Abstract
MafA is a key transcriptional activator of islet β cells, and its exclusive expression within β cells of the developing and adult pancreas is distinct among pancreatic regulators. Region 3 (base pairs −8118 to −7750 relative to the transcription start site), one of six conserved 5′ cis domains of the MafA promoter, is capable of directing β-cell-line-selective expression. Transgenic reporters of region 3 alone (R3), sequences spanning regions 1 to 6 (R1-6; base pairs −10428 to +230), and R1-6 lacking R3 (R1-6ΔR3) were generated. Only the R1-6 transgene was active in MafA+ insulin+ cells during development and in adult cells. R1-6 also mediated glucose-induced MafA expression. Conversely, pancreatic expression was not observed with the R3 or R1-6ΔR3 line, although much of the nonpancreatic expression pattern was shared between the R1-6 and R1-6ΔR3 lines. Further support for the importance of R3 was also shown, as the islet regulators Nkx6.1 and Pax6, but not NeuroD1, activated MafA in gel shift, chromatin immunoprecipitation (ChIP), and transfection assays and in vivo mouse knockout models. Lastly, ChIP demonstrated that Pax6 and Pdx-1 also bound to R1 and R6, potentially functioning in pancreatic and nonpancreatic expression. These data highlight the nature of the cis- and trans-acting factors controlling the β-cell-specific expression of MafA.
MafA is an essential transcriptional activator in adult pancreatic islet β cells, due in part to regulating genes associated with cell identity, including insulin. MafA is produced only in insulin+ cells within the pancreas, with production first detected around embryonic day 13.5 (E13.5), during the secondary and principal wave of insulin+ cell production (33). This expression pattern is not novel just among islet-enriched transcription factors but even for insulin, as both are produced earlier, with transcription factors also more broadly expressed. Thus, insulin is expressed prior to E13.5 in a distinct population of cells which lack important regulatory molecules necessary for islet β-cell function (40, 41). Adult islet MafA levels appear to be a sensitive barometer of β-cell function, since many key metabolic and cellular effectors, such as glucose (20, 26, 53, 58), fatty acids (18), and insulin (52), greatly impact MafA expression.
The functions of islet-enriched transcription factors in pancreatic function and formation have been examined in detail by use of gene knockouts in mice. For example, global Pdx-1 null mice are apancreatic because of the role of Pdx-1 in early endocrine and exocrine progenitor development (24, 39), while later β-cell-specific removal results in cell dysfunction and diabetes (1, 10). In contrast, all other factors act later and more specifically, as exemplified by the reduction in distinct islet cell populations in Pax6−/− (i.e., α and β cells) (47, 51) and Nkx6.1−/− (β cells) (48) mice and the general loss of these populations in NeuroD1−/− mice (36).
MafA is an atypical islet-enriched transcription factor in not being essential to β-cell development, presumably due to compensation by the closely related MafB protein, which is silenced in β cells and exclusively expressed in α (glucagon hormone+) cells soon after birth (2, 3). However, MafA is critical to adult β-cell function, with defects in knockout mice affecting multiple gene products associated with cell maturation (e.g., glucose transport, insulin transcription, glucose sensing, and the insulin secretory machinery [2a, 55, 57]). These results are further supported by the observation that human embryonic stem cells differentiated to produce insulin and many islet-enriched transcription factors were neither glucose responsive nor capable of protecting against streptozotocin-induced hyperglycemia until they became MafA+ (7, 28).
The cis-acting sequences directing β-cell-enriched transcription have been defined most extensively for the insulin and Pdx-1 genes. Control is mediated by sequences that are well conserved between mammalian genes, residing approximately between bp −250 and +1 (relative to the transcription start site) in the insulin gene and between bp −2761 and −2457 (termed area I) and bp −2153 and −1923 (area II) in Pdx-1. Notably, while each of these regions has a β-cell-line-specific activation pattern in vitro, none independently recapitulates the expression pattern of the endogenous gene in transgenic assays in vivo. Thus, insulin-transgenic reporters driven by roughly 700 bp of the proximal promoter are expressed selectively in islet β cells, but also in the brain (12). The proper β-cell-specific pattern was observed with a transgene containing approximately 8.5 kb of the insulin promoter (19). Similarly, only an area I/area II-driven transgene reiterated the endogenous Pdx-1 expression pattern in developing and adult islet β cells (54). Early exocrine and endocrine Pdx-1 expression is mediated by sequences within areas I, II, and III, with area III (bp −1879 to −1600) binding to the PTF1a transcription factor, a factor essential for acinar and endocrine progenitor cell development, contributing to activation (56).
There are six areas of high sequence identity within 10 kbp of the mammalian MafA gene (termed regions 1 through 6 [R1 to R6]), but just R3 (bp −8118 to −7750) is able to direct β-cell-line-selective reporter transcription (44). R3 is also the only conserved sequence domain in the chicken MafA promoter, with an 88% level of identity to the human gene over the 370-bp control domain. Interestingly, this identity is much greater than that in other islet β-cell control regions, such as insulin (63% identity between human and mouse I or mouse II genes [21]) or Pdx-1 (78% identity between area II of the human and mouse genes [14]). We first sought to determine the significance of R3 in directing MafA expression to insulin+ cells in vivo. Here we demonstrate that a bp −10428- to +230-driven transgene spanning R1 through R6 (termed R1-6:eGFP) recapitulated the endogenous pancreatic MafA expression pattern in mice during development and in adults, but transgenes driven by R3 alone or R1-6 lacking R3 (R1-6ΔR3) did not. Interestingly, although the nonpancreatic expression pattern of MafA has not been analyzed in mammals, R1-6:eGFP and R1-6ΔR3:eGFP were expressed in many tissues in the chicken (e.g., eye, nervous system, and limbs [29]). In addition, islet R1-6:eGFP activity was stimulated by glucose, the most important effecter of β-cell function. The essential role of R3 in driving MafA expression in β cells was also highlighted by our ability to link Pax6 and Nkx6.1, but not NeuroD1, to control in biochemical and transfection-based assays. Consistent with a direct and significant role in endogenous gene transcription, MafA was not present in the remaining insulin+ cells of Pax6 and Nkx6.1 mutant mice. In addition to R3 binding, Pdx-1 and Pax6 factors associated with other conserved regions of the MafA promoter in scanning chromatin immunoprecipitation (ChIP) assays. Collectively, these results indicate that correct spatial and inducible regulation of MafA involves cross talk between trans-acting factors binding to multiple conserved cis elements located within distinct regions of the promoter, with R3 acting to direct expression specifically to the β cell.
MATERIALS AND METHODS
DNA sequence analysis.
Transcription factor binding sites within conserved chicken, human, mouse, and rat R3 sequences of MafA were found using the TRANSFAC (22) and rVISTA (32) bioinformatic programs.
Transfection constructs.
Noncomplementary mutations (G to T and C to A) were produced in the mouse R3:pTk chloramphenicol acetyltransferase (CAT) reporter (44) by use of a QuikChange mutagenesis kit (Stratagene) and the following oligonucleotides (the mutated bases are underlined): Pax6 mutant, TCTCTGCGCGTTTGAGTGACAGCGAACCCCAC (positions −7917 to −7886); NeuroD1 mutant, CTGGAGAGAAACGAGTTTCCCTCTCC (positions −8046 to −8021); and Nkx6.1 mutant, CGAGGGCTGATTTCCTTAGAAAGCACCCAT (positions −8112 to −8083). Restriction enzyme digestion and partial DNA sequencing were used to verify each construct.
Transient transfections.
Monolayer cultures of pancreatic islet β (βTC3) cells were maintained as described previously (14). The day before transfection, 106 cells were plated in 35-mm-diameter plates and then transfected with 1 μg of R3:pTk CAT and 1 μg of pRSV-LUC (2 μg in total), using Lipofectamine reagent (Invitrogen Life Technologies). Cellular extracts were collected at 40 to 48 h posttransfection, and luciferase (LUC; Promega, Madison, WI) and CAT (38) assays were performed. The pRSV-LUC activity was used to normalize the CAT activity from R3:pTk. Each experiment was performed on several separate occasions, with at least two independently prepared plasmids.
Electrophoretic mobility shift assays.
Nuclear extracts were prepared from INS-1 and βTC3 cells as previously described (49). Gel shift reactions were performed at 4°C (20-μl final volume), with roughly 10 μg of βTC3 or INS-1 nuclear extract and 400 fmol of 32P-radiolabeled (via polynucleotide kinase) probe. The binding buffer varied between transcription factors, as follows: for Nkx6.1, 10 mM HEPES (pH 7.9), 75 mM KCl, 2.5 mM MgCl2, 0.1 mM EDTA, 1 mM dithiothreitol (DTT), 3% Ficoll, 1 μg of poly(dI-dC), and 10 μg of bovine serum albumin; for NeuroD1, 25 mM HEPES (pH 7.9), 9% glycerol, 100 mM NaCl, 0.2 mM EDTA, 5 mM DTT, and 1 μg of poly(dI-dC); and for Pax6, 20 mM Tris (pH 8), 200 mM NaCl, 2 mM EDTA, 20% glycerol, 4 mM DTT, 1 μg of poly(dA-dT), and 1 μg of poly(dI-dC). Competition analysis was performed with a molar excess of unlabeled competitor to labeled probe (for the wild type, 5- to 50-fold excess; and for mutants, 25- to 50-fold excess). Antibody supershift analyses were performed by preincubating nuclear extract protein with Pax6 (Covance Research)-, NeuroD1 (Santa Cruz Biotechnology)-, or Nkx6.1 (Beta Cell Biology Consortium)-specific antibodies for 20 min at 4°C prior to addition of the radiolabeled probe. Samples were electrophoresed in 6% nondenaturing polyacrylamide gels at 150 V for 2 h in 1× TGE buffer (50 mM Tris, 380 mM glycine, 2 mM EDTA). Gels were then dried and visualized by autoradiography.
The following double-stranded mouse R3 oligonucleotides (the mutated bases are underlined) were used for binding reactions: for the NeuroD1 site (positions −8046 to −8021), CTGGAGAGAACAGATGTTCCCTCTCC; for the NeuroD1 site mutant, CTGGAGAGAAACGAGTTTCCCTCTCC; for Pax6-like site 1 (positions −7993 to −7962), CTCCACTCAGCCTTGTTTAGGAGAGAAAAGAG; for Pax6-like site 2 (positions −7917 to −7886), TCTCTGCGCGTTTGAGTGACGATAAACCCCAC; for mutant site 2, TCTCTGCGCGTTTGAGTGACAGCGAACCCCAC; for Pax6-like site 3 (positions −7849 to −7820), TATCATTTTATTGTCATATTTCACGGCCGT; for Pax6-like site 4 (positions −7822 to −7793), CGTAACGTTAATGGAAGATGCTTGCTGCAG; for the Nkx6.1 site (positions −8112 to −8083), CGAGGGCTGATTTAATTAGAAAGCACCCAT; for the Nkx6.1 site mutant, CGAGGGCTGATTTCCTTAGAAAGCACCCAT; for the rat insulin 2 E1 element, TCTGGCCATCTGCTGATCC (50); for the Pax6 consensus, AATTCGTGAACTCAAGCGTGAAAATGG (46); and for the Nkx6.1 consensus, GATCTGACCATTTAATTACCCTTCGTTGACAAGG (35).
ChIP assays.
βTC3 cells (∼0.5 × 108) were formaldehyde cross-linked and the sonicated chromatin-DNA complexes isolated as described previously (14). Anti-Pax6, anti-NeuroD1, anti-Pdx-1 (a gift from Chris Wright, Vanderbilt University), anti-Nkx6.1 (Beta Cell Biology Consortium), normal rabbit IgG (Santa Cruz Biotechnology), or no antibody was added to the sonicated chromatin, and the antibody-protein-DNA complexes were isolated by incubation with salmon sperm DNA-protein A-agarose (Upstate). PCR was performed on 1/10 of the purified anti-Pax6-, anti-NeuroD1-, or anti-Pdx-1-immunoprecipitated DNA, using either Ready-to-Go PCR beads (Amersham Pharmacia Biotech, Piscataway, NJ) or a Master Mix PCR system (Eppendorf) with 15 pmol of mouse primers. The following primer sets were used: for region 1, −9471 (5′-TGGTGGGCAGTTTATAGGGTCAGT-3′) and −9124 (5′-GCCACCTACAGCTCACACAAACTT-3′); for R3, −8120 (5′-CACCCCAGCGAGGGCTGATTTAATT-3′) and −7750 (5′-AGCAAGCACTTCAGTGTGCTCAGTG-3′); for R4 and R5 −6348 (5′-TGTCCATTCCCTGTTCCTCTCCCT-3′), −6041 (5′-TGTGTGGTAGTCAAGACAGGCCAA-3′), −4661 (5′-ACCTCTTGCCCTATGGCTGATGAT-3′), −4330 (5′-TGCACATTGATCTGGTGAGGTGGA-3′), −1873 (5′-TGCCCAGACATGTAGCTCATCCTT-3′), and −1607 (5′-TGGTAGCCACAGCCATCAGTGTAA-3′); for R6, −506 (5′-GAATTCCTGAACCCATCCCAACCA-3′) and −251 (5′-AGACCAAGTGGCAGATTCTGAGGT-3′); and for phosphoenolpyruvate carboxykinase (PEPCK), −434 (5′-GAGTGACACCTCACAGCTGTGG-3′) and −96 (5′-GGCAGGCCTTTGGATCATAGCC-3′). Cycling parameters were 1 cycle of 95°C for 2 min and 30 cycles of 95°C for 30 s, 59°C for 30 s, and 72°C for 30 s. The PCR products were resolved in a 1.4% agarose gel in Tris-acetate-EDTA (TAE) buffer and then visualized by ethidium bromide staining. Each experiment was repeated on at least three independent occasions.
MafA transgene construction and generation of transgenic mice.
Sequences spanning positions −8120 to −7750 (R3), −10428 to +230 (R1-6), and −10428 to +230 lacking positions −8120 to −7750 (R1-6ΔR3) were cloned into hsp6811eGFPpA, which contains a tripartite nuclear localization signal (NLS) and the enhanced green fluorescent protein (EGFP) gene cloned into hsp6811pA (58). The minimal heat shock promoter was removed by restriction digestion, and R1-6 was inserted directly upstream of the mouse β-globin intron by homologous recombination (30, 44). The recombination arms were made with the following PCR primers: for the 5′ arm, −10393 (5′-GGACCCCTCTGCCAGGCTTTGTCC-3′) and −10194 (5′-ATCTCAAAATCATCCATGGTGCCA-3′); and for the 3′ arm, +57 (5′-CGGGCGGGAGAGCCCGGAGCGCGG-3′) and +230 (5′-CTGTGCTCAGGGGACGCCGCCGGC-3′). R3 was cloned upstream of the minimal heat shock promoter in hsp6811eGFPpA by use of PCR primers −8120 (5′-CACCCCAGCGAGGGCTGATTTAATT-3′) and −7750 (5′-AGCAAGCACTTCAGTGTGCTCAGTG-3′). To create the R1-6ΔR3:eGFP construct, positions −10393 to −8820 were amplified by PCR and cloned upstream of positions −7749 to +120 in the hsp6811eGFPpA construct lacking the minimal heat shock promoter. Primers used to amplify positions −10393 to −8820 were as follows: −10393, 5′-GCGCGCACTAGTGGACCCCTCTGCCAGGCTTTGTCC-3′; and −8820, 5′-GCGCGCGAATTCTGGTCTTCAAGGCAGCCGCTGCTG-3′. R1-6:eGFP, R1-6ΔR3:eGFP, and R3:eGFP constructs were verified by restriction digestion and DNA sequencing. Transgenic screening was performed by PCR, using DNA (∼1 μg) obtained from either neonatal or adult mouse tails by use of a Puregene DNA purification kit (Gentra Systems, Minneapolis, MN). The PCRs (1 cycle of 95°C for 3 min; 25 cycles of 95°C for 30 s, 58°C for 30 s, and 68°C for 1 min; and 1 cycle of 68°C for 5 min) were performed with primers spanning the unique NLS and eGFP sequences in the transgene (forward, 5′-AAAGAATTCGCCCTTCCATGGTGC; and reverse, 5′-TGTAGTTGCCGTCGTCCTTGAAGA).
Tissue preparation.
Embryonic pancreases were isolated from transcription factor mutant mice (NeuroD1−/− mice [6, 36] at E15.5 and Pax6Sey/Sey mice [47] at P2), while embryonic (E12.5, E13.5, E15.5, and E18.5) and adult (P33) samples were collected from R1-6:eGFP, R1-6ΔR3:eGFP, and R3:eGFP transgenic mice. The day of vaginal plug discovery was designated stage E0.5. The transgenic and NeuroD1−/− embryonic tissues were fixed in 4% paraformaldehyde for 2 h at 4°C, washed with phosphate-buffered saline (PBS), dehydrated in increasing concentrations of ethanol, and embedded in paraffin. The newborn Pax6Sey/Sey tissue was fixed overnight in 4% paraformaldehyde at 4°C, immersed in 30% sucrose in PBS overnight at 4°C, and embedded in tissue freezing medium (Tissue-Tek; Triangle Biomedical Sciences) for cryoprotection. Whole-mount and bright-field images of the digestive organs from R1-6:eGFP, R1-6ΔR3:eGFP, and R3:eGFP mice were taken on a Stemi 2000-C microscope (Zeiss) with a Nikon 5700 digital camera.
Immunofluorescence.
Confocal image analyses were performed on paraffin and frozen sections as described previously (34, 42). Briefly, paraffin (6 μm) and frozen (10 μm) block sections were mounted on glass slides. The slides were incubated with primary antibodies overnight at 4°C (rabbit anti-MafA [1:1,000; Bethyl Laboratories, Montgomery, TX], chicken anti-GFP [1:5,000; Abcam, Cambridge, MA], and guinea pig anti-insulin [1:1,000; Linco Research Immunoassay, St. Charles, MO]) and then with Cy2-, Cy3-, or Cy5-conjugated donkey anti-guinea pig, anti-chicken, and anti-rabbit IgG secondary antibodies (1:500; Jackson ImmunoResearch, West Grove, PA). Fluorescent images were captured on either a Zeiss LSM 510 confocal microscope, using an optical depth of 1.5 μm, or a Zeiss Axioskop 2 microscope. Nuclear counterstaining was performed with YoPro1 (Molecular Probes, Eugene, OR). Adobe Photoshop, version 7.0 (Adobe Systems, Inc.), was used to process the images. GFP- and MafA-stained cells were counted for GFP+, GFP+ MafA+, and MafA+ cells in transgenic sections from embryonic (for E12.5, every 24 μm; for E13.5, every 72 μm; and for E15.5 and E18.5, every 96 μm) and adult (every 250 μm) tissues. The transgenic analysis was performed on a minimum of three independent animals from each line for each time point.
Islet isolation and culture.
Islets were isolated from R1-6:eGFP mice as previously described (5). Briefly, mice were sacrificed by cervical dislocation, and collagenase P (Roche) was then injected at 0.6 mg/ml into the pancreas via the bile duct and islets were incubated for 10 to 20 min. Islets were then handpicked five times under a dissecting microscope and washed three times in RPMI 1640 containing 5 mM glucose, 1% penicillin-streptomycin, and 10% fetal bovine serum. Islets were maintained overnight in RPMI containing 5 mM glucose at 37°C. After being cultured overnight, approximately 100 islets were incubated for 24 h at 37°C in RPMI 1640 containing 5 or 11 mM glucose. Total RNA was isolated from the glucose-treated R1-6:eGFP mouse islets (∼180 islets per condition) by use of an RNeasy kit (Qiagen, Valencia, CA) and then subjected to DNase I treatment (Versagene RNA DNase kit; Gentra Systems, Minneapolis, MN). Reverse transcription PCR reagents (TaqMan; Applied Biosystems, Foster City, CA) were used to generate cDNAs from DNase I-treated RNA.
Real-time PCR.
SYBR green master mix reagents (Applied Biosystems) were used in the PCR mixes for R1-6:eGFP islet and Nkx6.1 ChIP DNAs, and reactions were performed and analyzed using an Applied Biosystems Prism 7900 sequence detection system and software. The following oligonucleotide primers were used for detection: for insulin, 5′-CCACCCAGGCTTTTGTCAAA-3′ (5′) and 5′-CCCAGCTCCAGTTGTTCCAC-3′ (3′); for MafA, 5′-CCTGTAGAGGAAGCCGAGGAA-3′ (5′) and 5′-CCTCCCCCAGTCGAGTATAGC-3′ (3′); for eGFP, 5′-ATCGAGCTGAAGGGCATCGACTT-3′ (5′) and 5′-TGTCGGCCATGATATAGACGTTGTGG-3′ (3′); for γ-actin, 5′-GCACCCGGTGCTTCTGAC-3′ (5′) and 5′-CCAGATGCATACAAGGAC-3′ (3′); for GAPDH, 5′-AACTTTGGCATTGTGGAAGG-3′ (5′) and 5′-GGATGCAGGGATGATGTTCT-3′ (3′); for 18S rRNA, 5′-AGTCCCTGCCCTTTGTACACA-3′ (5′) and 5′-GATCCGAGGGCCTCACTAAAC-3′ (3′); for MafA bp −9433 to −9319 (R1), 5′-CTGTATTTGGCCCTCCTCTCCG-3′ (5′) and 5′-GCGGGCAAAGTCAGGACACA-3′ (3′); for MafA bp −7988 to −7875 (R3), 5′-CACTCAGCCTTGTTTAGGAGAGAAAAGA-3′ (5′) and 5′-CCGAGCTGCGTGGGGTTTAT-3′ (3′); for MafA bp −6282 to −6186 (R5), 5′-GCACCCAGCCTCACTCTAGGACTT-3′ (5′) and 5′-CTGCCAATGCCACCACAGGA-3′ (3′); for MafA bp −4633 to −4541, 5′-GCCCTATGGCTGATGATCCACA-3′ (5′) and 5′-TGAGGGTGTGCTCGCTTGGA-3′ (3′); for MafA bp −1678 to −1560, 5′-CCAGTTGCTTTTCACGGCCTC-3′ (5′) and 5′-CGGGGAGCCATTGGAATGTC-3′ (3′); and for MafA bp −346 to −241 (R6), 5′-GAGACACCGAAGGAGTACCCTGGA-3′ (5′) and 5′-AGAGGGCGCGAGACCAAGTG-3′ (3′). Islet RNA levels were calculated by the comparative ΔCT method, in which γ-actin, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and 18S rRNA were used for normalization (31). ChIP data are expressed as the percent recovery of coimmunoprecipitated DNA relative to input DNA (before immunoprecipitation).
RESULTS
R3 is necessary but not sufficient for β-cell-specific expression in vivo.
To determine the significance of R3 in MafA expression in vivo, transgenic mouse eGFP reporter lines were created using R3 alone (bp −8120 to −7750), R1-6 (bp −10428 to +230), and R1-6 lacking R3 (Fig. 1A to C). The expected β-cell-selective activity pattern was found for each of the MafA:eGFP transgenes in cell-line-based experiments (data not shown).
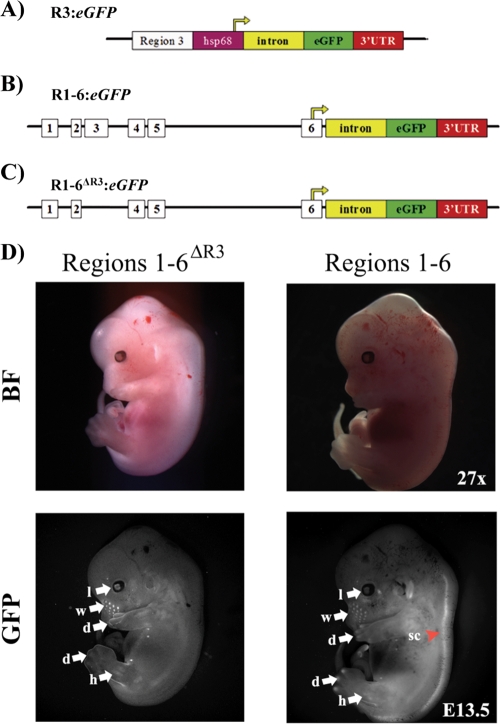
FIG. 1.
The MafA R1-6:eGFP and R1-6ΔR3:eGFP transgenes are expressed in similar tissue-specific manners. (A) Diagrams of the R3 (A), R1-6 (B), and R1-6ΔR3 (C) transgenic constructs. The start of transcription is from the hsp68 minimal heat shock promoter in R3:eGFP, while the endogenous start site in region 6 is utilized in the R1-6:eGFP and R1-6ΔR3:eGFP lines. Nuclear GFP was produced from all of the constructs (data not shown). (D) Whole-mount immunofluorescence of E13.5 embryos, performed with both R1-6:eGFP (E line is shown) and R1-6ΔR3:eGFP transgenic mouse lines. The expression patterns in these transgenic mouse lines were similar in most tissues, including the lens and limb buds (arrows), yet were absent in the spinal cord in R1-6ΔR3:eGFP mice (arrowhead). l, lens; d, digit tips; w, whisker hair shaft; h, hind limb; sc, spinal cord; BF, bright field; GFP, GFP fluorescence.
Whole-mount immunofluorescence was performed at E13.5 to determine the gross expression profiles of the R3:eGFP, R1-6:eGFP, and R1-6ΔR3:eGFP transgenes. Very similar expression patterns in the eyes, whiskers, and limbs were obtained between the R1-6:eGFP and R1-6ΔR3:eGFP lines (Fig. 1D), although expression was lost in the nervous system in R1-6ΔR3:eGFP mice. Such complementary results imply that this reflects endogenous MafA expression, which has not yet been determined for mammalian nonpancreatic tissues. Notably, the R1-6:eGFP and R1-6ΔR3:eGFP expression pattern corresponds well with that found for chicken MafA (29). In contrast, GFP fluorescence was not detected for the R3:eGFP lines (n = 3) in any organ at either embryonic or adult time points (data not shown).
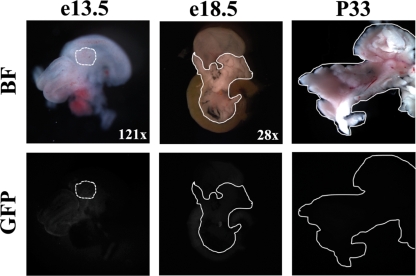
Light punctate staining in whole-mount images was first observed at E15.5 in R1-6:eGFP pancreases and became more pronounced by E18.5, as expected for a β-cell-expressed gene product (Fig. 2). Two R1-6:eGFP (labeled A and E) founder lines were established, and fluorescent nodes were also seen throughout the late developing and adult pancreases for both lines, with the E line having a stronger signal. In contrast to the case for R1-6:eGFP mice, GFP fluorescence was not observed in the adult pancreas for any of the R1-6ΔR3:eGFP lines (n = 3) (Fig. 3). These results demonstrate that R3 is essential for MafA expression in the pancreas, with other distinct sequences found between bp −10428 and +230 apparently necessary for expression in the pancreas and elsewhere.
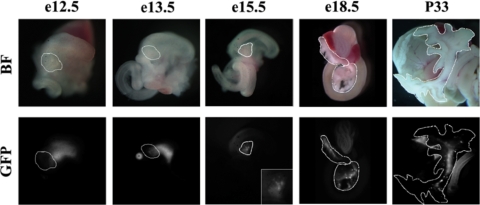
FIG. 2.
The MafA R1-6:eGFP transgene is active within the pancreas. Digestive organs, including the stomach, pancreas, spleen, and small intestine, were collected from the R1-6:eGFP A and E (shown) lines. The same developmental and adult GFP fluorescence patterns were obtained for both lines, with GFP not detected in the pancreatic buds until E15.5. The dorsal pancreatic bud is magnified (×4) in the E15.5 GFP inset. Pancreases are outlined in white. The punctate pattern in E15.5, E18.5, and P33 pancreases is characteristic of insulin+ cell expression. Low levels of EGFP were also detected in the E12.5 and E13.5 duodenum and stomach. BF, bright field; GFP, GFP fluorescence.
FIG. 3.
R3 is necessary for pancreatic transgenic expression. Gut tissues were collected from E13.5 and E18.5 embryos and P33 animals from the R-6ΔR3:eGFP line and examined via whole-mount immunofluorescence. No GFP expression was detected within the developing or adult pancreas, which is outlined in white. BF, bright field; GFP, GFP fluorescence.
Sequences spanning R1 to R6 recapitulate endogenous MafA expression in β cells.
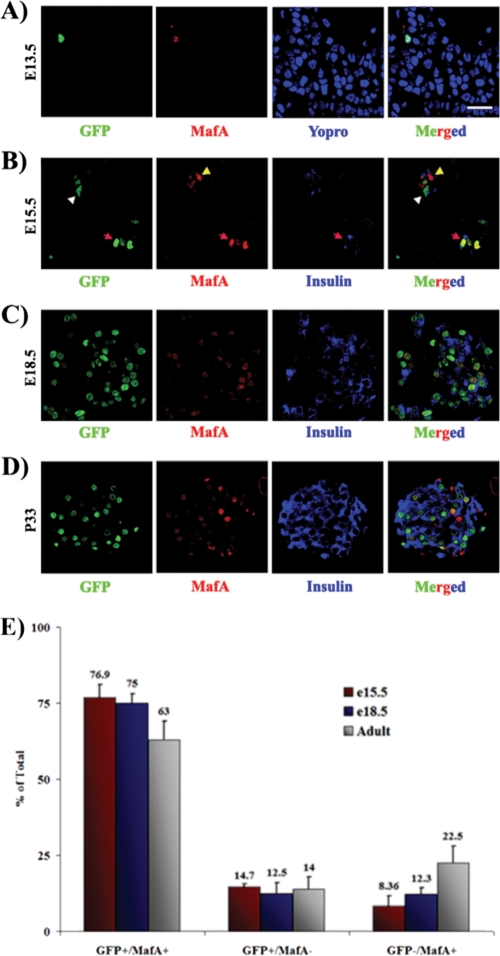
Pancreases were obtained from E12.5, E13.5, E15.5, E18.5, and P33 animals to precisely characterize the age-dependent expression pattern of the R1-6 transgene. GFP+ cells in the pancreatic epithelium were detected by antibody staining starting at E13.5, coincident with the onset of MafA expression (Fig. 4A). No GFP+ cells were detected at E12.5 (data not shown). The inability to detect GFP fluorescence in whole mounts presumably reflects the very few MafA+ cells normally produced at E13.5 (33). The GFP+ cell number increased during development, coinciding with the massive wave of β-cell formation during the secondary transition (Fig. 4B to D). Costaining of GFP with either insulin or MafA demonstrated that R1-6:eGFP activity was present in a majority of MafA+ cells throughout the lifetime of the animal and was localized exclusively to insulin+ cells (Fig. 4E) (33, 37). These results established that the correct temporal and spatial signals for MafA expression in the pancreas are contained within the roughly 10-kbp promoter region of the R1-6 transgene.
FIG. 4.
MafA R1-6 drives GFP transgene expression, predominantly in MafA+ cells, in the developing and adult pancreas. Sections from E13.5 (A), E15.5 (B), E18.5 (C), and P33 (D) mice were collected, and sections were stained for GFP (green), MafA (red), and/or insulin (blue). GFP was first detected in MafA+ cells at E13.5. GFP was usually coexpressed with MafA (red arrowheads), although the occasional single GFP+ (white arrowheads) or MafA+ (yellow arrowheads) cell was observed, as illustrated in panel B. These images are representative of both the R1-6:eGFP A and E lines. Nuclei in panel A were counterstained in blue. Bar, 20 μm. (E) The mean percentages ± standard deviations of pancreatic GFP+ MafA+, GFP+, and MafA+ cells to the total number of GFP+ and MafA+ cells at E15.5 (red bars), E18.5 (blue bars), and P33 (gray bars) are shown for both the A and E lines. Cells were counted from at least three independently isolated pancreases per time point.
MafA is regulated transcriptionally by glucose.
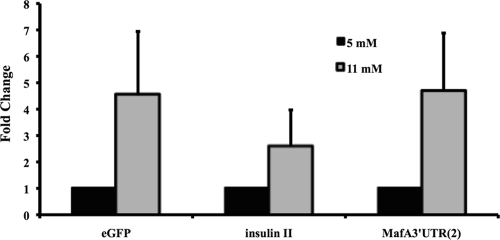
The most important regulator of insulin expression in islet β cells is glucose, which activates insulin gene transcription, protein synthesis, and secretion. Glucose-induced transcription is dependent upon MafA, in a process mediated by increased MafA mRNA and protein levels upon acute stimulation (20, 26, 53, 58). To determine if MafA transcription was also regulated by glucose, islets from 6-month-old R1-6:eGFP mice were cultured with stimulating (11 mM) and nonstimulating (5 mM) glucose concentrations for 24 h. Endogenous insulin mRNA levels were induced approximately 2.6-fold under these conditions, whereas MafA and eGFP reporter levels were both increased roughly 4.5-fold (Fig. 5). An identical regulatory pattern was obtained with 3-month-old R1-6:eGFP islets (data not shown). These collective results demonstrate that glucose signaling induces MafA transcription through factors binding within the 10 kb 5′ of the transcription start site.
FIG. 5.
MafA transcription is induced in response to stimulating glucose treatment. Mouse islets were collected from the R1-6:eGFP line and cultured at a high (11 mM) or low (5 mM) glucose concentration. Real-time PCR was performed with primers specific to eGFP coding sequences, insulin II coding sequences, or the MafA 3′-untranslated region (UTR). Normalized mRNA values ± standard deviations are presented as fold changes between low and high glucose stimulation.
Pax6, NeuroD1, and Nkx6.1 bind specifically to R3 sequences in vitro.
MafA is expressed exclusively in the wave of pancreatic insulin+ cells produced after E13.5 that eventually populate the mature islet (33). However, it is significant only to β-cell maturation and adult function, not to earlier cell development steps (2a, 57). We concluded from the R1-6:eGFP and R1-6ΔR3:eGFP transgenics and earlier MafA-driven cell line transfection studies (44) that R3 is the primary regulatory domain driving pancreas-specific expression. A TRANSFAC-based bioinformatic approach was previously used to identify potential R3 transcription regulators, with Pdx-1, FoxA2, and Nkx2.2 associated with activation (Fig. 6A) (44). Here we tested the role in R3 activity of Pax6, NeuroD1, and Nkx6.1, which are key regulators of β-cell development and function.
FIG. 6.
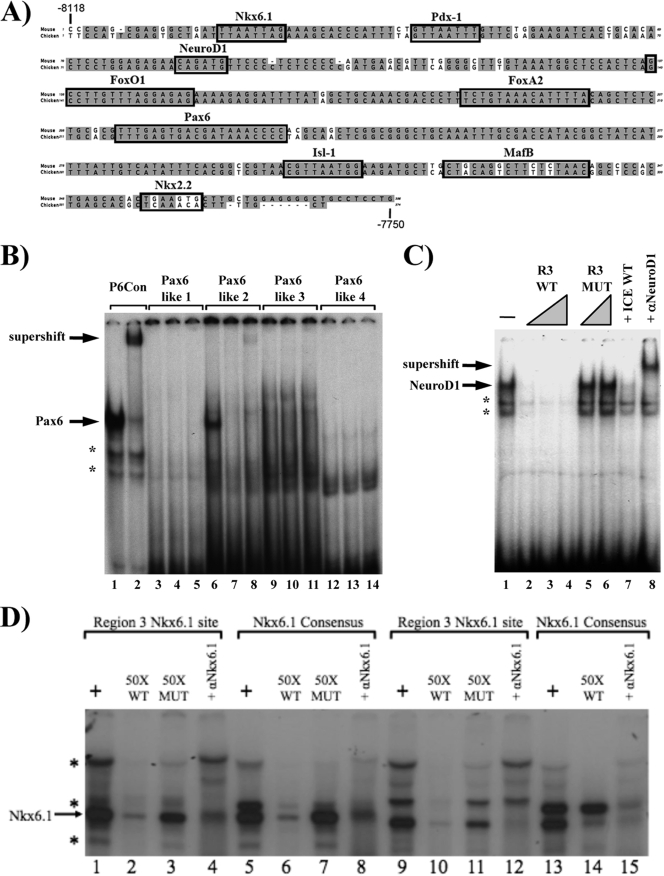
Identification of Pax6, NeuroD1, and Nkx6.1 binding sites within R3. (A) The gray highlighted sequences are identical between mouse and chicken R3 sequences. The locations of characterized and TRANSFAC-localized potential islet-enriched factor binding sites are superimposed on the R3 sequences. The Pdx-1, FoxA2, Nkx2.2, MafB, and Isl-1 binding sites (black boxes) have been described previously (2, 8, 27, 44). The Pax6, NeuroD1, and Nkx6.1 binding elements identified in the gel shift studies below are also denoted with black boxes. (B) Gel shifts were performed with probes corresponding to the consensus Pax6 site (lanes 1 and 2) and the four Pax6-like sites (lanes 3 to 14). The probes were incubated in the presence of βTC3 nuclear extract (alone; lanes 1, 3, 6, 9, and 12), and the specificity and composition of the βTC3 protein-DNA complexes were assayed with a 100-fold molar excess of the consensus Pax6 site (lanes 4, 7, 10, and 13) or by preincubation with an anti-Pax6 antibody (lanes 2, 5, 8, 11, and 14). (C) The βTC3 complexes formed with the NeuroD1-like site probe (bp −8046 to −8021; lane 1) were analyzed with a 10- to 50-fold molar excess of unlabeled wild-type competitor (R3 WT; lanes 2 to 4), a 25- to 50-fold molar excess of the R3 E-box mutant (R3 MT; lanes 5 and 6), or a 10-fold molar excess of the insulin NeuroD1 element (ICE WT; lane 7). Addition of anti-NeuroD1 antibody identified the specific location of the NeuroD1-containing binding complex (lane 8). (D) The Nkx6.1 consensus (lanes 5 to 8 and 13 to 15) and R3 Nkx6.1-like (lanes 1 to 4 and 9 to 12) sites were incubated in the presence of either βTC3 (lanes 1 to 8) or INS-1 (lanes 9 to 15) cell extracts. The Nkx6.1 complex was assessed with a 50-fold excess of either the wild-type R3 Nkx6.1 site (WT; lanes 2, 6, 10, and 14) or a probe for a R3 Nkx6.1 binding mutant (MUT; lanes 3, 7, and 11). The specificity of the Nkx6.1 complex was determined by incubation with an Nkx6.1 antibody (lanes 4, 8, 12, and 15). Asterisks denote nonspecific binding complexes in panels B to D.
R3 gel shift probes were made for each of the potential Pax6, NeuroD1, and Nkx6.1 binding sites (Fig. 6A). Only one of the four Pax6-like sites was found to bind to this factor in βTC3 nuclear extracts (bp −7917 to −7886), as demonstrated by both competition with a consensus Pax6 element and incubation with a Pax6 antibody (Fig. 6B). In addition, NeuroD1 was shown to interact with the basic helix-loop-helix (bHLH)-like site found within bp −8046 to −8021 by competition and antibody supershift analyses (Fig. 6C). Not only were competition experiments performed with wild-type and R3 mutant sites, but an identical pattern was obtained with the bHLH sites from the insulin and G6PC2 genes (data not shown). Several complexes were formed with the probe for bp −8112 to −8083 in R3, using βTC3 and INS-1 β-cell extracts (Fig. 6D), with binding of the principal specific complex disrupted by the Nkx6.1 antibody.
Pax6, NeuroD1, and Nkx6.1 bind to endogenous R3 sequences and function as positive regulators.
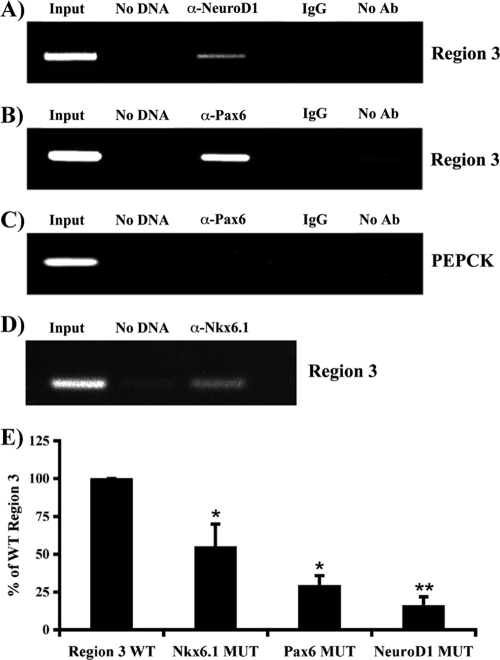
ChIP assays were used to determine whether Pax6, NeuroD1, and Nkx6.1 interactions with R3 could be observed in situ. R3 was selectively amplified by PCR from formaldehyde cross-linked β-cell chromatin precipitated with antibodies specific to NeuroD1, Pax6, and Nkx6.1 (Fig. 7) but not from chromatin treated with IgG or in the absence of antibody treatment. In contrast, no signal was detected with promoter sequences from the PEPCK gene, which is not transcribed in β cells. These results demonstrate that NeuroD1, Pax6, and Nkx6.1 bind within R3 in β cells.
FIG. 7.
Pax6, Nkx6.1, and NeuroD1 binding elements are necessary for R3 stimulation. Formaldehyde-cross-linked βTC3 chromatin was incubated with antibodies specific to NeuroD1 (A), Pax6 (B and C), and Nkx6.1 (D). Immunoprecipitated DNA was analyzed by PCR with primers specific to R3 (A, B, and D) or PEPCK (C). For controls, PCR was performed with total input DNA (1/100 dilution; input), no DNA, DNA immunoprecipitated with rabbit IgG (IgG), or DNA precipitated in the absence of antibody (no Ab). Only the anti-Pax6 results are shown for PEPCK, although the same pattern was seen for NeuroD1 and Nkx6.1. Each experiment was repeated with three separate chromatin preparations. (E) βTC3 cells were transfected with wild-type R3 (WT) and Pax6, NeuroD1, and Nkx6.1 binding-site mutants (MUT) of R3:pTk. Normalized region 3:pTk activity ± the standard error of the mean for each mutant is presented as a percentage of wild-type R3 activity. Wild-type and mutant R3:pTk constructs were active only in transfected β-cell lines, not in non-β cells (data not shown). Asterisks denote statistically significant differences between the mutant and wild-type activities, as assessed by Student's t test (*, P < 0.001; **, P < 0.005).
Element mutations were made in the mouse MafA region 3:pTk reporter to specifically disrupt Pax6, NeuroD1, or Nkx6.1 binding. The loss of binding led to a significant decrease in R3-driven reporter activity in transfected βTC3 cells (Fig. 7E). These results suggest that Pax6, NeuroD1, and Nkx6.1 directly activate MafA R3 in β cells.
Loss of only endogenous Pax6 and Nkx6.1 affects MafA expression in insulin+ cells in vivo.
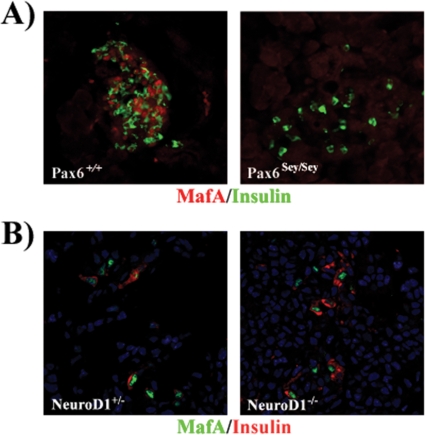
Pax6Sey/Sey mutant mice have a noticeable reduction in all islet cell types, but most significantly in glucagon- and insulin-producing cells (47). The Pax6 protein produced from the Pax6Sey allele lacks the homeo- and transactivation domains, effectively producing a null animal (15). Immunofluorescence staining revealed that MafA was not present in the insulin+ cells remaining at P2 in the Pax6Sey/Sey pancreas (Fig. 8A), although many other islet-enriched transcription factors were still present (e.g., Nkx2.2, Nkx6.1, and Pdx-1 [4, 51]). Similarly, MafA was not found in E18.5 insulin+ cells in the pancreases of Nkx6.1−/− mice (33), which specifically lack second-wave β cells (48).
FIG. 8.
MafA is not produced in insulin+ cells remaining in Pax6 mutant mice but is present in those in NeuroD1 mutant mice. MafA and insulin expression was examined by immunofluorescence analysis of pancreatic sections from P2 Pax6sey/sey (A) and E15.5 NeuroD1−/− (B) mice and either wild-type or NeuroD1+/− control mice. NeuroD1+/− and wild-type mice were phenotypically indistinguishable (36), enabling us to use NeuroD1+/− mice as the control in this analysis. MafA was undetectable in the Pax6 mutant and was present in NeuroD1−/− insulin-producing cells. Nuclei were counterstained in blue.
NeuroD1−/− mice are diabetic due to reduced β-cell numbers (36), although the severity is strain dependent (23). Interestingly, MafA was still detected in insulin+ cells of NeuroD1 mutants at E15.5 (Fig. 8B). Cumulatively, these results indicate that Pax6 and Nkx6.1, but not NeuroD1, are essential to MafA transcription in vivo.
Pax6 and Pdx-1 are also bound to non-R3 conserved sequences within the 10-kbp promoter region.
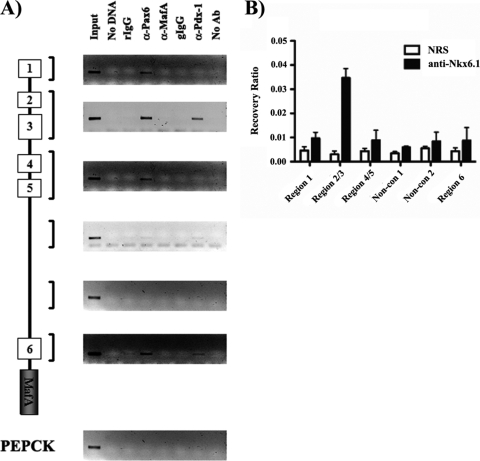
The R1-6:eGFP and R1-6ΔR3:eGFP transgenic experiments demonstrated that R3 was essential to β-cell-specific transcription in mice (Fig. 1 to 4). Moreover, MafA expression was severely and selectively compromised upon reducing the expression of various transcriptional effectors of β-cell development and function in vivo (Fig. 8) (44). These data raised the possibility that MafA expression was also mediated by the actions of such effectors on non-R3 promoter sequences. As a consequence, a scanning ChIP analysis was performed to assess Pdx-1, Pax6, and Nkx6.1 binding throughout the MafA promoter region. As expected, antibodies to Pdx-1, Nkx6.1, and Pax6 precipitated R3, not PEPCK, promoter sequences (Fig. 9). In addition, Pdx-1 binding was detected in R6, and Pax6 binding was detected in regions 1 and 6 (Fig. 9A). Taken together, these results imply that β-cell-specific transcription of the MafA gene in vivo is regulated principally by factors binding within R3, with further influence by (at least) Pax6 and Pdx-1 acting upon conserved cis-acting elements residing outside R3.
FIG. 9.
Nkx6.1 binds only within R3, whereas Pdx-1 and Pax6 bind to multiple conserved regions of the MafA promoter. Scanning ChIP analysis was performed on roughly 10 kb of MafA, using antibodies specific to Pdx-1, MafA, and Pax6 (A) and to Nkx6.1 (B). (A) Immunoprecipitated DNA was analyzed with MafA-specific (R1, R2 and R3, R4 and R5, for distinct nonconserved sequences 1 and 2 and R6) and PEPCK (control)-specific primers. The PCRs were performed with total input DNA (1:100 dilution), no DNA, nonspecific rabbit IgG (rIgG), anti-Pax6, anti-MafA, nonspecific goat IgG (gIgG), anti-Pdx-1, or no antibody (no Ab). (B) Real-time PCR was performed on Nkx6.1-immunoprecipitated DNA. DNA samples were incubated with either Nkx6.1 or control IgG (NRS), with the data presented as recovery of the indicated region (region 1, regions 2 and 3, regions 4 and 5, or region 6), as a percentage of the input level. Each experiment was repeated with at least three independently isolated chromatin preparations.
DISCUSSION
MafA is expressed late in pancreatic development and exclusively within insulin+ cells destined to populate the islet, a unique expression pattern in relation to all other islet transcription factors. Previously, MafA R3 was shown to be sufficient to direct β-cell-line-specific expression. Here we showed, upon analyzing the R1-6:eGFP, R1-6ΔR3:eGFP, and R3:eGFP transgenic constructs in mice, that R3 is essential for pancreatic MafA transcription. Hence, we found that only R1-6:eGFP recapitulated endogenous MafA expression in developing and adult pancreatic β cells, whose activity in islets was increased in response to glucose. Further support for a novel role for R3 in MafA expression came upon demonstrating that the β-cell developmental regulators Pax6, NeuroD1, and Nkx6.1 were associated with R3 activation in vitro and that MafA expression was lost in the insulin+ cell population remaining in Pax6 and Nkx6.1 mutant mice. The loss of MafA in these islet-enriched factor mutants is consistent with its late expression during β-cell formation and the absence of a developmental phenotype in MafA−/− mice. In addition, our ability to observe R3 regulators binding to other conserved promoter sequences indicates that functional interactions between factors acting in distinct cis-sequence domains are critical to β-cell expression.
Since R3 was the only mammalian conserved region capable of directing β-cell-line selectivity and the sole conserved MafA promoter control region in chickens (44), we tested if a transgene driven by R3 alone could direct expression in vivo. While the R3:eGFP lines were inactive in all tissues (data not shown), only Pdx-1 area II, not its area I and III control domains, independently directed transgenic reporter expression to a small fraction of Pdx-1+ β cells (11, 46). In a manner resembling that for MafA R1-6, the area I/II transgene recapitulated the comprehensive expression pattern of Pdx-1 in the secondary transition (E13.5 or later) (44), while area I/II/III lines were expressed in earlier pancreatic exocrine and endocrine Pdx-1+ progenitors (11). These results thus established that functional interactions between trans-acting factors of distinct control domains may be crucial in spatiotemporal gene expression within the pancreas. MafA R1-6 also mediated glucose-inducible expression (Fig. 5). In contrast, this critical metabolic effector of insulin transcription and β-cell function impacted Pdx-1 activation quite differently, by influencing nuclear localization (9, 43).
Interestingly, the R1-6:eGFP and R1-6ΔR3:eGFP transgenes retained expression in many extrapancreatic tissues at E13.5, yet R1-6ΔR3:eGFP was inactive in both the pancreas and the nervous system (Fig. 1 and 3). We presume that the sequences spanning R1 to R6 are critical for MafA transcription in these tissues, as the expression pattern for both transgenic lines was analogous to that described for MafA in chickens, the only species for which the distribution has been examined thoroughly (29). The apparent coordinating role for R3 in tissues of such distinct developmental origins as the pancreas and nervous system is not surprising, as many transcription factors have been shown to be shared between them (25). This includes Nkx6.1, Pax6, and NeuroD1, which were all found here to profoundly affect R3-mediated activation in vitro, with MafA lost in the insulin+ cells remaining in Nkx6.1 (33) and Pax6 mutant mice (Fig. 8).
In contrast to Nkx6.1, Pax6, and many other islet-enriched transcription factors linked to R3 control (e.g., Nkx2.2, Pdx-1, FoxO1, Isl-1, and MafB [2, 8, 27, 44]), MafA was still present in insulin+ cells of NeuroD1−/− mice (Fig. 8B). This result was unexpected, since NeuroD1 bound R3 endogenously and the binding-site mutant profoundly reduced transfected R3-driven activity (Fig. 7), although MafA mRNA levels were also recently found to be unaffected after conditional removal of NeuroD1 from islet β cells in vivo (17). Two interpretations of these results are (i) that NeuroD1 does not directly regulate MafA transcription and (ii) that a closely related bHLH family member acts in a compensatory manner. An example of the latter is the case of the MyoD and Myf5 bHLH proteins, which play functionally redundant roles in skeletal muscle formation. Hence, neither single mutant has a phenotype, while no skeletal muscle is formed in MyoD−/−; Myf5−/− mice (45). MafB is also thought to act in a functionally redundant manner relative to MafA during β-cell development (3). The presence of the closely related NeuroD2 protein in β cells suggests a compensatory role in NeuroD1−/− mice (13, 23), but studies on NeuroD1−/−; NeuroD2−/− mice need to be performed to test this hypothesis.
MafA is uniquely detected in the insulin+ cells produced during the secondary transition of pancreatic development, linking expression to β-cell maturation and function. The regulatory properties of R3 have been found to be fundamental to MafA transcription in the pancreas. This has been demonstrated using a variety of different transcriptional assays, and R3 appears to direct β-cell-specific MafA expression through coordinating actions with other conserved promoter regions. The activity of this transcription unit is exquisitely sensitive to the levels of glucose and many islet enriched-transcription factors, as, for example, expression was lost in the remaining insulin+ cells produced in all but NeuroD1 knockout mice (2, 8, 33, 44). It will be interesting to determine if R1-6 activity is also responsive to other extracellular signals that impact β cells, such as insulin (52) and BMP4 (16). Furthermore, since much of the conserved 370 bp of R3 is untested and few characterized islet-enriched transcription factors remain unassociated with control, an in-depth characterization may identify novel factors intimately involved in β-cell formation and function.
Acknowledgments
EL250 cells were kindly provided by D. Mortlock at Vanderbilt University Medical Center.
This work was supported by grants from the National Institutes of Health (DK50203 to R.S., DK60581 to R.G.M., VUMC9183 to B.S.-P., DK072504 to L.S., and DK083160 to C.S.H.), the American Lebanese Syrian Associated Charities (ALSAC) (to B.S.-P.), and the Vanderbilt Molecular Endocrinology Training Program (5T32 DK07563 to J.C.R.). The transgenic mice were developed with Vanderbilt Transgenic/ES shared resource grants CA68485 and DK20593. Partial support was also provided to the Molecular Biology Core Laboratory by the Vanderbilt University Diabetes Research and Training Center (Public Health Service grant P60 DK20593).
Footnotes
Published ahead of print on 28 June 2010.
REFERENCES
- 1.Ahlgren, U., J. Jonsson, L. Jonsson, K. Simu, and H. Edlund. 1998. Beta-cell-specific inactivation of the mouse Ipf1/Pdx1 gene results in loss of the beta-cell phenotype and maturity onset diabetes. Genes Dev. 12:1763-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Artner, I., B. Blanchi, J. C. Raum, M. Guo, T. Kaneko, S. Cordes, M. Sieweke, and R. Stein. 2007. MafB is required for islet beta cell maturation. Proc. Natl. Acad. Sci. U. S. A. 104:3853-3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2a.Artner, I., Y. Hang, M. Marzur, T. Yamamoto, M. Guo, J. Linder, M. Magnuson, and R. Stein. MafA and MafB regulate genes critical to β cells in a unique temporal manner. Diabetes, in press. [DOI] [PMC free article] [PubMed]
- 3.Artner, I., J. Le Lay, Y. Hang, L. Elghazi, J. C. Schisler, E. Henderson, B. Sosa-Pineda, and R. Stein. 2006. MafB: an activator of the glucagon gene expressed in developing islet alpha- and beta-cells. Diabetes 55:297-304. [DOI] [PubMed] [Google Scholar]
- 4.Ashery-Padan, R., X. Zhou, T. Marquardt, P. Herrera, L. Toube, A. Berry, and P. Gruss. 2004. Conditional inactivation of Pax6 in the pancreas causes early onset of diabetes. Dev. Biol. 269:479-488. [DOI] [PubMed] [Google Scholar]
- 5.Brissova, M., M. Fowler, P. Wiebe, A. Shostak, M. Shiota, A. Radhika, P. C. Lin, M. Gannon, and A. C. Powers. 2004. Intraislet endothelial cells contribute to revascularization of transplanted pancreatic islets. Diabetes 53:1318-1325. [DOI] [PubMed] [Google Scholar]
- 6.Chao, C. S., Z. L. Loomis, J. E. Lee, and L. Sussel. 2007. Genetic identification of a novel NeuroD1 function in the early differentiation of islet alpha, PP and epsilon cells. Dev. Biol. 312:523-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D'Amour, K. A., A. G. Bang, S. Eliazer, O. G. Kelly, A. D. Agulnick, N. G. Smart, M. A. Moorman, E. Kroon, M. K. Carpenter, and E. E. Baetge. 2006. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat. Biotechnol. 24:1392-1401. [DOI] [PubMed] [Google Scholar]
- 8.Du, A., C. S. Hunter, J. Murray, D. Noble, C. L. Cai, S. M. Evans, R. Stein, and C. L. May. 2009. Islet-1 is required for the maturation, proliferation, and survival of the endocrine pancreas. Diabetes 58:2059-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elrick, L. J., and K. Docherty. 2001. Phosphorylation-dependent nucleocytoplasmic shuttling of pancreatic duodenal homeobox-1. Diabetes 50:2244-2252. [DOI] [PubMed] [Google Scholar]
- 10.Gannon, M., E. T. Ables, L. Crawford, D. Lowe, M. F. Offield, M. A. Magnuson, and C. V. Wright. 2008. pdx-1 function is specifically required in embryonic beta cells to generate appropriate numbers of endocrine cell types and maintain glucose homeostasis. Dev. Biol. 314:406-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gannon, M., L. W. Gamer, and C. V. Wright. 2001. Regulatory regions driving developmental and tissue-specific expression of the essential pancreatic gene pdx1. Dev. Biol. 238:185-201. [DOI] [PubMed] [Google Scholar]
- 12.Gannon, M., C. Shiota, C. Postic, C. V. Wright, and M. Magnuson. 2000. Analysis of the Cre-mediated recombination driven by rat insulin promoter in embryonic and adult mouse pancreas. Genesis 26:139-142. [DOI] [PubMed] [Google Scholar]
- 13.Gasa, R., C. Mrejen, F. C. Lynn, P. Skewes-Cox, L. Sanchez, K. Y. Yang, C. H. Lin, R. Gomis, and M. S. German. 2008. Induction of pancreatic islet cell differentiation by the neurogenin-neuroD cascade. Differentiation 76:381-391. [DOI] [PubMed] [Google Scholar]
- 14.Gerrish, K., M. A. Cissell, and R. Stein. 2001. The role of hepatic nuclear factor 1 alpha and PDX-1 in transcriptional regulation of the pdx-1 gene. J. Biol. Chem. 276:47775-47784. [DOI] [PubMed] [Google Scholar]
- 15.Glaser, T., L. Jepeal, J. G. Edwards, S. R. Young, J. Favor, and R. L. Maas. 1994. PAX6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nat. Genet. 7:463-471. [DOI] [PubMed] [Google Scholar]
- 16.Goulley, J., U. Dahl, N. Baeza, Y. Mishina, and H. Edlund. 2007. BMP4-BMPR1A signaling in beta cells is required for and augments glucose-stimulated insulin secretion. Cell Metab. 5:207-219. [DOI] [PubMed] [Google Scholar]
- 17.Gu, C., G. H. Stein, N. Pan, S. Goebbels, H. Hornberg, K. A. Nave, P. Herrera, P. White, K. H. Kaestner, L. Sussel, and J. E. Lee. 2010. Pancreatic beta cells require NeuroD to achieve and maintain functional maturity. Cell Metab. 11:298-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagman, D. K., L. B. Hays, S. D. Parazzoli, and V. Poitout. 2005. Palmitate inhibits insulin gene expression by altering PDX-1 nuclear localization and reducing MafA expression in isolated rat islets of Langerhans. J. Biol. Chem. 280:32413-32418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hara, M., X. Wang, T. Kawamura, V. P. Bindokas, R. F. Dizon, S. Y. Alcoser, M. A. Magnuson, and G. I. Bell. 2003. Transgenic mice with green fluorescent protein-labeled pancreatic beta cells. Am. J. Physiol. Endocrinol. Metab. 284:E177-E183. [DOI] [PubMed] [Google Scholar]
- 20.Harmon, J. S., R. Stein, and R. P. Robertson. 2005. Oxidative stress-mediated, post-translational loss of MafA protein as a contributing mechanism to loss of insulin gene expression in glucotoxic beta cells. J. Biol. Chem. 280:11107-11113. [DOI] [PubMed] [Google Scholar]
- 21.Hay, C. W., and K. Docherty. 2006. Comparative analysis of insulin gene promoters: implications for diabetes research. Diabetes 55:3201-3213. [DOI] [PubMed] [Google Scholar]
- 22.Heinemeyer, T., X. Chen, H. Karas, A. E. Kel, O. V. Kel, I. Liebich, T. Meinhardt, I. Reuter, F. Schacherer, and E. Wingender. 1999. Expanding the TRANSFAC database towards an expert system of regulatory molecular mechanisms. Nucleic Acids Res. 27:318-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, H. P., K. Chu, E. Nemoz-Gaillard, D. Elberg, and M. J. Tsai. 2002. Neogenesis of beta-cells in adult BETA2/NeuroD-deficient mice. Mol. Endocrinol. 16:541-551. [DOI] [PubMed] [Google Scholar]
- 24.Jonsson, J., L. Carlsson, T. Edlund, and H. Edlund. 1994. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature 371:606-609. [DOI] [PubMed] [Google Scholar]
- 25.Jorgensen, M. C., J. Ahnfelt-Ronne, J. Hald, O. D. Madsen, P. Serup, and J. Hecksher-Sorensen. 2007. An illustrated review of early pancreas development in the mouse. Endocr. Rev. 28:685-705. [DOI] [PubMed] [Google Scholar]
- 26.Kataoka, K., S. I. Han, S. Shioda, M. Hirai, M. Nishizawa, and H. Handa. 2002. MafA is a glucose-regulated and pancreatic beta-cell-specific transcriptional activator for the insulin gene. J. Biol. Chem. 277:49903-49910. [DOI] [PubMed] [Google Scholar]
- 27.Kitamura, Y. I., T. Kitamura, J. P. Kruse, J. C. Raum, R. Stein, W. Gu, and D. Accili. 2005. FoxO1 protects against pancreatic beta cell failure through NeuroD and MafA induction. Cell Metab. 2:153-163. [DOI] [PubMed] [Google Scholar]
- 28.Kroon, E., L. A. Martinson, K. Kadoya, A. G. Bang, O. G. Kelly, S. Eliazer, H. Young, M. Richardson, N. G. Smart, J. Cunningham, A. D. Agulnick, K. A. D'Amour, M. K. Carpenter, and E. E. Baetge. 2008. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat. Biotechnol. 26:443-452. [DOI] [PubMed] [Google Scholar]
- 29.Lecoin, L., K. Sii-Felice, C. Pouponnot, A. Eychene, and M. P. Felder-Schmittbuhl. 2004. Comparison of maf gene expression patterns during chick embryo development. Gene Expr. Patterns 4:35-46. [DOI] [PubMed] [Google Scholar]
- 30.Lee, E. C., D. Yu, J. Martinez de Velasco, L. Tessarollo, D. A. Swing, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73:56-65. [DOI] [PubMed] [Google Scholar]
- 31.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 32.Loots, G. G., I. Ovcharenko, L. Pachter, I. Dubchak, and E. M. Rubin. 2002. rVista for comparative sequence-based discovery of functional transcription factor binding sites. Genome Res. 12:832-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuoka, T. A., I. Artner, E. Henderson, A. Means, M. Sander, and R. Stein. 2004. The MafA transcription factor appears to be responsible for tissue-specific expression of insulin. Proc. Natl. Acad. Sci. U. S. A. 101:2930-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsuoka, T. A., L. Zhao, I. Artner, H. W. Jarrett, D. Friedman, A. Means, and R. Stein. 2003. Members of the large Maf transcription family regulate insulin gene transcription in islet beta cells. Mol. Cell. Biol. 23:6049-6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mirmira, R. G., H. Watada, and M. S. German. 2000. Beta-cell differentiation factor Nkx6.1 contains distinct DNA binding interference and transcriptional repression domains. J. Biol. Chem. 275:14743-14751. [DOI] [PubMed] [Google Scholar]
- 36.Naya, F. J., H. P. Huang, Y. Qiu, H. Mutoh, F. J. DeMayo, A. B. Leiter, and M. J. Tsai. 1997. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/neuroD-deficient mice. Genes Dev. 11:2323-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishimura, W., T. Kondo, T. Salameh, I. El Khattabi, R. Dodge, S. Bonner-Weir, and A. Sharma. 2006. A switch from MafB to MafA expression accompanies differentiation to pancreatic beta-cells. Dev. Biol. 293:526-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nordeen, S. K., P. P. Green III, and D. M. Fowlkes. 1987. A rapid, sensitive, and inexpensive assay for chloramphenicol acetyltransferase. DNA 6:173-178. [DOI] [PubMed] [Google Scholar]
- 39.Offield, M. F., T. L. Jetton, P. A. Labosky, M. Ray, R. W. Stein, M. A. Magnuson, B. L. Hogan, and C. V. Wright. 1996. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development 122:983-995. [DOI] [PubMed] [Google Scholar]
- 40.Oster, A., J. Jensen, P. Serup, P. Galante, O. D. Madsen, and L. I. Larsson. 1998. Rat endocrine pancreatic development in relation to two homeobox gene products (Pdx-1 and Nkx 6.1). J. Histochem. Cytochem. 46:707-715. [DOI] [PubMed] [Google Scholar]
- 41.Pang, K., C. Mukonoweshuro, and G. G. Wong. 1994. Beta cells arise from glucose transporter type 2 (Glut2)-expressing epithelial cells of the developing rat pancreas. Proc. Natl. Acad. Sci. U. S. A. 91:9559-9563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prado, C. L., A. E. Pugh-Bernard, L. Elghazi, B. Sosa-Pineda, and L. Sussel. 2004. Ghrelin cells replace insulin-producing beta cells in two mouse models of pancreas development. Proc. Natl. Acad. Sci. U. S. A. 101:2924-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rafiq, I., H. J. Kennedy, and G. A. Rutter. 1998. Glucose-dependent translocation of insulin promoter factor-1 (IPF-1) between the nuclear periphery and the nucleoplasm of single MIN6 beta-cells. J. Biol. Chem. 273:23241-23247. [DOI] [PubMed] [Google Scholar]
- 44.Raum, J. C., K. Gerrish, I. Artner, E. Henderson, M. Guo, L. Sussel, J. C. Schisler, C. B. Newgard, and R. Stein. 2006. FoxA2, Nkx2.2, and PDX-1 regulate islet beta-cell-specific mafA expression through conserved sequences located between base pairs −8118 and −7750 upstream from the transcription start site. Mol. Cell. Biol. 26:5735-5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rudnicki, M. A., P. N. Schnegelsberg, R. H. Stead, T. Braun, H. H. Arnold, and R. Jaenisch. 1993. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell 75:1351-1359. [DOI] [PubMed] [Google Scholar]
- 46.Samaras, S. E., M. A. Cissell, K. Gerrish, C. V. Wright, M. Gannon, and R. Stein. 2002. Conserved sequences in a tissue-specific regulatory region of the pdx-1 gene mediate transcription in pancreatic beta cells: role for hepatocyte nuclear factor 3 beta and Pax6. Mol. Cell. Biol. 22:4702-4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sander, M., A. Neubuser, J. Kalamaras, H. C. Ee, G. R. Martin, and M. S. German. 1997. Genetic analysis reveals that PAX6 is required for normal transcription of pancreatic hormone genes and islet development. Genes Dev. 11:1662-1673. [DOI] [PubMed] [Google Scholar]
- 48.Sander, M., L. Sussel, J. Conners, D. Scheel, J. Kalamaras, F. Dela Cruz, V. Schwitzgebel, A. Hayes-Jordan, and M. German. 2000. Homeobox gene Nkx6.1 lies downstream of Nkx2.2 in the major pathway of beta-cell formation in the pancreas. Development 127:5533-5540. [DOI] [PubMed] [Google Scholar]
- 49.Schreiber, E., P. Matthias, M. M. Muller, and W. Schaffner. 1989. Rapid detection of octamer binding proteins with ‘mini-extracts,’ prepared from a small number of cells. Nucleic Acids Res. 17:6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sharma, A., and R. Stein. 1994. Glucose-induced transcription of the insulin gene is mediated by factors required for beta-cell-type-specific expression. Mol. Cell. Biol. 14:871-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.St.-Onge, L., B. Sosa-Pineda, K. Chowdhury, A. Mansouri, and P. Gruss. 1997. Pax6 is required for differentiation of glucagon-producing alpha-cells in mouse pancreas. Nature 387:406-409. [DOI] [PubMed] [Google Scholar]
- 52.Ueki, K., T. Okada, J. Hu, C. W. Liew, A. Assmann, G. M. Dahlgren, J. L. Peters, J. G. Shackman, M. Zhang, I. Artner, L. S. Satin, R. Stein, M. Holzenberger, R. T. Kennedy, C. R. Kahn, and R. N. Kulkarni. 2006. Total insulin and IGF-I resistance in pancreatic beta cells causes overt diabetes. Nat. Genet. 38:583-588. [DOI] [PubMed] [Google Scholar]
- 53.Vanderford, N. L., S. S. Andrali, and S. Ozcan. 2007. Glucose induces MafA expression in pancreatic beta cell lines via the hexosamine biosynthetic pathway. J. Biol. Chem. 282:1577-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Velkinburgh, J. C., S. E. Samaras, K. Gerrish, I. Artner, and R. Stein. 2005. Interactions between areas I and II direct pdx-1 expression specifically to islet cell types of the mature and developing pancreas. J. Biol. Chem. 280:38438-38444. [DOI] [PubMed] [Google Scholar]
- 55.Wang, H., T. Brun, K. Kataoka, A. J. Sharma, and C. B. Wollheim. 2007. MAFA controls genes implicated in insulin biosynthesis and secretion. Diabetologia 50:348-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wiebe, P. O., J. D. Kormish, V. T. Roper, Y. Fujitani, N. I. Alston, K. S. Zaret, C. V. Wright, R. W. Stein, and M. Gannon. 2007. Ptf1a binds to and activates area III, a highly conserved region of the Pdx1 promoter that mediates early pancreas-wide Pdx1 expression. Mol. Cell. Biol. 27:4093-4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, C., T. Moriguchi, M. Kajihara, R. Esaki, A. Harada, H. Shimohata, H. Oishi, M. Hamada, N. Morito, K. Hasegawa, T. Kudo, J. D. Engel, M. Yamamoto, and S. Takahashi. 2005. MafA is a key regulator of glucose-stimulated insulin secretion. Mol. Cell. Biol. 25:4969-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao, L., M. Guo, T. A. Matsuoka, D. K. Hagman, S. D. Parazzoli, V. Poitout, and R. Stein. 2005. The islet beta cell-enriched MafA activator is a key regulator of insulin gene transcription. J. Biol. Chem. 280:11887-11894. [DOI] [PubMed] [Google Scholar]