Abstract
Both transcription and RNA decay are critical for normal gene regulation. Arabidopsis mutants with defects in VARICOSE (VCS), a decapping complex scaffold protein, lack mRNA decapping and 5′-to-3′ decay. These mutants show either severe or suppressed phenotypes, depending on the Arabidopsis accession. Here, we show that the molecular basis for this variation is the SUPPRESSOR OF VARICOSE (SOV), a locus that encodes a conserved, cytoplasmically localized RRP44-like RNaseII-domain protein. In vivo RNA decay assays suggest that active forms of this protein carry out decay on mRNA substrates that overlap with those of the decapping complex. Members of this conserved gene family encode proteins lacking the PIN domain, suggesting that SOV is not a functional component of the RNA exosome.
Keywords: natural variation
The decay of mRNA allows rapid changes in mRNA populations and so enables fine-tuned cellular responses to new stimuli (1–3). Cytoplasmic mRNAs have two structural stability determinants: the 5′ cap, a 7-methyl guanine residue linked to the mRNA via a 5′-5′ triphosphate bond, and the 3′ poly(A)+ tail. Decay initiates by removal of one or both of these stability determinants or by internal cleavage. Typically shortening of the poly(A)+ tail is the earliest event (4), with the major deadenylase being the Ccr4–Caf1–Not complex (5). mRNAs can also be targeted for decay by a 3′ modification such as by addition of Us or CUCU, and these modifications appear to stimulate mRNA decapping (6, 7). Decapping hydrolyzes the 5′-5′ linkage, which causes the mRNA to be vulnerable to 5′-to-3′ exoribonucleases such as XRN1 in animals and yeast and XRN4 in plants (8, 9). Decapping is often carried out in cytoplasmic foci called processing bodies (10–12), although this localization is apparently dispensable because decapping also occurs in dispersed locations and on polysome-bound mRNAs (13–15). The decapping complex, RNA exosome, and deadenylases all play established roles in mRNA decay, although the composition of these complexes and their mechanisms for RNA substrate recognition are still not well understood. Moreover, the very wide range of mRNA stabilities (16, 17), and the specificity of mRNAs for particular decay pathways (18, 19), indicate a complex regulatory network governing mRNA decay that remains poorly understood. Here, we used natural variation among Arabidopsis accessions to identify a suppressor of vcs. The SUPPRESSOR OF VARICOSE (SOV) encodes a highly conserved cytoplasmic RNaseII domain protein. This protein shows mRNA decay activity in vivo, and its substrates overlap with those of the mRNA decapping enzyme.
Results
SOV Is a Dominant Suppressor of vcs Present in Many Arabidopsis Accessions.
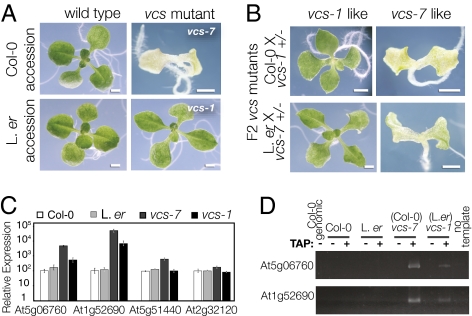
Forward genetic screens led to identification of Arabidopsis developmental mutants with defects in the mRNA decapping complex (18, 20). VCS encodes the decapping complex scaffold (homolog of Ge-1/HEDLS) (18, 20–22) and is also required for miRNA-mediated translational regulation (23). Mutants with vcs defects display two distinct phenotypes. A severe vcs phenotype, which includes root growth arrest, misshapen chlorotic cotyledons, loss of shoot stem cells, and failure to initiate leaves, occurs in vcs-6–vcs-8 (18, 24), whereas a mild phenotype, which includes apparently normal cotyledons, functional stem cells, and modestly defective leaves, occurs in vcs-1–vcs-5 (18, 20) (Fig. 1A). All mutants showing the severe phenotype are alleles in the Col-0 accession, whereas all mutants showing the mild phenotype were isolated in the Landsberg erecta (L. er) accession. We tested whether these vcs phenotypes depended on genetic background. Both vcs-1 (L. er) and vcs-7 (Col-0) produced indistinguishable suppressed and severe phenotypes in F2s after a cross to the complementary accession (Fig. 1B). This analysis confirmed that different phenotypes were accession-dependent, and it indicated that the L. er suppressor was unlinked. Moreover, this suppressor is dominant, as vcs-1/vcs-7 transheterozygotes showed a partially suppressed phenotype that was indistinguishable from vcs-1 (Fig. S1A). We call this locus SUPPRESSOR OF VARICOSE (SOV).
Fig. 1.
The L.er accession encodes a suppressor that modifies vcs mutant RNA accumulation defects. (A) The Col-0 and L.er accessions appear very similar, but vcs alleles isolated in these two genetic backgrounds have different phenotypes. In Col-0, the vcs-7 mutant is chlorotic and fails to make leaves, whereas the vcs-1 mutant, isolated in L.er, is green and produces broad leaves. (B) Both suppressed and severe vcs phenotypes are among the F2 after crosses of vcs-1 and vcs-7 mutants to Col-0 and L. er, respectively. (C) The relative expression of four RNAs in 3-d seedlings of both wild-type accessions and in vcs-1 (L.er) and vcs-7 (Col-0), determined by using quantitative real-time RT-PCR. These mRNAs accumulate to high levels in vcs-7, and in each case, the RNA accumulates to a lower level in the vcs-1 mutant. Error bars indicate SE (s.e.m.). (D) RNA ligation assay for the presence of a 5′ cap. Tobacco acid pyrophosphatase (TAP) addition indicated by a (+). Note that PCR products are produced in vcs-7 and vcs-1 mutants only after incubation with TAP, indicating that the abundant RNAs quantified in C retain their 5′ cap. (Scale bars: 1 mm.)
SOV Modifies vcs mRNA Accumulation Defects Independently of Decapping.
SOV appeared to carry out an RNA-related function because four RNAs that overaccumulate in vcs-7 (18) showed an intermediate expression level in vcs-1 (L. er accession) (Fig. 1C). We reasoned that SOVL.er might modify RNA levels in vcs mutants by restoring decapping. To test this idea, we carried out a ligation-based cap assay (25). High-abundance mRNAs in vcs-1 (L. er) retained their 5′-cap (Fig. 1D). This result suggested that SOVL.er modifies mRNA levels in vcs mutants by some different mechanism.
Molecular Identification of SOV.
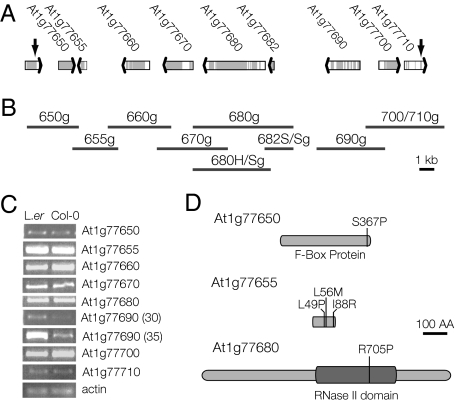
We mapped SOV to chromosome one by using codominant PCR markers and confirmed the map position by using recombinant inbred lines (Fig. S2) (26). The SOV interval contained nine candidate genes (Fig. 2A). We reasoned that SOV might confer suppression through either accession-specific differences in expression level or accession-specific differences in protein features. All nine of the candidate genes were expressed in young seedlings of both L. er and Col-0 (Fig. 2C); three showed differential Col-0/L.er expression levels (At1g77660, At1g77690, and At1g77700) and another three coded for slightly different proteins (At1g77650, At1g77655, and At1g77680) (Fig. 2D) (27, 28). To identify SOV, we generated rescue clones that included all candidate genes (L. er genomic DNA; Fig. 2B) and tested each for its ability to partially suppress the vcs-7 phenotype.
Fig. 2.
Molecular characterization of the SUPPRESSOR OF VARICOSE. (A) The mapping interval that contained SOV included nine genes (depicted by broad arrows) and was defined by recombination break points indicated by the heavy vertical black arrows. (B) The L.er genomic DNA used for complementation analysis is represented by the dark gray bars, which were positioned to correspond to the region shown above the bars in A. Each rescue construct contained ≈2-kb upstream sequences, 1-kb downstream sequences, and all introns and exons. Note that the 5-kb scale bar works for both A and B. (C) Semiquantitative RT-PCR analysis of candidate gene expression in 7-d seedlings. Three of the candidate genes showed different levels of expression in the two accessions. Note that for At1g77690, reactions with different cycle numbers are shown to confirm differential expression. (D) Cartoon representation of the protein polymorphisms found in three SOV candidate genes. (Scale bar: 100 amino acids.)
The severe vcs-7 phenotype was not affected in transgenic plants bearing clones 650g, 655g, 660g, 670g, 690g, and 700/710g; however, clone 680g conferred dominant partial suppression matching that of vcs-1 (Fig. 3A and Table S1). This result indicated that clone 680g contained SOVL.er. The genomic DNA included in this clone encoded two Arabidopsis genes: At1g77680 and At1g77682. These genes were independently tested by 680H/Sg (containing only At1g77680) and 682S/Sg (containing only At1g77682). Clone 680H/Sg, but not clone 682S/Sg, conferred the partially suppressed vcs-1 (L. er) phenotype to vcs-7 (Col-0; Table S1), indicating that At1g77680 was SOV.
Fig. 3.
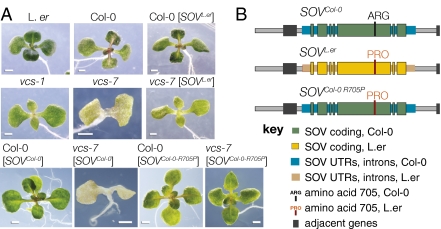
Suppression of vcs is conferred by At1g77680 encoding a proline at position 705. (A) The effect of SOV genomic clones transformed into the wild-type and vcs-7 mutants; only SOV encoding PRO-705 confer the partially rescued vcs phenotype. Transgenic wild-type carrying SOVL.er appear normal, and vcs-7 carrying SOVL.er appear indistinguishable from vcs-1. This partially suppressed vcs-1 phenotype is mostly easily seen by the presence of leaves and green organ color. The partial suppression required the L.er version of SOV because vcs-7 transgenic plants carrying an extra copy of SOVCol-0 were not suppressed unless SOVCol-0 had been modified to resemble the L.er version at amino acid 705. (B) Rescue constructs designed to test the importance of PRO at position 705 and used in the experiments shown in A.
SOV Encodes an RNase II Domain Protein.
At1g77680 encodes a 1,055-amino acid RNaseII domain protein with an L. er/Col-0 polymorphism at amino acid 705 (Fig. 2D) (27, 28). Among the 20 sequenced Arabidopsis accessions, only Col-0 encodes ARG-705, whereas L.er and the 18 other sequenced accessions encode PRO-705. To test whether PRO-705 was important for SOV activity, we analyzed 16 Arabidopsis accessions, all of which code for PRO at position 705, for SOV activity. Each accession was crossed to vcs-7 and, in every case, the F2 plants segregated for both severe and suppressed vcs mutant phenotypes (Table S2). These data were consistent with partial suppression arising from At1g77680 with a PRO at amino acid 705.
To directly test the importance of PRO-705, we generated SOVCol-0-R705P, a modified Col-0 genomic clone containing At1g77680, but mutagenized to encode PRO at position 705 instead of ARG (Fig. 3B). Simply increasing the SOV copy number by transforming vcs-7 with the unaltered Col-0 genomic clone did not alter the vcs-7 phenotype; however, vcs-7 mutants carrying SOVCol-0-R705P showed the partially suppressed vcs-1 phenotype (Fig. 3A). This result confirmed SOV’s identity as At1g77680 and highlighted the potential importance of the domain containing amino acid 705.
Evolutionary Conservation of SOV-Like Proteins.
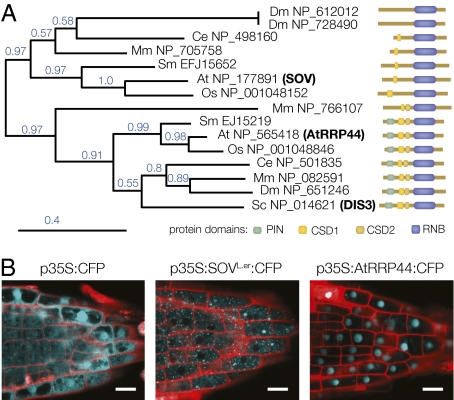
SOV/At1g77680, and its closest Arabidopsis homolog, At2g17510, both encode RNase II-domain proteins similar to RRP44/DIS3, which is the only known catalytic component of the yeast and human exosome (Fig. S3A) (29–31). The product of At2g17510 appears more likely to be the Arabidopsis RRP44 homolog; it shows 41% identity (58% similarity) to the yeast RRP44 protein, whereas SOV only shows 29% identity (44% similarity). To further evaluate the relationship between these two Arabidopsis proteins and RRP44, we identified SOV homologs from Mus musculus, Caenorhabditis elegans, Drosophila melanogaster, Oryza sativa, and Selaginella moellendorffii, and along with the yeast RRP44/DIS3, generated a phylogenetic tree (32) based on their RNA binding domains (RNB) (Fig. 4A). This analysis revealed two gene clusters: One cluster included the yeast exosome subunit RRP44/DIS3 along with proteins from each of the species examined, and the second cluster included At1g77680/SOV and uncharacterized proteins from each of these other species. Because the tree was based solely on the RNB, we tested the validity of these two groupings by comparing their domain organization (Fig. 4A). All proteins within the cluster that included RRP44/DIS3 contained the essential RRP44 domains, including a pilT N-terminal (PIN) domain and two cold shock domains (CSDs). By contrast, all proteins within the SOV cluster lacked the PIN domain and one or more of the CSD domains. The PIN domain is crucial for RRP44/DIS3 binding to the exosome core (33–35), thus its absence from the SOV cluster suggests that these proteins do not substitute for RRP44 in the exosome. Moreover, because SOV-like proteins have been conserved in these distant lineages, the SOV pathway might play important roles in RNA metabolism.
Fig. 4.
SOV encodes a conserved protein that localizes to the cytoplasm. (A) A phylogenetic tree, built using amino acid sequences of RNA binding domains (RNB) from SOV and SOV-like proteins. This analysis revealed that SOV as belonging to a conserved cohort that was distinct from RRP44 proteins. To the right, the domain organization of each analyzed protein is depicted. The RRP44 cohort all contained the typical RRP44 domains, whereas those of the SOV cohort all lacked the PIN domain and contained fewer CSD domains. Bootstrap values are indicated in blue. At, Arabidopsis; Sc, yeast; Mm, mouse; Ce, Caenorhabditis elegans; Dm, Drosophila; Os, rice; Sm, Selaginella. (B) Confocal image of CFP expression driven from the 35S promoter. CFP alone (Left) localizes throughout the cytoplasm and nucleus, whereas a translational fusion to SOV localizes to discrete cytoplasmic foci (Center) and a translational fusion to AtRRP44 largely localizes to the nucleus (Right). (Scale bars: 1 mm.)
SOV Functions in mRNA Decay.
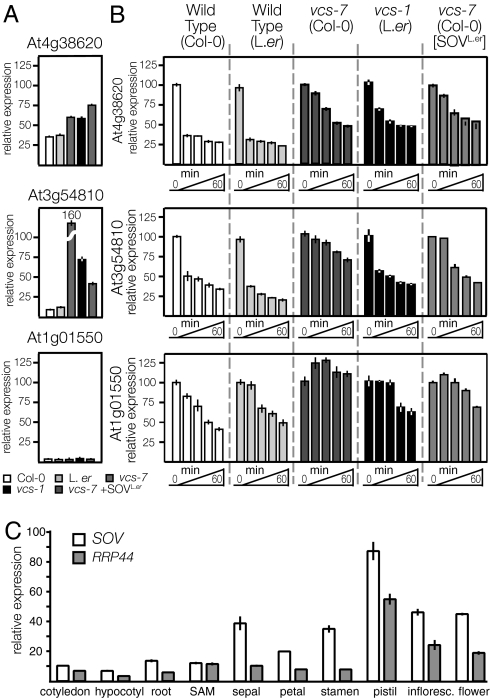
The conserved RNase II domain in SOV suggested that it catalyzed RNA decay (29). We compared the in vivo activity of SOVCol-0 and SOVL.er by measuring both the steady-state levels and the RNA decay kinetics of three seedling-expressed mRNAs (analyzed in ref. 18) in five genotypes: Col-0, L.er, vcs-7, vcs-1, and vcs-7 [SOVL.er] (Fig. 5 A and B). The RNA for At4g38620 is fairly abundant and accumulated to higher levels in all three genotypes that included vcs. We found similar exponential decay kinetics in the two wild-type accessions and similar reduced decay kinetics in vcs-1, vcs-7, and vcs-7 [SOVL.er]. Thus, this mRNA is normally a substrate for decapping because vcs mutants, which lack decapping and 5′-to-3′ decay, all showed reduced decay kinetics. However, because rapid decay still occurred in vcs mutants, and this residual decay rate was not affected by the addition of SOVL.er, this mRNA must also be a substrate for some other RNA decay pathway.
Fig. 5.
SOV functions in cytoplasmic mRNA decay. (A) Steady-state levels of three mRNAs, relative to the actin internal control. (B) RNA decay analysis comparing three mRNAs in five genotypes: wild-type (Col-0), wild-type (L.er), vcs mutants (vcs-7 in Col-0 and vcs-1 in L.er) and transgenic vcs-7 (Col-0) containing SOVL.er. (C) Relative expression of RRP44 and SOV in various organs of L.er plants. Both genes are expressed in all tissues analyzed; the relative abundance of SOV and AtRRP44 varied among the organs, but SOV expression was always greater than that of AtRRP44. All three analyzes used real-time RT-PCR. Error bars indicate SEM.
The abundance of At3g54810 was strongly increased in vcs mutants, especially in vcs-7, which lacks both decapping and SOV activity (Fig. 5A). Similar exponential decay kinetics was found for the two wild-type accessions (Fig. 5B). However, in vcs-7, which lacks both decapping and SOV activity, this mRNA's decay rate was strongly diminished. By contrast, both vcs genotypes containing an active SOV (vcs-1 and vcs-7 [SOVL.er]) showed intermediate decay kinetcs for this mRNA. The strong and additive effects of SOV and VCS loss on the steady-state level of At3g54810 mRNA indicated a pivotal role for RNA decay in its regulation. Consistent with the high steady state, decay rates were strongly affected in VCS and SOVL.er loss-of-function backgrounds, indicating that these two activities play major roles in this mRNA's decay.
The RNA for At1g01550 RNA is very low abundance (Fig. 5A). It showed similar decay kinetics in the two wild-type accessions; however, decay was undetectable in vcs-7, which lacks both decapping and SOV (Fig. 5B). By contrast, both vcs-1 and vcs-7 [SOVL.er] showed similar modest rates of decay of this mRNA. Because the addition of an active SOV stimulated decay, this mRNA is a substrate for SOV. Moreover, because decay was undetectable in vcs-7, decapping and SOV appear to be the only pathways for this RNA's degradation.
SOV and AtRRP44 Show Different Patterns of Expression and Localization.
To further understand the relationship between SOV and RRP44, we analyzed T-DNA insertion alleles for both genes (Fig. S1 A and D) (36). The sov-1 mutant, which had strongly diminished SOV RNA levels, was indistinguishable from Col-0. In contrast, insertional mutations in RRP44 disrupted female gametophyte development (Fig. S4A). Because insertional mutants were generated in the Col-0 accession, rrp44 mutants also lacked an active SOV. Therefore, to determine whether SOVL.er could compensate for the loss of RRP44, we generated SOVL.er/SOVL.er rrp44/+ plants and characterized their progeny. We reasoned that if the mRNA substrates of SOVL.er and RRP44 overlapped, then SOVL.er rrp44-1 female gametophytes might show a less severe phenotype. We tested this idea by using an endosperm marker (37) and found that despite the presence of SOVL.er, these plants still showed an equally penetrant female gametophyte defect (Fig. S4B). These data confirm other studies demonstrating that the Arabidopsis gametophyte has an essential requirement for the exosome (38) and indicate that, in female gametophytes, AtRRP44 carries out essential functions that cannot be supplied by SOV.
We also compared SOV and RRP44 intracellular localization and mRNA expression patterns. We generated transgenic plants with translational fusions to CFP that were expressed from the CaMV 35S promoter and that rescued the mutant phenotypes. The 35S:SOVL.er-CFP fusion construct showed strictly cytoplasmic localization (Fig. 4B). By contrast, 35S:AtRRP44-CFP showed predominantly nuclear localization. In L.er, both genes were broadly expressed and their relative transcript levels varied (e.g., nearly identical expression in the SAM, whereas in stamen tissues, SOV shows ≈3-fold higher expression; Fig. 5C). These distinct expression and localization patterns support distinct functions for SOV and AtRRP44.
The Col-0 variant of SOV, with an ARG at position 705 instead of PRO, appeared to lack activity, because vcs-7 sov-1 double mutants were indistinguishable from the vcs-7 single mutant (in Col-0) (Table S3). How the single amino acid change at position 705 affects activity is unknown. This residue is located within the RNase II domain and appears to be flanked by putative RNA contact sites (Fig. S3A) (33, 39). Although comparison with the RRP44 structure suggested that this R group projects outwards, away from the enzyme's active regions (Fig. S3 B–D), a PRO in this position appears well conserved. Among the amino acid sequences included in the phylogenetic analysis (Fig. 4A), the only sequences that lacked a PRO at this position were the RRP44 sequences from mouse and Drosophila, where SER is substituted for the PRO. In SOV, the loss of activity associated with an ARG substitution for PRO might be due to protein destabilization or it might interfere with assembly of a putative larger complex.
Discussion
The control of gene expression requires a balance between synthetic and degradative processes. The wide range of half-lives observed for mRNAs implies the existence of regulatory networks that initiate decay (e.g., by decapping), perhaps by selective targeting to degradative machinery. A complete understanding of this regulatory network requires the identification of mRNA decay machinery.
Our data suggest that SOVL.er encodes an RRP44-like protein, which is an active RNase. The importance of SOV was uncovered in Arabidopsis vcs mutants, which have mRNA decapping defects. For most Arabidopsis accessions, vcs mutants have a relatively mild phenotype; however, the Col-0 accession conditions a severe vcs phenotype because SOV is inactive. Phenotypic consequences of the loss of SOV in Col-0 appear to be largely masked by the activity of the exosome and decapping/XRN4.
The extent to which SOV and decapping activities overlap is revealed by the profound suppression of vcs-7 by SOVL.er. Suppressed phenotypes include root development, root hair formation, cotyledon vascular patterning, establishment of the shoot apical meristem, and leaf development. This broad range of tissues affected by SOVL.er suggests that SOV might be catalytically active on diverse mRNA substrates. In addition, SOV activity appears to be selective, because decay assays found that SOVL. er only enhanced decay of specific mRNAs.
By contrast, SOV function appears to be distinct from that of AtRRP44. Analysis of AtRRP44:CFP localization suggests that AtRRP44 might mostly function in the nucleus. This pattern is consistent with the effects of exosomal subunit knockdown plants, where the major primary defects were observed in nuclear RNAs, and only a relatively small subset of mRNAs was affected (38). Future work identifying the mRNAs substrates of different decay pathways, and the molecular mechanisms conferring specificity, will clarify this area.
Determining how SOV functions as a major cytoplasmic 3′-to-5′ exoribonuclease is a high priority for future research in this field. Although SOV is very similar to AtRRP44 (34% identity, 50% similarity), it lacks the PIN domain, which forms the binding interface between RRP44 and the exosome core (27–29). Thus, it is unlikely that SOV functions in association with the exosome core. However, SOV appears to function in the context of a larger macromolecular assemblage, because SOV:CFP translational fusions revealed microscopic cytoplasmic particles. Given the conservation of SOV with RRP44-like proteins of animals, we expect functionally relevant interaction partners to also be conserved in animals.
Materials and Methods
Plant Growth and Materials.
Photographed seedlings were grown on sterile growth medium (2.16 g of MS salts, 0.5 g of Mes, 10 g of sucrose per liter, 7.5% phytagar at pH 5.8) in a 16 °C growth chamber. Seedlings for analysis of steady-state mRNA levels (3-d) and for RNA decay (4-d) were grown in sterile conditions at 22 °C. To analyze SOV and RRP44 relative expression, vegetative organs (cotyledons, shoot apex, hypocotyl, and roots) were collected from 5-d seedlings, or for reproductive tissues from 5-wk plants, again grown at 22 °C.
The varicose (vcs) alleles were described (18, 20). The recombinant inbred lines (RILs) (26) were obtained from Nottingham Arabidopsis Stock Centre, and Arabidopsis accessions were obtained from Arabidopsis Biological Resource Center.
RNA Decay Analysis.
RNA decay assays were carried out as described (40). Four-day-old seedlings of Col-0, L.er, vcs-7, vcs-1, and vcs-7 [SOVL.er] were collected, pretreated for 30 min, and then supplied with cordycepin (Sigma) to a final concentration of 1 mM. Samples were collected before cordycepin addition (0 min), and 15, 30, 45, and 60 min after cordycepin addition. See SI Materials and Methods for controls carried out to test the efficacy of the cordycepin treatment (Fig. S5). Tissue for each time point was flash-frozen in liquid nitrogen and stored at −80 °C. RNA was purified by using Qiagen RNeasy kit, and cDNA was generated by using random primers. To quantify specific mRNAs, we carried out quantitative RT-PCR (qRT-PCR) by using the Lightcycler and FastStart DNA master SYBR Green 1 master mix (both by Roche). RNA quality was tested by using the Bioanalyzer 2100 (Agilent Technologies). cDNA quality was assessed by using different reference genes (detailed information listed in SI Materials and Methods). Each qRT-PCR analysis used a minimum of three biological replicas and two technical replicas (6 reactions total). Transcript abundance was expressed as a ratio relative to internal control for steady-state mRNAs (Fig. 5 A and C), relative to wild-type levels for individual mRNAs accumulation (Fig. 1C), or relative to pretreatment (−15 min) levels (for the cordycepin control, Fig. S1), or relative to the time 0 sample for RNA decay (Fig. 5B).
RACE Analysis to Assess Presence of 5′ Cap on vcs-1 mRNAs.
RNA ligase-mediated (RLM) RACE (Ambion) was carried out by using 10 μg of total RNA for samples isolated from Col-0, vcs-7, L.er, and vcs-1. Duplicate ligations were carried out for samples digested with Tobacco Acid Pyrophosphatase (TAP) to remove the 5′ cap and for untreated samples. Ligation efficiency was then analyzed by reverse transcription reactions using random primers followed by nested PCR primers.
Localization of SOV and AtRRP44.
SOV and AtRRP44 coding regions were cloned into Gateway vector pEarly102 (41). The constructs were transformed into Col-0 and vcs-7/+ by floral dipping (42). Confocal was performed on the roots of 4-d-old seedlings that grow at 22 °C.
Additional detailed methods are available in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the Arabidopsis Biological Resource Center for providing seed stocks; Martin Horvath for assistance with protein structure; Ed King for assistance with confocal microscopy; Richard Clark, Gary Drews, members of the L.E.S. and Drews laboratory groups, Dmitry Belostotsky, Marylou Spencer, Tanya Hooker, and Ravi Kumar for useful discussions; and anonymous reviewers for useful suggestions. This work was supported by National Science Foundation Grant NSF IOS 0642118 (to L.E.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1007060107/-/DCSupplemental.
References
- 1.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 2.Belostotsky DA, Sieburth LE. Kill the messenger: mRNA decay and plant development. Curr Opin Plant Biol. 2009;12:96–102. doi: 10.1016/j.pbi.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Wilusz CJ, Wilusz J. Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet. 2004;20:491–497. doi: 10.1016/j.tig.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 4.Couttet P, Fromont-Racine M, Steel D, Pictet R, Grange T. Messenger RNA deadenylylation precedes decapping in mammalian cells. Proc Natl Acad Sci USA. 1997;94:5628–5633. doi: 10.1073/pnas.94.11.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Decker CJ, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: Evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 6.Rissland OS, Norbury CJ. Decapping is preceded by 3′ uridylation in a novel pathway of bulk mRNA turnover. Nat Struct Mol Biol. 2009;16:616–623. doi: 10.1038/nsmb.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morozov IY, Jones MG, Razak AA, Rigden DJ, Caddick MX. CUCU modification of mRNA promotes decapping and transcript degradation in Aspergillus nidulans. Mol Cell Biol. 2010;30:460–469. doi: 10.1128/MCB.00997-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu CL, Stevens A. Yeast cells lacking 5′—>3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol Cell Biol. 1993;13:4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kastenmayer JP, Green PJ. Novel features of the XRN-family in Arabidopsis: Evidence that AtXRN4, one of several orthologs of nuclear Xrn2p/Rat1p, functions in the cytoplasm. Proc Natl Acad Sci USA. 2000;97:13985–13990. doi: 10.1073/pnas.97.25.13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cougot N, Babajko S, Séraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson P, Kedersha N. RNA granules. J Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu W, Sweet TJ, Chamnongpol S, Baker KE, Coller J. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature. 2009;461:225–229. doi: 10.1038/nature08265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu W, Petzold C, Coller J, Baker KE. Nonsense-mediated mRNA decapping occurs on polyribosomes in Saccharomyces cerevisiae. Nat Struct Mol Biol. 2010;17:244–247. doi: 10.1038/nsmb.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol. 2007;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narsai R, et al. Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana. Plant Cell. 2007;19:3418–3436. doi: 10.1105/tpc.107.055046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, et al. Precision and functional specificity in mRNA decay. Proc Natl Acad Sci USA. 2002;99:5860–5865. doi: 10.1073/pnas.092538799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goeres DC, et al. Components of the Arabidopsis mRNA decapping complex are required for early seedling development. Plant Cell. 2007;19:1549–1564. doi: 10.1105/tpc.106.047621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiao Y, Riechmann JL, Meyerowitz EM. Transcriptome-wide analysis of uncapped mRNAs in Arabidopsis reveals regulation of mRNA degradation. Plant Cell. 2008;20:2571–2585. doi: 10.1105/tpc.108.062786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deyholos MK, et al. VARICOSE, a WD-domain protein, is required for leaf blade development. Development. 2003;130:6577–6588. doi: 10.1242/dev.00909. [DOI] [PubMed] [Google Scholar]
- 21.Yu JH, Yang WH, Gulick T, Bloch KD, Bloch DB. Ge-1 is a central component of the mammalian cytoplasmic mRNA processing body. RNA. 2005;11:1795–1802. doi: 10.1261/rna.2142405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fenger-Grøn M, Fillman C, Norrild B, Lykke-Andersen J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 23.Brodersen P, et al. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–1190. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- 24.Xu J, Yang JY, Niu QW, Chua NH. Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell. 2006;18:3386–3398. doi: 10.1105/tpc.106.047605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gazzani S, Lawrenson T, Woodward C, Headon D, Sablowski R. A link between mRNA turnover and RNA interference in Arabidopsis. Science. 2004;306:1046–1048. doi: 10.1126/science.1101092. [DOI] [PubMed] [Google Scholar]
- 26.Lister C, Dean C. Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J. 1993;4:745–750. doi: 10.1046/j.1365-313x.1996.10040733.x. [DOI] [PubMed] [Google Scholar]
- 27.Clark RM, et al. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science. 2007;317:338–342. doi: 10.1126/science.1138632. [DOI] [PubMed] [Google Scholar]
- 28.Zeller G, et al. Detecting polymorphic regions in Arabidopsis thaliana with resequencing microarrays. Genome Res. 2008;18:918–929. doi: 10.1101/gr.070169.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frazão C, et al. Unravelling the dynamics of RNA degradation by ribonuclease II and its RNA-bound complex. Nature. 2006;443:110–114. doi: 10.1038/nature05080. [DOI] [PubMed] [Google Scholar]
- 30.Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 31.Dziembowski A, Lorentzen E, Conti E, Séraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- 32.Dereeper A, et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36(Web Server issue):W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang HW, et al. Architecture of the yeast Rrp44 exosome complex suggests routes of RNA recruitment for 3′ end processing. Proc Natl Acad Sci USA. 2007;104:16844–16849. doi: 10.1073/pnas.0705526104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schneider C, Leung E, Brown J, Tollervey D. The N-terminal PIN domain of the exosome subunit Rrp44 harbors endonuclease activity and tethers Rrp44 to the yeast core exosome. Nucleic Acids Res. 2009;37:1127–1140. doi: 10.1093/nar/gkn1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaeffer D, et al. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat Struct Mol Biol. 2009;16:56–62. doi: 10.1038/nsmb.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 37.Steffen JG, Kang IH, Macfarlane J, Drews GN. Identification of genes expressed in the Arabidopsis female gametophyte. Plant J. 2007;51:281–292. doi: 10.1111/j.1365-313X.2007.03137.x. [DOI] [PubMed] [Google Scholar]
- 38.Chekanova JA, et al. Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell. 2007;131:1340–1353. doi: 10.1016/j.cell.2007.10.056. [DOI] [PubMed] [Google Scholar]
- 39.Lorentzen E, Basquin J, Tomecki R, Dziembowski A, Conti E. Structure of the active subunit of the yeast exosome core, Rrp44: Diverse modes of substrate recruitment in the RNase II nuclease family. Mol Cell. 2008;29:717–728. doi: 10.1016/j.molcel.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 40.Gutierrez RA, Ewing RM, Cherry JM, Green PJ. Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: Rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc Natl Acad Sci USA. 2002;99:11513–11518. doi: 10.1073/pnas.152204099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Earley KW, et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- 42.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.