Abstract
Replication and transcription of viral RNA genomes rely on host-donated proteins. Qβ virus infects Escherichia coli and replicates and transcribes its own genomic RNA by Qβ replicase. Qβ replicase requires the virus-encoded RNA-dependent RNA polymerase (β-subunit), and the host-donated translational elongation factors EF-Tu and -Ts, as active core subunits for its RNA polymerization activity. Here, we present the crystal structure of the core Qβ replicase, comprising the β-subunit, EF-Tu and -Ts. The β-subunit has a right-handed structure, and the EF-Tu:Ts binary complex maintains the structure of the catalytic core crevasse of the β-subunit through hydrophobic interactions, between the finger and thumb domains of the β-subunit and domain-2 of EF-Tu and the coiled-coil motif of EF-Ts, respectively. These hydrophobic interactions are required for the expression and assembly of the Qβ replicase complex. Thus, EF-Tu and -Ts have chaperone-like functions in the maintenance of the structure of the active Qβ replicase. Modeling of the template RNA and the growing RNA in the catalytic site of the Qβ replicase structure also suggests that structural changes of the RNAs and EF-Tu:Ts should accompany processive RNA polymerization and that EF-Tu:Ts in the Qβ replicase could function to modulate the RNA folding and structure.
Keywords: complex structure, RNA replicase, translational factors
RNA viruses rely on host-donated factors for replication and transcription of their genomic RNAs. In some RNA viruses from eubacteria, plants, and animals, the virus-encoded RNA-dependent RNA polymerase (RdRp) forms a complex with host-donated translational factors (1). In eubacterial viruses, such as Qβ virus, the virus-encoded RdRp forms a replicative complex with the host-donated translational factors EF-Tu and -Ts (2, 3). The RdRps from plant viruses, such as brome mosaic virus and tobacco mosaic virus, form replicative complexes with the eukaryotic translation factor eIF-3 (4, 5). The RdRps from animal viruses, such as poliovirus and vesicular stomatitis virus, also reportedly form replicative complexes with eEF-1A (a eukaryotic counterpart of EF-Tu) or eEF-1A and 1B (a eukaryotic counterpart of EF-Ts) (6, 7). The formation of a complex between the virus-encoded RdRps and the host-donated translational factors is required for RNA replication and transcription of viral genomic RNAs in the host cells.
Qβ virus infects Escherichia coli and replicates and transcribes its own genomic RNA, using Qβ replicase (2). Qβ replicase comprises the virus-encoded RdRp (β-subunit) and the host-donated translational elongation factors EF-Tu, EF-Ts, and ribosomal protein S1 (2, 3, 8). The β-subunit is the catalytic core for the RNA polymerization activity, and the assembly of the β-subunit with EF-Tu and -Ts is required for the RNA polymerization activity (2, 3, 8, 9). The established roles of EF-Tu are to bind an aminoacyl-tRNA (aa-tRNA) and escort the aa-tRNA to the ribosome A-site in its GTP-bound form. After a codon-anticodon match is formed on the A-site, GTP is hydrolyzed and EF-Tu is released from the ribosome in its GDP-bound form (10, 11). EF-Ts acts as a guanine nucleotide exchange factor, binds GDP-bound EF-Tu, displaces the GDP, and recycles EF-Tu. However, the mechanism for the assembly of the β-subunit with the host-donated translational factors EF-Tu and -Ts, and the functions of the translational factors in the RNA polymerization activitiy of Qβ replicase, beyond their established functions in the protein synthesis cycle, have remained obscure. Although several structures of virus-encoded RdRps have been solved (12), no complex structure of viral RdRp with host-donated translational factors has been reported.
Here, we present the crystal structure of the active core Qβ replicase, comprising the β-subunit (RdRp), EF-Tu and -Ts. The β-subunit has a right-handed structure, and the catalytic palm domain structure is similar to that of the corresponding domain of other RdRps. The tight EF-Tu:EF-Ts binary complex maintains the structure of the catalytic core crevasse of the β-subunit. Thus, EF-Tu and -Ts have chaperone-like functions in the assembly and maintenance of the structure of the active core Qβ replicase. The structure of the core Qβ replicase revealed two distinct tunnels leading to the catalytic site of the β-subunit. One tunnel allows the access of an incoming nucleotide, and the other is for the access of the template RNA to the catalytic site. Modeling of the template RNA, the growing RNA, and the incoming nucleotide in the catalytic site of the core Qβ replicase, based on the structure and the biochemical studies, revealed that structural changes of the double-stranded RNA and EF-Tu:Ts should accompany the processive RNA polymerization. Thus, the host translational factors might be also required for processive RNA chain elongation during the replication and transcription of viral genomic RNAs.
Results
Determination of the Core Qβ Replicase Structure.
We crystallized the single chain, active Qβ replicase, comprising the β-subunit, EF-Tu and EF-Ts (13). The crystal belonged to the space group C2221 and contained one Qβ replicase complex in the asymmetric unit. The initial phase was calculated by MAD methods, using the selenomethionine derivative protein. Subsequently, the structures of the Qβ replicase in the presence of magnesium or calcium ion were determined. Finally, the structures of the Qβ replicase were refined to R factors of 22.0% (Rfree = 28.2%) at 2.8 Å resolution for the calcium ion-bound form (Ca-form), and 25.2% (Rfree = 31.5%) at 3.2 Å resolution for the magnesium ion-bound form (Mg-form) (Table S1). The stoichiometry of the β-subunit, EF-Tu, and EF-Ts in the asymmetric unit is 1∶1∶1. Thus, the structure reflects the biological unit of the Qβ replicase complex (2).
Overall Structure of the Core Qβ Replicase.
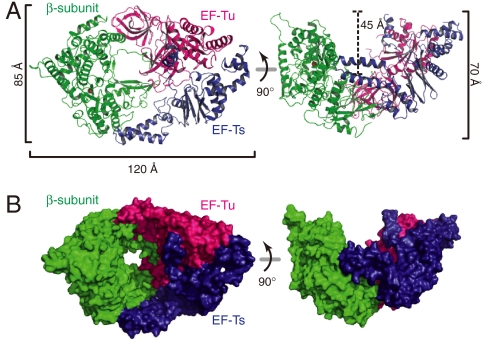
The three subunits in the Qβ replicase are tightly assembled together, and the overall structure of the Qβ replicase resembles a boat, with approximate dimensions of 85 Å × 120 Å × 70 Å (Fig. 1A). The convex curved surface of the β-subunit is covered by the concave surface of the EF-Tu:Ts binary complex (Fig. 1B), and the total interface area between the β-subunit and the EF-Tu:Ts binary complex is 2,287 Å2. The β-subunit has a right-handed structure and is composed of three functional domains—finger, thumb and palm domains—(Fig. 2A), as in other RdRps (12). EF-Tu and -Ts in the Qβ replicase closely interact with each other, as observed in the structure of the EF-Tu:Ts binary complex (14), and the complex structure of EF-Tu:Ts in the Qβ replicase is almost identical to that of the EF-Tu:Ts binary complex, with a root-mean-square-deviation of 0.7 Å for the Cα atoms (Fig. 3A). The interactions between the β-subunit and the EF-Tu:Ts binary complex maintain the structure of the catalytic core crevasse of the β-subunit, as described below in detail.
Fig. 1.
Overall structure of the Qβ replicase. (A) Ribbon and (B) surface models of Qβ replicase. The Qβ replicase adopts a boat-like structure. The β-subunit, EF-Tu and EF-Ts are colored green, red, and blue, respectively.
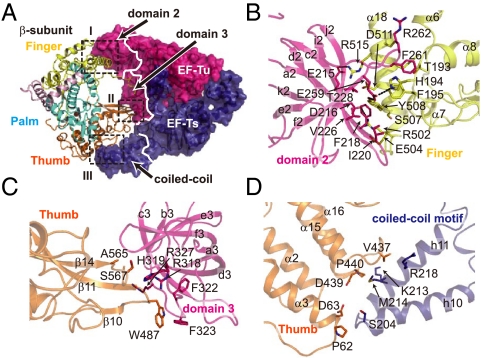
Fig. 2.
Interactions between the subunits of the Qβ replicase. (A) Interactions between the β-subunit and the EF-Tu:Ts complex (interfaces I, II, and III). The thumb, palm, and finger domains and the helix-loop-helix (HLH) of the β-subunit are depicted by ribbon models and are colored orange, cyan, yellow, and pink, respectively. EF-Tu and -Ts are shown in surface models and are colored as in Fig. 1A. Interfaces I, II, and III are depicted in boxes. (B–D) Detailed view of the interactions in interfaces I, II, and III in A. The numbering of the amino acid residues of EF-Tu and -Ts is according to the literature (14, 44).
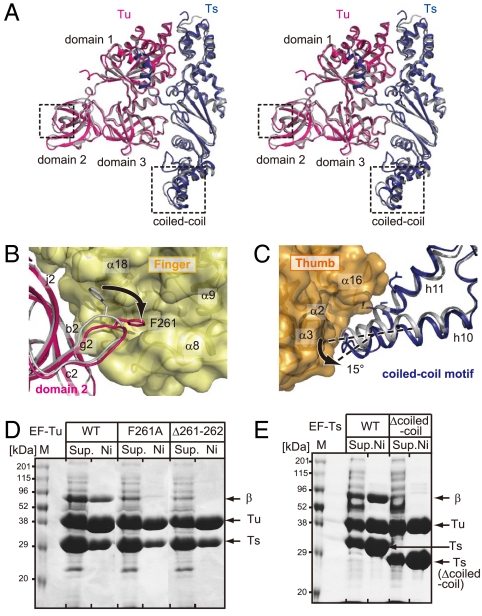
Fig. 3.
Structural differences between EF-Tu and -Ts. (A) A stereo-view comparison of the structures of EF-Tu:Ts in Qβ replicase (Tu and Ts are colored red and blue, respectively) and in the EF-Tu:Ts binary complex (14) (Tu and Ts are colored gray). The structural differences are observed in domain 2 of EF-Tu and a coiled-coil region in EF-Ts and are enclosed by boxes. (B and C) Detailed views of structural differences of EF-Tu and -Ts between the EF-Tu:Ts complex in Qβ replicase and the EF-Tu:Ts binary complex. The structures of EF-Tu and -Ts in the binary complex are colored gray. (D) The mutation or deletion of the amino acid residue(s) in the loop between f2 and i2 of EF-Tu reduced the complex formation and the expression of the Qβ replicase. (E) The deletion of the coiled-coil motif in EF-Ts reduced the complex formation and the expression of the Qβ replicase. The hexa-histidine tagged variants of EF-Tu (or EF-Ts) were coexpressed with wild-type EF-Ts (or EF-Tu) and the β-subunit and were purified by Ni-NTA column chromatography. The fractions eluted from the Ni-NTA column were separated by 10% (w/v) SDS/PAGE, and the gels were stained by Coomassie Brilliant Blue. “ Sup.” and “Ni” in the figures represent the total supernatant of the cell lysates and the eluted proteins from the Ni-NTA column, respectively.
Interactions Between the Subunits of Qβ Replicase.
Domains 2 and 3 in EF-Tu, which are involved in the recognition of the acceptor-TΨC helix and the TΨC loop of the aminoacyl-tRNA together with domain 1 (15, 16), interact with the finger and thumb domains of the β-subunit (Interfaces I and II) and are covered by the β-subunit (Fig. 2 A, B, and C). A coiled-coil motif in EF-Ts interacts with the thumb domain of the β-subunit (Interface III) (Fig. 2 A and D). The interactions between the finger domain of the β-subunit and domain 2 of EF-Tu (interface I, contact area of 976 Å2) are mainly hydrophobic. A loop between f2 and i2 in domain 2 of EF-Tu protrudes into the finger domain of the β-subunit, and Phe261 in the loop of EF-Tu is docked in the space between helices α18 and α8 in the finger domain of the β-subunit (Fig. 2B). Hydrogen bonds are also formed between the finger domain of the β-subunit and domain 2 of EF-Tu, contributing to the tight interactions. The interactions between domain 3 of EF-Tu and the thumb domain (interface II, contact area of 355 Å2) are also hydrophobic, and a β-turn (between β10 and β11) and the C-terminal part of β14 in the thumb domain interact with the loop between a3 and b3 in domain 3 of EF-Tu (Fig. 2C). The thumb domain of the β-subunit and the coiled-coil motif of EF-Ts contact each other through hydrophobic interactions (interface III, contact area of 588 Å2), where h10-h11 of EF-Ts interact with the N-terminal end of helix α3 and the loop between α15 and α16 of the thumb domain of the β-subunit (Fig. 2D). The conformational differences between EF-Tu:Ts in Qβ replicase and the EF-Tu:Ts binary complex (14) are observed in domain 2 of EF-Tu and the coiled-coil motif of EF-Ts (Fig. 3A). In the Qβ replicase complex, the loop between f2 and i2 in domain 2 of EF-Tu protrudes into the hydrophobic pocket (Fig. 3B), and the coiled-coil motif in EF-Ts rotates by about 15 ° (Fig. 3C). As a result, the finger and thumb domains of the β-subunit are tightly held by the EF-Tu:Ts binary complex. Thus, EF-Tu and -Ts have chaperone-like functions in the assembly and maintenance of the structure of the active Qβ replicase complex. Consistent with this, mutations or deletions of amino acid residues in the loop in domain 2 of EF-Tu and the deletion of the coiled-coil motif (17) in EF-Ts reduced the expression and assembly of the Qβ replicase complex in vivo (Fig. 3 D and E).
Structure of the β-Subunit of Qβ Replicase.
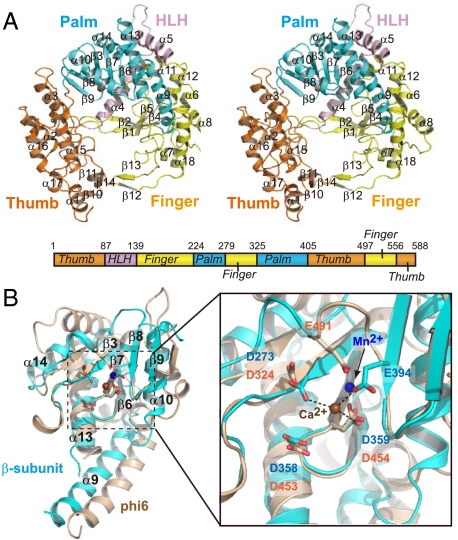
The domain organization of the β-subunit, which is right-handed, is similar to those of other RdRps (Fig. 4A) (12, 18). The finger domain of the β-subunit is composed of four antiparallel β sheets (β1, β2, β4, and β5) and six α helices (α6–α8, α11, α12, α18), and the thumb domain comprises a bundle of α helices (α1–α3, α15–α17) and three antiparallel β sheets (β10, β11, and β14). The finger and thumb domains of the β-subunit are connected by a long helix-loop-helix (HLH, α4–loop–α5) and antiparallel β strands (β12, β13), and as a result, the palm domain is surrounded by the finger and thumb domains. While the connection of the thumb and finger domains by the antiparallel β strands is commonly observed among viral RdRps (12), the other connection by the HLH is unique in the structure of the β-subunit of Qβ replicase and is conserved within the RdRps of the closely related RNA viruses (Fig. S1).
Fig. 4.
Structure of the β-subunit. (A) A stereo view of the β-subunit structure and a schematic representation of the β-subunit. The thumb, palm, and finger domains and the HLH are colored as in Fig. 2A. (B) Superposition of the palm domain of the β-subunit (Ca-form, colored cyan) onto that of phi-6 RdRp (colored beige, PDB code: 1hhs, 21) (Left). A close-up view of the superposed catalytic core regions (Right). The carboxylates in the catalytic sites are depicted by stick models, and the corresponding amino acid residues are labeled in blue for the β-subunit and orange for phi-6 RdRp. The calcium ion in the structure of the β-subunit is colored brown. Only one metal ion was observed in the structure of the β-subunit of Qβ replicase. The manganese ion in the structure of phi-6 RdRp is colored blue.
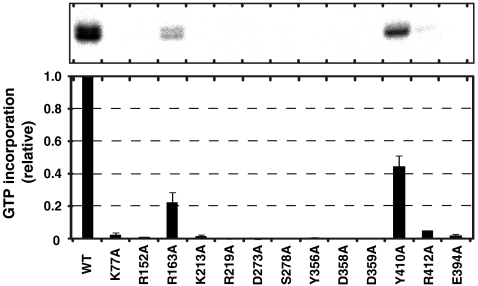
The catalytic palm domain of the β-subunit comprises five antiparallel β sheets (β3 and β6–β9), which are flanked by four α helices (α9, α10, α13, and α14), and the structure is similar to those of other RdRps (Fig. 4B) (18). Three carboxylates, Asp273, Asp358, and Asp359, in a conserved motif (Tyr-Gly-Asp-Asp), reside in the palm domain (Fig. S1), and the geometry of the three carboxylates is similar to those of the catalytic carboxylates of other RdRps, such as phi-6 RdRp (18, 19). Because conventional DNA/RNA polymerization proceeds by the two metal ion catalytic mechanism (20), two metals should be coordinated by these three carboxylates for the nucleotidyl transfer to proceed at the polymerization stage. The mutations of these carboxylates to Ala abolished the RNA polymerization activity of the Qβ replicase in vitro (Fig. 5).
Fig. 5.
RNA polymerization activities of core Qβ replicase variants. RNA replication activities of Qβ replicases containing β-subunit variants were assayed, using DN3 RNA as the template RNA (43). The activity (GMP incorporation activity) of the wild-type core Qβ replicase was defined as 1.0. The error bars in the graph indicate the standard deviations of more than two independent experiments.
In both the Ca-form and Mg-form β-subunit structures, only one respective metal ion was observed, at a different location from the expected catalytic positions (Fig. 4B and Fig. S2A). The fourth carboxylate, Glu394, is proximal to the catalytic carboxylates (Asp273, Asp358, and Asp359), and participates in the coordination of one calcium ion, together with Asp273 and Asp359, in the structure of the Ca-form β-subunit (Fig. 4B, right). In the structure of the Mg-form β-subunit, one magnesium ion was also located at the same position as the calcium ion in the structure of the Ca-form β-subunit (Fig. S2B). The mutation of Glu394 to Ala abolished the RNA polymerization activity of the Qβ replicase in vitro (Fig. 5). In the apo phi-6 RdRp structure (21), only one metal ion (manganese ion) is coordinated, at a different location from the expected catalytic positions (Fig. 4B Right), and the location is similar to that occupied by the calcium or magnesium ion in the β-subunit of Qβ replicase. The manganese ion at this location in phi-6 RdRp, which is different from the expected catalytic position, may be required for the efficient initiation of RNA polymerization, by capturing the triphosphate group of the first incoming nucleotide (21). Since Qβ replicase, like phi-6 RdRp, initiates RNA synthesis without using an RNA primer, the metal ion observed at the different location from the expected catalytic position may also be required for the initiation of RNA polymerization by Qβ replicase, as observed in the Phi-6 RdRp initiation complex.
A Model for RNA Polymerization by the Qβ Replicase.
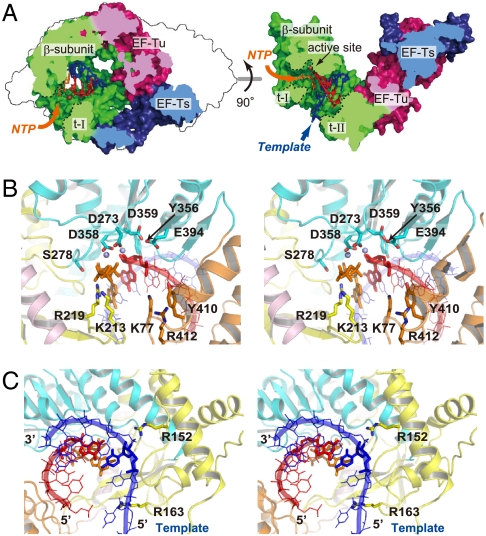
Two large tunnels were identified (tunnel-I and tunnel-II) in the Qβ replicase structure (Fig. 6A). Tunnel-I is located between the thumb and palm domains and leads to the catalytic site of the β-subunit from the outside of the Qβ replicase. Tunnel-II, with a diameter of approximately 15 Å, is surrounded by the three domains of the β-subunit and passes through the center of the catalytic crevasse of the β-subunit. Based on the structural similarity of the catalytic palm domains of the β-subunit to those of other RdRps (Fig. 4) (12, 18, 21), a template RNA, a growing RNA, and an incoming nucleotide were modeled in the catalytic crevasse of the Qβ replicase (Fig. 6A). The model represents the elongation stage of RNA polymerization and revealed that tunnel-I and tunnel-II are passages for incoming nucleotides and single-stranded template RNAs, respectively. The 3′-end of the template RNA enters tunnel-II first, from the bottom of the boat-like Qβ replicase. The 3′-terminal region of the template RNA reaches the catalytic site of the β-subunit to initiate RNA polymerization, and the elongation of the RNA proceeds.
Fig. 6.
RNA elongation model by Qβ replicase. (A) Substrate tunnels in Qβ replicase (tunnels I and II), shown in cross sections. Tunnel-I (t-I) and tunnel-II (t-II) are for the access of incoming nucleotides and a single-stranded template RNA, respectively. The template RNA, the growing RNA, and the incoming nucleotide are colored blue, red, and orange, respectively, and are shown in stick models. (B and C) Detailed view of the catalytic site of Qβ replicase, representing the elongation stage of RNA polymerization, in A. The incoming nucleotide and the 3′-nucleoside of the growing RNA are shown as stick models in B. Two magnesium ions are modeled in B using the structure of Norwalk virus RdRp (colored purple, PDB code: 3BSO), representing the RNA elongation stage (45). The nucleoside at the 3′-end of the growing RNA and the nucleoside of the template RNA that base-pairs with the nucleoside at the 3′-end of the growing RNA are shown in stick models in C.
In the catalytic site of the palm domain of the β-subunit (Fig. 6B), the triphosphate moiety of the incoming nucleotide is in the vicinity of Lys213, Arg219, and Ser278 and would hydrogen-bond with these amino acid residues. Lys213 and Arg219 are conserved among the other RdRps from positive-strand RNA viruses (Fig. S1). Lys77, Tyr410 and Arg412 are proximal to the phosphate backbone of the 3′-terminal nucleotide of the growing RNA and would hydrogen-bond with the phosphate backbone. The recognition of the phosphate backbone of the 3′-nucleoside of the growing RNA relocates the 3′-hydroxyl group of the acceptor nucleoside into the catalytic site for the nucleotidyl transfer reaction to proceed. Arg163 is located at the entrance of tunnel-II and is in the vicinity of the phosphate backbone of the template RNA (Fig. 6C). Arg152 resides close to the phosphate backbone in the template RNA, which is double-stranded with the growing RNA, and would fix the template nucleoside in the position opposite to that for the incoming nucleotide. Consistent with the model representing the elongation stage of RNA polymerization by the Qβ replicase, the mutations of the amino acid residues interacting with the incoming nucleotide, the 3′-end nucleotide of the nascent RNA, and the template RNA all reduced the RNA polymerization activity of the Qβ replicase in vitro (Fig. 5).
Discussion
The elongation factors EF-Tu and EF-Ts are ubiquitous and essential factors for the elongation cycle of protein synthesis in all organisms. In addition to the established function of EF-Tu in protein synthesis, alternative unexpected functions of EF-Tu and its eukaryotic counterpart eEF1A have emerged, including regulation of the actin cytoskeleton and cell morphology (22, 23) and protection of cells from heat shock and oxidative stress (24, 25). In the 1960s ∼ 1970s, EF-Tu and -Ts were found to be essential host-donated subunits of the Qβ replicase (2, 3). Since then, RdRps from animal, plant, and eubacterial viruses have also been reported to form replicative complexes with translational factors in infected cells (1, 4–7). Thus, the RdRPs from the RNA viruses, including animal, plant, and eubacterial viruses, might have evolved from a common ancestor, and share similar replication and transcription systems for their genomic RNA amplification. However, the detailed mechanisms for the assembly of virus-encoded RdRp with the host-donated translational factors and the functions of the translational factors in RNA replication and transcription activities have remained obscure.
In this study, we present a previously undescribed structure of the complex of a virus-encoded RdRp with the host-donated translational factors for the active replicative RNA polymerase. This structure, along with the biochemical analysis, provides mechanistic insights into the assembly of the active virus RdRp with the host-donated translational factors in the infected cells. The hydrophobic interactions between the β-subunit and EF-Tu and Ts are required for the maintenance of the structure of the catalytic core crevasse of Qβ replicase and for the expression and assembly of the complex in the infected cells (Figs. 1, 2, and 3). Eubacterial EF-Tu reportedly displays the properties of a chaperone (26, 27) by protecting proteins from heat-denaturation and facilitating the folding of unfolded proteins. Thus, EF-Tu and -Ts have chaperone-like functions in the assembly and maintenance of the structure of the active core Qβ replicase.
The present structure also provides mechanistic insight into other functions of translational factors beyond their established functions in protein synthesis during RNA polymerization by the viral RdRp complex with the translational factors. In the elongation stage of RNA polymerization by the Qβ replicase, the growing RNA forms a duplex with the template RNA immediately after polymerization, and the growing RNA duplex proceeds toward the EF-Tu:Ts complex in the Qβ replicase (Fig. 6A). Without the structural changes of the growing RNA duplex and (or) EF-Tu:Ts in the Qβ replicase, processive RNA polymerization would not occur. Thus, EF-Tu and -Ts in the Qβ replicase might have specific flexibility to influence the structures and (or) conformations of the growing RNA duplexes properly for processive RNA polymerization to occur. The EF-Tu:Ts in the Qβ replicase complex may modulate the RNA folding and structure by an as-yet-unknown mechanism, in a similar manner to the function of EF-Tu on proteins (26, 27). In vitro analyses using GTP analogues suggested that the GTPase activity of EF-Tu was not required for RNA polymerization by the Qβ replicase (Fig. S3A) (3, 28). Consistent with this, the mutations of the amino acid residues in EF-Tu involved in GTP binding or hydrolysis (11, 29–31) did not reduce the RNA polymerization activity in vitro (Fig. S3B). Thus, neither the large structural changes of EF-Tu, as observed between its GTP- and GDP-bound forms (30, 31) in translational cycles nor the GTP hydrolysis would occur during RNA polymerization by Qβ replicase.
For the replication and transcription of the genomic RNA of Qβ virus, the fourth component, host-donated ribosomal protein S1, is required (2). The present structure of the core Qβ replicase and the location of the template tunnel also suggest that the 3′-end of the Qβ genomic RNA should be relocated to the entrance of the template tunnel. The S1 protein might bind to the bottom of the boat-like structure of the core Qβ replicase and recruit the 3′-part of the genomic RNA to the positively charged area near the template tunnel entrance (Fig. S4). The binding of the S1 protein to specific internal sites of the viral genomic RNA would facilitate the initiation of the replication and transcription of the viral genomic RNA (32, 33).
Understanding the dynamic mechanism of the replication and transcription of Qβ virus genomic RNA by Qβ replicase awaits the determination of the complex structures of Qβ replicase with the template and growing RNAs, representing the entire RNA polymerization process.
Materials and Methods
Expression, Purification, and Crystallization of the Core Qβ Replicase.
The single chain core Qβ replicase was expressed in E. coli BL21 (DE3), using the pBAD33-Ts-Tu-β-3 plasmid (13). At about the midlog phase, the protein production was induced by the addition of arabinose to 0.2% (w/v). The cells were disrupted by sonication in 50 mM sodium phosphate, pH 8.0, 300 mM NaCl, 5 mM β-mercaptoethanol and 20% (v/v) glycerol. The protein was purified by successive chromatography steps on Ni-NTA (Qiagen, Japan), DEAE Sepharose, and Hi-Trap Heparin (GE Healthcare, Japan) columns. Finally, the protein was purified by chromatography on a Hi-Load Superdex 200 column (GE Healthcare, Japan), in 20 mM Tris-Cl, pH 8.0, 200 mM NaCl, and 5 mM β-mercaptoethanol. The protein was concentrated to 8–10 mg/ml using an Amicon filter (Millipore, Japan), and was stored at -80 °C until use. The crystals were obtained with a reservoir solution containing 100 mM HEPES, pH 5.6, 200 mM Ca(OAc)2, and 30% (v/v) PEG400, using the sitting-drop vapor diffusion method at 20 °C (Ca-form). For the phase determination, the crystal of the selenomethionine derivative was prepared with the same reservoir solution, except for the addition of 10 mM TCEP-HCl (Tris [2-carboxyethyl] phosphine hydrochloride) to the solution. The native crystal was also obtained with the reservoir solution containing 200 mM Mg(OAc)2, instead of 200 mM Ca(OAc)2, and the structure was determined (Mg-form).
Structure Determination of Qβ Replicase.
For structure determination, three datasets for the multiple wavelength anomalous dispersion (MAD) method with the selenomethionine derivative were collected at the beam-line PF-BL17A at KEK (Tsukuba, Japan). The dataset of the native crystals was also collected at the same beam-line. The crystals were cryoprotected by the reservoir solution and were flash-cooled in a 100 K nitrogen stream. All of the data were processed using the program HKL2000 (35).
The dataset at the peak wavelength was used to locate the selenium atoms with the program SnB (36), and 28 peaks were picked for the 28 atoms in the asymmetric unit. Refinement of the selenium sites, phase calculation, and density modification were performed with autoSHARP (37). Subsequent phase extension to the native data was performed with DM in CCP4i (38). The initial model was built using Buccaneer (39). Finally, manual model building was performed with Xtalview/Xfit (40). The model was refined with the programs CNS (41) and PHENIX (42).
Preparation of the Qβ Replicase Complex.
The core Qβ replicase complex, comprising the individual β-subunit, EF-Tu and -Ts, was expressed in BL21 Star (DE3) (Invitrogen, Japan) using the plasmids pBAD33-βH and pET-Ts-Tu (13). At about the midlog phase, the expression of EF-Ts and EF-Tu was induced by adding IPTG (isopropyl-β-D-thiogalactopyranoside) to a 0.01 mM final concentration, and then the β-subunit was induced by adding arabinose to 0.2% (w/v). The complex was purified by the same procedure as described above. For the preparation of the core Qβ replicase with mutations in either the β-subunit, EF-Tu or -Ts, the mutations were introduced by site-directed mutagenesis in the respective hexa-histidine tagged gene. The expression and purification of the Qβ replicase variants were the same as described.
Replication Activity Assay.
Replication activities of Qβ replicase and its variants were assayed, using DN3 (43) as the RNA template. The reaction solutions, containing 80 mM Tris-Cl, pH 8.0, 10 mM MgCl2, 1 mM DTT, 100 nM DN3 RNA, 100 μM NTP mix, 165 nM [α-32P] GTP (GE Healthcare, Japan, 3000 ci/mmol), and 380 nM Qβ replicase (or its variants), were incubated for 30 min at 37 °C. The reactions were stopped by adding EDTA (pH 8.0) to a final concentration of 25 mM. The reaction solutions were treated with phenol/chloroform, and the RNAs were ethanol-precipitated and dissolved in a buffer containing 90% (v/v) formamide and 10 mM EDTA, pH 8.0. The RNAs were heat-denatured at 90 °C for 5 min, chilled on ice, and separated by 10% (w/v) polyacrylamide gel electrophoresis under denaturing conditions. The intensity of the 32P-labeled products was analyzed and quantified by a BAS2500 Bio-Image Analyzer (Fuji Film, Japan).
Supplementary Material
Acknowledgments.
We thank Tetsuya Yomo for the plasmid encoding Qβ replicase and Azusa Hamada for technical assistance. We thank Shuya Fukai and Tsutomu Suzuki for comments on this paper and the beam-line staff of BL-17A (KEK, Tsukuba) for technical assistance during data collection. This work was supported by grants from the Precursory Research for Embryonic Science and Technology (PRESTO) program of the Japan Science and Technology Agency; the Ministry of Education, Culture, Sports, Science and Technology; the Japan Society for Promotion of Science, Toray Science Foundation, and Sumitomo Foundation to K.T.
Note Added in Proof.
During the review process of our manuscript, the crystal structure of the core Qβ replicase belonging to a different space group was reported (34).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1006559107/-/DCSupplemental.
Data deposition: The atomic coordinates and structural factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 3AGP and 3AGQ).
References
- 1.Lai MM. Cellular factors in the transcription and replication of viral RNA genomes: A parallel to DNA-dependent RNA transcription. Virology. 1998;244:1–12. doi: 10.1006/viro.1998.9098. [DOI] [PubMed] [Google Scholar]
- 2.Blumenthal T, Carmichael GG. RNA replication: Function and structure of Qbeta-replicase. Annu Rev Biochem. 1979;48:525–548. doi: 10.1146/annurev.bi.48.070179.002521. [DOI] [PubMed] [Google Scholar]
- 3.Blumenthal T, Landers TA, Weber K. Bacteriophage Q replicase contains the protein biosynthesis elongation factors EF Tu and EF Ts. Proc Natl Acad Sci USA. 1972;69:1313–1317. doi: 10.1073/pnas.69.5.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quadt R, Kao CC, Browning KS, Hershberger RP, Ahlquist P. Characterization of a host protein associated with brome mosaic virus RNA-dependent RNA polymerase. Proc Natl Acad Sci USA. 1993;90:1498–1502. doi: 10.1073/pnas.90.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Osaman TAM, Buck KW. The tobacco mosaic virus RNA polymerase complex contains a plant protein related to the RNA-binding subunit of yeast eIF-3. J Virol. 1997;71:6075–6082. doi: 10.1128/jvi.71.8.6075-6082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Das T, Mathur M, Gupta AK, Janssen GMC, Banerjee AK. RNA polymerase of vesicular stomatitis virus specifically associates with translation elongation factor-1 αβγ for its activity. Proc Natl Acad Sci USA. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harris KS, et al. Interaction of poliovirus polypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome: Identification of viral and cellular cofactors needed for efficient binding. J Biol Chem. 1994;269:27004–27014. [PubMed] [Google Scholar]
- 8.Wahba AJ, et al. Subunit I of Q beta replicase and 30 S ribosomal protein S1 of Escherichia coli. Evidence for the identity of the two proteins. J Biol Chem. 1974;249:3314–3316. [PubMed] [Google Scholar]
- 9.Kamen R, Kondo M, Romer W, Weissmann C. Reconstitution of Qβ :replicase lacking subunit α with protein-synthesis-interference factor i. Eur J Biochem. 1972;31:44–51. doi: 10.1111/j.1432-1033.1972.tb02498.x. [DOI] [PubMed] [Google Scholar]
- 10.Schuette JC, et al. GTPase activation of elongation factor EF-Tu by the ribosome during decoding. EMBO J. 2009;28:755–765. doi: 10.1038/emboj.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmeing TM, et al. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009;326:688–694. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrer-Orta C, Arias A, Escarmís C, Verdaguer N. A comparison of viral RNA-dependent RNA polymerases. Curr Opin Struct Biol. 2006;16:27–34. doi: 10.1016/j.sbi.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 13.Kita H, et al. Functional Qbeta replicase genetically fusing essential subunits EF-Ts and EF-Tu with beta-subunit. J Biosci Bioeng. 2006;101:421–426. doi: 10.1263/jbb.101.421. [DOI] [PubMed] [Google Scholar]
- 14.Kawashima T, Berthet-Colominas C, Wulff M, Cusack S, Leberman R. The structure of the Escherichia coli EF-Tu:EF-Ts complex at 2.5 Å resolution. Nature. 1996;379:511–518. doi: 10.1038/379511a0. [DOI] [PubMed] [Google Scholar]
- 15.Nissen P, et al. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- 16.Nissen P, Kjeldgaard M, Thirup S, Clark BF, Nyborg J. The ternary complex of aminoacylated tRNA and EF-Tu-GTP. Recognition of a bond and a fold. Biochimie. 1996;78:921–933. doi: 10.1016/s0300-9084(97)86714-4. [DOI] [PubMed] [Google Scholar]
- 17.Karring H, et al. Qβ-phage resistance by deletion of the coiled-coil motif in elongation factor Ts. J Biol Chem. 2004;279:1878–1884. doi: 10.1074/jbc.M306605200. [DOI] [PubMed] [Google Scholar]
- 18.O’Reilly EK, Kao CC. Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure. Virology. 1998;252:287–303. doi: 10.1006/viro.1998.9463. [DOI] [PubMed] [Google Scholar]
- 19.Koonin EV. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J Gen Virol. 1991;72:2197–2206. doi: 10.1099/0022-1317-72-9-2197. [DOI] [PubMed] [Google Scholar]
- 20.Brautigam CA, Steitz TA. Structural and functional insights provided by crystal structures of DNA polymerases and their substrate complexes. Curr Opin Struct Biol. 1998;8:54–63. doi: 10.1016/s0959-440x(98)80010-9. [DOI] [PubMed] [Google Scholar]
- 21.Butcher SJ, Grimes JM, Makeyev EV, Bamford DH, Stuart DI. A mechanism for initiating RNA-dependent RNA polymerization. Nature. 2001;410:235–240. doi: 10.1038/35065653. [DOI] [PubMed] [Google Scholar]
- 22.Yang F, Demma M, Warren V, Dharmawardhane S, Condeelis J. Identification of an actin-binding protein from Dictyostelium as elongation factor 1a. Nature. 1990;347:494–496. doi: 10.1038/347494a0. [DOI] [PubMed] [Google Scholar]
- 23.Gross SR, Kinzy TG. Translation elongation factor 1A is essential for regulation of the actin cytoskeleton and cell morphology. Nat Struct Mol Biol. 2005;12:772–778. doi: 10.1038/nsmb979. [DOI] [PubMed] [Google Scholar]
- 24.Shamovsky I, Ivannikov M, Kandel ES, Gershon D, Nudler E. RNA-mediated response to heat shock in mammalian cells. Nature. 2006;440:556–560. doi: 10.1038/nature04518. [DOI] [PubMed] [Google Scholar]
- 25.Chang R, Wang E. Mouse translation elongation factor eEF1A-2 interacts with Prdx-I to protect cells against apoptotic death induced by oxidative stress. J Cell Biochem. 2007;100:267–278. doi: 10.1002/jcb.20969. [DOI] [PubMed] [Google Scholar]
- 26.Caldas TD, Yaagoubi AE, Richarme G. Chaperone properties of bacterial elongation factor EF-Tu. J Biol Chem. 1998;273:11478–11482. doi: 10.1074/jbc.273.19.11478. [DOI] [PubMed] [Google Scholar]
- 27.Kudlicki W, Coffman A, Kramer G, Hardesty B. Renaturation of rhodanese by translational elongation factor (EF) Tu. J Biol Chem. 1997;272:32206–32210. doi: 10.1074/jbc.272.51.32206. [DOI] [PubMed] [Google Scholar]
- 28.Landers TA, Blumenthal T, Weber K. Function and structure in ribonucleic acid phage Qβ ribonucleic acid replicase. J Biol Chem. 1974;249:5801–5808. [PubMed] [Google Scholar]
- 29.Daviter T, Wieden HJ, Rodnina MV. Essential role of histidine 84 in elongation factor Tu for the chemical step of GTP hydrolysis on the ribosome. J Mol Biol. 2003;332:689–699. doi: 10.1016/s0022-2836(03)00947-1. [DOI] [PubMed] [Google Scholar]
- 30.Kjeldgaard M, Nissen P, Thirup S, Nyborg J. The crystal structure of elongation factor EF-Tu from Thermus aquaticus in the GTP conformation. Structure. 1993;1:35–50. doi: 10.1016/0969-2126(93)90007-4. [DOI] [PubMed] [Google Scholar]
- 31.Berchtold H, et al. Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature. 1993;365:126–132. doi: 10.1038/365126a0. [DOI] [PubMed] [Google Scholar]
- 32.Senear AW, Steitz JA. Site-specific interaction of Qbeta host factor and ribosomal protein S1 with Qbeta and R17 bacteriophage RNAs. J Biol Chem. 1976;251:1902–1912. [PubMed] [Google Scholar]
- 33.Miranda G, et al. Recognition of bacteriophage Qbeta plus strand RNA as a template by Qbeta replicase: Role of RNA interactions mediated by ribosomal proteins S1 and host factor. J Mol Biol. 1997;267:1089–1103. doi: 10.1006/jmbi.1997.0939. [DOI] [PubMed] [Google Scholar]
- 34.Kidmose RT, Vasiliev NN, Chetverin AB, Andersen GR, Knudsen CR. Structure of the Qbeta replicase, an RNA-dependent RNA polymerase consisting of viral and host proteins. Proc Natl Acad Sci USA. 2010;107:10884–10889. doi: 10.1073/pnas.1003015107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 36.Weeks CM, Miller R. The design and implementation of SnB v2.0. J Appl Crystallogr. 1999;32:120–124. [Google Scholar]
- 37.de La Fortelle E, Bricogne G. Maximum-likelihood heavy-atom parameter refinement for the multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Method Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 38.Cowtan KD. dm: An automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography. 1994;31:34–38. [Google Scholar]
- 39.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 40.McRee DE. XtalView/Xfit—A versatile program for manipulating atomic coordinates and electron density. J Struct Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 41.Brunger AT, et al. Crystallography & NMR System: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 42.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 43.Zamora H, Luce R, Biebricher CK. Design of artificial short-chained RNA species that are replicated by Q beta replicase. Biochemistry. 1995;34:1261–1266. doi: 10.1021/bi00004a020. [DOI] [PubMed] [Google Scholar]
- 44.Song H, Parsons MR, Rowsell S, Leonard G, Phillips SE. Crystal structure of intact elongation factor EF-Tu from Escherichia coli in GDP conformation at 2.05 Å resolution. J Mol Biol. 1999;285:1245–1256. doi: 10.1006/jmbi.1998.2387. [DOI] [PubMed] [Google Scholar]
- 45.Zamyatkin DF, et al. Structural insights into mechanisms of catalysis and inhibition in Norwalk virus polymerase. J Biol Chem. 2008;283:7705–7712. doi: 10.1074/jbc.M709563200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.