Abstract
The guts and casts of earthworms contain microbial assemblages that process large amounts of organic polymeric substrates from plant litter and soil; however, the enzymatic potential of these microbial communities remains largely unexplored. In the present work, we retrieved carbohydrate-modifying enzymes through the activity screening of metagenomic fosmid libraries from cellulose-depleting microbial communities established with the fresh casts of two earthworm species, Aporrectodea caliginosa and Lumbricus terrestris, as inocula. Eight glycosyl hydrolases (GHs) from the A. caliginosa-derived community were multidomain endo-β-glucanases, β-glucosidases, β-cellobiohydrolases, β-galactosidase, and β-xylosidases of known GH families. In contrast, two GHs derived from the L. terrestris microbiome had no similarity to any known GHs and represented two novel families of β-galactosidases/α-arabinopyranosidases. Members of these families were annotated in public databases as conserved hypothetical proteins, with one being structurally related to isomerases/dehydratases. This study provides insight into their biochemistry, domain structures, and active-site architecture. The two communities were similar in bacterial composition but significantly different with regard to their eukaryotic inhabitants. Further sequence analysis of fosmids and plasmids bearing the GH-encoding genes, along with oligonucleotide usage pattern analysis, suggested that those apparently originated from Gammaproteobacteria (pseudomonads and Cellvibrio-like organisms), Betaproteobacteria (Comamonadaceae), and Alphaproteobacteria (Rhizobiales).
Microorganisms producing diverse glycosyl hydrolases (GHs) are widespread and typically thrive in environments where plant materials tend to accumulate and deteriorate (42, 73). The habitats of microorganisms with great GH diversity are the ruminant animal rumen, mouse bowel, and rabbit cecum (10, 24, 26, 28, 49, 74). Microorganisms associated with soil invertebrates in general and with soil earthworms in particular carry out metabolic processes that contribute to element cycling and are essential in sustaining processes which their hosts are unable to perform (20, 52, 72, 76). Although some species of earthworms produce cellulases (15, 55), they generally rely on microbes inhabiting their gastrointestinal (GI) tracts to perform cellulose utilization processes (31, 47, 77). Casts are of special interest in this respect. Considering that the overall numbers of cellulolytic microbes in earthworm casts are greater than those in soil (57), earthworm casts seem to play an important role in the decomposition of plant litter, serving as an inoculum for cellulosic substrates (9). It is important to note that microorganisms from preingested substratum (soil or plant litter) are predominant in the gut lumen (20); however, microbial populations in earthworm casts differ from those in soil in terms of diversity and the relative abundance of different taxa (29, 57, 63). It is anticipated that the enzymatic repertoire of such microbial communities must be especially broad toward diverse sugar-based polymeric, oligomeric, and monomeric substrates; however, among approximately 115 families of GHs with thousands of members known to date (12), none of the GHs have been derived from microorganisms of earthworm-associated microbial communities.
The aim of the present work was therefore to examine the diversity of GHs in metagenome libraries derived from fresh casts of Aporrectodea caliginosa and Lumbricus terrestris earthworms via functional screening. Other important tasks of this work were to characterize individual enzymes and to gain insight into their structural-functional features. Finally, we performed sequence analysis of large contiguous DNA fragments of fosmids harboring the genes for GHs to associate them with the organism(s) that may produce them, which was complemented by conventional small-subunit (SSU) rRNA clone library sequencing analysis.
MATERIALS AND METHODS
Materials and strains.
Chemicals, biochemicals, and solvents were purchased from Sigma-Fluka-Aldrich Co. (St. Louis, MO) and were of pro analysi quality. Oligonucleotides for DNA amplification, mutagenesis, and sequencing were synthesized by Sigma Genosys Ltd. (Pampisford, Cambs, United Kingdom). Restriction and modifying enzymes were from New England Biolabs (Beverly, MA). Ni-nitrilotriacetic acid (NTA) His·Bind chromatographic medium was from Qiagen (Hilden, Germany). Escherichia coli EPI300-T1 for fosmid library construction and screening from Epicentre Biotechnologies (Madison, WI), XL10 Gold for site-directed mutagenesis from Invitrogen (Carlsbad, CA), and E. coli GigaSingles for cloning and BL21(DE3)pLysS for expression using the pET-30 Ek/LIC vector (Novagen, Darmstadt, Germany) were cultured and maintained according to the recommendations of the suppliers.
Earthworms and cellulose cultures.
Earthworms of the species L. terrestris and A. caliginosa were collected from the top lowed horizon (0 to 20 cm) of soddy-podzolic soil under crop rotation at the Ecological Soil Station of Lomonosov, Moscow State University (Solnechnogorskiy District, Moscow Region, Russia), as described earlier (53, 54). Worms were kept in terrariums with soil at 12 to 15°C for >3 months and fed sterile leaf grass and oak litter. Casts (ca. 0.5 g [wet weight]) of earthworms of each species were collected by keeping the animals on wet sterile filter paper at 15°C; the cast suspensions made of sterile distilled water (1/1 [wt/vol]) were briefly spun down at low speed (100 × g), and aqueous phases (0.5 ml) of suspension from each species were used to inoculate 1,000-ml Erlenmeyer flasks with 500 ml of Getchinson medium (1.3 g K2HPO2, 0.3 g MgSO2·7H2O, 0.1 g CaCl2·6H2O, 0.01 g FeCl2·6H2O, 2.5 g NaNO2, distilled water to 1,000.0 ml, pH 7.2 to 7.4) with a folded filter paper (Whatman 3MM) disk ca. 20 cm in diameter dipped into the medium. The flasks were sealed and incubated without agitation in the dark at 15°C for 10 days, long enough for the cellulose disk to be nearly fully degraded.
DNA extraction.
The liquid phase (50 ml) from each culture was centrifuged at 5,000 × g for 10 min at 4°C; total DNA was extracted from the pellet using the G'NOME DNA isolation kit (Qbiogene, Germany). Isolated DNA was quantified with the Quant-iT PicoGreen dsDNA assay kit (Invitrogen) and visualized via 0.8% agarose gel electrophoresis.
Metagenomic library construction and detection of GHs.
Fosmid libraries were established by using the pCCFOS vector and E. coli EPI300-T1 according to the instructions of Epicentre (WI). Fosmid clones (ca. 11,500 per library, each library harboring ca. 400 Mbp of community genomes) were picked with a QPix2 colony picker (Genetix Co., United Kingdom) and grown in 384-well microtiter plates containing Luria-Bertani broth (LB) with chloramphenicol (12.5 μg/ml) and 15% (vol/vol) glycerol and stored at −80°C. To screen for GH activity, the clones were replicated on large (22.5 by 22.5 cm) petri agar plates to give an array of 2,304 clones per plate. Subsets of 5,760 fosmids from each library were screened for the ability to hydrolyze p-nitrophenyl (pNP)-β-d-glucopyranoside (pNPβGlu) and pNP-α-l-arabinopyranoside (pNPαApyr). After overnight incubation on LB agar containing chloramphenicol (12.5 μg/ml) and fosmid copy number induction solution as recommended by the supplier (Epicentre), the plates were overlaid with 20 ml of 0.4% (wt/vol) agarose prepared in 100 mM sodium acetate buffer, pH 5.6, containing 10 mM pNP-α-d-arabinofuranoside (pNPαAfur) or pNPαApyr. Lichenan-active fosmids were screened in agar plates supplemented with 1% (wt/vol) lichenan, followed by incubation with a water solution of 1% (wt/vol) Congo red. A total of 55 positive colonies exhibiting a strong yellow color were obtained. Fosmid sequencing and assembly were done at the Göttingen Genomics Laboratory (Germany). Initially, each fosmid was partially digested with the endonuclease Sau3a and subcloned into the pBluescript SK+ vector (Stratagene, La Jolla, CA) to establish a clone library. Ninety-six clones of each library were grown overnight in a Greiner 96-deep-well block (Greiner Bio-one, Frickenhausen, Germany) and harvested. DNA extraction was done by using the Millipore Montage Plasmid MiniPrep 96 kit (Millipore, Schwalbach/Ts., Germany) on a Qiagen BioRobots 8000 (Qiagen, Hilden, Germany). End sequencing of plasmids was performed in a 96-well format on an ABI 3730XL (Applied Biosystems/Life Technology, Darmstadt, Germany) with, on average, a 550- to 750-bp read length using standard sequencing primers T3 and T7. Sequences were vector clipped and assembled using the Phred/Phrap program. PCR-based techniques were used to enhance sequence quality and to close gaps remaining after assembly using fosmid walking. The gene for GH in fosmid clone AcP3B3 was initially subcloned into the pUC19 vector and then Sanger sequenced using primer walking.
Cloning, expression, and purification of GHs.
Gene cloning was carried out using PCR with PfuTurbo DNA polymerase and custom oligonucleotide primers (see Table S2 in the supplemental material). To amplify the entire GH genes, the corresponding fosmid was used as the template with the pair of primers described in Table S2 in the supplemental material. The conditions were 95°C for 120 s; 30 cycles of 95°C for 45 s, 50°C for 60 s, and 72°C for 120 s; and 72°C for 500 s. The PCR products were purified from agarose gel after electrophoresis using the QiaExII gel extraction kit (Qiagen, Hilden, Germany) and cloned into pET-30-Ek/LIC (Novagen) according to the manufacturer's instructions. Plasmids were subsequently isolated and introduced into the nonexpressing E. coli GigaSingles host and further into E. coli BL21(DE3)pLysS for expression. The transformation mixtures were plated on LB agar supplemented with kanamycin (30 μg/ml). For enzyme expression and purification, the resulting cells were grown overnight at 37°C with shaking at 200 rpm in 100 ml of LB containing appropriate antibiotics. Each liter of medium was inoculated with 25 ml of culture, and the cells were grown for 4 h to an optical density at 600 nm of ∼0.6 before induction using 1 mM isopropyl-β-d-galactopyranoside (IPTG) for 5 to 6 h. The cells were harvested by centrifugation (5,000 × g) for 15 min to yield 2 to 3 g/liter pellet. The cell pellet was frozen at −80°C overnight and then thawed and resuspended at 10 ml/g of pellet in lysis buffer containing 20 mM HEPES (pH 7.0), 0.3 M NaCl 5 mM imidazole, and 2 μg/ml DNase. Lysozyme (1 mg/ml) was added, and the mixture was incubated for 1 h on ice with occasional mixing. The cell suspension was then sonicated for a total of 1.2 min and further spun at 15,000 × g for 15 min, and the supernatant was retained. His6-tagged enzymes were purified at 25°C after binding to Ni-NTA His·Bind resin. The columns were washed with 20 mM HEPES (pH 7.0)-0.3 M NaCl-5 mM imidazole, and enzymes were eluted with 20 mM HEPES (pH 7.0)-0.3 M NaCl-250 mM imidazole. Monitoring of enzyme elution was carried out by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and/or activity measurement using the standard assay. Purity was assessed as >95% using SDS-PAGE, which was performed with 12% (vol/vol) acrylamide gels as previously described (45), in a Bio-Rad Mini Protean system. Nondenaturing gel electrophoresis performed with 8 to 12% (vol/vol) acrylamide gels was used to determine the native molecular masses of the cloned proteins. Protein concentrations were determined according to Bradford (7), with bovine serum albumin as the standard.
Enzyme characterization.
Absorbance data were obtained on a BioTek Synergy HT spectrophotometer. The reaction conditions were [E]o = 0 to 12 nM, [substrate] ranging from 0 to 50 mM, 100 mM Tris-HCl (pH 8.5), and T = 40°C. For hydrolysis of pNP derivatives, a corresponding volume of a 120 mM pNP derivative stock solution in the corresponding buffer (Sigma) was incubated for 10 min with 12 nM enzyme diluted in 200 μl of 100 mM buffer and measured in a spectrophotometer at 405 nm in 96-well microtiter plates.
For cello- and xylooligosaccharides, released glucose and xylose were determined using glucose and xylose oxidase kits (Sigma). Initial rates were fitted to the Michaelis-Menten kinetic equation by nonlinear regression to extract the apparent Km and kcat. The concentrations used were 0 to 3.25 mM pNPβGlc (with 0.146 nM hydrolase), 0.35 to 5.06 mM cellobiose (with 0.243 nM hydrolase), 0.63 to 3.75 mM pNPαApyr (with 0.365 nM hydrolase), 0.50 to 3.75 mM pNP-β-d-cellobioside (pNPβCel) (with 0.183 nM hydrolase), and 0.50 to 3.75 mM pNPαAfur (with 0.183 nM hydrolase). All values were determined in triplicate and were corrected for the spontaneous hydrolysis of the substrate. The results shown are the averages of three independent assays ± the standard deviations.
The standard GH assay conditions were [E]o = 12 nM, [pNP derivative] 10 mM, 100 mM HEPES, a total volume 200 μl, pH 7.0, and T = 40°C.
The pH and temperature optima were determined in the ranges of pH 6.0 to 10.0 and 5 to 60°C. The buffers (100 mM) used were acetate (pH 5.0 to 6.0), morpholineethanesulfonic acid (MES; pH 6.0 to 7.0), HEPES (pH 7.0 to 8.0), Tris-HCl (pH 8.0 to 9.0), and glycine (pH 9.0 to 10.0). The pH was always adjusted at 25°C. The pH and temperature profiles were determined at 40°C and pH 7.0, respectively.
In silico analysis of proteins and three-dimensional (3D) modeling.
For gene calling in the cloned DNA fragments, the GeneMark.hmm prediction tool was used (http://exon.gatech.edu/genemark/) (50). Deduced proteins were screened via blastp and psi-blast (1) against the nonredundant database sourced from the nucleotide (nr/nt) collection, reference genomic sequences (refseq_genomic), whole-genome shotgun reads (wgs), and environmental samples (env_nt). The translation products were further searched for protein domains in the Pfam-A (5) and COG (67) databases. Multiple-sequence alignments were made using the ClustalW tool (http://www.ebi.ac.uk/clustalw/index.html) integrated in the BioEdit software (35). Structural alignments of proteins homologous to GHs obtained in this study were generated by GenTHREADER (39) and used to retrieve a model from the Swiss-Model server (32). The following Protein Data Bank (PDB) entries were used as templates: 2P5Y for G03-3, 2R4A for G05-26 and G05-27, 3bmx for G06-24, 1clc for G07-33, 2c0h for G08-17, 1qjw for G10-6, 1exg and 1tvn for G10-10, and 3cf7 for AcP3B3.
Oligonucleotide usage pattern analysis.
The frequencies of tetranucleotides, normalized by GC content, were calculated for the given sequences (58), and then the database was searched for the standard tetranucleotide usage patterns determined for 807 bacterial chromosomes and 503 plasmids (http://www.bi.up.ac.za/SeqWord/mhhapplet.php). The sequences of bacterial chromosomes and plasmids were downloaded from the NCBI public database (http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi). Inserts of mobile genetic elements characterized by alternative oligonucleotide usage were identified by the program SeqWord Sniffer available at http://www.bi.up.ac.za/SeqWord/sniffer/index.html (6, 59). Compositional sequence divergences were evaluated in distance values by the algorithm published previously (56, 57). Dendrograms were constructed based on the calculated distance matrix by the program fitch.exe of the Phylip package (http://evolution.genetics.washington.edu/phylip.html) using the Fitch-Margoliash algorithm. The likelihood that these DNA fragments originated in bacterial or plasmid genomes was analyzed by the in-house program MetaLingvo (www.bi.up.ac.za/SeqWord/metalingvo/index.html).
Construction of 16S and 28S rRNA gene clone libraries and clone sequencing.
PCR amplification was performed by serial dilution of template DNA using primers and PCR conditions described earlier (11). Bacterial 16S rRNA genes were amplified using bacterium-specific primers F27 (5′-AGAGTTTGATCMTGGCTCAG-3′) and R1492 (5′-CGGYTACCTTGTTACGACTT-3′) (11). Eukaryotic 28S rRNA gene fragments were amplified with primers NL-1 (5′-GCATATCAATAAGCGGAGGAAAA-3′) and NL-4 (5′-GGTCCGTGTTTCAAGACGG-3′). Amplification was done in a 20-μl reaction volume with recombinant Taq DNA polymerase (Invitrogen, Germany) and original reagents, according to the basic PCR protocol, with annealing temperatures of 45 and 50°C (bacterial and eukaryotic rRNA genes, respectively) for 30 cycles. PCR amplicons were purified by electrophoresis on 0.8% agarose gels, followed by isolation from excised bands using a QIAEX II gel extraction kit (Qiagen, Germany). The purified PCR products were ligated into plasmid vector pCRII-TOPO (TOPO TA cloning kit, Invitrogen, Germany) with subsequent transformation into electrocompetent cells of E. coli TOP10 (Invitrogen, Germany). After blue/white screening, randomly picked clones were resuspended in PCR lysis solution A without proteinase K (67 mM Tris-Cl [pH 8.8], 16 mM NH4SO4, 5 μM β-mercaptoethanol, 6.7 mM MgCl2, 6.7 μM EDTA [pH 8.0]) (62) and heated at 95°C for 5 min. The lysate (1 μl) was used as template DNA for PCR amplification using primers M13 forward (5′-GACGTTGTAAAACGACGGCCAG-3′) and M13 reverse (5′-GAGGAAACAGCTATGACCATG-3′). After verification on agarose gel, PCR products were purified with the MinElute 96 UF PCR purification kit (Qiagen, Germany); clones of bacterial and eukaryotic rRNA genes were sequenced using primers R1492 for bacteria and NL1 for eukaryotes according to the protocol for the BigDye Terminator v1.1 cycle sequencing kit from Applied Biosystems.
Phylogenetic analyses of SSU rRNA gene sequences.
All of the sequences obtained were analyzed with the MALLARD software (4). Sequences were scored against GenBank using the BLAST alignment software (http://www.ncbi.nlm.nih.gov/BLAST/) (1). Eukaryotic clone libraries from both enrichments were compared to each other using the webLIBSHUFF v0.96 software (64); bacterial clone libraries were compared with the LIBCOMPARE tool available at the Ribosomal Database Project II website (http://rdp.cme.msu.edu/comparison/comp.jsp) (14). Phylogenetic analysis was then performed with (i) Kimura's two-parameter algorithm for the neighbor-joining treeing method using the software MEGA version 4.0 (66); (ii) the maximum-likelihood method using the PHYML-aLRT program (3, 33) with the general time-reversible model, gamma-distributed rate heterogeneity, and a significant proportion of invariable sites; and (iii) MrBayes software package 3.1.2 (61) using the general time-reversible model of evolution. The Bayesian analysis was run in duplicate with four chains for 107 generations with a sampling frequency of 1,000 generations for bacterial clones and 106 generations with a sampling frequency of 100 generations for eukaryotic clones; the initial 2,500 of 104 generations were discarded as burn in.
Nucleotide sequence accession numbers.
The DNA sequences of the fosmid clones determined in this study were deposited in the GenBank/EMBL/DDBJ databases under accession numbers GQ996408 to GQ996414 (fosmid clones), GU045290 (pUC19 subclone encoding AcP3B3), and GQ902838 to GQ902937 (SSU rRNA gene sequences from clone libraries).
RESULTS
Screening of metagenomic libraries for GH activity.
Subsets of 5,760 fosmids from the L. terrestris and A. caliginosa libraries have been scored for the ability to hydrolyze pNPβGlu and pNPαApyr. Among the 55 positive clones obtained from both libraries, we have selected, essentially at random, 2 fosmid clones from the L. terrestris library (here, g03 and g04) and 6 (here, g05 to g10) from the A. caliginosa library. All cloned GH-encoding DNA fragments were fully sequenced, with the only exception of fosmid g09, which was subjected to shotgun subcloning into the pUC19 vector and consequent activity screening (the positive subclone was designated AcP3B3). The GHs characterized in the present work were named based on the source fosmid identification (ID) number and the number of the corresponding open reading frame (ORF) coding for the particular enzyme. Assignment of functions to the deduced proteins in each fosmid was also performed. See Fig. S1 and S2 in the supplemental material for maps of the genome fragments cloned into these fosmids and the corresponding arrangement of homologous ORFs within the genome of Cellvibrio japonicus (16), whose proteins were found to exhibit the highest level of homology to those predicted in fosmids g05, g07, g08, and g10 (see Table S1 in the supplemental material for details). The distribution of clusters of orthologous groups of deduced proteins varied significantly within cloned DNA fragments (see Fig. S3 and S4 and Table S1 in the supplemental material), with the fraction of deduced proteins of unknown function (hypothetical proteins) being between 10% and 50% of the total number of ORFs. Most frequently, the genes related to carbohydrate transport and metabolism occurred in fosmids g07 and g10, where they made up 16% and 29% of the total number of ORFs, respectively.
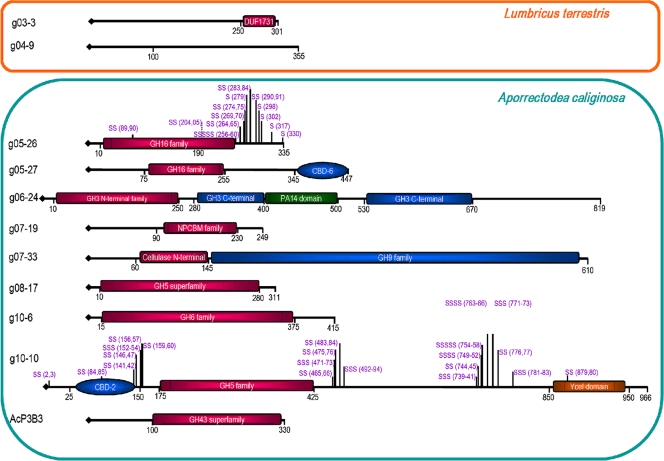
Further on, identified GHs were expressed in E. coli and purified (see Fig. S5 in the supplemental material) and their activities were tested with an array of substrates (Table 1) and temperature and pH optima were determined (see Fig. S6 in the supplemental material). The polypeptide sequences indicated significant differences between individual GHs, allowing their affiliation (with the exception of two GHs from the L. terrestris library) with certain GH classes (Fig. 1) and modeling of their 3D structures (see Fig. S7 in the supplemental material). Below, we provide a more detailed analysis of their structural and biochemical characteristics.
TABLE 1.
Kinetic parametersa and molecular masses of enzymes used in this study
| Enzyme (mol wt [monomer, native]) and substrate | Km (mM) | kcat (s−1) | kcat/Km (s−1 M−1) | Enzyme type |
|---|---|---|---|---|
| G03-3 (32,820.07, 66,000) | ||||
| pNPβGal | 1.01 ± 0.03 | 10.7 ± 1.1 | 1.1 × 104 | β-Galactosidase |
| pNPαApyr | 1.68 ± 0.35 | 7.5 ± 71.2 | 4.5 × 103 | |
| G04-9 (40,593.73, 80,000) | ||||
| pNPβGal | 0.27 ± 0.07 | 244.9 ± 9.3 | 9.1 × 105 | β-Galactosidase |
| pNPαApyr | 19.80 ± 1.0 | 193.2 ± 10.7 | 9.8 × 103 | |
| G05-26 (36,224.15, 37,000) | ||||
| pNPβGlu | 12.25 ± 2.1 | 360.8 ± 17.0 | 2.9 × 104 | β-Glucosidase (GHF16) |
| pNPβCel | 14.60 ± 1.76 | 43.4 ± 2.9 | 2.9 × 103 | |
| Cellobiose | 12.63 ± 1.6 | 24.4 ± 2.1 | 1.9 × 103 | |
| Cellotriose | 17.60 ± 1.7 | 12.6 ± 1.9 | 7.2 × 102 | |
| Cellotetraose | 23.80 ± 3.4 | 8.6 ± 2.3 | 3.6 × 102 | |
| Cellopentaose | 24.90 ± 4.8 | 1.8 ± 0.2 | 72.2 | |
| G05-27 (49,080.22, 50,000) | ||||
| pNPβGlu | 28.40 ± 4.9 | 43.5 ± 1.3 | 1.5 × 103 | Endo-β-1,3(4)-glucanase (GHF16) |
| Lichenanb | 0.17 ± 0.04 | 12.9 ± 0.2 | 76 | |
| G06-24 (88,300.54, 90,000) | ||||
| pNPβGal | 2.54 ± 0.17 | 87.4 ± 4.0 | 3.4 × 104 | β-Xylosidase (GHF3) |
| pNPβGlu | 0.37 ± 0.04 | 204.9 ± 9.5 | 5.5 × 105 | |
| pNPβXyl | 0.016 ± 0.001 | 379.0 ± 25.3 | 2.4 × 107 | |
| pNPαAfur | 1.36 ± 0.10 | 27.0 ± 1.0 | 1.9 × 104 | |
| Xylobiose | 0.16 ± 0.04 | 98.9 ± 4.4 | 6.2 × 105 | |
| Xylotriose | 0.46 ± 0.15 | 15.4 ± 3.9 | 3.3 × 104 | |
| Oat spelt xylanb | 4.6 ± 1.3 | 9.8 ± 2.9 | 2.13 | |
| G07-33 (64,791.79, 70,000) | ||||
| pNPβGlu | 27.75 ± 4.02 | 23.4 ± 1.1 | 0.8 × 103 | β-Glucosidase (GHF9) |
| CMCb | 2.18 ± 0.71 | 14.9 ± 1.6 | 6.8 × 103 | |
| Lichenanb | 1.67 ± 0.37 | 12.9 ± 2.5 | 7.72 | |
| G08-17 (35,785.76, 35,000) | ||||
| pNPβCel | 6.64 ± 0.83 | 119.0 ± 2.8 | 1.8 × 104 | Endo-β-1,4-glucanase (GHF5) |
| Cellobiose | 9.21 ± 1.40 | 59.6 ± 3.7 | 6.5 × 103 | |
| pNPβGlu | 16.84 ± 1.93 | 20.0 ± 0.9 | 1.2 × 103 | |
| Lichenanb | 0.68 ± 0.03 | 12.8 ± 6.3 | 19 | |
| CMC | 0.43 ± 0.02 | 3.1 ± 1.1 | 7 | |
| G10-6 (44,741.79, 85,000) | ||||
| pNPβCel | 0.041 ± 0.007 | 978.4 ± 56.4 | 2.4 × 107 | β-Cellobiohydrolase (GHF6) |
| Avicel | 2.28 ± 0.39 | 8.66 ± 2.4 | 3.79 | |
| G10-10 (101,532.54, 100,000) | ||||
| pNPβGlu | 10.52 ± 1.96 | 41.0 ± 1.0 | 3.9 × 103 | Endo-β-1,4-glucanase (GHF5) |
| pNPβCel | 17.61 ± 2.53 | 18.2 ± 1.3 | 1.0 × 103 | |
| Cellobiose | 16.75 ± 2.29 | 13.7 ± 0.8 | 8.2 × 102 | |
| Cellotetraose | 12.92 ± 3.62 | 10.0 ± 1.3 | 7.7 × 102 | |
| Cellopentaose | 29.87 ± 4.30 | 5.7 ± 0.8 | 1.9 × 102 | |
| CMCb | 0.64 ± 0.07 | 3.8 ± 0.3 | 6 | |
| AcP3B3 47,486.80, 200,000) | ||||
| pNPβGal | 0.19 ± 0.06 | 1,783.0 ± 48.7 | 9.4 × 106 | β-Galactosidase (GHF43) |
| pNPαApyr | 13.10 ± 1.7 | 1,837.7 ± 201.2 | 1.4 × 105 | |
| pNPαAfur | 13.10 ± 1.4 | 1,954.3 ± 58.6 | 1.5 × 105 | |
| pNPβXyl | 22.80 ± 3.7 | 2,850.7 ± 219.4 | 1.3 × 105 |
kcat and Km values were obtained as described in the supplemental material: [E]o = 0 to 12 nM, [substrate] ranging from 0 to 100 mM, 100 mM Tris-HCl, pH 8.5, T = 40°C.
Km values in mg/ml; kcat/Km values in s−1 mg−1 ml.
FIG. 1.
GH-related domains in functionally characterized GHs. Some proteins exhibit a multidomain architecture. Serine-rich regions are highlighted.
Two β-galactosidases from the L. terrestris library constitute two new families of GHs.
DNA fragments from fosmids g03 and g04 from the L. terrestris library were approximately 35.6 and 19.7 kbp in size, respectively, and analysis of their sequences predicted 37 and 19 ORFs. However, blastp and psi-blast analyses of all of the individual predicted polypeptide sequences could not suggest any candidate with reasonable similarity to any known GH. To identify the genes coding for the corresponding phenotype, clones were subjected to shotgun subcloning and activity screening.
We found that ORF3 in g03 and ORF9 in g04 were responsible for the GH activities and that the pure G03-3 and G04-9 enzymes (see Fig. S5 in the supplemental material) hydrolyze pNP-β-d-galactoside (pNPβGal) and pNPαApyr (Table 1). No other model (commercial) substrate tested was hydrolyzed, thus suggesting that their natural substrates should be elucidated. Approximately 90- and 2-fold higher catalytic efficiency with β-galactoside than with α-arabinopyranoside was observed for G04-9 and G03-3, respectively, due to their higher affinity for pNβGal. However, the polypeptides encoded by g03-3 and g04-9 exhibited high degrees of identity (63 and 75%) to conserved hypothetical proteins with no detectable GH domains (Fig. 1). Moreover, structural alignment and 3D modeling based on the crystal structure of the UDP-glucose epimerase of Thermus thermophilus (PDB 2P5Y; ca. 18% sequence identity) revealed that G03-3 is related to isomerases and dehydrogenases from COG1090 that actually have no relevance to the observed GH phenotype (see Fig. S7 in the supplemental material), whereas the G04-9 protein did not exhibit any conserved domains. Therefore, the sequence and experimental analyses suggested that these two enzymes belong to the two new families of functional GHs with β-galactosidase (EC 3.2.1.23) and α-arabinopyranosidase activities, whereas one of them (G03-3) is structurally related to isomerases/dehydrogenases. The absence of appropriate models did not allow us to suggest putative active sites. G03-3 and G04-9 consisted of two identical subunits with molecular masses of the native enzymes of 66 and 80 kDa (Table 1). The optima for the hydrolysis of pNβGal were 50°C and pH 8.0 to 10.0 for G04-9 and 40°C and pH 7.0 to 8.0 for G03-3 (see Fig. S6 in the supplemental material).
Additionally, analysis of the g04 fragment (see Fig. S3 in the supplemental material) showed that eight ORFs (g04-12 to g04-19) were arranged in a cluster presumably involved in the transport and biosynthesis of ectoine (2-methyl-1,4,5,6-tetrahydropyrimidine-4-carboxylic acid), a compatible solute whose biosynthesis is triggered under osmotic stress (43). In this context, the hydrolytic activity of G04-9 was assayed at elevated concentrations of NaCl. Strong activation (up to 3-fold in terms of kcat) was observed at NaCl concentrations between 0.2 and 2.5 M (see Fig. S8 in the supplemental material).
Functional heterogeneity of multidomain GHs from the A. caliginosa library. (i) g05 analysis.
The DNA insert in fosmid g05 encoded two proteins of GH family 16 (GHF16) most similar to β-glucanases from C. japonicus. G05-26 exhibited 78% and G05-27 exhibited 81% amino acid sequence identity to their counterparts, CJA_3705 and CJA_3706, from C. japonicus (see Table S1 in the supplemental material). Members of GHF16 hydrolyze and cleave β-1,4-glycosidic bonds precisely when β-1,3-glycosidic linkages are located prior to β-1,4-glycosidic linkages in lichenan or β-d-glucans (71). The same was proved for G05-27, which showed appreciable activity with pNPβGlu and lichenan {a β-[1→3(4)]-β-glucan} (with a 34-fold difference in terms of kcat/Km) (Table 1). It was therefore classified as a GHF16 endo-β-1,3(4)-glucanase. For the G05-26 protein, the highest hydrolytic activity was with pNPβGlu (19-fold higher than that shown by G05-27), whereas no longer polymers were used. pNPβCel and short cellooligosaccharides were also used but with much, 8- to 200-fold, lower catalytic efficiency. It was therefore classified as a GHF16 β-glucosidase. Both enzymes showed optimal activity over a broad range of temperatures (30 to 70°C) and in a narrow pH range (8.5 to 9.0) (see Fig. S6 in the supplemental material) and were found be to monomers of 37 (G05-26) and 50 (G05-27) kDa (Table 1).
The amino acid sequence identity of the two enzymes was about 77% (see Fig. S9 in the supplemental material). The 3D model analysis suggested that the two enzymes harbor quite similar catalytic domains (Fig. 1; see Fig. S7 in the supplemental material): an N-terminal catalytic domain with the general active-site motifs Glu67-Asp69-Glu71 (G05-26) and Glu137-Asp139-Glu141 (G05-27) with a β-propeller fold. However, substantial differences were observed in the C-terminal region: G05-27 contains a carbohydrate-binding domain (CBD) of family 6 that is probably involved in substrate recognition and binding, whereas that of G05-26 contains a serine-rich region distributed overall throughout the C terminus (Fig. 1), 24 residues in repeat units of 2 to 5 residues), with no functional role assigned to such regions linking catalytic and CBD modules in cellulases (21, 34, 37). The presence of a CBD may explain the capacity of G05-27 to hydrolyze soluble cellulosic substrates like lichenan.
(ii) Analysis of fosmid g06.
The DNA fragment in fosmid g06 encoded, inter alia, protein G06-24, affiliated with GHF3 and most similar to a β-glucosidase from Rhizobium etli CIAT 652 (61% identity). GHF3 contains 1,529 known enzymes (http://www.cazy.org), ranging from β-glucosidases (EC 3.2.1.21) to β-xylosidases (EC 3.2.1.37) that are able to remove successive β-d-glucose and β-d-xylose residues from the nonreducing termini, respectively (23). The pure enzyme showed 23- and 44-fold higher binding and catalytic efficiencies for pNP-β-d-xylopyranoside (pNPβXyl) than for pNPβGlu (Table 1). The enzyme was also able to hydrolyze xylobiose and xylotriose and the polymeric material xylan, although with low efficiency. The substrate specificity, together with sequence affiliation with GHF3, suggested the enzyme to be a β-d-xylosidase (EC 3.2.1.37). The enzyme was found to be a monomer of 90 kDa (Table 1) active over a broad range of pHs (5.5 to 9.0) and with an optimum temperature of 50°C (see Fig. S6 in the supplemental material).
The active nucleophile and acid/base that have been identified for various GHF3 β-glucosidases contained a conserved Ser-Asp-Trp segment possessing the nucleophile Asp, which was also found in the N-terminal sequence of G06-24 (Ser225-Asp226-Trp227) (Fig. 1; see Fig. S7 in the supplemental material). Moreover, this enzyme exhibited the most complex primary structure of any GH in this study, with four domains involved in catalysis and binding (Fig. 1; see Fig. S7 in the supplemental material).
(iii) Analysis of fosmid g07.
A genomic fragment from g07 encoded an enzyme of GHF9, named G07-33, that is most similar to the putative endo-1,4-β-glucanase CJA_1633 from C. japonicus (67% identity). Cellulase GHF9 has 424 entries in the CAZY database and includes endoglucanases (EC 3.2.1.4), cellobiohydrolases (EC 3.2.1.91), and β-glucosidases (EC 3.2.1.21). All of them catalyze the hydrolysis of β-1,4 linkages with inversion of the anomeric carbon configuration (36). Here we found that the G07-33 enzyme was highly specific for pNPβGlu, with an inability to hydrolyze any other pNP glycoside and therefore should be classified as a GHF9 β-glucosidase (EC 3.2.1.21). Carboxymethyl cellulose (CMC) and lichenan were used at similar turnover rates. The enzyme was a monomer of 70 kDa (Table 1) most active at 40 to 50°C and pH 8.0 to 9.0 (see Fig. S6 in the supplemental material). The enzyme contains a modular structure with a C-long-terminal catalytic domain with a putative Asp217, Asp220, and Glu588 catalytic core and a cellulose-binding N-terminal domain formed by β sheets (Fig. 1; see Fig. S7 in the supplemental material).
In addition to the above gene, the sequence analysis revealed five other ORFs (g07-18 to g07-22) arranged in a gene cluster encoding proteins that might be involved in sugar metabolism and possibly in cell wall biosynthesis. The deduced products of g07-18 to g07-22 showed a high level of homology (53 to 89% identity and 56 to 94% similarity) to glycosyltransferases, enzymes transferring sugar moieties from UDP-glucose, UDP-N-acetylgalactosamine, GDP-mannose, or CDP-abequose to a range of substrates, including cellulose, dolichol phosphate, dolichol-P-mannose, and teichoic acids. Moreover, the protein G07-19 exhibited significant relatedness (38% identity and 56% similarity) to the novel putative CBD named NPCBM (Pfam_08305) from Shewanella loihica PV-4. This domain is also known as NEW2, is found at the N termini of 18 different GHF98 proteins (see http://www.brenda-enzymes.info/), and serves for binding to carbohydrates (60). Taken together, these data suggest that the cloned fragment provides a set of genes for both the synthesis and hydrolysis of oligosaccharides.
(iv) Analysis of fosmid g08.
In fosmid g08, one of the deduced ORFs was predicted to code for protein G08-17 of GHF5, which has 75% sequence identity to the putative CJA_3369 protein of C. japonicus. GHF5 is one of the largest and most diverse GHFs (22), with the majority (approximately 60%) of the enzymes of bacterial origin and only a few proteins from archaea and higher eukaryotes, e.g., from plant-parasitic nematodes (17, 18, 44). Enzymes of GHF5 can degrade the β-1,4 linkages of cellulose, the most abundant plant polymer. Bacterial members of GHF5, as well as those from plants and fungi, contain a catalytic domain, a linker, and different types of CBDs of family 2, while currently known GHF5 endoglucanases of protists and beetles comprise no CBDs (44). Even though no evidence for a CBD-like domain was found in the G08-17 protein (Fig. 1; see Fig. S7 in the supplemental material), it was able to hydrolyze lichenan and, to a lesser extent, CMC. The pure enzyme, a monomer of 35 kDa, was also shown to act more efficiently (up to 950-fold in terms of kcat/Km) than pNPβCel, followed by pNPβGlu, with maximal efficiency shown at ∼55°C and pH 8.0 to 9.0 (see Fig. S6 in the supplemental material). According to its substrate specificity, the enzyme was classified as a GHF5 endo-1,4-β-d-glucanase.
Structurally, G08-17 was most similar to the mannase from Mytilus edulis (46), with a Glu141-and-Glu237 catalytic dyad and a (βα)8 barrel fold (see Fig. S7 in the supplemental material). In addition to that, ORF2, -4, and -16 encoded α/β fold hydrolases (esterase and feruloyl esterase-like) possibly involved in attack of the ester bond between hydroxycinnamic acids such as ferulic, p-coumaric, and sinnapic acids and sugars present in the intricate structure of the plant cell wall (70).
(v) Analysis of fosmid g010.
Analysis of fosmid g10 showed that putative genes for GHs were arranged in gene clusters probably involved in sugar binding and hydrolysis. ORF6 of g10 was predicted to encode a very hydrophilic 416-amino-acid (aa) protein, G10-6, with an estimated pI of 5.30 and without a signal peptide. The deduced protein belongs to GHF6, which comprises enzymes catalyzing the hydrolysis of crystalline cellulose by initiating their action from the ends of the cellulose chains and producing primarily cellobiose (41). G10-6 has obvious relatedness (78% identity and 87% similarity) to the predicted cellobiohydrolase CJA_2473 of C. japonicus. The affiliation of the enzyme with β-cellobiohydrolase GHF6 was confirmed by examining its substrate specificity and by showing that G10-6 was the only enzyme able to hydrolyze Avicel, although at much lower (112-fold, considering kcat values) rates than the pNPβCel. The enzyme, a dimer of 85 kDa, exhibits maximal activity at 55°C and pH 9.5 (see Fig. S6 in the supplemental material).
ORF10, also predicted in g10, encodes a product of 967 aa (G10-10) with a deduced molecular mass of 101,500 Da and an estimated pI value of 6.12 and with homology to known cellulases of GHF5 (73% identity to the predicted endo-1,4-β-glucanase CJA_2983 of C. japonicus). Sequence alignment revealed that this protein shows an identity of ca. 5% with the G08-17 endo-1,4-β-glucanase. Enzymatic analysis revealed that G10-10 showed a higher preference for both activity (4-fold) and binding (1.7-fold) to pNPβGlu over pNPβCel and that it was also able to hydrolyze CMC, albeit at lower rates (650-fold), and short cellooligosaccharides (up to 20-fold), as referred to G08-17. In contrast to G08-17, which also belongs to GHF5, this enzyme, functional as a monomer of 100 kDa, was unable to hydrolyze β-[1→3(4)]-β-glucan and thus should be considered an endo-1,4-beta-d-glucanase of GHF5. The maximal activity of this enzyme was found to occur at 35 to 55°C and pH 9.0 (see Fig. S6 in the supplemental material).
From a structural point of view, G10-6 exhibits a typical triosephosphate isomerase α/β fold with a putative active center formed by Asp79 and Asp128 (see Fig. S7 in the supplemental material), whereas the enzyme G10-10 showed much greater structural complexity (Fig. 1; see Fig. S7 in the supplemental material). The N-terminal domain is composed of an N-distal CBD (residues 35 to 150, named G10-10_NT) with a β-barrel fold and a number of β sheets and then a typical GHF5 cellulose domain (residues 170 to 450, named G10-10 CATDOM). The latter contains the catalytic dyad Asp79 and Asp128 with a (βα)8-barrel fold. The nature of the other two domains at the C terminus of this polypeptide, shown in Fig. 1, is unclear. Thus, the enzyme bears a long domain (residues 500 to 700) which, similar to the CATDOM, is located between two polyserine regions. This seemingly noncatalytic domain could be implicated in carbohydrate binding. Tandem Ser repeats were also found in the linker regions of two ORFs (ORF1 and -3) encoding peptides related to pectin esterases of the CBD2 and CBD6 families, as well as in three carbohydrate-binding proteins (ORF12, -15, and -16) that might be involved in sugar metabolism (from 20 to 33 repeats of two to five units) (for details, see in Table S1 in the supplemental material). Finally, the protein harbors a short C-distal domain of the YceI type whose homologues are related to lipid binding. Although several crystal structures have been solved, the degree of sequence identity of the protein in this region was too low to construct a model.
(vi) Analysis of plasmid AcP3B3.
Analysis of the 2,394 bp-long subcloned DNA fragment from plasmid clone pUC19-AcP3B3 revealed the presence of two ORFs. The first ORF (1,302 bp, positions 12 to 1313) encoded a putative 434-aa-long protein with a predicted molecular mass of 47 kDa and an estimated pI of 8.44. Sequence analysis revealed putative conserved domains for the sugar transporter superfamily and showed significant similarity to a predicted transport protein from C. japonicus (78% identity and 88% similarity). The second ORF (1,017 bp), located 58 bp upstream, encoded a 339-aa-long protein named AcP3B3 with a calculated molecular mass of 38,136.70 Da and an estimated pI of 5.04. The protein AcP3B3 was most homologous to GHF43 (pfam04617) and β-xylosidases and exhibited high similarity to the predicted α-N-arabinofuranosidase from Flavobacterium johnsoniae UW101 (72% identity). GHF43 is known to contain arabinanases, i.e., enzymes that hydrolyze the α-1,5-linked l-arabinofuranoside backbone of plant cell wall arabinans. Analysis of the pure enzyme, which formed five subunits with a total molecular mass of 200 kDa and worked optimally at 45 to 55°C and pH 7.5 to 10.0, revealed that, together with G06-24, it turned out to be the most promiscuous enzyme, able to hydrolyze pNPβXyl, pNPαAfur, pNPαApyr, and pNPβGal more than 3-fold more efficiently (see Fig. S4 in the supplemental material). The enzyme showed a binding preference for pNPβGal (up to 2 orders of magnitude), producing a 75-fold net positive effect on catalytic efficiency, and thus cataloguing it as a β-galactosidase (EC 3.2.1.23). No modular structure was detected, and the enzyme only contained a GHF43 domain at the C terminus of the polypeptide (Fig. 1). The Asp31 (base), Glu238 (acid), and Asp151 (substrate binding) residues may form the catalytic site of the enzyme (Fig. 1; see Fig. S7 in the supplemental material).
(vii) Analysis of metagenomic DNA fragments using a genome linguistic approach.
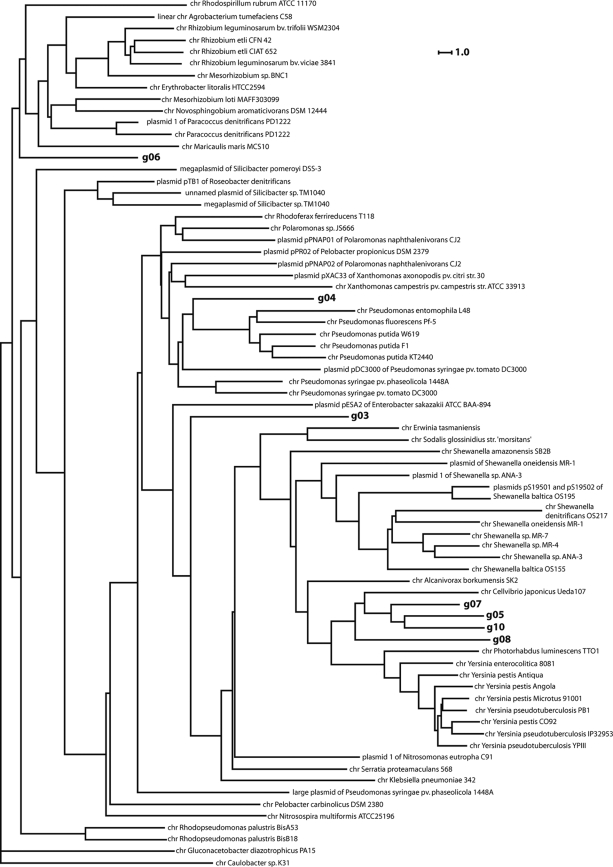
Compositional similarity between the metagenomic fragments and the sequences of related bacterial chromosomes and plasmids was analyzed by comparison of the frequencies of tetranucleotides in DNA sequences. The tetranucleotide usage patterns were calculated for the metagenomic clones, and the database of standard tetranucleotide usage patterns calculated for DNA sequences of bacterial chromosomes and plasmids was searched for similar patterns. For the selected sequences that showed compositional similarity to the metagenomic fragments, a dendrogram graph was built (Fig. 2). Below, the results of tetranucleotide usage, statistical comparison are summarized.
FIG. 2.
Dendrogram of compositional sequence similarities calculated by comparison of frequencies of tetranucleotides in DNA fragments from fosmids of L. terrestris and A. caliginosa libraries versus fully sequenced bacterial chromosomes (chr) and plasmids.
Fragment g03 showed some DNA compositional similarity to the chromosome of Rhodoferax ferrireducens T118; however, it was quite distant from this and all other organisms in the database. Therefore, one could assume only that the g03 fragment may most likely belong to Betaproteobacteria from the Rhodoferax or Polaromonas lineage.
The g04 fragment seems to be a part of a Pseudomonas chromosome. On the dendrogram in Fig. 2, this fragment clustered together with the chromosomes of P. entomophila and P. putida.
Fragments g05, g07, and g10 came from similar organisms, perhaps even from the same strain (fragments g07 and g10 overlap with zero nucleotide mismatches). The tetranucleotide usage pattern of g07 was more divergent, probably because it contains a horizontally transferred gene island (positions 18579 to 26761) (see Fig. S10 in the supplemental material). The gene island starts with several tandem repeats (TTTTCTTGTACTTCTTGTACTTCTTGTACTTCTTGTACTTCTTGTACTTCTTGTACTTCT) which are also known to flank a gene island in Methanothermobacter thermautotrophicus Delta H (positions 482301 to 482360; TCTTCTTGTACTTCTTGTACTTCTTGTACTTCTTGTACTTCTTGTACTTCTTGTACTTGT). A foreign origin of this gene island is consistent with the results of the annotation, as all of the genes of g07 showed homology with their counterparts from C. japonicus, but the homology of the gene island highlighted in Fig. S8 in the supplemental material remains obscure. The host organism of these genomic fragments is related to C. japonicus.
The g06 fragment likely belongs to an alphaproteobacterium related to Agrobacterium or Paracoccus chromosomes or plasmids.
Fragment g08 may have an origin in common with fragments g05, g07, and g10, but in contrast to the former genomic fragments, it most likely originated from a plasmid rather than from a chromosome. Its oligonucleotide usage shows a certain degree of similarity to several plasmids hosted by Shewanella spp.
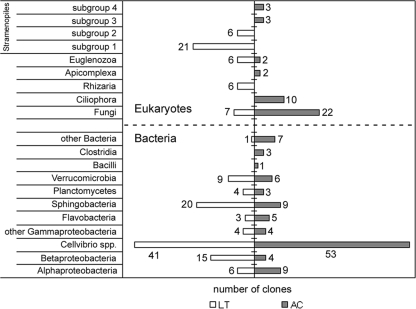
Taxonomic composition of bacterial communities.
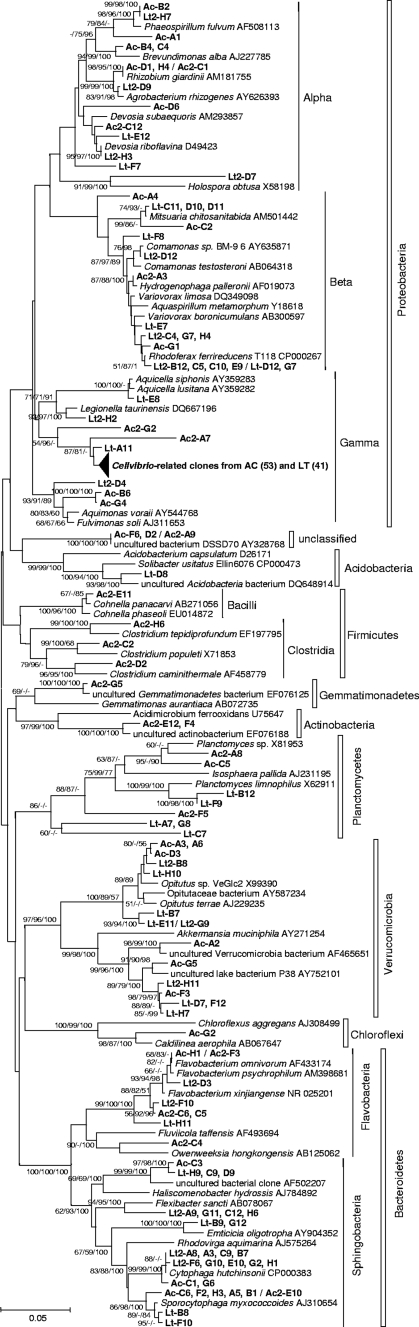
In total, 103 and 104 clones of 16S rRNA genes were sequenced from the L. terrestris and A. caliginosa clone libraries, respectively. Phylogenetic analysis revealed a relatively high diversity of bacterial ribotypes (Fig. 3 and 4): 35 and 38 different operational taxonomic units (OTUs) from 15 classes (eight phyla) were observed in the A. caliginosa and L. terrestris clone libraries, respectively. Clones derived from representatives of the phylum Proteobacteria (classes Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria) were predominant in both libraries: 64% (L. terrestris) and 67% (A. caliginosa). Among them, the clones showing 99.4 to 99.3% sequence identity (for a gene fragment of ca. 870 bp) with Cellvibrio fulvus (accession no. AF448514) and C. mixtus subsp. mixtus (accession no. AF448515), respectively, were the most abundant in both libraries, encompassing 40% (L. terrestris) and 51% (A. caliginosa) of the total number of clones (Fig. 3 and 4). Ribotypes related to the classes Flavobacteria and Sphingobacteria were also numerous in both clone libraries (Fig. 4). Even though few clones from certain bacterial groups were present only in the A. caliginosa library, comparative analysis with the LIBCOMPARE tool did not reveal any significant differences in clonal composition between the two libraries (see Fig. S12 in the supplemental material).
FIG. 3.
Phylogenetic analysis of bacterial 16S rRNA gene fragments ca. 600 to 620 bp in length from enrichments originating from A. caliginosa and L. terrestris. The tree was constructed by the neighbor-joining method. Clones sequenced in this work are in bold. Sequences with identities of >99.5% were taken as belonging to a single OTU and are shown as one ID number. Values in parentheses are numbers of clones. Numbers at nodes are bootstrap values greater than 50% and were calculated by the Kimura two-parameter, maximum-likelihood, and Bayesian treeing methods, respectively; branching topologies that are not supported by either method are marked by a dash at the corresponding node.
FIG. 4.
Phylogenetic affiliations of bacterial 16S and eukaryotic 28S rRNA clones in the libraries derived from cellulose enrichment cultures established with casts of L. terrestris and A. caliginosa as inocula.
Taxonomic diversity of eukaryotes.
PCR amplification of the DNA extracted from both communities with eukaryotic primers NL-1 and NL-4 generated amplicons of two different lengths, i.e., ca. 600 and of 800 bp, from each culture. Sublibraries were constructed from amplicons of both sizes; combined, 43 and 45 clones from L. terrestris and A. caliginosa, respectively, representing flagellate protists (Euglenozoa) and stramenopiles were found in short PCR amplicons, whereas alveolates, rhizaria, and fungi were found in long PCR products (Fig. 3; see Fig. S11 in the supplemental material). Two OTUs from fungi of the subphylum Mucoromycotina and a single OTU from Bodonidae (Euglenozoa) were detected in both libraries. Other eukaryotic microorganisms such as alveolates (three OTUs) and rhizaria (core Cercozoa; two OTUs) were detected in the A. caliginosa and L. terrestris communities, respectively (Fig. 3; see Fig. S11 in the supplemental material). Nine and five stramenopiles-related OTUs from different taxonomic groups were found in the L. terrestris and A. caliginosa clone libraries, respectively (Fig. 3; see Fig. S11 in the supplemental material). Statistical analysis of the A. caliginosa and L. terrestris libraries with the webLIBSUFF software (http://libshuff.mib.uga.edu/) established a significant difference between them (P = 0.001) (see Fig. S12 in the supplemental material).
DISCUSSION
Earthworms, as plant litter consumers, have a great impact on the mineralization of plant deposits and on soil formation (40). However, compared to other invertebrates, e.g., marine bivalves (21) or termites (19, 48, 56, 68, 69, 75), little is known about the enzymatic machinery of earthworm-associated microorganisms that allows them to deplete diverse polymeric lignocellulosic substrates at high rates. In this work, we established enrichment cultures in a minimal mineral medium with cellulose as the sole carbon and energy source and with fresh casts from earthworms as inocula. Metagenomic libraries constructed from the total DNA of enrichments were assayed for their enzymatic repertoire toward various glycosidic substrates, and taxonomic affiliations of GH-producing organisms were suggested using oligonucleotide word usage analysis of cloned genomic fragments, analysis of homology of deduced proteins, and sequence analysis of 16S rRNA gene libraries.
Microbial communities from enrichments were quite complex and encompassed the following microbial ecology groups: (i) common soil bacteria belonging to the lineages known to be able to degrade cellulose, namely, clostridia, sphingo- and flavobacteria, Cellvibrio spp. (most abundant in both clone libraries), and some other proteobacteria, (ii) bacterivorous protists, (iii) bacteria capable of symbiosis with protists, e.g., Legionella-related organisms, and (iv) quite surprisingly, highly diverse verrucomicrobia, planctomycetes, stramenopiles, and other microbes probably consuming products from fermentative processes. Previously we observed that cellvibrios were rather diverse in the soil substratum and enigmatic in the casts (54). Their abundance in cultures could indicate an ability to transiently flourish and quickly colonize cellulosic substrates upon their release with the cast. Interestingly, representatives of the sphingo- and flavobacteria and of the genus Pseudomonas capable of producing GHs were reported earlier either to maintain their densities during transit through the GI tract or even to become more numerous in the earthworm gut (11, 29, 38, 54, 63). These data are also consistent with an earlier report that cellulolytic microbes were more abundant in worm casts than in soil (57) and further support the importance of earthworm casts as inocula triggering organic matter decomposition processes in soil (9). Our previous study of soil substrata, gut content, and casts (the latter were used also in the present study as inocula to establish cellulose-depleting cultures) did not reveal any significant earthworm species-specific effects on bacterial populations passing through the GI tract (54), and therefore, differences between microbial populations from enrichments could be caused by some incidental factors rather than by physicochemical differences in the gut environments of the two earthworm species studied.
One of the most significant findings in the present work was the discovery of two novel GH protein families with β-galactosidase and α-arabinopyranosidase activities represented by G03-3 and G04-9, as their peptide sequences exhibited relatedness not to any known GH but to hypothetical proteins. These GHs were most probably derived from Betaproteobacteria and Gammaproteobacteria, respectively. This is additional clear evidence of the usefulness of activity-centered metagenomics for mining new enzymes which we were not able otherwise to predict in genome data sets and for attributing functions to hypothetical or conserved hypothetical proteins and protein families (25, 27). Furthermore, the G03-3 enzyme exhibited high 3D similarity to the isomerase/dehydrogenase family of enzymes frequently encoded in bacterial plasmids implicated in the biosynthesis of antibiotics and thus subject to fast evolution. Since β-galactoside and α-arabinopyranoside sugars are minor components of hemicellulose and intricate structures of the plant cell wall arabinans (8, 13), its significance in situ remains to be established. Interestingly, the analysis of the g04 fosmid clone derived from L. terrestris showed that it contained a gene cluster possibly involved in the biosynthesis and transport of compatible solutes (43) and the enzyme G04-9 from that gene cluster became active at elevated salt concentrations and had its optimum at a high pH. This observation suggests the capability of bacteria to be functionally active in plant polymer utilization even under conditions of temporary dehydration of soil or litter. This also correlates with our sequence analysis data (taxonomic distribution of best protein hits and genome linguistics) suggesting that the genome fragment may belong to the moderately halophilic species Pseudomonas mendocina or P. stutzeri.
Other enzymes showed significant degrees of similarity to specific protein domains of known GHs and CBDs and were mainly multimodular, with a broad spectrum of 3D structures and substrate specificities. An interesting example is that of the β-propeller fold G05-26 and G05-27 enzymes: G05-26 possessed a Ser-rich C-terminal domain (possibly acquired during the deletion of a CBD) (37), while G05-27 exhibited a type 6 CBD that conferred different hydrolytic capabilities. These results, together with the high degree of homology (77% identity) of the two enzymes, suggest a possible gene duplication event where the second copy was subjected to deleterious/beneficial mutations and positive selection where promiscuous protein intermediates emerged under the influence of environmental constraints and selection (51). This multidomain buildup suggests that genetic recombination leading to protein domain shuffling events may be of importance in GH evolution in a given ecosystem.
Psi-blastp analysis of individual deduced proteins revealed that the majority of the DNA fragments, including those from fosmids g05, g07, and g10, were derived from bacteria related to C. japonicus (the only strain with a fully sequenced genome among the Cellvibrio spp. so far); in turn, the sequence analysis suggested that bacteria related to C. fulvus and C. mixtus subsp. mixtus deliver the majority of 16S rRNA gene clones. Our data further suggested that the oligonucleotide usage pattern of g07 is most similar to that of the core genome of C. japonicus but is more diverse, as it contains a putative horizontally transferred gene island that starts with several tandem repeats. Exactly the same repetitive sequence was identified in the genome of Methanothermobacter thermautotrophicus strain Delta H (65). Interestingly, and similar to the situation in this fosmid, in the genome of M. thermautotrophicus, these repeats precede a gene island identified by the SeqWord genome browser (30). However, these gene islands do not have any genes in common. It is known that 8- to 12-nucleotide-long tandem repeats may play a role in the uptake of DNA sequences that facilitate genetic transformation between microorganisms by transduction and conjugation (2). An alien origin of this genome island is consistent with the results of the annotation, since all of the gene products of g07 showed homology with their counterparts from C. japonicus but not with those of the putative gene island localized at positions 18579 to 26761 (see Fig. S3C in the supplemental material).
Apart from two new GHs, the GH domains revealed in the present work were also found in microbial communities from lower and higher wood-feeding termites and Cryptocercus cockroaches (69, 75), whereas GHF6 from Cellvibrio spp. appeared to be specific to the earthworm-derived communities studied. The GH enzyme diversity characterized using activity screens of a bovine rumen expression library (28) differed from that reported in the present work; GHF3 and -5 domains were detected in both environments; however, these enzymes also have different microbial origins. Remarkably, the enzymes of GHF9 found in genomic fragments of Cellvibrio spp. were also reported earlier to be produced by earthworms, suggesting a cross-kingdom importance of this family in substrate-driven convergence (15, 55).
In conclusion, our study has led to the discovery of a series of new multidomain GHs of known families and two as-yet-unknown families, to the characterization of their biochemical and structural features, and to deduction of the corresponding producing microorganisms. This work has additionally underlined the utility of activity-centered metagenomics for mining proteins with new functions and emphasized the great potential of earthworm-associated microbial communities such as those in casts and likely the GI tract. The permanent exposure of microorganisms to a great variety of polymeric substrates is most likely to drive the evolution of cellulolytic enzymatic machinery toward the occurrence of enzymes composed of multiple GH domains or the emergence of as-yet-unknown enzymes able to hydrolyze unusual polymeric substrates. A mechanistic understanding of the processes involved in the degradation of organic matter will expand our knowledge of the contribution of microbes to global carbon cycling and expand our enzymatic toolbox for new biotechnological applications.
Supplementary Material
Acknowledgments
This research was supported by the Spanish MEC BIO2006-11738 and CSD2007-00005 projects.
A.B. thanks the Spanish MEC for an FPU fellowship. M.-E.G. thanks the CSIC for a JAE fellowship. T.Y.N., P.N.G., and O.V.G. acknowledge support by grant 0313751K from the Federal Ministry for Science and Education (BMBF) within the GenoMikPlus initiative.
Footnotes
Published ahead of print on 9 July 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambur, O. H., S. A. Frye, and T. Tønjum. 2007. New functional identity for the DNA uptake sequence in transformation and its presence in transcriptional terminators. J. Bacteriol. 189:2077-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anisimova, M., and O. Gascuel. 2006. Approximate likelihood ratio test for branches: a fast, accurate and powerful alternative. Syst. Biol. 55:539-552. [DOI] [PubMed] [Google Scholar]
- 4.Ashelford, K. E., N. A. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2006. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl. Environ. Microbiol. 72:5734-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bezuidt, O., G. Lima-Mendez, and O. N. Reva. 2009. SeqWord Gene Island Sniffer: a program to study the lateral genetic exchange among bacteria. World Acad. Sci. Eng. Technol. 58:1169-1174. [Google Scholar]
- 7.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 8.Bringhurst, R. M., Z. G. Cardon, and D. J. Gage. 2001. Galactosides in the rhizosphere: utilization by Sinorhizobium meliloti and development of a biosensor. Proc. Natl. Acad. Sci. U. S. A. 98:4540-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, G. G., and B. M. Doube. 2004. Functional interactions between earthworms, microorganisms, organic matter and plants, p. 213-240. In C. A. Edwards (ed.), Earthworm ecology, 2nd ed. CRC Press, Boca Raton, FL.
- 10.Brulc, J. M., D. A. Antonopoulos, M. E. Miller, M. K. Wilson, A. C. Yannarell, E. A. Dinsdale, R. E. Edwards, E. D. Frank, J. B. Emerson, P. Wacklin, P. M. Coutinho, B. Henrissat, K. E. Nelson, and B. A. White. 2009. Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases. Proc. Natl. Acad. Sci. U. S. A. 106:1948-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byzov, B. A., T. Y. Nechitaylo, B. K. Bumazhkin, S. A. Kharin, A. V. Kurakov, P. N. Golyshin, and D. G. Zvyagintsev. 2009. Cultivable microorganisms from digestive tract of earthworms. Mikrobiologiia 79:404-413. [PubMed] [Google Scholar]
- 12.Cantarel, B. L., P. M. Coutinho, C. Rancurel, T. Bernard, V. Lombard, and B. Henrissat. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37:D233-D238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Capek, P. 2008. An arabinogalactan containing 3-O-methyl-d-galactose residues isolated from the aerial parts of Salvia officinalis L. Carbohydr. Res. 343:1390-1393. [DOI] [PubMed] [Google Scholar]
- 14.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, A. M. Bandela, E. Cardenas, G. M. Garrity, and J. M. Tiedje. 2007. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 35:D169-D172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davison, A., and M. Blaxter. 2005. Ancient origin of glycosyl hydrolase family 9 cellulase genes. Mol. Biol. Evol. 22:1273-1284. [DOI] [PubMed] [Google Scholar]
- 16.DeBoy, R. T., E. F. Mongodin, D. E. Fouts, L. E. Tailford, H. Khouri, J. B. Emerson, Y. Mohamoud, K. Watkins, B. Henrissat, H. J. Gilbert, and K. E. Nelson. 2008. Insights into plant cell wall degradation from the genome sequence of the soil bacterium Cellvibrio japonicus. J. Bacteriol. 190:5455-5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dias, F. M., F. Vincent, G. Pell, J. A. Prates, M. S. Centeno, L. E. Tailford, L. M. Ferreira, C. M. Fontes, G. J. Davies, and H. L. Gilbert. 2004. Insights into the molecular determinants of substrate specificity in glycoside hydrolase family 5 revealed by the crystal structure and kinetics of Cellvibrio mixtus mannosidase 5A. J. Biol. Chem. 279:25517-25526. [DOI] [PubMed] [Google Scholar]
- 18.Domínguez, R., H. Souchon, M. Lascombe, and P. M. Alzari. 1996. The crystal structure of a family 5 endoglucanase mutant in complexed and uncomplexed forms reveals an induced fit activation mechanism. J. Mol. Biol. 257:1042-1051. [DOI] [PubMed] [Google Scholar]
- 19.Donovan, S. E., K. J. Purdy, M. D. Kane, and P. Eggleton. 2004. Comparison of Euryarchaea strains in the guts and food-soil of the soil-feeding termite Cubitermes fungifaber across different sol types. Appl. Environ. Microbiol. 70:3884-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drake, H. L., and M. A. Horn. 2007. As the worm turns: the earthworm gut as a transient habitat for soil microbial biomes. Annu. Rev. Microbiol. 61:169-189. [DOI] [PubMed] [Google Scholar]
- 21.Ekborg, N. A., W. Morrill, A. M. Burgoyne, L. Li, and D. L. Distel. 2007. CelAB, a multifunctional cellulase encoded by Teredinibacter turnerae T7902T, a culturable symbiont isolated from the wood-boring marine bivalve Lyrodus pedicellatus. Appl. Environ. Microbiol. 73:7785-7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elifantz, H., L. A. Waidner, V. K. Michelou, M. T. Cottrell, and D. L. Kirchman. 2008. Diversity and abundance of glycosyl hydrolase family 5 in the North Atlantic Ocean. FEMS Microbiol. Ecol. 63:316-327. [DOI] [PubMed] [Google Scholar]
- 23.Faure, D. 2002. The family 3 glycoside hydrolases: from housekeeping functions to host-microbe interactions. Appl. Environ. Microbiol. 68:1485-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng, Y., C. J. Duan, H. Pang, X. C. Mo, C. F. Wu, Y. Yu, Y. L. Hu, J. Wei, J. L. Tang, and J. X. Feng. 2007. Cloning and identification of novel cellulase genes from uncultured microorganisms in rabbit cecum and characterization of the expressed cellulases. Appl. Microbiol. Biotechnol. 75:319-328. [DOI] [PubMed] [Google Scholar]
- 25.Ferrer, M., A. Beloqui, K. N. Timmis, and P. N. Golyshin. 2009. Metagenomics for mining new genetic resources of microbial communities. J. Mol. Microbiol. Biotechnol. 16:109-123. [DOI] [PubMed] [Google Scholar]
- 26.Ferrer, M., A. Beloqui, O. V. Golyshina, F. J. Plou, T. N. Chernikova, L. Fernández-Arrojo, I. Ghazi, A. Ballesteros, K. Elborough, K. N. Timmis, and P. N. Golyshin. 2007. Biochemical and structural features of a novel cyclodextrinase from cow rumen metagenome. Biotechnol. J. 2:207-213. [DOI] [PubMed] [Google Scholar]
- 27.Ferrer, M., O. V. Golyshina, A. Beloqui, and P. N. Golyshin. 2007. Mining enzymes from extreme environments. Curr. Opin. Microbiol. 10:207-214. [DOI] [PubMed] [Google Scholar]
- 28.Ferrer, M., O. V. Golyshina, T. N. Chernikova, A. N. Khachane, D. Reyes-Duarte, V. A. Santos, C. Strompl, K. Elborough, G. Jarvis, A. Neef, M. M. Yakimov, K. N. Timmis, and P. N. Golyshin. 2005. Novel hydrolase diversity retrieved from a metagenome library of bovine rumen microflora. Environ. Microbiol. 7:1996-2010. [DOI] [PubMed] [Google Scholar]
- 29.Furlong, M. A., D. R. Singleton, D. C. Coleman, and W. B. Whitman. 2002. Molecular and culture-based analyses of prokaryotic communities from an agricultural soil and the burrows and casts of the earthworm Lumbricus rubellus. Appl. Environ. Microbiol. 68:1265-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ganesan, H., A. S. Rakitianskaia, C. F. Davenport, B. Tümmler, and O. N. Reva. 2008. The SeqWord Genome Browser: an online tool for the identification and visualization of atypical regions of bacterial genomes through oligonucleotide usage. BMC Bioinformatics 9:333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garvín, M. H., C. Lattaud, D. Trigo, and P. Lavelle. 2000. Activity of glycolytic enzymes in the gut of Hormogaster elisae (Oligochaeta, Hormogastridae). Soil Biol. Biochem. 32:929-934. [Google Scholar]
- 32.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modelling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 33.Guindon, S., and O. Gascuel. 2003. A simple, fast and accurate method to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 34.Hall, J., G. W. Black, L. M. Ferreira, S. J. Millward-Sadler, B. R. Ali, G. P. Hazlewood, and H. J. Gilbert. 1995. The non-catalytic cellulose-binding domain of a novel cellulase from Pseudomonas fluorescens subsp. cellulosa is important for the efficient hydrolysis of Avicel. Biochem. J. 309:749-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence aligment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 36.Han, S. O., H. Yukawa, M. Inui, and R. H. Doi. 2005. Molecular cloning and transcriptional and expression analysis of engO, encoding a new noncellulosomal family 9 enzyme, from Clostridium cellulovorans. J. Bacteriol. 187:4884-4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Howard, M. B., N. A. Ekborg, L. E. Taylor, S. W. Hutcheson, and R. M. Weiner. 2004. Identification and analysis of polyserine linker domains in prokaryotic proteins with emphasis on the marine bacterium Microbulbifer degradans. Protein Sci. 13:1422-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ihssen, J., M. A. Horn, C. Matthies, A. Gößner, A. Schramm, and H. L. Drake. 2003. N2O-producing microorganisms in the gut of the earthworm Aporrectodea caliginosa are indicative of ingested soil bacteria. Appl. Environ. Microbiol. 69:1655-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones, D. T. 1999. GenTHREADER: an efficient and reliable protein fold recognition method for genomic sequences. J. Mol. Biol. 287:797-815. [DOI] [PubMed] [Google Scholar]
- 40.Jouquet, P., J. Dauber, J. Lagerlöf, P. Lavelle, and M. Lepage. 2006. Soil invertebrates as ecosystem engineers: Intended and accidental effects on soil and feedback loops. Appl. Soil Ecol. 32:153-164. [Google Scholar]
- 41.Kanokratana, P., D. Chantasingh, V. Champreda, S. Tanapongpipat, K. Pootanakit, and L. Eurwilaichitr. 2008. Identification and expression of cellobiohydrolase (CBHI) gene from an endophytic fungus, Fusicoccum sp. (BCC4124) in Pichia pastoris. Protein Expr. Purif. 58:148-153. [DOI] [PubMed] [Google Scholar]
- 42.Kim, S. J., C. M. Lee, M. Y. Kim, Y. S. Yeo, S. H. Yoon, H. C. Kang, and B. S. Koo. 2007. Screening and characterization of an enzyme with beta-glucosidase activity from environmental DNA. J. Microbiol. Biotechnol. 17:905-912. [PubMed] [Google Scholar]
- 43.Kuhlmann, A. U., and E. Bremer. 2002. Osmotically regulated synthesis of the compatible solute ectoine in Bacillus pasteurii and related Bacillus spp. Appl. Environ. Microbiol. 68:772-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kyndt, T., A. Haegeman, and G. Gheysen. 2008. Evolution of GHF5 endoglucanase gene structure in plant-parasitic nematodes: no evidence for an early domain shuffling event. BMC Evol. Biol. 8:305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 46.Larsson, A. M., L. Anderson, B. Xu, I. G. Munoz, I. Uson, J. C. Janson, H. Stalbrand, and J. Stahlberg. 2006. Three-dimensional crystal structure and enzymic characterization of beta-mannanase Man5A from blue mussel Mytilus edulis. J. Mol. Biol. 357:1500-1510. [DOI] [PubMed] [Google Scholar]
- 47.Lattaud, C., S. Locati, P. Mora, and C. Rouland. 1997. Origin and activities of glycolytic enzymes in the gut of the tropical geophagous earthworm Millsonia anomala from Lamto (Côte d'Ivoire). Pedobiologia 41:242-251. [Google Scholar]
- 48.Li, L., J. Frölich, P. Pfeiffer, and H. Köning. 2003. Termite gut symbiotic archaezoa are becoming living metabolic fossils. Eukaryot. Cell 2:1091-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.López-Cortés, N., D. Reyes-Duarte, A. Beloqui, J. Polaina, I. Ghazi, O. V. Golyshina, A. Ballesteros, P. N. Golyshin, and M. Ferrer. 2007. Catalytic role of conserved HQGE motif in the CE6 carbohydrate esterase family. FEBS Lett. 581:4657-4662. [DOI] [PubMed] [Google Scholar]
- 50.Lukashin, A. V., and M. Borodovsky. 1998. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 26:1107-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lynch, M. 2002. Genomics, gene duplication and evolution. Science 297:945-947. [DOI] [PubMed] [Google Scholar]
- 52.Mohamed, N. M., A. S. Colman, Y. Tal, and R. T. Hill. 2008. Diversity and expression of nitrogen fixation genes in bacterial symbionts of marine sponges. Environ. Microbiol. 10:2910-2921. [DOI] [PubMed] [Google Scholar]
- 53.Nechitaylo, T. Y., K. N. Timmis, and P. N. Golyshin. 2009. ‘Candidatus Lumbricincola’, a novel lineage of uncultured Mollicutes from earthworms of family Lumbricidae. Environ. Microbiol. 11:1016-1026. [DOI] [PubMed] [Google Scholar]
- 54.Nechitaylo, T. Y., M. M. Yakimov, M. Godinho, K. N. Timmis, E. Belogolova, B. A. Byzov, A. V. Kurakov, D. L. Jones, and P. N. Golyshin. 2010. Effect of the earthworms Lumbricus terrestris and Aporrectodea caliginosa on bacterial diversity in soil. Microb. Ecol. 59:574-587. [DOI] [PubMed] [Google Scholar]
- 55.Nozaki, M., C. Miura, Y. Tozawa, and T. Miura. 2009. The contribution of endogenous cellulase to the cellulose digestion in the gut of earthworm (Pheretima hilgendorfi: Megascolecidae). Soil Biol. Biochem. 41:762-769. [Google Scholar]
- 56.Ohtoko, K., M. Ohkuma, S. Moriya, T. Inoue, R. Usami, and T. Kudo. 2000. Diverse genes of cellulase homologues of glycosyl hydrolase family 45 from the symbiotic protists in the hindgut of the termite Reticulitermes speratus. Extremophiles 4:343-349. [DOI] [PubMed] [Google Scholar]
- 57.Parle, J. N. 1963. A microbiological study of earthworm casts. J. Gen. Microbiol. 31:13-22. [Google Scholar]
- 58.Reva, O. N., and B. Tümmler. 2004. Global features of sequences of bacterial chromosomes, plasmids and phages revealed by analysis of oligonucleotide usage patterns. BMC Bioinformatics 5:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reva, O. N., and B. Tümmler. 2005. Differentiation of regions with atypical oligonucleotide composition in bacterial genomes. BMC Bioinformatics 6:251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rigden, D. J. 2005. Analysis of glycoside hydrolase family 98: catalytic machinery, mechanism and a novel putative carbohydrate binding module. FEBS Lett. 579:5466-5472. [DOI] [PubMed] [Google Scholar]
- 61.Ronquist, F., and J. P. Huelsenbeck. 2003. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 62.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed., p. 6.22. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 63.Schönholzer, F., D. Hahn, B. Zarda, and J. Zeyer. 2002. Automated image analysis and in situ hybridization as tools to study bacterial populations in food resources, gut and cast of Lumbricus terrestris L. J. Microbiol. Methods 48:53-68. [DOI] [PubMed] [Google Scholar]
- 64.Singleton, D. R., M. A. Furlong, S. L. Rathbun, and W. B. Whitman. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith, D. R., L. A. Doucette-Stamm, C. Deloughery, H. Lee, J. Dubois, T. Aldredge, R. Bashirzadeh, D. Blakely, R. Cook, K. Gilbert, D. Harrison, L. Hoang, P. Keagle, W. Lumm, B. Pothier, D. Qiu, R. Spadafora, R. Vicaire, Y. Wang, J. Wierzbowski, R. Gibson, N. Jiwani, A. Caruso, D. Bush, and J. N. Reeve. 1997. Complete genome sequence of Methanobacterium thermoautotrophicum deltaH: functional analysis and comparative genomics. J. Bacteriol. 179:7135-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 67.Tatusov, R. L., N. D. Fedorova, J. D. Jackson, A. R. Jacobs, B. Kiryutin, E. V. Koonin, D. M. Krylov, R. Mazumder, S. L. Mekhedov, A. N. Nikolskaya, B. S. Rao, S. Smirnov, A. V. Sverdlov, S. Vasudevan, Y. I. Wolf, J. J. Yin, and D. A. Natale. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Todaka, N., S. Moriya, K. Saita, T. Hondo, I. Kiuchi, H. Takasu, M. Ohkuma, C. Piero, Y. Hayashizaki, and T. Kudo. 2007. Environmental cDNA analysis of the genes involved in lignocellulose digestion in the symbiotic protist community of Reticulitermes speratus. FEMS Microbiol. Ecol. 59:592-599. [DOI] [PubMed] [Google Scholar]
- 69.Todaka, N., T. Inoue, K. Saita, M. Ohkuma, C. A. Nalepa, M. Lenz, T. Kudo, and S. Moriya. 2010. Phylogenetic analysis of cellulolytic enzyme genes from representative lineages of termites and a related cockroach. PLoS One 5:e8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Topakas, E., P. Christakopoulos, and C. B. Faulds. 2005. Comparison of mesophilic and thermophilic feruloyl esterases: characterization of their substrate specificity for methyl phenylalkanoates. J. Biotechnol. 115:355-366. [DOI] [PubMed] [Google Scholar]
- 71.Tsai, L. C., L. F. Shyur, Y. S. Cheng, S. H. Lee, L. C. Tsai, L. F. Shyur, Y. S. Cheng, and S. H. Lee. 2005. Crystal structure of truncated Fibrobacter succinogenes 1,3-1,4-beta-d-glucanase in complex with beta-1,3-1,4-cellotriose. J. Mol. Biol. 354:642-651. [DOI] [PubMed] [Google Scholar]
- 72.Vieites, J. M., M. E. Guazzaroni, A. Beloqui, P. N. Golyshin, and M. Ferrer. 2009. Metagenomics approaches in systems microbiology. FEMS Microbiol. Rev. 33:236-255. [DOI] [PubMed] [Google Scholar]
- 73.Voget, S., H. L. Steele, and W. R. Streit. 2006. Characterization of a metagenome-derived halotolerant cellulase. J. Biotechnol. 126:26-36. [DOI] [PubMed] [Google Scholar]
- 74.Walter, J., M. Mangold, and G. W. Tannock. 2005. Construction, analysis, and beta-glucanase screening of a bacterial artificial chromosome library from the large-bowel microbiota of mice. Appl. Environ. Microbiol. 71:2347-2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Warnecke, F., P. Luginbuhl, N. Ivanova, M. Ghassemian, T. H. Richardson, J. T. Stege, M. Cayouette, A. C. McHardy, G. Djordjevic, N. Aboushadi, R. Sorek, S. G. Tringe, M. Podar, H. G. Martin, V. Kunin, D. Dalevi, J. Madejska, E. Kirton, D. Platt, E. Szeto, A. Salamov, K. Barry, N. Mikhailova, N. C. Kyrpides, E. G. Matson, E. A. Ottesen, X. Zhang, M. Hernández, C. Murillo, L. G. Acosta, I. Rigoutsos, G. Tamayo, B. D. Green, C. Chang, E. M. Rubin, E. J. Mathur, D. E. Robertson, P. Hugenholtz, and J. R. Leadbetter. 2007. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450:560-565. [DOI] [PubMed] [Google Scholar]
- 76.Woyke, T., H. Teeling, N. N. Ivanova, M. Huntemann, M. Richter, F. O. Gloeckner, D. Boffelli, I. J. Anderson, K. W. Barry, H. J. Shapiro, E. Szeto, N. C. Kyrpides, M. Mussmann, R. Amann, C. Bergin, C. Ruehland, E. M. Rubin, and N. Dubilier. 2006. Symbiosis insights through metagenomic analysis of a microbial consortium. Nature 443:950-955. [DOI] [PubMed] [Google Scholar]
- 77.Zhang, B. G., C. Rouland, C. Lattaud, and P. Lavelle. 1993. Activity and origin of digestive enzymes in gut of the tropical earthworm Pontoscolex corethrurus. Eur. J. Soil Biol. 29:7-11. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.