Abstract
Biological macromolecules, and the complexes that they form, can be described in a variety of ways ranging from quantum mechanical and atomic chemical models, to coarser grained models of secondary structure and domains, to continuum models. At each of these levels, group theory can be used to describe both geometric symmetries and conformational motion. In this survey, a detailed account is provided of how group theory has been applied across computational structural biology to analyze the conformational shape and motion of macromolecules and complexes.
1. Introduction
Proteins and nucleic acids are fantastically complicated objects when viewed at the level of atomic detail. And yet, in comparison with man-made machines, they are tremendously robust to perturbations. As examples of their robustness, consider the following: (1) they operate under conditions dominated by Brownian motion; (2) mutation analysis of proteins shows that the overall protein shape and function is preserved under a wide variety of point substitutions of amino acid residue types; and (3) the overall double-helical structure of DNA is preserved under arbitrary substitutions of base-pair types and provides a robust method for encoding information.
Commonly used tools for analyzing the behavior of biomolecular structures are normal mode analysis [18, 19, 59] and molecular dynamics simulation [20, 79, 154]. It has become popular in recent years to describe large biomolecular structures as ‘machines’ by using coarse-grained models. In some coarse-grained models, the whole structure is sampled and spatially proximal sample points are connected with harmonic potentials [140–142, 4, 5, 88], or the collective motions in blocks of atoms obtained from examining normal modes of detailed models are used [137, 136, 99, 100, 138].
Some researchers have used the principles of statistical mechanics to extract parameters from large databases of structures for use in computations with coarse-grained models [102, 127]. Also, several papers have appeared recently that seek to match the parameters that define coarse-grained models of DNA to molecular dynamics simulations with full atomic detail [78, 42], as well as to experimental data [26].
In other models, a hierarchy of scales including full atomic (and even quantum mechanical detail) are used in regions that are deemed to be important, and the level of coarseness increases with distance away from them [114].
An alternative kind of coarse graining is the division of a biomolecular structure into rigid bodies [69, 70, 120, 121]. For example, individual bases, and sometimes whole base pairs, can be modeled as rigid ‘bricks’ in larger nucleic acid structures [91]. Furthermore, either short alpha helices or even whole domains within a large protein often can be taken as rigid without freezing out motions that are functionally critical. These substructures within coarse-grained models often have geometric symmetries (e.g., helices, indole rings, methyl groups) as do the assemblages that they form (e.g., viral capsids, GROEL-GROES, protein crystals).
As soon as one starts to talk about the geometric symmetry of individual objects, the symmetrical positioning of copies of objects in space, or the continuous motion of rigid bodies in space, group theory enters the picture. In some contexts in structural biology the relevant groups are finite (e.g., point groups describing rotational symmetries of objects), discrete but not finite (e.g., crystallographic space groups describing the symmetries of infinite lattices), or continuous (e.g., the group of rotations in three-dimensional space, described by three Euler angles, or the six-dimensional group of full rigid-body motions in space described by three translations along with the three rotational degrees of freedom). All of these cases are reviewed here in the context of the biomolecular applications in which they occur in practice. An overview of the biomolecular applications discussed in this paper is provided in section 1.2. But first, a very brief review of how group theory is used in other branches of physics is provided in section 1.1. The groups reviewed therein are generally different from those that we will be concerned with (which are groups of rigid-body motions and their subgroups). It is useful to put things in perspective by clarifying this difference in the presentation that follows.
1.1. Group theory in physics
Group theory is one of the cornerstone mathematical methods of modern physics. Wigner, Dirac, and other giants championed the use of group representation theory in the context of quantum mechanics [159, 155, 149]. Group theory also plays a role in modern particle physics [44, 51, 9], and in special relativity (Lorentz transformations) [158]. In solid-state physics the symmetry groups of crystals play a central role. In general when group theory is used in physics it is usually in the role of describing geometrical or dynamical symmetries of the phenomena under consideration. For example, it is well known that all of the equations of classical Newtonian mechanics are invariant under Galilean transformations. The fact that the rotation group is a subgroup of the Galilean group is one reason for the success of Cartesian tensors in mechanics.
In condensed matter physics at the molecular scale (and in particular in the biomolecular problems discussed here) the use of group theory is often somewhat disguised. This is because in these contexts Lie groups such as the rotation group, the Euclidean motion group, the torus group, and products thereof are not necessarily used to describe symmetries. Rather, they serves as the configuration space over which ensembles of conformations evolve. For example in the study of liquid crystals [87, 39, 63], as well as in the theory of rotational Brownian motion of rigid molecules more generally [110, 163, 96], partial differential equations that describe the evolution of ensemble orientation arise. Indeed, the role of group theory in these contexts is more akin to satellite control problems as articulated in [17], rather than the way that group theory is used in other branches of physics, since a group manifold serves as the configuration space for the system. Often the convenience of the Euler angles and other parameterizations of rotation obfuscate the underlying group-theoretic nature of these problems. In this review the aspects of group theory that are directly relevant to the description of biomolecular conformation, motion, and ensemble statistics are reviewed.
Recall that for counter-clockwise rotations about the natural unit basis vectors e3, e2, and e1 for ℝ3 are:
| (1) |
| (2) |
| (3) |
Each of these basic rotations can be written as the matrix exponential Ri(ϕ) = exp(ϕ Ei) where
| (4) |
Arbitrary rotations in 3D can be described using the ZXZ Euler angles
| (5) |
(Here and elsewhere the juxtaposition of two matrices or a matrix and a vector implies multiplication.) Similarly, the ZYZ Euler angles are defined by the product R3(α)R2(β)R3(γ). ZXZ and ZYZ Euler angles are two examples of parameterizations of the rotation group (which is also called SO(3), the group of 3 × 3 special orthogonal matrices).
Let T denote the transpose of a matrix or vector. Then, for example, ϕ = [α, β, γ]T denotes a column vector. One of the defining properties of rotations is that RRT = 𝕀3, the 3 × 3 identity matrix. This property is called orthogonality. The other defining property is that the determinant satisfies det R = +1, which restricts the discussion to ‘special’ or ‘proper’ orthogonal transformations. It is easy to show that: (a) the product of any two 3 × 3 rotation matrices is again a rotation matrix; (b) that every rotation R has a unique multiplicative inverse (i.e., R−1 = RT); (c) that the identity matrix satisfies the properties of a rotation; and (d) that the associative law (R1 R2) R3 = R1(R2 R3) holds. The abstraction of the properties (a)–(d) are exactly those that define a group. Namely, (a′) closure under a group operation (in this and every case in this paper, this will be matrix multiplication); (b′) the existence of a multiplicative inverse (which is guaranteed for invertible matrices); (c′) the existence of the identity (which will always be an identity matrix in this paper); (d′) the associative law holds (which is always true for multiplication of matrices with compatible dimensions).
Moreover, most of the groups that are relevant to biomolecular structure and conformational motions are Lie groups, which, roughly speaking, means that they have a differentiable structure and a continuum of elements. For example it takes a continuum of Euler angles in the range 0 ≤ α, γ < 2π and 0 ≤ β ≤ π to describe all possible rotations. Associated with any Lie group is a Lie algebra, which is a vector space together with an additional operation. For example, the matrices Ei in (4) form a basis for a vector space which is closed under the operation of matrix commutation [A, B] = AB − BA = − [B, A] since
The space spanned by linear combinations of these basis elements together with the commutator form a Lie algebra called so(3). The exponentiation of elements of so(3) produces rotation matrices in SO(3). This process is denoted as
When studying infinitesimal motions, the angular velocity vector can be written as
where ϕ̇ = dϕ/dt,
| (6) |
and
| (7) |
Here ωr is the description of this vector in a frame of reference fixed to the rotating rigid-body, and ωl is how it appears in inertial (space-fixed) coordinates. The reason for the choice of subscript names l and r is explained in [24], and are not standard. For example, if with respect to inertial coordinates the position of a classical particle is x(t) = R(ϕ(t))x0, then ẋ = ω1 × x. In contrast, the kinetic energy of a rigid-body would be where I is its body-fixed moment of inertia. Either form of angular velocity can be associated with an element of so(3) by defining Ω = ω1E1 + ω2E2 + ω3E3. A natural bijective linear transformation can be used to convert back and forth between 3 × 3 skew-symmetric matrices, Ω, and vectors, ω:
This is captured with the ‘vee–hat’ notation ω̂ = Ω and Ω∨ = ω.
If SO(3) were the only group of interest in applications, there would be no practical need for an abstract theory of groups. However, many different groups occur in applications, and in physics the value of group theory is well known. Multiple books on the application of group-theoretic methods in physics can be found, including [62, 116, 132]. For example, it is well known that in classical mechanics all of the equations of motion are invariant under Galilean transformations. That is, given f = ma, where f is the applied force, m is mass, and a = d2x/dt2, where x is the position as observed in an inertial reference frame, then the same equation will hold under the substitution x → x′ and f → f′ where
and
| (8) |
where R ∈ SO(3), v, b ∈ ℝ3 and a ∈ ℝ are all constant.
Group theory can also be used to reduce the complexity of equations of motion by observing transformations that preserve various quantities. For example, Euler’s equations of motion
| (9) |
are known to simplify when the rigid-body has an axis of symmetry. Suppose that it is the body-fixed z-axis that is the axis of symmetry. Then I = diag[I1, I1, I3]. Such symmetry is observed in the kinetic energy since
for arbitrary angle ϕ0. This is one example of a physical quantity that is preserved under a group action.
In classical mechanics it is known that problems can be simplified by separating out certain degrees of freedom corresponding to zero conjugate momenta, thereby reducing the resulting equations of motion. This reduction process can be extended within the framework of geometric mechanics [1], and has been applied in engineering contexts [24, 28].
In quantum mechanics, the concept of representations of groups enters prominently. This point of view was championed by Wigner [159] in the context of the rotation group. Indeed, for any 3 × 3 rotation matrix, R, the Wigner D-functions, Dl(R) are (2l + 1) × (2l + 1) matrices for l = 0, 1/2, 1, 3/2, 2, … such that
| (10) |
In other words, these are matrix-valued functions of rotation-valued arguments. Explicitly the matrix entries for −l ≤ m, n ≤ +l can be expressed in terms of ZXZ-Euler angles as [145, 50]
| (11) |
where
Other conventions define Dl(R) slightly differently1, but its essential properties are preserved under similarity transformations of the form Dl(R) → V Dl(R)V*, where VV* = 𝕀2l+1, with 𝕀2l+1 denoting the (2l + 1) × (2l + 1) identity and V* denoting the Hermitian conjugate of the constant (2l + 1) × (2l + 1) unitary matrix V.
In quantum mechanics, integer values of l are associated with angular momentum, and half-integer values are associated with spin.
The expression in (10) is reminiscent of the property ein(θ1+θ2) = einθ1 · einθ2 which, together with orthogonality and completeness, forms one of the cornerstones of Fourier analysis. Similarly, it can be shown that any well-behaved function on the rotation group can be expanded in a Fourier series of the form
| (12) |
Here tr(A) denotes the trace of the matrix A and the integral over SO(3) can be computed in terms of Euler angles by integrating over the range 0 ≤ α, γ ≤ 2π and 0 ≤ β ≤ π with respect to the volume element
| (13) |
where dϕ = dα dβ dγ and the matrices in the above determinants are those defined in (7). Equations (12) can be thought of as a specific case of the celebrated Peter–Weyl Theorem [111]. In recent years ‘fast transforms’ that evaluate (12) in computer codes have been developed [115, 93, 94, 92, 73]. Such computations are relevant in crystallography [74]2.
Similar Fourier series can be constructed on any compact Lie group such as U(n) and SU(n) [168]3. Roughly speaking, compactness in this context means closed and bounded, and as a result compact Lie groups have finite volume. In particular, the group manifold SU(2) is known to be the unit sphere in ℝ4, which is equivalent to the quaternion sphere, and forms a double cover of the rotation group. That is, there is a two-to-one mapping from SU(2) to SO(3). Higher-dimensional unitary groups are known in nuclear and particle physics [44, 51], but will not enter into the current formulation.
The irreducible unitary representations for a wide variety of groups, which have the relationship U(g1 ○ g2, λ) = U(g1, λ)U(g2, λ) have been worked out where g ∈ G. Here U(g,λ) is a matrix-valued function. The arguments of this function are group elements, g ∈ G, and elements of the so-called unitary dual of G, λ ∈ Ĝ. The structure of Ĝ varies widely from group to group. For compact Lie groups, this is a discrete space. For the noncompact commutative group G = (ℝn, +), Ĝ ≅ G. Below a noncompact noncommutative example is provided.
Whereas in the compact case the representation matrices are enumerated by some countable basis and written as a superscript, in the more general case the parameter λ may be discrete, continuous, or a hybrid. The actual matrix entries umn(g, λ) can be written explicitly and concretely for specific groups. For example, for the Euclidean motion group of the plane, SE(2), defined by the operation of matrix multiplication and elements of the form4
the corresponding Fourier transform pair are [24]
| (14) |
where dg = r dr dϕ dϕ and the group manifold SE(2) is defined by the parameter values 0 ≤ r ≤ ∞, 0 ≤ ϕ, θ ≤ 2π, and
Hence here Ĝ = ℝ≥0.
As with the case of SO(3), variations in the form of these irreducible unitary representations (IURs) for SE(2) exist in the literature. Note that SE(2) is not compact since r can extend to infinity. Though it does belong to a class of groups called unimodular.
A unimodular group is one with an integral that is invariant under both left and right shifts and inversions of its argument:
| (15) |
This is the first abstract statement in this paper. Here G denotes an arbitrary unimodular Lie group and dg is the volume element with which to perform the integral. Three examples are the rotation group SO(3), the special Euclidean group SE(2), and any finite group Γ (in which case the integral becomes a summation over the group and the volume element becomes the Dirac counting measure (and therefore dg disappears).
Similar relationships to (12) and (14) hold for other unimodular groups. The class of unimodular Lie groups includes all compact Lie groups, the Lie group of rigid-body motions in n-dimensional Euclidean space for any n, the groups SL(n, 𝔽) and GL(n, 𝔽) for 𝔽 = ℂ or ℝ, and the symplectic groups. The generalization of (12) and (14) for any unimodular Lie group is written more abstractly as
| (16) |
where Ĝ is called the ‘unitary dual of G’ and consists of all values of λ, d(λ) is an appropriately chosen integration measure on Ĝ, and U(g, λ) is a unitary matrix for any values of g ∈ G and λ ∈ Ĝ. The matrix-valued functions U(g, λ) are constructed such that
| (17) |
Using * to denote the Hermitian conjugate, it follows that
In words, (16) means that the entries in the matrices U(g, λ) form an orthonormal basis for the space of all square-integrable functions on G, which is denoted as L2(G). And furthermore, the mapping g → U(g, λ) with the property (17) is a called a homomorphism from G into a group of unitary matrices. Matrix-valued functions of the form of U(g, λ) are called unitary representations.
Note that in (12) the dimension of the representation matrix appears as a weight in the Fourier series whereas in (14) the continuous integration measure p dp appears. This is one of the fundamental differences between the representation theory and harmonic analysis on compact versus noncompact unimodular Lie groups. Advanced treatments of group theory and its relationship to the special functions of classical mathematical physics can be found in [98, 146].
While a wide variety of groups enter physics from a variety of sources, the application of group theory to biomolecular problems will be more restricted. The group of rigid-body motions in three-dimensional space will be of paramount interest. Its various subgroups (the group of pure rotations, proper crystallographic space groups, proper point symmetry groups, etc) will also be important. But more exotic groups such as those used in particle physics and relativity will not enter the picture.
1.2. Overview of the remainder of this paper
Since the group of rigid-body motions in 3D Euclidean space plays the central role in the analysis and modeling of biomolecular conformation, section 2 is devoted to reviewing its basic differential-geometric and algebraic properties. In section 3 group-theoretic models of DNA structure and conformational motions are reviewed. In these models, double-helical DNA is treated as a continuous elastic filament (or ‘elastica’) with a helical referential shape. Minimal energy conformations of this elastica subject to end constraints are examined, as is the ensemble of conformations that result from ambient Brownian motion forcing. The result is a generalization of the so-called Kratky–Porod and Yamakawa helical wormlike chain models of semi-flexible polymers. Section 4 addresses the proper way to normalize crossing angle data for pairs of alpha helices in proteins. Section 5 addresses the spatial relationships between essentially rigid domains of protein structures and complexes.
2. Group-theoretic properties of rigid-body motion in 3D
2.1. Defining properties, actions, and homogeneous matrix representation
An arbitrary rigid-body motion can be viewed as the pair g = (r, R) where R ∈ SO(3) (i.e., R is a 3 × 3 rotation matrix), and r ∈ ℝ3 is a translation vector in three-dimensional space. The composition law is g1 ○ g2 = (R1r2 + r1, R1R2) and the inverse of each element g is g−1 = (−RTr, RT). The action of the motion g on a position vector x ∈ ℝ3 is g •x = Rx+r. In other words, the motion group ‘acts on’ points x ∈ ℝ3 (which can be viewed as position vectors consisting of Cartesian coordinates) by moving them according to the rule x → Rx+r. (Note the distinction that ○ is used between group elements and · is used between a group element and a vector.) Any g describes the positional and orientational relationship between two reference frames. It is sometimes convenient to refer to the result of a rigid-body motion at a particular time as a ‘pose’, and to refer to a function of motion as a position and orientation distribution.
The collection of all rigid-body motions is denoted in this paper as G = SE(3). (The special (or proper) Euclidean motion group in three space.) Any g ∈ G can be faithfully represented with a 4 × 4 homogeneous transformation matrix of the form:
| (18) |
in the sense that H(g1 ○ g2) = H(g1)H(g2) (i.e., the matrix product of H(g1) and H(g2)). Here 0T = [0, 0, 0] and 1 is the number one. The structure of this bottom row is preserved under multiplication by matrices of the same kind.
Henceforth no distinction is made between G and the set of all 4 × 4 homogeneous transformation matrices with operation of matrix multiplication. That is, g and H(g) will be used interchangeably, and since the group operator can be viewed as matrix multiplication, it does not need to be written explicitly as ○.
2.2. Infinitesimal motions and Lie algebra
Given a one-parameter motion g(t), we can define the six-dimensional velocity of the rigid-body motion as observed in the moving frame as the nontrivial entries in the matrix
Here t can be thought of as time, and a dot denotes differentiation with respect to t.
Since RT Ṙ is skew symmetric as a result of R being orthogonal, it only has three independent nonzero entries. These can be extracted and used to form the dual vector ω(t), which is the angular velocity of the moving frame as seen in the moving frame. In some contexts it will be convenient to write this as ωr(t) to distinguish it from the dual vector of ṘRT, which we will call ωl (t). These are related as ωl (t) = Rωr(t).
The independent information in the matrix g−1ġ can be extracted and put in a six-dimensional vector defined as
| (19) |
The opposite operation of ∨ is:
where
Here E͂i is used for basis elements of se(3). The similarity between the first three of these and Ei in (4) is not coincidental; so(3) is a sub-algebra of se(3), and exponentiation of elements of this sub-algebra leads to 4 × 4 matrices that are isomorphic to SO(3).
2.3. Exponential, logarithm, and vee operation
In general an n-dimensional real matrix Lie algebra is defined by a basis consisting of real matrices {Xi} for i = 1, … , n that is closed under the matrix commutator5. That is,
for some real numbers , which are called the structure constants of the Lie algebra.
In a neighborhood around the identity of the corresponding Lie group, the parametrization
| (20) |
is always valid in a region around the identity in the corresponding Lie group. And in fact, for the examples discussed, this parametrization is good over almost the whole group, with the exception of a set of measure zero. Furthermore, exponentiation of scalar multiples of individual basis elements produces fundamental motions. For example, when G = SE(3),
and
The descriptive names rot(ei, θ) and trans(ei, z) respectively will be useful in the sequel to describe rigid-body rotations by θ clockwise around the axis defined by direction ei, and translation along that axis by z.
The logarithm map
(which is the inverse of the exponential) is valid for almost all g ∈ G for the case when G = SE(3) or SO(3). It will be convenient in the analysis to follow to identify a vector x ∈ ℝn as
| (21) |
Here {ei} is the natural basis for ℝn.
These correspond to infinitesimal rotations and translations about the 1, 2, and 3 axes and form a basis for the Lie algebra associated with G. Matrix exponentiation of any weighted sum of these basis elements produces elements of G. For example,
Furthermore, for small values of θ and z, the matrix exponential is approximated well as
2.4. Jacobians, integration, and Lie derivatives
It is clear from (20) that exponential coordinates can be used to parameterize the group SE(3), and from (18) it is clear that a combination of Euler angles and Cartesian coordinates can be used to respectively parameterize R and t. Depending on the application, some parameterizations are better than others. Furthermore, since in some of the calculations that follow the group of interest will be SE(3), and in other calculations the group of interest will be SO(3), a general description of Lie derivatives and how to construct the bi-invariant integration over Lie groups is presented here as they relate to Jacobian matrices analogous to those in (7).
For the moment it will be convenient to discuss Jacobians for general n-dimensional unimodular Lie groups, keeping the case of SO(3) (where n = 3) and SE(3) (with n = 6) in mind. Let q = [q1, q2, …, qn]T denote any parametrization of a unimodular Lie group G. Then Jacobian matrices Jr(q) and Jl(q) can be computed given g(q) and the definition of the ∨ operation in (21) as
and
This gives a hint as to why the subscripts l and r are used: if derivatives with respect to parameters appear on the ‘right’ of g−1, this is denoted with an r, and if they appear on the ‘left’ then a subscript l is used.
Given a function f(g), the left and right Lie derivatives are defined with respect to any basis element of the Lie algebra Xi ∈ 𝒢 as
| (22) |
The use of l and r mimicks the way that the subscripts were used in the Jacobians J1 and Jr in the sense that if exp(tXi) appears on the left/right then the corresponding derivative is given an l/r designation. This notation, while not standard in the mathematics literature, is useful in computations because when evaluating left/right Lie derivatives in coordinates g = g(q), the left/right Jacobians enter in the computation as [22, 24]
| (23) |
where , and ∇q = [∂/∂q1, … , ∂/∂qn]T is the gradient operator treating q like Cartesian coordinates. Here ‘–T’ denotes the inverse of the transpose (which is the same as the transpose of the inverse). An interesting fact that will be used later in the paper is that when q = x (i.e., the parametrization g(x) = exp(Σi xi Xi) is used), then Jr(0) = Jl(0) = 𝕀n, which means that
| (24) |
Since the two groups of paramount interest in the sequel are SO(3) and SE(3), it is important to distinguish between the Jacobian matrices for each of these cases. When considering SO(3), the notation Jr and J1 will be used, as it was in (21), to stand for these 3 × 3 matrices. In contrast, when considering SE(3), the resulting 6 × 6 Jacobian matrices will be denoted as 𝒥r and 𝒥1. In practice, the Jacobian matrices for SE(3) often will have embedded in them the Jacobian for SO(3).
Adjoint matrices Ad(g) and ad(X) are defined using Jacobians and the concept of the exponential and logarithm maps as
| (25) |
The dimensions of these square matrices is the same as the dimension of the Lie group, which can be very different from the dimensions of the matrices that are used to represent the elements of the group. The explicit forms of these matrices for SO(3) and SE(3) are given in [24, 28]. The function Δ(g) = det Ad(g) is called the modular function of G. For a unimodular Lie group, Δ(g) = 1, which is used in many texts as the defining property rather than (15). It follows that for unimodular Lie groups |det(Jl)| = |det(Jr)|, and so the subscripts l and r can be dropped when discussing Jacobian determinants.
3. Continuum models of DNA mechanics and statistical mechanics
The DNA double helix has been modeled at a variety of levels of coarseness. At the finest level of description, the Cartesian positions of all atomic nuclei are stored. At the next level up, the positions and orientations of individual bases are treated as rigid bodies that are paired and stacked with harmonic potentials. The model reviewed here is even coarser. The stiffness properties of DNA are averaged over several consecutive base pairs in the double helix. The result is a continuous elastic rod model with a minimal energy conformation that has a helical twist. A ‘backbone curve’ together with an arc-length-dependent stiffness matrix then describes the mechanical properties at this coarse level.
Such models have been used (not necessarily limited to the case of DNA) in the polymer physics literature for more than fifty years [37, 58, 75]. DNA is referred to as a ‘semiflexible’ or ‘stiff’ macromolecule when viewed as a serial chain, because it has resistance to local bending and twisting in addition to extension. As a result, even when there is ambient Brownian motion forcing, there is some persistence in correlations between the direction of the tangent to the backbone curve at nonzero separations in arc length.
The statistical mechanics of DNA has received substantial attention in the literature. (See, for example, [52, 56, 57, 65, 72, 76, 77, 85, 95, 109, 118, 123–125, 131, 147, 160–162, 164, 167]). Modeling semi-flexible polymer statistical mechanics using the theory of diffusion processes was studied in [11, 15, 84, 101].
Excluded-volume effects in polymer solutions have been studied extensively in [43, 47, 108]. Renormalization group concepts [38, 122] and mean field potentials [43, 139] are popular techniques to address this issue.
In theories of ring-closure probabilities, the probability density function describing the relative frequency of occurrence of positions and orientations of the distal end of the chain for a given position and orientation of the proximal end play an important role [40, 38, 43, 47, 55, 165]. Furthermore, a number of new theoretical models have been developed by the author’s group for generating this quantity from given stiffness models [22, 23, 169, 170, 25].
Experimental measurements of DNA stiffness parameters have been reported in [10, 61, 103, 104, 128, 165]. Efforts to characterize integrals of the joint positional and orientational probability density function over many of its arguments can be found in [160, 82], and the whole distribution in the case of the helical wormlike chain can be found in [165]. DNA elastic properties and experimental measurements of DNA elastic properties such as twist/stretch coupling have been reported in [21, 31, 64, 89, 90, 133, 151].
Elastic models of DNA mechanics has a long history [12, 13]. A number of recent studies on chiral and uncoupled end-constrained elastic rod models of DNA with circular cross-section have been presented [33, 135, 143, 34]. These models use the classical elasticity theory of continuum filaments with or without self-contact constraints to model the stable conformations of DNA in plasmids, in chromosomes, and during transcription. The remainder of this section addresses how group-theory enters in the mechanics of elastic filament models of DNA.
3.1. Elastic rod models of DNA expressed in Lie group terminology
A nonuniform extensible elastic filament with unstretched length L has elastic energy of the form [66]
| (26) |
where ξ(s) is exactly the 6D velocity defined in (19), with curve parameter s replacing time, and a quadratic (Hookean) elastic energy density is
| (27) |
Here ξ0(s) defines the local shape of the minimal energy conformation at each value of curve parameter s. Given ξ0(s), it is possible to integrate the matrix differential equation
subject to the initial condition g(0) = e (the identity element of the group, e, in this case is the identity matrix 𝕀4) for s ∈ [0, L] to obtain the minimal energy conformation rooted at the identity. In the case when ξ0(s) is a constant vector, this will produce a framed helix (with circular arcs and line segments as special cases).
Note that the independent variable is now a curve parameter, s, rather than time, t. Here the curve parameter s is taken to be the arc length of the filament in its undeformed (referential) conformation g0(s), but as the filament stretches or compresses s need not be arc length.
As a specific example, if the chain is uniform, inextensible and shearless, we have the constant stiffness matrix, K of the form
| (28) |
where si are very large numbers. As a result, the flexibility matrix (which, when measured in units of kBT will serve as a diffusion matrix in the sequel) is just the inverse of the stiffness matrix:
| (29) |
and if the minimal energy conformation is an arc-length-parameterized helix, we have the constant vector
| (30) |
In this case g(s) can be obtained in closed form as a framed helix.
As a specific example of (28) and (30) that has attracted attention in the recent literature is the Marko–Siggia DNA model [89]
Under the constraint that the molecule is inextensible, and all the frames of reference are attached to the backbone with their local z-axes pointing in the direction of the next frame, one observes
| (31) |
3.2. Minimal energy conformations of DNA loops with end constraints
A number of recent studies on chiral and uncoupled end-constrained elastic rod models of DNA with circular cross-section have been presented [33, 135, 143, 34]. These models use the classical elasticity theory of continuum filaments with or without self-contact constraints to model the stable conformations of DNA in plasmids, in chromosomes, and during transcription. That work is related to studies on DNA topology [48, 49, 61, 103, 113, 112, 148, 156] in the sense that the topological constraint of no self-interpenetration is enforced. In some works, Euler angles are used in parameterizing equations of the Kirchhoff elastic rod theory to obtain equilibrium conformations of DNA and determine its stability [166, 45, 46]. Also, the wormlike chain model has been used to model the equilibrium behavior of DNA [117]. More recent works involve the modeling of DNA as an anisotropic inextensible rod and also include the effect of electrostatic repulsion for describing the DNA loops bound to Lac repressor, etc [6, 7]. Another recent work includes sequence-dependent elastic properties of DNA [35]. All of these aforementioned works are based on Kirchhoff’s thin elastic rod theory [86]. This theory, as originally formulated, deals with non-chiral elastic rods with circular cross-section. Another example is the special Cosserat theory of rods [2], which can be viewed as an extension of Kirchhoff’s theory in that it includes extensible and shearable rods. Several researchers in elasticity have employed this rod theory to describe the static and dynamic characteristics of rods. For example, Simo and Vu-Quoc formulated a finite element method using rod theory [126]. Dichmann et al employed a Hamiltonian formulation using the special Cosserat theory of rods for the purpose of describing DNA [41]. Coleman et al reviewed dynamical equations in the theories of Kirchhoff and Clebsch [32]. Steigmann and Faulkner derived the equations of classical rod theory using parameter-dependent variational approach [130]. Recently, Gonzalez and Maddocks devised a method to extract sequence-dependent parameters for a rigid base-pair DNA model from molecular dynamics simulation [53]. In their paper, they used a force moment balance equation from Kirchhoff’s rod theory to extract stiffness and inertia parameters. Another recent work includes the application of Kirchhoff rod theory to marine cable loop formation and DNA loop formation [54]. In contrast to these uncoupled chiral models of DNA based on the elasticity of thin rod with isotropic or anisotropic cross-sectional properties, a number of stiffness models used in the statistical mechanics of semi-flexible polymers have been presented over the years [75, 147, 165]. These models address the chirality, anisotropic elasticity, and coupling between stiffnesses in semi-flexible polymers like DNA, though end-constrained equilibrium conformations for such models have not been obtained previously. Other models based on DNA structure [75, 109, 133, 125] and experimental measurements in which DNA is manipulated [10, 89, 105, 106] have also contributed to the development of anisotropic and coupled stiffness models of chiral macromolecules. Recently, Wiggins et al developed a theory based on nonlinear elasticity, called the kinkable wormlike chain model, for describing spontaneous kinking of polymers including DNA [157]. In subsequent sections of this chapter a model of elastic filaments that incorporates these stiffness properties is presented in which the theory of rotation and motion groups is used. The main differences between previous works and our approach are: (1) unlike previous works on DNA modeling which are based on rod theory (i.e., rods with uncoupled/diagonal stiffness tensor in a local frame of reference with one axis tangent to the filament in the shearless case), our approach applies to the chiral, anisotropic and coupled case. That is, we consider the most general small-strain, inextensible and shearless model, which is also the most accurate reflection of recent experimental measurements; (2) previous modeling works either use the balance equations for momentum and angular momentum from continuum mechanics and/or weak forms of these equations such as FEM/Galerkin methods. In contrast we use a Lie-group-based variational approach based on the Euler–Poincaré equation [60], which is completely different from previous works. Ideas from the theory of Lie groups have been applied in recent years in the fields of mechanics [3, 1] and robotics/systems theory (see, for example, the extensive reviews in [24, 28]). The material in section 3.2.1 is motivated by these previous works on applications of Lie theory, and is presented in a way so as to be directly applicable to the mechanics of end-constrained chiral and coupled rods.
The elastic energy in (26) is an example of a more general functional of the form
| (32) |
where g(t) is an element of a matrix Lie group G. If this is viewed as a cost that we seek to minimize subject to constraints hk(g) = 0 for k = 1,… , m, then the Euler–Poincaré equations (which are a coordinate-free version of the Euler–Lagrange equations for systems that evolve over group manifolds) can then be written in terms of the functions f and hk as [28]
| (33) |
The explicit form of (33) corresponding to the functionals in (26) is derived in the next sections (3.2.1 and 3.2.2), together with a method for solving for the values of the Lagrange multipliers that enforce the constraints.
3.2.1. Inextensible case
Considering the case when G = SO(3), the elastic energy density per unit length is of the form with the kinematic constraint of inextensibility (31), one writes (33) where G = SO(2) for i = 1, 2, 3 together as the vector equation
| (34) |
where a dot represents differentiation with respect to arc length s, λ ∈ ℝ3 is the vector of Lagrange multipliers necessary to enforce the vector constraint in equation (31), and the right-hand side of equation (34) results from the fact that
Equation (34) is solved iteratively subject to the initial conditions ω(0) = μ which are varied together with the Lagrange multipliers until a(L) and A(L) attain the desired values. A(s) is computed from ω(s) in equation (34) by integrating the matrix differential equation
and a(L) is then obtained from equation (31).
Numerical results using this model of DNA (and the one below) looped around histones is presented in [66].
3.2.2. Extensible rods
From equations (33) and (27) one can obtain the following Euler–Lagrange equation for the extensible case:
| (35) |
where ∧ is the product of infinitesimal rigid-body motions defined by
This wedge operator is related to the ad operator in (25) as
| (36) |
where , i = 1, 2 and the matrix of the ad operator for se(3) is defined as [28]
Equation (35) is solved subject to the initial conditions ξ(0) = η ∈ ℝ6. This, together with the kinematic condition
is integrated for 0 ≤ s ≤ L to define g(ξ, L). From this point everything follows in exactly the same way as for the inextensible case.
3.3. Conformational ensembles of DNA
Consider the equilibrium statistics of a stochastically forced elastic filament. Then instead of seeking one conformation that minimizes energy, the problem becomes that of characterizing the statistical mechanical ensemble of all conformations that fluctuate around the minimal energy conformation.
Let the evolution of the probability density of relative position and orientation of reference frames attached to a stochastically forced elastic filament at values of curve parameter 0 and s be denoted as f(g; 0, s). Since it is a probability density, by definition
| (37) |
Clearly f(g; 0, s) must be related in some way to the equilibrium shape of the filament, its stiffness, and the strength of the Brownian motion forcing from the ambient solvent. Also, the strength of this noise should be related in some way to the temperature. In fact, since f(g; 0, s) is the function describing the distribution of poses for a filament at equilibrium, it can be represented exactly as a path integral [24, 71, 165]:
| (38) |
Conceptually, this adds the contribution of the integrand over all possible paths g(σ) ∈ G for σ ∈ [0, s] that satisfy the end constraints g(0) = e (the identity, or ‘do nothing’ motion corresponding to the proximal end of the filament) and g(s) = g (the position and orientation of the frame attached to the distal end of the segment). The constant kB is Boltzmann’s constant and T is temperature measured in degrees kelvin. The integrand normalized by Z(s) is the Maxwell–Boltzmann distribution for the filament of length s. For conformations that are highly deformed relative to the referential shape (which is the shape that a filament would take as T → 0), the contribution to the path integral will be very small. Also, in this non-inertial theory, the statistical properties of any segment are independent of those of other concatenated segments that make up a longer chain.
3.3.1. DNA conformational statistics and diffusion on the Euclidean group
Equation (38) does not take into account the effects of excluded volume, which can be ignored for moderate values of filament length in the case when the DNA is not enclosed in a small compartment. The mathematical machinery associated with path integrals produces an evolution equation (i.e., a partial differential equation) for f(g; 0, s) of the form:
| (39) |
subject to the initial conditions
Here the diffusion matrix need not be limited to the special form in (29). This equation takes into account anisotropy and inhomogeneity of the elasticity (which has been observed in, e.g., [91]), as well as arbitrary minimal energy shape, and has essentially the same derivation as the homogeneous case presented in [22–24]. This equation can be written in classical terms using angular momentum operators and the gradient operator in Cartesian coordinates, as explained in [22]. The benefit of writing it in the group-theoretic form above is that then powerful tools from group theory can be applied to solve it either as a series or even in closed form, as explained in the sections that follow. As an alternative, the infinite continued fractions approach for generating the distribution of end-to-end distances for semi-flexible polymer chains is given in [97, 129].
Since stiffnesses are measured in terms of kBT, it follows that under the extreme condition T → 0, no diffusion would take place, and . For the biologically relevant case (T ≈ 300), (39) can be solved using the harmonic analysis approach in [22–24]. If we make the shorthand notation fs1,s2 (g) = f(g; s1, s2), then it will always be the case for s1 < s < s2 that6
| (40) |
This is the convolution of two position and orientation distributions. Here h is a dummy variable of integration, and the explicit form of the invariant integration measure, dh, is described in detail in [24]. While (40) will always hold for semi-flexible phantom chains, for the homogeneous rod there is the additional convenient properties that
| (41) |
The first of these says that for a uniform chain the position and orientation distribution only depends on the difference of arclength along the chain. The second provides a relationship between the position and orientation distribution for a uniform chain resulting from taking the frame at s1 to be fixed at the identity and recording the poses visited by s2, and the distribution of frames that results when s2 is fixed at the identity. However, neither of these nor (40) will hold when excluded-volume interactions are taken into account.
The density f(g; s1, s2) is a fundamental quantity for studying statistical problems. Even though it is defined for a chain/filament with freely moving ends, it can be used to obtain other important statistical properties. For example, given a chain with equilibrium (unstretched) length L, with proximal end fixed at the identity frame e, and distal end fixed at g, it is possible to write an expression for the probability of all framed filaments that satisfy these two end constraints as [27]
This is the probability density describing the relative likelihood that frame h ∈ G = SE(3) will be reached by some point s ∈ [0, L] along the filament when the ends of the filament are fixed at e and g. And f(h; 0, s) f(h−1 ○ g; s, L) is the relative likelihood that the frame attached to the base-pair at the specific value of s will reach the position and orientation h under the same end constraints.
Since a wide variety of quantities can be computed if f(g; s1, s2) is known, it makes sense to investigate methods for solving (39) to find it. Ideally, closed-form solutions analogous to Gaussian distributions would exist so that quantities of interest could be evaluated rapidly. In practice there are two cases in which f(g; s1, s2) can be evaluated rapidly: (1) when diffusion coefficients are very large (corresponding to a long polymer-like chain); (2) when the diffusion coefficients are very small (corresponding to a very stiff/short chain and/or very low temperature). In case 1, a group-Fourier solution can be evaluated rapidly, and in case 2 a Gaussian approximation can be written in closed form. This is analogous to solving the heat equation on a circular piece of metal; an initial delta function in temperature will diffuse as a Gaussian for small values of time until the tails meet, and at extremely large values of time Fourier series solutions that describe small deviations from uniformity can capture the situation well with a few terms. In the intermediate range either a folded Gaussian or Fourier series with more terms can be used.
3.3.2. Fourier solution of diffusion equations on the Euclidean group
The true benefit of the group-theoretic approach is realized when one observes that in coordinate form (39) is expressed as pages of complicated-looking (but essentially elementary) mathematical expressions. In contrast, it is possible to write out the solution very simply using results from group theory. One numerical approach that works well for dilute solutions of DNA of lengths in the range of 1/2–2 persistence lengths (60–300 basepairs at 300 K) is based on the group-Fourier transform for SE(3). The reason why this approach is most appropriate for this regime is that DNA of this length is flexible enough for Fourier methods (which work better for more spread out distributions than for highly focused ones) to be applicable, and it is short enough that the effects of self-contact can be neglected.
Equation (39) is a degenerate diffusion on SE(3) with constant coefficients. Methods for solving such equations are presented in [22–24]. These methods use the concept of the noncommutative Fourier transform for the Euclidean group. This builds on the work of Miller [98] and Vilenkin [146].
The SE(3) Fourier transform is completely analogous to those for SO(3) and SE(2) described in (12) and (14), respectively. The details, which are painstakingly reviewed in [24, 28], are much more involved. The essential feature that is used to solve (39) is that, in addition to the forward and inverse transform pair, Fourier transforms for Lie groups have operational properties, which for SE(3) are of the form
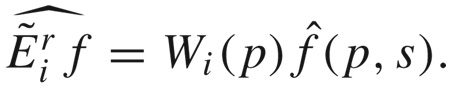 |
Here Wi(p) is an operator matrix. In other words, the group-Fourier transform converts Lie derivatives into matrix operations in Fourier space. This means that (39) can be written as
| (42) |
In the case of a referential configuration that has helical and stiffness parameters that are uniform (and therefore independent of s), then ℬ(s) = ℬ0 is constant and the solution can be written in Fourier space as f̂(p, s) = exp(sℬ0), and the inversion formula can be used to recover f(g, s). The details of this procedure have been discussed in a number of papers [22, 23, 25], together with the use of the convolution theorem for group-Fourier transforms to ‘stitch together’ the statistics of several segments of DNA connected by joints and/or kinks [169, 170]. In the case when ℬ(s) is not constant, the differential equation in (42), which is an ODE for each fixed value of 0 ≤ p ≤ ∞, can be solved either as a product of exponentials or by numerical integration.
3.3.3. Gaussian solution of diffusion equations on the Euclidean group
Here a new solution is presented for the case of very concentrated distributions (i.e., corresponding to lengths in the range of 5–50 base pairs). It uses the ‘chain rule for Lie groups’ reviewed in the appendix.
In addition to (39), we have another expression for the Fokker Planck equation:
| (43) |
where hi(s) = ξ0(s) • ei. The differential operators are the Lie derivatives:
| (44) |
Our goal here is to obtain an approximate functional form of f (g, s) that is efficient to compute in the case when ‖D‖ ≪ 1. In the case when D = 𝕆, the above SDE becomes the matrix ODE
| (45) |
with E͂i denoting the standard basis for se(3). Let the solution to the above ODE be denoted as m(s) ∈ SE(3). In the case when A is constant, m(s) = exp(As). More generally, if A and its integral commute then the solution can also be written as a matrix exponential:
| (46) |
However, if the above condition does not hold, then m(s) cannot be written as a single matrix exponential. In practice, if a baseline path is a circular arc or helix, then (46) will hold, because A will be constant.
In order to achieve the goal stated above, it will be useful to convert (43), which has potentially large values of hi, into an alternative form where all of the coefficients are small. An intuitive way to do this is to seek the new function F(g, s) such that
| (47) |
Here the notation m−1(s) is shorthand for [m(s)]−1. In other words, we will substitute (47) into (43) with the expectation that the large drift term will disappear and what remains can be described as a diffusion in a small neighborhood about m−1(s) ○ g.
To begin, we observe that the Lie algebra se(3) can be mapped bijectively to ℝ6 using the ∨ operation, where ∨ : E͂i → ei. Therefore, it is sometimes convenient to use the notation . Note that by the use of various notational substitutions,
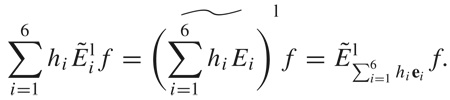 |
(48) |
This means that
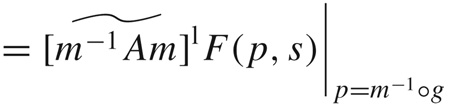 |
(49) |
| (50) |
| (51) |
Here (49)–(51) are just different ways of writing the same thing, each of which can be convenient in different contexts, where p ∈ SE(3) is a dummy variable, hi = hi (s), A = A(s) is defined in (45), and m = m(s). In the case when hi are all constant, we have m−1 Am = A. Therefore, in that case (49)–(51) can be avoided and
| (52) |
In the general case, the result in (51) can also be used to write
Direct substitution and using the fact that the 6 × 6 identity matrix can be written as then gives
| (53) |
When making the substitution (47) into (43) the left side of the equation becomes
When m(s) = exp(As) where A ∈ se(3) is constant, it can be shown that for m−1 (s) ○ g near the identity that
| (54) |
A detailed derivation of (54) is given in the appendix. Substituting (52)–(54) into (43) then gives
| (55) |
In other words, the drift term can be canceled, and we can study the diffusion (with time-varying diffusion matrix, D(s)) around the identity.
When motions are very close to the identity and m−1(s) ○ g = exp X, then the exponential coordinates {xi} are convenient, and The SE(3)-covariance defined in [152, 153] then is computed as
| (56) |
The resulting solution for the joint density on SE(3) for end-to-end position and orientation is then written in closed form as
| (57) |
From this joint density any quantities of interest can be extracted: end-to-end distance, ring-closure probabilities, end-to-end orientation distributions, etc.
4. Analysis of helix–helix interactions in proteins
In classical works on the analysis and prediction of how alpha helices within proteins should cross each other, the ‘knobs into holes’ [36] and ‘ridges into grooves’ [29] models predicted crossing angles that should be close to zero and 180°. This is because based on biophysical principles, hydrophobic side chains on alpha helices should want to bury as much area as possible from the surrounding solvent during the folding process [83]. However, a conundrum ensued when trying to observe this in tens of thousands of helix–helix pairs in the protein data bank. In fact, histograms of crossing angle showed peaks around 90°—in direct opposition to biophysical principles. Why should this be the case?
In a series of papers it was reasoned essentially that there must be ‘more room’ for helices to move in the range of crossing angles near 90° as opposed to 0 or 180. This is analogous to the fact that a strip of area of width 1° around the equator of a sphere has much more area than a 1° cap at the poles. The difficulty comes in quantifying this volumetric/entropic effect. For the only way to extract true preferences for helices to aggregate at particular angles is to first normalize by this effect (which is not accounted for when naively forming a histogram of crossing angle).
Bowie [16] argued that if β is the crossing angle then the integration factor should be sin β (just like the sphere, or the integration measure for SO(3) in Euler angles). Walther, Springer, and Cohen [150] argued that it should be sin2 β. The author and a collaborator clarified using group theory that there are actually three distinct cases [80]: (a) line-to-line interaction in which the common normal of the two finite helical axes intersects both on their interior; (b) end-to-line interaction in which the end of helix 1 is closer than any other point in helix 1 to the interior of helix 2; (c) end-to-end interaction in which the end points of the two helices are closer to each other than any points on the interior of either helix. Each of these cases parameterizes a subset of the group SE(3), and the Jacobian determinant for each provides the normalization factor required in order to extract true preferences from the underlying data. Below, the mathematical details are reviewed. For detailed analysis of the actual data see [80].
4.1. Case 1: line-to-line interaction
Given two finite line segments, the common normal of which intersects each on their interiors, the relative motion between these segments can be parameterized with six variables: the rotations around each axis α, γ, the translational displacement along each axis z1, z2, the distance between the axis measured along the common normal, r, and the angle between the axes measured around the common normal, β. It was reasoned in [80] that the position of a reference frame attached to one helix will be related that of the other by the rigid-body transformation
| (58) |
Performing the multiplications in equation (58),
| (59) |
Unlike the other two cases below, in this case three variables which appear in the translation part of the homogeneous transformation matrix do not appear in the rotation part. This gives a block structure to the Jacobian matrix, and makes the determinant easy to compute.
In particular, if we group the variables as q1 = (α, β, γ) and q2 = (r, z1, z2), then in this case the Jacobian will have the form
Here the matrix JR is [24]:
Due to the block lower diagonal form of this matrix, and the fact that A is a rotation matrix and therefore det A = +1, it is clear that
From (59) it is clear that
Therefore, a small computation shows that . And since |detJR|= sin β, it follows that
This verifies the result of Walther et al [150]. However, this is not the end of the story, because the line-on-line case is not the only way that helices can interact. The other two cases are reviewed below.
4.2. Case 2: end-to-line interaction
In this case spherical coordinates ϕ, θ, r from the tip of the first finite helical axis point to the interior of the second helical axis. The motion of the second helix is defined by translational motion along its axis, x, rotation around the vector connecting helix 1 and helix 2, α, and rotation of helix 2 around its axis, γ. This results in
| (60) |
Performing the multiplications in equation (61),
| (61) |
|det𝒥R| = r sin θ.
4.3. Case 3: end-to-end interaction
In this case there are essentially two spherical coordinate systems at the two tips of the helix axes, one which points from helix 1 to helix 2, and vice versa. They share a common radius, but the polar and azimuthal angles of each are ϕ, θ and α, β, respectively. A final angle, γ, describes twist around the radial vector. This leads to
| (62) |
Performing the multiplications in equation (62),
and
In short, basic Lie group-theoretic methods that lead to the computation of Jacobian determinants provide the geometric tools to evaluate biases in the naive analysis of helix–helix interactions. Accounting for these geometric effects allows for the extraction of the true underlying preferences that would correspond to potential energies of interaction of rigid-body models of alpha helices. This illustrates the usefulness of Lie theoretic methods in the analysis of secondary-structure interactions in proteins. However, it is not the end of the story, and work in this area continues. See, e.g., [144]. In addition, alpha helices are not the only structures that can be treated as rigid units within a protein. It is possible to analyze preferences in the relative positions and orientations of reference frames attached to alpha carbons of particular residue types, or at the ends of articulated sidechains. Effectively, this amounts to a 6D extension of the Ramachandran map [81]. At the other extreme, whole domains within multidomain proteins can be considered to be rigid, resulting in coarse-grained models with reduced degrees of freedom. That is the subject of section 5.
5. Coarse-grained models of protein structure and motion
Whereas static spatial relationships such as the crossing angle between pairs of alpha helices in contact within proteins can be studied from a statistical point of view as in section 4, another application of Lie group techniques occurs in studying the motion of proteins and complexes. This can be done at a variety of scales ranging from atomic, to coarse-grained alpha carbon traces, to rigid-body models of large domains within proteins. This section reviews these techniques.
5.1. Equilibrium statistical mechanics of elastic systems
In classical statistical mechanics, the partition function is defined as
| (63) |
where β = 1/kBT, pi = p • ei is the momentum conjugate to the ith generalized coordinate qi = q • ei, ℋ is the Hamiltonian for the system, and dpdq = dp1 ⋯ dpN dq1 ⋯ dqN for a system with N degrees of freedom. The range of integration is over all possible states of the system.
For any classical mechanical system the Hamiltonian is of the form
where V(q) is the potential energy and M(q) is the mass matrix.
For a macromolecule fluctuating closely about one conformation which globally minimizes its potential energy, the potential energy function can be expressed as
| (64) |
where the elements of the stiffness matrix K are
and q = 0 is defined to be the value for which V (0) = V0 is the minimum attainable potential energy. By appropriate choice of datum, one can take V0 = 0. Since q(t) never strays far from 0, it follows that M(q) is approximated well as the constant matrix M = M(0).
Therefore,
| (65) |
where M and K are constant matrices. Equation (65) holds for systems with one global minimum that is deeper than, and well separated from, any local energy minima. This equation holds regardless of whether Cartesian or internal coordinates are used to describe the motion. This simple formulation can in principle be used to extract effective stiffness parameters from observed data [26].
In section 5.2 an intuitive geometric method for constructing K is presented in which one or more stiffness parameters are left free. In a section 5.3 it is shown how these parameters can be fixed based on experimental measurements of molecular motion.
5.2. Cα-based elastic network models
As mentioned in section 1, a number of coarse-grained models of proteins have been developed [140, 4, 5, 88, 99] to infer possible harmonic motions from structures deposited in the protein data bank (PDB) [14]. In these papers, one point per amino acid residue is selected at the alpha carbon atom of each residue, and a contact map is used to store information about which residues are the nearest neighbors to each residue. In the elastic network model a Hookean spring (harmonic potential) is assigned to each pair of residues that are designated as neighbors by the contact map.
It should be noted that the purpose of the elastic network model is to examine small motions around a native state. Other methods can be used to explore larger deviations from this equilibrium. For example, Go-like models have been used to describe the folding of proteins, as well as the unfolding of proteins in numerical pulling experiments [30]. A survey of works pertaining to the force required to pull bead models of folded proteins apart is given in [134]. Numerical stretching experiments using bead models of DNA are presented in [107]. Also, it has been hypothesized that proteins form a state of matter that is distinct from solids and liquids, and that this state is characterized by excluded-volume effects and the overall serial nature of proteins, both of which are captured as a tube of finite thickness [8]. Therefore, if one has the goal of studying thermodynamic properties, folding pathways, etc, such models may be used. But that is not the purpose of the simple models reviewed here.
In the elastic network model, the initial PDB structure is assumed to be the minimal energy (equilibrium) conformation. While it is possible to assign different spring constants to each contact, usually they are taken to all be the same, as are the masses of each residue. The eigenvectors of the resulting stiffness matrix then correspond to small harmonic motions about equilibrium. The results corresponding to low-frequency motions have been shown to be very robust to changes in stiffness values, and match well to all-atom computations. In addition, they reproduce well experimentally observed crystallographic B-factors. These methods are reviewed here.
Given a set of Cα crystal structure coordinates for a protein, {xi(0)}, the Cartesian displacement of the i th α-carbon at time t can be written without loss of generality as
| (66) |
We define δi (t) to be a vector of small displacements.
The total kinetic energy in a network of n residues (each of which is treated as a point mass) then has the form
| (67) |
where the constant matrix M is the global mass matrix for the whole network and
| (68) |
In the current context, M is diagonal.
The total potential energy in a network of connected springs has the form
| (69) |
and
| (70) |
εi,j is a measure of the residual strain in the contact between residues i and j in the equilibrium conformation. ci,j is the (i, j) element of an n × n matrix called the linking or contact matrix. cij is equal to 1 if residues i and j are in contact, and zero otherwise. In coarse-grained models, one often sets κ[i],[j] = α, a single parameter. However, it is possible to partition the interactions in the macromolecule into several different types of interactions (e.g., covalent backbone interactions, disulfide bonds, hydrophobic contacts, solvent-mediated surface interactions, etc). That is, if contacting residue pairs (i, j) and (i′, j′) are in the same class, then κ[i],[j] = κ[i′],[j′]. The stiffnesses in each of these cases can either be all left as variables to be determined directly from experimental measurements, or some of them can be set using a priori knowledge of contact potentials.
Equation (69) is a non-quadratic function of the deformations even though the springs are linear. However, when we assume that the deformations are small, V becomes a classical quadratic potential energy function. In particular, using the Taylor series approximation
| (71) |
then for small deflections, the total potential energy (69) can be written in the form
| (72) |
where Gi,j ∈ ℝ3×3 is defined as
| (73) |
Note that the effects of residual strain do not appear in (72) in any material way (they do however appear in V0). Therefore, any residual strain effects are completely removed by setting a datum such that V0 = 0.
The stiffness matrix for the whole network is then the matrix K such that
| (74) |
where δ is defined in (68). Note that since a translation and rotation of all residues by the same rigid-body transformation does not change the value of V, six zero-frequency normal modes correspond to infinitesimal rigid-body motions about the equilibrium shape.
In the elastic network model, which is purely mechanical, the stiffness matrix K in (74) replaces the Hessian matrix in (64) and δ replaces q as the generalized coordinate. For more details regarding these models see [4, 67].
5.3. Coarse graining by rigid-body domains
In cases of very large structures such as GROEL-GROES, or even larger yet, viral capsids, even course-grained Cα-based models can contain tens or hundreds of thousands of degrees of freedom. One approach that has been taken is to sample even more coarsely, at every tenth, or even every hundredth, residue along the sequence of the protein [4]. However, not every degree-of-freedom is as important as others. If, for example, two structures have been obtained experimentally for the same protein (e.g., with/without a bound ligand), then a similarity analysis can be performed to assess which parts of the protein are preserved. Under the assumption that preserved regions (or domains) remain rigid as the protein undergoes functions such as binding, then very coarse models can be developed in which the protein is broken up into rigid bodies. This was done, for example, in [69], in which a viral capsid consisting of hundreds of copies of a capsomer protein were assembled into an icosahedral virus-the Hong Kong 97 (HK97) bacteriophage. It was determined that each capsomer essentially acted as two rigid bodies connected by a multi-degree-of-freedom hingelike connecting region. The result was a reduction of what would have been a 300 000 degree-of-freedom (dof) system to one which had several hundred rigid-body dof. And because all of the contact information between bodies is preserved in these models, some of the negative effects of decimation approaches are circumvented.
Under the same assumptions as in usual normal mode analysis (NMA) of small motions, NMA based on rigid-body-coarse-grained models of proteins is possible [119]. Essentially, the assumption of smallness is exactly the assumption that exp(εXi) = 𝕀 + εXi. It has been shown that this method reproduces well the low-frequency normal modes that are produced when using the residue-level coarse graining described above.
In addition to reducing degrees of freedom by freezing domains and treating them as rigid bodies, geometric symmetries in structures that result from the fact that they are composed of identical (or nearly identical) subunits, and arranged in a symmetrical pattern can be used to further reduce degrees of freedom when the conformational motions of these systems are assumed to preserve these symmetries. For example, in the analysis of the HK97 virus in [69], it was show that an additional factor of 60 (the number of elements in the icosahedral group, which is a finite subgroup of SO(3)) could be saved in the number of dof. This is significant because if the stiffness matrix is N × N, then operations such as matrix inversion and computation of eigenvalues/eigenvectors typically involve O(N3) operations. Reduction by a factor of 60 then reduces computation time by a factor of (60)3. The drawback of such symmetry methods is that they only apply when the structure remains symmetrical during its maturation.
Harmonic motions around an equilibrium clearly do not fully explain the structure-motion-function relationship. Sometimes large scale anharmonic rigid-body motions of domains are required to reorganize from one conformation to another. One method to animate these conformational transitions is elastic network interpolation [67, 68]. This can be implemented at the atomic, Cα, or rigid-body level. The rigid-body version is described in [70]. Another approach that involves the geometric/Lie group-theoretic tools reviewed earlier in this paper is that of iterative cluster NMA [120]. The basic idea is to perform rigid-body NMA, update the current conformation, and iterate. If two conformations are given, the iterations are updated starting with the first and driven to become the second. This method has been shown to work well with even very large motions of long protein loops [121].
6. Conclusions
This survey has reviewed the state of the art in how group-theoretic methods are applied to a variety of problems pertaining to biomolecular conformation. This includes computational analysis of structure and motion, and the reduction of the number of degrees of freedom required in modeling by taking into account geometric symmetries. Three specific areas were reviewed: (1) DNA models as continuum rods either subjected to end constraints or to ambient Brownian motion forcing; (2) geometric biases in observations of preferences in helix–helix crossing angles in proteins; (3) coarse-grained models of protein motions around the native state. It is hoped that by bringing these various topics together under one general framework that group theory will be used more widely by researchers on the more qualitative side of structural biology, and physicists who seek to enter the field will more readily understand how their tools can be brought to bear.
Acknowledgment
This work was performed under support from the NIH Grant R01 GM075310 ‘Group Theoretic Methods in Protein Structure Determination’ and its ARRA supplement R01 GM075310-04S1.
Appendix. The chain rule for functions on Lie groups
Given a mapping ϕ: ℝn × ℝ≥0 → ℝn and a function F: ℝn × ℝ≥0 → ℝ the classical chain rule states
| (75) |
or equivalently
| (76) |
Given a Lie group G, and defining x = (log g)∨, then one instance of the above that is relevant to the context of Lie groups is when
Though the logarithm map may not be defined for all g ∈ G, for the groups SO (3) and SE(3), which are of the most interest in the applications presented here, it will be defined for all g ∈ G except possibly a set of measure zero.
A function f: G × ℝ≥0 → ℝ can be expressed as one in exponential coordinates as
| (77) |
Now, if for each fixed value of t, the support of f(g, t) in G is confined to a small ball around m, then when computing integrals over G, only values for which d(m, g) = ‖log(m−1 ○ g) ‖ ≪ 1 will contribute. And so these are the only values of g ∈ G that really matter. This means that even though m(t) may not be small, ‖m−1 ○ g − 𝕀‖ will be small, and so
Therefore, since the ∨ and ∂/∂t operators are both linear and they commute, when the above approximation holds
If m(t) is defined by a system of ODEs of the form
where m(0) = m0 (as would be the case for a body-fixed or space-fixed description of free rigid-body motion) then
or
But if ‖log(m−1 ○ g)‖ is small, then the second term in each of the above two equations is insignificant compared to the first, and
| (78) |
As a special case, if A is constant of the form
then m−1 Am = A, and both expressions in (78) reduce to the same thing. Furthermore, if both ‖(log g)∨‖ and ‖(log m)∨‖ are small, then
and
which is consistent with, though not a necessary condition for, (78) to hold.
In any case, since (78) holds, and since near the identity e ∈ G
where the relationship between f and F is given in (77), it follows that (75) can be adapted to the Lie group setting involving concentrated functions as
| (79) |
or
| (80) |
Footnotes
Other common conventions have a multiplicative factor of (−1)m−n or im−n in front of each matrix element.
Part 2 of this paper is a detailed discussion of group-theoretic methods in experimental structure determination.
Recall that U(n) consists of all complex n × n matrices that satisfy the condition VV* = 𝕀n, and SU(n) consists of those entries of U(n) with det V = +1.
The notation SE(n) stands for a special Euclidean group acting on ℝn.
For the special Euclidean group SE(3) and its Lie algebra se(3), n = 6, and Xi = Ẽi. For the case of the rotation group SO(3), the Lie algebra is so(3), n = 3, and Xi = Ei. More abstractly, a Lie group G will have an associated Lie algebra 𝒢.
The notations fs1, s2 (g) and f(g; s1, s2) are used interchangeably.
References
- 1.Abraham R, Marsden JE. Foundations of Mechanics. 2nd edn. San Mateo, CA: Benjamin-Cummings; 1978. [Google Scholar]
- 2.Antman SS. Nonlinear Problems of Elasticity. New York: Springer; 1995. [Google Scholar]
- 3.Arnol’d VI. Mathematical Methods of Classical Mechanics. New York: Springer; 1978. [Google Scholar]
- 4.Atilgan AR, Durell SR, Jernigan RL, Demirel MC, Keskin O, Bahar I. Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophys. J. 2001;80:505–515. doi: 10.1016/S0006-3495(01)76033-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bahar I, Jernigan RL. Vibrational dynamics of transfer RNAs: comparison of the free and synthetase-bound forms. J. Mol. Biol. 1998;281:871–884. doi: 10.1006/jmbi.1998.1978. [DOI] [PubMed] [Google Scholar]
- 6.Balaeff A, Mahadevan L, Schulten K. Modeling DNA loops using the theory of elasticity arXiv.org. 2003. http://arxiv.org/abs/physics/0301006. [DOI] [PubMed] [Google Scholar]
- 7.Balaeff A, Mahadevan L, Schulten K. Structural basis for cooperative DNA binding by CAP and Lac repressor. Structure. 2004;12:123–132. doi: 10.1016/j.str.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 8.Banavar JR, Maritan A. Colloquium: geometrical approach to protein folding: a tube picture. Rev. Mod. Phys. 2003;75:23–34. [Google Scholar]
- 9.Barut AO, Raçzka R. Theory of Group Representations and Applications. Singapore: World Scientific; 1986. [Google Scholar]
- 10.Baumann CG, Smith SB, Bloomfield VA, Bustamante C. Ionic effects on the elasticity of single DNA molecules. Proc. Natl Acad. Sci. USA. 1997;94:6185–6190. doi: 10.1073/pnas.94.12.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bawendi MG, Karl FF. A wiener integral model for stiff polymer chains. J. Chem. Phys. 1985;83:2491–2496. [Google Scholar]
- 12.Benham CJ. Elastic model of the large-scale structure of duplex DNA. Biopolymers. 1979;18:609–623. doi: 10.1002/bip.1979.360180310. [DOI] [PubMed] [Google Scholar]
- 13.Benham CJ, Mielke SP. DNA mechanics. Annu. Rev. Biomed. Eng. 2005;7:21–53. doi: 10.1146/annurev.bioeng.6.062403.132016. [DOI] [PubMed] [Google Scholar]
- 14.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucl. Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhattacharjee SM, Muthukumar M. Statistical mechanics of solutions of semiflexible chains: a path integral formulation. J. Chem. Phys. 1987;86:411–418. [Google Scholar]
- 16.Bowie JU. Helix packing angle preferences. Nat. Struct. Biol. 1997;4:915–917. doi: 10.1038/nsb1197-915. [DOI] [PubMed] [Google Scholar]
- 17.Brockett RW. System theory on group manifolds and coset spaces. SIAM J. Control. 1972;10:265–284. [Google Scholar]
- 18.Brooks B, Karplus M. Harmonic dynamics of proteins: normal modes and fluctuations in bovine pancreatic typsin inhibitor. Proc. Natl Acad. Sci. USA. 1983;80:6571–6575. doi: 10.1073/pnas.80.21.6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brooks B, Karplus M. Normal modes for specific motions of macromolecules: application to the hinge-bending mode of lysozyme. Proc. Natl Acad. Sci. USA. 1985;82:4995–4999. doi: 10.1073/pnas.82.15.4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM—a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- 21.Buchiat C, Wang MD, Allemand JF, Strick T, Block SM, Croquette V. Estimating the persistence length of a worm-like chain molecule from force-extension measurements. Biophys. J. 1999;76:409–413. doi: 10.1016/s0006-3495(99)77207-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chirikjian GS, Wang YF. Conformational statistics of stiff macromolecules as solutions to PDEs on the rotation and motion groups. Phys. Rev. E. 2000;62:880–892. doi: 10.1103/physreve.62.880. [DOI] [PubMed] [Google Scholar]
- 23.Chirikjian GS, Kyatkin AB. An operational calculus for the Euclidean motion group with applications in robotics and polymer science. J. Fourier Anal. Appl. 2000;6:583–606. [Google Scholar]
- 24.Chirikjian GS, Kyatkin AB. Engineering Applications of Noncommutative Harmonic Analysis. Boca Raton, FL: CRC Press; 2001. [Google Scholar]
- 25.Chirikjian GS. Conformational statistics of macromolecules using generalized convolution. Comput. Theor. Polym. Sci. 2001;11:143–153. [Google Scholar]
- 26.Chirikjian GS. A methodology for determining mechanical properties of macromolecules from ensemble motion data. Trends Anal. Chem. 2003;22:549–553. [Google Scholar]
- 27.Chirikjian GS. The stochastic elastica and excluded-volume perturbations of DNA conformational ensembles. Int. J. Non-Linear Mech. 2008;43:1108–1120. doi: 10.1016/j.ijnonlinmec.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chirikjian GS. Stochastic Models, Information Theory, and Lie Groups. Basle: Birkhäuser; 2009. [Google Scholar]
- 29.Chothia C, Levitt M, Richardson D. Helix to helix packing in proteins. J. Mol. Biol. 1981;145:215–250. doi: 10.1016/0022-2836(81)90341-7. [DOI] [PubMed] [Google Scholar]
- 30.Cieplak M, Hoang TX, Robbins MO. Stretching of proteins in the entropic limit. Phys. Rev. E. 2004;69:011912. doi: 10.1103/PhysRevE.69.011912. [DOI] [PubMed] [Google Scholar]
- 31.Cluzel P, Lebrun A, Christoph H, Lavery R, Viovy JL, Chatenay D, Caron F. DNA: an extensible molecule. Science. 1996;271:792. doi: 10.1126/science.271.5250.792. [DOI] [PubMed] [Google Scholar]
- 32.Coleman BD, Dill EH, Lembo M, Lu Z, Tobias I. On the dynamics of rods in the theory of Kirchhoff and Clebsch. Arch. Ration. Mech. Anal. 1993;121:339–359. [Google Scholar]
- 33.Coleman BD, Tobias I, Swigon D. Theory of the influence of end conditions on self-contact in DNA loops. J. Chem. Phys. 1995;103:9101–9109. [Google Scholar]
- 34.Coleman BD, Swigon D, Tobias I. Elastic stability of DNA configurations. II: supercoiled plasmides with self-contact. Phys. Rev. E. 2000;61:759–770. doi: 10.1103/physreve.61.759. [DOI] [PubMed] [Google Scholar]
- 35.Coleman BD, Olson WK, Swigon D. Theory of sequence-dependent DNA elasticity. J. Chem. Phys. 2003;118:7127–7140. [Google Scholar]
- 36.Crick F. The packing of α-helices: simple coiled coils. Acta Crystallogr. 1953;6:689–697. [Google Scholar]
- 37.Daniels HE. The statistical theory of stiff chains. Proc. R. Soc. A. 1952;63:290–311. [Google Scholar]
- 38.de Gennes PG. Scaling Concepts in Polymer Physics. Ithaca, NY: Cornell University Press; 1979. [Google Scholar]
- 39.de Gennes PG, Prost J. The Physics of Liquid Crystals. 2nd edn. Oxford: Clarendon; 1998. [Google Scholar]
- 40.des Cloizeaux J, Jannink G. Polymers in Solution: Their Modelling and Structure. Oxford: Clarendon; 1990. [Google Scholar]
- 41.Dichmann DJ, Li Y, Maddocks JH. Hamiltonian formulations and symmetries in rod mechanics. In: Mesirov JP, Schulten K, Summers D, editors. Mathematical Approaches to Biomolecular Structure and Dynamics. New York: Springer; 1995. pp. 71–113. [Google Scholar]
- 42.Dixit SB, Beveridge DL, Case DA, Cheatham TE, Giudice E, Lankas F, Lavery R, Maddocks JH, Osman R, Sklenar H, Thayer KM, Varnai P. Molecular dynamics simulations of the 136 unique tetranucleotide sequences of DNA oligonucleotides. II: sequence context effects on the dynamical structures of the 10 unique dinucleotide steps. Biophys. J. 2005;89:3721–3740. doi: 10.1529/biophysj.105.067397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doi M, Edwards SF. The Theory of Polymer Dynamics. Oxford: Clarendon Press; 1986. [Google Scholar]
- 44.Dyson F. Selected Papers of Freeman Dyson with Commentary. Providence, RI: American Mathematical Society; 1996. [Google Scholar]
- 45.Fain B, Rudnick J. Conformations of closed DNA. Phys. Rev. E. 1999;60:7239–7252. doi: 10.1103/physreve.60.7239. [DOI] [PubMed] [Google Scholar]
- 46.Fain B, Rudnick J, Östlund S. Conformations of linear DNA. Phys. Rev. E. 1997;55:7364–7368. [Google Scholar]
- 47.Flory PJ. Statistical Mechanics of Chain Molecules. New York: Wiley-Interscience; 1969. [Google Scholar]
- 48.Fuller FB. Decomposition of the linking number of a closed ribbon: a problem from molecular biology. Proc. Natl Acad. Sci. USA. 1978;75:3557. doi: 10.1073/pnas.75.8.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fuller FB. The writhing number of a space curve. Proc. Natl Acad. Sci. USA. 1971;68:815. doi: 10.1073/pnas.68.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gel’fand IM, Minlos RA, Shapiro ZYa. Representations of the Rotation and Lorentz Groups and Their Applications. New York: Macmillan; 1963. [Google Scholar]
- 51.Gell-Mann M, Ne’eman Y. The Eightfold Way. Boulder, CO: Westview Press; 2000. [Google Scholar]
- 52.Gobush W, Yamakawa H, Stockmayer WH, Magee WS. Statistical mechanics of wormlike chains. I. Asymptotic behavior. J. Chem. Phys. 1972;57:2839–2843. [Google Scholar]
- 53.Gonzalez O, Maddocks JH. Extracting parameters for base-pair level models of DNA from molecular dynamics simulations. Theor. Chem. Acc. 2001;106:76–82. [Google Scholar]
- 54.Goyal S, Perkins NC, Lee CL. Nonlinear dynamics and loop formation in Kirchhoff rods with implications to the mechanics of DNA and cables. J. Comput. Phys. 2005;209:371–389. [Google Scholar]
- 55.Grosberg AYu, Khokhlov AR. Statistical Physics of Macromolecules. New York: American Institute of Physics; 1994. [Google Scholar]
- 56.Ha BY, Thirumalai D. Semiflexible chains under tension. J. Chem. Phys. 1997;106:4243–4247. [Google Scholar]
- 57.Hagerman PJ. Analysis of the ring-closure probabilities of isotropic wormlike chains: application to duplex DNA. Biopolymers. 1985;24:1881–1897. doi: 10.1002/bip.360241004. [DOI] [PubMed] [Google Scholar]
- 58.Hermans JJ, Ullman R. The statistics of stiff chains, with applications to light scattering. Physica. 1952;18:951–971. [Google Scholar]
- 59.Hinsen K. Analysis of domain motions by approximate normal mode calculations. Protein Struct. Function Genet. 1998;33:417–429. doi: 10.1002/(sici)1097-0134(19981115)33:3<417::aid-prot10>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 60.Holm DD, Marsden JE, Ratiu TS. The Euler-Poincaré equations and semidirect products with applications to continuum theories. Adv. Math. 1998;137:1. [Google Scholar]
- 61.Horowitz DS, Wang JC. Torsional rigidity of DNA and length dependence of the free energy of DNA supercoiling. J. Mol. Biol. 1984;173:75–91. doi: 10.1016/0022-2836(84)90404-2. [DOI] [PubMed] [Google Scholar]
- 62.Inui T, Tanabe Y, Onodera Y. Group Theory and its Applications in Physics. 2nd edn. Berlin: Springer; 1996. [Google Scholar]
- 63.Kalmykov YP, Coffey WT. Analytical solutions for rotational diffusion in the mean field potential: application to the theory of dielectric relaxation in nematic liquid crystals. Liq. Cryst. 1998;25:329–339. [Google Scholar]
- 64.Kamein RD, Lubensky TC, Nelson P, O’Hern CS. Direct determination of DNA twist-stretch coupling. Europhys. Lett. 1997;28:237–242. [Google Scholar]
- 65.Kholodenko AL. Statistical mechanics of semiflexible polymers: yesterday, today and tomorrow. J. Chem. Soc. Farady Trans. 1995;91:2473–2482. [Google Scholar]
- 66.Kim J-S, Chirikjian GS. Conformational analysis of stiff chiral polymers with end-constraints. Mol. Simul. 2006;32:1139–1154. doi: 10.1080/08927020601024137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim MK, Jernigan RL, Chirikjian GS. Efficient generation of feasible pathways for protein conformational transitions. Biophys. J. 2002;83:1620–1630. doi: 10.1016/S0006-3495(02)73931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim MK, Chirikjian GS, Jernigan RL. Elastic models of conformational transitions in macromolecules. J. Mol. Graph. Modeling. 2002;21:151–160. doi: 10.1016/s1093-3263(02)00143-2. [DOI] [PubMed] [Google Scholar]
- 69.Kim MK, Jernigan RL, Chirikjian GS. An elastic network model of HK97 capsid maturation. J. Struct. Biol. 2003;143:107–117. doi: 10.1016/s1047-8477(03)00126-6. [DOI] [PubMed] [Google Scholar]
- 70.Kim MK, Jernigan RL, Chirikjian GS. Rigid-cluster models of conformational transitions in macromolecular machines and assemblies. Biophys. J. 2005;89:43–55. doi: 10.1529/biophysj.104.044347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kleinert H. Path Integrals in Quantum Mechanics, Statistics and Polymer Physics. 2nd edn. Singapore: World Scientific; 1995. [Google Scholar]
- 72.Klenin K, Merlitz H, Langowski J. A Brownian dynamics program for the simulation of linear and circular DNA and other wormlike chain polyelectrolytes. Biophys. J. 1998;74:780–788. doi: 10.1016/S0006-3495(98)74003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kostelec PJ, Rockmore DN. FFTs on the Rotation Group (Santa Fe Institute Working Papers Series) 2003. Paper #03-11-060. [Google Scholar]
- 74.Kovacs JA, Wriggers W. Fast rotational matching. Acta Crystallogr. D. 2002;58:1282–1286. doi: 10.1107/s0907444902009794. [DOI] [PubMed] [Google Scholar]
- 75.Kratky O, Porod G. Röntgenuntersuchung Gelöster Fadenmoleküle. Recueil des Travaux Chimiques des Pays-Bas. 1949;68:1106–1122. [Google Scholar]
- 76.Kroy K, Frey E. Force-extension relation and plateau modulus for wormlike chains. Phys. Rev. Lett. 1996;77:306–309. doi: 10.1103/PhysRevLett.77.306. [DOI] [PubMed] [Google Scholar]
- 77.Lagowski JB, Noolandi J, Nickel B. Stiff chain model—functional integral approach. J. Chem. Phys. 1991;95:1266–1269. [Google Scholar]
- 78.Lavery R, Moakher M, Maddocks JH, Petkeviciute D, Zakrzewska K. Conformational analysis of nucleic acids revisited: curves+ Nucl. Acids Res. 2009;37:5917–5929. doi: 10.1093/nar/gkp608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Leach AR. Molecular Modelling: Principles and Applications. 2nd edn. Harlow: Longman; 2001. [Google Scholar]
- 80.Lee S, Chirikjian GS. Inter-helical angle and distance preferences in globular proteins. Biophys. J. 2004;86:1105–1117. doi: 10.1016/S0006-3495(04)74185-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee S, Chirikjian GS. Pose analysis of alpha-carbons in proteins. Int. J. Robot. Res. 2005;24:183–210. [Google Scholar]
- 82.Levene SD, Crothers DM. Ring closure probabilities for DNA fragments by Monte Carlo simulation. J. Mol. Biol. 1986;189:61–72. doi: 10.1016/0022-2836(86)90381-5. [DOI] [PubMed] [Google Scholar]
- 83.Lesk AM. Introduction to Protein Architecture. Oxford: Oxford University Press; 2001. [Google Scholar]
- 84.Liverpool TB, Edwards SF. Probability distribution of wormlike polymer loops. J. Chem. Phys. 1995;103:6716–6719. [Google Scholar]
- 85.Liverpool TB, Golestanian R, Kremer K. Statistical mechanics of double-stranded semiflexible polymers. Phys. Rev. Lett. 1998;80:405–408. [Google Scholar]
- 86.Love AEH. A Treatise on the Mathematical Theory of Elasticity. New York: Dover; 1944. [Google Scholar]
- 87.Luckhurst GR, Zannoni C, Nordio PL, Segre U. A molecular field theory for uniaxial nematic liquid crystals formed by non-cylindrically symmetric molecules. Mol. Phys. 1975;30:1345–1358. [Google Scholar]
- 88.Ma JP. New advances in normal mode analysis of supermolecular complexes and applications to structural refinement. Curr. Protein Peptide Sci. 2004;5:119–123. doi: 10.2174/1389203043486892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Marko JF, Siggia ED. Bending and twisting elasticity of DNA. Macromolecules. 1994;27:981–988. [Google Scholar]
- 90.Marko JF. DNA under high tension: overstretching, undertwisting, and relaxation dynamics. Phys. Rev. E. 1998;57:2134–2149. [Google Scholar]
- 91.Maroun RC, Olson WK. Base sequence effects in double-helical DNA. 2. Configurational statistics of rodlike chains. Biopolymers. 1988;27:561–584. doi: 10.1002/bip.360270403. [DOI] [PubMed] [Google Scholar]
- 92.Maslen DK. PhD Dissertation. Department of Mathematics, Harvard University; 1993. May, Fast transforms and sampling for compact groups. [Google Scholar]
- 93.Maslen DK, Rockmore DN. Generalized FFTs—a survey of some recent results. DIMACS Series in Discrete Mathematics and Theoretical Computer Science. 1997;28:183–237. [Google Scholar]
- 94.Maslen DK. Efficient computation of Fourier transforms on compact groups. J. Fourier Anal. Appl. 1998;4:19–52. [Google Scholar]
- 95.Matsutani S. Statistical mechanics of no-stretching elastica in three-dimensional space. J. Geom. Phys. 1999;29:243–259. [Google Scholar]
- 96.McConnell J. Rotational Brownian Motion and Dielectric Theory. New York: Academic; 1980. [Google Scholar]
- 97.Mehraeen S, Sudhanshu B, Koslover EF, Spakowitz AJ. End-to-end distribution for a wormlike chain in arbitrary dimensions. Phys. Rev. E. 2008;77:061803. doi: 10.1103/PhysRevE.77.061803. [DOI] [PubMed] [Google Scholar]
- 98.Miller W. Some applications of the representation theory of the Euclidean group in three-space. Commun. Pure Appl. Math. 1964;17:527–540. [Google Scholar]
- 99.Ming D, Kong YF, Lambert MA, Huang Z, Ma JP. How to describe protein motion without amino acid sequence and atomic coordinates. Proc. Natl Acad. Sci. 2002;99:8620–8625. doi: 10.1073/pnas.082148899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ming D, Kong YF, Wu YH, Ma JP. Substructure synthesis method for simulating large molecular complexes. Proc. Natl Acad. Sci. 2003;100:104–109. doi: 10.1073/pnas.232588999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Miyake A. Stiff-chain statistics in relation to the Brownian process. J. Phys. Soc. Japan. 1981;50:1676–1682. [Google Scholar]
- 102.Miyazawa S, Jernigan RL. Estimation of effective interresidue contact energies from protein crystal-structures—quasi-chemical approximation. Macromolecules. 1985;18:534–552. [Google Scholar]
- 103.Moroz JD, Nelson P. Torsional directed walks, entropic elasticity, and DNA twist stiffness. Proc. Natl Acad. Sci. USA. 1997;94:14418–14422. doi: 10.1073/pnas.94.26.14418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Moroz JD, Nelson P. Entropic elasticity of twist-storing polymers. Macromolecules. 1998;31:6333–6347. [Google Scholar]
- 105.Nelson P. Sequence-disorder effects on DNA entropic elasticity. Phys. Rev. Lett. 1998;80:5810. [Google Scholar]
- 106.Nelson P. New measurements of DNA twist elasticity. Biophys. J. 1998;74:2501. doi: 10.1016/S0006-3495(98)77958-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Niewierczerzam S, Cieplak M. Stretching and twisting of the DNA duplexes in coarse grained dynamical models arXiv:0901.0082v1. 2009. [DOI] [PubMed] [Google Scholar]
- 108.Norisuye T, Tsuboi A, Teramoto A. Remarks on excluded-volume effects in semiflexible polymer solutions. Polym. J. 1996;28:357–361. [Google Scholar]
- 109.Odijk T. Stiff chains and filaments under tension. Macromolecules. 1995;28:7016–7018. [Google Scholar]
- 110.Perrin F. Étude Mathématique du Mouvement Brownien de Rotation. Ann. Sci. L’ École Normale Supérieure. 1928;45:1–51. [Google Scholar]
- 111.Peter F, Weyl H. Die Vollständigkeit der primitiven Darstellungen einer geschlossenen kontinuierlichen Gruppe. Math. Ann. 1927;97:735–755. [Google Scholar]
- 112.Podtelezhnikov AA, Cozzarelli NR, Vologodskii AV. Equilibrium distributions of topological states in circular DNA: interplay of supercoling and knotting. Proc. Natl Acad. Sci. USA. 1999;96:12974. doi: 10.1073/pnas.96.23.12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pohl WF. The self-linking number of a closed space curve. J. Math. Mech. 1968;17:975. [Google Scholar]
- 114.Riccardi D, Schaefer P, Yang Y, Yu H, Ghosh N, Prat-Resina X, König P, Li G, Xu D, Guo H, Elstner M, Cui Q. Development of effective quantum mechanical/molecular mechanical (QM/MM) methods for complex biophysical processes. J. Phys. Chem. B. 2006;110:6458–6469. doi: 10.1021/jp056361o. [DOI] [PubMed] [Google Scholar]
- 115.Risbo T. Fourier transform summation of Legendre series and D-functions. J. Geodesy. 1996;70:383–396. [Google Scholar]
- 116.Sattinger DH, Weaver OL. Lie Groups and Algebras with Applications to Physics, Geometry, and Mechanics. New York: Springer; 1986. [Google Scholar]
- 117.Schiessel H, Rudnick J, Bruinsma R, Gelbart WM. Organized condensation of worm-like chains. Europhys. Lett. 2000;51:237–243. [Google Scholar]
- 118.Schmidt M, Stockmayer WH. Quasi-elastic light scattering by semiflexible chains. Macromolecules. 1984;17:509–514. [Google Scholar]
- 119.Schuyler AD, Chirikjian GS. Normal mode analysis of proteins: a comparison of rigid cluster modes and Cα coarse graining. J. Mol. Graph. Modelling. 2004;22:183–193. doi: 10.1016/S1093-3263(03)00158-X. [DOI] [PubMed] [Google Scholar]
- 120.Schuyler AD, Chirikjian GS. Efficient determination of low-frequency normal modes of large protein structures by cluster-NMA. J. Mol. Graph. Modelling. 2005;24:46–58. doi: 10.1016/j.jmgm.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 121.Schuyler AD, Jernigan RL, Qasba PK, Ramakrishnan B, Chirikjian GS. Iterative cluster-NMA (icNMA): a tool for generating conformational transitions in proteins. Prote. Struct. Function Bioinform. 2009;74:760–776. doi: 10.1002/prot.22200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shäfer L. Excluded Volume Effects in Polymer Solutions, as Explained by the Renormalization Group. New York: Springer; 1999. [Google Scholar]
- 123.Shi Y, He S, Hearst JE. Statistical mechanics of the extensible and shearable elastic rod and of DNA. J. Chem. Phys. 1996;105:714–731. [Google Scholar]
- 124.Shimada J, Yamakawa H. Statistical mechanics of DNA topoisomers. J. Mol. Biol. 1985;184:319–329. doi: 10.1016/0022-2836(85)90383-3. [DOI] [PubMed] [Google Scholar]
- 125.Shore D, Baldwin RL. Energetics of DNA twisting. J. Mol. Biol. 1983;170:957–981. doi: 10.1016/s0022-2836(83)80198-3. [DOI] [PubMed] [Google Scholar]
- 126.Simo JC, Vu-Quoc L. A three-dimensional finite-strain rod model. Part II: computational aspects. Comput. Methods Appl. Mech. Eng. 1986;58:79–116. [Google Scholar]
- 127.Sippl MJ. Knowledge-based potentials for proteins. Curr. Opin. Struct. Biol. 1995;5:229–235. doi: 10.1016/0959-440x(95)80081-6. [DOI] [PubMed] [Google Scholar]
- 128.Smith SB, Finzi L, Busmante C. Direct mechanical measurements of the elasticity of single DNA-molecules by using magnetic beads. Science. 1992;258:1122–1126. doi: 10.1126/science.1439819. [DOI] [PubMed] [Google Scholar]
- 129.Spakowitz AJ, Wang Z-G. End-to-end distance vector distribution with fixed end orientations for the wormlike chain model. Phys. Rev. E. 2005;72:041802. doi: 10.1103/PhysRevE.72.041802. [DOI] [PubMed] [Google Scholar]
- 130.Steigmann DJ, Faulkner MG. Variational theory for spatial rods. Arch. Ration. Mech. Anal. 1993;133:1–26. [Google Scholar]
- 131.Stepanow S. Kramer equation as a model for semiflexible polymers. Phys. Rev. E. 1996;54:R2209–R2211. doi: 10.1103/physreve.54.r2209. [DOI] [PubMed] [Google Scholar]
- 132.Sternberg S. Group Theory and Physics. New York: Cambridge University Press; 1994. [Google Scholar]
- 133.Strick TR, Allemand JF, Bensimon D, Bensimon A, Croquette V. The elasticity of a single supercoiled DNA molecule. Science. 1996;271:1835–1837. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 134.Sulkowska JI, Cieplak M. Mechanical stretching of proteins a theoretical survey of the protein data bank. J. Phys.: Condens. Matter. 2007;19:283201. [Google Scholar]
- 135.Swigon D, Coleman BD, Tobias I. The elastic rod model for DNA and its application to the tertiary structure of DNA minicircles in mononucleosomes. Biophys. J. 1998;74:2515–2530. doi: 10.1016/S0006-3495(98)77960-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tama F, Gadea FX, Marques O, Sanejouand YH. Building-block approach for determining low-frequency normal modes of macromolecules. Protein Struct. Function Genet. 2000;41:1–7. doi: 10.1002/1097-0134(20001001)41:1<1::aid-prot10>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 137.Tama F, Sanejouand YH. Conformational change of proteins arising from normal mode calculations. Protein Eng. 2001;14:1–6. doi: 10.1093/protein/14.1.1. [DOI] [PubMed] [Google Scholar]
- 138.Tama F, Wriggers W, Brooks CL. Exploring global distortions of biological macromolecules and assemblies from low-resolution structural information and elastic network theory. J. Mol. Biol. 2002;321:297–305. doi: 10.1016/s0022-2836(02)00627-7. [DOI] [PubMed] [Google Scholar]
- 139.Thirumalai D, Ha B-Y. Statistical mechanics of semiflexible chains: a mean field variational approach. In: Grosberg A, editor. Theoretical and Mathematical Models in Polymer Research. New York: Academic; 1998. pp. 1–35. [Google Scholar]
- 140.Tirion MM. Large amplitude elastic motions in proteins from a single-parameter, atomic analysis. Phys. Rev. Lett. 1996;77:1905–1908. doi: 10.1103/PhysRevLett.77.1905. [DOI] [PubMed] [Google Scholar]
- 141.Tirion MM, Ben-Avraham D. Normal mode analysis of G-actin. J. Mol. Biol. 1993;230:186–195. doi: 10.1006/jmbi.1993.1135. [DOI] [PubMed] [Google Scholar]
- 142.Tirion MM, Ben-Avraham D. Normal modes analyses of macromolecules. Physica A. 1998;249:415–423. [Google Scholar]
- 143.Tobias I, Swigon D, Coleman BD. Elastic stability of DNA configurations. I: general theory. Phys. Rev. E. 2000;61:747–758. doi: 10.1103/physreve.61.747. [DOI] [PubMed] [Google Scholar]
- 144.Trovato A, Seno F. A new perspective on the analysis of helix–helix packing preferences in globular proteins arXiv:cond-mat/0304429 v1. 2003. [DOI] [PubMed] [Google Scholar]
- 145.Varshalovich DA, Moskalev AN, Khersonskii VK. Quantum Theory of Angular Momentum. Singapore: World Scientific; 1998. [Google Scholar]
- 146.Vilenkin NJ, Akim EL, Levin AA. The matrix elements of irreducible unitary representations of the group of Euclidean three-dimensional space motions and their properties. Dokl. Akad. Nauk SSSR. 1957;112:987–989. (in Russian) [Google Scholar]; Vilenkin NJ, Klimyk AU. Representation of Lie Groups and Special Functions. 1–3. Dordrecht: Kluwer Academic; 1991. [Google Scholar]
- 147.Vologodskii AV, Anshelevich VV, Lukashin AV, Frank-Kamenetskii MD. Statistical mechanics of supercoils and the torsional stiffness of the DNA double helix. Nature. 1979;280:294–298. doi: 10.1038/280294a0. [DOI] [PubMed] [Google Scholar]
- 148.Vologodskii A. Topology and Physics of Circular DNA. Boca Raton, FL: CRC Press; 1992. [Google Scholar]
- 149.von Neumann J. Mathematical Foundations of Quantum Mechanics. Princeton, NJ: Princeton University Press; 1955. [Google Scholar]
- 150.Walther D, Springer C, Cohen FE. Helix–helix packing angle preferences for finite helix axes. Protein Struct. Funct. Genet. 1998;33:457–459. [PubMed] [Google Scholar]
- 151.Wang MD, Yin H, Landick R, Gelles J, Block SM. Stretching DNA with optical tweezers. Biophys. J. 1997;72:1335–1346. doi: 10.1016/S0006-3495(97)78780-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wang Y, Chirikjian GS. Error propagation on the Euclidean group with applications to manipulator kinematics. IEEE Trans. Robot. 2006;22:591–602. [Google Scholar]
- 153.Wang Y, Chirikjian GS. Nonparametric second-order theory of error propagation on the Euclidean group. Int. J. Robot. Res. 2008;27:1258–1273. doi: 10.1177/0278364908097583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Weiner PK, Kollman PA. AMBER: assisted model building with energy refinement. A general program for modeling molecules and their interactions. J. Comput. Chem. 1981;2:287–303. [Google Scholar]
- 155.Weyl H. The Theory of Groups and Quantum Mechanics. New York: Dover; 1950. [Google Scholar]
- 156.White JH, Bauer WR. Calculation of the twist and the writhe for representative models of DNA. J. Mol. Biol. 1986;189:329. doi: 10.1016/0022-2836(86)90513-9. [DOI] [PubMed] [Google Scholar]
- 157.Wiggins PA, Phillips R, Nelson PC. Exact theory of kinkable elastic polymers arXiv:cond-mat/0409003 v1. 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wigner EP. On unitary representations of the inhomogeneous Lorentz group. Ann. Math. 1939;40:149–204. [Google Scholar]
- 159.Wigner EP. Gruppentheorie und ihre Anwendungen auf die Quantenmechanik der Atomspektren. Braunschweig: Vieweg Verlag; 1931. [Google Scholar]; Griffin JJ. Group Theory and its Application to the Quantum Mechanics of Atomic Spectra. New York: Academic; 1959. (Engl. Transl.) [Google Scholar]
- 160.Wilhelm J, Frey E. Radial distribution function of semiflexible polymers. Phys. Rev. Lett. 1996;77:2581–2584. doi: 10.1103/PhysRevLett.77.2581. [DOI] [PubMed] [Google Scholar]
- 161.Winkler RG, Harnau L, Reineker P. Distribution functions and dynamical properties of stiff macromolecules. Macromol. Theory Simul. 1997;6:1007–1035. [Google Scholar]
- 162.Winkler RG. Analytical calculation of the relaxation dynamics of partially stretched flexible chain molecules: necessity of a wormlike chain descrption. Phys. Rev. Lett. 1999;82:1843–1846. [Google Scholar]
- 163.Wyllie G. Random motion and Brownian rotation. Phys. Rep. 1980;61:327–376. [Google Scholar]
- 164.Yamakawa H, Stockmayer WH. Statistical mechanics of wormlike chains. II. Excluded volume effects. J. Chem. Phys. 1972;57:2843–2854. [Google Scholar]
- 165.Yamakawa H. Helical Wormlike Chains in Polymer Solutions. Berlin: Springer; 1997. [Google Scholar]
- 166.Zandi R, Rudnick J. Constraints, histones, and 30-nm spiral. Phys. Rev. E. 2001;64:051918. doi: 10.1103/PhysRevE.64.051918. [DOI] [PubMed] [Google Scholar]
- 167.Zhao SR, Sun CP, Zhang WX. Statistics of wormlike chains. I. Properties of a single chain. J. Chem. Phys. 1997;106:2520–2529. [Google Scholar]
- 168.Želobenko DP. Compact Lie Groups and their Representations (Translations of Mathematical Monographs) Providence, RI: American Mathematical Society; 1973. [Google Scholar]
- 169.Zhou Y, Chirikjian GS. Conformational statistics of bent semiflexible polymers. J. Chem. Phys. 2003;119:4962–4970. [Google Scholar]
- 170.Zhou Y, Chirikjian GS. Conformational statistics of semi-flexible macromolecular chains with internal joints. Macromolecules. 2006;39:1950–1960. doi: 10.1021/ma0512556. [DOI] [PMC free article] [PubMed] [Google Scholar]


