SUMMARY
The normal aging process is a complex phenomenon associated with physiological alterations in the function of cells and organs over time. Although an attractive candidate for mediating transcriptional dysregulation, the contribution of epigenetic dysregulation to these progressive changes in cellular physiology remains unclear. In this study, we employed the genome-wide HELP assay to define patterns of cytosine methylation throughout the rat genome, and the LUMA assay to measure global levels of DNA methylation in the same samples. We studied both liver and visceral adipose tissue, and demonstrated significant differences in DNA methylation with age at >5% of sites analyzed. Furthermore, we showed that epigenetic dysregulation with age is a highly tissue-dependent phenomenon. The most distinctive loci were located at intergenic sequences and conserved non-coding elements, and not at promoters nor at CG-dinucleotide dense loci. Despite this, we found that there was a subset of genes at which cytosine methylation and gene expression changes were concordant. Finally, we demonstrated that changes in methylation occur consistently near genes that are involved in metabolism and metabolic regulation, implicating their potential role in the pathogenesis of age-related diseases. We conclude that different patterns of epigenetic dysregulation occur in each tissue over time and may cause some of the physiological changes associated with normal aging.
Keywords: aging, epigenetics, DNA methylation, tissue-specific, liver, visceral adiposity
INTRODUCTION
Aging is a complex phenomenon characterized by progressive loss of tissue homeostasis with decline of normal cellular functions and capacity for replication (Van Zant & Liang 2003; Chen 2004; Pelicci 2004; Sharpless & DePinho 2004; Campisi 2005). The liver exhibits age-dependent decay in overall structure, regenerative capacity, and function (Jackson et al. 1988; Schmucker 2005), while visceral fat depots expand with increasing age and contribute to the pathogenesis of late-onset diseases such as diabetes, dyslipidemias, and cardiovascular disease (Hayashi et al. 2003; Nieves et al. 2003; Carr et al. 2004; Huffman & Barzilai 2009). On both a tissue-specific and whole-organism level, aging is associated with accumulated genomic damage over time accompanied by progressive physiological decline (de Boer et al. 2002; Maslov & Vijg 2009).
Variation in an individual’s phenotypic age and lifespan is due in part to genetic influences (Gurland et al. 2004; Karasik et al. 2004; vB Hjelmborg et al. 2006), but epigenetics and the environment play major roles in determining physiological changes over a lifetime (Gartner 1990; Kennedy 2006; Whitelaw & Whitelaw 2006; Ordovas & Shen 2008; Vogt et al. 2008). Monozygotic twins show inherent epigenetic variability (Kaminsky et al. 2009) that increases with age (Fraga et al. 2005), a finding supported by longitudinal studies of DNA methylation in single individuals (Bjornsson et al. 2008). In animals, DNA methylation differences are acquired in multiple tissues with age (Golbus et al. 1990; Ahuja et al. 1998; Peng et al. 2001; Kwabi-Addo et al. 2007). Moreover, epigenetic dysregulation occurs in a locus-specific manner, with some hepatic genes showing age-dependent DNA hypermethylation, and other loci exhibiting no changes with age (Akintola et al. 2008; Jiang et al. 2008). It is now well-established that different tissues have specific patterns of epigenetic regulation (Song et al. 2005; Eckhardt et al. 2006b; Khulan et al. 2006; Lister et al. 2008; Yagi et al. 2008; Song et al. 2009), and there is some evidence that there exist genes whose methylation patterns do not change with age in liver but exhibit significant differences in methylation with age in other cell types, raising the possibility of a tissue-specific effect of age on the epigenome (Akintola et al. 2008).
In this study, we applied an epigenome-wide approach (the HELP assay (Khulan et al. 2006)) to interrogate DNA methylation patterns in aging F344*BN rats, and investigated whether significant changes in methylation status arise with increasing age. We used two tissues with distinct metabolic and cell growth characteristics (liver and visceral adipose tissue) to explore whether tissue-specific dysregulation of the epigenome occurs with age. We wanted to explore whether such changes were global and random, or whether certain loci emerged with specific patterns of hyper- or hypomethylation with age, and whether the patterns observed were globally or locally concordant between tissues.
RESULTS
Tissue-Specific Differences in DNA Methylation with Age
We performed a genome-wide analysis of cytosine methylation at almost 40,000 unique sites using the HELP assay (Khulan et al. 2006). We studied cytosine methylation in liver and adipose tissue isolated from young and old animals. Our interest in adipose tissue was prompted by our recent demonstration that removal of an adipose fat pad in young animals increases their longevity by ~30%, suggesting that the biology of adipose tissue maybe linked to aging, and could be in part the the explanation of how caloric restriction extends life (Muzumdar et al. 2008). Adipocytes have, however, a limited capacity to divide, so for comparison we chose liver as being representative of a more actively-dividing tissue that has been extensively studied for its changes with aging. We created four experimental groups: young liver (n=6), young adipose tissue (n=3), old liver (n=5), old adipose tissue (n=3). We confirmed overall experimental quality for each sample as described previously (Thompson et al. 2008). To detect global patterns of epigenomic change occurring with age, we compared one thousand randomly selected loci to generate a representative heatmap. This unsupervised clustering approach demonstrated that DNA methylation is globally comparable across all samples and tissues (not shown). Tissue-specific differences distinguishing liver and adipose tissue were present at ~5.5% of loci overall (using a Bonferroni correction for multiple testing with α=0.05).
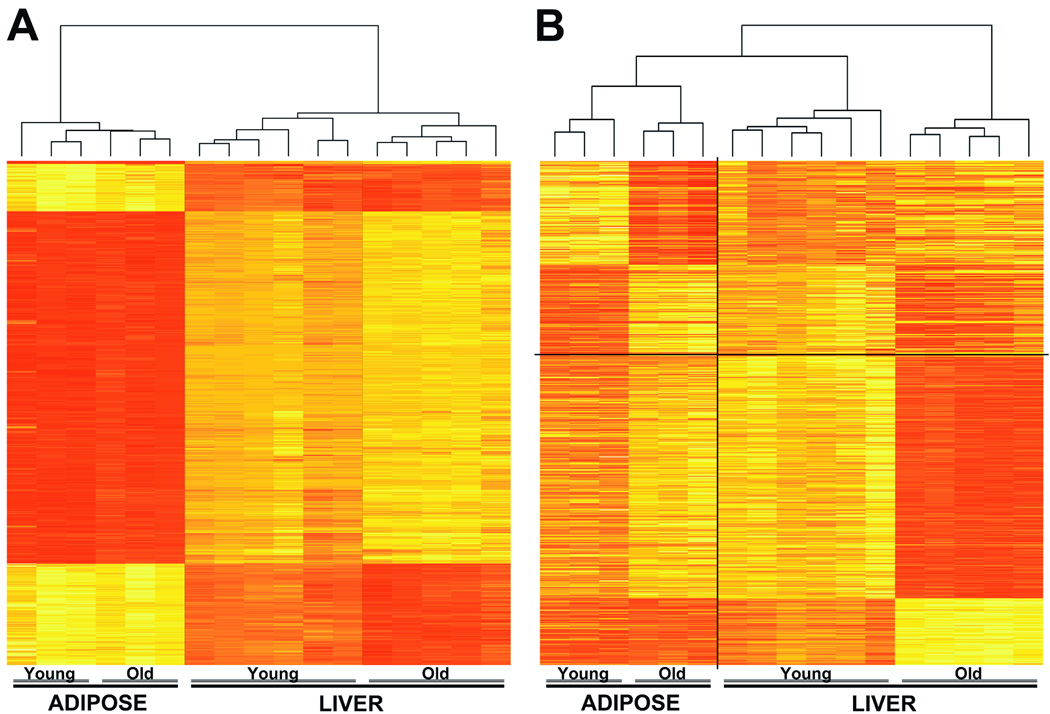
Each of the first three figures presents the results and analysis in complementary ways, with heatmaps for visualizing patterns of global DNA methylation (Figure 1), volcano plots for visualization of the statistical results (Figure 2) and the discordance from expected results demonstrated by a significance analysis of microarrays (SAM) study (Figure 3). We used the top 5% of loci at which tissue-specific differences in cytosine methylation had been identified for a subsequent unsupervised clustering analysis, revealing not only the clear differences between liver and adipose tissue but also age-related changes that appear to be more marked in liver than in fat (Figure 1A). As a complementary approach, we identified the loci at which the most marked changes in methylation are occurring with age and performed an unsupervised clustering analysis, again showing that the degree of difference of cytosine methylation due to tissue type exceeds that due to aging (Figure 1B).
Figure 1. Heatmap representation of global tissue-specific differences in DNA methylation in young and old animals.
(A) A heatmap of the top 5% of tissue-specific differences is shown, with each row corresponding to data from a single locus, and each column representing an organ sample (ADIPOSE and LIVER) obtained from a single rat (several young next to several old rats for each tissue). The branching dendrogram at the top represents the result of unsupervised clustering using these tissue-specific sites. Liver and adipose tissue show clear differences in methylation, with hyper- and hypomethylation shown on a continuum from red to yellow, respectively. While the profile of older adipose tissue is relatively similar to young, cytosine methylation in older liver tissue diverges more strongly from livers of young rats. (B) This panel shows a heatmap of the most significant age-related changes in DNA methylation identified in either liver (378 loci shown below the horizontal dividing line) or adipose tissue (240 loci shown above the horizontal dividing line). The loci included in this heatmap are identical to those identified in Figure 2 (red and blue datapoints). It is apparent that the large majority of changes in cytosine methylation with age occur distinctly in either liver or adipose tissue, and rarely in both tissues concordantly.
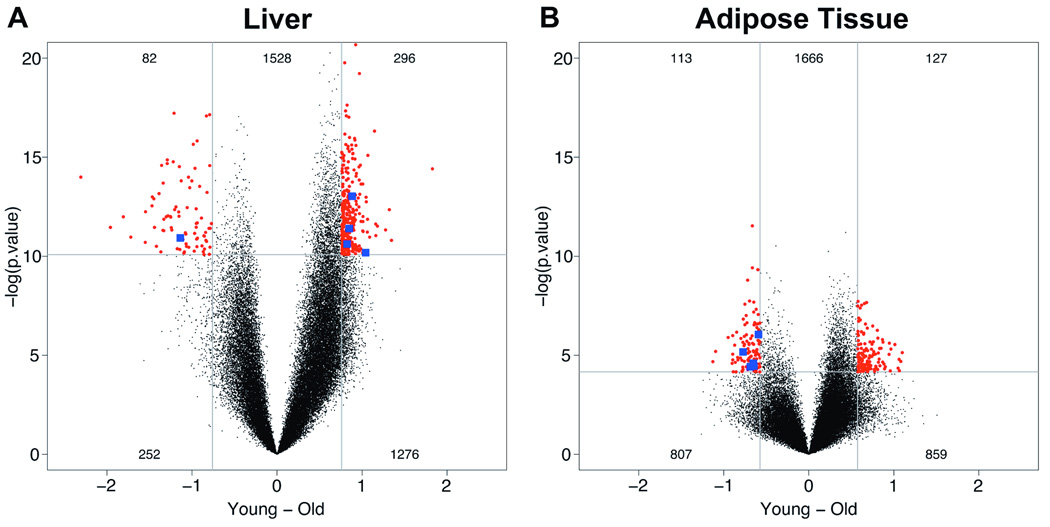
Figure 2. Volcano plot representation to assess age-related changes in global DNA methylation measured by HELP.
Volcano plots reveal differences in DNA methylation with age that are highly dependent upon the tissue in which they are found, liver (A) and adipose tissue (B). Every locus analyzed corresponds to a dot, showing the average differences between young and old (on x-axis, values below zero represent loss of methylation with increasing age, above zero hypermethylation with increasing age) with negative log-transformed significance (p-values) along the y-axis. Thresholds were determined from the magnitude of methylation or significance differences as ≥95th percentile of values. Red dots indicate those loci exceeding both the magnitude of difference and significance thresholds and therefore correspond with the most consistent differentially-methylated sites. Blue-labeled loci are those that exceed threshold values in both the liver and adipose tissue datasets.
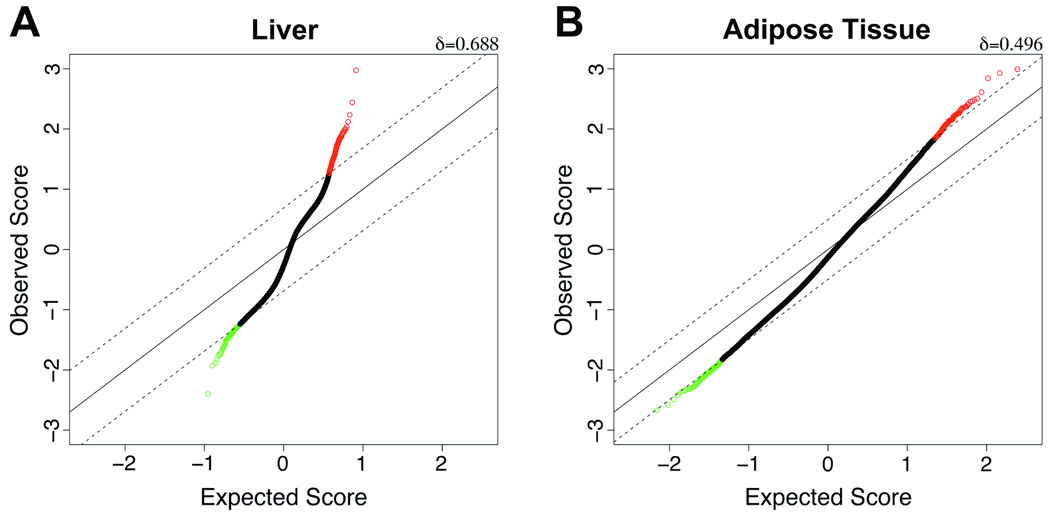
Figure 3. Significant age-related changes in DNA methylation in liver and adipose tissue.
Significance analysis of microarrays (SAM) was performed for young and old rats in two different tissues (liver and perinephric fat). (A) The panel shows a standard Q-Q plot with expected T statistic along the x-axis and observed T-statistic along the y-axis. Values were calculated using a two-class unpaired model comparing young and old liver data, with s0 (~0.59) automatically generated. The solid diagonal line indicates a 45-degree line of equivalent observed:expected ratios, and two dashed lines indicate thresholds of confidence corresponding to δ=0.688. Green and red datapoints represent significant hyper- and hypomethylation, with age, respectively. Thus, SAM identifies a large number of highly significant changes in DNA methylation that occur in liver specifically (380 total loci with estimated false discovery rate (FDR)<0.1%). (B) As before, a Q-Q plot generated from a comparison of young and old adipose tissue (two-class unpaired T statistic, s0~0.22) demonstrates the extent to which differences in DNA methylation occur beyond what one would expect to see by random chance alone (242 significant loci, δ=0.496). However, a direct comparison of both tissues demonstrates that the age-related changes observed in liver are much greater than those observed in perinephric fat, both in terms of their extent (380 in liver, with only 3 in fat, for δ=0.688 as shown in panel (A)) and significance (estimated FDR of 2.1% in liver compared to 46.1% in fat, for δ=0.496 as shown in panel (B)).
The distinct tissue-specific patterns observed using unsupervised clustering analysis of HELP data prompted us to investigate age-related differences in fat and liver separately. In Figure 2 we show the fat and liver-specific distributions of age-related methylation differences, respectively, along with the degree of significance associated with each of these changes (volcano plots). At stringent thresholds determining the most widely divergent and highly significant loci in each tissue, we find that liver has 50% more highly significant age-associated changes than visceral adipose tissue (378 compared with 240 sites, all of which are combined and shown as a heatmap in Figure 1B). Moreover, the degree, extent, and significance of age-related differences in liver are all much greater than the differences observed in perinephric fat (Figure 2), demonstrating that age preferentially induces epigenomic dysregulation in the liver with relatively fewer differences observed in adipose tissue. We confirmed these findings using the SAM approach (Tusher et al. 2001) (Figure 3). We estimate that the top 378 differentially methylated sites in liver (δ=0.688) are associated with a false discovery rate (FDR) less than 0.1%, indicating it is unlikely that any of these changes occur by chance alone, whereas the differences observed in visceral fat (240 identified with δ=0.496) are not comparably robust (FDR of 46.1%). Interestingly, the majority of epigenetic dysregulation in the liver skewed towards age-dependent hypermethylation (Figures 1B, 2A, and 3A).
We then asked whether any of the liver-specific and fat-specific differentially methylated loci were consistently dysregulated with age in both tissues. The large majority of changes appeared to be unique to each tissue (Figure 1B), with only five loci that were identified as differentially methylated in both tissues (Figure 2). However, contrary to any expectation that these sites may change methylation status concordantly, four out of the five loci become hypomethylated in fat but hypermethylated in the liver with increasing age (Figure 2, blue points). This observation is consistent with the overall tendency towards age-related hypermethylation in the liver (Figure 1B, 2A, and 3A).
Global Levels of DNA Methylation by Luminometric Methylation Assay
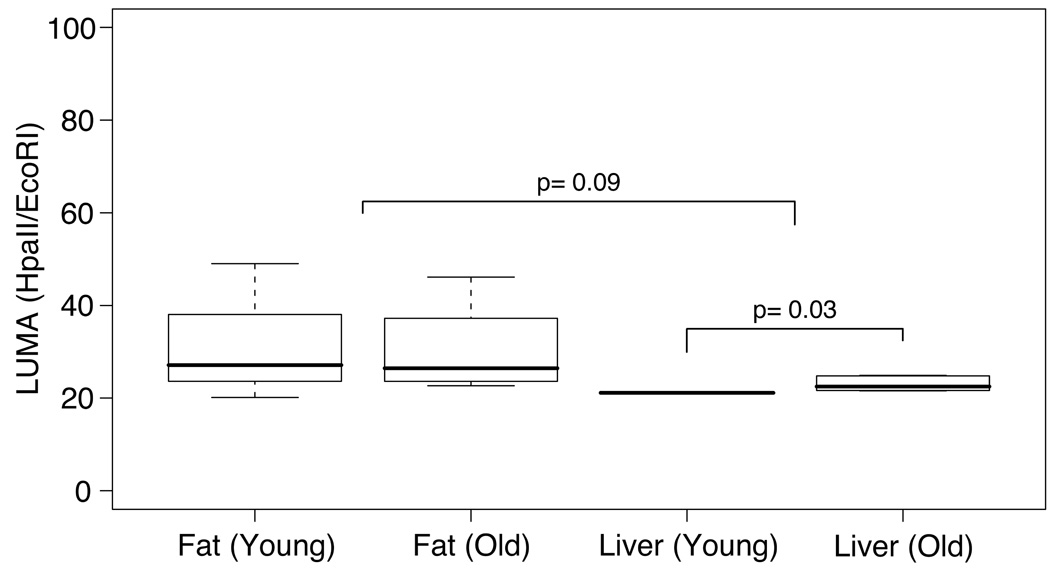
Using HELP, we demonstrated that liver exhibits global hypermethylation with age at the unique sequences interrogated using the microarray approach (Figure 2A), while adipose tissue shows no global tendency towards either hyper- or hypomethylation (Figure 2B). To test whether these changes reflect those of the genome as a whole, we measured global levels of DNA methylation in both tissues from young (age 3.2 ± 0.2 months) and old (age 18 months) F344*BN rats, a genetically homogeneous F1 hybrid strain, using the Luminometric Methylation Analysis (LUMA) assay (Karimi et al. 2006). As with HELP, this analysis revealed tissue-specific differences in methylation, with increased methylation in liver compared with visceral adipose tissue (p=0.09) (Figure 4). We estimated the percent of methylated HpaII sites by comparison of the data with corresponding MspI data, and found that 58.7±1.4% and 48.0±7.3% (± SEM) of HpaII sites were methylated in liver and fat, respectively. No aging-related effect on global DNA methylation levels was shown in adipose tissue, but we observed a small degree of hypomethylation in liver with age (p=0.03) (Figure 4). As LUMA should be more influenced by methylation status at repetitive elements than HELP (which predominantly samples unique sequences), the hypomethylation we observe is consistent with the demethylation and activation of specific repetitive elements seen in other aging rodents and in humans (Barbot et al. 2002; Bollati et al. 2009). The LUMA data therefore indicate that cytosine methylation changes in aging liver occur distinctively in different genomic sequence contexts.
Figure 4. LUMA as a technology to assess age-related changes in global DNA methylation.
DNA methylation was measured by the luminometric methylation assay (LUMA) in two tissues (Fat and Liver) from young and old rats. Each corresponding boxplot is a representation of the group-specific levels of methylation, shown along the y-axis (range from 0 to 100 with 0 indicating complete methylation). Within each boxplot, the solid black line indicates median methylation, with the upper and lower limits of each box corresponding to the 75% and 25% quantiles of the data. The bars associated with each box represent the extremes of the data. The distributions of methylation levels in each tissue were compared, demonstrating that liver tends to be more methylated than adipose tissue irrespective of age (p=0.09). Moreover, age-related differences were observed in liver, with relative hypomethylation in older animals (p=0.03). Note that p-values were obtained by two-group unpaired t-test.
Genomic Distributions of Cytosine Methylation
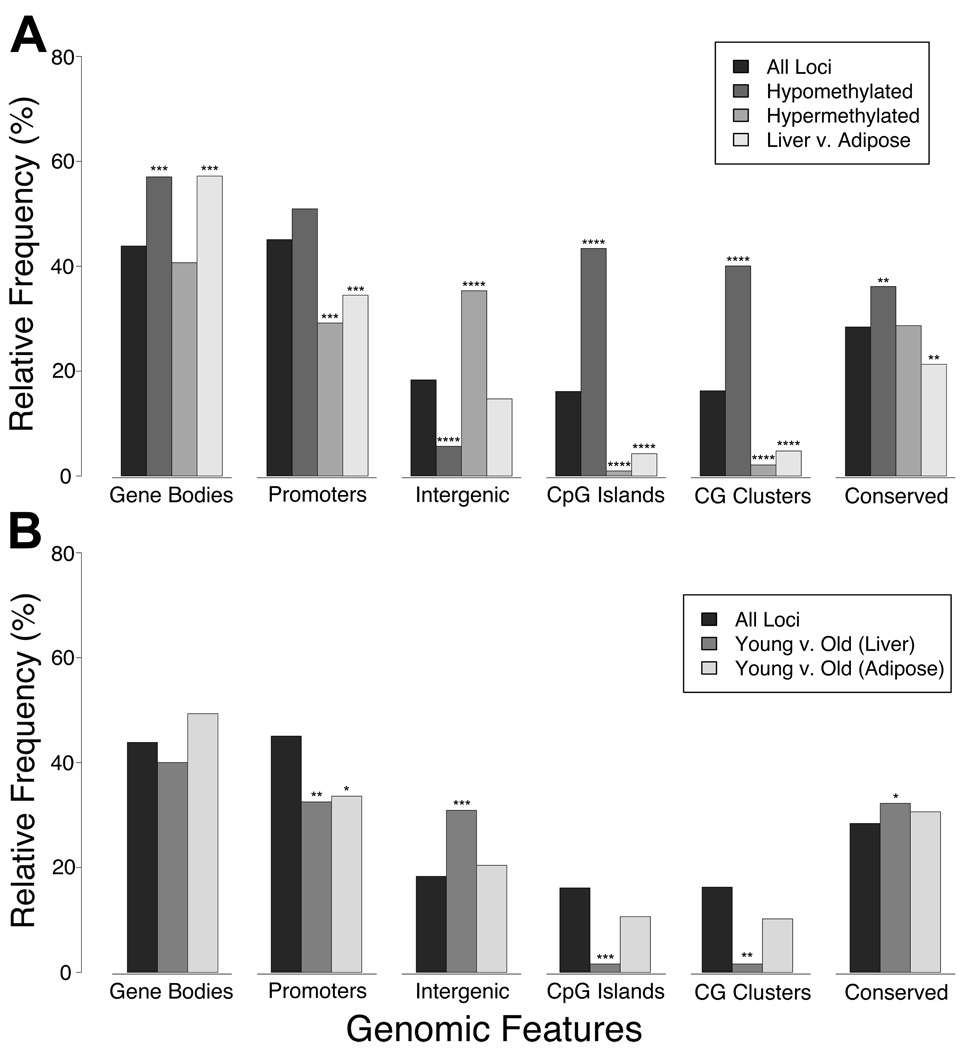
We therefore tested how our cytosine methylation data from the HELP experiments distributed by annotated DNA sequence features, studying both liver and adipose tissue at young and old timepoints. The data were partitioned into five non-overlapping subsets: 1) consistently hypomethylated sites in all animals; 2) consistently hypermethylated sites; 3) loci with tissue-specific differences in methylation; 4) loci with age-associated differences in methylation in liver; and 5) loci with age-associated differences in methylation in adipose tissue. The number of whole and partial sequence overlaps with CpG islands, CG clusters (Glass et al. 2007), conserved elements, repeat-masked sequences, gene bodies, and promoters (defined as the proximal 10 kb sequence upstream of RefSeq transcription start sites) were measured for each of these sets of loci (Supp. Table 2). The probability that each observed frequency could arise by random sampling of an equivalent number of loci from the array was calculated using the tailed hypergeometric distribution function (Supp. Table 2) (Johnson et al. 1992).
First we investigated the consistent patterns of cytosine methylation in both liver and adipose tissue, focusing on sites that are hypo- or hypermethylated in all samples. The epigenomic patterns we observed for these loci are concordant with prior expectations, with enrichment of hypomethylated sequences at CpG islands and CG clusters (Fig. 5A), a direct agreement with the generally hypomethylated nature of these CG-dense elements (Bird et al. 1985; Glass et al. 2007). Additionally, hypomethylated loci were enriched at promoters and conserved sequences, as well as in gene bodies (Fig. 5A). Repetitive elements by contrast tended to be hypermethylated (Supp. Table 2), as were intergenic sequences (Fig. 5A). Overall, these methylation patterns are typical of normal primary tissues from eutherian mammals.
Figure 5. Genomic distributions of DNA methylation in normal and aging tissues.
HELP data were divided into five different subsets representing constitutively hypomethylated loci, constitutively hypermethylated loci, and tissue-specific differentially-methylated regions (DMR, Liver compared with Adipose) in panel (A), as well as age-related DMRs (Young compared with Old) specific to either liver or adipose tissue in panel (B). Overlap of these subsets with six different genomic features (gene bodies, promoters including 10 kb upstream of transcription start sites, intergenic regions, CpG islands, CG clusters, and conserved non-coding elements) was measured and is shown from left to right in both panels. We also show genomic distributions for the whole microarray (All Loci, both panels), and are thus able to determine if a given subset of HELP data is enriched or depleted for any given genomic feature beyond what one might expect to see by random chance. Many of the differences shown are associated with negligible probability that they might occur with random sampling of the data (tailed hypergeometric distribution; *, **, ***, and **** indicate P<1%, P<0.01%, P<0.00000001%, and P~0, respectively) (Supp. Table 2).
We next investigated how these normal patterns of cytosine methylation differ between tissues. When we studied ~1,000 of the loci most distinctively methylated between liver and adipose tissue, (p<0.0001 and |liver – adipose| > 1.5), we found them to be highly depleted at CpG islands, CG clusters, and to a lesser extent, promoters and conserved sequences, but most strikingly enriched at gene bodies (Fig. 5A), which may be the consequence of transcriptional differences in gene expression between the two tissues (Zilberman et al. 2007; Ball et al. 2009).
Finally, we tested where age-related changes in methylation were occurring in each tissue. Overall, liver generated more significant data than adipose tissue, with a significant enrichment in dysregulated loci at intergenic and conserved sequences, and under-representation of dysregulated loci at promoters and CG-dense sequences (Fig. 5B). Similarly, conserved non-coding elements tend to be enriched among age-related differences in methylation in both tissues (Fig. 5B). The cytosine methylation data from these analyses are provided as a UCSC genome browser track available as an open-access resource at http://greallylab.aecom.yu.edu/ratAgeing/, and through the GEO database (accession number GSE17332).
Quantitative Validation of DNA Methylation States
We confirmed our HELP results with a quantitative validation approach, bisulphite MassArray (Ehrich et al. 2005), testing four loci representing pairs of constitutively-hypomethylated and constitutively-methylated sites identified by the HELP assay, and three loci at which tissue-specific differences in methylation were observed, two of which were hypomethylated and one of which was hypermethylated in liver compared to adipose tissue. Supplementary Figure 1 shows the expected inverse correlation between methylation values determined independently for these loci by HELP and MassArray, confirming our ability to discriminate methylation status in these samples, defining our hypermethylated category as greater than approximately 60% methylation, and hypomethylation as lower values.
Transcriptional profiling reveals concordant changes in gene expression at epigenetically-dysregulated loci
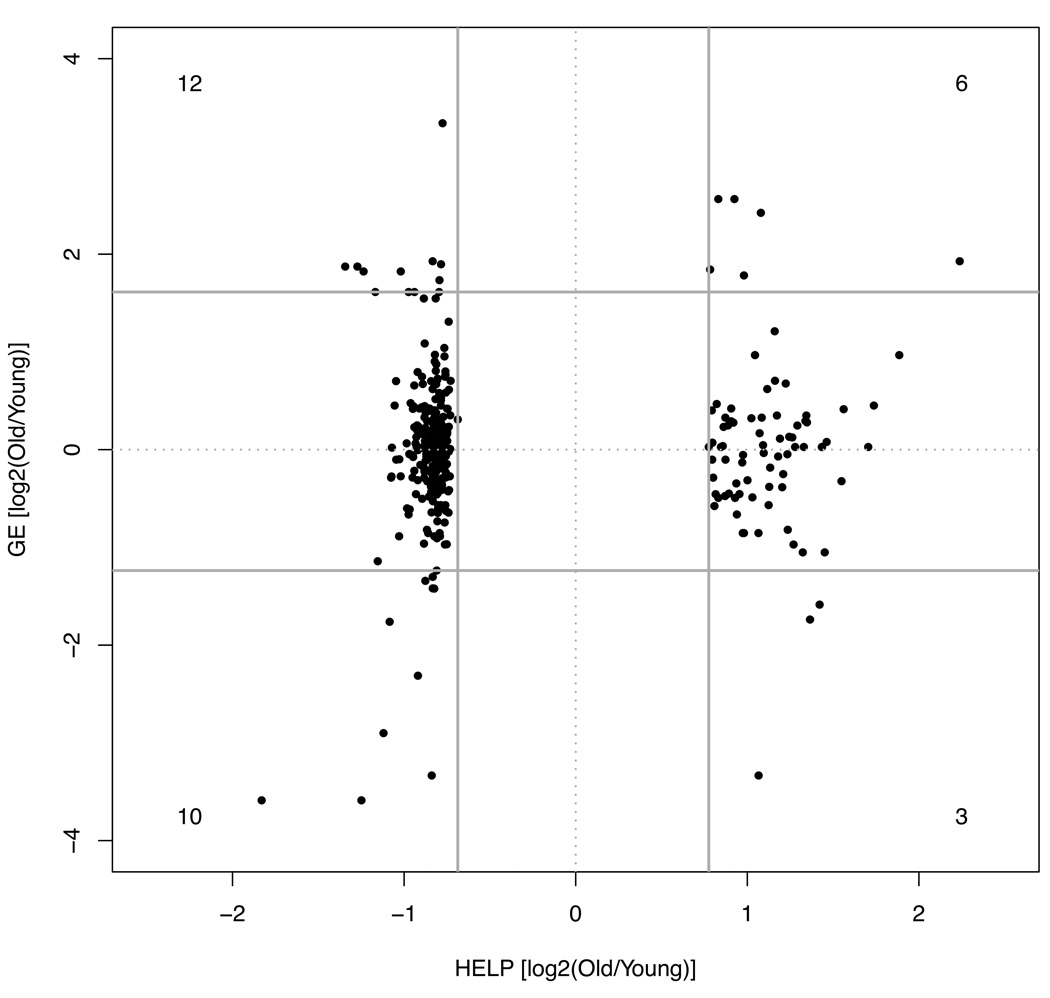
Gene expression studies were performed in liver tissue using a long oligonucleotide microarray approach, identifying genes with robust differences in expression with age. Our question was whether the epigenetic changes we observed, frequently located at non-promoter regions, were associated with changes in gene expression, implying a functional consequence to the underlying epigenetic dysregulation. We therefore compared loci with the most robust changes in cytosine methylation to corresponding gene expression differences with age. As demonstrated in Figure 6, a number of loci (31) exhibit robust changes in both DNA methylation and gene expression. A detailed description of these loci can be found in Table 1.
Figure 6. Identification of loci at which cytosine methylation and gene expression are both altered with aging.
We focused on the loci identified in Figure 2A, plotting HELP data along the x-axis and corresponding gene expression data for these loci along the y-axis. A total of 378 loci with significant differences in cytosine methylation were analyzed, 347 of which were mapped to RefSeq genes, and 31 of which demonstrated a corresponding robust difference in gene expression with age (> 2.5 SD from the mean of the overall distribution of gene expression data). Solid gray horizontal lines indicate the 2.5 SD cutoffs for these gene expression data, while solid gray vertical lines indicate the 95th percentile (approximately 2 SD) cutoffs for cytosine methylation data. Numerical labels appear in each of the four corners of the plot, corresponding to the number of datapoints meeting the defined criteria.
TABLE 1.
Loci at which cytosine methylation and gene expression both significantly dysregulated with aging.
| ACCESSION NUMBER |
GENE NAME |
GE [log2(old/y oung)] |
METH [log2(old/yo ung)] |
LOCATION [relative to TSS] |
DESCRIPTION |
|---|---|---|---|---|---|
| NM_012493 | Afp | − 3.5878254 13 |
−1.8301945 | Promoter | chloride intracellular channel 6 |
| NM_012493 | Afp | − 3.5878254 13 |
−1.2487887 | Promoter | chloride intracellular channel 6 |
| NM_031511 | Igf2 | − 3.3338100 79 |
− 0.839196979 |
25 kb downstream |
Insulin-like growth factor II |
| NM_031511 | Igf2 | − 3.3338100 79 |
1.06576529 | 22 kb downstream |
Insulin-like growth factor II |
| NM_012488 | A2m | − 2.9002281 63 |
− 1.120114809 |
Promoter | alpha-2-macroglobulin precursor |
|
NM_001013 083 |
Cpa2 | − 2.3129951 92 |
−0.919576 | 17 kb downstream |
carboxypeptidase A2 |
|
XM_001064 308 |
LOC68 5560 |
− 1.7613548 36 |
−1.084331 | 49 kb downstream |
similar to monoacylglycerol O- acyltransferase 2 |
|
NM_001024 369 |
Ypel4 | − 1.7382221 66 |
1.3655237 | 30 kb downstream |
yippee-like 4 |
|
XM_001080 259 |
E2f8 | − 1.5854266 39 |
1.4213172 | 24 kb downstream |
E2F transcription factor 8 |
|
XM_001070 878 |
Stfa2 | − 1.4214691 9 |
−0.824367 | 36 kb downstream |
stefin A2 |
| NM_053702 | Ccna2 | − 1.4193500 43 |
−0.832823 | 8 kb downstream |
cyclin A2 |
|
NM_001008 804 |
Krt10 | − 1.3422105 27 |
−0.876206 | 10 kb downstream |
type I keratin KA10 |
|
NM_001012 167 |
Pld3 | − 1.3017941 88 |
−0.833135 | 29 kb upstream | phospholipase D3 |
| NM_133547 |
Sult1c 2 |
3.3397381 99 |
−0.775869 | 38 kb upstream | sulfotransferase family, cytosolic, 1c |
| NM_053288 | Orm1 | 2.5637637 72 |
0.8309709 | Gene body | alpha-1-acid glycoprotein |
| NM_053288 | Orm1 | 2.5637637 72 |
0.9248454 | Gene body | alpha-1-acid glycoprotein |
| NM_177426 | Gstm2 | 2.4230501 84 |
1.0789278 | Gene body | Glutathione-S- transferase mu type 2 |
| NM_134350 | Mx2 | 1.9276835 46 |
−0.833708 | Gene body | myxovirus resistance 2 |
| NM_134350 | Mx2 | 1.9276835 46 |
2.237653 | 27 kb downstream |
myxovirus resistance 2 |
|
NM_001042 619 |
Hsd3b | 1.8970910 74 |
−0.78504 | Gene body | 3 beta-hydroxysteroid dehydrogenase |
| NM_053922 | Acacb | 1.8731377 25 |
− 1.343365164 |
Gene body | acetyl-Coenzyme A carboxylase 2 |
| NM_053922 | Acacb | 1.8731377 25 |
− 1.272142911 |
Gene body | acetyl-Coenzyme A carboxylase 2 |
| NM_173093 |
Cyp2d 13 |
1.8429318 03 |
0.7834656 | 12 kb downstream |
Cytochrome P450, subfamily IID3 |
| NM_130414 | Abcg8 | 1.8231229 78 |
−1.23628 | Gene body | ATP-binding cassette sub-family G, member 8 |
| NM_130414 | Abcg8 | 1.8231229 78 |
−1.0194304 | Gene body | ATP-binding cassette sub-family G, member 8 |
| NM_175578 | Rcan2 | 1.7824881 35 |
0.9803631 | 40 kb downstream |
regulator of calcineurin 2 |
| NM_052798 |
Zfp354 a |
1.7338909 21 |
−0.793965 | 45 kb upstream | zinc finger protein 354A |
| NM_031721 | Prss11 | 1.6127511 68 |
−1.167703822 | Gene body | protease, serine, 11 (Igf binding) |
| NM_031721 | Prss11 | 1.6127511 68 |
−0.97389659 | Gene body | protease, serine, 11 (Igf binding) |
| NM_031721 | Prss11 | 1.6127511 68 |
−0.939681764 | Gene body | protease, serine, 11 (Igf binding) |
| NM_031721 | Prss11 | 1.6127511 68 |
−0.796195907 | Gene body | protease, serine, 11 (Igf binding) |
|
Positive value: increased expression level in older animal |
Positive value: less methylated in older animal |
TSS: transcription start site.
GE: gene expression, microarray data
METH: methylation, HELP data
Molecular Interaction Network Analysis of Genes Associated with Age-Related Epigenetic Dysregulation
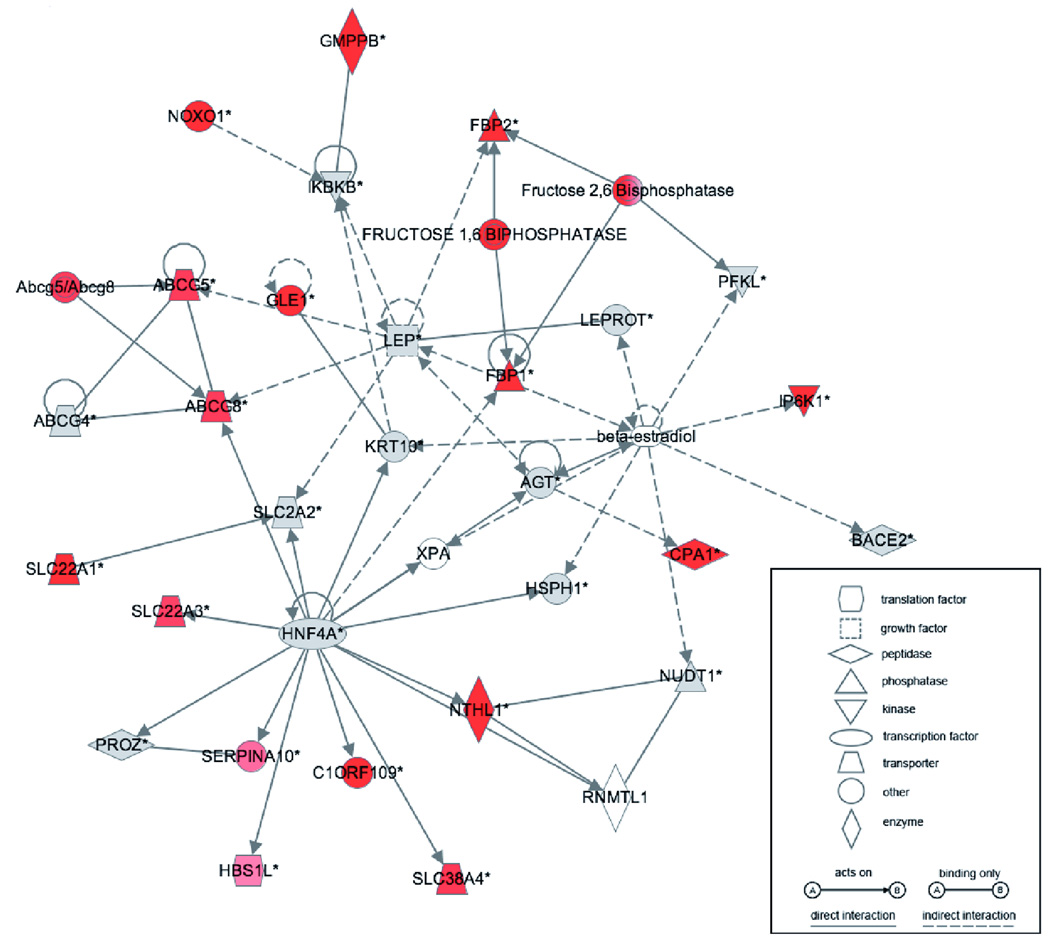
Using Ingenuity Pathway Analysis (IPA) software (Redwood City, CA), we carried out a network analysis to investigate whether genes associated with age-related epigenetic dysregulation shared any common functional roles or relationships. We linked loci with age-associated dysregulation of cytosine methylation to RefSeq genes if they mapped within 10 kb upstream of the transcription start site or anywhere within the body of the gene. We performed a filtered Ingenuity Pathway Analysis (IPA) on 102 genes (exclusively representing changes identified in liver, p<0.0000001) from an input list of 65,000 genes (p<0.37842) which provided a dataset-specific context for the enrichment analysis. Twenty-seven of the 102 input genes were removed from consideration as IPA possessed insufficient information to include them in the network analysis. The remaining 75 genes (Supp. Table 3) associated most strongly with a functional network relevant to metabolism (Figure 7). Among the nodes central to this network are Hnf4a and Leptin, the first being highly important in liver development and function (Duncan et al. 1994; Odom et al. 2004; Rhee et al. 2006), and the second an important adipokine mediator between visceral fat and liver metabolism (Szanto & Kahn 2000; Fishman et al. 2007). IPA further revealed that these 75 genes that have the most consistent epigenetic dysregulation with age are significantly enriched for functions in lipid metabolism (p<0.0001) and metabolic disease (p<0.0001).
Figure 7. Ingenuity Pathway Analysis (IPA) reveals that many of the most differentially-methylated genes form a molecular interaction network of particular relevance for metabolism.
RefSeq identifiers for 75 of the top 102 sites (filtered from an input list of 65,000, by p<0.0000001) that mapped to gene bodies and promoters were analyzed using the “Core Analysis” tool of IPA. This molecular interaction network was constructed with 35 nodes, 19 of which were among the top differentially methylated sites (red nodes). In alphabetical order, this network consists of ABCG4, ABCG5, ABCG8, Abcg5/Abcg8, AGT, BACE2, β-estradiol, C1ORF109, CPA1, FBP1, FBP2, Fructose 1,6 Bisphosphatase, Fructose 2,6 Bisphosphatase, GLE1, GMPPB, HBS1L, HNF4A, HSPH1, IKBKB, IP6K1, KRT10, LEP, LEPROT, NOXO1, NTHL1, NUDT1, PFKL, PROZ, RNMTL1, SERPINA10, SLC22A1, SLC22A3, SLC2A2, SLC38A4, XPA.
DISCUSSION
We investigated the effects of age on cytosine methylation throughout the genome using a genome-wide assay of a type not previously exploited in the study of aging metabolically-active tissues such as liver and visceral fat. This study represents one of the first such genome-wide studies of the aging epigenome, and is the first to demonstrate not only genome-wide but also locus-specific differences in cytosine methylation in both liver and adipose tissue. We find that normal aging in genetically identical rats exposed to the same environment throughout life causes consistent tissue-specific dysregulation of cytosine methylation, and that these changes skew globally towards hypermethylation of unique sequences in the liver. This increased methylation is accompanied by hypomethylation at a smaller set of loci, and less pronounced effects in perinephric visceral fat. The epigenomic dysregulation appears to be non-random in terms of genomic sequence context, preferentially affecting specific loci and genomic compartments such as intergenic and conserved sequences. These epigenetic changes may be adaptive and secondary to other alterations in cellular physiology, in which case they represent potential biomarkers of the aging process and indicators of the heterogeneity of the pathophysiology of aging between tissues. However, as many of these changes occur in proximity to genes with well-established roles in metabolism and metabolic dysregulation, epigenomic dysregulation is a clear candidate for being a primary mediator of the pathogenesis of age-related metabolic disease.
The extremely limited overlap that we observe between liver- and fat-specific epigenetic dysregulation is fundamentally due to the limited epigenetic variability observed in fat with age (Figures 3, 4), and results in an extremely small number of loci that are dysregulated in both tissues with age. Identification of loci that are jointly dysregulated in different tissues with age may suggest a common age-response mechanism operating in both tissues, but our results do little to shed light on such a mechanism with such a small number of loci involved and the high false-positive rate in adipose tissue in the current study. Furthermore, the preponderance of unique changes in each tissue suggests that a common mechanism might explain only a part of the story. Instead, we hypothesize that varying tissue environments with age, as well as varying mitotic activity and cellular susceptibilities to accumulated damage, are the hallmarks of tissue-specific epigenomic dysregulation with age.
Why liver as opposed to fat should be subject to large epigenomic changes (Figures 3, 4) is somewhat counterintuitive, given that visceral fat is centrally involved in the pathogenesis of age-related diseases (Muzumdar et al. 2008), and our prior expectation was that adipose tissue would have greater potential for epigenetic dysregulation due to the proximity of the nuclear DNA in adipocytes to free fatty acid flux and the accumulation of tissue macrophages, a phenomenon typical of aging (Einstein et al. 2008). However, the tissue-specific epigenetic differences may be related to the distinct cell proliferation properties of the tissues, analogous to the distinct tissue-specific DNA mutational rates previously observed in brain and small intestine (Busuttil et al. 2007). As liver is a relatively highly proliferative tissue type (Duncan et al. 2009), it may be more susceptible than adipose tissue to the accumulation of mutations, not just those of DNA but also those of epigenetic organization, or ‘epimutations’. DNA replication involves the propagation of cytosine methylation patterns to daughter chromatids, with restoration of symmetrical methylation from an initially hemimethylated state by DNA methyltransferase 1 (DNMT1), thus preserving the pattern of methylation present in the parental cell. However, DNMT1 has measurable de novo methylation activity and an estimated error rate of 0.3–5% (Vilkaitis et al. 2005; Goyal et al. 2006), thus epimutations are likely to occur with each cell division. A prior study has indicated that less mitotically-active cell types may be less prone to age-associated changes in cytosine methylation (Chu et al. 2007), results concordant with our data. The age-related epigenomic dysregulation that arises in a non-dividing cell type such as mature adipocytes (Neese et al. 2002) is more likely to reflect changes that occur within a single cell’s lifespan such as DNA repair-mediated loss of methylation (Barreto et al. 2007; Meulle et al. 2008).
While the locus-specific changes we observed are potentially valuable insights into the pathophysiology of aging, it is the striking tissue-specificity we observe that represents the most novel finding of our study. Because of these findings, we propose that epigenomic dysregulation in aging should be studied in a tissue-specific context, and that the lessons learned from one tissue cannot necessarily be applied to other tissues. The failure of a large-scale, quantitative study of cytosine methylation to find changes in DNA methylation with age (Eckhardt et al. 2006a) may be due to their measurement of age-related differences as average values across the many loci and tissue types they studied, whereas our results indicate that a locus and cell type-specific approach to the same dataset may yield different results. The changes we observe in our study are unlikely to be due to a random pattern of loss of epigenetic regulation, as might be concluded from a study of age effects on the epigenomes of twins (Fraga et al. 2005). Indeed, with an estimated false discovery rate of about 2.5% in liver and 21.3% in fat (δ=0.5), the epigenetic dysregulation we observe with age is likely due to reproducible, non-random biological differences.
The changes in methylation that we observed included but were not limited to promoter-proximal loci, making it difficult to predict whether these changes would have any consequences in terms of local transcription. We addressed this question by performing gene expression microarray studies on young and old rat livers, demonstrating a subset of loci at which changes were concordant. The role of intergenic loci in transcriptional regulation remains difficult to assess, but our data indicate that at least some such loci are potentially cis-reguatory and involved in cellular aging. Studies of the aging epigenome should not therefore be limited to promoter regions but should include other genomic contexts also.
We have to consider the possibility that fat- and more notably liver-specific dysregulation could be a product of a controlled tissue-specific response to accumulated stress with aging. This idea is supported by the results from our ontological and pathway analyses, which showed the enrichment of physiologically relevant loci in functional networks associated with metabolism and age-related diseases. The extremely limited overlap that we observe between liver- and fat-specific epigenetic dysregulation (Figure 3) suggests that each tissue responds differently to the damage and physiological insults that accumulate with age, with the potential additional contribution of distinct cellular environments during the aging process. We hypothesize that a combination of differences in mitotic activity, tissue environments and cellular susceptibilities to accumulated damage, define the reasons for cell type-specific differences in epigenomic dysregulation with age.
While this study provides us with new insights into the contribution of epigenetic dysregulation in aging, many questions remain unanswered. In prioritizing future directions, it is clear that the study in isolation of one epigenetic regulatory mechanism such as cytosine methylation is of less value than integrative studies of chromatin organization and gene expression, with orthogonal, quantitative single-locus studies to validate results at individual loci. Our results highlight the importance of testing a range of tissues and taking into account their replicative characteristics when assessing results. A focus on stem cells for analysis has the potential for greater insights than using differentiated cells, but carries the inherent problem of limited cell numbers for these genome-wide assays.
We conclude that the epigenomic dysregulation associated with aging is non-random and highly tissue-specific. Genome-wide assays focused on promoters or CG dinucleotide-dense regions would have failed to identify many of the changes we have found in this study, emphasizing the need for unbiased studies of the genome when exploring the role of cytosine methylation and the many other regulators of the epigenome in aging. More comprehensive studies of the epigenome integrated with transcriptomic assays performed on individual tissues has significant potential for identifying genes and pathways that are the targets for modification with aging, and thus insights into this aspect of the pathophysiology of aging in humans.
EXPERIMENTAL PROCEDURES
Animals
Young (3 months old, n=6) and old (18 months old, n=6) male Fischer 344/Brown Norway F1 Hybrid (F33XBN) rats (Harlan Worldwide, Somerville, NJ) were housed in individual cages and were subjected to a standard light (6:00 a.m. to 6:00 p.m)-dark (6:00 p.m to 6:00 a.m.) cycle. All rats were fed ad libitum using regular rat chow that consisted of 64% carbohydrate, 30% protein, and 6% fat with a physiological fuel value of 3.3 kcal/g chow. They were chronically catheterized 1 week before the study, recovered and were euthanized rapidly (by pentobarbital sodium, 60 mg/kg body weight intravenously) when unstressed and conscious in order to avoid prolonged severe stress that might potentially affect epigenetic characteristics (Barzilai et al. 1998). The abdomen was quickly opened, and adipose and liver tissues were freeze-clamped in situ with aluminum tongs pre-cooled in liquid nitrogen (Rossetti & Giaccari 1990). The study protocol was reviewed and approved by the Animal Care and Use Committee of the Albert Einstein College of Medicine.
HELP Assay
HELP (HpaII tiny fragment Enrichment by Ligation-mediated PCR) assays were performed as described previously (Khulan et al. 2006). High molecular weight genomic DNA was isolated from liver and perinephric fat tissues of young and old rats, digested to completion by HpaII and by MspI separately, and then amplified by ligation-mediated PCR (LM-PCR). Following PCR, the HpaII and MspI representations were labeled with different fluorophores using random priming and were cohybridized on a customized genomic microarray representing HpaII/MspI fragments of 200–2,000 bp in unique sequence. This microarray was designed specifically to target the 5’ regions of all known RefSeq genes in the rat genome as well as imprinted regions and a number of gene bodies, until the number of probes on the array was filled to the ~380,000 capacity.
HELP Microarray Data Analysis
Microarray data were pre-processed and subject to quality control and quantile normalization as previously described (Thompson et al. 2008). HpaII/MspI ratio values were subsequently normalized by RMA (Irizarry et al. 2003) for each of four distinct subgroups of rats/tissues: young liver, young fat, old liver, old fat (n=6, n=3, n=5, and n=3, respectively). This extra analytical step was used for inter-array normalization and to reduce within-group variability, ensuring a more conservative approach to interpretation of our data. Changes in methylation state were defined using a HpaII/MspI ratio threshold of zero, where methylated loci and hypomethylated loci had ratio values less than zero and greater than zero, respectively.
MassArray Validation
Target regions were amplified by PCR using the primers and cycling conditions described in Supplementary Table 1. Primers were selected with MethPrimer (http://www.urogene.org/methprimer/) using parameters as follows: 250–450 bp amplicon size, 56–60°C Tm, 24–30 bp length, and ≥1 CG in product. 50 µl PCR reactions were carried out using the Roche FastStart High Fidelity Kit. In cases where products showed primer-dimer or other contaminants, the bands of appropriate predicted size were excised from 2% agarose gels, purified by the Qiagen Gel Extraction Kit, and eluted with 1X Roche FastStart High Fidelity Reaction Buffer (+MgCl2). All PCR products (5 µl) were aliquotted onto 384-well microtiter plates and were treated with 2 µl Shrimp Alkaline Phosphatase (SAP) mix for 20 minutes at 37°C to dephosphorylate unincorporated dNTPs. Microtiter plates were processed by the MassARRAY Matrix Liquid Handler. A 2 µl volume of each SAP-treated sample was then heat-inactivated at 85°C for 5 minutes and subsequently incubated for 3 hours at 37°C with 5µl of Transcleave mix (T or C Cleavage Mix) for concurrent in vitro transcription and base-specific cleavage. Samples were transferred onto the spectroCHIP array by nanodispensation calibrated to ambient temperature and humidity, and analysis with the Sequenom MALDI-TOF MS Compact Unit following 4-point calibration with oligonucleotides of different mass provided in the Sequenom kit. Matched peak data were exported using EpiTYPER software and analyzed for quality and single nucleotide polymorphisms according to analytical tools that we have developed (Thompson et al. 2009).
Luminometric Methylation Assays
This protocol was adapted from that previously described (Karimi et al. 2006). Genomic DNA (1 µg) was cleaved with HpaII + EcoRI or MspI + EcoRI (5 µl each enzyme) in two separate 200 µl reactions containing 20 µl NEB buffers 1 and 2, respectively. The reactions were incubated at 37°C overnight, and purified by phenol-chloroform extraction and isopropanol precipitation, resuspended in 20 µl H2O. 20 µl of annealing buffer (20 mM Tris-acetate, 2 mM Mg-acetate pH 7.6) was added to the purified cleavage reactions, and samples were placed in a PSQ96™MA system (Biotage AB, Uppsala, Sweden). The instrument was programmed to add dNTPs in twelve consecutive steps: 1) dTTP, 2) dGTP, 3) dATP, 4) dCTP, 5) dATP, 6) dCTP, 7) dTTP, 8) dGTP, 9) dTTP, 10) dGTP, 11) dATP, and 12) dCTP. Peak heights were calculated using the PSQ96™A software. The HpaII/EcoRI and MspI/EcoRI ratios were calculated for each respective reaction as follows:
Gene expression microarray assays and analysis
Total RNA isolated from livers of rats of 2 weeks and 10.5 months of age was isolated and purified using Qiagen RNAEasy kit. The RNA was converted to cDNA and to dsDNA using the Superscript Double Stranded cDNA kit. These samples were labeled in our institutional Epigenomics Shared Facility using the Roche-Nimblegen One Color DNA labeling kit and co-hybridised in pairs to a rat gene expression microarray (Roche-Nimblegen design 090901 Rat HX12 expr HX12) which represents 26,419 genes with five long (60 nt) oligonucleotides per gene. The microarrays were scanned and analyzed using the Nimblegen Hybridization System. Genes showing robust differences in expression with age were identified following RMA normalization, and classified as those genes exhibiting a >2.5 SD increase or decrease in age-related expression beyond the average difference observed with age. Integration of this dataset with the HELP microarray dataset was performed by a genome-wide mapping approach, wherein all methylation loci found between 50 kb upstream and 50 kb downstream of the gene body were linked to the containing gene.
Ingenuity Pathway Analysis
The most significantly differentially methylated loci were mapped to RefSeq gene identifiers by chromosomal position (i.e. within 10 kb upstream of the transcription start site, or overlapping the gene body). The list of RefSeq identifiers was then uploaded to the Ingenuity Pathway Analysis program (Redwood City, CA), enabling exploration of ontology and molecular interaction networks. Each uploaded gene identifier was mapped to its corresponding gene object (focus genes) in the Ingenuity Pathways Knowledge Base. Core networks were constructed for both direct and indirect interactions using default parameters, and the focus genes with the highest connectivity to other focus genes were selected as seed elements for network generation. New focus genes with high specific connectivity (i.e. overlap between the initialized network and gene’s immediate connections) were added to the growing network until the network reached a default size of 35 nodes. Non-focus genes (i.e. those that were not among our differentially methylated input list) that contained a maximum number of links to the growing network were also incorporated.
The ranking score for each network was then computed by a right-tailed Fisher's exact test as the negative log of the probability that the number of focus genes in the network is not due to random chance. Similarly, significances for functional enrichment of specific genes were also determined by the right-tailed Fisher’s exact test, using all input genes as a reference set.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health (AG21654 and AG18381 to N.B., HG004401 and HD044078 to J.M.G.) and by the Core laboratories of the Albert Einstein Diabetes Research and Training Center (DK 20541) and by Einstein’s Center for Epigenomics. The contributions of Dr. Shahina Maqbool and Gael Westby of the Einstein Epigenomics Shared Facility are gratefully acknowledged. R.F.T was supported by a NIA T32 training grant.
Footnotes
AUTHOR CONTRIBUTIONS
J.M.G. and N.B. designed the experiments, which were performed by R.F.T., G.A., C.G., H.L., and C.L., with data analysis by R.F.T. and interpretation by R.F.T., J.M.G., and N.B. The manuscript was authored by R.F.T., J.M.G., and N.B. with contributions from G.A. and C.G.
REFERENCES
- Ahuja N, Li Q, Mohan AL, Baylin SB, Issa JP. Aging and DNA methylation in colorectal mucosa and cancer. Cancer Res. 1998;58:5489–5494. [PubMed] [Google Scholar]
- Akintola AD, Crislip ZL, Catania JM, Chen G, Zimmer WE, Burghardt RC, Parrish AR. Promoter methylation is associated with the age-dependent loss of N-cadherin in the rat kidney. Am J Physiol Renal Physiol. 2008;294:F170–F176. doi: 10.1152/ajprenal.00285.2007. [DOI] [PubMed] [Google Scholar]
- Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbot W, Dupressoir A, Lazar V, Heidmann T. Epigenetic regulation of an IAP retrotransposon in the aging mouse: progressive demethylation and de-silencing of the element by its repetitive induction. Nucleic Acids Res. 2002;30:2365–2373. doi: 10.1093/nar/30.11.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, Handa V, Doderlein G, Maltry N, Wu W, Lyko F, Niehrs C. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- Barzilai N, Banerjee S, Hawkins M, Chen W, Rossetti L. Caloric restriction reverses hepatic insulin resistance in aging rats by decreasing visceral fat. J Clin Invest. 1998;101:1353–1361. doi: 10.1172/JCI485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A, Taggart M, Frommer M, Miller OJ, Macleod D. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell. 1985;40:91–99. doi: 10.1016/0092-8674(85)90312-5. [DOI] [PubMed] [Google Scholar]
- Bjornsson HT, Sigurdsson MI, Fallin MD, Irizarry RA, Aspelund T, Cui H, Yu W, Rongione MA, Ekstrom TJ, Harris TB, Launer LJ, Eiriksdottir G, Leppert MF, Sapienza C, Gudnason V, Feinberg AP. Intra-individual change over time in DNA methylation with familial clustering. JAMA. 2008;299:2877–2883. doi: 10.1001/jama.299.24.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollati V, Schwartz J, Wright R, Litonjua A, Tarantini L, Suh H, Sparrow D, Vokonas P, Baccarelli A. Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mech Ageing Dev. 2009;130:234–239. doi: 10.1016/j.mad.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busuttil RA, Garcia AM, Reddick RL, Dolle ME, Calder RB, Nelson JF, Vijg J. Intra-organ variation in age-related mutation accumulation in the mouse. PLoS One. 2007;2:e876. doi: 10.1371/journal.pone.0000876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Carr DB, Utzschneider KM, Hull RL, Kodama K, Retzlaff BM, Brunzell JD, Shofer JB, Fish BE, Knopp RH, Kahn SE. Intra-abdominal fat is a major determinant of the National Cholesterol Education Program Adult Treatment Panel III criteria for the metabolic syndrome. Diabetes. 2004;53:2087–2094. doi: 10.2337/diabetes.53.8.2087. [DOI] [PubMed] [Google Scholar]
- Chen J. Senescence and functional failure in hematopoietic stem cells. Exp Hematol. 2004;32:1025–1032. doi: 10.1016/j.exphem.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Chu MW, Siegmund KD, Eckstam CL, Kim JY, Yang AS, Kanel GC, Tavare S, Shibata D. Lack of increases in methylation at three CpG-rich genomic loci in nonmitotic adult tissues during aging. BMC Med Genet. 2007;8:50. doi: 10.1186/1471-2350-8-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer J, Andressoo JO, de Wit J, Huijmans J, Beems RB, van Steeg H, Weeda G, van der Horst GT, van Leeuwen W, Themmen AP, Meradji M, Hoeijmakers JH. Premature aging in mice deficient in DNA repair and transcription. Science. 2002;296:1276–1279. doi: 10.1126/science.1070174. [DOI] [PubMed] [Google Scholar]
- Duncan AW, Dorrell C, Grompe M. Stem Cells and Liver Regeneration. Gastroenterology. 2009 Aug;137(2):466–481. doi: 10.1053/j.gastro.2009.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SA, Manova K, Chen WS, Hoodless P, Weinstein DC, Bachvarova RF, Darnell JE., Jr Expression of transcription factor HNF-4 in the extraembryonic endoderm, gut, and nephrogenic tissue of the developing mouse embryo: HNF-4 is a marker for primary endoderm in the implanting blastocyst. Proc Natl Acad Sci U S A. 1994;91:7598–7602. doi: 10.1073/pnas.91.16.7598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA, Haefliger C, Horton R, Howe K, Jackson DK, Kunde J, Koenig C, Liddle J, Niblett D, Otto T, Pettett R, Seemann S, Thompson C, West T, Rogers J, Olek A, Berlin K, Beck S. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrich M, Nelson MR, Stanssens P, Zabeau M, Liloglou T, Xinarianos G, Cantor CR, Field JK, van den Boom D. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci U S A. 2005;102:15785–15790. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einstein FH, Fishman S, Bauman J, Thompson RF, Huffman DM, Atzmon G, Barzilai N, Muzumdar RH. Enhanced activation of a "nutrient-sensing" pathway with age contributes to insulin resistance. FASEB J. 2008;22:3450–3457. doi: 10.1096/fj.08-109041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman S, Muzumdar RH, Atzmon G, Ma X, Yang X, Einstein FH, Barzilai N. Resistance to leptin action is the major determinant of hepatic triglyceride accumulation in vivo. FASEB J. 2007;21:53–60. doi: 10.1096/fj.06-6557com. [DOI] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suner D, Cigudosa JC, Urioste M, Benitez J, Boix-Chornet M, Sanchez-Aguilera A, Ling C, Carlsson E, Poulsen P, Vaag A, Stephan Z, Spector TD, Wu YZ, Plass C, Esteller M. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner K. A third component causing random variability beside environment and genotype. A reason for the limited success of a 30 year long effort to standardize laboratory animals? Lab Anim. 1990;24:71–77. doi: 10.1258/002367790780890347. [DOI] [PubMed] [Google Scholar]
- Glass JL, Thompson RF, Khulan B, Figueroa ME, Olivier EN, Oakley EJ, Van Zant G, Bouhassira EE, Melnick A, Golden A, Fazzari MJ, Greally JM. CG dinucleotide clustering is a species-specific property of the genome. Nucleic Acids Res. 2007;35:6798–6807. doi: 10.1093/nar/gkm489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golbus J, Palella TD, Richardson BC. Quantitative changes in T cell DNA methylation occur during differentiation and ageing. Eur J Immunol. 1990;20:1869–1872. doi: 10.1002/eji.1830200836. [DOI] [PubMed] [Google Scholar]
- Goyal R, Reinhardt R, Jeltsch A. Accuracy of DNA methylation pattern preservation by the Dnmt1 methyltransferase. Nucleic Acids Res. 2006;34:1182–1188. doi: 10.1093/nar/gkl002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurland BJ, Page WF, Plassman BL. A twin study of the genetic contribution to age-related functional impairment. J Gerontol A Biol Sci Med Sci. 2004;59:859–863. doi: 10.1093/gerona/59.8.m859. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Boyko EJ, Leonetti DL, McNeely MJ, Newell-Morris L, Kahn SE, Fujimoto WY. Visceral adiposity and the risk of impaired glucose tolerance: a prospective study among Japanese Americans. Diabetes Care. 2003;26:650–655. doi: 10.2337/diacare.26.3.650. [DOI] [PubMed] [Google Scholar]
- Huffman DM, Barzilai N. Role of visceral adipose tissue in aging. Biochim Biophys Acta. 2009 Oct;1790(10):1117–1123. doi: 10.1016/j.bbagen.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Jackson RA, Hawa MI, Roshania RD, Sim BM, DiSilvio L, Jaspan JB. Influence of aging on hepatic and peripheral glucose metabolism in humans. Diabetes. 1988;37:119–129. doi: 10.2337/diab.37.1.119. [DOI] [PubMed] [Google Scholar]
- Jiang MH, Fei J, Lan MS, Lu ZP, Liu M, Fan WW, Gao X, Lu DR. Hypermethylation of hepatic Gck promoter in ageing rats contributes to diabetogenic potential. Diabetologia. 2008;51:1525–1533. doi: 10.1007/s00125-008-1034-8. [DOI] [PubMed] [Google Scholar]
- Johnson NL, Kotz S, Kemp AW. Univariate Discrete Distributions. New York: Wiley; 1992. [Google Scholar]
- Kaminsky ZA, Tang T, Wang SC, Ptak C, Oh GH, Wong AH, Feldcamp LA, Virtanen C, Halfvarson J, Tysk C, McRae AF, Visscher PM, Montgomery GW, Gottesman, Martin NG, Petronis A. DNA methylation profiles in monozygotic and dizygotic twins. Nat Genet. 2009;41:240–245. doi: 10.1038/ng.286. [DOI] [PubMed] [Google Scholar]
- Karasik D, Hannan MT, Cupples LA, Felson DT, Kiel DP. Genetic contribution to biological aging: the Framingham Study. J Gerontol A Biol Sci Med Sci. 2004;59:218–226. doi: 10.1093/gerona/59.3.b218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Johansson S, Stach D, Corcoran M, Grander D, Schalling M, Bakalkin G, Lyko F, Larsson C, Ekstrom TJ. LUMA (LUminometric Methylation Assay)--a high throughput method to the analysis of genomic DNA methylation. Exp Cell Res. 2006;312:1989–1995. doi: 10.1016/j.yexcr.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Kennedy ET. Evidence for nutritional benefits in prolonging wellness. Am J Clin Nutr. 2006;83:410S–414S. doi: 10.1093/ajcn/83.2.410S. [DOI] [PubMed] [Google Scholar]
- Khulan B, Thompson RF, Ye K, Fazzari MJ, Suzuki M, Stasiek E, Figueroa ME, Glass JL, Chen Q, Montagna C, Hatchwell E, Selzer RR, Richmond TA, Green RD, Melnick A, Greally JM. Comparative isoschizomer profiling of cytosine methylation: the HELP assay. Genome Res. 2006;16:1046–1055. doi: 10.1101/gr.5273806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwabi-Addo B, Chung W, Shen L, Ittmann M, Wheeler T, Jelinek J, Issa JP. Age-related DNA methylation changes in normal human prostate tissues. Clin Cancer Res. 2007;13:3796–3802. doi: 10.1158/1078-0432.CCR-07-0085. [DOI] [PubMed] [Google Scholar]
- Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslov AY, Vijg J. Genome instability, cancer and aging. Biochim Biophys Acta. 2009 Oct;1790(10):963–969. doi: 10.1016/j.bbagen.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulle A, Salles B, Daviaud D, Valet P, Muller C. Positive regulation of DNA double strand break repair activity during differentiation of long life span cells: the example of adipogenesis. PLoS One. 2008;3:e3345. doi: 10.1371/journal.pone.0003345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar R, Allison DB, Huffman DM, Ma X, Atzmon G, Einstein FH, Fishman S, Poduval AD, McVei T, Keith SW, Barzilai N. Visceral adipose tissue modulates mammalian longevity. Aging Cell. 2008;7:438–440. doi: 10.1111/j.1474-9726.2008.00391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neese RA, Misell LM, Turner S, Chu A, Kim J, Cesar D, Hoh R, Antelo F, Strawford A, McCune JM, Christiansen M, Hellerstein MK. Measurement in vivo of proliferation rates of slow turnover cells by 2H2O labeling of the deoxyribose moiety of DNA. Proc Natl Acad Sci U S A. 2002;99:15345–15350. doi: 10.1073/pnas.232551499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieves DJ, Cnop M, Retzlaff B, Walden CE, Brunzell JD, Knopp RH, Kahn SE. The atherogenic lipoprotein profile associated with obesity and insulin resistance is largely attributable to intra-abdominal fat. Diabetes. 2003;52:172–179. doi: 10.2337/diabetes.52.1.172. [DOI] [PubMed] [Google Scholar]
- Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK, Fraenkel E, Bell GI, Young RA. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordovas JM, Shen J. Gene-environment interactions and susceptibility to metabolic syndrome and other chronic diseases. J Periodontol. 2008;79:1508–1513. doi: 10.1902/jop.2008.080232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelicci PG. Do tumor-suppressive mechanisms contribute to organism aging by inducing stem cell senescence? J Clin Invest. 2004;113:4–7. doi: 10.1172/JCI200420750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Shen N, Qian L, Sun XL, Koduru P, Goodwin LO, Issa JP, Broome JD. Hypermethylation of CpG islands in the mouse asparagine synthetase gene: relationship to asparaginase sensitivity in lymphoma cells. Partial methylation in normal cells. Br J Cancer. 2001;85:930–935. doi: 10.1054/bjoc.2001.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee J, Ge H, Yang W, Fan M, Handschin C, Cooper M, Lin J, Li C, Spiegelman BM. Partnership of PGC-1alpha and HNF4alpha in the regulation of lipoprotein metabolism. J Biol Chem. 2006;281:14683–14690. doi: 10.1074/jbc.M512636200. [DOI] [PubMed] [Google Scholar]
- Rossetti L, Giaccari A. Relative contribution of glycogen synthesis and glycolysis to insulin-mediated glucose uptake. A dose-response euglycemic clamp study in normal and diabetic rats. J Clin Invest. 1990;85:1785–1792. doi: 10.1172/JCI114636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmucker DL. Age-related changes in liver structure and function: Implications for disease? Exp Gerontol. 2005;40:650–659. doi: 10.1016/j.exger.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Sharpless NE, DePinho RA. Telomeres, stem cells, senescence, and cancer. J Clin Invest. 2004;113:160–168. doi: 10.1172/JCI20761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song F, Mahmood S, Ghosh S, Liang P, Smiraglia DJ, Nagase H, Held WA. Tissue specific differentially methylated regions (TDMR): Changes in DNA methylation during development. Genomics. 2009;93:130–139. doi: 10.1016/j.ygeno.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song F, Smith JF, Kimura MT, Morrow AD, Matsuyama T, Nagase H, Held WA. Association of tissue-specific differentially methylated regions (TDMs) with differential gene expression. Proc Natl Acad Sci U S A. 2005;102:3336–3341. doi: 10.1073/pnas.0408436102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szanto I, Kahn CR. Selective interaction between leptin and insulin signaling pathways in a hepatic cell line. Proc Natl Acad Sci U S A. 2000;97:2355–2360. doi: 10.1073/pnas.050580497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RF, Reimers M, Khulan B, Gissot M, Richmond TA, Chen Q, Zheng X, Kim K, Greally JM. An analytical pipeline for genomic representations used for cytosine methylation studies. Bioinformatics. 2008;24:1161–1167. doi: 10.1093/bioinformatics/btn096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RF, Suzuki M, Lau KW, Greally JM. A pipeline for the quantitative analysis of CG dinucleotide methylation using mass spectrometry. Bioinformatics. 2009 Sep 1;25(17):2164–2170. doi: 10.1093/bioinformatics/btp382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Zant G, Liang Y. The role of stem cells in aging. Exp Hematol. 2003;31:659–672. doi: 10.1016/s0301-472x(03)00088-2. [DOI] [PubMed] [Google Scholar]
- vB Hjelmborg J, Iachine I, Skytthe A, Vaupel JW, McGue M, Koskenvuo M, Kaprio J, Pedersen NL, Christensen K. Genetic influence on human lifespan and longevity. Hum Genet. 2006;119:312–321. doi: 10.1007/s00439-006-0144-y. [DOI] [PubMed] [Google Scholar]
- Vilkaitis G, Suetake I, Klimasauskas S, Tajima S. Processive methylation of hemimethylated CpG sites by mouse Dnmt1 DNA methyltransferase. J Biol Chem. 2005;280:64–72. doi: 10.1074/jbc.M411126200. [DOI] [PubMed] [Google Scholar]
- Vogt G, Huber M, Thiemann M, van den Boogaart G, Schmitz OJ, Schubart CD. Production of different phenotypes from the same genotype in the same environment by developmental variation. J Exp Biol. 2008;211:510–523. doi: 10.1242/jeb.008755. [DOI] [PubMed] [Google Scholar]
- Whitelaw NC, Whitelaw E. How lifetimes shape epigenotype within and across generations. Hum Mol Genet. 2006;15(Spec No 2):R131–R137. doi: 10.1093/hmg/ddl200. [DOI] [PubMed] [Google Scholar]
- Yagi S, Hirabayashi K, Sato S, Li W, Takahashi Y, Hirakawa T, Wu G, Hattori N, Ohgane J, Tanaka S, Liu XS, Shiota K. DNA methylation profile of tissue-dependent and differentially methylated regions (T-DMRs) in mouse promoter regions demonstrating tissue-specific gene expression. Genome Res. 2008;18:1969–1978. doi: 10.1101/gr.074070.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.