Abstract
Chlamydia trachomatis is a human pathogen of global importance. An obstacle to studying the pathophysiology of human chlamydial disease is the lack of a suitable murine model of C. trachomatis infection. Mice are less susceptible to infection with human isolates due in part to innate mouse-specific host defense mechanisms to which human strains are sensitive. Another possible factor that influences the susceptibility of mice to infection is that human isolates are commonly cultivated in vitro prior to infection of mice; therefore, virulence genes could be lost as a consequence of negative selective pressure. We tested this hypothesis by infecting innate immunity-deficient C3H/HeJ female mice intravaginally with a human serovar D urogenital isolate that had undergone multiple in vitro passages. We observed early and late infection clearance phenotypes. Strains of each phenotype were isolated and then used to reinfect naïve mice. Following infection, the late-clearance strain was significantly more virulent. It caused unvarying infections of much longer durations with greater infectious burdens that naturally ascended to the upper genital tract, causing salpingitis. Despite contrasting in vivo virulence characteristics, the strains exhibited no differences in the results of in vitro infectivity assays or sensitivities to gamma interferon. Genome sequencing of the strains revealed mutations that localized to a single gene (CT135), implicating it as a critical virulence factor. Mutations in CT135 were not unique to serovar D but were also found in multiple oculogenital reference strains. Our findings provide new information about the pathogenomics of chlamydial infection and insights for improving murine models of infection using human strains.
Chlamydia trachomatis is an obligate intracellular bacterial pathogen of humans. Infection of ocular and genital mucosal columnar epithelial cells causes blinding trachoma and sexually transmitted infection (STI), respectively. Trachoma is the leading cause of preventable blindness, afflicting an estimated 84 million individuals worldwide (29), and is recognized as one of the world's most neglected infectious diseases (15). Chlamydial STIs are epidemic globally, with an estimated 92 million new cases occurring each year (30). Chlamydial STI can result in pelvic inflammatory disease and tubal factor infertility and is an important risk factor in the transmission of HIV (11). Persistent infection and reinfection of human chlamydial strains drive irreversible inflammatory responses, fibrosis, and damaging scarring. Control of these diseases will require the development of a vaccine, for which there has been nominal progress. Regrettably, a vaccine will not be forthcoming until the pathophysiology of these globally important diseases is better understood.
Understanding the pathophysiology and immunology of human chlamydial infection and disease has been hampered by the lack of a suitable small-animal model for human C. trachomatis strains. There are excellent guinea pig and murine models that utilize naturally occurring chlamydial challenge strains. However, rodents are exceptionally resistant to genitourinary tract infection by human C. trachomatis isolates, where infections are characteristically acute and cleared rapidly by murine-specific innate immune mechanisms (17). Previous comparative pathogenomic studies highlighted the major differences between human and mouse gamma interferon (IFN-γ)-mediated antimicrobial effectors and portrayed how human and mouse chlamydial strains evade these host-specific defense mechanisms by utilizing a unique set of pathogen-specific genes that reside in the chromosome's plasticity zone (1, 4, 17). Little, however, is known about other C. trachomatis genes that influence virulence for the mouse. Human isolates are commonly cultivated in vitro prior to infection of animals; therefore, virulence genes might be lost as a consequence of negative selective pressure. Identifying such genes could lead to the development of a more practical murine model that utilizes human challenge isolates. We tested this hypothesis and identified human strains following in vivo infection of C3H/HeJ female mice that exhibited marked differences in virulence for the female mouse genital tract. We show that the in vivo-selected virulent strain exhibits superior infection and disease-causing characteristics that mimic those in human infection. A comprehensive genomic analysis of attenuated and virulent isolates shows that mutations in a single gene are responsible for altering the pathogenesis of C. trachomatis infection.
MATERIALS AND METHODS
Infection of mice and chlamydial strains.
Eight-week-old inbred female C3H/HeJ mice (Jackson Laboratories) were treated with 2.5 mg medroxyprogesterone acetate at 10 and 3 days prior to intravaginal inoculation with C. trachomatis. Swabbing of the vaginal vault was performed just prior to inoculation. In our initial study, 56 female mice were infected intravaginally with 1 × 105 inclusion-forming units (IFU) of our laboratory reference C. trachomatis strain D/UW-3/CX. This strain had been serially passed 10 times in embryonating hens' eggs and 17 times in HeLa 229 cells prior to infection of mice. We followed the individual infections by culture until the infections had spontaneously cleared. Chlamydiae were isolated from a single early-clearance mouse at day 10 postinfection (p.i.) and from a single late-clearance mouse at day 49 p.i. These strains were subsequently grown in McCoy cells and designated D-EC and D-LC, respectively. The strains were then used to reinfect naïve mice, and the infections were monitored over the entire infection period by culture. The 50% infective dose (ID50) of each strain was calculated using the Reed and Muench method (23). All animal protocols were reviewed and approved by the Animal Care and Use Committee at the Rocky Mountain Laboratories, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Histopathology.
Mice were infected with 105 IFU D-EC or D-LC or were mock infected with sucrose-phosphate glutamate (SPG) buffer, as described above. At different times p.i., mice were euthanized and their genital tracts were excised, fixed in 10% formalin, and embedded into paraffin blocks. Three to seven mice per time period per group were euthanized for tissue analysis. Tissue sections were stained with hematoxylin-eosin (H&E) to visualize inflammatory cell infiltration. Immunostaining was done with the anti-major outer membrane protein (MOMP) monoclonal antibody L2I-5 using methods described previously (16). Histological scoring of chronic inflammatory cellular infiltrates (lymphocytes, plasma cells, and macrophages) in H&E-stained tissue was performed blinded by a veterinary pathologist following previously described four-tiered semiquantitative criteria (8). The range of cellular infiltration scoring of 0 to 4 was defined as follows: 0 is none, 1 is minimal, 2 is mild, 3 is moderate, and 4 is severe.
In vitro characterization of strains.
The growth characteristics of D-EC and D-LC strains were studied in McCoy cells by performing one-step growth curve assays, plaque formation assays, and assays for susceptibility to growth inhibition by IFN-γ (BD Pharmingen), as described previously (7, 13, 24).
Genome sequencing.
C. trachomatis D-EC and D-LC genomic DNAs were isolated from purified elementary bodies (EBs), as described previously (6). Genomes were sequenced using Roche Applied Science's 454 Life Sciences GS Pyrosequencing platform. This high-throughput de novo sequencing method enabled an average 45-fold coverage and was performed and assembled into contigs by Beckman Coulter Genomics, Inc. Targeted sequencing across the six gaps between contigs resulted in two fully closed chromosomal and plasmid sequences. The entire CT456 gene (which encodes the translocated actin-recruiting phosphoprotein) was also PCR amplified and sequenced because of its highly repetitive sequence. The primers used for gap closing and CT456 sequencing are listed in Table S1 in the supplemental material. The sequences of the closed genomes were compared to each other and to the published serovar D sequence (26), identifying 62 loci that exhibited differences among the sequences. These loci were PCR amplified and capillary sequenced for confirmation. Primers for the 62 regions are listed in Table S2 in the supplemental material. Forty-two of these loci contained false-positive mutations that included frameshift mutations in long homopolymeric runs, a common 454 sequencing artifact (28). The remaining 20 loci had mutations in D-EC and D-LC compared to the published serovar D sequence (see Table 2).
TABLE 2.
Twenty mutations identified in genomes of D-EC and D-LC compared to C. trachomatis D/UW-3/CXa
| SNP locationb | Gene | Reference D/UW-3/CX |
D-EC |
D-LC |
|||
|---|---|---|---|---|---|---|---|
| Gene start position | Gene stop position | nt change | aa change | nt change | aa change | ||
| 30572 | CT025 | 29069 | 29941 | C → G | Thr → Ser | C → G | Thr → Ser |
| 55047 | CT049 | 54113 | 55585 | G → C | Gly → Ala | G → C | Gly → Ala |
| 55099 | CT049 | 54113 | 55585 | A → G | None | A → G | None |
| 152276 | CT135 | 152143 | 153225 | None | None | ΔT | Frameshift |
| 152686 | CT135 | 152143 | 153225 | ::T | Frameshift | None | None |
| 403784 | CT352 | 403499 | 403804 | A → C | Ile → Leu | A → C | Ile → Leu |
| 450477 | CT394 | 449841 | 451019 | A → G | Lys → Glu | A → G | Lys → Glu |
| 517109 | IGc (CT446-CT447) | 516946 | 519105 | G → T | None | G → T | None |
| 588168 | CT511 | 587942 | 588376 | G → T | Thr → Lys | G → T | Thr → Lys |
| 621054 | CT551 | 620037 | 621068 | ::G | Frameshift | ::G | Frameshift |
| 628883 | IG (CT556-CT557) | 628808 | 630414 | C → T | None | C → T | None |
| 706330 | CT621 | 704354 | 706852 | T → C | Thr → Ala | T → C | Thr → Ala |
| 717493 | CT630 | 717087 | 717770 | T → C | Gln → Arg | T → C | Gln → Arg |
| 720059 | IG (CT632-CT633) | 719973 | 721287 | A → T | None | A → T | None |
| 727542 | CT638 | 726792 | 727559 | ::G | Frameshift | ::G | Frameshift |
| 733706 | CT640 | 730899 | 733913 | T → C | Thr → Ala | T → C | Thr → Ala |
| 752502 | IG (CT655-CT656) | 752365 | 753143 | ::C | None | ::C | None |
| 858691 | 23S rRNA | 878039 | 880902 | G → A | None | G → A | None |
| 858694 | 23S rRNA | 878039 | 880902 | G → A | None | G → A | None |
| 858698 | 23S rRNA | 878039 | 880902 | ΔC | None | ΔC | None |
Boldface type indicates unique mutations identified in D-EC and D-LC. nt, nucleotide; aa, amino acid; ::, insertion; Δ, deletion.
SNP location refers to the location of the mutation in the reference D/UW-3/CX genome; for a single base pair insertion, the location of the preceding nucleotide is given.
IG, intergenic region and adjacent ORFs.
Targeted sequencing.
DNA was extracted from D-EC, D-LC, D/UW-3/CX, and clinical isolates from infected HeLa 229 cells by 0.5 M NaOH, followed by neutralization with 1 M Tris-HCl, pH 8.0. DNA was isolated from purified EBs from the 15 reference strains in the same way. The CT135 genes from these strains were PCR amplified using the flanking primers 5′-TGAGCTAAAGATCGTGATGGTC and 5′-CTATACCACAAATCCGCCGC. Capillary sequencing using BigDye chemistry (Applied Biosystems) was performed with the flanking primers and the four internal primers 5′-CTGCCTTCGCCCGTTTGAG, 5′-CTAGTCCAGCACTTGCTGC, 5′-CTTCTGCGGTAATCGCAGC, and 5′-CTCATCATCACAGTATCGCAAG.
CEL I digestion.
Intrastrain polymorphisms were analyzed by CEL I digestion of the PCR amplicons (18). PCR amplicons were heat denatured at 95°C and reannealed by very slow cooling (0.1°C/s) to allow heterodimers to form. The reannealed PCR products were digested with CEL I (Transgenomic, Inc.) at 42°C in a 20-μl reaction mixture, according to the manufacturer's instructions. After digestion, the samples were run on 1.5% agarose gels and the DNA bands were visualized by ethidium bromide staining.
Statistical analyses.
The Tukey-Kramer Studentized range test was performed to identify statistically significant differences among the inflammatory scores of mouse genital tract tissues at individual time points using SAS (version 9.2) software (SAS Institute, Inc.). Pairwise comparisons among the groups were made. P values of less than 0.05 were considered significant.
Nucleotide sequence accession numbers.
The closed genomes, associated plasmid sequences, and the original 454 sequencing reads for D-EC and D-LC have been submitted to GenBank under accession numbers CP002052, CP002054 (chromosomes) and CP002053 and CP002055 (plasmids) and to the NCBI Sequence Read Archive under study number SRP002534 (454 reads).
RESULTS
In vivo selection of C. trachomatis infection phenotypes differing in virulence. (i) Infection characteristics.
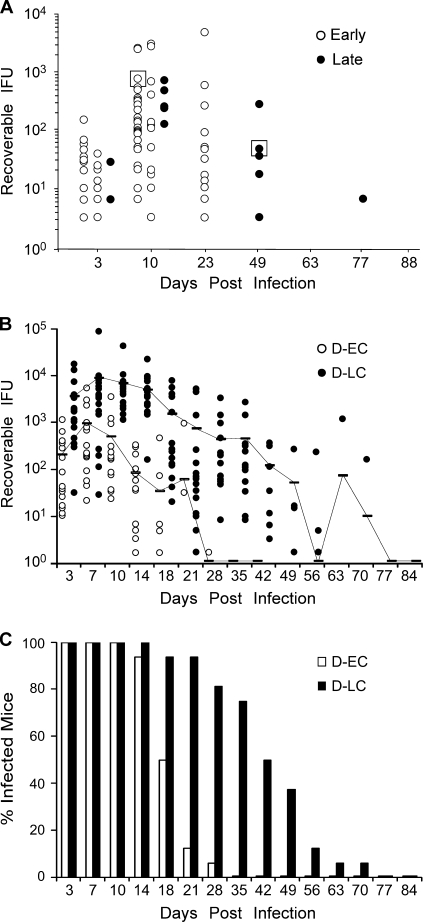
We infected 56 C3H/HeJ female mice with our C. trachomatis serovar D reference laboratory strain, and the clearance patterns were monitored over the entire course of the infection (Fig. 1A). Innate immunity-deficient C3H/HeJ mice were chosen for these experiments because they are known to be more susceptible to infection by human C. trachomatis strains (8). We observed infection durations that varied greatly among the mice: from 10 to 77 days p.i. The majority of the mice cleared their infection after 23 days p.i. In contrast, a small subset (five mice) remained infected for 49 to 77 days p.i. Organisms were isolated from one mouse at day 10 p.i. and another mouse at day 49 p.i. We designated these isolates early clearance (D-EC) and late clearance (D-LC) strains, respectively. The D-EC and D-LC strains were minimally propagated in vitro and were then used to infect naïve mice. Sixteen mice were challenged with each strain, and the course of infection was monitored by culture (Fig. 1B). Mice infected with either the D-EC or the D-LC strain exhibited a consistent and distinct clearance pattern. D-LC-infected mice had greater mean infectious burdens than D-EC-infected mice throughout the culture period (circa 10-fold), and D-LC infections persisted for a much longer period of time (70 versus 28 days for D-EC infections). In addition, a greater percentage of D-LC-challenged mice were culture positive at later time points p.i. (Fig. 1C). The ID50s of the D-EC and D-LC strains were 6.3 × 102 and 4.4 × 102 IFU, respectively (Table 1), indicating that the isolates did not differ in their ability to colonize the mouse urogenital epithelium. We concluded from these results that our serovar D parental seed stock was a mixture of organisms varying in virulence for the mouse.
FIG. 1.
In vivo isolation of C. trachomatis infection phenotypes. (A) Fifty-six C3H/HeJ mice were infected with 105 IFU parental strain D/UW-3/CX, and the course of infection was monitored by culture. The results for culture-positive mice are plotted. Infection durations varied from early (10 to 23 days; open circles) to late (49 to 77 days; closed circles). Chlamydiae were isolated from a single early-clearance (D-EC) or late-clearance (D-LC) strain-infected mouse (boxed) and serially passed in McCoy cells (five passages) to obtain sufficient amounts of organisms to rechallenge naïve animals. (B) Sixteen mice were infected with 105 IFU of D-EC or D-LC, and the course of the infection was monitored by culture. Each data point indicates the number of IFU cultured from a single mouse infected with either D-EC (open circle) or D-LC (filled circle). The results for culture-positive mice are plotted. Samples from mice were cultured until inclusion-negative samples were obtained at two successive time points. Contiguous lines indicate the 16-mouse mean IFU values for both groups and include the results for both positive and negative cultures. Mice infected with the individual isolates exhibited distinctly separate infection and clearance kinetics, suggesting that the in vivo-selected strains were clonal. (C) Data for the same groups whose results are shown in panel A plotted as the percentage of infected mice. The percentage of infected mice in the D-LC group was consistently greater in the later culture periods (days 18 to 42).
TABLE 1.
ID50s of C. trachomatis D-EC and D-LC isolates for the female mouse genital tract
| Infective dosea (IFU) | Result for the following C. trachomatis challenge isolate at the indicated day postinfectionb: |
|||
|---|---|---|---|---|
| D-EC |
D-LC |
|||
| 3 | 7 | 3 | 7 | |
| 106 | 5/5 | 5/5 | 5/5 | 5/5 |
| 105 | 5/5 | 5/5 | 5/5 | 5/5 |
| 104 | 8/10 | 8/10 | 15/15 | 15/15 |
| 103 | 8/15 | 10/15 | 8/15 | 11/15 |
| 102 | 0/15 | 1/15 | 0/15 | 1/15 |
| 101 | 0/5 | 0/5 | 0/5 | 0/5 |
Groups of medroxyprogesterone acetate-treated mice were infected cervicovaginally with 5 μl of EBs.
Results are shown as the number of mice yielding inclusion-positive cultures on the indicated day/total number of mice tested. ID50 calculations were performed by the method of Reed and Muench (23) with data for the day 7 culture results. C. trachomatis D-EC ID50, 6.3 × 102; C. trachomatis D-LC ID50, 4.4 × 102.
(ii) Disease characteristics.
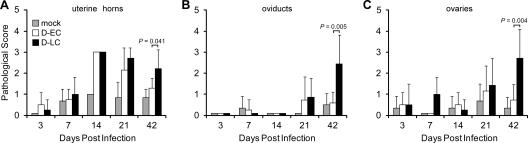
We next sought to determine if the different courses of infection caused by these strains were reflected in differences in pathology. Figure 2 shows the mean chronic inflammatory scores of the uterine horn, oviduct, and ovarian tissues of mock-, D-EC-, and D-LC-infected mice taken at different times p.i. The histopathological scores of the uterine horns (Fig. 2A) at days 14 and 21 p.i. were similar between D-EC- and D-LC-infected mice and were significantly higher relative to those of the mock-infected group (P < 0.0001). The inflammatory responses in the uterine horns of D-EC-infected mice diminished in intensity by day 42 p.i. In contrast, the inflammatory responses in the uterine horns of D-LC-infected mice remained significantly elevated (P = 0.041). A significantly greater inflammatory cell infiltrate was also observed in the oviducts (P = 0.005) and ovarian tissues (P = 0.004) of D-LC-infected mice than in those of D-EC-infected mice at the later time point. We observed minimal to no acute inflammatory infiltrates (neutrophils) in the same tissues in any of the infection groups (data not shown).
FIG. 2.
Inflammatory response in the upper genital tract of C. trachomatis-infected mice. Seventy-three C3H/HeJ mice were infected intravaginally with 105 IFU of D-EC, D-LC, or SPG buffer (mock infection). At five different times following infection, the genital tracts were excised and placed in 10% formalin, embedded into paraffin, sliced, and H&E stained. Upper genital tract tissues were examined for chronic inflammatory cellular infiltrates and scored accordingly: 0 for none, 1 for minimal, 2 for mild, 3 for moderate, and 4 for severe. Scoring was performed for the uterine horn (A), oviduct (B), and ovaries (C). The numbers of mock-, D-EC-, and D-LC-infected mice scored ranged from three to seven mice per group. Mean pathological scores are plotted, and error bars indicate the standard error of the mean. Tukey-Kramer pairwise statistical tests were performed at each time point. P values of less than 0.05 were considered significant and are indicated for comparisons of the results for D-EC and D-LC. D-EC and D-LC infections resulted in a similar inflammatory cell infiltrate in uterine horns at 14 and 21 days p.i. In contrast, only D-LC-infected mice produced a significant inflammatory response in the uterine horns, oviducts, and ovaries at 42 days p.i.
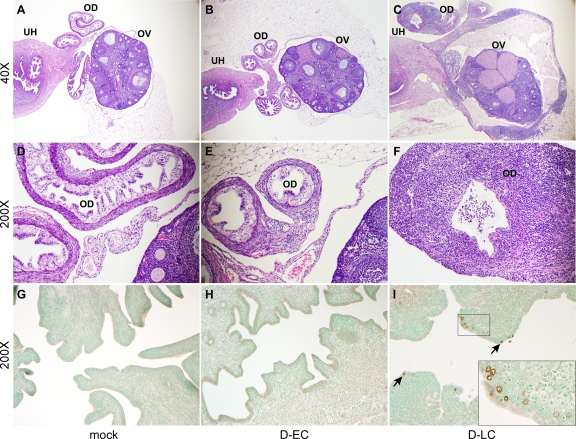
Figure 3A to F shows representative examples of H&E-stained uterine horns, oviducts, and ovaries of mock-, D-EC-, and D-LC-infected mice at 42 days p.i. A moderate to severe chronic inflammatory infiltrate was present within and surrounding the oviducts and ovaries of D-LC-infected mice (Fig. 3C and F), whereas the tissues of mock- and D-EC-infected mice were essentially within normal limits histologically (Fig. 3A, B, D, and E). The D-LC tissue infiltrates consisted of numerous lymphocytes and plasma cells and fewer macrophages that expanded the walls of the oviducts and the ovarian bursae. The lining epithelia of the ovarian bursae and oviducts were multifocally lost or attenuated. Immunostaining of tissues on day 42 p.i. detected chlamydial inclusions in the upper uterine horns of D-LC-infected mice but not those of D-EC- or mock-infected mice (Fig. 3G to I). Inclusions were sparse and localized to the epithelium.
FIG. 3.
Histopathological evaluation of oviduct and ovarian tissue from C. trachomatis-infected mice. (A to F) Micrographs are of representative examples of H&E-stained tissues evaluated at 42 days p.i. from mock-, D-EC-, and D-LC-infected mice at ×40 (A to C) and ×200 (D to F) magnifications. The indicated tissue types are uterine horns (UH), oviducts (OD), and ovaries (OV). The oviduct and ovarian tissues of mock-infected (A and D) and D-EC-infected (B and E) mice are of normal thickness and contain only minimal aggregates of perivascular lymphocytes. In contrast, the walls of the oviducts and ovarian bursa of D-LC-infected mice (C and F) are severely thickened by lymphocyte and plasma cell infiltrates. Proteinaceous fluid in the lumen of the oviducts and ovarian bursa primarily consisted of polymorphonuclear leukocytes and was observed in both mock-infected and infected mice. (G to I) Uterine horn tissues from mock-, D-EC-, and D-LC-infected mice stained at 42 days p.i. with anti-MOMP-specific monoclonal antibody L2I-5 (magnification, ×200). Chlamydiae were not detected in the tissues of mock-infected (G) or D-EC-infected (H) mice but were readily found in the tissues of D-LC-infected mice (I). Inclusions (arrows) were localized to the epithelial cells. (I, inset) Enlarged image of immunostained inclusions.
In vitro comparison of virulence phenotypes.
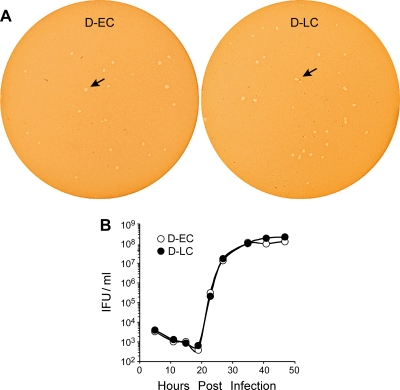
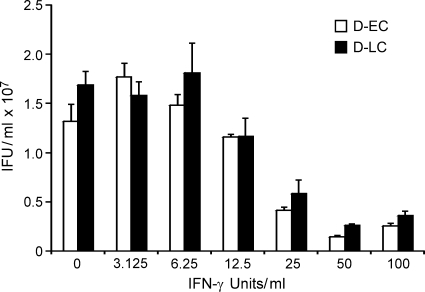
Interestingly, the D-EC and D-LC strains did not differ in their plaquing properties or growth rate in McCoy cells (Fig. 4). We next tested whether the strains exhibited differences in their sensitivities to growth inhibition by IFN-γ, as it is known that human strains are highly sensitive to murine IFN-γ-mediated effectors (19). We found no differences in the susceptibilities of D-EC and D-LC to IFN-γ-mediated inhibition of intracellular growth (Fig. 5).
FIG. 4.
Comparisons of in vitro growth of the strains. (A) Plaque formation of the D-EC and D-LC isolates in McCoy cells at day 9 p.i. (B) One-step growth curves of D-EC and D-LC in McCoy cells. The D-EC and D-LC strains did not differ in plaque size or growth rate.
FIG. 5.
Effect of IFN-γ on growth of the D-EC and D-LC strains in McCoy cells. McCoy cells were infected with D-EC and D-LC in the presence of IFN-γ and harvested at 42 h postinfection, and the numbers of recoverable IFUs were determined. The values are the means of triplicate samples. The growth characteristics of the D-EC and D-LC strains did not differ during in vitro IFN-γ treatment.
Genomic sequencing of D-EC and D-LC.
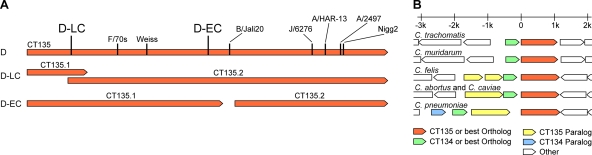
To determine the genetic basis for these important virulence differences, we sequenced the genomes of the D-EC and D-LC strains. We initially performed comparative genome hybridization sequencing (Nimblegen) and mutation mapping using the SOLiD platform (data not shown). Although the data from these experiments were in complete agreement with each other, the mutations identified between D-EC and D-LC were extremely limited. As both of these technologies identify mutations using a known reference sequence, it is not uncommon for mutations to be missed. To confidently attribute phenotypic differences to specific mutations, we performed 454 high-throughput de novo genomic sequencing on each of the strains. The assembly of sequencing reads and gap closings resulted in two fully closed chromosomal and plasmid sequences. Pairwise analyses between the D-EC and D-LC genomes as well as the published serovar D sequence were conducted (26). The genomic differences identified were PCR amplified and sequenced for confirmation. Polymorphisms were found at 20 chromosomal locations (Table 2); 18 of these were conserved between the D-EC and D-LC genomes compared to the sequence of the reference D genome sequence. The remaining two loci contained unique mutations, one in D-EC and one in D-LC. Remarkably, both of these mutations occurred in the same open reading frame (ORF), CT135. The mutations introduced frameshifts in both strains. The frame shift in the D-EC strain disrupts CT135 near the middle of the gene, generating two similarly sized predicted ORFs, while the frameshift in D-LC results in a slightly shorter predicted ORF than the annotated serovar D CT135 (Fig. 6A). The other 902 ORFs and all intergenic and structural RNA-coding regions of D-EC and D-LC were identical. C. trachomatis and C. muridarum contain a CT135 homolog that is predicted to form a bicistronic operon with CT134 (20) (Fig. 6B). All sequenced isolates of the family Chlamydiaceae have at least one CT134 and CT135 ortholog, although in the Chlamydophila species this gene pair is not predicted to form an operon due to a larger intergenic region. These results provide evidence that CT135 may be a key C. trachomatis virulence factor that influences the severity of infection and disease in the female mouse genital tract.
FIG. 6.
Schematic sequence comparisons and genome organization of CT135. (A) Frameshift mutations in D-EC, D-LC, and other C. trachomatis and C. muridarum strains (6, 10, 21, 25, 27) are shown mapped to CT135 in D/UW-3/CX. Depiction of predicted ORFs CT135.1 and CT135.2 (locus tags CTDEC_013501 and CTDEC_013502 in D-EC and CTDLC_013501 and CTDLC_013502 in D-LC under GenBank accession numbers CP002052 and CP002054 for the two strains, respectively) in D-EC and D-LC are also shown. (B) Regions of sequenced Chlamydiaceae genomes containing CT135 and CT134 orthologs and paralogs are drawn to scale, with the top bar indicating the kilobase distance relative to the start codons of the CT135 orthologs.
CT135 polymorphisms in other C. trachomatis reference strains and clinical isolates.
We investigated the possibility that CT135 intrastrain polymorphisms existed in other C. trachomatis reference strains by using the mismatch-specific nuclease CEL I. CEL I specifically cleaves single nucleotide polymorphisms (SNPs) and small insertion/deletion (in/del) mismatches in DNA heteroduplexes. To experimentally test the legitimacy of this procedure, we first generated CT135 PCR amplicons from strains D-EC and D-LC and the serovar D reference strain. The PCR products were heat denatured, reannealed to allow heteroduplex formation, digested with CEL I, and sized by gel electrophoresis (Fig. 7). Samples containing multiple CT135 genotypes are cleaved at the site of these polymorphisms and generate distinct DNA fragment sizes. A single CT135 product (1,217 bp, including the flanking regions) was observed for D-EC and D-LC, demonstrating the CT135 homogeneity of the strains. Conversely, the D-EC and D-LC mixture showed the expected fragments predicted from a single digestion and double digestions of D-EC and D-LC CT135 mutations (D-EC digestion, 628 bp and 589 bp; D-LC digestion, 1,001 bp and 216 bp; double digestions, 412 bp). Surprisingly, the multiply passaged serovar D reference strain exhibited a more complex digestion profile. Cleaved products corresponding to the D-LC mutation were significantly weaker or not detectable, suggesting a low frequency of the D-LC genotype. Two additional fragments (685 bp and 532 bp) were also detected, indicating the presence of additional CT135 genotypes. We characterized the CT135 genotypes in the serovar D reference strain by sequencing CT135 from plaque-cloned isolates and identified six distinct genotypes, including D-EC and D-LC. All six genotypes have disruptions in CT135 introduced by frameshift or nonsense mutations. Whether the other four genotypes produce distinct infection pathotypes in the murine model is unknown.
FIG. 7.
CEL I digestion of the D-EC, D-LC, and reference serovar D strains. CT135 PCR amplicons of D-EC, D-LC, a mixture of each strain, and the parental reference serovar D strain were digested with the mismatch-specific nuclease CEL I. Before digestion, the PCR products were heat denatured and reannealed to allow possible heteroduplexes to form. D-EC and D-LC have only the uncleaved 1,217-bp full-length CT135 PCR product. The 1:1 mixture of D-EC and D-LC exhibits five digestion products: 628 bp and 589 bp as a result of cleaving the D-EC mutation, 1,001 bp and 216 bp as a result of cleaving the D-LC mutation, and 412 bp as a result of cleaving both the D-EC and D-LC mutations (the complete digestion of a heteroduplex). In the parental serovar D strain, the 1,001-bp band is significantly weaker than that for the D-EC and D-LC mixture, and the 412-bp and 216-bp bands are not detectable, all of which are likely related to a low abundance of D-LC in the parental serovar D strain. The bands associated with D-EC are easily recognizable, as are two other bands (685 bp and 532 bp, indicated by asterisks), suggesting the presence of other genotypes in the parental serovar D strain.
We performed the same analysis on our other C. trachomatis laboratory reference strains to determine if they exhibited CT135 intrastrain polymorphisms. The majority of strains (strains A to J) were found to contain multiple genotypes for CT135; the exceptions were serovars K, L1, L2, and L3, which appeared to be homogeneous (Fig. 8A). Capillary sequencing of CT135 from serovars K, L1, L2, and L3 confirmed the presence of an intact ORF similar to one in the published serovar D sequence. A rigorous inspection of the chromatograms of the serovars showing intrastrain polymorphisms (Fig. 8A, lanes A to J) confirmed that these sequences indeed contain multiple CT135 genotypes with mutations that also produced frameshifts in CT135 that could reflect avirulent and virulent phenotypes. As a control, we conducted a similar analysis with ompA, the most polymorphic C. trachomatis gene (31). We observed no evidence for ompA polymorphisms in any of the 15 reference strains (Fig. 8B), whereas a mixture of serovar B and Ba CT135 amplicons revealed the presence of multiple ompA SNPs between the strains.
FIG. 8.
Polymorphism of CT135 in 15 reference laboratory strains. (A) CT135 PCR amplicons from 15 reference laboratory strains, representing the 15 major serovars, were digested with CEL I and run on an agarose gel. All strains except those of serovars K and L1, L2, and L3 exhibit digestion products indicating the presence of multiple genotypes of CT135 in these strains. (B) CEL I digestion of ompA from the same 15 strains. No digestion products were observed in any sample, indicating that all 15 strains contain only one genotype of ompA. A 1:1 mixture of serovars B and Ba was included as a positive control for CEL I digestion.
We also investigated the occurrence of CT135 polymorphisms in 55 clinical isolates that had undergone minimal in vitro passages in cell culture and that represented nine different urogenital serovars. Interestingly, sequencing of CT135 from 52 of these samples showed no CT135 polymorphisms. Two clinical samples differed by one SNP each and one differed by four SNPs compared to the published serovar D sequence (see Table S3 in the supplemental material). None of the 55 clinical samples exhibited disruptions in CT135 by either frameshift or nonsense mutations, nor did they have any sign of mixed genotypes after testing by both capillary sequencing and CEL I digestion.
DISCUSSION
The study described in this report shows that mutations in a single genetic locus (CT135) are sufficient to change the in vivo virulence of the human C. trachomatis urogenital strain D/UW-3/CX in the female genital tract of C3H/HeJ mice. Our findings support the conclusion that the D-LC strain but not the D-EC strain produced a naturally ascending infection in the female genital tract and evoked a chronic mononuclear inflammatory response, resulting in salpingitis. To our knowledge, this is the first demonstration of a naturally ascending infection in female mice with accompanying upper genital tract sequelae achieved using a C. trachomatis human STI isolate. We were successful in identifying the virulent human strain and mapping the virulence to CT135 by performing an in vivo infection screen that utilized a large cohort of mice infected with our laboratory reference strain, D/UW-3/CX, and de novo genome sequencing of the reisolated strains. The ID50s of D-EC and D-LC were similar, a finding consistent with the conclusion that CT135 is not critical for chlamydial colonization of urogenital epithelial cells. Strain virulence differences were discernible postcolonization and were characterized by significantly higher infectious burdens, durations of infection, and ascending infections, resulting in salpingitis. Our finding that CT135 mutations do not discriminate pathobiological differences in in vitro assays of infectivity, plaque morphology, or resistance to IFN-γ strongly support the conclusion that the function of CT135 is restricted to host-pathogen interactions within the in vivo infection environment.
Analysis of CT135 from our serovar D reference strain by CEL I digestion revealed numerous CT135 polymorphisms suggesting a mixture of phenotypes differing in virulence. The heterogeneity in CT135 was proven by direct sequencing from plaque-cloned isolates, also revealing that all genotypes had disruptions in CT135. Analysis of the 15 C. trachomatis reference strains by CEL I digestion showed that CT135 polymorphisms were not unique to serovar D. There was considerable CT135 diversity in most serovars, suggesting that similar disrupting mutations occur in these strains. The exception was the lack of CT135 polymorphisms in serovars K, L1, L2, and L3, the significance of which is unknown. Interestingly, sequencing of CT135 from clinical samples that had undergone minimal in vitro passages in cell culture showed no CT135 disruptions by either capillary sequencing or CEL I digestion. These findings suggest that most reference strains have lost their intact CT135, probably due to negative selection pressure, while there is an in vivo positive selection for intact CT135 in clinical isolates. A caveat, however, is that the clinical diagnosis associated with these clinical samples is not known. Therefore, future epidemiological genotyping studies of CT135 should minimally incorporate comparisons of symptomatic and asymptomatic subjects to more fully appreciate a potential role for CT135 in the pathogenesis of human infection.
CT135 encodes a hypothetical protein unique to the Chlamydiaceae. All sequenced C. trachomatis genomes and the C. muridarum genome contain a CT135 ortholog located immediately downstream from CT134, a gene that also encodes a hypothetical protein with orthologs in other Chlamydiaceae. The CT134 and CT135 ORFs are on the same DNA strand and are separated by only 58 bp in the C. trachomatis (26) and C. muridarum (22) genomes, suggesting that these genes might form a bicistronic operon with a related function (20). Consistent with this theory, it was shown that in C. trachomatis-infected human epithelial cells, both genes were coordinately upregulated at 3 h p.i. (3); in contrast, the adjacent genes CT133 and CT136 exhibited upregulation at 1 and 8 h p.i., respectively. However, in Chlamydophila species, the intergenic regions between CT134 and CT135 are larger than what would be predicted for a bicistronic operon. The conservation and organization of these genes, particularly among human C. trachomatis isolates, suggest that they could be common virulence factors that play important roles in the pathogenesis of chlamydial infection and disease. In support of the importance of CT135 in chlamydial pathogenesis, others have reported similar frameshift mutations in this gene in strains differing in in vitro or in vivo pathogenic properties (6, 10, 21, 25, 27). Unlike our findings, however, those studies identified mutations in multiple other genetic loci, making it impossible to associate a single gene with a pathogenic phenotype. Nevertheless, those studies collectively support our conclusion that CT135 is an important chlamydial virulence factor. Our CT135 sequence results are confounding, given that both the D-EC and D-LC strains contain mutations predicted to disrupt the ORF as initially annotated (26). Given the distinct in vivo phenotypic differences between the strains, it is likely that D-LC CT135 encodes a protein with enhanced virulence properties. Another possible explanation is that CT135 is an antivirulence factor and that it is functional in D-EC but not in D-LC (14). However, the fact that all clinical samples examined in this study possessed intact CT135 makes the second possibility less likely.
A fundamental question is how does CT135 function in the pathogenesis of in vivo chlamydial infection? The difference in virulence between the two strains was the ability of D-LC to sustain infections with greater organism burdens and infection durations and, importantly, to generate ascending infection of the upper reproductive tract. It is possible that a single metabolic, structural, or secreted protein could exhibit this phenotypic property, an argument supported by the critical role described for the C. trachomatis tryptophan synthase gene in the pathogenesis of human genital but not ocular infections (5, 9). It is also possible that CT135 might be a regulatory gene that affects the expression of multiple genes in response to in vivo environmental stimuli that are important to in vivo growth or survival. Interestingly, transcriptional profiling studies have shown that the levels of expression of CT135 and its putative bicistronic partner, CT134, are increased 2.7-fold when C. trachomatis is grown in IFN-γ-treated human epithelial cells (2). Under this growth condition, C. trachomatis exhibits differential gene expression that has been postulated to be part of a stress response stimulon. Our results did not show a difference in the susceptibilities of D-EC and D-LC strains to growth inhibition by IFN-γ in cultured mouse cells. However, the effects of IFN-γ in this in vitro environment may not be sufficiently selective or hostile to differentiate the strains.
Our findings could have important implications for the understanding of human disease and the utilization of the murine model for studying human infection and immunity. CT135 genotyping of C. trachomatis-infected humans might be useful for identifying individuals at greater risk for complicated postinfection sequelae. This will require sequencing of CT135 from patients with asymptomatic, symptomatic, or complicated infections. Also, identifying additional C. trachomatis strains from other serovars causing STIs that demonstrate an enhanced murine virulence phenotype would significantly improve the mouse genital tract small animal model to study the pathophysiology and immunity of human urogenital infection.
In summary, we have identified a single genetic locus that significantly changes the in vivo pathogenicity of a human clinical isolate for the female mouse genital tract. Future studies will focus on determining whether CT135 plays a similar role in other C. trachomatis serovars for the murine genital tract and defining the role of CT135 in the pathogenicity of human infection and disease.
Supplementary Material
Acknowledgments
This work was supported in part by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
We thank Frank DeLeo for critical review of the manuscript, Rebecca Rosenke and Jill Harmon for technical assistance, Mark VanRaden for performing statistical analyses, Anita Mora for assistance with graphic arts, and Kelly Matteson for manuscript preparation.
Editor: R. P. Morrison
Footnotes
Published ahead of print on 14 June 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Al-Zeer, M. A., H. M. Al-Younes, H. P. Braun, J. Zerrahn, and T. F. Meyer. 2009. IFN-gamma inducible Irga6 mediates host resistance against Chlamydia trachomatis via autophagy. PLoS One 4:e4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belland, R. J., D. E. Nelson, D. Virok, D. D. Crane, D. Hogan, D. Sturdevant, W. L. Beatty, and H. D. Caldwell. 2003. Transcriptome analysis of chlamydial growth during IFN-gamma-mediated persistence and reactivation. Proc. Natl. Acad. Sci. U. S. A. 100:15971-15976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belland, R. J., G. Zhong, D. D. Crane, D. Hogan, D. Sturdevant, J. Sharma, W. L. Beatty, and H. D. Caldwell. 2003. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc. Natl. Acad. Sci. U. S. A. 100:8478-8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernstein-Hanley, I., J. Coers, Z. R. Balsara, G. A. Taylor, and M. N. Starnbach. 2006. The p47 GTPases lgtp and lrgb10 map to the Chlamydia trachomatis susceptibility locus Ctrq-3 and mediate cellular resistance in mice. Proc. Natl. Acad. Sci. U. S. A. 103:14092-14097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caldwell, H. D., H. Wood, D. Crane, R. Bailey, R. B. Jones, D. Mabey, I. Maclean, Z. Mohammed, R. Peeling, C. Roshick, J. Schachter, A. W. Solomon, W. E. Stamm, R. J. Suchland, L. Taylor, S. K. West, T. C. Quinn, R. J. Belland, and G. McClarty. 2003. Polymorphisms in Chlamydia trachomatis tryptophan synthase genes differentiate between genital and ocular isolates. J. Clin. Invest. 111:1757-1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlson, J. H., S. F. Porcella, G. McClarty, and H. D. Caldwell. 2005. Comparative genomic analysis of Chlamydia trachomatis oculotropic and genitotropic strains. Infect. Immun. 73:6407-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlson, J. H., W. M. Whitmire, D. D. Crane, L. Wicke, K. Virtaneva, D. E. Sturdevant, J. J. Kupko, S. F. Porcella, N. Martinez-Orengo, R. A. Heinzen, L. Kari, and H. D. Caldwell. 2008. The Chlamydia trachomatis plasmid is a transcriptional regulator of chromosomal genes and a virulence factor. Infect. Immun. 76:2273-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darville, T., C. W. J. Andrews, K. K. Laffoon, W. Shymasani, L. R. Kishen, and R. G. Rank. 1997. Mouse strain-dependent variation in the course and outcome of chlamydial genital tract infection is associated with differences in host response. Infect. Immun. 65:3065-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fehlner-Gardiner, C., C. Roshick, J. H. Carlson, S. Hughes, R. J. Belland, H. D. Caldwell, and G. McClarty. 2002. Molecular basis defining human Chlamydia trachomatis tissue tropism. A possible role for tryptophan synthase. J. Biol. Chem. 277:26893-26903. [DOI] [PubMed] [Google Scholar]
- 10.Kari, L., W. M. Whitmire, J. H. Carlson, D. D. Crane, N. Reveneau, D. E. Nelson, D. C. Mabey, R. L. Bailey, M. J. Holland, G. McClarty, and H. D. Caldwell. 2008. Pathogenic diversity among Chlamydia trachomatis ocular strains in non-human primates is affected by subtle genomic variations. J. Infect. Dis. 197:449-456. [DOI] [PubMed] [Google Scholar]
- 11.Laga, M., A. Manoka, M. Kivuvu, B. Malele, M. Tuliza, N. Nzila, J. Goeman, F. Behets, V. Batter, M. Alary, et al. 1993. Non-ulcerative sexually transmitted diseases as risk factors for HIV-1 transmission in women: results from a cohort study. AIDS 7:95-102. [DOI] [PubMed] [Google Scholar]
- 12.Reference deleted.
- 13.Matsumoto, A., H. Izutsu, N. Miyashita, and M. Ohuchi. 1998. Plaque formation by and plaque cloning of Chlamydia trachomatis biovar trachoma. J. Clin. Microbiol. 36:3013-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maurelli, A. T. 2007. Black holes, antivirulence genes, and gene inactivation in the evolution of bacterial pathogens. FEMS Microbiol. Lett. 267:1-8. [DOI] [PubMed] [Google Scholar]
- 15.Molyneaux, D. H., P. J. Hotez, and A. Fenwick. 2005. “Rapid-impact interventions”: how a policy of integrated control for Africa's neglected tropical diseases could benefit the poor. PLoS Med. 2:e336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morrison, R. P., K. Feilzer, and D. B. Tumas. 1995. Gene knockout mice establish a primary protective role for major histocompatibility complex class II-restricted responses in Chlamydia trachomatis genital tract infection. Infect. Immun. 63:4661-4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson, D. E., D. P. Virok, H. Wood, C. Fehlner-Gardiner, R. M. Johnson, W. M. Whitmire, D. D. Crane, O. Steele-Mortimer, L. Kari, G. McClarty, and H. D. Caldwell. 2005. Chlamydial interferon gamma immune evasion is linked to host infection tropism. Proc. Natl. Acad. Sci. U. S. A. 102:10658-10663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oleykowski, C. A., C. R. Bronson Mullins, A. K. Godwin, and A. T. Yeung. 1998. Mutation detection using a novel plant endonuclease. Nucleic Acids Res. 26:4597-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perry, L. L., H. Su, K. Feilzer, R. Messer, S. Hughes, W. Whitmire, and H. D. Caldwell. 1999. Differential sensitivity of distinct Chlamydia trachomatis isolates to IFN-γ-mediated inhibition. J. Immunol. 162:3541-3548. [PubMed] [Google Scholar]
- 20.Price, M. N., K. H. Huang, E. J. Alm, and A. P. Arkin. 2005. A novel method for accurate operon predictions in all sequenced prokaryotes. Nucleic Acids Res. 33:880-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramsey, K. H., I. M. Sigar, J. H. Schripsema, C. J. Denman, A. K. Bowlin, G. A. Myers, and R. G. Rank. 2009. Strain and virulence diversity in the mouse pathogen Chlamydia muridarum. Infect. Immun. 77:3284-3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. DeBoy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reed, L. J., and H. Muench. 1938. A simple method of estimating 50 percent endpoints. Am. J. Hyg. (Lond.) 27:493-497. [Google Scholar]
- 24.Roshick, C., H. Wood, H. D. Caldwell, and G. McClarty. 2006. Comparison of gamma interferon-mediated antichlamydial defense mechanisms in human and mouse cells. Infect. Immun. 74:225-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seth-Smith, H. M., S. R. Harris, K. Persson, P. Marsh, A. Barron, A. Bignell, C. Bjartling, L. Clark, L. T. Cutcliffe, P. R. Lambden, N. Lennard, S. J. Lockey, M. A. Quail, O. Salim, R. J. Skilton, Y. Wang, M. J. Holland, J. Parkhill, N. R. Thomson, and I. N. Clarke. 2009. Co-evolution of genomes and plasmids within Chlamydia trachomatis and the emergence in Sweden of a new variant strain. BMC Genomics 10:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, L. Olinger, R. L. Tatusov, Q. Zhao, E. V. Koonin, and R. W. Davis. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754-759. [DOI] [PubMed] [Google Scholar]
- 27.Suchland, R. J., B. M. Jeffrey, M. Xia, A. Bhatia, H. G. Chu, D. D. Rockey, and W. E. Stamm. 2008. Identification of concomitant infection with Chlamydia trachomatis IncA-negative mutant and wild-type strains by genomic, transcriptional, and biological characterizations. Infect. Immun. 76:5438-5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomson, N. R., M. T. Holden, C. Carder, N. Lennard, S. J. Lockey, P. Marsh, P. Skipp, C. D. O'Conner, I. Goodhead, H. Norbertzake, B. Harris, D. Ormond, R. Rance, M. A. Quail, J. Parkhill, R. S. Stephens, and I. N. Clarke. 2008. Chlamydia trachomatis: genome sequence analysis of lymphogranuloma venereum isolates. Genome Res. 18:161-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.World Health Organization. 2003. Alliance for the global elimination of blinding trachoma by 2020. World Health Organization, Geneva, Switzerland.
- 30.World Health Organization. 2001. Global prevalence and incidence of selected curable sexually transmitted infections: overview and estimates. World Health Organization, Geneva, Switzerland.
- 31.Yuan, Y., Y.-X. Zhang, N. G. Watkins, and H. D. Caldwell. 1989. Nucleotide and deduced amino acid sequences for the four variable domains of the major outer membrane proteins of the 15 Chlamydia trachomatis serovars. Infect. Immun. 57:1040-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.