Abstract
Streptococcus mutans is known to be resistant to bacitracin, a cyclic polypeptide antibiotic produced by certain species of the genus Bacillus. This property is often exploited in the isolation of S. mutans strains from highly heterogeneous oral microflora. A genetic locus consisting of a four-gene operon, bceABRS (formerly mbrABCD), the component genes of which are homologous to Bacillus subtilis bceRS-bceAB (encoding a two-component system and an ABC transporter), is required for bacitracin resistance in S. mutans. Here we describe the identification of a DNA binding site for the BceR response regulator and its transcriptional control of the bceABRS operon in response to the presence of bacitracin. We provide evidence indicating that phosphorylated BceR binds directly to a conserved invert repeat located between bp −120 and −78 of the bceABRS promoter region and positively regulates expression of the bceABRS operon. We also demonstrate that sensing of bacitracin by the BceS histidine kinase requires the presence of an intact BceAB transporter, since deletion of either bceA or bceB abolishes BceRS-mediated bacitracin sensing. The results suggest that the BceAB transporter acts as a cosensor, together with the BceRS two-component system, for bacitracin perception in S. mutans. By searching the S. mutans genome databases, we have identified three additional genes that share the consensus BceR binding motif at their promoter regions. Our initial work has confirmed that expression of these genes is directly controlled by BceRS, indicating that the bceABRS operon, along with the three additional genes, forms the BceRS regulon in S. mutans. Taking these findings together, we conclude that BceABRS comprises a four-component system that plays an important role in stimulus sensing, signal transduction, the gene regulatory network, and substrate transport for the cell envelope stress response in S. mutans.
An important aspect of microbial life in natural ecosystems is the ubiquitous presence of antibiotics, which are a common threat to microorganisms (13). Their sensitive detection and the subsequent induction of appropriate resistance mechanisms are therefore prerequisites for microbial survival. For example, bacitracin, a cyclic polypeptide antibiotic that is produced by certain species of the genus Bacillus in the environment, can target essential components of the cell envelope and can suppress the biosynthesis of peptidoglycan in many Gram-positive bacteria (27, 31). Bacitracin binds to undecaprenyl pyrophosphate (UPP), the lipid carrier responsible for the translocation of cell envelope precursors from the cytosol to the extracellular side of the cytoplasmic membrane, preventing the dephosphorylation of UPP necessary for the recycling of the lipid carrier (6, 13, 29, 31). To survive threats represented by antibiotics such as bacitracin in the environment, bacteria must be able to sense, respond to, and cope with such threats. The first and major line of defense against the threats from the environment is the bacterial cell envelope, which provides a major sensory interface between a bacterial cell and its surroundings and plays important roles in signal sensing, transduction, and the adaptive response (13, 29). Thus, maintaining cell envelope integrity and an effective response to threats is crucial for microbial survival in natural ecosystems.
Dental plaque represents a natural biofilm community characterized by its vast diversity (>700 species) and high cell density (1011 cells/g of wet weight) (4, 15, 17). Many bacteria in dental biofilms produce bacteriocins, which kill competing species and are responsible for “chemical warfare” in the community (1, 16, 17). Thus, bacteria in dental biofilms often face life-threatening challenges by antibiotic compounds from competing species. S. mutans is a Gram-positive bacterium that depends on a “biofilm lifestyle” for survival and persistence in its natural ecosystem, dental plaque (4, 20, 25). During its natural life in dental biofilms, S. mutans is frequently exposed to various threats such as killing by antibiotics produced by other species. However, S. mutans is well known to be resistant to bacitracin, and this property is often exploited in the isolation of S. mutans strains from highly heterogeneous oral microflora (8, 23). Only a few studies have explored the molecular basis of bacitracin resistance in S. mutans, including the biosynthesis of cell wall rhamnose-glucose polysaccharides (37), altered expression of diacyglycerol kinase in diacyglycerol phosphorylation (24), and induction of a bacitracin-specific efflux ABC transporter (37). Recently, a three-component system, LiaFSR, was also found to play a role in bacitracin-induced stress response in S. mutans, although this system primarily responds to the damage caused by cell wall-acting antibiotics, including bacitracin (32).
By screening a high-density transposon mutant library, we have identified two transposon insertion mutants that are extremely sensitive to bacitracin (35). These mutants have been confirmed to have a transposon insertion in S. mutans SMU.1007 and SMU.1009 at the same genetic locus that consists of four open reading frames (ORFs). Our initial work confirmed that these four ORFs from SMU.1006-SMU.1009 are consistent with the mbrABCD locus previously described by Tsuda et al. (37), who reported the involvement of these genes in bacitracin resistance in S. mutans. However, the mechanism by which these genes and their gene products are regulated in response to bacitracin remains unclear. In this study, we set forth experiments to investigate this question. We have confirmed that the MbrABCD in S. mutans is homologous to the BceRS-AB in Bacillus subtilis, so we propose to redesignate these genes bceABRS to unify the names from the previous mbrABCD genes (37). We describe the identification of the DNA binding site for the BceR regulator, which binds directly to the upstream portion of the bceABRS promoter region and positively regulates expression of the bceABRS operon in response to bacitracin. We also provide evidence indicating that the BceAB transporter not only plays an important role in bacitracin resistance but also acts, together with the BceRS, as a cosensor to orchestrate bacitracin perception. Finally, we have identified three additional genes that share the conserved BceR binding motif at their promoter regions. Our initial work confirms that expression of these genes is directly controlled by BceRS, suggesting that bceABRS, together with those three additional genes, constitutes the BceRS regulon responsible for bacitracin resistance in S. mutans.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
Bacterial strains and plasmids and their characteristics are listed in Table 1. S. mutans wild-type (wt) strain UA159 was grown on Todd-Hewitt medium supplemented with 0.3% yeast extract (THYE), whereas all the mutants, complementation strains, and lacZ transcriptional reporter strains constructed from S. mutans UA159 were maintained on THYE supplemented with either erythromycin (10 μg/ml) or erythromycin plus spectinomycin (800 μg/ml) or kanamycin (800 μg/ml). Escherichia coli host strains were grown in Luria-Bertani (LB) medium supplemented with an appropriate antibiotic.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| S. mutans | ||
| UA159 (wt) | Wild type, Erys, Kans | 20 |
| SmΔbceA | UA159 ΔbceA::erm, Eryr | This study |
| SmΔbceB | UA159 ΔbceB::erm, Eryr | This study |
| SmΔbceR | UA159 ΔbceR::erm, Eryr | This study |
| SmΔbceS | UA159 ΔbceS::erm, Eryr | This study |
| Sm-pCpbceA | UA159 ΔbceA::erm harboring pCpbceA, Eryr, Sptr | This study |
| Sm-pCpbceR | UA159 ΔbceR::erm harboring pCpbceR, Eryr, Sptr | This study |
| Sm159-pPbceA | UA159 harboring pPbceA-lacZ, Kanr | This study |
| SmΔbceA-pPbceA | SmΔbceA::erm harboring pPbceA-lacZ, Eryr, Kanr | This study |
| SmΔbceB-pPbceA | SmΔbceB::erm harboring pPbceA-lacZ, Eryr, Kanr | This study |
| SmΔbceR-pPbceA | SmΔbceR::erm harboring pPbceA-lacZ, Eryr, Kanr | This study |
| SmΔbceS-pPbceA | SmΔbceS::erm harboring pPbceA-lacZ, Eryr, Kanr | This study |
| E. coli | ||
| BL21(DE3)pLysS | F−ompT hsdSB(rB−mB−)gal dcm (DE3)pLysS(Camr) | Novagen |
| BL21-pET41aBceR | BL21(DE3)pLysS harboring pET41aBceR, Kanr | This study |
| DH5α | E. coli supercompetence strain | Invitrogen |
| Plasmids | ||
| pET41a | E. coli expression vector, Kanr | Novagen |
| pET41aBceR | pET41a::bceR, Kanr | This study |
| pPbceA-lacZ | pSL-lacZ::PbceA399, Kanr | This study |
| pSL-lacZ | pDL276 containing a 3.45-kb HindIII fragment of a promoterless lacZ from pALH122, Kanr | 33 |
| pDL278 | Streptococcus-E. coli shuttle vector, Sptr | 19 |
| pCpbceA | pDL278::CpbceA-1372, Sptr | This study |
| pCpbceR | pDL278::CpbceR-1037, Sptr | This study |
Ery, erythromycin; Kan, kanamycin; Spt, spectinomycin.
Construction of gene deletion mutants.
To determine the effect of inactivation of each gene on bacitracin sensing and resistance, we constructed individual bceA, bceB, bceR, and bceS deletion mutants by an allelic replacement strategy involving PCR-ligation mutagenesis (18). The primers used to construct and confirm these mutants are listed in Table 2. To construct the bceA mutant, for example, a 670-bp fragment (bceA-up) upstream from an approximately 100-bp internal region of the bceA start codon was amplified in S. mutans UA159 by the use of primers bceA-KO-P1 and bceA-KO-P2 (AscI site at its 5′ end). Another amplicon, designated bceA-dw, consisting of 932 bp downstream from the approximately 100-bp internal region of the bceA stop codon, was amplified using primers bceA-KO-P3 (FseI site at the 5′ end) and bceA-KO-P4. An 860-bp erythromycin resistance cassette was amplified from mutant SmRr11 (21) by the use of primers Em-860-P1 and Em-860-P2. These amplicons were subjected to restriction digestion and ligation to produce a bceA-up::Em::bceA-dw fragment. The ligation product was directly transformed into S. mutans UA159. Following double-crossover recombination, the internal region of bceA gene was completely replaced by the erythromycin cassette. The transformants grown on THYE plus erythromycin (10 μg/ml) were genetically confirmed by a PCR strategy using primers P1 and P4 in combination with the primers designed for the ermr cassette (18, 22). The presence of the Erm marker with predicted sizes of the flanking region in the mutant but not in the parent strain indicated successful deletion and replacement by the ermr cassette at the desired locus (22). The same strategy was also used to construct the rest of the gene deletion mutants.
TABLE 2.
Primers used in this study
| Primer | Nucleotide sequencea (5′→3′) | Amplicon (bp) |
|---|---|---|
| ABrt-F | CTTGCAAGACGTTGACTTCA | 868 |
| ABrt-B | GGTAGCCAAACCTAGAGCAT | |
| BRrt-F | GACTCCTCTATGGCGATGATGAT | 618 |
| BRrt-B | GCCCTAAAGTTATCAACGC | |
| RSrt-F | GGTTGAAGATGATACAACC | 984 |
| RSrt-B | CAAGATCACTTGGTGCAG | |
| BceR-F | TGGTCCATGGTTATGCTAAAGC | 692 |
| BceR-B | GCGGGGAATTCTCTCAAATAAGATC | |
| bceA-qRT-F | CATTTGTATCGCCTTGCTCCG | 153 |
| bceA-qRT-B | TGCTGCTGTTGGTTCATCTGC | |
| bceB-qRT-F | GCAATCAAAGTCAAGGAGAACG | 144 |
| bceB-qRT-B | CCTGGAAAAAGCGTGTAATACC | |
| bceR-qRT-F | GGGAGGTGATGATTTTTTGAGC | 108 |
| bceR-qRT-B | GCCTTGCTTAGTGAACTCGTTAG | |
| bceR-qRT-F | TCTTTATCATCTGCCTCTCGTC | 104 |
| bceR-qRT-B | ATTTTTGGTGGAACCGCC | |
| 862-qRT-F | GGAACGATTGCCTCTTCAACTTTG | 110 |
| 862-qRT-B | GCAGACACCATAACCTTAGGATTGG | |
| 302-qRT-F | AAACCATTTCTGAAGCAGCGG | 246 |
| 302-qRT-B | AACAACCCAAGCCATCTCCAG | |
| 1856-qRT-F | GCGGCAGGATTGATGTTTTTG | 109 |
| 1856-qRT-B | TGGTAACTCCCAAGGCTAAGGTC | |
| gyrA-qRT-F | TTGCGACTATCTGCTATGTG | 111 |
| gyrA-qRT-B | CCAAGAATCTGCTGTCCG | |
| bceA-KO-P1 | GCCTTAATTGGTTGGCATGTTGT | 670 |
| bceA-KO-P2 | TATGGCGCGCCTCTCTGATTTTGGTGATGTCTTGG | |
| bceA-KO-P3 | ATAGGCCGGCCTATTCTGATGGTCACCCACTC | 932 |
| bceA-KO-P4 | GTTCTTACGACGACGTTTCA | |
| bceB-KO-P1 | GCCAAGACATCACCAAAATCAGAG | 707 |
| bceB-KO-P2 | GGCGCGCCGCATAAACTCCTGCCCCCATAG | |
| bceB-KO-P3 | GGCCGGCCCTGAAGGGTTGAGAATGACTGGC | 770 |
| bceB-KO-P4 | ATAGGCAGCGAGAAAGGCTTG | |
| bceR-KO-P1 | GAATGAGTCAGGCTAAGCATGG | 623 |
| bceR-KO-P2 | GGCGCGCCCCGCTTTTAACTTGATAGTGTTGTCC | |
| bceR-KO-P3 | GGCCGGCCCACGCTTAGTGTTAATATGACACG | 732 |
| bceR-KO-P4 | GGATTTTATTTCCCAATCGCC | |
| bceS-KO-P1 | GCGTTGATAACTTTAGGGCTATC | 688 |
| bceS-KO-P2 | GGCGCGCCCCGATAAAATAGACGAGAGGCAGATG | |
| bceS-KO-P3 | GGCCGGCCCGCCACTGGTCTTGGTCTTTATATGG | 687 |
| bceS-KO-P4 | GAAACGGGTCCTTATATCATCAGC | |
| Erm-860-P1 | TAGGCGCGCCCCGGGCCCAAAATTTGTTTGAT | 860 |
| Erm-860-P2 | GCTGGCCGGCCAGTCGGCAGCGACTCATAGAAT | |
| BceA-cp-F | ATAGAATTCGGTTGGCATGTTGTTGAAGG | 1,372 |
| BceA-cp-B | GCTAAGCTTCAAACCTAGAGCATAAACTCCTGC | |
| BceR-cp-F | ATAGAATTCCTTAAAAAGCTGCTCTTCCTG | 1,037 |
| BceR-cp-B | GCTAAGCTTGAACCGCCAGTATAAAATGAG | |
| PbceA399-F | TATCGTTATGCCAGTCA | 399 |
| PbceA399-B | AGCTTCTCCTTTTTATCAT | |
| PbceA187-F | GGCTTCTTTTTTGGCTG | 187 |
| PbceA187-B | AGCTTCTCCTTTTTATCAT | |
| PbceA160-F | TTAGGGTACCAAACAGAGCCA | 160 |
| PbceA160-B | AGCTTCTCCTTTTTATCAT | |
| PbceA120-F | TTAATGTCAATGCTTAC | 120 |
| PbceA120-B | AGCTTCTCCTTTTTATCAT | |
| PbceA78-F | GTGTAAGATATCTTTTC | 78 |
| PbceA78-B | AGCTTCTCCTTTTTATCAT | |
| PbceR389-F | GTAGGTACCGCCTTCTTACTTG | 389 |
| PbceR389-B | AACTTTATTCTACCAAAAAAGTG | |
| PΔIR1-A | TTAATGTCAATTTTTGTAAGCTACGAT | 112 |
| PbceA-B | GCTTCTCCTTTTTATCAT | |
| PbceA-C | TATCGTTATGCCAGTCA | 301 |
| PΔIR1-D | AGCTTACAAAAATTGACATTAATTTAATAG | |
| PΔIR2-A | TGCTTACAATTTACGATTCTTTAAGTG | 112 |
| PbceA-B | GCTTCTCCTTTTTATCAT | |
| PbceA-C | TATCGTTATGCCAGTCA | 311 |
| PΔIR2-D | AAAGAATCGTAAATTGTAAGCATTG | |
| PΔIR1 + 2-A | TTAATGTCAATTACGATTCTTTAAGTG | 102 |
| PbceA-B | GCTTCTCCTTTTTATCAT | |
| PbceA-C | TATCGTTATGCCAGTCA | 301 |
| PΔIR1 + 2-D | AAAGAATCGTAATTGACATTAATTTAAT | |
| PΔDR2-A | ATTCTTTAAGATATCTTTTC | 114 |
| PbceA-B | GCTTCTCCTTTTTATCAT | |
| PbceA-C | TATCGTTATGCCAGTCA | 332 |
| PΔDR2-D | GAAAAGATATCTTAAAGAATCGT | |
| P862-F | GATGTAGGTAAGAGGACAAACG | 231 |
| P862-B | CCTCAAAGTAAAGTCAAGAGCC | |
| P302-F | GGTATTGATTTGCGAGCG | 392 |
| P302-B | TTGAGTAAACTTCGTGCCTC | |
| P1856-F | AGTCTGCCCTCCTTTTCAC | 403 |
| P1856-B | CTTCGGCTAAGAATCAACG |
The engineered restriction sites are indicated by underlining as follows: CCATGG, NcoI; GAATTC, EcoRI; AAGCTT, HindIII; GGCGCGCC, AscI; and GGCCGGCC, FseI.
Construction of the complementation strains.
To rule out the possibility of a polar effect on the phenotype of these mutants, we constructed two S. mutans strains that harbored a low-copy-number Streptococcus-E. coli shuttle vector, pDL278 (19), carrying a wild-type copy of either bceA or bceR, for genetic complementation of the defects in trans. Briefly, we used PCR to amplify the entire bceA gene and its promoter region from S. mutans UA159 genomic DNA by the use of primers BceA-cp-F and BceA-cp-B. We also used primers BceR-cp-F and BceR-cp-B to amplify the entire bceR gene and its upstream sequence containing a putative ribosome-binding site preceded by a stop codon. The amplicons were then ligated into NcoI and EcoRI sites at the multiple cloning site of pDL278 and transformed into an E. coli host DH5α strain (Invitrogen). Transformants from LB agar plates containing spectinomycin (50 μg/ml) were confirmed by restriction digestion analysis. The confirmed plasmids, pCpbceA and pCpbceR, were transformed into mutants SmΔbceA and SmΔbceR, respectively. Transformants were selected from THYE plates containing both spectinomycin (800 μg/ml) and erythomycin (10 μg/ml). The resulting strains were designated Sm-pCpbceA and Sm-pCpbceR.
Antimicrobial susceptibility assays.
A modified broth dilution method was used with 96-well microtiter plates to determine the MICs of bacitracin and other cell wall-acting antibiotics, including vancomycin, nisin, penicillin-G, and lysozyme, for these mutants and the UA159 parent strain (34). Briefly, an aliquot of the mid-log-phase cells was inoculated into 200 μl/well (at an optical density at 600 nm [OD600] of approximately 0.02) of THYE broth containing a series of diluted antibiotics. The cultures were incubated at 37°C for 20 h, and the optical density was read at 590 nm using a microplate reader (Synergy HT; Biotek). All experiments were performed in triplicate.
RT-PCR.
To detect polycistronic transcripts of bceABRS genes, total RNA was isolated from mid-log-phase cells of S. mutans UA159 in the presence of bacitracin (0.2 units/ml) by the use of a FastPrep method (9). The resulting RNA was then treated with RNase-free DNase I to remove genomic DNA and further purified by passing through an RNeasy minicolumn (Qiagen). The purified RNA was converted to cDNA by reverse transcription (RT) using SuperScript II reverse transcriptase (Invitrogen). PCR was performed to amplify the cDNA by the use of three pairs of cross-gene primers (listed in Table 2). The same primers were also used for PCR amplifications of S. mutans UA159 genomic DNA (a positive control) and purified RNA (a negative control). RT-PCR and PCR products were then resolved and analyzed on a 1.2% agarose gel.
Cloning, expression, and purification of recombinant BceR protein.
To clone and express recombinant BceR protein, the entire bceR gene was amplified by PCR using primers BceR-F and BceR-B (Table 2) to introduce two restriction sites (NcoI and EcoRI). The resulting PCR product (692 bp) was digested, purified, and ligated to the same restriction sites of the cloning site of pET-41a(+) expression vector (Novagen), which contained a glutathione S-transferase (GST) tag facilitating subsequent identification, cleaving, and purification of the fusion protein. The ligation product was transformed into an E. coli host (NovaBlue), and the positive clones were confirmed by PCR. A confirmed clone (pET-41abceR) was then transformed into BL21(DE3)pLysS for overexpression of BceR protein by the use of the protocol recommended by the manufacturer. The strain was grown in LB broth to mid-log phase. The culture was added with 1.0 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and incubated at 26°C for 4 h to induce overexpression of a GST-BceR fusion protein. The cells were then harvested and washed and proteins were extracted by the use of a BugBuster GST-binding purification kit (Novagen). The GST-tagged BceR fusion protein was analyzed and identified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting using a monoclonal antibody against the GST tag (Novagen). The concentration of the purified BceR protein was estimated by a Bio-Rad protocol, and the purified protein was stored in 20% glycerol at −80°C until use.
EMSA.
To identify the DNA binding site for BceR, we used a two-color fluorescence-based electrophoretic mobility shift assay (EMSA) protocol (Invitrogen), which employs a fast, easy, and nonradioactive assay method, to detect a DNA-protein complex (12). A series of DNA fragments of decreasing length that contained putative BceR binding sites were amplified by PCR using the primers listed in Table 2. To activate the BceR protein for DNA binding, an aliquot of the purified protein was phosphorylated in a reaction buffer containing 25 mM acetyl phosphate, 50 mM Tris-HCl, 50 mM KCl, 10 mM MgCl2, and 4 mM dithiothreitol (pH 7.5) by a method previously described (7, 10). The phosphorylation reaction mixture was incubated at room temperature for 2 h before excess acetyl phosphate was removed by filtration. Phosphorylated BceR (BceR-P) was then serially diluted in a binding buffer (50 mM Tris-Cl, 10 mM MgCl2, 200 mM KCl, 0.5% Tween 20, 4 mM dithiothreitol, 50 μg of bovine serum albumen [BSA]/ml, 10% glycerol, pH 7.5) and incubated with 50 ng of a DNA fragment in each reaction mixture for an additional hour. The phosphorylation was assessed by determining the relative concentrations of BceR-P and BceR used to form a similar-density protein-DNA binding complex on nondenaturing polyacrylamide gel. An unrelated DNA fragment (248 bp) from plasmid pDL278 (19) was also included in the reaction as a negative control. The DNA-BceR-P complexes were resolved on 6% nondenaturing polyacrylamide gels and stained with SYBR green dye (Invitrogen). The DNA-BceR-P complexes were distinguished from unbound DNA or protein by mobility shift rates and differential staining. EMSA profiles were visualized and photographed using a FluorChem SP imaging system at the recommended excitation and emission settings (Alpha Innotech, CA). To further define the BceR binding site, we also generated several mutated DNA fragments, each of which had a site-directed deletion of a putative binding sequence, by an overlapped PCR extension strategy using specific sets of primers (Table 2) and a two-step PCR amplification method (9). The DNA-BceR-P binding and EMSA were performed by the methods described above.
Construction of lacZ transcriptional reporter strains.
To determine how the bceABRS operon was regulated, we constructed a lacZ transcriptional reporter fused to the promoter of the bceABRS operon. A 399-bp fragment containing the entire promoter region of bceABRS was amplified from S. mutans UA159 by PCR using primers PbceA-F and PbceA-B (Table 2). The resulting PCR product was digested with KpnI and XbaI, gel purified, and cloned into pSL-lacZ, a low-copy-number shuttle vector carrying a promoterless lacZ in the backbone of pDL276 (31). Positive clones grown on LB agar plates plus kanamycin (50 μg/ml) were screened for genetic confirmation by PCR. Two of the positive clones were selected for sequencing confirmation of the lacZ fusion site. The confirmed clone was named pPbceA-lacZ and was transformed into S. mutans UA159 and mutants SmΔbceA, SmΔbceB, SmΔbceS, and SmΔbceR to generate five lacZ transcriptional reporter strains (Table 1).
lacZ reporter assay.
Induction of bceABRS promoter activity by bacitracin was monitored using a lacZ reporter assay as previously described (36). When the culture reached the mid-log phase (OD600 of 0.5), it was divided into four tubes and added with bacitracin at a final concentration of 0, 0.1, 0.2, or 0.4 units/ml. The cultures were incubated at 37°C for half an hour before the cell pellets were collected by centrifugation at 4°C. Each cell pellet was resuspended in 1 ml of 50 mM Tris-HCl buffer (pH 7.5) supplemented with 0.27% β-mercaptoethanol. The cell suspensions were added with 50 μl of chloroform and 20 μl of 0.1% sodium dodecyl sulfate (SDS) and permeabilized using a FastPrep FP120 instrument (Thermo Electro Ltd.) at setting 6 for 30 s. The cell lysates were centrifuged, and 100 μl of the supernatant from each sample was transferred in triplicate to a 96-well microtiter plate for a β-galactosidase (β-Gal) activity assay. The reactions were initiated by adding 20 μl of ONPG (O-nitrophenyl-d-galactopyranoside) at a final concentration of 80 μM. The reaction mixtures were incubated for an hour, and then the reactions were terminated by adding 50 μl of 1 M Na2CO3. β-Gal activity was quantified by optical density readings at 420 nm using a multiplate reader (Synergy HT; Biotek). Protein concentrations of the supernatants were determined by a Bio-Rad protein assay. Specific β-Gal activity was calculated using triplicate samples from two independent experiments. The data determined from the β-Gal activity were normalized to a negative control before being plotted.
Gene expression analysis by real-time qRT-PCR.
Real-time quantitative RT-PCR (qRT-PCR) was performed to determine (i) the effect of bceA deletion on expression of bceABRS and (ii) whether BceRS controlled expression of newly identified genes that had the BceR binding motif at their promoter regions. S. mutans UA159, the SmΔbceA mutant and the corresponding complementation strain Sm-pCpbceA, and UA159 and SmΔbceR were used to isolate total RNA by the use of the same protocol as that described for RT-PCR. Real-time qRT-PCR was carried out in a Cepheid Smart Cycler using a QuantiTech SYBR green PCR kit (Qiagen). The primers for qRT-PCR analysis are listed in Table 2. Efficiency of amplification was confirmed by analyzing melting curves of each amplicon following amplification with template dilutions over 4 orders of magnitude. The expression levels were normalized to the gyrA gene of S. mutans as an internal standard, since it produced little variation in its levels of expression. A nontemplate control was also included to confirm the absence of primer-dimer formation. Each assay was performed in duplicate in two independent experiments, and severalfold changes in expression levels were calculated by the following equations: for each cDNA, ΔCT = CT (target gene) − CT (gyrA) and ΔΔCT = ΔCT (test cDNA) − ΔCT (reference cDNA); ratio = 2−ΔΔCT, where CT represents the threshold cycle (9).
RESULTS
bceABRS (formerly mbrABCD) forms a four-gene operon in S. mutans.
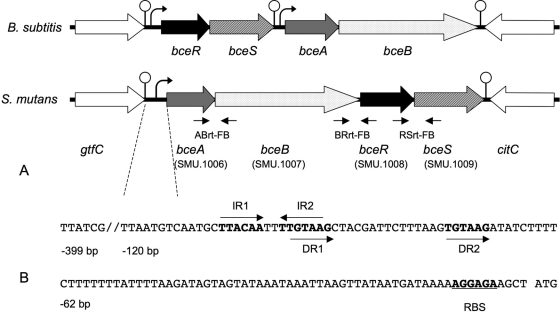
S. mutans SMU.1006 (mbrA), SMU.1007 (mbrB), SMU.1008 (mbrC), and SMU.1009 (mbrD) are located at the same genetic locus in the chromosome (from bp 960370 to 96479) (Fig. 1). The gene 273 bp immediately upstream from SMU.1006 is gtfC, which encodes a glucosyltransferase-SI responsible for the synthesis of a water-insoluble glucan, while the gene 149 bp downstream from this locus but in the opposite reading direction is citC, which encodes a citrate lyase synthetase. SMU.1006 (mbrA) and SMU.1007 (mbrB) encode an ABC transporter and a transporter permease, while SMU.1008 (mbrC) and SMU.1009 (mbrD) encode a two-component system (TCS). Sequence alignments reveal that these S. mutans proteins share high homology to B. subtilis BceA (67% similarity), BceB (45.5%), BceR (57.8%), and BceS (45.5%). To confirm the phenotype of these genes, we constructed individual gene deletion mutants, ΔSMU.1006 (SmΔbceA), ΔSMU.1007 (SmΔbceB), ΔSMU.1008 (SmΔbceR), and ΔSMU.1009 (SmΔbceS) (Table 1), by a PCR-ligation mutagenesis strategy involving an allelic replacement by an erythromycin resistance cassette (18). This method enables a target gene to be rapidly deleted and replaced by the ermr cassette without interruption of the downstream reading frame (18). We first tested the susceptibility of these mutants to bacitracin and several other cell wall-acting antibiotics (Table 3) and confirmed that all the mutants (MIC, ∼0.5 units/ml) were nearly 100-fold more sensitive to bacitracin than the parent UA159 strain (MIC, ∼48 units/ml). Introduction of a wild-type copy of one of these genes, for example, either bceA or bceR, into SmΔbceA or SmΔbceR completely restored their resistance to bacitracin (Table 3). Due to their similarities in protein sequences and phenotypes, we propose to redesignate these genes bceA (SMU.1006), bceB (SMU.1007), bceR (SMU.1008), and bceS (SMU.1009) to reflect their characteristics as bacitracin efflux genes as proposed by Ohki et al. (30) and to unify the names from the previous mbrA, mbrB, mbrC, and mbrD names designated by Tsuda et al. (37).
FIG. 1.
The genetic locus of bceABRS (SMU.1006-1009), together with the adjacent genes, is schematically presented. Figure 1A shows a comparison of the gene organization characteristics of B. subtilis and S. mutans. Sequence analysis of this genetic locus indicates two potential transcriptional terminators in the region between gtfC and bceA and downstream of bceS. Small horizontal arrows under the ORFs indicate the locations of three pairs of cross-gene primers (ABrt-FB, BRrt-FB, and RSrt-FB) for RT-PCR. Figure 1B shows the promoter sequence of bceABRS. Arrows indicate the locations and directions of orientation of a pair of invert repeats (IR1 and IR2) and a pair of direct repeats (DR1 and DR2), which may be putative binding motifs for BceR. The putative ribosome binding site (RBS) and the translation start codon (ATG) are also indicated.
TABLE 3.
Susceptibility of S. mutans strains to cell wall-acting antibiotics
| Strain | MIC |
||||
|---|---|---|---|---|---|
| Bacitracin (U/ml) | Nisin (μg/ml) | Vancomycin (μg/ml) | Penicillin G (μg/ml) | Lysozyme (mg/ml) | |
| UA159 | 48.0 | 0.50 | 1.25 | 0.40 | 2.5 |
| SmΔbceA | 0.50 | 0.50 | 1.25 | 0.40 | 2.5 |
| SmΔbceB | 0.50 | 0.50 | 1.25 | 0.40 | 2.5 |
| SmΔbceR | 0.50 | 0.50 | 1.25 | 0.40 | 2.5 |
| SmΔbceS | 0.50 | 0.50 | 1.25 | 0.40 | 2.5 |
| Sm-pCpbceA | 50.0 | 0.50 | 1.25 | 0.40 | 2.5 |
| Sm-pCpbceR | 50.0 | 0.50 | 1.25 | 0.40 | 2.5 |
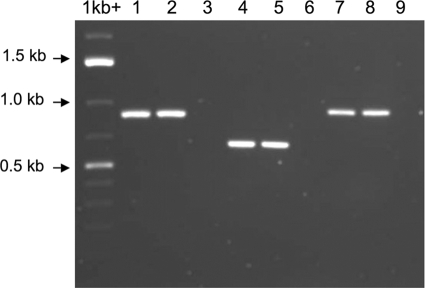
DNA sequence analysis of this genetic locus showed two potential transcriptional terminators, in the form of stem-loop structures followed by a poly(T) sequence, in the regions between gtfC and bceA and downstream of the last gene, bceS. There was no such clear transcriptional terminator identified between bceB and bceR, although a 41-bp intergenic spacer was present between these two genes (Fig. 1). To determine whether these genes were cotranscripted, we used RT-PCR to analyze the transcription products of these genes by using cross-gene primer pairs (Table 2). The RT-PCR results confirmed that these four genes were cotranscripted in a single polycistronic transcript in S. mutans UA159 (Fig. 2). The result was consistent with a previous report by Tsuda et al. (37), who used a different S. mutans strain. Thus, the evidence supports the idea that the bceABRS genes are organized as a four-gene operon and that their transcription is likely driven by the same promoter.
FIG. 2.
RT-PCR analysis of transcripts from the bceABRS operon in S. mutans UA159. Three pairs of cross-gene primers (also indicated by small horizontal arrows in Fig. 1) used for PCR amplification included ABrt-F and ABrt-B (lanes 1 to 3 [anticipated product size, 868 bp]), BRrt-F and BRrt-B (lanes 4 to 6 [618 bp]), and RSrt-F and RSrt-B (lanes 7 to 9 [984 bp]). Lanes 1, 4, and 7 represent positive controls amplified from the genomic DNA. Lanes 2, 5, and 8 represent RT-PCR products amplified from the cDNA. Lanes 3, 6, and 9 represent negative controls amplified from purified RNA, indicating no contamination with genomic DNA in the purified RNA samples.
Promoter analysis of the bceABRS operon.
In B. subtilis, the bceRS genes, which encode a two-component system, and the bceAB genes, which are their immediate downstream neighbors and encode an ABC transporter and a transporter permease, constitute two independent transcriptional units (30). BceR is known to bind directly to an invert repeat sequence (AAGCgTGTGACgaaaatGTCACAtGCTT) that is upstream of the bceAB promoter region and that positively regulates expression of bceAB in response to bacitracin (30). We used this sequence to search for the BceR binding site at the putative promoter region of bceAB or bceRS in S. mutans. However, we were unable to identify a similar BceR binding motif in these regions, suggesting that the DNA binding site for BceR in S. mutans might differ from that in B. subtilis. We then searched for putative binding motifs (invert and direct repeats) in these regions by the use of motif-searching tools such as MEME (2) and Virtual Footprint (http://prodoric.tu-bs.de/vfp/). By analyzing the region 400 bp upstream from the start codon of bceAB, we found a perfect invert repeat and an imperfect direct repeat located between the bp −120 and bp −70 regions (Fig. 1B). The invert repeat regions (IR1 and IR2) are separated by 2 nucleotides, while the direct repeat regions (DR1 and DR2) are separated by 12 nucleotides. Thus, IR2 has five nucleotides that overlap with DR1. Based on the sequence locations and features, we hypothesized that either the invert or the direct repeat might be the putative binding motif required for BceR binding in S. mutans.
Production and confirmation of the recombinant BceR protein.
To test the hypothesis that BceR might directly bind to one of the putative binding motifs upstream of the bceABRS promoter region, we used pET-41a(+) expression vector (Novagen) to express and produce the recombinant BceR protein. SDS-PAGE analysis of the resulting BceR protein demonstrated that we had successfully produced sufficient amounts of the BceR fusion protein, with a predicted molecular size of 56 kDa. The BceR fusion protein was further confirmed by Western blot analysis using a monoclonal antibody against the GST tag (data not shown). The purified recombinant BceR protein was then subjected to phosphorylation before being used for a gel mobility shift assay (EMSA).
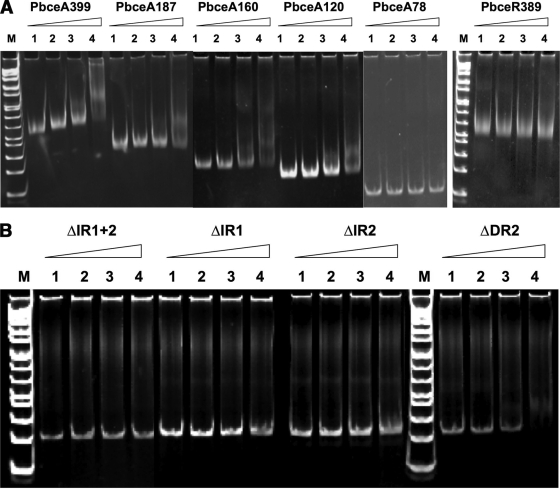
BceR binds directly upstream of the bceABRS promoter region.
To define the BceR binding site, we used a PCR strategy to generate a series of DNA fragments with decreasing lengths of 399 bp (PbceA399), 187 bp (PbceA187), 160 bp (PbceA160), 120 bp (PbceA120), and 78 bp (PbceA78) for EMSA. We also included a 389-bp PCR fragment (PbceR389) covering the upstream region from the start codon of bceRS in the EMSA. Before being mixed with the DNA fragments, the BceR protein was phosphorylated by a method that was confirmed to effectively phosphorylate the CovR response regulator in S. pyogenes (7, 11). The results revealed that the phosphorylated BceR (BceR-P) bound to the fragments of PbceA399, PbceA187, PbceA160, and PbceA120 in a concentration-dependent manner but not to the fragments of PbceA78 and PbceR389 (Fig. 3A). These results confirmed that BceR bound to the upstream of bceABRS and ruled out the possibility that BceR might bind upstream of bceRS. The results also suggested that the BceR binding site must be located between bp −120 and bp −78 of the bceABRS promoter region, since PbceA120 (rather than PbceA78) was the smallest fragment required for the BceR binding and gel mobility shift. Interestingly, this is exactly the location containing the invert and direct repeats (putative binding motifs). In addition, a comparison of the concentrations of BceR-P and BceR used for the DNA binding showed that the concentration of BceR-P required for the formation of a protein-DNA complex was about 20-fold lower than that of BceR required for the formation of a similar protein-DNA complex, suggesting that phosphorylation of BceR dramatically enhanced the affinity of binding to its target DNA. To further determine whether the invert or the direct repeat is the true BceR binding motif, we generated several PCR fragments with a deletion of one or two repeat sequences, including ΔIR1, ΔIR2, ΔIR1 plus ΔIR2, and ΔDR2, for binding analysis and EMSA. The results revealed that deletion of either IR1 or IR2 or of both (IR1 plus IR2) abolished the BceR binding but that a deletion of the DR2 alone did not affect the mobility shift profile (Fig. 3B). The results indicated that the invert repeat, rather than the direct repeat, is essential for BceR binding. This consensus invert repeat sequence (TTACAAnnTTGTAA) was then used to search for the BceR binding site of other genes in the S. mutans genome.
FIG. 3.
Electrophoresis mobility shift assay (EMSA) showing that phosphorylated BceR binds to the DNA fragments containing the putative binding motifs upstream of the becABRS promoter region. Panel A shows binding of BceR to the DNA fragments of PbceA399, PbceA187, PbceA160, and PbceA120 but not to PbceA78 and PbceR389. Panel B indicates that deletion of the inverse repeat(s) (i.e., of either ΔIR1 or ΔIR2 or both [ΔIR1+ΔIR2]) abolished the binding of BceR to the promoter region. However, a deletion of DR2 did not affect the mobility shift profile (still shift).
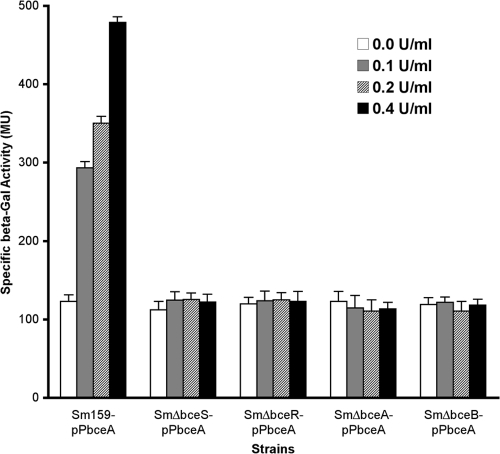
BceRS positively regulates the bceABRS operon in response to bacitracin.
Next, we investigated how the BceRS two-component system regulated expression of the bceABRS operon in response to bacitracin. To answer this question, we constructed several lacZ transcriptional reporter strains that harbored pSL-lacZ, a low-copy-number Streptococcus-E. coli shuttle vector that was engineered with a promoterless lacZ transcriptional reporter fused to the bceABRS promoter region, by using a strategy and method previously described (34). After genetic confirmation, the lacZ fusion construct (pPbceA-lacZ) was transformed into S. mutans UA159 and the mutants, SmΔbceS and SmΔbceR, to generate Sm159-pPbceA (the wild-type background), SmΔbceS-pPbceA, and SmΔbceR-pPbceA (Table 1). We then examined lacZ reporter activity in response to bacitracin in these strains. The results revealed that, in the absence of bacitracin, a baseline level of lacZ activity could be detected in all the strains. Upon addition of bacitracin, lacZ activity was significantly induced in Sm159-pPbceA (P < 0.01) and the activity increased with increasing concentrations of bacitracin (Fig. 4). However, the same concentrations of bacitracin did not significantly induce lacZ activity in the SmΔbceS-pPbceA and SmΔbceR-pPbceA strains, which lacked either the BceS histidine kinase or the BceR response regulator. The results suggest that induction of the bceABRS promoter by bacitracin is required for the BceRS signal transduction system, which positively regulates the bceABRS operon in response to bacitracin in a concentration-dependent manner.
FIG. 4.
Specific ß-galactosidase activity of the S. mutans strains in response to bacitracin. There was a dose-dependent induction of ß-galactosidase activity in Sm159-pPbceA, but no significant induction of ß-Gal activities was observed in other lacZ reporter strains that lacked bceS (SmΔbceS-pPbceA), bceR (SmΔbceR-pPbceA), bceA (SmΔbceA-pPbceA), or becB (SmΔbceB-pPbceA). MU, Miller units.
BceA and BceB are required for bacitracin sensing by BceRS.
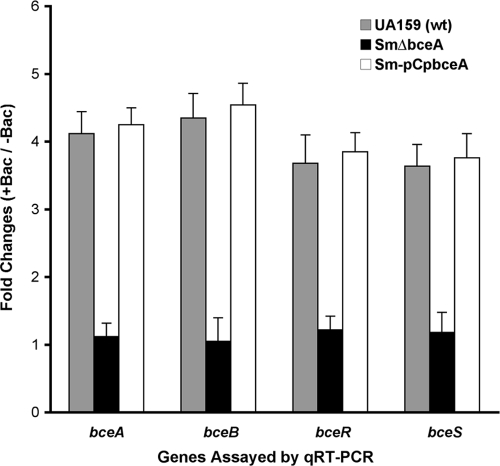
In B. subtilis, cells lacking BceA or BceB show no transcription from an intact bceAB promoter, despite the presence of bacitracin and the BceRS signal transduction system (3). This indicates that induction of the bceAB structural genes in B. subtilis requires the simultaneous presence of bacitracin, the BceRS signal transduction system, and the BceAB transporter. Thus, BceA and BceB actually act as cosensors for bacitracin perception in B. subtilis. To determine whether BceAB in S. mutans played a similar role in bacitracin sensing, we also transformed a pPbceA-lacZ fusion construct into the SmΔbceA and SmΔbceB mutants to generate two strains, SmΔbceA-pPbceA and SmΔbceB-pPbceA, which had a genetic background of either a bceA or a bceB deletion. We then assayed the lacZ reporter activity in response to bacitracin of Sm159-pPbceA and of these two strains. The results revealed that wild-type reporter strain Sm159-pPbceA was highly inducible by bacitracin and showed the expected lacZ reporter activity in response to the stimulus (Fig. 4). In contrast, neither SmΔbceA-pPbceA nor SmΔbceB-pPbceA showed an inducible response to bacitracin at the concentrations tested. The results suggest that both the BceA and BceB subunits are required for bacitracin perception in S. mutans. To further verify the results, we used real-time qRT-PCR to assay the relative expression levels of bceA, bceB, bceR, and bceS in wild-type UA159 and in the bceA deletion mutant SmΔbceA and its complementation strain, SmΔbceA-pCpbceA, in response to bacitracin. The results showed that expression of all the genes, including the bceA gene and the downstream bceB, bceR, and bceS genes, in wild type UA159 was highly inducible in the presence of bacitracin at the final concentration of 0.4 units/ml (Fig. 5). In contrast, no significant induction of expression levels of these genes by bacitracin was observed in the SmΔbceA mutant. However, bacitracin-dependent induction of these genes was fully restored in the bceA complementation strain SmΔbceA-pCpbceA (Fig. 5). Taken together, the results clearly demonstrated that both the BceA and BceB subunits of the BceAB transporter are required for bacitracin perception by the BceRS signal transduction system in S. mutans.
FIG. 5.
Real-time qRT-PCR showed the effects of bceA deletion and complementation on the relative expression levels of the bceA, bceB, bceR, and bceS genes in response to bacitracin. The severalfold changes represent the relative gene expression threshold cycle (CT) levels in the wild-type (wt) UA159 strain and the SmΔbceA mutant and the corresponding SmΔbceA-pCpbceA complementation strain grown in the presence (+bac [0.4 U/ml]) or absence (−bac) of bacitracin.
The bceABRS genes, along with other genes, constitute the BceRS regulon in S. mutans.
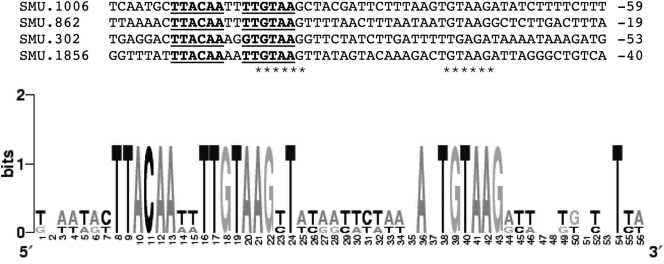
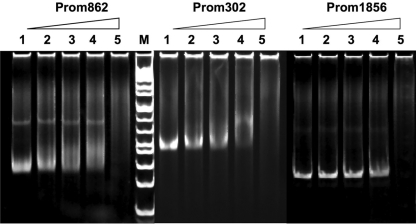
By searching for the BceR binding sequence (TTACAAnnTTGTAA) in the S. mutans genome, we found three additional genes, SMU.862, SMU.302, and SMU.1856, with the conserved BceR binding motif (BceR box) upstream of their promoter regions. The consensus sequence of the BceR box and its location at the promoter regions of these genes are presented in Fig. 6. As indicated in WebLogo graph, all of these genes shared this well-conserved binding sequence or BceR box at their promoter regions, with the exception of SMU.302, which had one nucleotide difference in the binding motif. These newly identified genes that encode hypothetical proteins of unknown function have not yet been characterized. However, the protein sequence alignments show that SMU.862 encodes a protein that is an ortholog (50% protein identity and 60% similarity) to an efflux transporter subunit of S. agalactiae (NCBI accession number YP_330002). SMU.302 encodes a protein that shares 39% identity and 58% similarity with a membrane protein of Pediococcus pentosaceus (NCBI accession number YP_803763), while SMU.1856 encodes a protein that shares 42% identity and 63% similarity with an unknown membrane protein of Enterococcus faecalis (NCBI accession number NP_815373). To determine whether BceR bound to the promoter regions of these genes, we examined BceR binding and EMSA profiles by using PCR fragments of P862-231, P302-392, and P1856-303, each of which contained its entire promoter region. The results confirmed that BceR-P bound to all of these DNA fragments, resulting in gel mobility shifts (Fig. 7) and suggesting that the BceR regulator did control the expression of these genes. The results also indicated that the imperfect binding sequence of SMU.302 did not appear to affect its gel mobility shift profile. To further determine how BceR regulated these genes, we used real-time qRT-PCR to analyze the relative expression levels of these genes in the wild-type UA159 strain and the bceR deletion mutant SmΔbceR in the presence or absence of bacitracin. We also included the Sm-pCpbceR complementation strain in the experiment to verify the role of BceR in the regulation of these genes. The results revealed that, in the absence of bacitracin, a baseline expression level of these three genes was observed for both the wild-type UA159 strain and the bceR deletion mutant SmΔbceR. Upon the addition of bacitracin, however, expression of all three genes was dramatically induced, resulting in about 4-fold to 5-fold increases in their expression levels in wild-type UA159 (Table 4). In contrast, the same concentration of bacitracin did not appear to significantly induce expression of these genes in the mutant SmΔbceR strain. Interestingly, introduction of a wild-type copy of the bceR gene in trans fully restored the wild-type expression levels of these genes (Table 4). The results suggest that the BceRS signal transduction system positively regulates expression of these genes in response to bacitracin, although their precise roles in bacitracin resistance remain to be elucidated. Thus, the evidence from this study supports the notion that the bceABRS genes and the genes discussed above may constitute the BceRS regulon responsible for the bacitracin-induced cell envelope stress response in S. mutans.
FIG. 6.
WebLog graph showing the invert repeat (IR, or BceR box) sequences (bold and underlined) at the promoter regions of SMU.1006 (bceA), SMU.862, SMU.302, and SMU.1856 in the S. mutans genome. The direct repeat (DR) sequences are also indicated by asterisks. Note that IR2 has five nucleotides that overlap with those of DR1.
FIG. 7.
Electrophoresis mobility shift assay (EMSA) showing that phosphorylated BceR also binds to the DNA fragments containing the imperfect invert repeat upstream of the promoter regions of newly identified SMU.862, SMU302, and SMU.1856 genes in the S. mutans genome.
TABLE 4.
Relative expression levels of the genes controlled by the BceRS signal transduction system in response to bacitracina
| Locus | Fold difference ± SD (+bacitracin vs −bacitracin) |
||
|---|---|---|---|
| UA159 (wt) | SmΔbceR | Sm-pCpbceR | |
| SMU.862 | 5.28 ± 0.42 | 1.65 ± 0.28 | 6.26 ± 0.53 |
| SMU.302 | 4.72 ± 0.39 | 1.84 ± 0.35 | 4.68 ± 0.48 |
| SMU.1856 | 6.45 ± 0.54 | 1.68 ± 0.32 | 6.53 ± 0.64 |
qRT-PCR values indicate severalfold changes in relative gene expression levels (CT) of cells grown in the presence (0.4 U/ml) or in the absence of bacitracin. The severalfold differences (ΔCT) in expression levels of the genes tested were calculated from triplicate experiments; the data shown represent comparisons to the CT values obtained for the constitutively expressed gene (gyrA of S. mutans UA159) as determined by the equations described in the text. Severalfold changes ≥ 2.0 are considered to represent significant induction. SD, standard deviation.
DISCUSSION
In this study, we investigated the transcription and regulatory mechanism of an S. mutans four-gene operon consisting of the bceABRS (formerly mbrABCD) genes, homologues of the B. subtilis bceRS-AB genes that encode a two-component system and an ABC transporter required for bacitracin resistance. In particular, we have described the identification of the BceR binding site at the promoter regions of the bceABRS operon and three additional genes in the S. mutans genome (1). We have shown by EMSA that the phosphorylated BceR regulator binds directly to a conserved invert repeat (BceR box) located between bp −120 and −78 at the promoter region of the bceABRS operon, positively regulating expression in response to bacitracin. Since binding of BceR to this region induces transcription of all four genes, the BceRS two-component system in S. mutans actually induces its own expression, forming an autoregulatory circuit in response to the presence of bacitracin. This regulatory mode differs from that found in B. subtilis, in which the bceRS genes and their immediate downstream neighbors, the bceAB genes, constitute two independent transcriptional units (3, 30). Although BceR binds directly to the bceAB promoter region and positively regulates expression of the genes, the BceRS two-component system does not control its own expression in B. subtilis. In fact, no self-induction of bceRS was observed upon addition of bacitracin or upon overexpression of bceR in B. subtilis (30). In contrast, incubation with bacitracin actually decreased the level of bceRS mRNA in B. subtilis. These findings imply that other factors may be involved in the regulation of expression of bceRS in B. subtilis. Despite this difference, both regulatory modules, BceRS-AB in B. subtilis and BceABRS in S. mutans, are topologically and functionally linked stimulus perception and detoxification systems. They have been confirmed to be the most sensitive and efficient bacitracin resistance determinants found in many Gram-positive bacteria (30, 31, 37, 38). We speculate that transcription of bceABRS in S. mutans might be highly inducible upon exposure to bacitracin. From the ecological point of view, the existence of such a regulatory module makes sense, because S. mutans does not produce bacitracin, and many known bacitracin producers, such as B. licheniformis and B. subtilis, are not common members in dental biofilms. Therefore, it is reasonable to assume that expression of bceABRS might be greatly induced when S. mutans needs to secure a defense against bacitracin producers that may transiently come into the oral cavity from contaminated food or water. Alternatively, S. mutans may require such a mechanism of such efficacy to cope not only with bacitracin but also with other peptide antibiotics such as bacteriocins from competing species in dental biofilms or host defense peptides in human saliva. However, there has been no report describing the effect of antimicrobial compounds other than bacitracin on the BceABRS system in S. mutans. A recent study by our group began to explore this issue and found that all the mutants, including SmΔbceA, SmΔbceB, SmΔbceR, and SmΔbceS, were more sensitive to human α- or β-defensin than the parent UA159 strain (unpublished data) but that no significant differences was observed in the levels of sensitivity of these strains to nisin, vancomycin, penicillin-G, and lysozyme (Table 3). We also found that α- and β-defensins induced expression of the bceABRS operon in a manner similar to that seen with bacitracin, although the mechanism whereby these peptides directly interact with BceAB or BceRS proteins is unknown. Further work may be required to elucidate the mechanism.
The evidence from this study has also shown that the cells lacking either BceA (SmΔbceA) or BceB (SmΔbceB) show no induction of transcription from the intact bceABRS promoter in S. mutans, despite the presence of bacitracin and the BceRS signal transduction system. This suggests that sensing of bacitracin by the BceRS two-component system requires the intact BceAB transporter. Thus, the BceAB transporter in S. mutans not only plays an important role in bacitracin resistance but also acts as a cosensor, together with the BceRS signal transduction system, to orchestrate bacitracin perception. This finding is highly consistent with the results of studies of B. subtilis, in which induction of the structural bceAB genes requires the simultaneous presence of bacitracin, the BceRS signal transduction system, and the BceAB transporter (3, 31). Such a regulatory interplay between a two-component system and an ABC transporter has recently drawn considerable attention, because employing transporters as cosensors appears to be a mechanism widely distributed in many bacteria (28, 29, 34). In Fimicutes such as Bacillus and Clostridium species, there are about 70 examples of genes that encode a two-component system (TCS) and its adjacent genes encoding ABC transporters (14, 28, 29). The BceRS-AB (TCS-ABC transporter) in B. subtilis is a typical example of such regulatory interplay for bacitracin perception and resistance (3). Although the direction of bacitracin transport by BceAB is still a matter of debate, it seems to be crucial that BceAB acts both as a bacitracin detoxification pump and as a cosensor of BceRS signal transduction system (3, 31). Thus, the coupling of transporter and stimulus perception is reasonable, because transporter proteins are well-suited information carriers, as they bind low-molecular-weight molecules in the external medium, allowing them to provide dynamic information on the metabolic flux (5, 34). Detoxification by the BceAB ABC transporter simultaneously removes the stimulus, thereby switching off the system once the bacitracin concentration decreases below the sensor threshold. At the same time, instant resistance is ensured, because the synthesis of the ABC transporter is regulated by the BceRS signal transduction system that is already activated at very low concentrations of bacitracin (34). We believe that the BceABRS module in S. mutans may be another good example of such regulatory interplay for stimulus perception and response. In particular, the four genes that encode BceAB and BceRS are cotranscripted, which may provide further control for regulation of bacitracin sensing and detoxification in S. mutans. Since all four proteins are required for bacitracin perception and response, we propose that BceABRS comprises a four-component system that plays an important role in stimulus sensing, signal transduction, and target gene activation and in their regulatory network in S. mutans in response to bacitracin. Further study that involves direct protein-protein and protein-substrate interactions may be required to elucidate detailed molecular mechanisms of stimulus perception.
The bacitracin stress response network in B. subtilis is known to consist of four signal transduction systems, the two-component systems BceRS, YvcPQ, and LiaRS and an extracytoplasmic function sigma factor, σM (6, 31). Each of these regulatory factors and pathways controls a set of target genes (regulon) in response to the stimulus (13, 27). In S. mutans, there are at least two signal transduction systems, BceRS (formerly mbrCD) and LiaSR (formerly HK/RR11) (21), which have been confirmed to regulate cell envelope stress response induced by lipid II cycle inhibitors such as bacitracin (32, 37). Suntharalingam et al. (32) reported that LiaF is an integral component of the LiaSR signal cascade in sensing of and response to bacitracin. In the absence of bacitracin, LiaF represses LiaSR-regulated expression, whereas bacitracin induces derepression of liaSR, resulting in transcription of several genes involved in membrane protein synthesis and peptidoglycan biosynthesis and with envelope chaperone-proteases and transcriptional regulators (32). However, the BceRS regulon in S. mutans has not yet been defined in the period since the first identification of bceABRS genes (37). In this report, we have identified three additional genes, SMU.862, SMU.302, and SMU.1856, which encode conserved hypothetical proteins of unknown functions and have the consensus BceR binding motif at their promoter regions. Our initial work revealed that BceR also binds to the promoter regions of these genes (Fig. 7). qRT-PCR analysis of relative expression levels of these genes in the UA159 strain and the bceR deletion mutant (SmΔbceR) confirmed that BceR appears to positively regulate these genes in response to bacitracin, although their precise roles in bacitracin-induced cell envelope stress response remain to be elucidated. Taken together, the bceABRS genes and at least three additional genes constitute the BceRS regulon in S. mutans. A genome-wide analysis of wild-type UA159 and the SmΔbceR mutant by the use of a DNA microarray may facilitate identification of additional genes in this regulon.
In summary, we have described the identification of the DNA binding site for the BceR response regulator and its transcription control of the bceABRS operon and three other genes in response to the presence of bacitracin. In contrast to the BceRS-BceAB TCS-ABC system in B. subtilis, the BceABRS system in S. mutans appears to constitute a more integrated regulatory unit whose components are genetically, topologically, and functionally linked to each other. In addition, this system forms an autoregulatory circuit in S. mutans, inducing expression of the bceABRS genes in response to bacitracin. Based on the results presented in this paper, we propose that BceABRS (formerly MbrABCD) comprises a four-component system that plays an important role in stimulus perception, signal transduction, the gene regulatory network, and substrate transport in the cell envelope stress response in S. mutans.
Acknowledgments
This work was supported in part by grant MOP-74487 from the Canadian Institutes for Health Research (CIHR) and by discovery grant RGPIN 311682-07 from the Natural Sciences and Engineering Research of Canada (NSERC). Y.-H. Li was a recipient of the Nova Scotia-CIHR Regional Partnership Program New Investigator Award. A. Wishart was a recipient of the CIHR Professional Studentship.
Footnotes
Published ahead of print on 6 July 2010.
REFERENCES
- 1.Ajdić, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. U. S. A. 99:14434-14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey, T. L., and C. Elkan. 1995. The value of prior knowledge in discovering motifs with MEME. Proc. Int. Conf. Intell. Syst. Mol. Biol. 3:21-29. [PubMed] [Google Scholar]
- 3.Bernard, R., A. Guiseppi, M. Chippaux, M. Foglino, and F. Denizot. 2007. Resistance to bacitracin in Bacillus subtilis: unexpected requirement of the BceAB ABC transporter in the control of expression of its own structural genes. J. Bacteriol. 189:8636-8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cvitkovitch, D. G., Y. H. Li, and R. P. Ellen. 2003. Quorum sensing and biofilm formation in streptococcal infections. J. Clin. Invest. 112:1626-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dawson, R. J., K. Hollenstein, and K. P. Locher. 2007. Uptake or extrusion: crystal structures of full ABC transporters suggest a common mechanism. Mol. Microbiol. 65:250-257. [DOI] [PubMed] [Google Scholar]
- 6.Eiamphungporn, W., and J. D. Helmann. 2008. The Bacillus subtilis σM regulon and its contribution to cell envelope stress responses. Mol. Microbiol. 67:830-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao, J., A. Gusa, J. R. Scott, and G. Churchward. 2005. Binding of the global response regulator protein CovR to the sag promoter of Streptococcus pyogenes reveals a new mode of CovR-DNA interaction. J. Biol. Chem. 280:38948-38956. [DOI] [PubMed] [Google Scholar]
- 8.Gold, O. G., H. V. Jordan, and J. van Houte. 1973. A selective medium for Streptococcus mutans. Arch. Oral Biol. 18:1357-1364. [DOI] [PubMed] [Google Scholar]
- 9.Gong, Y. X., X. L. Tian, T. Sutherland, G. Sisson, J. Mai, J. Ling, and Y. H. Li. 2009. Global transcriptional analysis of acid-inducible genes in Streptococcus mutans: multiple two-component systems involved in acid adaptation. Microbiology 155:3322-3332. [DOI] [PubMed] [Google Scholar]
- 10.Gusa, A. A., J. Gao, V. Stringer, G. Churchward, and J. R. Scott. 2006. Phosphorylation of the group A streptococci CovR response regulator causes dimerization and promoter-specific recruitment by RNA polymerase. J. Bacteriol. 188:4620-4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heckman, K. L., and L. R. Pease. 2007. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc. 2:924-932. [DOI] [PubMed] [Google Scholar]
- 12.Jing, D., J. Agnew, W. F. Patton, J. Hendrickson, and J. M. Beechem. 2003. A sensitive two-color electrophoresis mobility shift assay for detecting both nucleic acids and protein in gels. Proteomics 3:1172-1180. [DOI] [PubMed] [Google Scholar]
- 13.Jordan, S., M. I. Hutchings, and T. Mascher. 2008. Cell envelope stress response in Gram-positive bacteria. FEMS Microbiol. Rev. 32:107-146. [DOI] [PubMed] [Google Scholar]
- 14.Joseph, P., G. Fichant, Y. Quentin, and F. Denizot. 2002. Regulatory relationship of two-component and ABC transport systems and clustering of their genes in the Bacillus/Clostridium group, suggest a functional link between them. J. Mol. Microbiol. Biotechnol. 4:503-513. [PubMed] [Google Scholar]
- 15.Kolenbrander, P. E. 2000. Oral microbial communities: biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 54:413-437. [DOI] [PubMed] [Google Scholar]
- 16.Kreth, J., J. Merritt, W. Shi, and F. Qi. 2005. Competition and coexistence between Streptococcus mutans and Streptococcus sanguinins in the dental biofilm. J. Bacteriol. 187:7193-7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuramitsu, H. K., X. He, R. Lux, M. H. Anderson, and W. Shi. 2007. Interspecies interactions within oral microbial communities. Microbiol. Mol. Biol. Rev. 71:653-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lau, P. C. Y., C. K. Sung, J. H. Lee, D. A. Morrison, and D. G. Cvitkovitch. 2002. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J. Microbiol. Methods 49:193-205. [DOI] [PubMed] [Google Scholar]
- 19.LeBlanc, D. J., L. N. Lee, and A. Abu-Al-Jaibat. 1992. Molecular, genetic, and functional analysis of the basic replicon of pVA380-1, a plasmid of oral streptococcal origin. Plasmid 28:130-145. [DOI] [PubMed] [Google Scholar]
- 20.Li, Y. H., P. C. Y. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2001. Natural genetic transformation of Streptococcus mutans growing in biofilms. J. Bacteriol. 183:897-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, Y. H., P. C. Lau, N. Tang, G. Svensater, R. Ellen, and D. G. Cvitkovitch. 2002. Novel two-component regulatory system involved in biofilm formation and acid resistance in Streptococcus mutans. J. Bacteriol. 184:6333-6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, Y. H., X. L. Tian, G. Layton, C. Norgaard, and G. Sisson. 2008. Additive attenuation of virulence and cariogenic potential of Streptococcus mutans by simultaneous inactivation of the ComCDE quorum sensing system and HK/RR11 two-component regulatory system. Microbiology 154:3256-3265. [DOI] [PubMed] [Google Scholar]
- 23.Liljemark, W. F., D. H. Okrent, and C. G. Bloomquist. 1976. Differential recovery of Streptococcus mutans from various mitis-salivarius agar preparations. J. Clin. Microbiol. 4:108-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lis, M., and H. K. Kuramitsu. 2003. The stress-responsive dgk gene from Streptococcus mutans encodes a putative undecaprenol kinase activity. Infect. Immun. 71:1938-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marsh, P. D. 1994. Microbial ecology of dental plaque and its significance in health and disease. Adv. Dent. Res. 8:263-271. [DOI] [PubMed] [Google Scholar]
- 26.Martínez, B., A. L. Zomer, A. Rodriguez, J. Kok, and O. P. Kuipers. 2007. Cell envelope stress induced by the bacteriocin Lcn972 is sensed by the lactococcal two-component system CesSR. Mol. Microbiol. 64:473-486. [DOI] [PubMed] [Google Scholar]
- 27.Mascher, T., N. G. Margulis, T. Wang, R. W. Ye, and J. D. Helmann. 2003. Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol. Microbiol. 50:1591-1604. [DOI] [PubMed] [Google Scholar]
- 28.Mascher, T. 2006. Intramembrane-sensing histidine kinases: a new family of cell envelope stress sensors in Firmicutes bacteria. FEMS Microbiol. Lett. 264:133-144. [DOI] [PubMed] [Google Scholar]
- 29.Mascher, T., J. D. Helmann, and G. Unden. 2006. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol. Mol. Biol. Rev. 70:910-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohki, R., Giyanto, K. Tateno, W. Masuyama, S. Moriya, K. Kobayashi, and N. Ogasawara. 2003. The BceRS two-component regulatory system induces expression of the bacitracin transporter, BceAB, in Bacillus subtilis. Mol. Microbiol. 49:1135-1144. [DOI] [PubMed] [Google Scholar]
- 31.Rietkötter, E., D. Hoyer, and T. Mascher. 2008. Bacitracin sensing in Bacillus subtilis. Mol. Microbiol. 68:768-785. [DOI] [PubMed] [Google Scholar]
- 32.Suntharalingam, P., M. D. Senadheera, R. W. Mair, C. M. Levesque, and D. G. Cvitkovitch. 2009. The LiaFSR system regulates the cell envelope stress response in Streptococcus mutans. J. Bacteriol. 191:2973-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Syvitski, R. T., X. L. Tian, K. Sampara, A. Salman, S. F. Lee, D. L. Jakeman, and Y. H. Li. 2007. Structure-activity analysis of quorum sensing signaling peptides from Streptococcus mutans. J. Bacteriol. 189:1441-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tetsch, L., and K. Jung. 2009. The regulatory interplay between membrane-integrated sensors and transporter protein in bacteria. Mol. Microbiol. 73:982-992. [DOI] [PubMed] [Google Scholar]
- 35.Tian, X. L., D. Campbell, and Y. H. Li. 2007. Microarray-based transposon tracking to identify genes involved in bacitracin resistance and envelope stress response in Streptococcus mutans, abstr. K-010. Abstr. 107th Gen. Meet. Am. Soc. Microbiol., Toronto, Canada.
- 36.Tian, X. L., R. T. Syvitski, T. Liu, N. Livingstone, D. L. Jakeman, and Y. H. Li. 2009. A method for structure-activity analysis of quorum-sensing signaling peptides from naturally transformable streptococci. Biol. Proced. Online 11:207-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsuda, H., Y. Yamashita, Y. Shibata, Y. Nakano, and T. Koga. 2002. Genes involved in bacitracin resistance in Streptococcus mutans. Antimicrob. Agents Chemother. 46:3756-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wecke, T., B. Veith, A. Ehrenreich, and T. Mascher. 2006. Cell envelope stress response in Bacillus licheniformis: integrating comparative genomics, transcriptional profiling, and regulon mining to decipher a complex regulatory network. J. Bacteriol. 188:7500-7511. [DOI] [PMC free article] [PubMed] [Google Scholar]