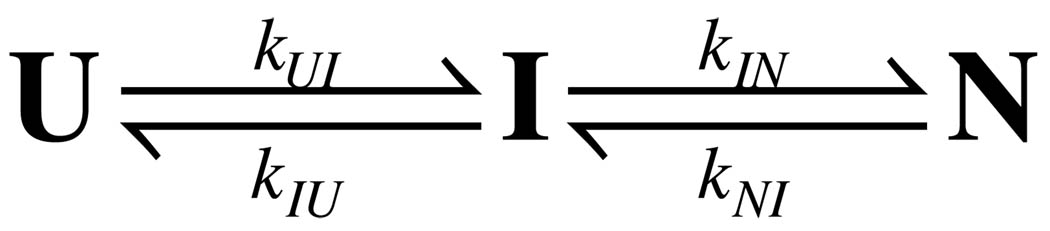Abstract
Understanding the interconversion between thermodynamically distinguishable states present in a protein folding pathway provides not only the kinetics and energetics of protein folding but also insights into the functional roles of these states in biological systems. The protein component of bacterial RNase P holoenzyme from Bacillus subtilis (P protein) was previously shown to be unfolded in the absence of its cognate RNA or other anionic ligands. P protein was used in the present study as a model system to explore general features of intrinsically disordered protein (IDP) folding mechanisms. The use of trimethylamine-N-oxide (TMAO), an osmolyte that stabilizes the unliganded folded form of the protein, enabled us to study the folding process of P protein in the absence of ligand. Transient stopped-flow kinetic traces at various final TMAO concentrations showed multiphasic kinetics. Equilibrium “cotitration” experiments were performed using both TMAO and urea during the titration to obtain a TMAO-urea titration surface of P protein. Both kinetic and equilibrium studies show evidence of a previously undetected intermediate state in the P protein folding process. The intermediate state is significantly populated and the folding rate constants involved in the reaction are relatively slow compared to intrinsically folded proteins of similar size and topology. The experiments and analysis described serve as a useful example for mechanistic folding studies of other IDPs.
Keywords: Protein folding, Osmolyte, Intrinsically disordered, RNase P protein
Protein folding kinetics has been extensively explored in many model protein systems (1, 2), but most previous studies were focused primarily on natively structured globular proteins. Recently, a biologically important class of proteins called intrinsically disordered proteins (IDP) have become widely recognized (3–6). It had long been accepted that the folded structure of a protein determines its function, but most IDPs lack definite structure in the absence of bound ligand(s). Many IDPs adopt active conformations through disorder-to-order transitions coupled to binding of other cellular components and this coupled folding and binding phenomenon is a major feature of the IDP family (7). Coupling a folding transition to a molecular interaction has been considered functionally advantageous because it may allow a protein to recognize different target substrates, could enhance the rate of intermolecular interactions, and could provide for large intermolecular interfaces with a relatively small protein (8). Recent studies have shown that many IDPs participate in transcriptional and translational regulation (9). Because of their structural flexibility, some IDPs may function as multimolecular hubs that interact with multiple binding partners (3).
Ribonuclease P holoenzyme (RNase P) is the endonuclease that catalyzes the maturation of pre-tRNA by removal of the 5′ leader sequence (10–12). The crystal structure of the 119 residue protein subunit of Bacillus subtilis RNase P (referred to hereafter as P protein) has been determined to 2.6Å resolution (13, 14). Our previous biophysical studies of P protein demonstrated that it is intrinsically disordered in the absence of ligand and at low ionic strength (15). The major functional role of P protein in the holoenzyme is to enhance substrate recognition (16) and stimulate the intrinsic ribonuclease activity of the catalytic RNA subunit (17). Several structural roles for P protein have also been proposed including stabilizing the structure of P RNA or compensating for charge repulsion (18). Because P protein is intrinsically disordered, holoenzyme assembly includes folding of the protein. Thus, the mechanism P protein folding is important to understanding the assembly of the RNase P ribonucleoprotein complex. Moreover, it is interesting to determine the sequence of events in multistep folding and RNA binding reactions.
Previous studies have demonstrated that many small proteins exhibit simple two-state folding kinetics (1). In this situation, the two thermodynamically distinguishable ensembles are in equilibrium and undergo cooperative conformational change as they interconvert. However, many proteins do not exhibit two-state folding but instead fold via one or more partially folded intermediates. These intermediates can be broadly divided into two classes (19). One class is obligatory folding intermediates, which usually, but not always, have partially formed folded structure. Under some circumstances, they can also accelerate the search for the folded state (20, 21). The second class is non-obligatory intermediates, which are usually kinetically trapped misfolded species (22). However, these intermediate states can also be steps in an alternative folding pathway (23). Although intermediate states can be crucial in the overall folding process, it is sometimes challenging to detect and characterize them. Many intermediates form rapidly, within the dead-time of a conventional stopped-flow instrument. Transient kinetic intermediates are generally not stable enough to be observed at equilibrium under strongly denaturing conditions. Intermediates may also be spectroscopically silent, making them difficult to detect. Thus, it is important to search for conditions that maximize the population and stability of possible intermediate states studying order to determine their thermodynamic, kinetic and even structural properties. In the present study, we detected a state that apparently is an obligatory, on-pathway intermediate in the folding of P protein.
There have been many studies of biological functions of IDPs (8), but fewer fundamental biochemical and biophysical studies of properties such as their folding mechanisms. Renaturation of IDPs and denaturation of natively folded proteins are two sides of the same experimental coin and both provide insights into the balance of forces and intrinsic steps of the folding/unfolding reaction. The theory and methods behind these two kinds of experiments are fundamentally the same. The main difference is the experimental design: whereas natively structured proteins require denaturant such as urea or guanidinium chloride to perturb the equilibrium, IDPs require “protection osmolyte” (24). In the present study, an osmolyte, trimethylamine-N oxide (TMAO), accompanied by the denaturant urea were used to perturb folding equilibrium so as to nearly fully deplete both the unfolded (high TMAO) and folded (high urea) states. Interestingly, as described below, we find no conditions that fully deplete the intermediate state. The highly basic amino acid composition (11 Arg, 19 Lys, and 3 His out of 119 residues) of P protein gives a net charge of +16 to +17 at neutral pH and a pI of 10.8. Electrostatic repulsion in the absence of the cognate polyanionic ligands may explain why folding to a compact folded state is unfavorable.
Osmolytes are small organic molecules, including certain amino acids, polyols, and methylamines, that are used in many organisms to maintain intracellular water level (25). This water stress can result from osmotic pressure, desiccation, extreme temperatures or high intracellular urea concentrations. Water stress resistant cells are evolved to use osmolytes to maintain macromolecular stability and function in the presence of these stresses. Many studies have used osmolytes to stabilize the folded state of proteins (26, 27) and RNA (28) secondary and tertiary structures. The major mechanism of protein stabilization by osmolyte is through unfavorable interaction with the backbone atoms, which increases the free energy of the denatured state (29). Most osmolytes are interchangeable and additive, which means that stabilization can be achieved in combination. Studies also show that the free energy of unfolding is linearly dependent on osmolyte concentration, analogous to the linear dependence of the unfolding free energy on chemical denaturant, albeit with a slope of opposite sign (30). This property is particularly useful for estimation of unfolding free energy of IDPs and partly folding proteins (31). IDPs can be titrated with osmolyte to generate a refolding curve from which the (positive) folding free energy can be estimated using the same linear extrapolation method used to analyze a urea or guanidine denaturation curve. The stability of a partly folded protein can be measured by mixing both urea and osmolyte solvent in protein sample to generate complete denatured or native state baselines in the titration curve (31).
In this study, P protein was used as a model system to elucidate the folding/unfolding process of intrinsically disordered proteins. Both kinetic and equilibrium data show the presence of an intermediate state in the P protein folding pathway. A sequential three-state mechanism was modeled to fit the experimental data. Kinetic and equilibrium data were fitted globally to extract the rate constants, thermodynamic parameters and the spectroscopic properties of these three states. To our knowledge, these are the first folding and unfolding rate constants determined by TMAO induced folding kinetics for an intrinsically disordered protein.
Experimental Procedures
Chemicals and reagents
Ultrapure urea was purchased from Nacalai Tesque Inc. Trimethylamine-N-oxide (TMAO) was from Fluka. Cacodylic acid was from Sigma. In order to remove the background fluorescence from TMAO, a 3 M stock TMAO solution was mixed with activated charcoal (0.025g/ml) for at least 5 hours. The charcoal was then remove by filtration using 0.22 µm syringe filter. Urea and TMAO concentrations were measured by refractive index using the equation from (32) and (26), respectively.
Expression and purification of P protein
The gene for the P protein variant F107W used in this study was constructed by the QuickChange site direct mutagenesis protocol (Strategene). The variant was overexpressed in E. coli and purified as described (13) with the following modifications. The pooled fractions of P protein that eluted from the second CM-Sepharose column were concentrated using Centriprep until the volume of the pooled fractions was down to 1~2 ml. The buffer of the protein stock was changed to 6 M Guanidine-HCl, 10 mM Tris at pH 7.5 during this concentrating step. The concentrated stock was then loaded on an S-100 column (GE Healthcare) equilibrated with 6 M Guanidine-HCl, 10 mM Tris (pH 7.5) to remove EDTA bound P protein. Fractions containing P protein were pooled and stored at −80 °C. Before equilibrium titration or stopped flow kinetic experiments, the P protein sample was dialyzed against water extensively, then 20 mM sodium cacodylate, pH 7. In all experiments the protein concentration was determined using the Edelhoch method using an extinction coefficient of 11,200 M−1 cm−1 at 276nm (33).
CD and fluorescence spectrum
An Aviv model 202 CD spectrometer was employed to collect CD wavelength spectra. Samples were scanned from 260nm to 190 nm in 1 nm increments with a bandwidth of 1.5 mm and averaged for 3s at each wavelength. Protein concentration was 10 µM in 20 mM sodium cacodylate, pH 7, in the presence or absence of TMAO using a cuvette with a 1 mm path length at 25 °C.
Fluorescence spectra were collected on a Fluorolog-3 spectrometer from HORIBA Jobin Yvon using a 1 cm pathlength cuvette. The sample was 5 µM P protein in 20 mM cacodylate, pH 7. The temperature was controlled at 25°C by a circulating water bath. The excitation wavelength was 285 nm with a 4 nm bandpass, and emission spectra were collected from 300 to 400 nm at 1nm increments.
CD and Fluorescence equilibrium cotitration experiments
The urea-TMAO equilibrium cotitration experiments were performed on an Applied Photophysics PiStar instrument equipped with both fluorescence and CD detection. A total of six renaturation equilibrium titration curves were collected in each set of experiments and all samples were in 20 mM sodium cacodylate buffer (pH 7.0) at 25 °C. The starting sample for the protein concentration was 5 µM in 0 M, 2 M, 4 M, 6 M urea. The two additional starting samples were 5 µM protein in 6M urea, 0.5 M TMAO or 6 M urea, 0.9M TMAO. The titrant solution is 5 µM protein in 1.4 M TMAO. An automated Hamilton titrator was used to perform all titrations. The CD signal was collected at 222 nm and fluorescence signal was collected using a 320 nm cut-off filter with an excitation wavelength of 285 nm and a 4 nm bandwidth. The CD and fluorescence signals were collected simultaneously from the same sample. Each urea-TMAO concentration pair data point was measured for 15 sec to obtain a mean signal. The equilibrium time between two titration points was 3 to 3.5 minutes.
Stopped-flow fluorescence kinetic experiments
The stopped-flow fluorescence kinetic experiments were performed on an Applied Photophysics SX20 instrument at 25 °C with a circulating water bath. The excitation wavelength was 285 nm with a slit width of 1 mm. The pathlength of the observation cell was 2 mm. The fluorescence signal was collected through a 320 nm high pass filter. For single mixing folding and unfolding experiments, a 6-fold or 11-fold dilution was used to initiate the reactions. In the refolding experiments, protein samples were 6 µM in 20 mM sodium cacodylate buffer (pH 7.0), and TMAO concentrations ranged from 0.15 M to 1.4 M in 20 mM sodium cacodylate buffer (pH 7.0). The final protein concentration was 1 µM with final TMAO concentrations ranging from 0.125–1.167 M. For unfolding experiments, protein samples were 11 µM in 1 M TMAO with 20 mM sodium cacodylate, and final TMAO concentrations ranged from 0 to 0.5 M. Following 11-fold dilution in the stopped flow instrument, the final protein concentration was 1 µM in 0.091 M to 0.55 M TMAO. The time traces of the fluorescence change for the reaction were collected for 20 s in a split time base mode. Five thousand points were collected for the first 10s, and 1000 points from were collected from 10 to 20 s.
Fluorescence signals of protein from equilibrium and kinetic experiments are normalized using signals between two experimental conditions. The signal is arbitrarily set to 1 when Protein is in 0M TMAO, 0M urea, and signal is set to 0 when protein is in 1.2M TMAO, 0M urea.
Data analysis
(a) Kinetic folding/unfolding data: TMAO induced folding/unfolding kinetic time traces from stopped flow experiments were fitted by the non-linear least-squares method with multiple the exponential function:
| (2) |
where F(t) and F(∞) are the observed fluorescence at time t and infinite time, respectively. Ai and λi are the fluorescence amplitudes and observed rate constants, respectively, of the different phases. The amplitudes and observed rate constants were plotted against the final TMAO concentration.
(b) Equilibrium titration data: Equilibrium urea-TMAO cotitration surface data were fit to a linear three-state model assuming that ΔGUI and ΔGUN are linearly dependent on urea and TMAO concentration (31), according to equations 3–7:
| (3) |
| (4) |
| (5) |
| (6) |
| (7) |
where and are the m values that describe the linear osmolyte concentration dependence of the free energy, ΔGij; Kij is the equilibrium constant of the i–j equilibrium; Fi is the fractional population of state i at a specific osmolyte concentration; and are the intensities of CD and fluorescence of the unfolded (U), intermediate (I), folded (N) states, respectively; SigCD and SigFL are the observed CD and fluorescence signals.
(c) Global fitting: The CD and fluorescence equilibrium titration surfaces were fit using equations 1–5. The amplitudes and observed rate constants were fit to a linear three-state model (scheme 1) using the analytical solutions described by Bujalowski (34). The TMAO dependence of the observed rate constants and associated amplitudes assumes a linear dependence of the logarithm of the microscopic rate constants in scheme 1 on TMAO concentration in the form:
| (8) |
where kij is the microscopic rate constant for the transition from state i to j at specific TMAO concentration. βij is the Tanford value (35) which is a measure of the average degree of solvent exposure of the transition state relative to the difference in solvent exposure between states i and j. Equilibrium data, kinetic amplitudes and observed rates were then fit simultaneously using Mathematica (Wolfram) to obtain the equilibrium and kinetic parameters for the model.
Scheme 1.
Results
Tryptophan variants of P protein as a probe to study kinetics and thermodynamics of P protein
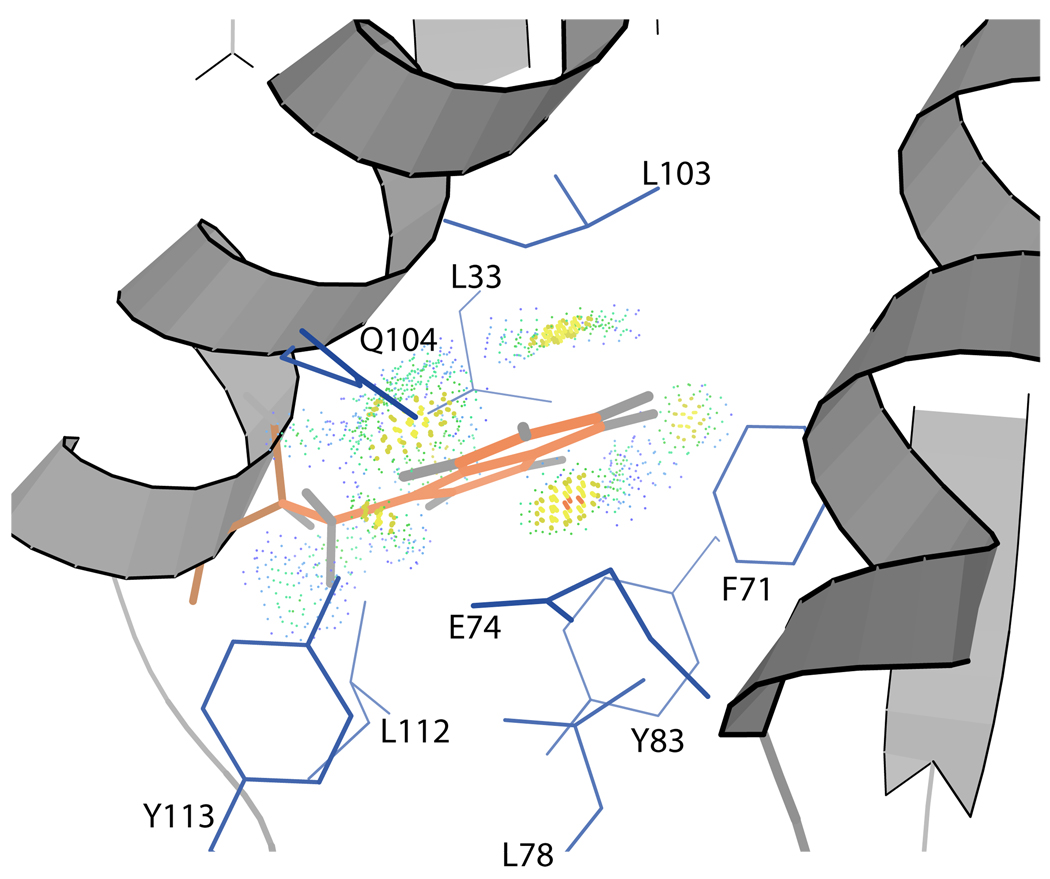
Wild type B. subtilis P protein does not contain any tryptophan residues, which are often used as intrinsic fluorescent probes to monitor protein conformational changes. To utilize this property for fluorescence-detected folding kinetic and equilibrium studies, a tryptophan containing variant of P protein, F107W, was generated. The site for mutagenesis was selected based on a sequence comparison between B. subtilis P protein and the highly homologous (similarity) E. coli P protein, C5. In E. coli P protein, tryptophan 107 is highly conserved among different gram-negative bacteria. The Trp residue fits in a hydrophobic pocket and has a fluorescence change upon urea unfolding (36). Figure 1 shows a model of the m0 rotamer of a tryptophan sidechain with −26° adjustment of χ2 that inserted into the crystal structure of wild type B. subtilis P protein (14). The Trp sidechain fits well in this conformation using Structural biology tool in software King (37) indicates good Van der Waals contacts with surrounding sidechains. The F107W variant shares the IDP property with wild type P protein and can be induced to fold upon small anion binding as observed in previous studies of wild type P protein (15). Addition of TMAO to the protein sample also shifts the folding equilibrium to the folded state. The fluorescence emission spectrum (285 nm excitation) of F107W in various TMAO concentrations from 0 M to 1 M is shown in Figure 2A. Unfolded F107W has a λmax at 358 nm, and the peak is blue-shifted to 308 nm when the protein folds to the native state, indicating that the tryptophan sidechain is located in a nonpolar region of the folded protein. (38, 39). The total fluorescence also decreases upon folding, suggesting that there is additional quenching in the native protein. The wild type P protein crystal structure supports these conclusions, but the Trp is not in a completely nonpolar environment. As shown in Figure 1, the modeled Trp sidechain is surrounded by 4 Leu, 1 Phe, 2 Tyr, 1 Gln, and 1 Glu residues. Adjacent aromatic sidechains are possible quenchers to Trp fluorescence. Figure 2B shows the CD spectrum of the variant also from 0M to 1 M TMAO. The appearance of distinct peaks around 220 and 208 nm indicates the formation of α-helical secondary structure at [TMAO] > 0.2 M. The CD spectrum of P protein in 1 M TMAO is similar to that found for 20 mM sulfate, in which P protein is predominantly folded (15). The NMR HSQC spectrum of F107W (Fig. 2C) under folded condition is similar to the wild type spectrum, and more than 90% of the peaks in the F107W spectrum can be assigned by comparing with the wild type spectrum. Most of the shifted resonances correspond to amides in close proximity to the site of the Trp substitution.
Figure 1.
Model of Trp 107 sidechain contacts with surrounding sidechains. The model was constructed using the crystal structure of B. subtilis P protein (PDB 1A6F) and the Sidechain Mutator Tool in King (50). The dots generated by Probe (51) indicate the contact surface of the Trp sidechain with the surrounding atoms. Red spikes represent steric clashes between atoms. The blue to green dots indicate a favorable Van der Waals contacts between each atom. The model shows that the engineered Trp fit well in this hydrophobic pocket and probably does not greatly perturb the overall structure. The modeled Trp sidechain rotamer is m0 with a minor adjustment of the chi2 angle.
Figure 2.
Fluorescence and circular dichroism spectra of the F107W variant of P protein in the presence of various TMAO concentrations. (A) Fluorescence spectrum of F107W variant of P protein (5µM, 1cm cuvette) in the same TMAO concentrations used for CD. The excitation wavelength was 285nm and the emission spectra were scanned from 400 to 300 nm in 1 nm increments. (B) Far-UV CD spectrum of F107W variant of P protein (10µM, 1mm cuvette) in 20mM cacodylate buffer in 0, 0.2, 0.4, 0.6, 0.8 and 1M TMAO. The spectra were scanned from 260 to 200 nm in 1 nm increments. (C) The HSQC spectrum of F107W (red) overlay on top of the wild type spectrum (black).
Taken together, these results suggest that the fluorescence of the Trp residue in F107W variant is sensitive to the structural change between folded and unfolded states and with minimum perturbation of the overall protein structure. For these reasons, the variant was used for the equilibrium and kinetic studies described below.
TMAO stopped flow kinetic studies and chevron plots
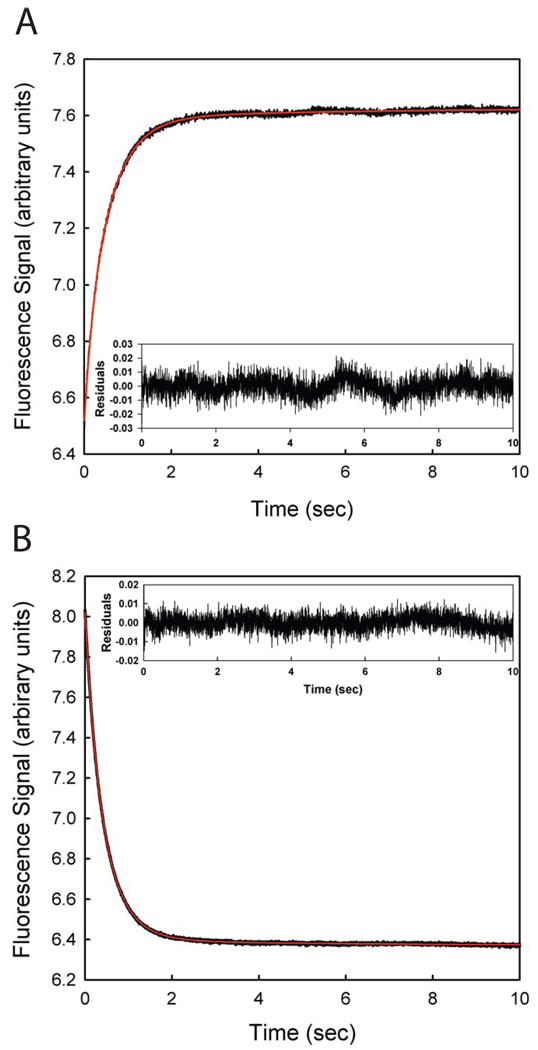
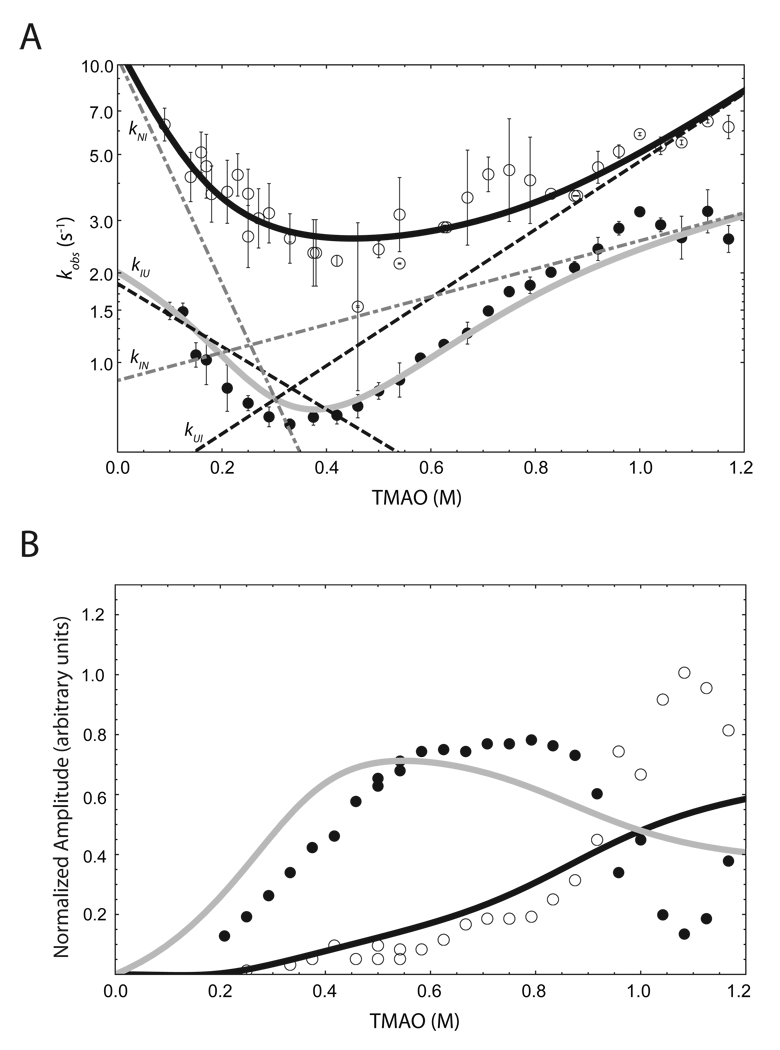
Transient fluorescence traces of P protein folding/unfolding reactions induced by changes in [TMAO] were collected using the stopped-flow method. As expected from the fluorescence emission spectra described above, the fluorescence signal decreases when folding is induced by mixing protein in buffer with TMAO solutions (TMAO up-jump) while the signal increases when unfolding is induced by mixing protein in TMAO with buffer alone (TMAO down-jump). Figures 3A and 3B show representative up-jump and down-jump traces, respectively. The kinetic traces were fitted to multiple exponential functions by nonlinear least squares method using Pro Data Viewer Software from Applied Photophysics. The minimum complexity model that fits both up-jump and down-jump kinetic traces is a sum of three exponentials (Equation 2 in materials and methods), so there are observable three phases in both the folding and unfolding reactions. Each fit was obtained by minimizing the residuals as shown in the inset panel of Figure 3A and 3B. The observed rate constants (kobs) of the fast phase (ranging from 2 to 7 s−1) and middle phase (ranging from 0.8 to 3 s−1) are dependent on TMAO concentration, as are the amplitudes associated with each phase, as shown in Figures 4A and 4B, respectively. Because the sum of the fast and the middle phase amplitudes account for more than 95% of the total raw amplitude, these two phases describe the major folding kinetics of P protein. The third phase, which is around 0.10 ~ 0.15 s−1 is independent of TMAO concentration and the amplitude of the phase is small (less than 5% of total raw amplitude). We have assigned this slowest phase to cis/trans isomerization of Pro 39 and/or Pro 90 based on the absence of the third phase in folding and unfolding of a P39A/P90A/F107W triple substitution variant of P protein (data not shown). All subsequent kinetic and equilibrium studies employed the prolinecontaining F107W variant, whose transients were fit to three exponentials but the slowest (third) phase was not included in subsequent data analysis.
Figure 3.
Folding/unfolding of P protein induced by changes in TMAO concentration and monitored by stopped-flow fluorescence. (A) Unfolding (down-jump) of P protein from 1 M to 0.09 M TMAO. The kinetic trace of the unfolding process monitored by fluorescence is the black line. The red line through the data points is the best fit to a triple exponential function. The inset shows the residuals of the curve fits. (B) Folding (up-jump) of P protein from 0 M to 0.54 M TMAO. The red line through the black data points is the best fit to a triple exponential function. The inset shows the residual of the fitting. Both data sets were collected at a rate of 500 points/sec.
Figure 4.
Folding and unfolding kinetics of F107W P protein as a function of final TMAO concentration. (A) TMAO concentration dependence of the natural logarithm of observed rate constants for both up-jump and down-jump experiments. The open circles and closed circles are the fast and slow observed rate constants obtained from the folding (up-jump) experiments, respectively. The black dashed lines are rate constants kUI and kIU as the function of TMAO concentration, and the gray dot-dashed lines are kIN and kNI as the function of TMAO concentration. (B) Raw amplitudes associated with fast and slow observed rate constants are plotted as a function of TMAO concentration. Only the amplitudes of folding (up-jump) experiments are shown. Smooth lines through the observed rate constants are calculated from the parameters determined in the global fit of kobs values and the equilibrium data (listed in Table 1) using the analytical solutions described by Bujalowski and Jezewska (34). The poor agreement between calculated and observed amplitudes at high [TMAO] is likely due to large uncertainties in the experimental amplitudes when the two phases have similar rate constants.
In many multiphase protein folding systems, the observed rate constants are usually at least one order of magnitude different from each other. Under these conditions, the nonlinear lease squares fitting method is robust and capable of accurately estimating both the rates and the associated amplitudes of each phase. However, an intrinsic challenge in kinetic data analysis of the P protein system originates from the similarity of the observed rate constants associated with the two major phases. The observed rate constants for the fast and medium phases in P protein folding processes are relatively close to each other (between 2 to 6 fold). When rate constants of two phases are close, the uncertainty of the parameters obtained from nonlinear square fits is large because the fitting parameters have a correlation coefficient close to 1. In order to best represent the uncertainty for each data point in Figure 4A, the same kinetic experiment was performed at least three times. The uncertainty was calculated for each condition from the fitting results of these independent experiments. Because there are two major folding phases, the simplest mechanism consistent with the data is a three-state reaction with an on-pathway intermediate state, as shown in Scheme 1. Four kinetic rate constants, kUI, kIU, kIN, and kNI are associated with this mechanism. To obtain the extrapolated values for all four rate constants, we attempted to fit their TMAO concentration dependence using Equation 8. The initial fitting analyses from the kinetic data alone did not yield convergent estimations of the parameters because the two observed rate constants were similar and the uncertainty in the fastest rate constant was high. Equilibrium titration experiments were performed to provide more constraints in the fitting process.
Urea-TMAO fluorescence and CD co-titration
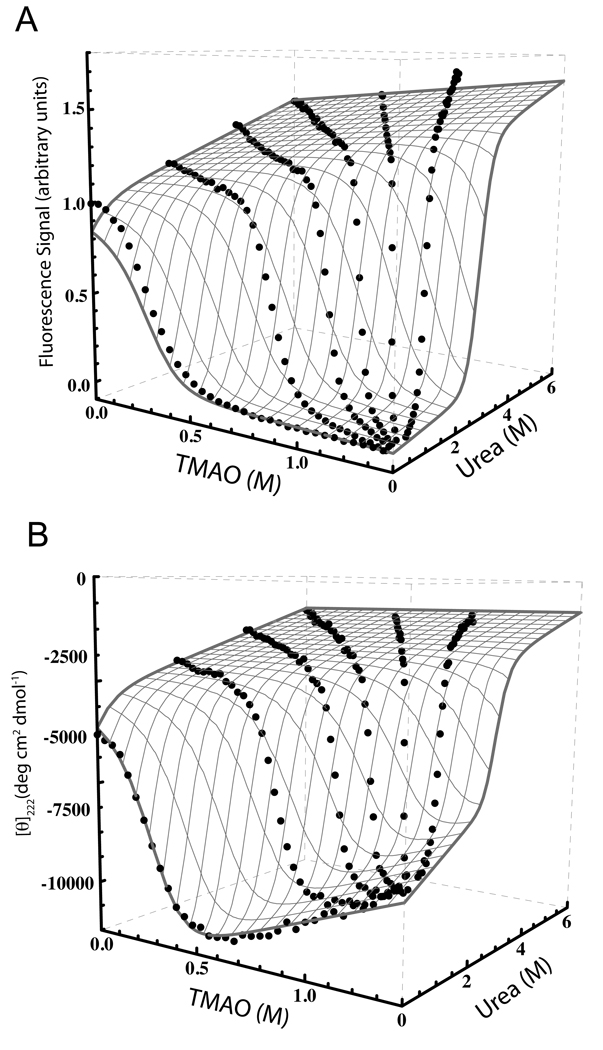
Urea and guanidinium chloride denaturation titrations are widely used to estimate the protein stability, but the method is suitable only for stable proteins with both folded and unfolded baselines are present in the titration curve. When applying this method to IDPs, the absence of a complete cooperative transition and folded baseline in the titration curve makes it impossible to estimate the thermodynamic parameters using denaturation titration. In order to estimate the stability of P protein, we used mixtures of urea and TMAO as the denaturant and renaturant, respectively, to populate the folded or unfolded states of P protein. Titrations were initiated with a starting sample consisting of protein in various urea or urea/TMAO mixtures. Protein at the same concentration, in 1.4 M TMAO was then added using an automated titrator, which resulted in progressive dilution of the starting urea concentration while the TMAO concentration increased. Six such titration curves allowed us to construct a three-dimensional urea-TMAO cotitration surface, which accessed a wide range of folded and unfolded populations. Each titration step yielded both fluorescence and CD data from the same protein sample using the same instrument. This procedure eliminated errors from discrepancies in protein, urea, or TMAO concentrations that might occur if the two types of data were collected separately. Figures 5A and 5B show the TMAO dependence of the equilibrium fluorescence signal (λex= 285 nm, λem > 320 nm) and CD signal (λ= 222 nm) for F107W variant, respectively. Taken separately, fluorescence and CD titration curves can be fit to a two state model but the resulting fits give statistically significant different C values. This observation indicates the presence of one or more significantly populated equilibrium intermediates in the folding process, consistent with the kinetic data. Our previous reports on P protein folding described the mechanism as two-state, based on statistically insignificant differences in C values (15). The small difference, combined with the fact that the near- and far-UV CD data were collected on different samples, led us to erroneously suggest that equilibrium intermediate is not present, when in fact it is (see below). However, by combining the equilibrium titration data with the stopped flow data, we can globally fit the equilibrium data to a model based on the three-state mechanism shown in scheme 1.
Figure 5.
Fluorescence and circular dichroism detected equilibrium co-titration surface of the P protein (5mM, 10mm path length cuvette). The gray meshed surfaces on the two data sets are the best global fit results using a three-state model described by equation 5. (A) The fluorescence titration surface constructed by six titration curves plotted as a function of [urea] and [TMAO]. The protein sample was excited at 285nm and the fluorescence signal was collected through a 320 nm high pass filter. (B) The CD titration surface constructed by six titration curves plotted as a function of [urea] and [TMAO]. The CD signal was monitored at 222 nm. The best-fit equilibrium parameters from the model were listed in Table 1.
The experimental data were analyzed using equations 3–7 (materials and methods), which assume that the free energy of each transition is linearly dependent on both urea and TMAO concentration. In general, the CD and fluorescence signals of each state may vary with the urea or TMAO concentration. In accordance with convention, we assumed that the CD and fluorescence signals of U, I and N states vary linearly with urea and TMAO concentrations:
| (1) |
where i = U, I, N; Sig is the CD or fluorescence signal; and are the slopes of the baseplanes describe the urea and TMAO dependence of the signal of each state. Therefore, nine parameters for the fluorescence signals and nine parameters for the CD signals for a total of 18 spectroscopic parameters were used in the model. Fluorescence and CD titration data were simultaneously fitted using a three-state equilibrium model. Six thermodynamic parameters, , and , were used as global fitting parameters between the fluorescence and CD data sets. Fitting of the equilibrium data alone did not yield well-determined results for every parameter, especially the spectroscopic parameters associated with the intermediate state. For this reason, the uncertainties of the free energy , and are large. The poor estimation of these parameters mainly results from the large parameter space (total of 24 parameters) and lack of two distinct transitions on the titration surfaces. This latter limitation indicates that either the intermediate state is poorly populated throughout the titration range or that its spectroscopic signals are very similar to U or N. In order to overcome these limitations in the data analysis, we estimated and from NMR spectra collected at various urea concentrations as described below. These parameters were held fixed in a global non-linear least squares fit of the remaining parameters to the combined kinetic and equilibrium data.
and from NMR urea titration experiments
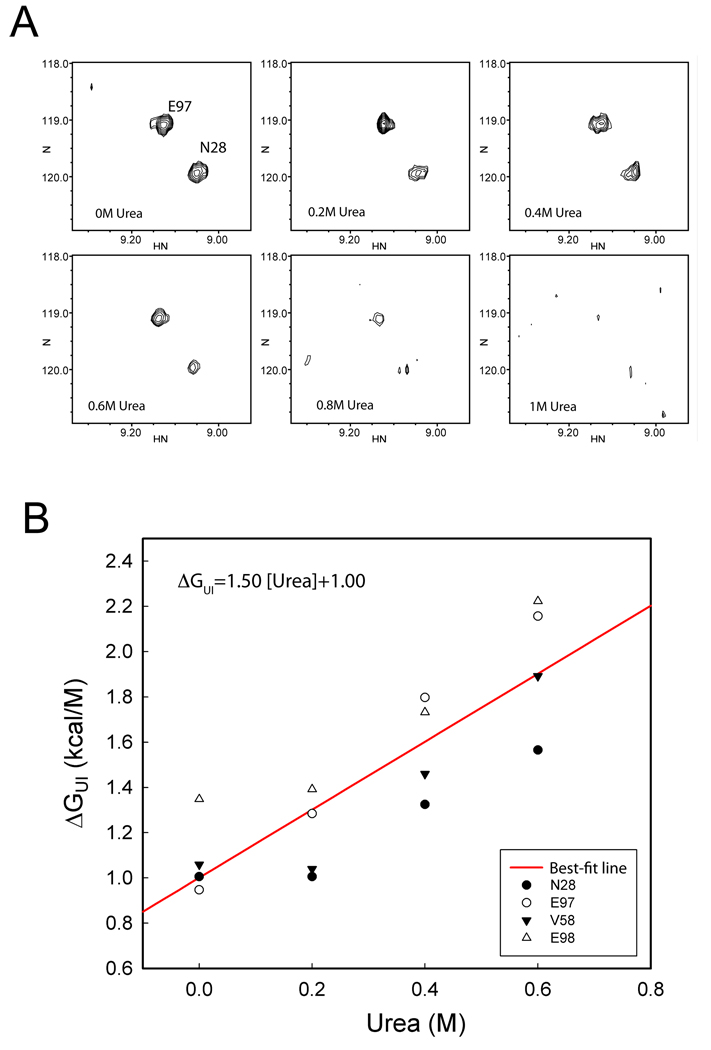
NMR experiments were performed to estimate the equilibrium constant between U and I states. More than 95% of the backbone amide 15N-1H resonances of the folded state P protein were assigned in a previous study (40). The presence of at least one alternative state was suggested by the observation of around 50 extra peaks in the 15N-1H HSQC spectrum of sulfate folded P protein. In addition, the 15N-1H HSQC spectrum of P protein in the absence of sulfate (20 mM sodium cacodylate buffer, pH 7) has some resonances whose positions are similar to those found in sulfate folded protein. Both of these results suggest the existence of an alternative state in the P protein folding transition, which gives rise to extra HSQC peaks under both folding and unfolding conditions. As a working hypothesis, we assumed that this alternative state corresponds to the intermediate state observed in the kinetic and equilibrium experiments. In addition, we assumed that the native state peaks observed in the sulfate-free sample correspond to residues that are in a native-like magnetic environment in the intermediate state. If correct, this interpretation suggests that the intermediate state has a significant population, comparable to that of the unfolded state. The equilibrium data suggest that the population of the native state is negligible under these conditions, so addition of urea alters the NMR spectrum by perturbing the equilibrium between the U and I state. Thus, 15N-1H HSQC spectra were collected at various urea concentrations. The relative populations of U and I states were estimated by peak intensity or volume change and used to determine the ΔGUI as a linear function of urea concentration.
Figure 6A shows a region of the unfolded P protein HSQC spectrum, which contains some of the observed crosspeaks of folded structure of P protein. These peaks were monitored in spectra obtained at urea concentrations ranging from 0 to 1 M. As urea concentration increases, the intensities of these peaks decrease, indicating a decrease in the population of the intermediate state to which they are tentatively assigned. To estimate the population of the intermediate from these spectra, the native state (in 20 mM sulfate, pH 7.0) HSQC spectrum was used as the reference to determine the intensity of each native-like peak in fully folded P protein. The peak intensities in all spectra were normalized to the intensity of the K119 amide peak, whose position is independent of sample conditions, making it an excellent internal standard. The normalized intensity of each intermediate peak was divided by the normalized intensity of the corresponding native spectrum peak to produce an estimate of the fractional population of the intermediate at each urea concentration. These populations were used to calculate KUI, which was converted to free energy using Equation 3. To avoid line shape artifacts, peak volume was used to estimate peak intensity. Figure 6B shows the free energy calculated in this fashion for each monitored peak plotted against urea concentration and fit to a linear function. The intercept of each line is the free energy , and the slope of urea m-value, , between U and I transition. The data points from 4 peaks gave ranging from 1.0 to 1.3 kcal/mol. The fact that urea m-values show larger variation between different peaks might be due to the large variation in estimation of the peak volume for different peaks. Combined estimates of (1.0 kcal mol−1) and (1.5 kcal mol−1 M−1) were obtained from a linear regression of all the data points, shown as the red line in the Figure 6B. These two values were used as fixed parameter values in the global fitting of the combined equilibrium and kinetic data.
Figure 6.
Urea titration of P protein in the absence of ligand (20mM cacodylate buffer, pH 7) monitored by HSQC spectrum and the estimation of free energy difference between U and I state. (A) A region of the HSQC spectrum is shown in the plot, and two peaks corresponding to N28 and R97 in a native like environment are labeled. The urea concentrations are shown in each plot. (B) The estimation of free energy between U and I state from different residues against urea concentration was plotted. The free energy was calculated from the population of U and I state from HSQC urea titration experiment. The line is the best-fit results of all the data points in the plot using the linear function.
Global fitting of kinetic and equilibrium data
Because the kinetic or equilibrium data alone cannot provide an accurate estimation of all of the parameters for the model, a global analysis of both data sets was performed to estimate the kinetic and spectroscopic parameters together. The experimental data included the two observed rate constants and the two associated amplitudes at 33 different TMAO concentrations and the fluorescence and CD titration surfaces. Because preliminary NMR studies (data not shown) indicated that the magnetic environment of the Trp residue in the intermediate state may be similar to that of the unfolded state, we further simplified the fit by assuming that the TMAO dependence of fluorescence of the intermediate state is the same as the unfolded state. The two solid lines in Figure 4A are the best-fit results for the observed rate constants. As depicted in Figure 4A, the usual chevron shape of kobs vs. [TMAO] is not observed because the two reaction steps in the scheme 1 have similar rate constants and are coupled to each other over most of the [TMAO] range. The dashed lines and dot-dashed lines in the plot are individual rate constants calculated as a function of TMAO concentration. In the high TMAO concentration (above 1.1M), the fast and slow phases approximately equal to kUI and kIN, respectively. The amplitudes were also analyzed using the model. The fitting results have the similarly general trends, but do not fit well to the data points. The reasons for this discrepancy might result from the large uncertainty of fluorescence signal of each state from the fitting (Table 1) and also likely due to large uncertainties in the experimental amplitudes when the two phases have similar rate constants. As shown in Figure 5, the best-fit model fit the equilibrium fluorescence and CD data quite well, although the denatured fluorescence baseplane did not fit as well as the CD. Inspection of the data in the denatured baseplane region shows differences in the denatured state fluorescence in different titrations. Although these differences could be caused by day-to-day variations in the fluorescence detection electronics, they also could result from small perturbations in the actual (not read) pH of the buffer caused by urea sensitivity of the pH electrode (41). The fluorescence of F107W variant is pH sensitive between 5 and 7 (data not shown), presumably because it is near the His 105 residue whose ionization state could affect the fluorescence signal of tryptophan fluorophore. Differences in the starting fluorescence of each titration curve could be due to slightly different pH values at each starting urea concentration.
Table 1.
Kinetic and equilibrium parameters of three-states folding and unfolding of P protein.
| g Kinetic Parameters | g Equilibrium Parameters | Spectroscopic Parameters | |||||
|---|---|---|---|---|---|---|---|
| akUI | 0.34±0.05 | 1.0±0.2 |
d |
−3899±339 | |||
| bkIU | 1.8±0.3 | 1.48±0.1 | −1 × 104±1017 | ||||
| kIN | 0.86±0.1 |
c |
−3±0.2 | −1.29 × 104±508 | |||
| bkNI | 10.5±0.3 | −5.8±0.3 |
e |
1±0.01 | |||
| βUI | 0.48±0.02 | 1.5±0.5 | 0.1±0.02 | ||||
| βIN | 0.89±0.02 | 2.14±0.2 | 0.05±0.01 | ||||
|
f |
0.34 | ||||||
| 0.41 | |||||||
Units are in s−1
Calculated using kIU/KUI and kIN/KIN, respectively.
Units are in kcal mol−1 M−1
Units are in [θ]222 (mean residue ellipticity)
Arbitrary units
Unit less
All kinetic and equilibrium parameters are the values in the absence of Urea and TMAO
The kinetic and equilibrium parameters for the best-fit model are listed in Table 1. The free energy differences between U and I and I and N are approximately the same and predict populations for unfolded, intermediate and native state populations of 83±5, 16±3, and 1±0.2%, respectively. The ratios of TMAO and urea m-values are −2.0±0.3 and −2.7±0.5 for U⇔I and I⇔N transitions, respectively. Auton and Bolen have proposed a method for calculating m-values based on differences in the backbone and sidechain solvent accessible surface areas (42). Because this calculation depends on side chain composition, there is no single expected ratio of TMAO to urea m-value for different equilibria. Previous studies have shown that Barnase and Nank1–7* have the ratio of −1.1 and −2.4, respectively (31). However, in general TMAO has a stronger opposite effect on protein stability than urea (larger absolute m-value). The best-fit values of both folding rate constants (kUI=0.34 s−1 and kIN=0.86 s−1 ) are relatively slow compared to other proteins of similar size (1). Further kinetic studies on other IDPs are necessary to determine if the slow refolding rate constant is a general property of the IDP folding reactions.
The best-fit model parameters include the intermediate state CD and fluorescence signals, which provide information about the structure of I. The best-fit value for the fluorescence signal of I is close to that of N, indicating that the environment surrounding Trp107 is similar in the intermediate and folded states. Moreover, the CD signal ([θ]222) of I is midway between U and N suggesting that approximately 60–70% of the native state secondary structure, particularly helix, is formed in the intermediate state.
Discussion
Comparison between intrinsically disordered protein and natively folded protein kinetics
These kinetic studies of P protein show that the folding rate constant is lower than the unfolding rate constant, as expected for an equilibrium that favors the unfolded state. Interestingly, the folding rate constant is quite low when compared to natively folded proteins of similar size and topology (1). Contact order is a measure of the topological complexity of a given protein’s structure and a measurement of the number of contacts a residue makes with other residues that are local in sequence, relative to the number of contacts with residues distant in sequence (43). Thus, the contact order is related to secondary structure, but is also influenced substantially by the tertiary fold. Plaxco et al. showed a correlation between the contact order and natural logarithm of the refolding rate constant and the position of the transition state. These correlations suggested that the lower the contact order the faster the folding rate constant and less folded-like the transition state. From this correlation, the contact order of the P protein (14.3) predicts a folding rate constant (1000 s−1) at least 2 to 3 orders of magnitude faster than our experimental estimates. This major discrepancy suggests that other factors besides topology, such as the charge-charge interactions in the protein, can be a major determinant of the actual folding kinetics. There are not many folding kinetic studies of IDPs in the literature, so it is too early to conclude whether slow folding (as opposed to fast unfolding) is a general property of such proteins. In the case of P protein, the low stability is at least partially due to unfavorable interactions in the more compact intermediate and folded state ensembles, many of which persist in the transition state ensembles , thus raising the barrier to folding. This conclusion is supported by the high βTanford values determined from the TMAO dependence of the rate constants (see Table 1), which indicate that the loss in solvent accessible surface area (ΔSASA) for is 48% of the ΔSASA for U→I and the ΔSASA for is 89% of that for I→N (see Table 1). Thus, both transition state ensembles are significantly more compact than the preceding ground states and unfavorable electrostatic interactions in these compact transition states are a likely explanation for slow folding.
Utility of TMAO (renaturant) stopped flow and cotitration experiments as a general method to determine IDP folding mechanisms
Various protective osmolytes have been used as co-solutes to stabilize proteins and favor the folded state (26, 44). Many studies have shown that some IDPs can fold to their folded states in the presence of their binding partners. Are the folded states of proteins induced by protective osmolytes the same as those induced by their binding partner? In this and previous studies (15), the CD and fluorescence spectrum of the ligand induced native state and the TMAO induced native state of P protein are very similar. Furthermore, most of the recognizable peaks in the 1H-15 N HSQC spectrum of the native state from these two conditions have similar chemical shifts indicating the structures of the protein under the two conditions are very similar. This conclusion is consistent with previous studies of the effect of TMAO on stabilities and structures of other proteins, which noted little perturbation on the folded structure (27, 45). Other studies have shown that TMAO enhances the enzymatic activity (46, 47), which makes large structural perturbations unlikely. For these reasons, addition of TMAO appears to be a useful way to shift the folding equilibrium of an IDP to favor the same folded conformation that is present in the absence of TMAO, albeit at much lower concentrations.
With the increasing number of reported IDPs and partially folded proteins, there is a need for systematic biophysical methods to measure the kinetic and thermodynamic properties of these proteins. Although some previous studies employed denaturant-induced protein unfolding kinetic experiments in the presence of various TMAO concentrations to investigate the stability effect (48, 49), there have been no previous kinetic studies using TMAO to initiate IDP refolding. The stopped-flow measurements of TMAO-induced IDP folding/unfolding described in the present study are the equivalent of traditional denaturant-induced unfolding/folding kinetics of natively structure proteins. The experimental approach described here should work well to study the folding mechanism of other IDPs.
Previous studies have demonstrated that TMAO combined with urea or guanidinium chloride denaturation can be used to properly estimate the free energy of partially folded protein (31). The cotitration stability measurement method we developed in this study should be applicable to other IDPs. The highest TMAO concentration in the titration was 1.4 M, but in many circumstances it could be used at much higher concentrations. The upper limit in this study was determined by the weak buffering capacity of our low affinity buffer (20 mM sodium cacodylate).. As shown in the Figure 6A and 6B, the six titration curves formed diagonal transects across the titration surface and well sampled the unfolded and folded baseplanes and the transition region between. Although the presence of an intermediate is not immediately apparent on these surfaces, the simultaneous detection of CD and fluorescence during the equilibrium titration made it possible for the global fitting procedure to detect an intermediate in the folding reaction, measure its population and provide information about its spectroscopic properties.
Role of partially folded intermediate in folding mechanism
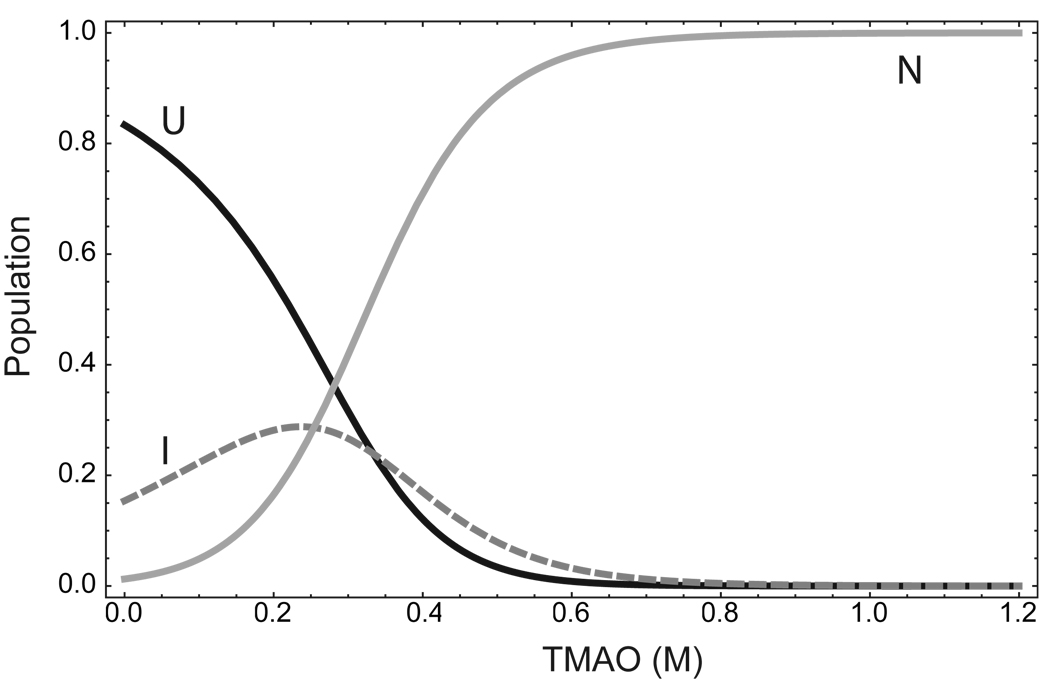
Based on the parameter values listed in Table 1, the population of the P protein intermediate state ensemble is around 16–20% of the total protein concentration in the absence of urea and TMAO. In addition, the mUI/mUN values for both urea and TMAO in Table 1 indicate that about 40% of the area buried in N is also buried in I, suggesting that I is significantly folded. The best-fit CD signal for I listed in Table 1 is significantly more negative than for U, indicating that I has significant secondary structure content. The secondary structure of the intermediate state of P protein explains the shape of the CD signal vs. [urea] along the 0 M TMAO line in Figure 2B. The curvature from 0 M to 2 M urea can be explained by the equilibrium transition between the unfolded state and the intermediate state of the P protein. The population of the three states as function of TMAO concentration is plotted in Figure 7 using the equilibrium parameters from the fitting. The population of the intermediate state reaches its maximum at 0.25 M TMAO. At that TMAO concentration, the population of I is comparable to U and N state, and the relative amounts of three states are approximately equal. By these criteria, I should be considered an important subensemble in the P protein energy landscape, distinguishable from U and N by the kinetic barriers between the three states.
Figure 7.
Population of U, I, and N state as a function of the TMAO concentration. (A) The population of each state was calculated using equation 4 and the equilibrium parameters listed in Table 1.
In addition to its role in the folding mechanism, the intermediate state ensemble may play a functional role in vivo such as protein-RNA recognition during the holoenzyme assembly process. Given the highly basic nature of P protein, there is almost certainly some non-specific binding of protein on the negatively charged RNA molecule. The partially folded P protein intermediate state may more efficiently search the binding pocket on the RNA than either fully folded or unfolded P protein leading to more productive binding events to form holoenzyme. Future studies will be aimed at determining the structure of this important species and whether it has significant affinity for the two anionic ligands that bind with high affinity to N. Kinetic studies on P protein with small anionic ligands and cognate binding partner, P RNA, will also provide insights about the holoenzyme assembly process.
Acknowledgement
Supported by National Institute of Health Grants 5RO1GM061367 and 5RO1GM081666 (to T.G.O.).
We thank Drs. K.V. Rajagopalan and Michael Fitzgerald for generously sharing their instruments. We also thank Dr. Gordon Hammes for critical comments and reading for the manuscript, and various members of the Oas laboratory for insightful discussion and advice.
Abbreviations
- CD
circular dichroism
Reference
- 1.Jackson SE. How do small single-domain proteins fold? Folding & design. 1998;3:R81–R91. doi: 10.1016/S1359-0278(98)00033-9. [DOI] [PubMed] [Google Scholar]
- 2.Clark AC. Protein folding: are we there yet? Arch Biochem Biophys. 2008;469:1–3. doi: 10.1016/j.abb.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunker A, Oldfield C, Meng J, Romero P, Yang J, Chen J, Vacic V, Obradovic Z, Uversky V. The unfoldomics decade: an update on intrinsically disordered proteins. BMC Genomics. 2008;9:S1. doi: 10.1186/1471-2164-9-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fink AL. Natively unfolded proteins. Curr Opin Struct Biol. 2005;15:35–41. doi: 10.1016/j.sbi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 5.Uversky V. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 2002;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wright P, Dyson H. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 7.Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol. 2002;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- 8.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 9.Wright PE, Dyson HJ. Linking folding and binding. Current Opinion in Structural Biology. 2009:1–8. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 11.Frank DN, Pace NR. Ribonuclease P: unity and diversity in a tRNA processing ribozyme. Annu Rev Biochem. 1998;67:153–180. doi: 10.1146/annurev.biochem.67.1.153. [DOI] [PubMed] [Google Scholar]
- 12.Kurz J, Fierke C. Ribonuclease P: a ribonucleoprotein enzyme. Curr Opin Chem Biol. 2000;4:553–558. doi: 10.1016/s1367-5931(00)00131-9. [DOI] [PubMed] [Google Scholar]
- 13.Niranjanakumari S, Kurz J, Fierke C. Expression, purification and characterization of the recombinant ribonuclease P protein component from Bacillus subtilis. Nucleic Acids Res. 1998;26:3090–3096. doi: 10.1093/nar/26.13.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stams T, Niranjanakumari S, Fierke C, Christianson D. Ribonuclease P protein structure: evolutionary origins in the translational apparatus. Science. 1998;280:752–755. doi: 10.1126/science.280.5364.752. [DOI] [PubMed] [Google Scholar]
- 15.Henkels CH, Kurz JC, Fierke CA, Oas TG. Linked folding and anion binding of the Bacillus subtilis ribonuclease P protein. Biochemistry. 2001;40:2777–2789. doi: 10.1021/bi002078y. [DOI] [PubMed] [Google Scholar]
- 16.Crary S, Niranjanakumari S, Fierke C. The protein component of Bacillus subtilis ribonuclease P increases catalytic efficiency by enhancing interactions with the 5' leader sequence of pre-tRNAAsp. Biochemistry. 1998;37:9409–9416. doi: 10.1021/bi980613c. [DOI] [PubMed] [Google Scholar]
- 17.Buck AH, Dalby AB, Poole AW, Kazantsev AV, Pace NR. Protein activation of a ribozyme: the role of bacterial RNase P protein. EMBO J. 2005;24:3360–3368. doi: 10.1038/sj.emboj.7600805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsieh J, Andrews A, Fierke C. Roles of protein subunits in RNA-protein complexes: lessons from ribonuclease P. Biopolymers. 2004;73:79–89. doi: 10.1002/bip.10521. [DOI] [PubMed] [Google Scholar]
- 19.Baldwin RL. On-pathway versus off-pathway folding intermediates. Folding & design. 1996;1:R1–R8. doi: 10.1016/S1359-0278(96)00003-X. [DOI] [PubMed] [Google Scholar]
- 20.Wagner C, Kiefhaber T. Intermediates can accelerate protein folding. Proc Natl Acad Sci USA. 1999;96:6716–6721. doi: 10.1073/pnas.96.12.6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Capaldi AP, Shastry MC, Kleanthous C, Roder H, Radford SE. Ultrarapid mixing experiments reveal that Im7 folds via an on-pathway intermediate. Nat Struct Biol. 2001;8:68–72. doi: 10.1038/83074. [DOI] [PubMed] [Google Scholar]
- 22.Kathuria SV, Day IJ, Wallace LA, Matthews CR. Kinetic traps in the folding of beta alpha-repeat proteins: CheY initially misfolds before accessing the native conformation. Journal of Molecular Biology. 2008;382:467–484. doi: 10.1016/j.jmb.2008.06.054. [DOI] [PubMed] [Google Scholar]
- 23.Kiefhaber T. Kinetic traps in lysozyme folding. Proc Natl Acad Sci USA. 1995;92:9029–9033. doi: 10.1073/pnas.92.20.9029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bolen DW, Baskakov IV. The osmophobic effect: natural selection of a thermodynamic force in protein folding1. J Mol Biol. 2001;310:955–963. doi: 10.1006/jmbi.2001.4819. [DOI] [PubMed] [Google Scholar]
- 25.Yancey PH, Clark ME, Hand SC, Bowlus RD, Somero GN. Living with water stress: evolution of osmolyte systems. Science. 1982;217:1214–1222. doi: 10.1126/science.7112124. [DOI] [PubMed] [Google Scholar]
- 26.Wang A, Bolen DW. A naturally occurring protective system in urea-rich cells: mechanism of osmolyte protection of proteins against urea denaturation. Biochemistry. 1997;36:9101–9108. doi: 10.1021/bi970247h. [DOI] [PubMed] [Google Scholar]
- 27.Lin SL, Zarrine-Afsar A, Davidson AR. The osmolyte trimethylamine-N-oxide stabilizes the Fyn SH3 domain without altering the structure of its folding transition state. Protein Sci. 2009;18:526–536. doi: 10.1002/pro.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lambert D, Draper DE. Effects of osmolytes on RNA secondary and tertiary structure stabilities and RNA-Mg2+ interactions. J Mol Biol. 2007;370:993–1005. doi: 10.1016/j.jmb.2007.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bolen DW, Rose GD. Structure and energetics of the hydrogen-bonded backbone in protein folding. Annu. Rev. Biochem. 2008;77:339–362. doi: 10.1146/annurev.biochem.77.061306.131357. [DOI] [PubMed] [Google Scholar]
- 30.Baskakov I, Bolen DW. Forcing thermodynamically unfolded proteins to fold. J Biol Chem. 1998;273:4831–4834. doi: 10.1074/jbc.273.9.4831. [DOI] [PubMed] [Google Scholar]
- 31.Mello CC, Barrick D. Measuring the stability of partly folded proteins using TMAO. Protein Sci. 2003;12:1522–1529. doi: 10.1110/ps.0372903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warren JR, Gordon JA. On the Refractive Indices of Aqueous Solutions of Urea. J Phys Chem. 1966;70:297–300. [Google Scholar]
- 33.Edelhoch H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry. 1967;6:1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- 34.Bujalowski W, Jezewska MJ. Kinetic mechanism of the single-stranded DNA recognition by Escherichia coli replicative helicase DnaB protein. Application of the matrix projection operator technique to analyze stopped-flow kinetics. J Mol Biol. 2000;295:831–852. doi: 10.1006/jmbi.1999.3378. [DOI] [PubMed] [Google Scholar]
- 35.Fersht A. Structure and mechanism in protein science : a guide to enzyme catalysis and protein folding. New York: W.H. Freeman; 1999. [Google Scholar]
- 36.Gopalan V, Golbik R, Schreiber G, Fersht AR, Altman S. Fluorescence properties of a tryptophan residue in an aromatic core of the protein subunit of ribonuclease P from Escherichia coli. Journal of Molecular Biology. 1997;267:765–769. doi: 10.1006/jmbi.1997.0907. [DOI] [PubMed] [Google Scholar]
- 37.Chen VB, Davis IW, Richardson DC. KING (Kinemage, Next Generation): a versatile interactive molecular and scientific visualization program. Protein Sci. 2009;18:2403–2409. doi: 10.1002/pro.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szabo AG, Stepanik TM, Wayner DM, Young NM. Conformational heterogeneity of the copper binding site in azurin. A time-resolved fluorescence study. Biophys J. 1983;41:233–244. doi: 10.1016/S0006-3495(83)84433-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lakowicz JR. Principles of fluorescence spectroscopy. 2nd ed. New York: Kluwer Academic/Plenum; 1999. [Google Scholar]
- 40.Henkels CH. Biophysical studies of Bacillus subtilis ribonuclease P protein. Duke University; 2005. p. xi. 303 leaves, [Google Scholar]
- 41.Acevedo O. pH Corrections in Chemical Denaturant Solutions. Analytical Biochemistry. 2002;306:158–161. doi: 10.1006/abio.2002.5668. [DOI] [PubMed] [Google Scholar]
- 42.Auton M, Bolen D. Application of the Transfer Model to Understand How Naturally Occurring Osmolytes Affect Protein Stability. Meth Enzymol. 2007;428:397–418. doi: 10.1016/S0076-6879(07)28023-1. [DOI] [PubMed] [Google Scholar]
- 43.Plaxco KW, Simons KT, Baker D. Contact order, transition state placement and the refolding rates of single domain proteins. J Mol Biol. 1998;277:985–994. doi: 10.1006/jmbi.1998.1645. [DOI] [PubMed] [Google Scholar]
- 44.Russo AT, Rosgen J, Bolen D. Osmolyte effects on kinetics of FKBP12 C22A folding coupled with prolyl isomerization. J Mol Biol. 2003;330:851–866. doi: 10.1016/s0022-2836(03)00626-0. [DOI] [PubMed] [Google Scholar]
- 45.He HL, Chen XL, Zhang XY, Sun CY, Zou BC, Zhang YZ. Novel Use for the Osmolyte Trimethylamine N-oxide: Retaining the Psychrophilic Characters of Cold-Adapted Protease Deseasin MCP-01 and Simultaneously Improving its Thermostability. Mar Biotechnol. 2009 doi: 10.1007/s10126-009-9185-2. [DOI] [PubMed] [Google Scholar]
- 46.Mashino T, Fridovich I. Effects of urea and trimethylamine-N-oxide on enzyme activity and stability. Arch Biochem Biophys. 1987;258:356–360. doi: 10.1016/0003-9861(87)90355-9. [DOI] [PubMed] [Google Scholar]
- 47.Kumar R, Serrette JM, Thompson EB. Osmolyte-induced folding enhances tryptic enzyme activity. Arch Biochem Biophys. 2005;436:78–82. doi: 10.1016/j.abb.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 48.Banks DD, Gloss LM. Folding mechanism of the (H3-H4)2 histone tetramer of the core nucleosome. Protein Sci. 2004;13:1304–1316. doi: 10.1110/ps.03535504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Topping TB, Gloss LM. Stability and folding mechanism of mesophilic, thermophilic and hyperthermophilic archael histones: the importance of folding intermediates. J Mol Biol. 2004;342:247–260. doi: 10.1016/j.jmb.2004.07.045. [DOI] [PubMed] [Google Scholar]
- 50.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Word JM, Lovell SC, LaBean TH, Taylor HC, Zalis ME, Presley BK, Richardson JS, Richardson DC. Visualizing and quantifying molecular goodness-of-fit: small-probe contact dots with explicit hydrogen atoms. J Mol Biol. 1999;285:1711–1733. doi: 10.1006/jmbi.1998.2400. [DOI] [PubMed] [Google Scholar]