The SIN pathway blocks inappropriate actin rearrangements during cytokinesis by preventing activation of the MOR pathway component Orb6.
Abstract
The mechanisms that regulate cytoskeletal remodeling during the transition between mitosis and interphase are poorly understood. In fission yeast the MOR pathway promotes actin polarization to cell tips in interphase, whereas the SIN signaling pathway drives actomyosin ring assembly and cytokinesis. We show that the SIN inhibits MOR signaling in mitosis by interfering with Nak1 kinase-mediated activation of the most downstream MOR component, the NDR family kinase Orb6. Inactivation of the MOR may be a key function of the SIN because attenuation of MOR signaling rescued the cytokinetic defects of SIN mutants and allowed weak SIN signaling to trigger ectopic cytokinesis. Furthermore, failure to inhibit the MOR is toxic when the cell division apparatus is compromised. Together, our results reveal a mutually antagonistic relationship between the SIN and MOR pathways, which is important for completion of cytokinesis and coordination of cytoskeletal remodeling at the mitosis-to-interphase transition.
Introduction
Eukaryotic cells undergo major reorganizations of their cytoskeleton during cell cycle transitions from interphase to mitosis and vice versa. Because cells reorganize many shared cytoskeletal elements such as actin in the transition between interphase and mitosis, they must have mechanisms to ensure both a rapid and decisive switch between the two states. Although there has been considerable study of how cells assemble the various cytoskeletal structures during each cell cycle stage, it remains unclear how the transition between stages is coordinated.
The fission yeast Schizosaccharomyces pombe has proven to be an excellent model to study cell cycle–dependent cytoskeletal arrangements (Roberts-Galbraith and Gould, 2008; Martin, 2009). This cylindrical eukaryote grows from both ends in interphase by polarization of actin patches and cables toward the growing tips of the cell (Snell and Nurse, 1993). However, the polarized actin distribution is lost with the onset of mitosis when growth ceases and actin is relocalized to the cell middle to form the actomyosin contractile ring. A conserved signaling network called the MOR is essential for polarized growth in interphase because MOR mutants have depolarized actin and become spherical in morphology. The MOR regulates cell polarity at least in part through control of spatial regulation of the Cdc42 GTPase (Das et al., 2009). The MOR pathway is comprised of several proteins including Pmo25 (MO25 in humans; Kanai et al., 2005; Mendoza et al., 2005; Kume et al., 2007), Nak1 (a GC kinase; Kanai et al., 2005; Leonhard and Nurse, 2005), and the homologue of the Drosophila Furry protein called Mor2 (Cong et al., 2001; Hirata et al., 2002), all of which are required for the activity of the NDR kinase Orb6 (Verde et al., 1998; Tamaskovic et al., 2003; Imai et al., 2004; Kanai et al., 2005), the most downstream component of the pathway.
Interestingly, Sid2, the other NDR family kinase in fission yeast, acts at the end of the SIN signaling pathway (Sparks et al., 1999). The SIN is essential for stable actomyosin ring formation, ring constriction, and septum formation during cytokinesis (Krapp and Simanis, 2008). Although the two NDR kinase pathways, MOR and SIN, are essential for separate cytoskeletal configurations, the mechanism that controls the switch between the two states remains unknown. Interestingly, an earlier study suggested that the SIN inhibits actin polarization toward cell tips (Mishra et al., 2004); however, the mechanism of SIN action was unclear.
In this study we show that the SIN can directly inhibit polarized growth by blocking MOR pathway signaling. Our data further suggest that this cross talk between the SIN and MOR pathways is important to coordinate cytoskeletal rearrangements during the transition from mitosis to interphase.
Results
SIN activation in interphase arrests nuclear division and inhibits the polarity machinery
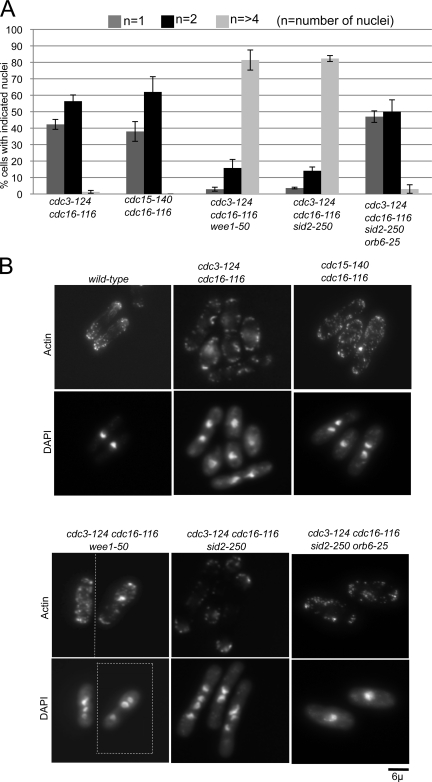
To examine how the SIN affects interphase polarity and cell cycle progression we ectopically activated the SIN in interphase. The SIN can be turned on by inactivation of Cdc16, which is the GAP for the Spg1 GTPase (Fankhauser et al., 1993; Schmidt et al., 1997). If the SIN is activated in asynchronous cells, the cells stop polarized growth because they cease to elongate, and arrest as mononucleate or binucleate cells with multiple septa (Minet et al., 1979; Fankhauser et al., 1993). The disruption of polarized growth in these cells could be an indirect effect caused by ectopic septa formation, which could interfere with growth at cell ends or cause cell death by cutting the nuclei in half. To remove potential complications due to formation of multiple septa, the SIN was activated using the cdc16-116 temperature-sensitive mutant in either the cdc3-124 or cdc15-140 temperature-sensitive mutant background. The cdc3-124 and cdc15-140 mutations block cytokinesis and septum formation by disrupting actomyosin ring assembly, but do not interfere with SIN signaling (Balasubramanian et al., 1994; Fankhauser et al., 1995; Sparks et al., 1999). The cdc16-116 cdc3-124 and cdc16-116 cdc15-140 strains were shifted to restrictive conditions for 4 h (almost two cell cycles) and were scored for cell length and number of nuclei. Because the cdc3-124 and cdc15-140 mutations block cytokinesis, the cdc16-116 cdc3-124 and cdc16-116 cdc15-140 cells should be tetranucleate and twice the length of normal wild-type cells if nuclear division and cell growth proceeded normally. Interestingly, the cdc16-116 cdc3-124 and cdc16-116 cdc15-140 double-mutant cells arrested as both mono- and binucleate cells and failed to elongate past the size of asynchronous wild-type cells (Fig. 1 A, Table I). These cells also displayed an increase in cell diameter consistent with a disruption in polarized growth (Table I). FACS analysis showed that the cells were arrested with 2C and 4C peaks of similar size to the mono- and binucleate peaks, consistent with each nuclei arresting with a G2 DNA content (Fig. S1 A). Thus, SIN activation blocked cell elongation and mitotic entry in interphase cells.
Figure 1.
SIN activation arrests nuclear division and inhibits the interphase polarity machinery. (A) Cells of the indicated genotypes were grown at 25°C and then shifted to 36°C for 4 h, methanol fixed, stained with DAPI, and scored for number of nuclei (n). The experiment was done in triplicate and error bars denote SD. (B) Cells of the indicated genotypes were shifted to 36°C for 4 h and then stained with Alexa Fluor 488–phalloidin and DAPI to visualize actin and DNA, respectively. Dashed dividing lines separate individual images for montage presentations.
Table I.
Activation of the SIN inhibits polarized cell growth
| Strain genotype | Number of nuclei (n) | Mean cell length ± SD | Mean cell diameter ± SD |
| (μm) | (μm) | ||
| wild type | n = 1 | 11.9 ± 1.8 | 2.93 ± 0.2 |
| n = 2 | 14.9 ± 0.817 | 2.95 ± 0.35 | |
| cdc3-124 cdc16-116 | n = 1 | 10.66 ± 2 | 4.62 ± 0.46 |
| n = 2 | 13.75 ± 2 | 4.7 ± 0.5 | |
| cdc15-140 cdc16-116 | n = 1 | 10.2 ± 2 | 6.46 ± 1.26 |
| n = 2 | 15 ± 1 | 6.086 ± 1.24 | |
| cdc3-124 cdc16-116 wee1-50 | n = 1 | 6.65 ± 0.85 | 4.81 ± 0.45 |
| n = 2 | 7.25 ± 1.4 | 4.43 ± 0.54 | |
| n = 4 | 9.6 ± 1.6 | 4.47 ± 0.61 | |
| cdc3-124 cdc16-116sid2-250 | n = 1 | N/A | N/A |
| n = 2 | 21.2 ± 1.75 | 3.47 ± 0.36 | |
| n = 4 | 26 ± 4 | 3.6 ± 0.434 | |
| cdc3-124 cdc16-116 sid2-250 orb6-25 | n = 1 | 7.86 ± 1.5 | 5.04 ± 0.41 |
| n = 2 | 13.23 ± 3 | 5.3 ± 0.365 | |
| n = 4 | 14.38 ± 1 | 8.23 ± 1.13 |
Average length and diameter measurements for cells with indicated genotype and the given number of nuclei are shown. SD, standard deviation. Measurements are not shown (N/A) if the strain did not arrest with a significant number of cells with the indicated number of nuclei. At least 100 cells were counted for each strain.
Cell elongation in interphase requires polarization of actin patches to the cell tips. To test if the block in cell elongation upon SIN activation is due to inhibition of the interphase polarity machinery, we looked at the actin distribution in the cdc3-124 cdc16-116 and cdc15-140 cdc16-116 cells. Upon shift to the restrictive temperature, the double-mutant cells had a dispersed actin cytoskeleton (Fig. 1 B and Fig. S1, B and C), suggesting that SIN signaling disrupted interphase polarity. To rule out the possibility that Cdc16 inactivation could trigger an alternative pathway besides the SIN or that the cdc3-124 ring mutation disrupts interphase polarity, we inactivated the most downstream SIN component Sid2 using the sid2-250 mutation. The cdc3-124 cdc16-116 sid2-250 cells showed no block in cell elongation and nuclear division at the restrictive temperature and became long and multi-nucleate with actin polarized at the cell tips (Fig. 1, A and B; Fig. S1, B and C; Table I), confirming that the effects of Cdc16 inactivation are caused by ectopic activation of SIN signaling.
The G2 arrest observed after SIN activation is presumably mediated by inhibitory phosphorylation on Cdk1 because the nuclear division arrest was lost when the Cdk1 inhibitory kinase Wee1 was inactivated (Fig. 1 A). Interestingly, examination of cdc3-124 cdc16-116 wee1-50 cells at the restrictive temperature showed that although the wee1-50 mutation overcame the nuclear division block caused by SIN signaling, these cells failed to elongate and had depolarized actin (Table I; Fig. 1 B; Fig. S1, B and C), showing that the wee1-50 mutation did not overcome the SIN-mediated inhibition of polarized growth. Thus, SIN inhibition of nuclear division can be uncoupled from its inhibition of interphase actin organization.
The SIN inhibits the activity of the MOR pathway kinase Orb6
The phenotype caused by ectopic activation of the SIN is very similar to that observed in MOR pathway loss of function mutants, which arrest with depolarized actin, fail to elongate, and undergo a Wee1-dependent G2 arrest (Hirata et al., 2002; Kanai et al., 2005). In fact, inactivation of the most downstream MOR pathway component Orb6 caused a block in cell elongation and nuclear division along with an increase in cell diameter in cdc3-124 cdc16-116 sid2-250 cells similar to that in the cdc3-124 cdc16-116 double mutant (Fig. 1 A and Table I). Consistent with the block in cell elongation, the actin cytoskeleton was depolarized in the cdc3-124 cdc16-116 sid2-250 orb6-25 cells (Fig. 1 B). These results suggested that the SIN might disrupt interphase polarity by inhibiting the MOR pathway.
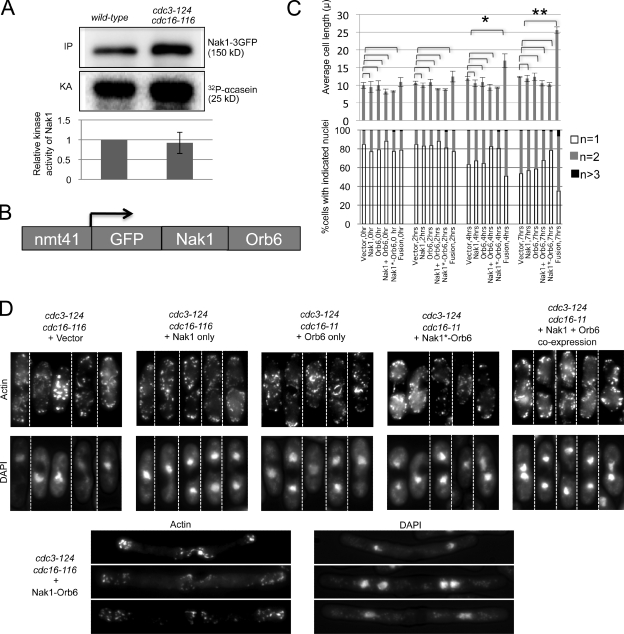
The most downstream component of the MOR pathway is the NDR family kinase Orb6, allowing the kinase activity of Orb6 to serve as a read-out for the functional status of the MOR pathway. To directly test if SIN activation blocks MOR signaling, we examined the kinase activity of Orb6 after ectopic activation of the SIN using the cdc16-116 mutation. As before, the cdc15-140 mutation was included to overcome complications due to constitutive septation in the cdc16-116 cells. In contrast to asynchronous wild-type and G2-arrested cdc25-22 cells, the cdc16-116 cells, which also arrest in G2, showed reduced Orb6 activity, with or without the cdc15-140 mutation (Fig. 2 A). We next examined if SIN activation had a similar effect in interphase-arrested cells. Interestingly, although cdc15-140 cdc25-22 cells maintained high Orb6 activity consistent with their bipolar growth in interphase, SIN activation in this mutant background using the cdc16-116 mutation inhibited Orb6 activity (Fig. 2 A). Hence, SIN activation in interphase results in loss of Orb6 kinase activity and blocks MOR signaling.
Figure 2.
The SIN has a dual role in regulation of the Orb6 kinase activity. (A) Ectopic activation of the SIN in interphase inhibits Orb6 kinase activity. All strains expressed mob2-13Myc from the chromosomal promoter. Cells were grown at 25°C, then shifted to 36°C for 3 h. Cell extracts were prepared and the Orb6–Mob2–13Myc complex was immunoprecipitated using anti-Myc antibodies. Immunoprecipitates were split, with one half immunoblotted with anti-Myc antibodies and the other half used to assay Orb6 kinase activity using MBP as an artificial substrate. The kinase activities (KA) were quantified using ImageJ software and normalized to the amount of Mob2–13Myc (IP) and the background kinase activity in untagged control cells. The activity relative to wild-type cells is shown. Error bars denote SD from three separate experiments. (B) The SIN inhibits Orb6 kinase activity in mitosis but promotes its full activity in the following interphase. Early G2 cells of the indicated strains expressing Mob2-13Myc from the chromosomal locus were grown at 25°C, synchronized by elutriation, and then shifted to 36°C at time zero, and portions were collected at the indicated times and assayed for Orb6 kinase activity. This experiment was repeated at least once with similar results.
We next examined the relationship between the SIN and Orb6 activity under more physiological circumstances. In wild-type cells, Orb6 kinase activity peaks in G2 phase and then decreases as cells go through mitosis and septation (Kanai et al., 2005). Interestingly, the interphase peak in Orb6 activity after cell division was shown to require the activity of some of the SIN components such as Cdc7 in the previous mitosis (Kanai et al., 2005), apparently contradicting our results showing that ectopic SIN activation inhibits Orb6. However, careful examination of Orb6 kinase activity in synchronous cdc7-24 mutant cells resolved the apparent discrepancy. Consistent with the earlier results, Cdc7 was required for the large peak in the Orb6 activity after completion of mitosis (Fig. 2 B, compare 200–240-min time points in wild-type and cdc7-24 cells). However, cdc7-24 cells maintained moderate levels of Orb6 activity, consistent with the continued polarized growth observed in cdc7-24 mutants. Thus, the SIN is not essential for Orb6 activity, per se, but is required for the peak in activity associated with the onset of bipolar growth in the following cell cycle. We next examined Orb6 activity in synchronous cdc15-140 mutants. The cdc15-140 mutant cells have defects in actomyosin ring assembly, which triggers the cytokinesis checkpoint, causing them to arrest as binucleate cells with activated SIN (Liu et al., 2000; Mishra et al., 2004). Interestingly, these cells maintained very low Orb6 kinase activity consistent with the SIN inhibiting Orb6 (Fig. 2, A and B; 160–240 min in cdc15-140). If Cdc7 is inactivated in these cells, they regained moderate levels of Orb6 kinase activity (Fig. 2 B, compare 160–240-min time points in cdc15-140 cdc7-24 and cdc15-140 cells) but lacked the peak in the following interphase (Fig. 2 B, compare 200–240-min time points in cdc15-140 cdc7-24 and wild-type cells). Thus, SIN has a dual role in regulation of the MOR pathway. The SIN suppresses Orb6 activity during cytokinesis but is required for its later increase as cells begin new end growth in the next cell cycle after cytokinesis.
The SIN inhibits the MOR pathway by blocking Nak1-mediated activation of Orb6
We next wanted to understand the mechanism by which the SIN inhibits Orb6 kinase activity. Similar to other members of the NDR kinase family, Orb6 kinase is activated by the GC kinase Nak1 (Kanai et al., 2005; Hergovich et al., 2006). Thus, the SIN could either inhibit the activity of Nak1, or it could inhibit the ability of Nak1 to activate Orb6. A previous study suggested that the SIN does not inhibit Nak1 because Nak1 activity remained high during cytokinesis when the SIN is active (Kanai et al., 2005). To test this possibility in another way, we examined Nak1 kinase activity in cdc3-124 cdc16-116 cells, which arrest with active SIN. Consistent with earlier results, upon SIN activation there was no significant reduction in Nak1 kinase activity (Fig. 3 A). This result suggested that the SIN did not alter Nak1 kinase activity, per se, but possibly disrupted the ability of Nak1 to activate Orb6.
Figure 3.
Fusion of Nak1 to Orb6 bypasses SIN inhibition of cell elongation. (A) The SIN does not inhibit Nak1 activity. Wild-type and cdc3-124 cdc16-116 cells expressing Nak1-3GFP from the chromosomal promoter were grown at 25°C and then shifted to 36°C for 3 h before harvesting. Cell extracts were prepared and Nak1-3GFP was immunoprecipitated with anti-GFP (Invitrogen) antibody. The immunoprecipitates were split, with one portion used for Western blotting using GFP antibodies (Santa Cruz Biotechnology, Inc.) (IP), and the other portion for in vitro kinase assays using α-casein (Sigma-Aldrich) as an artificial substrate (Leonhard and Nurse, 2005). The kinase activity (KA) was measured using a PhosphorImager (MDS Analytical Technologies), quantified using ImageQuant software, and normalized to the amount of Nak1 (IP). The activity relative to wild-type cells is shown. The difference in the normalized Nak1 kinase activity between the wild-type and the cdc3-124 cdc16-116 cells was not significant based on t test analysis from three different experiments. Error bars denote SD of the relative KA. (B) Schematic representation of the Nak1–Orb6 fusion construct expressed from the medium strength thiamine-repressible promoter, nmt41. (C) cdc3-124 cdc16-116 cells carrying the indicated plasmids were induced for 19 h in media lacking thiamine and shifted to 36°C; then cells were collected at 2, 4, and 7 h. The plasmids used were the pRep41 vector, or the pRep41 vector carrying Nak1, Orb6, the Nak1–Orb6 fusion (fusion), the Nak1 kinase-dead–Orb6 fusion (Nak1*-Orb6), or the Nak1 and Orb6 genes on separate plasmids (Nak1 + Orb6). Cells were then stained with DAPI and scored for cell length measurements (top panel) and nuclei count (n, bottom panel). Error bars in the cell length plot denote SD of the length measurements obtained from three separate experiments. Average cell length was compared between cells with vector control and those with the different transgenes as indicated in the figure at each time point. Statistically significant increase in cell length (based on P-value calculation by t test analysis) was observed only in cells expressing the fusion construct. *, P = 0.0118; **, P = 0.0001. At least 100 cells were analyzed for each time point. For clarity, the SD values for the nuclear count plot are shown in Fig. S4. (D) cdc3-124 cdc16-116 cells with the indicated transgenes were grown as in C and then processed for actin staining. Montage of representative cells (separated by dashed dividing lines) with actin (phalloidin staining) and nuclei (DAPI staining) staining are shown for the 7-h time point.
The exact mechanism by which Nak1 activates Orb6 remains unclear. Although interaction between the two proteins was not observed by coimmunoprecipitation, a two-hybrid interaction was observed (Kanai et al., 2005), suggesting a physical association between the two proteins. This raised the possibility that the SIN might interfere with Nak1-mediated activation of Orb6 by preventing association of the two proteins. To test this hypothesis, we constructed a Nak1–Orb6 fusion (Fig. 3 B). The Nak1–Orb6 fusion was able to rescue the growth defects of both the Nak1 and Orb6 single mutants (Table II), suggesting that the individual proteins in the fusion retained functionality. The fusion did not rescue a mutant in the upstream regulator Pmo25, consistent with the known dependence of Nak1 activity on Pmo25 (Kanai et al., 2005). Interestingly, the fusion, but not the Nak1 and Orb6 proteins individually or in combination, completely rescued a mutation in the scaffold protein Mor2, suggesting that the key function of Mor2 is to bring Nak1 and Orb6 together (Table II).
Table II.
Rescue of MOR pathway mutants by the Nak1–Orb6 fusion
| Strain | Vector | Nak1 | Orb6 | Nak1–Orb6 fusion |
| pmo25-35 | − | − | N/A | − |
| nak1-167 | − | + | − | + |
| orb6-25 | − | N/A | + | + |
| mor2-786 | − | − | − | + |
The ability of the Nak1–Orb6 fusion to rescue (+) or not rescue (−) MOR pathway mutants at the restrictive temperature of 36°C is shown. N/A indicates condition that was not tested.
To test whether the Nak1–Orb6 fusion could bypass the block in cell elongation when the SIN is activated, we expressed the Nak1–Orb6 fusion in cdc3-124 cdc16-116 background. Unlike cdc3-124 cdc16-116 cells carrying the vector control plasmid, which did not elongate, the cdc3-124 cdc16-116 cells containing the Nak1–Orb6 fusion plasmid were able to undergo significant elongation (Fig. 3 C). The cdc3-124 cdc16-116 cells expressing the Nak1–Orb6 fusion were able to polarize actin to the cell tips, but also showed medial actin distribution consistent with the cells trying to carry out both the SIN and MOR actin polarization programs (Fig. 3 D). Expression of Nak1 or Orb6 alone or coexpression of both Nak1 and Orb6 in the cdc3-124 cdc16-116 cells did not bypass the cell elongation and actin polarization defects observed in these cells (Fig. 3, C and D), showing that fusion of Nak1 and Orb6 was required to bypass the SIN-mediated inhibition of polarized growth. Furthermore, the kinase activity of Nak1 was required in the fusion because a kinase inactivating mutation in Nak1 blocked the ability of the fusion to drive polarized growth when the SIN is active (Fig. 3, C and D). Interestingly, the Nak1–Orb6 fusion also partially overrode the block in nuclear division caused by SIN activation, as seen by the reduction in mononucleate and increase in bi- and tetranucleate cells compared with controls at the 7-h time point (Fig. 3 C). Because either MOR inactivation or SIN activation blocks both nuclear division and cell elongation, the ability of the Nak1–Orb6 fusion to partially bypass both of these blocks when the SIN is active suggests that the SIN might block both nuclear division and cell growth by inhibiting the MOR. The failure of the fusion to completely bypass the SIN-mediated block in growth and nuclear division suggests that the SIN can still partially inhibit the MOR in the presence of the fusion, or the SIN can affect nuclear division and cell growth through an additional mechanism besides inhibition of the MOR.
Failure to inhibit MOR pathway signaling interferes with cytokinesis
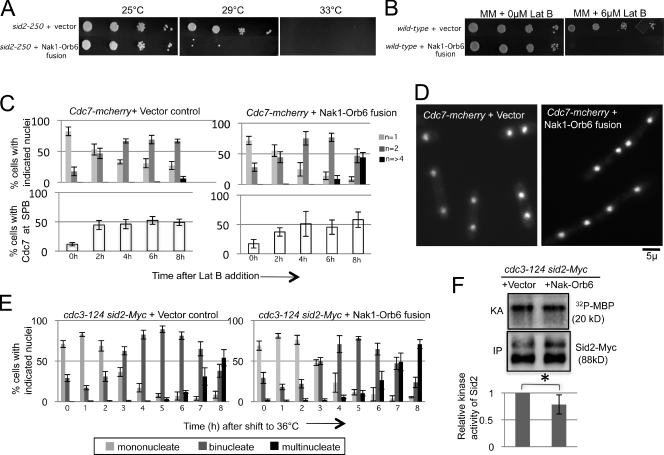
The previous results showed that the SIN blocks polarized growth during cytokinesis by inhibiting the MOR pathway. We next wanted to address why the SIN inhibits the MOR. One possibility is that the SIN might inhibit the MOR to keep it from interfering with cytokinesis by titrating shared cytoskeletal elements such as actin away from the cell division site and toward the cell tips. To test whether loss of SIN inhibition of the MOR caused defects in cytokinesis, we examined the effects of expressing the Nak1–Orb6 fusion in different S. pombe strains. The fusion protein caused a very slight increase in cell length (Fig. S5 C), and surprisingly, expression of the Nak1–Orb6 fusion had only mild effects on cytokinesis in wild-type cells (unpublished data), perhaps because expression of the fusion does not totally bypass SIN inhibition. To see if expression of the Nak1–Orb6 fusion might interfere with cytokinesis in a more sensitized background, we expressed the fusion in cells with compromised SIN signaling. Interestingly, expression of the fusion was lethal when expressed in the temperature-sensitive SIN mutant sid2-250 at the semi-permissive temperature of 29°C (Fig. 4 A). This result suggested that MOR inhibition becomes essential when cytokinesis is partially compromised. To further test this hypothesis we used an alternative way to interfere with the cell division machinery. Low doses of the actin-depolymerizing drug, latrunculin B (Lat B) causes a cell division delay in wild-type cells (Trautmann and McCollum, 2005). During the delay the SIN remains active, causing an arrest in polarized growth and nuclear division until cytokinesis is complete (Mishra et al., 2004). We found that expression of the Nak1–Orb6 fusion in wild-type cells treated with low doses of Lat B is lethal (Fig. 4 B). Examination of similarly treated wild-type cells in liquid culture showed that wild-type cells with the vector control initially accumulate binucleate cells because of the delay in cytokinesis, but are eventually able to divide as judged by their ability to maintain a population of mononucleate cells and failure to accumulate multinucleate cells (Fig. 4, C and D). In contrast, wild-type cells expressing the fusion protein are unable to complete cytokinesis as seen by the loss of mononucleate cells and the accumulation of multinucleate cells (Fig. 4, C and D). This phenotype could be caused by loss of SIN signaling, which also causes cells to fail cytokinesis and become multinucleate when cytokinesis is delayed (Mishra et al., 2004), or the Nak1–Orb6 fusion could be interfering with the cytokinetic apparatus, keeping cells from completing cytokinesis. We do not think that expression of the fusion protein is interfering with SIN signaling because the cells expressing the fusion maintained similar levels of SIN activity as those with the control vector, as judged by the presence of the SIN kinase Cdc7 at the SPB.
Figure 4.
MOR inhibition becomes essential when cytokinesis is perturbed. (A) sid2-250 cells expressing the vector alone or the Nak1–Orb6 fusion transgene were grown in medium lacking thiamine for 14 h at 25°C to induce expression of the fusion protein before spotting 10-fold serial dilutions on minimal media plates lacking thiamine at the indicated temperatures. (B) Wild-type cells carrying the indicated plasmids were grown as in A and then 10-fold serial dilutions were spotted on minimal media plates with or without 6 µM Lat B and tested for growth at 25°C. (C) Wild-type cells carrying integrated copies of Cdc7-mcherry were transformed with the indicated plasmids, and grown as in A for 19 h before being treated with a low dose (4 µM) of Lat B. Samples were collected every 2 h for 8 h after drug addition (0 h). Cells were fixed, stained with DAPI, and scored for number of nuclei (n, top panel) and Cdc7 localization at the SPB (bottom panel). At least 100 cells were analyzed for nuclei count and at least 50 for scoring Cdc7-mcherry localization. Error bars denote SD for three separate experiments. (D) Representative images of the Cdc7-mcherry cells expressing vector alone or the Nak1–Orb6 fusion at the 7-h time point are shown. (E) cdc3-124 sid2-13Myc cells expressing either the empty vector control or the fusion protein were grown in media lacking thiamine for 19 h at 25°C and then shifted to 36°C. Samples were collected for DAPI stain and nuclei count every hour for 8 h after shift to 36°C. Error bars denote SD for three separate experiments. (F) Cells with the indicated genotypes were grown as in E and shifted to 36°C for 3 h. Cell extracts were prepared and Sid2-13Myc was immunoprecipitated with anti-Myc (Santa Cruz Biotechnology, Inc.) antibody. The immunoprecipitates were split, with one portion used for Western blotting using Myc antibodies (Santa Cruz Biotechnology, Inc.) (IP) and the other portion used for in vitro kinase assays using myelin basic protein (MBP; Sigma-Aldrich) as an artificial substrate as described previously (Sparks et al., 1999) and in Materials and methods. The kinase activity (KA) was measured using a PhosphorImager (MDS Analytical Technologies), quantified using ImageQuant software, and normalized to the amount of Sid2 (IP). The activity relative to cells with the control plasmid is shown. Error bars denote SD of the relative KA. The difference in Sid2 kinase activity between cells expressing the vector and those expressing the fusion protein was not statistically significant based on t test analysis from three different experiments. *, P = 0.1047.
We also used another approach to test whether the Nak1–Orb6 fusion was affecting SIN signaling. We completely blocked cytokinesis using the cdc3-124 profilin mutant, which cannot form actomyosin rings. After shift to restrictive temperature, these cells undergo a prolonged SIN-dependent arrest as binucleates but eventually leak past the nuclear division arrest and become multinucleate (Trautmann et al., 2001). Expression of the Nak1–Orb6 fusion in these cells did not significantly interfere with the ability of these cells to arrest as binucleates, suggesting that it was not affecting SIN signaling (Fig. 4 E), which is required to maintain the nuclear division arrest. Furthermore, examination of the kinase activity of the most downstream component of the SIN pathway, Sid2, did not show a significant change when the Nak1–Orb6 fusion was expressed (Fig. 4 F). Along similar lines, we did not observe any evidence for persistent SIN signaling when the MOR was inhibited in orb6-25 mutants (Fig. S4). Together, these results suggest that the MOR pathway may interfere with actomyosin ring assembly and constriction downstream of the SIN, and that MOR inhibition is essential when the cell division machinery is compromised and cytokinesis is delayed.
Reduction in MOR activity allows weak SIN signaling to promote cytokinesis
If the MOR pathway interferes with the ability of the SIN to promote cytokinesis, then reduction in MOR pathway activity might be predicted to enhance cytokinesis in SIN mutants. Therefore, we tested the phenotype of double mutants between the MOR mutant orb6-25 and various SIN mutants. Interestingly, the orb6-25 temperature-sensitive mutant was able to rescue the growth defect of some temperature-sensitive SIN mutants at 29°C and 33°C, which are semi-permissive temperatures for orb6-25 (Fig. 5 A). Although the orb6-25 mutant dies at 36°C and was thus not able to rescue SIN mutants at this temperature, examination of double mutants between orb6-25 and various SIN mutants at 36°C showed that the orb6-25 mutation was able to partially rescue the septum formation defect in all SIN mutants tested (Fig. 5 B). We also examined this phenotype using sid2-250, orb6-25, and sid2-250 orb6-25 cells that had been synchronized in G2 and then shifted to restrictive temperature (Fig. S5 A). Consistent with results from asynchronous cells, the sid2-250 orb6-25 cells showed a substantial increase in septation compared with the sid2-250 single-mutant cells, which totally failed in septation. Although MOR inactivation allowed septation in the sid2-250 orb6-25 cells, absence of a completely functional SIN pathway could explain the reduced septation index in these cells relative to orb6-25 alone. Inactivation of orb6 was unable to promote septum formation in SIN double-mutant cells with mutations in two different SIN components (sid2-250 sid1-239 and sid2-250 cdc11-123), which presumably completely ablates SIN signaling (Fig. 5 B), suggesting that inactivation of orb6 does not bypass the requirement of the SIN in septum formation, but instead allows residual weak SIN signaling to promote cytokinesis.
Figure 5.
The MOR pathway mutant orb6-25 rescues growth and cell division defects of SIN mutants. (A) Cells of the indicated genotypes were grown at 25°C and their growth was tested at the indicated temperatures by spotting 10-fold serial dilutions on YE agar plates. (B) Cells of the indicated genotypes were shifted to 36°C for 4 h, methanol fixed, and stained with Calcofluor White (CW) to score for septum formation. Representative cdc7-24 (montage) and cdc7-24 orb6-25 cells show the septation status in respective cells and arrows mark the septum. Dashed dividing lines separate the individual images in the montage. Arrows mark the septum in the cdc7-24 orb6-25 cells. The septation index (percentage of cells with septa) for all the genotypes indicated is shown in the histogram plot. At least 100 cells were scored for each strain. Error bars denote the SD for three separate experiments. (C) Time-lapse images of the indicated strains were acquired using a spinning disc confocal microscope (TE 2000-E2; Nikon). Cells were grown at 25°C and then placed on the microscope stage that was maintained at 36°C during the entire time of imaging. Arrowheads indicate completion of ring constriction in representative cells. (D) Cells of the indicated genotypes were grown as in B and the proportion of cells with ectopic septa (either septa present in mononucleate cells, or multiple septa) was scored. At least 100 cells were scored for each genotype. The experiment was done in triplicate and error bars denote the SD values. Arrows in the montage image point to the ectopic septa found in representative cells. Dashed dividing lines separate the individual images in the montage.
To examine how loss of MOR activity rescued SIN mutants in more detail, we observed the dynamics of actomyosin ring assembly and septum formation in orb6-25 sid2-250 double-mutant cells. Previous studies have shown that SIN mutants form actomyosin rings that fall apart in anaphase and the cells do not form septa (Krapp and Simanis, 2008). Therefore, we examined actomyosin ring constriction and septum formation in the sid2-250 orb6-25 double-mutant cells expressing the GFP-tagged actomyosin ring component Rlc1 (Le Goff et al., 2000; Naqvi et al., 2000) at the restrictive temperature of 36°C using time-lapse microscopy. As expected, in wild-type cells, actomyosin rings formed then constricted (15/15 cells; Fig. 5 C). In contrast, actomyosin rings formed in sid2-250 single-mutant cells, but failed to constrict and then disassembled in 16 out of 21 cells observed (Fig. 5 C). Unlike sid2-250 mutant cells but similar to wild-type cells, actomyosin rings formed and constricted in sid2-250 orb6-25 double mutants (21/21 cells; Fig. 5 C). Thus, loss of Orb6 activity allows SIN mutants to maintain actomyosin ring stability and complete cytokinesis. Thus, loss of MOR activity allows weak SIN signaling to promote actomyosin ring constriction and septum formation.
Interestingly, MOR inactivation also enhanced the ability of weak SIN signaling to promote ectopic septation. The Spg1-GFP allele has a weakly activated SIN phenotype, which causes occasional formation of interphase septa, or additional rounds of septum formation after normal cytokinesis (García-Cortés and McCollum, 2009). When spg1-GFP was combined with orb6-25, or any other MOR mutant, the resulting double-mutant cells showed an increased rate of ectopic septum formation (Fig. 5 D). Together, these results show that reduction in MOR pathway activity enhances the ability of weak SIN signaling to promote cytokinesis.
Discussion
The SIN antagonizes interphase polarity through inhibition of the MOR pathway
The SIN is required for actomyosin ring assembly and septum synthesis (Balasubramanian et al., 1998; Simanis, 2003; Hachet and Simanis, 2008; Huang et al., 2008). Previous studies have shown that when perturbation of the actomyosin ring causes a delay in cytokinesis, SIN activity is maintained during the delay and is required for cells to continue to promote actomyosin ring assembly, block polarized growth, and arrest cells with two G2 phase nuclei in order to allow cells to complete cell division and maintain normal ploidy (Liu et al., 2000; Trautmann et al., 2001; Mishra et al., 2004). However, it was unclear how the effects of the SIN on cell polarity and nuclear division are mediated. Although part of the SIN inhibition of interphase polarity might be through competition for shared components, we demonstrated that SIN signaling clearly interferes with interphase polarity through inhibition of the MOR pathway. Interestingly, the ability of the SIN to inhibit G2/M transition might also be a consequence of inhibition of the MOR pathway because MOR pathway mutants, like cells with activated SIN, display a Wee1-dependent block in G2/M progression (Hirata et al., 2002; Kanai et al., 2005). Consistent with this idea, we found that blocking complete inhibition of the MOR by the SIN using the Nak1–Orb6 fusion could partially bypass the SIN-dependent block in nuclear division. The failure of the Nak1–Orb6 fusion to completely bypass the SIN inhibition of polarized growth and nuclear division could be because the SIN is still capable of partially inhibiting the MOR when the Nak1–Orb6 fusion is expressed.
Cross talk between the SIN and MOR pathways
The molecular details governing regulation of MOR/Orb6 activity by the SIN are likely to be complex. Orb6 kinase activity normally peaks at about the time when cells initiate bipolar growth in early G2 phase, and then decreases during mitosis (Kanai et al., 2005). We suspect that the decrease of Orb6 activity during mitosis may be biphasic. The initial reduction in Orb6 kinase activity at mitotic entry appears to be independent of the SIN because the SIN is not active in early mitosis (Guertin et al., 2000), and because Orb6 activity still dropped at mitotic entry in the SIN mutant cdc7-24 (Kanai et al., 2005; this paper). The initial drop in Orb6 activity may be triggered directly or indirectly by Cdk1 activation and mitotic entry, and then when Cdk1 activity drops in anaphase, the SIN becomes active and maintains inhibition of the MOR pathway until cytokinesis is complete. Intriguingly, some of the upstream SIN components are required for the increase in Orb6 activity that occurs in the following G2 phase when cells initiate growth at the new end (Kanai et al., 2005). The mechanistic details of how the SIN both activates and inhibits the MOR are as yet unclear. One explanation for these results could be that the SIN causes both inhibitory and activating modifications in the MOR pathway, with the inhibitory modifications being dominant. Removal of the inhibitory modifications after cytokinesis would then allow the MOR to become active in interphase. To address this issue, future studies will need to focus on identification of SIN-dependent changes in the levels or post-translational modifications of MOR components.
However, even without knowing the specific details of how the SIN affects the MOR pathway, our present data indicate that the SIN blocks signaling through the MOR pathway by inhibiting activation of the most downstream component of the pathway, the NDR family kinase Orb6. Our results showed that the SIN does not affect the kinase activity of the Orb6-activating kinase Nak1, suggesting that the SIN interferes with the ability of Nak1 to activate Orb6. Previous reports have suggested that the protein Mor2, which is required for Orb6 kinase activity, functions as a scaffold to bring together Orb6 with its activator, the GC family kinase Nak1 (Kanai et al., 2005). Consistent with this idea, we found that fusion of Nak1 to Orb6 could rescue the mor2-786 mutant. Interestingly, the SIN inhibition of polarized growth was also bypassed by expression of the Nak1–Orb6 fusion construct, suggesting that the SIN could inhibit Mor2 function, or the ability of Nak1 or Orb6 to associate with Mor2. A more detailed understanding of the molecular mechanism by which Orb6 is activated by Nak1 will be essential to uncover how the SIN interferes with the activation of Orb6 and the MOR pathway.
The MOR inhibits SIN-mediated actomyosin ring constriction and septum formation
So what is the purpose of the antagonistic relationship between the SIN and MOR pathways? Because each signaling pathway directs the actin cytoskeleton toward distinct processes, the mutual antagonism between them would keep each pathway from interfering with the functions of the other. The antagonism between the two pathways could be due to each pathway inhibiting signaling through the other, or by interfering with downstream functions such as actin organization. From our experiments, the SIN clearly inhibits signaling through the MOR pathway, although additional interference could come from competition over actin cytoskeletal components. In contrast, we found no evidence that the MOR pathway inhibited SIN signaling, but instead, the MOR appears to interfere with cytokinesis downstream of the SIN, most likely through competition for shared components. The antagonism between the two pathways may both enhance the efficiency of each pathway by removing a competitor, and ensure that cytoskeletal rearrangements occur at the correct point in the cell cycle. For example, the ability of the MOR to interfere with SIN functions might be important to keep weak or leaky SIN signaling from triggering multiple rounds of cytokinesis. Consistent with this, we observe that inactivation of Orb6 allows weak interphase SIN signaling caused by the spg1-GFP allele to trigger ectopic septum formation. Overall, our study identifies an antagonistic interaction between the two NDR pathways, the SIN and MOR, which is crucial for the ability to maintain the quite different cytoskeletal arrangements present during cytokinesis and interphase (Fig. 6).
Figure 6.
Model. Antagonistic interaction between the SIN and MOR is essential to inhibit bipolar growth until the completion of cytokinesis and to prevent ectopic cytokinesis in interphase. Actin and associated cytoskeletal elements at the cell ends and cell middle are shown in red.
Animal cells also undergo redistribution of cytoskeletal elements as they transition between interphase and mitosis. Interestingly, animals also have two pathways containing NDR kinases analogous to the SIN and MOR pathways called the Hippo/Lats and Ndr1/2 pathways, respectively. Although the functions of these pathways in animal cells appear complex, they have been reported to function in mitotic exit/cytokinesis and polarized growth/morphogenesis as in fission yeast (Preisinger et al., 2004; Bothos et al., 2005; Chan et al., 2005; Hergovich et al., 2005, 2008; Guo et al., 2007; ten Klooster et al., 2009). Additionally, evidence for cross talk between the homologous NDR pathways has been observed both in Drosophila melanogaster (Emoto et al., 2006) and in mammalian cells (Vichalkovski et al., 2008). Therefore, it is tempting to speculate that the cytoskeletal rearrangements that take place between anaphase/telophase and interphase might be regulated similarly in animal cells. Besides their role in polarized growth and mitotic exit, the animal homologues of the SIN and MOR pathway components are also involved in processes regulating cell proliferation and cell death. The homologues of the SIN kinase Sid2 in mammals, called Lats1/2, function as tumor suppressors (Takahashi et al., 2005; Hergovich et al., 2006; Seidel et al., 2007; Hao et al., 2008; Zhang et al., 2008). The role of Ndr1/2 in human diseases such as cancer is just beginning to be revealed (Hergovich et al., 2006, 2008). Thus, a detailed understanding of cross talk between NDR pathways will likely have important implications for our understanding of how cells regulate both growth and proliferation, as well as the cytoskeletal rearrangements that occur during the transitions between mitosis and interphase.
Materials and methods
Yeast strain culture and flow cytometry conditions
Fission yeast media, growth conditions, and manipulations were performed as described previously (Moreno et al., 1991). Except where noted, cells were grown in yeast extract (YE) medium. Flow cytometry analysis (FACS) was performed on isolated nuclei as described previously (Forsburg and Rhind, 2006) using a FACScan flow cytometer (BD).
Microscopy
GFP and fusion proteins were observed in cells that were grown in YE (unless otherwise mentioned) after fixation with −20°C methanol or in live cells for time-lapse studies. DNA and septum material were visualized by staining with DAPI (Sigma-Aldrich) and Calcofluor White (CW; Sigma-Aldrich), respectively. Cell staining with DAPI and Alexa Fluor 488–conjugated phalloidin was performed as described previously (Balasubramanian et al., 1997). Images were acquired at room temperature using a microscope (Eclipse E600; Nikon) equipped with a cooled charge-coupled device camera (Orca-ER; Hamamatsu Photonics) and IPlab Spectrum software (Scanalytics). Z-series of images were captured with a 100x oil (NA 1.3) objective lens and 3D stacked view was obtained using IP Laboratory software. For time-lapse studies, exponentially growing cells were concentrated and 1.8 µl of cell suspension (in YE) was placed on a microscope slide between a 2% YE-agar pad and a coverslip, which was sealed using VALAP. The cells were maintained at 36°C in a 20/20 Technologies micro-incubator, and images were acquired on an inverted microscope (TE 2000-E2; Nikon) equipped with a spinning disk confocal system (CSU10B; Solamere Technology Group) controlled by MetaMorph software. Time-lapse Z-series of images was captured with a 60x Plan Apo oil objective (NA 1.4; Nikon) using a camera (MGi EMCCD; Rolera) and processed using MetaMorph software to get the maximum projection of the z-stack images.
Plasmid construction
The fusion construct between the Nak1 and Orb6 genes was expressed in the pREP41-GFP vector system (Craven et al., 1998). The fusion products were sequenced to ensure there were no frame shift mutations. The kinase-dead allele of Nak1 was made by mutating the conserved lysine (K39) site in the proposed ATP binding region of the Nak1 kinase to arginine (R) using the Stratagene QuikChange Site-Directed Mutagenesis kit. The resulting constructs were sequenced to confirm the changes.
Immunoprecipitation and immunodetection
Preparation of cell extracts, immunoprecipitation, immunodetection, and kinase assays were performed as described previously (Sparks et al., 1999; Huang et al., 2003; Kanai et al., 2005). In brief, exponentially growing 2 x 108 cells were collected by centrifugation and cell pellets were frozen in liquid nitrogen. All subsequent steps were performed on ice or at 4°C. Cells were thawed in ice after addition of NP-40 Buffer (1% NP-40, 150 mM NaCl, 2 mM EDTA, 6 mM Na2HPO4, and 4 mM NaH2 PO4) supplemented with protease inhibitor cocktail for fungal and yeast extract (Sigma-Aldrich). After centrifugation for 3 min at 3,000 rpm (Beckman Coulter), 1.5 ml of glass beads (425–600 µm G8772 Sigma) were added to the cells and vortexed vigorously for 1 min. Another 1 ml of NP-40 buffer was added and the lysates were cleared by centrifugation at 8,000 rpm for 10 min in a microfuge and the supernatant was collected. The cell lysates were subject to a preclearing step by incubation with 20 µl of protein G–Agarose or 15 µl Dyna1 beads (as appropriate, see below) by rocking at 4°C for 45 min followed by centrifugation to collect precleared lysates. Relative protein amounts in the precleared lysates were estimated using the BCA Protein Assay kit (Thermo Fisher Scientific).
For immunoprecipitations of Myc-tagged Sid2, 0.2 µg anti-Myc monoclonal IgG (Santa Cruz Biotechnology, Inc.) was added to the NP-40 precleared lysates (described above) and incubated on a rocker for 1 h at 4°C. Immune complexes were prepared by adding 30 µl of a 1:1 slurry of protein G–Agarose beads (Sigma-Aldrich) and incubating for 1 h, followed by centrifugation in a microfuge for 1 min. Beads were washed three times with 1 ml NP-40 buffer. For immunoprecipitation of GFP-tagged Nak1 and Myc-tagged Mob2, precleared cell lysates in NP-40 buffer (as above) were added to anti-GFP monoclonal IgG (Invitrogen) or anti-Myc monoclonal IgG-bound magnetic protein G Dynabeads (DYNAL; Invitrogen) and incubated at 4°C on a roller for at least 3 h, after which beads were washed three times in NP-40 buffer as before.
For detection of the epitope-tagged proteins, cell lysates and immunoprecipitates were separated by SDS-PAGE (7%) and transferred to Immobilon-P nylon (Millipore) using a wet transfer apparatus (Bio-Rad Laboratories). Blots were probed with anti-Myc IgG (Santa Cruz Biotechnology, Inc.) or anti-GFP IgG (Santa Cruz Biotechnology, Inc.) at a 1:500 dilution, and developed using the HRP chemiluminescent detection system (Thermo Fisher Scientific).
In vitro kinase assays
Kinase assays were performed as described previously (Sparks et al., 1999; Huang et al., 2003; Leonhard and Nurse, 2005). In brief, immune complex bead preparations were washed twice in 1 ml kinase assay buffer (10 mM Tris-HCl, pH 7.4, 10 mM MgCl2, 0.1 mM sodium vanadate, and 1 mM DTT for Sid2 kinase assays; and 50 mM Tris-HCl, pH 7.4, 100 mM NaCl, 10 mM MgCl2, 1 mM MnCl2, and 1 mM DTT for Nak1 kinase assays) supplemented with a yeast protease inhibitor cocktail (Sigma-Aldrich). Washed immunoprecipitates were incubated at 30°C for 30 min in 20 µl respective kinase buffer containing 10 µg myelin basic protein (Sigma-Aldrich) for Sid2 kinase assays, or 5 µg α-casein (Sigma-Aldrich) for Nak1 kinase assays; 5 µCi of γ-[32P]ATP (PerkinElmer) and 50 µM unlabeled ATP per reaction. Reactions were stopped with the addition of 20 µl of 2x SDS sample buffer and subjected to SDS-PAGE. The gels were dried and quantified using a PhosphorImager.
Online supplemental material
Fig. S1 shows FACS plot and actin staining quantification data to show that SIN activation causes G2/M arrest and disrupts interphase-polarized actin distribution. Fig. S2 shows that immunoprecipitated untagged Mob2 does not have any kinase activity on MBP. Fig. S3 is the same as Fig. 3 C, with error bars that had been left out for clarity. Fig. S4 shows that the MOR pathway mutant orb6-25 does not cause persistent SIN signaling. Fig. S5 A shows septation counts after cell synchronization in G2 to support our conclusion in Fig. 5 B. Fig. S5, B and C, test the effects of Nak1–Orb6 fusion on septation and cell length in cdc16-116 and wild-type cells, respectively. Table S1 lists all the strains used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201002055/DC1.
Acknowledgments
We are grateful to Kathy Gould for providing strains and Takashi Toda for valuable discussions. We thank the McCollum laboratory members for discussions. We are thankful to Nick Rhind and his laboratory members for helping us with the FACS analysis and to Paul Furcinitti at the Spinning Disc Confocal Microscope Facility for help with the time-lapse movies. We would also like to thank Joshua Nordberg and Kip Sluder for help with the Slidebook Software for our actin staining quantifications.
This work was supported by National Institutes of Health grant to D. McCollum (GM058406-12), and by grants from the Ministry of Education, Science and Culture of Japan to D. Hirata (20380063) and K. Kume (21780097).
Footnotes
Abbreviation used in this paper:
- Lat B
- latrunculin B
References
- Balasubramanian M.K., Hirani B.R., Burke J.D., Gould K.L. 1994. The Schizosaccharomyces pombe cdc3+ gene encodes a profilin essential for cytokinesis. J. Cell Biol. 125:1289–1301 10.1083/jcb.125.6.1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian M.K., McCollum D., Gould K.L. 1997. Cytokinesis in fission yeast Schizosaccharomyces pombe. Methods Enzymol. 283:494–506 10.1016/S0076-6879(97)83039-X [DOI] [PubMed] [Google Scholar]
- Balasubramanian M.K., McCollum D., Chang L., Wong K.C., Naqvi N.I., He X., Sazer S., Gould K.L. 1998. Isolation and characterization of new fission yeast cytokinesis mutants. Genetics. 149:1265–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothos J., Tuttle R.L., Ottey M., Luca F.C., Halazonetis T.D. 2005. Human LATS1 is a mitotic exit network kinase. Cancer Res. 65:6568–6575 10.1158/0008-5472.CAN-05-0862 [DOI] [PubMed] [Google Scholar]
- Chan E.H., Nousiainen M., Chalamalasetty R.B., Schäfer A., Nigg E.A., Silljé H.H. 2005. The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene. 24:2076–2086 10.1038/sj.onc.1208445 [DOI] [PubMed] [Google Scholar]
- Cong J., Geng W., He B., Liu J., Charlton J., Adler P.N. 2001. The furry gene of Drosophila is important for maintaining the integrity of cellular extensions during morphogenesis. Development. 128:2793–2802 [DOI] [PubMed] [Google Scholar]
- Craven R.A., Griffiths D.J., Sheldrick K.S., Randall R.E., Hagan I.M., Carr A.M. 1998. Vectors for the expression of tagged proteins in Schizosaccharomyces pombe. Gene. 221:59–68 10.1016/S0378-1119(98)00434-X [DOI] [PubMed] [Google Scholar]
- Das M., Wiley D.J., Chen X., Shah K., Verde F. 2009. The conserved NDR kinase Orb6 controls polarized cell growth by spatial regulation of the small GTPase Cdc42. Curr. Biol. 19:1314–1319 10.1016/j.cub.2009.06.057 [DOI] [PubMed] [Google Scholar]
- Emoto K., Parrish J.Z., Jan L.Y., Jan Y.N. 2006. The tumour suppressor Hippo acts with the NDR kinases in dendritic tiling and maintenance. Nature. 443:210–213 10.1038/nature05090 [DOI] [PubMed] [Google Scholar]
- Fankhauser C., Marks J., Reymond A., Simanis V. 1993. The S. pombe cdc16 gene is required both for maintenance of p34cdc2 kinase activity and regulation of septum formation: a link between mitosis and cytokinesis? EMBO J. 12:2697–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C., Reymond A., Cerutti L., Utzig S., Hofmann K., Simanis V. 1995. The S. pombe cdc15 gene is a key element in the reorganization of F-actin at mitosis. Cell. 82:435–444 10.1016/0092-8674(95)90432-8 [DOI] [PubMed] [Google Scholar]
- Forsburg S.L., Rhind N. 2006. Basic methods for fission yeast. Yeast. 23:173–183 10.1002/yea.1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Cortés J.C., McCollum D. 2009. Proper timing of cytokinesis is regulated by Schizosaccharomyces pombe Etd1. J. Cell Biol. 186:739–753 10.1083/jcb.200902116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin D.A., Chang L., Irshad F., Gould K.L., McCollum D. 2000. The role of the sid1p kinase and cdc14p in regulating the onset of cytokinesis in fission yeast. EMBO J. 19:1803–1815 10.1093/emboj/19.8.1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C., Tommasi S., Liu L., Yee J.K., Dammann R., Pfeifer G.P. 2007. RASSF1A is part of a complex similar to the Drosophila Hippo/Salvador/Lats tumor-suppressor network. Curr. Biol. 17:700–705 10.1016/j.cub.2007.02.055 [DOI] [PubMed] [Google Scholar]
- Hachet O., Simanis V. 2008. Mid1p/anillin and the septation initiation network orchestrate contractile ring assembly for cytokinesis. Genes Dev. 22:3205–3216 10.1101/gad.1697208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y., Chun A., Cheung K., Rashidi B., Yang X. 2008. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J. Biol. Chem. 283:5496–5509 10.1074/jbc.M709037200 [DOI] [PubMed] [Google Scholar]
- Hergovich A., Bichsel S.J., Hemmings B.A. 2005. Human NDR kinases are rapidly activated by MOB proteins through recruitment to the plasma membrane and phosphorylation. Mol. Cell. Biol. 25:8259–8272 10.1128/MCB.25.18.8259-8272.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergovich A., Stegert M.R., Schmitz D., Hemmings B.A. 2006. NDR kinases regulate essential cell processes from yeast to humans. Nat. Rev. Mol. Cell Biol. 7:253–264 10.1038/nrm1891 [DOI] [PubMed] [Google Scholar]
- Hergovich A., Cornils H., Hemmings B.A. 2008. Mammalian NDR protein kinases: from regulation to a role in centrosome duplication. Biochim. Biophys. Acta. 1784:3–15 [DOI] [PubMed] [Google Scholar]
- Hirata D., Kishimoto N., Suda M., Sogabe Y., Nakagawa S., Yoshida Y., Sakai K., Mizunuma M., Miyakawa T., Ishiguro J., Toda T. 2002. Fission yeast Mor2/Cps12, a protein similar to Drosophila Furry, is essential for cell morphogenesis and its mutation induces Wee1-dependent G(2) delay. EMBO J. 21:4863–4874 10.1093/emboj/cdf495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T.Y., Markley N.A., Young D. 2003. Nak1, an essential germinal center (GC) kinase regulates cell morphology and growth in Schizosaccharomyces pombe. J. Biol. Chem. 278:991–997 10.1074/jbc.M208993200 [DOI] [PubMed] [Google Scholar]
- Huang Y., Yan H., Balasubramanian M.K. 2008. Assembly of normal actomyosin rings in the absence of Mid1p and cortical nodes in fission yeast. J. Cell Biol. 183:979–988 10.1083/jcb.200806151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T., Shimamura S., Kurosaka A., Yamagishi H., Terachi T. 2004. Cloning and characterization of a novel radish protein kinase which is homologous to fungal cot-I like and animal Ndr protein kinases. Genes Genet. Syst. 79:283–291 10.1266/ggs.79.283 [DOI] [PubMed] [Google Scholar]
- Kanai M., Kume K., Miyahara K., Sakai K., Nakamura K., Leonhard K., Wiley D.J., Verde F., Toda T., Hirata D. 2005. Fission yeast MO25 protein is localized at SPB and septum and is essential for cell morphogenesis. EMBO J. 24:3012–3025 10.1038/sj.emboj.7600782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp A., Simanis V. 2008. An overview of the fission yeast septation initiation network (SIN). Biochem. Soc. Trans. 36:411–415 10.1042/BST0360411 [DOI] [PubMed] [Google Scholar]
- Kume K., Goshima T., Miyahara K., Toda T., Hirata D. 2007. A method for Pmo25-associated kinase assay in fission yeast: the activity is dependent on two gC kinases Nak1 and Sid1. Biosci. Biotechnol. Biochem. 71:615–617 10.1271/bbb.60574 [DOI] [PubMed] [Google Scholar]
- Le Goff X., Motegi F., Salimova E., Mabuchi I., Simanis V. 2000. The S. pombe rlc1 gene encodes a putative myosin regulatory light chain that binds the type II myosins myo3p and myo2p. J. Cell Sci. 113:4157–4163 [DOI] [PubMed] [Google Scholar]
- Leonhard K., Nurse P. 2005. Ste20/GCK kinase Nak1/Orb3 polarizes the actin cytoskeleton in fission yeast during the cell cycle. J. Cell Sci. 118:1033–1044 10.1242/jcs.01690 [DOI] [PubMed] [Google Scholar]
- Liu J., Wang H., Balasubramanian M.K. 2000. A checkpoint that monitors cytokinesis in Schizosaccharomyces pombe. J. Cell Sci. 113:1223–1230 [DOI] [PubMed] [Google Scholar]
- Martin S.G. 2009. Microtubule-dependent cell morphogenesis in the fission yeast. Trends Cell Biol. 19:447–454 10.1016/j.tcb.2009.06.003 [DOI] [PubMed] [Google Scholar]
- Mendoza M., Redemann S., Brunner D. 2005. The fission yeast MO25 protein functions in polar growth and cell separation. Eur. J. Cell Biol. 84:915–926 10.1016/j.ejcb.2005.09.013 [DOI] [PubMed] [Google Scholar]
- Minet M., Nurse P., Thuriaux P., Mitchison J.M. 1979. Uncontrolled septation in a cell division cycle mutant of the fission yeast Schizosaccharomyces pombe. J. Bacteriol. 137:440–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra M., Karagiannis J., Trautmann S., Wang H., McCollum D., Balasubramanian M.K. 2004. The Clp1p/Flp1p phosphatase ensures completion of cytokinesis in response to minor perturbation of the cell division machinery in Schizosaccharomyces pombe. J. Cell Sci. 117:3897–3910 10.1242/jcs.01244 [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795–823 10.1016/0076-6879(91)94059-L [DOI] [PubMed] [Google Scholar]
- Naqvi N.I., Wong K.C., Tang X., Balasubramanian M.K. 2000. Type II myosin regulatory light chain relieves auto-inhibition of myosin-heavy-chain function. Nat. Cell Biol. 2:855–858 10.1038/35041107 [DOI] [PubMed] [Google Scholar]
- Preisinger C., Short B., De Corte V., Bruyneel E., Haas A., Kopajtich R., Gettemans J., Barr F.A. 2004. YSK1 is activated by the Golgi matrix protein GM130 and plays a role in cell migration through its substrate 14-3-3zeta. J. Cell Biol. 164:1009–1020 10.1083/jcb.200310061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts-Galbraith R.H., Gould K.L. 2008. Stepping into the ring: the SIN takes on contractile ring assembly. Genes Dev. 22:3082–3088 10.1101/gad.1748908 [DOI] [PubMed] [Google Scholar]
- Schmidt S., Sohrmann M., Hofmann K., Woollard A., Simanis V. 1997. The Spg1p GTPase is an essential, dosage-dependent inducer of septum formation in Schizosaccharomyces pombe. Genes Dev. 11:1519–1534 10.1101/gad.11.12.1519 [DOI] [PubMed] [Google Scholar]
- Seidel C., Schagdarsurengin U., Blümke K., Würl P., Pfeifer G.P., Hauptmann S., Taubert H., Dammann R. 2007. Frequent hypermethylation of MST1 and MST2 in soft tissue sarcoma. Mol. Carcinog. 46:865–871 10.1002/mc.20317 [DOI] [PubMed] [Google Scholar]
- Simanis V. 2003. Events at the end of mitosis in the budding and fission yeasts. J. Cell Sci. 116:4263–4275 10.1242/jcs.00807 [DOI] [PubMed] [Google Scholar]
- Snell V., Nurse P. 1993. Investigations into the control of cell form and polarity: the use of morphological mutants in fission yeast. Dev. Suppl. 1993:289–299 [PubMed] [Google Scholar]
- Sparks C.A., Morphew M., McCollum D. 1999. Sid2p, a spindle pole body kinase that regulates the onset of cytokinesis. J. Cell Biol. 146:777–790 10.1083/jcb.146.4.777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Miyoshi Y., Takahata C., Irahara N., Taguchi T., Tamaki Y., Noguchi S. 2005. Down-regulation of LATS1 and LATS2 mRNA expression by promoter hypermethylation and its association with biologically aggressive phenotype in human breast cancers. Clin. Cancer Res. 11:1380–1385 10.1158/1078-0432.CCR-04-1773 [DOI] [PubMed] [Google Scholar]
- Tamaskovic R., Bichsel S.J., Hemmings B.A. 2003. NDR family of AGC kinases—essential regulators of the cell cycle and morphogenesis. FEBS Lett. 546:73–80 10.1016/S0014-5793(03)00474-5 [DOI] [PubMed] [Google Scholar]
- ten Klooster J.P., Jansen M., Yuan J., Oorschot V., Begthel H., Di Giacomo V., Colland F., de Koning J., Maurice M.M., Hornbeck P., Clevers H. 2009. Mst4 and Ezrin induce brush borders downstream of the Lkb1/Strad/Mo25 polarization complex. Dev. Cell. 16:551–562 10.1016/j.devcel.2009.01.016 [DOI] [PubMed] [Google Scholar]
- Trautmann S., McCollum D. 2005. Distinct nuclear and cytoplasmic functions of the S. pombe Cdc14-like phosphatase Clp1p/Flp1p and a role for nuclear shuttling in its regulation. Curr. Biol. 15:1384–1389 10.1016/j.cub.2005.06.039 [DOI] [PubMed] [Google Scholar]
- Trautmann S., Wolfe B.A., Jorgensen P., Tyers M., Gould K.L., McCollum D. 2001. Fission yeast Clp1p phosphatase regulates G2/M transition and coordination of cytokinesis with cell cycle progression. Curr. Biol. 11:931–940 10.1016/S0960-9822(01)00268-8 [DOI] [PubMed] [Google Scholar]
- Verde F., Wiley D.J., Nurse P. 1998. Fission yeast orb6, a ser/thr protein kinase related to mammalian rho kinase and myotonic dystrophy kinase, is required for maintenance of cell polarity and coordinates cell morphogenesis with the cell cycle. Proc. Natl. Acad. Sci. USA. 95:7526–7531 10.1073/pnas.95.13.7526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vichalkovski A., Gresko E., Cornils H., Hergovich A., Schmitz D., Hemmings B.A. 2008. NDR kinase is activated by RASSF1A/MST1 in response to Fas receptor stimulation and promotes apoptosis. Curr. Biol. 18:1889–1895 10.1016/j.cub.2008.10.060 [DOI] [PubMed] [Google Scholar]
- Zhang J., Smolen G.A., Haber D.A. 2008. Negative regulation of YAP by LATS1 underscores evolutionary conservation of the Drosophila Hippo pathway. Cancer Res. 68:2789–2794 10.1158/0008-5472.CAN-07-6205 [DOI] [PubMed] [Google Scholar]