Abstract
Proteins with oxidizable thiols are essential to many functions of cell nuclei, including transcription, chromatin stability, nuclear protein import and export, and DNA replication and repair. Control of the nuclear thiol-disulfide redox states involves both the elimination of oxidants to prevent oxidation and the reduction of oxidized thiols to restore function. These processes depend on the common thiol reductants, glutathione (GSH) and thioredoxin-1 (Trx1). Recent evidence shows that these systems are controlled independent of the cytoplasmic counterparts. In addition, the GSH and Trx1 couples are not in redox equilibrium, indicating that these reductants have nonredundant functions in their support of proteins involved in transcriptional regulation, nuclear protein trafficking, and DNA repair. Specific isoforms of glutathione peroxidases, glutathione S-transferases, and peroxiredoxins are enriched in nuclei, further supporting the interpretation that functions of the thiol-dependent systems in nuclei are at least quantitatively distinct, and probably also qualitatively distinct, from similar processes in the cytoplasm. Elucidation of the distinct nuclear functions and regulation of the thiol redox pathways in nuclei can be expected to improve understanding of nuclear processes and also to provide the basis for novel approaches to treat aging and disease processes associated with oxidative stress in the nuclei. Antioxid. Redox Signal. 13, 489–509.
Introduction
The nucleus contains many of the same thiol/disulfide redox control systems as does the cytoplasm and other organelles, but accumulating evidence shows that the regulation and functions of the nuclear systems are distinct. This review summarizes current knowledge of the thiol/disulfide control systems in nuclei, their contents and redox states within nuclei, and their roles in supporting and controlling nuclear functions. The review is organized into five sections, with the first addressing the impact of changing concepts of oxidative stress on nuclear research. Important among these concepts are the recognition that nonequilibrium conditions exist in thiol/disulfide redox systems and that this necessitates inclusion of disruption of redox signaling and control as an important aspect of oxidative stress mechanisms. The second section includes current knowledge of the nuclear contents of redox control systems, principally covering glutathione, thioredoxin, and related proteins. This is followed by a section reviewing available knowledge about the steady-state redox balance of nuclear proteins. The nuclei appear to be relatively reduced and protected against oxidation, thereby emphasizing the critical nature of nuclear redox regulation. A fourth section addresses redox-dependent nuclear-import and -export mechanisms. Although studied in detail for only a small number of proteins, redox-dependent regulation of nuclear protein content may represent an important aspect of signaling for cell proliferation and differentiation as well as responses to stress. The final section covers the roles of nuclear redox systems in supporting and controlling nuclear functions. Substantial evidence is available to illustrate the importance of redox regulation of transcription, and additional research shows that studies to elucidate redox-dependent sites of DNA repair and chromatin remodeling are likely to clarify and enhance the understanding of these critical processes. The development of new methods to measure redox states of nuclear proteins under physiologic conditions provides considerable opportunity to advance further the understanding of redox control in cell nuclei.
Nuclear Redox Systems Function in Redox Signaling and Protection Against Two Types of Oxidative Stress
Nuclear redox reactions have been most frequently studied in the context of oxidative stress, but extensive research shows that central cell signaling and control systems also involve redox mechanisms. Many of these signaling pathways involve reversible oxidation of thiol-containing proteins. The redox states of these thiols are sensitive to two-electron (nonradical) oxidants and are controlled by reductant systems dependent on the thioredoxin (Trx) or glutathione (GSH). Nonradical oxidants include peroxides, aldehydes, quinones, epoxides, disulfides, peroxynitrites, and other species that can be generated enzymatically or as a consequence of nonenzymatic mechanisms. These can be quantitatively important in oxidative stress because free radical–scavenging mechanisms are efficient in maintaining free radicals at very low levels and because the nonradical oxidants are often produced at higher rates than free radicals are generated (83). Moreover, large-scale intervention trials with moderately high doses of free radical–scavenging antioxidants show little benefit in humans in terms of protection against cancer or age-related diseases. Consequently, the contemporary view of oxidative stress in disease has shifted away from a simple imbalance of prooxidants and antioxidants to disruption of redox signaling pathways dependent more on nonradical oxidants than on free radical mechanisms.
Definition of oxidative stress
Oxidative stress has been redefined as “an imbalance in prooxidants and antioxidants with associated disruption of redox circuitry and macromolecular damage” to emphasize the critical disruption of redox signaling mechanisms that occurs during oxidative stress and the importance of nonradical oxidants in the relevant mechanisms. This new definition of oxidative stress includes two different mechanistic outcomes (Fig. 1). The first of these is macromolecular damage, such as DNA damage, that has historically been a common focus of nuclear oxidative stress, and the more contemporary study of disruption of thiol redox systems, which are gaining increasing recognition as critical determinants of normal signaling and control of nuclear functions. Whereas free radical mechanisms are readily induced by ionizing radiation and metal-catalyzed oxidation by peroxides, nonradical oxidative mechanisms involving thiols appear to predominate under more usual physiologic conditions (83).
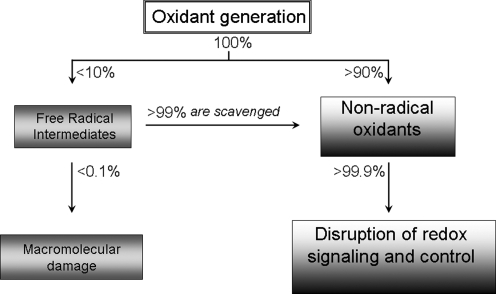
FIG. 1.
Mechanistic outcomes of oxidative stress include both macromolecular damage and disruption of redox signaling and control. Accumulating evidence indicates that both free radical (one-electron) and nonradical (two-electron) mechanisms contribute to damage from oxidative stress. Whereas superoxide anion radical and nitric oxide are free radicals, H2O2, lipid peroxides, quinones, epoxides, peroxynitrite, disulfides, and many other chemicals are nonradical oxidants. Based on model studies with xanthine oxidase, univalent reduction of O2 to superoxide anion radical is a minor fraction of the bivalent reduction to H2O2 (41), suggesting that two-electron oxidants predominate during oxidative stress. For an arbitrary rate of oxidant generation equal to 100%, perhaps 90% would go directly to two-electron oxidants, and only 10% would be one-electron oxidants. Because free radical–scavenging mechanisms are efficient and convert free radicals into nonradical oxidants, nonradical oxidants probably constitute >99.9% of all oxidants. Unlike reactive free radicals, which are nonspecific in their destruction of macromolecules, many of the nonradical oxidants selectively oxidize thiols. Many redox signaling and control mechanisms depend on critical cysteine residues. Consequently, disruption of redox signaling and control by nonradical oxidants may have a major role in oxidative stress as a mechanism of disease.
Quantification of the partitioning of univalent (1e−) radical oxidant production and bivalent (2e−) nonradical oxidant production is not available for in vivo conditions. However, model studies by Fridovich (41) using xanthine oxidase showed that under all conditions, production of the nonradical oxidant, H2O2, predominated over the rate of superoxide generation. Because very efficient fluorescent probes are available to detect free radical generation in cells, the extensive use of these indicators may have resulted in overinterpretation of results. For instance, if nonradical oxidants are important in toxicity mechanisms and oxidative stress produces proportionately more nonradical oxidants than free radical oxidants, then free radical–dependent macromolecular damage can correlate with pathologic insult but, nonetheless, be only a relatively minor contributor to the disease process.
Redox hypothesis of oxidative stress
To facilitate the consideration of nonradical mechanisms separately from free radical mechanisms, a “redox hypothesis of oxidative stress” has been delineated (83). This hypothesis contains four postulates:
All biologic systems contain redox elements (e.g., redox-sensitive cysteine residues) that function in cell signaling, macromolecular trafficking, and physiologic regulation.
Organization and coordination of the redox activity of these elements occurs through redox circuits dependent on common control nodes (e.g., thioredoxin, GSH).
The redox-sensitive elements are spatially and kinetically insulated so that “gated” redox circuits can be activated by translocation/aggregation or catalytic mechanisms or both.
Oxidative stress is a disruption of the function of these redox circuits caused by specific reaction with the redox-sensitive thiol elements, altered pathways of electron transfer, or interruption of the gating mechanisms controlling the flux through these pathways.
These postulates are largely untested with regard to nuclear functions. However, redox-sensitive thiols are critical for the control of replication and cell proliferation, chromatin structure, DNA repair, mechanisms of transcriptional activation, transcription factor binding to DNA, and transport of proteins into and out of nuclei. Similar to signaling proteins found in other compartments of the cell, these thiol-containing proteins are susceptible to oxidation or inactivation by nonradical oxidants. Accumulating data for cellular thiol systems show that many thiols are maintained under stable but nonequilibrium conditions because of continuous oxidation/reduction turnover at a rate of about 0.5% of the total thiol pool per minute (83). Although rates of oxidant production in nuclei under physiologic conditions are not known, evidence summarized later indicates that nonequilibrium conditions of thiol/disulfide couples also exist in nuclei. Nonequilibrium thiol/disulfide redox states of regulatory proteins mean that the altered abundance and distribution of redox enzymes provide a general mechanism for control of nuclear functions. Moreover, this means that nonradical mechanisms of oxidative stress are likely to contribute to altered cell proliferation, differentiated phenotype, or other fundamental properties associated with nuclei.
Distribution of Redox Systems in Nuclei
Nuclear glutathione system
Glutathione
Glutathione (GSH), the most abundant low-molecular-weight thiol-containing tripeptide (γ-glutamyl-cysteinyl-glycine), plays a major role in protecting cells from oxidant-induced toxicity. Cellular GSH at millimolar levels is widely distributed within cells and determines cellular redox state, based on the ratio of [GSH]2 to its disulfide form [GSSG]. Although a number of important functions of GSH are implicated in cell physiology and pathophysiology, relatively few studies have provided spatial or compartmental resolution to address specific functions in nuclei. Previously, important roles for GSH were shown for DNA synthesis and cell proliferation (71, 182), regulation of the nuclear matrix organization (29), and maintenance of Cys residues on zinc-finger DNA-binding motifs in a reduced and functional state (96). These results show that GSH plays a pivotal role in nuclear function. In this review, Cys-containing proteins regulating the GSH system (Fig. 2), including glutathione reductase, glutathione peroxidase, and glutaredoxin and glutathione S-transferase in nuclei, are discussed.
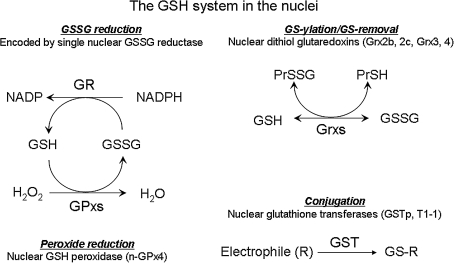
FIG. 2.
The GSH systems in the nuclei. Although systematic studies are limited, an extensive literature describes the GSH systems associated with nuclei. These include GSH, GSSG reductase (nuclear GSSG reductase), GSH peroxidase (n-Prx), glutaredoxins (Grx2b and Grx2c for human, Grx3 and Grx4 for yeast), and GST (GSTp and GST T1 are present in the nuclei). Many of these systems are identical or functionally equivalent to the cytoplasmic forms. However, the GSH systems exist under nonequilibrium redox conditions, so that function depends on abundance and distribution of individual redox components. In addition, changes in electron flow from NADPH or reduction of endogenously produced peroxide can change the steady-state redox potential and thereby control protein functions.
Glutaredoxins
Glutaredoxins (Grxs) are thiol-disulfide oxidoreductases that catalyze GSH/GSSG redox reactions with cysteine residues of target proteins (70). Grxs are classified into two main groups, dithiol Grxs and monothiol Grxs, depending on the active-site consensus sequences containing –Cys-Pro-Tyr-Cys– and –Cys-Gly-Phe-Ser–, respectively (106). Monothiol Grxs can be further categorized into single-domain monothiol Grxs containing one Grx domain and multidomain monothiol Grxs containing a Trx-like domain and one to three monothiol Grx domains.
All dithiol Grxs catalyze monothiol reactions, as shown by the hydroxyethyl disulfide assay, a general method used to measure Grx activity (106). Dithiol Grxs reduce protein disulfides through a mechanism involving two Cys in the active-site motif, whereas monothiol Grxs function to deglutathionylate GSH-protein mixed disulfides through a monothiol mechanism involving one Cys in the active site (36). Two monothiol Grxs, cytoplasmic Grx3 and mitochondrial Grx5, occur in human cells. Grx3 is a multidomain monothiol form, and Grx5 is a single-domain monothiol form. Four dithiol Grxs are found in human cells. These include Grx1 in cytoplasm, Grx2 (Grx2a) in mitochondria, and Grx2b and Grx2c in cytoplasm and nuclei. Grx2b and Grx2c are transcript variants derived from Grx2a, localized exclusively in mitochondria. Grx2c, but not Grx2b, forms an Fe-S cluster in an in vitro reconstitution assay (107). Grx2a and Grx2b expression was limited to testis in nontransformed cells, whereas expression was found in a broad range of transformed cancer tissues, including those of blood, breast, cervix, lung, and the nervous system (107). The pattern of distribution of Grxs among organelles and altered abundance in cancer suggests a complexity in functions of Grxs that is currently not understood.
Localization of Grxs in nuclei was recently identified. Specifically, Grx2b and Grx2c were found in nuclei of human cells, and Grx3 and Grx4 were found in nuclei of Saccharomyces cerevisiae cells (119). Compared with isoforms of Grxs found in other organelles, fewer functional studies of nuclear Grxs have been reported. Two research groups, Pujol-Carrion et al. (147) and Ojeda et al. (133), characterized a function for Grx3 and Grx4 in S. cerevisiae in cellular iron homeostasis. This occurs through the regulation of Aft1, a transcription factor regulating the high-affinity iron-uptake genes. The translocation of Aft1 from nuclei to cytoplasm was directly regulated by Grx3 and Grx4 as consequence of interacting with Aft1 (147). The glutaredoxin domain of Grx is essential for binding and control of Aft1 and suggests an important role for GSH in this mechanism (133).
A recent study suggests that critical nuclear functions of nuclear Grx can serve as a target for anticancer treatment. Message levels of Grx2b, and also of thioredoxin reductase-1, were examined with quantitative PCR and found to be significantly increased in leukemic cells from acute myeloid leukemia patients after treatment with selenite, a potent inhibitor of malignant cell growth (137). In principle, the increase in specific nuclear forms of Grx, which are expressed in cancer cells, could provide a target to kill rapidly proliferating cancer cells selectively.
GSH S-transferase
The mammalian glutathione S-transferases (GSTs) are phase II detoxification enzymes that function in elimination of drugs and other xenobiotics. GST catalyzes detoxification of reactive electrophilic compounds, including carcinogens, therapeutic drugs, environmental toxins, and products of oxidative stress, by conjugation with GSH. Increasing evidence supports a protective role of GST in oxidative stress and also a role in the regulation of ROS-mediated cell signaling. Importantly, the GST expression level shows significant correlation with cancer development. This increase is associated with growth and resistance of cancer cells; in addition, GST can provide a biomarker for early stages of cancer development.
Four major cytosolic GST families, including Alpha, Mu, Pi, and Theta (65), and four minor families, Zeta, Sigma, Kappa, and Omega, have been identified (179). Studies on subcellular localization show that most GSTs are localized in the cytoplasm. However, some of the alpha class GSTs are associated with the plasma membrane, and other forms from different families are found in mitochondria and nuclei. The endoplasmic reticulum contains microsomal GSTs (mGSTs), which are present as trimers and are structurally and immunologically distinct from the cytosolic GST (112), which are present as dimers of subunits (171).
GST alpha was detected in nuclei of human hepatocytes (15, 181), rat Purkinje cells, and neurons (81). The mGST not only was found in microsomal fractions but also was observed in nuclei of primary spermatocytes (139). The presence of the class theta GST T1-1 in the nucleus of mouse and human liver cells was confirmed with histochemical analysis (172). Nuclear expression of GSTpi (GSTp) occurs in cancer tissues (52, 85, 138), although it also was found in mitochondria (52). Increases in the expression of GSTp have been detected in precancerous cells and various cancer tissues, animal-type melanoma (138), hepatocarcinogenesis (165), prostate cancer (116), gynecologic cancers, urinary cancer cells (173), and glioma cells (2).
Nuclear expression of human GSTp is associated with anticancer drug resistance (53, 117). Nuclear accumulation of GSTp in cancer cell lines was observed in response to treatment with the anticancer drugs cisplatin, irinotecan hydrochloride, etoposide, and 5-fluorouracil (53). Inhibition of nuclear localization of GSTp-sensitized cells to cytotoxicity suggested that nuclear GSTp protected DNA against anticancer drug–induced damage (52, 53). Additional evidence supports the protective role of nuclear GSTp against H2O2-induced DNA damage (85). A negative relation between animal-type melanoma and nuclear GSTp levels also supports a critical role of nuclear GSTp in nuclear protection (138). Mechanistic studies of the regulation of GSTp in association with carcinogenesis showed that the GSTp gene expression was regulated by transcription factor C/EBP (CAAT enhancer–binding protein)α for suppression in normal liver, whereas the Nrf2 (NF-E2 p45-related factor 2)/MafK heterodimer was required for activation during hepatocarcinogenesis (165).
GSSG reductase
Glutathione reductase (GR) is a ubiquitous enzyme required for reduction of glutathione disulfide (GSSG) to GSH by NADPH. GR is a homodimeric flavoprotein consisting of subunits with a molecular mass of 55 kDa and containing two FAD molecules. The dimeric nature of this enzyme is maintained in the presence of GSH, which is critical for its catalytic activity. GR is found in most organisms from prokaryotes to eukaryotes. Biochemical activity of the GR present in the cytoplasm and mitochondria of mammalian cells is identical (46). Both the human and the mouse GR contain mitochondrial targeting sequences in the N-terminal region (89). As identified in a group of proteins, such as DNA-ligase and DNA-glycosylase, with different compartmental expressions of proteins encoded by a single nuclear gene (102, 140), GR is a product of a single nuclear gene that is expressed in cytoplasm, mitochondria, nuclei, and nucleoli. Truncations of the N-terminal domains of pea GR (pGR) resulted in significantly different organellar targeting patterns compared with wild-type pGR, suggesting that the N-terminal domain of the dual-targeted pGR signal peptide controls organellar targeting efficiency (162).
Earlier studies showed that GSH-dependent antioxidant defense mechanisms are significantly compartmentalized (176). Rogers et al. (158) studied hepatic nuclear and nucleolar GR, glutathione peroxidase (GPx) and GST expressions, and their activities in response to hepatic necrosis. They found that these distributions differed in female rats from those of male rats, suggesting significant roles for these enzymes in the different compartments. They also observed greater nuclear GR activities of CHO (Chinese hamster ovary) cells during S-phase than during other stages of the cell cycle, suggesting that a nuclear GR controls the cell cycle (159).
A significant number of studies support the critical function of GR in a variety of diseases. Expression levels and activities of GR and GPx were decreased in patients with glioblastoma multiforme (GBM) and transitional meningioma (TM), when compared with normal brain tissues. However, protein oxidation levels measured by protein carbonyl content were increased in GBM and TM, perhaps because of the decreases in other antioxidant systems (185). The role of GR in zinc-induced toxicity was shown by studies in human lung cell lines and rat astrocytes (6, 193). BCNU inactivation of GR sensitized cellular zinc toxicity by increasing the cellular GSSG level. This result suggests that GSSG itself is an important effector in zinc cytotoxicity, perhaps mediated by Grx reactions, as described earlier.
A role of GR in cardiovascular disease (CVD) also has been investigated (104, 149, 196) but specific nuclear functions have not been delineated. Decreased levels of GSH and GSH-related antioxidant enzymes, GR, GST, GPx, have been observed in human atherosclerotic lesions. Conversely, overexpression of GR targeted to cytoplasm or mitochondria in bone marrow–derived cells protected cells from oxidized LDL-induced mitochondrial hyperpolarization and reduced atherosclerotic lesion formation (149). Studies to define the role of GR in CVD have been extended to the regulation of signaling pathways. Laminar shear stress activated antiapoptotic signaling, including activation of Akt followed by eNOS activation, and increased NO production was regulated in a GR-dependent manner (195). This finding suggests that the more-reduced GSH redox system caused by laminar shear stress has a protective role in apoptosis, inflammation, and further CVD, by controlling cellular signaling.
Because of the large body of evidence showing a critical role of GR in cytotoxicity and diseases, additional studies focused on nuclear-localized GR are warranted. The redox potential of the nuclear GSH/GSSG couple is kinetically controlled and at least partially autonomous from the cytoplasmic compartment. Consequently, additional studies on the regulation of nuclear GR will be important to understand the functions and control of the nuclear GSH/GSSG system.
Glutathione peroxidase
Glutathione peroxidase (GPx) catalyzes the reduction of H2O2 or organic hydroperoxides to H2O and respective alcohols by using GSH as a reductant. Eight types of GPx (GPx1-8) have been characterized. The GPx family is one of the 25 known classes of selenoproteins (98), and all members, except for GPx5 in humans and mice, GPx7, and GPx8, contain a selenocysteine necessary for catalytic activity. Several of these selenoproteins, including GPx, selenoproteins P (67), W (191), and R (38), and thioredoxin reductases, have antioxidant functions, and deficiencies in these selenoproteins are associated with cancer. Most GPxs are tetramers. Each monomer of GPx1, GPx2, GPx3, GPx4, and GPx6 contains selenocysteine (98). In contrast, GPx5, GPx7, and GPx8 contain cysteine at the catalytic site. Although differing in the active site, GPx8 and GPx4 share structural similarities. Individual disruption of GPx1 and GPx2, alone or in combination, shows mild phenotypes (20, 33), whereas GPx4 deficiency resulted in embryonic lethality (74, 203). Mice deficient in GPx3 also have been reported to be viable, suggesting that GPx4 is a uniquely critical selenoprotein (22).
GPx4 has previously been described as a phospholipid hydroperoxide glutathione peroxidase (PHGPx) because of its unique biochemical functions toward phospholipid hydroperoxides, which are formed during lipid peroxidation (188). Despite its similarities to other GPxs in terms of structure and biochemical properties, many differences exist between GPx4 and other GPx members. A major structural difference is that GPx4 is a monomer, whereas the others are tetramers (187). In addition, GPx4 has a broad range of substrate specificities in addition to H2O2, including derivatives from cholesterol and cholesteroyl esters and thymine hydroperoxide (74). Previous findings also suggest that GPx4 functions as a structural protein and as a regulator of apoptosis and gene expression (168).
Examination of GPx4 expression shows unusual patterns in tissue distribution with a specific nuclear form. GPx4 is widely expressed in normal tissues but is especially high in the testis (11). GPx4 levels were normal even under selenium restriction, whereas others (e.g., GPx1) were more sensitive to the restriction (12, 167). Three different isoforms of GPx4 exist, differing in their 5' extension; these are derived from a single gene and include cytosolic (c-GPx4), mitochondrial (m-GPx4), and nuclear (n-GPx4) forms (144, 148). The m-GPx4 is higher than c-GPx4 in molecular mass; however, the mitochondrial targeting sequence is cleaved off in the mitochondria so that these two processed GPx4 forms are not distinguishable in their primary structures. The c-GPx4 is predominant in most mammalian tissues; however, in testis, the m-GPx4 expression was dominant. Independent of c-GPx4 and m-GPx4, the n-GPx4 (also called snGPx for sperm nucleus–specific GPx) was predominantly expressed in late spermatids and spermatozoa (24). The nuclear-targeting sequence was retained in this protein even after nuclear import, resulting in the higher molecular mass of n-GPx4 (34 kDa) than other isoforms (19 kDa, c-GPx4 and m-GPx4). In studies of n-GPx4 KO mice, Conrad et al. (24) concluded that n-GPx4 is not an indispensable player in spermatozoon physiology because KO mice develop normally, are fertile, and do not show any morphologic or functional defects. However, mice lacking n-GPx4 showed instability of the sperm nuclei, implying that n-GPx4 plays an important role in stabilizing nuclear structures in spermatozoa. To date, the mechanism for nuclear import of n-GPx4 has not been established, and the specific biologic function of n-GPx4 in spermatogenesis remains to be elucidated.
Nuclear thioredoxin system
Thioredoxin
Thioredoxins (Trxs) are small thiol-disulfide oxidoreductases (12 kDa) involved in a wide range of cellular redox processes (Fig. 3) and essential for the maintenance of cellular redox signaling and control (72). Trx is oxidized during the conditions of oxidative stress caused by chemical, physical, or biologic stimuli (18, 50, 57). Oxidized Trx is reversibly reduced by NADPH and thioredoxin reductase, whereas reduced Trx is used for reduction of peroxiredoxins (Prxs) (157).
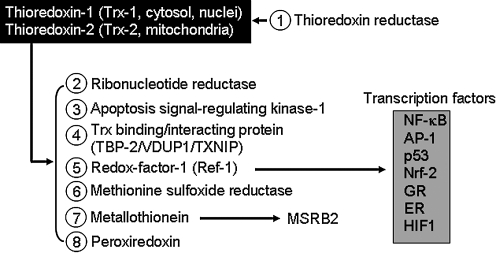
FIG. 3.
Thioredoxin-regulated cellular redox processes. (1) Thioredoxin (Trx) is reduced principally by thioredoxin reductases (TrxR). (2) Trx is an electron donor to support the activity of ribonucleotide reductase (RNR). (3) Trx binds to apoptosis signal-regulated kinase-1, inhibiting the kinase function. (4) Trx1 activity is inhibited by binding to Trx-binding protein-2 (TBP-2), also known as VDUP-1 and TXNIP. (5) Trx1 reduces redox factor-1 (Ref-1), a bifunctional protein with a redox activity and a separate DNA repair (APE-1, endonuclease) activity in nuclei; the redox activity maintains conserved Cys residues of transcription factors [NF-κB, AP-1, p53, Nrf2, glucocorticoid receptor (GR), estrogen receptor (ER), and HIF1] in the reduced form required for DNA binding. (6) Trx1 functions as a reductant for methionine sulfoxide reductases (MSRs). (7) Trx1 controls a pathway for MSRB2 reduction mediated by metallothionein. (8) Trx acts as a reductant for peroxiredoxin (Prx) activity to scavenge peroxides.
Two isoforms of mammalian Trx, Trx2 and Trx1, have been identified, and both are well studied. Trx2 is localized to mitochondria and has a protective function against mitochondrial oxidative stress. Similar to Trx1, Trx2 also functions as a thiol-disulfide oxidoreductase, which is controlled by thioredoxin reductase (TrxR2) and peroxidases (peroxiredoxin-3 and −5) in mitochondria. Trx2 protects against oxidant-induced cell death (19, 62), and recent evidence shows that increased mitochondrial Trx2 alters abundance of proteins controlled by nuclear gene expression (Hansen, et al., unpublished data). Interestingly, Prx5 is found in nuclei as well as mitochondria, but the functional significance of this is unclear.
Trx1 is found in cytoplasmic and nuclear compartments. The redox state of Trx1 in the cytoplasm and nuclei regulates cell death and survival signaling by controlling interactions with Trx1-binding proteins (164). Nuclear Trx1 is functionally autonomous from cytoplasmic Trx1. A distinct nuclear existence of Trx1 has been demonstrated by immunohistochemical studies, suggesting that Trx1 is imported into the nucleus during oxidative stress (69, 169, 177, 197). Spielberger et al. (177) found that cells in sparse culture have increased ROS level, and oxidation of both Trx1 and GSH redox states. In sparse cultures, nuclear localization of Trx1 was high relative to high-density cultures, indicating that the redox environments of the cytoplasm and the nucleus are distinct and that Trx1 has an important role of nuclear redox state in cell growth. A previous study examining the oxidation of the thiol-dependent antioxidant systems in subcellular compartments under conditions of limited energy supply of human colonic cancer cells showed that oxidative stress induced by depletion of glucose and glutamine affects the redox states of proteins in the cytoplasm and mitochondria more than those in the nucleus (Fig. 4). The results showed that the nuclear compartment has better protection against oxidative stress than does cytoplasm or mitochondria (50). Another study showed that nuclear GSH is more susceptible to localized oxidation than is nuclear Trx1 (56). This study examined the effect of nuclear H2O2 generation on the nuclear thiol/disulfide redox systems by targeting d-amino acid oxidase (DAO) to the nucleus. The results showed that nuclear Trx1, measured with redox Western blot analysis, was resistant to oxidation by DAO-induced H2O2. In contrast, the nuclear GSH system, measured with nuclear protein glutathionylation, was sensitive to oxidation. Under these conditions, neither cytoplasmic Trx1 nor cellular GSH/GSSG showed oxidation. Thus, the results show that the nuclear Trx1 and GSH systems are functionally distinct from the cytoplasmic systems and also respond independently to oxidant generation (Fig. 4).
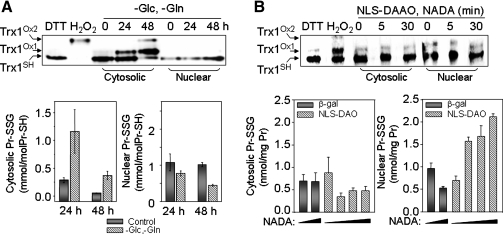
FIG. 4.
Quantification of nuclear redox state measured with Trx1 redox Western blotting and protein glutathionylation. Cells exposed to oxidative stress by depleting glucose (Glc) and glutamine (Gln) from culture (50) or by expressing nuclear-targeted d-amino acid oxidase (NLS-DAO) with variation of substrate amounts (N-acetyl-d-alanine, NADA) (56) were analyzed for cytoplasmic and nuclear redox-state measurements. (A) Nuclear Trx1 and GSH/GSSG were resistant to oxidation by Glc and Gln depletion, compared with cytoplasm. (B) Protein glutathionylation in nuclei was substantially increased by NLS-DAO expression, whereas no detectable change was seen in cytoplasmic protein glutathionylation or Trx1 oxidation in either cytoplasm or nuclei.
Another protein containing a conserved Trx domain is termed nucleoredoxin (42). This protein is more similar to tryparedoxin, a Trx family protein originally identified in parasitic trypanosomes, than to thioredoxins. The protein was originally localized in nuclei, but subsequent studies show that it functions to regulate the Wnt/β-catenin pathway and is principally cytoplasmic. The apparent contradiction appears to be due to formation of complexes with Dv1 and PP2A, which can shuttle between nuclei and cytoplasm under different signaling conditions (42).
Peroxiredoxins
Peroxiredoxins (Prxs) are antioxidant enzymes that were initially identified in yeast, based on their protection of glutamine synthetase against oxidation (91). Prxs undergo a cycle of peroxide-dependent oxidation and thiol-dependent reduction during their catalytic turnover. Oxidized Cys residues in the Prx are reduced by Trx specifically and not by GSH or Grx. The six isoforms of Prx are termed Prx1 through Prx6 in mammalian cells. These proteins are divided into three groups, including 2-Cys Prx (Prx1, Prx2, Prx3, and Prx4; each contain two conserved Cys in both N- and C-terminals), atypical 2-Cys Prx (Prx 5; contains only the N-terminal Cys and requires the other nonconserved Cys for activity), and 1-Cys Prx (Prx6; contains only the N-terminal Cys and requires only this conserved Cys for activity) (170). Prx1, 2, and 4 exist mainly in cytoplasm but also are found in nuclei, whereas Prx3 is localized exclusively in the mitochondria (86, 157). Prx5 is found in mitochondria, cytoplasm, nuclei, and peroxisomes (132, 170).
The catalytic functions of Prx involve multiple oxidized forms of the conserved Cys. Initial oxidation by peroxide involves oxidation of the thiol to a sulfenic acid form (200). This represents a two-electron equivalent oxidation and can be readily converted to a disulfide by reaction with another protein thiol, thereby releasing water. The disulfide is then reduced by Trx to regenerate thiols. The two conserved Cys residues in Prx1 through Prx4 were characterized to form intermolecular disulfides resulting in homodimers, but also exist in decameric forms on overoxidation (200). Overoxidation of the active site Cys to Cys-sulfinic (−SO2H) or Cys-sulfonic acid (−SO3H) can occur during peroxide oxidation (151) and result in inactivation of peroxidase activity. Oxidation-induced inactivation of Prx can be reversed by ATP-dependent reductases, termed sulfiredoxins (7, 16). The loss of peroxidase activity has been suggested to lead to a functional change from a peroxidase to a molecular chaperone under stress conditions (79, 120).
Structural studies show that, in addition to existing as a homodimer, Prx4 also interacts with small proteins such as Prx1 to form heterodimers. Prx4 is present in the cytoplasm and is also translocated to the extracellular space because of secretory signal sequences included in the N-terminal region (115). Prx5, localized in cytosol, mitochondria, and peroxisome, has been characterized to form two intermolecular disulfide bonds, which then rearrange to form intramolecular disulfides during peroxide-induced oxidation (34). The one N-terminal Cys of Prx6 is readily oxidized by peroxide, resulting in Cys-sulfenic acid (−SOH) (21).
The presence of multiple Prxs in various subcellular compartments suggests that the different Prx forms could have different functions in different compartments within cells. This possibility has been supported by studies with approaches including transient overexpression of Prx in cells (4, 60) and knockout Prx1 in mice (31). Egler et al. (31) showed that embryonic fibroblasts from Prx1 KO mice (Prx I−/−) have localized, increased ROS levels in the nucleus, resulting in DNA damage. This research provides evidence for a critical role of Prx1 for the regulation of ROS levels specifically in the nucleus (31). Overexpression of human Prx5 targeted to the nucleus confers resistance to hydrogen peroxide and tert-butylhydroperoxide–induced cell death and DNA damage (4). In addition, the use of nuclear- and cytoplasmic-targeted Prx1 to study effects on transcriptional activation, as described later, suggests that Prx1 governs the compartment-specific redox signaling and control mechanism during oxidative stress (60). This research specifically indicates that a dynamic regulatory mechanism can exist in nuclei in which a constitutive generation of H2O2 provides continuous oxidation of protein thiols, in opposition to reduction by the Trx1 system (Fig. 5). This mechanism provides an “off” mechanism for transcription factors that have a conserved Cys in the DNA-binding region that must be reduced for DNA binding (see later).
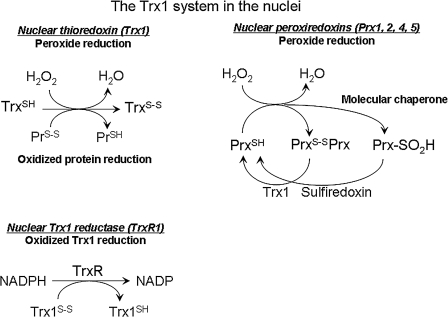
FIG. 5.
The thioredoxin-1 (Trx1) system in nuclei. The Trx system, including Trx1, thioredoxin reductase-1 (TrxR1), and Trx peroxidases (Prx1, Prx2, Prx4, and Prx5), is present in the nuclei. TrxR1 reduces oxidized Trx1 in the presence of NADPH. Trx1 functions to reduce peroxides and disulfide forms of oxidized proteins, such as Prx, which eliminates peroxides. Overoxidized Prx (Prx-SO2H) functions as a molecular chaperone and is reduced by sulfiredoxin.
Similar to the control of transcription-factor activity by Trx1, Prx-controlled regulation of transcriptional factors, including Yap1, NF-κB, Nrf-2, and c-Myc, has been demonstrated (60, 93, 136). Nuclear localization and activation of yeast transcription factor Yap1 was controlled by Tsa1, a ubiquitous Prx in yeast, by regulating intracellular reactive oxygen species (ROS) levels (136). In addition, direct interaction of Prx1 with c-Myc through MBII (Myc Box II) was shown to alter the c-Myc transcriptional activity, although the functional consequences of this interaction are complex and remain unclear (121). Expression of nuclear-targeted Prx1 in HeLa cells increases NF-κB binding to DNA and reporter expression (60). Prx2-deficient mouse embryonic fibroblasts have stimulated Ras, ERK, and NF-κB activation, suggesting that Prx II inhibits the pathway of Ras-ERK-NF-κB through controlling cellular ROS levels (59). Together, the data indicate that cytoplasmic and nuclear Prx distribution are part of a compartmentalized redox control mechanism allowing oxidative activation in the cytoplasm and oxidative inactivation in the nuclei.
Thioredoxin reductase
Thioredoxin reductases (TrxRs) catalyze the NADPH-dependent reduction of Trx and other oxidized cellular proteins. They belong to the pyridine nucleotide-disulfide oxidoreductase protein family and are ubiquitously found in mammalian tissues. TrxRs are homodimers, with each subunit containing a C-terminal selenocysteine necessary for catalytic activity (47). Three isoforms of mammalian TrxR have been identified: TrxR1 is localized to the cytoplasm and the nuclei; TrxR2 (also known as TR3) is localized to the mitochondria; and TrxR3 is expressed mainly in testis. TrxR3 is distinct, in that it has combined Trx and GSSG reductase activities (124). Cytosolic and nuclear TrxR1 and mitochondrial TrxR2 function independently (125, 143), and elimination of either TrxR1 or TrxR2 gene results in an embryonic lethal phenotype (23, 78).
Similar to the role of Trx in redox-sensitive signaling, TrxR also appears to regulate multiple downstream redox-sensitive proteins. Trx1 is translocated from cytoplasm to the nucleus, in association with the activation of NF-κB during oxidative stress induced by UVB irradiation or TNF-α treatment (69). A study from Karimpour et al. (87) suggests that TrxR1 has a role as a signaling factor that controls transcription factor activity, such as AP1 and subsequent gene expression by mediation of Trx1 nuclear localization. A recent study examining changes in expression level of TrxR1 under different growth rates shows higher nuclear expression of TrxR1 in proliferating cells than in nonproliferating cells (174). In CWR22 cells, which resemble androgen-dependent human prostate cancer, TrxR1 is expressed predominantly in the nucleus. Although the biologic function and reasons for the existence of TrxR1 in the nucleus are not firmly established, these findings suggest that the nuclear TrxR1 could be selectively targeted to enhance specificity in cancer therapy.
Quantification of Redox State in the Nucleus
Substantial advances have been made during the past several years in understanding the regulation and functions of thiol/disulfide systems in different subcellular compartments. One of the means to compare thiol/disulfide couples is to express the oxidation–reduction states in terms of the steady-state redox potentials, Eh. These potentials are calculated by using the Nernst equation and respective concentrations of reduced and oxidized components, taking into account the inherent tendency of the couple to accept or donate electrons, as expressed in the standard potential (Eo) for the respective couple. Analyses using currently available techniques show that the relative redox states from most reducing to most oxidizing are mitochondria (−330 mV) > nuclei (−280 mV) > cytoplasm (−250 mV) > endoplasmic reticulum > extracellular space (−150 mV) (62, 82), with some differences due to whether values are expressed for Trx or GSH. Data for different cell types are often similar, but five- to six-fold variation also can occur (62).
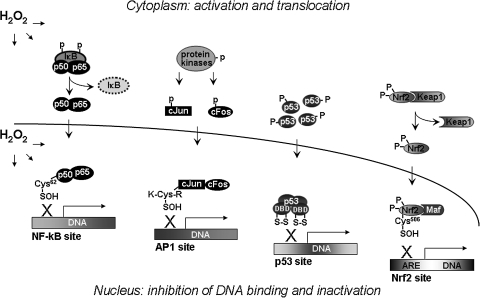
Approaches to measure steady-state redox potentials have shown that the nuclear compartment is maintained at a more-reduced value than is cytoplasm (48) and that the GSH/GSSG and Trx couples in different compartments are not in thermodynamic equilibrium (90). The nuclear redox systems dynamically interact with those in other redox compartments. This is evident in the regulation of transcription by nuclear thiol-disulfide redox-dependent mechanisms. Oxidants in the cytoplasm stimulate oxidative stress signaling, which allows transcriptional activation to enhance systems to protect from uncontrolled oxidative reactions involving lipid peroxidation, DNA crosslinking, and irreversible sulfur hyperoxidation. The fine-tuned redox control systems maintain relatively reduced intracellular redox states, despite responses to oxidative stimuli. A surprising and unexpected aspect of these responses is that major systems controlling cell fate, including signaling of cell survival, growth, and death, involve oxidants in the signaling mechanisms, including those for DNA synthesis, enzyme activation, cell-cycle regulation, and gene expression. However, this research also shows that oxidative stress–caused inactivation of transcription factors is critically important in cell processes, resulting in mutation of DNA and causing subsequent cell death (68). Thus, a general model has emerged (Fig. 6) in which cytoplasmic oxidants activate translocation of transcription factor proteins from the cytoplasm to the nuclei to activate transcription, whereas a nuclear oxidative mechanism, as described in Fig. 5, functions in opposition to turn off the system.
FIG. 6.
A model for transcription-factor regulation in the cytoplasm and nuclei by oxidants. Cytoplasmic oxidants stimulate phosphorylation and translocation of transcription-factor proteins (NF-κB, AP-1, p53, and Nrf2) from the cytoplasm to nuclei to activate transcription. A nuclear oxidative mechanism oxidizes critical Cys residues in the DNA-binding region of the transcription factors and inhibits binding to DNA. This oxidative reaction inactivates transcriptional functions to turn off the system. Thus, the nuclear Trx1 and GSH systems have two functions in the nuclei: maintenance of the oxidant tone, which turns off the transcription factors, and repeated reduction of the critical Cys residues through Ref1 to maintain DNA binding.
Generalizations must be made cautiously because the number of studies and variety of methods available are limited. However, current evidence indicates that, in addition to being relatively reduced, the nucleus is relatively resistant to oxidation by exogenously supplied oxidants. The most widely used methods are described later and involve redox Western blot analysis of Trx1 and quantification of protein S-glutathionylation. Newly available methods to image the GSH redox state by using a fusion of glutaredoxin-1 and a redox-sensitive green fluorescent protein provide a means to measure nuclear redox in live cells (55). In addition, a redox proteomics method provides the means to quantify thiol/disulfide redox changes in specific Cys in different nuclear proteins (49, 62).
Methods to measure nuclear redox states
A previous review summarizes approaches to distinguish nuclear redox states from those in the cytoplasm [see Table 2 and Fig. 2 in the review (48)]. These methods focus on thiol/disulfide redox states of Trx1 and GSH/GSSG because these are abundant molecules present in different subcellular compartments (48, 49); their oxidation provides direct evidence of compartment-specific oxidative stress. Previous studies show Trx1 translocation into the nucleus occurs in response to a variety of stimuli, including oxidants, UV irradiation, cytokines, and lipopolysaccharide (69, 198) in cell-culture and animal models (183). Translocation of Trx1 into nuclei in response to stress may support antioxidant and repair functions. Consequently, the increased Trx1 level in the nuclei may provide a more-reducing environment required for DNA binding by transcription factors. Existence of Trx1 in both the cytoplasm and nucleus enables redox states to be determined in these compartments by comparable redox measurement of Trx1 in cytosolic and nuclear fractions. In addition, cotransfection of cells with NLS-Trx1 (nuclear localization signal-linked, Myc epitope-tagged Trx1) and NES-Trx1 (nuclear export signal-linked, HA epitope-tagged Trx1) plasmid DNA provide tools to compare Trx1 redox state in cytoplasm and nuclei without fractionation (Y-M. Go and D.P. Jones, unpublished data). The latter has the benefit that measurement of redox states of compartment-targeted proteins, NLS-Trx1 for nuclei and NES-Trx1 for cytoplasm, reduces the possible artifactual oxidation in the Trx1 redox state during subcellular fractionation.
In parallel with Trx1 redox measurements, quantification of the GSH/GSSG redox state has been considered another tool to estimate nuclear redox. Technical difficulties in measuring GSH levels in the nuclear compartment have resulted in contradictory interpretations (5, 13, 27, 192). An alternative to measurement of GSH, which can diffuse out of nuclei during fractionation, is to quantify S-glutathionylated protein (Pr-SSG) in the nuclei, which is less subject to artifactual redistribution and provides an indirect estimation of nuclear GSH/GSSG. Pr-SSG can be formed from a reaction of GSSG with protein thiols (PrSH), as well as reaction of oxidized proteins with GSH by GSH S-transferases or thiol peroxidases. Glutathionylation of protein is a dynamic and reversible process thought to be involved in redox regulation of proteins in physiologic as well as pathologic conditions. Molecular mechanisms, quantitation methods, and the potential clinical significance of protein gluatathionylation were discussed in a recent review article (28). S-Glutathionylated hemoglobin has been analyzed as a marker of oxidative stress in some human diseases, including Friedreich ataxia, type I and type II diabetes mellitus, hyperlipidemia and uremia associated with hemodialysis or peritoneal dialysis, Down syndrome, and atherosclerosis (28). Increased glutathionylation of crystallins, human eye-lens proteins, was observed in aging human lenses and was correlated with nuclear color and opalescence, suggesting that Pr-SSG may play an important role in the cataractogenesis of aging (108).
Measurement of Pr-SSG concentrations in nuclei and cytoplasm can be obtained after nuclear isolation. Nuclear and cytoplasmic protein fractions are reduced with DTT, releasing GSH for HPLC analysis (105). Stimulated nuclear production of H2O2 by targeting d-amino acid oxidase to nuclei resulted in increased protein glutathionylation in the nuclei, but not in the cytosol (56). In contrast, culture of cells in media depleting glucose and glutamine resulted in increased protein S-glutathionylation in the cytosolic fraction but not in the nuclei (50). Thus, the data show that protein S-glutathionylation is independently controlled in the cytoplasmic and nuclear compartments and indicate that the GSH/GSSG redox potential is more reduced in nuclei than in cytoplasm.
Redox-dependent Nuclear-Import and -Export Mechanisms
Transport of molecules across the nuclear envelope is highly selective and regulated. Transported molecules include proteins, such as transcription factors that move from the cytoplasm to the nuclei and RNA that move toward to the cytoplasm. Generally, transport mechanisms begin with recognition of a protein by its cognate nuclear-transport signals, including nuclear-localization signal (NLS) (80) and nuclear-export signal (NES) (35) within the protein that mediate its nuclear import and export, respectively. Proteins containing NLS are recognized by a heterodimeric receptor, importin (or karyopherin) (51, 154); however, proteins that do not contain classic NLS, such as the mRNA-binding protein, hnRNPA1, do not interact with importin-α/β. The hnRNPA1 interacts with a different import receptor, a family of proteins related to importin-β [e.g., transportin, for nuclear import (10)]. For nuclear exit, proteins contain NES, which are short stretches of leucine-rich amino acid sequences. The NES regions interact with exportins such as CRM1, proteins that are also members of the importin-β family (39, 178).
Glucocorticoid receptor function is directly regulated by Trx1 through interaction with the DNA-binding domain (111). In addition, Okamoto et al. (135) used a chimeric human glucocorticoid receptor fused to green fluorescent protein and found that nuclear translocation of this receptor was impaired under oxidative conditions in living cells. The C481S mutant of the NLS was not sensitive to oxidative treatment, indicating that C481 is a target amino acid for redox regulation of import. This research demonstrates a direct redox dependence of import due to the presence of a redox-sensitive amino acid in the nuclear localization signal. A similar redox sensitivity is present in the nuclear-export signal (NES) for the fission yeast AP-1–like transcription factor (Pap1) (99, 100). Pap1 is negatively regulated by CRM1/exportin 1, the nuclear export factor. Deletion of the C-terminal cysteine-rich domain (CRD) resulted in nuclear accumulation of Pap1, and deletion and mutational analyses of the CRD showed that C532 was important for Pap1 export. Cys mutation-induced inhibition of nuclear export suggests that this NES provides a redox-sensitive mechanism for controlling nuclear export of Pap1. Consequently, oxidizable Cys residues in both NLS and NES sequences provide switchable mechanisms for redox control of nuclear import and export.
As discussed earlier in this review, Trx1 is present in the cytoplasm and in the nucleus. The mechanism for the oxidative stress–induced nuclear import of Trx1 has been studied (69, 169, 197). Schroeder et al. (169) found that under physiologic levels of H2O2 and nitric oxide, nuclear translocation of Trx1 was controlled by an importin (karyopherin)-α–dependent mechanism. Nuclear Trx1 then provides antiapoptotic properties by binding to several transcription factors and increasing transcription-factor binding to genes through the antioxidant response element, ARE (169). Thioredoxin-binding protein-2 (TBP-2), also known as vitamin D3-upregulated protein-1 or Trx-interacting protein (TXNIP), binds to Trx1 and negatively regulates Trx1 function (Fig. 7). TBP-2 bound to Trx1 accumulated in the nuclei on stress through direct interaction with Rch1, a member of the importin-α family (128). Several reports confirmed that TBP-2 inhibited the reducing activity of Trx1 (84, 129), and that its expression was downregulated in various tumor cells (14, 73). Additionally, Nishinaka et al. (128) found that nuclear localized TBP-2 was imported by interacting with Rch1, so that importin-α functioned as a growth suppressor. In the same study, the authors found that Rch1 inhibited redox-dependent interaction between Trx1 and TBP-2. Therefore, Trx1 may function to regulate nuclear import of TBP-2 by Rch1 and the tumor-suppressive activity of TBP-2.
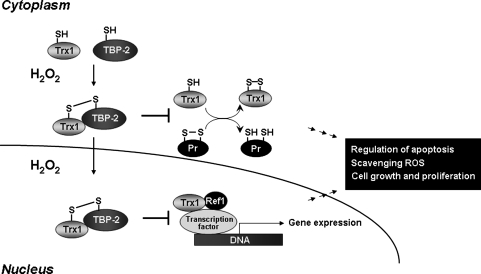
FIG. 7.
Inhibition of Trx1 functions by TBP-2. Trx1-binding protein (TBP)-2 (also called VDUP1 or TXNIP) binds to Trx1 and inhibits Trx1 functions in reduction of protein disulfides. Oxidative stress stimulates this binding and also stimulates accumulation of the complex in nuclei. The Trx1/TBP complex accumulated in the nucleus by oxidants inhibits Trx1 interaction with Ref1. This process inhibits Trx1 function for Ref1 reduction, which in turn affects Ref1 activity to maintain Cys residues of transcription factors for DNA binding and activation for gene expression required for cell growth and proliferation.
Other redox-dependent regulation of subcellular protein trafficking has been investigated. The SECIS (selenocysteine insertion sequence)-binding protein 2, named SBP2, shuttles between cytoplasm and nuclei through NLS and NES motifs. SBP2 regulates selenium incorporation into selenoproteins such as TrxR and Grx members by direct binding to SECIS elements (26). Compartmental trafficking of SBP2 from ribosome to nuclei and vice versa was regulated by NLS and NES, respectively (142). Nuclear accumulation of SBP2 was associated with concomitant decreases in selenoprotein expression levels. Oxidation of the SBP2 redox state resulted from direct Cys oxidation (142). In vitro studies showed that the oxidized form was efficiently reduced by Trx1, suggesting that SBP2 is a Trx1 substrate. In addition, glutathionylated SBP2 was formed during incubation with GSSG, and this formation was reversed by Grx1. However, because redox potentials of the in vitro experiments may not reflect in vivo conditions, it is not clear which of these mechanisms predominates in vivo. The decrease in oxidative modifications of SBP2 by Trx1 or Grx1 supports the idea that the nuclear Trx1 or GSH system plays a role in the nuclear export of SBP2. During cell recovery from oxidative stress, relocation to the ribosomes occurs as a result of reduction.
Redox regulation of the apurinic-apyrimidinic endonuclease 1/redox factor-1 (APE1/Ref-1) in DNA repair and tumorigenesis and neurodegerative diseases has been reported (43, 88, 189). APE1/Ref-1 is present predominantly in the nuclei, although its subcellular localization varies depending on cell types and stress. Nuclear localization of APE1/Ref-1 was dependent on NLS residing within this protein, and nuclear import was found to be mediated by importin (77). The modifications in critical Cys residues (Cys 93, Cys 310) in APE1/Ref-1 by nitrosative stress stimulated nuclear export of APE1/Ref-1 in a CRM1-independent manner, and this process was reversible (150). In addition, S-nitrosation of Cys 93 and Cys 310 suppressed importin-mediated nuclear import of APE1/Ref-1. This NO-stimulated, NO-dependent reaction further supports a critical role of a redox-controlled subcellular trafficking mechanism.
Depletion of glucose/glutamine (Glc/Gln) from cell-culture medium resulted in oxidation of the NADPH/NADP+ and resulted in redox regulation of Ref-1. Total Ref-1 contents in cytosol and nuclear fractions were unaffected by this treatment for 24 h, suggesting that nuclear import of Ref-1 did not occur in response to Glc/Gln depletion. However, quantification of the amount of reduced Ref-1 in each compartment showed that Ref-1 is more reduced in nuclei than in cytoplasm after Glc and Gln depletion. These results show that mechanisms are available to maintain the Ref-1 redox state in the nuclei during oxidative stress (50). Such protection can occur as a consequence of Trx1 translocation to nuclei or of variation in the content or activity of TBP-2, as indicated earlier.
Redox-dependent Regulation in Nuclei
Most nuclear proteins contain cysteine residues that are subject to oxidation during oxidative stress, and considerable evidence shows that GSH and Trx1 systems function in the protection of these Cys, either by elimination of oxidants or by reduction of oxidized residues. Such processes have been studied most extensively in terms of proliferative and growth-promoting effects in cancer and the in resistance of cancer cells to therapy (45, 113, 141, 146, 202, 204). More direct evidence for redox functions in regulation is provided by studies of transcription factors that contain a conserved Cys in the DNA-binding region. This research provides clear support for the interpretation that the thiol antioxidant systems serve to regulate signaling mechanisms in addition to providing the protective and repair functions. Furthermore, recent evidence shows that ROS generation occurs as a consequence of DNA damage, suggesting that nuclear generation of oxidants may also function in signaling. Redox-dependent transcriptional regulation, oxidative DNA damage and repair, and redox mechanisms in control of cell growth and proliferation have been extensively reviewed, so the current overview is intended to highlight some of the important redox-dependent characteristics.
Redox-sensitive transcriptional regulation
Modulation of DNA binding by redox-dependent transcription factors control expression of a wide range of genes in cell proliferation, differentiation, and apoptosis. These redox-dependent transcription factors including NF-κB, p53, AP-1, Nrf2, HIF, glucocorticoid receptor, estrogen receptor, and others. Each has a critical thiol moiety in a cysteine residue, which is controlled by the redox status of the nucleus and plays an important role in the transcriptional activity. For instance, the Cys 62 residue of the p50 subunit within the p50/p65 heterodimer plays a crucial role for the NF-κB activation and inactivation. The Cys 62 residue must be reduced for the p50 subunit to facilitate DNA binding; thus, the redox state of the nuclear compartment is critical for NF-κB activation (37). Both the Trx1 system (69) and the GSH system (161) modulate NF-κB transcriptional activity through Cys 62 of p50 subunit. The transcriptional activation was elevated by Trx1 overexpression, whereas mutations in active site C32 and C35 of Trx1 had significant inhibitory effect on NF-κB activation. In contrast, mutations in nonactive site Cys residues (Cys 62, 69, 73) of Trx1 did not have an effect on NF-κB binding to DNA and transcriptional activation.
The sensitivity of transcription factors to oxidant-induced inhibition was first recognized in studies of AP-1. AP-1 is activated by low levels of oxidative stress, but increased oxidative stress caused inhibition. In studies of the mechanism, a hepatic nuclear protein, identified as redox factor-1 (Ref-1) (201) was recognized to reduce c-Fos and c-Jun members of AP-1 (1). This redox modulation occurred at a conserved Cys residue (Lys-Cys-Arg) present in both c-Fos and c-Jun. Reductions of c-Fos and c-Jun by Ref-1 stimulated AP-1 DNA binding, and this stimulatory effect by nuclear protein was significantly elevated by reduced Trx1 (1). Numerous other studies show that AP-1 activation is redox dependent, regulated by the Trx1 system, including Ref-1 and TrxR, and also by the GSH system (87, 95, 197). DNA binding of AP-1 was shown to be inhibited by oxidized Trx1 and GSSG (45), and transcriptional activation was also shown to be regulated by the redox state of GSH/GSSG (95).
The p53 transcription factor controls the expression of a wide range of genes in response to DNA damage, chemotherapeutic drugs, growth factors, and oxidants (76). The p53 resembles other redox-dependent transcription factors, NF-κB and AP-1, in its susceptibility to redox regulation (155, 180). The p53 consists of three major domains, including a transactivation domain, a DNA-binding domain, and a tetramerization domain. Ten Cys residues are located within the DNA-binding domain of p53, and all are susceptible to oxidation, resulting in alteration of DNA-binding properties. The p53 forms a disulfide bond under mild oxidizing conditions by cis-diaminedichloroplatinum (II), and disulfide bond formation correlates with inhibition of the transcriptional activity of p53 (180). Trx1 enhances p53 DNA-binding activity directly or through Ref-1 mediation (186), whereas dominant-negative Trx1 inhibited p53 DNA-binding activity (156). It has been suggested that a protective role for the nuclear Trx1 in cisplatin-induced apoptosis is perhaps mediated by p53-dependent mechanism (17).
Evidence for the regulation of the DNA-binding function of p53 by GSH/GSSG has also been shown (190). Several Cys residues in the proximal DNA-binding domain are responsible for glutathionylation and have been suggested to regulate the structural dynamics of the DNA-binding domain. Increased glutathionylation of p53 was observed at a lower GSH/GSSG ratio, and its increase was associated with inhibition of DNA-binding function (190). In addition, increased glutathionylation of nuclear p53 was observed with diamide-treated cells, leading to reduction of DNA-binding ability. However, it has not been identified which of the 10 Cys residues in p53 are most reactive in regulation, and the functional consequences of alterations in each of the Cys residues remains unclear. Further investigations of biochemical mechanisms that affect the p53 functions during cellular stress are necessary to provide insight into the roles of these modifications in tumor biology and anticancer therapy.
NF-E2–related factor-2 (Nrf-2) is another redox-sensitive transcription factor that is implicated in cellular responses to oxidative stress. Nrf-2 regulates numerous genes through the antioxidant response element (ARE) as a heterodimer with a small Maf protein (126). Several important antioxidant genes are regulated, including Trx1 (92), glutathione synthesis enzymes (118, 199), GPx, GR (101), GSTs (163), and heme oxygenase-1 (75). Nrf-2 is normally sequestered in the cytoplasm by an inhibitor molecule, Kelch-like ECH-associated protein 1 (Keap-1). Oxidative stress stimulates Nrf-2 release and translocation to the nucleus (127). Similar to other redox-sensitive transcription factors, NF-κB, AP-1, and p53, a critical Cys residue (Cys 506), must be reduced in the nuclei for DNA binding (9). Nrf-2 function is controlled by the dissociation of the Nrf-2/Keap-1 complex. The Cys residues in Keap-1 (Cys 257, 273, 288, and 297) have a crucial role in the dissociation of this complex (30, 127). Compartment-specific Nrf-2 redox control has been shown with oxidative activation in the cytoplasm and oxidative inactivation in the nucleus (61). The data showed that oxidant-induced Nrf-2/Keap-1 dissociation in the cytoplasm was regulated by the GSH system, whereas the DNA-binding activity of Nrf-2 in the nucleus was controlled by nuclear Trx1 (61).
Redox sensitivity of ribonucleotide reductase and DNA damage/repair
Ribonucleotide reductase
Early studies of deoxyribonucleotide synthesis in Escherichia coli led to the discovery of Trx and Grx (131), and related research continues to be an important subject in DNA repair and cancer biology. Ribonucleotide reductase (RNR) catalyzes de novo synthesis of deoxyribonucleotides by reducing the corresponding ribonucleotides for DNA synthesis and repair (131). Mammalian RNR consists of a large R1 subunit containing the substrate-binding site, redox-active Cys residues and a small R2 subunit harboring a tyrosine free radical, which is important in catalysis (97). The RNR activity is cell cycle–dependent in mammalian cells (8). Proliferating cells require a large amount of deoxyribonucleotide during the S phase of the cell cycle. The R2 protein level is low in the G1 phase, but increases to the maximum level during the S phase. Conversely, the R1 protein level remains high and constant throughout the cell cycle. Thus, cell cycle–dependent RNR activity appears to be modulated by the R2 synthesis and degradation. More recently, the p53-dependent RNR small subunit (p53R2), a homologue of R2, was discovered and found to play a pivotal role in the p53-dependent cellular response to DNA damage (54, 184). The p53R2 induced in DNA-damaged cells also forms the RNR complex with the R1 subunit as a partner protein and supplies deoxyribonucleotides for DNA repair (54).
Because the reduction of ribonucleotides is a fundamental initial metabolic step for DNA synthesis, RNR has been considered an important target for cell-growth control and anticancer therapy. For instance, several RNR inhibitors have been used as chemotherapy agents (64, 97). More recent studies show the inhibitory regulation of p53R2 on the MAP kinase signaling pathway. This finding suggests a suppressive role of p53R2 in oncogenicity but also provides a new strategy for inhibiting tumor development (145).
The redox control of RNR in DNA repair is dependent on the Trx1 and GSH systems (3, 122, 160). Each enzymatic cycle results in oxidized RNR, with a disulfide in the C-terminal site of the R1 subunit. Trx1 in the Trx system and Grx1 in the GSH system reduce the oxidized RNR by donating electrons ultimately from NADPH. The study from Avval et al. (3) suggests that the redox control mechanisms of the Trx1 and GSH systems for the RNR reduction are different. Although Trx1 and Grx1 have similar catalytic efficiency in reduction of the C-terminal disulfide in RNR, these reductases appear to favor conditions depending on the levels of RNR (3). Additional in vivo research shows that mice deficient in GSH metabolism accumulate DNA damage in the organs. This finding indicates that GSH plays a critical role in DNA repair through RNR (160). Redox-dependent control of cell cycle also was found to be regulated by the Trx1 redox state. Oxidation of Trx1 under hypoxia decreased RNR activity and caused cell-cycle arrest, suggesting a key role of Trx1 in control of the cellular redox state without the involvement of ROS (122).
DNA damage and repair
DNA is susceptible to oxidative damage caused by ionizing radiation, environmental mutagenesis, and carcinogens. An increasing body of evidence indicates that significant damage to DNA contributes to the pathology of cancer, atherosclerosis, and neurodegenerative diseases (114, 194, 205). Most studies addressing thiol/disulfide systems in DNA damage and repair have focused on protection against reactive species and have not distinguished whether separate redox signaling and control events also function in regulation of the repair machinery. However, several studies show that redox-dependent mechanisms function in controlling the transcription of DNA-repair machinery (40).
A wide range of products are generated during oxidative DNA damage, such as sugar and base modifications and covalent crosslinks with single- and double-stranded breaks. In terms of oxidative damage to DNA, damage to bases and to the phosphodiester backbone are known to be the major causes. One of the most extensively investigated adducts from modifications to DNA bases is 8-oxo-2'-deoxy-guanosine (8-oxodG), an oxidative base lesion resulting in mismatches in the DNA strands (109, 114, 123, 134). Because it was hypothesized that age-related oxidative stress resulted from the loss of the physiologic repair function to the irreversible, oxidative damage in macromolecules and the accumulation of altered macromolecules, such as modified DNAs, a significant number of studies on 8-oxodG in aging also have been performed. Substantial debate about actual in vivo levels of 8-oxodG in DNA have occurred because an artificial increase in 8-oxodG can occur during subcellular fractionation and isolation. However, with extensive precautions in analysis, 8-oxodG levels are still observed to increase with aging in rat and mouse aging models (58). The 8-oxodG accumulates in both the nuclear and the mitochondrial DNA (58, 123, 134). Accumulation of 8-oxodG stimulates mutagenesis by pairing with adenine as well as cytosine (110). A recent study by Oka et al. (134) demonstrated that oxidative stress with menadione treatment stimulated 8-oxodG accumulation in both nuclei and mitochondria. They also identified distinct cell-death mechanisms associated with 8-oxodG in nuclear and mitochondrial DNA. The 8-oxodG in nuclear DNA caused poly-ADP-ribose polymerase (PPAR)-dependent translocation of apoptosis-inducing factor to nuclei, whereas that in mitochondrial DNA caused mitochondrial dysfunction by Ca2+ release and subsequent activation of calpain (134).
Indirect evidence for redox signaling in DNA repair comes from studies showing that reactive oxygen species are generated in response to DNA damage. The study by Salmon et al. (166) to determine the cellular effects of induced, oxidative DNA damage shows a relation between specific oxidative DNA damage levels and biologic consequences of DNA damage levels. They found that DNA damage and intracellular ROS levels are elevated under normal growth conditions (no exposure to exogenous oxidant) in cells deficient in repair enzymes, relative to that in the wild type. Also, cells with elevated levels of DNA damage and ROS exhibit altered abilities to resist exogenous oxidative stress. These findings suggest that the presence of DNA damage alone is sufficient to increase cellular ROS levels and provide insights into the contribution of increasing genetic instability to tumor progression and resistance to chemotherapeutic agents.
Oxidative stress to DNA damage leading to structural changes may be associated with subsequent repair of the lesion. The repair systems include base-excision repair (BER), nucleotide-excision repair (NER), and mismatch repair (109). The majority of adducts of DNA damage are repaired by BER. BER involves several types of DNA glycosylases, including thymine glycol DNA glycosylase, hydroxyl methyl uracil glycosylase, and 8-oxodG glycosylase (25, 63). In humans, hOgg1 and hOgg2, identified as 8-oxodG glycosylases, have a specificity for 8-oxodG in template DNA and misincorporated opposite C and G or A, respectively (66). The hMYH, a human homologue of the E. coli MUTY repair protein, excises A paired with 8-oxodG in template DNA (175). These three enzymes play a key role in BER-dependent suppression of spontaneous mutagenesis induced by oxidative DNA damage. Conversely, the NER-mediated repair of 8-oxodG results in the excision of a lesion-containing oligomer, 24 to 29 nucleotides in length, followed by subsequent reduction to 6- to 7-mers (44). GSH and Trx1 systems can protect against introduction of damage and also regulate expression of DNA-repair genes or modify activity of proteins by glutathionylation (103).
Several repair enzymes were upregulated under oxidative stress by atherosclerosis, providing evidence of DNA damage as a useful biomarker in atherosclerosis (114). These included enzymes regulating the BER system, Ref-1, and poly(ADP-ribose) polymerase 1, and proteins involved in nonspecific repair, p53 and DNA-dependent protein kinase. Urinary 8-oxodG amounts measured from patients with hypertension was correlated with the GSH/GSSG ratio and GSH and GSSG concentration, suggesting that detection of DNA damage could be a useful aspect in the oxidative stress–related diseases (32).
Because oxidative modifications of DNA bases in the nuclei and mitochondria have been suggested as a mechanism of aging and age-related diseases, studies have been performed to evaluate whether a moderate level of regular exercise could be beneficial to age-induced oxidative DNA damage. Results show that swimming and treadmill running attenuate the usual oxidative DNA modifications that are associated with aging, supporting a beneficial role of a moderate level of regular exercise in oxidative DNA damage (123, 152, 153). Moreover, investigation of the effect of a calorie-restricted diet on age-associated DNA damage by examining level of 8-oxodG in aged rodents showed that dietary restriction significantly prevented age-related increase in mitochondrial 8-oxodG, whereas it had little effect on nuclear 8-oxodG in liver and kidney. These results imply that dietary restriction had much greater effect on mitochondrial DNA damage than on nuclear DNA damage (58). The mitochondrial Trx2 system is much more sensitive to oxidation than the nuclear Trx1 system (18), but it is not known whether this difference contributes to the differences in DNA damage. Additional studies are needed to examine the dietary-restricted effect on other tissues, such as heart and brain of aged rodents, other than liver and kidney.
DNA replication, cell proliferation, chromatin remodeling, and other redox-dependent nuclear functions
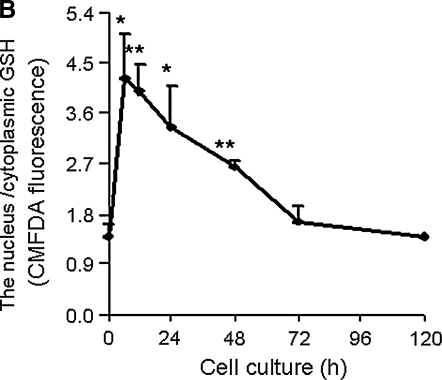
An extensive literature links the abundance and activity of GSH and Trx1-dependent redox systems to increased cell proliferation (45, 113, 130, 141, 204). A strong likelihood exists that nuclei-specific redox regulation is associated with these processes, just as such compartmentalization is recognized in transcriptional regulation. Such regulation is suggested by the redox dependence of the ribonucleotide reductase system. A number of studies show that GSH concentration is increased in rapidly proliferating cells and that decreased GSH limits cell proliferation. The data suggest that an increase in GSH occurs in nuclei of proliferating cells (Fig. 8) (113, 141, 177). This, in addition to the numerous studies showing elevated Trx1 in highly proliferative cancer cells and preferential localization of Trx1 in nuclei of highly proliferative cells (Fig 8), indicates that a highly reducing nuclear environment is critical for proliferation. At present, however, these data must be viewed as circumstantial evidence that a more reduced nuclear redox state occurs in the cell cycle. This subject is of considerable interest for cancer therapy targeting Trx1 and GSH systems (146, 202), and additional studies are needed to address this important subject.
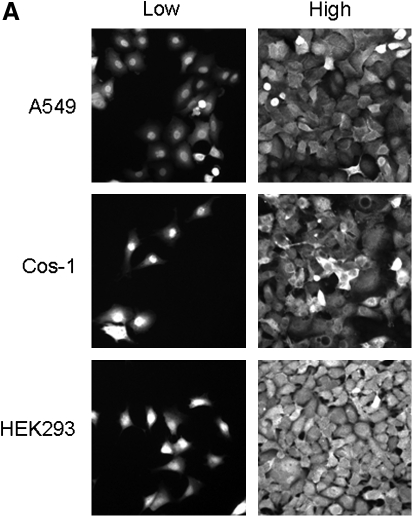
FIG. 8.
Nuclear localization of Trx1 and glutathione is associated with cell proliferation. (A) Low cell-culture density, which is associated with oxidation of thiols and increased cell proliferation, affects the localization of Trx1 in cells. Each cell line (A549, Cos-1, HEK293) was plated in six-well plates containing glass coverslips at low and high density, incubated for 2 days, and assayed for Trx1 localization with immunofluorescence microscopy by using the Trx1 antibody. Data are reproduced from reference (177) with permission. (B) The nucleus/cytoplasm-to-GSH ratio, quantified with CMFDA fluorescence, shows higher levels in proliferating cells than in confluence, suggesting that a reduced nuclear environment is required for cell growth. Data are redrawn from reference (113).
Redox regulation of histone deacetylases, known as sirtuins, has been linked to aging and longevity. This regulation involves NADH/NAD+ redox changes and is not known to be dependent on thiols. However, the processes functioning in chromatin structure and remodeling are very complex, and changes in cellular GSH/GSSG redox potential are known to occur in association with cell differentiation (94). Thus, a possibility exists that specific nuclear redox systems also have a role in regulation of chromatin structure and function. The critical roles of these processes in development and aging therefore warrant investigation of these possibilities.
Summary and Perspectives
Recent changes in the definition of oxidative stress have placed additional focus on the disruption of redox signaling and control as important subjects of investigation. Recognition that nonradical oxidants are quantitatively important in oxidative stress further emphasizes that thiol systems are central targets of injury. Methods are now available to examine specific thiols in cell nuclei, and available data indicate that the central GSH and Trx1 systems are more reduced than those in the cytoplasm and are also more resistant to exogenous oxidants. Despite the relative resistance of nuclear proteins to oxidation, the sensitivity to oxidation of transcription factor binding to DNA suggests that redox mechanisms are involved in a dynamic regulation of function. Additional research using newly developed methods to study nuclear protein oxidation and redox control pathways will aid in understanding complex regulatory events in the cell nucleus and can provide the basis for new therapeutic approaches targeting these critical redox-sensitive processes.
Abbreviations Used
- 8-oxodG
8-oxo-2'-deoxy-guanosine
- APE1
apurinic-apyrimidinic endonuclease 1
- BER
base excision repair
- Cys
cysteine
- DAO
d-amino acid oxidase
- Eh
redox potential
- Glc
glucose
- Gln
glutamine
- GPx
glutathione peroxidase
- GR
GSSG reductase
- Grx
glutaredoxin
- GSH
glutathione
- GSSG
glutathione disulfide
- GST
glutathione S-transferase
- NER
nucleotide-excision repair
- NES
nuclear-export signal
- NLS
nuclear-localization signal
- Nrf2
nuclear factor erythroid 2 (NF-E2)-related factor 2
- PrSH
protein thiol
- Pr-SSG
S-glutathionylated protein
- Prx
peroxiredoxin
- Ref-1
redox factor-1
- RNR
ribonucleotide reductase
- ROS
reactive oxygen species
- SBP2
selenocysteine insertion sequence–binding protein 2
- TBP-2
thioredoxin-binding protein-2
- Trx
thioredoxin
- TrxR
thioredoxin reductase
Acknowledgment
Research on nuclear redox signaling and control in the investigators' laboratory is supported by NIH grant ES011195.
References
- 1.Abate C. Patel L. Rauscher FJ., 3rd Curran T. Redox regulation of fos and jun DNA-binding activity in vitro. Science. 1990;249:1157–1161. doi: 10.1126/science.2118682. [DOI] [PubMed] [Google Scholar]
- 2.Ali-Osman F. Brunner JM. Kutluk TM. Hess K. Prognostic significance of glutathione S-transferase pi expression and subcellular localization in human gliomas. Clin Cancer Res. 1997;3:2253–2261. [PubMed] [Google Scholar]
- 3.Avval FZ. Holmgren A. Molecular mechanisms of thioredoxin and glutaredoxin as hydrogen donors for mammalian S phase ribonucleotide reductase. J Biol Chem. 2009;284:8233–8240. doi: 10.1074/jbc.M809338200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banmeyer I. Marchand C. Verhaeghe C. Vucic B. Rees JF. Knoops B. Overexpression of human peroxiredoxin 5 in subcellular compartments of Chinese hamster ovary cells: effects on cytotoxicity and DNA damage caused by peroxides. Free Radic Biol Med. 2004;36:65–77. doi: 10.1016/j.freeradbiomed.2003.10.019. [DOI] [PubMed] [Google Scholar]
- 5.Bellomo G. Vairetti M. Stivala L. Mirabelli F. Richelmi P. Orrenius S. Demonstration of nuclear compartmentalization of glutathione in hepatocytes. Proc Natl Acad Sci U S A. 1992;89:4412–4416. doi: 10.1073/pnas.89.10.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bishop GM. Dringen R. Robinson SR. Zinc stimulates the production of toxic reactive oxygen species (ROS) and inhibits glutathione reductase in astrocytes. Free Radic Biol Med. 2007;42:1222–1230. doi: 10.1016/j.freeradbiomed.2007.01.022. [DOI] [PubMed] [Google Scholar]
- 7.Biteau B. Labarre J. Toledano MB. ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature. 2003;425:980–984. doi: 10.1038/nature02075. [DOI] [PubMed] [Google Scholar]
- 8.Bjorklund S. Skog S. Tribukait B. Thelander L. S-phase-specific expression of mammalian ribonucleotide reductase R1 and R2 subunit mRNAs. Biochemistry. 1990;29:5452–5458. doi: 10.1021/bi00475a007. [DOI] [PubMed] [Google Scholar]
- 9.Bloom D. Dhakshinamoorthy S. Jaiswal AK. Site-directed mutagenesis of cysteine to serine in the DNA binding region of Nrf2 decreases its capacity to upregulate antioxidant response element-mediated expression and antioxidant induction of NAD(P)H:quinone oxidoreductase1 gene. Oncogene. 2002;21:2191–2200. doi: 10.1038/sj.onc.1205288. [DOI] [PubMed] [Google Scholar]
- 10.Bonifaci N. Moroianu J. Radu A. Blobel G. Karyopherin beta2 mediates nuclear import of a mRNA binding protein. Proc Natl Acad Sci U S A. 1997;94:5055–5060. doi: 10.1073/pnas.94.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brigelius-Flohe R. Tissue-specific functions of individual glutathione peroxidases. Free Radic Biol Med. 1999;27:951–965. doi: 10.1016/s0891-5849(99)00173-2. [DOI] [PubMed] [Google Scholar]
- 12.Brigelius-Flohe R. Muller C. Menard J. Florian S. Schmehl K. Wingler K. Functions of GI-GPx: lessons from selenium-dependent expression and intracellular localization. Biofactors. 2001;14:101–106. doi: 10.1002/biof.5520140114. [DOI] [PubMed] [Google Scholar]
- 13.Briviba K. Fraser G. Sies H. Ketterer B. Distribution of the monochlorobimane-glutathione conjugate between nucleus and cytosol in isolated hepatocytes. Biochem J. 1993;294:631–633. doi: 10.1042/bj2940631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butler LM. Zhou X. Xu WS. Scher HI. Rifkind RA. Marks PA. Richon VM. The histone deacetylase inhibitor SAHA arrests cancer cell growth, up-regulates thioredoxin-binding protein-2, and down-regulates thioredoxin. Proc Natl Acad Sci U S A. 2002;99:11700–11705. doi: 10.1073/pnas.182372299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell JA. Corrigall AV. Guy A. Kirsch RE. Immunohistologic localization of alpha, mu, and pi class glutathione S-transferases in human tissues. Cancer. 1991;67:1608–1613. doi: 10.1002/1097-0142(19910315)67:6<1608::aid-cncr2820670623>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 16.Chang TS. Jeong W. Woo HA. Lee SM. Park S. Rhee SG. Characterization of mammalian sulfiredoxin and its reactivation of hyperoxidized peroxiredoxin through reduction of cysteine sulfinic acid in the active site to cysteine. J Biol Chem. 2004;279:50994–51001. doi: 10.1074/jbc.M409482200. [DOI] [PubMed] [Google Scholar]
- 17.Chen XP. Liu S. Tang WX. Chen ZW. Nuclear thioredoxin-1 is required to suppress cisplatin-mediated apoptosis of MCF-7 cells. Biochem Biophys Res Commun. 2007;361:362–366. doi: 10.1016/j.bbrc.2007.07.033. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y. Cai J. Jones DP. Mitochondrial thioredoxin in regulation of oxidant-induced cell death. FEBS Lett. 2006;580:6596–6602. doi: 10.1016/j.febslet.2006.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y. Cai J. Murphy TJ. Jones DP. Overexpressed human mitochondrial thioredoxin confers resistance to oxidant-induced apoptosis in human osteosarcoma cells. J Biol Chem. 2002;277:33242–33248. doi: 10.1074/jbc.M202026200. [DOI] [PubMed] [Google Scholar]
- 20.Cheng WH. Ho YS. Ross DA. Valentine BA. Combs GF. Lei XG. Cellular glutathione peroxidase knockout mice express normal levels of selenium-dependent plasma and phospholipid hydroperoxide glutathione peroxidases in various tissues. J Nutr. 1997;127:1445–1450. doi: 10.1093/jn/127.8.1445. [DOI] [PubMed] [Google Scholar]
- 21.Choi HJ. Kang SW. Yang CH. Rhee SG. Ryu SE. Crystal structure of a novel human peroxidase enzyme at 2.0 A resolution. Nat Struct Biol. 1998;5:400–406. doi: 10.1038/nsb0598-400. [DOI] [PubMed] [Google Scholar]
- 22.Conrad M. Transgenic mouse models for the vital selenoenzymes cytosolic thioredoxin reductase, mitochondrial thioredoxin reductase and glutathione peroxidase 4. Biochim Biophys Acta. 2009;1790:1575–1585. doi: 10.1016/j.bbagen.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 23.Conrad M. Jakupoglu C. Moreno SG. Lippl S. Banjac A. Schneider M. Beck H. Hatzopoulos AK. Just U. Sinowatz F. Schmahl W. Chien KR. Wurst W. Bornkamm GW. Brielmeier M. Essential role for mitochondrial thioredoxin reductase in hematopoiesis, heart development, and heart function. Mol Cell Biol. 2004;24:9414–9423. doi: 10.1128/MCB.24.21.9414-9423.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conrad M. Moreno SG. Sinowatz F. Ursini F. Kolle S. Roveri A. Brielmeier M. Wurst W. Maiorino M. Bornkamm GW. The nuclear form of phospholipid hydroperoxide glutathione peroxidase is a protein thiol peroxidase contributing to sperm chromatin stability. Mol Cell Biol. 2005;25:7637–7644. doi: 10.1128/MCB.25.17.7637-7644.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cooke MS. Evans MD. Herbert KE. Lunec J. Urinary 8-oxo-2'-deoxyguanosine: source, significance and supplements. Free Radic Res. 2000;32:381–397. doi: 10.1080/10715760000300391. [DOI] [PubMed] [Google Scholar]
- 26.Copeland PR. Driscoll DM. Purification, redox sensitivity, and RNA binding properties of SECIS-binding protein 2, a protein involved in selenoprotein biosynthesis. J Biol Chem. 1999;274:25447–25454. doi: 10.1074/jbc.274.36.25447. [DOI] [PubMed] [Google Scholar]
- 27.Cotgreave IA. Analytical developments in the assay of intra- and extracellular GSH homeostasis: specific protein S-glutathionylation, cellular GSH and mixed disulphide compartmentalisation and interstitial GSH redox balance. Biofactors. 2003;17:269–277. doi: 10.1002/biof.5520170126. [DOI] [PubMed] [Google Scholar]
- 28.Dalle-Donne I. Milzani A. Gagliano N. Colombo R. Giustarini D. Rossi R. Molecular mechanisms and potential clinical significance of S-glutathionylation. Antioxid Redox Signal. 2008;10:445–473. doi: 10.1089/ars.2007.1716. [DOI] [PubMed] [Google Scholar]
- 29.Dijkwel PA. Wenink PW. Structural integrity of the nuclear matrix: differential effects of thiol agents and metal chelators. J Cell Sci. 1986;84:53–67. doi: 10.1242/jcs.84.1.53. [DOI] [PubMed] [Google Scholar]
- 30.Dinkova-Kostova AT. Holtzclaw WD. Cole RN. Itoh K. Wakabayashi N. Katoh Y. Yamamoto M. Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci U S A. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Egler RA. Fernandes E. Rothermund K. Sereika S. de Souza-Pinto N. Jaruga P. Dizdaroglu M. Prochownik EV. Regulation of reactive oxygen species, DNA damage, and c-Myc function by peroxiredoxin 1. Oncogene. 2005;24:8038–8050. doi: 10.1038/sj.onc.1208821. [DOI] [PubMed] [Google Scholar]
- 32.Espinosa O. Jimenez-Almazan J. Chaves FJ. Tormos MC. Clapes S. Iradi A. Salvador A. Fandos M. Redon J. Saez GT. Urinary 8-oxo-7,8-dihydro-2'-deoxyguanosine (8-oxo-dG), a reliable oxidative stress marker in hypertension. Free Radic Res. 2007;41:546–554. doi: 10.1080/10715760601164050. [DOI] [PubMed] [Google Scholar]
- 33.Esworthy RS. Aranda R. Martin MG. Doroshow JH. Binder SW. Chu FF. Mice with combined disruption of Gpx1 and Gpx2 genes have colitis. Am J Physiol Gastrointest Liver Physiol. 2001;281:G848–G855. doi: 10.1152/ajpgi.2001.281.3.G848. [DOI] [PubMed] [Google Scholar]
- 34.Evrard C. Capron A. Marchand C. Clippe A. Wattiez R. Soumillion P. Knoops B. Declercq JP. Crystal structure of a dimeric oxidized form of human peroxiredoxin 5. J Mol Biol. 2004;337:1079–1090. doi: 10.1016/j.jmb.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 35.Fabbro M. Henderson BR. Regulation of tumor suppressors by nuclear-cytoplasmic shuttling. Exp Cell Res. 2003;282:59–69. doi: 10.1016/s0014-4827(02)00019-8. [DOI] [PubMed] [Google Scholar]
- 36.Fernandes AP. Holmgren A. Glutaredoxins: glutathione-dependent redox enzymes with functions far beyond a simple thioredoxin backup system. Antioxid Redox Signal. 2004;6:63–74. doi: 10.1089/152308604771978354. [DOI] [PubMed] [Google Scholar]
- 37.Flohe L. Brigelius-Flohe R. Saliou C. Traber MG. Packer L. Redox regulation of NF-kappa B activation. Free Radic Biol Med. 1997;22:1115–1126. doi: 10.1016/s0891-5849(96)00501-1. [DOI] [PubMed] [Google Scholar]
- 38.Fomenko DE. Novoselov SV. Natarajan SK. Lee BC. Koc A. Carlson BA. Lee TH. Kim HY. Hatfield DL. Gladyshev VN. MsrB1 (methionine-R-sulfoxide reductase 1) knock-out mice: roles of MsrB1 in redox regulation and identification of a novel selenoprotein form. J Biol Chem. 2009;284:5986–5993. doi: 10.1074/jbc.M805770200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fornerod M. Ohno M. Yoshida M. Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 40.Fratelli M. Goodwin LO. Orom UA. Lombardi S. Tonelli R. Mengozzi M. Ghezzi P. Gene expression profiling reveals a signaling role of glutathione in redox regulation. Proc Natl Acad Sci U S A. 2005;102:13998–14003. doi: 10.1073/pnas.0504398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fridovich I. Quantitative aspects of the production of superoxide anion radical by milk xanthine oxidase. J Biol Chem. 1970;245:4053–4057. [PubMed] [Google Scholar]
- 42.Funato Y. Miki H. Nucleoredoxin, a novel thioredoxin family member involved in cell growth and differentiation. Antioxid Redox Signal. 2007;9:1035–1057. doi: 10.1089/ars.2007.1550. [DOI] [PubMed] [Google Scholar]
- 43.Gaiddon C. Moorthy NC. Prives C. Ref-1 regulates the transactivation and pro-apoptotic functions of p53 in vivo. EMBO J. 1999;18:5609–5621. doi: 10.1093/emboj/18.20.5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Galloway AM. Liuzzi M. Paterson MC. Metabolic processing of cyclobutyl pyrimidine dimers and (6-4) photoproducts in UV-treated human cells: evidence for distinct excision-repair pathways. J Biol Chem. 1994;269:974–980. [PubMed] [Google Scholar]
- 45.Galter D. Mihm S. Droge W. Distinct effects of glutathione disulphide on the nuclear transcription factor kappa B and the activator protein-1. Eur J Biochem. 1994;221:639–648. doi: 10.1111/j.1432-1033.1994.tb18776.x. [DOI] [PubMed] [Google Scholar]
- 46.George DL. Francke U. Gene dose effect: regional mapping of human glutathione reductase on chromosome 8. Cytogenet Cell Genet. 1976;17:282–286. doi: 10.1159/000130723. [DOI] [PubMed] [Google Scholar]
- 47.Gladyshev VN. Jeang KT. Stadtman TC. Selenocysteine, identified as the penultimate C-terminal residue in human T-cell thioredoxin reductase, corresponds to TGA in the human placental gene. Proc Natl Acad Sci U S A. 1996;93:6146–6151. doi: 10.1073/pnas.93.12.6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Go YM. Jones DP. Redox compartmentalization in eukaryotic cells. Biochim Biophys Acta. 2008;1780:1273–1290. doi: 10.1016/j.bbagen.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Go YM. Pohl J. Jones DP. Quantification of redox conditions in the nucleus. Methods Mol Biol. 2009;464:303–317. doi: 10.1007/978-1-60327-461-6_17. [DOI] [PubMed] [Google Scholar]
- 50.Go YM. Ziegler TR. Johnson JM. Gu L. Hansen JM. Jones DP. Selective protection of nuclear thioredoxin-1 and glutathione redox systems against oxidation during glucose and glutamine deficiency in human colonic epithelial cells. Free Radic Biol Med. 2007;42:363–370. doi: 10.1016/j.freeradbiomed.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gorlich D. Kostka S. Kraft R. Dingwall C. Laskey RA. Hartmann E. Prehn S. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr Biol. 1995;5:383–392. doi: 10.1016/s0960-9822(95)00079-0. [DOI] [PubMed] [Google Scholar]
- 52.Goto S. Ihara Y. Urata Y. Izumi S. Abe K. Koji T. Kondo T. Doxorubicin-induced DNA intercalation and scavenging by nuclear glutathione S-transferase pi. FASEB J. 2001;15:2702–2714. doi: 10.1096/fj.01-0376com. [DOI] [PubMed] [Google Scholar]
- 53.Goto S. Kamada K. Soh Y. Ihara Y. Kondo T. Significance of nuclear glutathione S-transferase pi in resistance to anti-cancer drugs. Jpn J Cancer Res. 2002;93:1047–1056. doi: 10.1111/j.1349-7006.2002.tb02482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guittet O. Hakansson P. Voevodskaya N. Fridd S. Graslund A. Arakawa H. Nakamura Y. Thelander L. Mammalian p53R2 protein forms an active ribonucleotide reductase in vitro with the R1 protein, which is expressed both in resting cells in response to DNA damage and in proliferating cells. J Biol Chem. 2001;276:40647–40651. doi: 10.1074/jbc.M106088200. [DOI] [PubMed] [Google Scholar]
- 55.Gutscher M. Pauleau AL. Marty L. Brach T. Wabnitz GH. Samstag Y. Meyer AJ. Dick TP. Real-time imaging of the intracellular glutathione redox potential. Nat Methods. 2008;5:553–559. doi: 10.1038/nmeth.1212. [DOI] [PubMed] [Google Scholar]
- 56.Halvey PJ. Hansen JM. Johnson JM. Go YM. Samali A. Jones DP. Selective oxidative stress in cell nuclei by nuclear-targeted D-amino acid oxidase. Antioxid Redox Signal. 2007;9:807–816. doi: 10.1089/ars.2007.1526. [DOI] [PubMed] [Google Scholar]
- 57.Halvey PJ. Watson WH. Hansen JM. Go YM. Samali A. Jones DP. Compartmental oxidation of thiol-disulphide redox couples during epidermal growth factor signalling. Biochem J. 2005;386:215–219. doi: 10.1042/BJ20041829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hamilton ML. Van Remmen H. Drake JA. Yang H. Guo ZM. Kewitt K. Walter CA. Richardson A. Does oxidative damage to DNA increase with age? Proc Natl Acad Sci U S A. 2001;98:10469–10474. doi: 10.1073/pnas.171202698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Han YH. Kwon JH. Yu DY. Moon EY. Inhibitory effect of peroxiredoxin II (Prx II) on Ras-ERK-NFkappaB pathway in mouse embryonic fibroblast (MEF) senescence. Free Radic Res. 2006;40:1182–1189. doi: 10.1080/10715760600868552. [DOI] [PubMed] [Google Scholar]
- 60.Hansen JM. Moriarty-Craige S. Jones DP. Nuclear and cytoplasmic peroxiredoxin-1 differentially regulate NF-kappaB activities. Free Radic Biol Med. 2007;43:282–288. doi: 10.1016/j.freeradbiomed.2007.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hansen JM. Watson WH. Jones DP. Compartmentation of Nrf-2 redox control: regulation of cytoplasmic activation by glutathione and DNA binding by thioredoxin-1. Toxicol Sci. 2004;82:308–3317. doi: 10.1093/toxsci/kfh231. [DOI] [PubMed] [Google Scholar]
- 62.Hansen JM. Zhang H. Jones DP. Mitochondrial thioredoxin-2 has a key role in determining tumor necrosis factor-alpha-induced reactive oxygen species generation, NF-kappaB activation, and apoptosis. Toxicol Sci. 2006;91:643–650. doi: 10.1093/toxsci/kfj175. [DOI] [PubMed] [Google Scholar]
- 63.Harrison L. Ascione AG. Wilson DM., 3rd Demple B. Characterization of the promoter region of the human apurinic endonuclease gene (APE) J Biol Chem. 1995;270:5556–5564. doi: 10.1074/jbc.270.10.5556. [DOI] [PubMed] [Google Scholar]
- 64.Hashemy SI. Ungerstedt JS. Zahedi Avval F. Holmgren A. Motexafin gadolinium, a tumor-selective drug targeting thioredoxin reductase and ribonucleotide reductase. J Biol Chem. 2006;281:10691–10697. doi: 10.1074/jbc.M511373200. [DOI] [PubMed] [Google Scholar]
- 65.Hayes JD. Pulford DJ. The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit Rev Biochem Mol Biol. 1995;30:445–600. doi: 10.3109/10409239509083491. [DOI] [PubMed] [Google Scholar]
- 66.Hazra TK. Izumi T. Maidt L. Floyd RA. Mitra S. The presence of two distinct 8-oxoguanine repair enzymes in human cells: their potential complementary roles in preventing mutation. Nucleic Acids Res. 1998;26:5116–5122. doi: 10.1093/nar/26.22.5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hill KE. Zhou J. McMahan WJ. Motley AK. Atkins JF. Gesteland RF. Burk RF. Deletion of selenoprotein P alters distribution of selenium in the mouse. J Biol Chem. 2003;278:13640–13646. doi: 10.1074/jbc.M300755200. [DOI] [PubMed] [Google Scholar]
- 68.Hirota K. Matsui M. Iwata S. Nishiyama A. Mori K. Yodoi J. AP-1 transcriptional activity is regulated by a direct association between thioredoxin and Ref-1. Proc Natl Acad Sci U S A. 1997;94:3633–3638. doi: 10.1073/pnas.94.8.3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hirota K. Murata M. Sachi Y. Nakamura H. Takeuchi J. Mori K. Yodoi J. Distinct roles of thioredoxin in the cytoplasm and in the nucleus: a two-step mechanism of redox regulation of transcription factor NF-kappaB. J Biol Chem. 1999;274:27891–27897. doi: 10.1074/jbc.274.39.27891. [DOI] [PubMed] [Google Scholar]
- 70.Holmgren A. Antioxidant function of thioredoxin and glutaredoxin systems. Antioxid Redox Signal. 2000;2:811–820. doi: 10.1089/ars.2000.2.4-811. [DOI] [PubMed] [Google Scholar]
- 71.Holmgren A. Hydrogen donor system for Escherichia coli ribonucleoside-diphosphate reductase dependent upon glutathione. Proc Natl Acad Sci U S A. 1976;73:2275–2279. doi: 10.1073/pnas.73.7.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Holmgren A. Johansson C. Berndt C. Lonn ME. Hudemann C. Lillig CH. Thiol redox control via thioredoxin and glutaredoxin systems. Biochem Soc Trans. 2005;33:1375–1377. doi: 10.1042/BST0331375. [DOI] [PubMed] [Google Scholar]
- 73.Ikarashi M. Takahashi Y. Ishii Y. Nagata T. Asai S. Ishikawa K. Vitamin D3 up-regulated protein 1 (VDUP1) expression in gastrointestinal cancer and its relation to stage of disease. Anticancer Res. 2002;22:4045–4048. [PubMed] [Google Scholar]
- 74.Imai H. Nakagawa Y. Biological significance of phospholipid hydroperoxide glutathione peroxidase (PHGPx, GPx4) in mammalian cells. Free Radic Biol Med. 2003;34:145–169. doi: 10.1016/s0891-5849(02)01197-8. [DOI] [PubMed] [Google Scholar]
- 75.Inamdar NM. Ahn YI. Alam J. The heme-responsive element of the mouse heme oxygenase-1 gene is an extended AP-1 binding site that resembles the recognition sequences for MAF and NF-E2 transcription factors. Biochem Biophys Res Commun. 1996;221:570–576. doi: 10.1006/bbrc.1996.0637. [DOI] [PubMed] [Google Scholar]
- 76.Inoue T. Wu L. Stuart J. Maki CG. Control of p53 nuclear accumulation in stressed cells. FEBS Lett. 2005;579:4978–4984. doi: 10.1016/j.febslet.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 77.Jackson EB. Theriot CA. Chattopadhyay R. Mitra S. Izumi T. Analysis of nuclear transport signals in the human apurinic/apyrimidinic endonuclease (APE1/Ref1) Nucleic Acids Res. 2005;33:3303–3312. doi: 10.1093/nar/gki641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jakupoglu C. Przemeck GK. Schneider M. Moreno SG. Mayr N. Hatzopoulos AK. de Angelis MH. Wurst W. Bornkamm GW. Brielmeier M. Conrad M. Cytoplasmic thioredoxin reductase is essential for embryogenesis but dispensable for cardiac development. Mol Cell Biol. 2005;25:1980–1988. doi: 10.1128/MCB.25.5.1980-1988.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jang HH. Lee KO. Chi YH. Jung BG. Park SK. Park JH. Lee JR. Lee SS. Moon JC. Yun JW. Choi YO. Kim WY. Kang JS. Cheong GW. Yun DJ. Rhee SG. Cho MJ. Lee SY. Two enzymes in one; two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell. 2004;117:625–635. doi: 10.1016/j.cell.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 80.Johnson HM. Subramaniam PS. Olsnes S. Jans DA. Trafficking and signaling pathways of nuclear localizing protein ligands and their receptors. Bioessays. 2004;26:993–1004. doi: 10.1002/bies.20086. [DOI] [PubMed] [Google Scholar]
- 81.Johnson JA. el Barbary A. Kornguth SE. Brugge JF. Siegel FL. Glutathione S-transferase isoenzymes in rat brain neurons and glia. J Neurosci. 1993;13:2013–2023. doi: 10.1523/JNEUROSCI.13-05-02013.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jones DP. Extracellular redox state: refining the definition of oxidative stress in aging. Rejuvenation Res. 2006;9:169–181. doi: 10.1089/rej.2006.9.169. [DOI] [PubMed] [Google Scholar]
- 83.Jones DP. Radical-free biology of oxidative stress. Am J Physiol Cell Physiol. 2008;295:C849–C868. doi: 10.1152/ajpcell.00283.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Junn E. Han SH. Im JY. Yang Y. Cho EW. Um HD. Kim DK. Lee KW. Han PL. Rhee SG. Choi I. Vitamin D3 up-regulated protein 1 mediates oxidative stress via suppressing the thioredoxin function. J Immunol. 2000;164:6287–6295. doi: 10.4049/jimmunol.164.12.6287. [DOI] [PubMed] [Google Scholar]
- 85.Kamada K. Goto S. Okunaga T. Ihara Y. Tsuji K. Kawai Y. Uchida K. Osawa T. Matsuo T. Nagata I. Kondo T. Nuclear glutathione S-transferase pi prevents apoptosis by reducing the oxidative stress-induced formation of exocyclic DNA products. Free Radic Biol Med. 2004;37:1875–1884. doi: 10.1016/j.freeradbiomed.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 86.Kang SW. Chae HZ. Seo MS. Kim K. Baines IC. Rhee SG. Mammalian peroxiredoxin isoforms can reduce hydrogen peroxide generated in response to growth factors and tumor necrosis factor-alpha. J Biol Chem. 1998;273:6297–6302. doi: 10.1074/jbc.273.11.6297. [DOI] [PubMed] [Google Scholar]
- 87.Karimpour S. Lou J. Lin LL. Rene LM. Lagunas L. Ma X. Karra S. Bradbury CM. Markovina S. Goswami PC. Spitz DR. Hirota K. Kalvakolanu DV. Yodoi J. Gius D. Thioredoxin reductase regulates AP-1 activity as well as thioredoxin nuclear localization via active cysteines in response to ionizing radiation. Oncogene. 2002;21:6317–6327. doi: 10.1038/sj.onc.1205749. [DOI] [PubMed] [Google Scholar]
- 88.Kelley MR. Parsons SH. Redox regulation of the DNA repair function of the human AP endonuclease Ape1/ref-1. Antioxid Redox Signal. 2001;3:671–683. doi: 10.1089/15230860152543014. [DOI] [PubMed] [Google Scholar]
- 89.Kelner MJ. Montoya MA. Structural organization of the human glutathione reductase gene: determination of correct cDNA sequence and identification of a mitochondrial leader sequence. Biochem Biophys Res Commun. 2000;269:366–368. doi: 10.1006/bbrc.2000.2267. [DOI] [PubMed] [Google Scholar]
- 90.Kemp M. Go YM. Jones DP. Nonequilibrium thermodynamics of thiol/disulfide redox systems: a perspective on redox systems biology. Free Radic Biol Med. 2008;44:921–937. doi: 10.1016/j.freeradbiomed.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kim K. Rhee SG. Stadtman ER. Nonenzymatic cleavage of proteins by reactive oxygen species generated by dithiothreitol and iron. J Biol Chem. 1985;260:15394–15397. [PubMed] [Google Scholar]
- 92.Kim YC. Yamaguchi Y. Kondo N. Masutani H. Yodoi J. Thioredoxin-dependent redox regulation of the antioxidant responsive element (ARE) in electrophile response. Oncogene. 2003;22:1860–1865. doi: 10.1038/sj.onc.1206369. [DOI] [PubMed] [Google Scholar]
- 93.Kim YJ. Ahn JY. Liang P. Ip C. Zhang Y. Park YM. Human prx1 gene is a target of Nrf2 and is up-regulated by hypoxia/reoxygenation: implication to tumor biology. Cancer Res. 2007;67:546–554. doi: 10.1158/0008-5472.CAN-06-2401. [DOI] [PubMed] [Google Scholar]
- 94.Kirlin WG. Cai J. Thompson SA. Diaz D. Kavanagh TJ. Jones DP. Glutathione redox potential in response to differentiation and enzyme inducers. Free Radic Biol Med. 1999;27:1208–1218. doi: 10.1016/s0891-5849(99)00145-8. [DOI] [PubMed] [Google Scholar]
- 95.Klatt P. Molina EP. De Lacoba MG. Padilla CA. Martinez-Galesteo E. Barcena JA. Lamas S. Redox regulation of c-Jun DNA binding by reversible S-glutathiolation. FASEB J. 1999;13:1481–1490. doi: 10.1096/fasebj.13.12.1481. [DOI] [PubMed] [Google Scholar]
- 96.Klug A. Rhodes D. Zinc fingers: a novel protein fold for nucleic acid recognition. Cold Spring Harb Symp Quant Biol. 1987;52:473–482. doi: 10.1101/sqb.1987.052.01.054. [DOI] [PubMed] [Google Scholar]
- 97.Kolberg M. Strand KR. Graff P. Andersson KK. Structure, function, and mechanism of ribonucleotide reductases. Biochim Biophys Acta. 2004;1699:1–34. doi: 10.1016/j.bbapap.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 98.Kryukov GV. Castellano S. Novoselov SV. Lobanov AV. Zehtab O. Guigo R. Gladyshev VN. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 99.Kudo N. Taoka H. Toda T. Yoshida M. Horinouchi S. A novel nuclear export signal sensitive to oxidative stress in the fission yeast transcription factor Pap1. J Biol Chem. 1999;274:15151–15158. doi: 10.1074/jbc.274.21.15151. [DOI] [PubMed] [Google Scholar]
- 100.Kudo N. Taoka H. Yoshida M. Horinouchi S. Identification of a novel nuclear export signal sensitive to oxidative stress in yeast AP-1-like transcription factor. Ann N Y Acad Sci. 1999;886:204–207. doi: 10.1111/j.1749-6632.1999.tb09417.x. [DOI] [PubMed] [Google Scholar]
- 101.Kwak MK. Wakabayashi N. Itoh K. Motohashi H. Yamamoto M. Kensler TW. Modulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1-Nrf2 pathway: identification of novel gene clusters for cell survival. J Biol Chem. 2003;278:8135–8145. doi: 10.1074/jbc.M211898200. [DOI] [PubMed] [Google Scholar]
- 102.Lakshmipathy U. Campbell C. The human DNA ligase III gene encodes nuclear and mitochondrial proteins. Mol Cell Biol. 1999;19:3869–3876. doi: 10.1128/mcb.19.5.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Langie SA. Knaapen AM. Houben JM. van Kempen FC. de Hoon JP. Gottschalk RW. Godschalk RW. van Schooten FJ. The role of glutathione in the regulation of nucleotide excision repair during oxidative stress. Toxicol Lett. 2007;168:302–309. doi: 10.1016/j.toxlet.2006.10.027. [DOI] [PubMed] [Google Scholar]
- 104.Lapenna D. de Gioia S. Ciofani G. Mezzetti A. Ucchino S. Calafiore AM. Napolitano AM. Di Ilio C. Cuccurullo F. Glutathione-related antioxidant defenses in human atherosclerotic plaques. Circulation. 1998;97:1930–1934. doi: 10.1161/01.cir.97.19.1930. [DOI] [PubMed] [Google Scholar]
- 105.Lash LH. Jones DP. Distribution of oxidized and reduced forms of glutathione and cysteine in rat plasma. Arch Biochem Biophys. 1985;240:583–592. doi: 10.1016/0003-9861(85)90065-7. [DOI] [PubMed] [Google Scholar]
- 106.Lillig CH. Berndt C. Holmgren A. Glutaredoxin systems. Biochim Biophys Acta. 2008;1780:1304–1317. doi: 10.1016/j.bbagen.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 107.Lonn ME. Hudemann C. Berndt C. Cherkasov V. Capani F. Holmgren A. Lillig CH. Expression pattern of human glutaredoxin 2 isoforms: identification and characterization of two testis/cancer cell-specific isoforms. Antioxid Redox Signal. 2008;10:547–557. doi: 10.1089/ars.2007.1821. [DOI] [PubMed] [Google Scholar]
- 108.Lou MF. Dickerson JE Jr. Tung WH. Wolfe JK. Chylack LT., Jr Correlation of nuclear color and opalescence with protein S-thiolation in human lenses. Exp Eye Res. 1999;68:547–552. doi: 10.1006/exer.1998.0638. [DOI] [PubMed] [Google Scholar]
- 109.Lunec J. Holloway KA. Cooke MS. Faux S. Griffiths HR. Evans MD. Urinary 8-oxo-2'-deoxyguanosine: redox regulation of DNA repair in vivo? Free Radic Biol Med. 2002;33:875–885. doi: 10.1016/s0891-5849(02)00882-1. [DOI] [PubMed] [Google Scholar]
- 110.Maki H. Origins of spontaneous mutations: specificity and directionality of base-substitution, frameshift, and sequence-substitution mutageneses. Annu Rev Genet. 2002;36:279–303. doi: 10.1146/annurev.genet.36.042602.094806. [DOI] [PubMed] [Google Scholar]
- 111.Makino Y. Yoshikawa N. Okamoto K. Hirota K. Yodoi J. Makino I. Tanaka H. Direct association with thioredoxin allows redox regulation of glucocorticoid receptor function. J Biol Chem. 1999;274:3182–3188. doi: 10.1074/jbc.274.5.3182. [DOI] [PubMed] [Google Scholar]
- 112.Mannervik B. Widersten M. Human Glutathione Transferases: Classification, Tissue Distribution, Structure, and Functional Properties. Luxembourg: Commission of the European Communities; 1995. [Google Scholar]
- 113.Markovic J. Borras C. Ortega A. Sastre J. Vina J. Pallardo FV. Glutathione is recruited into the nucleus in early phases of cell proliferation. J Biol Chem. 2007;282:20416–20424. doi: 10.1074/jbc.M609582200. [DOI] [PubMed] [Google Scholar]
- 114.Martinet W. Knaapen MW. De Meyer GR. Herman AG. Kockx MM. Elevated levels of oxidative DNA damage and DNA repair enzymes in human atherosclerotic plaques. Circulation. 2002;106:927–932. doi: 10.1161/01.cir.0000026393.47805.21. [DOI] [PubMed] [Google Scholar]
- 115.Matsumoto A. Okado A. Fujii T. Fujii J. Egashira M. Niikawa N. Taniguchi N. Cloning of the peroxiredoxin gene family in rats and characterization of the fourth member. FEBS Lett. 1999;443:246–250. doi: 10.1016/s0014-5793(98)01736-0. [DOI] [PubMed] [Google Scholar]
- 116.Meiers I. Shanks JH. Bostwick DG. Glutathione S-transferase pi (GSTP1) hypermethylation in prostate cancer: review. Pathology. 2007;39:299–304. doi: 10.1080/00313020701329906. [DOI] [PubMed] [Google Scholar]
- 117.Moffat GJ. McLaren AW. Wolf CR. Involvement of Jun and Fos proteins in regulating transcriptional activation of the human pi class glutathione S-transferase gene in multidrug-resistant MCF7 breast cancer cells. J Biol Chem. 1994;269:16397–16402. [PubMed] [Google Scholar]
- 118.Moinova HR. Mulcahy RT. Up-regulation of the human gamma-glutamylcysteine synthetase regulatory subunit gene involves binding of Nrf-2 to an electrophile responsive element. Biochem Biophys Res Commun. 1999;261:661–668. doi: 10.1006/bbrc.1999.1109. [DOI] [PubMed] [Google Scholar]
- 119.Molina MM. Belli G. de la Torre MA. Rodriguez-Manzaneque MT. Herrero E. Nuclear monothiol glutaredoxins of Saccharomyces cerevisiae can function as mitochondrial glutaredoxins. J Biol Chem. 2004;279:51923–51930. doi: 10.1074/jbc.M410219200. [DOI] [PubMed] [Google Scholar]
- 120.Moon JC. Hah YS. Kim WY. Jung BG. Jang HH. Lee JR. Kim SY. Lee YM. Jeon MG. Kim CW. Cho MJ. Lee SY. Oxidative stress-dependent structural and functional switching of a human 2-Cys peroxiredoxin isotype II that enhances HeLa cell resistance to H2O2-induced cell death. J Biol Chem. 2005;280:28775–28784. doi: 10.1074/jbc.M505362200. [DOI] [PubMed] [Google Scholar]
- 121.Mu ZM. Yin XY. Prochownik EV. Pag, a putative tumor suppressor, interacts with the Myc Box II domain of c-Myc and selectively alters its biological function and target gene expression. J Biol Chem. 2002;277:43175–43184. doi: 10.1074/jbc.M206066200. [DOI] [PubMed] [Google Scholar]
- 122.Muniyappa H. Song S. Mathews CK. Das KC. Reactive oxygen species-independent oxidation of thioredoxin in hypoxia: inactivation of ribonucleotide reductase and redox-mediated checkpoint control. J Biol Chem. 2009;284:17069–17081. doi: 10.1074/jbc.M109.008557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nakamoto H. Kaneko T. Tahara S. Hayashi E. Naito H. Radak Z. Goto S. Regular exercise reduces 8-oxodG in the nuclear and mitochondrial DNA and modulates the DNA repair activity in the liver of old rats. Exp Gerontol. 2007;42:287–295. doi: 10.1016/j.exger.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 124.Nakamura H. Thioredoxin and its related molecules: update 2005. Antioxid Redox Signal. 2005;7:823–828. doi: 10.1089/ars.2005.7.823. [DOI] [PubMed] [Google Scholar]
- 125.Nalvarte I. Damdimopoulos AE. Nystom C. Nordman T. Miranda-Vizuete A. Olsson JM. Eriksson L. Bjornstedt M. Arner ES. Spyrou G. Overexpression of enzymatically active human cytosolic and mitochondrial thioredoxin reductase in HEK-293 cells: effect on cell growth and differentiation. J Biol Chem. 2004;279:54510–54517. doi: 10.1074/jbc.M408494200. [DOI] [PubMed] [Google Scholar]
- 126.Nguyen T. Huang HC. Pickett CB. Transcriptional regulation of the antioxidant response element: activation by Nrf2 and repression by MafK. J Biol Chem. 2000;275:15466–15473. doi: 10.1074/jbc.M000361200. [DOI] [PubMed] [Google Scholar]
- 127.Nguyen T. Sherratt PJ. Pickett CB. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu Rev Pharmacol Toxicol. 2003;43:233–260. doi: 10.1146/annurev.pharmtox.43.100901.140229. [DOI] [PubMed] [Google Scholar]
- 128.Nishinaka Y. Masutani H. Oka S. Matsuo Y. Yamaguchi Y. Nishio K. Ishii Y. Yodoi J. Importin alpha1 (Rch1) mediates nuclear translocation of thioredoxin-binding protein-2/vitamin D(3)-up-regulated protein 1. J Biol Chem. 2004;279:37559–37565. doi: 10.1074/jbc.M405473200. [DOI] [PubMed] [Google Scholar]
- 129.Nishiyama A. Matsui M. Iwata S. Hirota K. Masutani H. Nakamura H. Takagi Y. Sono H. Gon Y. Yodoi J. Identification of thioredoxin-binding protein-2/vitamin D(3) up-regulated protein 1 as a negative regulator of thioredoxin function and expression. J Biol Chem. 1999;274:21645–21650. doi: 10.1074/jbc.274.31.21645. [DOI] [PubMed] [Google Scholar]
- 130.Nkabyo YS. Ziegler TR. Gu LH. Watson WH. Jones DP. Glutathione and thioredoxin redox during differentiation in human colon epithelial (Caco-2) cells. Am J Physiol Gastrointest Liver Physiol. 2002;283:G1352–G1359. doi: 10.1152/ajpgi.00183.2002. [DOI] [PubMed] [Google Scholar]
- 131.Nordlund P. Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 132.Oberley TD. Verwiebe E. Zhong W. Kang SW. Rhee SG. Localization of the thioredoxin system in normal rat kidney. Free Radic Biol Med. 2001;30:412–424. doi: 10.1016/s0891-5849(00)00486-x. [DOI] [PubMed] [Google Scholar]
- 133.Ojeda L. Keller G. Muhlenhoff U. Rutherford JC. Lill R. Winge DR. Role of glutaredoxin-3 and glutaredoxin-4 in the iron regulation of the Aft1 transcriptional activator in Saccharomyces cerevisiae. J Biol Chem. 2006;281:17661–17669. doi: 10.1074/jbc.M602165200. [DOI] [PubMed] [Google Scholar]
- 134.Oka S. Ohno M. Tsuchimoto D. Sakumi K. Furuichi M. Nakabeppu Y. Two distinct pathways of cell death triggered by oxidative damage to nuclear and mitochondrial DNAs. EMBO J. 2008;27:421–432. doi: 10.1038/sj.emboj.7601975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Okamoto K. Tanaka H. Ogawa H. Makino Y. Eguchi H. Hayashi S. Yoshikawa N. Poellinger L. Umesono K. Makino I. Redox-dependent regulation of nuclear import of the glucocorticoid receptor. J Biol Chem. 1999;274:10363–10371. doi: 10.1074/jbc.274.15.10363. [DOI] [PubMed] [Google Scholar]
- 136.Okazaki S. Naganuma A. Kuge S. Peroxiredoxin-mediated redox regulation of the nuclear localization of Yap1, a transcription factor in budding yeast. Antioxid Redox Signal. 2005;7:327–334. doi: 10.1089/ars.2005.7.327. [DOI] [PubMed] [Google Scholar]
- 137.Olm E. Jönsson-Videsäter K. Ribera-Cortada I. Fernandes AP. Eriksson LC. Lehmann S. Rundlöf A-K. Paul C. Björnstedt M. Selenite is a potent cytotoxic agent for human primary AML cells. Cancer Lett. 2009;282:116–123. doi: 10.1016/j.canlet.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 138.Orlandi A. Costantini S. Campione E. Ferlosio A. Amantea A. Bianchi L. Chimenti S. Spagnoli LG. Relation between animal-type melanoma and reduced nuclear expression of glutathione S-transferase pi. Arch Dermatol. 2009;145:55–62. doi: 10.1001/archderm.145.1.55. [DOI] [PubMed] [Google Scholar]
- 139.Otieno MA. Baggs RB. Hayes JD. Anders MW. Immunolocalization of microsomal glutathione S-transferase in rat tissues. Drug Metab Dispos. 1997;25:12–20. [PubMed] [Google Scholar]
- 140.Otterlei M. Haug T. Nagelhus TA. Slupphaug G. Lindmo T. Krokan HE. Nuclear and mitochondrial splice forms of human uracil-DNA glycosylase contain a complex nuclear localisation signal and a strong classical mitochondrial localisation signal, respectively. Nucleic Acids Res. 1998;26:4611–4617. doi: 10.1093/nar/26.20.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Pallardo FV. Markovic J. Garcia JL. Vina J. Role of nuclear glutathione as a key regulator of cell proliferation. Mol Aspects Med. 2009;30:77–85. doi: 10.1016/j.mam.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 142.Papp LV. Lu J. Striebel F. Kennedy D. Holmgren A. Khanna KK. The redox state of SECIS binding protein 2 controls its localization and selenocysteine incorporation function. Mol Cell Biol. 2006;26:4895–4910. doi: 10.1128/MCB.02284-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Patenaude A. Ven Murthy MR. Mirault ME. Mitochondrial thioredoxin system: effects of TrxR2 overexpression on redox balance, cell growth, and apoptosis. J Biol Chem. 2004;279:27302–27314. doi: 10.1074/jbc.M402496200. [DOI] [PubMed] [Google Scholar]
- 144.Pfeifer H. Conrad M. Roethlein D. Kyriakopoulos A. Brielmeier M. Bornkamm GW. Behne D. Identification of a specific sperm nuclei selenoenzyme necessary for protamine thiol cross-linking during sperm maturation. FASEB J. 2001;15:1236–1238. [PubMed] [Google Scholar]
- 145.Piao C. Jin M. Kim HB. Lee SM. Amatya PN. Hyun JW. Chang IY. You HJ. Ribonucleotide reductase small subunit p53R2 suppresses MEK-ERK activity by binding to ERK kinase 2. Oncogene. 2009;28:2173–2184. doi: 10.1038/onc.2009.84. [DOI] [PubMed] [Google Scholar]
- 146.Powis G. Kirkpatrick DL. Thioredoxin signaling as a target for cancer therapy. Curr Opin Pharmacol. 2007;7:392–397. doi: 10.1016/j.coph.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 147.Pujol-Carrion N. Belli G. Herrero E. Nogues A. de la Torre-Ruiz MA. Glutaredoxins Grx3 and Grx4 regulate nuclear localisation of Aft1 and the oxidative stress response in Saccharomyces cerevisiae. J Cell Sci. 2006;119:4554–4564. doi: 10.1242/jcs.03229. [DOI] [PubMed] [Google Scholar]
- 148.Pushpa-Rekha TR. Burdsall AL. Oleksa LM. Chisolm GM. Driscoll DM. Rat phospholipid-hydroperoxide glutathione peroxidase. cDNA cloning and identification of multiple transcription and translation start sites. J Biol Chem. 1995;270:26993–26999. doi: 10.1074/jbc.270.45.26993. [DOI] [PubMed] [Google Scholar]
- 149.Qiao M. Kisgati M. Cholewa JM. Zhu W. Smart EJ. Sulistio MS. Asmis R. Increased expression of glutathione reductase in macrophages decreases atherosclerotic lesion formation in low-density lipoprotein receptor-deficient mice. Arterioscler Thromb Vasc Biol. 2007;27:1375–1382. doi: 10.1161/ATVBAHA.107.142109. [DOI] [PubMed] [Google Scholar]
- 150.Qu J. Liu GH. Huang B. Chen C. Nitric oxide controls nuclear export of APE1/Ref-1 through S-nitrosation of cysteines 93 and 310. Nucleic Acids Res. 2007;35:2522–2532. doi: 10.1093/nar/gkl1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Rabilloud T. Heller M. Gasnier F. Luche S. Rey C. Aebersold R. Benahmed M. Louisot P. Lunardi J. Proteomics analysis of cellular response to oxidative stress: evidence for in vivo overoxidation of peroxiredoxins at their active site. J Biol Chem. 2002;277:19396–19401. doi: 10.1074/jbc.M106585200. [DOI] [PubMed] [Google Scholar]
- 152.Radak Z. Kaneko T. Tahara S. Nakamoto H. Ohno H. Sasvari M. Nyakas C. Goto S. The effect of exercise training on oxidative damage of lipids, proteins, and DNA in rat skeletal muscle: evidence for beneficial outcomes. Free Radic Biol Med. 1999;27:69–74. doi: 10.1016/s0891-5849(99)00038-6. [DOI] [PubMed] [Google Scholar]
- 153.Radak Z. Kaneko T. Tahara S. Nakamoto H. Pucsok J. Sasvari M. Nyakas C. Goto S. Regular exercise improves cognitive function and decreases oxidative damage in rat brain. Neurochem Int. 2001;38:17–23. doi: 10.1016/s0197-0186(00)00063-2. [DOI] [PubMed] [Google Scholar]
- 154.Radu A. Blobel G. Moore MS. Identification of a protein complex that is required for nuclear protein import and mediates docking of import substrate to distinct nucleoporins. Proc Natl Acad Sci U S A. 1995;92:1769–1773. doi: 10.1073/pnas.92.5.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Rainwater R. Parks D. Anderson ME. Tegtmeyer P. Mann K. Role of cysteine residues in regulation of p53 function. Mol Cell Biol. 1995;15:3892–3903. doi: 10.1128/mcb.15.7.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ravi D. Muniyappa H. Das KC. Endogenous thioredoxin is required for redox cycling of anthracyclines and p53-dependent apoptosis in cancer cells. J Biol Chem. 2005;280:40084–40096. doi: 10.1074/jbc.M507192200. [DOI] [PubMed] [Google Scholar]
- 157.Rhee SG. Chae HZ. Kim K. Peroxiredoxins: a historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic Biol Med. 2005;38:1543–1552. doi: 10.1016/j.freeradbiomed.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 158.Rogers LK. Gupta S. Welty SE. Hansen TN. Smith CV. Nuclear and nucleolar glutathione reductase, peroxidase, and transferase activities in livers of male and female Fischer-344 rats. Toxicol Sci. 2002;69:279–285. doi: 10.1093/toxsci/69.1.279. [DOI] [PubMed] [Google Scholar]
- 159.Rogers LK. Hansen TN. Welty SE. Smith CV. Nuclear glutathione reductase activities are increased during S phase of cell cycle in Chinese hamster ovary (CHO) cells. Toxicologist. 2002;66:143. [Google Scholar]
- 160.Rojas E. Valverde M. Kala SV. Kala G. Lieberman MW. Accumulation of DNA damage in the organs of mice deficient in gamma-glutamyltranspeptidase. Mutat Res. 2000;447:305–316. doi: 10.1016/s0027-5107(99)00191-8. [DOI] [PubMed] [Google Scholar]
- 161.Rokutan K. Teshima S. Miyoshi M. Kawai T. Nikawa T. Kishi K. Glutathione depletion inhibits oxidant-induced activation of nuclear factor-kappa B, AP-1, and c-Jun/ATF-2 in cultured guinea-pig gastric epithelial cells. J Gastroenterol. 1998;33:646–655. doi: 10.1007/s005350050151. [DOI] [PubMed] [Google Scholar]
- 162.Rudhe C. Clifton R. Whelan J. Glaser E. N-terminal domain of the dual-targeted pea glutathione reductase signal peptide controls organellar targeting efficiency. J Mol Biol. 2002;324:577–585. doi: 10.1016/s0022-2836(02)01133-6. [DOI] [PubMed] [Google Scholar]
- 163.Rushmore TH. Pickett CB. Glutathione S-transferases, structure, regulation, and therapeutic implications. J Biol Chem. 1993;268:11475–11478. [PubMed] [Google Scholar]
- 164.Saitoh M. Nishitoh H. Fujii M. Takeda K. Tobiume K. Sawada Y. Kawabata M. Miyazono K. Ichijo H. Mammalian thioredoxin is a direct inhibitor of apoptosis signal-regulating kinase (ASK) 1. EMBO J. 1998;17:2596–2606. doi: 10.1093/emboj/17.9.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sakai M. Muramatsu M. Regulation of glutathione transferase P: a tumor marker of hepatocarcinogenesis. Biochem Biophys Res Commun. 2007;357:575–578. doi: 10.1016/j.bbrc.2007.03.174. [DOI] [PubMed] [Google Scholar]
- 166.Salmon TB. Evert BA. Song B. Doetsch PW. Biological consequences of oxidative stress-induced DNA damage in Saccharomyces cerevisiae. Nucleic Acids Res. 2004;32:3712–3723. doi: 10.1093/nar/gkh696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Savaskan NE. Brauer AU. Kuhbacher M. Eyupoglu IY. Kyriakopoulos A. Ninnemann O. Behne D. Nitsch R. Selenium deficiency increases susceptibility to glutamate-induced excitotoxicity. FASEB J. 2003;17:112–114. doi: 10.1096/fj.02-0067fje. [DOI] [PubMed] [Google Scholar]
- 168.Savaskan NE. Ufer C. Kuhn H. Borchert A. Molecular biology of glutathione peroxidase 4: from genomic structure to developmental expression and neural function. Biol Chem. 2007;388:1007–1017. doi: 10.1515/BC.2007.126. [DOI] [PubMed] [Google Scholar]
- 169.Schroeder P. Popp R. Wiegand B. Altschmied J. Haendeler J. Nuclear redox-signaling is essential for apoptosis inhibition in endothelial cells: important role for nuclear thioredoxin-1. Arterioscler Thromb Vasc Biol. 2007;27:2325–2331. doi: 10.1161/ATVBAHA.107.149419. [DOI] [PubMed] [Google Scholar]
- 170.Seo MS. Kang SW. Kim K. Baines IC. Lee TH. Rhee SG. Identification of a new type of mammalian peroxiredoxin that forms an intramolecular disulfide as a reaction intermediate. J Biol Chem. 2000;275:20346–20354. doi: 10.1074/jbc.M001943200. [DOI] [PubMed] [Google Scholar]
- 171.Sharma R. Yang Y. Sharma A. Awasthi S. Awasthi YC. Antioxidant role of glutathione S-transferases: protection against oxidant toxicity and regulation of stress-mediated apoptosis. Antioxid Redox Signal. 2004;6:289–300. doi: 10.1089/152308604322899350. [DOI] [PubMed] [Google Scholar]
- 172.Sherratt PJ. Williams S. Foster J. Kernohan N. Green T. Hayes JD. Direct comparison of the nature of mouse and human GST T1-1 and the implications on dichloromethane carcinogenicity. Toxicol Appl Pharmacol. 2002;179:89–97. doi: 10.1006/taap.2002.9348. [DOI] [PubMed] [Google Scholar]
- 173.Shiratory Y. Soma Y. Maruyama H. Sato S. Takano A. Sato K. Immunohistochemical detection of the placental form of glutathione S-transferase in dysplastic and neoplastic human uterine cervix lesions. Cancer Res. 1987;47:1806–6809. [PubMed] [Google Scholar]
- 174.Singh SS. Li Y. Ford OH. Wrzosek CS. Mehedint DC. Titus MA. Mohler JL. Thioredoxin reductase 1 expression and castration-recurrent growth of prostate cancer. Transl Oncol. 2008;1:153–157. doi: 10.1593/tlo.08145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Slupska MM. Luther WM. Chiang JH. Yang H. Miller JH. Functional expression of hMYH, a human homolog of the Escherichia coli MutY protein. J Bacteriol. 1999;181:6210–6213. doi: 10.1128/jb.181.19.6210-6213.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Smith CV. Jones DP. Guenthner TM. Lash LH. Lauterburg BH. Compartmentation of glutathione: implications for the study of toxicity and disease. Toxicol Appl Pharmacol. 1996;140:1–12. doi: 10.1006/taap.1996.0191. [DOI] [PubMed] [Google Scholar]
- 177.Spielberger JC. Moody AD. Watson WH. Oxidation and nuclear localization of thioredoxin-1 in sparse cell cultures. J Cell Biochem. 2008;104:1879–1889. doi: 10.1002/jcb.21762. [DOI] [PubMed] [Google Scholar]
- 178.Stade K. Ford CS. Guthrie C. Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 179.Strange RC. Spiteri MA. Ramachandran S. Fryer AA. Glutathione-S-transferase family of enzymes. Mutat Res. 2001;482:21–26. doi: 10.1016/s0027-5107(01)00206-8. [DOI] [PubMed] [Google Scholar]
- 180.Sun XZ. Vinci C. Makmura L. Han S. Tran D. Nguyen J. Hamann M. Grazziani S. Sheppard S. Gutova M. Zhou F. Thomas J. Momand J. Formation of disulfide bond in p53 correlates with inhibition of DNA binding and tetramerization. Antioxid Redox Signal. 2003;5:655–665. doi: 10.1089/152308603770310338. [DOI] [PubMed] [Google Scholar]
- 181.Sundberg AG. Nilsson R. Appelkvist EL. Dallner G. Immunohistochemical localization of alpha and pi class glutathione transferases in normal human tissues. Pharmacol Toxicol. 1993;72:321–331. doi: 10.1111/j.1600-0773.1993.tb01658.x. [DOI] [PubMed] [Google Scholar]
- 182.Suthanthiran M. Anderson ME. Sharma VK. Meister A. Glutathione regulates activation-dependent DNA synthesis in highly purified normal human T lymphocytes stimulated via the CD2 and CD3 antigens. Proc Natl Acad Sci USA. 1990;87:3343–3347. doi: 10.1073/pnas.87.9.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Takagi Y. Mitsui A. Nishiyama A. Nozaki K. Sono H. Gon Y. Hashimoto N. Yodoi J. Overexpression of thioredoxin in transgenic mice attenuates focal ischemic brain damage. Proc Natl Acad Sci U S A. 1999;96:4131–4136. doi: 10.1073/pnas.96.7.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Tanaka H. Arakawa H. Yamaguchi T. Shiraishi K. Fukuda S. Matsui K. Takei Y. Nakamura Y. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404:42–49. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 185.Tanriverdi T. Hanimoglu H. Kacira T. Sanus GZ. Kemerdere R. Atukeren P. Gumustas K. Canbaz B. Kaynar MY. Glutathione peroxidase, glutathione reductase and protein oxidation in patients with glioblastoma multiforme and transitional meningioma. J Cancer Res Clin Oncol. 2007;133:627–633. doi: 10.1007/s00432-007-0212-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ueno M. Masutani H. Arai RJ. Yamauchi A. Hirota K. Sakai T. Inamoto T. Yamaoka Y. Yodoi J. Nikaido T. Thioredoxin-dependent redox regulation of p53-mediated p21 activation. J Biol Chem. 1999;274:35809–35815. doi: 10.1074/jbc.274.50.35809. [DOI] [PubMed] [Google Scholar]
- 187.Ursini F. Maiorino M. Gregolin C. The selenoenzyme phospholipid hydroperoxide glutathione peroxidase. Biochim Biophys Acta. 1985;839:62–70. doi: 10.1016/0304-4165(85)90182-5. [DOI] [PubMed] [Google Scholar]
- 188.Ursini F. Maiorino M. Valente M. Ferri L. Gregolin C. Purification from pig liver of a protein which protects liposomes and biomembranes from peroxidative degradation and exhibits glutathione peroxidase activity on phosphatidylcholine hydroperoxides. Biochim Biophys Acta. 1982;710:197–211. doi: 10.1016/0005-2760(82)90150-3. [DOI] [PubMed] [Google Scholar]
- 189.Vasko MR. Guo C. Kelley MR. The multifunctional DNA repair/redox enzyme Ape1/Ref-1 promotes survival of neurons after oxidative stress. DNA Repair (Amst) 2005;4:367–379. doi: 10.1016/j.dnarep.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 190.Velu CS. Niture SK. Doneanu CE. Pattabiraman N. Srivenugopal KS. Human p53 is inhibited by glutathionylation of cysteines present in the proximal DNA-binding domain during oxidative stress. Biochemistry. 2007;46:7765–7780. doi: 10.1021/bi700425y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Vendeland SC. Beilstein MA. Yeh JY. Ream W. Whanger PD. Rat skeletal muscle selenoprotein W: cDNA clone and mRNA modulation by dietary selenium. Proc Natl Acad Sci U S A. 1995;92:8749–8753. doi: 10.1073/pnas.92.19.8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Voehringer DW. McConkey DJ. McDonnell TJ. Brisbay S. Meyn RE. Bcl-2 expression causes redistribution of glutathione to the nucleus. Proc Natl Acad Sci U S A. 1998;95:2956–2960. doi: 10.1073/pnas.95.6.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Walther UI. Czermak A. Muckter H. Walther SC. Fichtl B. Decreased GSSG reductase activity enhances cellular zinc toxicity in three human lung cell lines. Arch Toxicol. 2003;77:131–137. doi: 10.1007/s00204-002-0421-z. [DOI] [PubMed] [Google Scholar]
- 194.Wang D. Kreutzer DA. Essigmann JM. Mutagenicity and repair of oxidative DNA damage: insights from studies using defined lesions. Mutat Res. 1998;400:99–115. doi: 10.1016/s0027-5107(98)00066-9. [DOI] [PubMed] [Google Scholar]
- 195.Wang J. Pan S. Berk BC. Glutaredoxin mediates Akt and eNOS activation by flow in a glutathione reductase-dependent manner. Arterioscler Thromb Vasc Biol. 2007;27:1283–1288. doi: 10.1161/ATVBAHA.107.144659. [DOI] [PubMed] [Google Scholar]
- 196.Wang Y. Qiao M. Mieyal JJ. Asmis LM. Asmis R. Molecular mechanism of glutathione-mediated protection from oxidized low-density lipoprotein-induced cell injury in human macrophages: role of glutathione reductase and glutaredoxin. Free Radic Biol Med. 2006;41:775–785. doi: 10.1016/j.freeradbiomed.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 197.Wei SJ. Botero A. Hirota K. Bradbury CM. Markovina S. Laszlo A. Spitz DR. Goswami PC. Yodoi J. Gius D. Thioredoxin nuclear translocation and interaction with redox factor-1 activates the activator protein-1 transcription factor in response to ionizing radiation. Cancer Res. 2000;60:6688–6695. [PubMed] [Google Scholar]
- 198.Wiesel P. Foster LC. Pellacani A. Layne MD. Hsieh CM. Huggins GS. Strauss P. Yet SF. Perrella MA. Thioredoxin facilitates the induction of heme oxygenase-1 in response to inflammatory mediators. J Biol Chem. 2000;275:24840–24846. doi: 10.1074/jbc.M000835200. [DOI] [PubMed] [Google Scholar]
- 199.Wild AC. Gipp JJ. Mulcahy T. Overlapping antioxidant response element and PMA response element sequences mediate basal and beta-naphthoflavone-induced expression of the human gamma-glutamylcysteine synthetase catalytic subunit gene. Biochem J. 1998;332:373–381. doi: 10.1042/bj3320373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Wood ZA. Schroder E. Robin Harris J. Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 201.Xanthoudakis S. Miao G. Wang F. Pan YC. Curran T. Redox activation of Fos-Jun DNA binding activity is mediated by a DNA repair enzyme. EMBO J. 1992;11:3323–3335. doi: 10.1002/j.1460-2075.1992.tb05411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Yang P. Mandrekar SJ. Hillman SH. Allen Ziegler KL. Sun Z. Wampfler JA. Cunningham JM. Sloan JA. Adjei AA. Perez E. Jett JR. Evaluation of glutathione metabolic genes on outcomes in advanced non-small cell lung cancer patients after initial treatment with platinum-based chemotherapy: an NCCTG-97-24-51 based study. J Thorac Oncol. 2009;4:479–485. doi: 10.1097/jto.0b013e31819c7a2c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Yant LJ. Ran Q. Rao L. Van Remmen H. Shibatani T. Belter JG. Motta L. Richardson A. Prolla TA. The selenoprotein GPX4 is essential for mouse development and protects from radiation and oxidative damage insults. Free Radic Biol Med. 2003;34:496–502. doi: 10.1016/s0891-5849(02)01360-6. [DOI] [PubMed] [Google Scholar]
- 204.Yoshida T. Nakamura H. Masutani H. Yodoi J. The involvement of thioredoxin and thioredoxin binding protein-2 on cellular proliferation and aging process. Ann N Y Acad Sci. 2005;1055:1–12. doi: 10.1196/annals.1323.002. [DOI] [PubMed] [Google Scholar]
- 205.Zhang J. Perry G. Smith MA. Robertson D. Olson SJ. Graham DG. Montine TJ. Parkinson's disease is associated with oxidative damage to cytoplasmic DNA and RNA in substantia nigra neurons. Am J Pathol. 1999;154:1423–1429. doi: 10.1016/S0002-9440(10)65396-5. [DOI] [PMC free article] [PubMed] [Google Scholar]