Abstract
When stem cells and multipotent progenitors differentiate, they undergo fate restriction, enabling a single fate and blocking differentiation along alternative routes. We herein present a mechanism whereby such unequivocal commitment is achieved, based on microRNA (miRNA)-dependent repression of an alternative cell fate. We show that the commitment of monocyte RAW264.7 progenitors to active macrophage differentiation involves rapid up-regulation of miR-155 expression, which leads to the suppression of the alternative pathway, namely RANK ligand-induced osteoclastogenesis, by repressing the expression of MITF, a transcription factor essential for osteoclast differentiation. A temporal asymmetry, whereby miR-155 expression precedes and overrides the activation of the osteoclast transcriptional program, provides the means for coherent macrophage differentiation, even in the presence of osteoclastogenic signals. Based on these findings, we propose that miRNA may provide a general mechanism for the unequivocal commitment underlying stem cell differentiation.
Keywords: microRNA, miR-155, differentiation, osteoclast, macrophage
The process whereby progenitor cells commit to one, and only one, fate occurs with remarkable spatial and temporal precision. Understanding the mechanism underlying this response is highly challenging since, in vivo, pluripotent progenitors often encounter diverse and even conflicting differentiation signals. For example, monocyte progenitors subjected to inflammatory stimuli are induced to differentiate into activated macrophages via the toll-like receptor (TLR) pathway (1), whereas the same progenitors can differentiate into bone-degrading osteoclasts, after stimulation with the receptor activator of nuclear factor κB ligand (RANKL) and macrophage colony-stimulating factor (M-CSF) (2–4).
Macrophages and osteoclasts serve distinct and essential physiological roles; therefore, once committed to a particular fate, each cell type acquires a unique network of genetic interactions. In this manner, a specific set of effector genes, whose expression is essential for a particular function, is expressed. Specifically, tissue-resident macrophages are induced by invading microorganisms and serve as the first line of defense by engulfing the pathogens. Additionally, macrophages express cytokines such as interleukin 1 (IL-1) (5, 6) and a variety of toxic products, including nitric oxide (NO) (7). On the other hand, osteoclasts derived from the same progenitors activate tartrate-resistant acid phosphatase (TRAP), cathepsin K (CtsK), and Na/K channels and undergo major morphological transformation, all important for their bone resorption activity. The osteoclast fate is induced by osteoblast-derived RANKL, which activates a signaling cascade that induces microphthalmia-associated transcription factor (MITF) and PU.1, two pivotal transcription factors involved in osteoclast differentiation (4).
In this study, we used a RAW264.7 monocyte cell line to test the hypothesis that the unequivocal commitment of the progenitor to a single pathway involves the activation of a regulatory genetic program that effectively blocks alternative cell fates and, in so doing, confers robustness on the fate determination process.
Our search for miRNAs involved in such fate restriction was inspired by growing evidence that miRNAs and their cognate target genes are often expressed in a mutually exclusive manner during development (8, 9) and that development is reinforced by miRNAs that repress genes belonging to earlier states (10, 11). We therefore hypothesized that a program for repressing alternative cell fates could be orchestrated by the activity of miRNA genes (12).
miRNAs are small, noncoding RNAs that are involved in posttranscriptional repression by binding to minimal binding sites on target mRNAs. Many miRNAs play a role in hematopoietic cells, including osteoclasts (13). Our findings, presented herein, suggest that the regulation of a specific miRNA gene, miR-155, is involved in the commitment of monocyte progenitors to macrophage differentiation and activation, by effectively interfering with the genetic network driving the alternative, osteoclast fate.
miR-155, a product of the BIC transcript that was first described as a frequent site of integration for the avian leukosis virus (14), is one of the most extensively studied miRNA genes. Its functions in the hematopoietic lineage are well-documented, and include its direct involvement in the normal proliferation of lymphocytes, in the repression of erythroid, myeloid, and megakaryocytic differentiation, and in the emergence of B cell lymphoma (15).
In the monocytic lineage, miR-155 was shown to be up-regulated by proinflammatory LPS and TNF-α (16–19), playing an important role in monocyte proliferation, and in macrophage and dendritic cell differentiation (15–21). However, the roles of miR-155 in the osteoclast trajectory, derived from the same monocytic precursor, were not known until now. Our data suggest that miR-155 represses osteoclast differentiation through inhibition of the osteoclast core transcriptional machinery, thus enabling robust acquisition of an activated macrophage fate.
Results
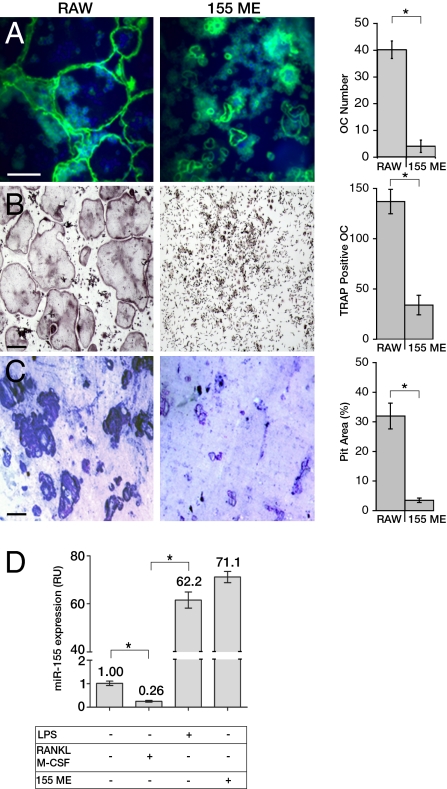
To assess the functional significance of miRNA expression for macrophage vs. osteoclast differentiation, we used the monocyte-derived cell line RAW 264.7, capable of differentiating in vitro into either activated macrophages, or osteoclasts (22, 23). First, we screened a list of genes involved in differentiation along the monocyte-macrophage lineage (detailed in Fig. S1), using a set of target prediction algorithms (24–26). This approach was used to identify candidate miRNAs that could potentially target more than one gene within this differentiation network. Based on this analysis, we carried out a secondary, functional screen as follows: Treatment of wild-type RAW 264.7 cells with RANKL and M-CSF induces massive osteoclastic differentiation, manifested by the formation of multinucleated syncytia displaying large (≈200 μm in diameter), actin-rich sealing zone-like peripheral rings. We used this trait to perform a preliminary survey of 15 candidate miRNA genes. In this screen, specific miRNA genes were misexpressed in RAW264.7 cells that were induced with RANKL and M-CSF to differentiate into osteoclasts. Of the 15 miRNA genes that were studied, 3 miRNA genes appeared to repress osteoclast differentiation (Fig. S1). This screen focused our attention on miR-155, whose misexpression blocked the formation of giant osteoclasts with sealing zone-like structures (Fig. 1A). miR-155 misexpression also suppressed TRAP expression, an early marker of osteoclast differentiation (Fig. 1B), and blocked the bone-resorbing activity of the treated cells (Fig. 1C). Intriguingly, these combined effects are rather specific to miR-155, because other miRNA genes, such as miR-200c, did not affect osteoclast differentiation; furthermore, miR-155 misexpression did not change the expression levels of other miRNAs. We also noted that this arrest of differentiation was not accompanied by changes in cell viability or proliferation (Fig. S1). miR-155 expression is dramatically up-regulated in activated macrophages (16, 27) and is down-regulated in osteoclasts (Fig. 1D). In fact, the levels of miR-155 misexpression in RAW264.7 cells are comparable to those normally found in differentiated macrophages after LPS stimulation (Fig. 1D), and miR-155 misexpression did not significantly affect LPS-induced macrophage phagocytotic capacity (Fig. S2).
Fig. 1.
miR-155 misexpression impairs osteoclast differentiation. (A–C) Wild-type (RAW) and RAW264.7 cells misexpressing miR-155 (155 ME), were analyzed 72 h after M-CSF and RANKL induction. (A) A representative micrograph of cultured RAW264.7 cells: sealing zone-like structures are delineated by phalloidin (green), and nuclei are labeled with DAPI (blue). The quantification of the average number of giant polarized multinucleated osteoclasts per well, is presented on the right. (B) Cells were assayed for TRAP activity, and the number of TRAP-positive, multinucleated osteoclasts per well was counted. (C) Bone resorption capacity was quantified by the relative pit area generated by cells plated on 0.5-cm2 sections of bovine bone for 6 d. (D) Expression levels of miR-155 were quantified by qPCR in untreated RAW264.7 cells stimulated with either M-CSF+RANKL or LPS, and in RAW264.7 cells misexpressing miR-155. Data from at least three independent experiments, performed in duplicates, are shown as mean ± SD. *P < 0.01. RU, relative units. (Scale bars: 150 μm.)
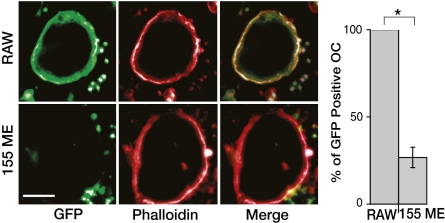
The formation of multinucleated osteoclasts is a biphasic process, initiated by the fusion of a few “lead” cells, followed by the rapid fusion of many mononuclear cells located in the vicinity of this “nucleus” (Movie S1). To determine whether miR-155 blocks the initial fusion event, or reduces the overall fusogenic capacity of the mononuclear cells, we used RANKL/M-CSF to stimulate a coculture of wild-type RAW264.7, and cells misexpressing miR-155. To visualize each cell population separately, either the wild-type or the miR-155-expressing cells were engineered to express actin-GFP. After 3 d of RANKL and M-CSF stimulation, the cocultures were fixed and labeled with TRITC phalloidin. When the cells expressing actin-GFP coexpressed miR-155, fusion was abrogated, as manifested by the absence of actin-GFP from the phalloidin-tagged rings. On the other hand, the wild-type controls were readily engaged in the formation of giant multinucleated cells. Actin-GFP expression was not attenuated by miR-155 misexpression, as revealed by a Western blot analysis (Fig. S3). Thus, RAW264.7 cells misexpressing miR-155 fail to initiate fusion and also do not merge into the syncytia as do wild-type osteoclasts (Fig. 2 and Movie S2). These results suggest that miR-155 interferes with RANK signaling upstream of the acquisition of fusogenic capacity, resulting in early arrest of osteoclastogenesis.
Fig. 2.
miR-155 arrests osteoclastogenesis before cell fusion. Wild-type RAW264.7 cells and cells expressing either miR-155 and actin-GFP (155 ME) or actin-GFP alone (RAW) were cocultured. The ability of control or miR-155 misexpressing cells to fuse into multinucleated cells upon RANKL and M-CSF induction was quantified by counting the relative number of giant polarized multinucleated osteoclasts expressing actin-GFP compared with the total number of osteoclasts in the well. Data from at least three independent experiments performed in triplicates are shown as mean ± SD. *P < 0.01. (Scale bar: 150 μm.)
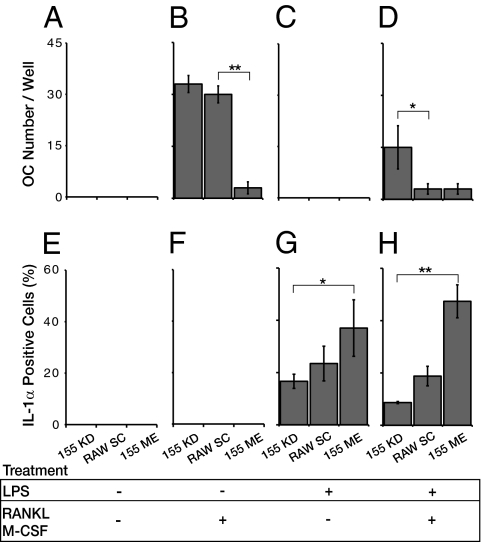
Because miR-155 appears to be an early repressor of osteoclast differentiation and is highly expressed in activated macrophages, we explored its role as a switch between the mutually exclusive osteoclast and macrophage fates. Specifically, we hypothesized that miR-155-based fate restriction allows coherent macrophage differentiation in the face of conflicting signals. To test this hypothesis experimentally, we simultaneously exposed RAW264.7 cells to conflicting osteoclastogenic (M-CSF+RANKL) and proinflammatory (LPS) signals. Cultures, which had been stimulated with a single cue, were propelled toward either an osteoclast or a macrophage fate (Fig. 3 B, C, F, and G). However, under combined (M-CSF+RANKL+LPS) stimulation, cultures that had not been subjected to any manipulation of miR-155 expression, yielded both activated macrophages and a small fraction of osteoclasts (Fig. 3 D and H).
Fig. 3.
miR-155 levels affect monocyte differentiation capacity when stimulated by conflicting proosteoclastic and inflammatory signals. The number of giant multinucleated osteoclasts per well (A–D) and the percentage of Il-1α+ macrophages (E–H) was quantified in cells transfected with scrambled, nontargeting oligonecleotides (RAW SC), cells misexpressing miR-155 (155 ME), or cells in which miR-155 was knocked down by antisense oligonucleotides (155 KD). (A and E) Untreated cells; (B and F) cells stimulated with RANKL and M-CSF; (C and G) with LPS alone; (D and H) simultaneously, with a mixture of RANKL, M-CSF, and LPS. Data from at least three independent experiments, replicated eight times, are shown as mean ± SD. *P < 0.05; **P < 0.01.
Under these conditions, miR-155 misexpression promoted macrophage differentiation, manifested by a 2.5-fold increase in Il-1α+ macrophages (Fig. 3H) and by comparable up-regulation of nitric oxide synthesis (Fig. S4). miR-155 knockdown by antisense oligos consistently reduced differentiation into macrophages by 2-fold (Fig. 3H) and increased osteoclast numbers ≈5-fold relative to control cultures (Fig. 3D). Of note, diminished osteoclast differentiation in this assay could not simply be attributed to a strong bias toward macrophage activation, because miR-155 misexpression, per se, is insufficient to induce the development of active macrophages if the progenitors are not challenged by an external inflammatory signal or if the cells are solely exposed to osteoclastogenic cues (Fig. 3 E and F). These results indicate that miR-155 favors a macrophage fate and represses osteoclast differentiation under combined (M-CSF+RANKL+LPS) stimulation.
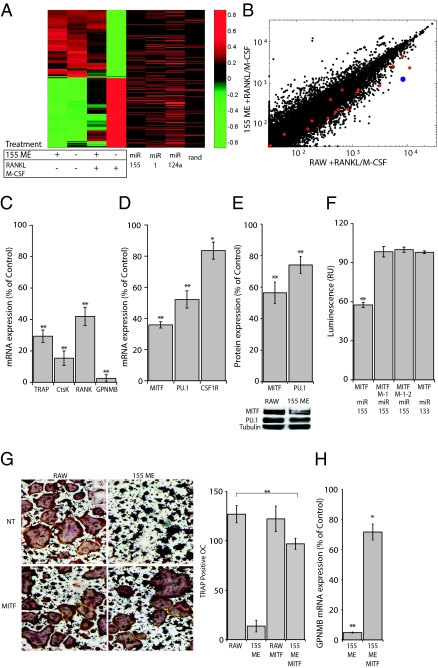
To address the molecular mechanism whereby miR-155 arrests osteoclastogenesis, we used Affymetrix arrays to compare gene expression profiles under four conditions: wild-type or miR-155 misexpression in RAW264.7 cells, which are either in the progenitor state, or subjected to RANKL/M-CSF osteoclastogenic stimulation. Analysis of the expression profiles obtained in the wild-type cells revealed a set of ≈500 genes that undergo major alterations upon osteoclast differentiation (Pearson correlation of −0.98). A “nearest neighbor” bioinformatics analysis demonstrated that RAW264.7 cells misexpressing miR-155 possess a genomic fingerprint remarkably similar to their undifferentiated precursors (Pearson correlation of 0.84). After RANKL and M-CSF stimulation, however, miR-155 misexpression induces an expression profile closer to that of undifferentiated progenitors (Pearson correlation of 0.47) than to that of differentiated osteoclasts (Pearson correlation of −0.59; Fig. 4A). Specifically, osteoclast effector genes such as MMP9, Ctsk, Acp5, and Clcn7 are only moderately up-regulated, whereas the expression of genes such as Lyz and Cybb that are associated with macrophage differentiation are not down-regulated (see list of additional genes in Table S1). These data indicate that miR-155 plays a major role in the regulation of macrophage vs. osteoclast differentiation, by down-regulating numerous osteoclast-related genes.
Fig. 4.
miR-155 misexpression blocks osteoclast differentiation by repressing MITF and PU.1 expression. mRNA extracted from wild-type and miR-155 misexpressing cells was analyzed on an Affymetrix microarray platform either before or after 72 h of stimulation with RANKL and M-CSF. (A) On the left, a gene expression heat map reveals the molecular fingerprint of M-CSF and RANKL-based osteoclast differentiation by depicting the 500 mRNAs whose log2 expression patterns have the most significant variance. For each mRNA, log2 expression levels are normalized to its mean log2 expression; expression levels are indicated on a green-to-red scale. The expression of 234 mRNAs is up-regulated (green-to-red), and the expression of 266 mRNAs is repressed (red-to-green), in comparison with unmanipulated cells. On the right, the distribution of potential targets (seed-matches) is given by red bars correspond to genes targeted miR-155, miR-1, miR-124a, or a random hexamer (rand). (B) A log2-scale scatter plot, presenting the expression of mRNAs from RANKL/M-CSF-treated wild-type RAW264.7 cells (RAW; x axis), and miR-155-expressing cells subjected to the same treatment (155 ME; y axis). MITF targets from ref. 38 are depicted in red; the expression of GPNMB is marked in blue. (C) mRNA expression levels of Acp5 (TRAP), Cathepsin K (CtsK), RANK, and GPNMB in miR-155 misexpressing cells. (D) qPCR analysis for the expression of MITF, PU.1, and CSF1R in cells misexpressing miR-155, shown as a percentage of control levels (mean ± SD). (E) Western blot analysis of MITF and PU.1 expression in cells misexpressing miR-155, shown as a percentage of control levels (mean ± SD). (F) Renilla luciferase reporters containing either the miR-155 complementary sites from mouse MITF 3′UTR (WT) or mutant versions of the UTR (M-1, M-2) were cotransfected with miR-155 (155) or miR-133 (133) expression vectors into HEK-293T cells. Luminescence was normalized to Firefly luciferase activity. (G) A representative micrograph of TRAP-stained RAW264.7 cells 72 h after induction with RANKL/M-CSF. Shown are control (NT) or misexpression of MITF (MITF) either in wild-type cells (RAW), or in cells that, in addition, misexpress miR-155 (155 ME). The number of TRAP-positive, multinucleated osteoclasts per well is quantified on the right. (H) GPNMB mRNA expression levels in RANKL/M-CSF-treated RAW cells that misexpress miR-155 (155 ME) alone or together with MITF (155 ME MITF). Data are presented as a percentage of GPNMB expression in RANKL/M-CSF-treated wild-type RAW264.7 cells (mean ± SD). Data from at least three independent experiments, performed in duplicates, are represented as mean ± SD. *P < 0.05; **P < 0.01. RU, relative units.
miRNA regulation may simultaneously repress multiple mRNAs. Therefore, one potential mechanism by which miR-155 could regulate the osteoclast program is through repression of a broad set of osteoclast genes. We searched for potential miR-155 binding sites in the 3′ untranslated region (3′UTR) of the mRNAs that undergo major alterations upon osteoclast differentiation. The distribution of potential miR-155 targets was completely random, and was not enriched in the sequences of down-regulated mRNAs, relative to two other miRNA genes (miR-1, miR-124a) or to a random sequence (Fig. 4A). In addition, the set of genes depicted in Fig. 4A does not show enrichment for any other miRNA binding sequence (Fig. S5). This analysis suggests that the impact of miR-155 on the osteoclast transcriptome might be conveyed through control of a few key regulators of osteoclast differentiation. For example, GPNMB is a transmembrane glycoprotein, essential for osteoclastogenesis (28). GPNMB is the most dramatically suppressed mRNA when miR-155 is misexpressed in progenitors (Fig. S6) and in cells subjected to osteoclast differentiation (Fig. 4 B and C). However, the GPNMB 3′UTR does not display potential miR-155 binding sites, implying that GPNMB is indirectly down-regulated by miR-155.
GPNMB expression is controlled by MITF (29). Furthermore, loss of MITF function (30) is reminiscent of the morphological and functional effects of miR-155. This similarity suggests that miR-155 may control GPNMB downstream of the MITF pathway. To determine whether the MITF pathway is specifically controlled by miR-155, our data were revisited, using a set of putative MITF targets described by Meadows et al. (31). All these MITF target genes possess an MITF binding sequence in their promoter and are also highly induced in RAW264.7 cells by MITF misexpression. Our analyses revealed a highly specific overlap between MITF targets and mRNAs repressed by miR-155 misexpression, under identical experimental conditions (P value <0.0001; Fig. 4B). Using quantitative PCR (qPCR), we further quantified a few MITF targets in osteoclasts, including TRAP/Acp5, CtsK, RANK, and GPNMB, and found that the expression of these genes is dramatically lower in RANKL/M-CSF-induced miR-155 misexpressing cells relative to controls (Fig. 4C). We therefore conclude that miR-155 interferes with the MITF pathway, including GPNMB, and may function as a direct regulator of MITF. Consistent with this hypothesis, we found that in cells misexpressing miR-155, MITF mRNA and protein levels decreased by ≈50% relative to controls (Fig. 4 D and E).
The core machinery in osteoclast differentiation involves a transcription factor complex composed of MITF and PU.1 (32), and miR-155 was reported to repress PU.1(18), suggesting that the repression of osteoclast differentiation by miR-155 may arise from its affect on PU.1 as well. We found that in cells misexpressing miR-155, PU.1 is also down-regulated, yet to a lesser extent (Fig. 4 D and E).
To address the direct regulation of miR-155 upstream of MITF, we verified that the mRNA isoform expressed in RAW 264.7-derived osteoclasts harbors miR-155 binding sites (Fig. S7) and then obtained a heterologous luciferase reporter assay to explore the functionality of two potential miR-155 binding sites at the 3′UTR of MITF. This analysis revealed that one of the predicted binding sites on the MITF 3′UTR is directly affected by miR-155. Moreover, once the binding site is mutated, these interactions are abolished (Fig. 4F).
If miR-155 epistatically regulates MITF, then the blockade conferred by miR-155 on osteoclastogenesis may be reversed by its coexpression with an MITF variant that lacks miR-155 binding sites. To experimentally assess this hypothesis, we infected miR-155 misexpressing RAW 264.7 cells and controls with a retroviral vector that coexpresses MITF and GFP. FACS-purified GFP+ cells were then induced to differentiate into osteoclasts with RANKL and M-CSF. Strikingly, MITF rescues osteoclastogenesis in the face of miR-155 misexpression, resulting in large, multinucleated TRAP-positive osteoclasts (Fig. 4G). In addition, we quantified the expression levels of GPNMB, because its expression was most significantly repressed by miR-155 misexpression (Fig. S6 and Fig. 4C). We found that MITF up-regulates GPNMB expression 15-fold, relative to the levels found in cells that only express miR-155, raising it to approximately the levels found in wild-type osteoclasts (Fig. 4H). These functional rescue experiments reveal the epistasis of miR-155 over MITF in the monocytic lineage.
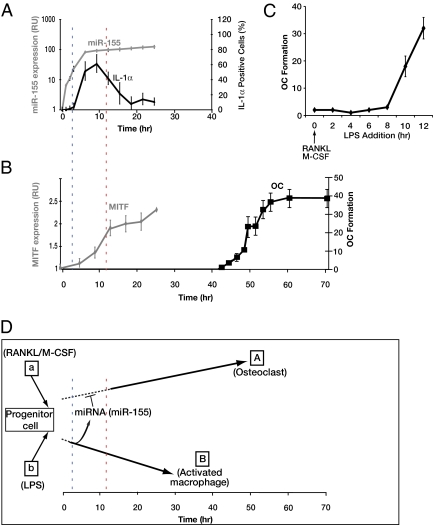
We then sought to compare the kinetics of miR-155 expression and its target MITF in wild-type RAW264.7 cells. As shown in Fig. 5, miR-155 is rapidly and extensively up-regulated (100-fold) after induction with LPS, approaching a plateau at ≈5 h. This rapid miR-155 up-regulation precedes stereotypical changes in the expression of Il-1α, an early and transient marker of activated macrophages (Figs. 5A and 6) and nitric oxide synthesis (Fig. S8). At the same time, osteoclast induction of parallel RAW264.7 cultures with RANKL/M-CSF caused MITF up-regulation, but the temporal scale of this reaction was significantly slower. Intriguingly, terminal differentiation of macrophages and osteoclasts follows a similar trend, revealing an intrinsic temporal asymmetry in these two related lineages. We suggest that the marked difference in miR-155 and MITF synthesis kinetics provides committed macrophages with the opportunity to block RANKL-dependent osteoclast differentiation. Thus, miR-155 is up-regulated much earlier than MITF and blocks it from approaching some critical level that is essential for initiation of osteoclast differentiation. To test whether the temporal asymmetry between miR-155 and MITF up-regulation represents a functional window, we induced osteoclast differentiation with RANKL/M-CSF and then added LPS to the culture at varying time points. Ten hours after stimulation with RANKL, LPS can no longer block osteoclastogenesis (Fig. 5C), consistent with the expression dynamics of miR-155/MITF and in line with the idea that irreversible osteoclast commitment is slower than macrophage commitment.
Fig. 5.
Temporal asymmetry in macrophage and osteoclast differentiation. (A) A detailed temporal analysis of miR-155 expression (gray, log10 scale; y axis on the left) and the relative number of RAW264.7-derived macrophages, expressing IL-1α (y axis on the right) quantified at various time points after LPS stimulation (x axis). (B) Temporal analysis of MITF expression (gray, linear scale; y axis on the left) and the emergence of osteoclast sealing zone-like structures (black, linear scale; y axis on the right) after stimulation of RAW264.7 cells with RANKL/M-CSF. Blue and red vertical dashed lines indicate the time for miR-155 and MITF half-maximal levels, respectively. Data from at least three independent experiments, performed in duplicates, are shown as mean ± SD. (C) The number of giant multinucleated osteoclasts per well, quantified in cells stimulated with RANKL/ M-CSF. LPS was added immediately (0 h) or in delayed intervals, as noted. Data from two independent experiments, replicated eight times, are shown as mean ± SD. **, P < 0.01. (D) A suggested model for an miRNA-based mechanism underlying the commitment of multipotent progenitors to a single cell fate. A progenitor may differentiate to either fate “A” or fate “B,” given specific induction with signal “a” or “b,” respectively. LPS induces differentiation along the macrophage lineage. Macrophage commitment, reflected by up-regulation of miR-155 expression, is depicted by a transition from a dashed to a solid black line, whereas osteoclast commitment is significantly slower. miR-155 plausibly represses osteoclast differentiation through direct repression of MITF within a ≈10-h window, denoted by the blue and red vertical dashed lines.
Discussion
The primary objective of this study was to gain insights into the molecular mechanism underlying the commitment of individual pluripotent progenitors to one and only one fate, despite the presence of two or more conflicting differentiation signals. We show that miRNAs, because of their capacity to block one pathway while enabling an alternative fate, may provide a general molecular mechanism for fate restriction. Specifically, we demonstrate that miR-155 drives the commitment of monocyte progenitors toward activated macrophages by interfering with the expression of MITF, which propels the same progenitors toward osteoclast differentiation (see model in Fig. 5D).
The differential expression kinetics of miR-155 and MITF appears to play a key role in regulating the macrophage vs. osteoclast commitment. Thus, within 2–3 h after LPS stimulation, transcriptional and posttranscriptional mechanisms up-regulate miR-155 ≈100-fold (Figs. 1 and 5A; refs. 19 and 33), whereas commitment to the osteoclast fate (approximated in Fig. 5B by the up-regulation of MITF), is a relatively slow process that becomes apparent in cultured RAW264.7 cells ≈10–12 h after RANKL induction (Fig. 5). We propose that the up-regulated miR-155 (Figs. 4 and 5) blocks MITF and, as a consequence, inhibits osteoclast development. Thus, a temporal asynchronous mechanism, based on rapid miR-155 up-regulation, represses MITF expression, thus blocking the osteoclast fate.
MITF is a nuclear effector that integrates M-CSF/RANKL signals during osteoclast differentiation to initiate the expression of osteoclast-specific effector genes. Although it was shown that MITF is repressed by inflammatory signals such as LPS (34), the mechanism underlying this inhibition was not clear. Our study suggests that miR-155 is a mediator of this suppression.
The repression of MITF by miR-155 is accompanied by corepression of PU.1 (Fig. 4), another key transcription factor in osteoclast development that was shown to synergize with MITF (35). Repression of PU.1 by miR-155 is well-documented (18, 33, 36, 37); yet because MITF expression alone is sufficient to reverse miR-155 activity, it likely constitutes the dominant player in this particular system.
It is noteworthy that the suppressive effect of miR-155 over osteoclast development is limited to a particular timeframe, after inflammatory or osteoclastogenic stimulation. Thus, if added simultaneously or up to ≈10 h after RANKL+M-CSF introduction, LPS effectively suppresses osteoclast induction through miR-155 activity. However, once MITF expression is established, naturally or artificially, using a vector that lacks the endogenous 3′UTR, LPS can no longer block osteoclast development (Figs. 4 and 5). Because mutually exclusive expression of miRNAs and their target genes is a prevailing theme in multiple developmental contexts (8, 9), it would be intriguing to learn whether the induction of miR-155 by LPS is equally suppressed, when the cells are first primed by RANKL+M-CSF.
miRNA may repress targets related to previous or alternative cell fates. Under these circumstances, the miRNA and target expression are mutually exclusive (8, 38). miRNA were also suggested to “fine-tune” the expression of targets coexpressed in the same cell (reviewed in refs. 12 and 39). The case of miR-155 may be distinguished from these patterns, because dramatic up-regulation of the miRNA that is coexpressed with its target blocks MITF from approaching the threshold level required for osteoclast differentiation.
A more general miRNA-based mechanism underlying the commitment of bi- or pluripotent progenitors to a single cell fate is suggested, considering the data presented herein on miR-155, along with other studies addressing the role of miRNAs in a variety of developmental contexts. For example, miR-203 promotes epidermal differentiation by restricting proliferative potential and inducing cell-cycle exit (10) and miR-150 expression in megakaryocytes represses erythrocyte differentiation (40).
Finally, miR-155 function is also intriguing within a clinical osteolytic/inflammatory context. For example, elevated miR-155 expression was documented in the synovial fluid of joints affected by rheumatoid arthritis (RA), where it may be involved in repressing the expression of the proteolytic metalloproteinases MMP-3 and MMP-1 (41). Therefore, it would be interesting to study whether miR-155 dysfunction contributes to the misregulation of conflicting inflammatory-osteolytic signals associated with RA etiology.
In summary, our study reveals a unique role for miR-155 in the monocytic lineage as a regulator of macrophage differentiation, where it is rapidly and dramatically up-regulated to repress the alternative osteoclastic cell fate. The rapid rate of the miRNA up-regulation, relative to the transcriptional mechanisms underlying osteoclast fate determination, provides the basis for coherent macrophage differentiation.
Materials and Methods
RAW264.7 cells were grown in DMEM/10% FBS, 2 mM L-glutamine, 100 U/mL penicillin/streptomycin or in α-MEM/20 ng/mL M-CSF and 20 ng/mL RANKL for 72 h to differentiate into osteoclast-like cells. Sealing zone-like structures were detected in cells stably expressing actin-GFP or by phalloidin staining (Sigma). TRAP-positive multinucleated osteoclasts (>3 nuclei) were identified by using a leukocyte acid phosphatase kit (Sigma). Bone resorption pits were identified by Coomassie Brilliant Blue stain 6 d after culturing RAW264.7-derived osteoclasts over bovine tibia bone slices. Additional methods are provided in detail in SI Materials and Methods and primers used in this study are described in Table S2.
Supplementary Material
Acknowledgments
We thank Zvi Kam, Shirley Horn-Saban, and Ari Elson for technical help and discussions, Barbara Morgenstern for editorial assistance, and Roni Apte (Ben-Gurion University of the Negev, Beer-Sheva, Israel) for anti-IL-1α Abs. B.G. holds the Erwin Neter Professorial Chair in Cell and Tumor Biology. E.H. is the incumbent of the Helen and Milton A. Kimmelman Career Development Chair. This work was supported by grants from the Volkswagen Foundation, the Nanotechnology Center for Mechanics in Regenerative Medicine (NIH Grant PN2 EY016586; part of the NIH Nanomedicine Development Center Network) and the National Institutes of Health Cell Migration Consortium Grant U54 GM64346 (to B.G.), the Israel Science Foundation (to E.H.), the Women’s Health Research Center (to E.H.), the Estate of Florence Blau (to E.H.) and the Wolfson Family Charitable Trust.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE16749).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0915022107/-/DCSupplemental.
References
- 1.Kawai T, Akira S. TLR signaling. Semin Immunol. 2007;19:24–32. doi: 10.1016/j.smim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 2.Lacey DL, et al. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell. 1998;93:165–176. doi: 10.1016/s0092-8674(00)81569-x. [DOI] [PubMed] [Google Scholar]
- 3.Yasuda H, et al. Osteoclast differentiation factor is a ligand for osteoprotegerin/osteoclastogenesis-inhibitory factor and is identical to TRANCE/RANKL. Proc Natl Acad Sci USA. 1998;95:3597–3602. doi: 10.1073/pnas.95.7.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature. 2003;423:337–342. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- 5.Cavaillon JM, Haeffner-Cavaillon N. Signals involved in interleukin 1 synthesis and release by lipopolysaccharide-stimulated monocytes/macrophages. Cytokine. 1990;2:313–329. doi: 10.1016/1043-4666(90)90061-w. [DOI] [PubMed] [Google Scholar]
- 6.Godambe SA, Chaplin DD, Bellone CJ. Regulation of IL-1 gene expression: Differential responsiveness of murine macrophage lines. Cytokine. 1993;5:327–335. doi: 10.1016/1043-4666(93)90064-c. [DOI] [PubMed] [Google Scholar]
- 7.Bogdan C. Nitric oxide and the immune response. Nat Immunol. 2001;2:907–916. doi: 10.1038/ni1001-907. [DOI] [PubMed] [Google Scholar]
- 8.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3’UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 9.Farh KK, et al. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 10.Yi R, Poy MN, Stoffel M, Fuchs E. A skin microRNA promotes differentiation by repressing ‘stemness’. Nature. 2008;452:225–229. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giraldez AJ, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 12.Hornstein E, Shomron N. Canalization of development by microRNAs. Nat Genet. 2006;38(Suppl):S20–S24. doi: 10.1038/ng1803. [DOI] [PubMed] [Google Scholar]
- 13.Sugatani T, Hruska KA. Impaired micro-RNA pathways diminish osteoclast differentiation and function. J Biol Chem. 2009;284:4667–4678. doi: 10.1074/jbc.M805777200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tam W. Identification and characterization of human BIC, a gene on chromosome 21 that encodes a noncoding RNA. Gene. 2001;274:157–167. doi: 10.1016/s0378-1119(01)00612-6. [DOI] [PubMed] [Google Scholar]
- 15.Teng G, Papavasiliou FN. Shhh! Silencing by microRNA-155. Philos Trans R Soc Lond B Biol Sci. 2009;364:631–637. doi: 10.1098/rstb.2008.0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tili E, et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 18.O’Connell RM, et al. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. 2008;205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruggiero T, et al. LPS induces KH-type splicing regulatory protein-dependent processing of microRNA-155 precursors in macrophages. FASEB J. 2009;23:2898–2908. doi: 10.1096/fj.09-131342. [DOI] [PubMed] [Google Scholar]
- 20.Rodriguez A, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci USA. 2009;106:7113–7118. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raschke WC, Baird S, Ralph P, Nakoinz I. Functional macrophage cell lines transformed by Abelson leukemia virus. Cell. 1978;15:261–267. doi: 10.1016/0092-8674(78)90101-0. [DOI] [PubMed] [Google Scholar]
- 23.Hsu H, et al. Tumor necrosis factor receptor family member RANK mediates osteoclast differentiation and activation induced by osteoprotegerin ligand. Proc Natl Acad Sci USA. 1999;96:3540–3545. doi: 10.1073/pnas.96.7.3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krek A, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 25.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: Targets and expression. Nucleic Acids Res. 2008;36(Database issue):D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 27.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheng MH, Wergedal JE, Mohan S, Lau KH. Osteoactivin is a novel osteoclastic protein and plays a key role in osteoclast differentiation and activity. FEBS Lett. 2008;582:1451–1458. doi: 10.1016/j.febslet.2008.03.030. [DOI] [PubMed] [Google Scholar]
- 29.Ripoll VM, et al. Microphthalmia transcription factor regulates the expression of the novel osteoclast factor GPNMB. Gene. 2008;413:32–41. doi: 10.1016/j.gene.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 30.Weilbaecher KN, et al. Linkage of M-CSF signaling to Mitf, TFE3, and the osteoclast defect in Mitf(mi/mi) mice. Mol Cell. 2001;8:749–758. doi: 10.1016/s1097-2765(01)00360-4. [DOI] [PubMed] [Google Scholar]
- 31.Meadows NA, et al. The expression of Clcn7 and Ostm1 in osteoclasts is coregulated by microphthalmia transcription factor. J Biol Chem. 2007;282:1891–1904. doi: 10.1074/jbc.M608572200. [DOI] [PubMed] [Google Scholar]
- 32.Sharma SM, et al. MITF and PU.1 recruit p38 MAPK and NFATc1 to target genes during osteoclast differentiation. J Biol Chem. 2007;282:15921–15929. doi: 10.1074/jbc.M609723200. [DOI] [PubMed] [Google Scholar]
- 33.Gatto G, et al. Epstein-Barr virus latent membrane protein 1 trans-activates miR-155 transcription through the NF-kappaB pathway. Nucleic Acids Res. 2008;36:6608–6619. doi: 10.1093/nar/gkn666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishii J, et al. Lipopolysaccharide suppresses RANK gene expression in macrophages by down-regulating PU.1 and MITF. J Cell Biochem. 2008;105:896–904. doi: 10.1002/jcb.21886. [DOI] [PubMed] [Google Scholar]
- 35.Luchin A, et al. Genetic and physical interactions between Microphthalmia transcription factor and PU.1 are necessary for osteoclast gene expression and differentiation. J Biol Chem. 2001;276:36703–36710. doi: 10.1074/jbc.M106418200. [DOI] [PubMed] [Google Scholar]
- 36.Martinez-Nunez RT, Louafi F, Friedmann PS, Sanchez-Elsner T. MicroRNA-155 modulates the pathogen binding ability of dendritic cells (DCs) by down-regulation of DC-specific intercellular adhesion molecule-3 grabbing non-integrin (DC-SIGN) J Biol Chem. 2009;284:16334–16342. doi: 10.1074/jbc.M109.011601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vigorito E, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hornstein E, et al. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature. 2005;438:671–674. doi: 10.1038/nature04138. [DOI] [PubMed] [Google Scholar]
- 39.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu J, et al. MicroRNA-mediated control of cell fate in megakaryocyte-erythrocyte progenitors. Dev Cell. 2008;14:843–853. doi: 10.1016/j.devcel.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stanczyk J, et al. Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 2008;58:1001–1009. doi: 10.1002/art.23386. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.