Abstract
Telomerase RNA is an essential component of telomerase, a ribonucleoprotein enzyme that maintains chromosome ends in most eukaryotes. Here we employ a novel approach, namely, RNA-guided RNA modification, to assess whether introducing 2′-O methylation into telomerase RNA can influence telomerase activity in vivo. We generate specific 2′-O methylation sites in and adjacent to the triple helix (within the conserved pseudoknot structure) of Saccharomyces cerevisiae telomerase RNA (TLC1). We show that 2′-O methylation at U809 reduces telomerase activity, resulting in telomere shortening, whereas 2′-O methylation at A804 or A805 leads to moderate telomere lengthening. Importantly, we also show that targeted 2′-O methylation does not affect TLC1 levels and that 2′-O-methylated TLC1 appears to be efficiently assembled into telomerase ribonucleoprotein. Our results demonstrate that RNA-guided RNA modification is a highly useful approach for modulating telomerase activity.
In eukaryotic cells, chromosomal ends are capped by telomeres, which are long tandem repeat sequences complexed with proteins (3, 34). Telomeres maintain the integrity and stability of chromosomes, which would otherwise undergo incomplete replication, fusion, or degradation with each cell division (6, 7). Telomerase is responsible for telomere elongation and maintenance of chromosomal ends in most eukaryotes (3, 14). It has long been known that telomerase is fundamental to cell survival, growth, and death (4). Telomerase malfunction is often associated with disease. For instance, most cancer cells have an unusually high level of telomerase activity (17). On the other hand, mutations in telomerase components have been linked to several degenerative diseases such as dyskeratosis congenita and aplastic anemia (4). Thus, to understand the molecular mechanisms of these diseases and to identify new treatments, it is desirable to regulate telomerase activity in vivo.
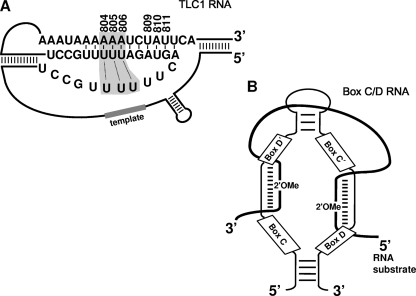
Telomerase is a ribonucleoprotein (RNP) complex (15) that consists of one noncoding RNA (known as TLC1 in Saccharomyces cerevisiae) (32) and several proteins, including a reverse transcriptase (Est2p) (22). The telomerase noncoding RNA not only folds into a structure that tethers proteins but also serves as a template for reverse transcription (39), which leads to the addition of a specific repeated sequence to the chromosome ends. S. cerevisiae TLC1 and its homologs in other organisms (including mammals) have been extensively studied. Several other possible functions (including catalysis) of telomerase RNA have been proposed (25, 27). Furthermore, nuclear magnetic resonance (NMR) studies and computational modeling coupled with functional analysis have revealed a conserved triple-helix structure within the pseudoknot region of human and Kluyveromyces lactis telomerase RNAs (31, 35). Recently, the Cech lab has presented experimental evidence for the presence of a similar triple-helix structure in yeast TLC1 RNA (27) (Fig. 1A). Changes of 2′-OH groups of nucleotides in and adjacent to the triple-helix region to 2′-H or 2′-OMe (2′-O methylated) lead to reduction of telomerase activity in yeast and mammalian in vitro systems (27).
FIG. 1.
Artificial box C/D RNA is expressed and functional. (A) The pseudoknot structure of S. cerevisiae TLC1 RNA is shown (27). Shaded nucleotides and dotted lines denote the triple helix. The template sequence is also indicated. Nucleotides in the triple-helix region that were evaluated in the current work are numbered. (B) The box C/D RNA structure is shown. Boxes C, D′, C′, and D are indicated. The sequence between box C and box D′ and the sequence between box C′ and box D function as guides that base pair with target RNA (thick line), as shown. 2′OMe shows the target nucleotide to be 2′-O methylated.
Box C/D RNPs are modifying enzymes that introduce 2′-O methylation into rRNAs and snRNAs at specific sites (38). Box C/D RNPs comprise one small RNA (box C/D RNA) and four core proteins (fibrillarin or Nop1p in S. cerevisiae, 15.5-kDa protein, Nop56, and Nop58) (38). A typical box C/D RNA folds into a unique secondary structure, leaving two short sequences—one between box C and box D′ and one between box C′ and box D—unpaired or single stranded (Fig. 1B). These single-stranded sequences function as guides that base pair with the natural rRNA and snRNA substrates, thereby directing 2′-O methylation at specific sites (1, 5, 20). Without exception, 2′-O methylation occurs at the target nucleotide in the substrate RNA that is base paired to the nucleotide in snoRNA precisely 5 nucleotides (nt) upstream from box D (or D′; Fig. 1B) (5, 20). Once the box C/D snoRNA finds its nucleotide target, fibrillarin, a methyltransferase associated with the box C/D guide RNA, delivers the methyl group to the target nucleotide at the 2′-O position. The “box D + 5 rule” for predicting the site of 2′-O methylation guided by box C/D RNAs has been verified in various organisms, including yeast, Xenopus laevis, and human, suggesting that RNA-guided 2′-O methylation of rRNA and snRNA is universal among eukaryotes (18, 19, 26, 33). Given the detailed mechanism of RNA-guided RNA 2′-O methylation, it is possible to design artificial box C/D RNAs to target telomerase RNA in and adjacent to the triple-helix region, thus offering an opportunity to manipulate telomerase activity in vivo.
Here we show that artificial box C/D RNAs can indeed target 2′-O methylation at specific sites in and adjacent to the triple-helix structure of TLC1, thereby affecting telomerase activity in vivo. 2′-O methylation did not affect the steady-state level of TLC1, and 2′-O-methylated TLC1 was incorporated into telomerase RNP. Our results indicate that telomerase activity can be manipulated in vivo.
MATERIALS AND METHODS
Plasmids and S. cerevisiae strains.
pSEC (snoRNA expression cassette) was constructed based on the parental yEPlac181 (a 2μm LEU2 vector kindly provided by E. M. Phizicky at the University of Rochester) (8). The glucose phosphate dehydrogenase (GPD) promoter region (a sequence corresponding to nucleotides −655 to 0 of TDH3) was inserted between EcoRI and BamHI. A 65-nt RNT1 element sequence (5′-TTTTTATTTCTTTCTAAGTGGGTACTGGCAGGAGTCGGGGCCTAGTTTAGAGAGAAGTAGACTCA-3′), corresponding to part of the 35S pre-rRNA 3′ external transcribed spacer (ETS) that is recognized by endonuclease RNT1 (RNase III activity) (11), was inserted between BamHI and SalI; a 55-nt snR13 termination sequence (5′-AGTAATCCTTCTTACATTGTATCGTAGCGCTGCATATATAATGCGTAAAATTTTC-3′), corresponding to nucleotides 26 to 80 downstream of snR13, was inserted between PstI and HindIII. The pSEC cassette thus constructed contains a pair of restriction sites (SalI and PstI) flanked by the RNT1 element on the 5′ side and the snR13 termination sequence on the 3′ side (11). These two restriction sites were then used for insertion of an snR52-based artificial box C/D RNA gene (in which one of the guide sequences was altered), resulting in the production of pSEC-gRNA-A804, pSEC-gRNA-A805, pSEC-gRNA-A806, pSEC-gRNA-U809, pSEC-gRNA-A810, and pSEC-gRNA-U811. Upon transformation into yeast cells, the mature artificial box C/D RNA (gRNA) was efficiently expressed (Fig. 2A).
FIG. 2.
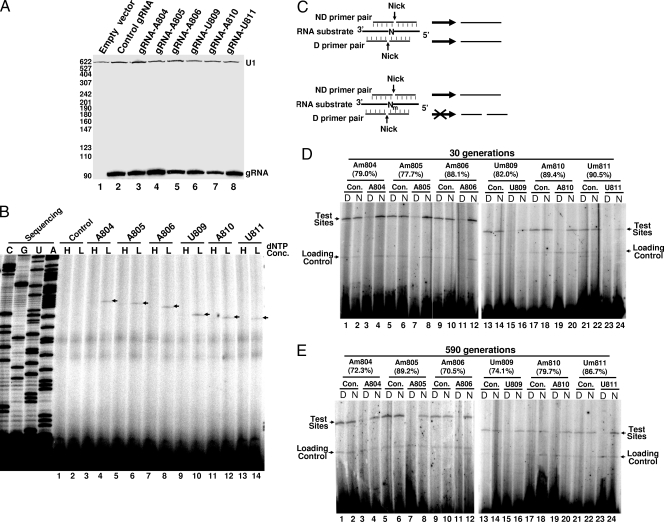
Artificial box C/D guide RNAs are expressed and functional. (A) Northern blot assay for gRNA expression. Total RNA isolated from cells (yCH-001) expressing no gRNA (empty vector), a random gRNA (Control), gRNA-A804, gRNA-A805, gRNA-A806, gRNA-U809, gRNA-A810, or gRNA-U811 was used for Northern blot analysis. Probes for U1 (loading control) and gRNAs were used, and signals corresponding to these RNAs are indicated. The levels of some gRNAs are slightly lower than the others, but they are all estimated (based on Northern blotting) as higher than the endogenous naturally occurring box C/D RNAs tested (data not shown). Numbers on the left are size markers (in nucleotides) of MspI-digested pBR322 DNA. (B) 2′-O-methylation mapping of individually modified nucleotides. Total RNA isolated from cells (yCH-002) expressing a random gRNA (Control), gRNA-A804 (lanes 3 and 4), gRNA-A805 (lanes 5 and 6), gRNA-A806 (lanes 7 and 8), gRNA-U809 (lanes 9 and 10), gRNA-A810 (lanes 11 and 12), or gRNA-U811 (lanes 13 and 14) was used for primer extension analysis in the presence of high (H; 1 mM) or low (L; 0.01 mM) dNTP concentrations. Arrows indicate the stop/pause signals corresponding to the 2′-O-methylated residues. The TLC1 sequencing ladder is shown on the left. (C) The strategy behind the ligation-based 2′-O-methylation assay (also see Materials and Methods and elsewhere in text). The thick lines represent the target RNA substrate, and the thin lines stand for primer pairs (ND, nondiscriminating; D, discriminating) used for ligation. N denotes a test nucleotide lacking 2′-O methylation; Nm represents a test nucleotide that is 2′-O methylated. Nicks (on the 5′ or 3′ side of the test nucleotide) are also shown. (D) Ligation-based quantification of 2′-O methylation. RNA that was isolated from cells after 30 generations expressed a random gRNA (Con.), gRNA-A804 (lanes 3 and 4), gRNA-A805 (lanes 7 and 8), gRNA-A806 (lanes 11 and 12), gRNA-U809 (lanes 15 and 16), gRNA-A810 (lanes 19 and 20), or gRNA-U811 (lanes 23 and 24) and was assayed for 2′-O methylation at the respective positions with position-specific primer pairs (A804, lanes 1 to 4; A805, lanes 5 to 8; A806, lanes 9 to 12; U809, lanes 13 to 16; A810, lanes 17 to 20; U811, lanes 21 to 24). D, discriminating primer pair; N, nondiscriminating primer pair. In all lanes, an additional pair of labeled primers was also included for a loading control. The relative modification efficiencies are calculated and shown in parentheses. (E) As in panel D, except that RNA was isolated from cells after 590 generations.
Strain yCH-001 (snR52Δ::URA3, BY4741 background) was used for analyzing the expression of snR52-based artificial gRNAs. In addition, using the kanMX4 cassette (23), we deleted RAD52 from another haploid strain, yHK53 (37) (BY4741 background; MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 Tel VII-URA3; kindly provided by X. Bi at the University of Rochester), generating a new strain (yCH-002) that no longer had the Rad52-mediated alternative telomere maintenance pathway. yCH-002 was then used for both the telomere length assay (see below) and the telomere position effect assay (see below).
A yeast strain, YKF103 (kindly provided by the Cech group), in which the chromosomal EST2 gene is fused with a protein A tag, was used for IgG immunoprecipitation assay (12) (see below).
Northern blot assay.
To examine gRNA expression, total RNAs isolated from cells (yCH-001) expressing no gRNA, a control gRNA, gRNA-A804, gRNA-A805, gRNA-A806, gRNA-U809, gRNA-A810, or gRNA-U811 were used for Northern blot analysis (Fig. 2A). The hybridization was performed using two 5′-end-radiolabeled DNA oligonucleotides: the first one was complementary to an snR52 sequence between box C′ and box D′ (5′-GTTTTTCTAATCCTAAAATCTTTCGATTTT-3′), and the second one was complementary to U1 snRNA (5′-AACGTCCTTCTACTATTGGAA-3′).
To check whether 2′-O methylation affects TLC1 levels, yeast strain yCH-002 was transformed with pSEC-gRNA-Control, pSEC-gRNA-A804, pSEC-gRNA-A805, pSEC-gRNA-A806, pSEC-gRNA-U809, pSEC-gRNA-A810, or pSEC-gRNA-U811. After 30, 310, or 590 generations, cells were collected, total RNAs were isolated, and Northern blotting was performed. In brief, upon electrophoresis on a 4% polyacrylamide-8 M urea gel, RNAs were transferred to a Hybond-N+ membrane (Amersham Pharmacia) and hybridized at room temperature with two 5′-end-labeled DNA oligonucleotide probes; one was complementary to TLC1 RNA (5′-TTCTCTGTCACATCGTTCGATGTAC-3′), and the other was complementary to U1 snRNA (5′-AACGTCCTTCTACTATTGGAA-3′). After an overnight hybridization, the membrane was extensively washed, and TLC1 and U1 signals were revealed by autoradiography.
Northern analysis was also carried out to check whether 2′-O-methylated TLC1 RNA associated with Est2p (or was assembled onto RNP) (see “Coimmunoprecipitation” and “Glycerol gradient assay” below).
Primer extension-based 2′-O-methylation assay.
A standard primer extension-based modification assay (with high and low deoxynucleoside triphosphate [dNTP] concentrations) was performed essentially as described previously (42) to detect RNA 2′-O methylation. Briefly, ∼6 μg of total RNA, or ∼200 ng of coimmunoprecipitated TLC1 RNA (see below), was mixed with 5′-radiolabeled DNA oligonucleotide Detect-TLC-2Ome (5′-TTCTCTGTCACATCGTTCGATGTAC-3′), and the primer extension reaction was carried out in the presence of either 1 mM dNTPs (high) or 0.01 mM dNTPs (low). The reaction mixtures were incubated at 42°C for 30 min and resolved on an 8% polyacryamide-8 M urea gel.
Ligation-based quantitative 2′-O-methylation assay.
Using biotinylated antisense TLC1 DNA oligonucleotide (5′-biotin-TCAATCCGAAATCCGACACTATCTC-3′) and biotin-streptavidin affinity chromatography (40, 41), we purified TLC1 RNA from 1 liter of yeast cells (optical density at 600 nm [OD600] of 7.0) that expressed no gRNA, a random gRNA, or gRNA targeting the nucleotides in and adjacent to the triple-helix region. Purified cellular TLC1 RNA served as the template in the subsequent ligation reaction.
To quantify the 2′-O methylation at any particular site of TLC1 RNA, two parallel ligation reactions (discriminating and nondiscriminating reactions) with two different site-specific pairs of oligodeoxynucleotides were carried out at 37°C for 30 min in the presence of 66 mM Tris-HCl (pH 7.6), 6.6 mM MgCl2, 10 mM dithiothreitol (DTT), 66 mM ATP, 15% dimethyl sulfoxide (DMSO), and 0.125 U/μl T4 DNA ligase (28).
In the discriminating ligation reaction, a pair of discriminating DNA oligonucleotides (D oligonucleotide pair) was used to hybridize with TLC1 RNA. Consequently, the two oligonucleotides were precisely aligned, placing a nick or the ligation junction (between the two oligonucleotides) on the 3′ side of U809. In the nondiscriminating ligation reaction, a pair of nondiscriminating DNA oligonucleotides (ND oligonucleotide pair) were used. Upon hybridization with TLC1 RNA, the oligonucleotides were aligned, leaving a nick or the ligation junction on the 5′ side of U809. It is well established that if the target nucleotide (U809) is 2′-O methylated, only the nondiscriminating oligonucleotide pair will be ligated; however, if U809 is not 2′-O methylated, both pairs of oligonucleotides will be ligated (28), thus providing a quantitative measurement of 2′-O methylation at U809.
For position 804 (A804), the D oligonucleotide pair were DF-804, 5′-GAAATTTCATCAGTAAGTTCAGTGAATAGATT-3′, and DA-804, 5′-[32P]TTTTATTTTACCTTTTTGTAGTGGG-3′; the ND oligonucleotide pair were NF-804, 5′-GAAATTTCATCAGTAAGTTCAGTGAATAGATTT-3′, and NA-804, 5′-[32P]TTTATTTTACCTTTTTGTAGTGGG-3′. Similarly, for positions 805, 806, 809, 810, and 811, the D oligonucleotide pairs were DF-805, 5′-GGAAATTTCATCAGTAAGTTCAGTGAATAGAT-3′, and DA-805, 5′-[32P]TTTTTATTTTACCTTTTTGTAGTGG-3′; DF-806, 5′-TGGAAATTTCATCAGTAAGTTCAGTGAATAGA-3′, and DA-806, 5′-[32P]TTTTTTATTTTACCTTTTTGTAGTG-3′; DF-809, 5′-AATTTCATCAGTAAGTTCAGTGAAT-3′, and DA-809, 5′-[32P]AGATTTTTTATTTTACCTTTTTGTA-3′; DF-810, 5′-AAATTTCATCAGTAAGTTCAGTGAA-3′, and DA-810, 5′-[32P]TAGATTTTTTATTTTACCTTTTTGT-3′; and DF-811, 5′-GAAATTTCATCAGTAAGTTCAGTGA-3′, and DA-811, 5′-[32P]ATAGATTTTTTATTTTACCTTTTTG-3′, respectively; the ND oligonucleotide pairs were NF-805, 5′-GGAAATTTCATCAGTAAGTTCAGTGAATAGATT-3′, and NA-805, 5′-[32P]TTTTATTTTACCTTTTTGTAGTGG-3′; NF-806, 5′-TGGAAATTTCATCAGTAAGTTCAGTGAATAGAT-3′, and NA-806, 5′-[32P]TTTTTATTTTACCTTTTTGTAGTG-3′; NF-809, 5′-AATTTCATCAGTAAGTTCAGTGAATA-3′, and NA-809, 5′-[32P]GATTTTTTATTTTACCTTTTTGTA-3′; NF-810, 5′-AAATTTCATCAGTAAGTTCAGTGAAT-3′, and NA-810, 5′-[32P]AGATTTTTTATTTTACCTTTTTGT-3′; and NF-811, 5′-GAAATTTCATCAGTAAGTTCAGTGAA-3′, and NA-811, 5′-[32P]TAGATTTTTTATTTTACCTTTTTG-3′, respectively. For sites U809, A810, and U811, the ligation products were 50 nucleotides long, and for sites A804, A805, and A806, the ligation products were 57 nucleotides long.
In all ligation reactions, another pair of DNA oligonucleotides (NF-1145, 5′-TTCCAAAAATTATCTAAA-3′; NA-1145, 5′-[32P]TGCATCGAAGGCATTAGGAGAAGTA-3′), which placed the ligation junction 5′ of U1145 (a nontargeting site) of TLC1 RNA, were used as a loading control.
After the ligation reaction, the radioactively labeled oligonucleotides (ligated and unligated) were resolved on an 8% polyacrylamide-8 M urea gel and quantified using a PhosphorImager (Molecular Dynamics).
Glycerol gradient assay.
The glycerol gradient assay procedure was essentially as described previously (22). Briefly, yeast cells from 2-liter overnight cultures (SD leucine dropout medium) were harvested at an OD600 of 1.0. Pelleted cells were suspended in 4 ml of extraction buffer (20 mM Tris-acetate [pH 7.5], 300 mM potassium glutamate, 1.1 mM MgCl2, 0.1 mM EDTA, 5% glycerol, 1 mM dithiothreitol [DTT], and 0.5 mM phenylmethylsulfonyl fluoride), placed in liquid nitrogen, and ground with a mortar and pestle for 30 min. The lysate was concentrated 3-fold in a Vivaspin ultrafiltration spin column and then loaded on the top of a 15 to 40% continuous glycerol gradient prepared with the extraction buffer. Ultracentrifugation (SW41 Ti rotor, 150,000 × g) was performed at 4°C for 18 h. Nineteen fractions were collected, and RNA was extracted from each fraction for Northern analysis (see above).
Coimmunoprecipitation.
Extracts were prepared from yeast cells (strain YKF103) expressing a control gRNA, gRNA-A804, gRNA-A805, gRNA-A806, gRNA-U809, gRNA-A810, or gRNA-U811, according to the glass bead lysis method (12). Briefly, 500 ml yeast cells (OD600, 1.0) was lysed, by glass bead beating, in TMG-300NaCl buffer containing 10 mM Tris-HCl, pH 8.0, 1 mM MgCl2, 10% glycerol, 0.1 mM DTT, and 300 mM NaCl. After clarification by brief centrifugation (15,000 × g at 4°C for 5 min, repeated 3 times), total protein concentration was adjusted to 5 mg/ml. Upon addition of Tween 20 (to a final concentration of 0.5%), 500 μl of extract was mixed with 10 μl IgG Sepharose 6 Fast Flow beads (GE Healthcare). After overnight nutation at 4°C, the beads were collected, washed three times with TMG-200NaCl-Tween buffer (10 mM Tris-HCl, pH 8.0, 1 mM MgCl2, 10% glycerol, 0.1 mM DTT, 200 mM NaCl, and 0.5% Tween) and one time with TMG-50NaCl buffer (10 mM Tris-HCl, pH 8.0, 1 mM MgCl2, 10% glycerol, 0.1 mM DTT, 50 mM NaCl), and then resuspended in 20 μl TMG-50NaCl buffer with 0.5 mM DTT. The beads were split into two equal aliquots of 10 μl each. One aliquot was used for in vitro telomerase activity assay (see below), and the other was used for TLC1 RNA extraction and analysis.
For TLC1 RNA extraction, 10 μl beads was treated with proteinase K at 37°C for 30 min in a 200-μl reaction mixture (10 mM Tris-HCl, pH 8.0, 0.5% SDS, 0.4 mg/ml proteinase K). RNA was then extracted with phenol-chloroform-isoamyl alcohol (PCA; 25:24:1) and precipitated with ethanol. Recovered TLC1 RNA was subsequently subjected to primer extension analysis (2′-O methylation assay) and Northern blot analysis (see above).
Western blot assay.
Western blotting was performed essentially as previously described (29). Briefly, 10 μl of immunoprecipitated beads was mixed with 10 μl 2× Laemmli loading buffer (125 mM Tris-HCl, pH 6.8, 4% SDS, 50% glycerol, 5% β-mercaptoethanol, 0.02% bromophenol blue). Upon incubation at 95°C for 5 min, the supernatant was resolved on a 4 to 15% Tris-glycine gel (Bio-Rad). Protein was then transferred to a Protran nitrocellulose membrane (Whatman) and probed with antibodies. The primary antibody was rabbit IgG (Sigma), and the secondary was goat anti-rabbit IgG-alkaline phosphatase (AP)-conjugated antibody (Bio-Rad).
Telomerase activity assay.
The in vitro telomerase activity assay was carried out as previously described (29). Briefly, 2 μl of Est2p-bound beads derived from immunoprecipitation (see above) was added to a final 10 μl of reaction mixture containing 40 mM Tris-HCl, pH 8.0, 50 mM NaCl, 5% glycerol, 2.5 mM MgCl2, 0.5 mM spermidine, 0.5 mM DTT, 2.5 μM telomerase substrate (5′-TGTGGTGTGTGTGGG-3′), 1 μl [α-32P]dGTP (10 μCi/μl), and 1 μl [α-32P]dTTP (10 μCi/μl). After incubation at 30°C for 20 min, the reaction was terminated by addition of 200 μl of proteinase K buffer (10 mM Tris-HCl, pH 8.0, 0.5% SDS) and 4 μl of proteinase K (20 mg/ml). The mixture was then incubated at 60°C for 30 min. Nucleic acids were recovered by PCA extraction and ethanol precipitation. The primer-extended DNA products were resolved on a 14% polyacrylamide denaturing gel and visualized by autoradiography.
Southern analysis to measure telomere length.
For Southern blotting, yeast cells were grown to saturation in 20 ml SD leucine dropout medium. The genomic DNA was subsequently extracted. Briefly, yeast cells were suspended in extraction buffer (10 mM Tris-HCl, pH 8.0, 2% Triton X-100, 1% SDS, 1 mM EDTA). Upon the addition of saturated phenol and glass beads, cells were broken by vigorous vortexing. Cell extracts were treated with proteinase K (∼0.4 mg/ml) at 55°C for 2 h, and genomic DNA was subsequently purified with PureLink genomic DNA spin columns (Invitrogen). The purified genomic DNA was digested with XhoI, resolved on a 0.8% agarose gel, transferred onto a Hybond-N+ membrane (Amersham Pharmacia), and hybridized with two radiolabeled probes, one against the telomeric repeat sequence probe (5′-TGTGGGTGTGGTGTGTGGGTGTGGTG-3′) and the other (as a control) against chromosome IV (nucleotides 31051 to 31075; 5′-GTCTGGCCTATGGTGCTAGTAGTAC-3′).
For detecting URA3 integrated into chromosome VII (yCH-002 strain), purified genomic DNA was digested with PstI and hybridized with a URA3-specific probe (5′-ACATTATTATTGTTGGAAGAGGACT-3′) and a control probe against chromosome IV (nucleotides 194700 to 194676; 5′-GATACACCTTCCGTTTCTGACCCAT-3′).
Telomere position effect assay.
After transformation with various pSEC-gRNA plasmids, cells were plated onto solid medium, and a single colony was picked and grown to saturation in 5 ml of SD leucine dropout medium. One microliter of the liquid culture was then withdrawn, placed in 5 ml of fresh SD leucine dropout medium, and grown to saturation again. This cycle was repeated until the cells reached 590 generations. At 310, 450, and 590 generations, an aliquot of cells was saved and stored at −80°C for future use. To assay growth phenotype, cells from different generations were first diluted to an OD600 of 0.1 and then plated with a series of 5-fold dilutions on the SD leucine dropout solid medium with or without 5-fluoroorotic acid (5-FOA) (1 mg/ml). The growth phenotype was monitored at 30°C.
RESULTS
Artificial box C/D RNA targeting TLC1 is efficiently expressed.
Qiao and Cech have recently shown that 2′-OH groups of the triple-helix (A804, A805, and A806) nucleotides and their adjacent nucleotides (U809, A810, and U811) within the conserved pseudoknot structure of TLC1 contribute to telomerase function in vitro (27) (Fig. 1A), thus offering an opportunity for an in vivo functional analysis of these 2′-OH groups using RNA-guided RNA 2′-O methylation. We designed six artificial box C/D guide RNAs (gRNA-A804, gRNA-A805, gRNA-A806, gRNA-U809, gRNA-A810, and gRNA-U811), each of which targeted one of the six nucleotides in the triple-helix region (Fig. 1A). These artificial guide RNAs were constructed based on snR52, a naturally occurring S. cerevisiae box C/D snoRNA that contains two guide sequences, one between box C and box D′ and the other between box C′ and box D (Fig. 1B). The short guide sequence between box C and box D′ or between box C′ and box D was altered to target the TLC1 nucleotides; all other nucleotide sequences of snR52 were left unchanged. The artificial box C/D RNA genes were separately inserted into a 2μm vector, with the expression of the box C/D guide RNAs under the control of the GPD promoter. Upon transformation, we measured the expression of guide RNA using Northern analysis. Our result showed that every guide RNA was efficiently expressed (Fig. 2A, lanes 3 to 8).
Artificial box C/D guide RNA is functionally active in vivo.
To ensure that the artificial box C/D guide RNA had a sufficient level of activity, we next carried out a primer extension-based 2′-O-methylation assay to detect 2′-O methylation of TLC1 at the target sites. It is well established that at low deoxynucleoside triphosphate (dNTP) concentrations, primer extension will stop/pause precisely one nucleotide before the 2′-O-methylated site (24). When a guide RNA was expressed, we clearly detected a stop/pause signal corresponding to its target site under low-dNTP conditions (Fig. 2B, lanes 4, 6, 8, 10, 12, and 14). As expected, when high dNTP concentrations were used, the stop/pause signal was barely detected (Fig. 2B, lanes 3, 5, 7, 9, 11, and 13). Thus, our results demonstrated that each guide RNA was capable of guiding TLC1 2′-O methylation at its target site.
Given that 2′-O-methylation efficiency is important for determining the degree to which the modification influences telomerase activity, we further quantified the level of 2′-O methylation at each target site. We took advantage of a recently developed ligation-based assay, in which a pair of DNA primers are aligned with the RNA substrate upon hybridization, leaving the ligation junction (nick) 5′ or 3′ of the test nucleotide in the RNA substrate (28) (Fig. 2C). If the test nucleotide is 2′-O methylated, the two primers will not be ligated if the ligation junction is placed 3′ of the modified nucleotide (discriminating or D primer pair), but they will be quantitatively ligated if the junction is placed 5′ of the modified nucleotide (nondiscriminating or ND primer pair). If the test nucleotide is not 2′-O methylated, the two primers (either D or ND primer pair) will be quantitatively ligated regardless of where the junction is (Fig. 2C). The ligation efficiency should correlate well with the modification level at the test site. Comparison of the ligation ratios will thus allow quantification of 2′-O methylation at the test nucleotide.
Figures 2D and E show the ligation experiments using TLC1 RNA harvested after 30 and 590 generations, respectively. When the ND primer pair was used, ligation efficiency (the ratio of the ligated product to the loading control) was about the same for all TLC1 RNAs at all sites tested, including untargeted TLC1 RNA (Con.) and gRNA-targeted TLC1 RNA (Fig. 2D and E, compare even-numbered lanes). When the D primer pair was used, a drastic reduction in ligation was, however, observed in reactions where TLC1 RNA was isolated from cells expressing gRNAs (Fig. 2D and E, compare lanes 3, 7, 11, 15, 19, and 23 with other odd-numbered lanes). Our results indicated a high level (70 to 90%) of 2′-O methylation at all target sites regardless of the number of cell generations (30 or 590 generations) (compare Fig. 2D and E, and compare targeted lanes with untargeted control lanes). Our results also indicated that 2′-O methylation was target specific, as reduced ligation was detected only in reactions where a target-specific gRNA was expressed and a D primer pair for the respective target site was used (Fig. 2D and E and data not shown).
2′-O methylation in the triple-helix region does not change TLC1 RNA levels.
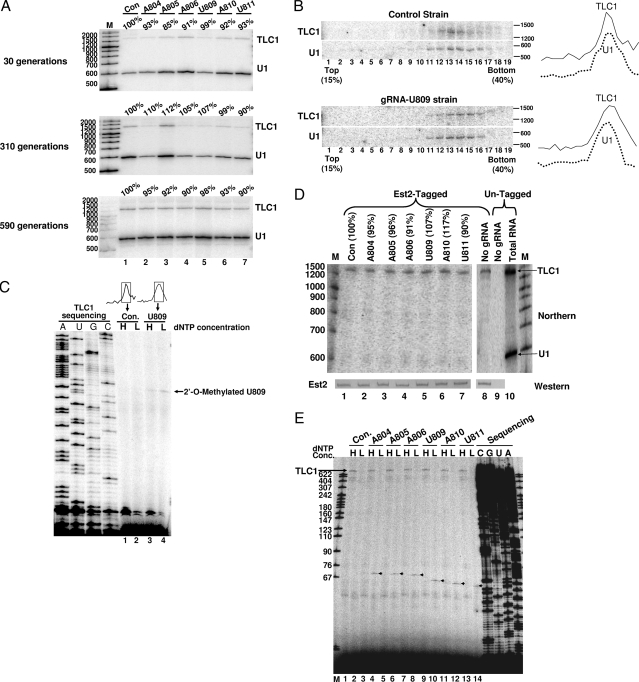
We next wished to test whether 2′-O methylation in the triple-helix region would affect TLC1 RNA levels. Using Northern analysis, we assessed the levels of TLC1 RNA in wild-type control cells and in cells expressing various artificial gRNAs. Total RNAs were isolated from these cells after different numbers of generations, and Northern analysis was carried out using a TLC1-specific probe and a U1-specific probe as an internal control. The signals of TLC1 RNA relative to U1 RNA were virtually identical in all the cells after 30, 310, and 590 generations (Fig. 3A), indicating that targeted 2′-O methylation had no effect on steady-state levels of TLC1 RNA.
FIG. 3.
2′-O-methylated TLC1 RNA is incorporated into telomerase RNP. (A) 2′-O methylation has no effect on the steady-state level of TLC1 RNA. Cells expressing a random gRNA (control [con]), gRNA-A804 (lane 2), gRNA-A805 (lane 3), gRNA-A806 (lane 4), gRNA-U809 (lane 5), gRNA-A810 (lane 6), or gRNA-U811 (lane 7) were grown for 30, 310, and 590 generations. RNAs were isolated from these cells and analyzed by Northern blotting with TLC1-specific and U1-specific probes. TLC1 RNA levels were calculated relative to U1 (in percentage). Lane M, size markers (indicated in nucleotides) of 100-bp-plus DNA ladder (Fermentas). (B) Incorporation of TLC1 RNA into telomerase RNP. Extracts prepared from cells expressing a random gRNA (Control) or gRNA-U809 were loaded on a 15 to 40% glycerol gradient. Nineteen fractions were collected. RNA from each fraction was analyzed by Northern blotting with TLC1-specific and U1-specific probes. On the right is a plot showing the TLC1 and U1 signal peaks. (C) Fractions 12 to 15 of the gradient described in panel B were pooled, RNAs were recovered, and primer extension was carried out in the presence of high (H) or low (L) dNTP concentrations. Lanes 1 and 2 are from cells expressing a random gRNA (Con.), and lanes 3 and 4 are from cells expressing gRNA-U809. A TLC1 sequencing ladder was electrophoresed in parallel on the left. A signal corresponding to 2′-O-methylated U809 is indicated. (D) Western and Northern analyses of immunoprecipitated Est2p complex. A yeast strain, in which the EST2 gene is fused with a protein A tag, was transformed with a plasmid containing no gRNA (lane 8), a random gRNA (lane 1), gRNA-A804 (lane 2), gRNA-A805 (lane 3), gRNA-A806 (lane 4), gRNA-U809 (lane 5), gRNA-A810 (lane 6), or gRNA-U811 (lane 7). IgG precipitation was subsequently carried out. As a control, an untagged yeast strain was also used for IgG precipitation (lane 9). Precipitated proteins were recovered, and Western analysis (with anti-protein A antibodies) was performed. Furthermore, RNA coprecipitated with protein A-Est2p was recovered, and Northern analysis was carried out. Probes for U1 (loading control) and TLC1 were used, and signals corresponding to these RNAs are indicated. As a control, total cellular RNA was also used (lane 10). Lane M, as in panel A. (E) 2′-O-methylation mapping was conducted as in panel C. All seven samples (corresponding to lanes 1 to 7 in panel D) were assayed. Lane M, size markers of MspI-digested pBR322 DNA.
2′-O-methylated TLC1 RNA is assembled into telomerase RNP.
To determine whether 2′-O-methylated TLC1 RNA was incorporated into telomerase RNP, we carried out a glycerol gradient analysis. TLC1 RNA from the control strain and that from the strain expressing gRNA-U809 peaked in the same fractions, just as U1 snRNP did in both strains (Fig. 3B). We then isolated the RNA from the telomerase RNP peaks and performed the 2′-O methylation assay. Although no 2′-O methylation signal was observed in control TLC1 RNA (Fig. 3C, lanes 1 and 2), a clear 2′-O methylation signal corresponding to U809 was detected in TLC1 RNA isolated from cells expressing gRNA-U809 (lanes 3 and 4). Thus, 2′-O-methylated TLC1 RNA appeared to be incorporated into the telomerase RNP.
To further confirm our gradient results, we took an independent approach, namely, the Est2p coprecipitation assay. We took advantage of the availability of a strain in which the telomerase reverse transcriptase gene EST2 is fused with a protein A tag (12). Upon transformation with the plasmid containing an artificial box C/D RNA gene, cells were harvested, and Est2p was pulled down through protein A-IgG precipitation. Western blotting was used to measure the precipitated Est2p. As shown in Fig. 3D, a nearly identical amount of Est2p was precipitated (compare the intensities of the Est2 bands in lanes 1 to 8 [Est2-tagged lanes]). As a control, when an untagged strain was used, no Est2p was detected (lane 9). RNA coprecipitated with Est2p was also recovered and analyzed. Northern blotting indicated that TLC1 RNA was efficiently coprecipitated with Est2p regardless of whether cells were transformed with a plasmid containing no gRNA gene (lane 8), a random box C/D RNA gene (lane 1), or any one of the six artificial box C/D RNA genes (lanes 2 to 7). 2′-O methylation was further assayed, and our experiments showed that Est2p-associated TLC1 RNA, isolated from cells expressing a gRNA, was efficiently 2′-O methylated at the expected target site (Fig. 3E). Thus, we conclude that 2′-O-methylated TLC1 RNA is efficiently incorporated into telomerase RNP (at least associated with Est2p).
In vitro functional assay indicates that 2′-O methylation at U809 (adjacent to the triple helix) reduces telomerase activity.
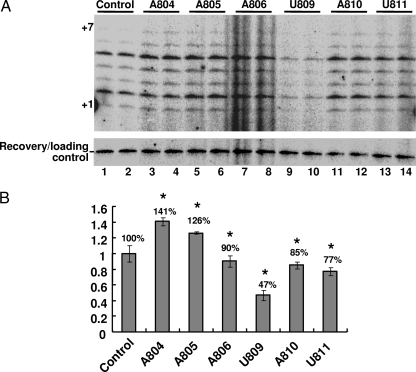
To assess whether 2′-O methylation at the nucleotides in and near the triple-helix structure affects function, we carried out the in vitro telomerase activity assay (12) using the Est2p-bound fractions described above. As shown in Fig. 4, targeted 2′-O methylation at position U809 resulted in a substantial reduction of telomerase activity (Fig. 4A, lanes 9 and 10, and Fig. 4B). When A804 or A805 was targeted by the artificial gRNAs, a relatively small but statistically significant enhancement of telomerase activity was observed (Fig. 4A, compare lanes 3 to 6 with lanes 1 and 2; Fig. 4B). 2′-O methylation at the other sites had no significant effect on telomerase activity (Fig. 4A and B).
FIG. 4.
In vitro telomerase activity assay. (A) The IgG-bound (protein A-Est2p) fraction described in the Fig. 3D legend was used directly for an in vitro telomerase activity assay (29). Three independent sets of experiments, each in duplicate, were carried out. Shown is one such experiment. Duplicate lanes represent two independent reactions for each strain. A control band (for recovery and loading control [30]) is also shown. +1 and +7, size markers corresponding to the first and seventh nucleotides incorporated into the products. (B) Relative telomerase activity of the IgG-bound fraction, derived from each strain, was quantified based on three independent sets of duplicate experiments. The quantification was performed by comparing the intensities of the bands (the sum of all seven bands) in each lane. Adjustment was made for every band by deducting a background area immediately above the band. The percentage of in vitro telomerase activity of each strain was calculated against control activity (set to 100%). The error bars represent the standard deviations of the measurements. Asterisks indicate that the P values are less than 0.05 (calculated using the Microsoft Excel t test software).
A804 and A805 are in the triple-helix structure, and U809 is in a nearby stem adjacent to the triple helix (Fig. 1A). The fact that 2′-O methylation at U809 reduced telomerase activity is consistent with the previous in vitro study by Qiao and Cech (27), who showed that combined substitution of the 2′-OH groups of all three adjacent nucleotides (U809, A810, and U811) with 2′-H groups resulted in diminished telomerase activity. In contrast, our targeted 2′-O methylation directed at the triple-helix nucleotide A804 or A805 resulted in no reduction of telomerase activity; rather, a slight increase in telomerase activity was observed. This observation is quite different from that made by Qiao and Cech, who show a substantial reduction of telomerase activity when the 2′-OH groups of all three triple-helix nucleotides are simultaneously changed to 2′-H groups. This apparent inconsistency is probably due to the fact that we changed the 2′-OH groups to 2′-O-methyl groups whereas they changed them to 2′-H groups (see Discussion).
A804, A805, and U809 in the triple-helix region are key 2′-O-methylation targets for influencing telomerase activity in vivo.
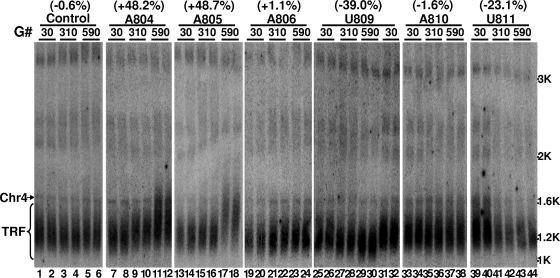
We next assessed the effect of the six artificial guide RNAs on chromosome end maintenance in vivo. Using Southern blotting, we monitored telomere length in cells expressing these artificial gRNAs. As shown in Fig. 5, when gRNA-U809 was expressed, chromosome ends progressively shortened with time (compare lanes 29 and 30 [590 generations] with lanes 27 and 28 [310 generations] and with lanes 25 and 26 [30 generations] or lanes 31 and 32 [30 generations]). Interestingly, the chromosome ends moderately lengthened when gRNA-A804 or gRNA-A805 was expressed (compare lanes 11 and 12 with lanes 9 and 10 and lanes 7 and 8; also compare lanes 17 and 18 with lanes 15 and 16 and lanes 13 and 14). In contrast, no apparent changes in telomere length were detected in cells expressing any other gRNAs (lanes 19 to 24 and 33 to 44) or no gRNA (data not shown). These results are consistent with our in vitro assay results described above (also see the Discussion), pinpointing three critical nucleotides: A804 and A805 for lengthening and U809 for shortening. The other sites appeared to be less important for telomerase activity when targeted individually.
FIG. 5.
Southern analysis of chromosome end length in cells expressing various artificial guide RNAs. Cells (yCH-002) expressing a random gRNA (Control), gRNA-A804 (lanes 7 to 12), gRNA-A805 (lanes 13 to 18), gRNA-A806 (lanes 19 to 24), gRNA-U809 (lanes 25 to 32), gRNA-A810 (lanes 33 to 38), or gRNA-U811 (lanes 39 to 44) were harvested after being grown for the indicated number of generations (G#). DNA was recovered, digested with XhoI, and hybridized with two radiolabeled probes, a telomere-specific one and a chromosome IV-specific one (as an internal control). The fragment of chromosome IV (Chr4) and the telomeres (TRF) are indicated. For each strain, two independent experiments are presented in duplicate lanes. The distances between the signal peaks of Chr4 and TRF were measured using ImageQuant software (Molecular Dynamics). Shown in parentheses at the top of each strain are the relative changes in distance between two time points (30 and 590 generations) (difference in average distance between the two time points, divided by the average distance at 30 generations). “+” indicates telomere lengthening, and “−” indicates telomere shortening. Numbers at the right indicate sites of double-stranded DNA.
Targeted 2′-O methylation influences telomerase activity as shown by the telomere position effect assay.
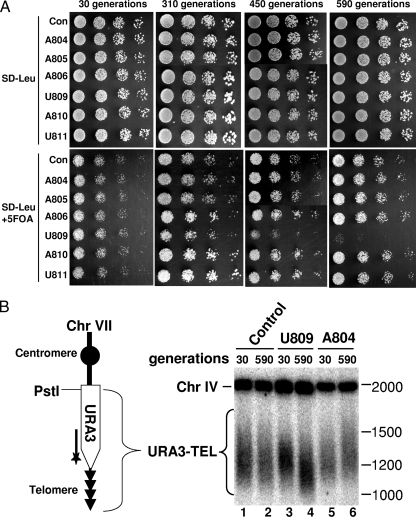
To further prove the effectiveness of our in vivo approach, we used an independent phenotypic assay involving the yeast strain (yHK53 rad52Δ::kanMX) (37) in which URA3 is inserted into chromosome VII near its end. Under normal conditions, the chromosome ends are stable and URA3 is silenced by the telomere position effect (13), thus allowing cells to grow on 5-fluoroorotic acid (5-FOA) medium. When the chromosome ends are progressively shortened, URA3 is, however, progressively activated, which thus results in cell death on 5-FOA-containing medium. Using this assay, we observed a clear progressive growth defect for cells expressing gRNA-U809. In contrast, no growth defect on 5-FOA medium was observed when cells were transformed with any other gRNAs targeting a single nucleotide in the triple-helix region or with a control box C/D guide RNA containing a random guide sequence (Fig. 6A).
FIG. 6.
Telomere position effect assay. (A) Cells (yCH-002) expressing a random gRNA (Con), gRNA-A804, gRNA-A805, gRNA-A806, gRNA-U809, gRNA-A810, or gRNA-U811 were grown for the indicated number of generations and then plated on SD-Leu medium with or without 5-FOA. In each panel, the four columns of cells are serial dilutions (5×) from left to right. (B) DNAs were recovered from the cells described in panel A, cleaved with PstI, and hybridized with a URA3-specific radiolabeled probe and a chromosome IV-specific radiolabeled probe. (Left panel) The schematic shows the modified chromosome VII with URA3 inserted near the telomere. A PstI site and the site of hybridization with the URA3 probe (a thick line with a star) are indicated. (Right panel) The signals corresponding to the chromosome IV fragment and the telomere of chromosome VII are indicated. Size markers of double-stranded DNA are on the right.
To confirm our observation, we further measured the telomere length of chromosome VII using Southern analysis. Whereas no change in telomere length was detected when cells expressed a random gRNA (Fig. 6B; compare lane 1 with lane 2), substantial telomere shortening was observed when cells were transformed with gRNA-U809 (compare lane 3 with lane 4). Interestingly, a moderate lengthening of the end of chromosome VII was observed when cells received gRNA-A804 (compare lane 5 with lane 6), which is consistent with what was observed earlier (Fig. 4 and 5). Taken together, these results indicate that targeted telomerase RNA 2′-O methylation is a very effective strategy for blocking telomerase activity in vivo.
DISCUSSION
Using RNA-guided RNA modification, we introduced 2′-O-methylation sites into specific nucleotides in or adjacent to the triple-helix structure of S. cerevisiae TLC1 RNA, thereby affecting telomerase activity in vivo. Specifically, 2′-O methylation at U809 resulted in telomere shortening, whereas 2′-O methylation at A804 and A805 led to moderate lengthening of telomeres. Although there may be some apparent inconsistencies between our data and recently published in vitro data (27) (discussed below), our results appear to be consistent with the notion that the 2′-OH groups of the nucleotides within the triple helix and the nearby stem contribute to function (directly or indirectly) (27). Our results also suggest that RNA-guided RNA modification can serve as an effective tool for regulating telomerase activity (and RNA function in general) in vivo.
With respect to function, the Cech lab demonstrated, using an in vitro system, that the 2′-OH groups of nucleotides in the triple-helix region, which are proximal to the catalytic active site, contribute to telomerase activity in vitro (27). Specifically, they show that the simultaneous changing of the 2′-OH groups of the triple-helix nucleotides (A804, A805, and A806) to 2′-H groups results in a substantial reduction in telomerase activity. Likewise, when the nucleotides (U809, A810, and U811) adjacent to the triple-helix structure are simultaneously changed to 2′-deoxynucleotides, a substantial reduction of in vitro telomerase activity is observed. Consistent with their results, we observed a clear reduction of telomerase activity both in vitro (Fig. 4) and in vivo (Fig. 5 and 6) when U809, one of the three nucleotides adjacent to the triple-helix structure, was targeted for 2′-O methylation, although targeting of either A810 or U811 did not lead to the reduction of telomerase activity. Our results thus argue that it is the 2′-OH group of U809, rather than the 2′-OH groups of A810 and U811, that contributes to function. However, given that our approach added a methyl group to the sugar ring, we cannot exclude the possibility that such a modification disrupted the local structure of the RNA, thereby resulting in the reduction of telomerase activity. This alternate possibility would then argue against the notion that the target 2′-hydroxyl group per se is critical. Caution should be taken in interpreting our results.
Rather surprisingly, we detected telomere lengthening when 2′-O methylation was introduced into either A804 or A805, two of the three triple-helix nucleotides (Fig. 5). This enhancing effect is in contrast to the inhibitory effect previously observed when the 2′-OH groups of the triple-helix nucleotides were changed to 2′-H groups (27). It is conceivable that if the 2′-OH group of A804 and/or A805 contributes directly to catalysis (as U809 does), changing the 2′-OH group to either 2′-H or 2′-O methyl should dramatically alter its chemical properties (e.g., preventing hydrogen bond donation), thus resulting in a reduction (rather than an enhancement) of telomerase activity. One possible explanation is that the 2′-OH groups of the triple-helix nucleotides may indirectly contribute to function (or catalysis): modifications at the 2′ position may influence the configuration of the sugar pucker, either C3′-endo or C2′-endo configuration, which may in turn affect its function. A sugar pucker with a 2′-OH favors the C3′-endo conformation compared with a sugar pucker with a 2′-H. However, when the 2′-OH of the sugar pucker is methylated (2′-O methyl), the C3′-endo conformation becomes even more favorable (16, 36). This order of preference (2′-O methyl > 2′-OH > 2′-H) for the C3′-endo conformation would be consistent with the observed effect of different 2′ moieties on telomerase activity if the C3′-endo (rather than C2′-endo) is the more functionally favorable conformation. Specifically, relative to 2′-OH, 2′-O methyl enhances, while 2′-H inhibits, telomerase activity.
This hypothesis (sugar pucker effect), however, does not explain the results obtained from the experiment with human telomerase RNA (27). Specifically, Qiao and Cech show that a change of 2′-OH to 2′-H or to 2′-O methyl at one of the nucleotides in the human telomerase RNA triple helix results in a similar inhibitory effect on telomerase activity. The apparent inconsistency between our yeast results and the human results (27) may be due to species or system differences. Along this line, we note that although qualitatively consistent, the effects on telomerase activity observed in vitro (Fig. 4) do not seem to correlate well with the effects observed in vivo (Fig. 5). For instance, when U809 was 2′-O methylated, we observed a relatively large reduction of telomerase activity in vitro (Fig. 4). However, the in vivo effect of this modification on telomere length was rather moderate (effect was detected only after many generations) (Fig. 5 and 6). On the other hand, while 2′-O methylation at either A804 or A805 showed a relatively small enhancing effect on telomerase activity when tested in vitro (Fig. 4), this effect seemed to be larger when analyzed in vivo (Fig. 5). While it is difficult to explain these phenomena at this point, we speculate that there are some other factors (or mechanisms) that are able to positively influence the overall telomerase activity in vivo and that these factors (mechanisms) are absent in the in vitro system. More work is necessary to clarify the mechanism.
We recognize the possibility that it is the extensive base-pairing interactions between the guide sequence of gRNA and the target sequence of TLC RNA, rather than 2′-O methylation per se, that have influenced telomerase activity. For instance, the base pairing may have altered TLC RNA folding, localization, and even telomerase RNP assembly. However, we regard this possibility as unlikely for at least two reasons. First, we detected clean and specific 2′-O methylation at target sites (Fig. 2B and 3E). Second, not all gRNAs, which targeted different nucleotides in the same region, exhibited an inhibitory or enhancing effect on telomerase activity. For instance, gRNA-U809 and gRNA-A810 targeted nucleotides adjacent to each other, but only gRNA-U809 exhibited an inhibitory effect on telomerase activity. Because both gRNAs maintained almost identical complementarity with TLC1 RNA, our results suggest that the observed inhibitory effect of targeting U809 was truly 2′-O methylation specific rather than an antisense effect that could impact TLC1 RNA folding, localization, or RNP assembly. Likewise, the same reasoning can be used to explain the enhancing effect of targeting A804 or A805 (comparing gRNA-A804 or gRNA-A805 with gRNA-A806).
It should also be noted that our artificial guide RNAs may have an unintended target(s), thus raising concerns about substrate specificity. To address this issue, we used the guide sequences (12 nt) to conduct a BLAST search against the yeast genome, with an attempt to identify other potential target RNAs. Our search generated only a few such candidates: six (STE24, PRM4, NAM7, SMI1, PUF3, and TUS1) for gRNA-A804 and gRNA-A805 and one (STB4) for U809. None of these potential targets have known functions in telomere maintenance. Thus, it is unlikely that the observed effects are due to the nonspecific effect of modifications of unintended off-targets.
With regard to the application of RNA-guided RNA modification, we have shown that an artificial box C/D guide RNA can efficiently target TLC1 2′-O methylation at specific sites. The artificial guide RNA can be constructed according to a naturally occurring box C/D snoRNA (e.g., snR52). According to the sequence/site to be targeted, only the short guide sequences of the original box C/D snoRNA need to be altered, and the remaining sequences do not have to be changed. Such an approach to target an RNA at a specific site appears to be straightforward and effective and could possibly be applied to many different RNA types.
Although any nucleotides of an RNA can, in theory, be targeted in vivo, control of RNA localization remains a major issue that must be addressed. It is known that box C/D snoRNA (or RNP) is localized to the nucleoli and/or Cajal bodies (18); however, some potential target RNAs (for example, mRNA) do not colocalize with snoRNA. The distinct localization of RNAs raises the question of whether all nuclear RNAs (including those that are temporarily present in the nucleus) can be targeted for modification. We note that although localization studies detect snoRNAs in the nucleoli and/or Cajal bodies (2, 9), such studies do not exclude the possibility that snoRNAs may exist in the nucleoplasm as well. Perhaps the failure to detect snoRNAs in the nucleoplasm merely reflects the fact that snoRNAs are too dilute to be detected in this subnuclear compartment. In this regard, two reports have suggested that a U2-specific guide RNA (40) and a guide RNA specific for spliced-leader RNA (a special type of spliceosomal snRNA involved in trans splicing) (21) may both reside within the nucleoplasm rather than within the nucleolus or Cajal bodies. Such conclusions are bolstered by a recent finding that suggests the presence of a number of Drosophila melanogaster snRNA-specific guide RNAs in the nucleoplasm as well as in Cajal bodies (10). In this regard, we and others have shown that mRNA as well as pre-mRNA can be targeted by artificial guide RNAs for modification in vivo (5, 42) (J. Karijolich and Y.-T. Yu, unpublished data). Thus, it appears that RNA-guided RNA modification can occur in one or a few different nuclear subcompartments, including the nucleolus, Cajal bodies, and/or even the nucleoplasm, and that RNA-guided RNA modification is a highly useful approach for regulating RNA function in eukaryotic cells.
Acknowledgments
We thank T. Cech, K. Friedman, and K. Goodrich (University of Colorado) for their generous gift of the YKF103 strain and X. Bi and H. Kuzmiak (University of Rochester, Department of Biology) for the yHK53 strain. We also thank the members (J. Karijolich, in particular) of the Yu laboratory for valuable discussions.
This work was supported by grant GM62937 (to Y.-T.Y.) from the National Institutes of Health.
Footnotes
Published ahead of print on 20 July 2010.
REFERENCES
- 1.Bachellerie, J. P., B. Michot, M. Nicoloso, A. Balakin, J. Ni, and M. J. Fournier. 1995. Antisense snoRNAs: a family of nucleolar RNAs with long complementarities to rRNA. Trends Biochem. Sci. 20:261-264. [DOI] [PubMed] [Google Scholar]
- 2.Balakin, A. G., L. Smith, and M. J. Fournier. 1996. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell 86:823-834. [DOI] [PubMed] [Google Scholar]
- 3.Blackburn, E. H., C. W. Greider, and J. W. Szostak. 2006. Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat. Med. 12:1133-1138. [DOI] [PubMed] [Google Scholar]
- 4.Blasco, M. A. 2007. Telomere length, stem cells and aging. Nat. Chem. Biol. 3:640-649. [DOI] [PubMed] [Google Scholar]
- 5.Cavaille, J., M. Nicoloso, and J. P. Bachellerie. 1996. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature 383:732-735. [DOI] [PubMed] [Google Scholar]
- 6.Cech, T. R. 2004. Beginning to understand the end of the chromosome. Cell 116:273-279. [DOI] [PubMed] [Google Scholar]
- 7.Collins, K. 2006. The biogenesis and regulation of telomerase holoenzymes. Nat. Rev. Mol. Cell Biol. 7:484-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Culver, G. M., S. M. McCraith, S. A. Consaul, D. R. Stanford, and E. M. Phizicky. 1997. A 2′-phosphotransferase implicated in tRNA splicing is essential in Saccharomyces cerevisiae. J. Biol. Chem. 272:13203-13210. [DOI] [PubMed] [Google Scholar]
- 9.Darzacq, X., B. E. Jady, C. Verheggen, A. M. Kiss, E. Bertrand, and T. Kiss. 2002. Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J. 21:2746-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deryusheva, S., and J. G. Gall. 2009. Small Cajal body-specific RNAs (scaRNAs) of Drosophila function in the absence of Cajal bodies. Mol. Biol. Cell 20:5250-5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fatica, A., M. Morlando, and I. Bozzoni. 2000. Yeast snoRNA accumulation relies on a cleavage-dependent/polyadenylation-independent 3′-processing apparatus. EMBO J. 19:6218-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedman, K. L., and T. R. Cech. 1999. Essential functions of amino-terminal domains in the yeast telomerase catalytic subunit revealed by selection for viable mutants. Genes Dev. 13:2863-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gottschling, D. E., O. M. Aparicio, B. L. Billington, and V. A. Zakian. 1990. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63:751-762. [DOI] [PubMed] [Google Scholar]
- 14.Greider, C. W., and E. H. Blackburn. 1985. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43:405-413. [DOI] [PubMed] [Google Scholar]
- 15.Greider, C. W., and E. H. Blackburn. 1989. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337:331-337. [DOI] [PubMed] [Google Scholar]
- 16.Hou, Y. M., X. Zhang, J. A. Holland, and D. R. Davis. 2001. An important 2′-OH group for an RNA-protein interaction. Nucleic Acids Res. 29:976-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, N. W., M. A. Piatyszek, K. R. Prowse, C. B. Harley, M. D. West, P. L. Ho, G. M. Coviello, W. E. Wright, S. L. Weinrich, and J. W. Shay. 1994. Specific association of human telomerase activity with immortal cells and cancer. Science 266:2011-2015. [DOI] [PubMed] [Google Scholar]
- 18.Kiss, T. 2001. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 20:3617-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiss, T. 2002. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 109:145-148. [DOI] [PubMed] [Google Scholar]
- 20.Kiss-Laszlo, Z., Y. Henry, J. P. Bachellerie, M. Caizergues-Ferrer, and T. Kiss. 1996. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell 85:1077-1088. [DOI] [PubMed] [Google Scholar]
- 21.Liang, X. H., Y. X. Xu, and S. Michaeli. 2002. The spliced leader-associated RNA is a trypanosome-specific sn(o) RNA that has the potential to guide pseudouridine formation on the SL RNA. RNA 8:237-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lingner, J., T. R. Hughes, A. Shevchenko, M. Mann, V. Lundblad, and T. R. Cech. 1997. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276:561-567. [DOI] [PubMed] [Google Scholar]
- 23.Ma, X., C. Yang, A. Alexandrov, E. J. Grayhack, I. Behm-Ansmant, and Y. T. Yu. 2005. Pseudouridylation of yeast U2 snRNA is catalyzed by either an RNA-guided or RNA-independent mechanism. EMBO J. 24:2403-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maden, B. E., M. E. Corbett, P. A. Heeney, K. Pugh, and P. M. Ajuh. 1995. Classical and novel approaches to the detection and localization of the numerous modified nucleotides in eukaryotic ribosomal RNA. Biochimie 77:22-29. [DOI] [PubMed] [Google Scholar]
- 25.Miller, M. C., and K. Collins. 2002. Telomerase recognizes its template by using an adjacent RNA motif. Proc. Natl. Acad. Sci. U. S. A. 99:6585-6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peculis, B. 1997. RNA processing: pocket guides to ribosomal RNA. Curr. Biol. 7:R480-R482. [DOI] [PubMed] [Google Scholar]
- 27.Qiao, F., and T. R. Cech. 2008. Triple-helix structure in telomerase RNA contributes to catalysis. Nat. Struct. Mol. Biol. 15:634-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saikia, M., Q. Dai, W. A. Decatur, M. J. Fournier, J. A. Piccirilli, and T. Pan. 2006. A systematic, ligation-based approach to study RNA modifications. RNA 12:2025-2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seto, A. G., K. Umansky, Y. Tzfati, A. J. Zaug, E. H. Blackburn, and T. R. Cech. 2003. A template-proximal RNA paired element contributes to Saccharomyces cerevisiae telomerase activity. RNA 9:1323-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seto, A. G., A. J. Zaug, S. G. Sobel, S. L. Wolin, and T. R. Cech. 1999. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature 401:177-180. [DOI] [PubMed] [Google Scholar]
- 31.Shefer, K., Y. Brown, V. Gorkovoy, T. Nussbaum, N. B. Ulyanov, and Y. Tzfati. 2007. A triple helix within a pseudoknot is a conserved and essential element of telomerase RNA. Mol. Cell. Biol. 27:2130-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singer, M. S., and D. E. Gottschling. 1994. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266:404-409. [DOI] [PubMed] [Google Scholar]
- 33.Smith, C. M., and J. A. Steitz. 1997. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell 89:669-672. [DOI] [PubMed] [Google Scholar]
- 34.Szostak, J. W., and E. H. Blackburn. 1982. Cloning yeast telomeres on linear plasmid vectors. Cell 29:245-255. [DOI] [PubMed] [Google Scholar]
- 35.Theimer, C. A., C. A. Blois, and J. Feigon. 2005. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol. Cell 17:671-682. [DOI] [PubMed] [Google Scholar]
- 36.Uesugi, S., H. Miki, M. Ikehara, H. Iwahashi, and Y. Kyogoku. 1979. A linear relationship between electronegativity of 2′-substituents and conformation of adenine nucleosides. Tetrahedron Lett. 20:4073. [Google Scholar]
- 37.Yu, Q., H. Kuzmiak, Y. Zou, L. Olsen, P. A. Defossez, and X. Bi. 2009. Saccharomyces cerevisiae linker histone Hho1p functionally interacts with core histone H4 and negatively regulates the establishment of transcriptionally silent chromatin. J. Biol. Chem. 284:740-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu, Y. T., R. M. Terns, and M. P. Terns. 2005. Mechanisms and functions of RNA-guided RNA modification, p. 223-262. In H. Grosjean (ed.), Fine-tuning of RNA functions by modification and editing, vol. 12. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 39.Zappulla, D. C., and T. R. Cech. 2004. Yeast telomerase RNA: a flexible scaffold for protein subunits. Proc. Natl. Acad. Sci. U. S. A. 101:10024-10029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao, X., Z. H. Li, R. M. Terns, M. P. Terns, and Y. T. Yu. 2002. An H/ACA guide RNA directs U2 pseudouridylation at two different sites in the branchpoint recognition region in Xenopus oocytes. RNA 8:1515-1525. [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao, X., and Y. T. Yu. 2004. Pseudouridines in and near the branch site recognition region of U2 snRNA are required for snRNP biogenesis and pre-mRNA splicing in Xenopus oocytes. RNA 10:681-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao, X., and Y. T. Yu. 2008. Targeted pre-mRNA modification for gene silencing and regulation. Nat. Methods 5:95-100. [DOI] [PubMed] [Google Scholar]