Abstract
Nedd4 and Itch are E3 ubiquitin ligases that, in vitro, ubiquitinate similar targets and thus are thought to function similarly. T cells lacking Itch show spontaneous activation and T helper type 2 (TH2) polarization. To test whether the loss of Nedd4 affects T cells in the same way, we generated Nedd4+/+ and Nedd4−/−fetal liver chimeras. Nedd4−/−T cells developed normally but proliferated less, produced less interleukin 2, and provided inadequate help to B cells. Nedd4−/−T cells contain increased amounts of Cbl-b protein, and Nedd4 was required for Cbl-b poly-ubquitination induced by CD28 co-stimulation. These data demonstrate that Nedd4 promotes the conversion of naive T cells into activated T cells. We propose that Nedd4 and Itch ubiquitinate distinct target proteins in vivo.
INTRODUCTION
The amount of protein present in a cell is determined by rates of production and degradation. Although the molecular mechanisms influencing protein production are relatively well understood, much less is known about how proteins are destroyed. However, it is known that the destructive process is initiated when ubiquitin is covalently attached to a target protein; accordingly, this process is regulated by a family of proteins known as ubiquitin ligases1,2.
Three groups of ubiquitin processing enzymes have been identified. The E1 and E2 enzymes process ubiquitin, preparing it for transfer to protein substrates. The E3 ubiquitin ligases are responsible for ‘selecting’ target proteins and directing the attachment of ubiquitin to these targets2. Of the E3 ubiquitin ligases, only members containing the homologous to the E6-AP carboxyl terminus (HECT) domain have intrinsic ubiquitin ligase activity; those of the really interesting new gene (RING) domain family do not, and therefore must remain associated with an E2 enzyme to function2. Both HECT-type and RING-type E3 ubiquitin ligases regulate protein degradation in T cells3.
Among the HECT-type E3 ubiquitin ligases, both neural precursor cell expressed developmentally down regulated 4 (Nedd4) (http://www.signaling-gateway.org/molecule/query?afcsid=A001622) and Itch (http://www.signaling-gateway.org/molecule/query?afcsid=A000959) are expressed in T cells4–6. Several lines of evidence suggest that Itch functions to maintain T cell tolerance.
First, expression of Itch is increased when T cells are primed in the absence of CD28 or ICOS co-stimulation6, 7. Second, mice with a genetic mutation that prevents Itch expression develop a severe inflammatory disease8,9. Itch can promote the ubiquitination and subsequent destruction of several proteins that regulate T cell responsiveness, including Bcl-10, JunB, Cbl-b (http://www.signaling-gateway.org/molecule/query?afcsid=A003923) and PKCθ, in vitro4,6,10,11. Interestingly, with the exception of JunB, these targets can also be ubiquitinated by Nedd4, suggesting that Itch and Nedd4 might function similarly12–14. Here, however, we present data inconsistent with this hypothesis.
As mice lacking Nedd4 die at birth (data not shown), we generated fetal liver chimeras that lack Nedd4 only in hematopoietic cells. Unlike Itch-deficient T cells, T cells lacking Nedd4 proliferated poorly and were less likely to produce cytokines after stimulation. Unlike Itch-deficient T cells, Nedd4-deficient T cells did not exhibit spontaneous activation or excessive T helper type 2 (TH2) polarization. The dissimilarities between the phenotypes of Itch- and Nedd4-deficient T cells suggest that Nedd4 and Itch ubiquitinate distinct targets. The TH2-mediated disease in Itch mutant mice can be explained by the loss of Itch-mediated degradation of JunB, a transcription factor that promotes TH2 polarization4. Here we showed that defective activation of Nedd4-deficient T cells can be explained, in part, by impaired ubiquitination and degradation of Cbl-b, a protein that facilitates destruction of T cell receptor (TCR) signaling components. Based on these data, we propose that Nedd4 ubiquitinates Cbl-b and promotes its destruction, thus allowing T cell activation to proceed.
Results
Nedd4−/−fetal liver chimeras
Embryonic stem (ES) cells harboring a disruption of the Nedd4 gene were obtained from BayGenomics (cell line codes XA209 and XB398). These targeted ES cells contain a gene trapping vector inserted within intron 25 or 12 (for XA209 and XB398, respectively) of Nedd4 (Supplementary Fig. 1a, online)16. The gene trap vector is composed of an artificial intron (En2), a splice acceptor site, and a β-Geo cassette. As Nedd4−/−mice are embryonic lethal, to study how the loss of Nedd4 affects T cell function, we generated fetal liver chimeras by transferring Nedd4+/+ and Nedd4−/− fetal liver cells into lethally irradiated C57BL/6 recipients. To confirm genotyping, detection of Nedd4 protein from fetal liver chimera T cells was analyzed by immunoblot (Supplementary Fig. 1b, online). Nedd4+/+ and Nedd4−/−fetal liver chimeras showed similar rates of death during the reconstitution process, suggesting that the number and viability of hematopoietic precursors within the fetal liver from Nedd4+/+ and Nedd4−/−embryos was similar.
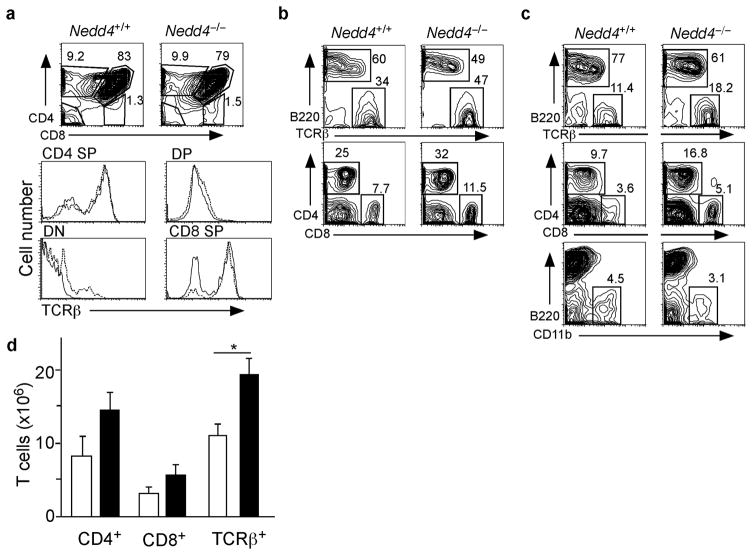
T cells in mice lacking Itch undergo normal development but, over time, become spontaneously activated in vivo4. We wished to determine whether the loss of Nedd4 similarly affected T cell development and activation. Nedd4+/+ and Nedd4−/−fetal liver chimeras showed no significant differences in the numbers or percentages of thymocytes at each developmental stage, or in the expression of TCRβ proteins on the thymocyte surface (Fig. 1a). From these findings we inferred that Nedd4 is not required for T cell development.
Figure 1.
Phenotypic analysis of Nedd4+/+ and Nedd4−/−cells isolated from thymus, lymph node and spleen. (a- c) Cells were isolated from (a) thymus, (b) lymph node, and (c) spleen and stained with antibodies that recognize indicated surface proteins on T cells (CD4, CD8, TCRβ), B cells (B220), or dendritic cells (CD11b). Representative FACS plots are shown. Numbers indicate percentages of each subset (boxed) among live cells. (d) Based on the FACS analysis, total numbers of T cells per spleen were enumerated using antibodies that recognize CD4+ T cells, CD8+ T cells or all TCRβ+ cells. White bars represent Nedd4+/+ cells and black bars represent Nedd4−/−cells. This phenotypic analysis of immune cell populations was performed on tissues from 6–8 fetal liver chimeras of each genotype. * P = 0.038
In the lymph nodes (LN) and spleen, the percentages of B cells and CD11b+ cells were slightly reduced, whereas the percentages of both CD4+ and CD8+ cells were increased, in Nedd4−/−fetal liver chimeras (Fig. 1b,c). In addition, we noted a modest but significant increase in the numbers of T cells in spleens of Nedd4−/−fetal liver chimeras (Fig. 1d). From these data we can infer that Nedd4 is not required for T cell development.
Impaired Nedd4−/−effector T cell proliferation
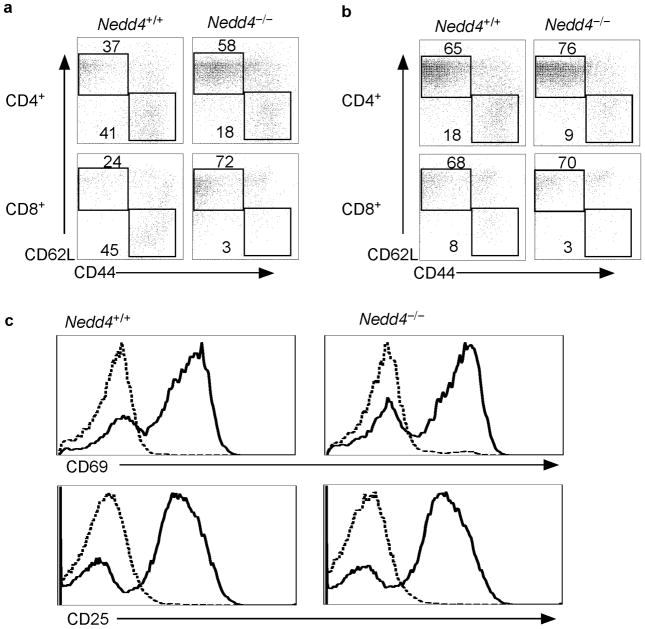
T cells from mice lacking Itch become activated as mice age and, as a consequence, display a surface phenotype characteristic of effector T cells (e.g. CD62LlowCD44hi)4. In contrast, compared to Nedd4+/+ fetal liver chimeras, a higher percentage of splenic and LN CD4+ and CD8+ T cells from Nedd4−/−fetal liver chimeras displayed a naïve T cell surface phenotype (CD62LhiCD44low) (Fig. 2a,b). These data imply that the loss of Nedd4 results in a defect in T cell activation.
Figure 2.
Fewer Nedd4−/−T cells express markers characteristic of an effector phenotype. (a, b) T cells were isolated from (a) spleen and (b) lymph node of Nedd4+/+ and Nedd4−/−fetal liver chimeras and were stained with antibodies specific for the indicated surface proteins. Representative FACS plots, gated on CD4+ or CD8+ populations as indicated, are shown. Numbers indicate the percentages of each subset (boxed) within the CD4+ or CD8+ population. (c) T cells were isolated from the spleen and lymph nodes of Nedd4+/+ and Nedd4−/−fetal liver chimeras and cultured in vitro without stimulation (dashed line) or with anti-CD3 and anti-CD28 (solid line). Expression of CD69 (a marker of B cell activation) and CD80 (a co-stimulatory marker) was analyzed by flow cytometry. Data are representative of three independent experiments.
When T cells are activated, their surface expression of CD69 and CD25 increases. Accordingly, to test whether T cells lacking Nedd4 have a defect in initial activation, we measured expression of these activation markers on the surface of Nedd4+/+ and Nedd4−/−T cells. Nedd4+/+ and Nedd4−/−T cells showed similar upregulation of CD69 and CD25, suggesting that Nedd4 is not required for initial TCR-mediated activation events (Fig. 2c).
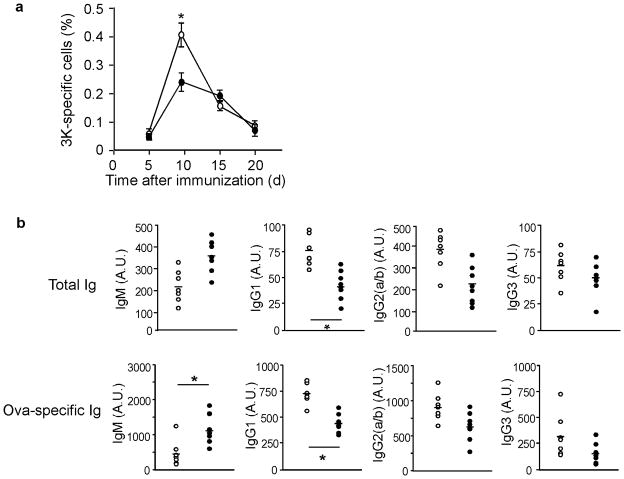
Having demonstrated that there were fewer effector T cells in Nedd4−/−mice, we suspected that T cells lacking Nedd4 might respond poorly to antigen stimulation. To test this hypothesis, we immunized Nedd4+/+ and Nedd4−/−fetal liver chimeras with the I-Ab-binding peptide 3K 17 and lipopolysaccharide (LPS), and measured the numbers of I-Ab–3K-specific T cells at various time points. In the LNs that drain the immunization site of Nedd4+/+ mice, I-Ab–3K-specific T cell populations had expanded from below the limit of detection to comprise approximately 0.41% of the total CD4+ T cell population (Fig. 3a). In contrast, I-Ab–3K-specific T cell populations lacking Nedd4 showed poor population expansion, accounting for only 0.23% of CD4+ T cells in the draining LN. This was not due to a delayed response by the I-Ab–3K-specific Nedd4−/−T cells, as both Nedd4+/+ and Nedd4−/−I-Ab–3K-specific T cell populations contracted after day 9. Thus, T cells lacking Nedd4 proliferated poorly in response to antigen immunization.
Figure 3.
T cells lacking Nedd4 expand poorly in response to antigen in vivo and do not provide adequate help to B cells. (a) 8 weeks after reconstitution, Nedd4+/+ and Nedd4−/−fetal liver chimeras were immunized with the 3K peptide and LPS. 3K-specific T cells were quantified by flow cytometer using an I-Ab-3K tetramer. *, P = 0.023. Open circles represent Nedd4+/+ and closed circles represent Nedd4−/−T cells. Error bars represent s.e.m. (b) Serum from Nedd4+/+ and Nedd4−/−fetal liver chimeras left unimmunized (Total Ig) or immunized with OVA in CFA and boosted with OVA in IFA (OVA-specific Ig) was measured by ELISA. *, P < 0.05. Open circles represent Nedd4−/−and closed circles represent Nedd4+/+ cells. Each dot represents the data point from one animal. Bar represents the mean of each sample population. This data is representative of three independent experiments.
Impaired Nedd4−/−T cell ‘help’ for B cells
T cell proliferation plays a critical role in the adaptive immune response, because T cell ‘help’ for B cells is a limiting factor. T cells provide important signals that promote B cell proliferation, as well as antibody affinity maturation and class switching18. Thus, we next measured whether the absence of Nedd4 in T cells prevented the T cells from delivering crucial signals to B cells. To analyze T cell help, we measured serum concentrations of immunoglobulins (Igs) from unimmunized and immunized Nedd4+/+ and Nedd4−/−fetal liver chimeras. We immunized mice with ovalbumin (OVA) protein and Complete Freund’s Adjuvant (CFA), and then boosted the mice 3 weeks later with OVA and Incomplete Freund’s Adjuvant (IFA). On day 4 after the second antigen dose (boost), we collected serum and measured total and OVA-specific Ig concentrations. In the absence of immunization, Nedd4−/−fetal liver chimeras contained increased concentrations of IgM, the non-class switched version of Ig, and lower concentrations of all class switched Ig isotypes analyzed (Fig. 3b). This same pattern was observed for OVA-specific Ig that developed after immunization. Taken together, these data suggested that Nedd4−/−T cells are not able to provide appropriate T cell help to B cells. However, it is also possible that B cells are intrinsically defective in the absence of Nedd4.
In an effort to determine whether Nedd4 is required for B cell function, we examined surface expression of the B cell receptor (BCR) and of CD21 and CD23, all of which promote B cell activation. These receptors were expressed in comparable amounts on Nedd4+/+ and Nedd4−/−B cells (Supplementary Fig. 2a, online). We then examined BCR-induced upregulation of the activation markers CD69 and CD80, the latter of which is of considerable interest as it facilitates the co-stimulation of T cells by binding to CD28 on the T cell surface. Neither CD69 nor CD80 expression was affected by the absence of Nedd4 (Supplementary Fig. 2b). These data suggest that Nedd4 is not required for B cells to become activated, at least not the initial phases of B cell activation measured here.
Nedd4−/−T cells produce less interleukin 2
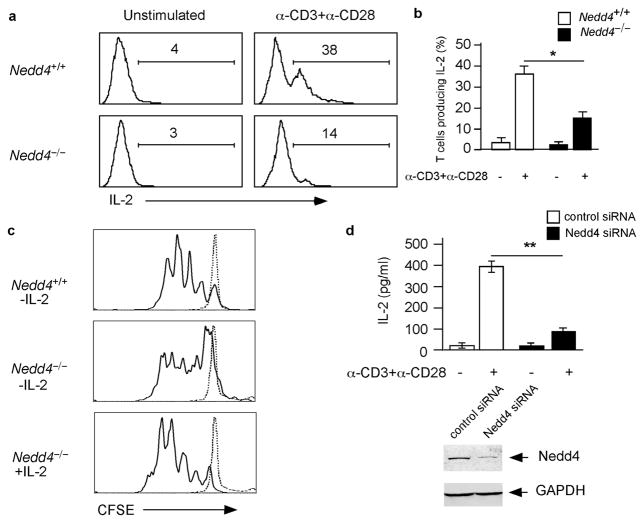
We next measured whether T cells lacking Nedd4 produced interleukin 2 (IL-2), a cytokine that promotes the proliferation of activated T cells. To do this we isolated T cells from Nedd4+/+ and Nedd4−/−fetal liver chimeras and cultured these cells for 20 hours in the absence or presence of plate bound anti-CD3 and anti-CD28, after which we measured IL-2 by intracellular cytokine staining. We chose this time point because it should allow examination of T cells at a time when they are producing IL-2 but have not yet undergone substantial proliferation. Using this protocol, we found that in the absence of stimulation, neither Nedd4+/+ nor Nedd4−/−T cells contained detectable amounts of IL-2 (Fig. 4a). On the other hand, after 20 h of activation, 38% of Nedd4+/+ CD4+ T cells produced IL-2, whereas only 14% of T cells lacking Nedd4 contained measurable amounts of IL-2 (Fig. 4a). Comparing data from multiple experiments, we found a significant decrease in IL-2 production by Nedd4−/−T cells (Fig. 4b). This difference could account for the poor proliferation of these cells.
Figure 4.
Fewer Nedd4−/−T cells produce IL-2. (a) T cells were isolated from Nedd4+/+ and Nedd4−/−fetal liver chimeras, stimulated or left unstimulated as indicated for 20 h, and intracellular IL-2 production was analyzed by FACS. Histograms are gated on CD4+ populations. Numbers indicate percentages of IL-2-producing T cells among total CD4+ T cells. (b) The percentages of IL-2-producing CD4+ T cells were quantified from multiple experiments and the data combined. *, P = 0.0092. (c) Nedd4+/+ and Nedd4−/−CD4+ T cells were isolated and labeled with CFSE. Cells were then cultured for 3 days in media (dashed line) or stimulated in the presence or absence of IL-2 (solid line). CFSE dilution was measured by flow cytometry. (d) D10 T cells were transfected with control siRNA or Nedd4-specific siRNA and then cultured in media or stimulated with anti-CD3 and anti-CD28. Top, after stimulation, IL-2 concentrations in the supernatants were measured by ELISA. Bottom, immunoblot of whole cell lysates to illustrate Nedd4 expression after siRNA treatment. These are representative data from four independent experiments. Error bars show s.e.m. **,P = 0.0002
We reasoned that if low IL-2 production by Nedd4−/−T cells caused the decreased proliferation of these cells, addition of exogenous IL-2 should restore proliferation. To test this hypothesis, we stimulated CFSE-labeled Nedd4−/−CD4+ T cells in vitro and assessed their ability to proliferate in the absence or presence of additional IL-2 (10 U/ml). In the absence of added IL-2, activated Nedd4−/−T cells underwent fewer divisions on average than Nedd4+/+ T cells (Fig. 4c). However, when cultured in the presence of exogenous IL-2, Nedd4−/−T cell proliferation was comparable to that of Nedd4+/+ T cells. This finding suggests that reduced IL-2 production accounts for the decreased proliferation of T cells lacking Nedd4.
Decreased IL-2 production by Nedd4−/−T cells could be due to a defect induced during development of the cells, or, alternatively, due to the absence of Nedd4 in the cells at the time they are stimulated. To distinguish between these possibilities we used siRNA to reduce the expression of Nedd4 in D10, a mature CD4+ T cell line, and then analyzed the ability of these cells to make IL-2. D10 T cells were transfected with either a control or Nedd4-specific siRNA. Forty-eight hours after transfection, Nedd4 protein expression was reduced >90% in cells transfected with Nedd4-specific siRNA, compared to mock transfected cells or cells transfected with control siRNA (Fig. 4d). T cells transfected with Nedd4 siRNA produced significantly less IL-2 than control siRNA-transfected cells (Fig. 4d). Thus, acute loss of Nedd4 in mature T cells is sufficient to reduce IL-2 production.
Taken together, these data suggest that Nedd4 plays an important role in T cell activation by promoting or facilitating signals that stimulate IL-2 production and proliferation. Previous data has shown that CD28 co-stimulation promotes both proliferation and IL-2 production19,20. T cells from mice lacking CD28 respond to antigen similarly to T cells lacking Nedd421–23. Thus, we reasoned that Nedd4 might regulate a protein that inhibits CD28 signaling.
Nedd4−/−T cells express excess Cbl-b
Nedd4 has been shown to ubiquitinate many proteins in studies performed in vitro6,10,11,24–26. These studies often found that the same target proteins were ubiquitinated by Nedd4 and Itch6,10, 11. However, these findings may reflect in vitro promiscuity of these E3 ubiquitin ligases rather than a true sharing of target protein specificity. Two lines of evidence support this hypothesis. First, neither of these E3 ubiquitin ligases is able to compensate for the loss of the other in vivo8. Second, T cells lacking Itch are hyperresponsive, whereas those lacking Nedd4 are hyporesponsive.
Our data suggest that Nedd4 regulates a protein that inhibits T cell activation. Thus, to identify the mechanism by which Nedd4 regulates T cell function we needed to focus on potential targets that would inhibit signals downstream of T cell activation. Phosphatase and tensin homologue deleted on chromosome 10 (PTEN) and Cbl-b are two such candidates. PTEN dephosphorylates PI(3,4,5)P3 thus opposing the effects of phosphatidyl inositol-3-kinase (PI3K) and negatively regulating signals downstream of CD28 co-stimulation27–29. Cbl-b is a ring-type ubiquitin ligase that is upregulated in a calcineurin-dependent manner to promote T cell anergy6. Both PTEN and Cbl-b have been shown to be ubiquitinated and degraded by Nedd4 in vitro9,25. Thus, we sought to determine whether expression of either of these proteins was altered in Nedd4−/−T cells.
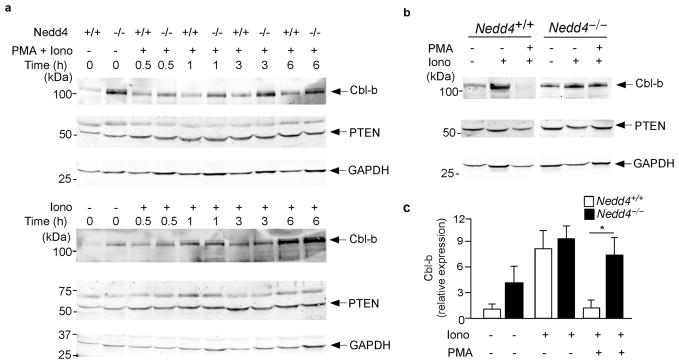
As many proteins targeted for ubiquitination by members of the Nedd4 family are subsequently degraded6,10,11,25, in the absence of Nedd4, target protein amounts would be expected to be increased in resting and/or activated T cells. We examined Nedd4 target gene expression in T cell lines generated from Nedd4+/+ and Nedd4−/−mice. These T cell lines were cultured alone or in the presence of ionomycin or a combination of PMA and ionomycin for 0–6 h, or for 24 h, after which Cbl-b and PTEN proteins were measured. In support of previously published data6, Cbl-b was expressed in Nedd4+/+ T cells and its expression increased when cells were treated with the anergizing stimulus of ionomycin alone (Fig. 5a,b). When these same cells were treated with PMA and ionomycin, Cbl-b protein expression remained low, again consistent with previously published data showing that CD28 signals promote the degradation of Cbl-b (Fig. 5a,b)30.
Figure 5.
Nedd4−/−CD4+ T cells express normal amounts of PTEN and increased amounts of Cbl-b. (a, b) T cell lines were generated from Nedd4+/+ and Nedd4−/−fetal liver chimeras and stimulated with PMA and ionomycin or ionomycin alone where indicated. Expression of PTEN, Cbl-b and GAPDH (loading control) was measured by immunoblot. Cells were analyzed (a) 0–6 h or (b) 24 h after activation. Data are representative of 3–5 different experiments. (c) The 24 h Cbl-b data from multiple experiments was quantified, normalized to that of the loading control (GAPDH) and expressed relative to basal amounts in Nedd4+/+ cells. Error bars show s.d. *, P = 0.0001
Although T cells lacking Nedd4 expressed higher basal amounts of Cbl-b, nonetheless, as in Nedd4+/+ T cells, Cbl-b expression increased further following ionomycin treatment. However, unlike Nedd4+/+ T cells, Nedd4−/−T cells stimulated with PMA and ionomycin, showed an initial decrease followed by a sustained increase in Cbl-b expression (Fig. 5a,b). Cbl-b amounts increased in Nedd4−/−T cells from 3 to 24 h after stimulation, suggesting that Cbl-b was not being appropriately degraded. Normalization of protein amounts at the 24 h time point revealed that compared to Nedd4+/+ T cells, Nedd4−/−T cells had significantly elevated amounts of Cbl-b when activated with both PMA and ionomycin (Fig. 5c). Taken together, these data support a role for Nedd4 in the ubiquitination and degradation of Cbl-b following T cell stimulation. However, based on these data, we could not rule out the possibility that a defect in Nedd4−/− T cell signaling alters Cbl-b transcription, rather than ubiquitin-mediated degradation. This might occur if these cells contained high amounts of PTEN.
T cells with increased PTEN protein would be less able to propagate signals downstream of the CD28 co-stimulatory receptor. We postulated that if Nedd4 regulated the ubiquitination and degradation of PTEN in T cells, than PTEN quantities might be increased and, thus, signaling diminished, in Nedd4−/− T cells. To address this possibility, we measured the relative expression of PTEN protein. Under these conditions, we were unable to detect any differences in PTEN protein expression in Nedd4+/+ and Nedd4−/− T cells (Fig. 5a,b). These data suggest that Nedd4 does not regulate PTEN protein expression in T cells under these conditions. However, these data did not reveal whether Nedd4 regulates Cbl-b by affecting transcription or ubiquitin-mediated degradation.
Nedd4 is an E3 ubiquitin ligase, so the most likely possibility was that Nedd4 affects the turnover of Cbl-b protein. To test this idea, we blocked protein production using cycloheximide, a potent inhibitor of translation (Supplementary Fig. 3a, online). Between 6 and 10 h after activation, Cbl-b degradation was evident in Nedd4+/+ but not Nedd4−/− T cells (Supplementary Fig. 3b). The results from several such experiments showed that very little Cbl-b degradation occurred in resting T cells, regardless of whether or not they contained Nedd4. After 5 h of activation, however, a considerable amount of Cbl-b was turned over in cells containing, but not in those lacking, Nedd4 (Supplementary Fig. 3c). Thus Nedd4 promotes Cbl-b degradation and this may account for the differences in Cbl-b amounts observed in activated Nedd4+/+ versus Nedd4−/−T cells.
Nedd4 binds Cbl-b and promotes its ubiquitination
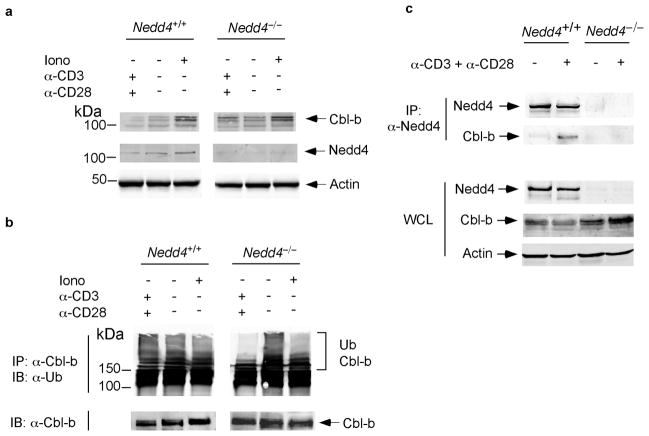
The generation of T cell lines requires one or more rounds of stimulation and propagation in the presence of IL-2. It is possible that the results described above are artifacts of cell lines and do not reflect a situation that would occur in primary T cells. To rule out this possibility, we measured Cbl-b protein expression in primary T cells isolated from Nedd4+/+ and Nedd4−/−fetal liver chimeras (Fig. 6a). Consistent with data shown for T cell lines, Cbl-b expression was higher in the Nedd4−/−T cells stimulated with anti-CD3 and anti-CD28. As an added precaution, we tested the Cbl-b antibody used in these experiments and found that it was specific for Cbl-b (Supplementary Fig. 4). Thus, Cbl-b was not ubiquitinated and degraded in primary T cells that lack Nedd4.
Figure 6.
Nedd4 binds to and promotes ubiquitination of Cbl-b. (a, b) Primary CD4+T cells were isolated from Nedd4+/+ and Nedd4−/−fetal liver chimeras. Cells were cultured for 24 h with or without stimulation as noted. Whole cell lysates were (a) immunoblotted with antibodies specific for Cbl-b, Nedd4 or actin (loading control), or (b) immunoprecipitated with anti-Cbl-b and immunoblotted with anti-ubiquitin. (c) Primary CD4+ T cells were isolated from Nedd4+/+ and Nedd4−/−fetal liver chimeras. Cells were cultured for 24 h in the absence or presence of plate-bound anti-CD3 and anti-CD28. Lysates were immunoprecipitated with anti-Nedd4 where indicated, and immunoblotted for Nedd4, Cbl-b and actin (loading control). WCL, whole cell lysate. These data are representative of 2–3 independent experiments.
We next wished to ascertain whether Nedd4 was required for Cbl-b ubiquitination. We immunoprecipitated Cbl-b from primary Nedd4+/+ and Nedd4−/−T cells and immunoblotted for ubiquitin (Fig. 6b). We noted markedly reduced polyubiquitination of Cbl-b immunoprecipitated from Nedd4−/−T cells stimulated with anti-CD3 and anti-CD28, compared to their stimulated Nedd4+/+ counterparts. This data supports a role for Nedd4 in ubiquitination of Cbl-b, and suggests that Nedd4 might be particularly relevant in the degradation of Cbl-b that occurs as a consequence of T cell activation. Based on this, we predicted that Nedd4 interacts with Cbl-b in activated T cells. Thus, we immunoprecipitated Nedd4 from Nedd4+/+ and Nedd4−/−T cells that were left unstimulated or were stimulated with anti-CD3 and anti-CD28 for 16 h (Fig. 6c). As expected, we were able to immunoprecipitate Nedd4 from Nedd4+/+ but not Nedd4−/−T cells.
Additionally, we found that Cbl-b co-immunoprecipitated with Nedd4 in stimulated but not unstimulated Nedd4+/+ T cells. Taken together, these data suggest that when wild- type T cells are stimulated, Nedd4 binds to and ubiquitinates, and thus promotes the degradation of, Cbl-b. Furthermore, our data suggest that the degradation of Cbl-b may be necessary for T cells to become fully activated, make IL-2 and proliferate to their full capacity.
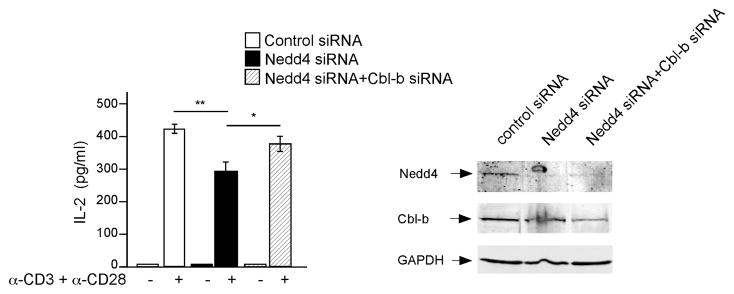
If this hypothesis is correct, then the reduction in IL-2 caused by loss of Nedd4 should not occur if Cbl-b is simultaneously reduced. To test this, siRNAs specific for Nedd4 and Cbl-b, alone and in combination, were transfected into activated primary CD4+ T cells. As a control, we used a pool of siRNAs that deplete irrelevant proteins. Protein expression measured 48 and 72 h after transfection showed that Nedd4 protein amounts were reduced approximately 90% by the Nedd4-specific siRNA, whereas Cbl-b expression was reduced 40% by the Cbl-b-specific siRNA, compared to control siRNA (Fig. 7). We tested several different Cbl-b siRNAs alone or in combination and, the 40% reduction shown here was the best we could achieve. Nevertheless, this reduction appeared to be effective as IL-2 production increased when Cbl-b siRNA was transfected into T cells (data not shown). As expected, Nedd4-specific siRNA significantly reduced T cell IL-2 production; however, this defect was restored by simultaneous transfection with Cbl-b-specific siRNA (Fig. 7). Taken together, our data indicate that Nedd4 promotes the degradation of Cbl-b, and thus regulates a checkpoint in T cell activation that ‘authorizes’ IL-2 production and subsequent proliferation after TCR and CD28 stimulation.
Figure 7.
Simultaneous depletion of both Nedd4 and Cbl-b restores IL-2 production. Primary CD4+ T cells were isolated and transfected with control siRNA (open bars), Nedd4-specific siRNA (filled bars) or a pool of Nedd4- and Cbl-b-specific siRNA (hatched bars). Cells were then stimulated with anti-CD3 and anti-CD28 and IL-2 production was measured by ELISA (left). Whole cell lysates were immunoblotted to illustrate siRNA-mediated suppression of Nedd4 and Cbl-b protein exprpession (right). T cells from 4 individual animals were tested in 4 independent experiments. Each test was replicated at least 2 times. Error bars represent s.e.m. *, P = 0.03, **, P = 0.0005 based on a paired sample, two tailed T test.
Discussion
By studying mice lacking Nedd4 in hematopoietic cells, we made several important observations. First, Nedd4−/−fetal liver chimeras show, by all measures analyzed in this study, a very different phenotype compared to mice that lack Itch. Mice lacking Itch have hyperresponsive T cells8 while mice lacking Nedd4 (in the hematopoietic compartment) had hyporesponsive T cells. One interpretation of this observation is that these two E3 ubiquitin ligases act on similar targets but in an opposing manner. This hypothesis seems unlikely considering that both of these E3 ligases appear to lead to similar outcomes, namely polyubiquitination and degradation, when tested on a given target protein in vitro6,9,10. Additionally, both of these E3 ubiquitin ligases catalyzed the production of lysine 48-linked polyubiquitin chains when tested on a synthetic substrate31. These data suggest that Nedd4 and Itch do not oppose each other by ubiquitinating the same target proteins in a functionally distinct manner.
A more likely scenario is that Nedd4 and Itch, although structurally similar, ubiquitinate different target proteins in vivo. This hypothesis presumes that in vitro ubiquitination assays and over-expression systems promote promiscuous behavior by these E3 ubiquitin ligases and thus, while useful for studying ligase activity, these assays are not able to accurately predict whether the E3 ubiquitin ligase interacts with a given target in vivo.
Nedd4 has been shown to ubiquitinate more than a dozen potential target proteins in vitro6,10,11, 24,26,32–35. At least 8 of these proteins are also ubiquitinated in vitro by Itch6,10,11,36. Targets that might be relevant to Nedd4 function in T cells include, Notch1, PKCθ, phospholipase C-γ1, PTEN, Cbl-b, c-Cbl, and Bcl1027,37–44. Among these, PTEN and Cbl-b stand out as potent inhibitors of T cell activation. Of these two proteins, Cbl-b has been shown to be ubiquitinated by both Itch and Nedd49, whereas ubiquitination of PTEN by Itch has not been reported. Although PTEN amounts were not increased in Nedd4−/−T cells, Cbl-b expression was much higher in Nedd4−/−T cells than in Nedd4+/+ controls. Unlike Nedd4+/+ controls, Nedd4−/−T cells stimulated with both PMA and ionomycin showed increased expression of Cbl-b, suggesting that Cbl-b is degraded after T cells are activated. These findings suggest that Nedd4 functions only under certain circumstances. Notably, Nedd4 protein expression does not change significantly after stimulation. Thus, it seems possible that Nedd4 relies on an adaptor protein that is only expressed in activated T cells.
Whereas Cbl-b degradation is only evident in activated T cells, Cbl-b was ubiquitinated in both stimulated and unstimulated Nedd4−/−T cells. This finding suggests that Cbl-b polyubiquitination in unstimulated T cells is non-degradative. A thorough analysis of Cbl-b ubiquitination, in both activated and naive T cells, will help to resolve this issue. It will also be important to determine how Cbl-b is ubiquitinated in unstimulated T cells, and whether the switch from non-degradative to degradative forms of ubiquitination involves a de-ubiquitinating enzyme.
Whereas the absence of Cbl-b leads to hyperresponsive T cells, elevated amounts of Cbl-b are coincident with T cell anergy. This suggests that high quantities of Cbl-b could inhibit T cell activation and cause the hypo-responsiveness of Nedd4−/−T cells. We confirmed this hypothesis by simultaneously depleting both Nedd4 and Cbl-b; lowered amounts of Cbl-b in T cells lacking Nedd4 restored IL-2 production. Whether impaired Cbl-b degradation is the cause of all the defects in Nedd4−/−T cells is not certain; nevertheless, the fact remains that Nedd4 expression is required for T cells to become fully activated.
Notably, Nedd4−/−fetal liver chimeras contained increased numbers of both CD4+ and CD8+ T cells. This could be due to abnormal development of T cell progenitors, or to altered T cell homeostasis. The latter option seems unlikely since these cells do not exhibit a CD44hiCD62Llow phenotype. Future studies will need to focus on whether these increased T cell numbers are due to altered amounts of Cbl-b or, alternatively, whether this is due to another Nedd4-regulated pathway.
Methods
Generation of Nedd4−/−fetal liver chimeras
Targeted ES cell lines with the mouse Nedd4 gene disrupted (XA209 and XB398) were obtained from BayGenomics and injected into mouse blastocysts for generation of chimeras as described previously5,45. Fetal liver cell suspensions from day14–16 Nedd4+/+ and Nedd4−/−embryos were transferred into lethally irradiated 6–10 week old C57BL/6 recipients (split dose 800 Rad, 2 h rest, 400 Rad) by tail vein injection.
Mice
C57BL/6J mice were purchased from The Jackson Laboratory. Unless otherwise specified, mice used in experiments were 8–10 weeks old. All mice were maintained in a specific pathogen free (SPF) barrier facility. Care of mice used in the experiments met the standards set forth by the National Institutes of Health in their guidelines for the care and use of experimental animals.
T cell isolation and stimulation, and production of T cell lines
T cells were isolated from LN and/or spleen by nylon wool. T cell lines were generated as previously described (ref 5), For short term in vitro stimulation, cells were cultured in the absence of IL-2, on plates bound with 50μg/ml anti-CD3 (clone 145-2C11) and anti-CD28 (clone 37.51), or with 1μM ionomycin (Calbiochem) with or without 50ng/ml PMA (Calbiochem).
Cytokine staining
Stimulated T cells were incubated for the final 4 h with Brefeldin A. The cells were surface stained with anti-CD4 (clone RM4–5), then fixed, and permeabilized (Cytofix/Cytoperm Plus Kit, BD Biosciences) and incubated with anti-IL-2 for 1 h. Data were acquired on a FACScalibur and analyzed using CellQuestPro software (Beckton Dickenson).
Immunization and analysis of serum Immunoglobulin isotypes
To measure T cell expansion in vivo, mice were immunized with 10μg 3K and 7μg LPS subcutaneously in the hind leg. Draining LN cells were harvested and stained with I-Ab–3K tetramer as previously described17. Data was acquired on a CYAN flow cytometer (Cytomation) and analysed by FlowJo software (Treestar). To measure serum immunoglobulins, mice were immunized with 100μg OVA and 7μg CFA emulsion subcutaneously and then boosted with OVA and IFA (as previously described5). Total or OVA-specific Ig was measured by ELISA (as described previously5).
Antibodies
For surface staining, cells were incubated with antibodies diluted in 2.4G2 conditioned supernatant (to block Fcγ receptors). Antibodies used for FACS analysis were purchased from BD Biosciences except anti-TCRβ (Ham597)46. Antibodies used for immunoblotting were as follows: Nedd4 (07-049, Upstate, 611480, BD Transduction Labs), GAPDH (MAB374, Chemicon International), Cbl-b (G-1, Santa Cruz Biotechnology), ubiquitin (P4D1, Cell Signaling Technology), and PTEN (#9552, Cell signaling Technology). Secondary antibodies were goat anti-mouse IgG1 (A680, Molecular Probes) and goat anti-rabbit IgG IRDye (800CW, Li-Cor Biosciences).
siRNA assay
CD4+ T cells were stimulated with anti-CD3 and anti-CD28 as described above and expanded for 2–4 days in IL-2. Approximately 5 × 106 cells were electroporated with 400 pmol of siRNA (mouse Nedd4 siGenome On-Target plus Smart Pool L-058562-00-0010, mouse Cbl-b siGenome On-Target plus Smart Pool L-051830-00-0010, Dharmacon RNA Technologies, mouse control siGenome On-Target plus Smart Pool) using a BTX ECM 830 Squarewave Electroporator, then cultured for 48 h in the presence of IL-2. Electroporated cells were re-stimulated without exogenous IL-2 for 24 h and IL-2 was measured using the Mouse IL-2 ELISA Ready-Set-Go! reagent set (eBioscience).
Cycloheximide treatment, immunoprecipitation and immunoblotting
Cells were incubated with 30 μg/ml Cycloheximide Ready Made (Sigma) for 4 h after the specified stimulation period and then lysed for immunoblot analyses. Cells were lysed as previously described5 and protein was quantified using a micro BCA kit (Pierce Protein Research). Whole cell lysates were run at 10–20μg/lane. Alternatively, 106 cells were lysed in 40μl sample buffer, briefly sonicated and heated to 100°C prior to loading.
For immunoprecipitation, lysates were precleared with protein A/G Sepharose beads (Pierce), then immunoprecipitated with anti-Nedd4 or anti-Cbl-b and protein A/G Sepharose beads for 1 hr at 4°C. Eluted protein was subjected to SDS-PAGE and transferred to immobilon-Fl (Bio-Rad). Membranes were blocked with Li-Cor blocking buffer, then immunoblotted and imaged using the Odyssey (Li-Cor Biosciences).
Supplementary Material
Anti-Cbl-b is specific for Cbl-b. Whole cell lysates from T cells purified from wild-type and Cbl-b-deficient mice were separated by SDS-PAGE. Blots were then analyzed to determine the specificity of the Cbl-b antibody used in our experiments. These data are representative of three independent experiments.
Generation of Nedd4−/−fetal liver chimeras. (a) The insertion point of the gene trapped vector (SA IRES β-GEO PLAP PolyA) was determined using 5′RACE and is shown for the two ES cells lines (XA209 and XA398). The following primers were used to genotype mice with Nedd4 gene disruption15, XA209f (5′ AGG TCA TCC ACT GGT TCT GG 3′) and XA209r (5′ TCT GAG AGC TCT GCA CAG GA 3′) amplified a 930 bp fragment from the wild type allele, whereas XA209f and XA209r2 (5′ TGT CCT CCA GTC TCC TCC AC 3′) amplified a 400 bp fragment from the gene trapped allele. For the ES line XA209, the insertion was mapped to an intron within the genomic region encoding the HECT domain. For XA398, the gene trap vector was inserted within an intron in the genomic region that encodes a protein sequence between the first two WW domains. Gray boxes depict domains within the Nedd4 protein. (b) Whole cell lysates of T cells isolated from Nedd4+/+ and Nedd4−/−fetal liver chimeras immunoblotted with anti-Nedd4.
B cells lacking Nedd4 are not markedly defective. (a) Bone marrow and spleen cells were isolated from Nedd4+/+ and Nedd4−/−fetal liver chimeras and stained with antibodies specific for the indicated surface proteins. Spleen cells are gated on B220+ cells. Gates show pre-B cells (B220+ IgM−), immature B cells (B220lowIgM+), and mature/recirculating B cells (B220hi IgM+) for bone marrow, and follicular B cells (CD23hiCD21low) or marginal zone B cells (CD21hiCD23low) for spleen. (b) B cells from the spleen were activated in vitro using anti-Ig (H+L) and expression of CD69 (a marker of B cell activation) and CD80 (a co-stimulatory molecule) was measured by FACS. Filled histograms represent unstimulated samples, while open histograms illustrate stimulated traces. Data are representative of three independent experiments.
Cbl-b degradation increases considerably after T cell activation. (a) Nedd4+/+ and Nedd4−/−T cell lines were stimulated for 6 h with anti-CD3 and anti-CD28 and then lysed (A) or treated with cycloheximide (CHX) for an additional 4 h (B). Cells were then lysed and Cbl-b protein expression was analyzed by immunoblot. (b) Cbl-b degradation was determined at various time points after the initiation of cycloheximide treatment as depicted in (a). Open bars represent Nedd4+/+ and filled bars are Nedd4−/−T cells.
Acknowledgments
We thank R. Schwartz for Cbl-b-deficient mice and M. Shaffer for help with siRNA experiments. We would also like to acknowledge J. Loomis for sorting cells.
References
- 1.Ciechanover A, Heller H, Elias S, Haas AL, Hershko A. ATP-dependent conjugation of reticulocyte proteins with the polypeptide required for protein degradation. Proc Natl Acad Sci U S A. 1980;77:1365–8. doi: 10.1073/pnas.77.3.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ciechanover A. Intracellular protein degradation: from a vague idea thru the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Cell Death Differ. 2005;12:1178–90. doi: 10.1038/sj.cdd.4401692. [DOI] [PubMed] [Google Scholar]
- 3.Liu YC. Ubiquitin ligases and the immune response. Annu Rev Immunol. 2004;22:81–127. doi: 10.1146/annurev.immunol.22.012703.104813. [DOI] [PubMed] [Google Scholar]
- 4.Fang D, et al. Dysregulation of T lymphocyte function in itchy mice: a role for Itch in TH2 differentiation. Nat Immunol. 2002;3:281–7. doi: 10.1038/ni763. [DOI] [PubMed] [Google Scholar]
- 5.Oliver PM, et al. Ndfip1 protein promotes the function of itch ubiquitin ligase to prevent T cell activation and T helper 2 cell-mediated inflammation. Immunity. 2006;25:929–40. doi: 10.1016/j.immuni.2006.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heissmeyer V, et al. Calcineurin imposes T cell unresponsiveness through targeted proteolysis of signaling proteins. Nat Immunol. 2004;5:255–65. doi: 10.1038/ni1047. [DOI] [PubMed] [Google Scholar]
- 7.Babu S, Blauvelt CP, Kumaraswami V, Nutman TB. Regulatory networks induced by live parasites impair both Th1 and Th2 pathways in patent lymphatic filariasis: implications for parasite persistence. J Immunol. 2006;176:3248–56. doi: 10.4049/jimmunol.176.5.3248. [DOI] [PubMed] [Google Scholar]
- 8.Perry WL, et al. The itchy locus encodes a novel ubiquitin protein ligase that is disrupted in a18H mice. Nat Genet. 1998;18:143–6. doi: 10.1038/ng0298-143. [DOI] [PubMed] [Google Scholar]
- 9.Liu YC. The E3 ubiquitin ligase Itch in T cell activation, differentiation, and tolerance. Semin Immunol. 2007;19:197–205. doi: 10.1016/j.smim.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magnifico A, et al. WW domain HECT E3s target Cbl RING finger E3s for proteasomal degradation. J Biol Chem. 2003;278:43169–77. doi: 10.1074/jbc.M308009200. [DOI] [PubMed] [Google Scholar]
- 11.Scharschmidt E, Wegener E, Heissmeyer V, Rao A, Krappmann D. Degradation of Bcl10 induced by T-cell activation negatively regulates NF-kappa B signaling. Mol Cell Biol. 2004;24:3860–73. doi: 10.1128/MCB.24.9.3860-3873.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonnevier JL, Zhang R, Mueller DL. E3 ubiquitin ligases and their control of T cell autoreactivity. Arthritis Res Ther. 2005;7:233–42. doi: 10.1186/ar1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mueller DL. E3 ubiquitin ligases as T cell anergy factors. Nat Immunol. 2004;5:883–90. doi: 10.1038/ni1106. [DOI] [PubMed] [Google Scholar]
- 13.Heissmeier V, Rao A. E3 ligases in T cell anergy--turning immune responses into tolerance. Sci STKE. 2004;241:pe29. doi: 10.1126/stke.2412004pe29. [DOI] [PubMed] [Google Scholar]
- 14.Ingham RJ, et al. WW domains provide a platform for the assembly of multiprotein networks. Mol Cell Biol. 2005;25:7092–106. doi: 10.1128/MCB.25.16.7092-7106.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao XR, et al. Nedd4 controls animal growth by regulating IGF-1 signaling. doi: 10.1126/scisignal.1160940. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stryke D, et al. BayGenomics: a resource of insertional mutations in mouse embryonic stem cells. Nucleic Acids Res. 2003;31:278–81. doi: 10.1093/nar/gkg064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rees W, et al. An inverse relationship between T cell receptor affinity and antigen dose during CD4(+) T cell responses in vivo and in vitro. Proc Natl Acad Sci U S A. 1999;96:9781–6. doi: 10.1073/pnas.96.17.9781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grewal IS, Flavell RA. The role of CD40 ligand in costimulation and T-cell activation. Immunol Rev. 1996;153:85–106. doi: 10.1111/j.1600-065x.1996.tb00921.x. [DOI] [PubMed] [Google Scholar]
- 19.Powell JD, Ragheb JA, Kitagawa-Sakakida S, Schwartz RH. Molecular regulation of interleukin-2 expression by CD28 co-stimulation and anergy. Immunol Rev. 1998;165:287–300. doi: 10.1111/j.1600-065x.1998.tb01246.x. [DOI] [PubMed] [Google Scholar]
- 20.Umlauf SW, Beverly B, Lantz O, Schwartz RH. Regulation of interleukin 2 gene expression by CD28 costimulation in mouse T-cell clones: both nuclear and cytoplasmic RNAs are regulated with complex kinetics. Mol Cell Biol. 1995;15:3197–205. doi: 10.1128/mcb.15.6.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Green JM, et al. Absence of B7-dependent responses in CD28-deficient mice. Immunity. 1994;1:501–8. doi: 10.1016/1074-7613(94)90092-2. [DOI] [PubMed] [Google Scholar]
- 22.Lucas PJ, Negishi I, Nakayama K, Fields LE, Loh DY. Naive CD28-deficient T cells can initiate but not sustain an in vitro antigen-specific immune response. J Immunol. 1995;154:5757–68. [PubMed] [Google Scholar]
- 23.Shahinian A, et al. Differential T cell costimulatory requirements in CD28-deficient mice. Science. 1993;261:609–12. doi: 10.1126/science.7688139. [DOI] [PubMed] [Google Scholar]
- 24.Trotman LC, et al. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell. 2007;128:141–56. doi: 10.1016/j.cell.2006.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, et al. NEDD4 is a proto-oncogenic ubiquitin ligase for PTEN. Cell. 2007;128:129–39. doi: 10.1016/j.cell.2006.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leykauf K, et al. Ubiquitin protein ligase Nedd4 binds to connexin43 by a phosphorylation-modulated process. J Cell Sci. 2006;119:3634–42. doi: 10.1242/jcs.03149. [DOI] [PubMed] [Google Scholar]
- 27.Seminario MC, Wange RL. Lipid phosphatases in the regulation of T cell activation: living up to their PTEN-tial. Immunol Rev. 2003;192:80–97. doi: 10.1034/j.1600-065x.2003.00013.x. [DOI] [PubMed] [Google Scholar]
- 28.Xu Z, Stokoe D, Kane LP, Weiss A. The inducible expression of the tumor suppressor gene PTEN promotes apoptosis and decreases cell size by inhibiting the PI3K/Akt pathway in Jurkat T cells. Cell Growth Differ. 2002;13:285–96. [PubMed] [Google Scholar]
- 29.Buckler JL, Walsh PT, Porrett PM, Choi Y, Turka LA. Cutting edge: T cell requirement for CD28 costimulation is due to negative regulation of TCR signals by PTEN. J Immunol. 2006;177:4262–6. doi: 10.4049/jimmunol.177.7.4262. [DOI] [PubMed] [Google Scholar]
- 30.Zhang J, et al. Cutting edge: regulation of T cell activation threshold by CD28 costimulation through targeting Cbl-b for ubiquitination. J Immunol. 2002;169:2236–40. doi: 10.4049/jimmunol.169.5.2236. [DOI] [PubMed] [Google Scholar]
- 31.Kim HT, et al. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J Biol Chem. 2007;282:17375–86. doi: 10.1074/jbc.M609659200. [DOI] [PubMed] [Google Scholar]
- 32.Levy F, et al. Ubiquitylation of a melanosomal protein by HECT-E3 ligases serves as sorting signal for lysosomal degradation. Mol Biol Cell. 2005;16:1777–87. doi: 10.1091/mbc.E04-09-0803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murdaca J, et al. Grb10 prevents Nedd4-mediated vascular endothelial growth factor receptor-2 degradation. J Biol Chem. 2004;279:26754–61. doi: 10.1074/jbc.M311802200. [DOI] [PubMed] [Google Scholar]
- 34.Vecchione A, Marchese A, Henry P, Rotin D, Morrione A. The Grb10/Nedd4 complex regulates ligand-induced ubiquitination and stability of the insulin-like growth factor I receptor. Mol Cell Biol. 2003;23:3363–72. doi: 10.1128/MCB.23.9.3363-3372.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakata T, et al. Drosophila Nedd4 regulates endocytosis of notch and suppresses its ligand-independent activation. Curr Biol. 2004;14:2228–36. doi: 10.1016/j.cub.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 36.Jennings MD, Blankley RT, Baron M, Golovanov AP, Avis JM. Specificity and autoregulation of Notch binding by tandem WW domains in suppressor of Deltex. J Biol Chem. 2007;282:29032–42. doi: 10.1074/jbc.M703453200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiang YJ, et al. Cbl-b regulates the CD28 dependence of T-cell activation. Nature. 2000;403:216–20. doi: 10.1038/35003235. [DOI] [PubMed] [Google Scholar]
- 38.Di Cristofano A, et al. Impaired Fas response and autoimmunity in Pten+/− mice. Science. 1999;285:2122–5. doi: 10.1126/science.285.5436.2122. [DOI] [PubMed] [Google Scholar]
- 39.Donovan JA, Wange RL, Langdon WY, Samelson LE. The protein product of the c-cbl protooncogene is the 120-kDa tyrosine-phosphorylated protein in Jurkat cells activated via the T cell antigen receptor. J Biol Chem. 1994;269:22921–4. [PubMed] [Google Scholar]
- 40.Gaide O, et al. CARMA1 is a critical lipid raft-associated regulator of TCR-induced NF-kappa B activation. Nat Immunol. 2002;3:836–43. doi: 10.1038/ni830. [DOI] [PubMed] [Google Scholar]
- 41.Robey E, et al. An activated form of Notch influences the choice between CD4 and CD8 T cell lineages. Cell. 1996;87:483–92. doi: 10.1016/s0092-8674(00)81368-9. [DOI] [PubMed] [Google Scholar]
- 42.Thome M, Tschopp J. TCR-induced NF-kappaB activation: a crucial role for Carma1, Bcl10 and MALT1. Trends Immunol. 2003;24:419–24. doi: 10.1016/s1471-4906(03)00177-7. [DOI] [PubMed] [Google Scholar]
- 43.Villalba M, et al. Protein kinase ctheta cooperates with calcineurin to induce Fas ligand expression during activation-induced T cell death. J Immunol. 1999;163:5813–9. [PubMed] [Google Scholar]
- 44.Washburn T, et al. Notch activity influences the alphabeta versus gammadelta T cell lineage decision. Cell. 1997;88:833–43. doi: 10.1016/s0092-8674(00)81929-7. [DOI] [PubMed] [Google Scholar]
- 45.McDonald FJ, et al. Disruption of the beta subunit of the epithelial Na+ channel in mice: hyperkalemia and neonatal death associated with a pseudohypoaldosteronism phenotype. Proc Natl Acad Sci U S A. 1999;96:1727–31. doi: 10.1073/pnas.96.4.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCormack JE, Kappler J, Marrack P. Stimulation with specific antigen can block superantigen-mediated deletion of T cells in vivo. Proc Natl Acad Sci U S A. 1994;91:2086–90. doi: 10.1073/pnas.91.6.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Anti-Cbl-b is specific for Cbl-b. Whole cell lysates from T cells purified from wild-type and Cbl-b-deficient mice were separated by SDS-PAGE. Blots were then analyzed to determine the specificity of the Cbl-b antibody used in our experiments. These data are representative of three independent experiments.
Generation of Nedd4−/−fetal liver chimeras. (a) The insertion point of the gene trapped vector (SA IRES β-GEO PLAP PolyA) was determined using 5′RACE and is shown for the two ES cells lines (XA209 and XA398). The following primers were used to genotype mice with Nedd4 gene disruption15, XA209f (5′ AGG TCA TCC ACT GGT TCT GG 3′) and XA209r (5′ TCT GAG AGC TCT GCA CAG GA 3′) amplified a 930 bp fragment from the wild type allele, whereas XA209f and XA209r2 (5′ TGT CCT CCA GTC TCC TCC AC 3′) amplified a 400 bp fragment from the gene trapped allele. For the ES line XA209, the insertion was mapped to an intron within the genomic region encoding the HECT domain. For XA398, the gene trap vector was inserted within an intron in the genomic region that encodes a protein sequence between the first two WW domains. Gray boxes depict domains within the Nedd4 protein. (b) Whole cell lysates of T cells isolated from Nedd4+/+ and Nedd4−/−fetal liver chimeras immunoblotted with anti-Nedd4.
B cells lacking Nedd4 are not markedly defective. (a) Bone marrow and spleen cells were isolated from Nedd4+/+ and Nedd4−/−fetal liver chimeras and stained with antibodies specific for the indicated surface proteins. Spleen cells are gated on B220+ cells. Gates show pre-B cells (B220+ IgM−), immature B cells (B220lowIgM+), and mature/recirculating B cells (B220hi IgM+) for bone marrow, and follicular B cells (CD23hiCD21low) or marginal zone B cells (CD21hiCD23low) for spleen. (b) B cells from the spleen were activated in vitro using anti-Ig (H+L) and expression of CD69 (a marker of B cell activation) and CD80 (a co-stimulatory molecule) was measured by FACS. Filled histograms represent unstimulated samples, while open histograms illustrate stimulated traces. Data are representative of three independent experiments.
Cbl-b degradation increases considerably after T cell activation. (a) Nedd4+/+ and Nedd4−/−T cell lines were stimulated for 6 h with anti-CD3 and anti-CD28 and then lysed (A) or treated with cycloheximide (CHX) for an additional 4 h (B). Cells were then lysed and Cbl-b protein expression was analyzed by immunoblot. (b) Cbl-b degradation was determined at various time points after the initiation of cycloheximide treatment as depicted in (a). Open bars represent Nedd4+/+ and filled bars are Nedd4−/−T cells.