Abstract
The blaTLA-1 gene encoding an extended-spectrum β-lactamase was identified in 11 enterobacterial isolates from Mexico City, Mexico. This gene was located on different plasmids and plasmid types with different sizes and incompatibility groups. It was associated with a novel insertion sequence, ISCR20, encoding a putative transposase that shared only 20% amino acid identity with the most closely related transposase of ISCR1. The ISCR20 element provided specific promoter sequences for expression of the blaTLA-1 gene.
Although the majority of Ambler class A extended-spectrum β-lactamases (ESBLs) identified in the family Enterobacteriaceae worldwide are mostly of the TEM, SHV, and CTX-M types, some “minor” ESBLs have been reported, such as VEB-like, GES-like, BES-1, SFO-1, and TLA-1 enzymes (5). The blaTLA-1 gene was first reported in 2000 from an Escherichia coli clinical isolate from Mexico (8). From April 2000 until February 2002, the blaTLA-1 gene was detected from an epidemic Klebsiella pneumoniae clone in Mexico, and both the blaTLA-1 and blaSHV-5 ESBL genes were located on the same plasmid (1). The blaTLA-1 gene was identified on a 150-kb conjugative plasmid named pRZA92, but its genetic context had not been further characterized (8). Considering that many ESBL genes may be transposon or integron associated, the aim of this study was to characterize the genetic elements at the possible origin of acquisition of the blaTLA-1 gene from isolates of members of the family Enterobacteriaceae recovered from several hospitals in Mexico City, Mexico.
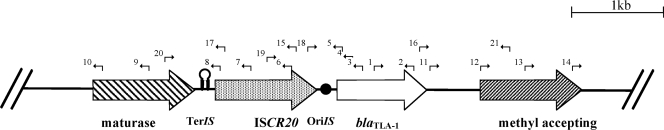
The 150-kb plasmid pRZA92 of E. coli R170 (8) was extracted by using the Qiafilter plasmid purification maxikit (Qiagen, Courtaboeuf, France) and partially sequenced using the primers indicated in Table 1. A sequence of 5,923 bp was identified through a primer-walking approach and contained several open reading frames (ORFs) (Fig. 1). Upstream of the blaTLA-1 gene, another gene (named orf432) was identified; this gene encoded a 432-amino-acid protein sharing 86% sequence identity with an uncharacterized orf430 gene (83% amino acid identity) located in a class 1 integron from a Proteus mirabilis isolate recovered from Mediterranean herring gulls in Italy (GenBank accession number DQ520941) (3). The orf432 gene encoded a putative transposase belonging to the IS91 family of transposases that includes the newly described ISCR elements. The Orf432 transposase shared 20%, 21%, and 18% amino acid identity with the transposases encoded by the ISCR1, ISCR2, and ISCR3 elements, respectively. Detailed analysis allowed us to identify the orf432 gene as part of a novel ISCR element, termed ISCR20 according to the official nomenclature (http://medicine.cf.ac.uk/en/research/research-groups/i3/research/antibacterial-agents/iscr-elements/). The ISCR20 element is 1,705 bp long and possesses a G+C content of 47.1% (Fig. 1). A putative oriIS sequence was identified 255 bp downstream of the stop codon of the transposase gene and shared 10 identical bp (5′ACTGATAGGAACTGTCATTTC3′ [the identical bases are shown in boldface type and underlined]) with the 19-bp consensus sequence reported for the other ISCRs (5′XXGTATAGGAAGTTCAAACGC3′) (10) (Fig. 1). A putative terIS sequence forming a hairpin structure (5′GGACCCGCACGCAGGGTGTT3′ [the boldfaced and underlined nucleotides indicate the complementary nucleo-tides forming the hairpin structure]) was also evidenced 136 bp upstream of the transposase start codon. The transposase of ISCR20 shared an overall significant degree of identity with members of the IS91 family transposases especially in the five functional domains previously described (Fig. 2). A tyrosine residue at position 218, which has been shown to be critical for the transposase activity of IS91 and speculated to be also critical for those of the ISCR elements, was present in motif IV of the ISCR20 transposase.
TABLE 1.
Sequences of primers used for detection of the blaTLA-1 gene and its circular form with ISCR20 and for mapping the genetic environment
| Primer name | Primer sequence | Positiona |
|---|---|---|
| TLA-1F | TGTGTGCTTTTTGCTTCTGC | 1 |
| TLA-1R | GCTTCCGGTTTTATGAGCAA | 2 |
| TLA-1-5′-TR1 | TACTGCTTTTAAGCGAATCCG | 3 |
| TLA-1-5′-TR2 | TTAGCCGCAAAAGCAGAAGC | 4 |
| TLA-1-5′-TR3 | AAGCAAAAAGCACACAAAATGC | 5 |
| IS91-5′-TR1 | ACAACCGACATTGACCGTGG | 6 |
| IS91-5′-TR4 | CTAAAACATGCCAGTGCGGG | 7 |
| IS91-5′-TR5 | AATGCTCTAGGCGGTCAAGG | 8 |
| IS91-5′-TR6 | GACATGGCTTTTTGCTGGCG | 9 |
| IS91-5′-TR7 | GATATTGCTTCGATGCAGCAC | 10 |
| TLA1-3′-TR1 | CATTGCTGTTTATGTGTCGG | 11 |
| TLA1-3′-TR4 | CATCGCGTCATTAATCCTGC | 12 |
| TLA1-3′-TR5 | GATCGAACAGATTTCCGCCT | 13 |
| TLA1-3′-TR6 | ACGAACAACAGAAAAGCGCC | 14 |
| IS91-3′extR | CTTTTGTAACAACCTTGCCC | 15 |
| TLA1-3′extF | GTATTATTACTCTGCCGAACGG | 16 |
| IS91-5′extR | TCTGAAGTACGAATGGCCAC | 17 |
| IS91-3′extF | ACCAAGATTATTGACTGGGC | 18 |
| ICR20F | CAAAGCTGATCGTGAAGCCC | 19 |
| Maturase F | AACATCATGCTCGACCCAC | 20 |
| Methyl R | CATCGAATTGAGCGAGTCG | 21 |
Positions of the primers indicated in Fig. 1. All the primers were designed in this study.
FIG. 1.
Schematic representation of the sequences surrounding the blaTLA-1 gene in E. coli X170. The positions of primers shown in Table 1 are indicated by arrows; primers 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, and 21 are primers TLA-1F, TLA-1R, TLA-1-5′-TR1, TLA-1-5′-TR2, TLA-1-5′-TR3, IS91-5′-TR1, IS91-5′-TR4, IS91-5′-TR5, IS91-5′-TR6, IS91-5′-TR7, TLA-1-3′-TR1, TLA-1-3′-TR4, TLA-1-3′-TR5, TLA-1-3′-TR6, IS91-3′-extR, TLA-1-3′extF, IS91-5′-extR, IS91-3′extF, ISCR20F, maturase F, and methyl R, respectively. Genes are shown by large arrows, and the transcription orientations of the genes are indicated. The putative origin of replication, oriIS, and termination of transposition, terIS, of the ISCR20 element are indicated by a solid black circle and a black hairpin, respectively.
FIG. 2.
Comparison of the sequence motifs of ISCR20 and those of the IS91 family transposases. The sequences (with their GenBank accession numbers shown in parentheses) shown are as follows: ISCR1 (FJ187822), ISCR2 (AY055428), ISCR3 group, including ISCR3, ISCR4, ISCR6, ISCR14, and ISCR16 (FJ183463), ISCR7 (AJ250371), ISCR8 (AF028594), ISCR15 (AM998375), and ISCR19 (EU503121). The five motifs found within IS91 group elements are indicated. The amino acid residues conserved in all IS91 group elements are shown boxed. Gaps introduced to maximize alignment are indicated by dashes.
The blaTLA-1 gene was followed by part of a gene encoding a methyl-accepting chemotaxis protein that shares 58% amino acid identity with that found in Desulfovibrio magneticus RS-1 (GenBank accession number YP_002952376). A group II intron, containing an orf458 gene, was identified at the left extremity of the ISCR20 element (Fig. 1). The orf458 gene encoded a putative reverse transcriptase and maturase sharing 69% amino acid identity with those of Vibrio cholerae (GenBank accession number ABV21790). Analysis of the nucleotide sequence surrounding the blaTLA-1 gene did not yield other insertion sequences that could be part of a transposon or any class 1 integron.
A collection of 11 TLA-1-producing Enterobacteriaceae clinical strains (3 E. coli strains [including E. coli R170], 1 K. pneumoniae strain, and 7 Enterobacter cloacae strains) isolated from different samples (urine, blood, and tracheal fluid samples) from 11 patients hospitalized over a 10-year period since 1991 from several hospitals in Mexico City, Mexico, were retrospectively analyzed. These isolates were selected among clinical ESBL-producing enterobacterial isolates on the basis of two criteria: their ESBL profile and their production of a β-lactamase with an isoelectric point of 9.0. By DNA restriction with XbaI endonuclease, followed by pulsed-field gel electrophoresis (PFGE) analysis, we showed that most of the isolates were not clonally related, with four distinct E. cloacae PFGE types and three distinct E. coli PFGE types, suggesting a diffusion of blaTLA-1-positive plasmids. The blaTLA-1 gene was transferred from seven of those distinct PFGE types to E. coli J53 by conjugation performed as described previously (6). Together with the expression of an ESBL phenotype, the E. coli transconjugants were resistant to trimethoprim, sulfamethoxazole, and tetracycline, and some of these transconjugants were resistant to amikacin (n = 5) or gentamicin (n = 1). Plasmid analysis performed by the Kieser technique (4) allowed us to visualize several plasmids in all the clinical isolates but only a single plasmid from each E. coli transconjugant. Southern blot hybridization with a blaTLA-1-specific probe indicated that this ESBL gene was located on a single plasmid, being of either ca. 110 or 150 kb. PCR-based replicon typing (PBRT) analysis performed on the E. coli transconjugants (2) showed that the plasmids carrying the blaTLA-1 gene belonged to the IncA/C, IncL/M, or IncN incompatibility group, corresponding to the 150-, 150-, and 110-kb plasmids, respectively, highlighting that three different plasmid types were at the origin of the diffusion of the blaTLA-1 gene.
Except for E. coli isolate R2915 and its transconjugant, PCR amplification identified in all cases the same ISCR20 element upstream of the blaTLA-1 gene. It has been suggested that the promoter of the blaTLA-1 gene was located 71 bp from the translational start site by researchers using a computer-based promoter analysis (8). However, by using a 5′ rapid amplification of cDNA end PCR system (Invitrogen, Cergy-Pontoise, France) and primers TLA-1-5′-TR1 to TLA-1-5′-TR3 (Table 1), the initiation site of transcription of the blaTLA-1 gene was mapped from E. coli R170. Promoter sequences were located 144 bp upstream of the translational start codon, corresponding to the −35 promoter sequence TTGACA separated by the 17-bp distance from the −10 promoter sequence TTAAAG. This result showed that the blaTLA-1 gene expression was driven by the ISCR20 element. One pair of outward primers chosen at each end of the ISCR20 insertion sequence was used to detect the circular form of this novel ISCR associated with the blaTLA-1 gene. The amplifications performed using strain R170 and its transconjugant as templates failed.
Nineteen ISCR elements with weak structural relationship (18 to 96%) have been found to be associated with different families of antibiotic resistance genes. As demonstrated with the transposition model of the IS1294 elements (9), it is proposed that a transposase encoded by the ISCR has the ability to cotranspose DNA adjacent to its terminal terIS sequence through a rolling-circle (RC) transposition mechanism (10). It has been suggested that the mobilization by ISCR elements of adjacent DNA sequences likely involving sequences located only at their left extremities and the frequent observation of resistance genes bracketed by two copies of ISCR may be the consequence of secondary recombination events (10). This has been recently shown with the identification of the blaVEB-1a ESBL gene flanked by two copies of ISCR2 in Acinetobacter baumannii (7). Here, no ISCR20 element was identified downstream of the blaTLA-1 gene, casting doubt on the involvement of that element in the mobilization process of the blaTLA-1 gene. Further work is needed to understand the process that gave rise to the mobilization of the blaTLA-1 gene.
Besides describing a novel ISCR element, our study indicates its involvement in the expression of the blaTLA-1 gene and its likely role in its acquisition. In addition, we showed here that the dissemination of the blaTLA-1 gene may have already occurred in Mexico and that it was related to different plasmid backbones. Further screening of TLA-1-positive isolates should be performed at least in neighboring countries, such as countries in Central America and in the southern United States. We showed a heterogeneity of plasmid backgrounds, supporting the hypothesis that the blaTLA-1 gene was very likely harbored by a mobile genetic structure.
Nucleotide sequence accession number.
The nucleotide sequence reported in this study has been deposited in the GenBank database under accession number GU441460.
Acknowledgments
This work was funded mostly by a grant from the INSERM (U914) and by grants from the European Community (TROCAR, HEALTH-F3-2008-223031) and from the Ministère de l'Education Nationale et de la Recherche (UPRES-EA3539), Université Paris XI, France.
We thank M. Toleman for providing us with an ISCR number.
Footnotes
Published ahead of print on 28 June 2010.
REFERENCES
- 1.Alcantar-Curiel, D., J. C. Tinoco, C. Gayosso, A. Carlos, C. Daza, M. C. Perez-Prado, L. Salcido, J. I. Santos, and C. M. Alpuche-Aranda. 2004. Nosocomial bacteremia and urinary tract infections caused by extended-spectrum β-lactamase-producing Klebsiella pneumoniae with plasmids carrying both SHV-5 and TLA-1 genes. Clin. Infect. Dis. 38:1067-1074. [DOI] [PubMed] [Google Scholar]
- 2.Carattoli, A., A. Bertini, L. Villa, V. Falbo, K. L. Hopkins, and E. J. Threlfall. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219-228. [DOI] [PubMed] [Google Scholar]
- 3.Gionechetti, F., P. Zucca, F. Gombac, C. Monti-Bragadin, C. Lagatolla, E. Tonin, E. Edalucci, L. A. Vitali, and L. Dolzani. 2008. Characterization of antimicrobial resistance and class 1 integrons in Enterobacteriaceae isolated from Mediterranean herring gulls (Larus cachinnans). Microb. Drug Resist. 14:93-99. [DOI] [PubMed] [Google Scholar]
- 4.Kieser, T. 1984. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid 12:19-36. [DOI] [PubMed] [Google Scholar]
- 5.Naas, T., L. Poirel, and P. Nordmann. 2008. Minor extended-spectrum β-lactamases. Clin. Microbiol. Infect. 14(Suppl. 1):42-52. [DOI] [PubMed] [Google Scholar]
- 6.Poirel, L., M. Guibert, S. Bellais, T. Naas, and P. Nordmann. 1999. Integron- and carbenicillinase-mediated reduced susceptibility to amoxicillin-clavulanic acid in isolates of multidrug-resistant Salmonella enterica serotype typhimurium DT104 from French patients. Antimicrob. Agents Chemother. 43:1098-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poirel, L., P. D. Mugnier, M. A. Toleman, T. R. Walsh, M. J. Rapoport, A. Petroni, and P. Nordmann. 2009. ISCR2, another vehicle for blaVEB gene acquisition. Antimicrob. Agents Chemother. 53:4940-4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Silva, J., C. Aguilar, G. Ayala, M. A. Estrada, U. Garza-Ramos, R. Lara-Lemus, and L. Ledezma. 2000. TLA-1: a new plasmid-mediated extended-spectrum β-lactamase from Escherichia coli. Antimicrob. Agents Chemother. 44:997-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tavakoli, N., A. Comanducci, H. M. Dodd, M. C. Lett, B. Albiger, and P. Bennett. 2000. IS1294, a DNA element that transposes by RC transposition. Plasmid 44:66-84. [DOI] [PubMed] [Google Scholar]
- 10.Toleman, M. A., P. M. Bennett, and T. R. Walsh. 2006. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol. Mol. Biol. Rev. 70:296-316. [DOI] [PMC free article] [PubMed] [Google Scholar]