Abstract
Myxococcus xanthus is a soil-dwelling bacterium that exhibits a complex life cycle comprising social behavior, morphogenesis, and differentiation. In order to successfully complete this life cycle, cells have to cope with changes in their environment, among which the presence of copper is remarkable. Copper is an essential transition metal for life, but an excess of copper provokes cellular damage by oxidative stress. This dual effect forces the cells to maintain a tight homeostasis. M. xanthus encodes a large number of genes with similarities to others reported previously to be involved in copper homeostasis, most of which are redundant. We have identified three genes that encode copper-translocating P1B-ATPases (designated copA, copB, and copC) that exhibit the sequence motifs and modular organizations of those that extrude Cu+. The expression of the ATPase copC has not been detected, but copA and copB are differentially regulated by the addition of external copper. However, while copB expression peaks at 2 h, copA is expressed at higher levels, and the maximum is reached much later. The fact that these expression profiles are nearly identical to those exhibited by the multicopper oxidases cuoA and cuoB suggests that the pairs CuoB-CopB and CuoA-CopA sequentially function to detoxify the cell. The deletion of any ATPase alters the expression profiles of other genes involved in copper homeostasis, such as the remaining ATPases or the Cus systems, yielding cells that are more resistant to the metal.
Copper is required as a cofactor for a number of enzymes involved in essential cellular processes. However, the two oxidation states of copper not only allow its participation in essential redox reactions but also form reactive oxygen species, leading to severe damage of cytoplasmic constituents (23). Copper homeostasis is a complicated process involving copper acquisition, sequestration, and efflux. These mechanisms are tightly regulated and are able to respond to changes in the extracellular bioavailability and intracellular demand for the metal. In order to prevent copper damage, sophisticated defense mechanisms have evolved, one of which is mediated by a group of P-type ATPases designated P1B-type ATPases (1).
P1B-type ATPases catalyze the transport of transition metal or heavy metal ions across cell membranes, coupling ATP hydrolysis to the transport of cations through a catalytic cycle involving the autophosphorylation of an Asp residue within a highly conserved DKTGT motif. The range of transported substrates is wide, including monovalent (Cu+, Ag+, and Au+) as well as divalent (Cu2+, Co2+, Zn2+, Cd2+, Hg2+, Pb2+, and Mn2+) cations. Structural features and conserved sequence motifs of transition-metal P-type pumps suggest a division into several subgroups with distinct substrate specificities. Two subgroups of P1B-ATPases export either Cu+ or Cu2+, which share a topological arrangement containing eight transmembrane domains (TMs) but differ in specific signature sequences in TMs 6, 7, and 8. Another structural characteristic of copper-dependent ATPases is the presence of cytoplasmic copper-binding domains in their N termini (1).
Myxococcus xanthus is considered a model to study multicellularity and differentiation in prokaryotes due to its unique life cycle. Under nutrient-rich conditions, M. xanthus cells glide on soil in search of organic matter, including other microorganisms, on which they feed. Starvation triggers a developmentally regulated signaling cascade that induces aggregation and the formation of multicellular fruiting bodies filled with environmentally resistant spores, which germinate when conditions become favorable for growth again (33). Because M. xanthus resides on soils, this complex cycle must be carried out in the presence of a variety of changing elements (21). One of these elements is copper, which is present in soils in concentrations that range between 2 and 100 ppm.
The elucidation of the copper response in M. xanthus has turned out to be interesting for several reasons. First, this metal induces the genes responsible for carotenogenesis (20). Second, the copper response can be studied during growth and development; in fact, growing cells exhibit around 15-fold-greater resistance to copper than do developing cells, while cells preadapted to this metal reach the same levels of resistance during both stages (27). Third, the concentration of metal required to induce the expression of some copper-dependent genes is around 10-fold lower during development than during growth (27).
The M. xanthus genome holds a plethora of gene products with sequence similarities to proteins known to be involved in copper homeostasis in other Gram-negative bacteria, most of which are redundant (19). Although the abundance of paralogous genes in myxobacteria seems to correlate with their large genome sizes (9, 25, 28), it is surprising to find such a large number of genes involved in copper homeostasis in M. xanthus, because this bacterium is not especially resistant to this metal. In order to clarify the physiological roles during the complete life cycle and the advantage of maintaining copper-related duplicated genes during evolution, we have previously characterized two families of paralogs: three multicopper oxidases (MCOs) and six CBA-type heavy metal efflux systems (19, 27).
In this work we have concentrated our efforts on the study of the three paralogs of the P1B-type ATPase family, named CopA, CopB, and CopC. We have found that copA and copB are differentially regulated by this metal, contributing to the adaptive response that allows the completion of this bacterium life cycle in a copper-fluctuating environment. The complexity of M. xanthus copper homeostasis is further illustrated by the interplay among the large number of components that participate in this process.
MATERIALS AND METHODS
Identification of copper-dependent P1B-type ATPases in the M. xanthus genome.
Genes encoding P-type ATPases were identified by BLASTP analysis against the M. xanthus genome by using the Pfam matrix for E1-E2_ATPase (Pfam accession number PF00122). The six sequences obtained with an E value of ≤0.1 were back-searched against the Pfam database (6) to confirm that they really encode P-type ATPases and to determine their modular organizations. The number of TMs was analyzed with the TMHMM (http://www.cbs.dtu.dk/services/TMHMM-2.0/) and SOSUI (http://bp.nuap.nagoya-u.ac.jp/sosui/) servers, and the conserved sequences defined previously by Argüello et al. (1) for copper-dependent ATPases were searched manually.
Bacterial strains, plasmids, and culture conditions.
Bacterial strains and plasmids used in this study are listed in Tables S1 and S2, respectively, in the supplemental material, together with their genotypes and origins. All M. xanthus strains were grown in CTT broth (11) with vigorous shaking (300 rpm) at 30°C. CTT agar plates contained 1.5% Bacto agar (Difco). When necessary, kanamycin (80 μg ml−1), X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (100 μg ml−1), or galactose (10 mg ml−1) was added. CF medium (10) was used to induce development. Cells were grown in CTT broth to 3.0 × 108 cells ml−1, corresponding to an optical density at 600 nm (OD600) of 1, and resuspended to 4.5 × 109 cells ml−1 (OD600 = 15) in TM buffer (10 mM Tris-HCl [pH 7.6], 1 mM MgSO4). Ten microliters of each suspension was spotted onto CF agar plates and incubated at 30°C, and the progression of development was monitored with a Wild Heerbrugg dissecting microscope. When required, nutrient-rich and starvation media were supplemented with metals at the concentrations indicated in the figures and tables. Escherichia coli strains were grown at 37°C in Luria-Bertani medium (26), which was supplemented with kanamycin (25 μg ml−1) or X-Gal (25 μg ml−1) when needed.
Nucleic acid manipulations.
Standard protocols were followed for nucleic acid manipulation (26). Total RNA was prepared by use of the High Pure RNA isolation kit provided by Roche. For these experiments, cells were grown on CTT or CF medium without the addition of external copper and in the same medium supplemented with 300 and 40 μM copper, respectively. Samples were treated with a DNA-free kit (Ambion) to completely remove chromosomal DNA. The RNA was first subjected to reverse transcription (RT) using primer CopCcDNA, and the cDNA thus obtained was amplified with internal copC primers CopCF and CopCR (see Table S3 in the supplemental material) by 30 cycles of PCR.
Construction and assay of strains harboring lacZ fusions.
The lacZ fusion plasmids were constructed by using vector pKY481 (4). Fragments encompassing the upstream copA, copB, and copC regions were amplified by PCR using wild-type (WT) chromosomal DNA as a template, the oligonucleotides listed in Table S3 in the supplemental material as primers, and the high-fidelity polymerase PrimeSTARHS (Takara). The BamHI site in the primers was introduced to create the fusion either at the start codon (for copB and copC) or inside the genes (for copA and copC) and in frame with the BamHI site existing in the lacZ gene of plasmid pKY481. PCR products were digested with KpnI and BamHI and ligated into vector pKY481 digested with the same enzymes. The resulting plasmids were introduced into M. xanthus cells (WT or mutants) by electroporation (13), and the kanamycin-resistant (Kmr) colonies were analyzed by Southern blotting.
Strains harboring lacZ fusions were incubated at 30°C on CTT and CF agar plates containing different metal concentrations. When necessary, the metal chelator bathocuproine sulfonate (BCS) (0.4 mM) or EDTA (0.4 mM) was added. For quantitative analysis of β-galactosidase activity, cell extracts were obtained at different times by sonication and analyzed as previously reported (20). Proteins in the supernatants were determined by using the Bio-Rad protein assay (Bio-Rad, Inc.) with bovine serum albumin as a standard. β-Galactosidase activity was determined as described previously by Kroos et al. (15). Specific activity is expressed as nmol o-nitrophenol produced per min and mg of protein. The results are the averages of data from three different experiments.
Construction of in-frame deletion mutants.
Sequences upstream and downstream of the copA, copB, and copC genes were amplified by PCR as described above, using the oligonucleotides listed in Table S3 in the supplemental material as primers. The PCR products were cloned into KpnI-HindIII-digested pBJ113 (12) to obtain plasmids harboring the in-frame deletion of the corresponding gene (Table S2). Those plasmids were introduced into M. xanthus by electroporation. The chromosomal integration of the plasmids was selected by plating cells onto CTT agar plates containing 80 μg ml−1 kanamycin. Several randomly chosen Kmr merodiploids were analyzed by Southern blot hybridization to confirm the proper recombination event. One positive strain was then grown in the absence of kanamycin and plated onto CTT plates containing 1% galactose. Southern blot analysis was used again to screen several kanamycin-sensitive (Kms) and galactose-resistant (Galr) colonies obtained to verify the loss of the WT allele.
Copper accumulation measurements.
For the determination of copper accumulation, cultures grown overnight in CTT medium were concentrated to an OD600 of 15 as described above, and 10 drops of 20 μl each were spotted onto CTT agar plates supplemented with 600 μM copper. After a 2- or 24-h incubation at 30°C, cells were harvested into a 15-ml sterile polypropylene centrifuge tube and washed three times with modified CTT medium (without MgSO4 and containing 1 mM EDTA). The pellets were dried at 80°C overnight and dissolved in 5 ml of trace-metal-grade nitric acid by heating at 80°C for 30 min according to a method described previously by Outten et al. (24). The metal content of the samples was measured by using the graphite furnace technique employing a Perkin-Elmer 5100ZL atomic absorption spectrophotometer. A Pd/Mg modifier was used to stabilize copper during the pyrolysis step. The calibration curve was constructed from four standards (100, 200, 400, and 600 ppb) and a blank. Measurements shown are the averages of data from triplicate experiments.
RESULTS
Three copper-dependent P1B-type ATPases in the M. xanthus genome.
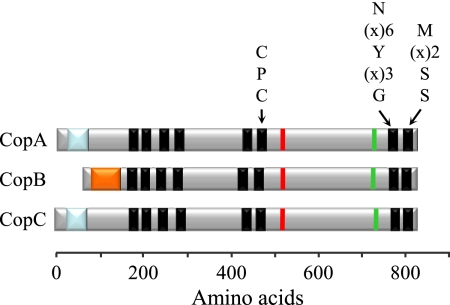
The in silico analysis of the M. xanthus genome showed that this bacterium holds six genes coding for proteins belonging to the family of P-type ATPases, three of which meet the criteria previously described for the subgroup of P1B-type ATPases involved in the export of Cu+ across the cytoplasmic membrane (1). We have named these genes copA, copB, and copC, which are found in the genome by the gene identifiers MXAN_3415, MXAN_3422, and MXAN_0979, respectively. Therefore, copC is located in copper region 1, while copA and copB are located in copper region 2, which also includes the three genes for the MCOs cuoA, cuoB, and cuoC (19). The three ATPases contain eight TMs and the conserved signature sequences in TMs 6, 7, and 8 (Fig. 1) present in most Cu-translocating ATPases. Additionally, they exhibit an N-terminal metal-binding domain. However, while CopA and CopC contain an uncharacterized YHS domain (Pfam accession number PF04945), which seems to be involved in copper binding, CopB contains an HMA (heavy metal-associated) domain, identified in the Pfam database as accession number PF00403 (8). Finally, CopA and CopC are very similar to one another, and their sequences exhibit 71.9% identity. On the contrary, CopB differs from CopA and CopC, exhibiting only 43.8 and 45.4% identities, respectively.
FIG. 1.
Relevant features of the three ATPases. The eight predicted TMs are shown as black boxes. The conserved sequences in TMs 6, 7, and 8 are indicated on the top of each box, where x represents any amino acid. The phosphorylation site (sequence DKTGT) and the nucleotide-binding domain are represented by a red line and a green line, respectively. The HMA domain of CopB is indicated by an orange box, whereas the YHS domains of CopA and CopC are represented by blue boxes.
Expression of the three cop genes.
In order to analyze the expression profiles of the three cop genes, M. xanthus strains harboring fusions between copA, copB, or copC and lacZ from E. coli were constructed. In the case of copB and copC, the fusions were designed at the start codon of each gene, while in the case of copA, the fusion was performed in a BamHI site located in the region that encodes the YHS domain of this ATPase, which is in frame with the same restriction site found in the lacZ gene of plasmid pKY481 (4).
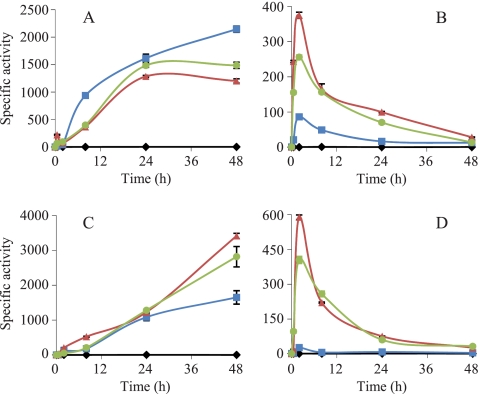
The analyses of the expression profiles of the three genes during growth indicated that the expression of the three ATPases was undetectable in the absence of metals. In contrast, the expression of copA and copB exhibited a high level of dependence on the addition of external copper. However, the expression levels of copA slowly increased in a time-dependent manner after copper supplementation (Fig. 2 A), while those of copB rose very rapidly, exhibiting a peak at 2 h, which subsequently decreased until basal levels were reached at 48 h (Fig. 2B).
FIG. 2.
Copper upregulation of copA and copB. (A and B) During growth, strains harboring the copA-lacZ (A) and copB-lacZ (B) fusions were incubated on CTT agar plates containing 0 μM (black lines), 300 μM (blue lines), 600 μM (red lines), or 800 μM (green lines) copper. (C and D) During development, strains harboring the copA-lacZ (C) and copB-lacZ (D) fusions were incubated on CF agar plates containing 0 μM (black lines), 20 μM (blue lines), 40 μM (red lines), or 60 μM (green lines) copper. The specific β-galactosidase activity in cell extracts was determined as described in Materials and Methods. The results are the averages of data from three different experiments. The error bars indicate standard deviations. Note the differences in the scales.
Surprisingly, the strain harboring the copC-lacZ fusion did not show expression in the presence or absence of copper (data not shown). To corroborate the lack of expression of copC, two other translational fusions were constructed to cover the two possible topologies of the protein in the membrane, one at the same location as that constructed for copA and another one in the region between the third and the fourth TMs. However, the expression of copC still remained undetectable in these two strains. Finally, we tested whether copper depletion could induce copC in order to increase metal uptake, as was reported previously for other bacteria (2, 30). To prove this hypothesis, the expression of copC was assayed in media containing BCS (an effective Cu+ chelator) and EDTA (which exhibits a broader range of chelating action on metal cations), but no expression was detected under those conditions either. The results obtained with the three translational fusions constructed indicated that copC could be expressed at very low levels, undetectable by using this methodology. Consequently, the expression of copC was approached by using the more sensitive RT-PCR technique. The total RNA from growing and developing cells incubated in the presence and absence of copper was used as a template for RT-PCR experiments. None of the reactions amplified the expected fragment, except for the positive control with genomic DNA as a template (data not shown).
The expression of the three ATPases during growth was also analyzed in the presence of other monovalent (Ag+ and Au+) and divalent (Ni2+, Co2+, Zn2+, and Cd2+) metals. Only Ni2+, Co2+, and Zn2+ increased the expression level of copA, although the levels reached with these other cations were significantly lower than those in the case of copper (Table 1). No induction of copB was observed when cations different from copper were added to the growth medium, suggesting a specificity of regulation by this metal. In the case of copC, the expression also remained undetectable with the other metals tested by using the three strains constructed (data not shown).
TABLE 1.
Response of the copA promoter to metals
| Inducer | Growth |
Development |
||||
|---|---|---|---|---|---|---|
| Metal concn (mM) | Avg β-galactosidase activity ± SDa | Fold inductionb | Metal concn (mM) | Avg β-galactosidase activity ± SDa | Fold inductionb | |
| None | 1.3 ± 0.3 | 0.5 ± 0.2 | ||||
| CuSO4 | 0.8 | 1,488 ± 58 | 1,145 | 0.040 | 3,414 ± 96 | 6,828 |
| Ni(NO3)2 | 1.0 | 73 ± 2 | 56 | 2.0 | 168 ± 1 | 336 |
| Zn(NO3)2 | 0.25 | 4 ± 0.1 | 3 | 0.1 | 64 ± 1 | 128 |
| Co(NO3)2 | 1.0 | 101 ± 10 | 78 | 1.0 | 9 ± 0.1 | 18 |
Data are averages and standard deviations of data from three independent experiments.
The fold induction was calculated by dividing the mean of each β-galactosidase activity by the average value for the cells without metal (none).
To analyze the cop promoter response to copper during development, 10-fold-lower concentrations of the metal were used to prevent cells from dying (27). The expression profiles and the levels of expression of copA and copB during development were similar to those exhibited during growth (Fig. 2C and D). Furthermore, the expression of copA was also induced by Ni2+, Co2+, and Zn2+ (Table 1), while copB did not respond to the addition of other cations to the starvation medium. In the case of copC, no expression was observed during development either in the presence or in the absence of metals with any of the strains that were constructed (data not shown).
Copper sensitivity of cop single-deletion mutants.
To study the physiological role of each ATPase, markerless in-frame deletion mutants for each gene were created. When the three mutants and the WT strain were cultured in liquid CTT medium without metals, no differences in growth were observed between the strains either in the generation times or in the cell densities reached at the stationary phase. Furthermore, the mutants' fruiting bodies were also similar to those of the WT strain on starvation medium (data not shown).
However, when the WT strain and the mutants were grown in media containing copper, some differences were observed. The deletion of copA or copC led to a very small decrease in copper tolerance. Surprisingly, the ΔcopB mutant turned out to be more resistant to the metal than the WT strain (Fig. 3 A). The fact that none of the mutants was very sensitive to copper and the ΔcopB mutant was even more resistant to the metal than the WT strain might be explained by the upregulation of other mechanisms involved in copper resistance. To check this possibility, we tested the tolerance of the cells when they were previously preincubated in the presence of a copper concentration high enough to induce the mechanisms involved in resistance to this metal. A preincubation period of 2 h in the presence of 300 μM copper was chosen for this experiment. After this preadaptation stage, cells were diluted into fresh CTT liquid medium supplemented with higher copper concentrations and incubated for 24 h. Under these conditions, the three mutants turned out to be more sensitive to copper than the WT strain, at least at high copper concentrations (Fig. 3B). Interestingly, the ΔcopB mutant now exhibited a greater sensitivity to the metal than did the other two mutants. Altogether, these two experiments indicate that the three ATPases confer copper tolerance to M. xanthus.
FIG. 3.
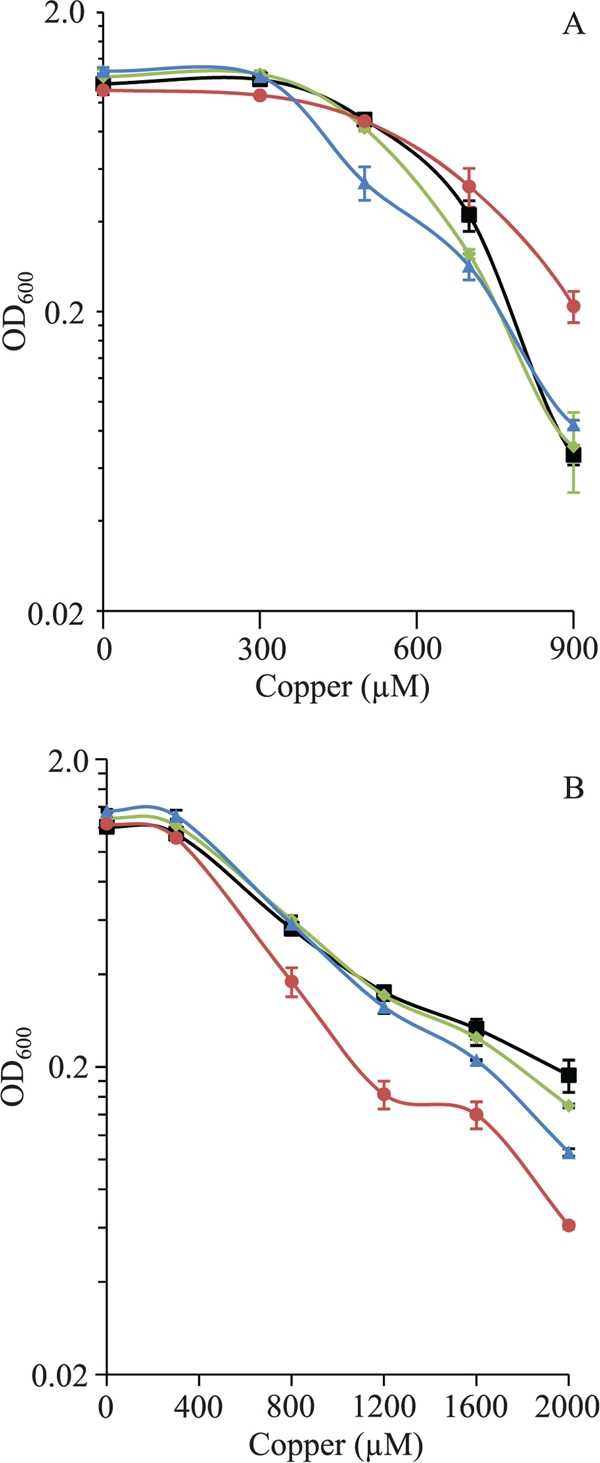
Copper sensitivity of the M. xanthus WT strain (black lines) and the ΔcopA (blue lines), ΔcopB (red lines), and ΔcopC (green lines) mutants during growth. (A) Copper tolerance of nonpreadapted cells to the metal. Cells grown in the absence of copper were diluted to an OD600 of 0.05 in fresh CTT liquid medium containing the indicated copper concentrations. The data shown indicate the OD600 monitored after 24 h of incubation. (B) Copper tolerance of cells adapted to copper. The strains were grown in the presence of 300 μM copper prior to dilution into fresh CTT liquid medium containing different copper concentrations. The data are expressed as mentioned above for A. Data are the averages of data from three experiments. The error bars indicate standard deviations.
Regarding development, the differences observed between the WT and the mutants in the presence of different copper concentrations in nonpreadapted cells were not significant, and only a slight delay in aggregation of about 2 h was detected in the case of the mutants. When cells were preadapted to the metal, the three mutants and the WT strain exhibited similar copper sensitivities and behaviors (data not shown).
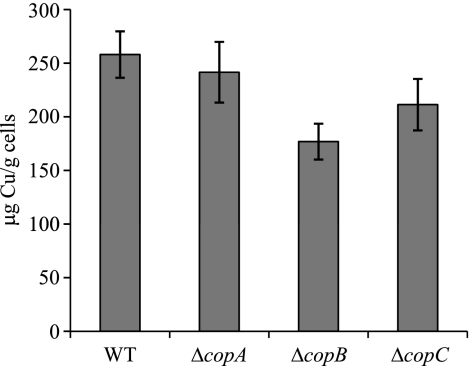
Copper accumulation in the cop deletion mutants.
As the copper sensitivity of nonpreadapted mutant cells was not as dramatic as expected, the level of copper accumulation was determined for the WT strain and the three deletion mutants after a 2-h incubation in the presence of metal. The results obtained showed that all the mutants accumulated a slightly smaller amount of metal than did the WT strain (Fig. 4), which might explain why the three cop mutants are not much more sensitive to copper than the WT strain (Fig. 3A). As the maximum expression levels of copA are not reached at 2 h after metal supplementation, copper accumulation was also determined for the WT strain and the ΔcopA mutant at 24 h. At this time, both strains also accumulated very similar amounts of copper (411 ± 37 μg Cu per g of cells in the case of the WT versus 387 ± 83 μg Cu per g of cells in the case of the mutant).
FIG. 4.
Copper accumulation of the WT strain and the cop deletion mutants during growth. Samples were taken after a 2-h incubation in the presence of 600 μM copper. The results are the averages of data from three different experiments. The error bars indicate standard deviations.
Expression of the cop genes in the deletion mutants.
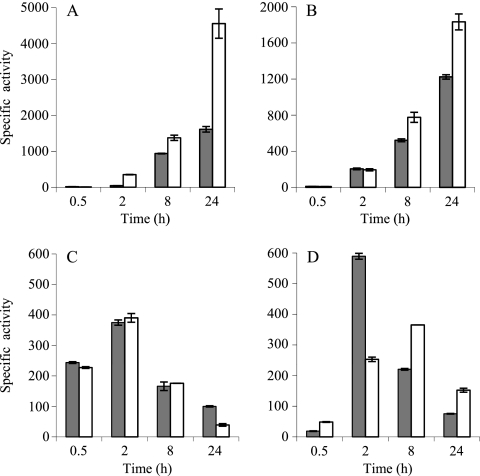
The fact that nonpreadapted mutant cells were not as sensitive to copper as one might expect and that they did not accumulate more copper than the WT strain could be explained by the upregulation of other mechanisms involved in copper resistance encoded in the genome. This possibility prompted us to test whether the P1B-type ATPases that still remained in each mutant exhibited an altered expression profile. To address this question, plasmids containing the fusions between the corresponding cop genes and lacZ were introduced into the different mutants. The ATPase copC remained undetectable in the ΔcopA and ΔcopB mutants. In contrast, the levels of copA in a ΔcopB background at 2 h (the time at which the level of copB peaks) were much higher than those in the WT strain during growth, similar to those of copB in the WT strain (compare Fig. 5 A to 2B). At 24 h, copA expression was more than 2-fold higher in the ΔcopB mutant than in the WT background (Fig. 5A). The levels of expression of copA in this mutant during development were also higher than those in the WT, although the increase observed was not as dramatic as in the case of growth (Fig. 5B).
FIG. 5.
(A and B) Expression of copA in WT (gray columns) and ΔcopB (white columns) backgrounds during growth (A) and development (B). Copper concentrations used were 300 μM during growth and 40 μM during development. (C and D) Expression of copB in WT (gray columns) and ΔcopA (white columns) backgrounds during growth (C) and development (D). Copper concentrations used were 600 μΜ during growth and 40 μM during development. The results are the averages of data from three different experiments. The error bars indicate standard deviations.
The influence of the deletion of copA on the expression of copB was different. The absence of CopA did not significantly affect the expression of copB during growth (Fig. 5C). In contrast, two major differences could be observed during development: (i) the maximum expression levels for copB in the ΔcopA mutant were lower than in the WT strain, and (ii) the peak appeared later, at 8 h instead of at 2 h (Fig. 5D).
Finally, when the copA and copB fusions were introduced into the ΔcopC mutant, it was found that the expression levels and profiles of these two ATPases were nearly identical to those determined in the WT background (data not shown).
Copper sensitivity of double- and triple-deletion mutants.
According to the data described above, it should be expected that the deletion of two or three cop genes yielded strains that were very sensitive to copper. Hence, we constructed the three reciprocal double mutants and the triple mutant and determined their sensitivities to copper during growth and development. The phenotypic characterization of the ΔcopA ΔcopC and ΔcopB ΔcopC double mutants revealed an increase in copper sensitivity compared to that of the WT strain during growth in both nonpreadapted and preadapted cells (Fig. 6). Contrary to what was expected, nonpreadapted cells of the ΔcopA ΔcopB mutant grew at higher rates than did the WT strain with copper concentrations as high as 800 μM (Fig. 6A). An even more pronounced resistance of these mutant cells was observed when they were preadapted to the metal (Fig. 6B). Regarding the ΔcopA ΔcopB ΔcopC triple mutant, the behavior of this strain during growth was almost identical to that of the ΔcopA ΔcopB mutant (Fig. 6).
FIG. 6.
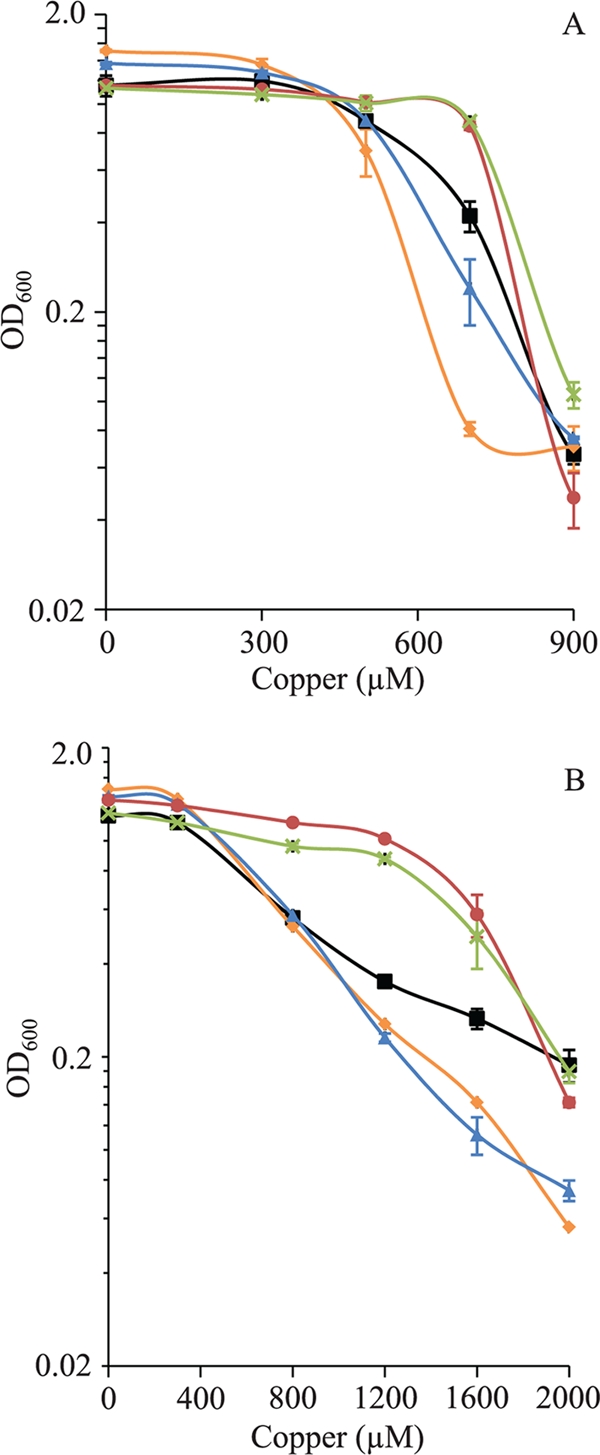
Copper sensitivities of the M. xanthus WT strain (black lines) and the ΔcopA ΔcopC (orange lines), ΔcopB ΔcopC (blue lines), ΔcopB ΔcopA (red lines), and ΔcopA ΔcopB ΔcopC (green lines) mutants during growth. (A) Copper tolerance of nonpreadapted cells to the metal. (B) Copper tolerance of cells adapted to copper. The experiment was carried out as indicated in the legend of Fig. 3.
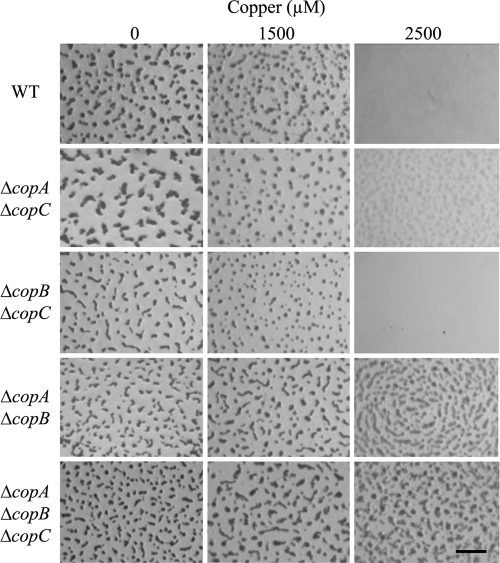
The analysis during development did not show significant differences between the WT strain and the double and triple mutants when cells were not preadapted to the metal. However, preadapted mutant cells exhibited a slight delay in aggregation when they were plated onto starvation medium with no copper. Moreover, the ΔcopB ΔcopC mutant exhibited copper sensitivities similar to those of the WT strain. In contrast, the ΔcopA ΔcopB and ΔcopA ΔcopC double mutants and the ΔcopA ΔcopB ΔcopC triple mutant turned out to be more resistant to the metal than the WT strain (Fig. 7).
FIG. 7.
Development of the preadapted M. xanthus WT strain and deletion mutants in the presence of different concentrations of copper. The bar represents 1 mm.
Expression of the cus systems in the ΔcopA ΔcopB mutant.
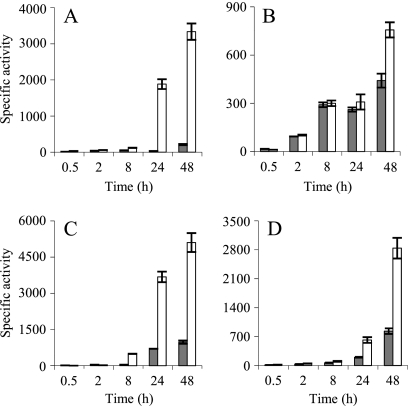
Since M. xanthus also encodes two Cus-like systems (19) that are clearly upregulated by copper (cus2 and cus3), their expression profiles in the ΔcopA ΔcopB mutant were analyzed. As shown in Fig. 8, these two cus systems were overinduced in the mutant. This induction was extremely marked in the case of the cus2 system during growth (Fig. 8A), where the level of activity was raised around 16-fold at 48 h in the double mutant compared to the WT. Similarly, at 48 h, the cus3 transporter increased its expression 5-fold during growth and 3-fold during development (Fig. 8C and D). The high level of upregulation of the cus systems in the ΔcopA ΔcopB mutant explains the unexpected copper resistance exhibited by this strain.
FIG. 8.
Expression of cus2 and cus3 in WT (gray columns) and ΔcopA ΔcopB (white columns) backgrounds during growth and development. (A and B) In the case of cus2, the expression was monitored by using 800 μM copper during growth (A), whereas 40 μM copper was added during development (B). (C and D) For cus3, 600 μM copper was used during growth (C), and 60 μM was used during development (D). The results are the averages of data from three different experiments. The error bars indicate standard deviations. Note the differences in the scales.
DISCUSSION
M. xanthus genome sequence analyses have revealed the presence of a variety of paralogous genes that encode proteins with similarities to others known to be involved in copper holding and trafficking, including MCOs, P1B-type ATPases, and CBA-like heavy metal efflux pumps (Cus and Czc systems). The question that arises from this observation is why this bacterium needs this abundance of redundant genes. To answer this intriguing question, the three MCOs and the six CBA-like heavy metal efflux transporters were first studied (19). Here, we continue the work with M. xanthus copper homeostasis with the characterization of three P-type ATPases that belong to the P1B-type subgroup proposed previously by Argüello et al. (1), which translocates Cu+ from the cytoplasm to the periplasmic space.
The results obtained reflect several similarities in the expression patterns of ATPases and MCOs. Thus, the timing and expression profile of copA are nearly identical to those of cuoA. In addition, the same metals mimic the copper effect on the expression of both genes. A similar correlation was observed between the expression patterns of copB and cuoB. The similarities in expression of cuoA-copA and cuoB-copB and their proximity in the genome suggest that each pair of genes must be forming part of the same operon or that they must be regulated by the same transcription factor. Moreover, the coincidence in the expression time of a P1B-type ATPase with an MCO indicates that they most likely will function together, with the Cop protein pumping Cu+ from the cytosol to the periplasmic space, where the cognate MCO will oxidize Cu+ to the less toxic form Cu2+ (27). This cooperative work will allow a proper detoxification of the whole cell. On the other hand, the timing of expression of each MCO/ATPase pair is different, pointing to the direction that M. xanthus possesses a primary copper-adaptive response consisting of CopB and CuoB, with a rapid gene induction after the external copper addition, while the ATPase CopA and the MCO CuoA are necessary for the maintenance of the adaptive response over time. Although it is tempting to speculate that the ATPase CopC and the constitutively expressed MCO CuoC (27) could also work together, representing a third detoxification mechanism at low copper concentrations, the fact that the expression of copC has not been detected by any of the methods and laboratory conditions tested does not allow the drawing of any conclusions.
Another interesting observation is that the copper induction of copA and copB cannot be mimicked by the monovalent metals, as was reported previously for E. coli for the ATPase CopA and the MCO CueO (3) and for M. xanthus for the Cus2 and Cus3 systems, whose promoters respond only to copper, Ag+, and Au+ (19). The specificity of the M. xanthus copA and copB genes for copper and divalent metals or only copper, respectively, indicates that metals most likely will be sensed in the exterior of the cytoplasmic membrane, where copper will be found mainly in the oxidized state instead of the reduced form that predominates in the cytoplasm (7, 31).
The function of P1B-type ATPases in copper homeostasis has been established for different organisms (5, 14, 17, 18, 29, 30, 32, 34, 35), but to our knowledge, only three bacteria have been reported to encode more than one ATPase involved in copper homeostasis (30). The need for so many elements involved in copper homeostasis in M. xanthus is not obvious, especially if we take into consideration that the level of copper resistance of this bacterium is low compared to those of other prokaryotes that carry only one gene of each type in their genomes. Nies et al. (22) previously suggested that paralogous genes involved in the copper response, including those that are silent, might provide an advantage that compensates for the cost of maintaining them during evolution. The possible benefit of this redundancy in the case of M. xanthus could be related to the life cycle of this bacterium. Growing M. xanthus cells are long rods with a cell envelope typical of Gram-negative bacteria although with several differences in chemical composition and structure that are required to allow conversion into round myxospores during development (33). These differences in the cell envelope might also make the cells more sensitive to the oxidative stress induced by copper. This notion is supported by the greater sensitivity exhibited by developing cells during the morphogenetic event. Undoubtedly, the participation of a large number of elements involved in copper detoxification that are differentially regulated by several types of environmental conditions will provide the cells with a great plasticity in the adaptation process, contributing to ensure survival in a changing metal environment during growth and development.
The phenotypic analysis of deletion mutants suggests that the three ATPases are implicated in the M. xanthus copper-adaptive response, although several results obtained for the copper sensitivity and accumulation of the mutants cannot be easily explained due to the intricate network of mechanisms involved in copper homeostasis in this bacterium and the interplay among all of them. Nevertheless, some of the data presented here provide, at least partially, an explanation for some of these observations. For instance, the earlier and higher level of expression of copA in the ΔcopB mutant seems to offer a good explanation about why this mutant is more resistant to copper than the WT strain during growth. As the same alteration was not observed during development, mutant and WT developing cells exhibit similar copper sensitivities. It must be taken into consideration that the lack of one ATPase (which is expressed in the WT strain at a specific time, along with an MCO after the copper addition) most likely will affect the allocation of the metal in the cell compartments (cytoplasm and periplasm) and its oxidation state. These differences may be sufficient to activate the transcriptional regulators that control the induction of the remaining ATPases, producing alterations in their expression profiles. Similarly, the structural and physiological differences between growing and developing cells might explain why the expression profiles of the cop genes do not exactly change in the same manner during both stages of the life cycle, given that the distribution and oxidation state of the metal most likely will not be identical in both cell types.
This explanation can also be valid for the upregulation of the cus systems in the ΔcopA ΔcopB mutant. The huge induction of the cus systems in this double mutant would compensate for the deficiency of the two P1B-type ATPases and might explain the contradictory copper-resistant phenotypes observed for this mutant. The complementation processes between Cus systems and other enzymes involved in copper homeostasis have also been described for other bacteria (16).
By compiling and analyzing all the data about MCOs (27), CBA systems (19), and ATPases, it can be concluded that M. xanthus requires the differential regulation of all these elements to ensure that copper homeostasis (and that of other metals) is tightly adjusted to prevent cell death by oxidative stress. The expression of this complex network of proteins must be controlled by different regulators that recognize the proper oxidation state of copper in different compartments. The identification and characterization of these regulators will undoubtedly help us to understand the complexity of copper handling and trafficking in M. xanthus throughout the entire life cycle.
Supplementary Material
Acknowledgments
This work has been supported by grants from the Junta de Andalucía (grant CVI-1377) and the Ministerio de Ciencia y Tecnología (grants BFU2006-00972/BMC and BFU2009-07565), Spain, 70% funded by FEDER, and funded by a grant from the Ministerio de Ciencia e Innovación, Spain, program Consolider-Ingenio 2010 (grant CSD2009-00006). A.M.-M. has been funded by a postdoctoral fellowship from the Plan Propio de la Universidad de Granada.
Footnotes
Published ahead of print on 23 July 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Argüello, J. M., E. Eren, and M. Gonzalez-Guerrero. 2007. The structure and function of heavy metal transport P1B-ATPases. Biometals 20:233-248. [DOI] [PubMed] [Google Scholar]
- 2.Cavet, J. S., G. P. Borrelly, and N. J. Robinson. 2003. Zn, Cu and Co in cyanobacteria: selective control of metal availability. FEMS Microbiol. Rev. 27:165-181. [DOI] [PubMed] [Google Scholar]
- 3.Changela, A., K. Chen, Y. Xue, J. Holschen, C. E. Outten, T. V. O'Halloran, and A. Mondragon. 2003. Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science 301:1383-1387. [DOI] [PubMed] [Google Scholar]
- 4.Cho, K., and D. R. Zusman. 1999. AsgD, a new two-component regulator required for A-signalling and nutrient sensing during early development of Myxococcus xanthus. Mol. Microbiol. 34:268-281. [DOI] [PubMed] [Google Scholar]
- 5.Espariz, M., S. K. Checa, M. E. Audero, L. B. Pontel, and F. C. Soncini. 2007. Dissecting the Salmonella response to copper. Microbiology 153:2989-2997. [DOI] [PubMed] [Google Scholar]
- 6.Finn, R. D., J. Tate, J. Mistry, P. C. Coggill, S. J. Sammut, H. R. Hotz, G. Ceric, K. Forslund, S. R. Eddy, E. L. Sonnhammer, and A. Bateman. 2008. The Pfam protein families database. Nucleic Acids Res. 36:D281-D288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finney, L. A., and T. V. O'Halloran. 2003. Transition metal speciation in the cell: insights from the chemistry of metal ion receptors. Science 300:931-936. [DOI] [PubMed] [Google Scholar]
- 8.Gitschier, J., B. Moffat, D. Reilly, W. I. Wood, and W. J. Fairbrother. 1998. Solution structure of the fourth metal-binding domain from the Menkes copper-transporting ATPase. Nat. Struct. Biol. 5:47-54. [DOI] [PubMed] [Google Scholar]
- 9.Goldman, B. S., W. C. Nierman, D. Kaiser, S. C. Slater, A. S. Durkin, J. A. Eisen, C. M. Ronning, W. B. Barbazuk, M. Blanchard, C. Field, C. Halling, G. Hinkle, O. Iartchuk, H. S. Kim, C. Mackenzie, R. Madupu, N. Miller, A. Shvartsbeyn, S. A. Sullivan, M. Vaudin, R. Wiegand, and H. B. Kaplan. 2006. Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Natl. Acad. Sci. U. S. A. 103:15200-15205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagen, D. C., A. P. Bretscher, and D. Kaiser. 1978. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev. Biol. 64:284-296. [DOI] [PubMed] [Google Scholar]
- 11.Hodgkin, J., and D. Kaiser. 1977. Cell-to-cell stimulation of movement in nonmotile mutants of Myxococcus. Proc. Natl. Acad. Sci. U. S. A. 74:2938-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Julien, B., A. D. Kaiser, and A. Garza. 2000. Spatial control of cell differentiation in Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 97:9098-9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kashefi, K., and P. L. Hartzell. 1995. Genetic suppression and phenotypic masking of a Myxococcus xanthus frzF− defect. Mol. Microbiol. 15:483-494. [DOI] [PubMed] [Google Scholar]
- 14.Kershaw, C. J., N. L. Brown, C. Constantinidou, M. D. Patel, and J. L. Hobman. 2005. The expression profile of Escherichia coli K-12 in response to minimal, optimal and excess copper concentrations. Microbiology 151:1187-1198. [DOI] [PubMed] [Google Scholar]
- 15.Kroos, L., A. Kuspa, and D. Kaiser. 1986. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev. Biol. 117:252-266. [DOI] [PubMed] [Google Scholar]
- 16.Legatzki, A., G. Grass, A. Anton, C. Rensing, and D. H. Nies. 2003. Interplay of the Czc system and two P-type ATPases in conferring metal resistance to Ralstonia metallidurans. J. Bacteriol. 185:4354-4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magnani, D., O. Barre, S. D. Gerber, and M. Solioz. 2008. Characterization of the CopR regulon of Lactococcus lactis IL1403. J. Bacteriol. 190:536-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monchy, S., M. A. Benotmane, R. Wattiez, S. van Aelst, V. Auquier, B. Borremans, M. Mergeay, S. Taghavi, D. van der Lelie, and T. Vallaeys. 2006. Transcriptomic and proteomic analyses of the pMOL30-encoded copper resistance in Cupriavidus metallidurans strain CH34. Microbiology 152:1765-1776. [DOI] [PubMed] [Google Scholar]
- 19.Moraleda-Muñoz, A., J. Pérez, A. L. Extremera, and J. Muñoz-Dorado. 2010. Differential regulation of six heavy metal efflux systems in the response of Myxococcus xanthus to copper. Appl. Environ. Microbiol. 76:6069-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moraleda-Muñoz, A., J. Perez, M. Fontes, F. J. Murillo, and J. Muñoz-Dorado. 2005. Copper induction of carotenoid synthesis in the bacterium Myxococcus xanthus. Mol. Microbiol. 56:1159-1168. [DOI] [PubMed] [Google Scholar]
- 21.Nies, D. H. 1999. Microbial heavy-metal resistance. Appl. Microbiol. Biotechnol. 51:730-750. [DOI] [PubMed] [Google Scholar]
- 22.Nies, D. H., G. Rehbein, T. Hoffmann, C. Baumann, and C. Grosse. 2006. Paralogs of genes encoding metal resistance proteins in Cupriavidus metallidurans strain CH34. J. Mol. Microbiol. Biotechnol. 11:82-93. [DOI] [PubMed] [Google Scholar]
- 23.Osman, D., and J. S. Cavet. 2008. Copper homeostasis in bacteria. Adv. Appl. Microbiol. 65:217-247. [DOI] [PubMed] [Google Scholar]
- 24.Outten, F. W., D. L. Huffman, J. A. Hale, and T. V. O'Halloran. 2001. The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J. Biol. Chem. 276:30670-30677. [DOI] [PubMed] [Google Scholar]
- 25.Perez, J., A. Castañeda-Garcia, H. Jenke-Kodama, R. Muller, and J. Muñoz-Dorado. 2008. Eukaryotic-like protein kinases in the prokaryotes and the myxobacterial kinome. Proc. Natl. Acad. Sci. U. S. A. 105:15950-15955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 27.Sanchez-Sutil, M. C., N. Gomez-Santos, A. Moraleda-Muñoz, L. O. Martins, J. Perez, and J. Muñoz-Dorado. 2007. Differential expression of the three multicopper oxidases from Myxococcus xanthus. J. Bacteriol. 189:4887-4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneiker, S., O. Perlova, O. Kaiser, K. Gerth, A. Alici, M. O. Altmeyer, D. Bartels, T. Bekel, S. Beyer, E. Bode, H. B. Bode, C. J. Bolten, J. V. Choudhuri, S. Doss, Y. A. Elnakady, B. Frank, L. Gaigalat, A. Goesmann, C. Groeger, F. Gross, L. Jelsbak, L. Jelsbak, J. Kalinowski, C. Kegler, T. Knauber, S. Konietzny, M. Kopp, L. Krause, D. Krug, B. Linke, T. Mahmud, R. Martinez-Arias, A. C. McHardy, M. Merai, F. Meyer, S. Mormann, J. Muñoz-Dorado, J. Perez, S. Pradella, S. Rachid, G. Raddatz, F. Rosenau, C. Ruckert, F. Sasse, M. Scharfe, S. C. Schuster, G. Suen, A. Treuner-Lange, G. J. Velicer, F. J. Vorholter, K. J. Weissman, R. D. Welch, S. C. Wenzel, D. E. Whitworth, S. Wilhelm, C. Wittmann, H. Blocker, A. Puhler, and R. Muller. 2007. Complete genome sequence of the myxobacterium Sorangium cellulosum. Nat. Biotechnol. 25:1281-1289. [DOI] [PubMed] [Google Scholar]
- 29.Solioz, M., and J. V. Stoyanov. 2003. Copper homeostasis in Enterococcus hirae. FEMS Microbiol. Rev. 27:183-195. [DOI] [PubMed] [Google Scholar]
- 30.Tottey, S., P. R. Rich, S. A. Rondet, and N. J. Robinson. 2001. Two Menkes-type ATPases supply copper for photosynthesis in Synechocystis PCC 6803. J. Biol. Chem. 276:19999-20004. [DOI] [PubMed] [Google Scholar]
- 31.Waldron, K. J., J. C. Rutherford, D. Ford, and N. J. Robinson. 2009. Metalloproteins and metal sensing. Nature 460:823-830. [DOI] [PubMed] [Google Scholar]
- 32.Ward, S. K., E. A. Hoye, and A. M. Talaat. 2008. The global responses of Mycobacterium tuberculosis to physiological levels of copper. J. Bacteriol. 190:2939-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitworth, D. E. (ed.). 2008. Myxobacteria. Multicellularity and differentiation. ASM Press, Washington, DC.
- 34.Yamamoto, K., and A. Ishihama. 2005. Transcriptional response of Escherichia coli to external copper. Mol. Microbiol. 56:215-227. [DOI] [PubMed] [Google Scholar]
- 35.Zhang, X. X., and P. B. Rainey. 2008. Regulation of copper homeostasis in Pseudomonas fluorescens SBW25. Environ. Microbiol. 10:3284-3294. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.