Abstract
Heparanase activity is highly implicated in cell dissemination associated with tumor metastasis, angiogenesis, and inflammation. Heparanase expression is induced in many hematological and solid tumors, associated with poor prognosis. Heparanase homolog, termed heparanase 2 (Hpa2), was cloned based on sequence homology. Detailed characterization of Hpa2 at the biochemical, cellular, and clinical levels has not been so far reported, and its role in normal physiology and pathological disorders is obscure. We provide evidence that unlike heparanase, Hpa2 is not subjected to proteolytic processing and exhibits no enzymatic activity typical of heparanase. Notably, the full-length Hpa2c protein inhibits heparanase enzymatic activity, likely due to its high affinity to heparin and heparan sulfate and its ability to associate physically with heparanase. Hpa2 expression was markedly elevated in head and neck carcinoma patients, correlating with prolonged time to disease recurrence (follow-up to failure; p = 0.006) and inversely correlating with tumor cell dissemination to regional lymph nodes (N-stage; p = 0.03). Hpa2 appears to restrain tumor metastasis, likely by attenuating heparanase enzymatic activity, conferring a favorable outcome of head and neck cancer patients.
Keywords: Enzymes, Extracellular Matrix Proteins, Heparin, Heparin-binding Protein, Immunochemistry, Heparanase, Head and Neck Carcinoma, Heparanase 2, Metastasis, Prognosis
Introduction
Enzymatic activity of the endo-β-d-glucuronidase heparanase has been recognized for nearly three decades (1, 2). Heparanase activity is considered a prerequisite for cellular invasion associated with tumor metastasis, inflammation, and angiogenesis, a consequence of heparan sulfate (HS)2 cleavage and remodeling of the subendothelial and subepithelial basement membrane and extracellular matrix (ECM) (3, 4). The clinical significance of heparanase activity critically emerges from numerous recent publications describing induced heparanase expression in human hematological and solid tumors, and its inverse correlation with post-operative patient survival, encouraging the development of heparanase inhibitors (5–9). Cloning of a single human heparanase cDNA sequence was independently reported by several groups (10–13) implying that one active heparanase enzyme exists in mammals. Further analysis of human genomic DNA led researchers to conclude that the heparanase gene is unique and that the existence of related proteins is unlikely (14). Based on amino acid sequences, McKenzie et al. (14) nonetheless reported the cloning of heparanase homolog termed heparanase 2 (Hpa2). Hpa2 is predicted to encode three proteins generated by alternative splicing (Hpa2a, Hpa2b, Hpa2c) and shares an overall identity of ∼40% with heparanase. Detailed characterization of Hpa2 at the biochemical, cellular, and clinical levels has not been so far reported, and its role in normal physiology and pathological disorders is obscure. Utilizing purified Hpa2c protein (wild type Hpa2) and newly generated Hpa2-specific polyclonal and monoclonal antibodies, we show that Hpa2c is secreted and exhibits high affinity toward heparin and HS but not hyaluronic acid (HA). Hpa2c binds cell membrane syndecan-1 and -4 but, unlike heparanase, failed to get internalized and remains on the cell surface for relatively long period of time. Notably, Hpa2c inhibits heparanase enzymatic activity, likely due to its high affinity to heparin and HS and its ability to associate physically with heparanase. Hpa2c expression correlates with reduced tumor metastasis in head and neck cancer patients.
MATERIALS AND METHODS
Antibodies and Reagents
Anti-syndecan-1 (sc-5632), anti-syndecan-4 (sc-12766), anti-bFGF (sc-79), anti-calnexin (sc-11397), and anti-Myc tag (sc-40) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Monoclonal antibody to heparanase was kindly provided by ImClone Systems Inc. (New York). Anti-actin monoclonal antibody, heparin, heparin-Sepharose, HA, wheat germ agglutinin-FITC conjugate, and tunicamycin were purchased from Sigma. Basic FGF was purchased from PeproTech (Rocky Hill, NJ). Anti-heparanase polyclonal antibodies 733 and 1453 were described previously (15). Antibody 58 was raised against a peptide (43DRRPLPVDRAAGLKEKT59) mapped at the N terminus of Hpa2. A cysteine residue was added after threonine 59 to enable efficient coupling of the peptide to maleimide-activated keyhole limpet hemocyanin, according to the manufacturer's (Pierce) instructions. This peptide was preferred because it exhibits minimal sequence homology with heparanase (14), and the respective antibody is expected to recognize the wild type and alternatively spliced Hpa2. The keyhole limpet hemocyanin-conjugated peptide was injected into rabbits, and antibody specificity was evaluated by ELISA, immunoblotting, and immunostaining. Monoclonal antibodies 1C7 and 20C5 were raised against Hpa2c protein purified from the conditioned medium of Hpa2c-transfected HEK293 cells, essentially as described (16). Hpa2a, Hpa2b, and Hpa2c cDNAs were kindly provided by Dr. Edward McKenzie (University of Manchester, Manchester, UK) (14) and subcloned into Myc-tagged mammalian expression vector pcDNA3.1. Heparanase and Hpa2c proteins were purified from the conditioned medium of stably transfected HEK293 cells essentially as described (17, 18).
Cell Culture, Transfection, Immunoprecipitation, and Immunoblotting
Human U87-MG glioma, Cal27 tongue carcinoma, and HEK293 cells were purchased from the American Type Culture Collection (ATCC). JSQ-3 nasal vestibule carcinoma cells were kindly provided by Dr. Ralph Weichselbaum (University of Chicago, Chicago, IL) (19). Cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with glutamine, pyruvate, antibiotics, and 10% fetal calf serum in a humidified atmosphere containing 5% CO2 at 37 °C. For stable transfection, cells were transfected with pcDNA3 vectors containing Myc-tagged full-length (Hpa2c) and splice variants (Hpa2a, 2b) using the FuGENE reagent according to the manufacturer's (Roche Applied Science) instructions, selected with G418 (400–800 μg/ml) for 2 weeks, expanded, and pooled. Protein expression and secretion were evaluated by immunoblotting, essentially as described (20, 21). Briefly, cells were incubated with the indicated concentrations of heparin, HS, or HA for 24 h under serum-free conditions. The medium was collected, and after three washes with ice-cold PBS, cell extracts were prepared using a lysis buffer containing 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 0.5% Triton X-100, supplemented with a mixture of protease inhibitors (Roche Applied Science). Protein concentration was determined (Bradford reagent, Bio-Rad), and 50 μg of protein and the equivalent volume of medium were resolved by SDS-PAGE under reducing conditions. After electrophoresis, proteins were transferred to PVDF membrane (Bio-Rad) and probed with the appropriate antibody followed by HRP-conjugated secondary antibody (Jackson ImmunoResearch, West Grove, PA) and an enhanced chemiluminescent substrate (Pierce) (20–22). Immunoprecipitation of heparanase and Hpa2c was carried out essentially as described (22) except that magnetic beads (30 μl; Dynabeads Protein G, Invitrogen) were applied to capture the antibody. Briefly, heparanase and Myc-tagged Hpa2c proteins (0.5 μg each in 0.5 ml of serum-free medium) were allowed to interact for 2 h at room temperature. Anti-heparanase monoclonal antibody coupled to protein G-magnetic beads was then added for 10 min; beads were collected by centrifugation and washed 3 times with PBS supplemented with 300 mm NaCl and 5% sucrose. Sample buffer was then added, and after boiling for 5 min, samples were subjected to electrophoresis and immunoblotting as described above.
Uptake Studies
Proteins (latent 65-kDa heparanase; constitutively active (GS3), Hpa2c, bFGF) were added to confluent cell cultures at a concentration of 1–5 μg/ml under serum-free conditions. At the indicated time points, the medium was aspirated, cells were washed twice with ice-cold PBS, and total cell lysates were prepared as described above. Protein uptake was analyzed by immunoblotting, applying the relevant antibody as described (15, 23). In acid wash studies, protein uptake was performed at 37 or 4 °C for 2 h. Cells were then washed with cold PBS and left untreated or subjected to three washes (1 min each) with serum-free medium adjusted to pH 2.5 (acid wash) to remove proteins tethered to the cell surface. Cell lysates were then prepared and subjected to immunoblotting as described above.
Purified proteins (3 μg) were incubated with heparin-Sepharose beads (30 μl) suspended in 1 ml of PBS for 30 min. Beads were then centrifuged (14,000 × g) and washed twice with cold PBS containing 0.3 m NaCl. Bound proteins were eluted stepwise by increased NaCl concentrations. Elution samples were then subjected to SDS-PAGE followed by Coomassie Blue staining.
Flow Cytometry
HEK293 cells were left untreated as a control or incubated with Myc-tagged Hpa2c (1 μg/ml, 30 min, 37 °C). Cells were detached with EDTA, centrifuged at 1000 rpm for 4 min, washed with PBS, and counted. Cells (2 × 105) were resuspended in PBS containing 1% FCS and incubated with anti-Myc or anti-Hpa2 monoclonal antibody for 30 min on ice. Cells were then washed twice with PBS, incubated with Cy2-conjugated secondary antibody, washed, and analyzed using a FACSCalibur fluorescent activated cell sorter and CellQuest software (BD Biosciences) as described (18).
Heparanase Enzymatic Activity
Preparation of ECM-coated 35-mm dishes and determination of heparanase activity were performed as described in detail elsewhere (20, 22). To evaluate inhibition by Hpa2c, active heparanase (50 ng) was applied onto 35S-labeled ECM without or with purified Hpa2c protein. The incubation medium (1 ml) containing sulfate-labeled degradation fragments was subjected to gel filtration on Sepharose CL-6B column. Fractions (0.2 ml) were eluted with PBS, and their radioactivity was counted in a β-scintillation counter. Degradation fragments of HS side chains are eluted at 0.5 < Kav < 0.8 (peak II, fractions 10–25). Nearly intact HS proteoglycans (HSPGs), liberated by proteolytic enzymes residing in the ECM and degrading the proteoglycan core protein, are eluted just after the V0 (Kav < 0.2, peak I). Each experiment was performed at least three times, and the variation in elution positions (Kav values) did not exceed ±15%.
Immunocytochemistry
Cells were left untreated or incubated with exogenously added heparanase or Hpa2c and subjected to indirect immunofluorescence staining, essentially as described (15, 23). Briefly, cells were fixed with cold methanol for 10 min. Cells were then washed with PBS and incubated in PBS containing 10% normal goat serum for 1 h at room temperature followed by a 2-h incubation with the indicated primary antibodies. Cells were then extensively washed with PBS and incubated with the relevant (Cy2/Cy3-conjugated) secondary antibody (Jackson ImmunoResearch) for 1 h, washed, and mounted (Vectashield; Vector, Burlingame, CA). Immunocytochemistry was similarly performed on live cells at 37 °C; cells were incubated with exogenously added Hpa2c protein for 30 min, medium was aspirated, and after washes with PBS cells were incubated with the appropriate antibody for 1 h. Cells were then washed with ice-cold PBS, fixed with 4% paraformaldehyde, and subjected Cy2/Cy3-conjugated secondary antibody, washed, and mounted as above. Staining was visualized by confocal microscopy, as described (21).
Immunohistochemistry
Staining of formalin-fixed, paraffin-embedded 5-μm sections for Hpa2 was performed essentially as described (24). Hpa2 staining was scored according to the intensity of staining (0, none; +1, weak-moderate; +2, strong) and the percentage (extent staining) of tumor cells that were stained. The extent of staining was further categorized as low (0, <10%), moderate (+1, 10–50%), and high (+2, >50%). Specimens that were similarly stained with pre-immune serum or in which the above procedure was applied but lacked the primary antibody yielded no detectable staining.
Statistical Analysis
Univariate association between Hpa2 immunostaining (intensity and extent) and clinical and pathological parameters as well as patient outcome were analyzed using χ2 tests (Pearson, Fisher exact test) as described (24).
RESULTS
Hpa2 Is Not Subjected to Proteolytic Processing and Lacks Enzymatic Activity Typical of Heparanase
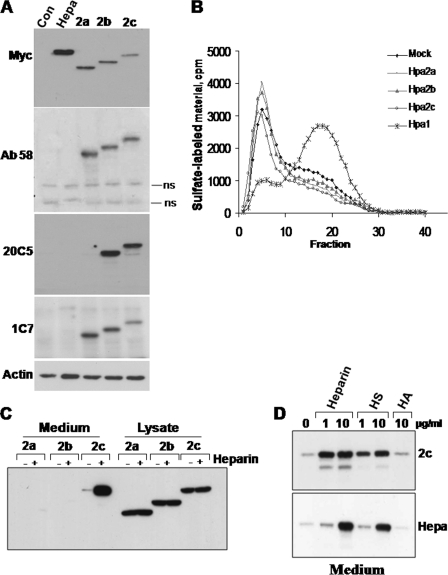
Sequence alignment analysis revealed 44% identity and 59% resemblance between wild type Hpa2 (Hpa2c) and heparanase (supplemental Fig. 1A). Heparin binding domains (Lys158–Asp171 and Pro271–Met278) critical for the interaction of heparanase with its HS substrate (20) are not well conserved (supplemental Fig. 1A). This and the questionable existence of a signal peptide required for Hpa2 protein secretion (14) suggest that heparanase and Hpa2 are functionally distinct. We subcloned each of the Hpa2 splice variants (2a, 2b and 2c; supplemental Fig. 1B) (14) in Myc-tagged mammalian expression vector pcDNA3.1 and examined protein expression after transfection of 293 cells. All three Hpa2 splice variants were readily detected and appeared as a single band at the expected molecular weight (Fig. 1A, upper panel). We have reported previously that heparanase is cleaved at its C terminus, resulting in loss of the introduced Myc-tag before its proteolytic processing and the formation of an active 8- and 50-kDa heterodimer (23, 25). Thus, Hpa2 processing may not be detected by anti-Myc antibodies. To verify this aspect, we generated a polyclonal antibody (Ab58) directed against a peptide (Asp43–Thr59) mapped at the protein N terminus and, hence, expected to recognize all three Hpa2 variants (supplemental Fig. 1B). In addition, the peptide exhibits minimal sequence homology with heparanase (27). Subjecting cell lysates to immunoblotting applying Ab58 confirmed its specificity for Hpa2 (i.e. no cross-reactivity with heparanase) and resulted in a band pattern similar to that obtained with the anti-Myc antibody (Fig. 1A, second panel). Furthermore, two monoclonal antibodies (20C5, 1C7) raised against Hpa2c yielded comparable results (Fig. 1A, third and fourth panels), indicating that Hpa2 proteins are not subjected to proteolytic processing. Corresponding lysate samples were evaluated for heparanase enzymatic activity. Although the release of 35S-labeled HS was markedly enhanced after transfection with heparanase (Hpa1; Fig. 1B), no such activity was evident in cells transfected with Hpa2 splice variants (Fig. 1B), indicating that Hpa2 is devoid of enzymatic activity typical of heparanase.
FIGURE 1.
Hpa2 expression and secretion. A, an immunoblot analysis is shown. HEK293 cells were stably transfected with the indicated Myc-tagged heparanase, Hpa2 variant, or control empty plasmid (Con), and total cell lysates were subjected to immunoblotting applying anti-Myc (upper panel), anti-Hpa2 (Ab58, second panel; 20C5, third panel; 1C7, fourth panel), or anti-actin (lower panel) antibodies. B, heparanase enzymatic activity is shown. Corresponding cell lysates were incubated with sulfate-labeled ECM. Labeled material released into the incubation medium was applied for gel filtration on Sepharose 6B and analyzed as described under “Materials and Methods.” Note the release of labeled degradation fragments eluted in fractions 15–30 after transfection with heparanase (×) but not Hpa2 cDNAs. C, secretion is shown. HEK293 cells stably expressing Hpa2 variants were cultured in serum-free medium without (−) or with heparin (+; 10 μg/ml) for 20 h. Cell lysates (50 μg) and corresponding volumes of conditioned medium samples were subjected to immunoblotting applying anti-Myc tag antibody. D, the effect of glycosaminoglycans is shown. HEK293 cells stably expressing Hpa2c (2c; upper panel) or heparanase (Hepa; lower panel) were incubated without (0) or with the indicated concentrations of heparin, HS, or HA, and the conditioned medium was subjected to immunoblotting as above. ns, nonspecific band.
Hpa2c Is Secreted, Exhibits High Affinity to Heparin and HS, and Inhibits Heparanase Activity
Treatment with tunicamycin resulted in decreased molecular weight of Hpa2 splice variants (not shown) and immunofluorescent staining localized Hpa2 in the endoplasmic reticulum (ER) (supplemental Fig. 2), suggesting that Hpa2 proteins are glycosylated and directed to the secretory pathway. To better appreciate Hpa2 secretion, conditioned medium and lysate samples were collected from stably transfected 293 cells and subjected to immunoblotting applying anti-Myc antibody. Although all three Hpa2 splice variants were abundantly expressed and readily detected in the cell lysates (Fig. 1C, Lysate), only Hpa2c protein was detected in the cell-conditioned medium (Fig. 1C, Medium, 2c, −). Notably, accumulation of Hpa2c extracellularly was markedly enhanced after the addition of heparin to the culture medium (Fig. 1C, Medium, 2c,+). This suggests that Hpa2c retains the capacity to interact with heparin and likely HS. To compare heparanase and Hpa2c for their affinity to heparin/HS, stably transfected cells were incubated with increasing concentrations of heparin, HS, or HA. Medium samples were collected and subjected to immunoblotting with anti-Myc antibody (Fig. 1D). As noted previously (20, 23), heparanase accumulated in the culture medium to high levels after the addition of 10 μg/ml heparin or HS but not HA (Fig. 1D, lower panel). Notably, Hpa2c accumulated in the culture medium to a similar extent already in the presence of a 10-fold lower concentration (1 μg/ml) of heparin or HS (Fig. 1D, upper panel). This finding suggests that Hpa2c exhibits a higher affinity toward heparin/HS than heparanase, as further demonstrated by the following experiments. Because only Hpa2c is found secreted, accumulates in the culture medium after the addition of low concentrations of heparin (1 μg/ml), and could be purified from the culture medium of stably transfected cell (see below), further studies mostly focused on this protein.
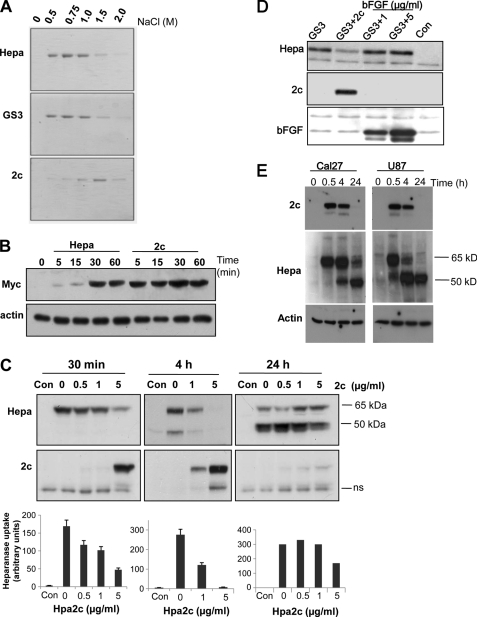
Purified heparanase or Hpa2c proteins were allowed to interact with heparin-Sepharose beads and were then eluted stepwise by increased salt concentrations. The latent (65 kDa) and constitutively active (GS3) (28) heparanase proteins were eluted with 0.5–1 m NaCl (Fig. 2A, upper and second panels), whereas higher NaCl concentrations (1.5 m) were required to elute Hpa2c from the heparin beads (Fig. 2A, lower panel). After exogenous addition to U87 glioma cells, heparanase was detected in the cell lysate after 30 min (Fig. 2B, upper panel, Hepa) (15, 20, 23, 29, 30). Intriguingly, cellular binding of Hpa2c was significantly faster than that of heparanase (Fig. 2B, 2c). Maximal Hpa2c binding was evident already 5 min after its addition, and cellular levels remained stable for 60 min (Fig. 2B, 2c). Similar binding kinetics was seen also in 293 cells (not shown). These results indicate that Hpa2c exhibits a higher affinity toward heparin/HS than heparanase and, thus, may interfere with the interaction of heparanase with its HS substrate.
FIGURE 2.
Hpa2c possesses higher affinity than heparanase toward heparin/HS. A, heparin affinity binding is shown. Purified latent (65 kDa) heparanase, constitutively active (GS3) heparanase, or Hpa2c proteins (3 μg/ml) were incubated with heparin-Sepharose beads for 30 min on ice. After washes with cold PBS containing 0.3 m NaCl, bound proteins were eluted with the indicated concentrations of NaCl. Eluted samples were resolved by SDS-PAGE and stained with Coomassie Blue. B, protein uptake is shown. U87 glioma cells were left untreated (0) or incubated with Myc-tagged heparanase (Hepa) or Hpa2c (2c) proteins (1 μg/ml) for the times indicated. Cell lysate samples were subjected to immunoblotting, applying anti-Myc (upper panel) or anti-actin (lower panel) antibodies. C and D, Hpa2c competes with heparanase uptake. C, Cal27 tongue carcinoma cells were incubated without (Con) or with latent 65-kDa heparanase (1 μg/ml) in the absence (0) or presence of the indicated concentrations of Myc-tag Hpa2c protein. Lysate samples were prepared after 30 min (left panels), 4 h (middle panels), or 24 h (right panels) and subjected to immunoblotting applying anti-heparanase (Hepa; upper panels) or anti-myc antibodies (2c; lower panels). Densitometry analysis of heparanase uptake is presented at the lower panels. D, U87 glioma cells were similarly left untreated (Con) or incubated (30 min, 37 °C) with constitutively active heparanase (1 μg/ml) without (GS3) or with Hpa2c (1 μg/ml; GS3 + 2c) or with the indicated concentration of bFGF (GS3 + 1, GS3 + 5). Lysate samples were subjected to immunoblotting with anti-heparanase (Hepa; upper panel), anti-Myc (2c; second panel), and anti-bFGF (third panel) antibodies. E, top panel, Hpa2c (1 μg/ml) was incubated with Cal27 tongue carcinoma (left panels) or U87 glioma cells (right panels) for the times indicated. Lysate samples were then prepared and subjected to immunoblotting applying anti-Hpa2 20C5 antibody. Cells were similarly incubated with heparanase (1 μg/ml), and lysate samples were immunoblotted with anti-heparanase (Hepa, middle panels) or anti-actin (lower panels) antibodies. ns, nonspecific band.
To examine this possibility, we evaluated cellular uptake of heparanase in the absence or presence of Hpa2c. Cal27 tongue carcinoma (Fig. 2C), JSQ-3 nasal carcinoma, U87 glioma, and 293 (supplemental Fig. 3B) cells were left untreated (Con) or were incubated with heparanase (1 μg/ml) in the absence (0) or presence on increasing concentrations of Hpa2c. At the times indicted, cells were extensively washed, and total cell lysates were subjected to immunoblotting. As demonstrated in Fig. 2C, Hpa2c inhibited uptake of heparanase in a dose-dependent manner. Reduced level of latent (65 kDa) (Fig. 2C, left panel; 30 min) and processed (50 kDa) heparanase (Fig. 2C, middle panel; 4 h) was evident already in the presence of equal concentration (1 μg/ml) of Hpa2c and even more so when Hpa2c concentration was elevated to 5 μg/ml (Fig. 2C, left and middle panels). Similarly, Hpa2c inhibited the uptake of constitutively active heparanase (GS3, Fig. 2D), whereas basic FGF, a typical heparin binding growth factor, did not compete with heparanase even at an ∼10-fold higher molar concentration (Fig. 2D, bFGF, 5 μg/ml). Importantly, the activity of purified recombinant heparanase (GS3) was markedly attenuated by Hpa2c (Fig. 3A, GS3 + 2c), whereas irrelevant protein such as BSA had no effect (Fig. 3A, GS3+BSA). Similarly, decreased activity of heparanase was evident after the addition of Hpa2c protein to Cal27 cells (Fig. 3A, right), likely due to its preferential interaction with HS and interference with uptake and processing of the endogenous enzyme (23). Notably, cell-associated Hpa2c levels were markedly decreased 24 h after its addition (Fig. 2C, right lower panel; 24 h). Concomitantly, its inhibitory capacity toward uptake of heparanase was markedly attenuated (Fig. 2C, right upper panel) evident by high levels of active (50 kDa) heparanase protein accumulating at this time point (Fig. 2C, right upper panel). Diminished levels of Hpa2c in Cal27 (Fig. 2, C, right lower panel, E, upper left panel) and U87 cells (Fig. 2E, upper right panel) 24 h after its addition was in contrast to accumulation of the processed 50-kDa heparanase at this time point (Fig. 2E, middle panels), suggesting a specific regulatory mechanism that dictates the levels of cell-associated Hpa2c.
FIGURE 3.
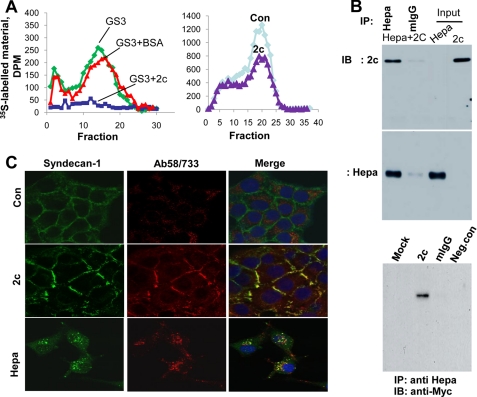
Hpa2c associates with heparanase and inhibits its enzymatic activity. A, heparanase activity is shown. Active heparanase (GS3, 50 ng) was applied onto culture dishes coated with 35S-labeled ECM for 4 h without (GS3) or with BSA or Hpa2c (1 μg/ml). Release of sulfate-labeled material eluted in fractions 10–25 was evaluated as a measure of heparanase activity, as described under “Materials and Methods” (left panel). Cal27 tongue carcinoma cells were grown for 2 days without (Con) or with Hpa2c protein (0.5 μg/ml); cells were subjected to three cycles of freeze/thaw, and heparanase activity in cell lysates was determined as above (right panel). B, co-immunoprecipitation is shown. Purified active heparanase (GS3) and Myc-tagged Hpa2c proteins (0.5 μg/0.5 ml) were incubated for 2 h at RT and were then immunoprecipitated with control mouse IgG (mIgG) or anti-heparanase monoclonal antibody coupled to magnetic beads. After three washes with PBS supplemented with 0.02% Tween 20, sample buffer was added, and after boiling the supernatant was subjected to immunoblotting applying anti-Myc (upper panel) or anti-heparanase (middle panel) monoclonal antibodies. Purified GS3 and Hpa2c (100 ng) were run in parallel as a reference to immune precipitation (IP) efficiency. 293 cells stably expressing heparanase were transiently transfected with Myc-tagged Hpa2c or with an empty vector (Mock). Lysate samples (200 μg) were immunoprecipitated with anti-heparanase antibody applying the ProFound co-immunoprecipitation kit according to the manufacturer's (Pierce) instructions followed by immunoblotting (IB) with anti-Myc antibody. Mouse IgG (mIgG) or uncoated beads (Neg. con) were included as controls for antibody specificity (lower panel). C, syndecan clustering. Cal27 tongue carcinoma cells were left untreated (Con, upper panels) or incubated with Hpa2c (second panels) or heparanase (lower panels) proteins for 2 h. Cells were then fixed and subjected to immunofluorescent staining applying anti-syndecan-1 (left panels, green) and anti-Hpa2 (Ab58; middle panel, red) or anti-heparanase (733; middle lower panel, red) antibodies. Merge images are shown in the right panels. Yellow designates co-localization. Note that Hpa2 clusters syndecan-1 but fails to get internalized, whereas heparanase appears in endocytic vesicles, co-localizing with syndecan-1.
We further suspected that Hpa2 may interfere with heparanase activity by means other than competition for substrate (HS) binding and examined a possible physical association between heparanase and Hpa2c. We first examined the association between purified heparanase and Hpa2c proteins applying immune precipitation. Indeed, after incubation of purified heparanase and Hpa2c proteins, immunoprecipitation with a monoclonal antibody to heparanase precipitated Myc-tagged Hpa2c, whereas control mouse IgG did not (Fig. 3B, upper panel). To examine the interaction between heparanase and Hpa2c in the context of live cells, 293 cells stably expressing heparanase were transiently transfected with an empty vector (mock) or Myc-tagged Hpa2c, and lysate samples were immunoprecipitated with anti-heparanase antibody followed by immunoblotting with anti-Myc antibody (Fig. 3B). Hpa2c was found to be associated with heparanase (Fig. 3B, lower panel). No reactivity was observed in control mock-transfected cells (Fig. 3B, lower panel; Mock) or when a control antibody was applied (Fig. 3B, lower panel; mIgG), ensuring the specificity of the assay. Thus, secreted Hpa2c can inhibit heparanase activity by competing for HS and can associate physically with heparanase and thereby modulate its enzymatic and possibly non-enzymatic functions.
Hpa2c Is Retained on the Cell Surface and Fails to Get Internalized
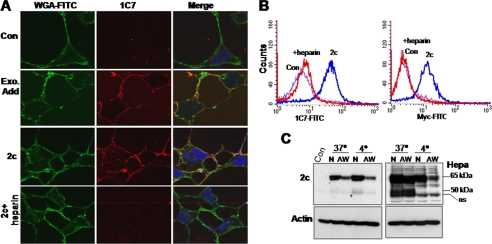
Immunofluorescent staining illustrates Hpa2c localization on the cell surface after its exogenous addition (Fig. 3C, Ab58), co-localizing with and clustering of syndecan-1, a class of cell membrane HSPGs (Fig. 3C, 2c, merge). Hpa2c similarly co-localized with and clustered syndecan-4 (supplemental Fig. 3A). Unlike heparanase (Fig. 3C, lower panel, Ab733), Hpa2c does not appear to get internalized into endocytic vesicles but, rather, remains on the cell surface (Fig. 3C, 2c). To ascertain membrane localization of Hpa2c, we applied the following experiments. We exogenously added Hpa2c protein to 293 cells and subjected live cells (i.e. without fixation and/or permeabilization) to immunofluorescent staining, applying newly generated anti-Hpa2 monoclonal antibody 1C7. The staining clearly shows peripheral Hpa2c (Fig. 4A, second panel, middle, red), partially co-localizing with wheat germ agglutinin-FITC (a cell membrane marker; Fig. 4A, second panel, merge). Membrane localization of Hpa2c was similarly demonstrated in stably transfected 293 cells (Fig. 4A, third panels), whereas no reactivity was observed in control mock-transfected cells (Fig. 4A, Con, upper panels). Furthermore, reactivity with antibody 1C7 is diminished in stably transfected cells incubated with heparin (Fig. 4A, lower panels). Membrane localization was further demonstrated by FACS analysis of cells incubated with purified Hpa2c applying anti-myc (Fig. 4B, right panel) or 1C7 (Fig. 4B, left panel) antibodies. Notably, cell-associated Hpa2c levels were markedly reduced after the addition of heparin (Fig. 4B, +heparin), further indicating that heparin competes with cell surface HS for Hpa2c binding. We next employed an acid wash to dissociate proteins tethered on the cell surface. 293 cells were incubated with purified heparanase or Hpa2c for 2 h at 37 or 4 °C and were then left untreated as control or washed 3 times (1 min each) with medium adjusted to pH 2.5 (acid wash). Total cell lysates were then subjected to immunoblotting applying anti-Hpa2 (Fig. 4C, upper left) or anti-heparanase (Fig. 4C, upper right) antibody. Hpa2c levels were markedly reduced after acid wash (Fig. 4C, upper panel, 2c). In contrast, heparanase levels were only modestly affected by acid wash once cells were incubated at 37 °C (Fig. 4C, upper right, Hepa) due to efficient internalization, evident by formation and accumulation of the processed 50-kDa active form (Fig. 4C, upper panel, Hepa, 37 °C). Heparanase levels were nonetheless significantly decreased by acid wash once uptake was carried out at 4 °C, when protein internalization is prevented, indicated by the lack of heparanase processing and absence of the 50-kDa subunit (Fig. 4C, Hepa, 4 °C). These results suggest that Hpa2c interacts with membrane HSPG and remains on the cell surface for relatively long periods of time. Hpa2c is not internalized but rather is being degraded on the cell surface or shed together with the associated HSPG (i.e. syndecan); the addition of heparin sequesters Hpa2c and prevents its degradation/shedding, resulting in accumulation of the protein in the cell media (Fig. 1, C and D).
FIGURE 4.
Hpa2c is localized to the plasma membrane. A, immunofluorescent staining of live cells is shown. 293 cells were left untreated as control (Con; upper panels) or incubated with Myc-tagged Hpa2c protein (1 μg/ml) for 30 min (second panels). After three washes with PBS, cells were incubated with anti-Hpa2 monoclonal antibody (1C7; 1 μg/ml, middle panel, red) for 60 min at 37 °C, washed, and incubated with wheat germ agglutinin (WGA)-FITC for 1 min to label the plasma membrane (left panels, green), fixed with 4% paraformaldehyde (15 min, RT), and incubated with Cy3-conjugated goat anti-mouse IgG secondary antibody. After three washes with PBS, slides were mounted and analyzed by confocal microscopy. Similar staining procedure was applied for 293 cells stably expressing Hpa2c and incubated without (third panels, 2c) or with heparin (fourth panels). Note that membrane localization of Hpa2c is diminished after incubation with heparin. B, FACS analysis is shown. 293 cells were left untreated as control (Con) or were incubated with Hpa2c protein (1 μg/ml, 30 min, 37 °C). Cells were then washed and left untreated (2c) or incubated with heparin (10 μg/ml) for 1 h. Cells were then dissociated by EDTA and subjected to FACS analysis applying anti-Hpa2 (1C7, left) or anti-Myc (right) antibodies. C, an acid wash is shown. 293 cells were left untreated as control (Con) or incubated with Hpa2c (2c, left) or heparanase (Hepa, right) for 2 h at 37 or at 4 °C. Cells were then left untreated (N) or washed three times (1 min each) with DMEM adjusted to pH 2.5 (AW). Cell lysates were prepared and subjected to immunoblotting applying anti-Hpa2 (2c; upper left), anti-heparanase (Hepa; upper right), or anti-actin (lower panel) antibodies.
Hpa2 Is Overexpressed in Head and Neck Carcinoma and Inversely Correlates with Tumor Metastasis
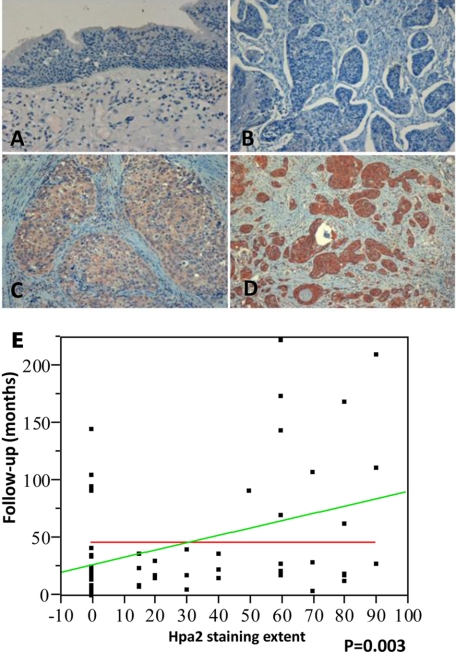
Having demonstrated the specificity (i.e. no cross-reactivity with heparanase; supplemental Fig. 3C) and suitability of Ab58 for immunohistochemical analysis, we next investigated the clinical significance of Hpa2. We have previously utilized head and neck tumor biopsies to study the clinical relevance of heparanase for this type of cancer. We reported (24) that heparanase expression is induced in the majority of cases (86%), inversely correlating with patient survival due, in part, to enhanced tumor metastasis to regional and distant lymph nodes (N-stage). We subjected this cohort of specimens to immunostaining with anti-Hpa2 antibody (Ab58) and correlated the intensity and extent (i.e. percent of positively stained cells) of staining with clinical parameters (24). Clinical description of the patients is presented in supplemental Table 1. Normal tissue adjacent to the tumor lesion stained negative for Hpa2 (Fig. 5A) in all of the specimens. Among the 58 biopsies available for staining, 25 tumor lesions (43%) stained negative for Hpa2 (Fig. 5B), and 33 (57%) were positive. The Hpa2-positive group was further categorized according to the intensity and extent of staining. Thus, weak staining (+1; Fig. 5C) was found in 60% (20/33) of positive specimens, whereas 40% (13/33) stained strongly (+2; Fig. 5D) for Hpa2. According to the extent criteria, 42% (14/33) of specimens that positively stained for Hpa2 were scored as low extent (+1), and 58% (19/33) were scored as a high extent (+2). No correlation was found between Hpa2 staining and tumor grade or tumor size (T-stage). Notably, an inverse correlation was found between Hpa2 staining extent and cell dissemination to lymph nodes (patient N-stage) (Table 1). Thus, 57% of patients that stained negative (0) for Hpa2 were diagnosed to have advanced metastases to lymph nodes (two or more infected lymph nodes; N2–3) compared with 13% of patients that were scored as high Hpa2 extent (+2), a decrease that is statistically significant (p = 0.03; Table 1). Likewise, an association was found between Hpa2 staining and the time to disease recurrence (follow-up to failure). Hence, while in patients stained negative for Hpa2, disease reappeared after 33.4 months on average, the time before disease reappearance was prolonged to 77.7 months for patients exhibiting a high Hpa2 staining extent, differences that are statistically highly significant (p = 0.006; Table 1). Furthermore, we found a linear association between Hpa2-staining extent and patient follow-up (=27.4 + 0.6 × Hpa2 extent (%); Fig. 5E), clearly pointing to Hpa2 expression levels as a favorable determinant in head and neck carcinoma.
FIGURE 5.
Immunohistochemical staining of Hpa2 in human head and neck tumor specimens. Formalin-fixed, paraffin-embedded 5-μm sections of 58 head and neck tumors were subjected to immunostaining, applying anti-Hpa2 polyclonal antibody (Ab58), as described under “Materials and Methods.” Shown are representative photomicrographs of normal-looking tissue adjacent the carcinoma lesion (A) and carcinoma specimens stained negative (B) or positive for Hpa2 and scored as weak (C; +1) and strong (D; +2) intensity. Original magnifications, ×50. A linear, statistically significant association (p = 0.003) between Hpa2 staining extent and patient survival is shown in E.
TABLE 1.
Hepa2 staining extent correlates with reduced lymph node metastasis and prolonged follow-up
| Parameter | HPA-2 staining extent |
||||
|---|---|---|---|---|---|
| 0 | 1 | 2 | Total | p | |
| Stage | 0.11 | ||||
| Stage II | 3 (33%) | 1 (11%) | 5 (56%) | 9 | |
| Stage III | 4 (24%) | 6 (35%) | 7 (41%) | 17 | |
| Stage IV | 18 (56%) | 7 (22%) | 7 (22%) | 32 | |
| N stage | 0.03 | ||||
| N0–1 | 12 (34%) | 7 (20%) | 16 (46%) | 35 | |
| N2–3 | 13 (57%) | 7 (30%) | 3 (13%) | 23 | |
| Follow-up time (months) | 33.4 | 26.9 | 77.7 | 0.006 | |
DISCUSSION
Enzymatic activity of heparanase is highly implicated in tumor metastasis, a notion that is now supported experimentally (31) and clinically (3, 4). Attempts to inhibit heparanase were initiated in parallel with the emergence clinical relevance of this activity, focusing on heparin and heparin mimetics, which bind heparanase with high affinity but serve as poor substrates (9, 32). Two types of endogenous inhibitors of heparanase have so far been reported, namely disaccharide end products of HS cleavage (33) and highly basic proteins such as eosinophil major basic protein (34). The relevance of this protein to human cancer progression is nonetheless questionable. Our results identify Hpa2 as a candidate endogenous inhibitor of heparanase activity. This conclusion emerges from our in vitro studies in which purified Hpa2c protein was capable of inhibiting the enzymatic activity of both recombinant and endogenous heparanase (Fig. 3A). The clinical relevance of Hpa2 decisively emerges from the inverse correlation found between Hpa2 staining extent and tumor metastasis (patient N-stage; Table 1). Although Hpa2 expression was undetected in normal epithelium adjacent to the tumor lesion (Fig. 5A), it appeared to be up-regulated in 57% of head and neck lesions. This is in agreement with a recent publication reporting increased Hpa2 expression in colorectal and ovarian carcinomas (35, 36). Clearly, Hpa2 expression was associated with reduced lymph nodes metastasis and prolonged follow-up time of head and neck cancer patients (Table 1; Fig. 5E), likely a consequence of heparanase inhibition. This notion is based on previous studies correlating high levels of heparanase with poor outcome of head and neck (24), nasopharynx (37), salivary gland (38), and tongue (39) cancer patients, among many other carcinomas (3). An anti-metastatic feature of Hpa2 is also supported by studies of the Polycomb protein EZH2. EZH2 is an important component of a multiprotein complex that methylates histone protein 3, leading to the repression of target genes (40). EZH2 is overexpressed in metastatic breast and prostate cancers and is a marker for aggressive disease (41). Thus, genes repressed by EZH2 are expected to function as tumor suppressors and to be associated with favorable prognosis. Interestingly, the Polycomb repression signature genes identified Hpa2 among the genes repressed by Polycomb proteins (42). Moreover, the Polycomb repression signature is thought to predict the clinical outcome of multiple solid tumors (42), suggesting that anti-metastatic features of Hpa2 are not limited to head and neck carcinoma. Studies examining Hpa2 levels in other human hematological and solid tumors are currently in progress.
Although the overall sequence resemblance between heparanase and Hpa2c is 59% (supplemental Fig. 1), elements critical for heparanase enzymatic activity are not well conserved. Particularly, complete removal of a linker segment (Ser110–Gln157) by cathepsin L is absolutely required to render the heparanase active site accessible for HS substrate binding (43, 44). Notably, this region shares the lowest sequence homology between heparanase and Hpa2c (supplemental Fig. 1) (14, 27). Moreover, amino acid residue (Tyr156) critical for heparanase processing, and removal of the linker segment by cathepsin L (43, 44) is not conserved in Hpa2c. Indeed, Hpa2 does not seem to undergo proteolytic processing. This was confirmed by applying antibodies directed against epitopes localized at the protein C (Myc tag) and N- (Ab58) terminus as well as novel monoclonal antibodies directed against Hpa2 (20C5, 1C7; Fig. 1A). As expected given the above consideration, Hpa2 lacks HS-degrading activity evaluated at acidic (pH 5.8; Fig. 1B) and physiological pH conditions (pH 7.4; data not shown) typical of heparanase and bacterial heparitinase, respectively. Heparanase and Hpa2 nonetheless share several biochemical properties. Like heparanase, Hpa2 is directed to the endoplasmic reticulum apparatus (supplemental Fig. 2) and gets glycosylated (evident by reduced molecular weight under SDS-PAGE analysis after tunicamycin treatment; not shown) (Fig. 6, 1). Only Hpa2c, however, is secreted, possibly due to extra glycosylation sites lost in the Hpa2a and Hpa2b splice variants (supplemental Fig. 1B) (14, 27). This possibility is supported by studies showing that altering glycosylation critically affects the secretion of heparanase (45). Despite the lack of intrinsic HS-degrading activity, the hallmark of heparanase, Hpa2c binds heparin/HS. In fact, Hpa2c exhibits even higher affinity toward heparin/HS than heparanase (Figs. 1 and 2), providing a rationale for heparanase inhibition by Hpa2. Unlike Hpa2c, bFGF, a characteristic heparin binding growth factor that orchestrates the assembly of a ternary complex (ligand/receptor/HS) to exert profound biological effects (46), did not interfere with heparanase uptake or activity (Fig. 2D and data not shown). This may imply that inhibition of heparanase activity by Hpa2c is not due solely to competition for HS and may involve other aspects. The observed physical association between Hpa2c and heparanase (Fig. 3B) suggests another mode by which Hpa2c can modulate heparanase functions. Such an interaction is thought to interfere with heparanase enzymatic and possibly non-enzymatic functions (47).
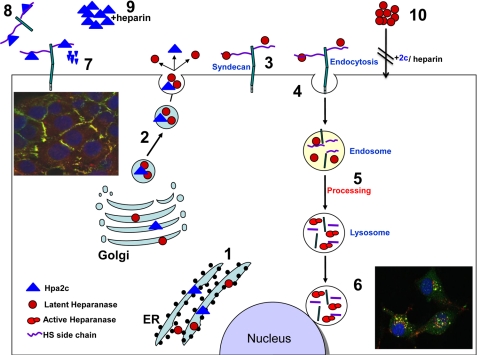
FIGURE 6.
Schematic representation of a proposed model for heparanase and Hpa2 biosynthesis and trafficking. Heparanase (Ο) and Hpa2 (Δ) are first targeted to the endoplasmic reticulum lumen via their signal peptide (1). Heparanase and Hpa2 are then shuttled to the Golgi apparatus and are subsequently secreted via vesicles that bud from the Golgi. Once secreted, heparanase and Hpa2 rapidly interacts with cell membrane HSPGs such as syndecan family members (3). Binding of heparanase is followed by rapid endocytosis of the heparanase-HSPG complex (4) that appears to accumulate in endocytic vesicles such as endosomes (5). Conversion of endosomes to lysosomes results in heparanase processing and activation (5, 6) and accumulation in perinuclear lysosomal vesicles (6; right inset, green, heparanase; red, syndecan-1; yellow, both colors). In contrast, Hpa2 is retained on the cell surface bound to syndecans and fail to get internalized (7; left inset, green, heparanase; red, syndecan-1; yellow, both colors). Instead, Hpa2 levels are regulated by local proteolysis, possibly by membrane-residing protease such as membrane-type metalloproteinase or shedding of the syndecan-Hpa2c complex 8). The addition of heparin sequesters Hpa2 from the cell surface and protects its degradation, resulting in its accumulation extracellularly (9). Heparin addition similarly results in accumulation of heparanase, and both heparin and Hpa2 inhibit uptake and proteolytic processing of heparanase 10).
Interestingly, despite its association with cell membrane HSPG and ability to cluster syndecans (Fig. 3C; supplemental Fig. 3A), Hpa2c does not appear to be internalized and was not detected in endocytic vesicles such as endosomes and lysosomes characteristic of heparanase uptake (Fig. 3) (15, 23, 48, 49) (Fig. 6, insets). Instead, Hpa2c is retained on the cell surface for a relatively long period of time. Membrane localization is confirmed by FACS analysis and immunofluorescent staining of live cells stably expressing Hpa2c or after its exogenous addition (Fig. 4, A and B) and by the release of Hpa2c but not heparanase from the cell surface by acid wash (Fig. 4C). These results clearly indicate that the rapid and efficient internalization of heparanase together with syndecans (Figs. 3C and 6) (21, 23) is unique and not purely a consequence of HS-ligand binding. Being not subjected to internalization, Hpa2c levels appear to be regulated by proteolysis (Fig. 2, C and E) by as yet unidentified membrane-residing protease(s), possibly membrane-type metalloproteinase. Alternatively, Hpa2c may be released from the cell surface by shedding together with the associated HSPG (i.e. syndecan; Fig. 6 and Ref. 8). Studies examining these possibilities are currently under way.
Because syndecans mediate the uptake of a large number of molecules including atherogenic lipoproteins (50, 51) and microorganisms such as bacteria and viruses (26), mechanisms that mediate internalization of syndecan ligands are of interest and clinical significance. Heparanase and Hpa2, thus, may serve as a good model system to study the association and fate of two closely related proteins with a dissimilar outcome. Although the biochemical and cellular consequences of syndecan clustering by Hpa2c is yet to be revealed, it may imply that the repertoire of Hpa2 functions resides beyond modulation of heparanase activity.
Taken together, our results expose for the first time the clinical significance of Hpa2 as a pro-survival protein in head and neck carcinomas. Whether mediated by inhibition of heparanase activity or by other mechanism(s), Hpa2 expression associates with favorable prognosis and prolonged follow-up. Hpa2 staining may, thus, aid in the diagnosis of patients with head and neck and possibly other carcinomas. In addition, elucidating the consequences of syndecan clustering by Hpa2c and the molecular mechanism underlying internalization of heparanase but not Hpa2c may shade light on cell entry by pathogens.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant RO1-CA106456 (NCI). This work was also supported by grants from the Israel Science Foundation (549/06), the Israel Cancer Research Fund, the DKFZ-MOST cooperation program in cancer research, and the Rappaport Family Institute Fund.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3 and Table 1.
- HS
- heparan sulfate
- ECM
- extracellular matrix
- Hpa2
- heparanase 2
- HA
- hyaluronic acid
- bFGF
- basic FGF
- HSPG
- HS proteoglycans.
REFERENCES
- 1.Nakajima M., Irimura T., Di Ferrante D., Di Ferrante N., Nicolson G. L. (1983) Science 220, 611–613 [DOI] [PubMed] [Google Scholar]
- 2.Vlodavsky I., Fuks Z., Bar-Ner M., Ariav Y., Schirrmacher V. (1983) Cancer Res. 43, 2704–2711 [PubMed] [Google Scholar]
- 3.Ilan N., Elkin M., Vlodavsky I. (2006) Int. J. Biochem. Cell Biol. 38, 2018–2039 [DOI] [PubMed] [Google Scholar]
- 4.Vlodavsky I., Ilan N., Naggi A., Casu B. (2007) Curr. Pharm. Des. 13, 2057–2073 [DOI] [PubMed] [Google Scholar]
- 5.Fairweather J. K., Hammond E., Johnstone K. D., Ferro V. (2008) Bioorg. Med. Chem. 16, 699–709 [DOI] [PubMed] [Google Scholar]
- 6.Ferro V., Hammond E., Fairweather J. K. (2004) Mini Rev. Med. Chem. 4, 693–702 [DOI] [PubMed] [Google Scholar]
- 7.McKenzie E. A. (2007) Br. J. Pharmacol. 151, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanderson R. D., Yang Y., Suva L. J., Kelly T. (2004) Matrix Biol. 23, 341–352 [DOI] [PubMed] [Google Scholar]
- 9.Vlodavsky I., Abboud-Jarrous G., Elkin M., Naggi A., Casu B., Sasisekharan R., Ilan N. (2006) Pathophysiol. Haemost. Thromb. 35, 116–127 [DOI] [PubMed] [Google Scholar]
- 10.Vlodavsky I., Friedmann Y., Elkin M., Aingorn H., Atzmon R., Ishai-Michaeli R., Bitan M., Pappo O., Peretz T., Michal I., Spector L., Pecker I. (1999) Nat. Med. 5, 793–802 [DOI] [PubMed] [Google Scholar]
- 11.Hulett M. D., Freeman C., Hamdorf B. J., Baker R. T., Harris M. J., Parish C. R. (1999) Nat. Med. 5, 803–809 [DOI] [PubMed] [Google Scholar]
- 12.Kussie P. H., Hulmes J. D., Ludwig D. L., Patel S., Navarro E. C., Seddon A. P., Giorgio N. A., Bohlen P. (1999) Biochem. Biophys. Res. Commun. 261, 183–187 [DOI] [PubMed] [Google Scholar]
- 13.Toyoshima M., Nakajima M. (1999) J. Biol. Chem. 274, 24153–24160 [DOI] [PubMed] [Google Scholar]
- 14.McKenzie E., Tyson K., Stamps A., Smith P., Turner P., Barry R., Hircock M., Patel S., Barry E., Stubberfield C., Terrett J., Page M. (2000) Biochem. Biophys. Res. Commun. 276, 1170–1177 [DOI] [PubMed] [Google Scholar]
- 15.Zetser A., Levy-Adam F., Kaplan V., Gingis-Velitski S., Bashenko Y., Schubert S., Flugelman M. Y., Vlodavsky I., Ilan N. (2004) J. Cell Sci. 117, 2249–2258 [DOI] [PubMed] [Google Scholar]
- 16.Gingis-Velitski S., Ishai-Michaeli R., Vlodavsky I., Ilan N. (2007) FASEB J. 21, 3986–3993 [DOI] [PubMed] [Google Scholar]
- 17.Zetser A., Bashenko Y., Edovitsky E., Levy-Adam F., Vlodavsky I., Ilan N. (2006) Cancer Res. 66, 1455–1463 [DOI] [PubMed] [Google Scholar]
- 18.Zetser A., Bashenko Y., Miao H. Q., Vlodavsky I., Ilan N. (2003) Cancer Res. 63, 7733–7741 [PubMed] [Google Scholar]
- 19.Weichselbaum R. R., Dunphy E. J., Beckett M. A., Tybor A. G., Moran W. J., Goldman M. E., Vokes E. E., Panje W. R. (1989) Head Neck 11, 437–442 [DOI] [PubMed] [Google Scholar]
- 20.Levy-Adam F., Abboud-Jarrous G., Guerrini M., Beccati D., Vlodavsky I., Ilan N. (2005) J. Biol. Chem. 280, 20457–20466 [DOI] [PubMed] [Google Scholar]
- 21.Levy-Adam F., Feld S., Suss-Toby E., Vlodavsky I., Ilan N. (2008) PLoS ONE 3, e2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levy-Adam F., Miao H. Q., Heinrikson R. L., Vlodavsky I., Ilan N. (2003) Biochem. Biophys. Res. Commun. 308, 885–891 [DOI] [PubMed] [Google Scholar]
- 23.Gingis-Velitski S., Zetser A., Kaplan V., Ben-Zaken O., Cohen E., Levy-Adam F., Bashenko Y., Flugelman M. Y., Vlodavsky I., Ilan N. (2004) J. Biol. Chem. 279, 44084–44092 [DOI] [PubMed] [Google Scholar]
- 24.Doweck I., Kaplan-Cohen V., Naroditsky I., Sabo E., Ilan N., Vlodavsky I. (2006) Neoplasia 8, 1055–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen E., Atzmon R., Vlodavsky I., Ilan N. (2005) FEBS Lett. 579, 2334–2338 [DOI] [PubMed] [Google Scholar]
- 26.Fears C. Y., Woods A. (2006) Matrix Biol. 25, 443–456 [DOI] [PubMed] [Google Scholar]
- 27.Vreys V., David G. (2007) J. Cell. Mol. Med. 11, 427–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nardella C., Lahm A., Pallaoro M., Brunetti M., Vannini A., Steinkühler C. (2004) Biochemistry 43, 1862–1873 [DOI] [PubMed] [Google Scholar]
- 29.Ben-Zaken O., Shafat I., Gingis-Velitski S., Bangio H., Kelson I. K., Alergand T., Amor Y., Maya R. B., Vlodavsky I., Ilan N. (2008) Int. J. Biochem. Cell Biol. 40, 530–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vreys V., Delande N., Zhang Z., Coomans C., Roebroek A., Dürr J., David G. (2005) J. Biol. Chem. 280, 33141–33148 [DOI] [PubMed] [Google Scholar]
- 31.Edovitsky E., Elkin M., Zcharia E., Peretz T., Vlodavsky I. (2004) J. Natl. Cancer Inst. 96, 1219–1230 [DOI] [PubMed] [Google Scholar]
- 32.Casu B., Guerrini M., Guglieri S., Naggi A., Perez M., Torri G., Cassinelli G., Ribatti D., Carminati P., Giannini G., Penco S., Pisano C., Belleri M., Rusnati M., Presta M. (2004) J. Med. Chem. 47, 838–848 [DOI] [PubMed] [Google Scholar]
- 33.Lider O., Cahalon L., Gilat D., Hershkoviz R., Siegel D., Margalit R., Shoseyov O., Cohen I. R. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 5037–5041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Temkin V., Aingorn H., Puxeddu I., Goldshmidt O., Zcharia E., Gleich G. J., Vlodavsky I., Levi-Schaffer F. (2004) J. Allergy Clin. Immunol. 113, 703–709 [DOI] [PubMed] [Google Scholar]
- 35.de Moura J. P., Jr., Nicolau S. M., Stávale J. N., da Silva Pinhal M. A., de Matos L. L., Baracat E. C., de Lima G. R. (2009) Int. J. Gynecol. Cancer 19, 1494–1500 [DOI] [PubMed] [Google Scholar]
- 36.Peretti T., Waisberg J., Mader A. M., de Matos L. L., da Costa R. B., Conceição G. M., Lopes A. C., Nader H. B., Pinhal M. A. (2008) Eur. J. Gastroenterol. Hepatol. 20, 756–765 [DOI] [PubMed] [Google Scholar]
- 37.Bar-Sela G., Kaplan-Cohen V., Ilan N., Vlodavsky I., Ben-Izhak O. (2006) Histopathology 49, 188–193 [DOI] [PubMed] [Google Scholar]
- 38.Ben-Izhak O., Kaplan-Cohen V., Ilan N., Gan S., Vlodavsky I., Nagler R. (2006) Neoplasia 8, 879–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagler R., Ben-Izhak O., Cohen-Kaplan V., Shafat I., Vlodavsky I., Akrish S., Ilan N. (2007) Cancer 110, 2732–2739 [DOI] [PubMed] [Google Scholar]
- 40.Sparmann A., van Lohuizen M. (2006) Nat. Rev. Cancer 6, 846–856 [DOI] [PubMed] [Google Scholar]
- 41.Kleer C. G., Cao Q., Varambally S., Shen R., Ota I., Tomlins S. A., Ghosh D., Sewalt R. G., Otte A. P., Hayes D. F., Sabel M. S., Livant D., Weiss S. J., Rubin M. A., Chinnaiyan A. M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 11606–11611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu J., Yu J., Rhodes D. R., Tomlins S. A., Cao X., Chen G., Mehra R., Wang X., Ghosh D., Shah R. B., Varambally S., Pienta K. J., Chinnaiyan A. M. (2007) Cancer Res. 67, 10657–10663 [DOI] [PubMed] [Google Scholar]
- 43.Abboud-Jarrous G., Atzmon R., Peretz T., Palermo C., Gadea B. B., Joyce J. A., Vlodavsky I. (2008) J. Biol. Chem. 283, 18167–18176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abboud-Jarrous G., Rangini-Guetta Z., Aingorn H., Atzmon R., Elgavish S., Peretz T., Vlodavsky I. (2005) J. Biol. Chem. 280, 13568–13575 [DOI] [PubMed] [Google Scholar]
- 45.Simizu S., Ishida K., Wierzba M. K., Osada H. (2004) J. Biol. Chem. 279, 2697–2703 [DOI] [PubMed] [Google Scholar]
- 46.Ornitz D. M. (2000) BioEssays 22, 108–112 [DOI] [PubMed] [Google Scholar]
- 47.Fux L., Ilan N., Sanderson R. D., Vlodavsky I. (2009) Trends Biochem. Sci 34, 511–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goldshmidt O., Nadav L., Aingorn H., Irit C., Feinstein N., Ilan N., Zamir E., Geiger B., Vlodavsky I., Katz B. Z. (2002) Exp. Cell Res. 281, 50–62 [DOI] [PubMed] [Google Scholar]
- 49.Nadav L., Eldor A., Yacoby-Zeevi O., Zamir E., Pecker I., Ilan N., Geiger B., Vlodavsky I., Katz B. Z. (2002) J. Cell Sci. 115, 2179–2187 [DOI] [PubMed] [Google Scholar]
- 50.Fuki I. V., Kuhn K. M., Lomazov I. R., Rothman V. L., Tuszynski G. P., Iozzo R. V., Swenson T. L., Fisher E. A., Williams K. J. (1997) J. Clin. Invest. 100, 1611–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stanford K. I., Bishop J. R., Foley E. M., Gonzales J. C., Niesman I. R., Witztum J. L., Esko J. D. (2009) J. Clin. Invest. 119, 3236–3245 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.