Abstract
Completion of the human genome project and rapid progress in genetics and bioinformatics have enabled the development of large public databases, which include genetic and genomic data linked to clinical health data. With the massive amount of information available, clinicians and researchers have the unique opportunity to complement and integrate their daily practice with the existing resources to clarify the underlying etiology of complex phenotypes such as allergic diseases. The genome itself is now often utilized as a starting point for many studies and multiple innovative approaches have emerged applying genetic/genomic strategies to key questions in the field of allergy and immunology. There have been several successes, which have uncovered new insights into the biologic underpinnings of allergic disorders. Herein, we will provide an in depth review of genomic approaches to identifying genes and biologic networks involved in allergic diseases. We will discuss genetic and phenotypic variation, statistical approaches for gene discovery, public databases, functional genomics, clinical implications, and the challenges that remain.
Keywords: gene, allergy, database, browser, genome, common variants, rare variants, HapMap, imputation
Human Genome Variation
Human genome variation encompasses all of the genetic characteristics observed within the human species. Genetic variation occurs both within and among populations and is the basis for natural selection. Insights regarding the distribution of genetic variants among human populations have recently become available1. Interestingly, human genetic diversity decreases in native populations as the migratory distance from Africa increases, presumably due to limitations in human migration2.
Nucleotide diversity is based on single mutations called single nucleotide polymorphisms (SNPs), which occur at a rate of 1 SNP per 1,000 base pairs3. Currently, there are more than 12 million SNPs deposited in GenBank, 6.5 million of which have been validated (http://www.ncbi.nih.gov/SNP). The bulk of variations at these nucleotide levels are not visible at the phenotypic level. A better understanding of the basis of genetic diversity was gained with the publication of full sequences of individuals genomes4, 5. The Human Genome Project and a parallel project by Celera Genomics yielded two haploid sequences, however, analysis of diploid sequences has revealed that non-SNP variation accounts for much more human genetic variation than single nucleotide diversity. Non-SNP variation includes copy number variation and results from deletions, inversions, insertions and duplications5. Copy number variation regions (CNVRs) have been found in 12% of the genome. CNVRs can be markedly different between populations and contain hundreds of genes, disease loci, functional elements and segmental duplications5. Taking into account this variation as well as SNPs, human to human genetic variation is estimated to be approximately 0.5%. This 0.5% difference amounts to a significant number of distinct genetic traits that uniquely distinguish the genome of every person and contribute to unique and distinct risks for diseases, responses to environmental exposures (including nutrition), and responses to pharmacologic treatment.
Epigenetic Variation in Allergic Disorders
Epigenetic variation does not affect the underlying DNA code, but rather modifies how it is expressed through covalent modifications including DNA methylation, histone modifications, and microRNAs. It is the structural adaptation of chromosomal regions so as to register, signal or perpetuate altered activity states6. Detailed analysis of methylation across several chromosomes has demonstrated that the promoter region of nearly 20% of genes are methylated, many of which influence transcription7. Progressive accumulation of phenotypic differences between genetically identical monozygotic (MZ) twins illustrates how pollution, smoking, mold, diet, habits or, in general, environment can shape phenotype and disease susceptibility. MZ are epigenetically indistinguishable early in life but, with age, exhibit substantial differences in particular when they have led different lifestyles and had spent less of their lives together8, 9. Therefore, MZ twin discordance for many common disorders could be interpreted as the result of external, environmental factors that modulate susceptibility through a change in the profile of epigenetic modifications that ultimately determine gene function. The field of epigenetics has emerged to explain how cells with the same DNA can differentiate into alternative cell types and how a phenotype can be passed from one cell to its daughter cells. It is now well established that epigenetic mechanisms are important to control the pattern of gene expression during development, the cell cycle, and in response to biological or environmental changes10–13. Unlike genetic alterations, which are permanent and usually affect all cells, epigenetic modifications are cell type specific14. Epigenetic regulation of the immune system occurs at many levels including the differentiation of T cells6, 15–19. Epigenetic effects on gene expression may persist even after the removal of the inducing agent, and can be passed on, through mitosis, to subsequent cell generations, constituting a heritable, epigenetic change. In a somatic cell, a heritable change can generate a dysfunctional clone of cells with phenotypic consequences (e.g. a tumor). In a germ-line cell, a heritable change may be transmitted to the germ cells themselves (sperm or ova) and potentially to the next generation. In this model, epialleles may be in linkage disequilibrium with SNPs that are genotyped in genome-wide association studies. The role of epigenetics in allergic disease is becoming increasingly evident. One recent study showed that epigenetic reprogramming involving aberrant DNA methylation of a 5′-CpG island in acyl-CoA synthetase long-chain family member 3 (ACSL3) was significantly associated with asthma risk in children born to mothers exposed to air pollutants such as traffic-related combustion emissions20. Another study found that neonates of allergic mothers are born with substantial changes in DNA methylation in their splenic dendritic cells and these dendritic cells show enhanced allergen-presentation activity in vitro21. Current knowledge of epigenetics in allergic diseases is limited and novel applications of epigenetic approaches including genome wide approaches to allergic diseases are necessary to uncover the role of epigenetics.
Defining Phenotypic Variation in Allergic Disease
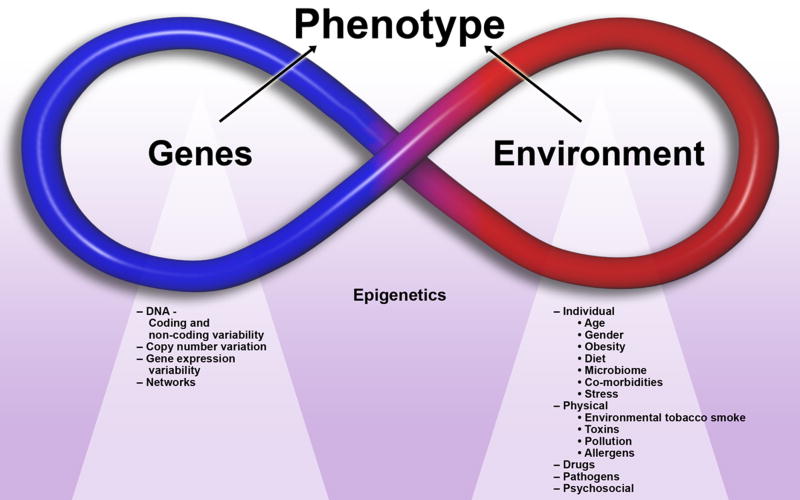
Phenotype is the observable characteristics of an organism, as determined by both genetic makeup and environmental influences, including individual, physical, psychosocial environmental exposures (Figure 1). Genotype is the descriptor of the genome which is the set of physical DNA molecules inherited from the organism’s parents, while phenotype is the descriptor of the phenome, the manifest physical properties of the organism including its physiology, morphology and behavior.
Figure 1.
Genetic and environmental factors, which interact to determine the overall phenotype.
Although single gene disorders in classical Mendelian inheritance result in direct genotype-phenotype correspondence, the relationship between genotype and phenotype in traits of multifactorial (complex) inheritance is complicated. In complex diseases with a multifaceted phenotype such as asthma, a given genotype can result in many different phenotypes and there are different genotypes corresponding to a given phenotype. While an individual’s genotype is fairly stable over a lifetime, an individual’s phenotype is dynamic, influenced by both the environment and the underlying genotype, including interactions between them22. The definition, measurement, and validity of phenotyping need to be standardized to increase the quality of research and the reproducibility of genetic studies22. Indeed, recently, NIH launched an initiative (PhenX) to address the need standardized phenotype and environmental exposures measures for cross-study comparison in genetics studies23. These measures do not include information for allergic diseases, however, the National Institute of Allergy and Infectious Diseases recently partnered with the National Heart, Lung, and Blood Institute, the National Institute of Environmental Health Sciences, the National Institute of Child Health and Human Development, the Agency for Healthcare Research and Quality, the Merck Childhood Asthma Network, and the Robert Wood Johnson Foundation to host a Asthma Outcomes Workshop. The objective of this workshop was to develop standardized definitions and data collection methodologies for established and validated asthma outcomes measures. The goal is that these outcomes will be broadly used in NIH-funded studies24.
There are several important variables to consider when defining a phenotype for studies of allergic disorders including disease definition, atopic status, comorbidities, and disease outcomes. For example, severe asthma is a recognized asthma phenotype defined by receiving ongoing treatment with high-dose inhaled corticosteroids, oral corticosteroids, or both for at least 6 months with persistent symptoms or exacerbations when the controller medications are tapered25. However, “severe asthma” is not a single phenotype. Population studies have revealed differences in severe asthma that begins in childhood versus adulthood26–28. Childhood-onset asthma is often “allergic”, while adulthood-onset asthma is more heterogeneous and often is not related to allergy, but rather to other influences including aspirin sensitivity, hormonal influences, and occupational exposures. This heterogeneity strongly supports the need for genetic studies aimed at uncovering the mechanistic bases for each distinct phenotype, rather than the mixed phenotype of asthma.
Age is an important factor in defining phenotypes for allergic disorders. As a population ages, it will be exposed to more environmental factors (e.g. environmental tobacco smoke, diesel exhaust, air pollution) that contribute to the pathogenesis of asthma and allergy, thus increasing sporadic (non-genetic) occurrences of these disorders. Thus, when studying a cohort of adults, there will be a proportion of individuals who could be classified as having asthma because of environmental exposures without a major genetic risk. Children on the other hand, may reduce the heterogeneity of the etiology of asthma because they have had minimal time to accumulate environment exposures, which would increase the risk of asthma. Given the risks of misclassification of asthma in the very young and the heterogeneity in the older groups, serious attention should be focused on the ages of participants. There has been a strong focus on powering genetic studies with very large sample sizes, however, large cohorts may not help improve our understanding of the genetic underpinnings of allergy phenotypes as much as precise phenotyping. Phenotypes can be defined through combinations of clinical information and individual biomarker and molecular data.
The phenotypic definition of controls is another important consideration, especially in studies of allergic disease where some features may be overlapping. For example, allergic sensitization may overlap with childhood asthma, so if a study aims to identify specifically childhood asthma genes, the control group should include sensitized subjects without asthma. The selection of the controls should be based on the goals of the research. With the availability of genotypic and phenotypic data through public resources such as dbGAP (http://www.ncbi.nlm.nih.gov/gap), it is enticing to consider the recruitment of controls as unnecessary. However, controls unselected with respect to phenotype increases the number of participants required to obtain similar power when using controls, which do not have the phenotype of interest. This is compounded by the fact that the publicly available controls are likely to be from a different population than the cases. When this situation occurs, researchers should consider applying genetic ancestry matching (discussed below) to minimize population stratification29.
Statistical Approaches to Finding Genetic Variation in Allergic Disorders
There are three main statistical approaches to gene discovery, linkage, association, and admixture mapping. Linkage analysis tests to determine whether a variant co-segregates with disease in families; association analysis tests to determine whether a genetic variant occurs more often in individuals with disease than without disease; and admixture mapping tests to determine whether there particular regions of the genome at which inheriting DNA from ancestors from a certain region of the world predisposes one to particular diseases. Linkage studies can be performed only in family-based studies, while association testing and admixture mapping can be performed in both population- or family-based studies. These approaches may appear to ask the same questions, but statistically these are independent tests, and the strategy affects the hypotheses that can be tested.
Linkage analysis is based on the assumption that the genetic marker and the disease variant are in close proximity and transmitted intact across generations30. Thus, markers in close proximity to the disease-causing gene segregate with disease in families. However, the resolution of linkage is poor with candidate regions encompassing hundreds of genes. Thus, linkage analysis only identifies regions not genes or variants. Further, as linkage is statistical evidence, replication is the gold standard to minimize the risk of false positives.
An alternative approach is an association study, which can utilize population or family based designs. It is important to recognize that association does not equal causation. Association studies simply measures statistical dependence between two or more variables. Significant associations can be due to one of several misleading factors including LD, population stratification, or random chance. Once significance is achieved, replication is required to ensure the validity31.
Admixture occurs when two or more genetically diverse populations merge to form a new population32. Localizing disease genes using an admixed population is called admixture mapping. In human admixture studies, researchers combine information about known population history with information from individuals’ measured genotypes using known ancestry informative markers (AIMs). Studies consistently show that allergic disorders such as asthma are more common in people of West African ancestry compared with people of European ancestry33. The African-American population is an admixed population for which about 20% of the genetic material traces to European ancestry34. The association between increased asthma risk and African ancestry and the admixed nature of the African-American population34 suggests that admixture mapping35 might be an important asthma gene-finding strategy to study genetically heterogeneous populations.
With current technology, it is not cost prohibitive to perform genome-wide linkage and association studies. An advantage of the genome-wide approach is that it requires no a priori evidence and, thus, has the ability to identify regions and variants in genes previously not implicated in allergic disorders and provide insights into the biologic underpinnings for these disorders. Researchers using genome wide approaches must adjust the level of significance to ensure that findings did not occur by chance; with the increased numbers of statistical tests, the likelihood of obtaining a p-value of 0.05 increases. For the current GWAS SNP chips (density 1M SNPs), significance thresholds of 10−8 are required31 to control for multiple comparisons. Given this level of significance, the number of samples required to obtain adequate power in a genome wide association study (GWAS) is in the thousands for a gene with modest effect. By limiting the analysis to those gene regions, which have promising a priori evidence of being involved with asthma, the severity of the correction for multiple testing becomes much less severe. A candidate gene study examining 1000 SNPs will require only 60.5% of the sample size required by a GWAS study examining 1 million SNPs to obtain the same statistical power of 80%. This reduced sample requirement may permit better phenotyping and reduced heterogeneity, which will also improve the power. Thus, there are benefits to both GWAS and candidate gene approaches.
Because asthma is a prevalent disorder, the classic population based sampling strategy is case-control. In this approach, the researcher collects individuals with disease (cases) and unrelated individuals without disease (controls). This method is very efficient; compared to a random sampling design, only 35% of the total sample would be required for equivalent power (assuming an asthma frequency of 10%). While this approach appears simple, the challenge is ensuring that the controls come from the same ancestrally homogeneous population as the cases. When cases and controls are not drawn from the same ancestral population, population stratification can result in spurious associations36. For example, suppose most people of African ancestry in a sample had brown eyes and also happened to have asthma, while most people of European ancestry were blue-eyed and asthma-free. A naïve analysis might conclude that the brown-eyes SNP is responsible for asthma, even if eye color and disease are completely unrelated. That is, the methods are likely to nab the wrong SNP suspects, due to “guilty by association”. This problem becomes more pronounced in studies surveying the entire genome because of the huge number of ancestry-related SNPs being tested. To address this genetic-mixing problem, researchers can test whether cases and controls differ over a large number of variants not expected to be associated with disease. If differences exist, adjustments can be made to minimize this effect37. Currently, three fundamentally different methods are used to correct for confounding in allergy genetic association studies37–39. These methods are (1) genomic control, (2) structured association, and (3) principal component analysis. Genomic control uses a set of non-candidate, unlinked loci to estimate an inflation factor, l, which was caused by the population structure present and then corrects the standard Chi-square test statistic for this inflation factor. The structured association method utilizes Bayesian techniques to assign individuals to “clusters” or subpopulation classes using information from a set of non-candidate, unlinked loci and then tests for an association within each “cluster” or subpopulation class. To control for population confounding by variations in background ancestry during structural association testing (SAT), ancestry informative markers (AIMs) panel can be used35. Therefore, AIMs can be also termed structure informative markers (SIMs). These markers exhibit differences in frequencies between population groups. Importantly, care should be taken in selecting which AIMs to use as some sets may be population specific40. Principal component analysis (PCA) involves a mathematical procedure that transforms a number of possibly correlated variables into a smaller number of uncorrelated variables called principal components. It can be used to identify and adjust for population substructure37. Family based association tests protection against stratification, a decided advantage of family based designs41.
Use of Public Databases to Inform Genetic Data
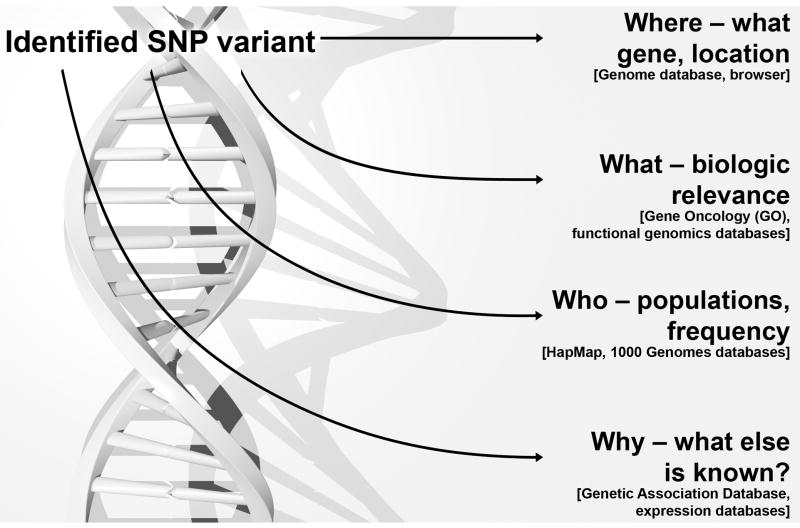
Publicly available databanks now contain billions of nucleotides sequence data collected from over 260,000 different organisms42. This proliferation of data from genome sequencing over the past decade has resulted in dramatic changes in the way the scientific community is communicating and carrying out genomic research. Once a genome wide or candidate gene study has been performed, the investigator can readily obtain information about an identified SNP, including where it is located, its potential functional significance, its frequency in different populations, and what else may already be known (Figure 2). A summary of available public resources is summarized in Table 1. The PUBMED (http://www.ncbi.nlm.nih.gov/sites/entrez) site will provide information on whether a SNP is in a gene and whether there are reported genotypic and allelic frequencies for major population groups. The database of genomic variants (http://projects.tcag.ca/variation/) is also a useful tool. This site permits the researcher to zoom out and get a broader view of the genomic region containing the SNP of interest including features such as newly reported genes, transcripts, and copy number variants. The website UCSC Genome Browser (http://genome.ucsc.edu) also provides excellent information about the features of the genome in a particular region. While each of these sites is an excellent tool to examine a small number of SNPs, a large number of SNPs can be investigated efficiently using a high throughput method, such as the SNP and CNV annotation database (http://genemem.bsd.uchicago.edu/newscan). Once the most promising SNPs have been identified, databases are available to provide estimates of putative functionality of the SNPs. FASTSNP (http://fastsnp.ibms.sinica.edu.tw/pages/input_CandidateGeneSearch.jsp) evaluates all SNPs in a gene region using the methodology proposed by Tabor and colleagues43, 44. If the SNP is non-synonymous then SNPeffect (http://snpeffect.vib.be/search.php) can provide additional information about the molecular properties of the variant. In order to determine what is already known about a specific SNP or genes in terms of disease associations, the Genetic Association Database (http://geneticassociationdb.nih.gov/) is a useful tool. It is an archive of genetic association studies. It is searchable by both disease and by gene45. A catalog of published GWAS is regularly updated and deposited at http://www.genome.gov/GWAStudies 46. Another resource available is the relationship between SNP variants and gene expression (http://www.scandb.org)47.
Figure 2.
Public databases can be utilized to rapidly provide key information about putative disease-associated genetic variants.
Table 1.
Summary of web based public databases and browsers of genetics/genomics tools
| TnQTable 1What do we know? | What is still unknown? |
|---|---|
| Genetic variation plays a large role in asthma and allergic disease risk. | Identified variants account for a small proportion of disease and the factors that contribute to the majority of the heritability of allergic diseases are still unknown. |
| Non-SNP variation accounts for much more human genetic variation than single nucleotide diversity. Copy number variation regions (CNVRs) have been found in 12% of the genome. | The impact of structural variation (including CNV) on asthma and allergic disease is unclear. Furthermore, the technical and statistical assessment of CNVs is still evolving. |
| Whole genome information and high-throughout tools are now available for high-resolution mapping. | Linkage of genetic variation to phenotypic variation and to translation into biological function is still at infancy. |
| Gene-environmental interactions play an important role in allergic diseases and have been relatively well studied in model organisms. | Rigorous quantitative assessment of environmental influences will be necessary to elucidate gene-environment in humans. |
| Epigenetic effects on gene expression may persist even after the removal of the inducing agent, and can be passed on, through mitosis, to subsequent cell generations, constituting a heritable, epigenetic change. | Approaches to efficiently dissecting the role of gene-gene and gene-environment interactions, epigenetics, and imprinting are lacking |
| There are three main statistical approaches to identify disease associated genes:, association, and admixture mapping. | A positive association does not imply causality or a direct effect on gene expression or protein function |
| Recent evidence has revealed that rare alleles with major phenotypic effects can contribute significantly to common traits in the general population. Sequencing of candidate genes or entire genomes is currently the optimal way to identify rare variants. | The role of rare variants unclear, Furthermore, while genetic association and linkage studies are well suited to find common variants for common diseases, they are not optimal for identification of rare variants. |
| Recent evidence has revealed that rare/private SNPs can contribute significantly to common traits in the general population. While genetic association and linkage studies are well suited to find common variants for common diseases, they are not optimal for identification of rare variants. Sequencing of candidate genes or entire genomes is currently the optimal way to identify rare variants. | Although rare and private SNPs are largely unknown, the 1000 Genomes Project, a deep-resequencing project will provide detailed genetic variation data on over 1000 genomes from 11 populations around the world. |
| Novel approaches to capture human genetic variation have integrated expression global gene expression arrays, DNA sequence variation arrays, and public databases. Variation in gene expression is an important mechanism underlying susceptibility to complex disease. An integrated genetic/genomic approach allows the mapping of the genetic factors that underpin individual differences in quantitative levels of expression (expression QTLs; eQTLs). | Genetic studies have identified hundreds of genetic variants associated with complex human diseases including 43 replicated genes for asthma. The variants identified so far confer relatively small increments in risk, and explain only a small proportion of disease heritability. The clinical implications, i.e., the contribution of the genetic variation to asthma subphenotypes, variations in treatment response, and different disease outcomes remain largely undetermined. |
HapMap, Tagging SNPs, and Imputation Analysis
To accelerate the identification of common disease alleles, the International HapMap Project in 2002 initiated the construction of a genome-wide SNP database of common variation (http://www.hapmap.org). In brief, the phase I and II project has genotyped over 3 million SNPs in 269 samples from 4 populations (90 Utah Residents (30 parent-offspring trios) with Northern and Western European Ancestry (CEU), 45 Han Chinese from Beijing, China (CHB), 44 Japanese from Tokyo, Japan (JPT), and 90 Yorubans (30 trios) from Ibadan, Nigeria (YRI). The average spacing of the map is one SNP per 1000 bp, and this vast resource is currently being used globally as a template for both LD-based candidate gene and genome-wide association studies in allergic disorders. To increase the sample size to over 1000 individuals in 11 populations, the HapMap phase III has recently released draft version of the dataset (http://www.hapmap.org). HapMap genotypic data, allele frequencies, LD data, phase information and sample documentation are publicly and freely available for download from HapMap website (http://www.hapmap.org).
While whole human genome sequencing is possible48, the cost and challenges with dealing with such a large quantity of data makes this approach untenable currently. However, SNPs that are physically close to one another on the chromosome are more likely to be inherited together than SNPs farther apart. Linkage disequilibrium (LD) is a measure of this non-random correlation between pairs of SNPs. Thus, if a causal variant is in LD with a marker SNP, then the marker will be associated with the phenotype proportional to the degree of LD between the two. Further, there are blocks of high LD conserved within populations49. The coinheritance between SNP alleles showing strong linkage disequilibrium, or LD enable most of the common genetic variations in a region to be captured by genotyping subsets of SNPs (termed haplotype-tagging SNPs, or tagSNPs) across a candidate gene or region of interest. Because redundant information can be reduced (thus reducing cost), many studies will often use the tagging SNP approach. A challenge is that tagging SNPs are not selected for their likelihood to be functional. However, recent work has shown that information from unmeasured SNPs can be imputed using tagging SNPs50, 51. Imputation requires use of a reference population in which genotype information is available for a large number of SNPs52. While some of these SNPs would overlap with the genotyped tagging SNPs in a given study, others would be untyped SNPs in LD with the genotyped SNPs. By delineating the genotype patterns in the reference set, researchers can make reasonable inferences about what genotypes are likely to be carried by individuals at untyped SNPs in their study. It is essential that the reference population is similar in ancestry to the population in which imputation will be performed. Fortunately, HapMap53 provides publicly available information on over 3 million SNPs in four major ancestry groups. Once imputation is performed then imputed SNPs can be tested for association with disease in the population of interest52. Since imputation interrogates all common variants, the likelihood of identifying biologically relevant associations (e.g with functional variants) is greater. Another advantage of imputation is that studies may not utilize the same SNPs in the original discovery phase. With imputation, even studies which have investigated different SNPs can be combined to determine the overall evidence for a given association52.
Rare Variants in Allergic Disorders
Most genetic studies, including GWAS, investigating common diseases have focused on common genetic variants on the assumption that common variants are mostly likely to contribute to common diseases (common disease/common variant hypothesis)54. There is emerging interest in association studies of rare variants and it is hypothesized that rare variants are more likely to be functional than common variants. Further, recent evidence supports that rare genetic variants can create synthetic associations that are credited to common variants55. While genetic association and linkage studies are well suited to find common variants for common diseases, they are not optimal for identification of rare variants56. Rare alleles with major phenotypic effects can contribute significantly to common traits in the general population57. Sequencing of candidate genes or entire genomes is the optimal way to identify rare variants. Unfortunately, most current studies are not designed or powered to identify and/or test the contributions of rare SNPs to common disease. Although current approaches are not optimal to elucidate rare variants, they can identify regions of interest, which harbor rare variants; these regions can then be further analyzed by deep resequencing (the determination of a new genome sequence relative to a reference genome is often referred to as “resequencing”).
Recently, approaches have been utilized to study the potential health impact of private SNPs, i.e. SNPs that have only been found in a given population58. In one study, investigators explored private SNPs in specific populations that may have phenotypic effects. They found that these SNPs contribute to variability in several cellular processes59. Such variability may provide clues regarding ethnicity-specific responses to diseases or drugs. Another recent study found that in African Americans, private SNPs were associated with asthma60. Investigation of rare and private SNPs requires deep sequencing approaches. The 1000 Genomes Project, a deep-resequencing project aimed at providing detailed genetic variation data on over 1000 genomes from 11 populations around the world, will aid these efforts (www.1000genomes.org). This project will identify over 95% of the variants with allele frequencies of more than 1% in human genome, substantially enhancing the HapMap data. Results from the 1000 Genome Project will provide data to allow evaluation of the common disease common variance (CD/CV) hypothesis versus the common disease many rare variants (CD/RV) hypothesis61.
Functional Genomics
Once a genetic study has been performed and allergy causing variants have been identified, the investigator can gain information to unify the biological function of gene products. Several groups have reported that genes involved in predisposing to a given polygenetic disease tend to share more commonalities (annotated by similar GO terms) in their molecular function or biological pathway than genes chosen at random or genes not involved in the same disease62–69. Gene Ontology (GO, http://www.geneontology.org) can be used to identify commonalities between gene products in the form of an agreed ontology. It provides a controlled vocabulary about genes and gene products based on known or predicted molecular function, cellular location, and biological process70. Because of the existing homologies between proteins among different taxa, the GO terms provides researchers with a powerful way to query and analyze functional genomic information in a way that is independent of species70, 71. Once genetic analyses determine which genes (among the thousands analyzed) may be related to the phenotypes, functional genomics experiments allow the scaling of the classical functional experiments to a genomic level72. The GO analysis could potentially be used to reduce the number of targets of a large group of correlated genes and to find biological functions potentially affected by multiple genes. In summary, GO annotation terms are enriched among genes linked to the trait, and such commonalities are often sufficient to narrow the list of candidate genes69.
Integration of Gene Expression and Sequence Variation Approaches in Allergic Disorders
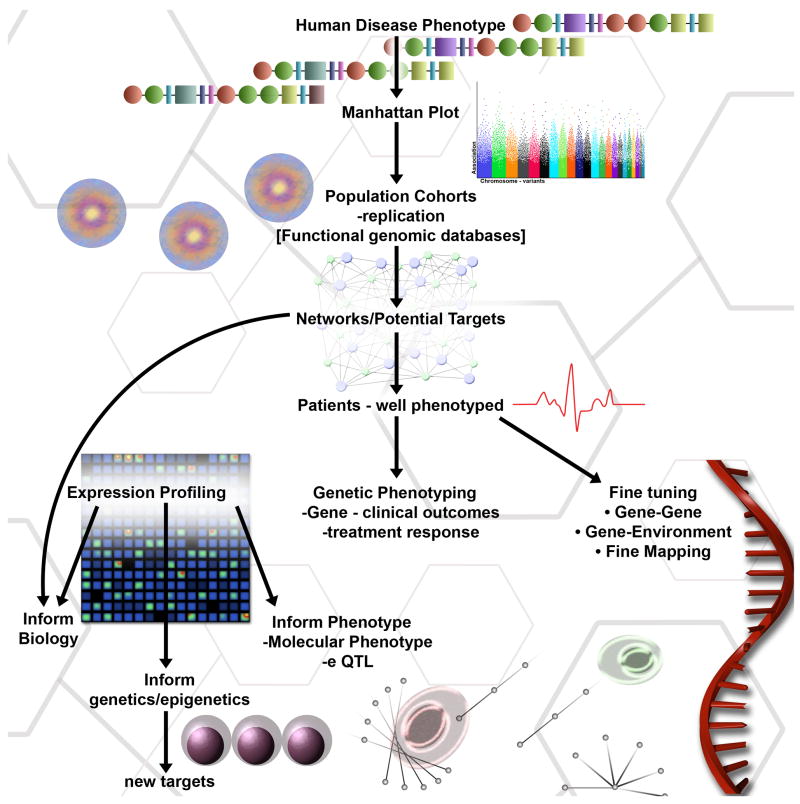
Both coding and non-coding variability contribute to genetic variation. Novel approaches to capture human genetic variation have integrated expression global gene expression arrays, DNA sequence variation arrays, and public databases (Figure 3)73. This strategy has been successfully applied to asthma74. In association studies, the investigators found markers on chromosome 17q21 to be reproducibly associated with childhood asthma. They then evaluated the relationships between the markers and transcript levels of genes in cell lines derived from children in the association study. The SNPs associated with childhood asthma were associated with transcript levels of ORMDL3, suggesting that genetic variants regulating ORMDL3 expression are determinants of susceptibility to childhood asthma. Thus, gene expression data informed the genetic data and provided insights regarding the biologic mechanisms that may be involved. Gene expression arrays can also be used in a discovery approach to identify dysregulated genes and pathways. The gene expression profiles can be used to identify key regulatory networks, to identify novel potential candidate genes, and to define phenotypes, which can then serve as quantitative traits for genetic studies. Variation in gene expression is an important mechanism underlying susceptibility to complex disease. An integrated genetic/genomic approach allows the mapping of the genetic factors that underpin individual differences in quantitative levels of expression (expression QTLs; eQTLs)75. The major public data repositories, ArrayExpress and Gene Expression Omnibus (GEO), house raw microarray data and serve as warehouses for processed experimental data, facilitating gene-based queries of multiple expression profiles. ArrayExpress (http://www.ebi.ac.uk/microarray-as/ae) is a public repository for experimental microarray data, queryable based on a range of gene annotations including gene symbols, GO terms and disease associations76. GEO (http://www.ncbi.nlm.nih.gov/geo) is a public repository that archives and freely distributes microarray, next-generation sequencing, and other forms of high-throughput functional genomic data.
Figure 3. From phenotypes to genotypes and back to phenotype.
An overview of how integrated genetic and genomics approaches can be used to inform current knowledge about allergic disorders.
Successes and Clinical Implications
Using a candidate gene approach, common mutations in filaggrin gene (FLG, 1q21) have been implicated in the causation of ichthyosis vulgaris77–79. Filaggrin80 (filament aggregation protein) is a major epidermal protein involved in maintaining the skin barrier81 and previous studies had demonstrated that filaggrin was absent or reduced in the skin cells of individuals with ichthyosis vulgaris82. Several independent replication studies have now provided convincing evidences of an association of FLG mutations with atopic dermatitis (AD)83–85. The estimated penetrance varies from 42% to 79%86, 87 i.e, between 42% and 79% of individuals with one or more FLG null mutations are likely to develop atopic dermatitis. The discovery that null mutations in FLG are associated with atopic eczema represents the single most significant breakthrough in understanding the genetic basis of this complex disorder. In addition, this association has yielded important insights into the biologic underpinnings of AD and support for the hypothesis that a barrier defect may be a contributory mechanism for the pathogenesis of AD and related atopic disorders83, 88. The exact contribution of FLG to atopic disorders remains to be delineated. The identification of patients with these FLG mutations may facilitate the targeting of novel therapies to repair or replace the defective epidermal barrier89.
Genome-wide association studies have also yielded successes. As discussed above, the association of ORMDL3 with asthma was first identified by GWAS74. Since the initial report, multiple groups have replicated the association between ORMDL3 variants and asthma90–96. Further, these variants have recently been found to associate not only with ORMDL3 expression, but with transcripts of multiple genes in this region92. Increased expression of ORMDL3 has been associated with the unfolded-protein response (UPR)97. There is still much work to be done in this area, but it further illustrates how genetic/genomic approaches can provide insights into novel biologic networks and potential disease mechanisms.
Missing Heritability and Future Directions
Genetic association including GWAS studies have identified hundreds of genetic variants associated with complex human diseases including 43 replicated genes for asthma98. Most variants identified so far confer relatively small increments in risk, and explain only a small proportion of disease heritability. This has lead to considerable speculation regarding the sources of the remaining, “missing heritability”99. Much of the speculation has focused on the possible contribution of rare variants (minor allele frequency 0.5% – 5.0%). Such variants are not sufficiently frequent to be captured by current genotyping arrays, nor do they carry sufficiently large effect sizes to be detected by current studies. With the completion of the human genome, more focus has gone into dense re-sequencing of regions. As the cost of sequencing is still high, researchers often sequence DNA pools to identify variants which that can be explored with additional genotyping100, 101. The pooled samples reliably detect variants at a frequency of 1% or greater with as little as 287 samples100. Further, if overlapping pools are used, these samples can be used to estimate allele frequencies101. Once variants are identified, the next challenge is how to proceed. Much larger samples are needed for the identification of associations with variants than those needed for the detection of the variants themselves. One technique that has been employed is to group rare variants such that the presence of any one of a number of rare variants is examined for disease association. However, this is complicated by the fact that the rare variants may have disparate effects on phenotype making this approach uninterpretable.
Structural variants, including copy number variants (including insertions and deletions) and copy neutral variation (including inversions and translocations) may account for some of the unexplained heritability102. While the variation affecting large chromosomal regions can result in large phenotypic perturbations, small/regional copy number variation can have minimal to severe effects on phenotype103. In 2006, the first comprehensive CNV map of the human genome was published104. Since then, CNVs have been associated with many different diseases including asthma105. The challenge for copy number variants is detection102. Furthermore, in a recent study, two copy number algorithms resulted in poor agreement106. Thus, while CNV analysis offers promise, the technical and statistical assessment of CNVs is still evolving107, 108.
The modest size of genetic effects detected thus far confirms the mulitfactorial etiology of these complex disorders. The next frontier of genetic studies will require innovative approaches to look for the sources of missing heritability. This will include application of whole genome sequencing to people with extreme phenotypes, use of expanded genome variation data provided by the 1,000 Genomes project, development of novel methods to detect additional sources of variation, improved phenotyping and use of eQTLs, expanded efforts in epigenetics and identification of epigenetic variation, rigorous assessment of environmental influences and gene-environment interactions, assessment of gene:gene interactions, and the design of meta-studies with well defined consistent phenotypes spanning across large population sets.
Acknowledgments
This work was supported by National Institutes of Health grants U19A170235 (GKKH and LJM) and P30HL10133 (TMB).
Abbreviations
- AM
Admixture mapping
- AIMs
ancestry informative markers
- CD/CV
common disease common variance
- CD/RV
common disease many rare variants
- CEU
Northern and Western European Ancestry
- CHB
Han Chinese from Beijing
- CNV
copy number variation
- CNVRs
copy number variation regions
- CpG
cytosine base followed immediately by a guanine base
- dbGAP
database of Genotypes and Phenotypes
- DNA
deoxyribonucleic acid
- ENCODE
Encyclopedia of DNA Elements (ENCODE)
- eQTL
expression quantitative trait loci
- FLG
Filaggrin
- GEO
Gene Expression Omnibus
- GO
gene ontology
- GWAS
genome wide association study
- HapMap
haplotype map
- JPT
Japanese from Tokyo
- LD
linkage disequilibrium
- MAF
minor allele frequency
- microRNA
small, ribonucleic acid
- MZ
monozygotic
- ORMDL3
ORM1-Like protein 3 gene
- PCA
Principal Component Analysis
- PhenX
consensus measures for Phenotypes and eXposures
- PUBMED
search engine for accessing the MEDLINE database of citations
- SAT
structural association testing
- SIMs
structural informative markers
- SNPs
single nucleotide polymorphisms
- tagSNPs
haplotype-tagging SNPs
- UCSC
University of California, Santa Cruz
- YRI
Yorubans from Ibadan
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tishkoff SA, Reed FA, Friedlaender FR, Ehret C, Ranciaro A, Froment A, et al. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–44. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manica A, Amos W, Balloux F, Hanihara T. The effect of ancient population bottlenecks on human phenotypic variation. Nature. 2007;448:346–8. doi: 10.1038/nature05951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.A haplotype map of the human genome. Nature. 2005;437:1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–54. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bird A. Perceptions of epigenetics. Nature. 2007;447:396–8. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 7.Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–85. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102:10604–9. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lara L, Calvanese V, Fraga MF. Epigenetic Drift and Aging. In: Tollefsbol TO, editor. Epigenetics of Aging. New York: Springer Science Business Media, LLC; 2010. pp. 257–73. [Google Scholar]
- 10.Brooks WH, Le Dantec C, Pers JO, Youinou P, Renaudineau Y. Epigenetics and autoimmunity. J Autoimmun. doi: 10.1016/j.jaut.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 11.Biniszkiewicz D, Gribnau J, Ramsahoye B, Gaudet F, Eggan K, Humpherys D, et al. Dnmt1 overexpression causes genomic hypermethylation, loss of imprinting, and embryonic lethality. Mol Cell Biol. 2002;22:2124–35. doi: 10.1128/MCB.22.7.2124-2135.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li E, Beard C, Forster AC, Bestor TH, Jaenisch R. DNA methylation, genomic imprinting, and mammalian development. Cold Spring Harb Symp Quant Biol. 1993;58:297–305. doi: 10.1101/sqb.1993.058.01.035. [DOI] [PubMed] [Google Scholar]
- 13.Sinclair KD, Allegrucci C, Singh R, Gardner DS, Sebastian S, Bispham J, et al. DNA methylation, insulin resistance, and blood pressure in offspring determined by maternal periconceptional B vitamin and methionine status. Proc Natl Acad Sci U S A. 2007;104:19351–6. doi: 10.1073/pnas.0707258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Renaudineau Y, Garaud S, Le Dantec C, Alonso-Ramirez R, Daridon C, Youinou P. Autoreactive B Cells and Epigenetics. Clin Rev Allergy Immunol. 2009 doi: 10.1007/s12016-009-8174-6. [DOI] [PubMed] [Google Scholar]
- 15.Nadeau JH. Transgenerational genetic effects on phenotypic variation and disease risk. Hum Mol Genet. 2009;18:R202–10. doi: 10.1093/hmg/ddp366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janson PC, Winerdal ME, Winqvist O. At the crossroads of T helper lineage commitment-Epigenetics points the way. Biochim Biophys Acta. 2009;1790:906–19. doi: 10.1016/j.bbagen.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 17.Lal G, Bromberg JS. Epigenetic mechanisms of regulation of Foxp3 expression. Blood. 2009;114:3727–35. doi: 10.1182/blood-2009-05-219584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Locksley RM. Nine lives: plasticity among T helper cell subsets. J Exp Med. 2009;206:1643–6. doi: 10.1084/jem.20091442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wells AD. New insights into the molecular basis of T cell anergy: anergy factors, avoidance sensors, and epigenetic imprinting. J Immunol. 2009;182:7331–41. doi: 10.4049/jimmunol.0803917. [DOI] [PubMed] [Google Scholar]
- 20.Perera F, Tang WY, Herbstman J, Tang D, Levin L, Miller R, et al. Relation of DNA methylation of 5′-CpG island of ACSL3 to transplacental exposure to airborne polycyclic aromatic hydrocarbons and childhood asthma. PLoS One. 2009;4:e4488. doi: 10.1371/journal.pone.0004488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li X, Yang A, Huang H, Zhang X, Town J, Davis B, et al. Induction of type 2 T helper cell allergen tolerance by IL-10-differentiated regulatory dendritic cells. Am J Respir Cell Mol Biol. 42:190–9. doi: 10.1165/rcmb.2009-0023OC. [DOI] [PubMed] [Google Scholar]
- 22.Schulze TG, McMahon FJ. Defining the phenotype in human genetic studies: forward genetics and reverse phenotyping. Hum Hered. 2004;58:131–8. doi: 10.1159/000083539. [DOI] [PubMed] [Google Scholar]
- 23.Stover PJ, Harlan WR, Hammond JA, Hendershot T, Hamilton CM. PhenX: a toolkit for interdisciplinary genetics research. Curr Opin Lipidol. doi: 10.1097/MOL.0b013e3283377395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Togias A, Fenton MJ, Gergen PJ, Rotrosen D, Fauci AS. Asthma in the inner city: the perspective of the National Institute of Allergy and Infectious Diseases. J Allergy Clin Immunol. 125:540–4. doi: 10.1016/j.jaci.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 25.American Thoracic Society. Proceedings of the ATS workshop on refractory asthma: current understanding, recommendations, and unanswered questions. Am J Respir Crit Care Med. 2000;162:2341–51. doi: 10.1164/ajrccm.162.6.ats9-00. [DOI] [PubMed] [Google Scholar]
- 26.Moore WC, Bleecker ER, Curran-Everett D, Erzurum SC, Ameredes BT, Bacharier L, et al. Characterization of the severe asthma phenotype by the National Heart, Lung, and Blood Institute’s Severe Asthma Research Program. J Allergy Clin Immunol. 2007;119:405–13. doi: 10.1016/j.jaci.2006.11.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levine SJ, Wenzel SE. Narrative review: the role of Th2 immune pathway modulation in the treatment of severe asthma and its phenotypes. Ann Intern Med. 152:232–7. doi: 10.1059/0003-4819-152-4-201002160-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haldar P, Pavord ID, Shaw DE, Berry MA, Thomas M, Brightling CE, et al. Cluster analysis and clinical asthma phenotypes. Am J Respir Crit Care Med. 2008;178:218–24. doi: 10.1164/rccm.200711-1754OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luca D, Ringquist S, Klei L, Lee AB, Gieger C, Wichmann HE, et al. On the use of general control samples for genome-wide association studies: genetic matching highlights causal variants. Am J Hum Genet. 2008;82:453–63. doi: 10.1016/j.ajhg.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bray MS. Genomics, genes, and environmental interaction: the role of exercise. J Appl Physiol. 2000;88:788–92. doi: 10.1152/jappl.2000.88.2.788. [DOI] [PubMed] [Google Scholar]
- 31.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–69. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 32.Chakraborty R. Gene Admixture in Human Populations: Models and Predictions. Yearbook of Physical Anthropol. 1986;29:1–43. [Google Scholar]
- 33.Higgins PS, Wakefield D, Cloutier MM. Risk factors for asthma and asthma severity in nonurban children in Connecticut. Chest. 2005;128:3846–53. doi: 10.1378/chest.128.6.3846. [DOI] [PubMed] [Google Scholar]
- 34.Parra EJ, Marcini A, Akey J, Martinson J, Batzer MA, Cooper R, et al. Estimating African American admixture proportions by use of population-specific alleles. Am J Hum Genet. 1998;63:1839–51. doi: 10.1086/302148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Redden DT, Divers J, Vaughan LK, Tiwari HK, Beasley TM, Fernandez JR, et al. Regional admixture mapping and structured association testing: conceptual unification and an extensible general linear model. PLoS Genet. 2006;2:e137. doi: 10.1371/journal.pgen.0020137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salmela E, Lappalainen T, Fransson I, Andersen PM, Dahlman-Wright K, Fiebig A, et al. Genome-wide analysis of single nucleotide polymorphisms uncovers population structure in Northern Europe. PLoS One. 2008;3:e3519. doi: 10.1371/journal.pone.0003519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 38.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 39.Pritchard JK, Stephens M, Rosenberg NA, Donnelly P. Association mapping in structured populations. Am J Hum Genet. 2000;67:170–81. doi: 10.1086/302959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Myles S, Stoneking M, Timpson N. An assessment of the portability of ancestry informative markers between human populations. BMC Med Genomics. 2009;2:45. doi: 10.1186/1755-8794-2-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spielman RS, Ewens WJ. The TDT and other family-based tests for linkage disequilibrium and association. Am J Hum Genet. 1996;59:983–9. [PMC free article] [PubMed] [Google Scholar]
- 42.Kulikova T, Akhtar R, Aldebert P, Althorpe N, Andersson M, Baldwin A, et al. EMBL Nucleotide Sequence Database in 2006. Nucleic Acids Res. 2007;35:D16–20. doi: 10.1093/nar/gkl913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuan HY, Chiou JJ, Tseng WH, Liu CH, Liu CK, Lin YJ, et al. FASTSNP: an always up-to-date and extendable service for SNP function analysis and prioritization. Nucleic Acids Res. 2006;34:W635–41. doi: 10.1093/nar/gkl236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tabor HK, Risch NJ, Myers RM. Candidate-gene approaches for studying complex genetic traits: practical considerations. Nat Rev Genet. 2002;3:391–7. doi: 10.1038/nrg796. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y, De S, Garner JR, Smith K, Wang SA, Becker KG. Systematic analysis, comparison, and integration of disease based human genetic association data and mouse genetic phenotypic information. BMC Med Genomics. 3:1. doi: 10.1186/1755-8794-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–7. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gamazon ER, Zhang W, Konkashbaev A, Duan S, Kistner EO, Nicolae DL, et al. SCAN: SNP and copy number annotation. Bioinformatics. 26:259–62. doi: 10.1093/bioinformatics/btp644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–6. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 49.Morton NE. Into the post-HapMap era. Adv Genet. 2008;60:727–42. doi: 10.1016/S0065-2660(07)00425-7. [DOI] [PubMed] [Google Scholar]
- 50.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–13. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 51.Servin B, Stephens M. Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet. 2007;3:e114. doi: 10.1371/journal.pgen.0030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Browning SR. Missing data imputation and haplotype phase inference for genome-wide association studies. Hum Genet. 2008;124:439–50. doi: 10.1007/s00439-008-0568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–7. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 55.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biol. 8:e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Visscher PM, Andrew T, Nyholt DR. Genome-wide association studies of quantitative traits with related individuals: little (power) lost but much to be gained. Eur J Hum Genet. 2008;16:387–90. doi: 10.1038/sj.ejhg.5201990. [DOI] [PubMed] [Google Scholar]
- 57.Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–72. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 58.Hinds DA, Stuve LL, Nilsen GB, Halperin E, Eskin E, Ballinger DG, et al. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–9. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 59.Baye TM, Wilke RA, Olivier M. Genomic and Geographic Distribution of Private SNPs and Pathways in Human Populations. Personalized Med. 2009;6:623–41. doi: 10.2217/pme.09.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haller G, Torgerson DG, Ober C, Thompson EE. Sequencing the IL4 locus in African Americans implicates rare noncoding variants in asthma susceptibility. J Allergy Clin Immunol. 2009;124:1204–9. e9. doi: 10.1016/j.jaci.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robinson R. Common Disease, Multiple Rare (and Distant) Variants. PLoS Biol. 2010;8:e1000293. doi: 10.1371/journal.pbio.1000293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lage K, Karlberg EO, Storling ZM, Olason PI, Pedersen AG, Rigina O, et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat Biotechnol. 2007;25:309–16. doi: 10.1038/nbt1295. [DOI] [PubMed] [Google Scholar]
- 63.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–13. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 64.Badano JL, Katsanis N. Beyond Mendel: an evolving view of human genetic disease transmission. Nat Rev Genet. 2002;3:779–89. doi: 10.1038/nrg910. [DOI] [PubMed] [Google Scholar]
- 65.Huang D, Wei P, Pan W. Combining gene annotations and gene expression data in model-based clustering: weighted method. OMICS. 2006;10:28–39. doi: 10.1089/omi.2006.10.28. [DOI] [PubMed] [Google Scholar]
- 66.Huang TH, Perry MR, Laux DE. Methylation profiling of CpG islands in human breast cancer cells. Hum Mol Genet. 1999;8:459–70. doi: 10.1093/hmg/8.3.459. [DOI] [PubMed] [Google Scholar]
- 67.Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, et al. The genetic landscape of a cell. Science. 327:425–31. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shriner D, Baye TM, Padilla MA, Zhang S, Vaughan LK, Loraine AE. Commonality of functional annotation: a method for prioritization of candidate genes from genome-wide linkage studies. Nucleic Acids Res. 2008;36:e26. doi: 10.1093/nar/gkn007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oti M, van Reeuwijk J, Huynen MA, Brunner HG. Conserved co-expression for candidate disease gene prioritization. BMC Bioinformatics. 2008;9:208. doi: 10.1186/1471-2105-9-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Clark JI, Brooksbank C, Lomax J. It’s all GO for plant scientists. Plant Physiol. 2005;138:1268–79. doi: 10.1104/pp.104.058529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jones AR, Miller M, Aebersold R, Apweiler R, Ball CA, Brazma A, et al. The Functional Genomics Experiment model (FuGE): an extensible framework for standards in functional genomics. Nat Biotechnol. 2007;25:1127–33. doi: 10.1038/nbt1347. [DOI] [PubMed] [Google Scholar]
- 73.Franke L, Jansen RC. eQTL analysis in humans. Methods Mol Biol. 2009;573:311–28. doi: 10.1007/978-1-60761-247-6_17. [DOI] [PubMed] [Google Scholar]
- 74.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–3. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 75.Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M. Mapping complex disease traits with global gene expression. Nat Rev Genet. 2009;10:184–94. doi: 10.1038/nrg2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brazma A, Parkinson H, Sarkans U, Shojatalab M, Vilo J, Abeygunawardena N, et al. ArrayExpress--a public repository for microarray gene expression data at the EBI. Nucleic Acids Res. 2003;31:68–71. doi: 10.1093/nar/gkg091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McGrath JA. Filaggrin and the great epidermal barrier grief. Australas J Dermatol. 2008;49:67–73. doi: 10.1111/j.1440-0960.2008.00443.x. quiz-4. [DOI] [PubMed] [Google Scholar]
- 78.Henderson J, Northstone K, Lee SP, Liao H, Zhao Y, Pembrey M, et al. The burden of disease associated with filaggrin mutations: a population-based, longitudinal birth cohort study. J Allergy Clin Immunol. 2008;121:872–7. e9. doi: 10.1016/j.jaci.2008.01.026. [DOI] [PubMed] [Google Scholar]
- 79.Palmer CN, Irvine AD, Terron-Kwiatkowski A, Zhao Y, Liao H, Lee SP, et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet. 2006;38:441–6. doi: 10.1038/ng1767. [DOI] [PubMed] [Google Scholar]
- 80.Elias PM, Steinhoff M. “Outside-to-inside” (and now back to “outside”) pathogenic mechanisms in atopic dermatitis. J Invest Dermatol. 2008;128:1067–70. doi: 10.1038/jid.2008.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McGrath JA, Uitto J. The filaggrin story: novel insights into skin-barrier function and disease. Trends Mol Med. 2008;14:20–7. doi: 10.1016/j.molmed.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 82.List K, Szabo R, Wertz PW, Segre J, Haudenschild CC, Kim SY, et al. Loss of proteolytically processed filaggrin caused by epidermal deletion of Matriptase/MT-SP1. J Cell Biol. 2003;163:901–10. doi: 10.1083/jcb.200304161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.O’Regan GM, Sandilands A, McLean WH, Irvine AD. Filaggrin in atopic dermatitis. J Allergy Clin Immunol. 2009;124:R2–6. doi: 10.1016/j.jaci.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 84.Weidinger S, O’Sullivan M, Illig T, Baurecht H, Depner M, Rodriguez E, et al. Filaggrin mutations, atopic eczema, hay fever, and asthma in children. J Allergy Clin Immunol. 2008;121:1203–9. e1. doi: 10.1016/j.jaci.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 85.Cookson WO, Moffatt MF. The genetics of atopic dermatitis. Curr Opin Allergy Clin Immunol. 2002;2:383–7. doi: 10.1097/00130832-200210000-00003. [DOI] [PubMed] [Google Scholar]
- 86.Marenholz I, Nickel R, Ruschendorf F, Schulz F, Esparza-Gordillo J, Kerscher T, et al. Filaggrin loss-of-function mutations predispose to phenotypes involved in the atopic march. J Allergy Clin Immunol. 2006;118:866–71. doi: 10.1016/j.jaci.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 87.Morar N, Cookson WO, Harper JI, Moffatt MF. Filaggrin mutations in children with severe atopic dermatitis. J Invest Dermatol. 2007;127:1667–72. doi: 10.1038/sj.jid.5700739. [DOI] [PubMed] [Google Scholar]
- 88.Bieber T. Atopic dermatitis. N Engl J Med. 2008;358:1483–94. doi: 10.1056/NEJMra074081. [DOI] [PubMed] [Google Scholar]
- 89.Brown SJ, Irvine AD. Atopic eczema and the filaggrin story. Semin Cutan Med Surg. 2008;27:128–37. doi: 10.1016/j.sder.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 90.Sleiman PM, Annaiah K, Imielinski M, Bradfield JP, Kim CE, Frackelton EC, et al. ORMDL3 variants associated with asthma susceptibility in North Americans of European ancestry. J Allergy Clin Immunol. 2008;122:1225–7. doi: 10.1016/j.jaci.2008.06.041. [DOI] [PubMed] [Google Scholar]
- 91.Galanter J, Choudhry S, Eng C, Nazario S, Rodriguez-Santana JR, Casal J, et al. ORMDL3 gene is associated with asthma in three ethnically diverse populations. Am J Respir Crit Care Med. 2008;177:1194–200. doi: 10.1164/rccm.200711-1644OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Verlaan DJ, Berlivet S, Hunninghake GM, Madore AM, Lariviere M, Moussette S, et al. Allele-specific chromatin remodeling in the ZPBP2/GSDMB/ORMDL3 locus associated with the risk of asthma and autoimmune disease. Am J Hum Genet. 2009;85:377–93. doi: 10.1016/j.ajhg.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Leung TF, Sy HY, Ng MC, Chan IH, Wong GW, Tang NL, et al. Asthma and atopy are associated with chromosome 17q21 markers in Chinese children. Allergy. 2009;64:621–8. doi: 10.1111/j.1398-9995.2008.01873.x. [DOI] [PubMed] [Google Scholar]
- 94.Wu H, Romieu I, Sienra-Monge JJ, Li H, del Rio-Navarro BE, London SJ. Genetic variation in ORM1-like 3 (ORMDL3) and gasdermin-like (GSDML) and childhood asthma. Allergy. 2009;64:629–35. doi: 10.1111/j.1398-9995.2008.01912.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Flory JH, Sleiman PM, Christie JD, Annaiah K, Bradfield J, Kim CE, et al. 17q12–21 variants interact with smoke exposure as a risk factor for pediatric asthma but are equally associated with early-onset versus late-onset asthma in North Americans of European ancestry. J Allergy Clin Immunol. 2009;124:605–7. doi: 10.1016/j.jaci.2009.05.047. [DOI] [PubMed] [Google Scholar]
- 96.Hirota T, Harada M, Sakashita M, Doi S, Miyatake A, Fujita K, et al. Genetic polymorphism regulating ORM1-like 3 (Saccharomyces cerevisiae) expression is associated with childhood atopic asthma in a Japanese population. J Allergy Clin Immunol. 2008;121:769–70. doi: 10.1016/j.jaci.2007.09.038. [DOI] [PubMed] [Google Scholar]
- 97.Cantero-Recasens G, Fandos C, Rubio-Moscardo F, Valverde MA, Vicente R. The asthma-associated ORMDL3 gene product regulates endoplasmic reticulum-mediated calcium signaling and cellular stress. Hum Mol Genet. 19:111–21. doi: 10.1093/hmg/ddp471. [DOI] [PubMed] [Google Scholar]
- 98.Weiss ST, Raby BA, Rogers A. Asthma genetics and genomics 2009. Curr Opin Genet Dev. 2009;19:279–82. doi: 10.1016/j.gde.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 99.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Out AA, van Minderhout IJ, Goeman JJ, Ariyurek Y, Ossowski S, Schneeberger K, et al. Deep sequencing to reveal new variants in pooled DNA samples. Hum Mutat. 2009;30:1703–12. doi: 10.1002/humu.21122. [DOI] [PubMed] [Google Scholar]
- 101.Prabhu S, Pe’er I. Overlapping pools for high-throughput targeted resequencing. Genome Res. 2009;19:1254–61. doi: 10.1101/gr.088559.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fanciulli M, Petretto E, Aitman TJ. Gene copy number variation and common human disease. Clin Genet. 2009 doi: 10.1111/j.1399-0004.2009.01342.x. [DOI] [PubMed] [Google Scholar]
- 103.Wain LV, Armour JA, Tobin MD. Genomic copy number variation, human health, and disease. Lancet. 2009;374:340–50. doi: 10.1016/S0140-6736(09)60249-X. [DOI] [PubMed] [Google Scholar]
- 104.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–54. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ionita-Laza I, Perry GH, Raby BA, Klanderman B, Lee C, Laird NM, et al. On the analysis of copy-number variations in genome-wide association studies: a translation of the family-based association test. Genet Epidemiol. 2008;32:273–84. doi: 10.1002/gepi.20302. [DOI] [PubMed] [Google Scholar]
- 106.Shtir C, Pique-Regi R, Siegmund K, Morrison J, Schumacher F, Marjoram P. Copy number variation in the Framingham Heart Study. BMC Proc. 2009;3 (Suppl 7):S133. doi: 10.1186/1753-6561-3-s7-s133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Oldridge DA, Banerjee S, Setlur SR, Sboner A, Demichelis F. Optimizing copy number variation analysis using genome-wide short sequence oligonucleotide arrays. Nucleic Acids Res. doi: 10.1093/nar/gkq073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cooper GM, Zerr T, Kidd JM, Eichler EE, Nickerson DA. Systematic assessment of copy number variant detection via genome-wide SNP genotyping. Nat Genet. 2008;40:1199–203. doi: 10.1038/ng.236. [DOI] [PMC free article] [PubMed] [Google Scholar]