Abstract
The bones of the vertebrate limb form by the process of endochondral ossification, whereby limb mesenchyme condenses to form an intermediate cartilage scaffold that is then replaced by bone. Although Indian hedgehog (IHH) is known to control hypertophic differentiation of chondrocytes during this process, the role of hedgehog signaling in the earlier stages of chondrogenesis is less clear. We have conditionally inactivated the hedgehog receptor Ptc1 in undifferentiated limb mesenchyme of the mouse limb using Prx1-Cre, thus inducing constitutively active ligand-independent hedgehog signaling. In addition to major patterning defects, we observed a marked disruption to the cartilage elements in the limbs of Prx1-Cre:Ptc1c/c embryos. Using an in vitro micromass culture system we show that this defect lies downstream of mesenchymal cell condensation and likely upstream of chondrocyte differentiation. Despite early increases in levels of chondrogenic genes, soon after mesenchymal condensation the stromal layer of Prx1-Cre:Ptc1c/c-derived micromass cultures is characterized by a loss of cell integrity, which is associated with increased cell death and a striking decrease in Alcian blue staining cartilage nodules. Furthermore, inhibition of the hedgehog pathway activation using cyclopamine was sufficient to essentially overcome this chondrogenic defect in both micromass and ex vivo explant assays of Prx1-Cre:Ptc1c/c limbs. These data demonstrate for the first time the inhibitory effect of cell autonomously activated hedgehog signaling on chondrogenesis, and stress the importance of PTC1 in maintaining strict control of signaling levels during this phase of skeletal development.
Keywords: Apoptosis, Cell Death, Development, Embryo, Mouse Genetics, Chondrogenesis, Limb Development, Patched1, prx1-cre, Sonic Hedgehog Pathway
Introduction
There are currently close to 400 recognized human genetic disorders with a skeletal component (1). A detailed understanding of skeletal patterning during development will aid in the treatment and diagnosis of such disorders, as well as in formulating approaches to skeletal regeneration and repair. With the exception of some flat bones of the skull, most bones form by the process of endochondral ossification, whereby mesenchymal cells differentiate into a cartilage intermediate that is then replaced by bone. The onset of chondrogenesis, the process by which cartilage is formed, is marked by the aggregation of proliferating undifferentiated mesenchymal cells into pre-cartilage condensations. The increased cell density in these condensations is thought to result from cell-cell interactions, consistent with the expression of specific cell adhesion molecules such as N-cadherin and neural cell adhesion molecule (2). Condensed precursor cells subsequently differentiate into chondrocytes that initially continue to proliferate. One of the earliest markers of chondrogenesis is the transcription factor SOX9, which is expressed in mesenchymal condensations and proliferating chondrocytes, and is necessary for chondrogenesis (3, 4). SOX9 directly activates expression of the cartilage-specific extracellular matrix protein COL2A1 (5), as well as controlling SOX5 and SOX6, which are also essential for cartilage formation (6). A complex network of interactions between members of the fibroblast growth factor (7), bone morphogenetic protein (4, 8, 9), and RHO (10, 11) families of signaling molecules is also required to regulate these early stages of chondrogenesis.
As condensed mensenchymal cells differentiate into chondrocytes, these aggregated condensations become surrounded by a layer of fibroblastic cells known as the perichondrium. The chondrocytes in the center of the condensation subsequently stop proliferating and become hypertrophic. Bone forming progenitors first occur in the perichondrium adjacent to the hypertrophic chondrocytes and vascularization allows invasion of osteoblasts into the cartilage, ultimately leading to replacement by bone (reviewed in Ref. 12). Ihh4 is first expressed in the mouse limb bud at 12.0 days post-coitum (dpc), in the prehypertrophic and hypertrophic chondrocytes of the developing growth plate (13). IHH controls differentiation of hypertrophic chondrocytes through a feedback loop involving the parathyroid-related protein (14, 15). Analysis of Ihh−/− null mice, which display shortened long bones, further revealed roles for IHH in controlling chondrocyte proliferation and osteoblast development in a parathyroid-related protein independent manner (15, 16). Although Sonic hedgehog (SHH) has a well established role during anterior-posterior patterning of the limb bud prior to Ihh expression, its role in regulating early chondrogenesis is less clear. Micromass studies suggest that this ligand is not required for the initiation of mesenchymal cell condensation and chondrocyte differentiation, although Shh overexpression in this system promotes chondrocyte hypertrophy (17). However, a number of in vitro and in vivo studies have suggested that SHH acts at a stage prior to chondrocyte differentiation to positively regulate chondrogenic cell differentiation and cartilage formation (18–21). In particular, in tracheal cartilage development SHH was shown to be involved in mesenchymal proliferation, condensation, and chondrocyte differentiation through regulation of Sox9 expression (21).
Hedgehog signaling plays a central role in the development of virtually all vertebrate tissues, and has been implicated in a number of human disorders associated with skeletal defects (reviewed in Ref. 22). Patched1 (PTC1, also known as PTCH1) is a transmembrane receptor that both negatively regulates and is transcriptionally activated by hedgehog signaling, thus creating a negative autoregulatory feedback loop. In the absence of hedgehog ligand (Sonic, Indian, or Desert), PTC1 constitutively represses the signaling molecule smoothened. This inhibition is relieved on ligand binding, leading to downstream transcriptional regulation. The strict requirement for controlled repression of hedgehog signaling has been revealed in Ptc1 null mice, which display severe developmental defects and lethality at 9.5 dpc (23). To overcome the early lethality of these mice, we used a Ptc1 conditional allele (24) to inactivate Ptc1 specifically in the early limb mesenchyme using the Prx1-Cre transgenic driver (25). This strategy leads to high level ligand-independent activation of hedgehog signaling across the limb, and provides the unique opportunity to interrogate the mechanistic effects of hedgehog signaling during early limb development. We have recently undertaken a detailed phenotypic and molecular analysis of the early patterning defects observed in the limbs of these embryos (26). However, in addition to effects on patterning, we uncovered a novel skeletal defect in these mice, as revealed by a severe disruption to cartilage elements in the embryonic limb. We have further characterized this phenotype using in vitro micromass and ex vivo limb explant cultures, and show a likely defect in the differentiation of condensed mesenchyme into chondrocytes, a step not previously associated with negative hedgehog control.
EXPERIMENTAL PROCEDURES
Animal Breeding
Breeding and genotyping of the Prx1-Cre:Ptc1c/c line has been previously described (26). Briefly, Prx1-Cre (25) males harboring one Ptc1 conditional allele (24) were crossed to Ptc1c/c females to produce embryos designated as wild-type (WT; Prx1-Cre:Ptc1w/w), heterozygous (Prx1-Cre:Ptc1c/wt), or homozygous (Prx1-Cre:Ptc1c/c). For all timed matings, 0.5 dpc was designated noon on the day after mating. All experimentation involving animals was approved by a University of Queensland Animal Ethics Committee and conformed to relevant ethical guidelines.
RNA in Situ Hybridization
Whole mount (27) and section (28) in situ hybridization was performed as previously described. The Sox9 probe was a kind gift of Peter Koopman (The University of Queensland, Australia) (29), and Col2a1 was from Kathy Cheah (University of Hong Kong).
Micromass Culture
Micromass cultures were prepared as previously described (30–32) from samples of pooled limbs. Forelimbs and hindlimbs were obtained from 11.5 dpc Prx1-Cre:Ptc1c/c mutants and from heterozygous and wild-type embryos. Although homozygous mutant embryos were easily identifiable phenotypically, it was not possible to distinguish between heterozygous and wild-type embryos within the time period required to pool limbs and establish cultures. Given that we have observed no molecular or phenotypic differences between heterozygous and wild-type limbs (26), these limbs were pooled to produce “control cultures.” Limbs were dissociated in 1 unit/ml of dispase II (Roche Diagnostics) solution containing 10% fetal bovine serum (FBS)/Puck's saline A buffer for 1.5 h at 37 °C. Digested limb solutions were triturated in 2:3 DMEM/F-12 medium containing 10% FBS (Invitrogen), passed through a 40-μm cell strainer to obtain a single cell suspension, and briefly centrifuged. Cells were resuspended in growth medium at a concentration of 2 × 107 cells/ml and spotted in 10-μl droplets on Nunclon 4-well Delta surface culture dishes. After cells adhered to culture dishes for 1.5 h at 37 °C in a humidified atmosphere containing 5% CO2, 500 μl of 2:3 DMEM/F-12 medium containing 10% FBS, 0.5 mm glutamine, 25 units/ml of penicillin, and 25 μg/ml of streptomycin was added. Growth medium was replaced every second day. Cyclopamine (Sigma, number C4116) was resuspended in dimethyl sulfoxide and added to micromass cultures at the time of the initial seeding and during medium replacement to a final concentration 5–10 μm. Recombinant mouse Sonic hedgehog (rSHH; R&D 461-SH) was reconstituted in sterile PBS containing 0.1% bovine serum albumin and stored following the manufacturer's instructions. Recombinant protein was added at the time of micromass seeding and every second day during medium replacement to a final concentration 10–100 ng/ml.
Alcian Blue, Lectin Peanut Agglutinin (PNA) and SOX9 Staining
Micromass cultures were fixed in 4% paraformaldehyde for 30 min. For Alcian blue staining cultures were washed briefly in PBS then incubated in 0.1% Alcian blue GX (Sigma) in 0.1 N HCl, pH 1.0, overnight at room temperature (the method was modified from Ref. 33). Cultures were cleared in 70% ethanol before images were captured on a stereomicroscope (Olympus SZX12). ImageJ software (Rasband, WS, ImageJ, rsb.info.nih.gov/ij) was used to quantify differences in Alcian blue staining between samples. To standardize measurements, all images were taken immediately after staining using identical camera settings (Olympus SZX12). The ImageJ automatic thresholding routine was used to avoid bias introduced by an observer manually setting the image threshold. Analysis was undertaken on 2–3 individual micromass experiments. Representative staining results were expressed as a fraction of total area. Alcian blue staining of cartilage in whole (34) and sectioned (35) 13.5 dpc limbs was performed according to published protocols.
For PNA staining, fixed micromass cultures were incubated with biotinylated peanut agglutinin overnight at 4 °C (Sigma; 100 g/ml). PNA binding was detected with the ABC Elite kit and colorimetric substrate (Vector Laboratories) as described by the manufacturer. For detection of SOX protein a SOX9-specific antibody (36) was used to stain fixed micromass cultures.
Limb Explant Culture and Skeletal Staining
Autopods were isolated from 11.5 dpc Prx1-Cre:Ptc1c/c embryos and compared with either wild-type or heterozygous controls. The tissue was washed in Puck's saline solution A and placed on a Nuclepore Polycarbonate membrane (Track-Etch Whatman 25-mm 0.1 UM). BGJ culture medium (Invitrogen 12591) containing 10% FBS, 0.5 mm glutamine, 25 units/ml of penicillin, and 25 μg/ml of streptomycin was changed every second day. Cyclopamine was added as described for micromass experiments.
Skeletal staining was undertaken to visualize bone (Alizarin red) and cartilaginous (Alcian blue) elements using a method modified from Ref. 37. Briefly, embryos were fixed in 95% (v/v) ethanol, stained with 70% (v/v) ethanol containing 0.005% (w/v) Alizarin red S, 0.015% (w/v) Alcian blue, and 5% (v/v) acetic acid, cleared in 20% (v/v) aqueous glycerin containing 1% (w/v) KOH, and stored in 100% glycerin. Images were captured on an Olympus BX-51 microscope using a DP-70 camera (Olympus).
Quantitative Real Time RT-PCR (qRT-PCR)
Total RNA was extracted using TRIzol (Invitrogen), and RNA quantity and quality were determined using an Agilent 2100 BioAnalyzer. cDNA was synthesized from 2 μg of DNase I-treated total RNA using Superscript III (Invitrogen) and oligo(dT)12–15 (Promega) according to the manufacturer's instructions. qRT-PCR was carried out using the Applied Biosystems SYBR Green dye system and 7500 Real Time Cycler in 96-well plates. Cycling variables were as follows: 50 °C for 2 min, 95 °C for 10 min, then 40 cycles of 15-s denaturation at 95 °C and 1-min at appropriate annealing temperatures, optimized for each set of primers based on dissociation curves. Primer sequences are available on request. Expression levels were normalized to hypoxanthine-guanine phosphoribosyltransferase as determined from the ratio of δCT values. For the whole autopod study (9.5–12.5 dpc) the mean ± S.D. values were determined from 2–6 biological replicates, each replicate consisting of two paired autopods. For the micromass studies, the mean of relative expression ± S.D. was determined from two biological replicates. Importantly, each biological replicate contained between 8 and 20 autopods pooled from somite and phenotypically matched embryos. A Student's t test was used to compare the experimental conditions. Sequencing of gel-purified amplicons was performed to ensure correct product amplification for each primer pair.
Time Lapse Live Cell Imaging
Micromass cultures were prepared as described above with minor modifications. Briefly, following 2 days of culture on Lab-Tek II Chamber slides (Nalge Nunc International) the medium was removed, cultures were washed in PBS and then incubated in CO2-independent medium (Invitrogen, number 18045). Still images were captured every 2 min for 20 h on an Olympus IX-81 inverted microscope (×10 objective). All images were processed using Cell® application software (Olympus). Video files were saved at 32 FPS. Cells were maintained at 37 °C in the absence of CO2 throughout the imaging period, fixed as described, and stained with Alcian blue.
Analysis of Apoptosis
For caspase-3 staining, micromass cultures were seeded as described above onto 8-well glass chamber slides (Lab-Tek II, Nalge Nunc International). Following 4 days of culture the wells were washed and the cultures fixed for 30 min in 4% paraformaldehyde at room temperature. Cultures were rinsed in 100% methanol for 10 min at −20 °C, blocked in 1:50 normal goat serum, 1% BSA for 60 min then incubated in 1:200 goat anti-rabbit cleaved caspase-3 (Asp175) (Cell Signaling number 9661) overnight at 4 °C. The following day the cultures were washed in PBS and incubated in DAPI (1:5000) (Sigma) for 10 min, washed, and then mounted using Prolong-Gold antifade reagent (Invitrogen). Confocal images were captured using an Axivert 200M SP LSM 510 META confocal laser-scanning microscope (Zeiss). Data were processed using LSM510 Meta software (Zeiss).
In vivo apoptosis was assessed using the TUNEL method to detect fragmented DNA on 4% paraformaldehyde-fixed paraffin-embedded 8-μm sections of whole limbs (11.5 and 12.5 dpc), according to the manufacturer's instructions (Roche Applied Science In situ cell death detection kit, fluorescein). Images were captured using an Olympus BX-51 microscope and DP-70 12Mp color camera.
RESULTS
Ptc1 Inactivation Inhibits Cartilage Formation in the Prx1-Cre:Ptc1c/c Forelimb and Hindlimb
We have previously described a differential oligodactyly/polydactyly phenotype in forelimbs and hindlimbs of Prx1-Cre:Ptc1c/c embryos (26). Here we describe an additional and severe defect in chondrogenesis which, unlike the patterning defect, is equivalent in both forelimbs and hindlimbs. In our previous study we confirmed that hedgehog signaling was activated in Prx1-Cre:Ptc1c/c mutant forelimbs from 10.5 dpc and hindlimbs from 11.5 dpc by qRT-PCR analysis of universal downstream targets Ptc1 and Gli1 (26, 38, 39). Here, we show that hedgehog signaling levels increase further in Prx1-Cre:Ptc1c/c forelimbs and hindlimbs at 12.5 dpc, a stage when early condensations are visible in control limbs (supplemental Fig. S1, A and B).
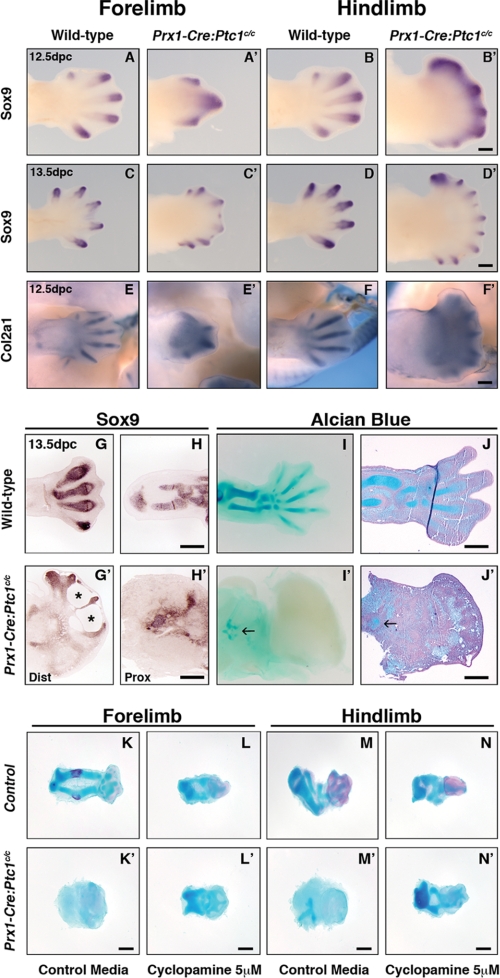
Whole mount in situ hybridization revealed altered expression of Sox9 in the Prx1-Cre:Ptc1c/c autopod relative to control. At 12.5 and 13.5 dpc Sox9 marks the digital rays in wild-type limbs, but in Prx1-Cre:Ptc1c/c limbs Sox9 expression is primarily restricted to the distal tips of the condensing digits (Fig. 1, A–D′). Compared to controls, ectopic interdigital Sox9 expression was also seen in distal 12.5 dpc Prx1-Cre:Ptc1c/c limbs (Fig. 1, A–B′). Similarly, expression of Col2a1 was restricted to the distal tips of skeletal elements in Prx1-Cre:Ptc1c/c limbs, with diffuse staining evident throughout the mesenchyme (Fig. 1, E and F′). Section in situ hybridization at 13.5 dpc revealed disorganized and hypoplastic cartilage precursor condensations marked by Sox9 expression, in both the autopod and zeugopod of Prx1-Cre:Ptc1c/c limbs (Fig. 1, G and G′, distal limb or autopod, and H and H′, proximal limb or zeugopod). A loss of cell integrity or blistering phenotype is also evident in the interdigital mesenchyme in these limbs (Fig. 1G′, asterisk). Most strikingly, sulfated proteoglycan produced by differentiated chondrocytes and detected by Alcian blue staining in whole and sectioned limbs is markedly reduced in Prx1-Cre:Ptc1c/c limbs compared with controls (Fig. 1, I and I′, and J and J′). Differentiated cartilage was consistently only visible in the proximal limb likely corresponding to the zeugopod region (Fig. 1, I′ and J′, arrows), and within the distal autopod (data not shown). However, these severely truncated cartilage elements do not resemble the highly organized condensations in the wild-type limb (Fig. 1, I and J). In some cases more extensive cartilage elements staining weakly for Alcian blue were visible in the mutant autopod by whole mount staining (data not shown), but again these were not equivalent to wild-type structures. To quantitate the gene expression changes observed by in situ hybridization, whole autopods were dissected from somite-matched 9.5–12.5 dpc embryos and analyzed by qRT-PCR.
FIGURE 1.
Altered chondrogenic gene expression and structural abnormalities in Prx1-Cre:Ptc1c/c limbs. Compared to wild type (A, B, C, and D), whole mount in situ hybridization for Sox9 revealed expression at distal tips of Prx1-Cre:Ptc1c/c digits at 12.5 dpc (A′ and B′) and 13.5 dpc (C′ and D′), along with ectopic inter-digital expression at 12.5 dpc. Localized expression of Col2a1 was also disrupted in the Prx1-Cre:Ptc1c/c limb at 12.5 dpc (E–F′). Scale bar, 500 μm. At 13.5 dpc, section in situ hybridization for Sox9 revealed disorganized and reduced expression in the distal autopod (dist) and proximal zeugopod (prox) of the limb (G, G′, H, and H′). Whole mount (I and I′) and section (J and J′) Alcian blue staining confirmed disrupted cartilage development. Arrows in I′ and J′ show small regions of Alcian blue staining in mutant limbs. Scale bar, 200 μm. Inhibition of hedgehog signaling with cyclopamine rescues the chondrogenic defect in Prx1-Cre:Ptc1c/c limbs cultured ex vivo. Whole autopods were harvested at 11.5 dpc and cultured for 6 days. Addition of cyclopamine perturbed Alcian blue staining in control limbs (K–N), but partially rescued the phenotype in limbs derived from Prx1-Cre:Ptc1c/c embryos (K′–N′). Images are representative of four individual experiments (see supplemental Figs. 2 and 3). Scale bar, 200 μm.
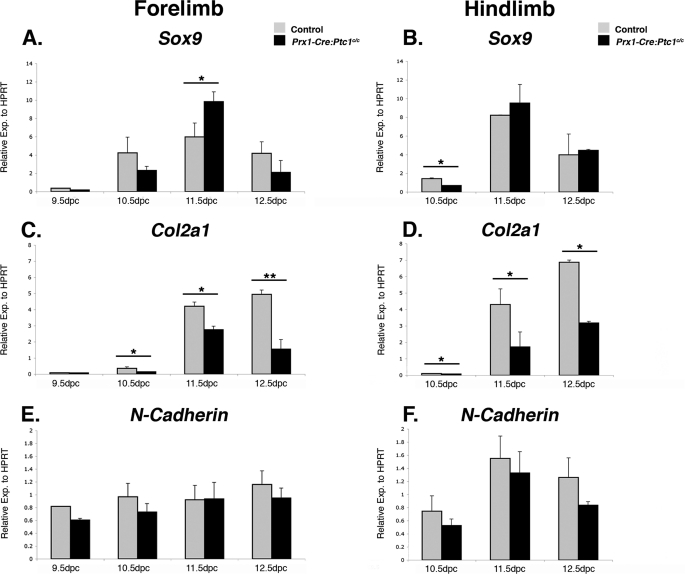
Sox9 expression in control and mutant limbs was highest at 11.5 dpc, with expression decreasing at 12.5 dpc (Fig. 2, A and B). Mutant forelimbs showed increased expression at 11.5 dpc but other stages revealed no significant difference between mutant and control samples. Importantly, significant down-regulation of Col2a1 expression was observed at all time points from 10.5 dpc in the Prx1-Cre:Ptc1c/c limbs (Fig. 2, C and D). We also analyzed expression of N-Cadherin (N-Cad) and saw no difference in expression in Prx1-Cre:Ptc1c/c versus control limbs at the time points tested (Fig. 2, E and F), suggesting that altered cellular adhesion in mesenchymal condensations is not the primary cause of the chondrogenic defects observed.
FIGURE 2.
Expression of chondrogenic markers in control versus Prx1-Cre:Ptc1c/c autopods. Quantitative real time RT-PCR revealed up-regulated Sox9 in Prx1-Cre:Ptc1c/c forelimb autopods at 11.5 dpc but no significant difference at other time points in forelimb or hindlimbs (A and B). Col2a1 was significantly down-regulated in Prx1-Cre:Ptc1c/c autopods from 10.5 dpc (C and D). N-Cadherin expression was unaffected in mutant limbs (E and F). Gli1 and Ptc1 data for 12.5 dpc autopods is available under supplemental Fig. S1. *, p < 0.05; **, p < 0.01. Error bars represent the S.D.
Chondrogenic Defects in the Prx1-Cre:Ptc1c/c Limbs Are Recapitulated in Culture and Partially Rescued by Inhibition of Hedgehog Signaling
Because Prx1-Cre:Ptc1c/c embryos do not survive past 14.0 dpc (26), we used ex vivo explant limb culture to ensure that the chondrogenic defect is not the result of developmental delay. Autopods were dissected from stage matched 11.5 dpc embryos of each phenotype and cultured for 6–7 days at an air-liquid interface. During this period, significant growth was observed in all control limbs, with striking development of zeugopod elements initiated from the cultured autopods (Fig. 1, K-N′, and supplemental Figs. 2 and 3). Over the culture period, control forelimb cultures developed cartilage elements that stained intensely with Alcian blue, and also showed evidence of bone deposition in the zeugopod elements, as visualized by Alizarin red staining (Fig. 1K). Although the ex vivo limb culture period (11.5 dpc + 6 days) cannot be directly compared with the equivalent in vivo maturation, digits and elements resembling both the radius and ulna were clearly evident (Fig. 1K). Furthermore, the presence of Alizarin red positive tissue suggests that these control cultures have progressed beyond the normal stage of lethality of Prx1-Cre:Ptc1c/c embryos (14.0 dpc). In contrast, Alcian blue staining was virtually undetectable in Prx1-Cre:Ptc1c/c cultured limbs compared with those from littermate controls, and no organized zeugopod elements were evident (Fig. 1, K and K′ and M and M′, and supplemental Figs. 2 and 3). Overall, the size and shape of Prx1-Cre:Ptc1c/c cultured limbs appeared altered relative to control limbs, partly due to the lack of zeugopod outgrowth. In addition, cultured mutant limbs also adopted a flatter morphology along the dorso-ventral axis compared with the more three-dimensional growth of control limbs, likely due to the lack of structural reinforcement.
To confirm that the observed defects were due to hedgehog signaling, limb bud explants were treated with cyclopamine, a steroidal alkaloid that antagonizes hedgehog signaling through direct interaction with signaling molecule smoothened (40, 41). Based on dose-response testing of cyclopamine in the micromass system (supplemental Fig. S4), 5 μm was chosen as the optimal concentration for use in subsequent experiments. Addition of cyclopamine to the culture medium from the time of initial seeding resulted in retarded growth and ossification of control limbs, compared with untreated controls (Fig. 1, L and N). Importantly, Prx1-Cre:Ptc1c/c limb explants cultured in the presence of cyclopamine showed dramatic improvement in the elongation and definition of Alcian blue staining elements (Fig. 1, L′ and N′). In the majority of cultures, cyclopamine treatment effectively restored the staining pattern in mutant cultures to a level similar to that observed in control cyclopamine-treated limbs (Fig. 1, L and L′ and N and N′, and supplemental Figs. 2 and 3). These data suggest that whereas a certain level of hedgehog signaling is required for optimal chondrogenesis, pathway activation can lead to inhibition of chondrogenesis in certain contexts.
Micromass Culture of Prx1-Cre:Ptc1c/c Limb Cells Reveals a Decrease in Formation of Cartilage Nodules
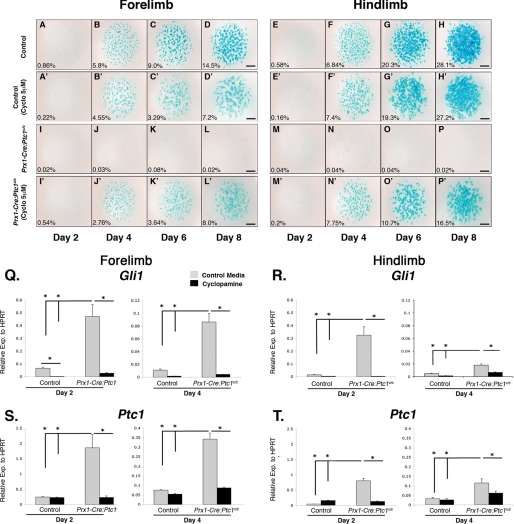
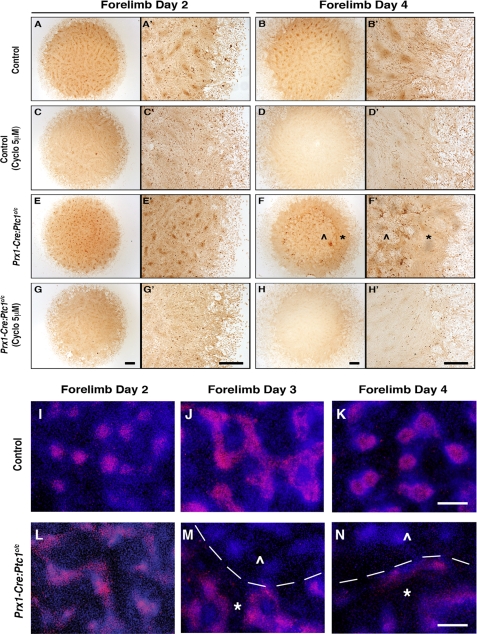
We next employed the in vitro micromass assay to further define the underlying chondrogenic defect identified in the in vivo and ex vivo analyses. Micromass has been used extensively as a model to analyze chondrogenesis in primary cells of the limb and face (e.g. Ref. 42). Mesenchymal cells plated at high density condense and undergo a process analogous to endochondral ossification in a sequence that can be monitored by staining with appropriate markers (43). Micromass cultures were established from somite-matched 11.5 dpc mutant and control autopods, collected at 2-day intervals and stained with Alcian blue to determine the level of chondrocyte differentiation. Micromass from control forelimbs and hindlimbs showed Alcian blue staining nodules from day 4 onwards, with a progressive increase in cartilaginous nodule size and boundary definition with time (Fig. 3, A–H). In accordance with in vivo lack of cartilage, Alcian blue staining cartilage nodules were not detected in micromass cultures established from Prx1-Cre:Ptc1c/c limbs (Fig. 3, I–P). Addition of cyclopamine to control micromass cultures from day 1 caused some disruption to Alcian blue-stained nodules, primarily in forelimb derived cultures, as quantified by ImageJ (Fig. 3, A′–H′). More significantly, cyclopamine treatment altered the gross morphology of control cartilage nodules in both forelimb and hindlimb cultures, with more diffusely aggregated cells, reduced boundary definition, and evidence of ectopic Alcian blue staining between nodules (Fig. 3, A′–H′). This was more evident when cultures were treated with higher concentrations (10 μm) of cyclopamine (supplemental Fig. S4). Strikingly, addition of cyclopamine to Prx1-Cre:Ptc1c/c cultures dramatically rescued the formation of Alcian blue staining nodules (Fig. 3, I′–P′). Although nodule morphology was not restored completely, staining in cyclopamine-treated Prx1-Cre:Ptc1c/c micromass cultures closely resembled that in cyclopamine-treated control cultures. Again, this confirms that the chondrogenic defects observed in the Prx1-Cre:Ptc1c/c limbs are hedgehog specific, and shows that the defect can be significantly rescued up until at least 11.5 dpc, a stage when limb mesenchymal cells are beginning to condense in vivo.
FIGURE 3.
Inhibition of hedgehog signaling with cyclopamine rescues the chondrogenic defect in Prx1-Cre:Ptc1c/c micromass. For micromass experiments, cells were harvested from 11.5 dpc autopods and maintained in culture for up to 8 days. Micromass cultures derived from Prx1-Cre:Ptc1c/c limbs (I–P) lacked the Alcian blue staining profile observed in control cultures (A–H). Addition of cyclopamine disrupted nodule morphology in control cultures (A′–H′) and restored Alcian blue staining in Prx1-Cre:Ptc1c/c micromass (I′–P′). ImageJ analysis shows the Alcian blue-stained area as a percentage of the total micromass area. Results are representative of three individual micromass experiments. Scale bar, 1 mm. Addition of cyclopamine (5 μm) significantly reduced both Gli1 and Ptc1 expression levels in day 2 and 4 micromass cultures from Prx1-Cre:Ptc1c/c limbs (Q–T) (*, p < 0.05). Error bars represent S.D.
In parallel with Alcian blue staining, qRT-PCR analysis was undertaken to measure the expression level of key hedgehog and chondrogenesis related genes in micromass cultures from Prx1-Cre:Ptc1c/c and control autopods. RNA samples were prepared from micromass cultures harvested at days 2 and 4, a time period during which rapid cellular condensation and early differentiation is occurring. As expected, Gli1 and Ptc1 expression was significantly elevated in Prx1-Cre:Ptc1c/c micromass compared with control cultures at days 2 and 4, in both forelimb (Fig. 3, Q and S, gray bars) and hindlimb (Fig. 3, R and T) cultures. Importantly, the addition of cyclopamine (5 μm) significantly reduced Gli1 and Ptc1 expression in all samples to levels approaching those in control cultures (Fig. 3, Q–T, black bars).
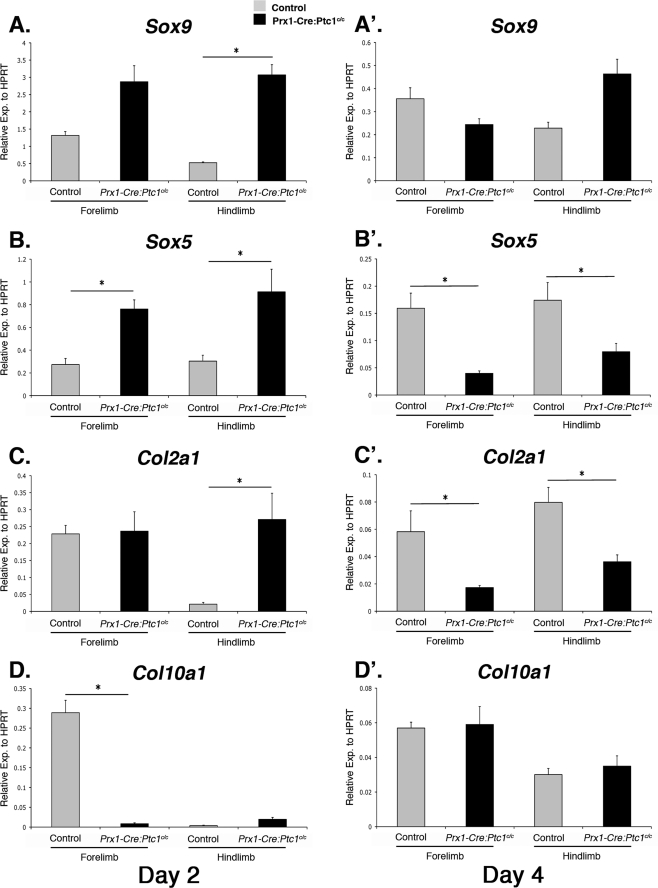
At day 2, when cells begin to undergo condensation in control cultures, expression of Sox9, Sox5, and Col2a1 was significantly up-regulated in Prx1-Cre:Ptc1c/c hindlimb micromass cultures compared with controls (Fig. 4, A, B, and C). By day 4, no significant difference in expression was identified for Sox9 (Fig. 4A′), but both Sox5 and Col2a1 showed decreased expression in Prx1-Cre:Ptc1c/c cultures from both forelimbs and hindlimbs compared with controls (Fig. 4, B′ and C′). Although up-regulation of Col2a1 in day 2 hindlimb Prx1-Cre:Ptc1c/c micromass cultures suggests early enhanced chondrogenesis, we detected no such increase in expression in whole limb buds. Specific differences between expression profiles in in vivo limb and micromass cultures are likely to be context dependent, but importantly day 4 micromass cultures show the same trend in expression of Sox9 and Col2a1 compared with 12.5 dpc whole limbs. Expression of Col10a1, a marker of hypertrophic differentiation was decreased in Prx1-Cre:Ptc1c/c forelimb cultures at day 2 (Fig. 4D), but showed no significant difference in forelimb cultures at day 2 or in day 4 cultures from either limb (Fig. 4, D and D′). We also analyzed expression of the gene encoding the proteoglycan aggrecan that is expressed in mature chondrocytes, but expression was low in control cultures and undetectable in Prx1-Cre:Ptc1c/c mutant cultures (data not shown). Because aggrecan is regulated by SOX9 and SOX5 (44), we may have expected aggrecan expression to more closely parallel these genes at early time points. However, low aggrecan expression is consistent with lack of differentiated chondrocytes and Alcian blue staining in these cultures at all stages. Consistent with the data from whole limbs, expression of N-Cad was not significantly altered in Prx1-Cre:Ptc1c/c compared with control cultures at either time point analyzed (data not shown).
FIGURE 4.
The chondrogenic gene expression profile is altered in Prx1-Cre:Ptc1c/c micromass. By day 2 expression, chondrogenic markers Sox9, Sox5, and Col2a1 were significantly up-regulated in hindlimb Prx1-Cre:Ptc1c/c micromass cultures (A–C). By day 4 expression, Sox5 and Col2a1 were significantly lower in Prx1-Cre:Ptc1c/c forelimb and hindlimb micromass cultures (B′ and C′). Col10a1 expression was significantly lower at day 2 in Prx1-Cre:Ptc1c/c forelimb-derived cultures, but was unchanged in all other samples analyzed (D and D′)(*, p < 0.05). Error bars represent S.D.
Analyses of micromass cultures support in vivo and limb explant data, suggesting that constitutively active hedgehog signaling within the embryonic limb mesenchyme of Prx1-Cre:Ptc1c/c embryos leads to defects in the formation of mature cartilage elements. Chemical interference targeted specifically at the hedgehog signaling pathway using cyclopamine was sufficient to restore the appearance of cartilage nodules in culture. Interestingly, expression of chondrogenic genes such as Sox9, Sox5, and Col2a1 was increased at early time points in Prx1-Cre:Ptc1c/c micromass cultures, suggesting an initial enhancement of the chondrogenic program that is never realized as mature cartilage nodules. To investigate this further we analyzed the early stages of mesenchymal condensation in wild-type and Prx1-Cre:Ptc1c/c micromass cultures.
PNA Staining and Live Imaging of Micromass Reveals Significant Cell Disruption between Days 2 and 4 in Prx1-Cre:Ptc1c/c Cultures
To determine how early the chondrogenic program is disrupted in Prx1-Cre:Ptc1c/c micromass cultures, we stained with the lectin PNA, which binds condensed mesenchymal chondroprogenitors (8). Similar trends were seen in cultures derived from forelimbs and hindlimbs, and data are presented from forelimb cultures only. In control cultures, well defined mesenchymal condensations were evident by day 2, consistent with the time frame for mesenchymal condensation (Fig. 5, A and A′). Staining persisted at day 4 (Fig. 5, B and B′). This defined staining pattern was significantly reduced in control cultures treated with cyclopamine (Fig. 5, C–D′), likely correlating with the diffuse Alcian blue staining observed in control cultures at later stages (Fig. 3, B′–D′ and supplemental Fig. S4). The most striking changes were seen in micromass derived from Prx1-Cre:Ptc1c/c limbs. At day 2, condensing nodules formed in approximately equivalent numbers to control cultures, although they appeared smaller and more tightly organized and distinct compared with control cultures (Fig. 5, compare E and E′ with A and A′). This suggests that mesenchymal condensation occurs in Prx1-Cre:Ptc1c/c limbs. Although difficult to quantify from the PNA staining, the early increase in chondrogenic gene expression described earlier suggests that this may be enhanced. However, by day 4, a time at which control nodules normally stain with Alcian blue, cells within the Prx1-Cre:Ptc1c/c micromass core were dramatically disrupted with no detectable nodules remaining (see ⋀ in Fig. 5, F and F′). In mutant cultures, the only organized cell populations were at the outer rim of the original micromass droplet (see * in Fig. 5, F and F′; n = 5). Cyclopamine treatment of Prx1-Cre:Ptc1c/c micromass cultures rescued the cell disruption at day 4 (Fig. 5, H and H′), although nodule formation was generally only restored to that seen in cyclopamine-treated control cultures (Fig. 5, D and D′).
FIGURE 5.
Disrupted cell integrity and loss of Sox9 in Prx1-Cre:Ptc1c/c micromass cultures. At day 2, cell condensations formed in approximately equivalent numbers in Prx1-Cre:Ptc1c/c micromass cultures but were smaller and stained with greater intensity (E and E′) when compared with control cultures (A and A′). By day 4 the Prx1-Cre:Ptc1c/c micromass had undergone dramatic remodeling (F and F′) creating a “central zone” showing significant cell disruption (⋀) surrounded by an outer “intact zone” of cells (*). Addition of cyclopamine (5 μm) to control cultures decreased localized PNA staining (C–D′). In contrast, cyclopamine inhibited the cell disruption observed in Prx1-Cre:Ptc1c/c micromass cultures (H and H′). Scale bar, 500 μm. Immunofluorescent staining revealed a distinct loss of SOX9 protein in the disrupted zone (⋀) compared with the intact zone (*) in Prx1-Cre:Ptc1c/c micromass cultures from 3 days (M and N). (Margin defined by dotted line; M and N.) Scale bar, 100 μm.
Time lapse microscopy was used to further characterize the cellular disruption that occurs between days 2 and 4 in Prx1-Cre:Ptc1c/c micromass culture. Cultures were imaged for 20 h following 3 days of culture. Time lapse imaging of control micromass cultures revealed the dynamic complexity of chondrogenesis, with condensations growing via recruitment of cells from the underlying stroma, and the culture overlaid with a dynamic population of myoblast progenitors (supplemental Movie S1). In contrast, condensations formed in micromass cultures from Prx1-Cre:Ptc1c/c limbs at the equivalent time points appeared smaller (supplemental Movie S2), consistent with the PNA staining observed in static cultures (Fig. 5, E and E′). Interestingly, Prx1-Cre:Ptc1c/c micromass cultures almost completely lack the myoblast population observed in control cultures, suggesting these progenitors have not migrated into the 11.5 dpc mutant autopod at the time of collection. Strikingly, in Prx1-Cre:Ptc1c/c cultures, disruption to the central cell layer occurs 3–4 days after initial micromass seeding. Tearing of the stromal layer disrupts all cell condensations, consistent with the lack of Alcian blue and PNA staining in day 4 Prx1-Cre:Ptc1c/c micromass (Figs. 3, J and N, and 5, F and F′, respectively). Following this event, the underlying stromal cells repopulated the disrupted area but failed to condense during the remaining time frame examined. In support of our previous observations, this cell disruption was not observed in Prx1-Cre:Ptc1c/c cultures treated with 5 μm cyclopamine (supplemental Movie S3).
To investigate the changes in chondrogenic gene expression at different stages of micromass culture we stained cultures with a SOX9 antibody to correlate gene expression with protein levels. This was also instructive in understanding the dynamic changes in expression in a spatio-temporal sense. In day 2 wild-type cultures SOX9 staining was confined to discrete condensed mesenchymal cell nodules (Fig. 5I). Staining increased at day 3 in tracts between nodules, and subsequently became confined to the core of cartilage nodules post chondrocyte differentiation at day 4 (Fig. 5, J and K). In Prx1-Cre:Ptc1c/c mutant cultures staining at day 2 appeared more reminiscent of day 3 control cultures, consistent with the increased gene expression at this stage (Fig. 5L). However, by days 3 and 4 the disrupted core of the Prx1-Cre:Ptc1c/c micromass cultures showed no evidence of SOX9 antibody staining, with staining persisting only in the undisturbed outer rim of these cultures (Fig. 5, M and N). Although this does not correlate with the unchanged Sox9 mRNA levels observed at this stage (Fig. 4A′), it is more consistent with the decreased expression of Col2a1 and Sox5 (Fig. 4, B′ and C′).
Taken together, these data suggest that mesenchymal cells derived from Prx1-Cre:Ptc1c/c limbs are able to undergo initial condensation in micromass culture, and at this stage express enhanced levels of chondrogenic genes. However, cells within these condensed nodules either do not progress to differentiated chondrocytes, or are eliminated soon after differentiation. Although the lack of Alcian blue staining suggests the former, we cannot exclude the latter possibility, especially given the early up-regulation of chondrogenic gene expression. Regardless of this, the defect appears to be related to disruption of the integrity of the cell layer within the central core of the micromass, resulting in ablation of the nodules prior to formation of mature cartilage nodules.
Cell Death Is Elevated in Prx1-Cre:Ptc1c/c Micromass Cultures
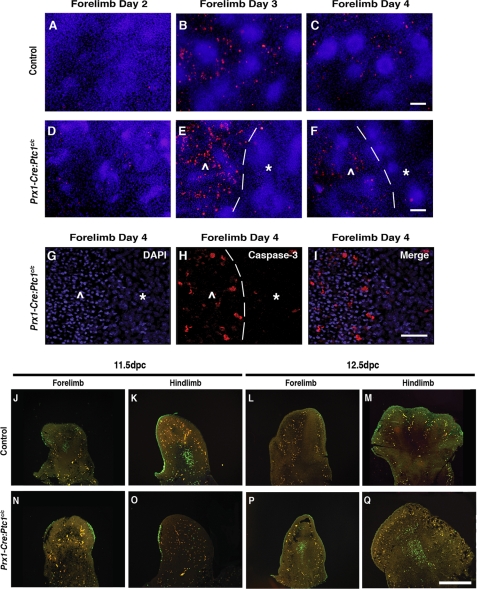
To investigate possible mechanisms for the morphological disruption within the Prx1-Cre:Ptc1c/c micromass, we next analyzed cultures for evidence of cell death based on immunofluorescent imaging of cleaved caspase-3. In control day 3 and 4 micromass cultures, caspase-3 positive cells were found throughout the stromal layer interspersed between nodules (Fig. 6, B and C). Time-matched Prx1-Cre:Ptc1c/c micromass cultures displayed elevated caspase-3 levels (Fig. 6, D–I) and abnormal nuclei morphology (Fig. 6, G–I) specifically within cells of the disrupted core. The leading edge of the phenotypic change was easily distinguishable and always consistent with elevated caspase-3 staining (Fig. 6, E, F, and H, dotted line, and supplemental Fig. S5). Notably, the outer rim of the mutant cultures, where cell integrity remains intact, was not characterized by increased apoptotic cells. As expected, no obvious nodules were identified within the disrupted area by day 4 either morphologically or by PNA staining. These observations suggest that the disrupted core of Prx1-Cre:Ptc1c/c micromass cultures is likely to result from increased cell death at a time corresponding to chondrocyte differentiation in control cultures.
FIGURE 6.
Increased cell death in Prx1-Cre:Ptc1c/c micromass cultures and limbs. Caspase-3 staining identified increased apoptotic cell death within the inner disrupted zone (⋀) compared to the outer intact zone (*) in Prx1-Cre:Ptc1c/c micromass from day 3 (margin defined by dotted line; E, F, and H). Scale bar in A–F, 200 μm. DAPI staining revealed a distinct morphological change in nuclei within the disrupted zone (⋀) in day 4 Prx1-Cre:Ptc1c/c micromass (G–I). Scale bar in G–I, 100 μm. TUNEL staining of a section through 11.5 dpc Prx1-Cre:Ptc1c/c autopods (N and O) revealed a lack of the typical apoptotic pattern observed in time-matched control tissues (J and K). In contrast, apoptotic cells were not detected in the central mesenchyme of control autopods at 12.5 dpc (L and M), but easily identified in Prx1-Cre:Ptc1c/c tissue (P and Q).
The disruption to the core of Prx1-Cre:Ptc1c/c micromass cultures is reminiscent of the loss of tissue integrity observed in the distal region between the digit condensations in the Prx1-Cre:Ptc1c/c limbs from 13.5 dpc (Fig. 1G′). We therefore performed TUNEL staining on whole autopods to investigate cell death in vivo. We have previously shown that apoptosis is not altered in Prx1-Cre:Ptc1c/c limbs at 10.5 dpc, and that normal interdigital cell death and digit separation is inhibited at 13.5 dpc (26). Here we present analysis at 11.5 and 12.5 dpc. In 11.5 dpc control autopods, we detected clusters of apoptotic cells within the proximal forelimb and hindlimb mesenchyme (Fig. 6, J and K), but these were not detected in 11.5 dpc Prx1-Cre:Ptc1c/c limbs (Fig. 6, N and O). These clusters, termed the opaque patch, have been described previously in 11.5 dpc, but not 12.5-dpc wild-type limbs (45). In contrast, apoptotic cells were readily detectable in the central mesenchyme of Prx1-Cre:Ptc1c/c limbs at 12.5 dpc, the time corresponding to mesenchymal condensation in control limbs and to the appearance of disrupted cell integrity in the Prx1-Cre:Ptc1c/c limbs (Fig. 6, P and Q). Although the ectopic cell death does not extend to the distal edge of the limb where loss of cell integrity is most pronounced, it is possible that it contributes to the chondrogenic phenotype given the dynamic tearing phenotype observed in micromass.
Ligand-dependent Activation of the Hedgehog Pathway Promotes Chondrogenic Differentiation in Micromass Culture
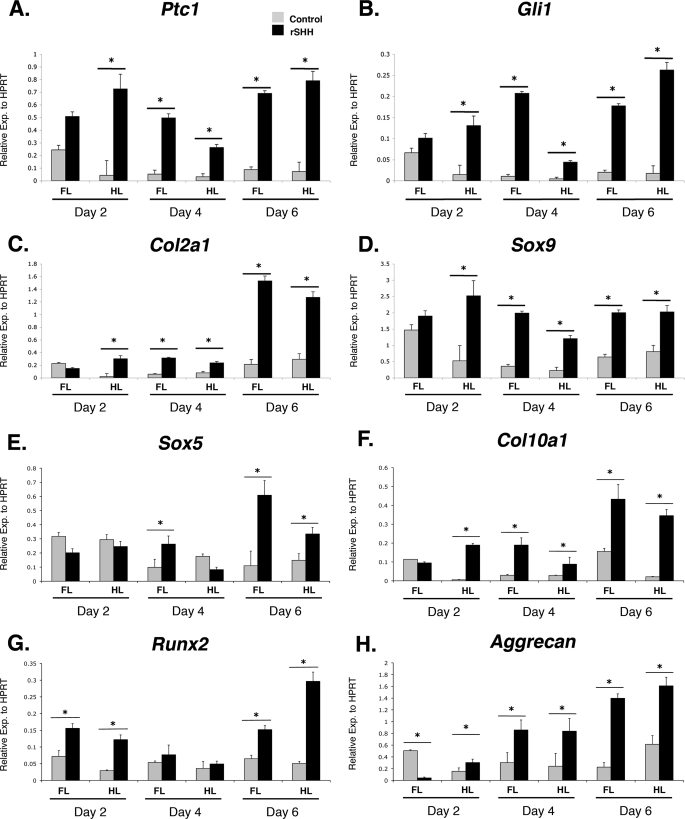
A number of studies have suggested that hedgehog signaling enhances the early stages of chondrogenesis (18–21). This seems contradictory to the disruption of chondrogenesis evident in the Prx1-Cre:Ptc1c/c limbs, which are characterized by constitutive ligand-independent activation of the hedgehog pathway. However, our micromass studies suggest that cultures derived from Prx1-Cre:Ptc1c/c limbs show evidence of early enhanced expression of chondrogenic genes, despite a failure to form mature cartilage nodules. We therefore treated control micromass cultures with rSHH protein (10 ng/ml) to allow a comparative study of ligand-dependent versus ligand-independent activation of the hedgehog pathway, with the proviso that activation will occur earlier and to a higher level in the Prx1-Cre:Ptc1c/c cultures. Gene expression levels were determined at 2, 4, and 6 days of culture using qRT-PCR (Fig. 7, A–H). As expected, Ptc1 and Gli1 were shown to be significantly up-regulated at all time points with the exception of forelimb cultures at day 2 (Fig. 7, A and B), supporting hedgehog pathway activation. Consistent with Prx1-Cre:Ptc1c/c mutant micromass cultures, Sox9 and Col2a1 were also significantly induced in the presence of rSHH from day 2 hindlimb cultures (Fig. 7, C and D). By day 6 expression of Sox9, Col2a1, and Sox5 was enhanced in rSHH exposed cultures from both forelimbs and hindlimbs (Fig. 7, C–E). This is in contrast to Prx1-Cre:Ptc1c/c mutant cultures that show down-regulation of Col2a1 and Sox5 by day 4 when the cellular integrity of the cultures is disrupted. N-Cad remained unchanged at all time points (data not shown). Expression of Col10A1 was increased in all Prx1-Cre:Ptc1c/c-derived cultures analyzed, with the exception of day 2 forelimb cultures (Fig. 7F). This is consistent with a previous study showing evidence of enhanced chondrocyte hypertrophy in retrovirally transfected SHH micromass cultures (17). Runx2, which encodes a transcription factor required for bone formation and osteoblast differentiation, was significantly elevated in rSHH-treated micromass at days 2 and 6 (Fig. 7G). In contrast to Prx1-Cre:Ptc1c/c mutant micromass cultures where aggrecan expression was undetectable, rSHH-treated cultures showed increased expression at all stages with the exception of day 2 forelimb cultures (Fig. 7H). All expression changes are consistent with enhanced chondrogenesis in rSHH-treated micromass cultures, however, cultures treated with 0, 10, or 100 ng/ml of rSHH revealed a dose-dependent reduction in Alcian blue staining intensity by day 6 as quantified using ImageJ (supplemental Fig. S6, A–C). It is possible that this is also indicative of an enhancement of the chondrogenic program in these cultures.
FIGURE 7.
Exogenous application of rSHH accelerates the chondrogenic program in micromass culture. Addition of rSHH (10 ng/ml) to control micromass cultures resulted in significantly up-regulated expression of Ptc1 and Gli1 at all time points (with the exception of day 2 forelimb cultures) compared with control medium (A and B). Col2a1, Sox9, and aggrecan expression was also up-regulated in the same cultures, as was the hypertrophy marker Col10a1 (C, D, F, and H). The pre-hypertrophic chondrogenic marker Sox5, and osteoblast marker Runx2, showed a consistent elevated response to rSHH by day 6 of culture (E and G). *, p < 0.05. Error bars represent S.D. FL, forelimb; HL, hindlimb.
DISCUSSION
Although many studies have characterized the requirement for hedgehog signaling during the late stages of chondrogenesis, little is known about its role during the initial stages of condensation and chondrocyte differentiation. For the first time, we have demonstrated defects in the formation of cartilage elements following ligand-independent up-regulation of the hedgehog pathway. Micromass studies suggest that chondrogenic gene expression is initially enhanced in this system, but decreases at later stages as disruption to cellular integrity prevents progression to Alcian blue staining cartilage nodules. In contrast, addition of exogenous SHH to micromass cultures causes sustained increases in expression of chondrogenic and osteogenic markers accompanied by the formation of mature cartilage nodules. This suggests that hedgehog regulation of chondrogenesis involves competing positive and negative inputs, most likely related to the timing and level of pathway activation.
Prx1-Cre:Ptc1c/c Limbs Show Disrupted Chondrogenesis in Vivo and in Vitro
It is well accepted that members of the vertebrate hedgehog family are important in skeletal patterning and chondrogenesis. In the limb, the role of SHH in skeletal patterning is mediated through effects on post-translational modification of the GLI3 transcription factor (46, 47). Together these molecules are the primary determinants of digit number and identity in the developing limb. However, the observation that skeletal elements form in both the Shh null and the Shh;Gli3 double mutant embryos suggests that SHH is not absolutely required for the actual process of chondrogenesis (48–50). However, analysis of tracheal cartilage development in Shh null mice suggests that, at least in this system, SHH contributes to mesenchymal cell proliferation, condensation, and chondrocyte differentiation through regulation of Sox9 (21). In contrast, whereas IHH is not involved in the early stages of chondrogenesis, it plays a role in regulating the proliferation and subsequent hypertrophy of chondrocytes, as well as in osteoblast differentiation (15). Prx1-Cre:Ptc1c/c embryos are characterized by high level ligand-independent activation of hedgehog signaling in the early limb mesenchyme. The patterning defects in these limbs have been presented previously (26), but the striking skeletal defects observed in both the forelimbs and hindlimbs prompted us to assess a possible effect on the earliest stages of mesenchymal condensation and chondrocyte differentiation.
Our data suggest that mesenchymal condensation occurs in Prx1-Cre:Ptc1c/c embryos, but differentiation to cartilage, as judged by Alcian blue staining, seems to be severely impaired. Importantly, we were able to culture limbs ex vivo to a stage beyond that achieved in vivo and thus demonstrate that the chondrogenic defect is unlikely to simply reflect a delay in this process in the Prx1-Cre:Ptc1c/c limb. This is significant because we have previously detected a delay in chondrogenesis in the anterior limb of the Gli3 extra-toes mouse (51). To further study this defect we moved to in vitro micromass culture. Although micromass culture does not maintain the precise cell-cell interactions seen in vivo and in limb culture, it has successfully been used to study the process of chondrogenesis in vitro (30). We showed that the formation of Alcian blue-stained cartilage nodules was severely impaired in cultures derived from both forelimbs and hindlimbs of Prx1-Cre:Ptc1c/c embryos. This defect was rescued by the addition of cyclopamine, which inhibits hedgehog signaling at the level of smoothened (40). It should be noted that morphologically the nodules in cyclopamine-treated Prx1-Cre:Ptc1c/c cultures resembled those observed in control cultures treated with cyclopamine. This is consistent with the fact that inhibition of HH signaling with cyclopamine alters chondrogenesis in control cultures. The effect of cyclopamine treatment on both control and Prx1-Cre:Ptc1c/c cultures will likely depend on the ultimate level of signaling achieved. Overall, the cyclopamine studies confirm that the effects on chondrogenesis in Prx1-Cre:Ptc1c/c mutants are hedgehog-dependent. These rescue data also suggest that the condensed mesenchymal cells do not transdifferentiate to other cell types, at least up to the time we commence exposure to cyclopamine.
Time-lapse imaging of micromass cultures allowed visualization of the condensation process in real time. As expected from our static analyses, condensations were initially evident in Prx1-Cre:Ptc1c/c-derived cultures, but the stromal layer was subsequently disrupted. We propose that this disruption prevents the maturation of condensed cells into mature cartilage nodules, although we cannot rule out the possibility that cells do differentiate just prior to disruption of the micromass. A similar live cell imaging approach has been used previously to visualize chondrogenesis in micromass cultures, showing that bone morphogenetic protein signaling regulates early compaction events in the undifferentiated mesenchyme, whereas SOX9 determines cell shape during chondrocyte differentiation (4). Our data fully support the usefulness of this approach as a tool to visualize the dynamic processes occurring during the early stages of chondrogenesis in micromass cultures.
The nature of the tearing in the mutant micromass cultures suggested either a defect in cellular adhesion or enhanced cell death. N-Cadherin has previously been shown to be important in cellular condensation and chondrocyte differentiation in chick limb micromass cultures (52). We found no difference in the expression of N-Cad in Prx1-Cre:Ptc1c/c mutant limbs or micromass cultures. This is consistent with normal condensation in these limbs, although we cannot rule out adhesion or cytoskeletal effects at later stages, or those not mediated by N-Cadherin.
The disrupted core of the Prx1-Cre:Ptc1c/c mutant micromass cultures was consistently characterized by enhanced cell death relative to the outer intact rim of cells. Although some level of cell death is observed in the stromal cell layer between condensed nodules in control cultures, this is not equivalent to the enhanced cell death observed in the mutant micromass core that is devoid of recognizable nodules. We suggest that this increased cell death contributes to the disruption seen in this region, and hence to the inability of Prx1-Cre:Ptc1c/c micromass cultures to form Alcian blue-positive cartilage nodules. Our previous studies have shown no increase in early apoptosis in the 10.5 dpc Prx1-Cre:Ptc1c/c limb buds, and a decrease in interdigital cell death and digit separation at later stages (26). Our analysis at 11.5 dpc confirmed a lack of apoptosis in the mutant limbs at this stage. However, at 12.5 dpc, a stage when apoptosis is not observed within the central mesenchyme of control limbs (Ref. 45, and our data), we observed evidence of apoptotic cells in this region in the Prx1-Cre:Ptc1c/c limbs. The precise contribution of this in vivo cell death to the final chondrogenic phenotype in the mutant limbs remains to be determined. As a general phenomenon, increased cell death appears at odds with enhanced hedgehog pathway activity, because SHH is thought to protect against apoptosis in the limb, with Shh null limbs characterized by massive cell death (48). In addition, PTC1 has been shown to act as a dependence receptor, mediating cell death in the absence of bound ligand through direct recruitment of a caspase-activating complex (53). However, although PTC1 is required to mediate this process, there is no evidence that its absence would decrease apoptosis levels, especially in regions where hedgehog ligands are normally active. We propose that effects on cell death are likely to be context dependent, and in the case of the Prx1-Cre:Ptc1c/c limb may relate to the timing, level, or site of up-regulation of the hedgehog pathway. It is also likely that the altered apoptosis observed in this mutant is not due to a direct effect on the cell death pathway, but that a more indirect mechanism is involved.
Hedgehog Gain of Function Models Suggest Timing, Level, and Context-dependent Effects on Chondrogenesis
Both the in vivo and in vitro data from our Prx1-Cre:Ptc1c/c model shows that widespread ligand-independent up-regulation of hedgehog signaling in undifferentiated limb mesenchyme leads ultimately to the failure to form mature cartilage. In a previous study deletion of Ptc1 specifically in Col2a1 expressing chondrocytes in vivo resulted in impaired chondrocyte hypertrophy, but also lead to enhanced ectopic osteoblast differentiation (54). Phenotypic differences between this model and the Prx1-Cre:Ptc1c/c mouse are likely related to timing of enhanced hedgehog activation, because Ptc1 deletion occurs earlier in our model. This is further supported by the fact that overexpression of Shh specifically in mouse chondrocytes in vivo also leads to inhibition of chondrocyte hypertrophy (55), as does application of exogenous SHH or IHH to limb explant cultures (14). Transgenic overexpression of Shh in mice using the same Prx1 enhancer element employed in this study did not give rise to the severe chondrogenic defects observed in the Prx1-Cre:Ptc1c/c embryos, but rather resulted in mature skeletal elements in the limb (56). This suggests that the level of hedgehog signaling may also be important in determining the final chondrogenic outcome. This is supported by the fact that cell autonomous activation of the hedgehog pathway in the Prx1-Cre:Ptc1c/c model is likely to result in higher overall signaling levels than other non-cell autonomous approaches.
A number of other studies have suggested a more positive role for hedgehog signaling in chondrogenesis. Implantation of SHH expressing fibroblasts into nude mice was shown to induce both cartilage and bone, and treatment of chondrogenic precursor cells with recombinant SHH or IHH stimulated chondrogenic differentiation (19). Likewise, it has been shown that transient SHH treatment of presomitic mesoderm cultures is required to generate precursor cells that are competent to differentiate to chondrocytes in response to bone morphogenetic protein signals (20). Closer to our own model, a study employing retroviral transduction of SHH in micromass cultures derived from limb bud cells showed decreased Alcian blue staining but an increase in hypertrophic markers (17). Our own data support and extend this conclusion by showing a consistent and sustained increase in markers of chondrogenesis, hypertrophy, and osteogenesis upon treatment of micromass cultures with exogenous SHH. Alcian blue is also disrupted by this treatment. Importantly, this is the first study to undertake an extensive quantitative analysis of relevant markers in this system. In the case of micromass cultures derived from Prx1-Cre:Ptc1c/c mutant mouse limbs we saw a similar increase in expression of chondrogenic markers at early stages prior to chondrocyte differentiation, however, these increases were curtailed at later stages by the disruption to cell integrity specifically in the core of the micromass cultures. Although levels of Sox9 mRNA in mutant cultures did not correlate closely with downstream chondrogenic genes such as Col2a1 and Sox5, protein distribution was more closely aligned with these changes. An observed discrepancy between Sox9 mRNA levels and that of downstream genes has previously been attributed to changes in protein activity in the micromass system (10).
Our data suggest that Prx1-Cre:Ptc1c/c cultures initially undergo changes similar to rSHH-treated cultures, but condensed mesenchymal cells do not subsequently differentiate to cartilage, likely due in part to increased cell death. It should be noted that unlike rSHH-treated cultures, Col10a1 levels were not enhanced in early Prx1-Cre:Ptc1c/c micromass cultures. Although it was surprising to detect expression of Col10a1 in early wild-type micromass cultures, it is possible that this represents basal levels of expression. The important point from these analyses is that unlike rSHH-treated cultures, hypertrophy is not accelerated in the Prx1-Cre:Ptc1c/c cultures, or at least this is not detectable up until the time of cell disruption. Although not tested directly, the differences between the two micromass models almost certainly arise from variation in timing and level of hedgehog pathway activation. This factor may be important in considering the use of SHH as a growth factor in in vitro approaches to skeletal regeneration.
Interestingly, the role of hedgehog signaling in adult bone homeostasis is also thought to depend on the level and timing or context of signaling. Haploinsufficiency for Ptc1 leads to an increase in bone mass due to enhanced osteoblast differentiation (57). In contrast, whereas complete conditional Ptc1 deletion in mature osteoblasts results in enhanced bone formation, the bone is porous due to excessive bone resorption (58). It is not possible to distinguish between effects of signaling levels or cell context in these studies, but it is likely that both are involved.
Taken together, our data and that of others suggest that the final contribution of hedgehog signaling to chondrogenesis in the embryo and to adult bone homeostasis is a fine balance between positive and negative inputs. The striking defect in the Prx1-Cre:Ptc1c/c model both in vivo and in vitro implies that timing and level of hedgehog pathway activation are likely to be important parameters in early chondrogenesis, and reinforce the importance of PTC1-dependent negative control of the hedgehog pathway at early stages in the chondrogenic process.
Supplementary Material
Acknowledgments
We thank Brandon Wainwright for providing the Ptc1 conditional mouse, Malcolm Logan for the Prx1-Cre mouse line, and Dagmar Wilhelm and Peter Koopman for the SOX9 antibody. We also thank Anne Hardacre, Julie Conway, and Tara Davidson for assistance with mouse husbandry, and Joy Richman and Kerry Manton for help establishing the micromass technology. Confocal microscopy was performed at the ACRF Cancer Biology Imaging Facility at the IMB, established with the generous support of the Australian Cancer Research Foundation.
This work was supported by National Health and Medical Research Council (NHMRC) of Australia Project Grant 569713.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and Movies S1–S3.
- Ihh
- Indian hedgehog
- dpc
- days post-coitum
- SHH
- Sonic hedgehog
- PTC1
- patched 1
- PNA
- peanut agglutinin
- rSHH
- recombinant Sonic hedgehog.
REFERENCES
- 1.Superti-Furga A., Unger S. (2007) Am. J. Med. Genet. A 143, 1–18 [DOI] [PubMed] [Google Scholar]
- 2.Hall B. K., Miyake T. (2000) Bioessays 22, 138–147 [DOI] [PubMed] [Google Scholar]
- 3.Bi W., Deng J. M., Zhang Z., Behringer R. R., de Crombrugghe B. (1999) Nat. Genet. 22, 85–89 [DOI] [PubMed] [Google Scholar]
- 4.Barna M., Niswander L. (2007) Dev. Cell 12, 931–941 [DOI] [PubMed] [Google Scholar]
- 5.Bell D. M., Leung K. K., Wheatley S. C., Ng L. J., Zhou S., Ling K. W., Sham M. H., Koopman P., Tam P. P., Cheah K. S. (1997) Nat. Genet. 16, 174–178 [DOI] [PubMed] [Google Scholar]
- 6.Smits P., Li P., Mandel J., Zhang Z., Deng J. M., Behringer R. R., de Crombrugghe B., Lefebvre V. (2001) Dev. Cell 1, 277–290 [DOI] [PubMed] [Google Scholar]
- 7.Yu K., Ornitz D. M. (2008) Development 135, 483–491 [DOI] [PubMed] [Google Scholar]
- 8.Pizette S., Niswander L. (2000) Dev. Biol. 219, 237–249 [DOI] [PubMed] [Google Scholar]
- 9.Yoon B. S., Ovchinnikov D. A., Yoshii I., Mishina Y., Behringer R. R., Lyons K. M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 5062–5067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woods A., Beier F. (2006) J. Biol. Chem. 281, 13134–13140 [DOI] [PubMed] [Google Scholar]
- 11.Montero J. A., Zuzarte-Luis V., Garcia-Martinez V., Hurle J. M. (2007) Dev. Biol. 303, 325–335 [DOI] [PubMed] [Google Scholar]
- 12.Goldring M. B., Tsuchimochi K., Ijiri K. (2006) J. Cell. Biochem. 97, 33–44 [DOI] [PubMed] [Google Scholar]
- 13.Bitgood M. J., McMahon A. P. (1995) Dev. Biol. 172, 126–138 [DOI] [PubMed] [Google Scholar]
- 14.Vortkamp A., Lee K., Lanske B., Segre G. V., Kronenberg H. M., Tabin C. J. (1996) Science 273, 613–622 [DOI] [PubMed] [Google Scholar]
- 15.St-Jacques B., Hammerschmidt M., McMahon A. P. (1999) Genes Dev. 13, 2072–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karp S. J., Schipani E., St-Jacques B., Hunzelman J., Kronenberg H., McMahon A. P. (2000) Development 127, 543–548 [DOI] [PubMed] [Google Scholar]
- 17.Stott N. S., Chuong C. M. (1997) J. Cell Sci. 110, 2691–2701 [DOI] [PubMed] [Google Scholar]
- 18.Warzecha J., Göttig S., Brüning C., Lindhorst E., Arabmothlagh M., Kurth A. (2006) J. Orthop. Sci. 11, 491–496 [DOI] [PubMed] [Google Scholar]
- 19.Enomoto-Iwamoto M., Nakamura T., Aikawa T., Higuchi Y., Yuasa T., Yamaguchi A., Nohno T., Noji S., Matsuya T., Kurisu K., Koyama E., Pacifici M., Iwamoto M. (2000) J. Bone Miner. Res. 15, 1659–1668 [DOI] [PubMed] [Google Scholar]
- 20.Murtaugh L. C., Chyung J. H., Lassar A. B. (1999) Genes Dev. 13, 225–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park J., Zhang J. J., Moro A., Kushida M., Wegner M., Kim P. C. (2010) Dev. Dyn. 239, 514–526 [DOI] [PubMed] [Google Scholar]
- 22.Nieuwenhuis E., Hui C. C. (2005) Clin. Genet. 67, 193–208 [DOI] [PubMed] [Google Scholar]
- 23.Goodrich L. V., Milenković L., Higgins K. M., Scott M. P. (1997) Science 277, 1109–1113 [DOI] [PubMed] [Google Scholar]
- 24.Ellis T., Smyth I., Riley E., Graham S., Elliot K., Narang M., Kay G. F., Wicking C., Wainwright B. (2003) Genesis 36, 158–161 [DOI] [PubMed] [Google Scholar]
- 25.Logan M., Martin J. F., Nagy A., Lobe C., Olson E. N., Tabin C. J. (2002) Genesis 33, 77–80 [DOI] [PubMed] [Google Scholar]
- 26.Butterfield N. C., Metzis V., McGlinn E., Bruce S. J., Wainwright B. J., Wicking C. (2009) Development 136, 3515–3524 [DOI] [PubMed] [Google Scholar]
- 27.Fowles L. F., Bennetts J. S., Berkman J. L., Williams E., Koopman P., Teasdale R. D., Wicking C. (2003) Genesis 35, 73–87 [DOI] [PubMed] [Google Scholar]
- 28.Wilhelm D., Hiramatsu R., Mizusaki H., Widjaja L., Combes A. N., Kanai Y., Koopman P. (2007) J. Biol. Chem. 282, 10553–10560 [DOI] [PubMed] [Google Scholar]
- 29.Wright E., Hargrave M. R., Christiansen J., Cooper L., Kun J., Evans T., Gangadharan U., Greenfield A., Koopman P. (1995) Nat. Genet. 9, 15–20 [DOI] [PubMed] [Google Scholar]
- 30.Ahrens P. B., Solursh M., Reiter R. S. (1977) Dev. Biol. 60, 69–82 [DOI] [PubMed] [Google Scholar]
- 31.Stanton L. A., Sabari S., Sampaio A. V., Underhill T. M., Beier F. (2004) Biochem. J. 378, 53–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.James C. G., Appleton C. T., Ulici V., Underhill T. M., Beier F. (2005) Mol. Biol. Cell 16, 5316–5333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cash D. E., Bock C. B., Schughart K., Linney E., Underhill T. M. (1997) J. Cell Biol. 136, 445–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagy A., Gertsenstein M., Vintersten K., Behringer R. (2003) Manipulating the Mouse Embryo: A Laboratory Manual, Cold Spring Harbor Laboratory Press, New York [Google Scholar]
- 35.Bancroft J., Stevens A. (1996) Theory and Practice of Histological Techiques, 4th Ed., Churchill Livingstone, Edinburgh, UK [Google Scholar]
- 36.Wilhelm D., Martinson F., Bradford S., Wilson M. J., Combes A. N., Beverdam A., Bowles J., Mizusaki H., Koopman P. (2005) Dev. Biol. 287, 111–124 [DOI] [PubMed] [Google Scholar]
- 37.Wallin J., Wilting J., Koseki H., Fritsch R., Christ B., Balling R. (1994) Development 120, 1109–1121 [DOI] [PubMed] [Google Scholar]
- 38.Goodrich L. V., Johnson R. L., Milenkovic L., McMahon J. A., Scott M. P. (1996) Genes Dev. 10, 301–312 [DOI] [PubMed] [Google Scholar]
- 39.Marigo V., Johnson R. L., Vortkamp A., Tabin C. J. (1996) Dev. Biol. 180, 273–283 [DOI] [PubMed] [Google Scholar]
- 40.Chen J. K., Taipale J., Cooper M. K., Beachy P. A. (2002) Genes Dev. 16, 2743–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Incardona J. P., Gaffield W., Kapur R. P., Roelink H. (1998) Development 125, 3553–3562 [DOI] [PubMed] [Google Scholar]
- 42.Bobick B. E., Thornhill T. M., Kulyk W. M. (2007) J. Cell. Physiol. 211, 233–243 [DOI] [PubMed] [Google Scholar]
- 43.Lee Y., Stott N., Jiang T., Widelitz R., Chuong C. (1998) Cells Materials 8, 19–32 [Google Scholar]
- 44.Han Y., Lefebvre V. (2008) Mol. Cell. Biol. 28, 4999–5013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fernández-Terán M. A., Hinchliffe J. R., Ros M. A. (2006) Dev. Dyn. 235, 2521–2537 [DOI] [PubMed] [Google Scholar]
- 46.Wang B., Fallon J. F., Beachy P. A. (2000) Cell 100, 423–434 [DOI] [PubMed] [Google Scholar]
- 47.Wang C., Rüther U., Wang B. (2007) Dev. Biol. 305, 460–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chiang C., Litingtung Y., Harris M. P., Simandl B. K., Li Y., Beachy P. A., Fallon J. F. (2001) Dev. Biol. 236, 421–435 [DOI] [PubMed] [Google Scholar]
- 49.Litingtung Y., Dahn R. D., Li Y., Fallon J. F., Chiang C. (2002) Nature 418, 979–983 [DOI] [PubMed] [Google Scholar]
- 50.te Welscher P., Zuniga A., Kuijper S., Drenth T., Goedemans H. J., Meijlink F., Zeller R. (2002) Science 298, 827–830 [DOI] [PubMed] [Google Scholar]
- 51.McGlinn E., van Bueren K. L., Fiorenza S., Mo R., Poh A. M., Forrest A., Soares M. B., Bonaldo Mde F., Grimmond S., Hui C. C., Wainwright B., Wicking C. (2005) Mech. Dev. 122, 1218–1233 [DOI] [PubMed] [Google Scholar]
- 52.Delise A. M., Tuan R. S. (2002) Dev. Dyn. 225, 195–204 [DOI] [PubMed] [Google Scholar]
- 53.Mille F., Thibert C., Fombonne J., Rama N., Guix C., Hayashi H., Corset V., Reed J. C., Mehlen P. (2009) Nat. Cell Biol. 11, 739–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mak K. K., Chen M. H., Day T. F., Chuang P. T., Yang Y. (2006) Development 133, 3695–3707 [DOI] [PubMed] [Google Scholar]
- 55.Tavella S., Biticchi R., Schito A., Minina E., Di Martino D., Pagano A., Vortkamp A., Horton W. A., Cancedda R., Garofalo S. (2004) J. Bone Miner. Res. 19, 1678–1688 [DOI] [PubMed] [Google Scholar]
- 56.Chen M. H., Li Y. J., Kawakami T., Xu S. M., Chuang P. T. (2004) Genes Dev. 18, 641–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ohba S., Kawaguchi H., Kugimiya F., Ogasawara T., Kawamura N., Saito T., Ikeda T., Fujii K., Miyajima T., Kuramochi A., Miyashita T., Oda H., Nakamura K., Takato T., Chung U. I. (2008) Dev Cell 14, 689–699 [DOI] [PubMed] [Google Scholar]
- 58.Mak K. K., Bi Y., Wan C., Chuang P. T., Clemens T., Young M., Yang Y. (2008) Dev. Cell 14, 674–688 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.