Abstract
The genus Listeria includes (i) the opportunistic pathogens L. monocytogenes and L. ivanovii, (ii) the saprotrophs L. innocua, L. marthii, and L. welshimeri, and (iii) L. seeligeri, an apparent saprotroph that nevertheless typically contains the prfA virulence gene cluster. A novel 10-loci multilocus sequence typing scheme was developed and used to characterize 67 isolates representing six Listeria spp. (excluding L. grayi) in order to (i) provide an improved understanding of the phylogeny and evolution of the genus Listeria and (ii) use Listeria as a model to study the evolution of pathogenicity in opportunistic environmental pathogens. Phylogenetic analyses identified six well-supported Listeria species that group into two main subdivisions, with each subdivision containing strains with and without the prfA virulence gene cluster. Stochastic character mapping and phylogenetic analysis of hly, a gene in the prfA cluster, suggest that the common ancestor of the genus Listeria contained the prfA virulence gene cluster and that this cluster was lost at least five times during the evolution of Listeria, yielding multiple distinct saprotrophic clades. L. welshimeri, which appears to represent the most ancient clade that arose from an ancestor with a prfA cluster deletion, shows a considerably lower average sequence divergence than other Listeria species, suggesting a population bottleneck and a putatively different ecology than other saprotrophic Listeria species. Overall, our data suggest that, for some pathogens, loss of virulence genes may represent a selective advantage, possibly by facilitating adaptation to a specific ecological niche.
Population genetics-based and phylogenetic studies have greatly contributed to the understanding of the evolutionary history and ecology of bacterial pathogens. In particular, multilocus sequence analyses (MLSA) and single-nucleotide polymorphism (SNP)-based population genetics research have revealed the microevolutionary patterns of species complexes like the Bacillus cereus complex (12) or the microevolution of well-known pathogens like Yersinia pestis (2), Salmonella enterica serovar Typhi (57), and Mycobacterium tuberculosis (18). One of the common findings of these studies is that obligate pathogens generally have a genetically clonal population structure as inferred by MLSA (1), while the population structure of free-living facultative pathogenic bacteria is characterized by relatively high genetic variability (12, 70). It has been hypothesized that these differences in population structure are related to the fact that some obligate pathogens represent epidemic clones (38), i.e., clonal lineages whose members have an epidemiological advantage compared to other lineages and are therefore able to quickly spread within the population. Because this dispersal of the members of an epidemic clone occurs rapidly, there is not enough time to accumulate mutations.
In this paper we present a phylogenetic and population genetics study of the genus Listeria. This genus consists of six closely related pathogenic (L. monocytogenes and L. ivanovii) and nonpathogenic (L. innocua, L. welshimeri, L. seeligeri, and a newly described species, L. marthii) species as well as a distantly related species, L. grayi (22). Another new species, L. rocourtiae, has been recently reported (33), but isolates were not available for inclusion in the study reported here. Because of the distant phylogenetic relatedness of L. grayi to the other Listeria species, it has been suggested that this species should be put in a separate genus, Murraya (63); L. grayi was thus not included in our study reported here. L. monocytogenes and L. ivanovii are facultative pathogens of warm-blooded animals and are the causative agents of a severe infectious disease, listeriosis (67). While L. monocytogenes has a wide host range, including humans, the host range of L. ivanovii seems to be largely restricted to ruminants, in particular sheep (13), even though some human listeriosis cases caused by L. ivanovii have been reported (34).
Key virulence genes in Listeria include (i) six genes (prfA, plcA, hly, mpl, actA, and plcB) clustered in a genomic element, designated the prfA virulence cluster or the Listeria pathogenicity island (LiPI), and (ii) members of the internalin family (61). Genes in the prfA cluster encode functions that that are necessary for inter- and intracellular motility and intracellular survival in the host cell. While some internalin genes encode proteins essential for host cell invasion (e.g., inlA and inlB) (3), inlC has recently been shown to encode a protein critical for cell-to-cell spread (52), and the functions of a number of other internalin proteins still remain to be elucidated (40). A number of internalin genes are also organized in clusters, including the inlAB operon, the inlGHE operon (which can also be present as an inlGC2DE or as an inlC2DE operon), which is found in L. monocytogenes and an L. ivanovii species-specific pathogenicity island encoding sphingomyelinase and numerous internalins (13). Importantly, the presence or absence of the prfA cluster and virulence characteristics can also be used to classify Listeria species and clades into three groups, including (i) species that do contain the prfA virulence cluster and are known pathogens, like L. monocytogenes and L. ivanovii, (ii) species that lack the prfA virulence cluster and are nonpathogenic (L. marthii and L. welshimeri), and (iii) species in which the presence of the prfA virulence cluster varies by strain. The last group contains L. seeligeri, which is nonpathogenic, although the majority of strains in the population contain the prfA virulence cluster (69), and L. innocua, which is also nonpathogenic, and although most strains lack the prfA virulence cluster, a small proportion of strains do carry this cluster (31, 68). The facts that the genus Listeria contains closely related nonpathogenic and pathogenic species and that strains with and without the prfA cluster within the same species make this genus an interesting model system for studies on the evolution of pathogenicity in opportunistic environmental pathogens. In addition, an improved understanding of the phylogeny and evolution of pathogenic and nonpathogenic Listeria spp. will also help in the development of appropriate assays for the specific detection and identification of human and animal pathogenic Listeria strains as well as regulations and intervention strategies that specifically target pathogenic species and strains.
MATERIALS AND METHODS
Isolates.
A total of 50 Listeria spp. isolates were selected for characterization by multilocus sequence typing (MLST), including L. monocytogenes (n = 15), L. innocua (n = 11), L. marthii (n = 3), L. welshimeri (n = 6), L. seeligeri (n = 7), and L. ivanovii (n = 8). Except for L. ivanovii, isolates were selected from the Cornell University Food Safety Laboratory (CUFSL) culture collection. As the CUFSL culture collection only included a limited number of L. ivanovii subsp. ivanovii isolates, an additional three L. ivanovii subsp. ivanovii and three L. ivanovii subsp. londoniensis isolates were obtained from ATCC (Table 1 ). Selection of isolates other than L. ivanovii was made using a sigB-based phylogeny (see the supplemental material for the sigB tree and phylogenetic analysis) based on sigB sequence data for 676 Listeria spp. isolates. sigB was used as a phylogenetic marker for isolate selection, as (i) it had previously been shown to provide no evidence for positive selection and recombination in L. monocytogenes (45), (ii) allows for reliable classification into Listeria spp., and (iii) sequence data for a large isolate set were available. Isolates were specifically selected to (i) represent the main genetic lineages (based on a sigB-based phylogeny) in each species and (ii) represent sigB genetic diversity within each species and lineage (see Fig. S1 in the supplemental material).
TABLE 1.
Isolates used
| Species and isolate identifiera | Additional information |
|---|---|
| L. innocua isolates | |
| FSL S4-045 | Isolated from algae along lake shore, NY |
| FSL S4-051 | Isolated from lake water, NY |
| FSL S4-176† | Isolated from pond water, NY |
| FSL S4-235 | Isolated from sidewalk, urban environment, NY |
| FSL S4-378† | Isolated from water puddle, NY |
| FSL R2-609 | Isolated from food processing plant environment, USA |
| FSL S4-846 | Isolated from sidewalk, urban environment, NY |
| FSL R2-604 | Isolated from soil, urban environment, NY |
| FSL R6-556 | Isolated from food related environment, NE |
| FSL W3-075 | Hemolytic, isolated from food, USA |
| FSL J1-023† | Hemolytic, origin unknown |
| CLIP 11262* | Isolated from food, Morocco |
| L. ivanovii subsp. ivanovii | |
| FSL C2-010† | Bovine isolate, obtained from USDA-ARS, IA |
| FSL C2-011† | Bovine fetus isolate, obtained from USDA-ARS, IA |
| FSL F6-600 | ATCC 19119, sheep isolate, Bulgaria |
| FSL F6-599 | ATCC BAA-678, clinical specimen, sheep fetus, Spain |
| FSL F6-597 | ATCC 700402, quality control strain for API products, origin unknown |
| L. ivanovii subsp. londoniensis | |
| FSL F6-598b | ATCC BAA-139, isolated from washing water, Switzerland |
| FSL F6-595 | ATCC 49953, goat isolate, Belgium |
| FSL F6-596 | ATCC 49954, food isolate, France |
| L. monocytogenes | |
| FSL S4-440 | Lineage I, serotype 1/2b, isolated from sidewalk, urban environment NY |
| FSL C1-406† | Lineage I, serotype 1/2b, isolated from food, NY |
| FSL S4-643 | Lineage I, serotype 4b, isolated from surface bench, NY |
| FSL E1-039 | Lineage I, serotype 1/2b, isolated from bovine clinical specimen, NY |
| FSL J1-194* | Lineage I, serotype 1/2b, isolated from human sporadic case, NY |
| FSL R2-503* | Lineage I, serotype 1/2b, human isolate, human outbreak, IL |
| HPB2262* | Lineage I, serotype 4b, human isolate, human outbreak, Italy |
| FSL N1-017* | Lineage I, serotype 4b, isolated from trout brine, NY |
| FSL J1-175* | Lineage I, serotype 1/2b, isolated from water, NY |
| FSL J2-064* | Lineage I, serotype 1/2b, isolated from animal clinical specimen, NY |
| CDC F2365* | Lineage I, serotype 4b, food isolate, human outbreak, CA |
| FSL F2-553† | Lineage II, serotype 1/2a, isolated from human sporadic case, NY |
| FSL N4-290 | Lineage II, serotype 1/2a, isolated from animal clinical specimen, NY |
| CDC J0161* | Lineage II, serotype 1/2a, human isolate, human outbreak, USA |
| 10403S* | Lineage II, serotype 1/2a, isolated from human skin lesion, USA |
| CDC J2818* | Lineage II, serotype 1/2a, food isolate, human outbreak, USA |
| FSL N3-165* | Lineage II, serotype 1/2a, isolated from soil, farm environment, NY |
| CDC F6900* | Lineage II, serotype 1/2a, isolated from human sporadic case, USA |
| EGD-e* | Lineage II, serotype 1/2a, strain derived from animal case, Great Britain |
| FSL S4-465 | Lineage IIIA, serotype 4b, isolated from water puddle, NY |
| FSL S4-839 | Lineage IIIA, serotype 4c, isolated from sidewalk, urban environment, NY |
| FSL F2-695† | Lineage IIIA, serotype 4a, isolated from human sporadic case, NY |
| FSL F2-525† | Lineage IIIA, serotype 4b, isolated from human sporadic case, NY |
| FSL J2-071* | Lineage IIIA, serotype 4c, isolated from animal clinical specimen, NY |
| FSL F2-208† | Lineage IIIC, serotype 4a, isolated from human sporadic case, OH |
| FSL W1-111† | Lineage IV, serotype 4c, origin unknown |
| FSL R2-142 | Lineage IV, serotype 4c, isolated from food, NY |
| FSL W1-112 | Lineage IV, serotype 4a, origin unknown |
| FSL M2-030 | Lineage IV, serotype 4b, origin unknown |
| L. seeligeri | |
| FSL S4-003 | hly positive,c isolated from leaves/debris, natural environment, NY |
| FSL S4-009 | hly positive, isolated from soil, natural environment, NY |
| FSL L5-054† | hly positive, isolated from soil, natural environment, NY |
| FSL S4-171 | hly negative, isolated from urban environment, NY |
| FSL N1-067† | hly positive, isolated from food related environment, NY |
| FSL S4-015 | hly positive, isolated from water, natural environment, NY |
| FSL S4-039 | hly positive, isolated from soil, urban environment, NY |
| L. marthii | |
| FSL S4-120 | ATCC BAA-1595, isolated from soil, natural environment, NY |
| FSL S4-710 | BEIR NR-9581, isolated from water, natural environment, NY |
| FSL S4-965† | BEIR NR-9582, isolated from water, natural environment, NY |
| L. welshimeri | |
| FSL S4-059† | Isolated from sidewalk, urban environment, NY |
| FSL S4-126† | Isolated from algae in a pond, natural environment, NY |
| FSL S4-182 | Isolated from soil, natural environment, NY |
| FSL S4-027 | Isolated from floor, urban environment, NY |
| FSL C2-006 | Isolated from food-related environment, NY |
| FSL S4-145 | Isolated from leaves/debris, natural environment, NY |
| SLCC 5334* | Isolated from decaying vegetation, USA |
Isolates listed included 50 isolates for which genes sequences for the 10 MLST loci were determined here as well as 16 isolates for which sequence data were derived from existing genome sequences (these isolates are marked with a *); isolates that were used for initial MLSA development are marked with a †.
This isolate was originally deposited with ATCC as L. ivanovii, and it is listed by ATCC as L. ivanovii subsp. ivanovii; trees for all 10 loci group this isolate into the L. ivanovii subsp. londoniensis clade (see Fig. 3), and this isolate is thus listed here as L. ivanovii subsp. londoniensis.
hly positive and negative notations indicate L. seeligeri isolates that were positive or negative by PCR for the gene encoding the L. seeligeri hemolysin.
In addition to the 50 Listeria spp. isolates that were characterized by MLST here, DNA sequence data for the 10 genes targeted by MLST were also obtained from genome sequences for Listeria spp. isolates for which appropriate genome sequence data were available (Table 1), including L. monocytogenes (two genome sequences [F2365 and EGD-e] from references 12, 21, and 42; genome sequences available at http://www.broadinstitute.org/annotation/genome/listeria_group/), L. innocua (one genome [21]), and L. welshimeri (one genome [28]).
Selection of MLST target loci and primer design.
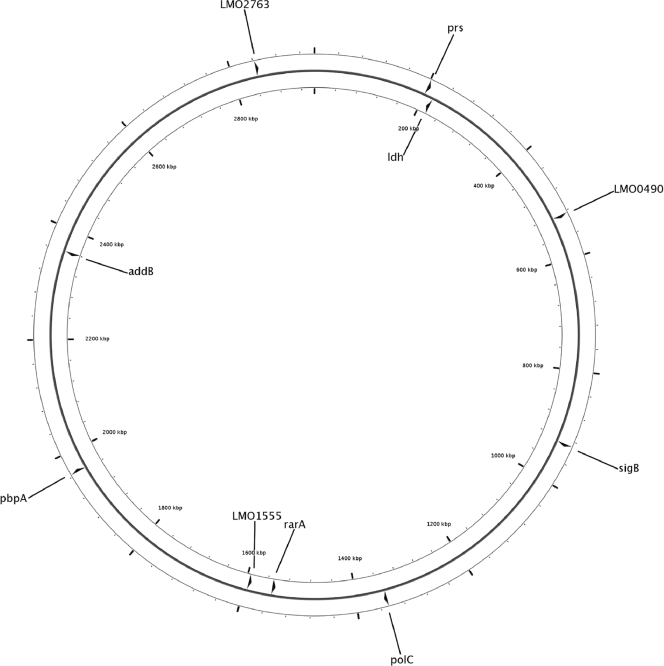
Loci for the MLST scheme used here were selected to fulfill five criteria: (i) no evidence for positive selection, (ii) no previous evidence for homologous recombination, (iii) present in all targeted Listeria species (meaning that they are part of the Listeria core genome), (iv) a dispersed distribution of target loci on the chromosome, and (v) inter- and intraspecific variability (i.e., no extreme conservation). Positive selection and recombination analyses performed on core genes found in four L. monocytogenes genomes and one L. innocua genome by Orsi et al. (46) were used to initially identify genes that showed (i) no evidence for positive selection and (ii) no detectable homologous recombination in these genomes. As the members of the genus Listeria show a high level of genome synteny (27), loci were selected to be dispersed throughout the EGD-e genome (Fig. 1). For loci that were initially selected as possible target loci for a 10-gene MLST scheme, the presence of the loci in all six targeted Listeria spp. was confirmed by initial PCR amplification and sequencing of the targeted loci in a subset of 16 isolates (Table 1).
FIG. 1.
Position of 10 MLST loci on the L. monocytogenes EGD-e chromosome (GenBank accession number AL591824).
PCR primers were designed with the online version of PriFi (19) and alignments of the target gene sequences, which were created using sequence data obtained from the genome sequences for L. monocytogenes isolates EGD-e (GenBank accession number AL591824) and F2365 (GenBank accession number AE017262), L. innocua CLIP 11262 (GenBank accession number AL592022), and L. welshimeri SLCC5334 (GenBank accession number AM263198).
PCR amplification and sequencing.
PCR amplification of the targeted gene fragments was performed using either genomic DNA prepared with the QIAamp DNA minikit (Qiagen, Valencia, CA) or a cell lysate; cell lysates were prepared in 100 μl 1× PCR buffer using lysozyme (2 mg/ml final concentration) and proteinase K (200 μg/ml final concentration), similar to a procedure described by Furrer et al. (20). PCR was performed in 50-μl reaction mixtures containing 0.5 μl of template, 10 μl of 5× PCR buffer, MgCl2 at a final concentration of 1.5 mM, deoxynucleotide triphosphates at a final concentration of 200 μM each, forward and reverse primers at a final concentration of 0.5 μM each, and 0.5 μl of Taq polymerase (5 U/μl; GoTaq Flexi; Promega, Madison, WI). PCR conditions for all loci were similar and included (i) one cycle at 95°C for 5 min; (ii) 20 touchdown cycles with 95°C for 30 s, 65°C for 30 s with a decrease of the annealing temperature by 0.5°C per cycle, and 72°C for 1 min; (iii) 20 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min; and (iv) one cycle at 72°C for 5 min. Some loci were amplified using a hot start protocol (see Table 2 for further details and primer sequences).
TABLE 2.
List of loci and primers used for PCR amplification
| Locus | Expected product size (bp)a | Hot startb | Primer name and sequence (5′-3′) |
|
|---|---|---|---|---|
| Forward | Reverse | |||
| ldh | 921 | No | HdB35ldhF, CAAAAAATTATTTTAGTTGGCGACGGAGCAGTTGG | HdB36ldhR, TTGTTTCATTGCRTCGTCWAG |
| lmo0490 | 791 | Yes | HdB37LMO490F, GCCACTCCAATCAGACACAGTTTATCACCAAC | HdB38LMO490R, ACTGGCATTTCTTTGTGTGTCCAAATTTCGAAAGC |
| prs | 809 | No | HdB40prsF, CCAAATTAACATTGAAGAAAGTATCCGTGGTTGTC | HdB41prsR, GAACTTACAGAWGCATTYTCATGWAC |
| sigB | 841 | Yes | Lm sigB15, AATATATTAATGAAAAGCAGGTGGAG | Lm sigB16, ATAAATTATTTGATTCAACTGCCTT |
| polC | 968 | No | HdB14polCF, CAAGCWGCTGATTGGGGWCAYAAAGC | HdB15polCR, GCMGCTTGCATYCCTTTTTGCATCATYGCTTCA |
| rarA | 860 | No | HdB30rarAF, ATGGCAATTCAACCTTTAGCTTACCGRATGCG | HdB31rarAR, TCCTCATAWGCCATAACDAGCATTCTCC |
| LMO1555 | 712 | No | HdB32LMO1555F, ATGAKTAAAAAARTTRTTTTAACAAGAGARGC | HdB33LMO1555R, CCTCCTGTATTATCAAATCAGCTARGTGCTTCATG |
| pbpA | 1,240 | Yes | HdB20pbpAF, TCGCAGAAGTTGGAACCGAACGGCGGG | HdB21pbpAR, GCAGCATARGCWCCRGCCATTTGCATTGG |
| addB | 845 | No | HdB26addBF, TGGAAATGGGAAAAAGARGGMGAYTGGAT | HdB27addBR, GCTTGYTTAGARCGAATRTAACTACTTTGTTC |
| LMO2763 | 1,216 | Yes | HdB39LMO2763F: GAACACGGCATGGAACGTGTTTTAGTACCAG | HdB40LMO2763R, AGCCGACCAGAAGCGCGAGCCAGTTTCCTCCTG |
The expected product size was calculated based on the respective gene sequence for L. monocytogenes EGD-e.
This column indicates whether hot start PCR was performed (hot start PCR was done by manual addition of the Taq polymerase after the initial denaturation step).
Sequence alignments.
Alignments were constructed in MacClade 4.08 (35). Since all of the loci are protein-coding genes without insertions or deletions, the pair-wise alignment option in MacClade was used for automatic alignment.
Descriptive analyses.
DnaSP 4.90.1 (58) was used to calculate the average number of pair-wise nucleotide differences per site (π), the average number of pair-wise nucleotide differences per sequence (k), the number of polymorphic sites, the number of mutations, the number of alleles, the GC content, Tajima's D value (test for neutrality of the data [65]), the number of synonymous and nonsynonymous mutations, and the rate of nonsynonymous-to-synonymous changes with a Jukes-Cantor correction.
Recombination analyses.
A Sawyer test (60), implemented in GENECONV, and the PHI (pair-wise homoplasy index) statistic (7), implemented in PhiPack, were used to test for evidence of intragenic recombination; the default settings were used for both methods. Extensive simulation studies (7, 49) have shown that these methods are less prone to type I errors, as opposed to neighbor similarity score (NSS) (29) and maximum chi-square (37) statistics.
Structure analysis.
To infer the ancestry of different clades identified among the sequence types (STs) in our data set, we performed an analysis using the linkage model of the program STRUCTURE (16); these analyses were performed essentially as described previously (11).
Positive selection analyses.
PAML 4.1 (75) was used to test for evidence of positive selection within the 10 loci. Specifically, two tests for positive selection were performed, including (i) a likelihood ratio test for the overall presence of positive selection within a locus, based on model M1a (nearly neutral) compared to model M2a (positive selection) (74), and (ii) the branch site test 2 as described by Zhang et al. (77), which is a likelihood ratio test for the presence of positive selection in a specific branch within a phylogeny. In most cases the branches leading to the individual species were tested; however, in cases where a species could be subdivided into several distinct branches (e.g., lineages I, II, III, and IV in L. monocytogenes) these branches were also tested for evidence of positive selection (branches tested in these analyses represented the actual branches found in a given gene tree; these did not necessarily always correspond to major phylogenetic lineages [see. Fig. S2 in the supplemental material]). For both tests, the gene trees used were constructed with the neighbor-joining algorithm in PAUP* 4.010b (64) using sequences for all unique allelic types.
Phylogenetic reconstruction.
BEAST 1.5.2 (14, 15) was used to construct separate phylogenies with a relaxed molecular clock model for each individual locus and to infer the position of the root of the tree in the absence of a suitable (closely related) outgroup. An advantage of the use of the relaxed molecular clock model is that it gives a summary statistic of the clockliness of the data, σr. If σr is 0 then the sequence data are perfectly clocklike, i.e., there is no variation in the branch rates. Larger values of σr correspond to increased rates of heterogeneity among branches. If the 95% highest posterior probability density of σr contained 0, a comparison with a strict clock model was performed using the Bayes factor as calculated using Tracer 1.5 (available from A. Rambaut at http://tree.bio.ed.ac.uk/software/tracer/). The analyses were performed assuming a coalescent process with a constant population as a prior for intraspecific relationships and a Yule model (76) of speciation as a prior for the interspecific phylogenetic relationships.
PHYML version 3.0 (25) was used to infer maximum likelihood (ML) phylogeny and bootstrap values based on concatenated sequences for the 10 loci. The hierarchical likelihood ratio test in Modeltest 3.7 (50) was used to select the model of nucleotide evolution for this analysis; a general time-reversible nucleotide substitution model with variant sites assumed to follow a gamma distribution and a probability I of sites being invariant (GTR+G+I) was identified as the best model and was used. ML bootstrap support (17) was inferred from 100 bootstrap replicates.
Reconstruction of evolutionary history of the prfA cluster in Listeria.
To infer the evolutionary history of the prfA virulence gene cluster in Listeria, we used both parsimony-based (35) and stochastic/maximum likelihood-based (36) criteria to map the character evolution of the cluster on the phylogeny of Listeria. To map the character evolution on the phylogeny using the parsimony criterion, MacClade 4.08 (35) was used. Stochastic character mapping was performed with Mesquite (36).
To allow for these analyses, all strains were tested for the presence or absence of hemolytic activity using the Christie, Atkins, Munch, Petersen (CAMP) test (24). Hemolytic activity is associated with the presence of the hemolysin gene, which is found in the prfA virulence cluster. As the CAMP test for a number of L. seeligeri isolates was inconclusive (due to very weak hemolysis), a PCR assay was performed to test for the presence or absence of the L. seeligeri hemolysin gene (forward primer, 5′-GGGATCCGCATAGGAAAAATAATGGAGTAAACAGC-3′; reverse primer, 5′-GCGGCCGCTTATTTTATGGTGTGTGTGTTAAGCG-3′). The presence of the hemolysin gene in the atypical CAMP-positive L. innocua isolates (FSL W3-075 and FSL J1-023) was further confirmed by PCR and sequencing of part of the hemolysin gene, and strain identity was confirmed by resequencing part of the sigB locus in these isolates.
To further probe the evolutionary history of the prfA cluster, we also constructed a phylogeny for the hemolysin gene (hly, which is located in the prfA cluster) for a subset of isolates that were found to be hly positive. These analyses were performed with 10 hly gene sequences that were available from publicly available genome sequences (L. monocytogenes EGD-e, F2365, and FSL J1-072 [see Table 1 for GenBank accession numbers]) and the following individual sequences (GenBank accession numbers in parentheses): L. monocytogenes FSL F2-208 (GU810924), L. ivanovii subsp. ivanovii (AY510072), L. ivanovii subsp. londoniensis (AY510073), L. seeligeri FSL S4-039 (EU755301), L. seeligeri FSL N1-067 (GU810921), L. innocua FSL J1-023 (GU810922), L. innocua FSL W3-075 (GU810923). The coding nucleotide sequences for hly were (manually) aligned in MacClade 4.08. Maximum likelihood analyses were performed using PAUP* version 4.010b (64). The hierarchical likelihood ratio test in Modeltest 3.7 (50) identified the Tamura Nei model with variant sites assumed to follow a gamma distribution (TrN+G) as the appropriate model to infer the hly phylogeny; ML bootstrap support (17) was inferred from 100 bootstrap replicates. The resulting hly phylogenetic tree was compared to the phylogenetic tree inferred from the 10 gene MLSA for the same isolates.
Nucleotide sequence accession numbers.
Sequences and alignments for all 10 loci (ldh, lmo0490, prs, sigB, polC, rarA, LMO1555, pbpA, addB, and LMO2763) have been deposited in GenBank (accession numbers GU475501 to GU475964).
RESULTS
Descriptive analysis of sequence data.
Among the 66 isolates (including 50 isolates sequenced here and sequence data obtained from 16 genome sequences [Table 1]), the 10 MLST loci yielded between 36 (prs) and 50 (lmo0490) allelic types (Table 3 ). The overall divergence between allelic types, as measured by π (the average number of pair-wise nucleotide differences per site) ranged from 0.0779 for lmo2763 to 0.2457 for lmo1555 (Table 3). π was chosen as a measure of sequence diversity, as this value is less affected by gene length or the number of sequences. L. monocytogenes was the most diverse species, as it showed the highest average π value for all 10 loci (0.0484), which is likely a reflection of the fact that this species is represented in four distinct lineages (see below). L. welshimeri showed the lowest diversity (with an average π for all 10 loci of 0.0050), and the corresponding average π values for the other species were 0.0261 (for L. ivanovii), 0.0177 (for L. seeligeri), 0.0168 (for L. innocua), and 0.0122 (for L. marthii).
TABLE 3.
Descriptive analysis of nucleotide sequence data
| Species (n) and gene | Length in bp (% of full ORF) | No. of variable sites | No. of mutations | No. of alleles | GC content | π/siteb | kc | Tajima's Dd | No. of mutations |
dN/dSe | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Synonymous | Nonsynonymous | ||||||||||
| All sequences (66)a | |||||||||||
| addB | 669 (20.1) | 283 | 418 | 43 | 35.10 | 0.1715 | 114.75 | NA | NA | NA | 0.08 |
| ldh | 711 (75.4) | 201 | 277 | 48 | 42.20 | 0.0970 | 68.95 | NA | NA | NA | 0.01 |
| lmo0490 | 611 (69.7) | 155 | 203 | 50 | 39.50 | 0.0810 | 49.50 | NA | NA | NA | 0.02 |
| prs | 511 (53.4) | 143 | 180 | 36 | 41.40 | 0.0808 | 46.48 | NA | NA | NA | 0.02 |
| polC | 695 (16.0) | 222 | 329 | 41 | 36.90 | 0.1312 | 90.43 | NA | NA | NA | 0.02 |
| pbpA | 871 (35.1) | 223 | 330 | 41 | 37.40 | 0.1254 | 91.32 | NA | NA | NA | 0.04 |
| LMO2763 | 958 (70.8) | 232 | 297 | 48 | 39.60 | 0.0779 | 74.40 | NA | NA | NA | 0.02 |
| lmo1555 | 495 (68.6) | 282 | 442 | 39 | 33.60 | 0.2457 | 116.15 | NA | NA | NA | 0.21 |
| rarA | 574 (44.7) | 188 | 277 | 39 | 39.60 | 0.1276 | 71.82 | NA | NA | NA | 0.03 |
| sigB | 660 (84.6) | 161 | 218 | 48 | 38.20 | 0.0866 | 57.07 | NA | NA | NA | 0.01 |
| L. monocytogenes (29) | |||||||||||
| addB | 669 (20.1) | 115 | 112 | 18 | 36.30 | 0.0716 | 47.91 | 2.09* | 97 | 18 | 0.07 |
| ldh | 711 (75.4) | 91 | 97 | 19 | 41.60 | 0.0447 | 31.77 | 1.10 | 92 | 5 | 0.01 |
| lmo0490 | 611 (69.7) | 66 | 71 | 21 | 39.80 | 0.0303 | 18.53 | 0.10 | 66 | 5 | 0.01 |
| prs | 511 (53.4) | 42 | 42 | 12 | 41.40 | 0.0240 | 13.26 | 0.89 | 42 | 0 | NA |
| polC | 695 (16.0) | 92 | 99 | 13 | 36.90 | 0.0523 | 36.25 | 1.63 | 98 | 1 | 0.00 |
| pbpA | 871 (35.1) | 115 | 130 | 13 | 38.40 | 0.0530 | 46.08 | 1.51 | 113 | 12 | 0.03 |
| LMO2763 | 958 (70.8) | 96 | 108 | 19 | 40.40 | 0.0286 | 27.28 | −0.03 | 106 | 2 | 0.00 |
| lmo1555 | 495 (68.6) | 135 | 154 | 16 | 33.90 | 0.1143 | 56.12 | 1.67 | 72 | 55 | 0.22 |
| rarA | 574 (44.7) | 51 | 56 | 13 | 40.60 | 0.0298 | 16.79 | 0.67 | 50 | 4 | 0.01 |
| sigB | 660 (84.6) | 74 | 80 | 17 | 38.80 | 0.0355 | 23.40 | 0.57 | 74 | 3 | 0.02 |
| L. innocua (12) | |||||||||||
| addB | 669 | 30 | 30 | 9 | 34.00 | 0.0120 | 8.02 | −0.87 | 37 | 2 | 0.02 |
| ldh | 711 | 55 | 57 | 10 | 43.60 | 0.0291 | 20.71 | 0.45 | 26 | 4 | 0.03 |
| lmo490 | 611 | 45 | 46 | 9 | 39.80 | 0.0214 | 13.09 | −0.64 | 54 | 3 | 0.02 |
| prs | 511 | 28 | 28 | 9 | 42.70 | 0.0126 | 6.95 | −1.12 | 45 | 1 | 0.00 |
| polC | 695 | 39 | 41 | 10 | 38.00 | 0.0150 | 10.39 | −1.07 | 28 | 0 | NA |
| pbpA | 871 | 40 | 40 | 10 | 37.20 | 0.0141 | 12.26 | −0.34 | 36 | 5 | 0.03 |
| lmo2763 | 958 | 39 | 39 | 9 | 39.30 | 0.0137 | 13.15 | 0.08 | 38 | 2 | 0.01 |
| lmo1555 | 495 | 30 | 30 | 6 | 32.80 | 0.0150 | 7.45 | −1.12 | 38 | 0 | NA |
| rarA | 574 | 36 | 38 | 9 | 38.70 | 0.0221 | 12.68 | 0.04 | 17 | 13 | 0.21 |
| sigB | 660 | 21 | 22 | 10 | 37.20 | 0.0128 | 8.42 | 0.69 | 19 | 3 | 0.05 |
| L. marthii (3) | |||||||||||
| addB | 669 | 1 | 1 | 2 | 36.00 | 0.0010 | 0.66 | NA | 0 | 1 | NA |
| ldh | 711 | 20 | 20 | 3 | 43.70 | 0.0188 | 13.33 | NA | 19 | 1 | 0.02 |
| lmo490 | 611 | 24 | 24 | 3 | 41.20 | 0.0262 | 16.00 | NA | 21 | 3 | 0.04 |
| prs | 633 | 9 | 9 | 3 | 42.90 | 0.0109 | 6.00 | NA | 9 | 0 | NA |
| polC | 695 | 28 | 28 | 3 | 39.20 | 0.0268 | 18.67 | NA | 28 | 0 | NA |
| pbpA | 871 | 8 | 8 | 3 | 38.30 | 0.0061 | 5.33 | NA | 8 | 0 | NA |
| lmo2763 | 958 | 13 | 13 | 3 | 40.30 | 0.0091 | 8.67 | NA | 13 | 0 | NA |
| lm01555 | 495 | 4 | 4 | 3 | 33.50 | 0.0054 | 2.67 | NA | 2 | 2 | 0.26 |
| rarA | 574 | 7 | 7 | 3 | 40.20 | 0.0081 | 4.67 | NA | 5 | 2 | 0.12 |
| sigB | 660 | 10 | 10 | 3 | 37.30 | 0.0101 | 6.67 | NA | 10 | 0 | NA |
| L. welshimeri (7) | |||||||||||
| addB | 669 | 4 | 4 | 4 | 33.50 | 0.0017 | 1.14 | −1.43 | 2 | 2 | 0.27 |
| ldh | 711 | 12 | 12 | 5 | 42.40 | 0.0071 | 5.05 | 0.17 | 12 | 0 | NA |
| lmo490 | 611 | 10 | 10 | 7 | 37.20 | 0.0053 | 3.24 | −1.11 | 10 | 0 | NA |
| prs | 511 | 4 | 4 | 4 | 40.00 | 0.0024 | 1.33 | −0.88 | 4 | 0 | NA |
| polC | 695 | 11 | 11 | 6 | 36.40 | 0.0069 | 4.76 | 0.33 | 10 | 1 | 0.02 |
| pbpA | 871 | 11 | 11 | 7 | 38.10 | 0.0058 | 4.67 | 0.21 | 8 | 3 | 0.12 |
| lmo2763 | 958 | 16 | 16 | 7 | 39.40 | 0.0056 | 5.33 | −1.02 | 15 | 1 | 0.02 |
| lmo1555 | 495 | 7 | 7 | 6 | 30.90 | 0.0048 | 2.38 | −0.86 | 3 | 4 | 0.48 |
| rarA | 574 | 8 | 8 | 7 | 39.20 | 0.0060 | 3.43 | 0.26 | 7 | 1 | 0.03 |
| sigB | 660 | 8 | 8 | 7 | 36.90 | 0.0040 | 2.67 | −0.96 | 6 | 2 | 0.08 |
| L. seeligeri (7) | |||||||||||
| addB | 669 | 22 | 22 | 7 | 33.70 | 0.0110 | 7.33 | −1.03 | 15 | 7 | 0.16 |
| ldh | 711 | 17 | 19 | 7 | 42.00 | 0.0118 | 8.38 | 0.45 | 19 | 0 | NA |
| lmo490 | 611 | 68 | 74 | 7 | 38.60 | 0.0481 | 29.38 | −0.16 | 69 | 5 | 0.02 |
| prs | 553 | 13 | 15 | 5 | 42.20 | 0.0084 | 4.67 | −1.31 | 15 | 0 | NA |
| polC | 695 | 30 | 32 | 7 | 37.60 | 0.0143 | 9.90 | −1.38 | 30 | 2 | 0.02 |
| pbpA | 871 | 31 | 31 | 6 | 38.10 | 0.0108 | 9.43 | −1.46 | 30 | 1 | 0.01 |
| lmo2763 | 958 | 31 | 31 | 7 | 37.70 | 0.0121 | 11.62 | −0.47 | 31 | 0 | NA |
| lmo1555 | 495 | 32 | 32 | 5 | 33.20 | 0.0204 | 10.10 | −1.30 | 22 | 10 | 0.14 |
| rarA | 574 | 43 | 45 | 5 | 38.90 | 0.0226 | 12.95 | −1.70* | 37 | 8 | 0.07 |
| sigB | 660 | 30 | 31 | 7 | 38.60 | 0.0174 | 11.48 | −0.53 | 29 | 2 | 0.02 |
| L. ivanovii (8) | |||||||||||
| addB | 669 | 33 | 33 | 3 | 36.10 | 0.0258 | 17.29 | 1.90† | 27 | 6 | 0.07 |
| ldh | 711 | 33 | 33 | 4 | 40.70 | 0.0241 | 17.11 | 1.83† | 31 | 2 | 0.01 |
| lmo490 | 611 | 46 | 46 | 3 | 38.90 | 0.0394 | 24.07 | 1.92† | 41 | 5 | 0.03 |
| prs | 553 | 30 | 30 | 3 | 40.90 | 0.0287 | 15.86 | 1.96† | 30 | 0 | NA |
| polC | 695 | 26 | 26 | 2 | 37.00 | 0.0200 | 13.93 | 2.05* | 23 | 3 | 0.04 |
| pbpA | 871 | 38 | 38 | 2 | 37.20 | 0.0234 | 20.36 | 2.08* | 35 | 3 | 0.02 |
| lmo2763 | 958 | 67 | 67 | 3 | 38.80 | 0.0372 | 35.61 | 2.05* | 63 | 4 | 0.02 |
| lmo1555 | 495 | 21 | 21 | 3 | 35.10 | 0.0219 | 10.86 | 1.78† | 10 | 11 | 0.31 |
| rarA | 574 | 22 | 22 | 2 | 38.50 | 0.0205 | 11.79 | 2.04* | 16 | 6 | 0.11 |
| sigB | 660 | 25 | 25 | 4 | 39.50 | 0.0197 | 13.00 | 1.83† | 23 | 2 | 0.02 |
These sequences were obtained from 50 isolates as well as 16 publicly available genomes (see Table 1 for more information).
Average number of pair-wise nucleotide differences per site.
Average number of pair-wise nucleotide differences per sequence.
Tajima's D was not inferred for (i) all isolates (because it is a population genetics statistic) or for (ii) L. marthii (because of the small number of isolates), and such cases are marked as not available (NA). *; P < 0.05; †, 0.05 < P < 0.10.
If the number of synonymous or nonsynonymous mutations could not be unambigiously inferred, the result is reported as not available (NA).
Tajima's D values for the individual loci were positive for all 10 loci for L. ivanovii (with 4 loci showing values significantly [P < 0.05] higher than 0 and the other 6 showing P values between 0.05 and 0.10) and for 9/10 loci for L. monocytogenes (with only addB showing values significantly [P < 0.05] higher than 0). A significantly positive value for Tajima's D is indicative of a decrease in population size, balancing selection (43), or subdivision of the population (62); these findings are consistent with the fact that both L. monocytogenes and L. ivanovii contain clearly distinct subspecies and lineages. L. innocua and L. welshimeri predominantly showed Tajima's D values that were negative or positive but close to zero; none of the values for these species was significantly different from 0. For L. seeligeri, Tajima's D values for 9 of 10 genes were negative, including 1 gene (rarA) which showed a negative value that was significantly different from 0. Negative values for Tajima's D are indicative of a population bottleneck or a selective sweep (43).
Recombination.
Overall, 4 of the 10 loci showed statistical evidence for intragenic homologous recombination in at least one of the two recombination tests used (i.e., Sawyer Test and PHI) (Table 4). Only one locus, lmo0490 (encoding shikimate 5-dehydrogenase), showed significant (P < 0.01) evidence of intragenic recombination with both the Sawyer test and the PHI statistic. The output for the Sawyer test indicated that two separate fragments of this sequence were involved in recombination, including (i) a 174-bp fragment near the 5′ end of the alignment, which appears to represent a recombination event between the lmo0490 allele found in the hemolytic L. innocua FSL J1-023 (acceptor) and the allele found in L. marthii FSL S4-710 (donor), and (ii) a 163-bp fragment, which appears to represent a recombination event between the lmo0490 allele found in another hemolytic L. innocua isolate, FSL W3-075 (acceptor), and the allele found in L. monocytogenes lineage IIIA isolate FSL F2-525 (donor).
TABLE 4.
Recombination statistics and models of substitution nucleotides inferred for the individual loci
| Locus | Geneconv result (no. of fragments)a | PHI statisticb | Nucleotide substitution model |
|---|---|---|---|
| ldh | 0.4854 | 0.0268* | GTR+I+G |
| lmo0490 | 0.0078 (2)** | 0.00106** | TrN+I+G |
| prs | 0.0019 (6)** | 0.0792 | TrN+I+G |
| sigB | 0.4262 | 0.8720 | TrN+I+G |
| polC | 0.1055 | 0.0155* | TrN+I+G |
| rarA | 0.1776 | 0.3940 | TrN+I+G |
| LMO1555 | 0.2525 | 0.5530 | HKY+I+G |
| pbpA | 0.2856 | 0.5780 | TrN+I+G |
| addB | 0.3446 | 0.1640 | TrN+I+G |
| LMO2763 | 0.1104 | 0.4810 | HKY+I+G |
**, P < 0.01.
*, P < 0.01; **, P < 0.01.
The Sawyer test further found significant (P < 0.01) evidence for recombination within the prs locus, while the PHI statistic found no significant signal of recombination for this gene; this may indicate that the prs locus has a region with significant clustering of substitutions (a signal of recombination for the Sawyer test [49]); however, these substitutions are not (phylogenetically) incompatible and are thus not considered a signal of recombination by the PHI statistic (7). The six recombinant sequence fragments in prs identified by the Sawyer test were located in a 176-bp section in the 5′ end of the locus sequenced and likely represent a single recombination event (as these fragments share 5′ and 3′ breakpoints) (45); this event involved alleles found in three L. innocua isolates (FSL S4-045, FSL S4-176, and FSL S4-235) as possible acceptors and alleles found in two L. monocytogenes lineage IIIA isolates (FSL F2-525 and FSL S4-465) as donors.
The PHI statistic also found statistically significant evidence (with P values between 0.01 and 0.05) for recombination in ldh and polC (with no significant evidence for recombination in these genes based on Sawyer's test); as recombination tests were performed for 10 loci and no correction for multiple comparisons was performed, we deemed these findings as likely false positives (a Bonferroni corrected P value cutoff would have been <0.005).
Structure analysis.
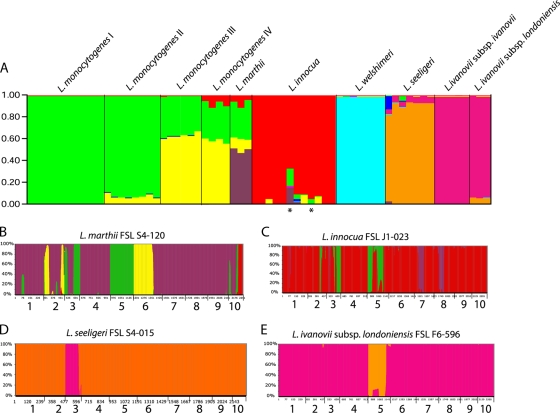
Structure analysis was performed to identify (i) putative recombination between different Listeria species and (ii) recombination between different lineages (for L. monocytogenes lineages I, II, III, and IV) or subspecies (for L. ivanovii subsp. ivanovii and L. ivanovii subsp. londoniensis). Structure analysis at the species level indicates very distinct allelic compositions between species, with a few notable exceptions. Specifically, the structure analysis showed that a considerable part of the loci of the three L. marthii isolates is derived from L. monocytogenes lineages I, II, and III and L. innocua. It appears that most of the L. monocytogenes and L. innocua gene content in L. marthii represents different recombination events, including (i) acquisition, by L. marthii, of part of sigB from L. innocua and (ii) acquisition of a complete locus (lmo2763) and an almost-complete locus (pbpA) from L. monocytogenes (Fig. 2 B). These events would not have been detected by the Sawyer test or the PHI statistic; however, the recombination event in lmo2763 was supported by phylogenetic analyses of the individual loci (see below). Structure analysis also showed that the lmo0490 locus in L. seeligeri includes a sequence fragment introduced from L. ivanovii by homologous recombination. In addition, selected L. innocua sequence types showed evidence for recombination with other species, most notably the ST of the hemolytic L. innocua isolate FSL J1-023, which includes parts of several loci (lmo1555, polC, and prs) that appear to have been introduced into the genome from L. monocytogenes and L. marthii.
FIG. 2.
Mixture of ancestry as inferred by the program STRUCTURE. (A) Proportions of ancestry from ancestral L. monocytogenes lineages I and II population (green), ancestral L. monocytogenes lineages III and IV population (yellow), ancestral L. marthii population (purple), the ancestral L. innocua population (red), the ancestral L. welshimeri population (light blue), the ancestral L. seeligeri population (orange), and the ancestral L. ivanovii population) as inferred by STRUCTURE assuming K = 8 ancestral populations. The asterisks mark hemolytic L. innocua isolates. Each vertical column represents an isolate and is colored according to the inferred proportion of single-nucleotide alleles that were derived from one of the ancestral subpopulations. (B to E) Posterior probabilities that an individual SNP allele is derived from one of the ancestral subpopulations within an individual isolate, including L. marthii FSL S4-120 (B), L. innocua FSL J1-023 (C), L. seeligeri FSL S4-015 (D), and L. ivanovii subsp. londoniensis FSL F6-596 (E). Colors of the columns indicate the ancestral subpopulation of a given SNP (see the color legend, above); the height of the color indicates the posterior probability (indicated on the y axis) that a given SNP is derived from a given ancestor. The numbers on the x axis represent the loci where these SNP alleles are found (1, addB; 2, ldh; 3, lmo0490; 4, lmo1555; 5, lmo2763; 6, pbpA; 7, polC; 8, prs; 9, rarA; 10, sigB).
Structure analysis of the four L. monocytogenes lineages (72) showed a considerable amount of shared alleles as well as a considerable allelic composition that is unique to lineages III and IV (Fig. 2); lineages I and II and lineages III and IV share a considerable proportion of their alleles. In addition, lineage IV (which has also previously has been described as lineage IIIB [55]) contains some L. innocua alleles, which appear to largely represent horizontal gene transfer of part of sigB from L. innocua to L. monocytogenes lineage IV. The two L. ivanovii subspecies show >90% shared alleles, a considerably higher shared proportion of alleles than for the alleles shared between L. monocytogenes lineages I/II and lineages III/IV. L. ivanovii subsp. londoniensis shows some lmo2763 alleles that appear to be derived from L. seeligeri through a homologous recombination event involving this locus.
Positive selection.
None of the 10 loci analyzed showed evidence for positive selection with the overall test for positive selection (75) or background on the positive selection analysis, indicating that the loci evolved neutrally or under negative selection (see File S1 in the supplemental material for results). Weak evidence for branch-specific positive selection was only found for pbpA in the L. seeligeri clade (P = 0.04); as no correction of P values for multiple comparisons was performed, this is likely a false positive (the Bonferroni corrected P value cutoff would have been <0.007, corrected for the fact that seven branch-specific analyses were performed for pbpA).
Phylogenetic reconstruction.
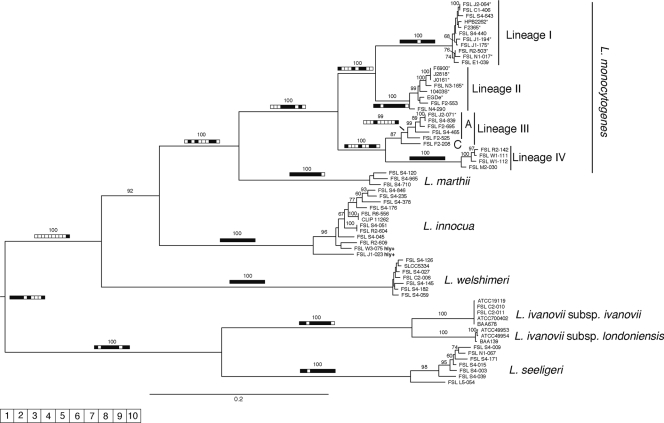
The ML analysis of the concatenated sequences of the 10 loci analyzed resulted in a well-supported and highly resolved phylogenetic tree of Listeria (Fig. 3). All six species included here formed well-supported clades (bootstrap support [BS], 100%) in the ML tree based on the concatenated gene and were generally well supported by the individual gene trees. For example, the L. innocua and L. welshimeri clades received significant Bayesian support in all 10 individual gene trees, while the L. seeligeri and L. marthii clades received significant Bayesian support in 9 out of 10 gene trees. Interestingly, while isolate ATCC BAA-139, which was originally deposited with ATCC as L. ivanovii, is now listed by ATCC as L. ivanovii subsp. ivanovii, trees for all 10 loci group this isolate into the L. ivanovii subsp. londoniensis clade (Fig. 3), suggesting that this isolate represents L. ivanovii subsp. londoniensis. The phylogenetic placement of atypical hemolytic L. innocua isolates in the L. innocua clade was confirmed by our analysis and is consistent with previous studies (31, 68).
FIG. 3.
Phylogram inferred by the maximum likelihood based on 10 concatenated loci. Bootstrap values of >60% are indicated above the branches. The bars summarize Bayesian support for the individual branches as found with the BEAST analysis of the individual loci, depicted as individual compartments of the bar (1, prs; 2, ldh; 3, lmo490; 4, sigB; 5, polC; 6, rarA; 7, lmo1555; 8, pbpA; 9, addB; 10, lmo2763); white indicates <95% PP and black indiates >95% PP.
L. monocytogenes was the species that was the least well supported by the individual gene trees; while no contradicting gene trees were found, the monophyly of this species was only significantly supported (posterior probability [PP], >95%) by 5 out of 10 gene trees (sigB, polC, rarA, lmo1555, and addB). Significant Bayesian support for branches that interrupted the monophyly of L. monocytogenes was found for two gene trees, including (i) the sigB gene tree (see Fig. S3a in the supplemental material), which places L. monocytogenes lineage IV in a sister group position to L. innocua, suggesting an ancient recombination event between the ancestor of lineage IV and the ancestor of L. innocua, and (ii) the lmo2763 gene tree (see Fig. S3b), which nested L. marthii sequences within L. monocytogenes lineage II sequences, suggesting a recent recombination event (as the divergence between the L. marthii allelic types and the phylogenetically most closely related L. monocytogenes lineage II allelic types is lower than the overall divergence between L. monocytogenes lineage II allelic types). A high bootstrap support (>85%) was found for all L. monocytogenes lineages (lineages I, II, III, and IV), which further supports the observation that L. monocytogenes can be subdivided into four separate evolutionary lineages (72).
In the lmo0490 tree (see Fig. S3c in the supplemental material), L. ivanovii subsp. ivanovii and L. ivanovii subsp. londoniensis form two distinct clusters which are nested between L. seeligeri isolates, suggesting two individual recombination events between L. seeligeri and L. ivanovii. The STRUCTURE analysis, however, suggested that only one recombination event took place between the most recent common ancestor (MRCA) of L. ivanovii subsp. londoniensis and L. seeligeri.
MLST data also allowed for characterization of phylogenetic relationships between the different Listeria spp. Although the phylogram constructed (Fig. 3) is midpoint rooted and therefore may have an arbitrary rooting position, the majority of the BEAST analyses (i.e., the analyses for 6 out of 10 loci: prs, ldh, lmo0490, sigB, rarA, and lmo2763) placed the root between the L. ivanovii/L. seeligeri clade and a clade that contains the species L. welshimeri, L. innocua, L. marthii, and L. monocytogenes. The sister group relationship of L. seeligeri and L. ivanovii is well supported (100% BS), with a PP of >95% in 8 out of 10 gene trees. The L. welshimeri/L. innocua/L. marthii/L. monocytogenes clade also received high bootstrap support (100%); however, only one individual gene tree (lmo2763) showed a significant PP for this clade. The interspecific phylogenetic relationships within the L. welshimeri/L. innocua/L. marthii/L. monocytogenes clade all received high ML BSs (92 to 100%); however, significant PP values for these relationships within individual gene trees were scarce (i.e., for the sister group relationship of L. marthii and L. monocytogenes) or completely absent (i.e., for the sister group relationship of L. innocua and the L. marthii/L. monocytogenes clade). While L. grayi was not included in our study reported here, due to its extremely distant relationship to all other Listeria spp., we also performed a phylogenetic analysis which used the sequence data for nine loci for all isolates included here as well as for one L. grayi isolate for which a genome sequence has recently become available (see Fig. S4 in the supplemental material); the lmo0490 locus was not included in this analysis because it was not present in the available L. grayi genome sequence. The resulting phylogeny clearly showed the distant relationship between L. grayi and all other Listeria spp. (see. Fig. S3 in the supplemental material).
The σr statistic showed that 7 of the 10 loci did not evolve in a clocklike manner, which indicates that these genes did not evolve at the same approximate mutation rate in the entire tree (14). Three loci (sigB, polC, and rarA) have a posterior probability density that includes 0, which indicates that these genes possibly have a clocklike mutation rate. A clocklike mutation rate could only be confirmed, through comparison with a strict clock model using the Bayes factor, for sigB. A putative reason for a nonclocklike mutation rate in some loci could be the presence of (homologous) recombination in all loci but sigB. As only a few of the 10 loci showed evidence for recombination (e.g., lmo0490), more likely explanations for this finding include (i) differences in generation time between clades and (ii) differences in mutation rates between clades. Differences in mutation rates between clades are usually associated with changes in biological function or selective pressure (73); hence, a possible explanation for clocklike evolution of sigB may be that this gene is essential in the stress response and survival and thus is under negative selection in all clades (signals for positive selection may have been too weak to be detected by the analyses performed here).
Reconstruction of the evolutionary history of the prfA cluster in Listeria.
The isolates tested here included 29 L. monocytogenes isolates (all with the prfA cluster), 12 L. innocua isolates (2 with and 10 without the prfA cluster), 3 L. marthii isolates (all without the prfA cluster), 7 L. welshimeri isolates (all without the prfA cluster), 7 L. seeligeri isolates (6 with and 1 without the prfA cluster), and 8 L. ivanovii isolates (all with the prfA cluster). For all L. seeligeri isolates, presence of the prfA cluster was determined using a PCR assay for the L. seeligeri hly gene. For the other isolates, the presence of the hly gene could be inferred by a positive CAMP reaction or by examination of the genome sequence. Information on prfA cluster presence/absence and the 10-gene ML tree was used to reconstruct the evolutionary history of the prfA cluster in Listeria. Based on parsimony criteria the observed contemporary pattern of prfA cluster distribution (Fig. 4) could be explained by either five gains or five losses of this cluster. An approach using parsimony criteria did, thus, not provide an unambiguous scenario for the evolutionary history of the prfA cluster in Listeria. Stochastic character mapping (Fig. 4), an approach that takes branch lengths (an approximate of evolutionary time and therefore the probability of a character to change) into account, suggests a scenario where the MRCA of Listeria had a prfA cluster as the most likely scenario; in this scenario, the MRCA of L. marthii and L. welshimeri lost the prfA cluster, while the MRCA of L. innocua had the prfA cluster, which was subsequently lost at two occasions during the evolutionary history of L. innocua. Under this scenario, the nonhemolytic L. seeligeri would have lost the prfA cluster recently.
FIG. 4.
Stochastic character mapping of gain/loss of the prfA cluster over the evolutionary history of Listeria. Solid branches indicate the presence of the prfA cluster, and white branches indicate the absence of the prfA cluster. This analysis shows that the most likely scenario for Listeria is one in which the MRCA of Listeria had the prfA cluster, and the cluster was lost on five separate occasions, including (i) once in the ancestral lineage of L. welshimeri, (ii) once in the ancestral lineage of L. marthii, (iii) twice within L. innocua, and (iv) a recent loss in L. seeligeri.
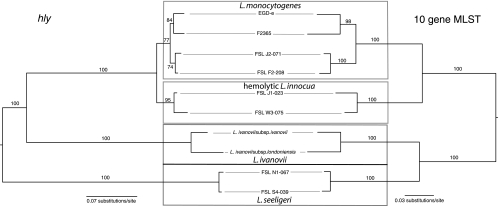
To further probe the evolution of the prfA cluster, we also constructed an ML tree based on the hly sequences available for 10 isolates included in our analyses reported here (four L. monocytogenes isolates, two hemolytic L. innocua isolates, a representative of each of the subspecies of L. ivanovii, and two hemolytic L. seeligeri isolates; these analyses resulted in a tree with a likelihood score of −ln5,702.1579 (Fig. 5). The topology of the hly tree is highly similar to the 10-gene MLST tree; the L. monocytogenes isolates are found in a moderately supported (77% BS) clade, while the hemolytic L. innocua isolates form a well-supported (95% BS) clade. The L. innocua isolates have a well-supported (100% BS) sister group relationship to L. monocytogenes. The hly phylogeny does not support the sister group relation of L. ivanovii and L. seeligeri found in the 10-gene MLST analyses. Instead, the hly phylogeny places L. ivanovii in a sister group position to the L. monocytogenes/L. innocua clade, while L. seeligeri seems to represent an early split from the rest of the “core” Listeria species. This is consistent with the suggestion that the prfA virulence cluster in L. seeligeri may represent an ancestral form of this cluster (66). Overall, these data support the hypothesis that the prfA cluster in the hemolytic L. innocua strains was not obtained through horizontal gene transfer but was instead vertically transmitted.
FIG. 5.
Comparison of hly (left) and 10 concatenated loci (right) ML phylograms of a subset of isolates of species with the prfA cluster. Values above the individual branches are bootstrap values based on 100 ML bootstrap replicates.
DISCUSSION
A new 10-gene MLST scheme was developed and applied to an initial isolate set representing the overall genetic diversity of the core species in the genus Listeria in order to probe the phylogeny and evolution of this genus, which contains a number of pathogenic and nonpathogenic clades. Overall, our data indicate that (i) the genus Listeria includes at least six phylogenetically well-supported species, including well-supported subspecies-like clades in at least L. ivanovii and L. monocytogenes, and (ii) while the common ancestor of the genus Listeria contained the prfA virulence gene cluster, this cluster was most likely lost at least five times during the evolution of Listeria, yielding multiple distinct saprotrophic clades.
The genus Listeria includes at least six phylogenetically well-supported species, including well-supported subspecies-like clades in at least L. ivanovii and L. monocytogenes.
Despite the availability of a number of MLST schemes for L. monocytogenes (45, 51), an MLST scheme that can be used on all Listeria spp. has not previously been available. While the main species in the genus Listeria (i.e., L. innocua, L. monocytogenes, L. seeligeri, L. ivanovii, and L. welshimeri) have been well defined for a few decades based on phenotypic and some genetic methods (5, 39, 56), development of the new MLST scheme reported here provided a new opportunity to further probe the phylogeny and taxonomy of the genus Listeria. Overall, our MLST-based phylogeny not only supported that the previously known Listeria species represent phylogenetically well-supported units but also provided further clear and unambiguous support that L. marthii, which was first described in 2009 (22), represents a distinct species. Our data thus showed that the MLST scheme reported here will have considerable utility by facilitating clear speciation of unusual Listeria isolates, as also supported by clear classification, into the respective species clades of hemolytic L. innocua (31, 68) and nonhemolytic L. seeligeri (69) isolates included in our isolate set. Interestingly, even though the 10 genes included in our MLST were preselected to represent genes that showed no evidence for recombination (based on genome sequence analysis for L. monocytogenes and L. innocua [46]), a few loci showed a low number of recombination events (summarized in Table 5) involving part of (lmo490, prs, and polC) or the complete locus sequenced (lmo0490, lmo2763, and sigB). We specifically found evidence for interspecific recombination between (i) L. innocua and L. monocytogenes, (ii) L. monocytogenes and L. marthii, (iii) L. innocua and L. marthii, and (iv) L. ivanovii and L. seeligeri. While bacterial species are often defined as clonal groups that show no/limited horizontal gene transfer and homologous recombination of a core gene (32), our data suggest that genetic exchange between closely related Listeria spp. still occurs, possible reflecting (i) relatively limited diversification between species (e.g., between L. seeligeri and L. ivanovii, which showed evidence for horizontal transfer) and/or (ii) overlapping or shared niches for individual Listeria species, consistent with previous studies in other pathogenic species (6).
TABLE 5.
Summary of homologous recombination events identified for the 10 genes characterized in this studya
| Gene | Evidence for recombination within locus identified (species or strain[s] involved)b | Evidence for recombination of complete locusb,c |
|---|---|---|
| lmo0490 | Two events identified by GENECONV and PHI (event 1: D, L. marthii; A, L. innocua; event 2: D, L. monocytogenes; A, L. innocua) | One event identified by STRUCTURE and GENE TREE (D, L. ivanovii; A, L. seeligeri) |
| LMO2763 | One event identified by STRUCTURE and GENE TREE (D, L. monocytogenes lineage II; A, L. marthii) | |
| sigB | One event identified by STRUCTURE and GENE TREE (D, L. innocua; A, L. monocytogenes lineage IV) | |
| polC | One event identified by STRUCTURE (D, L. marthii; A, L. innocua) | |
| prs | One event identified by GENECONV (D, L. monocytogenes lineage III; A, L. innocua); one event identified by STRUCTURE (D, L. marthii; A, L. innocua) |
Genes which showed evidence for recombination only by PHI with P values between 0.01 and 0.05 (Table 4) are not included here, as they likely represent false positives (due to borderline P values with no Bonferroni correction).
D, donor; A, acceptor.
This column lists recombination events that involved the full locus sequenced; the events were typically identified in the STRUCTURE analysis (Fig. 2) and/or through inspection of the gene trees (see Fig. S2a, b, and c in the supplemental material).
Importantly, the MLST data and their phylogenetic analyses also allowed for further insights into the possible subspecies or subspecies-like clades within some Listeria spp. (i.e., L. ivanovii and L. monocytogenes). Our data clearly support that L. ivanovii represents two highly divergent subspecies, consistent with the initial description of this subspecies by Boerlin et al. (4), with very limited genetic variation within a given subspecies (particularly in L. ivanovii subsp. ivanovii). Limited genetic variation, based on pulsed-field gel electrophoresis patterns, in L. ivanovii subsp. ivanovii was previously reported based on 38 isolates from various sources and geographic localities from the United Kingdom (53). As L. ivanovii is characterized by an apparent stringent host specificity for ruminants, where it predominantly causes abortions, its genetically homogeneous population structure seems to point toward L. ivanovii as being a highly specialized, clonal pathogen, comparable in population structure to known obligate pathogens like Yersinia pestis and Mycobacterium tuberculosis (1). Interestingly, the genetically homogeneous population structure of L. ivanovii is in clear contrast to the genetically heterogeneous population structure of L. monocytogenes, suggesting that the pathogenic life style probably plays a greater role in the ecology of L. ivanovii than L. monocytogenes. For L. monocytogenes, our data also support previous findings that this species represents multiple distinct species-like lineages. In addition to L. monocytogenes lineages I and II, which have been well documented in a large number of studies (11, 45, 48, 51, 54), our data also allowed further insights into the taxonomy and evolution of L. monocytogenes strains that have previously been classified as either lineage III (including IIIA, B, and C) (30, 53, 54) or lineages III and IV (72). Strains classified into these lineages (i) are, overall, rare and appear to be associated with animal hosts, even though they are also isolated from humans (23, 30, 55); (ii) represent serotypes 4a and 4c as well as 4b (even though these strains are distinct from the more common lineage I 4b strains [44, 55]); (iii) are genetically diverse; and (iv) show evidence for more common recombination compared to lineage I and II strains. Our MLST data show that isolates previously grouped into these isolates clearly represent two distinct lineages, which were named III (representing lineages previously designated as IIIA and C [55]) and IV (previously designated as IIIB [55]), consistent with a previous study by Ward et al. (72). Our findings thus overall support that L. monocytogenes represents at least four distinct subspecies-like lineages which, based on a variety of previous studies (23, 30, 71), may also differ in their ecology. Even though isolates were selected to represent diverse and distinct strains within each species (based on sigB allelic type data for >650 isolates), no clear subspecies-like clades were identified in our study for the other Listeria spp.; characterization of larger and more geographically diverse isolate sets by MLST may reveal new clades and subspecies in other species, however, particularly if isolates are obtained from sources and hosts not represented or underrepresented in our isolate collection. For example, while we did not identify distinct lineages within L. seeligeri, a recent study on isolates obtained in Upper Franconia, Germany, identified two distinct L. seeligeri clades (41).
Our data clearly support a basal split of the core species in the genus Listeria into two main clades, including (i) a clade consisting of L. seeligeri and L. ivanovii and (ii) a clade consisting of L. welshimeri, L. innocua, L. marthii, and L. monocytogenes. Phylogenetic analyses based on 9 of the 10 genes analyzed here, furthermore, showed that L. grayi is very distinct from the other Listeria spp. and represents a very distant separate clade, consistent with a number of other studies (61, 69), including a proposal that L. grayi should be reclassified as a separate genus (63). While a number of previous analyses (61, 69) have supported that the core Listeria spp. represent two main clades, consistent with those defined here, most of these studies were only based on a few genes or genes that are in close chromosomal proximity (e.g., 16S and 23S, open reading frames [ORFs] of the virulence cluster, and prs and ldh) or only studied very few isolates. For example, Schmid et al. (61) only used six isolates in their study. While our analyses clearly put L. welshimeri into a clade with L. innocua, L. marthii, and L. monocytogenes and indicate that L. welshimeri represents a lineage that diverged from the common ancestor of L. innocua/L. marthii/L. monocytogenes early during the evolutionary history of Listeria, previous studies were inconclusive about the position of L. welshimeri. For example, a study reporting a 16S rRNA sequence-based phylogeny (69) showed weak support (<60%) for the grouping of L. welshimeri in the L. innocua and L. monocytogenes clade. On the other hand, a gene tree based on iap (69) provided strong support for placement of L. welshimeri in the L. seeligeri/L. ivanovii clade, and another study (61) also showed strong support for grouping of L. welshimeri with L. seeligeri and L. ivanovii, based on a phylogenetic analysis of 16S rRNA, 23S rRNA, iap, prs, vclB, and ldh. However, the majority of loci in these studies (61, 69) either flank the prfA virulence cluster (prs, vclB, and ldh) or represent a potential virulence gene (iap), which also appears to show a considerable amount of deletions (8), and thus they may not be reliable phylogenetic markers. We thus conclude that our results from a phylogeny with 10 genes with no evidence for positive selection and only a few genes with evidence of recombination likely allowed for accurate reconstruction of the phylogenetic position of L. welshimeri, particularly since the grouping of L. welshimeri found here is also supported by a 100-gene phylogeny constructed using genome sequences for six Listeria spp. isolates representing all six species studied here (unpublished data).
While the common ancestor of the genus Listeria contained the prfA virulence gene cluster, this cluster was lost at least five times during the evolution of Listeria, yielding multiple distinct saprotrophic clades.
Construction of a robust phylogeny for the genus Listeria also allowed us to reconstruct the evolutionary history of the prfA pathogenicity cluster in this genus. While a parsimony criterion suggests that a scenario of five losses and a scenario of five gains of the prfA cluster are equally likely, the hypothesis that the MRCA of the contemporary core Listeria spp. already contained the prfA virulence gene cluster and that this cluster was lost in (at least) five separate events in the evolutionary history of Listeria is supported by (i) maximum likelihood-based stochastic character mapping (Fig. 4) and (ii) phylogenetic analysis of hly sequences (Fig. 5). This hypothesis of an MRCA with the prfA cluster is also consistent with the observation that the prfA cluster does not contain the features typical for classical pathogenicity islands, which generally show evidence for horizontal gene transfer (26). While features that suggest mobility of pathogenicity islands include the presence of direct repeats, association with tRNA genes, or insertion sequence elements, integrases, or transposases, and an aberrant GC content compared to the rest of the chromosomal DNA (9), the GC content of the prfA cluster is comparable to the rest of the chromosome and this cluster does not seem to be associated with mobile genetic elements. However, it cannot be excluded that the prfA cluster once was associated with mobile elements; two ORFs found in the cluster show similarity to viral proteins (66), and a putative conjugative transposon insertion junction has been reported between the prfA cluster and the adjacent prs gene (9). This suggests that the prfA cluster may once have been a mobile element but lost its mobility after integration into the chromosome of the ancestor of Listeria (66).
While the hypothesis of an MRCA that possessed the prfA cluster with subsequent losses of the cluster in nonpathogenic species has also been suggested by Schmid et al. (61), no rigorous phylogenetic tests of this hypothesis have previously been reported. Interestingly, our data also indicate that deletions of the prfA cluster occurred at different times in the evolutionary history of Listeria, yielding some monophyletic prfA cluster-negative clades that represent nonpathogenic Listeria spp. (e.g., L. welshimeri and L. marthii) as well as apparently more recent losses of this cluster, e.g., in L. seeligeri, where this loss yields a prfA cluster-negative clade within an otherwise-prfA-positive species. Interestingly, an evolutionary trend toward the loss of virulence-associated characteristics has also been observed in L. monocytogenes, where a large number of strains and clades are characterized by different premature stop codon mutations in the virulence gene inlA which yield invasion-attenuated strains (51). These observations suggested that losses of the prfA cluster and other virulence factors are not necessarily deleterious to Listeria but may even represent a selective advantage under some circumstances and/or in some lineages. As Listeria species are, even in the case of facultative pathogenic species, commonly found in the environment and considered saprotrophs (59), it is tempting to speculate that loss of virulence-associated characteristics may accompany, or even facilitate, a transition to organisms with improved environmental survival, consistent with the observation that L. innocua appears to sometimes outcompete L. monocytogenes in non-host-associated environments, including in certain enrichment media (10, 47). Further experimental efforts are needed, however, to test this hypothesis.
Interestingly, our data indicate that L. welshimeri, which appears to represent the most ancient clade that arose from an ancestor with a prfA cluster deletion, shows a considerably lower average sequence divergence than other Listeria spp. Although this low within-population divergence could be explained by the small and geographically limited sample size, this seems to be unlikely, since other species in this study with a similarly limited sampling (L. innocua and L. seeligeri) showed an average sequence divergence that was approximately 3.5 times higher than that observed in L. welshimeri. Another argument against the low diversity of L. welshimeri being a sampling artifact is the fact that an extensive sampling of sigB (see Fig. S1 in the supplemental material) showed the same pattern of low within-population divergence in L. welshimeri (π, 0.00531) versus a comparably higher within-sequence divergence in L. innocua (π, 0.01213) and L. seeligeri (π, 0.01963). A putative biological explanation for this observation is either (i) a lower mutation rate or (ii) a population bottleneck. In our relaxed molecular clock analyses, no evidence was found for L. welshimeri having a lower mutation rate, as neither the median relative mutation rate nor the 95% posterior probability density of the relative mutation rate (data not shown) proved to be lower than the relative mutation rates inferred for other species/branches. The pattern observed is more consistent with a scenario of a population bottleneck or a scenario in which the contemporary population descended from a highly successful fast-spreading clone. Both scenarios leave the same population genetics fingerprint and can therefore not be distinguished from each other. A population bottleneck is generally characterized by a (significantly) negative value of Tajima's D (43). Although negative values of Tajima's D were found for 5 out of the 10 genes examined in L. welshimeri, none of these values was significantly different from 0 and thus cannot be seen as convincing evidence for the occurrence of a population bottleneck in the recent history of L. welshimeri. Regardless of the actual evolutionary or biological mechanisms that are responsible for the relatively low genetic diversity in L. welshimeri, our data do suggest that L. welshimeri has a unique natural history in the genus Listeria, which warrants further investigations, including additional diversity data for isolates collected from other regions.
Conclusions.
Overall, our data indicate that the genus Listeria represents a group of well-supported species and also support the hypothesis that these species have evolved from an ancestor with a prfA virulence gene cluster. Over time these species have diversified into highly clonal pathogens (e.g., L. ivanovii), nonpathogens (L. welshimeri and L. marthii), and clades that include isolates with and without the prfA virulence cluster (L. innocua and L. seeligeri). Gene loss, lateral gene transfer, and recombination, including in housekeeping genes, as well as positive selection (46) clearly have contributed to the evolution of the genus Listeria into closely related species and clades that have adapted to different niches. Further application of the Listeria genus MLST scheme, which was developed and used here, to characterize additional isolates will not only help to further improve our understanding of the evolution and population genetics of the genus Listeria but also will help in rapid and unambiguous classification of unusual Listeria isolates, which are increasingly being described (31, 68, 69).
Supplementary Material
Acknowledgments
We thank Sherry Roof for her help with sequencing.
This work was supported by USDA Special Research grants 2005-34459-15625 and 2006-34459-16952.
Footnotes
Published ahead of print on 23 July 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Achtman, M. 2008. Evolution, population structure, and phylogeography of genetically monomorphic bacterial pathogens. Annu. Rev. Microbiol. 62:53-70. [DOI] [PubMed] [Google Scholar]
- 2.Achtman, M., G. Morelli, P. Zhu, T. Wirth, I. Diehl, B. Kusecek, A. J. Vogler, D. Wagner, C. J. Allender, W. R. Easterday, V. Chenal-Francisque, P. Worsham, N. R. Thomson, J. Parkhill, L. E. Lindler, E. Carniel, and P. Keim. 2004. Microevolution and history of the plague bacillus, Yersinia pestis. Proc. Natl. Acad. Sci. U. S. A. 101:17837-17842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bierne, H., C. Sabet, N. Personnic, and P. Cossart. 2007. Internalins: a complex family of leucine-rich repeat-containing proteins in Listeria monocytogenes. Microbes Infect. 9:1156-1166. [DOI] [PubMed] [Google Scholar]
- 4.Boerlin, P., J. Rocourt, F. Grimont, P. Grimont, C. Jacquet, and J. C. Piffaretti. 1992. Listeria ivanovii subsp. londoniensis subsp. nov. Int. J. Syst. Bacteriol. 42:69-73. [DOI] [PubMed] [Google Scholar]
- 5.Boerlin, P., J. Rocourt, and J. C. Piffaretti. 1991. Taxonomy of the genus Listeria by using multilocus enzyme electrophoresis. Int. J. Syst. Bacteriol. 41:59-64. [DOI] [PubMed] [Google Scholar]
- 6.Brown, E. W., M. K. Mammel, J. E. LeClerc, and T. A. Cebula. 2003. Limited boundaries for extensive horizontal gene transfer among Salmonella pathogens. Proc. Natl. Acad. Sci. U. S. A. 100:15676-15681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruen, T. C., H. Philippe, and D. Bryant. 2006. A simple and robust statistical test for detecting the presence of recombination. Genetics 172:2665-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bubert, A., S. Köhler, and W. Goebel. 1992. The homologous and heterologous regions within the iap gene allow genus- and species-specific identification of Listeria spp. by polymerase chain reaction. Appl. Environ. Microbiol. 58:2625-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai, S., and M. Wiedmann. 2001. Characterization of the prfA virulence gene cluster insertion site in non-hemolytic Listeria spp.: probing the evolution of the Listeria virulence gene island. Curr. Microbiol. 43:271-277. [DOI] [PubMed] [Google Scholar]
- 10.Curiale, M., and C. Lewus. 1994. Detection of Listeria monocytogenes in samples containing Listeria innocua. J. Food Prot. 57:1048-1051. [DOI] [PubMed] [Google Scholar]
- 11.den Bakker, H. C., X. Didelot, E. Fortes, K. K. Nightingale, and M. Wiedmann. 2008. Lineage specific recombination rates and microevolution in Listeria monocytogenes. BMC Evol. Biol. 8:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Didelot, X., M. Barker, D. Falush, and F. G. Priest. 2009. Evolution of pathogenicity in the Bacillus cereus group. Syst. Appl. Microbiol. 32:81-90. [DOI] [PubMed] [Google Scholar]
- 13.Domínguez-Bernal, G., S. Müller-Altrock, B. González-Zorn, M. Scortti, P. Herrmann, H. J. Monzó, L. Lacharme, J. Kreft, and J. A. Vázquez-Boland. 2006. A spontaneous genomic deletion in Listeria ivanovii identifies LIPI-2, a species-specific pathogenicity island encoding sphingomyelinase and numerous internalins. Mol. Microbiol. 59:415-432. [DOI] [PubMed] [Google Scholar]
- 14.Drummond, A. J., S. Y. M. Ho, M. J. Phillips, and A. Rambaut. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4:e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drummond, A. J., and A. Rambaut. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falush, D., M. Stephens, and J. Pritchard. 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felsenstein, J. 1985. Confidence-limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 18.Filliol, I., A. S. Motiwala, M. Cavatore, W. Qi, M. H. Hazbón, M. Bobadilla del Valle, J. Fyfe, L. García-García, N. Rastogi, C. Sola, T. Zozio, M. I. Guerrero, C. I. León, J. Crabtree, S. Angiuoli, K. D. Eisenach, R. Durmaz, M. L. Joloba, A. Rendón, J. Sifuentes-Osornio, A. Ponce de León, M. D. Cave, R. Fleischmann, T. S. Whittam, and D. Alland. 2006. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J. Bacteriol. 188:759-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fredslund, J., L. Schauser, L. H. Madsen, N. Sandal, and J. Stougaard. 2005. PriFi: using a multiple alignment of related sequences to find primers for amplification of homologs. Nucleic Acids Res. 33:W516-W520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furrer, B., U. Candrian, C. Hoefelein, and J. Luethy. 1991. Detection and identification of Listeria monocytogenes in cooked sausage products and in milk by in vitro amplification of haemolysin gene fragments. J. Appl. Bacteriol. 70:372-379. [DOI] [PubMed] [Google Scholar]
- 21.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. Garcia-del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 22.Graves, L. M., L. O. Helsel, A. G. Steigerwalt, R. E. Morey, M. I. Daneshvar, S. E. Roof, R. H. Orsi, E. D. Fortes, S. R. Millilo, H. C. den Bakker, M. Wiedmann, B. Swaminathan, and B. D. Sauders. 10 August 2009, posting date. Listeria marthii sp. nov., isolated from the natural environment, Finger Lakes National Forest. Int. J. Syst. Evol. Microbiol. 60:1280-1288. [DOI] [PubMed] [Google Scholar]
- 23.Gray, M., R. Zadoks, E. Fortes, B. Dogan, S. Cai, Y. Chen, V. Scott, D. Gombas, K. Boor, and M. Wiedmann. 2004. Listeria monocytogenes isolates from foods and humans form distinct but overlapping populations. Appl. Environ. Microbiol. 70:5833-5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Groves, R. D., and H. J. Welshimer. 1977. Separation of pathogenic from apathogenic Listeria monocytogenes by three in vitro reactions. J. Clin. Microbiol. 5:559-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 26.Hacker, J., G. Blum-Oehler, I. Mühldorfer, and H. Tschäpe. 1997. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol. Microbiol. 23:1089-1097. [DOI] [PubMed] [Google Scholar]
- 27.Hain, T., C. Steinweg, and T. Chakraborty. 2006. Comparative and functional genomics of Listeria spp. J. Biotechnol. 126:37-51. [DOI] [PubMed] [Google Scholar]
- 28.Hain, T., C. Steinweg, C. T. Kuenne, A. Billion, R. Ghai, S. S. Chatterjee, E. Domann, U. Karst, A. Goesmann, T. Bekel, D. Bartels, O. Kaiser, F. Meyer, A. Puhler, B. Weisshaar, J. Wehland, C. Liang, T. Dandekar, R. Lampidis, J. Kreft, W. Goebel, and T. Chakraborty. 2006. Whole-genome sequence of Listeria welshimeri reveals common steps in genome reduction with Listeria innocua as compared to Listeria monocytogenes. J. Bacteriol. 188:7405-7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jakobsen, I. B., and S. Easteal. 1996. A program for calculating and displaying compatibility matrices as an aid in determining reticulate evolution in molecular sequences. Comput. Appl. Biosci. 12:291-295. [DOI] [PubMed] [Google Scholar]
- 30.Jeffers, G., J. Bruce, P. McDonough, J. Scarlett, K. Boor, and M. Wiedmann. 2001. Comparative genetic characterization of Listeria monocytogenes isolates from human and animal listeriosis cases. Microbiology 147:1095-1104. [DOI] [PubMed] [Google Scholar]
- 31.Johnson, J., K. Jinneman, G. Stelma, B. G. Smith, D. Lye, J. Messer, J. Ulaszek, L. Evsen, S. Gendel, R. W. Bennett, B. Swaminathan, J. Pruckler, A. Steigerwalt, S. Kathariou, S. Yildirim, D. Volokhov, A. Rasooly, V. Chizhikov, M. Wiedmann, E. Fortes, R. E. Duvall, and A. D. Hitchins. 2004. Natural atypical Listeria innocua strains with Listeria monocytogenes pathogenicity island 1 genes. Appl. Environ. Microbiol. 70:4256-4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lan, R., and P. R. Reeves. 2001. When does a clone deserve a name? A perspective on bacterial species based on population genetics. Trends Microbiol. 9:419-424. [DOI] [PubMed] [Google Scholar]
- 33.Leclercq, A., D. Clermont, C. Bizet, P. Grimont, A. Le Flèche-Matéos, S. Roche, C. Buchrieser, V. Cadet-Daniel, A. Le Monnier, M. Lecuit, and F. Allerberger. 13 November 2009, posting date. Listeria rocourtiae sp. nov. Int. J. Syst. Evol. Microbiol. [Epub ahead of print.] [DOI] [PubMed]
- 34.Lessing, M. P., G. D. Curtis, and I. C. Bowler. 1994. Listeria ivanovii infection. J. Infect. 29:230-231. [DOI] [PubMed] [Google Scholar]
- 35.Maddison, D. R., and W. P. Maddison. 2005. MACCLADE version 4.08. Sinauer Associates Inc., Sunderland, MA.
- 36.Maddison, W. P., and D. R. Maddison. 2009. Mesquite: a modular system for evolutionary analysis, 2.72 ed.
- 37.Maynard Smith, J. 1992. Analyzing the mosaic structure of genes. J. Mol. Evol. 34:126-129. [DOI] [PubMed] [Google Scholar]
- 38.Maynard Smith, J., N. H. Smith, M. O'Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. U. S. A. 90:4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McLauchlin, J. 1997. The identification of Listeria species. Int. J. Food Microbiol. 38:77-81. [DOI] [PubMed] [Google Scholar]
- 40.Milillo, S. R., and M. Wiedmann. 2009. Contributions of six lineage-specific internalin-like genes to invasion efficiency of Listeria monocytogenes. Foodborne Pathog. Dis. 6:57-70. [DOI] [PubMed] [Google Scholar]
- 41.Müller, A. A., M. W. Schmid, O. Meyer, and F. G. Meussdoerffer. 2010. Listeria seeligeri isolates from food processing environments form two phylogenetic lineages. Appl. Environ. Microbiol. 76:3044-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nelson, K. E., D. E. Fouts, E. F. Mongodin, J. Ravel, R. T. DeBoy, J. F. Kolonay, D. A. Rasko, S. V. Angiuoli, S. R. Gill, I. T. Paulsen, J. Peterson, O. White, W. C. Nelson, W. Nierman, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, D. H. Haft, J. Selengut, S. Van Aken, H. Khouri, N. Fedorova, H. Forberger, B. Tran, S. Kathariou, L. D. Wonderling, G. A. Uhlich, D. O. Bayles, J. B. Luchansky, and C. M. Fraser. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 32:2386-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nielsen, R. 2001. Statistical tests of selective neutrality in the age of genomics. Heredity 86:641-647. [DOI] [PubMed] [Google Scholar]
- 44.Nightingale, K., L. Bovell, A. Grajczyk, and M. Wiedmann. 2007. Combined sigB allelic typing and multiplex PCR provide improved discriminatory power and reliability for Listeria monocytogenes molecular serotyping. J. Microbiol. Methods 68:52-59. [DOI] [PubMed] [Google Scholar]
- 45.Nightingale, K., K. Windham, and M. Wiedmann. 2005. Evolution and molecular phylogeny of Listeria monocytogenes isolated from human and animal listeriosis cases and foods. J. Bacteriol. 187:5537-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Orsi, R. H., Q. Sun, and M. Wiedmann. 2008. Genome-wide analyses reveal lineage specific contributions of positive selection and recombination to the evolution of Listeria monocytogenes. BMC Evol. Biol. 8:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petran, R., and K. Swanson. 1993. Simultaneous growth of Listeria monocytogenes and Listeria innocua. J. Food Prot. 56:616-618. [DOI] [PubMed] [Google Scholar]
- 48.Piffaretti, J. C., H. Kressebuch, M. Aeschbacher, J. Bille, E. Bannerman, J. M. Musser, R. K. Selander, and J. Rocourt. 1989. Genetic characterization of clones of the bacterium Listeria monocytogenes causing epidemic disease. Proc. Natl. Acad. Sci. U. S. A. 86:3818-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Posada, D., and K. A. Crandall. 2001. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc. Natl. Acad. Sci. U. S. A. 98:13757-13762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 51.Ragon, M., T. Wirth, F. Hollandt, R. Lavenir, M. Lecuit, A. L. Monnier, and S. Brisse. 2008. A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 4:e1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rajabian, T., B. Gavicherla, M. Heisig, S. Müller-Altrock, W. Goebel, S. Gray-Owen, and K. Ireton. 2009. The bacterial virulence factor InlC perturbs apical cell junctions and promotes cell-to-cell spread of Listeria. Nat. Cell Biol. 11:1212-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramage, C. P., J. C. Low, J. McLauchlin, and W. Donachie. 1999. Characterisation of Listeria ivanovii isolates from the UK using pulsed-field gel electrophoresis. FEMS Microbiol. Lett. 170:349-353. [DOI] [PubMed] [Google Scholar]
- 54.Rasmussen, O. F., P. Skouboe, L. Dons, L. Rossen, and J. E. Olsen. 1995. Listeria monocytogenes exists in at least three evolutionary lines: evidence from flagellin, invasive associated protein and listeriolysin O genes. Microbiology 141:2053-2061. [DOI] [PubMed] [Google Scholar]
- 55.Roberts, A., K. Nightingale, G. Jeffers, E. Fortes, J. M. Kongo, and M. Wiedmann. 2006. Genetic and phenotypic characterization of Listeria monocytogenes lineage III. Microbiology 152:685-693. [DOI] [PubMed] [Google Scholar]
- 56.Rocourt, J., F. Grimont, P. Grimont, and H. Seeliger. 1982. DNA relatedness among serovars of Listeria monocytogenes sensu lato. Curr. Microbiol. 7:383-388. [Google Scholar]
- 57.Roumagnac, P., F. X. Weill, C. Dolecek, S. Baker, S. Brisse, N. T. Chinh, T. A. Le, C. J. Acosta, J. Farrar, G. Dougan, and M. Achtman. 2006. Evolutionary history of Salmonella typhi. Science 314:1301-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rozas, J., J. Sánchez-DelBarrio, X. Messeguer, and R. Rozas. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496-2497. [DOI] [PubMed] [Google Scholar]
- 59.Sauders, B. D., M. Z. Durak, E. Fortes, K. Windham, Y. Schukken, A. J. Lembo, Jr., B. Akey, K. K. Nightingale, and M. Wiedmann. 2006. Molecular characterization of Listeria monocytogenes from natural and urban environments. J. Food Prot. 69:93-105. [DOI] [PubMed] [Google Scholar]
- 60.Sawyer, S. A. 1999. GENECONV: a computer package for the statistical detection of gene conversion. Department of Mathematics, Washington University in St. Louis, St. Louis, MO. http://www.math.wustl.edu/∼sawyer/geneconv/.
- 61.Schmid, M., E. Ng, R. Lampidis, M. Emmerth, M. Walcher, J. Kreft, W. Goebel, M. Wagner, and K. Schleifer. 2005. Evolutionary history of the genus Listeria and its virulence genes. Syst. Appl. Microbiol. 28:1-18. [DOI] [PubMed] [Google Scholar]
- 62.Simonsen, K., G. Churchill, and C. Aquadro. 1995. Properties of statistical tests of neutrality for DNA polymorphism data. Genetics 141:413-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stuart, S., and H. Welshimer. 1974. Taxonomic reexamination of Listeria Pirie and transfer of Listeria grayi and Listeria murrayi to a new genus, Murraya. Int. J. Syst. Evol. Microbiol. 24:177-185. [Google Scholar]
- 64.Swofford, D. L. 2002. PAUP*: Phylogenetic Analysis Using Parsimony (* and other methods), version 4.0. Sinauer Associates Inc., Sunderland, MA.
- 65.Tajima, F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vazquez-Boland, J. A., G. Dominguez-Bernal, B. Gonzalez-Zorn, J. Kreft, and W. Goebel. 2001. Pathogenicity islands and virulence evolution in Listeria. Microbes Infect. 3:571-584. [DOI] [PubMed] [Google Scholar]
- 67.Vazquez-Boland, J. A., M. Kuhn, P. Berche, T. Chakraborty, G. Dominguez-Bernal, W. Goebel, B. Gonzalez-Zorn, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14:584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Volokhov, D., S. Duperrier, A. Neverov, J. George, C. Buchrieser, and A. Hitchins. 2007. The presence of the internalin gene in natural atypically hemolytic Listeria innocua strains suggests descent from L. monocytogenes. Appl. Environ. Microbiol. 73:1928-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Volokhov, D., J. George, C. Anderson, R. Duvall, and A. Hitchins. 2006. Discovery of natural atypical nonhemolytic Listeria seeligeri isolates. Appl. Environ. Microbiol. 72:2439-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Walk, S. T., E. W. Alm, L. M. Calhoun, J. M. Mladonicky, and T. S. Whittam. 2007. Genetic diversity and population structure of Escherichia coli isolated from freshwater beaches. Environ. Microbiol. 9:2274-2288. [DOI] [PubMed] [Google Scholar]
- 71.Ward, T., L. Gorski, M. Borucki, R. Mandrell, J. Hutchins, and K. Pupedis. 2004. Intraspecific phylogeny and lineage group identification based on the prfA virulence gene cluster of Listeria monocytogenes. J. Bacteriol. 186:4994-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ward, T. J., T. F. Ducey, T. Usgaard, K. A. Dunn, and J. P. Bielawski. 2008. Multilocus genotyping assays for single nucleotide polymorphism-based subtyping of Listeria monocytogenes isolates. Appl. Environ. Microbiol. 74:7629-7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Whelan, S. 2008. Spatial and temporal heterogeneity in nucleotide sequence evolution. Mol. Biol. Evol. 25:1683-1694. [DOI] [PubMed] [Google Scholar]
- 74.Wong, W. S., Z. Yang, N. Goldman, and R. Nielsen. 2004. Accuracy and power of statistical methods for detecting adaptive evolution in protein coding sequences and for identifying positively selected sites. Genetics 168:1041-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang, Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24:1586-1591. [DOI] [PubMed] [Google Scholar]
- 76.Yule, G. U. 1925. A mathematical theory of evolution, based on the conclusions of Dr. JC Willis, FRS. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1925:21-87. [Google Scholar]
- 77.Zhang, J., R. Nielsen, and Z. Yang. 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 22:2472-2479. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.