Abstract
Human peripheral blood dendritic cells (PBDC) are a rare population comprised of several distinctive subsets. Analysis of these cells has been hindered by their low frequency. In this study, we report a novel direct ex vivo 11-color flow cytometric assay that combines subset identification with analysis of activation status and endocytic ability of three major PBDC subsets (CD1c+CD11c+ “MDC1,” CD141+CD11c+ “MDC2,” and CD303+CD11c− “PDC”) within a single platform. This method eliminates the need for DC enrichment, isolation, or prolonged culture. Human peripheral blood mononuclear cells (PBMC) from healthy donors are incubated with FITC-dextran directly ex vivo, prior to cell surface staining with various markers. As expected, PBDC identified by this assay express low levels of CD40 and CD86 directly ex vivo, and significantly upregulate expression of these molecules upon stimulation with toll-like receptor ligands LPS and CpG oligonucleotides. In addition, PDC internalize FITC-labeled dextran poorly in comparison to MDC1 and MDC2 subsets. Specificity of FITC-dextran endocytosis is further verified by imaging flow cytometry. Furthermore, the combination of surface markers used in this assay reveals a previously unreported CD4+CD11c+CD303−CD1c-CD141− cell population. Taken together, this assay is a rapid and cost-effective method that avoids manipulation of PBDC while providing direct ex vivo high-dimensional flow cytometry data for PBDC studies.
1. Introduction
Dendritic cells (DC) are arguably the most important professional antigen-presenting cells (APC) in the immune system. They play a critical role in adaptive immunity by priming naïve T cells, maintaining tolerance to self-antigens, and bridging the innate and adaptive response (Steinman, 2007). DC undergo a series of phenotypic changes and become functionally mature in response to a variety of stimuli such as microbial products, inflammatory cytokines, and CD40 ligand. Subsequent to stimulation, DC upregulate surface expression of major histocompatibility complex (MHC) molecules as well as co-stimulatory molecules CD40, CD80, and CD86 and decrease endocytic activity for antigens (Reis e Sousa, 2006). Given the paramount role DC play in regulating various aspects of the immune response and the potential impact in areas such as vaccine development, tumor immunity, autoimmune disorders, and organ transplantations, DC biology has attracted great interest from researchers in both basic and clinical science. However, research on DC has been hampered by the scarcity of these cells in vivo, particularly for human DC studies where accessible tissue sites are very limited. Thus, many researchers rely heavily on the in vitro generation of DC from precursor blood cells such as CD14+ monocytes (Romani et al., 1994; Sallusto and Lanzavecchia, 1994) or CD34+ hematopoietic progenitor cells (Caux et al., 1992; Reid et al., 1992) for human studies. Although these in vitro cultures provide a way to obtain sufficient quantities of DC with relative ease, the plasticity of these DC precursors in many cases could present a serious drawback for studying DC biology. Phenotypes of in vitro cultured DC can vary greatly depending on culture conditions. In particular, DC are highly sensitive to the concentration and type of cytokines as well as the specific batch of serum used to supplement the culture medium. Variation in DC phenotypes has caused serious confusion in the field, as numerous studies report contradictory results when using seemingly similar or identical experimental protocols (Schuler-Thurner et al., 2002; Banerjee et al., 2006). Of central importance is that these in vitro-generated DC may not truly reflect the phenotypes and functions of DC in vivo, particularly for clinical studies where pre-existing conditions such as infections, cancers or other immune perturbations are present. Under such circumstances, the analysis of circulating DC in readily accessible human peripheral blood is an attractive alternative to in vitro generated DC. Therefore, a method to detect these rare DC populations (about 1% or less of total PBMC) in the circulation and to simultaneously assess DC function and activation status directly ex vivo will be an invaluable tool for the examination of DC biology in various diseases.
The origin of human blood DC has yet to be fully elucidated. Based on experimental results from mouse models where purified CD11c+ DC were found to migrate into the spleen, lung, and liver within hours after intravenous adoptive transfer (Cavanagh et al., 2005), it is hypothesized that circulating DC may be immature DC emerging from the bone marrow en route to peripheral tissues and/or secondary lymphoid organs (Bonasio and von Andrian, 2006). Alternatively, circulating DC may be tissue-resident DC re-entering circulation through the thoracic duct en route to other organs, since small numbers of DC have been found in the thoracic duct and DC injected subcutaneously can be found 24-48 hours later in murine spleen and bone marrow (Cavanagh et al., 2005).
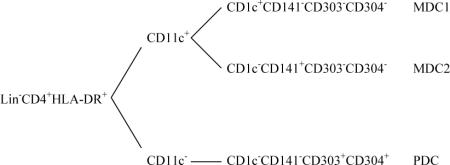
Several DC subset populations have been identified in human blood. In general, human peripheral blood DC (PBDC) are negative for lineage markers CD3, CD14, CD16, CD19, CD34, and CD56 but positive for CD4 and HLA-DR (Dzionek et al., 2000). It is noteworthy that some HLA-DR+CD16+ or HLA-DR+CD34+ cells with DC characteristics have been reported, though whether they represent distinctive DC subsets has yet to be fully elucidated (MacDonald et al., 2002). PBDC can be divided into CD11c+ and CD11c− subsets. The CD11c+ DC population is the conventional “myeloid” DC (MDC) possessing a typical DC morphology with short cell protrusions (dendrites) on the surface. Human blood CD11c+ DC express a broader range of toll-like receptors (TLRs) and higher levels of HLA-DR compared to CD11c− DC (Kadowaki et al., 2001). It is now known that the CD11c+ DC are actually a heterogeneous population composed of at least two subsets: a major CD1c+ subset and a minor CD1c− subset (Dzionek et al., 2000). On the other hand, CD11c− DC referred to as “plasmacytoid DC” appear to be a more homogeneous population expressing high levels of CD123 and exhibiting a plasmacytoid morphology (Grouard et al., 1996; Olweus et al., 1997). Plasmacytoid DC (PDC) are a major source of type I interferons (IFNα/β) when stimulated by TLR ligands such as CpG oligodeoxynucleotide (a TLR9 ligand) (Kadowaki et al., 2001). In addition, it has been found that PDC may also play an important role in tolerance, either directly suppressing T cell proliferation through the production of indoleamine 2,3-dioxegenase (IDO) or indirectly by promoting differentiation of inducible regulatory T cells (iTreg) (Munn et al., 2002; Moseman et al., 2004). Further definition and characterization of PBDC subsets have been facilitated by three recently identified PBDC subset-specific markers CD141 (BDCA-3), CD303 (BDCA-2), and CD304 (BDCA-4). Under steady state (non-inflammatory conditions), CD303 and CD304 are co-expressed on CD11c− plasmacytoid DC in blood, whereas both CD1c+ and CD141+ markers are associated with CD11c+ “myeloid DC” and can be used to further define myeloid DC subsets, CD11c+CD1c+CD141− (MDC1) and CD11c+CD1c−CD141+ (MDC2), respectively. Importantly, in freshly isolated PBMC from healthy donors both MDC1 and MDC2 are devoid of CD303 and CD304 expression, whereas there is no CD1c or CD141 expression on PDC (Dzionek et al., 2000). The patterns of surface marker expression by various PBDC subsets are summarized below.
The primary functions of DC are processing protein antigens into peptide fragments for presentation in the form of peptide:MHC complexes to T cells and secretion of cytokines upon sensing the presence of pathogens (through pattern recognition receptors such as TLRs) as well as other stimuli such as inflammatory mediators. Under steady state conditions, both MDC and PDC exhibit immature phenotypes with low expression of MHC molecules and the costimulatory molecules CD40, CD80, and CD86 (Sallusto and Lanzavecchia, 1994; Dzionek et al., 2000; MacDonald et al., 2002). Once immature DC are exposed to “danger signals” such as microbial products and/or inflammatory mediators, they will express high levels of MHC and costimulatory molecules and differentiate to a mature phenotype. In contrast to mature MDC which downregulate endocytic function, immature MDC demonstrate great capacity to endocytose extracellular antigens (Kohrgruber et al., 1999; Robinson et al., 1999). On the other hand, freshly isolated PDC showed low but positive endocytic activity for Lucifer Yellow and FITC-labeled dextran (Dzionek et al., 2000), whereas PDC cultured in vitro in the presence or absence of CD40 ligand did not endocytose FITC-dextran (Grouard et al., 1997).
The endocytic capability of DC may be evaluated in multiple ways. One frequently used method is to mix fluorescent chromophore-labeled molecules such as FITC-dextran with DC and measure the uptake of the fluorescent molecules by DC using flow cytometry. Here, we report a method to simultaneously identify all three PBDC subsets and assess their phenotypes and function directly from PBMC by combining the multicolor surface marker staining and the FITC-dextran uptake assay. Since this method allows the identification and characterization of rare PBDC subpopulations in one single 11-color flow cytometry-based assay with minimal prior manipulation, it has advantages over other methods in terms of required sample volume and cost. The application of this method will provide a valuable tool for both basic science and clinical research where it is now possible to assess circulating PBDC from small blood volumes.
2. Materials and methods
2.1. Blood Specimen Collection and Preparation
Peripheral venous blood samples were obtained from healthy adult donors with IRB approval following informed consent and collected in CPT tubes (BD, Franklin Lakes, NJ) with heparin. PBMC were isolated following the manufacture's protocol. Briefly, CPT tubes were centrifuged at 1500 × g for 30 minutes at room temperature. Mononuclear cells were harvested and washed twice with 1% BSA (Celliance, Kankakee, IL) / HBSS (Cellgro, Herndon, VA) by centrifugation at 300 × g for 10 minutes at 4°C. After resuspension in 1% BSA/HBSS, PBMC were adjusted to a final concentration of 20 × 106 cells/ml and kept on ice until stained.
In some experiments, cryopreserved normal control PBMC obtained from one subject with IRB approval were used. In brief, vials containing frozen cells were quickly thawed at 37°C for 65-70 seconds, and cell suspension was transferred to a 15 ml polypropylene at room temperature and diluted by slowly adding 1% BSA / HBSS at room temperature while gently swirling the tube to mix cells thoroughly, then washed twice with 1% BSA / HBSS by centrifugation at 300 × g for 10 minutes at 20°C.
2.2. Antibodies used for flow cytometric analysis
The following monoclonal antibodies were used in this study: CD1c-PE (clone AD5-8F7, Miltenyi Biotec, Auburn, CA), CD3-PE-Cy5.5 (clone S4.1, Caltag/Invitrogen, Carlsbad, CA), CD4-APC-Alexa-fluor 750 (clone RPA-T4, eBioscience, San Diego, CA), CD11c-PE-Cy7 (clone 3.9, Biolegend, San Diego, CA), CD14-Alexa-fluor 700 (clone M5E2, BD Biosciences, San Jose, CA), CD16-PerCp-Cy5.5 (clone 3G8, BD Biosciences, San Jose, CA), CD19-PerCp-Cy5.5 (clone SJ25C1, BD Biosciences, San Jose, CA), CD34-PerCp-Cy5.5 (clone 8G12, BD Biosciences, San Jose, CA), CD40-PE-Cy5 (clone 5C3, BD Biosciences, San Jose, CA), CD86-Pacific Blue (clone IT2.2, Biolegend, San Diego, CA), CD141-biotin (clone AD5-14H12, Miltenyi Biotec, Auburn, CA), CD303-APC (clone AC144, Miltenyi Biotec, Auburn, CA), HLA-DR-PE-Texas Red (clone TU36, Caltag/Invitrogen, Carlsbad, CA). Streptavadin-Pacific Orange (Molecular Probes/Invitrogen, Carlsbad, CA) was used as secondary staining reagent for CD141-biotin. 7-Amino-Actinomycin D (7-AAD) (BD Biosciences, San Jose, CA) was included in the antibody cocktails as a vital dye to exclude dead cells. The following isotype controls were also used: mIgG2a-PE (clone eBM2a, eBioscience, San Diego, CA), mIgG2b-PE-Texas Red (Caltag/Invitrogen, Carlsbad, CA), mIgG1-PE-Cy5 (clone P3, eBioscience, San Diego, CA), mIgG1-PE-Cy7 (clone MOPC-21, Biolegend, San Diego, CA), mIgG2b-Pacific Blue (clone MPC-11, Biolegend, San Diego, CA), mIgG1-Biotin (Caltag/Invitrogen, Carlsbad, CA), mIgG1-APC (clone P3, eBioscience, San Diego, CA), mIgG2a-Alexa-fluor 700 (clone eBM2a, eBioscience, San Diego, CA), and mIgG1-APC-Alexa-fluor 750 (clone P3, eBioscience, San Diego, CA). The antibody cocktails were made by mixing antigen-specific antibodies at optimal concentrations determined by prior titration. Isotype control antibodies were individually substituted for the specific antibody-fluorochrome in the cocktail at a similar antibody concentration. The configuration of lasers and optical filters and the compatible PMT channels for fluorochromes used in the instrument for this study is based on the “6-Blue 2 Violet 0-UV 3-Red Configuration” of the “Common Special Order Configurations” in the BD LSR II User's Guide, with the following minor modifications: we used a 735 LP longpass dichroic mirror for PE-Cy7, a 665 LP longpass dichroic mirror and a 670/14 bandpass filter for PE-Cy5, the Violet PMT A for Pacific Orange, a 440/40 bandpass filter for Pacific Blue, a 735 LP longpass dichroic mirror for APC-Alexa fluor 750, a 685 LP longpass dichroic mirror and a 720/40 longpass filter for Alexa fluor 700. The BD LSR II User's Guide can be obtained from the BD website.
2.3. FITC-dextran uptake assay
The FITC-dextran uptake assay was setup by incubating cells with FITC-dextran in duplicate plates at 4°C and 37°C, respectively. Briefly, 50 μl of PBMC (1 × 106 cells) in 1% BSA/HBSS were added to triplicate wells on each of the two 96-well V-bottom plates (BD, Franklin Lakes, NJ) before adding 4 μl of FITC-dextran (molecular weight = 40,000; Molecular Probes/Invitrogen, Carlsbad, CA) at 12.5 mg/ml for a final concentration of FITC-dextran of 1 mg/ml. The FITC-dextran solution was vortexed for 30 seconds and sonicated for an additional 30 seconds immediately before use. One plate was incubated at 37°C and the second was incubated at 4°C (to determine baseline FITCdextran uptake level) for 30 minutes. Both plates were gently tapped every 5 to 10 minutes to ensure adequate mixing. Following FITC-dextran incubation, 200 μl of 1% BSA/HBSS was added into each well and the plates were spun at 400 × g at 4°C for 6 minutes, decanted supernatant, washed one more time with 250 μl of 1% BSA/HBSS, and followed by cell surface marker staining (see below).
2.4. Cell surface marker staining for flow cytometry analysis
The staining for cell surface markers in this study was done by a “micromethod” that significantly reduces the amount of antibodies and staining volume compared to the standard staining protocol that usually stains one million cells in 100 μl. Briefly, 1 × 106 PBMC per well in the v-bottom plates (BD, Franklin Lakes, NJ) were resuspended with 50 μl of 5% normal mouse serum (Sigma-Aldrich, St. Louis, MO) diluted in 1% BSA/HBSS and incubated at room temperature for 10 minutes to pre-block non-specific staining. After pre-blocking, cells were centrifuged at 400 × g for 6 minutes at 4°C, supernatant was decanted, plate was gently blotted on paper towels, and cell pellets were resuspended with 15 μl of antibody-fluorochrome cocktails containing 20% of Fc receptor blocker (Miltenyi Biotec, Auburn, CA) by thorough but gentle pipetting. Cells and cocktail were incubated on ice for 20 minutes in the dark. Afterwards, cells were washed twice with 200 μl of 1% BSA/HBSS and centrifuged at 400 × g for 6 minutes at 4°C. Cells were then stained with 10 μl of Streptavadin-Pacific Orange at 1 mg/ml, and incubated on ice for 20 minutes in the dark. Following the SA-Pacific orange staining, cells were washed twice as above and resuspended in 200 μl of fixation buffer (2% formaldehyde in DPBS). The staining plate containing fixed cells was loaded onto an 11-color LSR II flow cytometer (BD Biosciences, San Jose, CA) through the High Throughput Sampler (HTS) for data acquisition. The unstained and single stained compensation controls were setup by staining antibody-capturing compensation beads (Simply Cellular anti-mouse compensation standard, Bangs Laboratories, Fishers, IN) with staining buffer alone or the optimal dilution of antibody-fluorochrome conjugate. In average, we acquired minimum 1.5 × 106 total events from triplicate wells for each sample, and about 0.8 × 106 to 1 × 106 leukocytes (based on FSC-SSC gating) minimum were collected, whereas 5000 total events were acquired for compensation beads controls. All flow cytometric data acquired on LSR II were analyzed using FlowJo software (Treestar, Ashland, OR). The raw data files from each of the triplicate samples were concatenated to one single fcs file for analysis. Note: staining was performed using 1 million cells in triplicate wells rather than 3 million cells in one single well following the standard operating procedure established in our laboratory.
2.5. MDC1 and PDC isolation by FACS and in vitro stimulation
To isolate pure MDC1 and PDC subsets for in vitro stimulation, PBMC were stained with an antibody cocktail of CD1c-PE, CD3-PE-Cy5.5, CD4-APC-Alexa Fluor 750, CD11c-PE-Cy7, CD14-PE-Texas Red (clone TUK4, Caltag/Invitrogen, Carlsbad, CA), CD16-PerCP-Cy5.5, CD19-PerCP-Cy5.5, CD34-PerCP-Cy5.5, CD304-APC (clone AD5-17F6, Miltenyi Biotec, Auburn, CA) plus 7-AAD and sorted by FACS on a 7-color FACSAria cell sorter (BD Biosciences, San Jose, CA). Five to six thousand sorted MDC1 (Lin−CD4+CD11c+CD1c+) and PDC (Lin−CD4+CD11c−CD304+) were resuspended in 100 μl of tissue culture medium (RPMI-1640 (Cellgro, Herndon, VA) with 8% of FCS (Sigma-Aldrich, St. Louis, MO) and antibiotic/antimycotic solution) per well in a 96-well v-bottom plate in the presence of 100 ng/ml of LPS from E. coli O55:B5 (Sigma-Aldrich, St. Louis, MO) or 20 μg/ml of CpG oligodeoxynucleotides 2006 (Coley Pharmaceutical Group, Wellesley, MA) and incubated at 37°C for 18 hours, followed by staining with 7-AAD, CD40-PE-Cy5, and CD86-Pacific Blue. Cells were then fixed and analyzed on a LSR II flow cytometer.
2.6. Visualization of FITC-dextran uptake by imaging flow cytometry (ImageStream)
PBDC were enriched by magnetic beads using the MACS Human Blood Dendritic Cell Isolation Kit II (Miltenyi Biotec, Auburn, CA). The isolated PBDC (a mixture of all three PBDC subsets with purity greater than 93%) were then used to perform the FITC-dextran uptake assay, followed by staining with CD1c-PE or CD303-PE. Cells were analyzed by ImageStream imaging flow cytometry with IDEAS software (Amnis, Seattle, WA). Nuclear dye DRAQ-5 was added to samples 3 minutes before loading samples into the flow cytometer to exclude dead cells.
3. Results
3.1 Identification of PBDC subsets from freshly isolated human PBMC
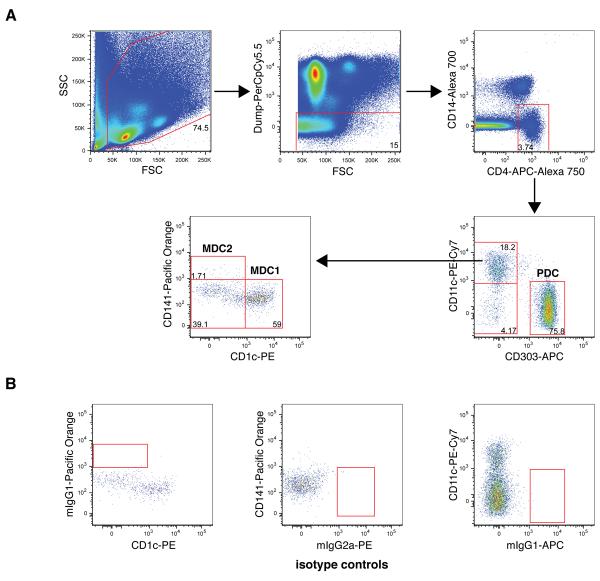
Fresh PBMC were isolated from healthy donors and stained with antibody-fluorochrome cocktails of CD1c, CD3, CD4, CD11c, CD14, CD16, CD19, CD34, CD40, CD86, CD141 and CD303 using the method described in section 2.4. Three major PBDC subsets MDC1, MDC2, and PDC as defined by Dzionek et al. (Dzionek et al., 2000) were identified along with two new additional subpopulations among Lin−CD4+CD14− cells. All the PBDC subsets share a common phenotype characterized by negative staining for CD3, CD14, CD16, CD19, CD34 and positive staining for CD4. PBDC subsets were identified using the bi-variate gating as shown in Fig. 1A for a typical healthy donor sample. A FSC and SSC leukocyte gate was applied to exclude contaminating red blood cells and cell debris prior to applying a “dump gate” to exclude Lin+ (CD3/CD16/CD19/CD34) cells as well as apoptotic (7-AAD+) cells. The CD4+CD14− cells were selected from live Lin− cells and further divided based on surface expression of CD11c and CD303 into CD11c+CD303− MDC and CD11c−CD303+ PDC subsets. Consistent with a previous report (Dzionek et al., 2000), PDC in freshly isolated PBMC from healthy donors did not express CD1c or CD141 (data not shown). In addition to CD11c+CD303− MDC and CD11c−CD303+ PDC subsets, a third subset of CD11c−CD303− cells was also seen in the CD4+CD14− population. The identity of this CD11c−CD303− has yet to be fully investigated. The MDC population was further subdivided based on surface expression of CD1c and CD141 into CD1c+CD141− “MDC1” and CD1c−CD141+ “MDC2” subpopulations. Interestingly, an additional subpopulation negative for CD1c and CD141 was defined in the CD11c+CD303− MDC gate that to our knowledge has not been previously reported. This new subpopulation merits further characterization to determine whether it may be a new type of DC.
Fig. 1. Identifying PBDC subsets.
The analysis for one representative specimen from the 18 healthy adult donor specimens is shown. (A) First, the leukocytes in total PBMC were gated based on FSC and SSC, followed by a “dump” gate to exclude Lin+ (CD3/CD16/CD19/CD34) as well as 7-AAD+ cells. Live Lin− cells were selected on the CD4+CD14− population and gated for CD11c−CD303+ PDC cells, CD11c+CD303− conventional MDC cells and a new subpopulation of CD11c−CD303− cells. CD11c+CD303− cells were further gated for CD1c+CD141− and CD1c−CD141+ (MDC1 and MDC2, respectively) and a new subpopulation of CD1c−CD141− cells. The number in each subset indicates the percentage of cells in its parent gate. Isotype control staining for CD1c, CD141, and CD303-fluorochromes is shown in (B).
To confirm the specificity of PBDC subset-specific marker staining in our assay, isotype-fluorochrome controls were individually substituted in the antibody-fluorochrome cocktail for the CD1c, CD141 and CD303. As shown in Fig. 1B, there was no staining of cell populations when the isotype control reagents were substituted. Gates shown in Fig. 1 are based on the isotype controls and are consistent with gate placement with fluorescent-minus-one (FMO) controls (data not shown).
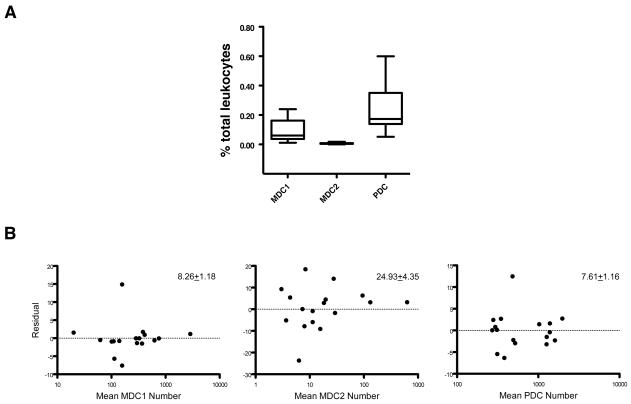
PBDC subset populations were consistently detected in all healthy donor specimens tested with the specific cocktail panel. The box plot in Fig. 2A shows the 25th and 75th quartile distributions obtained for the MDC1, MDC2, and PDC subset populations in specimens from 18 different subjects. For these 18 subjects, the CD11c-CD303− and CD11c+CD303−CD1c−CD141− subpopulations were also consistently present in all specimens (data not shown). The within day precision (repeatability) of this assay was determined from triplicate measurements of PBDC subsets for the 18 subject samples. The mean % CV ± S.D. was 8.26 ± 1.18 for MDC1, 24.93% ± 4.35 for MDC2 and 7.61 ± 1.16 for PDC subsets. The residual plots of the difference from the mean % CV versus the cell number for the 18 sets of triplicates are shown in Fig. 2B. The intermediate precision over time of this assay was determined by measuring the PBDC subset frequency in different vials of a batch of cryopreserved PBMC from a single donor on three consecutive days. The % CV of MDC1, MDC2, and PDC frequency were 10.33%, 24.85%, and 6.25%, respectively.
Fig. 2. Distributions of MDC1, MDC2, and PDC in healthy subjects.
(A) MDC1, MDC2, and PDC in specimens obtained from adult healthy donors (n = 18) were analyzed based on the gating strategy shown in Fig. 1. Boxes shown represent 25th and 75th percentiles. (B) Intra-assay repeatability of PBDC phenotyping assay. The residual plots of difference from the mean %CV of PBDC subset frequency against mean of PBDC subset cell number from triplicate wells are shown. The mean %CV ± SD for each PBDC subset are shown at the upper right corner of each plot.
3.2. Phenotypic characterization of PBDC subsets
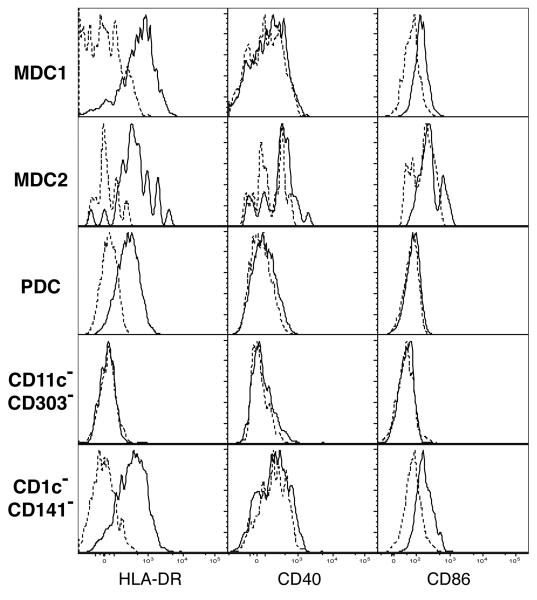
In addition to lineage and DC subset markers, the antibody cocktail used to stain PBDC also contained markers HLA-DR, CD40, and CD86 to further characterize the PBDC phenotypes. The typical expression levels for these markers with one PBMC specimen is shown in Fig. 3. Among the fresh PBDC subsets, MDC1 exhibited the highest level of HLA-DR expression, followed by MDC2 and then with significantly less by PDC (Fig. 3). This is consistent with previous reports that CD11c+ PBDC (MDCs) express a higher level of HLA-DR than CD11c− PBDC (PDC) (Kohrgruber et al., 1999; Robinson et al., 1999). In addition, it was observed that MDC1, MDC2 and PDC subsets obtained from healthy adult donors exhibited an immature phenotype; only a small percentage of cells in each DC subset were CD40 or CD86 positive (Fig. 3).
Fig. 3. Immunophenotypic characterization of MDC1, MDC2, PDC, CD4+CD14−CD11c−CD303−, and CD11c+CD303−CD1c−CD141− cells.
A representative analysis of HLA-DR, CD40, and CD86 expression on MDC1, MDC2, PDC, CD4+CD14−CD11c−CD303− and CD11c+CD303−CD1c−CD141− cells in samples collected from healthy adult donors (n = 18) is shown. Solid lines, staining with anti-HLA-DR, anti-CD40, or CD86 antibody; dashed lines, staining with relevant isotype control.
The additional CD11c−CD303− and CD11c+CD303−CD1c−CD141− subsets were similarly characterized for HLA-DR, CD40 and CD86 expression and are shown in Fig. 3. The CD11c−CD303− subset was negative for HLA-DR, CD40, and CD86 coinciding with the isotype control. For the CD11c+CD303−CD1c−CD141− subset, on the other hand, HLA-DR expression was intermediate between that of MDC1 and PDC while expression of CD40 and CD86 was similar to the MDC1 population.
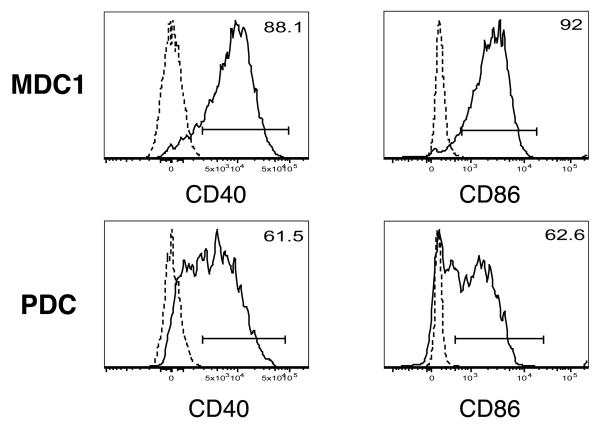
Given the scarcity of CD40 and CD86 positive PBDC and the low level of CD40 and CD86 staining in PBDC subsets directly ex vivo, the ability to detect CD40+ and CD86+ PBDC was confirmed by measuring these co-stimulatory molecules on PBDC stimulated with TLR ligands. Previous studies show that such stimulation can substantially upregulate the expression of CD40 and CD86 on PBDC (Takeda et al., 2003). Since it has also been reported that the expression of PBDC subset-specific markers (CD141 and CD303) may change once DC undergo maturation in culture (Dzionek et al., 2000), it was necessary to isolate each DC subset prior to in vitro stimulation. MDC1 and PDC were sorted by flow cytometry and cultured overnight in the presence of LPS (for MDC1) or CpG DNA (for PDC). Cell surface expression of CD40 and CD86 was measured by flow cytometry the following day. As shown in Fig. 4, the majority of stimulated MDC1 and PDC (ranging from 61.5% to 92%) expressed detectable levels of CD40 and CD86 over isotype controls, verifying the sensitivity of anti-CD40 and anti-CD86 staining in this panel. In separate experiments, the expression of surface markers other than CD40 and CD86 on sorted MDC1 and PDC stimulated in vitro was also monitored. It was found that on both MDC1 and PDC, CD4 expression became undetectable among about half of the cells. In addition, CD141 expression was slightly up on stimulated MDC1, whereas CD303 was downregulated on stimulated PDC. No gain or loss of other surface marker expression was observed (data not shown).
Fig. 4. CD40 and CD86 expression on TLR ligand-stimulated MDC1 and PDC.
MDC1 and PDC were sorted by FACS and stimulated with 100 ng/ml of LPS from E.coli O155:B5 (for MDC1) or 20 μg/ml of CpG 2006 oligonucleotides (for PDC) for 18 hours. Solid lines, staining with anti-HLA-DR, anti-CD40, or anti-CD86 monoclonal antibodies; dashed lines, staining with the relevant isotype controls. The percentages of MDC1 and PDC positive for CD40 or CD86 are shown at the upper right corner of each histogram.
3.3. Assessment of endocytosis capability of PBDC subsets by FITC-dextran uptake
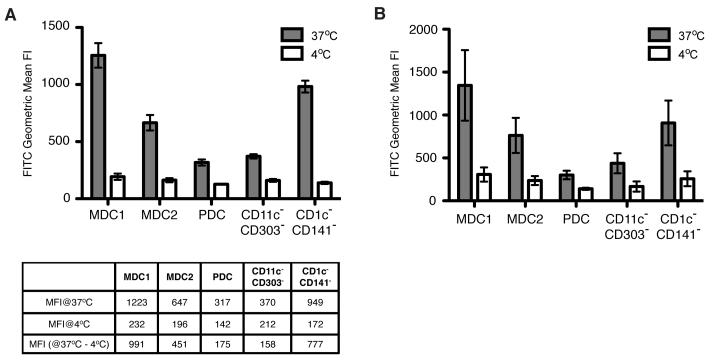
In addition to phenotypic characterization of PBDC subsets, the ability of PBDC to endocytose extracellular antigens was assessed in a FITC-dextran uptake assay. Given the vast majority of PBDC from healthy donors have an immature phenotype and immature MDC are able to endocytose foreign antigens effectively, a significant amount of FITC-dextran was expected to be taken up by MDC1 and MDC2. As shown in Fig. 5, all of the subset populations endocytosed FITC-dextran greater at 37°C than 4°C. The geometric mean fluorescent intensity difference between 37°C and 4°C was 991, 451, 175, 158, 777 for MDC1, MDC2, PDC, CD11C−CD303−, and CD11c+CD303−CD1c-CD141− subsets, respectively. The CD11c−CD303− subset exhibited levels of endocytic activity for FITC-dextran at 37°C similar to PDC, whereas the CD11c+CD303−CD1c-CD141− subset showed similar endocytic activity as MDC1.
Fig. 5. FITC-dextran uptake by the five CD4+CD14− subset populations.
(A) Freshly isolated whole PBMC were incubated with FITC-dextran at either 37°C or 4°C for 30 minutes, washed extensively to remove excess FITC-dextran, followed by staining with a panel of antibody cocktail to identify each subset. Filled bars and open bars represent geometric mean fluorescence intensity of FITC from cells incubated at 37°C and 4°C, respectively. Data shown are from triplicate wells of one representative of four separate experiments. (B) The mean ± SD from four separate experiments with one subject per day are shown; N= 4 subjects.
3.4. Validation of FITC-dextran uptake by ImageStream
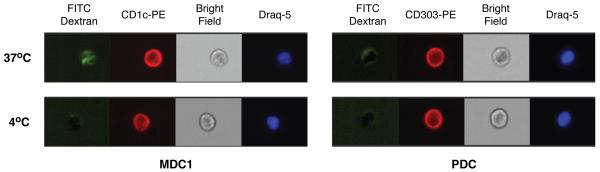
The readout of a flow cytometry-based endocytosis assay is the fluorescence intensity of the dye conjugated to the particles being taken up by cells. To assure the FITC fluorescence intensity detected in this assay was indeed from endocytosed dextran rather than non-specific adherence of FITC-dextran particles on the surface of cells, we examined the pattern of fluorescence emission using Amnis's ImageStream, a technology that combines microscopy and flow cytometry. PBDC were pre-enriched by Miltenyi's Blood DC isolation kit (which positively selects all three subsets of PBDC), followed by FITC-dextran uptake assay and counterstained with CD1c or CD303 to identify MDC1 and PDC, respectively. As shown in Fig. 6, it is clear that there was no green fluorescence inside either MDC1 or PDC when cells were incubated with FITC-dextran at 4°C, whereas at 37°C incubation green fluorescence was clearly visible inside the MDC1 but was very dim in PDC. This pattern is consistent with the flow cytometry data shown in Fig. 5 as well as findings from literature that MDC endocytose FITC-dextran efficiently whereas PDC are less efficient than MDC (Dzionek et al., 2000). In addition, a discernible FITC signal on the surface rather than inside cells was only seen at a frequency < 1 in 500 sorted MDC1 or PDC cells analyzed (data not shown). The low non-specific FITC signal and visual internalization of FITC in these cells in this imaging flow cytometry assay verifies specific FITC-dextran uptake by MDC1 and PDC.
Fig. 6. Validation of FITC-dextran uptake assay by ImageStream analysis.
PBDC enriched by MACS magnetic beads were incubated with FITC-dextran for 30 minutes at either 37°C or 4°C, followed by staining with anti-CD1c or anti-CD303 to identify MDC1 and PDC and nuclear morphology dye Draq-5 to exclude dead cells during data analysis. Stained cells were analyzed by an Amnis ImageStream flow cytometer.
4. Discussions
DC, the “master regulators” of immunity, are critical players in the pathogenesis of various diseases, ranging from infections to cancers to autoimmune diseases (Steinman and Banchereau, 2007; Ueno et al., 2007). Differences in the phenotypes or frequencies of PBDC in patients with SLE, RA, and liver cancer as well as malaria infection have been reported (Beckebaum et al., 2004; Migita et al., 2005; Jongbloed et al., 2006; Ormandy et al., 2006; Urban et al., 2006). Therefore, PBDC may prove useful as an indicator of immune status in vivo. Phenotypic characterization of PBDC subsets ex vivo has potential applications in the diagnosis and treatment in different disease states. Most exciting is the potential use of the multichromatic flow method reported here in clinical and translational studies. Such studies are now made feasible since peripheral blood is a readily available source of PBDC for direct ex vivo analysis.
In this study, we report a multichromatic flow cytometric method that combines identification, phenotypic analysis, and effector function assessment of PBDC subsets into one single assay without the need for a pre-enrichment step (either by depletion of non-DC populations or positive selection on DCs) other than PBMC isolation from whole blood. This eliminates any potential bias (i.e. selective enrichment or loss of a particular cell population) from being introduced during an enrichment step. Given the low frequency of PBDC, even a small error in enumeration can have significant impact on the integrity of data for identification, quantitation, or characterization of DC subsets and subsequent study conclusions. Perhaps the most significant advantage for the 11-color over 3- or 4-color flow methods is the ability to include definitive lineage exclusion and inclusion markers within a single assay tube. In a typical 3 or 4-color experiment, it is impossible to include all the positive and negative markers required to identify the three PBDC subsets (MDC1, MDC2 and PDC). This common 3/4-color immunofluorescence approach takes multiple “snapshots” to identify individual PBDC subsets one at a time without further characterization and may increase cell requirements by 3 fold over the 11-color method. Thus the 11-color method may relieve constraints in cell numbers for clinical studies where the volume of blood or other tissue is limited. From an economic point, operating costs in terms of reagents, labor, and time are greatly increased when each PBDC subset is analyzed individually. In addition, the multichromatic micromethod used in this protocol further reduces reagent and handling costs. Therefore, not only does our 11-color PBDC assay provide a cost-efficient way to study PBDC phenotype and function, it also makes such analysis possible where it is otherwise prohibited by limited sample availability.
It should be emphasized that the method presented here is flexible. Functional characterization markers may be varied and additional markers can be added where equipment is available with greater than 11 colors. The DC multichromatic identification scheme illustrated in this report is an excellent platform that may be modified for other research needs. For instance, the FITC-dextran uptake may be replaced with other functional measures such as intracellular staining of cytokine production by PBDC subsets. Similarly, CD40 and CD86 detection can be substituted with other markers such as CD83, CD205, CD209 or chemokine receptors depending on research interests. Even the lineage markers can be modified to include other DC subsets such as CD16+CD11c+ DCs or CD34+ DC, which have distinctive features from the PBDC (MDC1, MDC2, PDC) (Almeida et al., 2001; MacDonald et al., 2002). Importantly, while the use of peripheral blood is emphasized here, this panel has been used by us to characterize DCs from other tissues including tonsils, lymph nodes, bone marrow and tumors and can be readily extended to multiple types of tissues and body fluids.
The last decade has witnessed the rapid evolution of flow cytometry, especially in terms of the number of markers that can be measured simultaneously. One major impact has been the revelation of the heterogeneity within immune cell populations. Some cell subsets that were first thought to be homogeneous such as memory T cells and regulatory T cells are now recognized to be composed of a variety of subpopulations with diverse function and ontogeny. In fact, by using the 11-color method, we found a new Lin−CD14−CD4+CD11c−CD303− cell population as well as an additional cell population with surface marker expression pattern similar to MDC (Lin−CD14−CD4+CD11c+HLA-DR+) but not fitting the definition for MDC1 or MDC2 subsets, as this cell population was negative in all three PBDC subset markers CD1c, CD141, and CD303. The Lin−CD14−CD4+CD11c−CD303− cells were HLA-DR−, suggesting these cells may lack antigen presentation function. Further characterization is needed to determine whether this cell population is within a DC lineage or is another cell type. Examination of HLA-DR and co-stimulatory molecule upregulation and cytokine production upon TLR ligands stimulation in sorted Lin−CD14−CD4+CD11c−CD303− cells will be of interest. On the other hand, the Lin−CD14−CD4+CD11c+CD1c−CD141−CD303− cell population expressed HLA-DR and showed FITC-dextran uptake similar to MDC1. While these cells share some characteristics with defined MDC, whether they truly are of DC lineage or other cell type is yet to be definitively determined. If they belong to the DC family, then careful characterization is required to demonstrate whether they are a novel DC subset or they represent a distinctive stage in MDC differentiation. In either scenario, it is of interest to determine the role of these cells in both steady state and pathological conditions. It has been reported using 3 or 4-color flow methods that the number of blood circulating MDC subsets in RA and SLE patients is lower than normal subjects (Migita et al., 2005; Jongbloed et al., 2006). One possibility is that some MDC1 and MDC2 downregulate CD1c and CD141 and may no longer be identified as MDC subsets using prior methods. A concurrent increase of frequency in the CD4+CD11c+CD303−CD1c−CD141− population in this case could provide support to this explanation. In addition, since in vitro activated MDC1 and PDC downregulated expression of CD4 (as well as CD303 on PDC), it is of interest to include CD4−CD14− and CD11c−CD303− cell populations for analysis when examining samples from patients suspected with increased numbers of activated DC or whenever significant difference of PBDC subset frequency is seen compared to normal controls, though we have not found cells with activated phenotype (e.g. elevated CD40 and CD86 expression) in these two populations in PBMC directly ex vivo from healthy donors. Nevertheless, the power of multichromatic flow cytometry for identifying new cell populations becomes evident, as the CD4+CD11c+CD303−CD1c−CD141− population is visible only when all PBDC subset markers are used simultaneously to stain the cells.
In summary, we report here a novel multichromatic flow cytometric assay that allows a rapid, cost-efficient, and flexible multi-dimensional analysis of PBDC subsets. Although the data shown in this paper were obtained from samples of healthy donors, the real potential and value of this assay lies in the applications onto various diseases as a tool to monitor DC activation status and functions directly ex vivo. In fact, this assay method has been adapted by our collaborators to study PBDC in RA patients and lymphoma patients (data not shown). Furthermore, in addition to the direct comparisons of frequency, phenotype, and effector function between healthy donors and patients, we can further explore the relationship between costimulatory molecule expression and effector functions of PBDC subsets under pathological conditions. It is our hope this assay will be an invaluable tool to both DC biology research and translational clinical studies.
Acknowledgements
The authors wish to thank Nathan Laniewski for technical assistance in FACS sorting of PBDC subsets. We are also grateful to Dr. Tim Bushnell's expert assistance in performing and analyzing data of imaging flow cytometry and Dr. Alexandra Livingstone for her critical reading and comments on this manuscript. The CpG oligodeoxynucleotides 2006 was a generous gift from Dr. Iñaki Sanz. This study was funded by NIH grants R24 AI054953, U19 AI56390, and NIH contract N01-AI50029.
Abbreviations
- PBDC
peripheral blood dendritic cells
- MDC
myeloid dendritic cells
- PDC
plasmacytoid dendritic cells
- PBMC
peripheral blood mononuclear cells
- APC
antigen-presenting cells
- TLR
Toll-like receptors
- BSA
bovine serum albumin
- HBSS
Hanks' balanced salt solution
- FACS
fluorescence-activated cell sorting
- LPS
lipopolysaccharide
- SLE
systemic lupus erythematosus
- RA
rheumatoid arthritis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Almeida J, Bueno C, Alguero MC, Sanchez ML, de Santiago M, Escribano L, Diaz-Agustin B, Vaquero JM, Laso FJ, San Miguel JF, Orfao A. Comparative analysis of the morphological, cytochemical, immunophenotypical, and functional characteristics of normal human peripheral blood lineage(−)/CD16(+)/HLA-DR(+)/CD14(−/lo) cells, CD14(+) monocytes, and CD16(−) dendritic cells. Clin Immunol. 2001;100:325–38. doi: 10.1006/clim.2001.5072. [DOI] [PubMed] [Google Scholar]
- Banerjee DK, Dhodapkar MV, Matayeva E, Steinman RM, Dhodapkar KM. Expansion of FOXP3high regulatory T cells by human dendritic cells (DCs) in vitro and after injection of cytokine-matured DCs in myeloma patients. Blood. 2006;108:2655–61. doi: 10.1182/blood-2006-03-011353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckebaum S, Zhang X, Chen X, Yu Z, Frilling A, Dworacki G, Grosse-Wilde H, Broelsch CE, Gerken G, Cicinnati VR. Increased levels of interleukin-10 in serum from patients with hepatocellular carcinoma correlate with profound numerical deficiencies and immature phenotype of circulating dendritic cell subsets. Clin Cancer Res. 2004;10:7260–9. doi: 10.1158/1078-0432.CCR-04-0872. [DOI] [PubMed] [Google Scholar]
- Bonasio R, von Andrian UH. Generation, migration and function of circulating dendritic cells. Curr Opin Immunol. 2006;18:503–11. doi: 10.1016/j.coi.2006.05.011. [DOI] [PubMed] [Google Scholar]
- Caux C, Dezutter-Dambuyant C, Schmitt D, Banchereau J. GM-CSF and TNF-alpha cooperate in the generation of dendritic Langerhans cells. Nature. 1992;360:258–61. doi: 10.1038/360258a0. [DOI] [PubMed] [Google Scholar]
- Cavanagh LL, Bonasio R, Mazo IB, Halin C, Cheng G, van der Velden AW, Cariappa A, Chase C, Russell P, Starnbach MN, Koni PA, Pillai S, Weninger W, von Andrian UH. Activation of bone marrow-resident memory T cells by circulating, antigen-bearing dendritic cells. Nat Immunol. 2005;6:1029–37. doi: 10.1038/ni1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzionek A, Fuchs A, Schmidt P, Cremer S, Zysk M, Miltenyi S, Buck DW, Schmitz J. BDCA-2, BDCA-3, and BDCA-4: three markers for distinct subsets of dendritic cells in human peripheral blood. J Immunol. 2000;165:6037–46. doi: 10.4049/jimmunol.165.11.6037. [DOI] [PubMed] [Google Scholar]
- Grouard G, Durand I, Filgueira L, Banchereau J, Liu YJ. Dendritic cells capable of stimulating T cells in germinal centres. Nature. 1996;384:364–7. doi: 10.1038/384364a0. [DOI] [PubMed] [Google Scholar]
- Grouard G, Rissoan MC, Filgueira L, Durand I, Banchereau J, Liu YJ. The enigmatic plasmacytoid T cells develop into dendritic cells with interleukin (IL)-3 and CD40-ligand. J Exp Med. 1997;185:1101–11. doi: 10.1084/jem.185.6.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongbloed SL, Lebre MC, Fraser AR, Gracie JA, Sturrock RD, Tak PP, McInnes IB. Enumeration and phenotypical analysis of distinct dendritic cell subsets in psoriatic arthritis and rheumatoid arthritis. Arthritis Res Ther. 2006;8:R15. doi: 10.1186/ar1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki N, Ho S, Antonenko S, Malefyt RW, Kastelein RA, Bazan F, Liu YJ. Subsets of human dendritic cell precursors express different toll-like receptors and respond to different microbial antigens. J Exp Med. 2001;194:863–9. doi: 10.1084/jem.194.6.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohrgruber N, Halanek N, Groger M, Winter D, Rappersberger K, Schmitt-Egenolf M, Stingl G, Maurer D. Survival, maturation, and function of CD11c- and CD11c+ peripheral blood dendritic cells are differentially regulated by cytokines. J Immunol. 1999;163:3250–9. [PubMed] [Google Scholar]
- MacDonald KP, Munster DJ, Clark GJ, Dzionek A, Schmitz J, Hart DN. Characterization of human blood dendritic cell subsets. Blood. 2002;100:4512–20. doi: 10.1182/blood-2001-11-0097. [DOI] [PubMed] [Google Scholar]
- Migita K, Miyashita T, Maeda Y, Kimura H, Nakamura M, Yatsuhashi H, Ishibashi H, Eguchi K. Reduced blood BDCA-2+ (lymphoid) and CD11c+ (myeloid) dendritic cells in systemic lupus erythematosus. Clin Exp Immunol. 2005;142:84–91. doi: 10.1111/j.1365-2249.2005.02897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseman EA, Liang X, Dawson AJ, Panoskaltsis-Mortari A, Krieg AM, Liu YJ, Blazar BR, Chen W. Human plasmacytoid dendritic cells activated by CpG oligodeoxynucleotides induce the generation of CD4+CD25+ regulatory T cells. J Immunol. 2004;173:4433–42. doi: 10.4049/jimmunol.173.7.4433. [DOI] [PubMed] [Google Scholar]
- Munn DH, Sharma MD, Lee JR, Jhaver KG, Johnson TS, Keskin DB, Marshall B, Chandler P, Antonia SJ, Burgess R, Slingluff CL, Jr., Mellor AL. Potential regulatory function of human dendritic cells expressing indoleamine 2,3-dioxygenase. Science. 2002;297:1867–70. doi: 10.1126/science.1073514. [DOI] [PubMed] [Google Scholar]
- Olweus J, BitMansour A, Warnke R, Thompson PA, Carballido J, Picker LJ, Lund-Johansen F. Dendritic cell ontogeny: a human dendritic cell lineage of myeloid origin. Proc Natl Acad Sci U S A. 1997;94:12551–6. doi: 10.1073/pnas.94.23.12551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormandy LA, Farber A, Cantz T, Petrykowska S, Wedemeyer H, Horning M, Lehner F, Manns MP, Korangy F, Greten TF. Direct ex vivo analysis of dendritic cells in patients with hepatocellular carcinoma. World J Gastroenterol. 2006;12:3275–82. doi: 10.3748/wjg.v12.i20.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid CD, Stackpoole A, Meager A, Tikerpae J. Interactions of tumor necrosis factor with granulocyte-macrophage colony-stimulating factor and other cytokines in the regulation of dendritic cell growth in vitro from early bipotent CD34+ progenitors in human bone marrow. J Immunol. 1992;149:2681–8. [PubMed] [Google Scholar]
- Reis e Sousa C. Dendritic cells in a mature age. Nat Rev Immunol. 2006;6:476–83. doi: 10.1038/nri1845. [DOI] [PubMed] [Google Scholar]
- Robinson SP, Patterson S, English N, Davies D, Knight SC, Reid CD. Human peripheral blood contains two distinct lineages of dendritic cells. Eur J Immunol. 1999;29:2769–78. doi: 10.1002/(SICI)1521-4141(199909)29:09<2769::AID-IMMU2769>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Romani N, Gruner S, Brang D, Kampgen E, Lenz A, Trockenbacher B, Konwalinka G, Fritsch PO, Steinman RM, Schuler G. Proliferating dendritic cell progenitors in human blood. J Exp Med. 1994;180:83–93. doi: 10.1084/jem.180.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallusto F, Lanzavecchia A. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor alpha. J Exp Med. 1994;179:1109–18. doi: 10.1084/jem.179.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler-Thurner B, Schultz ES, Berger TG, Weinlich G, Ebner S, Woerl P, Bender A, Feuerstein B, Fritsch PO, Romani N, Schuler G. Rapid induction of tumor-specific type 1 T helper cells in metastatic melanoma patients by vaccination with mature, cryopreserved, peptide-loaded monocyte-derived dendritic cells. J Exp Med. 2002;195:1279–88. doi: 10.1084/jem.20012100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman RM. Lasker Basic Medical Research Award. Dendritic cells: versatile controllers of the immune system. Nat Med. 2007;13:1155–9. doi: 10.1038/nm1643. [DOI] [PubMed] [Google Scholar]
- Steinman RM, Banchereau J. Taking dendritic cells into medicine. Nature. 2007;449:419–26. doi: 10.1038/nature06175. [DOI] [PubMed] [Google Scholar]
- Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–76. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- Ueno H, Klechevsky E, Morita R, Aspord C, Cao T, Matsui T, Di Pucchio T, Connolly J, Fay JW, Pascual V, Palucka AK, Banchereau J. Dendritic cell subsets in health and disease. Immunol Rev. 2007;219:118–42. doi: 10.1111/j.1600-065X.2007.00551.x. [DOI] [PubMed] [Google Scholar]
- Urban BC, Cordery D, Shafi MJ, Bull PC, Newbold CI, Williams TN, Marsh K. The frequency of BDCA3-positive dendritic cells is increased in the peripheral circulation of Kenyan children with severe malaria. Infect Immun. 2006;74:6700–6. doi: 10.1128/IAI.00861-06. [DOI] [PMC free article] [PubMed] [Google Scholar]