Abstract
Over the past 5 years, large-scale sequencing has been revolutionized by the development of several so-called next-generation sequencing (NGS) technologies. These have drastically increased the number of bases obtained per sequencing run while at the same time decreasing the costs per base. Compared to Sanger sequencing, NGS technologies yield shorter read lengths; however, despite this drawback, they have greatly facilitated genome sequencing, first for prokaryotic genomes and within the last year also for eukaryotic ones. This advance was possible due to a concomitant development of software that allows the de novo assembly of draft genomes from large numbers of short reads. In addition, NGS can be used for metagenomics studies as well as for the detection of sequence variations within individual genomes, e.g., single-nucleotide polymorphisms (SNPs), insertions/deletions (indels), or structural variants. Furthermore, NGS technologies have quickly been adopted for other high-throughput studies that were previously performed mostly by hybridization-based methods like microarrays. This includes the use of NGS for transcriptomics (RNA-seq) or the genome-wide analysis of DNA/protein interactions (ChIP-seq). This review provides an overview of NGS technologies that are currently available and the bioinformatics analyses that are necessary to obtain information from the flood of sequencing data as well as applications of NGS to address biological questions in eukaryotic microorganisms.
The first report on the sequence of 10 consecutive bases in a DNA strand was published in 1968 (117), but methods to reliably obtain longer DNA sequences, namely, Sanger and Maxam-Gilbert sequencing, were not available until 1977 (71, 96). Of these, only Sanger sequencing underwent improvements that led to automation and therefore, for the next 30 years, large-scale sequencing projects, e.g., whole-genome sequencing for various species, relied on this technology (41). However, despite (or indeed because of) much progress in the area of genome sequencing, it became clear that even more information was to be gained not only from sequencing one genome per species but rather from sequencing and comparing the genomes of different individuals or strains/lines from the same species. This would enable a better grasp of genetic diversity and, in the case of humans, allow “personalized medicine” approaches. To make this feasible, novel techniques were needed that overcame current limitations of Sanger sequencing with respect to throughput and costs (98), and in the last decade, a number of different methods were developed that not only have revolutionized the field of genome sequencing but also can be applied to other biological questions not previously addressed by sequencing-based approaches. This review provides an overview of these so-called second-generation or next-generation sequencing (NGS) technologies and their applications with a special focus on addressing questions relevant to the biology of eukaryotic microorganisms.
NEXT-GENERATION SEQUENCING TECHNOLOGIES
All commercially available NGS technologies differ from automated Sanger sequencing in that they do not require cloning of template DNA into bacterial vectors. Apart from being less labor-intensive, this has the distinct advantage that cloning biases, e.g., due to difficulties with cloning of AT-rich regions or genes that are toxic for Escherichia coli, are avoided. In most NGS approaches, template DNA is fragmented, bound to a substrate, and amplified by PCR to generate clonal representations of the original fragments that are spatially separated for subsequent sequencing (75, 99). A current exception to this is the Helicos system, which does not require template amplification but rather sequences single-template molecules (9, 36). The sequencing itself is achieved by a number of methods that make use of different enzymes (polymerases or ligases) and chemistries to generate light signals that are recorded by highly sensitive detection methods. A common theme of all NGS technologies is the high degree of parallelization, in which millions to billions of sequencing reactions take place at the same time in small reaction volumes, thereby allowing a much higher throughput than automated Sanger sequencing. The following section provides a brief overview of NGS technologies; for a detailed description of the currently available NGS systems, the reader is referred to several comprehensive reviews (66, 75, 99).
The first NGS technology that became commercially available was the Roche/454 genome sequencer (454 Life Sciences, Branford, CT) (68). While the first machines were capable of delivering read lengths of ∼100 bases, the current generation of instruments and chemistry reaches read lengths of >400 bases. These read lengths are the longest that can currently be achieved by any NGS system; however, this is offset by lower throughput and higher per-base costs than for other systems (Table 1). Two other widely used NGS platforms are the Illumina/Solexa genome analyzer (Illumina, San Diego, CA) (6) and the ABI SOLiD system (Applied Biosystems, Carlsbad, CA). While they have a higher throughput than the 454 genome sequencer, they yield much shorter read lengths (Table 1). Another platform that is commercially available is the Polonator (http://www.polonator.org) (100). It is the only open-source system among the NGS technologies but currently also the system with the shortest read lengths (Table 1).
Table 1.
Overview of sequencing techniques that are currently commercially availablea
| NGS technology | Sequencing principle | Read length in bases | % raw read accuracy | Gbc |
|---|---|---|---|---|
| Sanger | Dideoxy sequencing | ∼1,000 | 99.999 | 0.0003 |
| Roche/454 | Pyrosequencing | ∼450 | ≥99 | 0.6 |
| Illumina/Solexa | Reversible terminator chemistry | 36–100 | ≥98–99 | 3–20 |
| ABI/SOLiD | Sequencing by ligation | 35–50 | ≥99.94 | 50–100 |
| Polonator | Sequencing by ligation | 26 | ≥98 | 4–5 |
| Helicos | Single-molecule sequencing | 25–55 | ≥97–99.8b | 21–35 |
Due to rapid progress in this field, error rates are likely to decrease while sequencing lengths will increase. A detailed description of NGS technologies can be found in several recent reviews (66, 75, 99).
Error rates depend on type of error (substitutions, insertions, deletions).
Gb, gigabases.
So far, the only platform that uses single-molecule sequencing and therefore avoids amplification biases is the HeliScope sequencer (Helicos, Cambridge, MA). The technique of single-molecule sequencing has recently been reported to be applicable not only to DNA but also to RNA (86). A robust, commercially available single-molecule system that directly sequences RNA would constitute another major innovation since this would circumvent not only the need for cloning and/or amplification but also the need for reverse transcription of RNA to generate cDNA, thereby avoiding a number of experimental steps that can introduce biases and misrepresentations of the original RNA molecules.
Yet another NGS technology that is scheduled to become available in 2010 is real-time DNA sequencing with single polymerase molecules (28) (Pacific Biosciences, Menlo Park, CA). This method makes use of the high processivity of DNA polymerase, i.e., the fact that DNA polymerase can add thousands of nucleotides to a growing DNA strand without releasing its substrate in between. Consequently, read lengths of >1,000 bases are theoretically possible from a single DNA polymerase molecule and have been achieved under test conditions (28). Further developments in the area of NGS can be expected in the near future, reducing the costs of genome sequencing even more. One example is DNA nanoball sequencing, which was reported to bring the costs for a human genome sequence down to $4,400 (24). Several methods that no longer rely on DNA polymerases or ligases but use nanopore or scanning probe techniques may be emerging novel sequencing technologies. However, these technologies are in a preliminary state, and it has yet to be demonstrated that they can deliver DNA sequences at single-nucleotide resolution (8, 62).
One general problem with all current NGS reads is their shorter length and higher error rate than those of Sanger sequence reads (Table 1). The short read lengths in particular present significant hurdles when it comes to assembling large sequence stretches, e.g., during whole-genome shotgun sequencing. The development of new algorithms to handle NGS data has helped to overcome this problem (see below). Another important improvement is the ability to sequence both ends from a DNA fragment (paired-end sequencing) now implemented for most of the commercially available NGS platforms. Paired-end data allow the scaffolding of contigs (contiguous sequences) in the absence of contiguous coverage of intervening sequences (33). Paired ends can be obtained from the ends of random, usually small DNA fragments. In addition, methods like mate-pair sequencing have been devised, where random DNA fragments are circularized, thereby combining previously distant ends. This DNA is then sheared to generate linear fragments as templates for sequencing. This procedure makes it possible to obtain sequence information from the ends of long fragments (6, 48).
Another issue that has prevented the use of NGS in many cases is that for projects where a large number of samples need to be sequenced, the costs of NGS can be prohibitive. Furthermore, for many purposes the sequence depth delivered by NGS is not needed. However, NGS can still be of use in these cases, because multiplexing, i.e., the sequencing of many individual samples in a single sequencing run, has been developed for a number of NGS platforms (19, 52, 76, 103). Multiplexing can be achieved by labeling individual samples using bar-coded primers during preparation of the sequencing library. The libraries are pooled prior to sequencing, and afterwards, sequence reads are computationally assigned to different samples.
APPLICATIONS OF NGS TECHNIQUES
Genome sequencing.
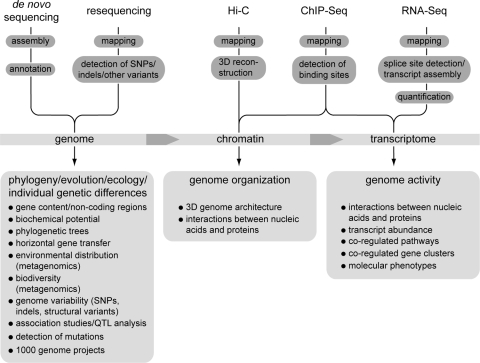
One of the most obvious applications of sequencing techniques with high throughput at low costs is genome sequencing. Here, one can distinguish between resequencing, i.e., sequencing of genomes from a species for which a reference genome is already available, and de novo sequencing (Fig. 1). Resequencing is currently one of the major areas of application for NGS. Here, platforms with short reads but high throughput can reach their full potential, because with a reference genome available even relatively short reads can be mapped with high confidence to the reference sequence. This approach works well even for large genomes, like those of mammals, and a number of individual genomes, e.g., from humans and cattle, have already been sequenced using NGS technologies (1, 6, 26, 47, 72, 91, 97, 111, 114). Sequence reads that are mapped to a reference genome can be used to identify single-nucleotide polymorphisms (SNPs), small insertions or deletions (indels), and copy number variations (CNV) or other structural variants, thereby helping to better understand the genetic basis of phenotypic differences.
Fig. 1.
Next-generation sequencing to address biological questions. SNP, single-nucleotide polymorphism; indel, insertion/deletion; QTL, quantitative trait loci.
The widespread application of NGS to the de novo sequencing and assembly of genomes is an important goal (Fig. 1). However, the short read lengths make the assembly difficult, especially for the larger, more complex, and often repeat-rich genomes of eukaryotes. Therefore, the first genomes that were sequenced with NGS technologies were from bacteria (32, 68, 93). For eukaryotic de novo genome sequencing, NGS was first used in combination with Sanger sequencing, thereby reducing the overall costs because a lower coverage of the (more expensive) Sanger reads was needed. This combined approach was used for sequencing the genomes of, e.g., the filamentous fungus Grosmannia clavigera (21) and the cucumber Cucumis sativus (39). However, a combination of improvements in read lengths, the ability to sequence paired-end reads, and novel assembly algorithms that can deal with large numbers of short reads have now made possible the assembly of eukaryotic genomes from NGS reads. The first two eukaryotic genomes to be assembled solely from NGS reads are those of the giant panda, assembled from Solexa reads (56), and the filamentous fungus Sordaria macrospora, assembled from a combination of Solexa and 454 reads (85).
Analysis of genome organization.
NGS technologies can be used not only for determining the DNA sequence of a genome but also for resolving higher-order structures within the eukaryotic nucleus. Examples are methods like Hi-C, ChIP-(chromatin immunoprecipitation)-seq, and methyl-seq (Fig. 1). Hi-C allows identification of genomic regions that are not necessarily close in terms of consecutive sequence but are brought into spatial proximity within the nucleus as a consequence of the three-dimensional organization of chromatin (59). Analysis of this spatial organization is achieved by first cross-linking DNA and proteins in the cell, then digesting the DNA with a restriction enzyme, end labeling it with biotin, and then ligating under diluted conditions that promote self-ligation of fragments that are close to each other, i.e., cross-linked fragments. The resulting fragments are sheared, and biotinylated fragments are isolated and sequenced using paired-end NGS strategies. The sequence reads are mapped to a reference genome, and three-dimensional maps of DNA organization at the nuclear, chromosomal, or megabase (intrachromosomal interactions) scale can be established. This was first reported for the human genome at a resolution of 1 Mb (59), and a similar technique was reported for yeast with a resolution in the kilobase range (25).
In eukaryotes, nuclear DNA is packed with proteins into chromatin, and the chromatin structure is highly dynamic and influences a range of cellular processes, e.g., the transcriptional activity in a genomic region (38). Changes in chromatin structure, e.g., nucleosome positioning, are mediated by a number of proteins that interact directly with DNA or with one another. One recently developed method to study the genome-wide distribution of DNA-interacting proteins is ChIP-seq. This approach also starts with cross-linking DNA and proteins within the cell and fragmenting the DNA; then the protein of choice in a complex with bound DNA is immunoprecipitated with a specific antibody. Next, the cross-linking is reversed and the DNA is isolated and sequenced (89). For both Hi-C and ChIP-seq, short-read/high-throughput methods are suitable because, usually, these methods are used when a reference genome is available. ChIP-seq offers a number of advantages over its predecessor ChIP-chip because the array-based ChIP-chip method requires microarrays ideally covering a complete genome, and due to the high costs of producing such arrays, they are available for only a few species (89). ChIP-seq experiments were first performed with the yeast Saccharomyces cerevisiae and several mammalian cell types and helped to shed light on the genome-wide distribution of nucleosomes, transcription factor binding sites, and methylation states of chromatin proteins that influence genome activity and hence cellular development (2, 5, 45, 77, 95). Similar types of experiments, including methyl-seq or MeDIP-(methylated DNA immunoprecipitation)-seq, can be conducted to determine the genome-wide distribution of methylated cytosine bases in DNA that provide another layer of epigenetic control in eukaryotic cells (50).
Analysis of gene expression.
Next to the (re)sequencing of genomes, sequencing of cDNAs is one field to which NGS was applied quickly and successfully. The use of NGS to obtain transcriptomics data is collectively known as RNA deep sequencing or RNA-seq (112) (Fig. 1). The first studies were published in 2008 for the yeasts S. cerevisiae (81) and Schizosaccharomyces pombe (115) and the plant Arabidopsis thaliana (60), as well as mammalian cells (17, 80), and since then for a number of other organisms (67).
RNA-seq has several advantages over other methods, e.g., expressed sequence tag (EST) sequencing, serial analysis of gene expression (SAGE), and microarray hybridizations to obtain transcriptomics data (84). Compared to EST sequencing and SAGE, both of which also rely on the sequencing of cDNAs or cDNA tags, the much higher throughput and consequently the increased sequencing depth of RNA-seq provides a much better resolution that allows not only the quantification of expression levels but the identification of transcript boundaries at the single-nucleotide level (80, 115). During the last decade, microarrays were the most widely used method to determine transcript levels at a whole-genome scale, and they have greatly contributed to our knowledge about gene expression in eukaryotic microbes (10, 27, 83). However, using microarrays, transcripts can be detected only when there is a corresponding probe on the chip, and arrays for most organisms were designed to cover only the annotated coding (or, if known, transcribed) regions of a genome. Later generations of arrays, so-called tiling arrays, can cover complete genomes at various resolution levels (7); however, they are more expensive and consequently available for only a few organisms. Nevertheless, the use of tiling arrays has already allowed a glimpse at the transcriptional landscape of eukaryotic genomes—a landscape that is much more complex than previously thought (46). RNA-seq can be used to extend this picture because transcribed regions of a genome can be determined at single-nucleotide resolution without having to rely on prior annotation or indeed any prior knowledge concerning transcribed regions within a genome. The use of tiling arrays and RNA-seq led to the discovery that, contrary to what was expected, most of the eukaryotic genome is transcribed; i.e., large numbers of (presumably) noncoding RNAs are present in those organisms that have been analyzed so far (43, 60, 81).
Apart from the discovery of novel transcripts, RNA-seq data can be used to map transcript boundaries, e.g., the ends of 5′ and 3′ untranslated regions, as well as exon/intron boundaries. For eukaryotic microorganisms, this will be extremely useful in combination with de novo genome sequencing since most sequenced genomes can only be annotated automatically, with manual annotation projects being extremely labor- and cost-intensive and therefore restricted to a few select model organisms. This leads to automatically predicted gene models that are often not correct, especially with respect to intron distribution. The inclusion of RNA-seq data in a genome sequencing project allows much better, evidence-based gene predictions during automated annotation without greatly increasing costs. This was the case even for the well-annotated genomes of S. cerevisiae and Schizosaccharomyces pombe (81, 115), and RNA-seq data were also used to improve genome annotation of the filamentous fungus Aspergillus oryzae (110).
To fully exploit the information contained in RNA-seq reads, one has to consider how to identify and map reads that span splice junctions and therefore cannot be mapped directly to the genome sequence. A number of novel algorithms have been developed to deal with this problem, and some of them also allow quantification of mapped reads (see below). This is important when RNA-seq data are to be used for the genome-wide quantitative analysis of gene expression, similar to microarray hybridization data. Among eukaryotic microorganisms, RNA-seq has already been used to quantify gene expression for, e.g., yeasts (81, 115), A. oryzae (110), and the truffle Tuber melanosporum (69). Compared to microarrays, RNA-seq is still more expensive when large numbers of samples need to be analyzed to compare gene expression under multiple different conditions. However, for organisms where no microarrays are currently available or where transcriptomics data are to be used primarily for annotation or general screening purposes, RNA-seq is an attractive alternative. Furthermore, the development of multiplexing approaches where different samples can be sequenced in one sequencing run as well as the steady increase of reads per run and decrease in costs per base will make RNA-seq an important tool for future transcriptomics studies.
RNA-seq can be extended to sequence only specific subsets of RNAs. One example for this is ribosome profiling, a method for quantitatively monitoring those parts of mRNAs that are translated (42). For this purpose, mRNA fragments that are bound by ribosomes and thereby protected from nuclease digestion are isolated, reverse transcribed, and sequenced. Studies with S. cerevisiae indicated that the data obtained by ribosome profiling correlate well with protein abundance as measured with mass spectrometry (42). Thus, RNA-seq and its variants provide a means for detailed, genomewide studies of gene expression at various levels.
BIOINFORMATICS CHALLENGES
As outlined above, NGS can be used for a wide variety of highly interesting and promising experimental approaches, and additional NGS-based applications will certainly be devised within the coming years. However, the most significant hurdle that researchers face with any NGS approach is the analysis of the huge amounts of data generated by these experiments. This starts with the fact that there are currently no unified data formats, moves on to problems associated with de novo assembly or mapping of millions to billions of reads, assembly of spliced transcripts, or quantification of RNA-seq data, and at some point leads to the questions of where and how to store the large files associated with different steps of NGS analysis. These challenges and some solutions will be described in this section.
Data formats.
Both Sanger and NGS techniques result in light signals that have to be decoded to determine the base sequence in the DNA. This process is named “base-calling” and depends on the sequencing platform used; however, in general each base call results in the base and a respective quality score that describes how likely it is that the called base is the correct one. A widely used base-calling software for Sanger sequencing reads is phred, and the corresponding quality scores are called phred scores (30, 31). A number of file formats to represent sequence data and/or quality scores have already been developed for Sanger reads, and one format for the combined base sequence and phred scores that has also been adopted for NGS reads is the FASTQ format (18). However, apart from the original FASTQ format developed at the Wellcome Trust Sanger Institute, two FASTQ variants are used by Illumina/Solexa that calculate quality scores using equations different from the ones used by phred (18). Therefore, there are now at least three FASTQ versions that differ in the way the quality scores are calculated, and it is usually up to the user to determine the format for any given file. In addition to FASTQ, there are a number of other file formats, most of them specific to different NGS platforms or applications. As most software packages currently accept only a limited number of input file formats, the first step in dealing with NGS data is often the conversion of the original files into a format that can be used as input for downstream processing, e.g., assembly, mapping, etc. A number of freely available software tools have been developed to deal with this problem (e.g., http://bioinf.comav.upv.es/sff_extract/index.html or http://maq.sourceforge.net/fq_all2std.pl); however, the constant development of new technologies accompanied by new file formats as well as the lack of standard formats for downstream applications requires that users pay close attention to data format changes.
Downstream analyses involving NGS data also generate large files. Many NGS applications start with the mapping of reads to a reference genome (see below and Fig. 1), and, similar to the case with the sequence data, there are also a number of formats in which mapping data can be stored. Most of these were developed for a specific mapping software and cannot directly be used by other programs. However, there are efforts under way to standardize mapping data formats, thereby making them more interchangeable among programs. For example, mapping data in the Sequence/Alignment Map (SAM) format or its compressed equivalent BAM can now be used by a number of downstream applications, including several genome viewers (54).
Because the field of NGS is new and advancing rapidly, there are no “start to finish” software suites available for dealing with NGS data. Rather, users have to piece together their own analysis pipelines according to their experimental questions. A distinct advantage when dealing with NGS data is the ability to write small scripts in one of the programming languages commonly used for bioinformatics tasks (http://www.open-bio.org), because this allows users to perform data transformations and extractions tailored to their specific needs.
De novo assembly.
The de novo assembly of genomes from NGS reads is not a trivial task, and assembly programs that were developed for Sanger reads usually are not suitable for this purpose. One reason for this is the number of reads that are required for NGS assemblies. Whereas a 3- to 10-fold coverage can yield high-quality draft assemblies from whole-genome shotgun Sanger data for eukaryotic genomes, assemblies from shorter reads require a much higher coverage. To date, two eukaryotic genome sequences have been published, both of which were assembled solely from NGS reads. The genome of the giant panda was assembled from a 73-fold coverage of 52-nucleotide (nt) Solexa reads (56), and the genome of the filamentous fungus Sordaria macrospora was assembled from an 85-fold coverage of 36-nt Solexa reads and an additional 10-fold 454 coverage (85). There are two main reasons why such high coverages are necessary for these assemblies: (i) the short read lengths require more reads within a region to confidently assemble contigs and (ii) the NGS reads have a higher error rate. With enough coverage, the higher error rate is not a problem, because the higher number of reads effectively “quenches” errors in single reads and leads to overall high accuracy in the final assembly. The short read length, however, causes difficulties that cannot be overcome by increasing the number of reads alone because repeat regions, which are widespread especially in the genomes of higher eukaryotes, cannot be assembled from reads that are shorter than the lengths of the repeats. One solution to improve an assembly by scaffolding reads known to be physically linked is the generation of paired-end data (see above). This approach was used for sequencing both the giant panda and Sordaria macrospora genomes (56, 85). Another solution is the combination of shorter and longer reads, e.g., Solexa and 454 reads (21, 85). During the last years, a number of programs were developed specifically for the de novo assembly of NGS reads (Table 2). These programs incorporate new types of algorithms that are able to deal with the huge number of reads (79). Most of them allow combinations of reads from different sequencing platforms and incorporate paired-end data. This makes the assembly of large, complex genomes from NGS data possible; e.g., the assembly of the giant panda genome was performed with SOAPdenovo (58) while the Sordaria macrospora genome was assembled with Velvet (118).
Table 2.
Some freely available software tools for the analysis of NGS dataa
| Area of application | Software | Reference(s) |
|---|---|---|
| De novo assembly | ABySS | 102 |
| ALLPATHS | 12 | |
| Celera/CABOG | 78 | |
| Edena | 37 | |
| EULER-SR | 15 | |
| QSRA | 11 | |
| SHARCGS | 23 | |
| SOAPdenovo | 58 | |
| SSAKE | 113 | |
| VCAKE | 44 | |
| Velvet | 118 | |
| Assembly visualization | ABySS-Explorer | 82 |
| EagleView | 40 | |
| Mapping to reference genome | Bowtie | 51 |
| BWA | 53 | |
| Maq | 55 | |
| SOAP2 | 57 | |
| SNP/indel identification, consensus building | Maq | 55 |
| SAMtools | 54 | |
| Slider II | 65 | |
| SOAPsnp | http://soap.genomics.org.cn | |
| Graphical view of mapped reads | Artemis/BamView | 13, 14 |
| IGV | http://www.broadinstitute.org/igv | |
| SAMtools | 54 | |
| Identification of splice junctions, transcript assembly/quantification | Tophat, Cufflinks | 107, 109 |
| Oases | http://www.ebi.ac.uk/∼zerbino/oases |
This (noncomprehensive) list gives programs for different stages of NGS analysis. Software packages specifically designed for the analysis of ChIP-seq data are not included here, but an overview can be found in reference 90. Further information is available in several recent reviews (79, 90, 108) or at http://www.seqanswers.com.
Mapping of reads to a reference genome, quantification, and detection of sequence variants.
With the exception of de novo assembly, NGS applications usually require mapping of reads to a reference genome prior to downstream analyses (Fig. 1; Table 2). Similar to de novo assembly, mapping NGS reads requires algorithms that can deal with the huge number of short reads—a computational problem for which algorithms developed for a usually much smaller number of Sanger reads are not well suited. Therefore, a number of novel algorithms specifically adapted to NGS read mapping were developed in the last years (90, 108) (Table 2). These programs are designed to deal with a number of issues that have to be considered when mapping NGS reads: some mapping algorithms use quality score data to calculate a mapping quality for individual reads. This is especially important because of the combination of short read lengths and higher error rates of NGS reads compared to Sanger reads. A related problem is the number of mismatches that are allowed for a read to still be mapped to a genomic position. Mismatches can originate from sequencing errors; however, when genomes from different individuals/strains/lines are sequenced, some mismatches will be caused by SNPs or other sequence variants and therefore are of biological interest. As in de novo assembly, repeats are one of the major concerns in mapping, because one read might be mapped to several genomic regions. There is no single “right” solution to this problem, and the behavior of mapping algorithms varies from completely excluding such reads to mapping them to the best hit to mapping them to all hits. Especially in applications where the quantification of mapped reads is the desired readout, e.g., in ChIP-seq or RNA-seq experiments, the question of how to map such “multireads” is important. For example, using only uniquely mapped reads increases specificity whereas using multireads increases sensitivity (90).
A specific mapping problem occurs with RNA-seq reads because reads that span splice junctions cannot be mapped to a genomic site. Several programs were developed to identify splice junctions either during the mapping or by first mapping all “mappable” reads and then identifying those reads that connect the transcribed regions that were identified in the previous mapping step (90) (Table 2). Thus, RNA-seq can be used to determine transcribed regions and splice sites at the single-nucleotide level (81, 115), but another area of application is the quantification of transcript levels across samples. The number of reads for each gene is a first measure of the gene's expression level; however, the number of reads is dependent not only on the transcript amount in the original sample but also on the length of the gene's mRNA. To normalize the read count by the mRNA length, expression values can be given as mapped reads per kilobase per million reads (RPKM) (80). Similar to the mapping of genomic reads, the multiread problem also occurs during mapping of RNA-seq reads. Here, it not only is due to repeat regions including paralogous genes but can also arise from, e.g., alternative splice forms and thereby cause not only mapping but also quantification problems. As with genomic mapping, there is no single solution to these concerns, but users can choose between a number of programs that offer different solutions to these problems (90, 108). Downstream analyses of RNA-seq data include normalization and validation steps similar to those for microarrays. By now, procedures for the analysis of microarray data are well established and include sophisticated statistical analyses that can be adapted for use with RNA-seq data (27, 35, 83).
One of the major aims of genome resequencing is to detect genomic variability between individuals or different strains/lines. This requires the ability to detect differences between the reference genome and the resequenced DNA. These differences can take the form of SNPs or indels of few bases but also include larger insertions or deletions and more complex forms of rearrangements, e.g., structural variants (48). A number of tools can identify SNPs and small indels in mapped read data (Table 2), but these tools usually do not find extensive insertions, deletions, or rearrangements. These types of sequence differences can be extracted, e.g., by using paired-end data, because insertions or deletions will shift the position of one end of the pair to a distance farther from or closer to its companion read. When the size of the sequenced fragments and therefore the length of the distance between ends is known, deviations of the mapping distance of reads on the reference genome from the expected distance indicate either deletions (mapping distance is larger than expected) or insertions (mapping distance is smaller than expected). More complex types of rearrangements can, for example, be determined by taking read orientation into account (48).
Data storage.
NGS has made large-scale sequencing projects possible even for small research groups, whereas genome centers are now gearing up for even higher throughput than was previously achieved with automated Sanger sequencing (74). However, one problem for all researchers conducting NGS projects is how to store the resulting data. The large number of sequence reads delivered by NGS platforms results in file sizes, e.g., for FASTQ files, that are well in the gigabyte range, and downstream applications like mapping yield files of a similar size so that the amount of data even from a single project often reaches the terabyte range. Therefore, especially for individual researchers, storage and backup schemes that were quite sufficient for Sanger sequencing projects can quickly run out of storage space with even a few NGS-based experiments.
The International Nucleotide Sequence Database Collaboration (INSDC) (http://www.insdc.org/), which includes the National Center for Biotechnology Information (NCBI), the European Bioinformatics Institute (EBI), and the DNA Data Bank of Japan (DDBJ), has established a database for NGS data, the SRA (Sequence Read Archive) (101). The SRA contains not only the actual sequences and quality scores but also metadata about the sequencing project to which individual data files belong. However, the large number of reads makes searching those data with established algorithms like BLAST (3) impractical in terms of search time and would in most cases not yield informative results anyway due to the short read lengths. Therefore, researchers can download the data, but the convenient online searches and other tools that we have become accustomed to are currently not available for NGS data. One major challenge is therefore how to make NGS data accessible to the research community.
NGS TO STUDY EUKARYOTIC MICROORGANISMS
NGS technologies hold great potential for the study of eukaryotic microorganisms. One reason for this is that because of their mostly small genome sizes, high sequence coverage can already be achieved with a moderate amount of NGS and therefore projects like genome sequencing that were previously restricted to large genome centers are now possible even for small groups. Filamentous fungi were among the first eukaryotes for which de novo genome sequencing projects included or were solely based on NGS reads (21, 70, 85), and yeasts were among the first organisms for which RNA-seq was established (81, 115). For other eukaryotic microorganisms, NGS has been so far mainly used in transcriptomics studies in combination with Sanger sequencing to establish EST libraries, e.g., for the oomycete Pythium ultimum (16) or the charophyte algae Coleochaete orbicularis and Spirogyra pratensis (106). However, with 65 sequenced genomes and more than 70 genome sequences in progress from green algae to a number of species that belong to diverse phylogenetic groups summarized as protists (http://www.ncbi.nlm.nih.gov/genomes/leuks.cgi), it is only a matter of time until NGS-based approaches are applied to eukaryotic microorganisms on a broad scale. Several areas of (potential) application of NGS to help answer questions concerning eukaryotic microbiology are described below.
Metagenomics.
One area where higher sequencing capacities might shed light on fundamental issues is biodiversity research. It has been known for decades that the number of microorganisms that cannot be cultured under laboratory conditions by far exceeds the number of those which can be cultured (20). One way to gain access to these uncultured organisms (archaea, bacteria, and eukaryotes) was developed in the 1980s and involves environmental surveys of rRNAs; i.e., partial rRNA genes are cloned directly or (mostly) after PCR amplification from environmental samples and sequenced (87). This approach still has some limitations, one of which is that because of the high degree of conservation of the rRNA sequences, they cannot be used to differentiate between closely related species.
One further step along the road to a broader glimpse at biodiversity was taken by metagenomics approaches (49, 116). Metagenomics involves the cloning and sequencing of nucleic acids isolated directly from environmental samples and is therefore not restricted to the rRNA genes. Moreover, this approach not only can be used to study microbial biodiversity in general but also allows the characterization of the environmental distribution of microorganisms as a means to characterize specific biotopes. First, metagenomic studies were performed using Sanger sequencing; however, the use of NGS techniques further improves the possibilities offered by metagenomics because NGS eliminates the need for cloning, thereby excluding cloning biases. Additionally, it has a higher sensitivity due to increased sequencing depths at lower costs (20, 22). It has long been known that eukaryotic microorganisms make up most of the eukaryotic biodiversity (29, 88), and metagenomic approaches using NGS are one way to increase our knowledge about these little-known groups. Moreover, metagenomics using NGS can be taken a step further to include not only environmental DNA but also RNA, as was recently demonstrated in a study of gene expression in ocean waters (34). Furthermore, tree-of-life projects, e.g., for the fungal tree of life (73), will benefit from increased sequencing capabilities.
Analysis of individual genetic variation.
While sequencing of one genome per species already allows insights into the species' biology, genetic variation that leads to phenotypic variation between individuals cannot be deduced from one genome sequence alone. Information about individual genetic variation is of pivotal interest, as it helps to address a number of fundamental biological questions. For example, such information can be used for evolutionary analyses as well as, especially in the case of microorganisms, identifying mutations and differences between pathogenic and apathogenic strains, toxin producers and nontoxinogenic variants, or wild-type strains and strains optimized for industrial production. Individual genetic variations range from SNPs to small indels and rearrangements to large structural variants. The most comprehensive overview of such variations is of course to be gained by sequencing not only candidate regions but whole genomes, and with the advent of NGS, sequencing of a large number of genomes from one species becomes feasible. The availability of NGS technologies has led to “1,000 genomes” projects, where the genomes for ∼1,000 individuals from one species are to be sequenced, e.g., for humans (http://www.1000genomes.org/), Drosophila melanogaster (http://www.dpgp.org/), and Arabidopsis thaliana (http://www.1001genomes.org/). Eukaryotic microorganisms with their usually smaller genomes are also good candidates for such projects, and for several species, there is already more than one genome available or the sequencing is under way, e.g., for S. cerevisiae (61) and other yeasts as well as filamentous fungi (e.g., http://www.jgi.doe.gov/genome-projects/, http://www.broadinstitute.org/science/projects/fungal-genome-initiative).
Genomic comparisons can be used to address open questions like mutation rates in evolving populations and their correlation to organismic adaptation. This was, for example, studied by whole-genome sequencing of yeast strains that evolved under controlled laboratory conditions (4, 64). Another study with yeast used whole-genome sequences of parental strains and their offspring to characterize sites of meiotic recombination at the nucleotide level (92). This combination of genetic and genomic analyses can even be used to determine mutations that are responsible for mutant phenotypes or, in the case of humans, cause disease. This was shown in two cases of inheritable disorders where the affected genes were identified by whole-genome sequencing of several family members (94) or of only one genome from an affected individual and subsequent genotyping of family members for putative candidate genes (63).
Another application where whole-genome sequencing is of particular interest is the analysis of industrial production strains. Such strains are mostly generated by repeated rounds of conventional mutagenesis, and therefore the number and nature of the mutations that contribute to the desired phenotype are usually not known. Genome sequencing of production strains as well as the original strains and/or intermediates of the mutation/selection process can help to narrow down candidate genes/genomic regions that are targets of mutations that improve strain properties. A study of a Pichia stipitis strain selected for high ethanol conversion from xylose showed that a 10× to 15× sequence coverage with any one of three NGS platforms (Solexa, SOLiD, or 454) was sufficient to detect single-nucleotide mutations with high sensitivity in the strain (104).
Genomewide dynamics and expression studies.
The yeasts S. cerevisiae and Schizosaccharomyces pombe were among the first species for which RNA-seq studies were performed (81, 115); similar studies of filamentous fungi were not long to follow (69, 110). In the oomycete Pythium ultimum as well as in the charophyte algae Coleochaete orbicularis and Spirogyra pratensis, NGS was used in combination with Sanger sequencing to establish EST databases (16, 106). Especially for species for which microarrays have not yet been established, it will in most cases be easier to start directly with sequencing-based approaches for high-throughput studies.
Apart from unraveling genome activity in a single species, with more and more transcriptomics data available, comparative functional genomics will become even more powerful. Comparative functional genomics uses the conservation of coexpression in different species as a means to identify those genes whose expression might be of functional significance under the conditions investigated (105). Thus, gene expression patterns can be used as molecular phenotypes, similarly to other phenotypic traits that are used for phylogenetic analysis.
Over the last years, high-throughput analyses have demonstrated genomewide links between genome organization at the nucleosome level and genome activity, e.g., regulation of gene expression. Thus, a systems biology approach using methods like ChIP-seq, methyl-seq, and RNA-seq to simultaneously study genome conformation and transcriptional activity will greatly enhance our understanding of eukaryotic biology. For the human genome, this integrated approach is ongoing in the ENCODE (Encyclopedia of DNA elements) project (29a), and similar projects for filamentous fungi are currently under way (e.g., http://projectreporter.nih.gov/project_info_description.cfm?aid=7633564&icde=0).
CONCLUSIONS
NGS technologies offer a promising new avenue for discovery in many fields of research, including several that, so far, have mainly used approaches not based on sequencing. Although there is still room for improvement, especially with respect to read lengths and error rates, NGS platforms as well as analysis software tools have evolved to already allow, for example, the de novo sequencing and assembly of eukaryotic genomes solely from NGS reads. However, one major concern about NGS is the question of how to deal with the flood of data that NGS platforms produce. Therefore, the development of bioinformatics tools and databases to better cope with these types of data will be one of the main factors determining how useful NGS will be for a wider research community. Nevertheless, the advent of NGS is already a major breakthrough in molecular biology, genetics, and beyond, as well as a great leap forward for genomics and systems biology analyses.
ACKNOWLEDGMENTS
I would like to thank Ulrich Kück for his support and Ines Engh, Birgit Hoff, and Jens Kamerewerd for critical reading of the manuscript.
Funding for my work comes from the German Science Foundation (Deutsche Forschungsgemeinschaft, DFG, grant no. NO 407/2-1) and the Protein Research Department of the Ruhr-University Bochum.
Biography
Minou Nowrousian studied biology at the Ruhr-Universität Bochum, Germany. In 1998, she completed her Ph.D. on fungal morphogenesis under the supervision of Ulrich Kück. In 1999, she started a postdoctoral position working on circadian rhythmicity in the lab of Jay Dunlap and Jennifer Loros at Dartmouth Medical School. In 2002, she returned to Bochum, where she completed a habilitation in botany in 2007 and currently leads a research group working on functional genomics of fungi. Her research focuses on comparative genomics and transcriptomics of sexual development in ascomycetes to identify evolutionary conserved factors involved in fungal morphogenesis.
Footnotes
Published ahead of print on 2 July 2010.
REFERENCES
- 1.Ahn S.-M., Kim T.-H., Lee S., Kim D., Ghang H., Kim D.-S., Kim B.-C., Kim S.-Y., Kim W.-Y., Kim C., Park D., Lee Y. S., Kim S., Reja R., Jho S., Kim C. G., Cha J.-Y., Kim K.-H., Lee B., Bhak J., Kim S.-J. 2009. The first Korean genome sequence and analysis: full genome sequencing for a socio-ethnic group. Genome Res. 19:1622–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albert I., Mavrich T. N., Tomsho L. P., Qi J., Zanton S. J., Schuster S. C., Pugh B. F. 2007. Translational and rotational settings of H2A. Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446:572–576 [DOI] [PubMed] [Google Scholar]
- 3.Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Araya C. L., Payen C., Dunham M. J., Fields S. 2010. Whole-genome sequencing of a laboratory-evolved yeast strain. BMC Genomics 11:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barski A., Cuddapah S., Cui K., Roh T. Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129:823–837 [DOI] [PubMed] [Google Scholar]
- 6.Bentley D. R., Balasubramanian S., Swerdlow H. P., Smith G. P., Milton J., Brown C. G., Hall K. P., Evers D. J., Barnes C. L., Bignell H. R., Boutell J. M., Bryant J., Carter R. J., Keira Cheetham R., Cox A. J., Ellis D. J., Flatbush M. R., Gormley N. A., Humphray S. J., Irving L. J., Karbelashvili M. S., Kirk S. M., Li H., Liu X., Maisinger K. S., Murray L. J., Obradovic B., Ost T., Parkinson M. L., Pratt M. R., Rasolonjatovo I. M. J., Reed M. T., Rigatti R., Rodighiero C., Ross M. T., Sabot A., Sankar S. V., Scally A., Schroth G. P., Smith M. E., Smith V. P., Spiridou A., Torrance P. E., Tzonev S. S., Vermaas E. H., Walter K., Wu X., Zhang L., Alam M. D., Anastasi C., Aniebo I. C., Bailey D. M. D., Bancarz I. R., Banerjee S., Barbour S. G., Baybayan P. A., Benoit V. A., Benson K. F., Bevis C., Black P. J., Boodhun A., Brennan J. S., Bridgham J. A., Brown R. C., Brown A. A., Buermann D. H., Bundu A. A., Burrows J. C., Carter N. P., Castillo N., Chiara M., Catenazzi E., Chang S., Neil Cooley R., Crake N. R., Dada O. O., Diakoumakos K. D., Dominguez-Fernandez B., Earnshaw D. J., Egbujor U. C., Elmore D. W., Etchin S. S., Ewan M. R., Fedurco M., Fraser L. J., Fuentes Fajardo K. V., Scott Furey W., George D., Gietzen K. J., Goddard C. P., Golda G. S., Granieri P. A., Green D. E., Gustafson D. L., Hansen N. F., Harnish K., Haudenschild C. D., Heyer N. I., Hims M. M., Ho J. T., Horgan A. M., et al. 2008. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456:53–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertone P., Gerstein M., Snyder M. 2005. Applications of DNA tiling arrays to experimental genome annotation and regulatory pathway discovery. Chromosome Res. 13:259–274 [DOI] [PubMed] [Google Scholar]
- 8.Branton D., Deamer D. W., Marziali A., Bayley H., Benner S. A., Butler T., Di Ventra M., Garaj S., Hibbs A., Huang X., Jovanovich S. B., Krstic P. S., Lindsay S., Ling X. S., Mastrangelo C. H., Meller A., Oliver J. S., Pershin Y. V., Ramsey J. M., Riehn R., Soni G. V., Tabard-Cossa V., Wanunu M., Wiggin M., Schloss J. A. 2008. The potential and challenges of nanopore sequencing. Nat. Biotechnol. 26:1146–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braslavsky I., Hebert B., Kartalov E., Quake S. R. 2003. Sequence information can be obtained from single DNA molecules. Proc. Natl. Acad. Sci. U. S. A. 100:3960–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breakspear A., Momany M. 2007. The first fifty microarray studies in filamentous fungi. Microbiology 153:7–15 [DOI] [PubMed] [Google Scholar]
- 11.Bryant D., Wong W.-K., Mockler T. 2009. QSRA—a quality-value guided de novo short read assembler. BMC Bioinformatics 10:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butler J., MacCallum I., Kleber M., Shlyakhter I. A., Belmonte M. K., Lander E. S., Nusbaum C., Jaffe D. B. 2008. ALLPATHS: de novo assembly of whole-genome shotgun microreads. Genome Res. 18:810–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carver T., Berriman M., Tivey A., Patel C., Bohme U., Barrell B. G., Parkhill J., Rajandream M.-A. 2008. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24:2672–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carver T., Bohme U., Otto T. D., Parkhill J., Berriman M. 2010. BamView: viewing mapped read alignment data in the context of the reference sequence. Bioinformatics 26:676–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaisson M. J., Pevzner P. A. 2008. Short read fragment assembly of bacterial genomes. Genome Res. 18:324–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheung F., Win J., Lang J. M., Hamilton J., Vuong H., Leach J. E., Kamoun S., Lévesque C. A., Tisserat N., Buell C. R. 2008. Analysis of the Pythium ultimum transcriptome using Sanger and pyrosequencing approaches. BMC Genomics 9:541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cloonan N., Forrest A. R. R., Kolle G., Gardiner B. B. A., Faulkner G. J., Brown M. K., Taylor D. F., Steptoe A. L., Wani S., Bethel G., Robertson A. J., Perkins A. C., Bruce S. J., Lee C. C., Ranade S. S., Peckham H. E., Manning J. M., McKernan K. J., Grimmond S. M. 2008. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat. Methods 5:613–619 [DOI] [PubMed] [Google Scholar]
- 18.Cock P. J. A., Fields C., Goto N., Heuer M. L., Rice P. M. 2010. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 38:1767–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Craig D. W., Person J. V., Szelinger S., Sekar A., Redman M., Corneveaux J. J., Pawlowski T. L., Laub T., Nunn G., Stephan D. A., Homer N., Huentelman M. J. 2008. Identification of genetic variants using bar-coded multiplexed sequencing. Nat. Methods 5:887–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeLong E. F. 2009. The microbial ocean from genomes to biomes. Nature 459:200–206 [DOI] [PubMed] [Google Scholar]
- 21.DiGuistini S., Liao N., Platt D., Robertson G., Seidel M., Chan S., Docking T. R., Birol I., Holt R., Hirst M., Mardis E., Marra M., Hamelin R., Bohlmann J., Breuil C., Jones S. 2009. De novo genome sequence assembly of a filamentous fungus using Sanger, 454 and Illumina sequence data. Genome Biol. 10:R94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dinsdale E. A., Edwards R. A., Hall D., Angly F., Breitbart M., Brulc J. M., Furlan M., Desnues C., Haynes M., Li L., McDaniel L., Moran M. A., Nelson K. E., Nilsson C., Olson R., Paul J., Brito B. R., Ruan Y., Swan B. K., Stevens R., Valentine D. L., Thurber R. V., Wegley L., White B. A., Rohwer F. 2008. Functional metagenomic profiling of nine biomes. Nature 452:629–632 [DOI] [PubMed] [Google Scholar]
- 23.Dohm J. C., Lottaz C., Borodina T., Himmelbauer H. 2007. SHARCGS, a fast and highly accurate short-read assembly algorithm for de novo genomic sequencing. Genome Res. 17:1697–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drmanac R., Sparks A. B., Callow M. J., Halpern A. L., Burns N. L., Kermani B. G., Carnevali P., Nazarenko I., Nilsen G. B., Yeung G., Dahl F., Fernandez A., Staker B., Pant K. P., Baccash J., Borcherding A. P., Brownley A., Cedeno R., Chen L., Chernikoff D., Cheung A., Chirita R., Curson B., Ebert J. C., Hacker C. R., Hartlage R., Hauser B., Huang S., Jiang Y., Karpinchyk V., Koenig M., Kong C., Landers T., Le C., Liu J., McBride C. E., Morenzoni M., Morey R. E., Mutch K., Perazich H., Perry K., Peters B. A., Peterson J., Pethiyagoda C. L., Pothuraju K., Richter C., Rosenbaum A. M., Roy S., Shafto J., Sharanhovich U., Shannon K. W., Sheppy C. G., Sun M., Thakuria J. V., Tran A., Vu D., Zaranek A. W., Wu X., Drmanac S., Oliphant A. R., Banyai W. C., Martin B., Ballinger D. G., Church G. M., Reid C. A. 2010. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science 327:78–81 [DOI] [PubMed] [Google Scholar]
- 25.Duan Z., Andronescu M., Schutz K., McIlwain S., Kim Y. J., Lee C., Shendure J., Fields S., Blau C. A., Noble W. S. 2010. A three-dimensional model of the yeast genome. Nature 465:363–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eck S. H., Benet-Pagès A., Flisikowski K., Meitinger T., Fries R., Strom T. M. 2009. Whole genome sequencing of a single Bos taurus animal for single nucleotide polymorphism discovery. Genome Biol. 10:R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ehrenreich A. 2006. DNA microarray technology for the microbiologist: an overview. Appl. Microbiol. Biotechnol. 73:255–273 [DOI] [PubMed] [Google Scholar]
- 28.Eid J., Fehr A., Gray J., Luong K., Lyle J., Otto G., Peluso P., Rank D., Baybayan P., Bettman B., Bibillo A., Bjornson K., Chaudhuri B., Christians F., Cicero R., Clark S., Dalal R., deWinter A., Dixon J., Foquet M., Gaertner A., Hardenbol P., Heiner C., Hester K., Holden D., Kearns G., Kong X., Kuse R., Lacroix Y., Lin S., Lundquist P., Ma C., Marks P., Maxham M., Murphy D., Park I., Pham T., Phillips M., Roy J., Sebra R., Shen G., Sorenson J., Tomaney A., Travers K., Trulson M., Vieceli J., Wegener J., Wu D., Yang A., Zaccarin D., Zhao P., Zhong F., Korlach J., Turner S. 2009. Real-time DNA sequencing from single polymerase molecules. Science 323:133–138 [DOI] [PubMed] [Google Scholar]
- 29.Embley T. M., Martin W. 2006. Eukaryotic evolution, changes and challenges. Nature 440:623–630 [DOI] [PubMed] [Google Scholar]
- 29a.ENCODE Project Consortium 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447:799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ewing B., Green P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186–194 [PubMed] [Google Scholar]
- 31.Ewing B., Hillier L., Wendl M. C., Green P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175–185 [DOI] [PubMed] [Google Scholar]
- 32.Farrer R. A., Kemen E., Jones J. D. G., Studholme D. J. 2009. De novo assembly of the Pseudomonas syringae pv. syringae B728a genome using Illumina/Solexa short sequence reads. FEMS Microbiol. Lett. 291:103–111 [DOI] [PubMed] [Google Scholar]
- 33.Fleischmann R. D., Adams M. D., White O., Clayton R. A., Kirkness E. F., Kerlavage A. R., Bult C. J., Tomb J. F., Dougherty B. A., Merrick J. M., McKenney K., Sutton G., FitzHugh W., Fields C., Gocayne J. D., Scott J., Shirley R., Liu L. I., Glodek A., Kelley J. M. M., Weidman J. F., Phillips C. A., Spriggs T., Hedblom E., Cotton M. D., Utterback T. R., Hanna M. C., Nguyen D. T., Saudek D. M., Brandon R. C., Fine L. D., Fritchman J. L., Fuhrmann J. L., Geoghagen N. S. M., Gnehm C. L., McDonald L. A., Small K. V., Fraser C. M., Smith H. O., Venter J. C. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496–512 [DOI] [PubMed] [Google Scholar]
- 34.Frias-Lopez J., Shi Y., Tyson G. W., Coleman M. L., Schuster S. C., Chisholm S. W., DeLong E. F. 2008. Microbial community gene expression in ocean surface waters. Proc. Natl. Acad. Sci. U. S. A. 105:3805–3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gentleman R., Carey V. J., Huber W., Irizarry R. A., Dudoit S.(ed.). 2005. Bioinformatics and computational biology solutions using R and Bioconductor. Springer, Heidelberg, Germany [Google Scholar]
- 36.Harris T. D., Buzby P. R., Babcock H., Beer E., Bowers J., Braslavsky I., Causey M., Colonell J., DiMeo J., Efcavitch J. W., Giladi E., Gill J., Healy J., Jarosz M., Lapen D., Moulton K., Quake S. R., Steinmann K., Thayer E., Tyurina A., Ward R., Weiss H., Xie Z. 2008. Single-molecule DNA sequencing of a viral genome. Science 320:106–109 [DOI] [PubMed] [Google Scholar]
- 37.Hernandez D., Francois P., Farinelli L., Osteras M., Schrenzel J. 2008. De novo bacterial genome sequencing: millions of very short reads assembled on a desktop computer. Genome Res. 18:802–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ho L., Crabtree G. R. 2010. Chromatin remodelling during development. Nature 463:474–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang S., Li R., Zhang Z., Li L., Gu X., Fan W., Lucas W. J., Wang X., Xie B., Ni P., Ren Y., Zhu H., Li J., Lin K., Jin W., Fei Z., Li G., Staub J., Kilian A., van der Vossen E. A. G., Wu Y., Guo J., He J., Jia Z., Ren Y., Tian G., Lu Y., Ruan J., Qian W., Wang M., Huang Q., Li B., Xuan Z., Cao J., Asan, Wu Z., Zhang J., Cai Q., Bai Y., Zhao B., Han Y., Li Y., Li X., Wang S., Shi Q., Liu S., Cho W. K., Kim J.-Y., Xu Y., Heller-Uszynska K., Miao H., Cheng Z., Zhang S., Wu J., Yang Y., Kang H., Li M., Liang H., Ren X., Shi Z., Wen M., Jian M., Yang H., Zhang G., Yang Z., Chen R., Liu S., Li J., Ma L., Liu H., Zhou Y., Zhao J., Fang X., Li G., Fang L., Li Y., Liu D., Zheng H., Zhang Y., Qin N., Li Z., Yang G., Yang S., Bolund L., Kristiansen K., Zheng H., Li S., Zhang X., Yang H., Wang J., Sun R., Zhang B., Jiang S., Wang J., Du Y., Li S. 2009. The genome of the cucumber, Cucumis sativus L. Nat. Genet. 41:1275–1281 [DOI] [PubMed] [Google Scholar]
- 40.Huang W., Marth G. 2008. EagleView: a genome assembly viewer for next-generation sequencing technologies. Genome Res. 18:1538–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hutchison C. A., III 2007. DNA sequencing: bench to bedside and beyond. Nucleic Acids Res. 35:6227–6237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ingolia N. T., Ghaemmaghami S., Newman J. R. S., Weissman J. S. 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324:218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jacquier A. 2009. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat. Rev. Genet. 10:833–844 [DOI] [PubMed] [Google Scholar]
- 44.Jeck W. R., Reinhardt J. A., Baltrus D. A., Hickenbotham M. T., Magrini V., Mardis E. R., Dangl J. L., Jones C. D. 2007. Extending assembly of short DNA sequences to handle error. Bioinformatics 23:2942–2944 [DOI] [PubMed] [Google Scholar]
- 45.Johnson D. S., Mortazavi A., Myers R. M., Wold B. 2007. Genome-wide mapping of in vivo protein-DNA interactions. Science 316:1497–1502 [DOI] [PubMed] [Google Scholar]
- 46.Kapranov P., Willingham A. T., Gingeras T. R. 2007. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 8:413–423 [DOI] [PubMed] [Google Scholar]
- 47.Kim J.-I., Ju Y. S., Park H., Kim S., Lee S., Yi J.-H., Mudge J., Miller N. A., Hong D., Bell C. J., Kim H.-S., Chung I.-S., Lee W.-C., Lee J.-S., Seo S.-H., Yun J.-Y., Woo H. N., Lee H., Suh D., Lee S., Kim H.-J., Yavartanoo M., Kwak M., Zheng Y., Lee M. K., Park H., Kim J. Y., Gokcumen O., Mills R. E., Zaranek A. W., Thakuria J., Wu X., Kim R. W., Huntley J. J., Luo S., Schroth G. P., Wu T. D., Kim H., Yang K.-S., Park W.-Y., Kim H., Church G. M., Lee C., Kingsmore S. F., Seo J.-S. 2009. A highly annotated whole-genome sequence of a Korean individual. Nature 460:1011–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Korbel J. O., Urban A. E., Affourtit J. P., Godwin B., Grubert F., Simons J. F., Kim P. M., Palejev D., Carriero N. J., Du L., Taillon B. E., Chen Z., Tanzer A., Saunders A. C. E., Chi J., Yang F., Carter N. P., Hurles M. E., Weissman S. M., Harkins T. T., Gerstein M. B., Egholm M., Snyder M. 2007. Paired-end mapping reveals extensive structural variation in the human genome. Science 318:420–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kunin V., Copeland A., Lapidus A., Mavromatis K., Hugenholtz P. 2008. A bioinformatician's guide to metagenomics. Microbiol. Mol. Biol. Rev. 72:557–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laird P. W. 2010. Principles and challenges of genome-wide DNA methylation analysis. Nat. Rev. Genet. 11:191–203 [DOI] [PubMed] [Google Scholar]
- 51.Langmead B., Trapnell C., Pop M., Salzberg S. L. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lefrancois P., Euskirchen G. M., Auerbach R. K., Rozowsky J., Gibson T. J., Yellman C. M., Gerstein M., Snyder M. 2009. Efficient yeast ChIP-Seq using multiplex short-read DNA sequencing. BMC Genomics 10:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H., Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H., Ruan J., Durbin R. 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18:1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li R., Fan W., Tian G., Zhu H., He L., Cai J., Huang Q., Cai Q., Li B., Bai Y., Zhang Z., Zhang Y., Wang W., Li J., Wei F., Li H., Jian M., Li J., Zhang Z., Nielsen R., Li D., Gu W., Yang Z., Xuan Z., Ryder O. A., Leung F. C.-C., Zhou Y., Cao J., Sun X., Fu Y., Fang X., Guo X., Wang B., Hou R., Shen F., Mu B., Ni P., Lin R., Qian W., Wang G., Yu C., Nie W., Wang J., Wu Z., Liang H., Min J., Wu Q., Cheng S., Ruan J., Wang M., Shi Z., Wen M., Liu B., Ren X., Zheng H., Dong D., Cook K., Shan G., Zhang H., Kosiol C., Xie X., Lu Z., Zheng H., Li Y., Steiner C. C., Lam T. T.-Y., Lin S., Zhang Q., Li G., Tian J., Gong T., Liu H., Zhang D., Fang L., Ye C., Zhang J., Hu W., Xu A., Ren Y., Zhang G., Bruford M. W., Li Q., Ma L., Guo Y., An N., Hu Y., Zheng Y., Shi Y., Li Z., Liu Q., Chen Y., Zhao J., Qu N., Zhao S., Tian F., Wang X., Wang H., Xu L., Liu X., Vinar T., et al. 2010. The sequence and de novo assembly of the giant panda genome. Nature 463:311–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li R., Yu C., Li Y., Lam T.-W., Yiu S.-M., Kristiansen K., Wang J. 2009. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25:1966–1967 [DOI] [PubMed] [Google Scholar]
- 58.Li R., Zhu H., Ruan J., Qian W., Fang X., Shi Z., Li Y., Li S., Shan G., Kristiansen K., Li S., Yang H., Wang J., Wang J. 2010. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 20:265–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lieberman-Aiden E., van Berkum N. L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B. R., Sabo P. J., Dorschner M. O., Sandstrom R., Bernstein B., Bender M. A., Groudine M., Gnirke A., Stamatoyannopoulos J., Mirny L. A., Lander E. S., Dekker J. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326:289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lister R., O'Malley R. C., Tonti-Filippini J., Gregory B. D., Berry C. C., Millar A. H., Ecker J. R. 2008. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133:523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liti G., Carter D. M., Moses A. M., Warringer J., Parts L., James S. A., Davey R. P., Roberts I. N., Burt A., Koufopanou V., Tsai I. J., Bergman C. M., Bensasson D., O'Kelly M. J. T., van Oudenaarden A., Barton D. B. H., Bailes E., Nguyen A. N., Jones M., Quail M. A., Goodhead I., Sims S., Smith F., Blomberg A., Durbin R., Louis E. J. 2009. Population genomics of domestic and wild yeasts. Nature 458:337–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lund J., Parviz B. A. 2009. Scanning probe and nanopore DNA sequencing: core techniques and possibilities. Methods Mol. Biol. 578:113–122 [DOI] [PubMed] [Google Scholar]
- 63.Lupski J. R., Reid J. G., Gonzaga-Jauregui C., Rio Deiros D., Chen D. C., Nazareth L., Bainbridge M., Dinh H., Jing C., Wheeler D. A., McGuire A. L., Zhang F., Stankiewicz P., Halperin J. J., Yang C., Gehman C., Guo D., Irikat R. K., Tom W., Fantin N. J., Muzny D. M., Gibbs R. A. 2010. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N. Engl. J. Med. 362:1181–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lynch M., Sung W., Morris K., Coffey N., Landry C. R., Dopman E. B., Dickinson W. J., Okamoto K., Kulkarni S., Hartl D. L., Thomas W. K. 2008. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc. Natl. Acad. Sci. U. S. A. 105:9272–9277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Malhis N., Jones S. J. M. 2010. High quality SNP calling using Illumina data at shallow coverage. Bioinformatics 26:1029–1035 [DOI] [PubMed] [Google Scholar]
- 66.Mardis E. R. 2008. Next-generation DNA sequencing methods. Annu. Rev. Genomics Hum. Genet. 9:387–402 [DOI] [PubMed] [Google Scholar]
- 67.Marguerat S., Bähler J. 2010. RNA-seq: from technology to biology. Cell. Mol. Life Sci. 67:569–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Margulies M., Egholm M., Altman W. E., Attiya S., Bader J. S., Bemben L. A., Berka J., Braverman M. S., Chen Y.-J., Chen Z., Dewell S. B., Du L., Fierro J. M., Gomes X. V., Godwin B. C., He W., Helgesen S., Ho C. H., Irzyk G. P., Jando S. C., Alenquer M. L. I., Jarvie T. P., Jirage K. B., Kim J.-B., Knight J. R., Lanza J. R., Leamon J. H., Lefkowitz S. M., Lei M., Li J., Lohman K. L., Lu H., Makhijani V. B., McDade K. E., McKenna M. P., Myers E. W., Nickerson E., Nobile J. R., Plant R., Puc B. P., Ronan M. T., Roth G. T., Sarkis G. J., Simons J. F., Simpson J. W., Srinivasan M., Tartaro K. R., Tomasz A., Vogt K. A., Volkmer G. A., Wang S. H., Wang Y., Weiner M. P., Yu P., Begley R. F., Rothberg J. M. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Martin F., Kohler A., Murat C., Balestrini R., Coutinho P. M., Jaillon O., Montanini B., Morin E., Noel B., Percudani R., Porcel B., Rubini A., Amicucci A., Amselem J., Anthouard V., Arcioni S., Artiguenave F., Aury J.-M., Ballario P., Bolchi A., Brenna A., Brun A., Buee M., Cantarel B., Chevalier G., Couloux A., Da Silva C., Denoeud F., Duplessis S., Ghignone S., Hilselberger B., Iotti M., Marcais B., Mello A., Miranda M., Pacioni G., Quesneville H., Riccioni C., Ruotolo R., Splivallo R., Stocchi V., Tisserant E., Viscomi A. R., Zambonelli A., Zampieri E., Henrissat B., Lebrun M.-H., Paolocci F., Bonfante P., Ottonello S., Wincker P. 2010. Périgord black truffle genome uncovers evolutionary origins and mechanisms of symbiosis. Nature 464:1033–1039 [DOI] [PubMed] [Google Scholar]
- 70.Martinez D. A., Nelson M. A. 2010. The next generation becomes the now generation. PLoS Genet. 6:e1000906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maxam A. M., Gilbert W. 1977. A new method for sequencing DNA. Proc. Natl. Acad. Sci. U. S. A. 74:560–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McKernan K. J., Peckham H. E., Costa G. L., McLaughlin S. F., Fu Y., Tsung E. F., Clouser C. R., Duncan C., Ichikawa J. K., Lee C. C., Zhang Z., Ranade S. S., Dimalanta E. T., Hyland F. C., Sokolsky T. D., Zhang L., Sheridan A., Fu H., Hendrickson C. L., Li B., Kotler L., Stuart J. R., Malek J. A., Manning J. M., Antipova A. A., Perez D. S., Moore M. P., Hayashibara K. C., Lyons M. R., Beaudoin R. E., Coleman B. E., Laptewicz M. W., Sannicandro A. E., Rhodes M. D., Gottimukkala R. K., Yang S., Bafna V., Bashir A., MacBride A., Alkan C., Kidd J. M., Eichler E. E., Reese M. G., De La Vega F. M., Blanchard A. P. 2009. Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome Res. 19:1527–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McLaughlin D. J., Hibbett D. S., Lutzoni F., Spatafora J. W., Vilgalys R. 2009. The search for the fungal tree of life. Trends Microbiol. 17:488–497 [DOI] [PubMed] [Google Scholar]
- 74.McPherson J. D. 2009. Next-generation gap. Nat. Methods 6:S2–S5 [DOI] [PubMed] [Google Scholar]
- 75.Metzker M. L. 2010. Sequencing technologies—the next generation. Nat. Rev. Genet. 11:31–46 [DOI] [PubMed] [Google Scholar]
- 76.Meyer M., Stenzel U., Myles S., Prüfer K., Hofreiter M. 2007. Targeted high-throughput sequencing of tagged nucleic acid samples. Nucleic Acids Res. 35:e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mikkelsen T. S., Ku M., Jaffe D. B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.-K., Koche R. P., Lee W., Mendenhall E., O'Donovan A., Presser A., Russ C., Xie X., Meissner A., Wernig M., Jaenisch R., Nusbaum C., Lander E. S., Bernstein B. E. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448:553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Miller J. R., Delcher A. L., Koren S., Venter E., Walenz B. P., Brownley A., Johnson J., Li K., Mobarry C., Sutton G. 2008. Aggressive assembly of pyrosequencing reads with mates. Bioinformatics 24:2818–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Miller J. R., Koren S., Sutton G. 2010. Assembly algorithms for next-generation sequencing data. Genomics 95:315–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5:621–628 [DOI] [PubMed] [Google Scholar]
- 81.Nagalakshmi U., Wang Z., Waern K., Shou C., Raha D., Gerstein M., Snyder M. 2008. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320:1344–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nielsen C. B., Jackman S. D., Birol I., Jones S. J. 2009. ABySS-Explorer: visualizing genome sequence assemblies. IEEE Trans. Vis. Comput. Graph. 15:881–888 [DOI] [PubMed] [Google Scholar]
- 83.Nowrousian M. 2007. Of patterns and pathways: microarray technologies for the analysis of filamentous fungi. Fungal Biol. Rev. 21:171–178 [Google Scholar]
- 84.Nowrousian M., Dunlap J. C., Nelson M. A. 2004. Functional genomics in fungi, p. 115–128InKück U.(ed.), The Mycota, vol. 2, 2nd ed.Springer, Berlin, Germany [Google Scholar]
- 85.Nowrousian M., Stajich J. E., Chu M., Engh I., Espagne E., Halliday K., Kamerewerd J., Kempken F., Knab B., Kuo H. C., Osiewacz H. D., Pöggeler S., Read N. D., Seiler S., Smith K. M., Zickler D., Kück U., Freitag M. 2010. De novo assembly of a 40 Mb eukaryotic genome from short sequence reads: Sordaria macrospora, a model organism for fungal morphogenesis. PLoS Genet. 6:e1000891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ozsolak F., Platt A. R., Jones D. R., Reifenberger J. G., Sass L. E., McInerney P., Thompson J. F., Bowers J., Jarosz M., Milos P. M. 2009. Direct RNA sequencing. Nature 461:814–818 [DOI] [PubMed] [Google Scholar]
- 87.Pace N. R. 2009. Mapping the tree of life: progress and prospects. Microbiol. Mol. Biol. Rev. 73:565–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pace N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734–740 [DOI] [PubMed] [Google Scholar]
- 89.Park P. J. 2009. ChIP-seq: advantages and challenges of a maturing technology. Nat. Rev. Genet. 10:669–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pepke S., Wold B., Mortazavi A. 2009. Computation for ChIP-seq and RNA-seq studies. Nat. Methods 6:S22–S32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pushkarev D., Neff N. F., Quake S. R. 2009. Single-molecule sequencing of an individual human genome. Nat. Biotech. 27:847–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Qi J., Wijeratne A. J., Tomsho L. P., Hu Y., Schuster S. C., Ma H. 2009. Characterization of meiotic crossovers and gene conversion by whole-genome sequencing in Saccharomyces cerevisiae. BMC Genomics 10:475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Reinhardt J. A., Baltrus D. A., Nishimura M. T., Jeck W. R., Jones C. D., Dangl J. L. 2009. De novo assembly using low-coverage short read sequence data from the rice pathogen Pseudomonas syringae pv. oryzae. Genome Res. 19:294–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Roach J. C., Glusman G., Smit A. F. A., Huff C. D., Hubley R., Shannon P. T., Rowen L., Pant K. P., Goodman N., Bamshad M., Shendure J., Drmanac R., Jorde L. B., Hood L., Galas D. J. 2010. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science 328:636–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Robertson G., Hirst M., Bainbridge M., Bilenky M., Zhao Y., Zeng T., Euskirchen G., Bernier B., Varhol R., Delaney A., Thiessen N., Griffith O. L., He A., Marra M., Snyder M., Jones S. J. 2007. Genome-wide profiles of STAT1 DNA association using chromatin immunopreciptation and massively parallel sequencing. Nat. Methods 4:651–657 [DOI] [PubMed] [Google Scholar]
- 96.Sanger F., Nicklen S., Coulson A. R. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U. S. A. 74:5463–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schuster S. C., Miller W., Ratan A., Tomsho L. P., Giardine B., Kasson L. R., Harris R. S., Petersen D. C., Zhao F., Qi J., Alkan C., Kidd J. M., Sun Y., Drautz D. I., Bouffard P., Muzny D. M., Reid J. G., Nazareth L. V., Wang Q., Burhans R., Riemer C., Wittekindt N. E., Moorjani P., Tindall E. A., Danko C. G., Teo W. S., Buboltz A. M., Zhang Z., Ma Q., Oosthuysen A., Steenkamp A. W., Oostuisen H., Venter P., Gajewski J., Zhang Y., Pugh B. F., Makova K. D., Nekrutenko A., Mardis E. R., Patterson N., Pringle T. H., Chiaromonte F., Mullikin J. C., Eichler E. E., Hardison R. C., Gibbs R. A., Harkins T. T., Hayes V. M. 2010. Complete Khoisan and Bantu genomes from southern Africa. Nature 463:943–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Service R. F. 2006. Gene sequencing. The race for the $1000 genome. Science 311:1544–1546 [DOI] [PubMed] [Google Scholar]
- 99.Shendure J., Li H. 2008. Next-generation DNA sequencing. Nat. Biotechnol. 26:1135–1145 [DOI] [PubMed] [Google Scholar]
- 100.Shendure J., Porreca G. J., Reppas N. B., Lin X., McCutcheon J. P., Rosenbaum A. M., Wang M. D., Zhang K., Mitra R. D., Church G. M. 2005. Accurate multiplex polony sequencing of an evolved bacterial genome. Science 309:1728–1732 [DOI] [PubMed] [Google Scholar]
- 101.Shumway M., Cochrane G., Sugawara H. 2010. Archiving next generation sequencing data. Nucleic Acids Res. 38:D870–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Simpson J. T., Wong K., Jackman S. D., Schein J. E., Jones S. J. M., Birol I. 2009. ABySS: a parallel assembler for short read sequence data. Genome Res. 19:1117–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Smith A. M., Heisler L. E., Mellor J., Kaper F., Thompson M. J., Chee M., Roth F. P., Giaever G., Nislow C. 2009. Quantitative phenotyping via deep barcode sequencing. Genome Res. 19:1836–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Smith D. R., Quinlan A. R., Peckham H. E., Makowsky K., Tao W., Woolf B., Shen L., Donahue W. F., Tusneem N., Stromberg M. P., Stewart D. A., Zhang L., Ranade S. S., Warner J. B., Lee C. C., Coleman B. E., Zhang Z., McLaughlin S. F., Malek J. A., Sorenson J. M., Blanchard A. P., Chapman J., Hillman D., Chen F., Rokhsar D. S., McKernan K. J., Jeffries T. W., Marth G. T., Richardson P. M. 2008. Rapid whole-genome mutational profiling using next-generation sequencing technologies. Genome Res. 18:1638–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Stuart J. M., Segal E., Koller D., Kim S. K. 2003. A gene-coexpression network for global discovery of conserved genetic modules. Science 302:249–255 [DOI] [PubMed] [Google Scholar]
- 106.Timme R. E., Delwiche C. F. 2010. Uncovering the evolutionary origin of plant molecular processes: comparison of Coleochaete (Coleochaetales) and Spirogyra (Zygnematales) transcriptomes. BMC Plant Biol. 10:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Trapnell C., Pachter L., Salzberg S. L. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Trapnell C., Salzberg S. L. 2009. How to map billions of short reads onto genomes. Nat. Biotechnol. 27:455–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., van Baren M. J., Salzberg S. L., Wold B. J., Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28:511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang B., Guo G., Wang C., Lin Y., Wang X., Zhao M., Guo Y., He M., Zhang Y., Pan L. 14April2010, posting date Survey of the transcriptome of Aspergillus oryzae via massively parallel mRNA sequencing. Nucleic Acids Res. [Epub ahead of print.] doi:10.1093/nar/gkq256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang J., Wang W., Li R., Li Y., Tian G., Goodman L., Fan W., Zhang J., Li J., Zhang J., Guo Y., Feng B., Li H., Lu Y., Fang X., Liang H., Du Z., Li D., Zhao Y., Hu Y., Yang Z., Zheng H., Hellmann I., Inouye M., Pool J., Yi X., Zhao J., Duan J., Zhou Y., Qin J., Ma L., Li G., Yang Z., Zhang G., Yang B., Yu C., Liang F., Li W., Li S., Li D., Ni P., Ruan J., Li Q., Zhu H., Liu D., Lu Z., Li N., Guo G., Zhang J., Ye J., Fang L., Hao Q., Chen Q., Liang Y., Su Y., San A., Ping C., Yang S., Chen F., Li L., Zhou K., Zheng H., Ren Y., Yang L., Gao Y., Yang G., Li Z., Feng X., Kristiansen K., Wong G. K.-S., Nielsen R., Durbin R., Bolund L., Zhang X., Li S., Yang H., Wang J. 2008. The diploid genome sequence of an Asian individual. Nature 456:60–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang Z., Gerstein M., Snyder M. 2009. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10:57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Warren R. L., Sutton G. G., Jones S. J. M., Holt R. A. 2007. Assembling millions of short DNA sequences using SSAKE. Bioinformatics 23:500–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wheeler D. A., Srinivasan M., Egholm M., Shen Y., Chen L., McGuire A., He W., Chen Y.-J., Makhijani V., Roth G. T., Gomes X., Tartaro K., Niazi F., Turcotte C. L., Irzyk G. P., Lupski J. R., Chinault C., Song X.-Z., Liu Y., Yuan Y., Nazareth L., Qin X., Muzny D. M., Margulies M., Weinstock G. M., Gibbs R. A., Rothberg J. M. 2008. The complete genome of an individual by massively parallel DNA sequencing. Nature 452:872–876 [DOI] [PubMed] [Google Scholar]
- 115.Wilhelm B. T., Marguerat S., Watt S., Schubert F., Wood V., Goodhead I., Penkett C. J., Rogers J., Bähler J. 2008. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 453:1239–1243 [DOI] [PubMed] [Google Scholar]
- 116.Wooley J. C., Godzik A., Friedberg I. 2010. A primer on metagenomics. PLoS Comput. Biol. 6:e1000667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wu R., Kaiser A. D. 1968. Structure and base sequence in the cohesive ends of bacteriophage lambda DNA. J. Mol. Biol. 35:523–537 [DOI] [PubMed] [Google Scholar]
- 118.Zerbino D. R., Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]