Abstract
The zinc finger transcription factor, Krüppel-like factor 4 (KLF4), regulates numerous biological processes, including proliferation, differentiation, and embryonic stem cell self-renewal. Although the DNA sequence to which KLF4 binds is established, the mechanism by which KLF4 controls transcription is not well defined. Small ubiquitin-related modifier (SUMO) is an important regulator of transcription. Here we show that KLF4 is both SUMOylated at a single lysine residue and physically interacts with SUMO-1 in a region that matches an acidic and hydrophobic residue-rich SUMO-interacting motif (SIM) consensus. The SIM in KLF4 is required for transactivation of target promoters in a SUMO-1-dependent manner. Mutation of either the acidic or hydrophobic residues in the SIM significantly impairs the ability of KLF4 to interact with SUMO-1, activate transcription, and inhibit cell proliferation. Our study provides direct evidence that SIM in KLF4 functions as a transcriptional activation domain. A survey of transcription factor sequences reveals that established transactivation domains of many transcription factors contain sequences highly related to SIM. These results, therefore, illustrate a novel mechanism by which SUMO interaction modulates the activity of transcription factors.
Keywords: Promoters, Protein Domains, Protein-Protein Interactions, Transcription Regulation, Yeast, KLF4, SIM, SUMO, Transactivation, Transcription Factor
Introduction
Eukaryotic gene transcription is controlled by transcription factors, which contain domains that recognize specific target DNA sequences. Transcription factors also contain distinct domains that regulate the transcriptional machinery by interacting with both general and specific transcription factors and regulators (1–4). The acidic activation domain, for example, represents the largest class of transcriptional activation domains and contains acidic and hydrophobic amino acid residues crucial for protein-protein interaction (1–4). Given that transcriptional control is fundamental to a cell, the mechanism by which acidic activation domains regulate transcription is an important question.
SUMO2 (small ubiquitin-related modifier), which is structurally related to but functionally distinct from ubiquitin, has emerged as an essential regulator in transcriptional control. There are two types of regulatory effects related to SUMO, non-covalent binding and covalent modification (5–8). SUMO can either bind in a reversible manner to a SUMO-interacting motif (SIM) (5, 6) or be covalently conjugated to lysine residues within a target protein by an enzymatic process known as SUMOylation (7, 8). The lysine residue often resides in a SUMOylation motif (SM) with the consensus sequence of ψKXE, where Ψ is hydrophobic, and X is any residue (7, 8). There are three functional forms of SUMO. SUMO-1 exists predominantly in conjugated forms, whereas SUMO-2 and -3, which are nearly identical to each other, are both free and conjugated (7, 8). A typical SIM contains a core of hydrophobic residues with juxtaposed acidic residues (6, 9–12). The presence of the SIM in a vast number of proteins, including transcription factors, implies its importance in the control of eukaryotic gene expression (6, 13). However, how the SIM regulates transcription is not well defined (6).
Krüppel-like factor 4 (KLF4), a zinc finger-containing transcription factor, has been subjected to intense investigation in recent years. It is one of the four original factors that induce the formation of pluripotent stem cells by the reprogramming of somatic cells (14, 15). KLF4 also plays crucial roles in numerous physiological and pathophysiological conditions (16–21). For example, KLF4 transactivates the C/EBPβ and Lefty1 promoters to stimulate adipogenesis and embryonic stem cell self-renewal, respectively (22, 23). KLF4 is essential for terminal differentiation of the epidermis and intestinal epithelium (16, 24, 25). It is also a potent inhibitor of axon outgrowth (26, 27). In pathological states, KLF4 plays a role in tumorigenesis (16, 18, 20, 28) and cardiovascular (21, 29) and inflammatory disorders (29, 30). Thus, identifying a common mechanism that regulates KLF4 transcriptional activity may aid in the development of novel therapeutic strategies against various disorders involving KLF4.
Here, we report that KLF4 is both associated with SUMO-1 via a SIM and SUMOylated at a single site. The KLF4 SIM acts as a transcriptional activation domain in both yeast and mammalian systems, and SUMO-1 binding is crucial for this activity. Hence, SUMO can directly regulate transcription through a SIM. A survey of transcription factor sequences reveals that established transactivation domains of many transcription factors contain sequences that are highly related to SIM. Our study, therefore, identified a novel and perhaps common mechanism by which SUMO interaction modulates the transcriptional activity of transcription factors.
EXPERIMENTAL PROCEDURES
Plasmids
Various SUMOylation and SIM mutants of KLF4 were constructed with QuikChange site-directed mutagenesis kit (Stratagene, catalog #200521). Mammalian and yeast plasmids expressing GAL4 DNA binding domain (GAL4-DBD) and green fluorescence protein (GFP) fusion proteins were prepared by inserting cDNA fragments encoding wild type or mutated KLF4 sequences into the NcoI-EcoRI site of pGBKT7 (Clontech, catalog #630443) or the EcoRI-SalI site of pEGFPC1 (Clontech, catalog #6084-1). The amounts of plasmids were equally adjusted for transfection in all experiments. The plasmids GFP-SUMO-1, Myc-SUMO-1ΔGG, His-SUMO-1, FLAG-PIAS1, pMT3-KLF4, pMT3-KLF4-EEE, pMT3-KLF4-DDD, pCMV-Myc-KLF4, pLefty1-luc, and pGL3-C/EBPβ-luc (B3K) have been described previously, many of which were generously provided by other investigators (22, 31–37).
SUMOylation Assay
SUMOylation assays were performed as previously described (38). Briefly, COS-1 cells were transfected with the indicated plasmids and disrupted in lysis buffer (20 mm Tris-HCl, pH 7.4, 1% Nonidet P-40, 135 mm NaCl, 20 mm N-ethylmaleimide (Sigma, catalog #1271) and complete protease inhibitor mixture (Roche Applied Science, catalog #11-836-153-001)) followed by immunoprecipitation and Western blotting with the indicated antibodies, including rabbit Myc (Chemicon, catalog #06-549), mouse Myc (Sigma, clone 9E10, catalog #M4439), GFP (Roche Applied Science, catalog #1814460), and His (Qiagen, catalog #34660) antibodies. Alternatively, KLF4 was immunoprecipitated from cells with rabbit anti-KLF4 (Santa Cruz, H180, catalog #SC-20691) followed by Western blotting with mouse anti-GFP or either goat (Santa Cruz, M19, catalog #SC-1905) or rabbit anti-KLF4 (Santa Cruz, H180, catalog #SC-20691).
Glutathione S-Transferase (GST) Pulldown Assay
To detect binding of SUMO-1 to the KLF4 SIM, HEK293T cells were transfected with the indicated GFP-SIM fusion constructs, and lysates were prepared using lysis buffer containing phosphatase inhibitors (Active Motif, catalog #54001) according to the manufacturer's instructions. Purified recombinant GST-SUMO-1 (Biomol/Enzo Life Sciences, catalog #UW0160-0500) was incubated with the lysates overnight, and bovine serum albumin (BSA)-blocked glutathione-Sepharose beads (GE Biosciences, catalog #17-0756-01) were then added. One hour later, the beads were washed twice with lysis buffer supplemented with 5% glycerol, 0.05% Nonidet P-40, and 1 mg/ml BSA and twice with wash buffer without BSA. The precipitates were blotted with mouse GFP (Roche Applied Science, catalog #1814460) and SUMO-1 (Zymed Laboratories Inc., catalog #33-2400) antibodies. To detect binding of SUMO-1 to Myc-tagged KLF4, cells were transfected with the indicated KLF4 or mutant construct and lysed in the lysis buffer containing phosphatase inhibitors, and the supernatants were incubated with purified recombinant GST-SUMO-1 for 5 h. BSA-blocked glutathione-Sepharose beads were added for 1 h and then washed twice with lysis buffer supplemented with 5% glycerol, 0.05% Nonidet P-40, and 1 mg/ml BSA and twice with wash buffer without BSA. The precipitated proteins were detected with mouse anti-Myc and mouse anti-SUMO-1.
Co-immunoprecipitation
To independently detect binding of SUMO-1 to KLF4, Klf4-null (Klf4−/−), mouse embryonic fibroblasts (39) were co-transfected with the indicated KLF4 construct and Myc-tagged SUMO-1 lacking two carboxyl-terminal glycine residues that support SUMOylation (Myc-SUMO-1ΔGG). Three days later, cells were lysed in lysis buffer containing phosphatase inhibitors (Active Motif, catalog #54001). The supernatant was incubated with a rabbit KLF4 antibody (H180, Santa Cruz), rocked for 5 h, and followed by incubation with BSA/salmon sperm DNA-blocked protein A beads (Upstate Biotechnology, catalog #16-157) overnight. The immune complexes were washed 3 times with lysis buffer supplemented with 5% glycerol, 0.05% Nonidet P-40, and 1 mg/ml BSA and twice with lysis buffer without BSA. The immunoprecipitates were probed with mouse anti-Myc and rabbit anti-KLF4.
In Vitro Binding Assay
Twenty μg of His6-tagged SIM or mutant peptide (GenScript, Peptide Services) was mixed with either 5 μg of purified human SUMO-1 (LAE Biotechnology, catalog #P012) in binding buffer (20 mm Tris-HCl, pH 6.8, 5% glycerol, and 0.05% Nonidet P-40) or buffer alone as a control and incubated overnight at 4 °C. Fifteen mm imidazole was then added followed by incubation with 40 μl of BSA-blocked His-Select Nickel Affinity Gel (Sigma, catalog #P6611) for 15 min. The beads were washed 6 times for 10, 10, 5, 2, 1, and 1 min sequentially with binding buffer containing progressively reduced amounts of imidazole and subjected to immunoblotting with a mouse SUMO-1 antibody (Zymed Laboratories Inc.).
Yeast Transactivation Assay
Saccharomyces cerevisiae strain AH109 was transformed with plasmid pGBKT7 (Clontech) or pGBKT7 inserted with the indicated SIM or SIM mutants and streaked on synthetic dropout medium lacking either adenine or both adenine and histidine to test transactivation of adenine (Ade) and histidine (His) reporters. Alternatively, strain Y187 was transformed with the indicated SIM or mutant constructs and cultured in liquid synthetic dropout medium lacking tryptophan. A colorimetric α-galactosidase assay was performed as instructed (Clontech, Yeast Protocols Handbook, PT3024-1) with the medium, which contained secreted α-galactosidase expressed from MEL1 reporter, with p-nitrophenyl-α-d-galactoside as the substrate. In addition, cells were lysed by glass bead disruption and immunoblotted with a mouse Myc antibody to detect GAL4-DBD-Myc-SIM fusion proteins. Both GAL4-DBD and fusion proteins expressed from the pGBKT7 vector contain a Myc tag near their carboxyl terminus (Clontech).
Luciferase Reporter Assay
To determine transcriptional activity of wild type KLF4 and a hydrophobic core SIM mutant in mammalian cells, 0.1 μg of pCMV-Script, pCMV-Myc-KLF4, or pCMV-Myc-KLF4-L101A/I106A (LI) was co-transfected into HEK293T cells with 0.4 μg of pLefty1-luc plus 2 ng of control renilla luciferase plasmid (for Lefty1 reporter) or with 0.25 μg of pGL3-C/EBPβ-luciferase plus 4 ng of renilla luciferase plasmid (for C/EBPβ reporter). Alternatively, pCMV-Myc-KLF4 was substituted with pMT3-KLF4 or the corresponding pMT3-KLF4-EEE or pMT3-KLF4-DDD mutant to detect the transcriptional activity of KLF4 SIM acidic stretch mutants.
Small Interfering RNA
Small interfering RNA (siRNA) against SUMO-1, in the form of either a mixture of two siRNAs targeting different regions of SUMO-1 (Santa Cruz Biotechnology, catalog #SC-29498), two individual siRNAs (Santa Cruz Biotechnology, #SC-29498a or #SC-29498b), or a Universal Negative Control siRNA (Invitrogen, catalog #12935300) (40) was transfected into 40% confluent HEK293T cells with Lipofectamine RNAiMAX (Invitrogen, catalog #13778-150) according to the manufacturer's instructions. On the next day, 0.1 μg of pCMV-Script or pCMV-Myc-KLF4 was co-transfected in the cells with 0.4 μg of pLefty1-luc plus 2 ng of renilla luciferase control (for Lefty1 reporter) or with 0.25 μg of pGL3-C/EBPβ-luc plus 4 ng of renilla luciferase control (for C/EBPβ reporter) using Lipofectamine 2000 (Invitrogen, catalog #11668-019). Two days posttransfection, dual luciferase reporter assay (Promega, catalog #E1960) was conducted, and a portion of the lysate was immunoblotted with a mouse antibody against SUMO-1 (Zymed Laboratories Inc., catalog #33-2400), SUMO-2/3 (MBL, clone 1E7, catalog #M114-3), ubiquitin (Santa Cruz, clone P4D1, catalog #SC-8017), or β-actin (Sigma, clone AC15, catalog #A1978).
Quantitative Reverse Transcription-PCR
siRNA against SUMO-1 or the Universal Negative Control siRNA was transfected into 40% confluent HEK293T cells with Lipofectamine RNAiMAX. Three days later, total RNA was isolated with TRIzol (Invitrogen; catalog #15596-018), and quantitative real-time RT-PCR was performed in triplicate with primer sets specific for human SUMO-1, C/EBPβ, Lefty1, and the control gene β-actin (Qiagen; QT00014280, QT00237580, QT01667421, and QT00095431). Products were amplified and detected with Power SYBR Green RNA-to-CT 1-Step kit (Applied Biosystems; catalog #4389986) on an Eppendorf REALPLEX epgradient S real-time PCR Mastercycler according to the manufacturer's instructions. Relative changes in expression were calculated based on the CT (−ΔΔCT) method after normalization with the actin control. Reactions without the reverse transcriptase (-RT Enzyme mix) were also performed as negative controls.
Cycloheximide Chase Assay
The cycloheximide chase assay was performed as described (81, 82). Briefly, HEK293T cells were transfected with either pCMV-Myc-KLF4 or pCMV-Myc-KLF4-K275R, treated with 100 μg/ml cycloheximide, and lysed at the indicated time points. Total proteins were subjected to Western blotting for Myc (Sigma, 9E10, catalog #M4439) and β-actin (Sigma, AC15, catalog #A1978). The chemiluminescence signals were quantitatively measured with a Typhoon 9200 Variable Mode Imager from GE Healthcare and normalized with the β-actin control.
Bromodeoxyuridine (BrdU) Incorporation Assay
The assay was performed as previously described (41). Briefly, cells were split to 30% confluence and transfected with the indicated plasmids. Cells were then labeled for 24 h with BrdU, fixed with methanol, and treated with HCl to denature the DNA. Cells were stained for immunofluorescence with mouse anti-BrdU (BD Pharmingen, clone 3D4, catalog #555627) and rabbit Myc (Chemicon) or KLF4 (H180, Santa Cruz) antibodies. Cells were also stained with Hoechst dye to reveal nuclei.
RESULTS
KLF4 Is SUMOylated at a Single Lysine Residue
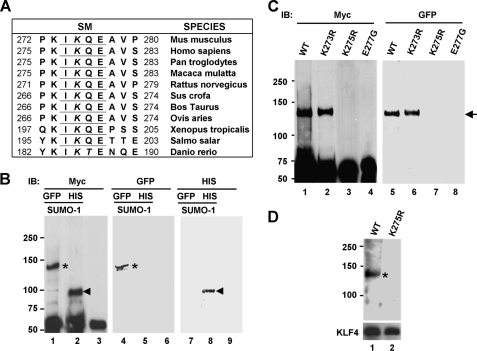
KLF4 contains a highly conserved sequence that resembles the SM, ψKXE (7, 8, 42) (Fig. 1A). We first determined whether KLF4 is SUMOylated within this motif. COS-1 cells were co-transfected with expression constructs containing Myc-tagged KLF4 (Myc-KLF4) and GFP-tagged SUMO-1 (GFP-SUMO-1), and cell lysates were prepared in the presence of a deSUMOylation inhibitor, N-ethylmaleimide (7). Myc-KLF4 was immunoprecipitated from the lysates with a rabbit Myc antibody followed by Western blotting with a mouse Myc or GFP antibody. As seen in Fig. 1B, a single Myc-KLF4 species was detected at ∼130 kDa (lane 1; asterisk). This form was SUMOylated as confirmed by blotting with the GFP antibody (lane 4; asterisk). The size of the protein is consistent with conjugation of a single GFP-SUMO-1 molecule (∼45-kDa apparent molecular mass) to Myc-KLF4 (65-kDa apparent molecular mass), taking into account a slowed migration due to the branching effect seen in SUMOylated proteins upon conjugation of SUMO near the center of the protein (38, 43–45).
FIGURE 1.
The mouse KLF4 is SUMOylated at lysine residue 275. A, conservation across species of the SM (underlines) in KLF4 is shown. The SM is located between amino acid residues 274 and 277 of the mouse KLF4. The conserved SUMO target lysine residues in the SM are italicized. B, SUMOylation of KLF4 by GFP-tagged or His-tagged SUMO-1 is shown. pCMV-Myc-KLF4 was co-transfected into COS-1 cells with GFP-SUMO-1 (lanes 1, 4, and 7), His-SUMO-1 (lanes 2, 5, and 8), or vector alone (lanes 3, 6, and 9). Lysates were immunoprecipitated with a rabbit Myc antibody followed by Western blotting with mouse Myc (lanes 1–3), GFP (lanes 4–6), or His (lanes 7–9) antibody. Asterisks indicate GFP-SUMO-1-conjugated Myc-KLF4, and arrowheads indicate His-SUMO-1-conjugated Myc-KLF4. The positions of the molecular mass markers in kDa are shown to the left of the blots. IB, immunoblot. C, identification of the SUMOylation site within KLF4 is shown. COS-1 cells were co-transfected with GFP-SUMO-1 and one of the following constructs: pCMV-Myc-KLF4 (WT), pCMV-Myc-KLF4-K273R (K273R), pCMV-Myc-KLF4-K275R (K275R), or pCMV-Myc-KLF4-E277G (E277G). Lysates were immunoprecipitated with a rabbit Myc antibody followed by Western blotting with a mouse Myc (lanes 1–4) or GFP (lanes 5–8) antibody. Arrow, GFP-SUMO-1-conjugated Myc-KLF4 or Myc-KLF4-K273R. D, SUMOylation of untagged KLF4. COS-1 cells, which express negligible amount of endogenous KLF4, were co-transfected with GFP-SUMO-1 and either pMT3-KLF4 (WT) or pMT3-KLF4-K275R (K275R). Lysates were immunoprecipitated with a rabbit KLF4 antibody and blotted with a mouse GFP antibody. SUMOylated KLF4 is indicated by the asterisk on the top panel. The bottom panel shows levels of KLF4 or KLF4-K275R in the immune complexes as probed by a goat KLF4 antibody.
As an alternative method to detect SUMOylation, we co-transfected COS-1 cells with Myc-KLF4 and His-tagged SUMO-1 followed by immunoprecipitation of Myc-KLF4. Upon Western blotting with Myc and His antibodies, SUMOylated Myc-KLF4 at ∼90 kDa was apparent in the immunoprecipitates (Fig. 1B, lanes 2 and 8; arrowheads). Because of the smaller size of the His tag, His-SUMO-1 (∼18 kDa) conjugated to Myc-KLF4 migrated faster than the GFP-SUMO-1-modified counterpart. These results confirm that KLF4 is SUMOylated.
We next attempted to identify the SUMOylation site within KLF4. The lysine residue within the putative KLF4 SUMOylation motif is located at amino acid position 275 of the mouse sequence (Fig. 1A). We substituted arginine for this lysine and determined the effect of the K275R mutation on the ability of KLF4 to become SUMOylated. The SUMOylated form (Fig. 1C, lanes 1 and 5; arrow) was completely absent from cells transfected with Myc-KLF4-K275R (Fig. 1C, lanes 3 and 7), indicating that Lys-275 is the site of SUMOylation. Reinforcing this conclusion, loss in SUMOylation was fully reproduced by mutating the conserved glutamate residue within the ψKXE SUMOylation motif, Glu-277, as the E277G mutant could not be SUMOylated (Fig. 1C, lanes 4 and 8). In contrast, the K273R mutant, which contains a mutation at a nearby lysine residue 273, was SUMOylatable (Fig. 1C, lanes 2 and 6). Lending further support, SUMOylation was revealed when untagged KLF4 was co-transfected with GFP-SUMO-1 into COS-1 cells, immunoprecipitated with a KLF4 antibody, and probed with a GFP antibody (Fig. 1D, lane 1; asterisk). Again, the SUMOylated form of KLF4 was absent when the Lys-275 SUMOylation site was mutated to arginine (Fig. 1D, lane 2). These results demonstrate that the KLF4 SUMOylation motif represents a bona fide site of SUMOylation.
In addition to the consensus SM between residues 274 and 277, an inverted SM (EPKP) is located between residues 382 and 385 in mouse KLF4 sequence (supplemental Fig. 1C). However, lysine 384 was SUMOylated, as KLF4 with a mutation at this site (K384R) was SUMOylated normally (supplemental Fig. 1A, lanes 2 and 5), but an additional mutation at Lys-275 (K275R/K384R) completely abolished SUMOylation (supplemental Fig. 1A, lanes 3 and 6). Moreover, other conserved lysine residues within the amino terminus (residues 1–100) and carboxyl-terminal zinc finger DNA binding domain (residues 350–483) of KLF4 were not SUMOylated (data not shown). Thus, multiple lines of evidence identified lysine residue 275 in the mouse KLF4 as the sole site of SUMOylation.
Although KLF4 is SUMOylated and mutations in the consensus SUMOylation motif block this activity, neither the K275R nor the E277G mutant showed gross alternation in nuclear localization or transcriptional activity with various target promoter reporters (data not shown). This could be attributed to the relatively small fraction of KLF4 that is SUMOylated at any given time (Fig. 1 and supplemental Fig. 1).
SUMOylation did not affect the stability of KLF4 as the protein level of neither the K275R nor E277G mutant in transfected cells was considerably different from that of wild type KLF4 (Fig. 2A, compare lanes 4 and 5 with lane 2). SUMO-1 co-expression did not reduce the level of either KLF4 (Fig. 2B, lane 2) or the SUMOylatable K273R mutant (Fig. 2B, lane 3). In contrast, although PIAS1, the proposed SUMO E3 ligase for KLF4 (46), reduced KLF4 levels (compare Fig. 2, A, lane 2, and C, lane 2), the level of the SUMOylatable K273R mutant was not affected (compare Fig. 2, A, lane 3, and C, lane 3), suggesting that the destabilizing effect of PIAS1 co-expression was not due to direct SUMOylation of KLF4. Furthermore, cycloheximide chase assays showed that the rate of degradation between wild type KLF4 and the SUMOylation site mutant K275R was identical (Fig. 2, D and E). These results indicate that SUMOylation does not directly trigger KLF4 degradation.
FIGURE 2.
SUMOylation of KLF4 does not reduce its protein levels. pCMV-Script (Vector), pCMV-Myc-KLF4 (WT), pCMV-Myc-KLF4-K273R (K273R), pCMV-Myc-KLF4-K275R (K275R), or pCMV-Myc-KLF4-E277G (E277G) were co-transfected into COS-1 cells with either vector alone (A), His-SUMO-1 (B), or PIAS1 (C). Lysates were immunoblotted with mouse Myc (upper panel) or β-actin (lower panel) antibody. D and E, pCMV-Myc-KLF4 (WT) or pCMV-Myc-KLF4-K275R (K275R) was transfected into HEK293T cells and subjected to cycloheximide chase assay for up to 3 h. D, shown is an immunoblot with Myc (upper panel) and β-actin (lower panel) antibody. E, corresponding quantitative chemiluminescence measurements after normalization with actin control are shown. The relative protein levels at 0 h were set as 1, and the average protein levels and S.D. (n = 3) at the other time points relative to those at 0 h are shown.
KLF4 Physically Interacts with SUMO-1 through a SIM
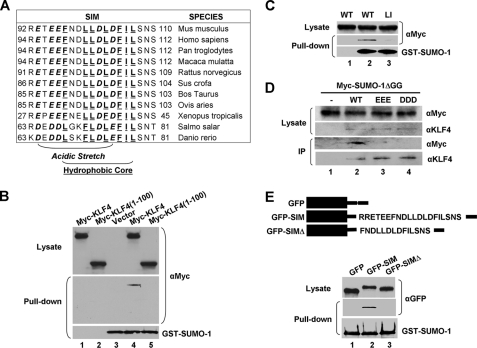
Although we have yet to identify a functional role for the covalent modification of KLF4 by SUMO, we hypothesized that non-covalent interaction between KLF4 and SUMO may have a more apparent effect as any protein that possesses a SIM could potentially bind and be regulated by SUMO. KLF4 contains a putative SIM that consists of an acidic stretch and a hydrophobic core between amino acid residues 92 and 110 in the mouse sequence (Fig. 3A). This putative SIM matches established SIMs in a host of other proteins (supplemental Fig. 2) (5, 6, 9–12, 47–51), suggesting that KLF4 may physically interact with SUMO. To investigate this possibility, we transfected cells with Myc-KLF4 or a truncated mutant, Myc-KLF4 (1–100), the latter containing only the amino terminus of KLF4 without an intact SIM, and incubated the lysates with purified recombinant GST-SUMO-1. A GST pulldown assay followed by Western blotting with anti-Myc indicated that SUMO-1 was physically associated with KLF4 (Fig. 3, B, lane 4, and C, lane 2) but not with its amino-terminal 100 amino acids (Fig. 3B, lane 5). The association between KLF4 and SUMO-1 was further demonstrated by co-transfecting KLF4 and Myc-SUMO-1ΔGG, which supports SUMO binding but not SUMOylation, into mouse embryonic fibroblasts null for Klf4 (Klf4−/−) (39) followed by immunoprecipitation with a rabbit KLF4 antibody (Fig. 3D, lane 2). These results indicate that KLF4 physically interacts with SUMO.
FIGURE 3.
KLF4 physically interacts with SUMO-1 through a SIM. A, conservation across species of the putative SIM in KLF4 is shown. The acidic stretch is shown in bold and italics, and the hydrophobic core is shown in bold and underlines. B, HEK293T cells were transfected with vector only, pCMV-Myc-KLF4, or pCMV-Myc-KLF4(1–100), a truncated mutant containing only the amino-terminal 100 residues. The corresponding lysates were either incubated with purified recombinant GST-SUMO-1 (lanes 3–5) or not (lanes 1 and 2). The GST pulldown assay was conducted followed by Western blotting with mouse Myc (upper and middle panels) and SUMO-1 (lower panel) antibodies. C, lysates from HEK293T cells transfected with pCMV-Myc-KLF4 (WT) (lane 2) or pCMV-Myc-KLF4-L101A/I106A (LI) (lane 3) were incubated with purified GST-SUMO-1. An equal portion of WT lysate not incubated with GST-SUMO-1 (lane 1) served as a control. GST pulldown assay was followed by Western blotting with mouse anti-Myc and SUMO-1. D, lysates from Klf4-null (Klf4−/−) mouse embryonic fibroblasts (39) co-transfected with Myc-SUMO-1ΔGG and pMT3 (-), pMT3-KLF4 (WT), pMT3-KLF4-E93V/E95V/E96V (EEE), or pMT3-KLF4-D99V/D102V/D104V (DDD) were immunoprecipitated (IP) with a rabbit KLF4 antibody followed by Western blotting with mouse Myc or rabbit KLF4 antibody. E, GFP or GFP linked to full-length KLF4 SIM (GFP-SIM) or the carboxyl terminus of SIM (GFP-SIMΔ) (upper panel) were transfected into HEK293T cells, and lysates were incubated with purified GST-SUMO-1 followed by a GST pulldown assay and Western blotting with mouse GFP or SUMO-1 antibody.
To demonstrate that the putative SIM in KLF4 is the site of interaction between SUMO-1 and KLF4, we transfected cells with a GFP fusion construct containing residues 92–110 (GFP-SIM; Fig. 3E) or 97–100 (GFP-SIMΔ; Fig. 3E), which has a truncated acidic stretch, and incubated the lysates with purified GST-SUMO-1 followed by pulldown assay. As seen in Fig. 3E, GST-SUMO-1 was co-precipitated with GFP-SIM (lane 2) but not with GFP-SIMΔ (lane 3) or GFP alone (lane 1). In agreement with these findings, co-immunoprecipitation and yeast two-hybrid assays showed that neither the amino-terminal residues (1–100) nor carboxyl-terminal residues (350–483) of KLF4 bound SUMO-1 (data not shown). These results demonstrate that the sequence encompassing residues 92–110 in KLF4 indeed acts as a SIM.
To further corroborate the above findings, mutations at the critical hydrophobic and acidic residues within the SIM in KLF4 were tested for their ability to affect SUMO binding. First, mutations at Leu-101 and Ile-106 within the hydrophobic core significantly reduced KLF4 binding to SUMO-1 in a GST pulldown assay, as demonstrated by the dramatically reduced ability of the Myc-KLF4-L101A/I106A (LI) mutant to co-precipitate GFP-SUMO-1 (Fig. 3C, lane 3). Second, KLF4 containing mutations at the three glutamates (EEE) or aspartates (DDD) within the acidic stretch (34) was unable to co-precipitate Myc-SUMO-1ΔGG when compared with wild type KLF4 (Fig. 3D, lanes 2–4). In addition, in vitro binding assay of a His-tagged SIM peptide, but not an EEE, DD, or LI mutant peptide, bound purified SUMO-1 (supplemental Fig. 3B, lanes 2–5). The results of these studies indicate that KLF4 interacts with SUMO-1 in a manner that is dependent on both the acidic stretch and hydrophobic core in its SIM.
The KLF4 SIM Is a Transcriptional Activation Domain
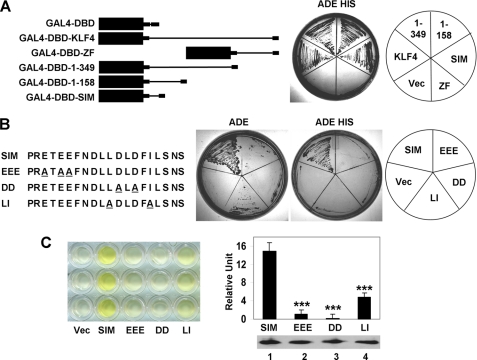
Previous studies indicated that the EEE and DDD KLF4 mutants were unable to activate transcription of target genes (34). This coupled with the inability of the two mutants to bind SUMO-1 raised the question of whether the SIM is a transcriptional activation domain. To explore this possibility, we first mapped the minimal transcriptional activation domain in KLF4 using a classical transactivation assay in yeast (52–54). Here we determined whether a series of truncated KLF4 proteins fused to the DBD of the yeast transcription factor, GAL4, could drive expression of the Ade and His reporters in the yeast strain AH109. As seen in Fig. 4A, full-length KLF4 fused to the GAL4-DBD transactivated the Ade and His reporters in a manner consistent with other previously characterized mammalian transcription factors (2, 3, 55–58). Likewise, KLF4 with carboxyl-terminal truncations (1–349 and 1–158) fused to the GAL4-DBD transactivated the reporters (Fig. 4A), a result consistent with the previous study in mammalian cells showing that the first 109 residues of KLF4 served as a transcriptional activation domain (34). In contrast, neither GAL-DBD alone nor GAL4-DBD fused to the carboxyl-terminal zinc finger region of KLF4 (GAL4-DBD-ZF) transactivated the reporters. Remarkably, transactivation can be fully capitulated by fusing KLF4s SIM to the GAL4-DBD (Fig. 4A; GAL4-DBD-SIM). These results indicate that SIM is the minimal transcriptional activation domain responsible for the transcriptional activity of KLF4.
FIGURE 4.
The KLF4 SIM is a transcriptional activation domain. A, localization of the KLF4 transcriptional activation domain is shown. Left, shown is a schematic of GAL4-DBD fused to full-length KLF4 (GAL4-DBD-KLF4), the KLF4 carboxyl terminus (residues 350–483) that contains the zinc finger DNA binding domain (GAL4-DBD-ZF), KLF4 amino-terminal 349 residues (GAL4-DBD-1–349), the KLF4 first 158 residues (GAL4-DBD-1–158), and KLF4 SIM (residues 92–110) (GAL4-DBD-SIM). Right, the GAL4-DBD constructs indicated on the left were transformed into AH109 yeast strain, and their ability to transactivate both Ade and His reporters was detected by growth in synthetic dropout medium lacking adenine and histidine (ADE HIS). Vec, vector (GAL4-DBD) alone. ZF, zinc finger. B, shown is localization of the transactivation domain of KLF4 to it SIM. Left, a schematic of wild type or mutated SIMs fused to GAL4-DBD is shown. The alanine residues used to substitute for the acidic or hydrophobic residues are underlined. Right, the GAL4-DBD-SIM constructs indicated on the left were transformed into AH109 yeast strain, and their ability to transactivate the single reporter Ade or both Ade and His reporters was detected by growth in synthetic dropout medium lacking either only adenine (ADE) or both adenine and histidine (ADE HIS). C, shown is quantification of the transcriptional activation activity of KLF4 SIM or SIM mutants fused to the GAL4-DBD by α-galactosidase assay using MEL1 as a reporter. Left, a representative experiment in triplicate is shown. The intensity of yellow color reflects the strength of transactivation. The residual yellowish color from vector alone was due to endogenous α-galactosidase in yeast host and served as a blank control. Right, quantitative results from the α-galactosidase assay are shown on the left. All GAL4-DBD-SIM fusion proteins were expressed at similar levels, as judged by immunoblotting against the pre-existing Myc tag (bottom right). ***, p < 0.001 compared with wild type SIM by two-tailed t test.
To further corroborate the finding that the SIM is the transactivation domain for KLF4, all three mutant constructs of SIM (EEE, DD, and LI) that failed to bind SUMO exhibited reduced transactivation of the Ade reporter and abrogation of transactivation by the more stringent Ade/His selection when compared with the wild type SIM (Fig. 4B). Moreover, an independent colorimetric assay using MEL1 as a reporter resulted in similar results (Fig. 4C). These findings demonstrate unequivocally that the SIM in KLF4 is its transactivation domain.
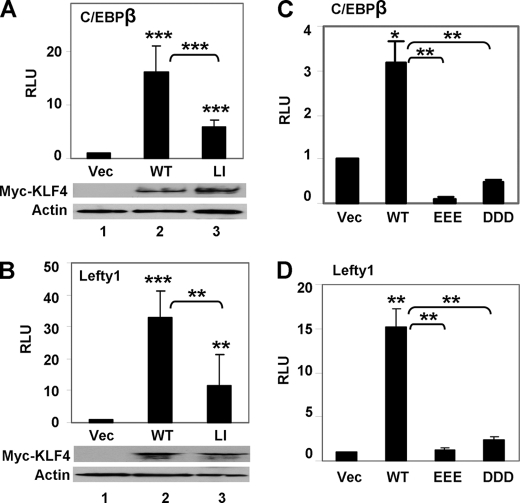
The requirement of the KLF4 SIM for transcriptional control was also observed in mammalian cells. We performed luciferase reporter assays to measure the transcriptional activity of KLF4 and its SUMO binding-deficient mutants by co-transfection in HEK293T cells. We studied two distinct KLF4 target promoters, C/EBPβ and Lefty1, which regulate the processes of adipogenesis and pluripotency, respectively, and have been shown to be robustly transactivated by KLF4 (22, 35). As shown in Fig. 5, wild type KLF4 cloned in two different expression constructs, pCMV (Figs. 5, A and B) and pMT3 (Figs. 5, C and D), transactivated both C/EBPβ and Lefty1 promoters. In contrast, the KLF4 LI mutant had significantly reduced activity toward both promoters compared with wild type KLF4 (Fig. 5, A and B), and the EEE and DDD mutants essentially lost the ability to transactivate (Fig. 5, C and D). These phenotypes are reminiscent of those from the yeast assay as, for example, the LI mutant lost approximately two-thirds of wild type activity in both yeast α-galactosidase and mammalian luciferase reporter assays (Figs. 4C and 5, A and B), whereas the acidic mutants suffered an even greater reduction of activity in both assays (Figs. 4C and 5, C and D). Combining the results of these studies, it is apparent that the transcriptional activity of KLF4 depends on the interaction between its SIM and SUMO.
FIGURE 5.
The KLF4 SIM is crucial for transcriptional activation in mammalian cells. Dual luciferase reporter assays were performed with C/EBPβ or Lefty1-luciferase reporter plasmids and expression constructs of KLF4 or its mutants in HEK293T cells as described under “Experimental Procedures”. A and B, shown is co-transfection of vector (Vec) alone, pCMV-Myc-KLF4 (WT), or pCMV-Myc-L101A/I106A (LI) with C/EBPβ-luciferase (A) or Lefty1-luciferase (B). The expression levels of Myc-KLF4 (WT) and Myc-KLF4-L101A/I106A (LI) were shown by Western blotting against Myc and β-actin. C and D, shown is co-transfection of vector (Vec) alone, pMT3-KLF4 (WT), pMT3-KLF4-E93V/E95V/E96V (EEE), or pMT3-D99V/D102V/D104V (DDD) with C/EBPβ-luciferase (C) and Lefty1-luciferase (D). The expression levels of both the EEE and DDD mutants have previously been documented (34). Shown are the means and S.D. of four independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001; by two-tailed t test. Asterisks not associated with brackets are comparisons to vector alone. RLU is relative luciferase unit.
SUMO-1 Depletion Inhibits Transactivation by KLF4
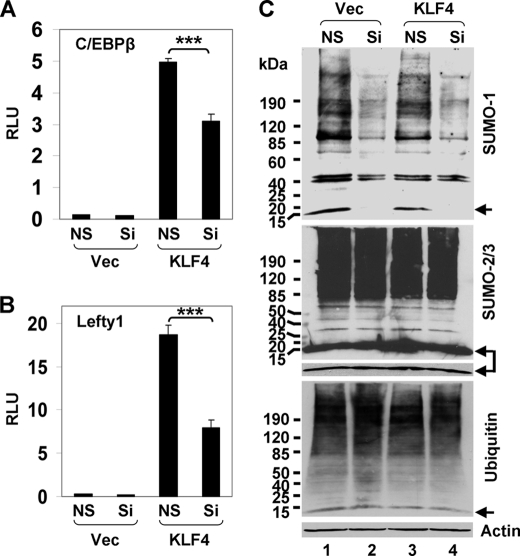
To examine whether SUMO-1 is necessary for transcriptional activation by KLF4, HEK293T cells were treated with SUMO-1-specific siRNA, and the effect of SUMO-1 deficiency on KLF4 transcriptional activity was investigated. As shown in Fig. 6, knockdown of endogenous SUMO-1 by a dual siRNA mixture commonly used to specifically deplete SUMO-1 (79, 80) led to a significant reduction in the ability of KLF4 to transactivate both the C/EBPβ and Lefty1 promoters compared with controls (Fig. 6, A and B). The effect of knockdown was highly specific to SUMO-1 (79, 80), as levels of both conjugated SUMO-1 and free SUMO-1 were significantly reduced (Fig. 6C, upper panel) but not those of SUMO-2/3 (Fig. 6C, middle panel) or ubiquitin (Fig. 6C, lower panel). Furthermore, the transcriptional inhibition by SUMO-1 depletion was recapitulated when two individual siRNAs were used separately, and the inhibition in transactivation correlated with the relative knockdown efficiency of these two siRNAs (supplemental Fig. 4). These results demonstrate that SUMO-1 depletion inhibits the transactivation by KLF4.
FIGURE 6.
Reduction of SUMO-1 inhibits KLF4 transcriptional activity. HEK293T cells were transfected with the C/EBPβ-luciferase (A) or Lefty1-luciferase (B) reporter, vector (Vec) or pCMV-Myc-KLF4 (KLF4), nonspecific siRNA (NS) or the dual siRNA mixture against SUMO-1 (Si), and the control renilla luciferase plasmid. Dual luciferase assays were performed, and the normalized luciferase activity presented as relative luciferase units (RLU). Shown are the means and S.D. of three independent experiments. ***, p < 0.001 by two-tailed t test. C, a fraction of lysates from the corresponding cells was subjected to Western blotting with mouse antibodies against SUMO-1 (upper panel), SUMO-2/3 (middle panel), and ubiquitin (lower panel). β-actin was used as a loading control. The arrows indicate free SUMO-1 (upper panel), SUMO-2/3 (middle panel), and ubiquitin (lower panel). A short exposure of free SUMO-2/3 in the middle panel is also provided.
As endogenous C/EBPβ and Lefty1 are expressed in various differentiated cells (35, 83), we further determined whether SUMO-1 depletion affects endogenous C/EBPβ and Lefty1 mRNA levels in HEK293T cells by quantitative RT-PCR. As shown in supplemental Fig. 5, knockdown with the specific SUMO-1 siRNA not only drastically reduced the relative expression of SUMO-1 but also decreased the expression of both C/EBPβ and Lefty1, reinforcing the observation that SUMO-1 is important for transcriptional activation of these genes. Altogether, these results provide additional evidence that transactivation by KLF4 requires SUMO-1.
The KLF4 SIM Is Crucial for Its Anti-proliferative Activity
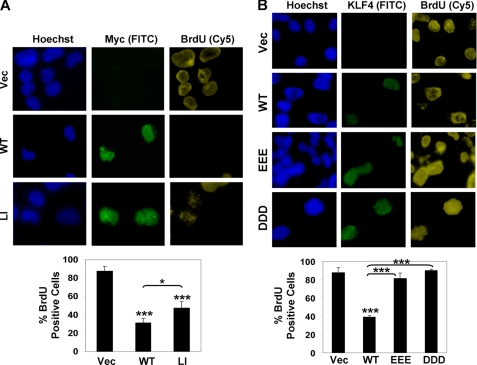
KLF4 has been shown to inhibit proliferation (41). To investigate whether the deficiencies in SUMO binding and transcriptional activation impair the ability of KLF4 to suppress cell growth, BrdU incorporation assays were performed in COS-1 cells transfected with various KLF4 constructs. As shown in Fig. 7, cells transfected with wild type pCMV-Myc-KLF4 or pMT3-KLF4 had significantly reduced proliferation compared with vector alone-transfected cell as demonstrated by a reduction in BrdU uptake in cells stained positive for KLF4 (Figs. 7, A and B, WT versus Vec). In contrast, all three KLF4 SIM mutants (Fig. 7, A, LI, and B, EEE and DDD) were less able or unable to inhibit BrdU incorporation when compared with wild type KLF4. Moreover, the degree of “disinhibition” of proliferation by the three mutants correlated with their relative ability to transactivate reporters (Figs. 4 and 5). These results indicate that the SIM in KLF4 is critical for the ability of KLF4 to modulate transcriptional activation and cellular proliferation.
FIGURE 7.
KLF4 SIM is crucial for its anti-proliferative activity. A, COS-1 cells were transfected with vector alone (Vec), pCMV-Myc-KLF4 (WT), or pCMV-Myc-KLF4-L101A/I106A (LI), labeled with BrdU, and stained with mouse BrdU and rabbit Myc antibodies. Hoechst dye was used to reveal the nuclei. B, cells were transfected with pMT3 (Vec), pMT3-KLF4 (WT), pMT3-KLF4-E93V/E95V/E96V (EEE), or pMT3-KLF4-D99V/D102V/D104V (DDD), labeled with BrdU, and stained with mouse BrdU and rabbit KLF4 antibodies. Shown are several representative cells in each panel. Between 100 and 300 cells were observed for each construct, and the percentages of KLF4-positive cells (green) that were also positive for BrdU (yellow) are shown in the charts. *, p < 0.05 and ***, p < 0.001; by two-tailed t test. Asterisks not associated with brackets are comparisons to vector alone.
DISCUSSION
Similar to ubiquitin, SUMO is a universal regulator in eukaryotes (6–8, 42). A prevailing role for SUMO is transcriptional control (7, 8, 42). However, the exact mechanism by which SUMO regulates transcription remains largely undefined. In this study we show that KLF4 is not only SUMOylated but binds SUMO-1 via a SIM. This SIM coincides with the transcriptional activation domain of KLF4. Although SIM co-localizes with the activation domain of a transcription co-activator, Sizn1 (48), our study is the first to demonstrate that SIM serves as the transcriptional activation domain of a known transcription factor (59).
Our results may help explain the often perplexing biophysical properties observed for the transcriptional activation domains of transcription factors. A transcriptional activation domain is intrinsically unstructured and mutations typically have little effect on its secondary structure (53, 60, 61). As such, a transcriptional activation domain can fold into either an α-helix (62) or a β-pleated sheet (1, 63, 64). Moreover, an unstructured activation domain may become structured upon binding to partners (1, 3, 65). It is, therefore, of interest to note that although an activation domain of a transcription factor becomes α-helical upon binding to basal transcription factors such as TATA-binding protein (TBP) (1), SIM usually assumes a β-sheet structure (6, 9, 10, 66) and can bind in either a parallel or antiparallel orientation to the β2-strand of SUMO (6, 10). Because an activation domain often has multiple binding surfaces with several possible orientations (1, 53, 67–69), it is conceivable that a transcriptional activation domain is induced to become α-helical upon binding to basal transcription factors but becomes a β-sheet upon binding to SUMO. Thus, a SIM may influence protein conformation (6). In this way a switch between the basal transcription factor binding and SUMO-binding modes may be important for the cyclic transcriptional initiation, elongation, and termination required for efficient transcription (1, 2). The dynamic levels of SUMO may tilt this balance toward either transcription activation or inhibition.
It is remarkable that a short SIM alone is sufficient to function as an activation domain. A shared feature between a SIM and an activation domain is the presence of hydrophobic and acidic residues (1, 6). Both require acidic residues, typically multiple, although on occasions positively charged residues may act as efficiently when interacting with basal transcription factors (63). In vivo, SUMO binding to SIM involves specifically orientated acidic residues (10). Our results also showed that mutation of the triple acidic residues in the KLF4 SIM to three valine residues, E93V/E95V/E96V (EEE) and D99V/D102V/D104V (DDD), abolishes SUMO-1 binding. This is consistent with the previous findings that electrostatic interaction provided by acidic or phosphorylated residues in a SIM play important roles in the affinity and orientation of SUMO-SIM interaction (6, 10, 66). We, therefore, demonstrated the coexistence of a SIM and transcriptional activation domain as a dual function motif.
Our results also provide a rationale for the often puzzling effects conferred by SUMOylation. As a major type of posttranslational modification, SUMOylation, is commonly thought to act by interaction between the conjugated SUMO in a SUMOylated protein and a SIM in an effector protein, such as a transcriptional co-repressor (6, 7). Thus, SUMO binding is pivotal in achieving the effect of SUMOylation. The recruitment of distinct co-repressors contributes to transcriptional regulation by SUMOylation (7, 70–74), but additional mechanisms, although not yet resolved, could exist (7, 74). For example, no single class of co-repressors can provide all of the suppressive effects in a transcription factor. In addition, although SUMOylation tends to suppress transcription, it can also be stimulatory (7, 8, 42). Here, we propose a new hypothesis wherein SUMOylation may inhibit a SIM-containing transcriptional activation domain through competitive interaction of the conjugated SUMO moiety with SIM. Our results are consistent with the transcriptional suppressive property of GAL4-DBD-SUMO in model systems (70–73), because sequestering additional SUMO to a promoter will compete with the interaction between a SIM and endogenously SUMOylated partners, especially when the SIM is involved in transactivation. Our results are also consistent with the localization of the above repressive activity to key hydrophobic and basic residues within a small surface on SUMO that are responsible for SIM binding (51, 75, 76). In addition, our results may explain why structures enriched in SUMOylation, such as promyelocyte leukemia nuclear bodies, are also implicated in stimulating transcription (8, 42).
It is well known that SUMO-1 mainly exists in conjugated forms, whereas there are more free forms of SUMO-2/3 (7, 8). In the current study, deficiency in SUMO-1 is sufficient to suppress KLF4 transcriptional activity, and additional deficiency in SUMO-2/3 does not lead to further inhibition (data not shown). These results suggest a functional diversity between SUMO-1 and SUMO-2/3 in transcriptional regulation and raise the question of whether this diversity is related to the SUMOylation capacity of the two SUMO classes or whether free SUMO may compete with conjugated SUMO. It is noteworthy that in various clinical disorders such as neurodegeneration and neoplasm the levels of SUMO and/or enzymes catalyzing reversible SUMOylation are significantly altered (42, 77). We predict that these alterations may result in dysregulated transcription and subsequent pathogenesis.
Another major difference between SUMO-1 and SUMO-2/3 lies in their ability to bind SIM. Some SIMs preferentially bind SUMO-1 and some primarily interact with SUMO-2/3, whereas others bind both (6, 9, 10, 66). One underlying molecular basis may be that negatively charged residues in a SIM are critical for SUMO-1 binding but less involved in SUMO-2/3 binding, although SUMO-2/3 binding does not appear to be exclusively mediated by the hydrophobic core (6, 10). Thus, the observation that knocking down SUMO-1 is sufficient to suppress KLF4 could also be attributable to preferential interaction of KLF4 SIM with SUMO-1 over SUMO-2/3.
A survey of transcription factors reveals that a considerable number of transcriptional activation domains contain consensus SIM sequences. A few such examples are listed in supplemental Fig. 6. For example, activation domains from KLF4 and v-Myb possess classic SIM consensus sequences. The activation domain of peroxisome proliferator-activated receptor α (15EADDLESPLSEEFLQEMGNIQEISQSIGEE44) (78) contains a sequence similar to the second SIM of thymine DNA glycosylase with a core sequence of VQEV (47). Mutation of Met-31 to leucine resulted in an increase in transcriptional activation, whereas M31G decreased transactivation (78). Remarkably, the M31L mutation created LQEL, another VQEV type of SIM consensus sequence, whereas M31G destroyed this consensus. AH (amphipathic α-helix), a synthetic peptide (ELQELQELQALLQQQ) extensively studied in biochemical studies of transcriptional activation (52), contains at least two patches matching thymine DNA glycosylase SIM (supplemental Fig. 6). As SUMO can bind in opposite orientations (66), the minimal VP16 activation domain (DALDDFDLDML) has considerable similarity to both the classical and VQEV types of SIMs when read from both the sense and inverted orientations (supplemental Fig. 6). The activation domain of v-Myb (TDEDPEKEKRIKELELLLMSTENEL) (58) contains typical consensuses for four divergent types of SIM sequences (supplemental Fig. 6), and it will not be surprising if this domain turns out to be a bona fide SIM. This knowledge may, thus, be useful in predicting new SIMs and transcriptional domains. As SIMs have a loose consensus and structural flexibility and variants are plentiful (6, 9, 66), there is also a high likelihood of identifying additional transcriptional domains as SIMs. The close relationship revealed by this survey strongly favors the hypothesis that a major mechanism for SUMO to regulate eukaryotic transcription is directly controlling a transcriptional domain by binding SIM-containing transcription factors.
SUMOylation of KLF4 was recently reported, but the site of SUMOylation in KLF4 was not identified (46). Although PIAS1 promotes KLF4 degradation (46), our investigation indicates that this effect is not caused by direct SUMOylation of KLF4. SUMOylated KLF4 was also reported by the same study to exist in two forms (46), the major form of which is identical to SUMOylated KLF4 revealed in our study. However, we failed to observe the minor form. This minor form, which migrates at ∼130 kDa after co-transfection of HA-tagged SUMO-1 and FLAG-tagged KLF4, was barely present in FLAG immunoblot but became almost as intensely detectable as the major SUMOylated form in HA immunoblot (46). This drastic increase in detection by HA antibody over FLAG antibody and the shift in size from ∼90 kDa for the major HA-SUMO-1-SUMOylated form to 130-kDa indicate that this minor form may be di- or tri-SUMOylated KLF4. Because SUMO-1 lacks a motif that supports poly-SUMOylation, this di-/tri-SUMOylation almost has to occur at two or three lysine sites in KLF4. However, deletion and site-directed mutagenesis studies have essentially ruled out such an additional site(s). The reason behind this discrepancy between the two studies is, therefore, not clear.
Known to play a critical role in development, differentiation, metabolism, and physiology, KLF4 has been extensively investigated in recent years, especially after it was reported to be one of the four transcription factors that induce pluripotent stem cells (14, 23, 26, 39). Thus, unveiling a general mechanism that regulates KLF4 transcriptional activity will greatly help understand the many basic events orchestrated by this multifunctional transcription factor and design better therapeutic strategies. Here, we show that KLF4 is SUMOylated at a single site and physically associated with SUMO-1. Importantly, we identify a novel SIM in KLF4 that behaves as a transcriptional activation domain. Altering SUMO interaction by both mutations of various acidic and hydrophobic residues within the SIM and siRNA against SUMO-1 critically affects KLF4 transcriptional activity in both yeast and mammalian systems. This study provides the first direct evidence that a SIM is also a transcriptional activation domain in a transcription factor, which could be a common mechanism that controls transcription and uncovers a novel role by which SUMO regulates transcription.
Supplementary Material
Acknowledgments
We are indebted to F. Claessens, A. Dejean, M. D. Lane, K. Morohashi, H. Niwa, and H. Will for providing plasmids and M. Nandan for expert technical advice in quantitative PCR.
This work was supported, in whole or in part, by National Institutes of Health Grants DK52230, DK64399, DK76742, DK77381, and CA84197.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–6.
- SUMO
- small ubiquitin-related modifier
- SM
- SUMOylation motif
- Ade
- adenine
- DBD
- DNA binding domain
- KLF4
- Krüppel-like factor 4
- SIM
- SUMO-interacting motif
- AH
- amphipathic α-helix.
REFERENCES
- 1.Triezenberg S. J. (1995) Curr. Opin. Genet. Dev. 5, 190–196 [DOI] [PubMed] [Google Scholar]
- 2.Xiao H., Friesen J. D., Lis J. T. (1994) Mol. Cell. Biol. 14, 7507–7516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang J., Kim D. H., Lee S. W., Choi K. Y., Sung Y. C. (1995) J. Biol. Chem. 270, 25014–25019 [DOI] [PubMed] [Google Scholar]
- 4.Seipel K., Georgiev O., Schaffner W. (1994) Biol. Chem. Hoppe Seyler 375, 463–470 [DOI] [PubMed] [Google Scholar]
- 5.Sekiyama N., Ikegami T., Yamane T., Ikeguchi M., Uchimura Y., Baba D., Ariyoshi M., Tochio H., Saitoh H., Shirakawa M. (2008) J. Biol. Chem. 283, 35966–35975 [DOI] [PubMed] [Google Scholar]
- 6.Kerscher O. (2007) EMBO Rep. 8, 550–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gill G. (2005) Curr. Opin. Genet. Dev. 15, 536–541 [DOI] [PubMed] [Google Scholar]
- 8.Heun P. (2007) Curr. Opin. Cell Biol. 19, 350–355 [DOI] [PubMed] [Google Scholar]
- 9.Song J., Durrin L. K., Wilkinson T. A., Krontiris T. G., Chen Y. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 14373–14378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hecker C. M., Rabiller M., Haglund K., Bayer P., Dikic I. (2006) J. Biol. Chem. 281, 16117–16127 [DOI] [PubMed] [Google Scholar]
- 11.Hannich J. T., Lewis A., Kroetz M. B., Li S. J., Heide H., Emili A., Hochstrasser M. (2005) J. Biol. Chem. 280, 4102–4110 [DOI] [PubMed] [Google Scholar]
- 12.Minty A., Dumont X., Kaghad M., Caput D. (2000) J. Biol. Chem. 275, 36316–36323 [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Dominguez M., Reyes J. C. (2009) Biochim. Biophys. Acta. 1789, 451–459 [DOI] [PubMed] [Google Scholar]
- 14.Takahashi K., Yamanaka S. (2006) Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 15.Yamanaka S. (2008) Cell Prolif. 41, 51–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McConnell B. B., Ghaleb A. M., Nandan M. O., Yang V. W. (2007) BioEssays 29, 549–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghaleb A. M., Nandan M. O., Chanchevalap S., Dalton W. B., Hisamuddin I. M., Yang V. W. (2005) Cell Res. 15, 92–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghaleb A. M., Yang V. W. (2008) Curr. Colorectal Cancer Rep. 4, 59–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nandan M. O., Yang V. W. (2009) Histol. Histopathol. 24, 1343–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evans P. M., Liu C. (2008) Acta Biochim. Biophys. Sin. (Shanghai) 40, 554–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzuki T., Aizawa K., Matsumura T., Nagai R. (2005) Arterioscler. Thromb. Vasc. Biol. 25, 1135–1141 [DOI] [PubMed] [Google Scholar]
- 22.Nakatake Y., Fukui N., Iwamatsu Y., Masui S., Takahashi K., Yagi R., Yagi K., Miyazaki J., Matoba R., Ko M. S., Niwa H. (2006) Mol. Cell. Biol. 26, 7772–7782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Birsoy K., Chen Z., Friedman J. (2008) Cell Metab. 7, 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katz J. P., Perreault N., Goldstein B. G., Lee C. S., Labosky P. A., Yang V. W., Kaestner K. H. (2002) Development 129, 2619–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Segre J. A., Bauer C., Fuchs E. (1999) Nat. Genet. 22, 356–360 [DOI] [PubMed] [Google Scholar]
- 26.Moore D. L., Blackmore M. G., Hu Y., Kaestner K. H., Bixby J. L., Lemmon V. P., Goldberg J. L. (2009) Science 326, 298–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subang M. C., Richardson P. M. (2009) Science 326, 238–239 [DOI] [PubMed] [Google Scholar]
- 28.Wei D., Kanai M., Huang S., Xie K. (2006) Carcinogenesis 27, 23–31 [DOI] [PubMed] [Google Scholar]
- 29.Atkins G. B., Jain M. K. (2007) Circ. Res. 100, 1686–1695 [DOI] [PubMed] [Google Scholar]
- 30.Feinberg M. W., Wara A. K., Cao Z., Lebedeva M. A., Rosenbauer F., Iwasaki H., Hirai H., Katz J. P., Haspel R. L., Gray S., Akashi K., Segre J., Kaestner K. H., Tenen D. G., Jain M. K. (2007) EMBO J. 26, 4138–4148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghaleb A. M., Katz J. P., Kaestner K. H., Du J. X., Yang V. W. (2007) Oncogene 26, 2365–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Callewaert L., Verrijdt G., Haelens A., Claessens F. (2004) Mol. Endocrinol. 18, 1438–1449 [DOI] [PubMed] [Google Scholar]
- 33.Komatsu T., Mizusaki H., Mukai T., Ogawa H., Baba D., Shirakawa M., Hatakeyama S., Nakayama K. I., Yamamoto H., Kikuchi A., Morohashi K. (2004) Mol. Endocrinol. 18, 2451–2462 [DOI] [PubMed] [Google Scholar]
- 34.Geiman D. E., Ton-That H., Johnson J. M., Yang V. W. (2000) Nucleic Acids Res. 28, 1106–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J. W., Klemm D. J., Vinson C., Lane M. D. (2004) J. Biol. Chem. 279, 4471–4478 [DOI] [PubMed] [Google Scholar]
- 36.Fogal V., Gostissa M., Sandy P., Zacchi P., Sternsdorf T., Jensen K., Pandolfi P. P., Will H., Schneider C., Del Sal G. (2000) EMBO J. 19, 6185–6195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muller S., Berger M., Lehembre F., Seeler J. S., Haupt Y., Dejean A. (2000) J. Biol. Chem. 275, 13321–13329 [DOI] [PubMed] [Google Scholar]
- 38.Du J. X., Bialkowska A. B., McConnell B. B., Yang V. W. (2008) J. Biol. Chem. 283, 31991–32002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hagos E. G., Ghaleb A. M., Dalton W. B., Bialkowska A. B., Yang V. W. (2009) Oncogene 28, 1197–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nandan M. O., McConnell B. B., Ghaleb A. M., Bialkowska A. B., Sheng H., Shao J., Babbin B. A., Robine S., Yang V. W. (2008) Gastroenterology 134, 120–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shields J. M., Christy R. J., Yang V. W. (1996) J. Biol. Chem. 271, 20009–20017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Müller S., Ledl A., Schmidt D. (2004) Oncogene. 23, 1998–2008 [DOI] [PubMed] [Google Scholar]
- 43.Perdomo J., Verger A., Turner J., Crossley M. (2005) Mol. Cell. Biol. 25, 1549–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hietakangas V., Ahlskog J. K., Jakobsson A. M., Hellesuo M., Sahlberg N. M., Holmberg C. I., Mikhailov A., Palvimo J. J., Pirkkala L., Sistonen L. (2003) Mol. Cell. Biol. 23, 2953–2968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hsu Y. H., Sarker K. P., Pot I., Chan A., Netherton S. J., Bonni S. (2006) J. Biol. Chem. 281, 33008–33018 [DOI] [PubMed] [Google Scholar]
- 46.Kawai-Kowase K., Ohshima T., Matsui H., Tanaka T., Shimizu T., Iso T., Arai M., Owens G. K., Kurabayashi M. (2009) Arterioscler. Thromb. Vasc. Biol. 29, 99–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mohan R. D., Rao A., Gagliardi J., Tini M. (2007) Mol. Cell. Biol. 27, 229–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cho G., Lim Y., Golden J. A. (2009) J. Biol. Chem. 284, 19592–19600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin D. Y., Huang Y. S., Jeng J. C., Kuo H. Y., Chang C. C., Chao T. T., Ho C. C., Chen Y. C., Lin T. P., Fang H. I., Hung C. C., Suen C. S., Hwang M. J., Chang K. S., Maul G. G., Shih H. M. (2006) Mol. Cell 24, 341–354 [DOI] [PubMed] [Google Scholar]
- 50.Shen T. H., Lin H. K., Scaglioni P. P., Yung T. M., Pandolfi P. P. (2006) Mol. Cell 24, 331–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ouyang J., Shi Y., Valin A., Xuan Y., Gill G. (2009) Mol. Cell 34, 145–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Giniger E., Ptashne M. (1987) Nature 330, 670–672 [DOI] [PubMed] [Google Scholar]
- 53.Sainz M. B., Goff S. A., Chandler V. L. (1997) Mol. Cell. Biol. 17, 115–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Breiding D. E., Grossel M. J., Androphy E. J. (1996) Virology 221, 34–43 [DOI] [PubMed] [Google Scholar]
- 55.Defossez P. A., Baert J. L., Monnot M., de Launoit Y. (1997) Nucleic Acids Res. 25, 4455–4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Remacle J. E., Albrecht G., Brys R., Braus G. H., Huylebroeck D. (1997) EMBO J. 16, 5722–5729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tell G., Perrone L., Fabbro D., Pellizzari L., Pucillo C., De Felice M., Acquaviva R., Formisano S., Damante G. (1998) Biochem. J. 329, 395–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang D. M., Lipsick J. S. (2002) Oncogene 21, 1611–1615 [DOI] [PubMed] [Google Scholar]
- 59.Shields J. M., Yang V. W. (1998) Nucleic Acids Res. 26, 796–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rajagopalan S., Andreeva A., Teufel D. P., Freund S. M., Fersht A. R. (2009) J. Biol. Chem. 284, 21728–21737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O'Hare P., Williams G. (1992) Biochemistry 31, 4150–4156 [DOI] [PubMed] [Google Scholar]
- 62.Donaldson L., Capone J. P. (1992) J. Biol. Chem. 267, 1411–1414 [PubMed] [Google Scholar]
- 63.Leuther K. K., Salmeron J. M., Johnston S. A. (1993) Cell 72, 575–585 [DOI] [PubMed] [Google Scholar]
- 64.Van Hoy M., Leuther K. K., Kodadek T., Johnston S. A. (1993) Cell 72, 587–594 [DOI] [PubMed] [Google Scholar]
- 65.Kim D. H., Lee S. H., Nam K. H., Chi S. W., Chang I., Han K. H. (2009) BMB Rep. 42, 411–417 [DOI] [PubMed] [Google Scholar]
- 66.Song J., Zhang Z., Hu W., Chen Y. (2005) J. Biol. Chem. 280, 40122–40129 [DOI] [PubMed] [Google Scholar]
- 67.Jonker H. R., Wechselberger R. W., Boelens R., Folkers G. E., Kaptein R. (2005) Biochemistry 44, 827–839 [DOI] [PubMed] [Google Scholar]
- 68.McAndrew P. C., Svaren J., Martin S. R., Hörz W., Goding C. R. (1998) Mol. Cell. Biol. 18, 5818–5827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Drysdale C. M., Jackson B. M., McVeigh R., Klebanow E. R., Bai Y., Kokubo T., Swanson M., Nakatani Y., Weil P. A., Hinnebusch A. G. (1998) Mol. Cell. Biol. 18, 1711–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Holmstrom S., Van Antwerp M. E., Iñiguez-Lluhi J. A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 15758–15763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ross S., Best J. L., Zon L. I., Gill G. (2002) Mol. Cell 10, 831–842 [DOI] [PubMed] [Google Scholar]
- 72.Yang S. H., Jaffray E., Hay R. T., Sharrocks A. D. (2003) Mol. Cell 12, 63–74 [DOI] [PubMed] [Google Scholar]
- 73.Yang S. H., Sharrocks A. D. (2004) Mol. Cell 13, 611–617 [DOI] [PubMed] [Google Scholar]
- 74.Kuo H. Y., Chang C. C., Jeng J. C., Hu H. M., Lin D. Y., Maul G. G., Kwok R. P., Shih H. M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 16973–16978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chupreta S., Holmstrom S., Subramanian L., Iñiguez-Lluhí J. A. (2005) Mol. Cell. Biol. 25, 4272–4282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rosendorff A., Sakakibara S., Lu S., Kieff E., Xuan Y., DiBacco A., Shi Y., Shi Y., Gill G. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 5308–5313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pountney D. L., Huang Y., Burns R. J., Haan E., Thompson P. D., Blumbergs P. C., Gai W. P. (2003) Exp. Neurol. 184, 436–446 [DOI] [PubMed] [Google Scholar]
- 78.Hi R., Osada S., Yumoto N., Osumi T. (1999) J. Biol. Chem. 274, 35152–35158 [DOI] [PubMed] [Google Scholar]
- 79.Venteclef N., Jakobsson T., Ehrlund A., Damdimopoulos A., Mikkonen L., Ellis E., Nilsson L. M., Parini P., Jänne O. A., Gustafsson J. A., Steffensen K. R., Treuter E. (2010) Genes. Dev. 24, 381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chalkiadaki A., Talianidis I. (2005) Mol. Cell. Biol. 25, 5095–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu R., Zheng H. Q., Zhou Z., Dong J. T., Chen C. (2009) J. Biol. Chem. 284, 16791–16798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen Z. Y., Wang X., Zhou Y., Offner G., Tseng C. C. (2005) Cancer Res. 65, 10394–10400 [DOI] [PubMed] [Google Scholar]
- 83.Tabibzadeh S., Hemmati-Brivanlou A. (2006) Stem Cells 24, 1998–2006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.