Abstract
Staphylococcus saprophyticus is a common cause of uncomplicated urinary tract infections in women. S. saprophyticus strain ATCC 15305 carries two staphylococcal cassette chromosome genetic elements, SCC15305RM and SCC15305cap. The SCC15305cap element carries 13 open reading frames (ORFs) involved in capsular polysaccharide (CP) biosynthesis, and its G+C content (26.7%) is lower than the average G+C content (33.2%) for the whole genome. S. saprophyticus strain ATCC 15305 capD, capL, and capK (capDSsp, capLSsp, and capKSsp) are homologous to genes encoding UDP-FucNAc biosynthesis, and gtaB and capISsp show homology to genes involved in UDP-glucuronic acid synthesis. S. saprophyticus ATCC 15305 CP, visualized by immunoelectron microscopy, was extracted and purified using anionic-exchange and size exclusion chromatography. Analysis of the purified CP by 1H and 13C nuclear magnetic resonance (NMR) spectroscopy and gas-liquid chromatography revealed two types of branched tetrasaccharide repeating units composed of the following:
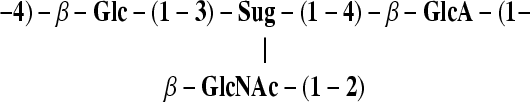 |
Sug represents two stereoisomers of 2-acetamido-2,6-dideoxy-hexos-4-ulose residues, one of which has an arabino configuration. The encapsulated ATCC 15305 strain was resistant to complement-mediated opsonophagocytic killing by human neutrophils, whereas the acapsular mutant C1 was susceptible. None of 14 clinical isolates reacted with antibodies to the ATCC 15305 CP. However, 11 of the 14 S. saprophyticus isolates were phenotypically encapsulated based on their resistance to complement-mediated opsonophagocytic killing and their failure to hemagglutinate when cultivated aerobically. Ten of the 14 clinical strains carried homologues of the conserved staphylococcal capD gene or the S. saprophyticus gtaB gene, or both. Our results suggest that some strains of S. saprophyticus are encapsulated and that more than one capsular serotype exists.
Approximately 13 million women develop urinary tract infections (UTIs) annually in the United States, with a recurrence rate between 25% and 44% (45). Staphylococcus saprophyticus is second only to Escherichia coli as a cause of uncomplicated UTI in young women (45, 46). A novobiocin-resistant member of the coagulase-negative staphylococci (60), S. saprophyticus has rarely exhibited resistance to other antibiotics (25). However, a recent report (19) indicated that methicillin-resistant S. saprophyticus isolates have emerged in Japan. The gastrointestinal tract and the vagina are the major reservoirs of S. saprophyticus (18, 30) and the likely sources of recurrent infection (20, 37, 49). Approximately 40% of patients with S. saprophyticus UTI present with acute pyelonephritis (22, 30). These patients experience symptoms more severe than those of patients infected by E. coli (24), and they are more likely to develop recurrent infections (21).
A number of potential virulence factors have been identified in S. saprophyticus. Gatermann et al. showed that in a rodent model of ascending UTI, the production of urease contributes to S. saprophyticus growth and pathogenicity in the bladder (10, 12). Other putative virulence factors of S. saprophyticus include a surface-associated lipase (11, 51, 53), the collagen binding protein SdrI (52), and a cell wall-anchored hemagglutinin protein that mediates the binding of S. saprophyticus to sheep erythrocytes, fibronectin, and human uroepithelial cells (14, 29, 34, 35). The hemagglutinin was dubbed UafA in the sequenced ATCC 15305 strain, and deletion of the uafA gene resulted in reduced S. saprophyticus hemagglutination (HA) and adherence to human bladder carcinoma cells (29). Kuroda et al. noted that UafA-mediated adherence of S. saprophyticus to the T24 cell line was inhibited by the presence of the ATCC 15305 polysaccharide capsule (29).
Staphylococcal species produce a variety of extracellular glycopolymers that contribute to the surface properties and virulence of the bacterium, such as capsular polysaccharides (CP), teichoic acids, and poly-N-acetylglucosamine (PNAG). CP production renders Staphylococcus aureus resistant to opsonophagocytic killing; alanine modifications of teichoic acids promote bacterial resistance to antimicrobial peptides (40); and PNAG is involved in biofilm formation (4). Recently, the secretion of another anionic polymer (poly-γ-dl-glutamic acid) by certain other coagulase-negative staphylococci was reported (28). Polyglutamic acid production is enhanced under high-salt conditions and may contribute to the survival of Staphylococcus epidermidis on human skin.
S. saprophyticus strain 15305 does not produce PNAG or polyglutamic acid (28, 29), but this uropathogenic species is encapsulated. CP are lacking in isolates of S. epidermidis, the most common of the coagulase-negative species, but genomic evidence indicates that Staphylococcus haemolyticus (7, 57), S. saprophyticus (29), and Staphylococcus carnosus (47) carry capsule loci with genetic similarity to the Staphylococcus aureus cap5 (cap8) gene locus. In this study, we purified and characterized the CP produced by S. saprophyticus ATCC 15305 and investigated the CP phenotype of S. saprophyticus clinical isolates.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The S. saprophyticus strains used in this study are listed in Table 1. Strain ATCC 15305 was obtained from the American Type Culture Collection (ATCC, Manassas, VA). S. saprophyticus C1, kindly provided by Makoto Kuroda, is an ATCC 15305 mutant (29) with an excised SCC15305cap region (deletion of nucleotides [nt] 50103 to 83737). Andrew Onderdonk provided clinical isolates of S. saprophyticus from patients with urinary tract infections treated at Brigham and Women's Hospital, Boston, MA. S. saprophyticus strains 7108 and 9325 were provided by Sören Gatermann, Ruhr-Universität, Bochum, Germany. Unless otherwise noted, the bacteria were cultivated aerobically for 24 h at 37°C in tryptic soy broth (TSB) or tryptic soy agar (TSA).
TABLE 1.
Presence of S. saprophyticus capsule genes in clinical isolates
| S. saprophyticus strain | Presence or absence of the following gene within SCC15305capa: |
|||
|---|---|---|---|---|
| SSP0056b | gtaB |
capD |
||
| PCR | Southern blotting | |||
| ATCC 15305 | + | + | + | + |
| C1 | − | − | − | − |
| BWH2 | − | + | + | + |
| 9777 | − | + | + | + |
| 1815 | − | + | − | + |
| 7108 | − | + | − | + |
| 9325 | − | + | − | + |
| 3201 | − | − | + | + |
| B2 | − | − | − | + |
| 1146 | − | + | − | − |
| 2262 | − | + | − | − |
| 9556 | − | + | − | − |
| BWH3 | − | − | − | − |
| 889 | − | − | − | − |
| 0937 | − | − | − | − |
| 3751 | − | − | − | − |
+, present; −, absent. The presence of the SSP0056 and gtaB genes was determined by PCR. Mutant C1 and all of the clinical isolates gave negative PCRs with primers to capABCDSsp, capEFGSsp, capHIJSsp, and capKLSsp. In addition, DNA fragments containing capEFGSsp or capKLSsp did not hybridize to DNA from any S. saprophyticus clinical isolates.
The SSP0056 gene is upstream of the capSsp locus within the SCC15305cap element.
Anti-CP antibodies and serotyping.
An antiserum to the S. saprophyticus CP was prepared by immunizing Swiss Webster mice or a New Zealand White rabbit with UV-killed cells of strain ATCC 15305 as described previously (61). A capsule-specific antiserum was obtained by absorption of immune serum with the acapsular mutant C1. The reactivities of S. saprophyticus clinical isolates with the ATCC 15305 CP-specific antiserum were determined by a colony immunoblotting method (31), and their reactivities with an S. aureus CP-specific antiserum were determined by double immunodiffusion of capsular extracts.
Immunogold electron microscopy.
Immunogold labeling of the CP produced by S. saprophyticus cells was performed on Formvar carbon-coated copper grids. The samples were blocked for 10 min in 0.5% fish skin gelatin in phosphate-buffered saline (PBS) with 0.1% Tween 20. The samples were incubated for 30 min at ambient temperature with the CP-specific antiserum. The grids were washed four times in PBS and were incubated with protein A-gold particles (diameter, 20 nm). After four washes in deionized water, the samples were air dried and examined with a JEOL 1200EX transmission electron microscope (TEM).
CP purification.
CP was purified by methods similar to those we have described previously (7, 59). Briefly, S. saprophyticus ATCC 15305 was cultivated on TSA medium overlaid with a dialysis membrane with a molecular size cutoff of 12,000 to 14,000 Da. Autoclaved aqueous bacterial extracts were treated with DNase and RNase for 6 h at 37°C, followed by overnight treatment with pronase. The dialyzed material was passed over a column of DEAE-Sephacel (Amersham Biosciences, Uppsala, Sweden) equilibrated with 0.05 M sodium acetate-0.05 M NaCl (pH 6.0). Fractions that eluted in a 2-liter gradient (0.05 M sodium acetate with 0.05 to 0.5 M NaCl) were analyzed for absorbance at 206 nm and for serologic activity with antibodies to S. saprophyticus CP. Serologically active fractions were pooled, dialyzed, and lyophilized. The CP was subjected to chromatography on a Sephacryl S-300 (Amersham Biosciences) column equilibrated in 0.2 M NaCl, and the fractions were monitored as described above. Serologically active fractions were pooled, dialyzed, and lyophilized. The purity of the polysaccharide was assessed by UV (260 and 280 nm) absorption spectra, chemical assays for protein and phosphate, and nuclear magnetic resonance (NMR) spectroscopy.
Structural characterization of the S. saprophyticus CP.
1H and 13C NMR spectra were recorded using a 500-MHz Varian Inova spectrometer in D2O solutions at 60°C with an acetone standard (2.225 ppm for 1H and 31.5 ppm for 13C) using standard pulse sequences for correlation spectroscopy (COSY), total correlation spectroscopy (TOCSY) (mixing time, 120 ms), nuclear Overhauser effect spectroscopy (NOESY) (mixing time, 200 ms), heteronuclear single-quantum correlation (HSQC), and heteronuclear multiple-bond correlation (HMBC) (optimized for a 5-Hz coupling constant). For monosaccharide analysis, the CP (0.5 mg) was hydrolyzed (0.2 ml of 3 M TFA, 120°C, 2 h) and evaporated to dryness under a stream of air. The residue was dissolved in water (0.5 ml), reduced with NaBH4 (∼5 mg, 1 h), neutralized with acetic acid (0.3 ml), and dried, and methanol (1 ml) was added. The mixture was dried twice with the addition of methanol, and the residue was acetylated with acetic anhydride (0.5 ml, 100°C, 30 min), dried, and analyzed by gas-liquid chromatography (GLC) on an HP1 capillary column (30 m by 0.25 mm) with a flame ionization detector (Agilent 6850 chromatograph) in a temperature gradient of 170°C (4 min) to 260°C at 4°C/min.
DNA and RNA preparations.
S. saprophyticus cells were lysed with lysozyme and lysostaphin (1 mg/ml each) for 1 h at 37°C, and genomic DNA was isolated with the Wizard genomic purification kit (Promega, Madison, WI). PCRs were performed with Taq DNA polymerase (Invitrogen, Carlsbad, CA) and an annealing temperature of 50°C. The S. saprophyticus recA gene served as a positive PCR control. Primer sequences and the lengths of the amplicons are given in Table S1 in the supplemental material. Total RNA was prepared from S. saprophyticus cells lysed with 0.5 ml zirconia-silicon beads (Fisher Scientific, Pittsburgh, PA) in a dental amalgamator (62), and RNA was purified with the RNeasy Mini kit (Qiagen, Valencia, CA). RNA was treated with RNase OUT (Invitrogen) and DNase I (Invitrogen) before amplification with the Access RT-PCR system kit (Promega). A control sample without reverse transcriptase was included for each RNA sample to confirm the absence of contaminating DNA. For Southern blotting, chromosomal DNA was digested with HindIII and separated on a 0.8% agarose gel. DNA was blotted on GeneScreenPlus (Perkin-Elmer, Boston, MA) membranes and hybridized with capDSsp, capEFGSsp, or capIJKLSsp. The probes were labeled with the ECL direct nucleic acid labeling and detection system (Amersham) according to the manufacturer's recommendations. After overnight hybridization with the probe at 42°C, the membranes were washed under low-stringency conditions: two washes for 5 min each at 42°C in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). These washes were followed by two additional washes at 42°C in 6 M urea-0.4% sodium dodecyl sulfate-0.5× SSC and two washes at room temperature in 2× SSC. The detection reagents included with the kit were used as directed, and positive signals were detected by exposure to radiographic film.
In vitro opsonophagocytic killing assay.
The opsonophagocytic killing assay is a modification of the method of Xu et al. (61). S. saprophyticus strains, cultivated on TSA plates for 24 h at 37°C, were suspended in a minimal essential medium (MEM; Invitrogen) containing 1% bovine serum albumin (BSA; Sigma, St. Louis, MO). Human neutrophils were isolated from the blood of healthy male donors with Mono-Poly resolving medium (MP Biomedicals, Solon, OH) and were washed once with MEM-BSA. The opsonophagocytic assay was performed in polypropylene tubes containing 2.5 × 106 neutrophils, 5 × 105 CFU of S. saprophyticus, 0.25% heat-inactivated rabbit antiserum against S. saprophyticus CP, and 1% guinea pig serum (as a complement source) in a total volume of 500 μl MEM-BSA. Control samples included (i) S. saprophyticus and neutrophils with either complement or antiserum and (ii) S. saprophyticus incubated with complement and antibodies but no neutrophils. The tubes were rotated end over end (12 rpm) for 2 h at 37°C. Bacterial killing was estimated by diluting the samples in sterile water and plating the samples in duplicate on TSA plates at time zero and after 2 h. The percentage of reduction in CFU per milliliter for each sample at 2 h was compared to the input inoculum at time zero. Assays with the clinical isolates were performed in identical fashion except that the samples included S. saprophyticus, neutrophils, and complement but no antiserum.
HA assay.
S. saprophyticus strains were cultivated either aerobically for 24 h in TSB at 37°C on a rotator or anaerobically for 48 h under static conditions in a Coy anaerobic chamber containing 10% CO2, 80% N2, and 10% H2. Bacterial cells were harvested, washed in PBS, and suspended to an optical density at 600 nm (OD600) of 1.0. The CFU per milliliter was determined on each suspension, and serial 2-fold dilutions (50 μl per well) of the bacterial suspensions were prepared in 96-well U-bottom microtiter plates. An equal volume of 0.4% sheep erythrocytes (Hemostat Laboratories, Dixon, CA) was added to each well, and the microtiter plate was incubated at ambient temperature for 4 h. The titer (the highest dilution of the bacterial suspension that resulted in hemagglutination [HA]) was determined for the anaerobic and aerobic cultures, and the HA index for each S. saprophyticus strain was calculated by dividing the HA titer of the anaerobic culture by the HA titer of the aerobic culture. Clinical isolates with an HA index of ≥8 were scored as positive for CP production, and isolates with an HA index of ≤2 were scored as negative.
RESULTS
The S. saprophyticus capsule genes.
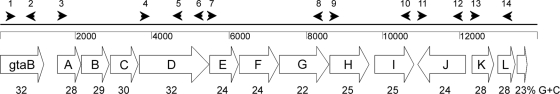
The genome of S. saprophyticus ATCC 15305 was sequenced by Kuroda et al. (29), and a capsule operon similar to the S. aureus cap1 gene locus (32) was identified within a staphylococcal cassette chromosome (SCC) genetic element named SCC15305cap. The capsule locus comprises 13 open reading frames (ORFs) (SSP0059 to SSP0072) spanning nucleotides 69784 to 83251 on the S. saprophyticus chromosome. The capASsp through capDSsp genes are homologous to S. aureus cap1A through cap1D, with DNA identities between 69% and 74% and amino acid similarities between 73% and 85% (Table 2). The putative gene products of capDSsp and capLSsp show homology to bacterial proteins involved in UDP-d-FucNAc biosynthesis, whereas CapKSsp is a putative UDP-FucNAc transferase. The gtaB gene upstream of the capSsp locus (encoding a putative UTP-glucose-1-phosphate uridyltransferase) and capISsp (encoding a putative UDP-glucose-6-dehydrogenase) are homologous to bacterial genes involved in UDP-glucuronic acid synthesis (6, 56). The putative gene products of capESsp, capFSsp, capGSsp, capHSsp, and capJSsp share only limited homology with proteins in the databases and include putative glycosyltransferases, as well as proteins involved in CP transport and polymerization. Of note, the orientation of capJSsp is opposite to that of the other capSsp genes (Fig. 1), and the last two ORFs appear to be truncated versions of the same gene (capLSsp). The overall G+C content in the coding regions of the S. saprophyticus cap region is 26.7% (Fig. 1), which is lower than that of the total genome (33.2%), suggesting that these genes may have been acquired by horizontal gene transfer. A putative promoter and a Rho-independent transcriptional terminator were located upstream of capASsp and downstream of capLSsp, respectively.
TABLE 2.
Putative functions of S. saprophyticus cap genes based on sequence homologies
| Genea | % Amino acid identity/similarity | Organism and gene | Putative function |
|---|---|---|---|
| gtaB | 76/89 | S. aureus gtaB | UTP-glucose-1-phosphate uridyltransferase |
| capASsp | 62/81 | S. aureus cap1A | Chain length determination |
| capBSsp | 65/78 | S. aureus cap1B | Chain length determination |
| capCSsp | 54/73 | S. aureus cap1C | Tyrosine protein phosphatase |
| capDSsp | 73/85 | S. aureus cap1D | UDP-GlcNAc 4,6-dehydratase |
| capESsp | 42/65 | Bacillus thuringiensis RBTH_06178 | Glycosyl transferase |
| capFSsp | 32/51 | Desulfuromonas acetoxidans Dace_1590 | Glycosyl transferase |
| capGSsp | 29/51 | S. aureus cap5K | Flippase |
| capHSsp | 27/48 | Hahella chejuensis HCH_02393 | Glycosyl transferase |
| capISsp | 60/77 | Escherichia coli ugd | UDP-glucose-6-dehydrogenase |
| capJSsp | 22/44 | Bacillus cereus wzy | Polysaccharide polymerase |
| capKSsp | 78/85 | S. aureus cap5M | UDP-FucNAc transferase |
| capLSsp | 50/68 | S. aureus cap5N | UDP-2-acetamido-2,6-dideoxy-d-xylo-4-hexulose reductase |
| capLSsp | 44/68 | S. aureus cap5N |
The last two genes in the capsule operon are truncated versions of the same gene (capLSsp).
FIG. 1.
S. saprophyticus capsule gene cluster. Distinguishing features include the gtaB gene upstream of the cap locus, the reverse orientation of the capSsp J gene, and the truncated capSsp L gene. The capABCD genes are conserved among S. aureus, S. haemolyticus, and S. saprophyticus. The G+C content for individual genes, expressed as a percentage, is shown below each ORF. The arrowheads indicate the relative positions of the primers described in Table S1 in the supplemental material.
To determine whether the capsule genes were expressed by S. saprophyticus ATCC 15305, we performed reverse transcription-PCR (RT-PCR) experiments with primers that yielded products ranging from 0.875 kb to 3.51 kb and spanned the length of the capsule operon from capASsp through capLSsp (Fig. 1). Amplicons corresponding to capABCDSsp, capEFGSsp, capHISsp, capJSsp, and capKLSsp were obtained (data not shown), indicating that the capsule genes were transcribed by the sequenced S. saprophyticus strain.
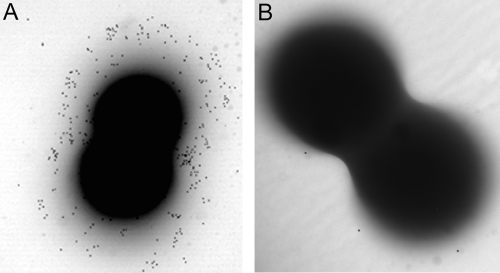
We used immunogold labeling experiments with a capsule-specific mouse antiserum to verify CP production by S. saprophyticus ATCC 15305. Electron micrographs of strain ATCC 15305 showed cells decorated with gold particles, indicating CP production. In contrast, mutant C1, a derivative of ATCC 15305 with a deletion of the capSsp locus, showed minimal gold labeling (Fig. 2). CP was not visualized on strain ATCC 15305 incubated with preimmune serum (not shown). We compared CP production by S. saprophyticus ATCC 15305 on different growth media. CP, visualized by electron microscopy and immunogold labeling, was abundant when strain ATCC 15305 was grown aerobically in tryptic soy medium with or without 1% glucose (Fig. 2 and data not shown). Little CP was observed on bacteria harvested from logarithmic-phase cultures or from broth cultures incubated under anaerobic conditions (not shown). In contrast to the observations with S. aureus (42, 43), cultivation on Columbia medium supplemented with 2% NaCl was suboptimal for CP production by S. saprophyticus (not shown). The addition of as much as 0.4 M urea to the culture medium did not markedly influence CP production (not shown).
FIG. 2.
Immunogold labeling of the S. saprophyticus capsule. (A) Strain ATCC 15305; (B) acapsular mutant C1. Both strains were harvested from tryptic soy agar plates and were then incubated with an antiserum specific for the S. saprophyticus capsule in order to visualize the capsule. A protein A-gold conjugate was used as the labeling reagent.
Purification of S. saprophyticus CP.
Capsules were extracted and purified from strain ATCC 15305 cells. A crude CP preparation was partially separated from contaminating teichoic acids by ion-exchange chromatography. The CP was eluted from a DEAE column with 0.18 to 0.30 M NaCl and was further purified by size exclusion chromatography. The purified CP (∼30 mg) eluted near the void volume of an S-300 Sephacryl column with a distribution coefficient (Kav) of 0.024. The capsule preparations used for biochemical characterization contained <0.2% protein, <1% nucleic acid, and <0.16% phosphorus.
Structural characterization of S. saprophyticus CP.
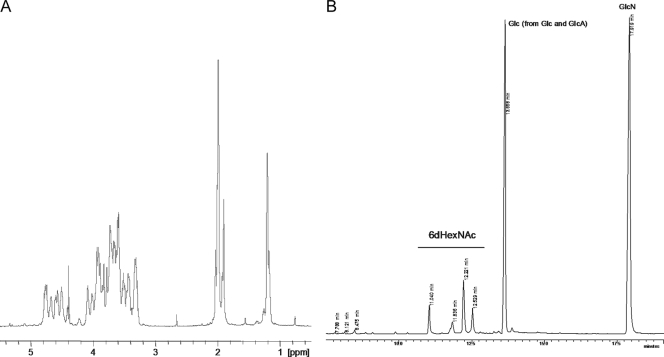
The NMR spectra of the ATCC 15305 CP were complex and included a number of signals of different intensities (Fig. 3A). Monosaccharide analysis showed the presence of GlcA, Glc, and GlcN, and the visible amount of GlcN was several times more than that of Glc. Reduction of the CP with NaBH4 and subsequent monosaccharide analysis revealed four isomers of 2-acetamido-2,6-dideoxy alditols (Fig. 3B). Two of these isomers were identified as derivatives of QuiN and FucN, expected to form from residue D (Fig. 4). Two others were not identified because of the absence of appropriate standards. The high-orifice-voltage mass spectrum of the S. saprophyticus CP (see Fig. S1 in the supplemental material) contained peaks that were consistent overall with fragments comprising the monosaccharides noted above.
FIG. 3.
Structural analysis of the S. saprophyticus CP. (A) 1H NMR spectrum (60°C) of the purified CP. (B) GC analysis of alditol acetates obtained from the reduced CP. Four isomers of 2-acetamido-2,6-dideoxy hexose are apparent.
FIG. 4.
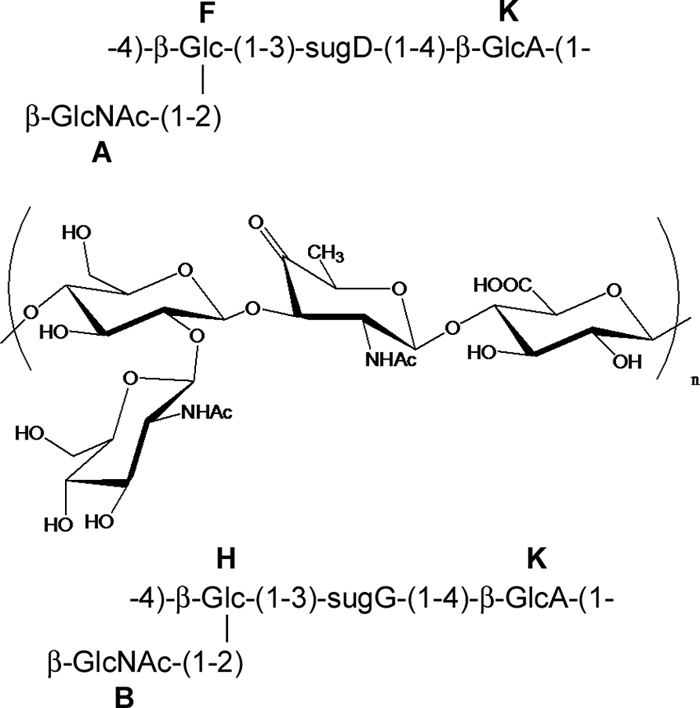
Proposed structure of the S. saprophyticus ATCC 15305 CP. The configuration of sugG is unknown. There is one more minor unidentified 6-deoxy sugar. 1H and 13C NMR chemical shifts for sugar residues A, B, D, F, G, H, and K are provided in Table S2 in the supplemental material.
Most of the signals in 2-dimensional (2D) NMR spectra were assigned. Glucose, glucosamine, and glucuronic acid residues were identified, as well as two types of 2-acetamido-2,6-dideoxy-4-keto-hexoses. The configuration of one of these (residue D) is shown in Fig. 4. Residue D had large (8- to 10-Hz) vicinal proton-coupling constants between H-1, H-2, and H-3, and assuming 4C1 conformation, it had a β-arabino configuration, which agrees with the detection of FucN and QuiN in the hydrolysate of the NaBH4-reduced polymer. The other 2-acetamido-2,6-dideoxy-4-keto-hexose (residue G) had overlapping H-2 and H-3 signals (see Fig. S2 and Table S2 in the supplemental material); thus, no configuration could be assigned on the basis of NMR data. The polymer contained two types of repeating units, differing in the nature of the 4-keto sugar. On the basis of signal positions, coupling constants, NOE correlation (see Fig. S2 in the supplemental material), and HMBC, the structures presented in Fig. 4 were deduced. There were some minor unidentified signals. Interestingly, the H-6 of GlcNAc showed large differences in chemical shifts, and the C-6 signal was slightly shifted downfield, which may indicate substitution by a keto sugar, but no substituent was identified. The spectra of the NaBH4-reduced polymer were even harder to interpret than that of the intact polymer and provided no new information.
Attempts to perform methylation analysis of the polymer were not successful. Mild methanolysis (0.5 M HCl in methanol, 50°C, 3 h) was used in order to prepare oligosaccharides, but it gave methyl β-GlcNAc and no other identifiable product.
Influence of the CP on in vitro opsonophagocytic killing of S. saprophyticus.
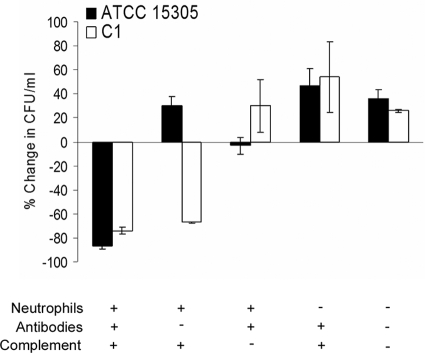
To determine whether there were differences in resistance to phagocytosis attributable to CP production by S. saprophyticus, we measured the killing of strains ATCC 15305 and the acapsular mutant C1 by human neutrophils. As shown in Fig. 5, ≥75% of wild-type and mutant bacterial cells were killed by neutrophils in the presence of S. saprophyticus CP-specific antibodies and complement. Similarly, 68% of the mutant C1 (lacking CP) inoculum was effectively opsonized for phagocytic killing by complement alone. In contrast, ATCC 15305 cells increased in number when incubated with neutrophils and complement but no CP-specific antibodies. In serum with no complement activity, rabbit CP-specific antibodies were not opsonic for either strain. No killing was observed in control samples lacking neutrophils (Fig. 5) or in samples with neutrophils but no opsonin (not shown). These results confirm the predicted antiphagocytic activity of the capsule produced by S. saprophyticus.
FIG. 5.
Assay of opsonophagocytic killing of S. saprophyticus ATCC 15305 and acapsular mutant C1 by human neutrophils. The percentage of reduction from the CFU per milliliter in each sample at time zero was calculated after 2 h at 37°C. The results shown are the means ± standard errors of the means for 14 independent experiments.
CP genes of clinical isolates of S. saprophyticus.
To investigate CP production by clinical isolates of S. saprophyticus, we tested their reactivities with antibodies specific to the CP produced by S. saprophyticus ATCC 15305. In contrast to strain 15305, neither the C1 mutant (Fig. 6) nor any of the 14 clinical isolates (not shown) reacted with the anti-CP antiserum by colony immunoblotting. Likewise, none of the S. saprophyticus strains reacted with antibodies to S. aureus capsule type 1, 5, or 8 (not shown).
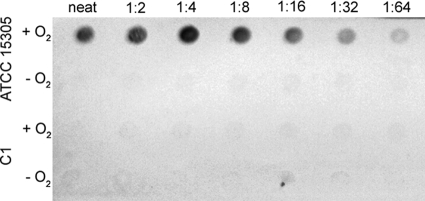
FIG. 6.
CP production by S. saprophyticus ATCC 15305 and mutant C1 cultivated under aerobic (+O2) or anaerobic (−O2) conditions. The bacteria were serially diluted in PBS, starting with an undiluted (neat) sample equivalent to 1.1 × 108 CFU (aerobic) or 2.6 ×107 CFU (anaerobic). The filters were incubated in a CP-specific antiserum, and colorimetric detection was achieved with a protein A-horseradish peroxidase conjugate and an appropriate substrate.
To determine whether the clinical isolates carried capsule genes similar to those expressed by ATCC 15305, we designed primers (see Table S1 in the supplemental material) to amplify specific regions of the S. saprophyticus capsule locus: capABCDSsp, capEFGSsp, capHIJSsp, and capKLSsp. Genomic DNA from S. saprophyticus ATCC 15305 yielded amplicons of the expected sizes with each primer pair, whereas the genomic DNA preparations from deletion mutant C1 and the clinical isolates were negative by PCR. Likewise, none of the isolates gave a positive reaction with primers to SSP0056 (an ORF encoding a hypothetical protein), which is upstream of the capsule locus but still within the SCC15305cap element (Table 1). All of the strains yielded amplicons with primers for S. saprophyticus recA, used as a positive control. These results suggest that none of the clinical isolates carry the SCC15305cap element.
To determine whether the clinical strains might carry heterologous capsule genes, we performed Southern blot hybridization analysis using a DNA fragment comprising the central region of the capD gene, which is conserved among the S. aureus cap1, cap5, and cap8 loci, as well as within the S. haemolyticus JCSC1435 cap (capSh) (57) and S. saprophyticus ATCC 15305 cap (capSsp) (29) loci. Chromosomal digests from 7 (7108, 9325, 3201, B2, BWH2, 1815, and 9777) of the 14 clinical isolates hybridized with the capD probe, and 3 of these yielded PCR products with primers (capD-f and capD-r2 [see Table S1 in the supplemental material]) designed to amplify the conserved capD region. Probes made to several of the downstream genes (capEFGSsp and capKLSsp) hybridized only to DNA digests from ATCC 15305, not to those from mutant C1 or any S. saprophyticus clinical isolates. Eight of the clinical isolates yielded PCR products with primers to gtaB, which is upstream of capASsp and is critical for the synthesis of the GlcA component of bacterial CP (2), and 5 of these 8 isolates also hybridized to the capD probe (Table 1). Five strains carried either gtaB or capD, and four strains carried neither. These data suggest that certain clinical isolates of S. saprophyticus may carry capsule genes distinct from those of the sequenced strain ATCC 15305.
Functional assays of S. saprophyticus clinical isolates.
S. saprophyticus HA has been shown to correlate with its in vitro adherence to human ureteral epithelial cells (9, 13). S. saprophyticus ATCC 15305 expresses a protein (named UafA to indicate its role as a uroadherence factor) that correlates with HA activity and mediates the adherence of S. saprophyticus to human T24 bladder carcinoma cells in vitro (29). UafA is encoded by a large ORF (SSP0135; 2,316 amino acids) that represents the only cell wall-anchored protein in ATCC 15305 (29). The acapsular mutant C1 showed greater HA activity and adherence to T24 cells than ATCC 15305, and Kuroda et al. postulated that CP might envelop the bacterial surface, resulting in the inhibition of UafA-mediated bacterial adherence (29).
Gatermann and Meyer observed that S. saprophyticus strain 9325 was phenotypically HA positive when grown under anaerobic conditions but HA negative after growth under aerobic conditions (16). Similarly, we observed that ATCC 15305 cells were HA positive when grown under anaerobic, but not under aerobic, conditions. Mutant C1 showed a positive HA phenotype when it was cultivated aerobically or anaerobically (not shown). We verified by colony immunoblotting (Fig. 6) and transmission electron microscopy (not shown) that S. saprophyticus ATCC 15305 produces CP under aerobic, but not anaerobic, culture conditions, and this finding is consistent with the minimal HA activity associated with aerobic S. saprophyticus cultures. We considered that HA and complement-mediated opsonophagocytic killing assays might be useful functional assays for screening clinical isolates for CP production.
HA assays were performed on serial 2-fold dilutions of suspensions of each S. saprophyticus clinical isolate harvested from TSB cultures cultivated either aerobically or anaerobically. The HA results were expressed as the minimum titer of each suspension that resulted in HA, and the HA index was calculated as the HA titer of the anaerobic culture divided by the HA titer of the aerobic culture. All of the clinical isolates were HA positive when cultivated anaerobically except for isolates 1815 and 2262. Unlike the other clinical isolates, these two strains did not generate PCR amplicons with uafA gene primers (see Table S1 in the supplemental material). As noted above, aerobic cultures of S. saprophyticus ATCC 15305 did not show HA, resulting in an average HA index of 32, whereas the acapsular mutant C1 had an average HA index of 2 (similar titers for anaerobic and aerobic cultures). The HA indices of most of the clinical isolates were high (≥8), suggesting that CP produced by these S. saprophyticus strains under aerobic conditions interfered with HA. The HA indices of S. saprophyticus strains 3751 and 7108 were both ≤2, suggesting that these clinical isolates were CP negative.
Because encapsulated staphylococci are resistant to complement-mediated opsonophagocytic killing by human neutrophils (7, 38, 41, 58, 61), we used this assay to further assess the capsular phenotypes of clinical isolates of S. saprophyticus cultivated under aerobic conditions. As a control, we measured the susceptibility to complement-mediated opsonophagocytic killing of ATCC 15305 cultivated aerobically or anaerobically in TSB. ATCC 15305 cells cultivated aerobically were resistant to complement-mediated opsonophagocytic killing (29% ± 7% increase in CFU per milliliter after 2 h). In contrast, anaerobic cultures of ATCC 15305 cells were susceptible to complement-mediated opsonophagocytic killing by human neutrophils (30% ± 11% reduction in the inoculum after 2 h). This finding is consistent with serologic evidence of poor CP expression under anaerobic conditions (Fig. 6).
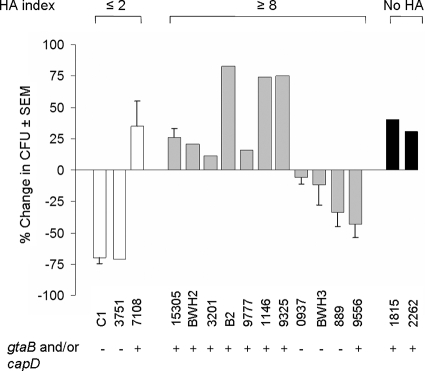
The majority of clinical isolates with HA indices of ≥8 resisted killing by neutrophils when opsonized with complement (Fig. 7), suggesting that CP (or other antiphagocytic factors) was produced by these strains. In contrast, 67% and 71% of the inocula of the acapsular mutant C1 and clinical isolate 3751, respectively, were killed by neutrophils following opsonization by complement (without CP antibodies). A few of the clinical isolates did not fit our paradigm. Strains 889 and 9556 showed modest killing by neutrophils (34% ± 11% and 43% ± 11% of the inocula were killed, respectively), despite HA indices of ≥8. Likewise, strain 7108 was not killed by complement-mediated opsonophagocytosis, suggesting that it produced CP; however, strain 7108 exhibited HA under both anaerobic and aerobic conditions (HA index, ≤2). Besides UafA, S. saprophyticus 7108 produces several additional surface-associated proteins: Ssp (a lipase) (53), SdrI (a collagen binding protein) (52), and Aas (a multifunctional protein with autolysin activity, adherence to fibronectin and uroepithelial cells, and HA activity) (14, 15, 17, 35). The presence of a second protein with HA activity (lacking in ATCC 15305) may explain the discrepancies observed between the phagocytic assay and HA for strain 7108. Of note, most of the clinical isolates with HA indices of ≥8 that were resistant to complement-mediated opsonophagocytic killing were positive for the CP-associated genes gtaB and capD (Fig. 7).
FIG. 7.
Phenotypic analysis of S. saprophyticus clinical isolates by HA index and complement-mediated opsonophagocytic killing assays. HA indices were calculated by dividing the HA titer of the anaerobic culture by the HA titer of the aerobic culture. If the HA index was ≤2, the strain was scored as CP negative, and if the HA index was ≥8, the strain was scored as CP positive. Cultures of strains 1815 and 2262 did not hemagglutinate under either aerobic or anaerobic growth conditions, and both strains were uafA negative by PCR.
DISCUSSION
S. saprophyticus is second only to E. coli as the most frequent cause of uncomplicated urinary tract infection in women (25, 45, 46), and most infections occur in young, sexually active women. Unique features of S. saprophyticus include its ability to adhere to uroepithelial cells and its high urease activity compared with that of other staphylococcal species (29). CP production is an important determinant of microbial virulence and is a trait common to many invasive bacterial pathogens. Encapsulated pathogens resist uptake and killing by professional phagocytes in the absence of specific anticapsular antibodies. CP production by S. aureus has been extensively characterized, but CP production by coagulase-negative staphylococci was not reported until recently. Our group showed that the sequenced S. haemolyticus strain produces a CP that is serologically cross-reactive with CP produced by several clinical isolates (7). Likewise, genes with homology to those involved in capsule biosynthesis have been identified in the genome of a recently sequenced Staphylococcus carnosus strain (47). Here we report that S. saprophyticus ATCC 15305 produces a CP that protects the organism from uptake and killing by neutrophils. We isolated and purified the CP, and we provide a preliminary biochemical structure. We investigated the presence of CP genes among S. saprophyticus clinical isolates from patients with urinary tract infections, and we provide the initial evidence that some of the isolates are encapsulated.
When the S. saprophyticus genome sequence was published, two SCCs were identified: SCC15305RM and SCC15305cap (29). Similarly to the location of SCCmec elements in S. aureus (5, 39), SCC15305RM was inserted within orfX. SCC15305cap is integrated into the potential attachment site on SCC15305RM as a nested structure, and it carries an intact and functional CP gene locus, reminiscent of the cap1 locus identified within the SCCcap1 element of the serotype 1 S. aureus strain M (32). The S. saprophyticus composite island encodes cassette chromosome recombinases (ccrA, ccrB, and ccrC) and restriction-modification system enzymes (hsdS, hsdM, and hsdR) but no antibiotic resistance genes. The existence of SCC elements lacking the mec determinant has been reported for other coagulase-negative staphylococcal species (26, 36).
The S. saprophyticus capsule locus comprises 13 ORFs, although the last two (capLSsp) appear to be truncated versions of the same gene (Fig. 1). Based on their putative functions and homologies to other genes in the databases, we predicted correctly that the S. saprophyticus CP would contain glucuronic acid (gtaB and capISsp) and a 2,6-dideoxy sugar (capDSsp). In fact, the CP polymer was quite complex and contained two types of repeating units differing in the nature of the 2,6-dideoxy-4-keto sugar. Two-dimensional NMR spectra could resolve one of the keto sugars (residue D), but the other keto sugar (residue G) had overlapping H-2 and H-3 signals, and thus, assignment of its precise configuration was impossible. NMR analysis of the reduced polymer and methylation analyses failed to provide meaningful data to enable elucidation of the final structure. We postulate that the unusual keto sugars that characterize the S. saprophyticus CP structure result from the truncated and presumably dysfunctional capLSsp gene, encoding a putative reductase enzyme. Similarly, a symbiotic mutant of Rhizobium etli carrying a Tn5 insertion in a putative reductase gene expressed a 2-acetamido-2,6-dideoxy-4-keto-hexose rather than N-acetylquinovosamine in the outer core region of its lipopolysaccharide (8). Other examples of bacterial polysaccharides containing a 4-keto hexose (2-acetamido-2,6-dideoxy-d-xylo-hexos-4-ulose) have been reported (8, 23, 27, 33, 50, 55).
None of 14 S. saprophyticus clinical isolates reacted with antisera specific for the ATCC 15305 CP or yielded amplicons from primers designed to amplify the ATCC 15305 capsule genes. However, 7 of the 14 isolates did react by Southern blotting with a gene probe made to a region of capD that is conserved within encapsulated strains of S. aureus, S. haemolyticus, and S. saprophyticus. This result suggested that other strains of S. saprophyticus might produce a CP unrelated to that of strain ATCC 15305.
Encapsulated S. saprophyticus strains cultivated aerobically do not hemagglutinate, presumably because the CP masks the UafA protein, which mediates HA. The same strains cultivated anaerobically do hemagglutinate, since CP is not produced under these conditions. Acapsular strains hemagglutinate when grown under aerobic or anaerobic conditions. This assay provided us with a simple screen for S. saprophyticus strains that synthesize a CP that does not cross-react serologically or share genetic homology with the ATCC 15305 capsule locus. We also took advantage of the fact that encapsulated staphylococci resist complement-mediated uptake and killing by neutrophils. Characterization of 14 clinical isolates of S. saprophyticus revealed that 11 are likely encapsulated, since they were HA negative when grown aerobically (but not anaerobically), and they resisted complement-mediated opsonophagocytic killing by neutrophils.
CP expression interferes with the binding of bacteria to neutrophils, but also to uroepithelial cells, as demonstrated by Kuroda et al. (29). Nonetheless, there is a precedent for CP enhancing the virulence of uropathogenic bacteria. Intact K2 and K15 capsule loci of E. coli have been shown to be critical for virulence in a murine model of ascending UTI (3, 54). Likewise, Bahrani-Mougeot et al. (1) used signature-tagged mutagenesis to identify genes that are essential for the survival of uropathogenic E. coli within the murine urinary tract. Among 19 survival-defective mutants, 3 had insertions in a type II capsule biosynthesis locus on a pathogenicity island. The three mutants were impaired in capsule production in vivo and were avirulent in a mouse model of UTI. Not all E. coli capsule types promote urovirulence, however, since Russo et al. (48) found no difference between the abilities of a K54 blood isolate and an isogenic acapsular mutant to cause UTI in a mouse model.
We hypothesize that the S. saprophyticus CP plays a unique role in survival within the urinary tract and that CP production, as well as the production of urease and specific adhesins, enhances the urovirulence of the organism in vivo. Our immunoelectron microscopy studies indicated that, like the S. aureus and S. haemolyticus CP, S. saprophyticus produced little CP during the exponential phase of bacterial growth. Under these conditions, the bacterium would be maximally adherent but also susceptible to killing by neutrophils. Poutrel et al. (44) reported that CP production by individual S. aureus isolates varied greatly and that the number of CP-positive bacteria ranged from 22% to 95% of the population. Thus, in any given population, staphylococci lacking a capsule could adhere to (and possibly invade) mammalian cells, whereas bacterial cells expressing CP would not adhere but would resist uptake and killing by phagocytes recruited to the site of infection. It is likely that for coagulase-negative species, such as S. saprophyticus, the situation is similar, i.e., at any given time the bacterial population would include both encapsulated and unencapsulated cells. The proportion of each would differ according to the microenvironment and the bacterial growth phase, and selection for the fittest bacterial cells would occur in vivo. Further evaluation of the role of CP expression in S. saprophyticus virulence, using an animal model of UTI, is warranted.
Supplementary Material
Acknowledgments
This work was supported by Public Health Service grant AI29040 (to J.C.L.). S.P. was supported by NIH Infectious Disease Training Grant T32 AI07061.
Makoto Kuroda, Sören Gatermann, and Andy Onderdonk kindly provided isolates of S. saprophyticus. We thank Kevin Liu for technical assistance.
Footnotes
Published ahead of print on 16 July 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bahrani-Mougeot, F. K., E. L. Buckles, C. V. Lockatell, J. R. Hebel, D. E. Johnson, C. M. Tang, and M. S. Donnenberg. 2002. Type 1 fimbriae and extracellular polysaccharides are preeminent uropathogenic Escherichia coli virulence determinants in the murine urinary tract. Mol. Microbiol. 45:1079-1093. [DOI] [PubMed] [Google Scholar]
- 2.Bonofiglio, L., E. Garcia, and M. Mollerach. 2005. Biochemical characterization of the pneumococcal glucose 1-phosphate uridylyltransferase (GalU) essential for capsule biosynthesis. Curr. Microbiol. 51:217-221. [DOI] [PubMed] [Google Scholar]
- 3.Buckles, E. L., X. Wang, M. C. Lane, C. V. Lockatell, D. E. Johnson, D. A. Rasko, H. L. Mobley, and M. S. Donnenberg. 2009. Role of the K2 capsule in Escherichia coli urinary tract infection and serum resistance. J. Infect. Dis. 199:1689-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cramton, S. E., C. Gerke, N. F. Schnell, W. W. Nichols, and F. Gotz. 1999. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 67:5427-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deurenberg, R. H., C. Vink, S. Kalenic, A. W. Friedrich, C. A. Bruggeman, and E. E. Stobberingh. 2007. The molecular evolution of methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 13:222-235. [DOI] [PubMed] [Google Scholar]
- 6.Drummelsmith, J., P. A. Amor, and C. Whitfield. 1997. Polymorphism, duplication, and IS1-mediated rearrangement in the chromosomal his-rfb-gnd region of Escherichia coli strains with group IA and capsular K antigens. J. Bacteriol. 179:3232-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flahaut, S., E. Vinogradov, K. A. Kelley, S. Brennan, K. Hiramatsu, and J. C. Lee. 2008. Structural and biological characterization of a capsular polysaccharide produced by Staphylococcus haemolyticus. J. Bacteriol. 190:1649-1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forsberg, L. S., K. D. Noel, J. Box, and R. W. Carlson. 2003. Genetic locus and structural characterization of the biochemical defect in the O-antigenic polysaccharide of the symbiotically deficient Rhizobium etli mutant, CE166. Replacement of N-acetylquinovosamine with its hexosyl-4-ulose precursor. J. Biol. Chem. 278:51347-51359. [DOI] [PubMed] [Google Scholar]
- 9.Fujita, K., T. Yokota, T. Oguri, M. Fujime, and R. Kitagawa. 1992. In vitro adherence of Staphylococcus saprophyticus, Staphylococcus epidermidis, Staphylococcus haemolyticus, and Staphylococcus aureus to human ureter. Urol. Res. 20:399-402. [DOI] [PubMed] [Google Scholar]
- 10.Gatermann, S., J. John, and R. Marre. 1989. Staphylococcus saprophyticus urease: characterization and contribution to uropathogenicity in unobstructed urinary tract infection of rats. Infect. Immun. 57:110-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gatermann, S., B. Kreft, R. Marre, and G. Wanner. 1992. Identification and characterization of a surface-associated protein (Ssp) of Staphylococcus saprophyticus. Infect. Immun. 60:1055-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gatermann, S., and R. Marre. 1989. Cloning and expression of Staphylococcus saprophyticus urease gene sequences in Staphylococcus carnosus and contribution of the enzyme to virulence. Infect. Immun. 57:2998-3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gatermann, S., R. Marre, J. Heesemann, and W. Henkel. 1988. Hemagglutinating and adherence properties of Staphylococcus saprophyticus: epidemiology and virulence in experimental urinary tract infection of rats. FEMS Microbiol. Immunol. 1:179-185. [DOI] [PubMed] [Google Scholar]
- 14.Gatermann, S., and H. G. Meyer. 1994. Staphylococcus saprophyticus hemagglutinin binds fibronectin. Infect. Immun. 62:4556-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gatermann, S., H. G. Meyer, and G. Wanner. 1992. Staphylococcus saprophyticus hemagglutinin is a 160-kilodalton surface polypeptide. Infect. Immun. 60:4127-4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gatermann, S. G., and H. G. Meyer. 1995. Expression of Staphylococcus saprophyticus surface properties is modulated by composition of the atmosphere. Med. Microbiol. Immunol. (Berl.) 184:81-85. [DOI] [PubMed] [Google Scholar]
- 17.Hell, W., H. G. Meyer, and S. G. Gatermann. 1998. Cloning of aas, a gene encoding a Staphylococcus saprophyticus surface protein with adhesive and autolytic properties. Mol. Microbiol. 29:871-881. [DOI] [PubMed] [Google Scholar]
- 18.Higashide, M., M. Kuroda, S. Ohkawa, and T. Ohta. 2006. Evaluation of a cefoxitin disk diffusion test for the detection of mecA-positive methicillin-resistant Staphylococcus saprophyticus. Int. J. Antimicrob. Agents 27:500-504. [DOI] [PubMed] [Google Scholar]
- 19.Higashide, M., M. Kuroda, C. T. Omura, M. Kumano, S. Ohkawa, S. Ichimura, and T. Ohta. 2008. Methicillin-resistant Staphylococcus saprophyticus isolates carrying staphylococcal cassette chromosome mec have emerged in urogenital tract infections. Antimicrob. Agents Chemother. 52:2061-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hooton, T. M. 2001. Recurrent urinary tract infection in women. Int. J. Antimicrob. Agents 17:259-268. [DOI] [PubMed] [Google Scholar]
- 21.Hovelius, B., and P. A. Mardh. 1979. Haemagglutination by Staphylococcus saprophyticus and other staphylococcal species. Acta Pathol. Microbiol. Scand. B 87B:45-50. [DOI] [PubMed] [Google Scholar]
- 22.Hovelius, B., and P. A. Mardh. 1984. Staphylococcus saprophyticus as a common cause of urinary tract infections. Rev. Infect. Dis. 6:328-337. [DOI] [PubMed] [Google Scholar]
- 23.Jansson, P. E., B. Lindberg, and U. Lindquist. 1985. Structural studies of the capsular polysaccharide from Streptococcus pneumoniae type 5. Carbohydr. Res. 140:101-110. [DOI] [PubMed] [Google Scholar]
- 24.Jellheden, B., R. S. Norrby, and T. Sandberg. 1996. Symptomatic urinary tract infection in women in primary health care. Bacteriological, clinical and diagnostic aspects in relation to host response to infection. Scand. J. Prim. Health Care 14:122-128. [DOI] [PubMed] [Google Scholar]
- 25.Kahlmeter, G. 2003. An international survey of the antimicrobial susceptibility of pathogens from uncomplicated urinary tract infections: the ECO.SENS Project. J. Antimicrob. Chemother. 51:69-76. [DOI] [PubMed] [Google Scholar]
- 26.Katayama, Y., F. Takeuchi, T. Ito, X. X. Ma, Y. Ui-Mizutani, I. Kobayashi, and K. Hiramatsu. 2003. Identification in methicillin-susceptible Staphylococcus hominis of an active primordial mobile genetic element for the staphylococcal cassette chromosome mec of methicillin-resistant Staphylococcus aureus. J. Bacteriol. 185:2711-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kilcoyne, M., A. S. Shashkov, Y. A. Knirel, R. P. Gorshkova, E. L. Nazarenko, E. P. Ivanova, N. M. Gorshkova, S. N. Senchenkova, and A. V. Savage. 2005. The structure of the O-polysaccharide of the Pseudoalteromonas rubra ATCC 29570T lipopolysaccharide containing a keto sugar. Carbohydr. Res. 340:2369-2375. [DOI] [PubMed] [Google Scholar]
- 28.Kocianova, S., C. Vuong, Y. Yao, J. M. Voyich, E. R. Fischer, F. R. DeLeo, and M. Otto. 2005. Key role of poly-γ-dl-glutamic acid in immune evasion and virulence of Staphylococcus epidermidis. J. Clin. Invest. 115:688-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuroda, M., A. Yamashita, H. Hirakawa, M. Kumano, K. Morikawa, M. Higashide, A. Maruyama, Y. Inose, K. Matoba, H. Toh, S. Kuhara, M. Hattori, and T. Ohta. 2005. Whole genome sequence of Staphylococcus saprophyticus reveals the pathogenesis of uncomplicated urinary tract infection. Proc. Natl. Acad. Sci. U. S. A. 102:13272-13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Latham, R. H., K. Running, and W. E. Stamm. 1983. Urinary tract infections in young adult women caused by Staphylococcus saprophyticus. JAMA 250:3063-3066. [PubMed] [Google Scholar]
- 31.Lee, J. C., M. J. Liu, J. Parsonnet, and R. D. Arbeit. 1990. Expression of type-8 capsular polysaccharide and production of toxic shock syndrome toxin-1 are associated among vaginal isolates of Staphylococcus aureus. J. Clin. Microbiol. 28:2612-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luong, T. T., S. Ouyang, K. Bush, and C. Y. Lee. 2002. Type 1 capsule genes of Staphylococcus aureus are carried in a staphylococcal cassette chromosome genetic element. J. Bacteriol. 184:3623-3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacLean, L. L., M. B. Perry, E. M. Crump, and W. W. Kay. 2003. Structural characterization of the lipopolysaccharide O-polysaccharide antigen produced by Flavobacterium columnare ATCC 43622. Eur. J. Biochem. 270:3440-3446. [DOI] [PubMed] [Google Scholar]
- 34.Meyer, H. G., J. Muthing, and S. G. Gatermann. 1997. The hemagglutinin of Staphylococcus saprophyticus binds to a protein receptor on sheep erythrocytes. Med. Microbiol. Immunol. (Berl.) 186:37-43. [DOI] [PubMed] [Google Scholar]
- 35.Meyer, H. G., U. Wengler-Becker, and S. G. Gatermann. 1996. The hemagglutinin of Staphylococcus saprophyticus is a major adhesin for uroepithelial cells. Infect. Immun. 64:3893-3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mongkolrattanothai, K., S. Boyle, T. V. Murphy, and R. S. Daum. 2004. Novel non-mecA-containing staphylococcal chromosomal cassette composite island containing pbp4 and tagF genes in a commensal staphylococcal species: a possible reservoir for antibiotic resistance islands in Staphylococcus aureus. Antimicrob. Agents Chemother. 48:1823-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mulvey, M. A., Y. S. Lopez-Boado, C. L. Wilson, R. Roth, W. C. Parks, J. Heuser, and S. J. Hultgren. 1998. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science 282:1494-1497. [DOI] [PubMed] [Google Scholar]
- 38.Nilsson, I.-M., J. C. Lee, T. Bremell, C. Ryden, and A. Tarkowski. 1997. The role of staphylococcal polysaccharide microcapsule expression in septicemia and septic arthritis. Infect. Immun. 65:4216-4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noto, M. J., B. N. Kreiswirth, A. B. Monk, and G. L. Archer. 2008. Gene acquisition at the insertion site for SCCmec, the genomic island conferring methicillin resistance in Staphylococcus aureus. J. Bacteriol. 190:1276-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peschel, A., M. Otto, R. W. Jack, H. Kalbacher, G. Jung, and F. Gotz. 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274:8405-8410. [DOI] [PubMed] [Google Scholar]
- 41.Peterson, P. K., B. J. Wilkinson, Y. Kim, D. Schmeling, S. D. Douglas, and P. G. Quie. 1978. The key role of peptidoglycan in the opsonization of Staphylococcus aureus. J. Clin. Invest. 61:597-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pöhlmann-Dietze, P., M. Ulrich, K. B. Kiser, G. Doring, J. C. Lee, J. M. Fournier, K. Botzenhart, and C. Wolz. 2000. Adherence of Staphylococcus aureus to endothelial cells: influence of the capsular polysaccharide, the global regulator agr, and the bacterial growth phase. Infect. Immun. 68:4865-4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poutrel, B., F. B. Gilbert, and M. Lebrun. 1995. Effects of culture conditions on production of type 5 capsular polysaccharide by human and bovine Staphylococcus aureus strains. Clin. Diagn. Lab. Immunol. 2:166-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poutrel, B., P. Rainard, and P. Sarradin. 1997. Heterogeneity of cell-associated CP5 expression on Staphylococcus aureus strains demonstrated by flow cytometry. Clin. Diagn. Lab. Immunol. 4:275-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raz, R., R. Colodner, and C. M. Kunin. 2005. Who are you—Staphylococcus saprophyticus? Clin. Infect. Dis. 40:896-898. [DOI] [PubMed] [Google Scholar]
- 46.Ronald, A. 2002. The etiology of urinary tract infection: traditional and emerging pathogens. Am. J. Med. 113(Suppl. 1A):14S-19S. [DOI] [PubMed] [Google Scholar]
- 47.Rosenstein, R., C. Nerz, L. Biswas, A. Resch, G. Raddatz, S. C. Schuster, and F. Gotz. 2009. Genome analysis of the meat starter culture bacterium Staphylococcus carnosus TM300. Appl. Environ. Microbiol. 75:811-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Russo, T., J. J. Brown, S. T. Jodush, and J. R. Johnson. 1996. The O4 specific antigen moiety of lipopolysaccharide but not the K54 group 2 capsule is important for urovirulence of an extraintestinal isolate of Escherichia coli. Infect. Immun. 64:2343-2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Russo, T. A., A. Stapleton, S. Wenderoth, T. M. Hooton, and W. E. Stamm. 1995. Chromosomal restriction fragment length polymorphism analysis of Escherichia coli strains causing recurrent urinary tract infections in young women. J. Infect. Dis. 172:440-445. [DOI] [PubMed] [Google Scholar]
- 50.Sadovskaya, I., J. R. Brisson, N. H. Khieu, L. M. Mutharia, and E. Altman. 1998. Structural characterization of the lipopolysaccharide O-antigen and capsular polysaccharide of Vibrio ordalii serotype O:2. Eur. J. Biochem. 253:319-327. [DOI] [PubMed] [Google Scholar]
- 51.Sakinc, T., B. Kleine, and S. G. Gatermann. 2007. Biochemical characterization of the surface-associated lipase of Staphylococcus saprophyticus. FEMS Microbiol. Lett. 274:335-341. [DOI] [PubMed] [Google Scholar]
- 52.Sakinc, T., B. Kleine, and S. G. Gatermann. 2006. SdrI, a serine-aspartate repeat protein identified in Staphylococcus saprophyticus strain 7108, is a collagen-binding protein. Infect. Immun. 74:4615-4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sakinc, T., M. Woznowski, M. Ebsen, and S. G. Gatermann. 2005. The surface-associated protein of Staphylococcus saprophyticus is a lipase. Infect. Immun. 73:6419-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schneider, G., U. Dobrindt, H. Bruggemann, G. Nagy, B. Janke, G. Blum-Oehler, C. Buchrieser, G. Gottschalk, L. Emody, and J. Hacker. 2004. The pathogenicity island-associated K15 capsule determinant exhibits a novel genetic structure and correlates with virulence in uropathogenic Escherichia coli strain 536. Infect. Immun. 72:5993-6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schoenhofen, I. C., V. V. Lunin, J. P. Julien, Y. Li, E. Ajamian, A. Matte, M. Cygler, J. R. Brisson, A. Aubry, S. M. Logan, S. Bhatia, W. W. Wakarchuk, and N. M. Young. 2006. Structural and functional characterization of PseC, an aminotransferase involved in the biosynthesis of pseudaminic acid, an essential flagellar modification in Helicobacter pylori. J. Biol. Chem. 281:8907-8916. [DOI] [PubMed] [Google Scholar]
- 56.Stevenson, G., K. Andrianopoulos, M. Hobbs, and P. R. Reeves. 1996. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J. Bacteriol. 178:4885-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takeuchi, F., S. Watanabe, T. Baba, H. Yuzawa, T. Ito, Y. Morimoto, M. Kuroda, L. Cui, M. Takahashi, A. Ankai, S. Baba, S. Fukui, J. C. Lee, and K. Hiramatsu. 2005. Whole-genome sequencing of Staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. J. Bacteriol. 187:7292-7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thakker, M., J.-S. Park, V. Carey, and J. C. Lee. 1998. Staphylococcus aureus serotype 5 capsular polysaccharide is antiphagocytic and enhances bacterial virulence in a murine bacteremia model. Infect. Immun. 66:5183-5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tzianabos, A. O., J. Y. Wang, and J. C. Lee. 2001. Structural rationale for the modulation of abscess formation by Staphylococcus aureus capsular polysaccharides. Proc. Natl. Acad. Sci. U. S. A. 98:9365-9370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.von Eiff, C., G. Peters, and C. Heilmann. 2002. Pathogenesis of infections due to coagulase-negative staphylococci. Lancet Infect. Dis. 2:677-685. [DOI] [PubMed] [Google Scholar]
- 61.Xu, S., R. D. Arbeit, and J. C. Lee. 1992. Phagocytic killing of encapsulated and microencapsulated Staphylococcus aureus by human polymorphonuclear leukocytes. Infect. Immun. 60:1358-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yim, H. H., and C. E. Rubens. 1997. Use of a dental amalgamator to extract RNA from the gram-positive bacterium Streptococcus agalactiae. Biotechniques 23:229-231. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.