Abstract
Nucleosome positioning has been the subject of intense study for many years. The properties of micrococcal nuclease, the enzyme central to these studies, are discussed. The various methods used to determine nucleosome positions in vitro and in vivo are reviewed critically. These include the traditional low resolution method of indirect end-labelling, high resolution methods such as primer extension, monomer extension and nucleosome sequencing, and the high throughput methods for genome-wide analysis (microarray hybridisation and parallel sequencing). It is established that low resolution mapping yields an averaged chromatin structure, whereas high resolution mapping reveals the weighted superposition of all the chromatin states in a cell population. Mapping studies suggest that yeast DNA contains information specifying the positions of nucleosomes and that this code is made use of by the cell. It is proposed that the positioning code facilitates nucleosome spacing by encoding information for multiple alternative overlapping nucleosomal arrays. Such a code might facilitate the shunting of nucleosomes from one array to another by ATP-dependent chromatin remodelling machines.
Introduction
Nuclease digestion has been a central tool of chromatin research since its inception. Hewish & Burgoyne (1) demonstrated the essentially ordered structure of chromatin when they observed that digestion of chromatin in nuclei by endogenous nucleases gave rise to a series of discrete DNA fragments, rather than the DNA smear that might have been expected. They observed that the sizes of the DNA fragments were multiples of a constant length, later called the “repeat length” of the chromatin. Subsequently, chromatin researchers have made use of purified nucleases, primarily micrococcal nuclease (MNase), but also pancreatic DNase I. The repeat length is a characteristic of particular tissues and organisms. Most somatic tissues have a repeat length of ~195 bp, but neuronal chromatin has a repeat of only ~165 bp, similar to that of budding yeast.
Early studies identified the nucleosome as the basic structural repeat unit of chromatin. It is composed of a nucleosome core containing 147 bp of DNA wrapped around a central histone octamer containing two molecules each of the four core histones (H2A, H2B, H3 and H4), and a “linker” DNA of characteristic length, which connects one nucleosome to the next. A single molecule of histone H1 (linker histone) is bound to the nucleosome at the point where the DNA enters and exits the core, and to the linker DNA. The DNA within the nucleosome core is protected from nucleases by the core histones, whereas the linker DNA is vulnerable to digestion. Thus, chromatin is composed of arrays of regularly spaced nucleosomes. Excellent reviews of the early work are available (2, 3).
Digestion of DNA by MNase
MNase digests both single- and double-stranded polynucleotides, yielding fragments with a 5′-hydroxyl and a 3′-phosphate. The enzyme is dependent on calcium for its activity and therefore digestion can be halted with EDTA. MNase has both endonuclease and exonuclease activities: it cuts DNA and then trims it from the exposed ends. The ideal nuclease for use in chromatin studies would cut DNA solely where it is accessible and would be unaffected by DNA sequence. However, MNase is far from ideal in this regard: it cuts DNA primarily at runs of alternating dA and dT that are preceded by dG or dC (CATA is a particularly good site), but it ignores runs of dA or dT (4–6). Once cleaved at a preferred site, the exonuclease activity rapidly removes dA and dT, but proceeds much more slowly when confronted with dC and dG (4). The exonuclease is a powerful enzyme at 37°C, but it is much weaker at 4°C (7).
Digestion of Chromatin by MNase
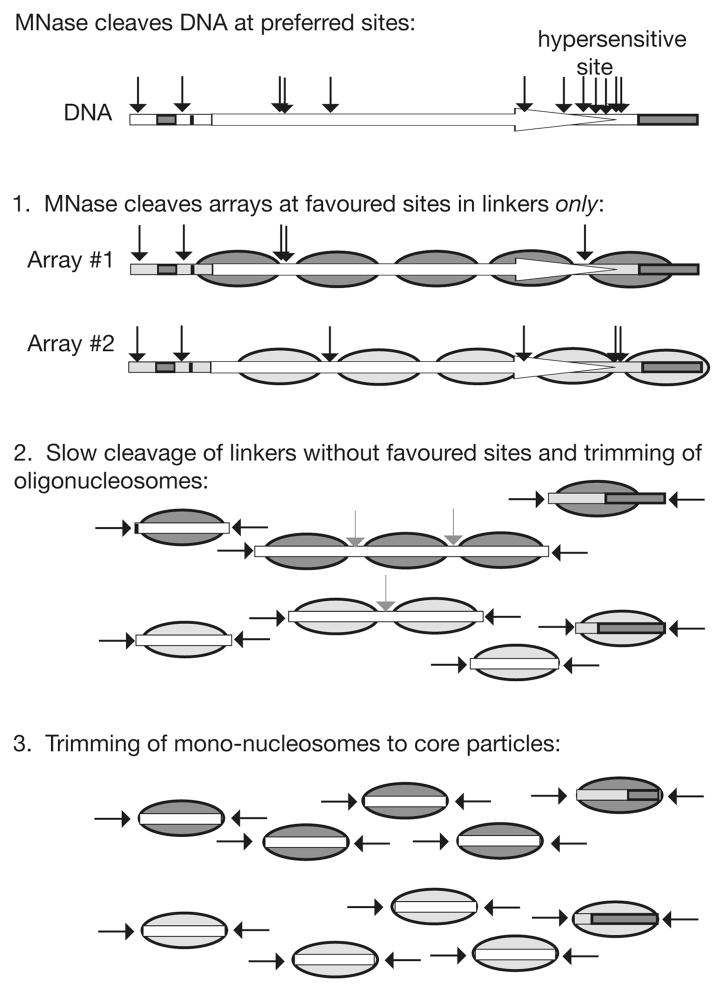
The digestion of chromatin is much slower than that of protein-free DNA. It proceeds through several stages, each involving metastable intermediates stabilised by bound histones (Figure 1). Initial digestion involves endonucleolytic cleavage of the linker DNA between nucleosomes, resulting in the characteristic “ladder” pattern (see Figure 3C for an example). The presence of H1 slows digestion of linker DNA significantly, but H1 protects DNA much less strongly than the core histones do (7). As digestion proceeds, the average number of nucleosomes per fragment decreases. The average fragment size for a given number of nucleosomes also decreases, because the trimming activity slowly shortens the cut linker at each end of the nucleosomal oligomer. The slope of a plot of average DNA fragment size versus nucleosome number yields the repeat length (simply dividing the DNA size by the number of nucleosomes will give an incorrrect result).
Figure 1.
Digestion of chromatin by MNase. The three main stages of digestion are illustrated, indicating the dominant role of the sequence preference of MNase. As an example, the distribution of CATA sites in the yeast HIS3 gene is shown (black arrows). A cluster of six such sites accounts for the MNase hypersensitive site observed at the 3′-end of HIS3. Two different nucleosomal arrays are shown: they are predicted to be digested differently even though they contain the same DNA sequence (the arrows indicate CATA sites in linkers). Grey arrows indicate the much slower cutting of linkers lacking CATA. Note that in mammalian chromatin, the linkers are usually longer than in yeast and so most are likely to contain a preferred site, resulting in a more even digestion of the chromatin.
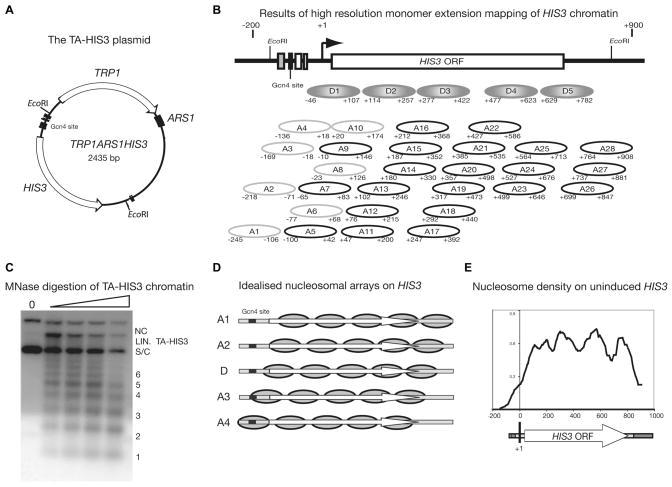
Figure 3.
Yeast HIS3 chromatin is organised into multiple alternative nucleosomal arrays. (A) Map of TA-HIS3. This plasmid contains the HIS3 and TRP1 genes and an origin of replication (ARS1). It was purified as chromatin from cells in which HIS3 is expressed at basal levels (i.e., lacking the activator, Gcn4p) or from cells in which HIS3 is active. (B) Comparison of the chromatin structures of active and basal HIS3 as revealed by high resolution monomer extension mapping: in gcn4Δ and snf2Δ cells, about one third of HIS3 chromatin is organised into a dominant nucleosomal array (D1–D5), which is similar to that observed by indirect end-labelling. The rest of the nucleosomes adopt alternative positions (A1–A28). In active chromatin, the D1–D5 positions are present, but no longer dominant. (C) Digestion of TA-HIS3 chromatin with MNase shows that it is organised into nucleosomal arrays with a typical spacing of ~165 bp (a Southern blot probed with TA-HIS3 DNA is shown). S/C = supercoiled; NC = nicked circle; LIN. = linear TA-HIS3. (D) Idealised nucleosomal arrays on HIS3: it is proposed that the nucleosomes mapped by monomer extension exist in multiple overlapping arrays (D, A1–A4), each with a spacing of ~165 bp. The black box indicates the binding site for Gcn4p. (E) Nucleosome density map for uninduced HIS3 obtained by quantification of monomer extension data. Data are from (18).
The first major histone-dependent block to MNase is the “chromatosome” (8), which contains 160–168 bp of DNA. It reflects protection of ~20 bp of linker DNA protruding from the nucleosome core, primarily by H1. There is an additional stabilising contribution from the core histones, since chromatin depleted of H1 and then digested with MNase shows some weak chromatosome-like protection. There is controversy over whether the ~20 bp of linker DNA in the chromatosome is all on one side of the nucleosome core, or whether ~10 bp projects from each side (9, 10). This relates to the question of the precise nature of the H1 binding site (11, 12). Chromatosomes can be isolated in a careful limited digestion, but they are relatively unstable. Continued trimming removes residual linker DNA from the chromatosome, resulting in eviction of H1, and halts at the second major histone-dependent block to digestion: the nucleosome core particle. The core particle contains 145–147 bp of DNA and is very stable, if digestion is stopped with EDTA. If allowed to continue, MNase will destroy it, by gradually nicking and cutting the DNA at ~10 bp intervals where it is most exposed on the outer surface of the core particle (13, 14).
Nucleosome Mapping in vivo by Indirect End-labelling
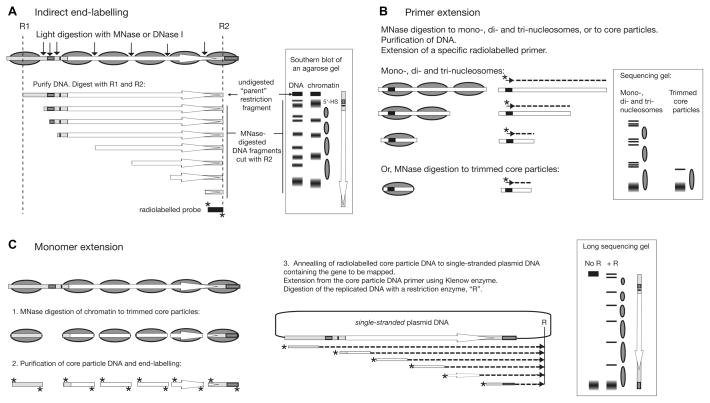
The traditional method for determining nucleosome positions with respect to DNA sequence in vivo is indirect end-labelling (Figure 2A) (15), which was originally developed to map DNase I hypersensitive sites (16). Chromatin in nuclei is digested with different amounts of MNase such that a range of nucleosome ladders is obtained (e g., Figure 3C). The purified DNA is digested using a restriction enzyme(s) with sites on either side of the region of interest and electrophoresed in a long agarose gel. A blot is probed with a radio-labelled DNA fragment abutting one end of the restriction fragment of interest. A series of defined bands is revealed, the sizes of which indicate the sites of MNase cleavage with respect to the probe end of the restriction fragment. These sites are located in the accessible linker DNA between nucleosomes. The digestion pattern of chromatin is compared with that of protein-free genomic DNA digested to the same extent. If the spacing between two neighbouring strong bands in chromatin is approximately the size of a nucleosome (147 bp) and there are bands in between these bands in the DNA sample (i.e., protected from cleavage in chromatin), then the presence of a nucleosome positioned between these cut sites is inferred (15). The interpretation of the band pattern to obtain a nucleosome map can be complicated by the following problems:
Figure 2.
Principles of nucleosome mapping techniques used in vivo. Outlines of the methods are shown with idealised results, assuming a single nucleosomal array on a gene of interest. (A) Indirect end-labelling. In this example, the major MNase or DNase I cleavage sites in chromatin and DNA are mapped with respect to restriction site R2 using a radiolabelled probe (*). 5′-HS: 5′-hypersensitive site. (B) Primer extension. The distance from the specific primer (indicated by a short arrow) to the site of MNase cleavage in the linker is mapped. Trimmed nucleosome core particles give the most accurate position, but only one nucleosome can be mapped. Good (but less accurate) data can be obtained by using oligo-nucleosomes. Several primers would be needed to map the entire gene shown. (C) Monomer extension. Core particle DNA is used as primer in a primer extension reaction with single-stranded DNA as template (the use of double-stranded DNA would result in extension of both core particle DNA strands in opposite directions). The bands observed correspond to the distance from the far border of the nucleosome to the unique restriction site, R. The entire gene can be mapped in one lane, giving accurate positions and relative occupancies. Overlapping positions are easily detected by primer extension or monomer extension: bands closer together than the allowed 147-bp will be observed. The same is true for indirect end-label maps, but in this case interpretation is problematic because the method relies on the observation of nucleosome-sized protected regions. See text for more details.
The band pattern observed for protein-free DNA is often disturbingly similar to that of the corresponding chromatin. Many of the bands are the same or almost the same, although free DNA often gives rise to additional bands. However, the intensities of the bands are usually different. A fair comparison of DNA and chromatin requires that they are digested to the same extent (indicated by similar amounts of intact parent band, assuming equal loadings of DNA in the gel). In many studies, a nucleosome map is derived simply by assuming that gaps in the band pattern that are about the size of a nucleosome correspond to positioned nucleosomes. This is reasonable only if there is some evidence of protection (i.e., a band is present in DNA, but absent within the postulated nucleosome). The DNA control is missing in a surprisingly large number of studies.
The band patterns of DNA and chromatin change as digestion proceeds. The question then arises as to which samples are the correct ones to compare. This problem derives from the following issues: (i) Digestion at other sites influences the amount of DNA in a particular band. For example, the rates of appearance and disappearance of a particular band during digestion would be affected indirectly by rapid cleavage at a strong site located nearer to the probe. Thus, the intensity of a band does not necessarily reflect just the accessibility of the DNA at that site. This problem can be circumvented if “one-hit kinetics” are used, i.e., if each parent DNA fragment is cut only once by MNase (statistically, this requires that most of the parent fragments remain intact). In many studies, this is not the case. (ii) Trimming of chromatin fragments by MNase shortens cut linkers, shifting bands to shorter sizes. Since trimming is slower with dG/dC than dA/dT, a new band might appear when MNase encounters a GC-rich sequence in a linker. New bands might also reflect digestion of unstable nucleosomes (perhaps of remodelled nucleosomes), or the presence of alternative arrangements of nucleosomes (15). Our own high resolution mapping data support the latter interpretation (see below).
The spacing between two neighbouring bands is sometimes inconsistent with the size of one nucleosome, being either too small or too large. The presence of a nucleosome cannot be inferred if the bands are less than 145 bp apart, because at this stage in digestion the nucleosome is resistant to MNase. The interpretation of a gap between two bands that is much larger than one nucleosome but smaller than two nucleosomes (especially if it exceeds the repeat length of the chromatin) is also problematic, because a canonical nucleosome can protect only 147 bp. Examples include one of the yeast ARS1 nucleosomes (180 bp) (15) and the yeast PHO8 gene (17). In the latter case, two alternative overlapping nucleosome positions were proposed. Indeed, complex indirect end-labelling patterns are often observed, in which there are numerous bands and few nucleosome-sized gaps; these are difficult to interpret.
The Sequence Preference of MNase Dominates the Early Stages of Chromatin Digestion
The digestion of protein-free DNA always gives rise to a discrete set of bands, indicating that the sequence preference of the enzyme dominates the early stages of digestion (Figure 1). For example, our own studies have shown that the digestion pattern of protein-free DNA containing the yeast HIS3 gene is predicted quite well by the distribution of CATA sequences. Clusters of preferred sequences might be expected to result in MNase hypersensitive sites, as is the case at the 3′ end of HIS3 (18). In chromatin, the rate at which a particular linker is cut is likely to depend on the number of preferred sites it contains. Linkers with preferred sites will be cut much faster than those which lack them. Most linkers are likely to possess such a site, because a typical MNase site has only ~4 base pairs, but the probability will depend on linker length. Consequently, chromatin with short linkers (yeast and neuronal chromatin) should give rise to a wider range of linker digestion rates. Thus, linkers will be cut at different rates, even if they have the same accessibility to MNase. Conversely, it is important to realise that if a preferred MNase site is present in a nucleosome, it is resistant to cleavage at this stage of digestion. For example, there is a CATA sequence near the dyad of the X. borealis 5S rRNA gene, a relatively strong nucleosome positioning sequence that is popular in nucleosome reconstitution studies. Another example is the D5 nucleosome on the yeast HIS3 gene (Figure 3B), which contains a cluster of five CATA sites.
In summary, the aim of the indirect end-labelling experiment is to infer the positions of nucleosomes by identifying accessible and protected regions in chromatin. The sequence specificity of MNase presents significant problems for interpretation of the map. The bands observed primarily reflect: (i) the accessibility of each favoured cleavage site (if present in a linker it is cut; if nucleosomal, it is resistant); (ii) the number and distribution of favoured cleavage sites; (iii) the rate and degree of trimming of cut linkers to the actual border of the nucleosome. Finally and most importantly, because of these difficulties, indirect end-label studies usually admit only one of two possible interpretations: either there is an array of positioned nucleosomes, or there is not. The true situation is usually more complex, as revealed by high resolution mapping studies. In conclusion, nucleosome maps obtained using the indirect end-labelling method should be considered low resolution (i.e., relatively imprecise and simplified). It is fair to say that indirect end-label maps are often heavily over-interpreted.
High Resolution Mapping Methods
The problems of indirect end-labelling reflect the marked sequence specificity of MNase, which complicates interpretation of what is essentially a kinetic experiment. They can be largely avoided by preparing nucleosome core particles and analysing their DNA. Methods involving core particles differ from indirect end-labelling in that they identify sequences within nucleosomes, rather than determining the rate of digestion of the linkers between them.
High resolution methods include primer extension, hydroxyl radical or DNase I footprinting, exonuclease III digestion and restriction mapping of core particle DNA (19–21). These are all good, accurate methods, but they are usually limited in scope to just one or two nucleosomes, for various reasons. Primer extension is the method most commonly used in vivo: oligo-nucleosomal DNA is purified and used as a template for primer extension using a radio-labelled primer; the length of the run-off product defines the distance from the primer to the nucleosome border (Figure 2B). Multiple products could indicate overlapping positions, or incomplete trimming. To avoid this problem, it is better to use core particle DNA as the template, but then the information obtained is limited to a single nucleosome. Another method, monomer extension (22), has a long range (similar to that of indirect end-labelling), high precision and, moreover, can provide quantitative information concerning the relative amounts of each nucleosome. For monomer extension (Figure 2C), radio-labelled core particle DNA is used as primer in a primer extension experiment with single-stranded plasmid containing the target sequence as the template (which can be several kilobases). Single-stranded plasmid DNA is used because a double-stranded template would allow both strands of the end-labelled core particle DNA to anneal and permit subsequent extension in both directions. The replicated DNA is digested with a suitable restriction enzyme and the DNA fragments are resolved in a long sequencing gel. The length of each DNA fragment is equal to the distance from the far border of the nucleosome to the restriction site. The positions and relative amounts of each nucleosome are revealed. Importantly, this method can resolve overlapping positions. The main disadvantage of monomer extension when applied to native chromatin is that the chromatin of interest has to be separated from the rest of the chromatin, or the background will be too high (23).
High resolution mapping typically provides positions with a precision of a few base pairs. The accuracy of the measured position depends on the extent to which the nucleosomes have been fully trimmed to core particles. Although “mono-nucleosome” and “core particle” are often used interchangeably, it should be noted that the core particle is fully trimmed to 145–150 bp, whereas mono-nucleosomes might not be fully trimmed and may contain H1. This is important because the accuracy of the position is critically dependent on full trimming. For example, the position of a mono-nucleosome containing 165 bp and therefore incompletely trimmed, cannot be determined more accurately than within ~20 bp.
A secondary problem is that by the time the chromatin has been completely digested to core particles, some of them have been nicked or even cut, with some sequence bias (13), perhaps resulting in under-representation of some nucleosomes in the core particle population. The traditional method for preparing core particles from the chromatin of higher eukaryotes involves two separate digestions with MNase: nuclei are first digested to oligomeric chromatin, the H1 is removed and then the chromatin is digested again to core particles. This provides high quality core particles, although the removal of H1 has to be done carefully to avoid nucleosome sliding. Yeast chromatin lacks a canonical linker histone, but a careful MNase titration is required to obtain fully trimmed core particles. A compromise is usually necessary, involving the analysis of either incompletely digested chromatin (some di- and tri-nucleosomes are still present and trimming is incomplete), or slightly over-digested chromatin (fully trimmed core particles with some nicking and perhaps a small amount of double-stranded cutting). The extent of trimming and nicking of purified core particle DNA should be determined routinely by analysis of radio-labelled DNA in native and denaturing polyacrylamide gels. The problem of nicked core particle DNA can be significant if the DNA is to be amplified by PCR. It can be solved by repairing the nicks using DNA repair enzymes. However, double-stranded cuts cannot be repaired.
High Resolution Mapping of Reconstituted Nucleosomes: Overlapping Positions
Numerous mapping studies in vitro indicate that uniquely positioned nucleosomes are very rare. A unique position is one in which the nucleosome forms on the same sequence on all DNA molecules in the population. Instead, nucleosomes adopt multiple overlapping positions with different occupancies. That is, the chromatin is structurally heterogeneous, because two canonical nucleosomes cannot physically overlap on the same DNA molecule. Examples include the 5S RNA positioning sequence (21) and the Drosophila hsp70 promoter (24). The only exception is the 601 sequence and its relatives. The 601-sequence is a synthetic DNA that was selected for its high affinity for the histone octamer in vitro (25). The 601-sequence has such a high affinity for the octamer that essentially all nucleosomes reconstituted on a short DNA fragment form on the 601-sequence. This is an invaluable property for experiments in vitro where the fate of the nucleosome is in question (e.g., after transcription or remodelling). Consequently, most groups now use 601 for their in vitro studies. However, 601 is exceptional in its positioning properties and it is not a natural sequence.
The general observation of overlapping positions necessitates a refinement of the definition of nucleosome positioning, distinct from the traditional concept of unique positions. It may be imagined that during nucleosome assembly, histone octamers must choose from all possible 147-bp DNA sequences in the DNA that is presented to them. Those 147-bp sequences with the highest affinity for the histone octamer will be bound preferentially. In vitro, the distribution of nucleosomes on a DNA fragment is therefore determined by the relative affinities of all possible 147-bp sites for the histone octamer (26). With the exception of 601, none of these sequences is so powerful that it can completely out-compete all the other sequences to yield a uniquely positioned nucleosome.
This analysis suggests that two properties should be defined for nucleosomes in chromatin: (i) their positions (i.e., the DNA sequence they contain); (ii) the occupancy of each position (the fraction of nucleosomes occupying this precise position relative to all possible overlapping positions). From this, it follows that all nucleosomes are precisely positioned, by definition. It is more useful to consider the occupancy of each possible position (i.e., 147-bp window) along a DNA sequence. Some positions will have high occupancies (the 601-sequence will have an occupancy close to 1 in vitro); others will be occupied rarely, if at all, having occupancies close to zero.
High Resolution Mapping of Nucleosomes in Native Chromatin
Early high resolution mapping of native chromatin focussed on repetitive sequences such as the X. borealis 5S RNA gene (27) or satellite DNA (28), using various ingenious methods to map them, which depend on the repetitive nature of the DNA. Most recent high resolution mapping studies have employed primer extension (e.g., the mouse mammary tumour virus (MMTV) promoter (29) and the H. polymorpha MOX promoter (30)), or nucleosome sequencing (e.g., SV40 chromatin (31)). In all of these cases, clusters of overlapping positions were revealed, perhaps rotationally related, rather than the unique positions expected from indirect end-labelling. The clearest exception in yeast is at silenced regions, where arrays of strongly positioned nucleosomes have been mapped (32). These might constitute a special case in view of their heterochromatin-like structure.
We have used monomer extension to map nucleosomes in plasmid chromatin carrying a functional CUP1 or HIS3 gene purified from yeast cells (Figure 3A) (18, 33). Purification of plasmid chromatin facilitates the removal of chromosomal chromatin, which would otherwise interfere with the mapping. Plasmid chromatin carrying either the CUP1 or the HIS3 gene is heterogeneous: nucleosomes are observed in many overlapping positions (Figure 3B). HIS3 chromatin is more ordered in the absence of the activator (Gcn4p), or of the SWI/SNF remodelling complex, in that a dominant nucleosomal array is apparent. But the overall impression is of a disordered structure, particularly for activated chromatin. However, a MNase titration confirms that the chromatin is organised into regular nucleosomal arrays independently of gene activation, with the average spacing of ~165 bp that is typical of yeast (Figure 3C). Therefore, the chromatin must be highly ordered, even though there are many overlapping positions (Figure 3B). To account for these observations, we proposed that HIS3 can exist in one of several alternative nucleosomal arrays (Figure 3D). Thus, in an individual cell, the HIS3 gene might undergo transitions from one array to another as events unfold; the monomer extension map (Figure 3B) represents the sum of all nucleosome positions within these arrays.
In summary, high resolution mapping has revealed a more complex picture of chromatin in vivo than that derived from indirect end-labelling. Low resolution mapping yields an averaged chromatin structure, whereas high resolution mapping reveals the weighted superposition of all the chromatin states in the cell population. The representation of a gene, especially an active gene, as an array of uniquely positioned nucleosomes is appealing in its simplicity, but is likely to be a major over-simplification for most genes (34). The complexity of the nucleosome maps of the CUP1 and HIS3 genes and the MOX and MMTV promoters, for example, have been a source of dismay to some in the field. However, such a complex structure should be expected, once events occurring on an active gene are considered. At a given moment in a cell population, there will be many possible chromatin states, including cells in which RNA polymerase II is initiating transcription at a nucleosome-free promoter, cells in which elongating RNA polymerase II is present at different places on the gene, causing local disruptions, and cells in which the gene is transiently in a non-transcribed state, or in the process of being remodelled. Thus, the combined effects of transcription and remodelling would be expected to result in different chromatin structures at different times on the same gene. A nucleosome mapping study will record the weighted average of these chromatin structures in the cell population.
Genome-wide Mapping using Microarrays and Nucleosome Sequencing
More recently, methods for mapping nucleosomes on a genome-wide scale have been developed. The first approach to be described was the hybridisation of yeast nucleosomal DNA to microarrays carrying oligonucleotides representative of an entire yeast chromosome (35). The method has a resolution equivalent to that of indirect end-labelling, because the borders of the nucleosome cannot be defined. Microarray data are best understood as measurements of nucleosome density or occupancy, i.e., the relative probability of each oligonucleotide on the array being found in a nucleosome. Many genes exhibit a sinusoidal nucleosome density profile, with peaks interpreted as positioned nucleosomes and troughs as linkers; many other genes exhibit more complex patterns that are difficult to interpret (see below) (35–37).
The most important finding from the microarray studies is that the old observation that active promoters are much less likely to be nucleosomal than coding regions is generally true. This is not to say that promoters are nucleosome-free - a critical point! It is illustrated by many studies in which the accessibility of a restriction site in a promoter is measured in nuclei. The assay is based on the observation that the nucleosome affords essentially complete protection of a restriction site from digestion in vitro (e.g., (38)). It is possible for a restriction enzyme to cut a site within a nucleosome, particularly if it is close to the edge, but high concentrations of enzyme are required (39). In most studies, digestion of a restriction site in nuclei reaches a plateau in the region of 50% (e.g., the yeast PHO5 promoter (40) and the chicken β-globin enhancer (41)); very few studies record complete accessibility. A conceptually weak point of the assay when applied in vivo is that proteins other than nucleosomes might also protect against restriction enzymes, although such proteins would have to be unusually strongly bound to DNA. Our own studies provide direct evidence for nucleosomes on two active yeast promoters in vivo, measured both by monomer extension mapping and by restriction enzyme accessibility (18, 33, 42). Thus, the active promoter is much less likely to be nucleosomal than the neighbouring coding region, but it is not always nucleosome-free. This can be understood in terms of a dynamic chromatin structure: at some points in the transcription cycle, there is a nucleosome on the promoter, at other times it is nucleosome-free (i.e., some arrays have a nucleosome on the promoter; others do not (Figure 3D)).
The latest breakthough in the field is parallel sequencing of nucleosomal DNA (43–47). Nucleosome sequencing is the ultimate high resolution mapping technique, because the nominal resolution is one base pair. Nucleosome sequencing using traditional methods has been described previously, leading to important insights into how DNA interacts with the histone octamer (48, 49). It is the scale of the new sequencing experiments that is breathtaking: the new machines can sequence millions of nucleosomes! For parallel sequencing, adaptors containing suitable primer sequences are ligated to purified nucleosomal DNA, which is then amplified by PCR and sequenced in parallel (50). Two different technologies are available: the Illumina-Solexa system which yields millions of short reads (~40 bases), and the Roche 454 system, which yields >100 times fewer reads, but of greater length (~250 bases). The advantage of the latter is that the length of the nucleosome is obtained from the sequence and therefore the degree of trimming and the accuracy of the position data should be apparent. However, most studies have used the Illumina system because it yields much higher genome coverage. Short sequences corresponding to one end of each nucleosomal DNA molecule are obtained and identified. To infer the position of the nucleosome, forward and reverse reads are paired assuming that they should be ~150 bp apart and so can be attributed to the same nucleosome (50). However, it is difficult to distinguish between different degrees of trimming of the same nucleosome and a cluster of overlapping positions (18). These difficulties can be resolved experimentally using Illumina paired end sequencing, in which both ends of the same DNA molecule are sequenced, yielding the length of the nucleosomal DNA. All the caveats concerning high resolution mapping mentioned above also apply to nucleosome sequencing, particularly the requirement for full trimming of core particles to 145–150 bp. An additional issue is the potential for amplification bias.
Most high throughput studies differ from traditional studies in two ways: (i) An attempt is made to prevent nucleosome sliding during core particle preparation by prior fixation of the cells with formaldehyde. However, it is unlikely that cross-linking of the histones to DNA is complete and therefore effective. It is unclear whether formaldehyde fixation is appropriate, given that it might introduce artefacts resulting from modification of DNA-binding lysine residues and DNA bases. (ii) An attempt is made to correct for the sequence bias of MNase by comparing nucleosomal DNA with protein-free genomic DNA digested by MNase to about the same size. This seems inappropriate given that protein-free DNA is destroyed long before mono-nucleosomes appear in the digestion (discussed above). A different kind of bias might be introduced if some core particles are more susceptible to digestion by MNase than others (13), resulting in under-representation of particular nucleosomes (34, 51).
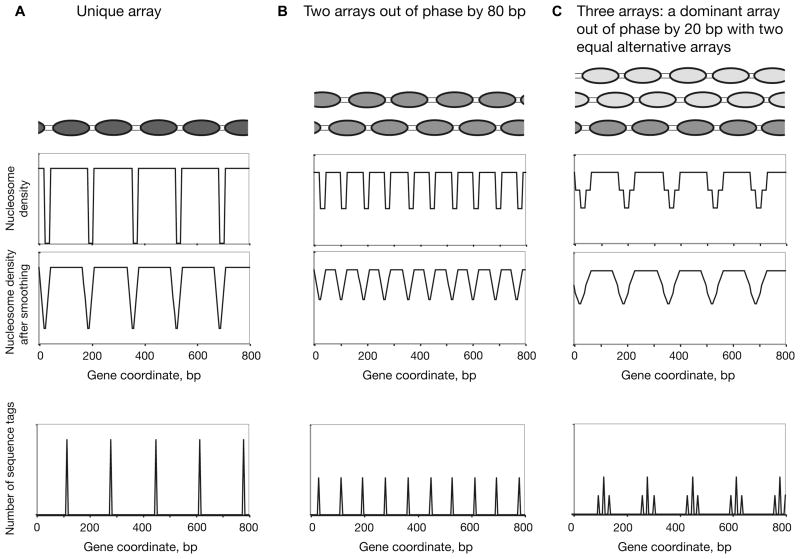
Are the genome-wide data consistent with multiple alternative arrays (Figure 3D)? Nucleosome sequencing has also revealed overlapping nucleosomes in yeast (43), although the genes given as examples exhibit more obvious dominant arrays than our data have indicated for CUP1 and HIS3 (18, 33). Overlapping positions appear to be a general feature of C. elegans chromatin (47). Data from genome-wide studies are typically presented as nucleosome occupancy maps for specific genes, with peaks and troughs interpreted as nucleosomes and linkers, respectively. These represent an averaged chromatin structure. Quantitative conversion of our monomer extension data for HIS3 chromatin to nucleosome density revealed a similar sinusoidal variation (Figure 3E), even though multiple arrays are present (18). We proposed that such patterns correspond to “interference” between the nucleosome density signals from overlapping nucleosomal arrays. Different patterns can be obtained depending on the phase and relative occupancy of each array (Figure 4). Such analysis suggests an explanation for the apparently “fuzzy” positioning of a large fraction of nucleosomes genome-wide, even though the majority of them must be present in ordered nucleosomal arrays.
Figure 4.
Model nucleosome density/occupancy profiles for unique and multiple nucleosomal arrays. Three different model array sets: (A) Unique array. (B) Two arrays with equal occupancies that are out of phase by 80 bp. (C) A set of three arrays (a dominant array accounting for 50% of the nucleosomes and two alternative arrays out of phase by +20 bp or −20 bp relative to the dominant array, each corresponding to 25% of the nucleosomes). Upper middle panels: High resolution nucleosome occupancy profiles calculated for each model, assuming 145 bp per nucleosome and 20 bp per linker, using a 5 bp window. Lower middle panels: Simulated low resolution occupancy profiles obtained by smoothing the high resolution profiles using a moving average of 25 bp. Note that two equal arrays (B) give twice as many peaks as the unique array (A) and that multiple arrays (C) can give a pattern similar to that for a unique array. Clearly, different combinations of arrays can give rise to a wide variety of nucleosome occupancy profiles (i.e., averaged chromatin structures). Lower panels: Simulated nucleosome sequencing profiles for the same arrays; each peak represents the inferred midpoint of a nucleosome. It is proposed that overlapping arrays such as those in B and C can account for the nucleosome occupancy profiles observed in high throughput analyses (often referred to as “fuzzy” positioning).
Nucleosome Spacing and a Nucleosome Positioning Code in Yeast
Is the positioning information or “code” inherent in DNA sequence utilised in vivo? Many studies have addressed this question, but they have generally relied on indirect end-labelling to map nucleosomes in vivo, which does not provide data with sufficient precision. We have mapped nucleosomes on the yeast CUP1 gene at high resolution both in vivo and in vitro (33, 52): the same overlapping positions are present, but their occupancies are different, indicating that DNA sequence does play a major role in determining nucleosome positions in vivo. The idea that genomic DNA might contain a positioning code has gained a major boost from genomewide studies (the analysis of microarray data is problematic given that nucleosome borders cannot be defined, but nucleosome sequencing is ideal). By comparing the positions of nucleosomes reconstituted on yeast genomic DNA in vitro with those observed in vivo, several studies have demonstrated that DNA sequence makes a major contribution to positioning genome-wide (45, 46, 49).
There is disagreement over the extent to which DNA sequence determines nucleosome positioning in vivo, based on the observation that genome-wide nucleosome occupancies in reconstituted chromatin (which contains only nucleosomes) differ significantly from those in chromatin in vivo (45, 46). This is not surprising: major differences in position occupancies should be expected, given the presence of many additional proteins in native chromatin. These include transcription factors, remodelling activities and RNA polymerases, which will strongly influence the local distribution of nucleosomes. Sequence-specific transcription factors bound to their cognate sites might act as barriers which sterically occlude nucleosomes, causing arrays to form to either side, which would be phased with respect to the DNA sequence. Theoretically, this “statistical positioning” (53) can occur even if sequence does not contribute to positioning; it depends instead on barrier locations and nucleosome spacing factors. However, it seems unlikely that nucleosomes would be assembled in total disregard of local sequence information. Instead, it may be imagined that as an array is formed by a nucleosome spacing factor, each histone octamer will adopt the local position with the highest affinity within the short length of DNA that is consistent with maintaining an average repeat length of 165 bp.
A question of major importance is the extent to which chromatin structure is dynamic. Consider the barriers involved in phasing: promoters usually contain several binding sites for sequence-specific factors, but their binding is reversible. Such barriers are therefore transient and will shift as different sites are occupied or vacated. The neighbouring nucleosomes, on the other hand, are bound irreversibly and mobilised only by chromatin remodelling activities. However, nucleosomal arrays might also be highly dynamic, if spacing factors can respond rapidly to alternating barriers, making use of the overlapping positions specified by the DNA sequence to re-position the nucleosomes in an array. For example, we may speculate that the binding of Gcn4p at its site in the HIS3 promoter might preclude the formation of arrays A3 and A4, allowing only arrays A1, A2 and D (Figure 3D). More generally, as gene expression patterns change in a cell, so will the locations of the barriers, with consequences for position occupancies.
We suggest that the nucleosome spacing factors responsible for forming arrays on either side of each barrier exploit the degeneracy of the nucleosome code, in that it specifies multiple overlapping positions. Some support for this hypothesis comes from experiments in vitro indicating that chicken β-globin DNA encodes positions with a spacing similar to that measured in native chicken chromatin (54) and that nucleosomes mapped on yeast CUP1 DNA (52) can be arranged into overlapping arrays with a spacing of ~165 bp. In the latter case, a comparison of the positions of reconstituted nucleosomes with those adopted in native chromatin revealed that all of the overlapping positions are encoded in CUP1 DNA; the positions are the same, but the occupancies are different (52). Furthermore, prokaryotic DNA yields a poor nucleosomal ladder in yeast cells (55), perhaps because it does not contain appropriate positioning signals (45).
In conclusion, it is proposed that the function of the nucleosome code is to facilitate nucleosome spacing, by encoding information specifying multiple overlapping arrays. Each nucleosomal array would utilise the set of encoded positions which gives a spacing closest to the required repeat length of ~160–165 bp. Such a code might facilitate the shunting of nucleosomes from one array to another by ATP-dependent remodelling machines (18).
Acknowledgments
I thank Gary Felsenfeld, Jeff Hayes and Rohinton Kamakaka for helpful comments on the manuscript. This research was supported by the Intramural Research Program of the NIH (NICHD).
Footnotes
This research was reported by the author in part at Albany 2009: The 16th Conversation (56).
References and Footnotes
- 1.Hewish DR, Burgoyne LA. Biochem Biophys Res Comm. 1973;52:504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- 2.van Holde KE. Springer Series in Molecular Biology: Chromatin. Springer-Verlag; 1988. [Google Scholar]
- 3.Wolffe A. Structure and Function. Academic Press; 1995. Chromatin. [Google Scholar]
- 4.Hörz W, Altenburger W. Nucl Acids Res. 1981;9:2643–2658. doi: 10.1093/nar/9.12.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dingwall C, Lomonossof GP, Laskey RA. Nucl Acids Res. 1981;9:2659–2673. doi: 10.1093/nar/9.12.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flick JT, Eissenberg JC, Elgin SCR. J Mol Biol. 1986;190:619–633. doi: 10.1016/0022-2836(86)90247-0. [DOI] [PubMed] [Google Scholar]
- 7.Noll M, Kornberg RD. J Mol Biol. 1977;109:393–404. doi: 10.1016/s0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- 8.Simpson RT. Biochem. 1978;17:5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- 9.An W, Leuba SH, van Holde KE, Zlatanova J. Proc Natl Acad Sci USA. 1998;95:3396–3401. doi: 10.1073/pnas.95.7.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nikitina T, Ghosh RP, Horowitz-Sherer RA, Hansen JC, Grigoryev SA, Woodcock CL. J Biol Chem. 2007;282:28237–28245. doi: 10.1074/jbc.M704304200. [DOI] [PubMed] [Google Scholar]
- 11.Pruss D, Hayes JJ, Wolffe AP. BioEssays. 1995;17:161–170. doi: 10.1002/bies.950170211. [DOI] [PubMed] [Google Scholar]
- 12.Thomas JO. Curr Opin Cell Biol. 1999;11:312–317. doi: 10.1016/S0955-0674(99)80042-8. [DOI] [PubMed] [Google Scholar]
- 13.McGhee JD, Felsenfeld G. Cell. 1983;32:1205–1215. doi: 10.1016/0092-8674(83)90303-3. [DOI] [PubMed] [Google Scholar]
- 14.Cockell M, Rhodes D, Klug A. J Mol Biol. 1983;170:423–446. doi: 10.1016/s0022-2836(83)80156-9. [DOI] [PubMed] [Google Scholar]
- 15.Thoma F, Bergman LW, Simpson RT. J Mol Biol. 1984;177:715–733. doi: 10.1016/0022-2836(84)90046-9. [DOI] [PubMed] [Google Scholar]
- 16.Wu C. Nature. 1980;286:854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- 17.Gregory PD, Schmid A, Zavari M, Münsterkötter M, Hörz W. EMBO J. 1999;18:6407–6414. doi: 10.1093/emboj/18.22.6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim Y, McLaughlin N, Lindstrom K, Tsukiyama T, Clark DJ. Mol Cell Biol. 2006;26:8607–8622. doi: 10.1128/MCB.00678-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimizu M, Roth SY, Szent-Gyorgyi C, Simpson RT. EMBO J. 1991;10:3033–3041. doi: 10.1002/j.1460-2075.1991.tb07854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayes JJ, Clark DJ, Felsenfeld G. Proc Nat Acad Sci USA. 1991;88:6829–6833. doi: 10.1073/pnas.88.15.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clark DJ, Felsenfeld G. Cell. 1992;71:11–22. doi: 10.1016/0092-8674(92)90262-b. [DOI] [PubMed] [Google Scholar]
- 22.Yenidunya A, Davey C, Clark DJ, Felsenfeld G, Allan J. J Mol Biol. 1994;237:401–414. doi: 10.1006/jmbi.1994.1243. [DOI] [PubMed] [Google Scholar]
- 23.Kim Y, Shen CH, Clark DJ. Methods. 2004;33:59–67. doi: 10.1016/j.ymeth.2003.10.021. [DOI] [PubMed] [Google Scholar]
- 24.Hamiche A, Sandaltzopoulos R, Gdula DA, Wu C. Cell. 1999;97:833. doi: 10.1016/s0092-8674(00)80796-5. [DOI] [PubMed] [Google Scholar]
- 25.Lowary PT, Widom J. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 26.Lowary PT, Widom J. Proc Natl Acad Sci USA. 1997;94:1183–1188. doi: 10.1073/pnas.94.4.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gottesfeld JM, Bloomer LS. Cell. 1980;21:751–760. doi: 10.1016/0092-8674(80)90438-9. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X, Hörz W. J Mol Biol. 1984;176:105–129. doi: 10.1016/0022-2836(84)90384-x. [DOI] [PubMed] [Google Scholar]
- 29.Fragoso GS, John S, Roberts MS, Hager GL. Genes Dev. 1995;9:1933–1947. doi: 10.1101/gad.9.15.1933. [DOI] [PubMed] [Google Scholar]
- 30.Costanzo G, di Mauro E, Negri R, Pereira G, Hollenberg C. J Biol Chem. 1995;270:11091–11097. doi: 10.1074/jbc.270.19.11091. [DOI] [PubMed] [Google Scholar]
- 31.Ambrose C, Lowman H, Rajadhyaksha A, Blasquez V, Bina M. J Mol Biol. 1990;214:875–884. doi: 10.1016/0022-2836(90)90342-J. [DOI] [PubMed] [Google Scholar]
- 32.Ravindra A, Weiss K, Simpson RT. Mol Cell Biol. 1999;19:7944–7950. doi: 10.1128/mcb.19.12.7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen CH, Leblanc BP, Alfieri JA, Clark DJ. Mol Cell Biol. 2001;21:534–547. doi: 10.1128/MCB.21.2.534-547.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Travers A, Caserta M, Churcher M, Hiriart E, Di Mauro E. Mol Biosyst. 2009:10.1039. doi: 10.1039/b907227f. [DOI] [PubMed] [Google Scholar]
- 35.Yuan G, Liu Y, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 36.Whitehouse I, Rando OJ, Delrow J, Tsukiyama T. Nature. 2007;450:1031–1036. doi: 10.1038/nature06391. [DOI] [PubMed] [Google Scholar]
- 37.Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C. Nature Gen. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 38.Studitsky V, Clark DJ, Felsenfeld G. Cell. 1994;76:371–382. doi: 10.1016/0092-8674(94)90343-3. [DOI] [PubMed] [Google Scholar]
- 39.Polach KJ, Widom J. J Mol Biol. 1995;254:130–149. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- 40.Fascher KD, Schmitz J, Hörz W. EMBO J. 1990;9:2523–2528. doi: 10.1002/j.1460-2075.1990.tb07432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boyes J, Felsenfeld G. EMBO J. 1996;15:2496–2507. [PMC free article] [PubMed] [Google Scholar]
- 42.Kim Y, Clark DJ. Proc Natl Acad Sci USA. 2002;99:15381–15386. doi: 10.1073/pnas.242536699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Field Y, Kaplan N, Fondufe-Mittendorf Y, Moore IK, Sharon E, Lubling Y, Widom J, Segal E. PLoS Comp Biol. 2008;4:1–25. doi: 10.1371/journal.pcbi.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Albert I, Mavrich TN, Tomsho LP, Zanton SJ, Schuster SC, Pugh BF. Nature. 2008;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y, Moqtaderi Z, Rattner BP, Euskirchen G, Snyder M, Struhl K. Nat Struc Mol Biol. 2009;16:847–852. doi: 10.1038/nsmb.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, Segal E. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Valouev A, Ichikawa J, Tonthat T, Stuart J, Ranade S, Peckham H, Zeng K, Malek JA, Costa G, McKernan K, Sidow A, Fire A, Johnson SM. Gen Res. 2008;18:1051–1063. doi: 10.1101/gr.076463.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Satchwell SC, Drew HR, Travers AA. J Mol Biol. 1986;191:659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- 49.Segal E, Fondufe-Mittendorf Y, Chen L, Thåström A, Field Y, Moore IK, Wang JZ, Widom J. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiang C, Pugh BF. Nat Rev Genet. 2009 doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weiner A, Hughes A, Yassour M, Rando OJ, Friedman N. Gen Res. 2009 doi: 10.1101/gr.098509.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shen CH, Clark DJ. J Biol Chem. 2001;276:35209–35216. doi: 10.1074/jbc.M104733200. [DOI] [PubMed] [Google Scholar]
- 53.Fedor MJ, Lue NF, Kornberg RD. J Mol Biol. 1988;204:109–127. doi: 10.1016/0022-2836(88)90603-1. [DOI] [PubMed] [Google Scholar]
- 54.Davey C, Pennings S, Meersseman G, Wess TJ, Allan J. Proc Natl Acad Sci USA. 1995;92:11210–11214. doi: 10.1073/pnas.92.24.11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tong W, Kulaeva OI, Clark DJ, Lutter LC. J Mol Biol. 2006;361:813–822. doi: 10.1016/j.jmb.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 56.Clark DJ. The Dynamic Chromatin Structure of Transcriptionally Active Yeast Genes. Abstracts of Albany 2009: 16th Conversation; June 16–20; Albany, New York, USA. Abstract Date Received: November 12, 2009. [Google Scholar]