Abstract
Vibrio cholerae is a Gram-negative bacillus that is the causative agent of cholera. Pathogenesis in vivo occurs through a series of spatiotemporally controlled events under the control of a gene cascade termed the ToxR regulon. Major genes in the ToxR regulon include the master regulators toxRS and tcpPH, the downstream regulator toxT, and virulence factors, the ctxAB and tcpA operons. Our current understanding of the dynamics of virulence gene expression is limited to microarray analyses of expression at selected time points. To better understand this process, we utilized a systems biology approach to examine the temporal regulation of gene expression in El Tor V. cholerae grown under virulence-inducing conditions in vitro (AKI medium), using high-resolution time series genomic profiling. Results showed that overall gene expression in AKI medium mimics that of in vivo studies but with less clear temporal separation between upstream regulators and downstream targets. Expression of toxRS was unaffected by growth under virulence-inducing conditions, but expression of toxT was activated shortly after switching from stationary to aerating conditions. The tcpA operon was also activated early during mid-exponential-phase growth, while the ctxAB operon was turned on later, after the rise in toxT expression. Expression of ctxAB continued to rise despite an eventual decrease in toxT. Cluster analysis of gene expression highlighted 15 hypothetical genes and six genes related to environmental information processing that represent potential new members of the ToxR regulon. This study applies systems biology tools to analysis of gene expression of V. cholerae in vitro and provides an important comparator for future studies done in vivo.
Vibrio cholerae is a Gram-negative, motile bacillus that is the pathogenic agent of the diarrheal disease cholera. The WHO estimates that there are 5 million cases of cholera worldwide a year, with more than 100,000 deaths (22). Infection occurs when bacteria pass from the external environment into humans through fecal-oral transmission (19). Once inside the body, V. cholerae causes a severe, secretory diarrhea that can lead to death through dehydration. There are over 200 serogroups of V. cholerae that are distinguished by the O antigen of the lipopolysaccharide. The only serogroups that are known to cause epidemic or pandemic cholera are serogroups O1 and O139. The O1 serogroup can be further broken down into the classical and El Tor biotypes, with the latter representing the dominant strain of the ongoing global pandemic. Both biotypes can be further subdivided into the Ogawa and Inaba serotypes.
Organisms that survive passage through the acidic stomach enter the small intestine, where they produce cholera toxin (CT), a prototypical A1B5 subunit ADP-ribosylating enterotoxin. The B pentamer of the exotoxin binds to the ganglioside GM1 on the surfaces of absorptive intestinal epithelial cells and translocates the A subunit into the host cytoplasm. The enzymatically active A subunit activates adenylate cyclase through ADP-ribosylation of its G protein, resulting in an increase in cyclic AMP levels and subsequent efflux of chloride ions into the intestinal lumen. The resulting osmotic shift in the lumen results in poor water absorption and copious, watery diarrhea (14). The other essential virulence factor of V. cholerae is the toxin-coregulated pilus (TCP), a type IV pilus that (i) facilitates colonization of the gastrointestinal tract by mediating bacterial-bacterial interactions to protect them from the shear stresses of the intestine (20, 28) and (ii) serves as the receptor for the bacteriophage encoding the cholera toxin genes (41).
The genes for cholera toxin are encoded in the filamentous bacteriophage CTXΦ, which enters bacterial cells by binding to TCP and then integrating into the bacterial chromosome and converting a nontoxigenic strain into a toxigenic strain (41). TCP is assembled by a set of genes located within the tcpA operon on a 40-kb pathogenicity island that also contains other virulence genes that are jointly regulated (23). Both the ctxAB and tcpA operons are activated primarily by the ToxT transcription factor, a member of the AraC protein family (16). ToxT also positively regulates expression of accessory colonization factor (acf) genes, the gene for aldehyde dehydrogenase (aldA), and ToxR-activated gene A (tagA) (29). Expression of toxT is itself positively regulated by two sets of dimeric transcription factors, ToxRS and TcpPH. ToxR is a transmembrane protein with a cytoplasmic DNA binding domain (35) whose activity is greatly enhanced by the presence of another transmembrane protein, ToxS. TcpP is a homologue of ToxR and is similarly localized to the membrane, with a DNA binding cytoplasmic domain; a second protein, TcpH, enhances the stability of TcpP (3). Both ToxR and TcpP bind the toxT promoter; however, it appears that TcpP is more directly responsible for the transcription of toxT, with ToxR playing a more indirect role (25). ToxR is known to regulate at least 17 genes besides toxT, including the outer membrane porin genes ompU and ompT (8). ToxR has also been shown to directly stimulate ctxAB expression, independently of ToxT (7). These ToxR-regulated virulence genes are collectively called the ToxR regulon, after the first discovered master regulator (35).
The sequencing of the genome of the V. cholerae O1 El Tor strain N16961 in 2000 opened the door to using high-throughput technologies to analyze this bacterium (15). The major advantage of these approaches is that they give researchers the opportunity to examine the effects of genetic and/or environmental perturbations on the expression of thousands of genes or proteins simultaneously without observation bias. DNA microarrays have been used to identify members of the rpoH regulon required for the V. cholerae heat shock response (39). Other work has elucidated the changes in the transcriptional profile of V. cholerae from early to late infection directly in humans (27). Two recent studies used analysis of the in vivo transcriptome of V. cholerae to highlight the existence of an “escape” response to help the bacteria detach from the intestinal epithelial surface and express a “preenvironmental” response to help prepare the organism for exit from the host into the marine environment (34, 38).
Studies using animal and human models have highlighted the temporal nature of virulence gene expression in V. cholerae (28); however, studies of global gene expression in vivo are impractical due to the technical difficulties of creating and resolving reporter gene constructs for all 3,890 open reading frames (ORFs) of the V. cholerae genome. Furthermore, genome profiling studies of humans are able to capture the V. cholerae transcriptome in early and late infection, using vomit and stool samples of infected patients for these phases (27), but miss gene expression in the small intestine, the site of active colonization.
To overcome these difficulties, we utilized a systems biology approach to assay gene expression at multiple time points during the growth of V. cholerae under virulence-inducing conditions to allow for high temporal resolution of the entire genome in parallel as virulence gene expression occurs over time. Previous studies have shown that El Tor V. cholerae strains grown in bicarbonate (1, 17) or in bicarbonate-containing AKI medium (18) activate virulence gene expression in vitro; we utilized growth in AKI medium in this study for analysis of gene expression at multiple time points. We assessed when the major ToxR regulon genes were expressed in relation to one another and then used the expression profiles of known virulence genes to identify additional genes regulated in parallel that might play a yet-unidentified role in pathogenesis. These potential virulence genes may enhance our understanding of the infectious process.
MATERIALS AND METHODS
Media and bacterial growth.
V. cholerae strain N16961 was streaked on LB agar containing streptomycin (100 μg/ml). After overnight incubation, a single colony was inoculated into 25 ml of LB broth with streptomycin (100 μg/ml) and grown overnight. A 10-ml inoculum of the overnight culture was then transferred to 1 liter of either AKI medium (1.5% Bacto peptone, 0.4% yeast extract, 0.5% NaCl, 0.3% NaHCO3) to induce cholera virulence gene expression or LB medium, both of which were preheated to 37°C to minimize the cold shock transcriptional response. Cultures in AKI medium were grown initially under stationary microaerophilic conditions at 37°C with a flask-to-medium volume ratio of 1:1. Once cultures reached mid-log phase (optical density at 600 nm [OD600] of ∼0.300), the culture was switched to aerobic growth conditions, with a flask-to-medium volume ratio of 2:1 and shaking at 220 rpm (18). LB medium cultures were incubated aerobically with a flask-to-medium volume ratio of 2:1 and shaking at 220 rpm at 37°C for the entire growth curve. OD600 measurements of growth in AKI medium were done in triplicate, while the LB medium growth curve was done once.
Measurement of cholera toxin production.
Detection of cholera toxin was done to confirm activation of the virulence cascade as previously described (5). One-milliliter samples of culture supernatants were taken from each time point and centrifuged at 3,000 × g for 5 min to remove bacterial cells and debris. Purified supernatants were then serially diluted 2-fold on enzyme-linked immunosorbent assay (ELISA) plates coated with GM1-ganglioside. Goat anti-cholera toxin subunit B (List Biological Laboratories) was used as the primary antibody, with rabbit anti-goat conjugated to horseradish peroxidase used as the secondary antibody and hydrogen peroxide as the detecting reagent. The cholera toxin B subunit at 1 mg/ml was used to generate the standard curve.
Isolation of bacterial RNA for transcriptional analysis.
Thirteen time points were taken every 25 min starting at 1 h and continuing until 6 h after the beginning of growth in either LB or AKI medium. A solution of 50% ethanol-50% acetone (chilled to −20°C) plus rifampin (200 μg/ml; Sigma-Aldrich) was added 1:1 to an aliquot of the growth medium at each time point to halt transcription and preserve RNA (37). Samples were immediately stored at −80°C overnight. The volume of sample used for transcriptional analysis varied inversely with the OD600 of the culture aliquot, with lower optical densities requiring larger volumes of sample to generate enough signal for subsequent microarray hybridization (OD600 of 0.000 to 0.060, 100-ml aliquot; OD600 of 0.061 to 0.100, 50-ml aliquot; OD600 of 0.101 to 0.250, 25-ml aliquot; OD600 of >0.250, 10-ml aliquot). Culture aliquots were thawed at room temperature and spun at 3,000 rpm at 4°C for 10 min to pellet cells. The supernatant was removed, and the cells were resuspended in 1 ml of lysis buffer. The RNeasy minicolumn kit was used for mRNA purification and on-the-column DNase treatment according to the manufacturer's specifications (Qiagen). The concentration of RNA in samples was quantitated using spectrophotometry, and the RNA was visualized on a 1% agarose gel to confirm RNA integrity.
Microarray hybridization.
The V. cholerae microarray (J. Craig Venter Institute, http://www.jcvi.org) contains all 3,890 annotated ORFs of V. cholerae El Tor strain N16961. Seventy-base-pair oligomers from the ORF of each gene are spotted on each slide in triplicate. mRNA from time points following growth in AKI and LB media was purified, labeled, hybridized, and quantitated as previously described (4, 10). Genomic DNA from V. cholerae O1 El Tor strain N16961 was isolated using the Easy-DNA kit (Invitrogen) according to the manufacturer's specifications and was used as a universal internal control to normalize the quality of the array and to allow for comparison between time points and conditions.
Microarray data analysis.
Microarray slides were scanned and spot intensities quantitated using the ScanArray Express (PerkinElmer; version 4.0) software. All spots were checked manually for artifacts. Data were analyzed using the software package Limma, within the “R” computing environment (40). Limma is part of the Bioconductor project (http://bioconductor.org [13]) and has been validated across a range of microarray data sets. Data normalization within and between arrays was performed using standard techniques (42), and log fold changes in expression of each gene were estimated using linear modeling. Fold changes were first calculated for LB medium and AKI medium separately. The final expression profile represents the expression of each gene in AKI medium at each time point after subtracting expression of that gene at that time point in LB medium, with expression given relative to that at the 1-h time point. We did the expression profiling in AKI medium in triplicate at each time point and then averaged those results before subtracting the results for growth in LB medium. For genes that had little change in expression in LB medium, this had little effect on the profile other than to average the fold changes among the AKI medium replicates.
Clustering analysis of gene expression.
For the clustering analysis, we used the cluster analysis of gene expression dynamics (CAGED) algorithm, a Bayesian clustering method that is ideally suited for time series microarray data (36). The algorithm models the time series profile of each gene using a polynomial or autoregressive model and then iteratively merges the time series into clusters of genes with similar expression patterns based on a user-defined similarity measure and the overall posterior probability of the model. The algorithm stops when remaining mergers no longer increase the model's posterior probability. Theoretically, each cluster represents a separate biologic process, generating a unique time series profile. For this analysis, we used fourth-order polynomial equations and Euclidean distance as our model parameter and similarity measure, respectively. Both the prior precision (the sample size of imaginary observations that builds the prior distribution) and the Bayes factor threshold (the minimum value that must be met for two series to be merged) were set to the default value of 1.
RESULTS
V. cholerae produces significant amounts of cholera toxin under inducing conditions.
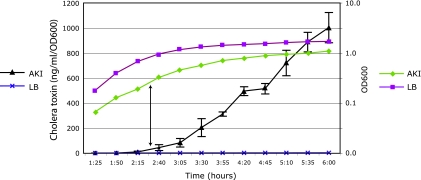
El Tor V. cholerae strains grown in vitro produce toxin under AKI medium conditions (18). We utilized this property, as well as the inability of V. cholerae to produce toxin in standard LB medium, to evaluate the impact of toxin-inducing and noninducing conditions on gene expression. Figure 1 compares the growth curves of V. cholerae El Tor strain N16961 grown in AKI medium and LB medium, superimposed with levels of toxin production over time. V. cholerae grew more rapidly and reached a higher steady-state cell density in LB medium than in AKI medium. Expression of cholera toxin was seen specifically under AKI medium conditions, as expected. Bacteria were already in the exponential phase of growth by 1 h postinoculation in both media and reached stationary phase by 3 h in LB and 4 h in AKI medium. The delayed growth in AKI medium likely reflects the more limited nutrients than in LB medium and the growth initially under microaerophilic conditions. Cholera toxin was detectable in the supernatants of cultures grown in AKI medium shortly after the cultures were switched to aerating conditions at ca. 2.5 hours and increased linearly through the 6-h time point.
FIG. 1.
V. cholerae O1 El Tor N16961 growth curves and toxin production in LB and AKI media. Average AKI medium and LB medium growth curves and toxin production are shown. Toxin concentrations in ng/ml/OD600 appear on the left y axis. The OD600 log scale for growth curves appears on the right y axis. The arrow represents the time at which AKI medium cultures were switched to shaking (aerobic) growth conditions. LB medium cultures were grown aerobically throughout the growth curve.
V. cholerae virulence genes are upregulated in AKI medium versus in LB medium.
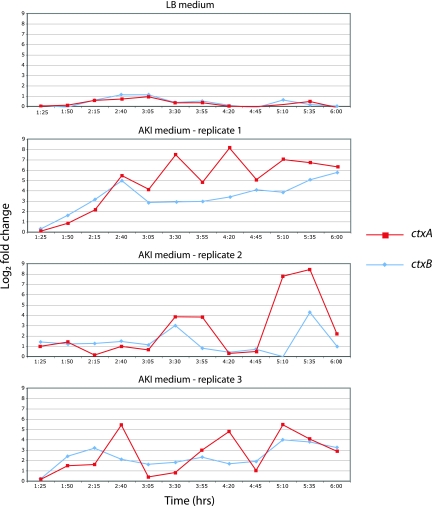
In order to investigate the effect of growth under AKI medium conditions on V. cholerae global gene expression, we compared the dynamic transcriptomes of V. cholerae grown in LB and AKI media at multiple time points during the exponential and stationary phases of growth. ctxA had the highest peak fold change of all genes and also exhibited large fluctuations in expression over a range of 7 log2 at the various time points. There was the suggestion of periodicity in ctxA expression, but the three AKI medium replicates had differing amplitudes and frequencies of these periodic swings (Fig. 2). The time series expression profile of ctxB followed the general trend of ctxA's but lacked the periodicity and large fluctuations. Expression of both genes in LB medium remained near baseline over the entire growth curve. The full data sets of expression of all genes are available as supplementary material via the Gene Expression Omnibus website (http://www.ncbi.nlm.nih.gov/geo/).
FIG. 2.
Comparison of levels of cholera toxin gene expression in LB medium and AKI medium replicates. Each data point represents the log2 fold change from the expression level at 1 h. Cultures were switched to aerating conditions at 2 h 30 min.
Major virulence genes and regulators display differing levels of gene expression over time.
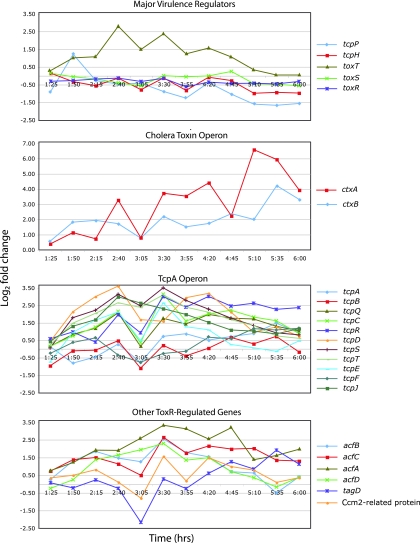
Of the genes known to be involved in regulation of virulence gene expression in V. cholerae (toxRS, tcpPH, and toxT), there was a brief spike in tcpP expression at 1 h 50 min, followed by significant upregulation of toxT during growth under AKI medium conditions (Fig. 3), suggesting that growth under AKI medium conditions upregulates toxT independently of expression of ToxRS. All of these genes (toxRS, tcpPH, and toxT) remained at baseline during growth in LB medium throughout the time course (data not shown).
FIG. 3.
Time series expression profiles of major virulence genes and gene regulators. Each data point represents the log2 fold change averaged from three AKI medium replicates with respect to expression in LB medium at 1 h. Cultures were switched to aerating conditions at 2 h 30 min.
The genes of the tcpA operon were also expressed at significantly higher levels in AKI medium. In contrast to what occurred with ctxAB, whose expression did not increase until after the expression of toxT spiked at 2 h 40 min, the time course profiles of the tcpA operon increased simultaneously with the increase in toxT. Nearly all tcpA operon members increased in expression from 1 h 25 min until 2 h 40 min and then transiently decreased shortly after the point at which the cultures were switched to aerating conditions; they increased again at 3 h 30 min (Fig. 3). There was considerable variation in expression between the operon members within a replicate, with less interreplicate variation. tcpA showed a negative fold change earlier in the time series in all AKI medium replicates, but then expression tended upward during the stationary phase of growth. Expression of the tcpA operon in LB medium was generally close to baseline throughout, except with tcpE and tcpB, which were expressed at levels 0.5- to 1.7-log2-fold above baseline throughout, and tcpR, whose expression remained below −1.2 log2 after 3 h 30 min (data not shown).
We examined genes for similarities in their time series profiles during growth under toxin-inducing conditions. The overall shape of the toxT profile was quite similar to the shapes of profiles of its downstream targets ctxA, tcpB, tcpC, tcpE, tcpQ, tcpR, tcpS, and tcpT, at least to 4 h 20 min. The time expression profile of toxT was less similar to those of other downstream targets, ctxB, tcpA, tcpD, tcpF, and tcpJ, although the reason for the difference from other members of the same ctxAB and tcpA operons is unknown (data not shown).
Expression profiles of the accessory colonization factors (acfABCD) increased early in the time course, peaked at 3 h 30 min, and then gradually decreased as bacteria entered stationary phase. Expression of the gene for the Ccm2-related protein (VCA1042), as well as of acfD and tagD, oscillated in phase with toxT between 2 h 40 min and 4 h 20 min, before expression decreased to baseline by 6 h. Expression of tagD was negatively regulated initially, reaching a minimum of −2.2 log2 below baseline at 3 h 5 min before rebounding and remaining near 1.0 log2 above baseline by the end of growth (Fig. 3). None of these genes were upregulated in LB medium (data not shown).
Potential new members of the V. cholerae virulence regulon identified through cluster analysis.
We used the CAGED algorithm on our time series data as an unsupervised method of identifying potential new members of the virulence regulon. The analysis was limited to the 127 genes that exhibited more than ±2-log2-fold changes in AKI medium at any point in the time series and that also did not have a similar fold change during growth in LB medium. Presumably, these are the genes that are most closely associated with toxin production and other virulence attributes. This assumption is supported by the fact that this subset contains 10 of the 13 genes identified through transcriptome analysis to be downregulated in a toxT mutant (2) and also contains the cholera toxin operon, toxT itself, and the acf operon.
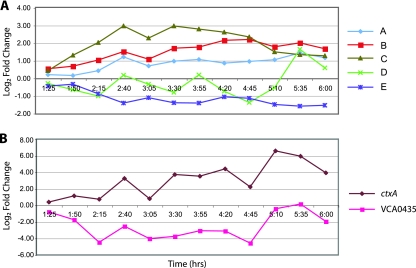
Using the model parameters described in Materials and Methods, the CAGED analysis generated five distinct clusters, labeled A to E, suggesting five different biologic processes associated with pathogenesis. Cluster A contains 41 genes, cluster B 32 genes, cluster C 6 genes, cluster D 5 genes, and cluster E 41 genes. All known virulence genes except for ctxA grouped into clusters A to C. The series profile shown in Fig. 4A represents the average levels of expression of the genes over time and provides a visual representation of the expression pattern of each cluster. The series profile for clusters A and B have similar patterns, peaking late in the growth curve, with cluster B genes expressed at a higher average value. Cluster C expression peaked earlier and genes were expressed to a greater degree than with cluster A and B genes, reaching 3.0 log2 above baseline between 2 h 40 min and 3 h 30 min. Cluster D was negatively regulated, oscillating between −1.4 and 0 log2 until 5 h 10 min, when expression rose ∼2-log2-fold in 25 min to 1.6 log2. The expression of cluster E tended downward and reached a plateau around −1.4 log2 below baseline from 2 h 40 min until 6 h. ctxA and locus VCA0435 did not cluster with any other genes due to their very large positive and negative fold changes, respectively (Fig. 4B).
FIG. 4.
(A) Cluster time series expression profiles. Each data point represents the average expression of all genes within the cluster, normalized to expression in LB medium and to time at 1 h. Cluster A contains 41 genes, cluster B 32 genes, cluster C 6 genes, cluster D 5 genes, and cluster E 41 genes. (B) Time series expression profiles of ctxA and VCA0435. These two genes did not cluster with any genes due to the sizes of their positive and negative fold changes, respectively.
Gene clusters were further divided into functional categories: virulence genes, transcription/translation genes, metabolism genes, miscellaneous genes, and unknown genes (Table 1 ). Cluster A, which contained toxT, tcpE, acfB, acfD, tagD, and tagE, also included 11 hypothetical genes of unknown function. Cluster B contained tcpC, tcpQ, tcpR, ctxB, and acfC, along with four hypothetical genes, and cluster C contained tcpD, tcpJ, tcpS, tcpT, and acfA. Other genes of interest in clusters A to C included three ATP-binding cassette active-transport genes (VC1092, VC1882, VCA0759), an ion transporter (VCA1093), and two signal transduction genes (VC0200, VCA0232). A subunit of SecY (VC2576), a member of the protein complex involved in cholera toxin secretion, was included in cluster A. A large number of housekeeping genes associated with transcription/translation and cellular metabolism also showed a time course of expression identical to that of genes specific to virulence.
TABLE 1.
Results of cluster analysis
| Cluster | Category | Common name or characteristic | Locus |
|---|---|---|---|
| A | Virulence | TagD protein | VC0824 |
| Toxin-coregulated pilus biosynthesis protein E | VC0836 | ||
| TCP pilus virulence regulatory protein | VC0838 | ||
| Accessory colonization factor AcfB | VC0840 | ||
| TagE protein | VC0843 | ||
| Accessory colonization factor AcfD | VC0845 | ||
| Unknown | Hypothetical protein | VC0712 | |
| Hypothetical protein | VC0821 | ||
| Conserved hypothetical protein | VC0842 | ||
| Hypothetical protein | VC1221 | ||
| Conserved hypothetical protein | VC2278 | ||
| Conserved hypothetical protein | VCA0006 | ||
| Hypothetical protein | VCA0393 | ||
| Hypothetical protein | VCA0394 | ||
| Hypothetical protein | VCA0480 | ||
| Hypothetical protein | VCA0817 | ||
| Hypothetical protein | VCA0932 | ||
| Transcription/translation | DNA polymerase III, beta chain | VC0013 | |
| Ribosomal protein L11 | VC0324 | ||
| Ribosomal protein L10 | VC0326 | ||
| Elongation factor G | VC0361 | ||
| Ribosomal protein L13 | VC0570 | ||
| Elongation factor G | VC2342 | ||
| Ribosomal protein L36 | VC2575 | ||
| Ribosomal protein L24 | VC2585 | ||
| Ribosomal protein S3 | VC2590 | ||
| Ribosomal protein L3 | VC2596 | ||
| Metabolism | Citrate lyase, gamma subunit | VC0797 | |
| Cytochrome d ubiquinol oxidase, subunit II | VC1843 | ||
| NADH:ubiquinone oxidoreductase, Na translocating, alpha subunit | VC2295 | ||
| UDP-N-acetylmuramate:l-alanyl-gamma-d-glutamyl-meso-diaminopimelate ligase | VC2542 | ||
| Aspartate ammonia-lyase | VC2698 | ||
| C4-dicarboxylate transporter, anaerobic | VC2699 | ||
| ATP synthase Fo, B subunit | VC2768 | ||
| ATP synthase Fo, A subunit | VC2770 | ||
| 3,4-Dihydroxy-2-butanone 4-phosphate synthase | VCA1060 | ||
| Other | LysE/YggA family protein | VC0481 | |
| Oligopeptide ABC transporter, permease protein | VC1092 | ||
| Lipoprotein-releasing system permease protein | VC1882 | ||
| Preprotein translocase, SecY subunit | VC2576 | ||
| Na+/H+ antiporter, putative | VCA0193 | ||
| B | Virulence | Toxin-coregulated pilus biosynthesis protein Q | VC0830 |
| Toxin-coregulated pilus biosynthesis outer membrane protein C | VC0831 | ||
| Toxin-coregulated pilus biosynthesis protein R | VC0832 | ||
| Accessory colonization factor AcfC | VC0841 | ||
| Cholera enterotoxin, B subunit | VC1456 | ||
| Unknown | Hypothetical protein | VC2098 | |
| Hypothetical protein | VCA0408 | ||
| Hypothetical protein | VCA0619 | ||
| Conserved hypothetical protein | VCA0689 | ||
| Transcription/translation | Transcription antitermination protein NusG | VC0323 | |
| DNA-directed RNA polymerase, beta subunit | VC0328 | ||
| Ribosomal protein S7 | VC0360 | ||
| Ribosomal protein L21 | VC0435 | ||
| Cell division protein FtsH | VC0637 | ||
| Ribosomal protein L25 | VC1640 | ||
| Ribosomal protein S11 | VC2573 | ||
| Ribosomal protein L14 | VC2586 | ||
| Ribosomal protein S17 | VC2587 | ||
| Ribosomal protein S19 | VC2592 | ||
| Ribosomal protein L31 | VC2679 | ||
| Metabolism | Aldehyde dehydrogenase | VC0819 | |
| Long-chain-fatty acid transport protein | VC1043 | ||
| Phosphoserine aminotransferase | VC1159 | ||
| Glyceraldehyde 3-phosphate dehydrogenase | VC2000 | ||
| Enolase | VC2447 | ||
| Other | Iron(III) compound receptor | VC0200 | |
| NifR3/Smm1 family protein | VC0291 | ||
| Antioxidant, AhpC/Tsa family | VC0731 | ||
| Enterobactin synthetase component F-related protein | VC1579 | ||
| Enterobactin receptor VctA, authentic frameshift | VCA0232 | ||
| Arginine ABC transporter, periplasmic arginine-binding protein | VCA0759 | ||
| DnaJ-related protein | VCA0788 | ||
| C | Virulence | Toxin-coregulated pilus biosynthesis protein D | VC0833 |
| Toxin-coregulated pilus biosynthesis protein S | VC0834 | ||
| Toxin-coregulated pilus biosynthesis protein T | VC0835 | ||
| Leader peptidase TcpJ | VC0839 | ||
| Accessory colonization factor AcfA | VC0844 | ||
| Transcription/translation | Ribosomal protein L19 | VC0564 | |
| D | Unknown | Hypothetical protein | VC0507 |
| Hypothetical protein | VCA0306 | ||
| Other | RstB1 protein | VC1453 | |
| Chaperonin, 60-kDa subunit | VC2664 | ||
| C4-dicarboxylate transporter, anaerobic | VCA0205 | ||
| E | Unknown | Conserved hypothetical protein | VC0340 |
| Hypothetical protein | VC0546 | ||
| Hypothetical protein | VC0569 | ||
| Conserved hypothetical protein | VC0758 | ||
| Conserved hypothetical protein | VC0770 | ||
| Hypothetical protein | VC0967 | ||
| Conserved hypothetical protein | VC1153 | ||
| Conserved hypothetical protein | VC1354 | ||
| Hypothetical protein | VC1794 | ||
| Hypothetical protein | VC1924 | ||
| Hypothetical protein | VC1999 | ||
| Hypothetical protein | VC2149 | ||
| Conserved hypothetical protein | VC2216 | ||
| Hypothetical protein | VC2306 | ||
| Hypothetical protein | VC2351 | ||
| Conserved hypothetical protein | VC2523 | ||
| Conserved hypothetical protein | VCA0835 | ||
| Transcription/translation | RNase P protein component | VC0006 | |
| Ribosomal protein L9 | VC0369 | ||
| Alanyl-tRNA synthetase | VC0545 | ||
| Peptidyl-tRNA hydrolase | VC2184 | ||
| Elongation factor Ts | VC2259 | ||
| Site-specific recombinase IntI4 | VCA0291 | ||
| Metabolic | Pyrroline-5-carboxylate reductase | VC0460 | |
| Glutathione synthetase | VC0468 | ||
| Hypoxanthine phosphoribosyltransferase | VC0585 | ||
| Inositol monophosphate family protein | VC0745 | ||
| Cytidine deaminase | VC1231 | ||
| 3-Hydroxydecanoyl-(acyl carrier protein) dehydratase | VC1483 | ||
| Mannose-6-phosphate isomerase | VC1827 | ||
| l-Asparaginase I | VC1995 | ||
| 3-Oxoacyl-(acyl carrier protein) reductase | VC2021 | ||
| Long-chain fatty acid-coenzyme A ligase, putative | VC2484 | ||
| Periplasmic nitrate reductase | VCA0678 | ||
| Other | ATP-dependent DNA helicase RecQ | VC0196 | |
| Factor for inversion stimulation protein | VC0290 | ||
| Cellobiose/cellodextrin-phosphorylase, putative | VC0612 | ||
| Heat shock protein HslJ | VC1663 | ||
| Heme exporter protein C | VC2055 | ||
| MinD-related protein | VC2067 | ||
| Aerobic respiration control protein FexA | VC2368 | ||
| None (unclustered) | Virulence | Cholera enterotoxin, A subunit | VC1457 |
| Unknown | Hypothetical protein | VCA0435 |
The negatively regulated clusters D and E contained 2 and 17 hypothetical genes, respectively. Among the other genes belonging to these groups were the oxygen sensor fexA (VC2368), an anaerobic C4-dicarboxylate transporter (VCA0205), the phage protein RstB1 (VC1453), a factor-for-inversion stimulation protein (VC0290), a heat shock gene (VC1663), and the flagellar assembly protein MinD (VC2067).
DISCUSSION
Previous studies of V. cholerae virulence gene regulation have focused on a limited number of individual genes or time points, comparison of expression levels between two conditions or strains, or expression in two different samples from humans (4, 27, 28). This work represents the first attempt to utilize systems biology tools to study expression at the whole-genome level, with high temporal resolution, during growth of the organism in vitro under virulence-inducing conditions. We show that virulence genes of El Tor V. cholerae O1 are expressed at much higher levels under toxin-inducing conditions than in standard medium. We found that tcpP and toxT were expressed early during mid-log-phase growth in AKI medium despite no increase in toxRS expression at that point. The fact that we saw only a brief spike in tcpP expression may reflect the very short half-life of the tcpPH transcript (ca. 2 min), shown previously (33). Members of the tcpA operon were also upregulated early, while ctxAB was expressed after the increase in expression of toxT and shortly after the switch to shaking conditions. Once initiated, however, ctxAB expression continued to increase independently of toxT levels. Using our time series clustering method, we distilled gene expression profiles into five distinct clusters containing known virulence genes and identified 15 hypothetical genes positively associated with virulence gene expression that are potential new members of the ToxR regulon and represent future targets of study.
The temporal nature of virulence gene expression in V. cholerae is of critical importance in understanding disease pathogenesis. Lee et al. showed in a murine model that expression of tcpA increased early as the bacteria began microcolonization of the upper intestine, while expression of ctxAB occurred later, after bacteria had attached to intestinal epithelial cells. They hypothesized that tcpA is induced before ctxAB to ensure that only bacteria that are adherent to cells secrete toxin, thereby increasing the efficiency of intoxication (28). Merrell et al. and Schild et al. described the activation of hyperinfectious and preenvironmental genetic programs late in infection, as the bacteria prepare to exit the intestine and reenter the external environment (31, 38). These results point to a highly evolved sequence of adaptive responses in V. cholerae as the organism colonizes the intestine, causes infection, and then escapes back into the environment to repeat the process again. Although growth in AKI medium cannot mimic the full complexity of the spatiotemporal interactions between host and pathogen, we present evidence that the major ToxR regulon members show temporal variations during in vitro growth in AKI medium similar to those that occur during intestinal colonization. Activation of the tcpA operon occurred during the exponential growth phase, while expression of cholera toxin genes increased later (Fig. 3). This suggests that although ToxT is responsible for tcpA and ctxAB operon expression, its ability to act on their respective promoters in vitro is dependent on different factors. The tcpA operon appears to be more responsive to cell density signals, perhaps relayed via quorum-sensing molecules, whereas the ctxAB operon may be triggered by different environmental conditions. The temporally distinct expression of the tcpA operon early and ctxAB later is analogous to the in vivo hypothesis put forth by Lee et al. (28) but in response to different signals.
Growth in AKI medium did not appear to influence toxRS expression, and there was only a brief spike of tcpP expression seen at 1 h 50 min. The low levels of tcpPH expression contrast with prior work by Murley et al. and Kovacikova and Skorupski, who showed upregulation of tcpPH in El Tor strains in AKI medium at the end of static phase and after overnight growth, respectively (24, 33). If we reexamine our data without normalizing to LB medium or to time at 1 h, we can reconcile these results, since tcpPH transcript levels were found to tend upward at the end of 6 h of growth (data not shown). Also, the tcpPH message has a very short half-life (33), and results seen previously with promoter fusions may give results quite different from gene microarray results, as demonstrated here to detect the presence of specific mRNA at individual time points. The constitutive level of toxR expression in both media is consistent with prior research by Medrano et al., who demonstrated that ToxR appears early during static growth in AKI medium and is present constitutively thereafter (30). Our results suggest that higher levels of toxT expression in AKI medium occur at a time when the proximal regulators, ToxRS, are not upregulated. Activation of toxT expression at this point could be the result of an unknown positive regulator or of specific environmental signals during growth in AKI medium.
The cholera toxin operon responds in phase to toxT expression starting at 2 h 40 min but, interestingly, continues to increase in expression even as toxT expression declines. This behavior suggests that ToxT is required for the initiation of cholera toxin gene expression but perhaps not for its continued expression in vitro. These results are consistent with the work of Medrano et al., who demonstrated that cholera toxin mRNA continues to increase linearly through 10 h of growth despite the disappearance of toxT after hour 5 (30). Those authors proposed that ToxR is present constitutively but that it is able to bind to the promoter region of toxT only during the early stationary phase of growth, when it receives the proper signals to switch to the “on” conformation. There are at least three possible explanations for continued ctxAB transcription despite decreasing levels of toxT expression. The first is that ToxR directly binds to the ctxAB promoter, bypassing toxT. Direct activation of ctxAB by ToxR has been shown before (7), but Dirita et al. showed that ToxT was the primary activator of ctxAB in vitro (9). Another possibility is that toxT is present at later time points mainly as part of a larger transcript of the tcpA operon, in which toxT resides, but that the oligonucleotide array did not detect this multigene transcript as efficiently. This possibility is supported by footprinting studies that have shown the toxT transcript attached to a tcpA-dependent message (6). A third possibility is that ToxT activates a feed-forward loop that stimulates ctxAB expression, which after initiation no longer requires ToxT for further stimulation. Possible mediators of this feed-forward loop remain to be identified.
The overall dynamics of the gene expression profiles under AKI medium conditions showed oscillation of expression across multiple virulence operons beginning at 2 h 15 min and dampening by 4 h 20 min. Genes exhibiting this behavior include toxT, the ctxAB operon, most (but not all) members of the tcpA operon, acfC, acfD, tagD, and the Ccm2-related protein gene. The oscillatory behavior of nodes in a complex system has been well documented in eukaryotic and prokaryotic genetic networks (11, 26) and reflects the dynamic balance achieved between positive- and negative-feedback loops on downstream elements after perturbation (2). In agreement with previous work on biologic networks, the timing of these oscillations is more precise than the amplitude (26, 32). It has been posited that variations in amplitude are secondary to noise generated during protein production, a property inherent in all biochemical circuits (2). Notably, the ctxA transcript showed variation in both the amplitudes and frequencies of its oscillations in AKI medium among three replicates (Fig. 2). This behavior has been associated with negative-feedback loops containing multiple regulators, termed repressilators, which have been shown to be noisy with regard to both the timing and amplitude of oscillations (12). Potential candidates for regulators are suggested through cluster analysis and are the targets of future investigation.
The CAGED algorithm clusters time series profiles together on the assumption that similarly shaped profiles are generated by genes involved in the same biological process. Though our results suggest five unique gene transcription profiles during growth in AKI medium, it is more likely that there is only one positive and one negative module. The existence of three different clusters all containing genes from the tcpA operon is likely the result of the same fundamental process being split up due to differing transcriptional dynamics and the message stability of the genes. We were unable to conclusively generate sets of genes that correspond to “early” and “late” regulators. This may be because our time intervals were too broad to capture the necessary changes in transcription at precise time intervals, or it may be that the sequence of gene activation seen in animal/human models of cholera is not fully replicated in vitro.
The primary significance of the cluster analysis is the inclusion, based on a systems biology approach to expression profiling, of potential new genes in the ToxR regulon which were not previously suggested to be related to the expression of virulence genes. The majority of these candidates, particularly the metabolic and transcription-related genes, may have similar expression profiles but not be related to virulence. An example is aldehyde dehydrogenase (aldA, VC0819), a recognized downstream target of ToxT that was included in cluster B. Although its presence in a cluster with other ToxT-regulated genes helps to validate our results, its broad role in metabolism may make unlikely the possibility that it also plays a major role in pathogenesis. Among the remaining genes, the presence of several ABC transporters, an ion transporter, and two signal transducers suggests an important role for processing of information about the environment; these represent better candidates for genes important in the pathogenic process. Their interactions with known members of the ToxR regulon, along with functional characterization of the 11 hypothetical genes in cluster A and four hypothetical genes in cluster B, are the subjects of future research.
This study is limited by the fact that it was performed in vitro. Work by Lee and colleagues has shown that the gene-regulatory relationships seen in the laboratory are not the same as those seen in natural infection, likely due to the absence of essential environmental signals from the gut (28). Although our data show clear temporal activation of the ToxR regulon genes in AKI medium, the complex sequence of gene expression described in vivo was not as clearly present, for example, in the separation in time between expression of key regulators and expression of downstream effectors. There could be two reasons for this. The first is that a temporal resolution of 25 min between time points is not sufficiently fine to detect the changes. The second explanation is that the milieu of factors during growth in AKI medium does not fully capture the intestinal environment; this is more likely the case. Despite this, in vitro study of V. cholerae gene expression clearly still has relevance to clinical disease, as many experiments cannot be performed in animal models or directly in humans and thus require an alternative, albeit imperfect, surrogate.
A systems biology approach to identifying potential pathogenesis genes using time series microarrays is based on the principle that genes that show similar dynamic expression profiles as known virulence genes are likely to be related to the pathogenetic process. The ultimate goal is to build an accurate topology of the genetic network responsible for pathogenesis, thus allowing rational therapeutic and vaccine design. One of the disadvantages of this current approach, however, is illustrated by the fact that not all genes known to be parts of the same operon were upregulated to the same extent in the time series profiles. One possible explanation for this is that the oligonucleotides on the array specific to each gene in a multigenic operon may have differing affinities for binding the multigenic transcript and therefore give differing levels of up- or downregulated expression of genes in the same operon. For example, expression of ctxB was clearly not as significantly upregulated as ctxA, and some of the genes in the tcpA operon were less upregulated than others, including tcpA itself. In interpreting a time series profile of gene expression as in this paper, it may be more prudent to look at the overall expression of genes in an operon, rather than the expression of specific individual genes within each operon.
In this study, we analyzed the global gene expression profile of V. cholerae in vitro. By analyzing time-dependent profiles, we have gained insight into the mechanisms of information processing that are common to many different types of genetic networks of prokaryotes and eukaryotes. Our data show appropriate temporal expression of major virulence genes, suggesting that growth in AKI medium, coupled with a high-resolution time series design, allows for the preliminary characterization of the dynamic interactions between known and newly identified genes that are associated with virulence. Further studies of these interactions in appropriate in vivo situations will contribute to our understanding of pathogenesis.
Acknowledgments
This work was supported by the Medical Fellows Program at the Howard Hughes Medical Institute (S.K.), grant U01 AI058935 from the National Institute of Allergy and Infectious Diseases (S.B.C.), grant K01 TW07144 from the Fogarty International Center (R.C.L.), a Physician-Scientist Early Career Award from the Howard Hughes Medical Institute (R.C.L.), and grant R01 HG003354 from the National Human Genome Research Institute (M.F.R.).
Footnotes
Published ahead of print on 2 July 2010.
REFERENCES
- 1.Abuaita, B. H., and J. H. Withey. 2009. Bicarbonate induces Vibrio cholerae virulence gene expression by enhancing ToxT activity. Infect. Immun. 77:4111-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alon, U. 2007. Network motifs in developmental, signal transduction, and neuronal networks, p. 97-124. In U. Alon (ed.), An introduction to systems biology: design principles of biological circuits. Chapman & Hall/CRC, Boca Raton, FL.
- 3.Beck, N. A., E. S. Krukonis, and V. J. DiRita. 2004. TcpH influences virulence gene expression in Vibrio cholerae by inhibiting degradation of the transcription activator TcpP. J. Bacteriol. 186:8309-8316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bina, J., J. Zhu, M. Dziejman, S. Faruque, S. Calderwood, and J. J. Mekalanos. 2003. ToxR regulon of Vibrio cholerae and its expression in vibrios shed by cholera patients. Proc. Natl. Acad. Sci. U. S. A. 100:2801-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bina, J. E., D. Provenzano, C. Wang, X. R. Bina, and J. J. Mekalanos. 2006. Characterization of the Vibrio cholerae vexAB and vexCD efflux systems. Arch. Microbiol. 186:171-181. [DOI] [PubMed] [Google Scholar]
- 6.Brown, R. C., and R. K. Taylor. 1995. Organization of tcp, acf, and toxT genes within a ToxT-dependent operon. Mol. Microbiol. 16:425-439. [DOI] [PubMed] [Google Scholar]
- 7.Champion, G. A., M. N. Neely, M. A. Brennan, and V. J. DiRita. 1997. A branch in the ToxR regulatory cascade of Vibrio cholerae revealed by characterization of toxT mutant strains. Mol. Microbiol. 23:323-331. [DOI] [PubMed] [Google Scholar]
- 8.DiRita, V. J., C. Parsot, G. Jander, and J. J. Mekalanos. 1991. Regulatory cascade controls virulence in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 88:5403-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DiRita, V. J., M. Neely, R. K. Taylor, and P. M. Bruss. 1996. Differential expression of the ToxR regulon in classical and El Tor biotypes of Vibrio cholerae is due to biotype-specific control over toxT expression. Proc. Natl. Acad. Sci. U. S. A. 93:7991-7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dziejman, M., E. Balon, D. Boyd, C. M. Fraser, J. F. Heidelberg, and J. J. Mekalanos. 2002. Comparative genomic analysis of Vibrio cholerae: genes that correlate with cholera endemic and pandemic disease. Proc. Natl. Acad. Sci. U. S. A. 93:1556-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eichenberger, P., M. Fujita, S. T. Jensen, E. M. Conlon, D. Z. Rudner, S. T. Wang, C. Ferguson, K. Haga, T. Sato, J. S. Liu, and R. Losick. 2004. The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol. 2:e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elowitz, M. B., and S. Leibler. 2000. A synthetic oscillatory network of transcriptional regulators. Nature 403:335-338. [DOI] [PubMed] [Google Scholar]
- 13.Gentleman, R. C., V. J. Carey, D. M. Bates, B. Bolstad, M. Dettling, S. Dudoit, B. Ellis, L. Gautier, Y. Ge, J. Gentry, K. Hornik, T. Hothorn, W. Huber, S. Iacus, R. Irizarry, F. Leisch, C. Li, M. Maechler, A. J. Rossini, G. Sawitzki, C. Smith, G. Smyth, L. Tierney, J. Y. H. Yang, and J. Zhang. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gill, D. M., and C. A. King. 1975. The mechanism of action of cholera toxin in pigeon erythrocyte lysates. J. Biol. Chem. 250:6424-6432. [PubMed] [Google Scholar]
- 15.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Higgins, D. E., E. Nazareno, and V. J. DiRita. 1992. The virulence gene activator ToxT from Vibrio cholerae is a member of the AraC family of transcriptional activators. J. Bacteriol. 174:6974-6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwanaga, M., and K. Yamamoto. 1985. New medium for the production of cholera toxin by Vibrio cholerae O1 biotype El Tor. J. Clin. Microbiol. 22:405-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwanaga, M., K. Yamamoto, N. Higa, Y. Ichinose, N. Nakasone, and M. Tanabe. 1986. Culture conditions for stimulating cholera toxin production by Vibrio cholerae O1 El Tor. Microbiol. Immunol. 30:1075-1083. [DOI] [PubMed] [Google Scholar]
- 19.Kaper, J. B., J. G. Morris, and M. M. Levine. 1995. Cholera. Clin. Microbiol. Rev. 8:48-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirn, T. J., M. J. Lafferty, C. M. Sando, and R. K. Taylor. 2000. Delineation of pilin domains required for bacterial association into microcolonies and intestinal colonization by Vibrio cholerae. Mol. Microbiol. 35:896-910. [DOI] [PubMed] [Google Scholar]
- 21.Reference deleted.
- 22.Kosek, M., C. Bern, and R. L. Guerrant. 2003. The global burden of diarrhoeal disease, as estimated from studies published between 1992 and 2000. Bull. World Health Organ. 81:197-204. [PMC free article] [PubMed] [Google Scholar]
- 23.Kovach, M. E., M. D. Shaffer, and K. M. Peterson. 1996. A putative integrase gene defines the distal end of a large cluster of ToxR-regulated colonization genes in Vibrio cholerae. Microbiology (Reading) 142:2165-2174. [DOI] [PubMed] [Google Scholar]
- 24.Kovacikova, G., and K. Skorupski. 1999. A Vibrio cholerae LysR homolog, AphB, cooperates with AphA at the tcpPH promoter to activate expression of the ToxR virulence cascade. J. Bacteriol. 181:4250-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krukonis, E. S., R. R. Yu, and V. J. DiRita. 2000. The Vibrio cholerae ToxR/TcpP/ToxT virulence cascade: distinct roles for two membrane-localized transcriptional activators on a single promoter. Mol. Microbiol. 38:67-84. [DOI] [PubMed] [Google Scholar]
- 26.Lahav, G., N. Rosenfeld, A. Sigal, N. Geva-Zatorsky, A. J. Levine, M. B. Elowitz, and U. Alon. 2004. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat. Genet. 36:147-150. [DOI] [PubMed] [Google Scholar]
- 27.Larocque, R. C., J. B. Harris, M. Dziejman, X. Li, A. I. Khan, A. S. G. Faruque, S. M. Faruque, G. B. Nair, E. T. Ryan, F. Qadri, J. J. Mekalanos, and S. B. Calderwood. 2005. Transcriptional profiling of Vibrio cholerae recovered directly from patient specimens during early and late stages of human infection. Infect. Immun. 73:4488-4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, S. H., D. L. Hava, M. K. Waldor, and A. Camilli. 1999. Regulation and temporal expression patterns of Vibrio cholerae virulence genes during infection. Cell 99:625-634. [DOI] [PubMed] [Google Scholar]
- 29.Matson, J. S., J. H. Withey, and V. J. DiRita. 2007. Regulatory networks controlling Vibrio cholerae virulence gene expression. Infect. Immun. 75:5542-5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Medrano, A. I., V. J. DiRita, G. Castillo, and J. Sanchez. 1999. Transient transcriptional activation of the Vibrio cholerae El Tor virulence regulator toxT in response to culture conditions. Infect. Immun. 67:2178-2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merrell, D. S., S. M. Butler, F. Qadri, N. A. Dolganov, A. Alam, M. B. Cohen, S. B. Calderwood, G. K. Schoolnik, and A. Camilli. 2002. Host-induced epidemic spread of the cholera bacterium. Nature 417:642-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mihalcescu, I., W. Hsing, and S. Leibler. 2004. Resilient circadian oscillator revealed in individual cyanobacteria. Nature 430:81-85. [DOI] [PubMed] [Google Scholar]
- 33.Murley, Y. M., J. Behari, R. Griffin, and S. B. Calderwood. 2000. Classical and El Tor biotypes of Vibrio cholerae differ in timing of transcription of tcpPH during growth in inducing conditions. Infect. Immun. 68:3010-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nielsen, A. T., N. A. Dolganov, G. Otto, M. C. Miller, C. Y. Wu, and G. K. Schoolnik. 2006. RpoS controls the Vibrio cholerae mucosal escape response. PLoS Pathog. 2:e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peterson, K. M., and J. J. Mekalanos. 1988. Characterization of the Vibrio cholerae ToxR regulon: identification of novel genes involved in intestinal colonization. Infect. Immun. 56:2822-2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramoni, M. F., P. Sebastiani, and I. S. Kohane. 2002. Cluster analysis of gene expression dynamics. Proc. Natl. Acad. Sci. U. S. A. 99:9121-9126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts, C., K. L. Anderson, E. Murphy, S. J. Projan, W. Mounts, B. Hurlburt, M. Smeltzer, R. Overbeek, T. Disz, and P. M. Dunman. 2006. Characterizing the effect of the Staphylococcus aureus virulence factor regulator, SarA, on log-phase mRNA half-lives. J. Bacteriol. 188:2593-2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schild, S., R. Tamayo, E. Nelson, F. Qadri, S. B. Calderwood, and A. Camilli. 2007. Genes induced late in infection increase fitness of Vibrio cholerae after release into the environment. Cell Host Microbe 2:264-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Slamti, L., J. Livny, and M. K. Waldor. 2007. Global gene expression and phenotypic analysis of a Vibrio cholerae rpoH deletion mutant. J. Bacteriol. 189:351-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smyth, G. K., Y. H. Yang, and T. Speed. 2003. Statistical issues in cDNA microarray data analysis. Methods Mol. Biol. 224:111-136. [DOI] [PubMed] [Google Scholar]
- 41.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910-1914. [DOI] [PubMed] [Google Scholar]
- 42.Yang, Y. H., S. Dudoit, P. Luu, D. M. Lin, V. Peng, J. Ngai, and T. P. Speed. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]