Abstract
There is a significant potential to improve the plant-beneficial effects of root-colonizing pseudomonads by breeding wheat genotypes with a greater capacity to sustain interactions with these bacteria. However, the interaction between pseudomonads and crop plants at the cultivar level, as well as the conditions which favor the accumulation of beneficial microorganisms in the wheat rhizosphere, is largely unknown. Therefore, we characterized the three Swiss winter wheat (Triticum aestivum) cultivars Arina, Zinal, and Cimetta for their ability to accumulate naturally occurring plant-beneficial pseudomonads in the rhizosphere. Cultivar performance was measured also by the ability to select for specific genotypes of 2,4-diacetylphloroglucinol (DAPG) producers in two different soils. Cultivar-specific differences were found; however, these were strongly influenced by the soil type. Denaturing gradient gel electrophoresis (DGGE) analysis of fragments of the DAPG biosynthetic gene phlD amplified from natural Pseudomonas rhizosphere populations revealed that phlD diversity substantially varied between the two soils and that there was a cultivar-specific accumulation of certain phlD genotypes in one soil but not in the other. Furthermore, the three cultivars were tested for their ability to benefit from Pseudomonas inoculants. Interestingly, Arina, which was best protected against Pythium ultimum infection by inoculation with Pseudomonas fluorescens biocontrol strain CHA0, was the cultivar which profited the least from the bacterial inoculant in terms of plant growth promotion in the absence of the pathogen. Knowledge gained of the interactions between wheat cultivars, beneficial pseudomonads, and soil types allows us to optimize cultivar-soil combinations for the promotion of growth through beneficial pseudomonads. Additionally, this information can be implemented by breeders into a new and unique breeding strategy for low-input and organic conditions.
Improvement of plant fitness and yield by root-colonizing microorganisms is of special value in low-input or organic wheat production. Beneficial soil bacteria, such as certain Pseudomonas strains, are known to promote plant growth, which might help to circumvent potential negative consequences of low-input cropping systems, such as the limited supply of nutrients and higher disease pressure. A wide range of traits in Pseudomonas spp. are responsible for plant-beneficial effects. Many pseudomonads are capable of solubilizing poorly soluble or insoluble mineral phosphates, thereby rendering this element available for the plant and promoting plant growth (25, 43). Root-colonizing pseudomonads are also able to indirectly promote plant growth by providing protection against plant diseases. The most important mechanisms for plant protection against attacking pathogens are the induction of systemic resistance in plants (3) and the direct suppression of soilborne pathogens through the production of antimicrobial metabolites (16). The protection of wheat plants against Gaeumannomyces graminis var. tritici by naturally occurring pseudomonads in take-all decline soils is a well-described phenomenon and highlights the importance of these bacteria in a successful and environmentally friendly wheat production (53). Interestingly, in many naturally disease-suppressive soils a specific group of fluorescent pseudomonads is enriched, which is able to produce the antimicrobial compound 2,4-diacetylphloroglucinol (DAPG) (6, 38, 53). The production of the polyketide DAPG, which has broad-spectrum activity against bacteria, plants, fungi, and nematodes (8, 9, 21, 28, 33, 45), has been shown to be a key factor in the suppression of soilborne plant diseases by various Pseudomonas biocontrol strains (16).
The degree of plant protection and plant growth promotion provided by root-colonizing pseudomonads is highly dependent on different environmental factors. For example, the expression of important biocontrol genes such as DAPG or HCN biosynthetic genes in the rhizosphere is modulated by biotic factors such as fungi and other bacteria present in the rhizosphere and the secondary metabolites they release (7, 19, 27, 29, 32). Moreover, it has been observed that the plant species and cultivar as well as the physiological stage of the plant can influence the expression of biocontrol genes and the production of antimicrobial metabolites (4, 7, 19, 32, 35). In addition to the production of DAPG and other antimicrobial metabolites, efficient colonization of roots is a prerequisite for beneficial plant-Pseudomonas interactions. Root colonization is dependent not only upon specific characteristics of the bacterium itself but also on root morphology and root exudates that vary between host plant species and even between cultivars of the same species (5, 34). The host species/cultivar also influences the abundance and diversity of naturally occurring pseudomonads (13). This has been shown in particular for DAPG-producing populations (4, 5, 26, 30, 36).
Wheat is a crop known to benefit strongly from naturally occurring DAPG-producing pseudomonad populations (52). It has been shown that the size and composition of DAPG-producing populations in the wheat rhizosphere and also the amount of DAPG produced by these populations may vary substantially between different cultivars (4, 35). However, holistic studies which evaluate specific wheat cultivars for both their ability to benefit from plant growth-promoting pseudomonads and their influence on bacterial populations and production of biocontrol compounds are missing. A comprehensive characterization of different cultivars is needed in order to better understand which cultivars promote beneficial interactions with the pseudomonads. This knowledge has potential in future breeding strategies to be used for selection of new cultivars that optimally attract and respond to these bacteria.
In order to address this gap in knowledge, this study evaluated three Swiss winter wheat (Triticum aestivum) cultivars for several characteristics considered important in a successful wheat-pseudomonas interplay: (i) the ability to accumulate pseudomonads and phlD+ pseudomonads in two different Swiss soils, (ii) the ability to select for individual phlD+ genotypes in two different soils, (iii) the ability to benefit from the two model biocontrol strains, Pseudomonas fluorescens strain CHA0 (a DAPG producer) and P. putida KD (a DAPG nonproducer), in terms of direct plant growth promotion and disease suppression, and finally (iv) the level of biocontrol gene expression (DAPG-biosynthetic gene phlA) in the rhizosphere.
MATERIALS AND METHODS
Wheat cultivars.
Three different Swiss winter wheat cultivars were used in this study: Arina, Zinal, and Cimetta. They were bred in the Swiss breeding program at Agroscope ACW in Changins, Switzerland. Arina is one of the most prominent cultivars used in Switzerland. Arina and Zinal have a common ancestor, the cultivar Zenith. Arina is a first-generation descendant and Zinal a second-generation descendant of Zenith. Cimetta has no near relationship with the other cultivars.
Microorganisms used in this study.
P. fluorescens CHA0 is a DAPG-producing strain isolated from the rhizosphere of tobacco grown in a Swiss soil naturally suppressive to Thielaviopsis basicola (48). P. fluorescens KD was isolated in China from the rhizosphere of wheat (46). Strain KD shows an outstanding biocontrol performance against both damping-off disease of cucumber caused by Pythium ultimum and crown and root rot of tomato caused by Fusarium oxysporum f. sp. radicis-lycopersici (46), although it does not produce the biocontrol compound DAPG, which is often associated with effective biocontrol in pseudomonads. Strain CHA0/pME7100 is a derivative of P. fluorescens CHA0 carrying a phlA-gfp reporter fusion (2). All strains were routinely cultivated at 27°C on King's B agar (KB) (23) and in Luria broth (LB) (44). For cultivation of CHA0/pME7100, tetracycline hydrochloride was added to the medium at 125 μg/ml.
Pythium ultimum strain 67-1 (obtained from Allelix Agriculture, Mississaugua, Ontario, Canada) was cultured on 1.5% malt agar plates (Oxoid, Basingstoke, England) at 20°C for 7 days. For disease suppression assays, a 0.7-cm plug of an actively growing Pythium ultimum culture was transferred to a petri dish containing 25 g of autoclaved millet seeds (Biofarm Kleindietwil, Switzerland) and 10 ml of autoclaved double-distilled water. After 7 days of incubation at 20°C the mycelium-covered millet was sieved and particles of 1 mm in diameter were used to infest soil.
Quantification of total pseudomonads and phlD+ pseudomonads on the roots of different wheat cultivars grown in two different soils.
Soil samples were taken in April 2005 from two fields in Switzerland. One site was located near Moudon (Ecole Cantonale d'Agriculture de Grange-Verney, Moudon, Switzerland, 46°40′43″N, 6°57′29″E); the other site was located at Delley (Delley Semences et Plantes SA, Delley, Switzerland, 46°40′34″N, 6°48′22″E). On these fields, the performance of different wheat cultivars was tested. Soil was collected from three plots each planted with one of the winter wheat cultivars Arina, Zinal, or Cimetta. For experiments evaluating Pseudomonas accumulation and phlD diversity, pots (10-cm diameter, 420-cm3 volume) were filled with the collected soils. Into each pot five seeds of the respective wheat cultivar were sown. Three replicates per cultivar and soil were prepared. The plants were grown in a growth chamber with 70% relative humidity and 12 h of light (160 μE/m2/s) at 18°C, followed by a 12-h dark period at 15°C. In order to accumulate Pseudomonas spp. adapted to each cultivar, pots were resown monthly to achieve continuous wheat cultures.
After seven successive cycles of 1 month each, the Pseudomonas populations in three different locations were investigated: in nonrhizosphere soil, on the root surface, and in the root interior. To this end, 0.1-g samples of nonrhizosphere soil were collected from each pot and suspended in 10 ml 0.9% NaCl solution. One-ml samples of the resulting suspensions were collected for quantification of pseudomonads in the soil. In order to quantify pseudomonads on the roots, plants were harvested carefully, and the roots were cut off and washed with tap water. Approximately 500 mg of roots from each sample were placed into a 50-ml Erlenmeyer flask containing 10 ml of a 0.9% sterile NaCl solution. The flasks were vigorously shaken for 20 min at 400 rpm. From the root washes, 1-ml samples were taken to estimate the numbers of Pseudomonas spp. closely adhering to the roots. Subsequently, the roots were removed from the flasks, washed with 0.9% NaCl solution, and placed for 45 s into 70% ethanol for surface sterilization. The roots were washed again and then macerated in 5 ml 0.9% NaCl solution with a homogenizer (Homex 6; Bioreba AG, Reinach, Switzerland) for 45 s. From the resulting suspensions, samples were taken to estimate Pseudomonas populations inside the roots. In order to evaluate the efficiency of surface sterilization, subsamples of roots, which had their surfaces sterilized with ethanol, were not macerated but were shaken again in 0.9% sterile NaCl solution for 20 min at 400 rpm. Afterwards the number of pseudomonads was determined as described below. The obtained results showed that 99.99% of the pseudomonads on the root surface were removed by washing with NaCl solution and subsequent ethanol treatment.
From all samples (soil suspensions, root washes, and macerated root suspensions), aliquots of 20 μl were transferred to 96-well microtiter plates containing 180 μl of the Pseudomonas selective medium KB+++ per well. The KB+++ medium consists of King's medium B (23), amended with actidione (100 μg/ml), chloramphenicol (13 μg/ml), and ampicillin (40 μg/ml) (37). Tenfold serial dilutions up to a final dilution of 10−6 were made in the microtiter plates. Four replicates were prepared per sample. The plates were incubated for 4 days at 24°C with continuous shaking at 150 rpm. Bacterial growth was estimated visually, and the highest dilution showing growth was used to calculate the total Pseudomonas population size of a sample by the most probable number (MPN) technique (1, 14). Sterile glycerol was added to all wells to achieve a final concentration of 50%, and microtiter plates were sealed with aluminum tape (Greiner Biotech, Germany) for storage at −80°C.
The proportion of rhizosphere pseudomonads carrying the phlD gene was examined using the MPN-PCR approach described by Ramette et al. (40). Briefly, 20 μl of bacterial suspensions from each dilution in the microplate were transferred to a 96-well PCR plate (Simport Plastics, Beloeil, Canada) and incubated for 10 min at 99°C. The heat-lysed suspensions were then frozen at −20°C for 30 min. After thawing, 4-μl aliquots of the suspensions were used as templates for a PCR with forward primer B2BF (25-mer 5′-ACCCACCGCAGCATCGTTTATGAGC-3′) and reverse primer BPR4 (26-mer 5′-CCGCCGGTATGGAAGATGAAAAAGTC-3′) in order to amplify a fragment of phlD (31). Primers were synthesized by MWG Biotech, Basel, Switzerland. Amplifications were carried out in 12-μl reaction mixtures containing 4 μl of lysed bacterial suspension, 1× PCR buffer (Amersham Pharmacia, Uppsala, Sweden), bovine serum albumin (0.5 g/liter; Fluka, Buchs SG, Switzerland), 5% dimethyl sulfoxide (Fluka), 100 μM (each) dATP, dCTP, dGTP, and dTTP (Amersham Pharmacia), 0.40 μM each primer, and 1.4 U of Taq DNA polymerase (Amersham Pharmacia). The initial denaturation (2 min at 94°C) was followed by 35 cycles of 94°C for 30 s, 60°C for 30 s, and 7°C for 60 s with a final extension at 72°C for 10 min. The terminal dilution showing phlD amplification was used to calculate the population size of phlD-positive pseudomonads by MPN as described above.
phlD amplification for denaturing gradient gel electrophoresis (DGGE) analysis.
phlD diversity was studied in root surface samples. In order to obtain satisfactory phlD amplification in certain samples, a nested PCR approach was needed. In the first step, phlD was amplified using primers B2BF and BPR4. In the second step, the 10-fold diluted product from the first PCR was used as a template to perform a PCR with primers Phl2b1GC (21-mer, 5′-ACC GCA GCA TCG TGT ATG AGC-3′) containing a 40-bp GC clamp (5′-CGCCCGCCGCGCCCCGCGCCCGTCCCGCCGCCCCCGCCCG-3′) and Mir1a (21-mer, 5′-GGA GTT CAT GAC CGC CTT GTC-3′).
PCR was carried out in 20-μl reaction mixtures containing 1 μl of DNA template, 2 μl of PCR buffer (New England Biolabs, Beverly, MA), 0.5 g/liter 100× bovine serum albumin (New England Biolabs), 5% dimethyl sulfoxide (Fluka), 200 μM each dATP, dCTP, dGTP, and dTTP (New England Biolabs), 0.50 μM each primer, and 1.4 U of Taq DNA polymerase (New England Biolabs). The initial denaturation (3 min at 94°C) was followed by 40 cycles of 94°C for 30 s, 60°C for 60 s, and 72°C for 60 s with a final extension at 72°C for 10 min. PCR products were analyzed on a 1% agarose gel to check the presence of 670-bp bands for the first and 571-bp bands for the second PCR.
DGGE analysis.
DGGE analysis was performed using the DCode universal mutation detection system (Bio-Rad, Hercules, CA). DGGE gels for both types of PCR products were cast using a double gradient ranging from 7% to 12% of acrylamide and from 30% to 60% denaturants (100% denaturant corresponds to 7 M urea and 40% deionized formamide). The marker consisted of individually amplified phlD fragments from 10 reference Pseudomonas strains (Table 1 and Fig. 1) belonging to different genotypic groups of phlD+ pseudomonads as described by Frapolli et al. (10). The samples were run for 14 h at 140 V in 1× TAE buffer (40 mM Tris base, 20 mM acetic acid, 1 mM EDTA, pH 8) preheated at 60°C. The gels were stained with SYBR Gold (Molecular Probes, Eugene, OR) for 1 h and visualized with a UV transilluminator.
TABLE 1.
Fluorescent Pseudomonas strains used in this study
| Strain | phlD genotypea | Reference |
|---|---|---|
| CHA0 | 9 | 48 |
| CHA0/pME7100 (phlA-gfp) | 2 | |
| KD | 46 | |
| Pf-5 | 9 | 18 |
| Q2-87 | 7 | 49 |
| Q65c-80 | 5 | 17 |
| CM1′A2 | 5 | 12 |
| F113 | 9 | |
| PITR2 | 6 | 22 |
| P97.38 | 8 | 50 |
| F96.27 | 8 | 50 |
| K94.37 | 2 | 50 |
| Q37.87 | 4 | 22 |
| P12 | 1 | 22 |
As determined by cluster analysis of partial phlD sequences by Frapolli et al. (10).
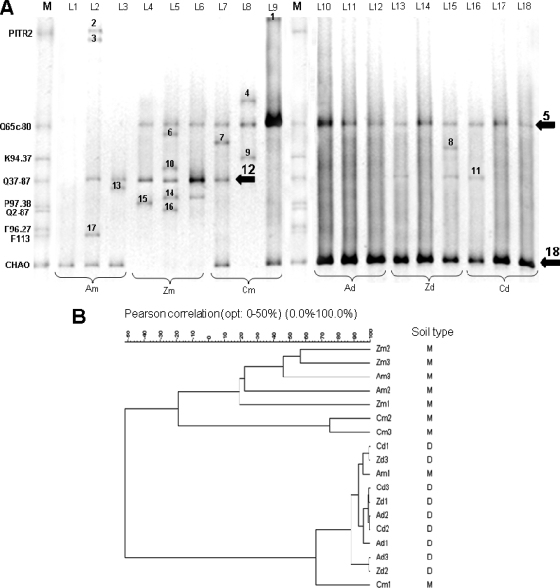
FIG. 1.
(A) Comparison of DGGE patterns of phlD fragments amplified from Pseudomonas populations isolated from the wheat cultivars Arina (A), Zinal (Z), and Cimetta (C) grown in Moudon soil (lanes L1 to L9) or Delley soil (lanes L10 to L18); M, marker composed from P. fluorescens strains PITR2, Q65c-80, K94.37, Q37-87, P97.38, Q2-87, F96.27, and CHA0, belonging to the phlD cluster groups 6, 5, 2, 4, 8, 7, 8, and 9, respectively, as described by Frapolli et al. (10), as well as F113. The three lanes per cultivar and soil represent DGGE analysis of root samples from three replicate pots. Arrows indicate major bands showing differences between soil type and wheat cultivar. (B) The hierarchical cluster analysis is based on Pearson's correlation index (band intensities) and the Ward algorithm (51). A, C, and Z, Arina, Cimetta, and Zinal, respectively. M or m, Moudon soil; D or d, Delley soil. 1, 2, and 3, replicate numbers.
Sequencing of DGGE bands.
Dominant DGGE bands were characterized as follows. The central part of DGGE bands was cut out using sterile pipette tips. The gel pieces were then washed with 100 μl sterile double-distilled water at room temperature for 1 h and subsequently used as a template for a 40-μl PCR with primers Mir1a and Phl2b1GC with the conditions described above. The PCR products were run on a DGGE gel to check migration and presence of a single band and then sequenced using the Applied Biosystems 3130 genetic analyzer. Sequences were edited using the Sequencher software (version 4.8; Gene Codes Corporation) and aligned with phlD sequences from known pseudomonad reference strains.
Analysis of DGGE band patterns.
DGGE pictures were analyzed using the GelCompar II software (version 5.1; Applied Maths). Band patterns of the samples were normalized using the DGGE marker as a reference and analyzed using a rolling background subtraction of 10% disk size. For cluster analysis, the Pearson correlation index, which takes the band intensities into account, and the Ward algorithm were used. To test for significant differences (P ≤ 0.05) between the two soil types and the wheat cultivars, the similarity matrix obtained with the Pearson's correlation coefficient was submitted to 1,000,000 unrestricted permutations using the Permtest software (24).
Diversity of phlD+ pseudomonad community.
To compare the phlD+ Pseudomonas sp. community diversity between soils and wheat cultivars, the Shannon index (H′) was calculated from the band abundance values retrieved from the GelCompar II program. This diversity index takes into account both abundance and distribution (evenness) of band patterns present in a sample and is calculated using the formula H′ = 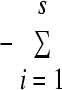 pilnpi, where pi is the relative abundance of a given DGGE band expressed as a ratio of its intensity to the total DGGE band intensity present in the sample i.
pilnpi, where pi is the relative abundance of a given DGGE band expressed as a ratio of its intensity to the total DGGE band intensity present in the sample i.
Ability of different wheat cultivars to profit from beneficial P. fluorescens strains.
Seeds of the winter wheat cultivars Arina, Zinal, and Cimetta were surface disinfected for 15 min in 7% sodium hypochlorite (vol/vol) and then thoroughly rinsed with sterile double-distilled water. Seeds were pregerminated for 3 days on 0.85% water agar at 24°C in darkness. Plants were grown in artificial soil containing vermiculite and quartz sand of different fractions (20). The soil was moistened with double-distilled water (100 ml per 1 kg of soil). One-liter Erlenmeyer flasks were partly filled with 300 g of artificial soil, plugged with cotton wool stoppers, and autoclaved for 30 min at 121°C. Overnight LB cultures of CHA0 and KD were washed with 0.9% NaCl solution and diluted to give a final concentration of 107 CFU/ml. To each flask, 6 ml of this bacterial suspension was added. Millet powder completely colonized by Pythium ultimum (see above) was mixed into the soil at a rate of 100 mg per flask. Control flasks without the pathogen received the same amount of autoclaved millet powder. Five seedlings were transferred to each flask, covered with soil, and incubated in a growth chamber with 16 h of light at 18°C (160 μE/m2/s) followed by 8 h of darkness at 13°C. The flasks were arranged in a randomized complete block design. Three weeks later, plants were removed from the flasks and gently shaken to discard loosely adhering soil. Roots with tightly adhering soil (in this article defined as rhizosphere) were shaken in sterile 0.9% NaCl solution at 450 rpm for 20 min. Rhizosphere colonization with bacteria was determined from the resulting suspension by plating serial dilutions on KB agar. Plants were washed with tap water, blotted dry, and weighed. The experiment was performed twice with four replicate flasks (each flask containing five plants) per treatment. The obtained results are separated in two parts for presentation: Table 2 shows the disease suppression, and Table 3 shows the plant growth promotion in the absence of the pathogen. In order to account for the growth differences between the individual wheat cultivars, fresh weights presented in Table 2 and Table 3 are expressed as percentages of the fresh weights of untreated control plants.
TABLE 2.
Protection of Swiss wheat cultivars against the root pathogen Pythium ultimum by P. fluorescensa
| Cultivar | Strain(s) used for treatment | Fresh wt (%)b |
|
|---|---|---|---|
| Root | Plant | ||
| Arina | Pythium | 11.5 cd | 27.9 d |
| Pythium + CHA0 | 60.7 a | 82.9 a | |
| Pythium + KD | 15.9 c | 46.6 c | |
| Zinal | Pythium | 9.6 cd | 33.8 d |
| Pythium + CHA0 | 40.9 b | 65.7 b | |
| Pythium + KD | 16.9 c | 51.3 c | |
| Cimetta | Pythium | 6.9 d | 28.7 d |
| Pythium + CHA0 | 47.0 b | 67.2 b | |
| Pythium + KD | 16.3 c | 50.4 c | |
Plants grown under gnotobiotic conditions were inoculated with bacteria and on the same day infested with Pythium ultimum. After 3 weeks of growth in artificial soil under sterile conditions, plants were harvested and assessed for fresh weights.
Plant fresh weights are expressed as percentages of weights of corresponding untreated control plants (total plant weights: Arina, 3.04 g; Zinal, 2.94 g; Cimetta, 2.75 g; root weights: Arina, 1.44 g; Zinal, 1.46 g; Cimetta, 1.42 g). Values are the means of two experiments with four replicates each. Values within the same column followed by the same letter are not significantly different according to Fisher's protected LSD (P ≤ 0.05).
TABLE 3.
Plant fresh weight of Swiss wheat cultivars grown in the presence or absence of P. fluorescens in artificial soil under sterile conditions
| Cultivar | Plant fresh wt (%)a in presence of strain |
||
|---|---|---|---|
| None | CHA0 | KD | |
| Arina | 100.00 c | 112.84 b | 98.12 c |
| Zinal | 100.00 c | 127.43 a | 95.50 c |
| Cimetta | 100.00 c | 129.73 a | 102.90 c |
Total plant fresh weights are expressed as percentages of weights of corresponding untreated control plants (Arina, 3.04 g; Zinal, 2.94 g; Cimetta, 2.75 g). Values are the means of two experiments with four replicates each. Values marked with the same letter are not significantly different according to Fisher's protected LSD (P ≤ 0.05).
phlA expression in P. fluorescens CHA0 on wheat roots.
The expression of the DAPG biosynthetic gene phlA by CHA0 on the roots of the three wheat cultivars was measured using a phlA-gfp reporter system and a flow cytometry-based approach as described in detail by De Werra et al. (7). Briefly, surface-sterilized seeds of the wheat cultivars Arina, Zinal, and Cimetta were germinated on soft agar for 2 days and placed into plant growth pouches (Mega International, West St. Paul, MN) at three seedlings per pouch. Growth pouches were amended with 15 ml of modified Knop plant nutrition solution (20). P. fluorescens suspensions containing 108 cells/ml were prepared as described above, and 1 ml was added to each seedling in the growth pouch. Plants were grown for 10 days in a growth chamber under the same conditions as described above. The experiment consisted of the following treatments: control without added bacteria, inoculation with wild-type CHA0 (for setting the background for flow cytometric analysis), and inoculation with CHA0/pME7100 carrying a phlA-gfp reporter fusion. Six replications per treatment were made, and the experiment was carried out twice. After a 10-day incubation period, plants were removed from the pouches. Roots of each pouch were placed into 7 ml of sterile distilled water and were vigorously shaken at 400 rpm for 20 min. From the resulting suspensions, samples of 2 ml were taken, transferred on ice, and immediately analyzed by fluorescence-activated cell sorting (FACS) as described by De Werra et al. (7) using a FACSCalibur flow cytometer equipped with a 15-mW, air-cooled argon ion laser excitation light source (488 nm) (Becton Dickinson, San Jose, CA). After subtracting background fluorescence, the green fluorescence measured by FACS corresponding to phlA-gfp expression was calculated as mean fluorescence per expressing cell and per gram of analyzed roots. Roots were assessed for fresh weight, and bacterial root colonization was determined by plating serial dilutions of root suspensions on KB agar.
Statistical analysis.
Statistical analyses were performed using the statistics program Systat, version 10.0 (Systat Inc., Evanston, IL). For each isolation site, the data for the two soils were pooled and soil versus cultivar interactions were analyzed by a two-way analysis of variance (see Table 5). Subsequently, means for one soil and one isolation site and also for one cultivar and isolation site were separated using Fisher's protected least significant difference (LSD) test (P ≤ 0.05) (see Table 5). As shown in Tables 2 to 4, results of independent repetitions over time (disease suppression assay and phlA expression assay) were first analyzed by a two-way analysis of variance to examine repetition versus treatment interactions. If this analysis revealed no significant repetition-treatment interactions, individual repetitions over time were pooled for statistical analysis and means were separated using Fisher's protected least significant difference (LSD) test (P ≤ 0.05).
TABLE 4.
Root colonization by P. fluorescens of Swiss wheat cultivars in the presence or absence of Pythium ultimuma
| Cultivar | Root colonizationb (log CFU/g root) |
|||
|---|---|---|---|---|
| CHA0 |
KD |
|||
| With Pythium | Without Pythium | With Pythium | Without Pythium | |
| Arina | 7.73 a | 7.10 a* | 8.13 a | 8.20 a |
| Zinal | 7.82 a | 7.26 a* | 7.74 b | 7.81 b |
| Cimetta | 7.68 a | 7.50 a | 8.30 a | 8.08 a |
Plants grown under gnotobiotic conditions were inoculated with bacteria and on the same day infested with Pythium ultimum. After 3 weeks of growth in artificial soil under sterile conditions, plants were harvested and assessed for bacterial root colonization.
Values are the means of two experiments with four replicates each. Values in the same column followed by the same letter are not significantly different according to Fisher's protected LSD (P ≤ 0.05). Values in the same row followed by an asterisk indicate a significant difference between results for treatments with and without Pythium ultimum for the same cultivar and bacterial inoculant according to Fisher's protected LSD (P ≤ 0.05).
RESULTS
Accumulation of total pseudomonads and phlD+ pseudomonads by different wheat cultivars in two different soils.
To assess the ability of different wheat cultivars to accumulate Pseudomonas spp. in distinct soils, wheat plants were grown for seven cycles in pots filled with field soils of two different geographic origins. Pseudomonads were isolated from the soil, from the root surface, and from inside the roots. Between 5.37 log and 5.98 log CFU/g soil of pseudomonads were detected in the soil originating from Moudon after seven successive cycles (1 month each) of wheat growing (Table 5). The Pseudomonas population was slightly smaller in the soil originating from Delley. In both soils, no significant differences between the cultivars Arina, Zinal, and Cimetta were found. A much higher Pseudomonas concentration was found on the root surface, ranging from 6.6 to 8.1 log CFU/g root fresh weight. When the plants were grown in the soil from Moudon no significant differences between cultivars were detected. In the Delley soil, however, the cultivar Arina was able to accumulate 15 to 30 times more pseudomonads on its roots than Cimetta and Zinal. Compared to the root surface, considerably fewer pseudomonads were found inside the roots of plants grown in the Moudon soil. Colonization ranged between 4.37 log and 4.52 log CFU/g root fresh weight with no significant differences between cultivars. The situation differed in the soil from Delley. The root interior of plants grown in the Delley soil was generally colonized by more pseudomonads (5.25 log to 7.09 log CFU/g root fresh weight) than that of plants grown in the Moudon soil. In this soil, the cultivar Cimetta accumulated significantly higher Pseudomonas numbers inside the plant roots than the other two cultivars.
TABLE 5.
Accumulation of Pseudomonas spp. and phlD+ Pseudomonas spp. by Swiss wheat cultivars in different soilsa
| Pseudomonas spp. isolated from: | Wheat cultivar | Colonization (log CFU/g)b by: |
ANOVA Pc |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
Pseudomonas spp. |
phlD+ Pseudomonas spp. |
Total Pseudomonas spp. |
phlD+ Pseudomonas spp. |
||||||||
| Moudon | Delley | Moudon | Delley | Cultivar | Soil | Cultivar × soil | Cultivar | Soil | Cultivar × soil | ||
| Soil | Arina | 5.53 a | 4.68 a | BD | 4.24, BD, BD | 0.231 | ≤0.05 | 0.984 | 0.642 | 0.382 | 0.402 |
| Zinal | 5.98 a | 5.21 a | BD | 3.76, BD, BD | |||||||
| Cimetta | 5.37 a | 4.46 a | BD | 3.10, BD, BD | |||||||
| Root surface | Arina | 6.78 a | 8.11 a* | 4.40 a | 5.71 a* | 0.208 | 0.353 | 0.034 | 0.248 | 0.679 | ≤0.05 |
| Zinal | 6.75 a | 6.93 b | 5.49 ab | 4.66 b | |||||||
| Cimetta | 7.31 a | 6.61 b | 5.94 b | 5.14 ab* | |||||||
| Root interior | Arina | 4.52 a | 6.07 a* | 4.43, BD, BD | 5.73 a | ≤0.05 | ≤0.05 | ≤0.05 | ≤0.05 | 0.223 | 0.209 |
| Zinal | 4.37 a | 5.25 a* | 3.93, BD, BD | 4.99 a | |||||||
| Cimetta | 4.43 a | 7.09 b* | 4.40, BD, BD | 6.80 b | |||||||
Wheat cultivars were grown in a growth chamber in pots filled with Moudon or Delley soil in seven successive cycles of 1 month each.
Means of three replicates are presented. In cases in which bacterial populations were below the detection limit (log 3.1 CFU/g for soil and root surface and 2.1 log CFU/g for root interior) in some of the replicates, results for individual replicates are presented. BD, below detection limit. Values within the same column for the same isolation site, followed by the same letter, are not significantly different according to Fisher's protected LSD (P ≤ 0.05). Values in the same row followed by an asterisk indicate a significant difference between results for soils for the same cultivar and isolation site according to Fisher's protected LSD (P ≤ 0.05).
Data for the two soils were pooled and analyzed by two-way analysis of variance (Systat, version 10.0; Systat Inc., Evanston, IL).
In the same experimental setup, the accumulation of Pseudomonas spp. carrying the phlD gene was assessed (Table 5). In the Moudon soil the population size of phlD+ pseudomonads was below the detection limit of 3.1 log CFU/g soil. In the Delley soil the phlD+ population sizes were below the detection limit in two out of three replicates for all cultivars and below 4.3 log CFU/g soil in the third replicate. The numbers of phlD+ pseudomonads adhering to the roots differed between the wheat cultivars tested. In the Moudon soil, Cimetta (5.94 log CFU/g root) accumulated significantly higher numbers of phlD+ pseudomonads on its roots than Arina (4.40 log CFU/g root). In Delley soil, the colonization of roots by phlD+ pseudomonads was significantly superior on the cultivar Arina than on Zinal. The accumulation of phlD+ pseudomonads in the root interior differed completely between the two soils. In the Delley soil, wheat plants accumulated 4.99 log to 6.80 log CFU/g root fresh weight inside their roots, with the cultivar Cimetta accumulating significantly more phlD+ pseudomonads than the other two cultivars. In the Moudon soil, however, no phlD+ pseudomonads were detected inside the roots of two out of three replicates (all cultivars). In the third replicate, the colonization of the root interior was around one log unit above the detection limit.
Resolution power of the new primers used for DGGE analysis.
In a previous study (10), a DGGE approach was developed to analyze communities of phlD+ pseudomonads. However, for the soils analyzed in this study the previous approach was not effective enough, since the amplification of phlD was insufficient in many samples (data not shown). In contrast, with the new nested PCR approach, good phlD amplification was obtained in all samples. Twelve reference strains (Table 1) that represent the known diversity of DAPG-producing pseudomonads were used to test the resolution power of the new nested method by DGGE. The approach presented here produced 10 different migration patterns (see Fig. S1 in the supplemental material), whereas the approach of Frapolli et al. (10) produced 11 different migration patterns.
phlD diversity of Pseudomonas populations enriched by different wheat cultivars in two different soils.
DGGE analysis was performed to study the diversity of phlD+ pseudomonads in the rhizosphere of the wheat cultivars Arina, Zinal, and Cimetta grown in Moudon and Delley soil (Fig. 1A). In the Delley soil, where only four migration patterns were found, the Shannon index was lower (H′ = 0.82) than in the Moudon soil (H′ = 1.19), where a total of 16 different migration patterns were detected. In each sample of the Delley soil, there were two bands, band 5, corresponding to the migration pattern of Q65c-80, and the more intense band 18, corresponding to the migration pattern of strain CHA0 (Fig. 1A). In Moudon soil samples, there were three prominent bands, i.e., the same two found in Delley soil (bands 5 and 18) and one (band 12) with the same migration pattern as strain Q37-87. The Moudon soil was characterized by the presence of other nondominant bands, in contrast to the Delley soil, where only very few additional weak bands were present. When the three different wheat cultivars were compared for the presence of the three dominant bands, it appeared that (i) in the Delley soil there were no differences between the cultivars in that bands 5 and 18 were always present, (ii) in Moudon soil Cimetta also harbored those two bands, (iii) in Moudon soil Zinal was characterized by the presence of band 5 and 12, and (iv) in Moudon soil Arina enriched band 12 and the even stronger band 18. The community diversity found on the roots of Zinal was higher than that of the other two cultivars in both types of soil, with H′ = 1.55 in Moudon and H′ = 1.03 in Delley soil, whereas Cimetta and Arina, respectively, displayed a diversity of H′ = 1.19 and H′ = 0.86 in Moudon soil and of H′ = 0.79 and H′ = 0.65 in Delley soil. The DGGE fingerprints (Fig. 1A) were then subjected to Pierson cluster analysis and converted into a dendrogram (Fig. 1B) that shows a clear differentiation between the samples of Moudon and those of Delley soils and displayed two main clusters. The first cluster contained only samples from Moudon soil, whereas the second cluster contained all Delley samples and two Moudon samples (Arina Moudon 1 and Cimetta Moudon 1). The difference between soil types was found to be significant (P ≤ 0.05) when submitted to the permutation test. In the Delley soil, phlD profiles from all three wheat cultivars and replicates were quite similar. In the Moudon soil, DGGE profiles from different wheat cultivars and different replicates of the same cultivar shared lower similarity compared to profiles from the Delley soil. However, as in the Delley soil, no statistically significant differences were found between the different wheat cultivars.
Sequencing of dominant phlD bands found in Moudon and Delley soils.
The three DGGE bands corresponding to the dominant phlD genotypes were sequenced in at least four samples from different cultivars and soils, and sequences were compared to NCBI sequences of P. fluorescens reference strains. Band number 5, with the same migration pattern as Q65c-80, had 100% identity (based on 460 bp) with the sequences of reference strains S7-42 (GenBank accession no. EF554345) and Q8r1-96 (GenBank accession no. AF207693) except in samples Cimetta Moudon 3 (with 1 bp difference) and Cimetta Deley 1 (with 2 bp difference). Band number 12, with the same migration pattern as Q37-87, had 100% identity (based on 472 bp) with the sequence of strain Q37-87 (GenBank accession no. AY928641) except in sample Zinal Moudon 2 (1 bp difference). The most dominant band, number 18, shared the same migration pattern and always had 100% sequence identity (based on 437 bp) with CHA0 (GenBank accession no. AJ278806).
Ability of different wheat cultivars to profit from beneficial P. fluorescens strains.
The potential of the two distinct P. fluorescens strains CHA0 and KD to protect three different wheat cultivars against Pythium ultimum was assessed in a gnotobiotic system. After 3 weeks the total fresh weights of the untreated control plants were 3.04 g (Arina), 2.94 g (Zinal), and 2.75 g (Cimetta) and the root fresh weights were 1.44 g, 1.46 g, and 1.42 g, respectively. Pythium ultimum strongly reduced root weights (by 89 to 93%) and total plant weights (by 66 to 72%) of all cultivars (Table 2). P. fluorescens KD was able to protect the plants only to a limited extent, with no differences between wheat cultivars. The root weights of plants inoculated with KD and Pythium ultimum ranged between 15.9 and 16.9% of those of noninoculated control plants and were not significantly different (for two out of three cultivars) from those of plants inoculated with the pathogen alone. Total weights of plants protected by KD, however, were significantly different from those of plants inoculated with Pythium only and ranged between 46.6 and 51.3% of the weights of the respective controls. P. fluorescens CHA0 provided significantly better disease suppression than strain KD (Table 2). In the presence of strain CHA0, root weights of plants growing in Pythium-infested soil were significantly enhanced and reached 41 to 61% of those of untreated controls. CHA0-treated pathogen-exposed plants reached between 66 and 83% of the total plant weight of untreated controls. Comparing the three wheat cultivars, strain CHA0 had the best protective effect on the cultivar Arina. In the presence of the oomycete pathogen, the total weight of Arina treated with CHA0 was around 25% higher than those of Zinal and Cimetta.
Interestingly, in the absence of the pathogen, the other two cultivars profited more than Arina from the presence of CHA0 (Table 3). Here, P. fluorescens KD had no impact on total plant weight of any of the three tested wheat cultivars. Strain CHA0, however, significantly enhanced the growth of all cultivars. The plant growth-promoting effect on Arina, however, was quite small (13%), whereas it was much better for Zinal (30%) and Cimetta (27%).
The effect of the wheat cultivars on root colonization by plant-beneficial pseudomonads was also evaluated. KD proved generally to be a better colonizer than CHA0 (Table 4). Root colonization by strain KD ranged between 7.74 log and 8.30 log CFU/g fresh weight, while colonization by CHA0 ranged between 7.1 log and 7.82 log CFU/g. The roots of Zinal were significantly less colonized by strain KD than the roots of Arina and Cimetta, independent of the presence or absence of Pythium. This cultivar-specific difference was not observed for root colonization by CHA0. In treatments that included inoculation with CHA0, the presence of Pythium had a significant enhancing effect (half a log unit) on the colonization of the roots of Arina and Zinal but not on that of Cimetta (Table 4).
phlA expression on wheat roots.
In order to test the influence of wheat cultivar on expression of an important biocontrol gene, Arina, Zinal, and Cimetta were grown in growth pouches inoculated with CHA0 carrying a phlA-gfp reporter fusion. The green fluorescence emitted by CHA0 cells, corresponding to the expression of phlA, was measured by FACS. After 10 days, the relative fluorescence per phlA-expressing CHA0 cell did not differ among the cultivars and ranged between 88.6 on the roots of Cimetta and 98.1 on the roots of Arina. Similarly, no differences were found for total phlA expression per root fresh weight. The obtained values were 1.24 × 1011 (Arina), 9.34 × 1010 (Zinal), and 9.02 × 1010 (Cimetta) relative fluorescence per gram of roots.
DISCUSSION
There is great potential for exploitation of host response to beneficial microorganisms (reviewed in reference 54), which could allow breeders to select for traits that encourage interaction with plant-beneficial pseudomonads. This is of interest especially in breeding programs aimed at low-input agriculture. A quality cultivar should to be able to promote colonization and profit from naturally occurring beneficial organisms in the soil. A prerequisite for breeders to exploit cultivar-driven beneficial interactions with microorganisms is specific knowledge pertaining to the ability of each cultivar to promote or support beneficial interactions. This study represents the first attempt to gather such information for three Swiss winter wheat cultivars currently being used in a Swiss governmental breeding program.
The influence of these three Swiss wheat cultivars on accumulation of naturally occurring pseudomonads and phlD+ Pseudomonas spp. was assessed in two soils after seven cycles of continuous wheat cultivation in pots. In the Moudon soil, no cultivar-specific differences in the accumulation of total pseudomonads in any of the tested compartments (soil, root surface, root interior) were detected. In contrast, the three cultivars accumulated different numbers of pseudomonads in the Delley soil. In both soils there were cultivar-specific differences in the accumulation of phlD+ pseudomonads on the roots. One cultivar, Cimetta, was superior in building up phlD+ populations in the Moudon soil, whereas another cultivar, Arina, was found to be superior in the Delley soil. These findings indicate the importance that soil has in the interaction between the plant and pseudomonads. It has been shown previously that fluorescent pseudomonads play an important role in take-all decline soils or against soilborne diseases in general. Raaijmakers et al. (39) reported a threshold of 105 CFU/g of root of phlD+ pseudomonads required for a successful suppression of Gaeumannomyces graminis var. tritici. In the Moudon soil, colonization of Arina by phlD+ Pseudomonas spp. was slightly below this threshold level, in contrast to the Delley soil, where Arina displayed the highest root colonization by phlD+ pseudomonads of all three tested cultivars (Table 5). This indicates that a wheat cultivar might in one soil accumulate enough phlD+ pseudomonads to be protected against soilborne diseases, while the same cultivar in another soil may not reach the threshold level needed for protection. Gu and Mazzola (15) tested different wheat cultivars for the ability to build up suppressiveness against replant disease in apple. They observed that the severity of apple replant disease was reduced after the preculture of some wheat cultivars but not others. As Cimetta proved to be a good attractor of phlD+ pseudomonads in both tested soils, it might be worthwhile to test this cultivar as a preculture with apple production or other perennial crops.
Interestingly, the soil type seems to have a greater impact on the accumulation of phlD+ pseudomonads in the root interior than the cultivar. In the Delley soil, all cultivars accumulated high levels of phlD+ pseudomonads inside the roots. In sharp contrast, in the Moudon soil, no phlD+ pseudomonads could be detected inside the roots in two out of three replicates. The importance of soil in determining the population sizes of plant-beneficial pseudomonads has already been shown in previous studies where soils suppressive to black root rot of tobacco were compared to nonsuppressive soils (11, 41).
Soil was also found to be a strong determining factor in the diversity of phlD+ pseudomonads on the different cultivars. Whereas in the Delley soil the wheat cultivar had no impact on diversity, in the Moudon soil individual cultivars selected for specific bacterial genotypes (Fig. 1). In general, the phlD diversity was much higher in the Moudon soil (16 bands detected) than in the Delley soil, where in the majority of the samples only two bands (number 5 and 18) were detected. The same two bands and additionally a third band (number 12) were the most prominent in the Moudon soil. Sequencing the three major bands 5, 12, and 18 detected by the DGGE analysis showed that they share 99.6 to 100% identity with the well-described P. fluorescens biocontrol reference strains S7-42 (41) and Q8r1-96 (38) (band 5), Q37-87 (22) (band 12), and CHA0 (band 18), which belong to the phlD genotypes (clusters) 5, 4, and 9 as defined by cluster analysis of phlD sequences by Frapolli et al. (10) and, respectively, to phlD genotypes D, E, and A as defined by De la Fuente et al. (5). We therefore suggest that in the Delley soil phlD+ pseudomonads of genotypes 5 and 9 are predominant. The high intensity of the two bands on DGGE gels made with samples from Delley soil suggests that the population sizes of genotypes 5 and 9 in the Delley soil are higher than that of any genotype present in Moudon soil. The great abundance of genotypes 5 and 9 in the Delley soil may explain the similarity between the two soils in the rhizosphere population level of phlD+ pseudomonads (Table 5) despite the large difference in the numbers of genotypes present.
Interestingly, the cultivar Arina seems to enrich phlD genotype 9 (band 18) in both soils (Fig. 1A). We speculate that the cultivar's ability to enrich the most dominant genotype in the Delley soil may have enabled this cultivar to accumulate the highest numbers of phlD+ pseudomonads and also of total pseudomonads in this soil (Table 5). We therefore suggest that a wheat cultivar with strong affinity for a phlD genotype that is present in abundance in the soil could have an advantage over other cultivars that do not possess the same affinity for that particular genotype. For an optimal cultivar-soil combination with respect to phlD+ pseudomonad enrichment it would therefore be necessary to know the preferences of different wheat cultivars for individual phlD+ genotypes and, additionally, the most prominent phlD genotypes present in the soil of interest.
In this study, we demonstrated that the three wheat cultivars differ in their ability to profit from a bacterial inoculant in a gnotobiotic system (Tables 2 and 3). The cultivar Arina was significantly better protected against Pythium damping off by P. fluorescens CHA0 than the other cultivars tested. Interestingly, the cultivar Arina enriched pseudomonads with phlD sequences identical to that of strain CHA0 in all samples from both Moudon and Delley soils (Fig. 1), suggesting that the better biocontrol activity of CHA0 on Arina could result from better adaptation of this cultivar to CHA0-like pseudomonads. However, CHA0 displayed neither a higher root colonization level (Table 4) nor a higher expression of the biocontrol gene phlA (in the absence of the pathogen) on Arina compared to the other varieties. We therefore suppose that in this simplified system further mechanisms of disease suppression were responsible for the better protection of Arina against Pythium ultimum.
Although it has been shown that P. fluorescens strain KD is a very good biocontrol agent in the control of Pythium on cress and cucumber (42, 46), this strain protected all tested wheat cultivars significantly less than CHA0 against Pythium ultimum (Table 2). Since biocontrol activity of strain KD is dependent on the presence of the type III secretion system (42), it might be possible that on monocotyledonous plants, such as wheat, type III secretion system-dependent mechanisms of disease suppression are of less importance in the control of Pythium.
To our best knowledge, we have shown for the first time that the plant growth promotion effects of a Pseudomonas inoculant, in the absence of a pathogen, can be wheat cultivar dependent. Growth promotion in the absence of the pathogen was lowest on Arina. Either this cultivar is less responsive to plant growth-promoting factors secreted by CHA0, or it is more sensitive to potential phytotoxic metabolites produced by this strain. Some secondary metabolites secreted by CHA0 (e.g., DAPG and pyoluteorin) can negatively affect plant growth (28). Direct growth promotion was lowest on Arina; however, protection, in the presence of the pathogen, was found to be the highest. This indicates a separation between the plant traits which favor plant-bacterium interactions that protect against disease and those that promote growth. If genetically controlled, it might thus be possible, through breeding, to combine these traits in a single cultivar.
For many of the investigated traits, differences between wheat cultivars were found. It is not surprising that the cultivar Cimetta differs from Arina and Zinal, since it originates from a distinct genetic background (Dario Fossatti, Agroscope ACW Changins, Nyon, Switzerland, personal communication). However, Zinal and Arina, which share an ancestor, also differ in several of the investigated traits. This genetic diversity could be exploited to breed a cultivar combining as many beneficial traits as possible. Quantitative trait locus (QTL) analyses in the progeny of crosses between these cultivars could be used to identify some genetic regions responsible for positive interactions with beneficial pseudomonads. Such an approach has been used to explore the genetic basis for interactions of tomato and the disease-suppressive bacterium Bacillus cereus (47). Smith and coworkers (47) suggest that the discovery of a genetic basis for plant-beneficial interactions with microorganisms opens up new opportunities to exploit natural genetic variation in host species. Furthermore, studies of this kind will enhance the current understanding of beneficial plant-microbe interactions and promote the development of ecologically sound strategies for disease control in agriculture.
Interactions between cultivars and soil might complicate efforts to exploit the genetic basis of beneficial wheat-bacterium interactions. Nevertheless, our study indicates that there is a great, unexplored potential for better exploitation of plant-bacterium interactions through the breeding of cultivars which are specifically adapted to accumulate and to profit from beneficial microbes.
Supplementary Material
Acknowledgments
We gratefully acknowledge Dario Fossatti and Fabio Mascher, Agroscope ACW, Changins, Switzerland, for helpful discussions and providing seed material.
This research was supported by the State Secretariat for Education and Research (project C04.0200; COST action 860) and the Swiss National Science Foundation (project 3100A0-105881 and NRP-59 project 405940-115596).
Footnotes
Published ahead of print on 30 July 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alexander, M. 1982. Most probable number method for microbial populations, p. 815-820. In S. Segoe (ed.), Methods of soil analysis, part 2, chemical and microbiological properties—agronomy monograph no. 9. American Society of Agronomy-Soil Science Society of America, Madison, WI.
- 2.Baehler, E., M. Bottiglieri, M. Péchy-Tarr, M. Maurhofer, and C. Keel. 2005. Use of green fluorescent protein-based reporters to monitor balanced production of antifungal compounds in the biocontrol agent Pseudomonas fluorescens CHA0. J. Appl. Microbiol. 99:24-38. [DOI] [PubMed] [Google Scholar]
- 3.Bakker, P. A. H. M., C. M. J. Pieterse, and L. C. van Loon. 2007. Induced systemic resistance by fluorescent Pseudomonas spp. Phytopathology 97:239-243. [DOI] [PubMed] [Google Scholar]
- 4.Bergsma-Vlami, M., M. E. Prins, and J. M. Raaijmakers. 2005. Influence of plant species on population dynamics, genotypic diversity and antibiotic production in the rhizosphere by indigenous Pseudomonas spp. FEMS Microbiol. Ecol. 52:59-69. [DOI] [PubMed] [Google Scholar]
- 5.De La Fuente, L., B. Landa, and D. M. Weller. 2006. Host crop affects rhizosphere colonization and competitiveness of 2,4-diacetylphloroglucinol-producing Pseudomonas fluorescens. Phytopathology 96:751-762. [DOI] [PubMed] [Google Scholar]
- 6.De Souza, J. T., D. M. Weller, and J. M. Raaijmakers. 2003. Frequency, diversity, and activity of 2,4-diacetylphloroglucinol-producing fluorescent Pseudomonas spp. in Dutch take-all decline soils. Phytopathology 93:54-63. [DOI] [PubMed] [Google Scholar]
- 7.De Werra, P., E. Baehler, A. Huser, C. Keel, and M. Maurhofer. 2008. Detection of plant-modulated alterations in antifungal gene expression in Pseudomonas fluorescens CHA0 on roots by flow cytometry. Appl. Environ. Microbiol. 74:1339-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dowling, D. N., and F. O'Gara. 1994. Metabolites of Pseudomonas involved in the biocontrol of plant disease. Trends Biotechnol. 12:133-141. [Google Scholar]
- 9.Fenton, A. M., P. M. Stephens, J. Crowley, M. O'Callaghan, and F. O'Gara. 1992. Exploitation of gene(s) involved in 2,4-diacetylphloroglucinol biosynthesis to confer a new biocontrol capability to a Pseudomonas strain. Appl. Environ. Microbiol. 58:3873-3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frapolli, M., Y. Moënne-Loccoz, J. Meyer, and G. Défago. 2008. A new DGGE protocol targeting 2,4-diacetylphloroglucinol biosynthetic gene phlD from phylogenetically contrasted biocontrol pseudomonads for assessment of disease-suppressive soils. FEMS Microbiol. Ecol. 64:469-481. [DOI] [PubMed] [Google Scholar]
- 11.Frapolli, M., G. Défago, and Y. Moënne-Loccoz. 2010. Denaturing gradient gel electrophoretic analysis of dominant 2,4-diacetylphloroglucinol biosynthetic phlD alleles in fluorescent Pseudomonas from soils suppressive or conducive to black root rot of tobacco. Soil Biol. Biochem. 42:649-656. [Google Scholar]
- 12.Fuchs, J., and G. Défago. 1991. Protection of cucumber plants against black root rot caused by Phomopsis sclerotioides with rhizobacteria, p. 57-62. In C. Keel, B. Koller, and G. Défago (ed.), Plant growth-promoting rhizobacteria. Progress and prospects. IOBC/WPRS Bulletin XIV/8, Interlaken, Switzerland.
- 13.Garbeva, O., J. D. van Elsas, and J. A. van Veen. 2008. Rhizosphere microbial community and its response to plant species and soil history. Plant Soil 302:19-32. [Google Scholar]
- 14.Garthright, W. E., and R. J. Blodgett. 2003. FDA's preferred MPN methods for standard, large or unusual tests, with a spreadsheet. Food Microbiol. 20:439-445. [Google Scholar]
- 15.Gu, Y.-H., and M. Mazzola. 2003. Modification of fluorescent pseudomonad community and control of apple replant disease induced in a wheat cultivar-specific manner. Appl. Soil Ecol. 24:57-72. [Google Scholar]
- 16.Haas, D., and C. Keel. 2003. Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease. Annu. Rev. Phytopathol. 41:117-153. [DOI] [PubMed] [Google Scholar]
- 17.Harrison, L. A., L. Letendre, P. A. Kovacevich, E. A. Pierson, and D. M. Weller. 1993. Purification of an antibiotic effective against Gaeumannomyces graminis var. tritici produced by a biocontrol agent Pseudomonas aureofaciens. Soil Biol. Biochem. 25:215-221. [Google Scholar]
- 18.Howell, C. R., and R. D. Stipanovich. 1979. Control of Rhizoctonia solani on cotton seedlings with Pseudomonas fluorescens and with an antibiotic produced by the bacterium. Phytopathology 69:480-482. [Google Scholar]
- 19.Jamali, F., A. Sharifi-Tehrani, M. P. Lutz, and M. Maurhofer. 2009. Influence of host plant genotype, presence of a pathogen and coinoculation with Pseudomonas fluorescens strains on the rhizosphere expression of hydrogen cyanide- and 2,4-diacetylphloroglucinol biosynthetic genes in Pseudomonas fluorescens biocontrol strain CHA0. Microb. Ecol. 57:267-275. [DOI] [PubMed] [Google Scholar]
- 20.Keel, C., C. Voisard, C. H. Berling, G. Kahr, and G. Défago. 1989. Iron sufficiency, a prerequisite for the suppression of tobacco black root rot by Pseudomonas fluorescens strain CHA0 under gnotobiotic conditions. Phytopathology 79:584-589. [Google Scholar]
- 21.Keel, C., U. Schnider, M. Maurhofer, C. Voisard, J. Laville, U. Burger, P. Wirthner, D. Haas, and G. Défago. 1992. Suppression of root diseases by Pseudomonas fluorescens CHA0: importance of the bacterial secondary metabolite 2,4-diacetylphloroglucinol. Mol. Plant Microbe Interact. 5:4-13. [Google Scholar]
- 22.Keel, C., D. M. Weller, A. Natsch, G. Défago, R. J. Cook, and L. S. Thomashow. 1996. Conservation of the 2,4-diacetylphloroglucinol biosynthesis locus among fluorescent Pseudomonas strains from diverse geographic locations. Appl. Environ. Microbiol. 62:552-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.King, E. O., M. K. Ward, and D. E. Raney. 1954. Two simple media for the demonstration of pyocyanin and fluorescein. J. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 24.Kropf, S., H. Heuer, M. Gruning, and K. Smalla. 2004. Significance test for comparing complex microbial community fingerprints using pairwise similarity measures. J. Microbiol. Methods 57:187-195. [DOI] [PubMed] [Google Scholar]
- 25.Kucey, R. M. N., H. H. Jenzen, and M. E. Leggett. 1989. Microbially mediated increases in plant available phosphorus. Adv. Agron. 42:199-228. [Google Scholar]
- 26.Landa, B. B., O. V. Mavrodi, K. L. Schroeder, R. Allende-Molar, and D. M. Weller. 2006. Enrichment and genotypic diversity of phlD-containing fluorescent Pseudomonas spp. in two soils after a century of wheat and flax monoculture. FEMS Microbiol. Ecol. 55:351-368. [DOI] [PubMed] [Google Scholar]
- 27.Lutz, M. P., S. Wenger, M. Maurhofer, G. Défago, and B. Duffy. 2004. Signalling between bacterial and fungal biocontrol agents in a strain mixture. FEMS Microbiol. Ecol. 48:447-455. [DOI] [PubMed] [Google Scholar]
- 28.Maurhofer, M., C. Keel, U. Schnider, C. Voisard, D. Haas, and G. Défago. 1992. Influence of enhanced antibiotic production in Pseudomonas fluorescens strain CHA0 on its disease suppressive capacity. Phytopathology 82:190-195. [Google Scholar]
- 29.Maurhofer, M., E. Bähler, R. Notz, V. Martinez, and C. Keel. 2004. Crosstalk between 2,4-diacetylphloroglucinol-producing biocontrol pseudomonads on wheat roots. Appl. Environ. Microbiol. 70:1990-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mazzola, M., D. L. Funnell, and J. M. Raaijmakers. 2004. Wheat cultivar-specific selection of 2,4-diacetylphloroglucinol-producing fluorescent Pseudomonas species from resident soil populations. Microb. Ecol. 48:338-348. [DOI] [PubMed] [Google Scholar]
- 31.McSpadden Gardener, B. B., D. V. Mavrodi, L. S. Thomashow, and D. M. Weller. 2001. A rapid polymerase chain reaction-based assay characterizing rhizosphere populations of 2,4-diacetylphloroglucinol-producing bacteria. Phytopathology 91:44-54. [DOI] [PubMed] [Google Scholar]
- 32.Notz, R., M. Maurhofer, U. Schnider-Keel, B. Duffy, D. Haas, and G. Défago. 2001. Biotic factors affecting expression of the 2,4-diacetylphloroglucinol biosynthesis gene phlA in Pseudomonas fluorescens biocontrol strain CHA0 in the rhizosphere. Phytopathology 91:873-881. [DOI] [PubMed] [Google Scholar]
- 33.Nowak-Thompson, B., S. J. Gould, J. Kraus, and J. E. Loper. 1994. Production of 2,4-diacetylphloroglucinol by the biocontrol agent Pseudomonas fluorescens PF-5. Can. J. Microbiol. 40:1064-1066. [Google Scholar]
- 34.Okubara, P. A., J. P. Kornoely, and B. B. Landa. 2004. Rhizosphere colonization of hexaploid wheat by Pseuodmonas fluorescens strains Q8r1-96 and Q2-87 is cultivar-variable and associated with changes in gross root morphology. Biol. Control 30:392-403. [Google Scholar]
- 35.Okubara, P. A., and R. F. Bonsall. 2008. Accumulation of Pseudomonas-derived 2,4-diacetylphloroglucinol on wheat seedlings roots is influenced by host cultivar. Biol. Control 46:322-331. [Google Scholar]
- 36.Picard, C., and M. Bosco. 2006. Heterozygosis drives maize hybrids to select elite 2,4-diacetylphloroglucinol-producing Pseudomonas strains among resident soil populations. FEMS Microbiol. Ecol. 58:193-204. [DOI] [PubMed] [Google Scholar]
- 37.Raaijmakers, J. M., D. M. Weller, and L. S. Thomashow. 1997. Frequency of antibiotic-producing Pseudomonas spp. in natural environment. Appl. Environ. Microbiol. 63:881-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raaijmakers, J. M., and D. M. Weller. 1998. Natural plant protection by 2,4-diacetylphloroglucinol-producing Pseudomonas spp. in take-all decline soils. Mol. Plant Microbe Interact. 11:144-152. [Google Scholar]
- 39.Raaijmakers, J. M., R. F. Bonsall, and D. M. Weller. 1999. Effect of population density of Pseudomonas fluorescens on production of 2,4-diacetylphloroglucinol in the rhizosphere of wheat. Phytopathology 89:470-475. [DOI] [PubMed] [Google Scholar]
- 40.Ramette, A., J. J. LiPuma, and J. M. Tiedje. 2005. Species abundance and diversity of Burkholderia cepacia complex in the environment. Appl. Environ. Microbiol. 71:1193-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramette, A., Y. Moënne-Loccoz, and G. Défago. 2006. Genetic diversity and biocontrol potential of fluorescent pseudomonads producing phloroglucinols and hydrogen cyanide from Swiss soils naturally suppressive or conducive to Thielaviopsis basicola-mediated black root rot of tobacco. FEMS Microbiol. Ecol. 55:369-381. [DOI] [PubMed] [Google Scholar]
- 42.Rezzonico, F., C. Binder, G. Défago, and Y. Moënne-Loccoz. 2005. The type III secretion system of biocontrol Pseudomonas fluorescens KD targets the phytopathogenic chromista Pythium ultimum and promotes cucumber protection. Mol. Plant Microbe Interact. 18:991-1001. [DOI] [PubMed] [Google Scholar]
- 43.Rodríguez, H., and R. Fraga. 1999. Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol. Adv. 17:319-339. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 45.Shanahan, P., D. J. O'Sullivan, P. Simpson, J. D. Glennon, and F. O'Gara. 1992. Isolation of 2,4-diacetylphloroglucinol from a fluorescent pseudomonad and investigation of physiological parameters influencing its production. Appl. Environ. Microbiol. 58:353-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharifi-Tehrani, A., M. Zala, A. Natsch, Y. Moenne-Loccoz, and G. Défago. 1998. Biocontrol of soil-borne fungal plant diseases by 2,4-diacetylphloroglucinol-producing fluorescent pseudomonads with different restriction profiles of amplified 16S rDNA. Eur. J. Plant Pathol. 104:631-643. [Google Scholar]
- 47.Smith, K. P., J. Handelsman, and R. M. Goodman. 1999. Genetic basis in plants for interactions with disease-suppressive bacteria. Proc. Natl. Acad. Sci. U. S. A. 96:4786-4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stutz, E. W., G. Défago, and H. Kern. 1986. Naturally occurring fluorescent pseudomonads involved in suppression of black root rot of tobacco. Phytopathology 76:181-185. [Google Scholar]
- 49.Vincent, M. N., L. A. Harrison, J. M. Brackin, P. A. Kovacevich, P. Mukerji, D. M. Weller, and E. A. Pierson. 1991. Genetic analysis of the antifungal activity of a soilborne Pseudomonas aureofaciens strain. Appl. Environ. Microbiol. 57:2928-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, C. X., A. Ramette, P. Punjasamarnwong, M. Zala, A. Natsch, Y. Moënne-Loccoz, and G. Défago. 2001. Cosmopolitan distribution of phlD-containing dicotyledonous crop-associated biocontrol pseudomonads of worldwide origin. FEMS Microbiol. Ecol. 37:105-116. [Google Scholar]
- 51.Ward, J. H. 1963. Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 58:236-244. [Google Scholar]
- 52.Weller, D. J., B. B. Landa, O. V. Mavrodi, K. L. Schroeder, L. De La Fuente, S. Blouin Bankhead, R. Allende Molar, R. F. Bonsall, D. V. Mavrodi, and L. S. Thomashow. 2007. Role of 2,4-diacetylphloroglucinol-producing fluorescent Pseudomonas spp. in the defense of plant roots. Plant Biol. 9:4-20. [DOI] [PubMed] [Google Scholar]
- 53.Weller, D. J. 2007. Pseudomonas biocontrol agents of soilborne pathogens: looking back over 30 years. Phytopathology 97:250-256. [DOI] [PubMed] [Google Scholar]
- 54.Wissuwa, M., M. Mazzola, and C. Picard. 2009. Novel approaches in plant breeding for rhizosphere-related traits. Plant Soil 321:409-430. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.