Abstract
Lim3 encodes an RNA polymerase II transcription factor with a key role in neuron specification. It was also identified as a candidate gene that affects lifespan. These pleiotropic effects indicate the fundamental significance of the potential interplay between neural development and lifespan control. The goal of this study was to analyze the causal relationships between Lim3 structural variations, and gene expression and lifespan changes, and to provide insights into regulatory pathways controlling lifespan. Fifty substitution lines containing second chromosomes from a Drosophila natural population were used to analyze the association between lifespan and sequence variation in the 5′-regulatory region, and first exon and intron of Lim3A, in which we discovered multiple transcription start sites (TSS). The core and proximal promoter organization for Lim3A and a previously unknown mRNA named Lim3C were described. A haplotype of two markers in the Lim3A regulatory region was significantly associated with variation in lifespan. We propose that polymorphisms in the regulatory region affect gene transcription, and consequently lifespan. Indeed, five polymorphic markers located within 380 to 680 bp of the Lim3A major TSS, including two markers associated with lifespan variation, were significantly associated with the level of Lim3A transcript, as evaluated by real time RT-PCR in embryos, adult heads, and testes. A naturally occurring polymorphism caused a six-fold change in gene transcription and a 25% change in lifespan. Markers associated with long lifespan and intermediate Lim3A transcription were present in the population at high frequencies. We hypothesize that polymorphic markers associated with Lim3A expression are located within the binding sites for proteins that regulate gene function, and provide general rather than tissue-specific regulation of transcription, and that intermediate levels of Lim3A expression confer a selective advantage and longer lifespan.
Introduction
Lifespan is determined by a complex interplay between environmental and genetic factors. Temperature, air pollution, nutrition, and other factors affect multiple processes through various signaling and metabolic pathways. Many genes are involved in these pathways, and therefore control lifespan. Indeed, hundreds of genes are known to affect lifespan in model organisms [1]–[3]. However, many aspects of the genetic control of lifespan remain unclear. One that is especially interesting for us is how naturally occurring structural and functional variations in a gene can affect this phenotypic trait. Recent studies of natural nucleotide divergence in a variety of Drosophila genes demonstrated associations between structural polymorphisms in several genes and quantitative traits, including lifespan [4]–[6]. However, the causal relation of these structural variations and gene expression changes and phenotype alterations remains poorly understood.
Several candidate genes affecting lifespan have been revealed using recombination mapping followed by quantitative complementation tests with deficiencies and mutations at candidate loci [7]. Among others, Lim3 was identified as a candidate gene affecting lifespan [8]. Recent data show that this gene is also associated with locomotion behavior [9].
Lim3 is located in cytological region 37B13-37C1 of the second chromosome, and is a homeobox gene that encodes an RNA polymerase II transcription factor (TF) required for development and function of neurons. Lim3 is involved in complicated motor neuron specification networks, and is activated by Nkx6 and repressed by Even skipped (Eve) [10]. Lim3 may regulate axon extension and fasciculation through its downstream target, FasciclinIII [11]. With Islet and Drifter, Lim3 constitutes a “combinatorial code” that generates distinct motor neuron identities [10], [12]. The Lim3 protein contains two LIM domains, a carboxyterminal homeodomain, and a highly conserved 22-amino acid region called the Lim3-specific domain (LSD). Lim3 is highly homologous to the vertebrate LHX3/4 subclass of LIM-homeodomain proteins, with 95% and 98% identity to human LHX3 and LHX4 in the homeodomain region, 89% identity in the LIM domains, and 45% identity in the LSD [13]. Like Lim3, human LHX3/4 are TFs required for pituitary development and motor neuron specification. Mutations in LHX3/4 are associated with combined pituitary hormone deficiency, rigid cervical spine, or short stature [14]–[16].
The involvement of Lim3 in both the regulation of neuron development and lifespan control could be of fundamental significance. The effect of Drosophila Lim3 on lifespan control could be conserved in multicellular eucaryotes, including humans, similar to its role in neuron identification. Analysis of the causal relationships between Lim3 structure, transcription level, and lifespan will provide insight into conserved regulatory pathways controlling lifespan. In this paper, we demonstrate the potential of naturally occurring polymorphisms in the Lim3 5′-regulatory region to modulate gene expression and fly lifespan.
Results
The exact mechanisms of Lim3A transcription, and the structure of its potential regulatory region were unknown. To characterize and evaluate the functional role of naturally occurring polymorphisms of the Lim3 5′-regulatory region, we first analyzed initiation of Lim3A transcription and determined the exact border between the regulatory and structural parts of the gene, and outlined proximal promoter region and potential binding sites for regulatory proteins within the regulatory region.
Analysis of Lim3A transcription initiation and proximal promoter region
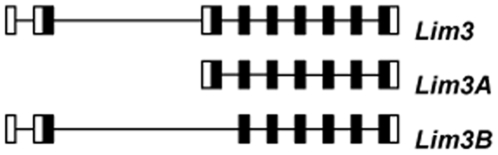
Lim3 was found to produce two mRNAs: Lim3A and Lim3B (Gen Bank accession nos. NM_057258 and NM_165277), with the same structure, except that the first exon of Lim3A is replaced by two different exons in Lim3B (Figure 1). We focused on Lim3A, which has been shown to have a function in Drosophila neuron development [10].
Figure 1. The structure of Drosophila Lim3 gene.
Exons are depicted by rectangles, white rectangles correspond to untranslated regions; introns are indicated by black lines.
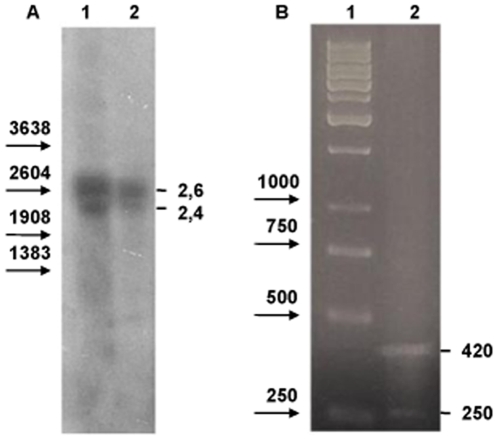
Northern blot using a Lim3A-specific probe revealed two transcripts (Figure 2A). The larger 2.6 kb major transcript was identical in size to Lim3A; the minor 2.4 kb transcript, which we called Lim3C, was new. 5′-RACE analysis (Figure 2B) confirmed the additional Lim3C mRNA. Sequences of 47 clones obtained by 5′-RACE (GenBank accession no. GU814523–GU814569) demonstrated that each transcript had an array of closely located transcriptional start sites (TSSs) with different initiation rates. The major Lim3A TSS (Figure 3) was at −6 nucleotides (18 clones), and the minor TSSs were at −16 (3 clones), −2 (8 clones), and +14 (4 clones) relative to the earlier annotated start site. The major Lim3C TSS (Figure 3) was at +184 (8 clones), and the minor TSSs were at +169 (3 clones), and +179 (3 clones) relative to the earlier annotated start site. TSSs located downstream of the major TSS might correspond to accidentally truncated fragments of full-length RNA molecules, so only TSSs represented by three or more clones were considered. Lim3C appeared to be 190 bp shorter than Lim3A because of the reduced length of the untranslated region (UTR). Seven identical exons were present in both transcripts (data not shown).
Figure 2. Molecular analyses of Lim3A.
(A). Northern blot analysis of Lim3A. Lanes 1 and 2 present RNA from 12 hr embryos of two different homozygous substitution Drosophila lines. PCR fragment including the 5′ region and the first exon of Lim3A was used as a probe. The mobilities of standard DNA markers (Promega) are depicted by arrows on the left. The approximate sizes of the transcripts are indicated in kilobases on the right. (B). 5′ RACE analysis of Lim3A. Lane 1 is a 1-kb DNA ladder from Fermentas. Lane 2 represents 5′ RACE products, sizes are indicated in base pairs on the right.
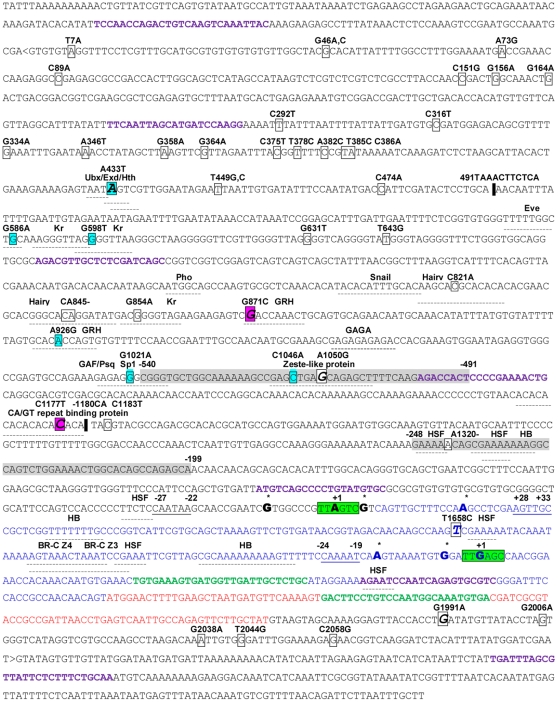
Figure 3. The structure and nucleotide sequence of the 5′ end of Lim3A and Lim3C.
Letters in blue correspond to 5′ UTR of Lim3A, letters in red – to translated region of the first exon of Lim3A, Lim3C. Letters in violet correspond to primers for PCR and sequence analysis. Letters in green correspond to primers for Real-time RT-PCR. The borders of the analysed sequence are depicted by </>. The promoter sequences predicted by the Neural Network Promoter Prediction database, version 2.2 are depicted by light grey color. Sequences marked with green rectangles are initiation regions. TSSs are indicated by large bold letters and asterisks. The underlined sequences are putative core promoter elements. Letters outlined by squares are polymorphic sites which were present in the sample with the frequency 0.06 and higher, insertions are depicted by black rectangles. Large, bold letters in italics outlined by squares are lifespan associated SNPs. Large, bold letters in italics outlined by pink squares are SNPs composing haplotype significantly associated with lifespan. Letters outlined by blue squares are SNPs significantly associated with Lim3A mRNA amount. The transcription factor binding sites are denoted by the dotted lines.
Lim3A and Lim3C TSSs were located within the large 12-kb intron of Lim3B (Figure 3). Several bioinformatic resources were used to determine regulatory elements present in the core and proximal promoter regions of Lim3A and Lim3C. The Lim3A transcript start region (initiator) was a close match to the consensus sequence of the D. melanogaster initiator T-C-A+1-G/T-T-T/C [17], [18], and appeared to be TTA+1GTC. Almost identical initiators were found in 13.2% of genes in the Drosophila Core Promoter Database. Most of these (63%) contained downstream core promoter elements (DPEs), and mainly had similar functions, specifically RNA polymerase II TF activity, which correlated with the Lim3 function.
The Eukaryotic Promoter Database, Current Release 100, and Drosophila Core Promoter Database were used to detect Drosophila core promoter elements in the Lim3A regulatory region. A DPE was identified at +28 to +33 nucleotides relative to the major TSS of Lim3A (Figure 3). The DPE sequence, AGTTGC, was a reasonable match to the consensus DPE sequence (A/G/T+28-C/G-A/T-C/T-A/C/G-C/T) [17], [18], and was encountered in 0.8% of 1926 genes included in the Eukaryotic Promoter Database.
No TATA box was found in the Lim3A regulatory region. However, the sequence CAATAA, found at −27 to −22 nucleotides upstream of the Lim3A TSS, often occurs at positions from −36 to −21 nucleotides in the regulatory regions of D. melanogaster genes (0.6% of 1926 promoter sequences in the Eukaryotic Promoter Database). For example, CAATAA was found in the regulatory regions of five Enhancer of split [E(spl)] genes (HLHm3, HLHm5, HLHm8, HLHmβ, HLHmγ) [19], which encode basic helix-loop-helix transcriptional repressors that are expressed mainly during the embryonic stage, and function in neuronal development, similar to Lim3. Thus, the CAATAA sequence is common to genes with overlapping expression patterns during embryogenesis [20].
In contrast to Lim3A, the Lim3C transcription start region TTG+1AGC was less similar to the consensus. Promoter elements were not found in the regulatory region of this transcript. The sequence CAAAAT, at −24 to −19 nucleotides relative to the Lim3C major TSS, has been found in the regulatory regions of 0.6% of 1926 genes of the Eukaryotic Promoter Database, at a position of −36- to −18 nucleotides, relative to the TSSs.
In addition to the two major and some minor TSSs mentioned above, 5′-RACE analysis revealed TSSs represented by a single clone each, located approximately 250 bp upstream of the Lim3A major TSS. These rare long transcripts might use promoters predicted by the Neural Network Promoter Prediction database (Figure 3) at −534 to −485 (score 0.98), and −242 to −193 (score 1.00), relative to the Lim3A major TSS or, more likely, are “slippery promoters” typical of both TATA-containing and TATA-less Drosophila genes with multiple TSSs [21].
To identify potential TF-binding sites within the proximal regulatory regions of Lim3A and Lim3C, TFSEARCH version 1.3, MOTIF Search, and other bioinformatic resources (see Materials and Methods) were used. Potential TF binding sites were found for heat-shock factor, which also controls the expression of non-heat shock protein genes, for example, eve, in Drosophila embryonic development [22]; Hunchback (HB) which is necessary and sufficient for specifying early-born temporal identity in multiple neuroblast lineages [23]; and broad-complex Z3 and broad-complex Z4 (BR-C Z3/ Z4), which are essential for metamorphic reorganization of the central nervous system [24] (Figure 3). HB and BR-C Z3/Z4 are specialized TFs participating in Drosophila nervous system morphogenesis that might take part in Lim3 transcription regulation, which is also essential for neuron development.
Naturally occurring polymorphisms at Lim3
To determine if Lim3 function is associated with molecular variation in natural populations of Drosophila, we sequenced 2094 bp from 50 alleles from the Raleigh natural population, including 1557 bp of the Lim3A regulatory region, 300 bp of the 5′ UTR, 109 bp of the translated region from the first Lim3A exon, and 128 bp from the first intron (Table 1, GenBank accession no. 9GU814570, 33GU814571, 40GU814572, 44GU814573, 49GU814574, 58GU814575, 74GU814576, 76GU814577, 77GU814578, 87GU814579, 89GU814580, 98-21GU814581, 98-5GU814582, 100GU814583, 113GU814584, 115GU814585, 122GU814586, 161GU814587, 166GU814588, 180GU814589, 183GU814590, 200GU814591, 201GU814592, 207GU814593, 215GU814594, 226GU814595, 266GU814596, 273GU814597, 284GU814598, 285GU814599, 311GU814600, 316GU814601, 317GU814602, 325GU814603, 327GU814604, 336GU814605, 345GU814606, 351GU814607, 354GU814608, 361GU814609, 369GU814610, 376GU814611, 382GU814612, 407GU814613, 429GU814614, 434GU814615, 444GU814616, 461GU814617, 472GU814618, 473GU814619).
Table 1. Parameters of nucleotide diversity in the regulatory region and the beginning of the structural part of the Lim3A.
| Region | Nucleotide position | Number of Indels +SNPs | π (s. d.) | θ (s. d.) |
| All sequence | 1–2094 | 16+74 | 0.00709 (0.00046) | 0.00864 (0.00098) |
| Regulatory region | 1–1557 | 14+58 | 0.00795 (0.00046) | 0.00875 (0.00112) |
| 1–779 | 8+39 | 0.01168 (0.00069) | 0.01118 (0.00179) | |
| 780–1557 | 6+19 | 0.00421 (0.00043) | 0.00631 (0.00135) | |
| Exon | 1558–1966 | 1+7 | 0.00152 (0.00059) | 0.00600 (0.00181) |
| 5′UTR | 1558–1857 | 1+5 | 0.00181 (0.00078) | 0.00670 (0.00223) |
| Translated region | 1858–1966 | 0+2 | 0.00073 (0.00070) | 0.00410 (0.00290) |
| Intron | 1967–2094 | 1+9 | 0.01443 (0.00131) | 0.01570 (0.00523) |
Numbers of nucleotides in the second column correspond to the standard sequence, regardless indel variation in the natural population.
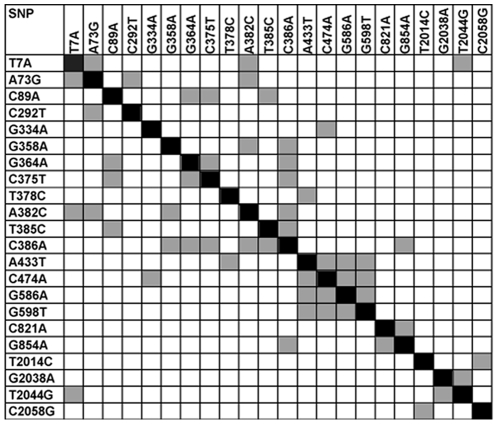
In total, 90 polymorphic markers were found, including 74 single nucleotide polymorphisms (SNPs) and 16 insertions and deletions (indels). Estimates of nucleotide diversity based on the number of differences between pairs of sites (π, [25]) and the number of segregating sites (θ, [26]) were within the range observed for D. melanogaster [27]: π = 0.00709±0.00046 and θ = 0.00864±0.00098. The highest level of variation was in the intron. However, when the regulatory region was divided arbitrarily into two equal parts, the distal section had approximately the same level of variation as the intron, while the proximal section closest to the 5′ UTR was much more conserved (Table 1). Not surprisingly, the most conserved was the translated part of the exon (Table 1), where only two nonsynonymous substitutions were found, each with a frequency of 0.02. Little significant linkage disequilibrium (LD) was observed between polymorphic markers (Figure 4), and the pattern of linked loci was as expected under assumptions of normal recombination, with few exceptions. This result was favorable for the identification of casual associations between molecular and phenotypic variations.
Figure 4. LD at the Lim3 locus.
Only markers with significant (corrected for multiple tests, DnaSP 4.0 [55]) LDs are included. LDs significant according both to Fishers's exact test and χ2 test are depicted in grey.
Molecular population genetic tests for selection were used to determine whether evolutionary forces might be regulating nucleotide variation at Lim3 locus. Significant negative values for D [28], D* and F* [29] were observed for the first exon (Table 2). Most parameters were also significant for the 5′ UTR alone, and for the translated region of the exon alone (Table 2). Only two nonsynonymous polymorphisms were found in our sample, so other neutrality tests were not applied. Overall, our results indicated less variation in the Lim3A first exon than expected under neutral expectations, and the action of purifying selection on this region. To understand in more detail the biological significance of molecular variation observed in the Raleigh natural population, we tested effects of nucleotide diversity on gene expression and fly phenotype.
Table 2. Neutrality tests in the regulatory region and the beginning of the structural part of the Lim3A.
| Region | D | D* | F* | D*, simulans | F*, simulans | D*, yakuba | F*, yakuba |
| All sequence | −0.63216 | −1.75232 | −1.59703 | −1.99481 | −1.89078 | ||
| Regulatory region | −0.31834 | −1.03457 | −0.92083 | −1.35484 | −1.28096 | ||
| 0.15386 | −0.88538 | −0.60895 | −1.11283 | −0.91096 | |||
| −1.07509 | −1.02695 | −1.23886 | −1.29958 | −1.51835 | −1.55057 | −1.67083 | |
| Exon | −2.10512 | −3.73109 | −3.79399 | −3.40290 | −3.56259 | −2.78912 | −3.07972 |
| 5′UTR | −2.04396 | −3.26405 | −3.37500 | −2.78749 | −3.02202 | −2.08900 | −2.46833 |
| Translated region | −1.46443 | −2.53305 | −2.57464 | −2.58770 | −2.63091 | −2.58770 | −2.63091 |
| Intron | −0.22559 | −1.28572 | −1.10719 | −0.44091 | −0.35893 | −1.99059 | −1.81791 |
Significant D, D*, F* are in bold case (P<0.05 according to DnaSP 4.0 [55]), in bold case and underlined (P<0.02). Sequences from D. simulans and D. yakuba have GenBank accession no. XM_002079849 and NT_167063, respectively.
Association between molecular variation at Lim3 locus and lifespan
Association studies used 44 polymorphic markers that were present in our sample at a frequency of 0.06 (in three lines out of 50) and higher. This restriction allowed us to concentrate on polymorphisms that were truly segregating in nature. Lifespan measurements were published in [5], [6].
Analysis of variance (ANOVA) revealed six polymorphic markers significantly associated with lifespan, while no significant association with lifespan was seen for sex or marker by sex interaction. Based on these results and the restricted sample size of sequenced alleles, we combined data on the sexes for nonparametric, distribution-free Wilcoxon tests, to assess association between molecular variation at Lim3 and lifespan.
The same six markers showed significant association with lifespan (Figure 3, Table 3, Table S1): four were located in the regulatory region (A433T; G871C; A1050G; C1177T), one in the 5′ UTR (T1658C), and one in the first intron (G1991A). None were in significant LD with each other. We also checked association of lifespan with several haplotypes composed of combinations of significant markers from the regulatory region that were most likely to influence lifespan through transcription alteration. Haplotypes composed of four makers, A433T, G871C, A1050G, and C1177T, three proximal markers adjacent to the structural gene, G871C, A1050G, and C1177T, and two markers with minimal P-values for individual association with lifespan, G871C and C1177T, were significantly associated with lifespan (Table 3, Table S1). In total, we carried out 47 association tests. Only lifespan association with haplotype G871C+C1177T survived Bonferroni correction, and lifespan associations with two other haplotypes and the G1991A marker survived a less conservative false discovery rate (FDR) correction. We concluded that the combination of two markers in the regulatory region, G871C+C1177T, which were present in all haplotypes, and the single marker in the first intron of Lim3A were important for lifespan.
Table 3. Genotype-phenotype associations at the Lim3 locus.
| SNP, haplotype | Numbers of lines with alternative alleles | Trait | P value3 | Mean (s. e.) |
| A433T | 38/121; 14/22 | Lifespan | 0.0357 | |
| RNA, embryos | 0.0030/0.1384 | 0.7(0.06)/2.0(0.37) | ||
| RNA, heads | 0.0682/0.1543 | |||
| RNA, testes | 0.0325/0.3327 | |||
| G586A, G598T | 36/141; 12/42 | Lifespan | 0.0992 | |
| RNA, embryos | 0.0131/0.0896 | 0.7(0.06)/1.4(0.29) | ||
| RNA, heads | 0.1773/0.2230 | |||
| RNA, testes | 0.0198/0.0896 | |||
| G871C | 46/41; 13/32 | Lifespan | 0.0151 | |
| RNA, embryos | 0.0002/0.0059 | 0.6(0.06)/1.8(0.27) | ||
| RNA, heads | 0.0105/0.0180 | 0.4(0.05)/0.7(0.12) | ||
| RNA, testes | 0.0138/0.1924 | |||
| A926G | 47/31; 14/22 | Lifespan | 0.0891 | |
| RNA, embryos | 0.0021/0.0682 | 0.7(0.06)/2.0(0.37) | ||
| RNA, heads | 0.0052/0.0025 | 0.4(0.05)/0.8(0.07) | ||
| RNA, testes | 0.0209/0.4588 | |||
| G1021A, | 40/101; 10/62, | Lifespan | 0.7528, 0.6801 | |
| C1046A | 39/111; 10/62 | RNA, embryos | 0.0018/0.0195 | 1.1(0.20)/0.5(0.09) |
| RNA, heads | 0.0356/0.0091 | |||
| RNA, testes | 0.0091/0.0014 | |||
| A1050G | 26/241; 11/52 | Lifespan | 0.0226 | |
| RNA, embryos | 0.5153/0.8548 | |||
| RNA, heads | 0.9029/0.2550 | |||
| RNA, testes | 0.9190/0.1039 | |||
| C1177T | 47/31; 14/22 | Lifespan | 0.0084 | |
| RNA, embryos | 0.0033/0.0037 | 0.9(0.11)/0.3(0.04) | ||
| RNA, heads | 0.0021/0.0021 | 0.5(0.05)/0.1(0.02) | ||
| RNA, testes | 0.0121/0.0044 | |||
| G1991A | 46/41; 13/32 | Lifespan | 0.0028 | 37(1)/29(2) |
| RNA, embryos | 0.2992/0.2005 | |||
| RNA, heads | 0.2183/0.2274 | |||
| RNA, testes | 0.4115/0.2333 | |||
| T1658C | 45/51; 13/32 | Lifespan | 0.0195 | |
| RNA, embryos | 0.1842/0.2881 | |||
| RNA, heads | 0.1111/0.1475 | |||
| RNA, testes | 0.1407/0.0535 | |||
| 871+1177, | 4/43/31; 3/11/22 | Lifespan | 0.0010 | 31(2)/38(1)/29(2) |
| CC/GC/GT | RNA, embryos | 0.0001/0.0014 | 1.8(0.27)/0.7(0.06)/0.3(0.04) | |
| RNA, heads | 0.0011/0.0016 | 0.7(0.12)/0.4(0.04)/0.1(0.02) | ||
| RNA, testes | 0.0053/0.0125 |
Data for the sample of 50 lines.
Data for the sample of 16 lines.
For associations with lifespan, P values of Wilkoxon test of line means, and for associations with Lim3 transcription, P values of Wilkoxon test of mRNA amounts/C(t) are shown, see text for details.
Significant P values are in italics; P values surviving FDR correction are in italics and bold case; P values surviving Bonferroni correction are in italics, bold case and underlined.
Markers and haplotypes significantly associated with lifespan are in bold case.
One of the alleles at each polymorphic site composing the significant haplotype had a low population frequency (pC = 0.08 for C871G; pT = 0.06 for C1177T), and was associated with short lifespan (Table 3). Of four possible combinations of alleles, only three were present in the population. Their frequencies were in good agreement with those expected from the frequencies of single alleles (χ2 = 0.0055), which confirmed the absence of LD between the markers. Multiple comparisons of means allowed us to divide the GC, CC, and GT haplotype variants of the G871C and C1177T markers into two groups that significantly differed in lifespan (P<0.05). The first group included 86% of lines and was characterized by the GC haplotype and a mean lifespan of 38 (±1) days. The second group included lines with the rare CC (8%) and GT (6%) haplotype, and mean lifespans of 31 (±2) and 29 (±2) days.
We proposed that polymorphisms in the regulatory region of the gene affect its expression, and thus a phenotypic trait such as lifespan. Our next goal was to test this hypothesis experimentally.
Association between molecular variation in Lim3A regulatory region and Lim3A expression
Lim3A and Lim3C differ in their 5′ UTR region, with Lim3C shorter by 190 bp. Therefore, the amount of either Lim3A alone, or both transcripts could be detected and measured. As Lim3A was more abundant (Figure 2A), and has functional significance for neuron development [10], we focused our analysis on Lim3A. To assess association between molecular variation in the Lim3A regulatory region and its transcript level, 16 lines with different G871C and C1177T haplotypes were selected. According to the information available [http://flyatlas.org, accession no. FBgn0002023], Lim3 transcription is predominantly observed in embryos, and in adult brains and testes. Guided by this information, we evaluated the amount of Lim3A in embryos, heads (Table 4), and testes of selected lines using real time RT-PCR.
Table 4. Polymorphism in the TFs binding sites and Lim3A transcription.
| Line | Nucleotide in the putative binding site for Grh | Nucleotide in the putative binding site for CA/TG – repeat binding protein | Lim3A mRNA amount in embryos (s. e.) | Lim3A mRNA amount in heads (s. e.) | Mean lifespan, males and females combined from [5], days |
| 284 | C | C | 2.578 (0.241) | 0.910 (0.078) | 27.5 |
| 207 | C | C | 1.369 (0.116) | 0.774 (0.195) | 41.5 |
| 285 | C | C | 1.361 (0.119) | 0.514 (0.077) | 27.5 |
| 472 | G | C | 1.068 (0.364) | 0.232 (0.027) | 37.0 |
| 74 | G | C | 1.045 (0.043) | 0.301 (0.067) | 38.0 |
| 100 | G | C | 0.876 (0.097) | 0.432 (0.061) | 48.5 |
| 40 | G | C | 0.847 (0.028) | 0.402 (0.059) | 51,0 |
| 115 | G | C | 0.729 (0.085) | 0.490 (0.034) | 51.5 |
| 201 | G | C | 0.676 (0.033) | 0.757 (0.085) | 30.0 |
| 325 | G | C | 0.615 (0.148) | 0.712 (0.057) | 42.5 |
| 336 | G | C | 0.481 (0.130) | 0.420 (0.144) | 40.0 |
| 200 | G | C | 0.457 (0.138) | 0.349 (0.009) | 36.5 |
| 461 | G | C | 0.452 (0.043) | 0.394 (0.022) | 30.5 |
| 180 | G | C | 0.439 (0.134) | 0.193 (0.022) | 46.5 |
| 76 | G | T | 0.358 (0.039) | 0.127 (0.005) | 31.0 |
| 226 | G | T | 0.236 (0.049) | 0.161 (0.049) | 23.5 |
Alleles corresponding to the standard sequence are in italics and bold case.
Correlations between independent measurements of Lim3A transcripts were highly significant across the 16 lines in both embryos (P<0.0001) and in heads (P = 0.0074), strengthening reliability of the results. The correlation between independent measurements in the testes was not significant (P = 0.1082), probably because of the substantially smaller amount of detected Lim3A mRNA. The amount of Lim3A mRNA was also correlated in embryos and heads (P = 0.0064), in embryos and testes (P = 0.0579), and in heads and testes (P = 0.0204) across the 16 lines.
In total, 30 of the 44 markers segregated in these lines, and eight were in complete LD with the others: 24 association tests with 22 markers and two haplotypes were performed. According to the distribution-free Wilcoxon test, significant association was seen between Lim3A levels in embryos for 14 polymorphic markers. For four markers (G871C, A926G, G1021A, C1046A), this held after Bonferroni correction, and another four (A433T, G586A, G598T, C1177T) held after FDR correction (Table 3, Table S1). Markers G1021A and C1046A, G586A and G598T were in complete LD in the 16 lines. Another method [30] based on the analysis of direct C(t) measurements proportional to the logarithm of the substrate quantity was used for verification. Significant associations surviving FDR corrections were confirmed for G871C and C1177T (Table 3, Table S1). Finally, REST [31], a program that accounts for different PCR efficiencies for target and reference genes, confirmed associations of G871C and C1177T (P = 0.0001 for both).
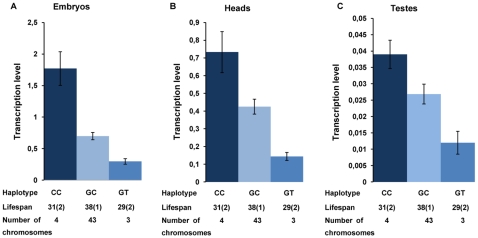
G871C and C1177T are the two polymorphic markers that form the haplotype that is significantly associated with lifespan. Association with Lim3A levels in embryos was highly significant for this haplotype, by all methods of analysis (Table 3), including pairwise comparisons using REST software (P = 0.0001 for each comparison). Multiple comparisons of means allowed us to categorize lines with different haplotype variants of the G871C and C1177T markers, specifically CC, GC, and GT, into three groups with an approximately six-fold significant difference (P<0.05) in the amount of Lim3A in embryos (CC: 1.8±0.27; GC: 0.7±0.06; GT: 0.3±0.04; Figure 5).
Figure 5. Lim3A transcription level in lines with different haplotype variants.
(A). Lim3A transcription level in embryos. (B). Lim3A transcription level in adult heads. (C). Lim3A transcription level in adult testes. Haplotypes composed of segregating markers G871C and C1177T, haplotype mean lifespan in days (s. e.) and numbers of chromosomes with corresponding haplotypes sampled from Raleigh population are given below the diagrams.
According to the Wilcoxon test, significant associations were found for Lim3A levels in adult heads for five polymorphic markers. One marker (C1177T) survived Bonferroni correction and another two (G871C, A926G) survived FDR correction (Table 3, Table S1). Analysis of direct C(t) measurements revealed significant associations surviving FDR correction for A926G and C1177T (Table 3, Table S1). These results were not confirmed using REST. Association with Lim3A levels in adult heads was highly significant for the G871C+C1177T haplotype by both nonparametric analysis methods (Table 3), and only one of the three pair-wise comparisons was significant (REST, P = 0.023 for GT compared to CC). Multiple comparisons of means allowed us to categorize lines with the CC, GC, and GT haplotype variants of the G871C and C1177T markers, into three groups with approximately six-fold significant differences (P<0.05) in Lim3A levels in adult heads (CC: 0.7±0.12; GC: 0.4±0.04; GT: 0.1±0.02; Figure 5).
According to the Wilcoxon test, significant associations were seen between the amount of Lim3A in testes and 16 polymorphic markers, although none survived Bonferroni or FDR correction (Table 3, Table S1). Association was also significant for the G871C+C1177T haplotype, according to both nonparametric analysis methods, but these also did not survive Bonferroni or FDR correction (Table 3). Multiple comparisons of means showed that Lim3A transcription in testes was significantly different (P<0.05) between lines with the CC (0.04±0.004) and GT (0.01±0.003) haplotype variants of the G871C and C1177T markers (Figure 5).
Many polymorphic markers appeared to be significantly associated with the amount of Lim3A in different tissues. Different methods of analysis and different P-value corrections gave slightly different, though not contradictory results (Table 3). The most notable polymorphic markers were G871C and C1177T, which formed a haplotype significantly associated with lifespan (P = 0.0010), and with transcription in embryos (P = 0.0001), adult heads (P = 0.0011), and testes (P = 0.0053). Each of the two markers alone was also significantly associated with transcription in embryos (G871C: P = 0.0002; C1177T: P = 0.0033), adult heads (P = 0.0105, P = 0.0021), and testes (P = 0.0138, P = 0.0121), as well as lifespan (P = 0.0151, P = 0.0084). The polymorphic markers A926G and G1021A+C1046A (linked in the sample of 16 lines), located between G871C and C1177T, were also significantly associated with the Lim3 transcription level in embryos (A926G: P = 0.0021; G1021A+C1046A: P = 0.0018), adult heads (P = 0.0052, P = 0.0356), and testes (P = 0.0209, P = 0.0091), as well as the haplotype composed of all five markers G871C+A926G+(G1021A+C1046A)+C1177T (P = 0.0005, P = 0.0058, P = 0.0053, for embryos, heads and testes, Table S1). We propose that the entire region from 380 to 686 bp upstream of the Lim3A major TSS is important for gene expression, while only two markers within this region are important for lifespan.
All five polymorphic markers mentioned above are in potential TF-binding sites: G871C and A926G are in the Grainy Head (Grh) binding site consensus sequence, G1021A is in the specificity protein-1 (Sp1)/Krüppel-like factor (KLF) binding-site consensus sequence, C1046A in the Zeste-like motif, and C1177T is in the (CA/TG)9 repeat (Figure 3).
G871C and C1177T appeared to be the most essential markers for Drosophila Lim3A expression and lifespan. When C, a frequent allele in the Raleigh population, was present at the 1177 position, Lim3A transcription was intermediate and Drosophila lifespan was high. When C was substituted for T, a rare allele, the expression and Drosophila lifespan were low (Table 4). Hence, we suggest that this site normally functions in Lim3 activation, as an activator-binding site. The (CA/TG)9 repeat where the C1177T polymorphic site is located is a a cis-regulatory element [32], however, nothing is known about the proteins that bind this repeat [33].
In a background of C at the 1177 position, Lim3A transcription and Drosophila lifespan was dependent on the G871C marker (Table 4). When G, a frequent allele in Raleigh population, was present at the 871 position, Lim3A transcription was intermediate and Drosophila lifespan was high. When G was substituted for C, a rare allele, the expression increased, and Drosophila lifespan was short (Table 4). Hence, we suggest that normally this site is involved in Lim3 repression as a repressor binding site. Indeed, Grh, which presumably interacts with G871C as part of its specific binding site, cooperates with Polycomb-group (PcG) proteins that inactivate genes by chromatin remodeling, and Grh-binding sites are often encountered in Polycomb response elements (PREs) [34]–[36].
Both intermediate level of Lim3A expression and longer lifespan are associated with the same polymorphic haplotype. Square regression (with mean lifespan as a dependent variable and Lim3A mRNA amount as independent variable) is a better approximation for our data (R2 = 0.062 for embryos; R2 = 0.011 for heads; R2 = 0.007 for testes) than linear regression (R2 = 0.0029 for embryos; R2 = 0.0006 for heads; R2 = 0.000 for testes). For embryos, square regression is significant (P = 0.0472), indicating that the model accounts for a low but significant portion of variation in the data; linear regression is not significant (P = 0.2814). This result is in agreement with the hypothesis that intermediate levels of Lim3A expression confer longer lifespan.
Discussion
We found that Lim3 produces three mRNAs. In addition to the already known Lim3A and Lim3B transcript, we discovered the additional Lim3C mRNA. The promoter region of Lim3A is DPE-containing, but lacks a TATA box, and possesses multiple start sites with one major initiation site, and additional nearby minor ones. The distance between the Lim3A DPE and initiator is appropriate for TFIID binding, which is essential for transcription [37]. Reduced expression of Lim3C compared to Lim3A is most likely explained by the lack of a strong initiator, TATA-box, or other core promoter elements. However, other elements such as CAAAAT, and other different mechanisms of initiation may be used to regulate Lim3C transcription. The alternative promoters of Lim3A and Lim3C may provide a mechanism for tissue- and developmental stage-specific Lim3 activation. TATA box-containing promoters are activated after embryonic development, and TATA-less promoters of the same genes are active during early embryo development [38], [39]. Mammalian LHX3a and LHX3b, which are homologues of Lim3A and Lim3B, are transcribed from two alternative TATA-less, GC-rich promoters [40], have distinct temporal expression profiles, and have different regulatory roles in the development of the distinct cell types [41].
Statistical analyses demonstrated that the first exon of Lim3A (Lim3C) is affected by purifying selection. The normal recombination found in this region suggests that the selection should be highly effective against deleterious alleles, removing them from the population [42]. Indeed, only two polymorphisms with minimal detectable frequency were found in the translated region of the first exon. Thus, the conserved structure of the Lim3A (Lim3C) protein can be assumed to be essential for its proper function, and therefore maintained by selection. An alternative explanation is that the Raleigh population recently experienced a bottleneck. However, this is not confirmed by analysis of selection forces acting on other regions of the gene, or on other genes whose molecular variation was analyzed using the same sample of second chromosomes from the Raleigh population (Dopa dcarboxilase [5]; Catecholamines Up [6]; shuttle craft, Simonenko, Pasyukova, unpublished results).
Regulatory regions can have crucial roles in evolution, and modifications in these regions have mainly adaptive evolutionary effects [43], [44]. Statistical analysis did not reveal any evidence for natural selection in the Lim3A regulatory region. Nevertheless, the significance of the regulatory region for transcription and phenotype was demonstrated by the finding that nucleotide substitutions within this region that segregated in the Raleigh population appeared to result in differences as large as six-fold in gene transcription, and 1.3-to-1.5-fold in lifespan. No significant associations were found between markers located outside the regulatory region, (i.e. in the 5′ UTR or the Lim3A structural gene) and Lim3A levels, and markers significantly associated with Lim3A expression were not in LD with each other or with markers within the gene in a sample of 50 alleles. Therefore, we have likely identified actual casual relationships between natural polymorphisms and gene function. Haplotype variants of the G871C and C1177T polymorphic markers associated with short lifespan, and either high or low Lim3A transcription (CC, GT) were found in the Raleigh population at low frequencies. The haplotype variant associated with long lifespan and intermediate Lim3A transcription (GC) was present at high frequency. Thus, association analysis predicted that an intermediate level of Lim3A expression provided longer lifespan, and a selective advantage. Statistical tests were possibly not sensitive enough to detect this selection, however. Even when the fitness effects of mutations are in the nearly neutral range, natural selection is still able to influence transcriptional phenotype [45].
A possible general explanation for the absence of selection on the regulatory region is that nucleotide substitutions in a single, or in several TF binding sites might affect gene expression only in the tissues where these TFs are active, so the impact of the substitutions on phenotypic traits would be small. However, as mentioned above, this was not true for several polymorphisms within the Lim3A 5′-regulatory region, which significantly affected expression and phenotype. Moreover, it is difficult to point to polymorphic markers within Lim3A regulatory region which have tissue-specific effects. Rather, most polymorphic markers that were significantly associated with transcript abundance seemed to be important in all tissues, and the exact significance level of the effect depended on the reliability of measurements in a particular tissue and on methods of analysis. Thus, nucleotide substitutions found in the Lim3A regulatory region in the Raleigh natural population must be located within sites that regulate transcription in a general, rather than a tissue-specific manner.
Most polymorphic markers significantly associated with transcription were located in the compact region that was 380–680 bps upstream of the Lim3A major TSS, and were within binding sites for important transcriptional regulators. For example, Grh is involved in many regulatory networks, including the complex regulation of neuroblast specification and neuron apoptosis [46], [47]. Sp1 mediates transcription of the LHX3 gene, the human homologue of Lim3 [40]. Grh and Sp1/KLF are members of the PcG and trxG complexes. Binding sites for other members of these complexes (Pho, GAGA or GAF/Psq), were also found in the Lim3A regulatory region, suggesting that PRE-TRE sites for PcG and trxG complexes are present in the region.
We presume that both repressor and activator proteins bind the essential sites for Lim3 transcription and fly lifespan in which the polymorphic markers are located. We hypothesize that the repressor protein Grh and the unknown activator protein that binds the (CA/TG)9 repeat might provide negative and positive transcriptional regulation of Lim3A, and consequently affect Drosophila lifespan. Disrupting the balance between negative and positive regulation would result in deviations in Lim3A transcription, and a decrease in Drosophila lifespan An intermediate expression based on a balance between activation and repression of the gene and favorable for long lifespan could be provided by the combined activity of PcG and trxG protein complexes through maintenance of a silent or active transcriptional state of their target genes. The PcG and trxG complexes bind to genes encoding transcription factors, including homeodomain-containing proteins such as Lim3, and are implicated in the regulation of various transcriptional pathways [48].
Overexpression or RNAi knockdown of a number of Drosophila genes showed the involvement of these genes in lifespan control [for example, 49–51]. Direct proof of Lim3 involvement in lifespan control is required, however, gene overexpression or RNAi knockdown are not applicable in this particular case. We are considering site-specific integration of a Lim3 transgene using carefully chosen sets of landing sites, transgene constructs, and drivers as a possible approach to verify the results presented here. Experimental manipulations with Lim3 expression levels are also necessary to prove that intermediate levels of Lim3A expression confer longer lifespan.
The mechanism underlying Drosophila lifespan variation through alteration of Lim3 expression is not understood. Molecular variation at the Lim3 regulatory region most strongly affected Lim3 expression in embryos. Previously, Lim3 was found to be active in the Drosophila embryonic nervous system and to take part in regulatory networks leading to the specification of motor neuron subclass identity, axon pathfinding, and finally, proper muscle innervation [12]. Lim3 was reported to be expressed in the Drosophila ring gland [10], but later studies failed to confirm Lim3 expression in the embryonic Drosophila endocrine system [52]. Whether these Lim3 functions are sufficient to explain the lifespan variations caused by alterations in Lim3 expression in embryos, and other mechanisms that might explain lifespan effects initiated during early development are unknown. Recently, however, genes responsible for sex determination during early Drosophila development that also affect lifespan were found [53].
The role of Lim3 in adult flies is not known, and we do not possess any information about alterations of Lim3 transcription level with age. Lim3 was first discovered as a male-specific candidate lifespan gene [8]. Thus, Lim3 expression in testes is assumed to affect lifespan and even to have a main casual relation to lifespan variation. However, we failed to find a strong association between Lim3 transcription in testes and lifespan, probably because of insufficient sensitivity in measuring of small amounts of mRNA. We demonstrated that Lim3 is substantially expressed in adult heads. This confirms that Lim3 expression in adults is tissue-specific, and probably associated with the nervous system. Lim3 function in the adult brain may be involved in lifespan regulation. We intend to ascertain the Lim3 function in the nervous and neuroendocrine systems of adult flies, to move closer to understanding the mechanisms underlying Lim3 involvement in Drosophila lifespan control.
Materials and Methods
Drosophila stocks
We used 50 substitution D. melanogaster lines containing second chromosomes from the Raleigh (USA) population in homozygous Samarkand genetic background and differing in lifespan (22–62 days, P<0, 0001; [5]). All lines were reared in glass vials with wheat-sugar-agar medium, at 25°C.
Nucleic acids isolation
DNA was extracted from 50 lines according to the standard procedures [54]. Total RNA for Northern and 5′RACE analyses was isolated using the SV Total RNA Isolation System (Promega) according to the manufacturer's instructions. Total RNA for real-time quantitative PCR was extracted from 50 12-hour embryos and from 20 heads (10 males and 10 females) or 50 pairs of testes of 15-day old adult flies using Trizol reagent (Invitrogen) and DNase I Kit (TURBO DNA-free™, Ambion) according to the manufacturers' instructions.
DNA sequencing and analysis
Isolated DNA was used in PCR reaction with forward primer TCC AAC CAG ACT GTC AAG TCA AAT TAC and reverse primer TTG CAG AAA GAG AAT AAC GCT AAA TCA. Then PCR products were sequenced with Big Dye Terminator V. 3.1. Kit (Applied Biosystems), according to the manufacturer's protocol, on ABI PRIZM 310 Genetic Analyser (Applied Biosystems). Six primers for sequencing were used: TCC AAC CAG ACT GTC AAG TCA AAT TAC, TTC AAT TAG CAT GAT CCA AGG, AGA CGT TGC TCT CGA TCA GC, AGA CCA CTC CCC GAA AAC TG, ATG TCA GCC CCT GTA TGT GC, AGA ATC CAA TCA GAG TGC GTC.
The putative RNA Polymerase II promoter sites were predicted with Neural Network Promoter Prediction database, version 2.2 (http://www.fruitfly.org/seq_tools/promoter.html), Eukaryotic Promoter Database Current Release 100 (http://www.epd.isb-sib.ch/seq_download.html) and the Drosophila core promoter database (http://www.biology.ucsd.edu/labs/kadonaga/DCPD). To identify possible binding sites of transcription factors TFSEARCH at http://www.cbrc.jp/research/db/TFSEARCH.htm; MOTIF Search at http://motif.genome.jp/; ConSite at http://asp.ii.uib.no:8090/cgi-bin/CONSITE/consite/; TESS at http://www.cbil.upenn.edu/cgi-bin/tess/tess; Match™ at http://www.gene-regulation.com/cgi-bin/pub/programs/match/bin/match.cgi; Drosophila Melanogaster Major Position Matrix Motifs at http://line.imb.ac.ru/DMMPMM/ were used.
Northern blot
Total RNA was quantified by absorbance at 260 nm, and 8–14 mg of total RNA were resolved by 1.5% denaturing agarose-formaldehyde gel, blotted onto Hybond-N+ membrane (Amersham) and fixed by UV cross-linking. Pre-hybridization and hybridization were performed at 42°C overnight in 25ml of the solution containing 50% formamide, 10× Denhardt solution, 5×SSC, 0.5% SDS, 100mg/ml tRNA. After hybridization, membrane was washed twice (10 min each) in 2×SSC, 0.1% SDS at 42°C, three times (5 min each) in 0.1×SSC, 0.1% SDS at 55°C, and exposed on a phosphor-imager (Storm, Amersham) using Image Quant Version 5.2 computational tool.
A 1640 bp Lim3 PCR fragment was amplified (forward primer: TTC AAT TAG CAT GAT CCA AGG, reverse primer: TCA CAT TTG CCA TTG GAC AGG AAG TC) and used as a probe to detect Lim3 transcripts. DNA probes (5–10×106 cpm) added to the hybridization mixture were labeled by Hexa Label™ DNA Labeling Kit (Fermentas) with 20–40 µCi [α-32P] and then purified with CentriSep columns (Princeton Separations).
Rapid amplification of 5′cDNA end (5′RACE) analysis
Transcription start sites of Lim3A mRNA in D. melanogaster were identified with the rapid amplification of the cDNA ends (RACE) technique using Smart™ RACE cDNA Amplification Kit (Clontech) for the first-strand cDNA synthesis. The touchdown PCR of the first-strand cDNA was then performed by using the gene-specific reverse primer, TCA CAT TTG CCA TTG GAC AGG AAG TC and the manufacturer's Abridged Anchor primer (Smart™ RACE cDNA Amplification Kit, Advantage 2 Polymerase Mix, Clontech). The annealing was performed at 64°C for 30 sec and extension at 68°C for 3 min, other parameters of the touchdown PCR were selected according to the manufacturer's recommendations. PCR products were gel-purified (Wizard® PCR Preps DNA Purification System, Promega) and cloned into pGEM-T EasyVector (Promega). Plasmid DNA was isolated (Wizard® Plus Minipreps DNA Purification Systems, Promega) and sequenced.
Real-time RT-PCR
The first strand of cDNA was synthesized using Super Script™ II Reverse Transcriptase (Invitrogen) with oligo(dT) primer, according to the manufacturer's instructions. cDNA amount was analyzed by real-time quantitative PCR using SYBR Green I/Rox in Chromo4 Real-Time PCR Detector (Bio-Rad). Equal amounts of mRNA and cDNA for real time RT-PCR analysis were used to evaluate the Lim3A expression in various tissues and life stages.
Gdh, a housekeeping gene located on the chromosome 3 which was common to all the substitution lines and characterized by relatively low expression level comparable with expression level of Lim3 was used as a reference gene to normalize for differences in total cDNA between samples. Forward and reverse primer sequences were: Lim3-RA: TGT GAA AGT GAT GGT TGA TTG CTC TGC, TCA CAT TTG CCA TTG GAC AGG AAG TC; Gdh: TAT GCC ACC GAG CAC CAG ATT CC, GGA TGC CCT TCA CCT TCT GCT TCT T.
MJ Opticon Monitor™ Analysis Software V. 3.1. 32 (Bio-Rad laboratories Inc., 2004–2005) was used to evaluate C(t) value and relative Lim3A mRNA amount which was considered as a measure of Lim3 transcription level in each Drosophila line.
Statistical analyses
The nucleotide diversity was analyzed as the pairwise distance between alleles (π) and the average number of segregating sites (θ) using DnaSP 4.0 [55]. This software was also used to assess linkage disequilibrium (LD) between polymorphic sites, and selective neutrality of observed polymorphisms (D, D* and F*, D* and F* with outgroup, [28], [29]).
Association between molecular polymorphisms and lifespan was assessed by two-way fixed effects ANOVA of line means, with polymorphic marker and sex as main effects, and by nonparametric distribution free Wilkoxon test of line means. Association between molecular polymorphisms and Lim3 transcription was assessed by nonparametric distribution free Wilkoxon test of mRNA amount or C(t)s [30]. REST V2.0.7 program [31], with the number of randomizations equal to 10,000, was used to verify the results. Multiple comparison of means (Tukey's test) was used to compare lifespan and Lim3 expression in groups of lines with different molecular haplotypes. Regression analysis with mean lifespan as a dependent variable and Lim3A mRNA amount as independent variable was used to assess association between lifespan and Lim3 transcription. Bonferroni and False Discovery Rate (FDR, [56]) corrections for multiple analyses were used when appropriate.
Supporting Information
Genotype-phenotype associations at the Lim3 locus.
(0.17 MB DOC)
Acknowledgments
We are grateful to T. Mackay who provided us with substitution Drosophila lines, to T. Kapelinskaya for help with 5′RACE analysis, to A. Krementcova for help with regression analysis, and to D. Mukha and O. Bylino for valuable discussions.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Russian Fund of Basic Research (RFBR) grant #09-04-01181-a for EGP; Grant form Russian Academy of Sciences (RAS) Program “Biodiversity” for EGP; Ministry of Education and Science of RF Program “Scientific and Educational Human Resources of Innovative Russia” Contract #Π317. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Pérez VI, Bokov A, Van Remmen H, Mele J, Ran Q, et al. Is the oxidative stress theory of aging dead? Biochim Biophys Acta. 2009;1790:1005–1014. doi: 10.1016/j.bbagen.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Broughton S, Partridge L. Insulin/IGF-like signalling, the central nervous system and aging. Biochem J 2009. 2009;418:1–12. doi: 10.1042/BJ20082102. [DOI] [PubMed] [Google Scholar]
- 3.Stanfel MN, Shamieh LS, Kaeberlein M, Kennedy BK. The TOR pathway comes of age. Biochim Biophys Acta. 2009;1790:1067–1074. doi: 10.1016/j.bbagen.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmidt PS, Duvernell DD, Eanes WF. Adaptive evolution of a candidate gene for aging in Drosophila. Proc Natl Acad Sci U S A. 2000;97:10861–10865. doi: 10.1073/pnas.190338897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Luca M, Roshina NV, Geiger-Thornsberry GL, Lyman RF, Pasyukova EG, et al. Dopa-decarboxylase affects variation in Drosophila longevity. Nat Genet. 2003;34:429–433. doi: 10.1038/ng1218. [DOI] [PubMed] [Google Scholar]
- 6.Carbonne MA, Jordan KW, Lyman RF, Harbison ST, Leips J, et al. Phenotypic variation and natural selection at Catsup, a pleiotropic quantitative trait gene in Drosophila. Current Biology. 2006;16:912–919. doi: 10.1016/j.cub.2006.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mackay TFC, Roshina NV, Leips JW, Pasyukova EG. Complex genetic architecture of Drosophila longevity. In: Masoro E, Austad S, editors. Handbook on the Biology of Ageing. Elsevier; 2005. pp. 181–216. [Google Scholar]
- 8.Roshina NV, Pasyukova EG. Genes regulating the development and functioning of the nervous system determine life span in Drosophila melanogaster. Russian J Genet. 2007;43:275–280. [PubMed] [Google Scholar]
- 9.Jordan KW, Morgan TJ, Mackay TFC. Quantitative Trait Loci for Locomotor Behavior in Drosophila melanogaster. Genetics. 2006;174:271–284. doi: 10.1534/genetics.106.058099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thor S, Andersson SGE, Tomlinson A, Thomas JB. A LIM-homodomain combinatorial code for motorneuron pathway selection. Nature. 1999;397:76–80. doi: 10.1038/16275. [DOI] [PubMed] [Google Scholar]
- 11.Landgraf M, Thor S. Development and structure of motoneurons. Inernat Rev Neurobiology. 2006;75:33–53. doi: 10.1016/S0074-7742(06)75002-4. [DOI] [PubMed] [Google Scholar]
- 12.Certel SJ, Thor S. Specification of Drosophila motoneuron identity by the combinatorial action of POU and LIM-HD factors. Development. 2004;131:5429–5439. doi: 10.1242/dev.01418. [DOI] [PubMed] [Google Scholar]
- 13.Mullen RD, Colvin SC, Hunter CS, Savage JJ, Walvoord EC, et al. Roles of the LHX3 and LHX4 LIM-homeodomain factors in pituitary development. Mol Cell Endocrinol. 2007;265–266:190–195. doi: 10.1016/j.mce.2006.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Netchine I, Sobrier ML, Krude H, Schnabel D, Maghnie M, et al. Mutations in LHX3 result in a new syndrome revealed by combined pituitary hormone deficiency. Nat Genet. 2000;25:182–186. doi: 10.1038/76041. [DOI] [PubMed] [Google Scholar]
- 15.Tajima T, Hattori T, Nakajima T, Okuhara K, Tsubaki J, Fujieda K. A novel missense mutation (P366T) of the LHX4 gene causes severe combined pituitary hormone deficiency with pituitary hypoplasia, ectopic posterior lobe and a poorly developed sella turcica. Endocr J. 2007;54:637–641. doi: 10.1507/endocrj.k06-200. [DOI] [PubMed] [Google Scholar]
- 16.Pfaeffle RW, Savage JJ, Hunter CS, Palme C, Ahlmann M, et al. Four novel mutations of the LHX3 gene cause combined pituitary hormone deficiencies with or without limited neck rotation. J Clin Endocrinol Metab. 2007;92:1909–191. doi: 10.1210/jc.2006-2177. [DOI] [PubMed] [Google Scholar]
- 17.Kutach AK, Kadonaga JT. The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters. Mol Cell Biol. 2000;20:4754–4764. doi: 10.1128/mcb.20.13.4754-4764.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butler JEF, Kadonaga JT. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 2002;16:2583–2592. doi: 10.1101/gad.1026202. [DOI] [PubMed] [Google Scholar]
- 19.Maeder ML, Polansky BJ, Robson BE, Eastman DA. Phylogenetic footprinting analysis in the upstream regulatory regions of the Drosophila Enhancer of split Genes. Genetics. 2007;177:1377–1394. doi: 10.1534/genetics.107.070425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wech IS, Delidakis BC, Preiss A. Distinct expression patterns of different enhancer of split bHLH genes during embryogenesis of Drosophila melanogaster. Dev Genes Evol. 1999;209:370–375. doi: 10.1007/s004270050266. [DOI] [PubMed] [Google Scholar]
- 21.Yasuhara JC, DeCrase CH, Wakimoto BT. Evolution of heterochromatic genes of Drosophila. Proc Natl Acad Sci USA. 2005;102:10958–10963. doi: 10.1073/pnas.0503424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuchar J, McDonough C, Sackerson C. Heat shock factor controls expression of a non-heat shock protein gene in Drosophila embryos. BIOS. 2007;78:62–68. [Google Scholar]
- 23.Isshiki T, Pearson B, Holbrook S, Doe CQ. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell. 2001;106:511–521. doi: 10.1016/s0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- 24.Spokony RF, Restifo LL. Broad complex isoforms have unique distributions during central nervous system metamorphosis in Drosophila melanogaster. J Comp Neurol. 2009;517:15–36. doi: 10.1002/cne.22119. [DOI] [PubMed] [Google Scholar]
- 25.Nei M, Tajima F. DNA polymorphism detectable by restriction endonucleases. Genetics. 1981;97:145–163. doi: 10.1093/genetics/97.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watterson GA. On the number of segregating sites in genetical models without recombination. Theor Popul Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- 27.Moriyama EN, Powell JR. Intraspecific nuclear DNA variation in Drosophila. Mol Biol Evol. 1996;13:261–277. doi: 10.1093/oxfordjournals.molbev.a025563. [DOI] [PubMed] [Google Scholar]
- 28.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan JS, Reed A, Chen F, Stewart CN., Jr Statistical analysis of real-time PCR data. BMC Bioinformatics. 2006;7:85–97. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST(C)) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucl Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma VK, Kumar N, Brahmachari SK, Ramachandran S. Abundance of dinucleotide repeats and gene expression are inversely correlated: a role for gene function in addition to intron length. Physiol Genomics. 2007;31:96–103. doi: 10.1152/physiolgenomics.00183.2006. [DOI] [PubMed] [Google Scholar]
- 33.Papatsenko DA, Makeev VJ, Lifanov AP, Régnier M, Nazina AG, et al. Extraction of Functional Binding Sites from Unique Regulatory Regions: The Drosophila Early Developmental Enhancers. Genome Res. 2002;12:470–481. doi: 10.1101/gr.212502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tuckfield A, Clouston DR, Wilanowski TM, Zhao LL, Cunningham JM, et al. Binding of the RING Polycomb Proteins to Specific Target Genes in Complex with the grainyhead-Like Family of Developmental Transcription Factors. Mol Cell Biol. 2002;22:1936–1946. doi: 10.1128/MCB.22.6.1936-1946.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blastya'k A, Mishra RK, Karch F, Gyurkovics H. Efficient and Specific Targeting of Polycomb Group Proteins Requires Cooperative Interaction between Grainyhead and Pleiohomeotic. Mol Cell Biol. 2006;26:1434–1444. doi: 10.1128/MCB.26.4.1434-1444.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muller J, Kassis JA. Polycomb response elements and targeting of Polycomb group proteins in Drosophila. Curr Opin Genet Dev. 2006;16:476–484. doi: 10.1016/j.gde.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 37.Burke TW, Kadonaga JT. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes& Dev. 1997;11:3020–3031. doi: 10.1101/gad.11.22.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davis W, Jr, Schultz RM. Developmental change in TATA-box utilization during preimplantation mouse development. Dev Biol. 2000;218:275–83. doi: 10.1006/dbio.1999.9486. [DOI] [PubMed] [Google Scholar]
- 39.Duan ZJ, Fang X, Rohde A, Han H, Stamatoyannopoulos G, et al. Developmental specificity of recruitment of TBP to the TATA box of the human gamma-globin gene. Proc Natl Acad Sci U S A. 2002;99:5509–5514. doi: 10.1073/pnas.072084499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yaden BC, Garcia M, 3rd, Smith TP, Rhodes SJ. Two promoters mediate transcription from the human LHX3 gene: involvement of nuclear factor I and specificity protein 1. Endocrinology. 2006;147:324–337. doi: 10.1210/en.2005-0970. [DOI] [PubMed] [Google Scholar]
- 41.Sloop KW, Meier BC, Bridwell JL, Parker GE, Schiller AM, Rhodes SJ. Differential activation of pituitary hormone genes by human Lhx3 isoforms with distinct DNA binding properties. Mol Endocrinol. 1999;13:2212–25. doi: 10.1210/mend.13.12.0395. [DOI] [PubMed] [Google Scholar]
- 42.Hill WG, Robertson A. The effect of linkage on limits to artificial selection. Genet Res. 1966;8:269–294. [PubMed] [Google Scholar]
- 43.Andolfatto P. Adaptive evolution of non-coding DNA in Drosophila. Nature. 2005;437:1149–1152. doi: 10.1038/nature04107. [DOI] [PubMed] [Google Scholar]
- 44.Madan Babu M, Balaji S, Aravind L. General trends in the evolution of prokaryotic transcriptional regulatory networks. Genome Dyn. 2007;3:66–80. doi: 10.1159/000107604. [DOI] [PubMed] [Google Scholar]
- 45.Bedford T, Hartl DL. Optimization of gene expression by natural selection. Proc Natl Acad Sci USA. 2009;106:1133–1138. doi: 10.1073/pnas.0812009106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brody T, Odenwald WF. Programmed transformations in neuroblast gene expression during Drosophila CNS lineage development. Dev Biol. 2000;226:34–44. doi: 10.1006/dbio.2000.9829. [DOI] [PubMed] [Google Scholar]
- 47.Cenci C, Gould AP. Drosophila Grainyhead specifies late programmes of neural proliferation by regulating the mitotic activity and Hox-dependent apoptosis of neuroblasts. Development. 2005;132:3835–3845. doi: 10.1242/dev.01932. [DOI] [PubMed] [Google Scholar]
- 48.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–45. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 49.Parkes T, Elia AJ, Dickinson D, Hilliker AJ, Phillips JP, et al. Extension of Drosophila lifespan by overexpression of human SOD1 in motoneurons. Nature Genetics. 1998;19:171–174. doi: 10.1038/534. [DOI] [PubMed] [Google Scholar]
- 50.Chavous DA, Jackson FR, O'Connor CM. Extension of the Drosophila lifespan by overexpression of a protein repair methyltransferase. Proc Natl Acad Sci U S A. 2001;98:14814–14818. doi: 10.1073/pnas.251446498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morrow G, Samson M, Michaud S, Tanguay RM. Overexpression of the small mitochondrial Hsp22 extends Drosophila life span and increases resistance to oxidative stress. FASEB J. 2004;18:598–599. doi: 10.1096/fj.03-0860fje. [DOI] [PubMed] [Google Scholar]
- 52.De Velasco B, Shen J, Go S, Hartenstein V. Embryonic development of the Drosophila corpus cardiacum, a neuroendocrine gland with similarity to the vertebrate pituitary, is controlled by sine oculis and glass. Dev Biol. 2004;274:280–294. doi: 10.1016/j.ydbio.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 53.Shen J, Ford D, Landis GN, Tower J. Identifying sexual differentiation genes that affect Drosophila lifespan. BMC Geriatr. 2009;9:56. doi: 10.1186/1471-2318-9-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sambrook J, Maniatis T, Fritsch EF. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 55.Rozas J, Rozas R. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics. 1999;15:174–175. doi: 10.1093/bioinformatics/15.2.174. [DOI] [PubMed] [Google Scholar]
- 56.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genotype-phenotype associations at the Lim3 locus.
(0.17 MB DOC)