Abstract
The ability to count bacteria associated with reef-building corals in a rapid, reliable, and cost-effective manner has been hindered by the viscous and highly autofluorescent nature of the coral mucus layer (CML) in which they live. We present a new method that disperses bacterial cells by trypsinization prior to 4′,6-diamidino-2-phenylindole (DAPI) staining and quantification by epifluorescence microscopy. We sampled seawater and coral mucus from Porites lobata from 6 reef sites influenced by wastewater intrusion and 2 reef sites unaffected by wastewater in Hawaii. Bacterial and zooxanthella abundances and cell sizes were quantified for each sample. Bacteria were more abundant in coral mucus (ranging from 5.3 × 105 ± 1.0 × 105 cells ml−1 to 1.8 × 106 ± 0.2 × 106 cells ml−1) than in the surrounding seawater (1.9 × 105 ± 0.1 × 105 cells ml−1 to 4.2 × 105 ± 0.2 × 105 cells ml−1), and the mucus-associated cells were significantly smaller than their seawater counterparts at all sites (P < 0.0001). The difference in cell size between mucus- and seawater-associated bacteria decreased at wastewater-influenced sites, where simultaneously mucus bacteria were larger and seawater bacteria were smaller than those at uninfluenced sites. The abundance of zooxanthellae in mucus ranged from 1.1 × 105 ± 0.1 × 105 cells ml−1 to 3.4 × 105 ± 0.3 × 105 cells ml−1. The frequency of dividing cells (FDC) was higher in the surrounding seawater than in mucus, despite finding that a 1,000-fold-higher zooxanthella biovolume than bacterial biovolume existed in the CML. Establishment of a standardized protocol for enumeration will provide the field of coral microbial ecology with the urgently needed ability to compare observations across studies and regions.
The extremely viscous and highly autofluorescent nature of coral mucus has been a major challenge in developing enumeration techniques and has limited our ability to study the ecological interactions among coral mucus layer (CML)-associated microbial communities. Only a few studies have used direct counts to quantify bacteria in the CML, and the methods and subsequent results vary widely. The techniques have included scanning electron microscopy (SEM) (34), phase-contrast microscopy (27), and epifluorescent microscopy using a variety of stains (acridine orange staining [8], SYBR gold [20], and 4′,6-diamidino-2-phenylindole [DAPI] [3]). Bacterial abundances reported from these studies spanned more than 5 orders of magnitude (from 1.6 × 102 cells [cm2]−1 using acridine orange [8] to 6.2 × 107 cells [cm2]−1 using SYBR gold [20]), and some of the studies are difficult to compare to each other because different units were used, such as cells ml−1 of mucus and cells (cm2)−1 of coral. Some variation in abundance is likely due to differences in mucus sampling methods and differences among coral species. However, the enormous quantity of autofluorescence emitted in green and red wavelengths found in most coral species creates a substantial challenge for reliably counting fluorescently stained cells in that portion of the spectrum, because many of the particles are bacterium sized. Many of these same particles could be visible with phase-contrast microscopy as well. Thus far, researchers quantifying CML-associated bacteria using epifluorescence microscopy have prepared their samples by following well-established protocols that were developed for seawater. We suggest that the viscous and autofluorescent nature of coral mucus may require some modifications from standard seawater protocols for epifluorescence microscopy to be most effective.
SEM is an alternative to fluorescence-dependent techniques. It has the advantage of acquiring images with sufficient detail to distinguish among particles and cells, but this method is time-consuming, visualizes only the surface of the sample, and is not widely available or affordable enough for it to be a standard field protocol. An additional limitation is that most studies that have employed SEM for CML observation have found bacteria to be too dispersed to count in a reasonable number of micrographs (8, 19).
Here we present a new method that disperses bacterial cells by enzymatically digesting the mucus with trypsin (an adaptation of routine cellular biology cell line culture procedures) and subsequently staining the cells with DAPI for rapid quantification using epifluorescence microscopy. DAPI fluoresces in the blue end of the spectrum, and its emission does not overlap with the autofluorescence of the mucus samples. This method is rapid, uses reagents and equipment readily available in microbial ecology laboratories, and can provide necessary information for studies of the ecology of microbial cells associated with mucus. It may also be helpful for studies of other aquatic gel-associated microbial communities.
This visualization capability revealed that bacteria living with the reef-building coral Porites lobata were significantly smaller than their water-associated counterparts and that this difference is reduced in reefs heavily influenced by anthropogenic impacts. There is only one other report that we are aware of that observed small bacterial cell size in mucus from corals (of the genus Fungia), but that study did not quantify cell size (34). Given that mucus is a carbon-rich environment (6, 11, 12, 18, 24, 25, 31), this discovery is counterintuitive. It highlights questions regarding the ecological interactions that must occur in situ to select for small cell size in such a rich environment (3, 4, 7, 8, 11, 25, 34).
MATERIALS AND METHODS
Sampling sites.
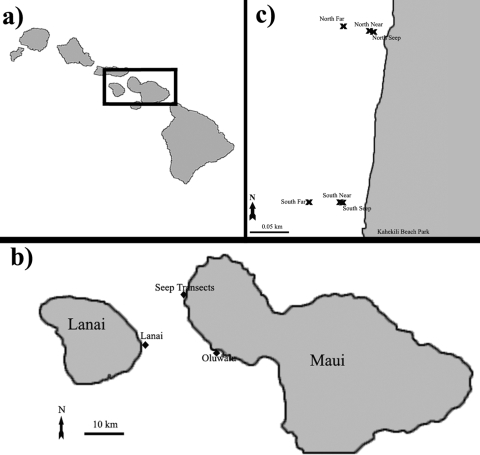
In July 2009, samples were collected from 7 different sites on the west side of Maui, HI, and 1 site on the east side of Lanai, HI (Fig. 1). There were two transects (north and south; 3 sites each) that began at freshwater seeps known to carry wastewater treatment plant effluent to the reef (10). On both transects, no living coral existed directly at the seep sites, so the “near” sites were chosen as the closest living coral cover offshore of the seep (∼2 m away from each seep), and the “far” sites were ∼50 m west (offshore) of the near sites. All 6 sites (seep, near, and far sites for both the north and south transects) showed the influence of wastewater in nitrogen isotope fractionation throughout the year (10). The two control sites that did not show evidence of wastewater influence were at Oluwalu and Lanai, HI (Fig. 1) (10). Samples were also collected from flowthrough seawater aquaria maintained at Scripps Institution of Oceanography (SIO), La Jolla, CA, for comparison.
FIG. 1.
(a) Overview of the Hawaiian Islands. (b) Our 3 major sampling locations on Maui and Lanai. (c) Close-up view of the two wastewater-influenced transects.
Sample collection.
Triplicate samples were collected by scuba diving at each site of seawater 0.5 m above the substrate (using 4-liter Cubitainers) and of coral mucus from Porites lobata (using 10-ml syringes) all between 1 to 5 m in depth. The two exceptions were at the seep sites, where only seawater was collected because no live coral cover existed. Samples were kept at ambient seawater temperature in the dark until arrival at the lab (<1 h), where they were fixed with 2% 0.02-μm-filtered formaldehyde. The mucus samples formed a compact mucus ball in the syringe, and that ball was removed with a sterile pipette tip, rinsed briefly in autoclaved 0.02-μm-filtered seawater to removed loosely associated seawater bacteria, and then fixed. All samples were allowed to fix at 4°C for at least 4 h. Mucus samples were stored frozen without filtering, while water samples were filtered on 0.2-μm polycarbonate filters (17), dried, wrapped in foil, and stored at −20°C until processing.
For the coral species Acropora yongei, maintained in aquaria at SIO, two mucus collection methods were compared. Triplicate mucus samples were collected using a 10-ml syringe, as done in the field, and an additional set of samples were collected by quickly rinsing the coral with 0.02-μm-filtered autoclaved seawater (FASW) to remove any loosely associated bacteria and then exposing the fragments to air for 3 min over a sterile 1.5-ml tube. Triplicate seawater samples were collected from the tank using sterile 15-ml Falcon tubes from the inflow valve and from directly above the coral samples. All samples were fixed as described above.
Seawater carryover experiment.
To quantify the number of water-associated bacteria remaining in coral mucus samples, A. yongei fragments were sampled in triplicate using the two methods described above (air exposure and syringe). A total of 200 μl of each mucus sample was removed with a cut, sterile 1,000-μl tip and put into 1 ml of FASW with 2% 0.02-μm-filtered formaldehyde. (Cut tips were used to reduce physical sheering of the viscous sample during transfer.) Samples were gently inverted three times and incubated at room temperature for 10 min to allow any loosely associated cells to disassociate. Samples were spun for 45 s at 4,000 × g to gently pellet the mucus. A total of 1 ml of supernatant was removed, fixed at 4°C overnight, and filtered onto 0.2-μm polycarbonate filters to enumerate (see below) as “carryover,” while the mucus pellets had another 2% 0.02-μm-filtered formaldehyde added to them before being put at 4°C overnight. Mucus samples were then treated and enumerated by following the trypsin protocol described below. Tank seawater was collected (as described above) and enumerated (as described below) at the same time, and FASW controls were carried along as well.
Mucus-associated bacterial dispersal treatment experiments.
Fixed mucus from three Porites lobata colonies was used for dispersal experiments. No-treatment controls for each colony were included in each trial. Three different methods were initially tried on 1 ml of 1:10 dilutions of mucus in 0.02 μm filtered autoclaved Milli-Q water, as follows: (i) 10% methanol incubated at 35°C for 15 min and sonicated on ice for three rounds of 1 min at 50% power and 30 s rest (adapted from reference 22), (ii) 20 μg ml−1 proteinase K digestion at room temperature for 10 min (adapted from reference 3), and (iii) 10 μM EDTA incubation on ice, followed by 0.4% trypsin incubation for 10 min at 37°C (quenched on ice) (adapted from reference 26). All samples (including untreated controls) were incubated for 8 min at 4°C with 5 μg ml−1 DAPI before being filtered onto black 0.2-μm polycarbonate filters. Filters were dried in the dark, wrapped in foil, and stored at −20°C until prepared for counting (see below).
To find the best incubation length for 0.4% trypsin at 37°C, the subsamples of mucus from each of the three corals were incubated for 0, 5, 10, 15, 20, 25, and 30 min (see the supplemental material for additional optimization experiments). All other parameters were maintained as described above. The following three different protocols were tried after the 15 min of trypsin incubation to quench autofluorescence present in the mucus samples: (i) 2 mg ml−1 of crystal violet was added at the DAPI staining step (adapted from reference 15), (ii) 500 μl 1× acidic Alcian blue was incubated on the filter (after DAPI staining) while the sample was in the filter tower for 45 s before filtering through (adapted from reference 2), and (iii) 1 mg ml−1 FeSO4 incubated on ice for 30 min before DAPI staining (adapted from reference 1). Three trials examined the effect of prefiltration after trypsin treatment with 10-μm-, 3-μm-, and 1-μm-pore-size filters on the abundances of bacteria, zooxanthellae, and autofluorescent particles in subsequent mucus samples collected on 0.2-μm polycarbonate filters (17).
Dispersal procedure.
The final procedure used for obtaining all results presented after the trials described above was dilution of fixed mucus at 1:10 in 0.02 μm filtered autoclaved Milli-Q water using a cut, sterile 1-ml tip, vortexing for 10 s at maximum speed, incubation for 30 min on ice with 10 μM EDTA (to chelate cations that inhibit the enzymatic activity of trypsin), addition of 0.4% trypsin, incubation for 15 min at 37°C, quenching on the ice-water slurry, addition of 5 μg ml−1 DAPI, incubation for 8 min in the dark at 4°C, immediate filtering onto a 0.2-μm black polycarbonate filter, drying in the dark, and store wrapping in foil at −20°C until slide preparation.
Counting and sizing of cells.
Stored DAPI-stained 0.2-μm filters were thawed, and one-quarter of each filter was mounted onto glass slides using Vectashield mounting medium (Vector Labs, CA) mixed with 2 μg ml−1 of the lipid stain 10-N-nonyl-acridine orange (NAO; Sigma-Aldrich, St. Louis, MO), while the rest of the filter was archived at −20°C. NAO provides a secondary stain to confirm the DAPI counts. Only dual-stained DAPI-NAO cells were enumerated. Eubacterial fluorescence in situ hybridization (FISH) probes were also used to confirm DAPI counts (Cy5-labeled EUB-388 I to III and a non-EUB-388 control) (protocol adapted from reference 13). Zooxanthellae were counted at 400× magnification, and bacterial cells and the frequency of dividing cells (FDC) (14) were counted at 1,000× magnification on a Nikon Eclipse TE-2000U. Images were taken using the NIS Elements software program. The dimensions of 100 to 200 bacterial cells and 20 zooxanthella cells were measured from each sample using the NIS-Elements measurement function in the DAPI channel for bacteria and the tetramethyl rhodamine isocyanate (TRITC) channel for zooxanthellae. Biovolume was estimated by the formula V = (π/4)W2 × (L − W/3), where L is the length and W is the width (5). DAPI, FITC, TRITC, and 5-cyano-2,3-ditoyl tetrazolium chloride (CTC) filter sets were used to quantify fluorescent particles from the quenching trials. To verify the abundances counted with the epifluorescence microscope, two other microscopy techniques were employed. The same slides were enumerated using laser scanning confocal microscopy z-stacks (Nikon Eclipse TE-2000U) to include the full z-axis dimension of each sample. Scanning electron microscopy (SEM) was done by following an established protocol (34) for half of the archived portion of filters.
Statistics.
The software program JMP was used for statistical analysis of cell length data. Cell lengths from water and coral mucus bacteria at all sites were transformed using Box-Cox analysis and tested for normality using a Shapiro-Wilk test. Analysis of variance (ANOVA) was performed to quantify the effect of “site” and “source” (cells from coral mucus or water) on cell length. A post hoc Tukey-Kramer honest significant difference (HSD) test was performed to determine which sets of sites were significantly different from each other for each source.
RESULTS
Cell counting methods.
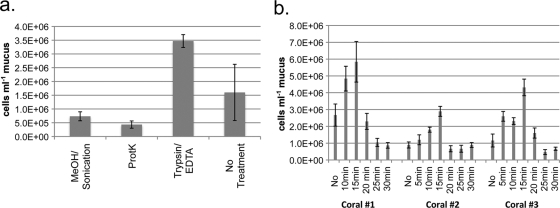
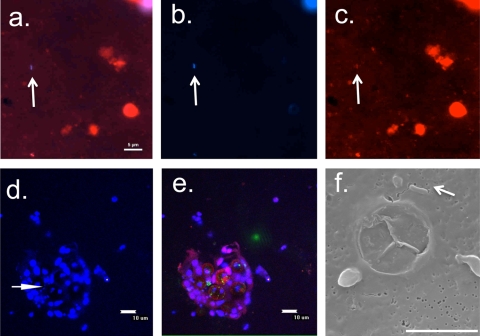
Our initial attempts to adapt methanol, sonication, and proteinase K protocols yielded lower counts than the no-treatment control protocol (Fig. 2 a). Dissociation of cells with a trypsin digestion was more successful than use of other treatments and the no-treatment controls. The error in counting samples from P. lobata was reduced by more than 9-fold compared to that in untreated samples when digested with trypsin, while total cell counts were >2.5 times higher in trypsin-treated A. yongei samples than in untreated samples. We found that 15 min of 0.4% trypsin incubation maximized the number of DAPI-stained cells visualized by epifluorescent microscopy (Fig. 2b). Attempts to quench autofluorescent particles in the coral mucus were marginally successful (see Fig. S1 in the supplemental material); however, these treatments also reduced our ability to count zooxanthella cells (see Fig. S2 in the supplemental material). Prefiltration experiments to physically remove autofluorescent particles after trypsin treatment reduced final bacterial cell counts by a minimum of 2-fold (with a 10-μm prefilter) or removed almost all cells (3-μm and 1-μm prefilters). DAPI cell counts were successfully verified with a secondary label using both NAO and Eub388 fluorescence in situ hybridization (FISH) probes. The final dispersal procedure is described above but, in short, involved 15 min of trypsin incubation, no quenching of autofluorescence, 5 μg ml−1 DAPI staining, filtration, and mounting the slide with Vectashield mixed with NAO to stain lipids as a secondary stain—only dual NAO-DAPI-stained cells were enumerated (Fig. 3 a to c).
FIG. 2.
(a) Average bacterial cell counts obtained from 3 P. lobata mucus samples, either untreated (“No Treatment”) or treated with methanol and sonication, subjected to proteinase K digest, or treated with EDTA followed by 10 min of trypsin incubation. (b) Bacterial cell counts obtained from P. lobata mucus treated with 30 min of EDTA incubation followed by varied trypsin incubation durations.
FIG. 3.
Representative images of P. lobata mucus treated with the trypsin protocol. (a to c) Single images of a mucus sample, each with an arrow pointing toward a bacterial cell (the scale bar is 5 μm). Epifluorescence images of a dual-stained sample showing red (NAO and chlorophyll) and blue (DAPI) channels overlaid (a), the blue channel alone (b), and the red channel alone (c). (d and e) Integrated three-dimensional stacks displayed in two dimensions of a single field of view taken with laser scanning confocal microscopy, with an arrow pointing toward several bacterial cells (the scale bar is 10 μm). (d) Blue channel shown from a laser scanning confocal image, showing the DAPI signal. The arrow points toward bacterial cells in a cluster of zooxanthella cells. (e) Red, green, and blue channels from the same image as shown in panel d, showing the autofluorescence detected in the red and green channels. (f) SEM image of a bacterium (arrow) in the mucus, with a 5-μm scale bar.
The intrusion level of bacteria from surrounding seawater was low with both the syringe and air exposure mucus collection methods (see Fig. S3 in the supplemental material). Air-exposed samples carried over 3.9 to 6.8% of total cell counts from entrained seawater in the mucus, while syringe-collected mucus brought in 5.8 to 11.5% of total counts. These percentages are within the error of any given count.
Abundances.
Bacterial abundances in P. lobata mucus ranged from 5.3 × 105 ± 1.0 × 105 cells ml−1 at Oluwalu to 1.8 × 106 ± 0.2 × 106 cells ml−1 at the north near site (Table 1). The surrounding seawater at each site had concentrations ranging from 1.9 × 105 ± 0.1 × 105 cells ml−1 at Oluwalu to 4.1 × 105 ± 0.2 × 105 cells ml−1 at the south near site (Table 1). Zooxanthella cell abundance in P. lobata mucus ranged from 1.1 × 105 ± 0.1 × 105 cells ml−1 at the north far site to 3.4 × 105 ± 0.3 × 105 cells ml−1 at Oluwalu (Table 1). The frequency of dividing cells (FDC) was higher in the surrounding seawater than in the coral mucus (Table 1).
TABLE 1.
Seawater and coral mucus cell data from all sitesa
| Site | Bacterial abundance (105 cells ml−1) |
Zooxanthella abundance in mucus (105 cells ml−1) | Bacterial FDC (% of DAPI count) |
Avg cell length of bacteria (μm) |
|||
|---|---|---|---|---|---|---|---|
| Mucus | Seawater | Mucus | Seawater | Mucus | Seawater | ||
| Lanai | 13.6 ± 0.2 | 3.3 ± 0.1 | 1.1 ± 0.2 | 3 | 7 | 0.36 ± 0.01 | 0.78 ± 0.04 |
| Oluwalu | 5.3 ± 0.1 | 1.9 ± 0.1 | 3.4 ± 0.3 | 4 | 11 | 0.44 ± 0.02 | 0.86 ± 0.03 |
| North far | 15.3 ± 0.3 | 2.6 ± 0.1 | 1.1 ± 0.1 | 7 | 6 | 0.49 ± 0.02 | 0.62 ± 0.02 |
| North near | 13.8 ± 0.3 | 4.1 ± 0.2 | 1.4 ± 0.1 | 5 | 12 | 0.52 ± 0.02 | 0.73 ± 0.03 |
| North seep | NA | 3.4 ± 0.3 | NA | NA | 12 | NA | 0.67 ± 0.02 |
| South far | 12.8 ± 0.2 | 3.0 ± 0.1 | 2.3 ± 0.2 | 6 | 9 | 0.54 ± 0.02 | 0.76 ± 0.03 |
| South near | 17.9 ± 0.2 | 3.6 ± 0.1 | 1.8 ± 0.2 | 5 | 11 | 0.58 ± 0.02 | 0.75 ± 0.03 |
| South seep | NA | 3.8 ± 0.1 | NA | NA | 11 | NA | 0.69 ± 0.02 |
NA, not applicable.
The bacterial abundances assessed by laser scanning confocal microscopy using z-stacks to image the full three dimensions of each sample were the same, within error, as the epifluorescence counts for both water and trypsin-treated coral mucus samples (Fig. 3d and e). Tenfold fewer bacterial cells (a maximum of 3.7 × 105 cells ml−1) were observed by scanning electron microscopy (SEM) than by either of the other methods in P. lobata mucus (Fig. 3f).
Cell size.
Bacterial cells from P. lobata mucus were significantly (P < 0.0001) shorter in length than their seawater counterparts at every site (Table 1). Seawater bacteria were 2.0 to 2.2 times longer than mucus-associated bacteria at sites uninfluenced by wastewater effluent (Lanai and Oluwalu) and 1.3 to 1.4 times longer at effluent-receiving sites (all north and south sites). The smallest length measured for any zooxanthella cell was 6.92 μm, which is more than 12 times longer than the longest average cell length from any mucus-associated bacterial community (0.58 μm; south near site). Bacterial cells associated with P. lobata mucus had an average volume of 5.2 × 10−2 ± 0.6 × 10−2 μm3, while zooxanthella cells were more than 4 orders of magnitude larger, with an average volume of 8.7 × 102 ± 3.0 × 102 μm3.
DISCUSSION
Digestion of the mucus sample before filtration dramatically improved our ability to visualize and enumerate coral-associated bacterial cells. Trypsin is a serine protease that specifically cleaves peptide chains on the carboxyl side of lysine or arginine. Hydrolysis is slowed if an acidic residue is on either side of the cleavage site, and cleavage does not occur at all if a proline residue is located on the C-terminal side of the site (9). These characteristics contribute to the variation in trypsin sensitivity that has been observed among different cell lines (29). In the case of marine bacteria, we observed at least one culture that appears insensitive to trypsinization and two distinct natural assemblages (CML associated and seawater associated) that had different levels of sensitivity (see the supplemental material). The protocol we have proposed here may be useful for microbial ecology studies of any aquatic gel-associated community, but the trypsinization step should be optimized for each new type of sample. Our results provide a practical starting place for designing optimization experiments for other marine samples.
Observed cell counts are always considered to be a minimum estimate of actual abundance. The two main things that we wanted to verify during the development of this method were (i) that all “cells” counted were indeed actual cells and (ii) that the sample preparation sufficiently dispersed the cells so that they could be reliably counted in two dimensions using epifluorescence microscopy. To address the first concern, we used eubacterial FISH probes as well as a lipid stain (NAO) to confirm the DAPI counts. The difference between DAPI-only cell abundance and dual-stained cell abundance was within the errors of the counts (<2.5%). In the interest of creating a streamlined protocol, we used the equally effective NAO dual-staining method in place of FISH for routine counting. Unstained mucus samples did not fluoresce in the blue channel, which also supported the use of DAPI rather than other stains.
We used laser scanning confocal microscopy and SEM to address the second point. Confocal microscopy has the advantage of being able to optically slice the sample and create three-dimensional images that include the insides of potentially thick mucus particles. The fact that confocal counts were within errors (±<2.5%) of epifluorescence counts suggests that the trypsin treatment and subsequent volume of mucus filtered sufficiently dispersed the sample such that very few cells were out of the primary plane of focus. The disadvantage of confocal microscopy is that it remains a fluorescence-dependent technique. SEM can image cell shape independently of fluorescence but lacks the ability to see beyond the surface of the sample. The fact that we generally observed 10-fold fewer cells with SEM than with fluorescence-based techniques, by following an established protocol (34), could be due to cell loss during preparation or gold-palladium sputtering that obscured the morphology of cells still embedded in a thin layer of mucus. It is also possible that some of the particles we counted were, in fact, not cells. However, given that both FISH probes and a lipid stain confirmed the DAPI cell counts, it is unlikely that this is the case. Our observations are in accord with two other studies that found that SEM underestimated bacteria associated with coral mucus (8, 19).
Two general sources of potential error in coral-associated bacterial counts are as follows: (i) missed cells that were too deeply entrained in mucus to either be stained or be visualized and (ii) the incorrect enumeration of bacterium-sized autofluorescent particles in green and red areas of the spectrum. We observed an average of 9.3 × 106 ± 0.7 × 106 green (FITC) particles per milliliter of untreated mucus in P. lobata samples, while average DAPI-stained bacterial concentrations were never greater than 1.8 × 106 ± 0.2 × 106 cells ml−1 of mucus. If a stain with emission in the green portion of the spectrum was used, such as SYBR green I or acridine orange, bacterial abundance would have been overestimated by at least 1 order of magnitude.
We found that mucus collection by either air exposure or syringe suction is appropriate. The bacterial cell abundances we observed are within the range of other reported values. Some degree of species-to-species variability is expected (as we observed between A. yongei and P. lobata), and thus, standardization of a method becomes even more important. Given that there are less than 10 published studies (3, 8, 16, 21, 27, 28, 34) reporting directly counted bacterial concentrations in the mucus layer, and none of those studies examined the same species of coral from the same geographic region nor used the same methods, we are at the very early stages of acquiring a baseline as a field. This is an ideal time to develop a standard method on which all subsequent studies can build. This protocol is a useful starting point in the development of a toolbox of consistent methods that will allow microbial ecology hypotheses to be tested in the viscous, autofluorescent mucus environment.
This is among the first reports of zooxanthella concentrations in coral mucus. They were far more abundant in P. lobata than expected. Cell biovolume measurements show that zooxanthellae are more than 4 orders of magnitude larger than bacteria. These cell volumes are likely an underestimate of the true volume, due to shrinking that occurs during fixation (see reference 23 for a full discussion). Zooxanthella abundance was only 1 order of magnitude less than bacterial abundance, indicating that 1,000-fold more algal biomass existed in the mucus layer than bacterial biomass. Given that dinoflagellates tend to exude photosynthate, and bacteria readily grow using that carbon source (3, 11, 31), why are mucus-associated bacteria so much smaller than their seawater counterparts? The opposite might be expected in an organically enriched environment. The higher frequency of dividing cells (FDC) in surrounding seawater than in mucus may suggest that CML bacteria are not capitalizing on enrichment from zooxanthella exudate for growth or that the zooxanthellae are less “leaky” than other dinoflagellates. It is also possible that the CML environment is distinct enough from the surrounding seawater that the FDC cannot be used to directly compare the bacterial growth rates between these two communities. Torreton and Dufour (33) found that the frequency of dividing-divided cells (FDDC) in tropical reef waters could not be directly interpreted in terms of growth rates of the assemblage using temperate water conversion factors (14) without severely overestimating bacterial production (by comparing the FDDC-based calculations to thymidine and leucine incorporation rates in the same samples). Those authors reinforce the necessity of calibrating FDC and FDDC conversion factors when working in any new ecosystem. It is possible that in the CML, the conversion factors for the FDC to bacterial production would be significantly different from the surrounding seawater.
Another question raised by this study concerns why wastewater effluent would encourage seawater-associated bacteria to be smaller than cells on uninfluenced reefs. The observation of significantly larger and more abundant mucus-associated bacteria at wastewater-influenced sites (P < 0. 0001) could be explained by excess surrounding nutrients stimulating growth or by a higher proportion of water-associated bacteria invading into the mucus layer. Perhaps mucus bacteria are generally small due to size-selective grazing pressure in the CML (by coral polyps or other microbes), antagonistic interactions among bacteria (30, 32), or a limiting nutrient or micronutrient (thus encouraging a higher surface area/volume ratio). While our study cannot answer these questions, the described method for visualizing and enumerating bacteria should assist in future resolution of such questions for coral microbial ecology.
Supplementary Material
Acknowledgments
This work was supported by grants from the Gordon and Betty Moore Foundation MMI, National Science Foundation (NSF) grants OCE06-48116 and OCE09-62721 to F. Azam, and NSF IGERT grant 0333444. M. Garren was supported by an NSF Graduate Research Fellowship and the Robert and Patricia Switzer Foundation.
We thank the residents of Maui, the University of Hawaii at Manoa, Celia Smith, Meghan Dailer, Robin Knox, and Aaron Hartmann for support and guidance in the field. We are grateful to Maui Community College, Peter Fisher, and Sally Irwin for their generous logistical and laboratory support. Thank you to Penny Dockry for logistical support at SIO. We thank Fernando Nosratpour and the Birch Aquarium at Scripps for the A. yongei fragments. We thank Francesca Malfatti, Krystal Rypien, and Steven Smriga for helpful discussions. We also greatly appreciate the statistical support and coral samples provided by K. Rypien and the SWAT-3 culture provided by F. Malfatti. We thank two anonymous reviewers who helped to greatly improve the manuscript.
Footnotes
Published ahead of print on 23 July 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ali, A., Q. Zhang, J. S. Dai, and X. Huang. 2003. Calcein as a fluorescent iron chemosensor for the determination of low molecular weight iron in biological fluids. Biometals 16:285-293. [DOI] [PubMed] [Google Scholar]
- 2.Alldredge, A. L., U. Passow, and B. E. Logan. 1993. The abundance and significance of a class of large, transparent organic particles in the ocean. Deep Sea Res. A 40:1131-1140. [Google Scholar]
- 3.Allers, E., C. Niesner, C. Wild, and J. Pernthaler. 2008. Microbes enriched in seawater after addition of coral mucus. Appl. Environ. Microbiol. 74:3274-3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benson, A. A., and L. Muscatin. 1974. Wax in coral mucus—energy-transfer from corals to reef fishes. Limnol. Oceanogr. 19:810-814. [Google Scholar]
- 5.Bratbak, G. 1985. Bacterial biovolume and biomass estimations. Appl. Environ. Microbiol. 49:1488-1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, B. E., and J. C. Bythell. 2005. Perspectives on mucus secretion in reef corals. Mar. Ecol. Prog. Ser. 296:291-309. [Google Scholar]
- 7.Coffroth, M. A. 1984. Ingestion and incorporation of coral mucus aggregates by a gorgonian soft coral. Mar. Ecol. Prog. Ser. 17:193-199. [Google Scholar]
- 8.Coffroth, M. A. 1990. Mucous sheet formation on poritid corals—an evaluation of coral mucus as a nutrient source on reefs. Mar. Biol. 105:39-49. [Google Scholar]
- 9.Coligan, J., B. Dunn, D. Speicher, and P. Wingfield (ed.). 2003. Short protocols in protein science. Wiley, New York, NY.
- 10.Dailer, M., R. Knox, J. Smith, M. Napier, and C. Smith. 2010. Using δ15N values in algal tissue to map locations and potential sources of anthropogenic nutrient inputs on the island of Maui, Hawai'i, USA. Mar. Pollut. Bull. 60:655-671. [DOI] [PubMed] [Google Scholar]
- 11.Ducklow, H. W., and R. Mitchell. 1979. Bacterial-populations and adaptations in the mucus layers on living corals. Limnol. Oceanogr. 24:715-725. [Google Scholar]
- 12.Ducklow, H. W., and R. Mitchell. 1979. Composition of mucus released by coral-reef coelenterates. Limnol. Oceanogr. 24:706-714. [Google Scholar]
- 13.Glockner, F. O., R. Amann, A. Alfreider, J. Pernthaler, R. Psenner, K. Trebesius, and K.-H. Schleifer. 1996. An in situ hybridization protocol for detection and identification of planktonic bacteria. Syst. Appl. Microbiol. 19:403-406. [Google Scholar]
- 14.Hagstrom, A., U. Larsson, P. Horstedt, and S. Normark. 1979. Frequency of dividing cells, a new approach to the determination of bacterial-growth rates in aquatic environments. Appl. Environ. Microbiol. 37:805-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hed, J., C. Dahlgren, and I. Rundquist. 1983. A simple fluorescence technique to stain the plasma-membrane of human-neutrophils. Histochemistry 79:105-110. [DOI] [PubMed] [Google Scholar]
- 16.Herndl, G. J., and B. Velimirov. 1986. Microheterotrophic utilization of mucus released by the Mediterranean coral Cladocora cespitosa. Mar. Biol. 90:363-369. [Google Scholar]
- 17.Hobbie, J. E., R. J. Daley, and S. Jasper. 1977. Use of Nuclepore filters for counting bacteria by fluorescence microscopy. Appl. Environ. Microbiol. 33:1225-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huettel, M., C. Wild, and S. Gonelli. 2006. Mucus trap in coral reefs: formation and temporal evolution of particle aggregates caused by coral mucus. Mar. Ecol. Prog. Ser. 307:69-84. [Google Scholar]
- 19.Johnston, I. S., and F. Rohwer. 2007. Microbial landscapes on the outer tissue surfaces of the reef-building coral Porites compressa. Coral Reefs 26:375-383. [Google Scholar]
- 20.Koren, O., and E. Rosenberg. 2006. Bacteria associated with mucus and tissues of the coral Oculina patagonica in summer and winter. Appl. Environ. Microbiol. 72:5254-5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koren, O., and E. Rosenberg. 2008. Bacteria associated with the bleached and cave coral Oculina patagonica. Microb. Ecol. 55:523-529. [DOI] [PubMed] [Google Scholar]
- 22.Lunau, M., A. Lemke, K. Walther, W. Martens-Habbena, and M. Simon. 2005. An improved method for counting bacteria from sediments and turbid environments by epifluorescence microscopy. Environ. Microbiol. 7:961-968. [DOI] [PubMed] [Google Scholar]
- 23.Malfatti, F., T. J. Samo, and F. Azam. 2010. High-resolution imaging of pelagic bacteria by atomic force microscopy and implications for carbon cycling. ISME J. 4:427-439. [DOI] [PubMed] [Google Scholar]
- 24.Meikle, P., G. N. Richards, and D. Yellowlees. 1987. Structural determination of the oligosaccharide side-chains from a glycoprotein isolated from the mucus of the coral Acropora formosa. J. Biol. Chem. 262:16941-16947. [PubMed] [Google Scholar]
- 25.Meikle, P., G. N. Richards, and D. Yellowlees. 1988. Structural investigations on the mucus from 6 species of coral. Mar. Biol. 99:187-193. [Google Scholar]
- 26.Minnikin, S. M., and A. Allen. 1973. Cell-surface mucosubstances from trypsin disaggregation of normal and virus-transformed lines of baby hamster kidney-cells. Biochem. J. 134:1123-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pascal, H., and E. Vacelet. 1982. Bacterial utilization of mucus on the coral reef of Aqaba (Red Sea), vol. 1, p. 669-677. In E. D. Gomez, C. E. Birkeland, R. W. Buddemeier, R. E. Johannes, J. A. Marsh, Jr., and R. T. Tsuda (ed.), Proceedings of the 4th International Coral Reef Symposium. Marine Science Center, University of the Philippines, Manila, Philippines. [Google Scholar]
- 28.Paul, J. H., M. F. Deflaun, and W. H. Jeffrey. 1986. Elevated levels of microbial activity in the coral surface microlayer. Mar. Ecol. Prog. Ser. 33:29-40. [Google Scholar]
- 29.Pumper, R. W., P. Fagan, and W. G. Taylor. 1971. Trypsin sensitivity of mammalian cells grown in a serum-free medium. In Vitro 6:266-268. [DOI] [PubMed] [Google Scholar]
- 30.Ritchie, K. B. 2006. Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar. Ecol. Prog. Ser. 322:1-14. [Google Scholar]
- 31.Ritchie, K. B., and G. W. Smith. 1995. Preferential carbon utilization by surface bacterial communities from water mass, normal, and white-band diseased Acropora cervicornis. Mol. Mar. Biol. Biotechnol. 4:345-352. [Google Scholar]
- 32.Rypien, K. L., J. R. Ward, and F. Azam. 2010. Antagonistic interactions among coral-associated bacteria. Environ. Microbiol. 12:28-39. [DOI] [PubMed] [Google Scholar]
- 33.Torreton, J. P., and P. Dufour. 1996. Bacterioplankton production determined by DNA synthesis, protein synthesis, and frequency of dividing cells Tuamotu atoll lagoons and surrounding ocean. Microb. Ecol. 32:185-202. [DOI] [PubMed] [Google Scholar]
- 34.Vacelet, E., and B. A. Thomassin. 1991. Microbial utilization of coral mucus in long-term in situ incubation over a coral reef. Hydrobiologia 211:19-32. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.