Abstract
During DNA polymerase switching, the Xenopus laevis Cip/Kip-type cyclin-dependent kinase inhibitor Xic1 associates with trimeric proliferating cell nuclear antigen (PCNA) and is recruited to chromatin, where it is ubiquitinated and degraded. In this study, we show that the predominant E3 for Xic1 in the egg is the Cul4-DDB1-XCdt2 (Xenopus Cdt2) (CRL4Cdt2) ubiquitin ligase. The addition of full-length XCdt2 to the Xenopus extract promotes Xic1 turnover, while the N-terminal domain of XCdt2 (residues 1 to 400) cannot promote Xic1 turnover, despite its ability to bind both Xic1 and DDB1. Further analysis demonstrated that XCdt2 binds directly to PCNA through its C-terminal domain (residues 401 to 710), indicating that this interaction is important for promoting Xic1 turnover. We also identify the cis-acting sequences required for Xic1 binding to Cdt2. Xic1 binds to Cdt2 through two domains (residues 161 to 170 and 179 to 190) directly flanking the Xic1 PCNA binding domain (PIP box) but does not require PIP box sequences (residues 171 to 178). Similarly, human p21 binds to human Cdt2 through residues 156 to 161, adjacent to the p21 PIP box. In addition, we identify five lysine residues (K180, K182, K183, K188, and K193) immediately downstream of the Xic1 PIP box and within the second Cdt2 binding domain as critical sites for Xic1 ubiquitination. Our studies suggest a model in which both the CRL4Cdt2 E3- and PIP box-containing substrates, like Xic1, are recruited to chromatin through independent direct associations with PCNA.
The eukaryotic cell cycle is positively regulated by cyclin-dependent kinases (CDKs) and negatively regulated by CDK inhibitors (CKIs) (22, 25, 27, 28). A complete knockout of all CDK inhibitor function, although as yet not attained in mammalian cells, has been accomplished in Saccharomyces cerevisiae and is shown to result in genomic instability due to premature entry into S phase (19). Conversely, the overexpression of cyclin E in mammalian cells has also been observed to induce chromosome instability (31). These studies suggest that CDK inhibitor function can play a critical role in maintaining genomic stability through the proper regulation of DNA replication initiation. Mammalian Cip/Kip-type CDK inhibitors p27 and p21 are stoichiometric inhibitors of CDK2-cyclins that regulate the entry into S phase and are targeted by ubiquitin- and proteasome-dependent proteolysis during the G1-to-S-phase transition (4, 5, 33, 35). In the frog, Xenopus laevis, three types of CDK inhibitors have been identified that share sequence and functional similarities with mammalian p27 and p21. The first type of CDK inhibitor includes the Xenopus inhibitor of CDK (p27Xic1 or Xic1) and kinase inhibitor from Xenopus (p28Kix1 or Kix1), which share ∼90% amino acid sequence identity with each other, preferentially inhibit the activity of CDK2-cyclin E or A and bind all CDK-cyclins and proliferating cell nuclear antigen (PCNAs) (30, 32). The second and third types of Xenopus CDK inhibitors are p16Xic2 and p17Xic3, which share sequence homology with p21 and p27, respectively, and exhibit restricted developmental expression but have not been extensively characterized biochemically (9).
In an effort to study the molecular mechanism of Cip/Kip-type CDK inhibitor proteolysis in the context of the temporal events of DNA replication initiation, we utilize the biochemically tractable Xenopus egg extract system. This extract can recapitulate all of the events of semiconservative DNA replication and fully support protein ubiquitination and degradation in the context of DNA replication initiation (3, 36). Using this system, we have shown that during DNA polymerase switching, Xic1 is recruited to sites of DNA replication initiation through its association with proliferating cell nuclear antigen (PCNA) and is targeted for ubiquitination and degradation (6). Using a strategy of PCNA reconstitution to PCNA-depleted extracts, our studies showed that Xic1 ubiquitination and turnover required not only PCNA binding but also the ability of PCNA to be loaded at a site of DNA replication initiation by replication factor C (RFC) (6). Our previous study indicated that like mammalian p27 and p21, Xic1 could be ubiquitinated in vitro by SCFXSkp2 (21), but our subsequent studies suggested that Xenopus Skp2 (XSkp2) levels were very low in the early embryo, and XSkp2 immunodepletion did not stabilize Xic1 in the Xenopus egg extract (our unpublished observations). Therefore, we postulated that in the interphase egg extract, Xic1 was targeted for ubiquitination by an alternate ubiquitin ligase.
In this study, we identify Cul4-DDB1-XCdt2 (CRL4Cdt2) as the ubiquitin ligase for Xic1 in the egg. We also identify both the critical residues of Xic1 required for association to Cdt2 and the critical lysine residues of Xic1 ubiquitinated by CRL4Cdt2. Importantly, we report a direct interaction between the C-terminal domain of Cdt2 and PCNA and show that the C-terminal domain of Cdt2 is required to promote the proteolysis of Xic1. Our studies suggest a model for Xic1 ubiquitination and proteolysis which requires the Xic1 PIP box for association with PCNA and Xic1 chromatin recruitment, the Xic1 sequences flanking the PIP box for association with Cdt2, specific lysine residues within the Cdt2 binding domain of Xic1 for efficient Xic1 ubiquitination, and a direct association between the Cdt2 C terminus and PCNA.
MATERIALS AND METHODS
Cloning of Xenopus Cdt2.
To identify the putative full-length cDNA sequence for Xenopus Cdt2, a BLAST search of the National Center for Biotechnology Information (NCBI) database was performed using the human Cdt2 (hCdt2) amino acid sequence (gi 7012714). This resulted in the identification of an mRNA sequence (gi 147904833) encoding the Xenopus laevis hypothetical protein MGC114697, which exhibited an overall 55% amino acid identity and 65% amino acid similarity to the human Cdt2 protein. The entire open reading frame encoding amino acids 1 to 710 was then PCR amplified from a Xenopus cDNA library (embryonic stage 11.5) using Pfu DNA polymerase and primers 5′-ATAAGCTTGGCCGGCCACCATGTTGTTTCGCTCTGTGATG and 5′-CCCTCGAGGGCGCGCCTTACTCAGACTTCTTGAAAAAGTAGGTGC. The PCR product was subcloned into the FseI/AscI sites of the pCS2+FA vector (kindly provided by Ethan Lee, Vanderbilt University) and was verified by DNA sequencing.
Generation of the XCdt2, Xic1, and p21 mutants.
XCdt2 deletion mutants XCdt21-400 (residues 1 to 400 of XCdt2) and XCdt2401-710 were generated by PCR mutagenesis using pCS2+/XCdt2 as the template plasmid, followed by subcloning into pCS2+FA and pGEX-FA. For the generation of Xic1 and p21 deletion mutants, pCS2+/Xic1, pCS2+FA/XCdt2, pCS2+/p21, and pGEM2/p21Δ156-161 were used as the template for PCR, and the products were cloned into the BamHI/EcoRI or BamHI/XhoI sites of pCS2+ and pGEX-4T. The following point mutants Xic1 (K2R, K2R-2, K5R, K6R, K8R, K11R, and K13R), XCdt2R247A, and p21F150A were generated using the pCS2+/wild-type Xic1 (Xic1-WT) and pCS2+/Xic1-NLS2 (also called Xic1-K3R), pCS2+FA/XCdt2, or pCS2+/p21 plasmids, respectively, and the QuikChange site-directed mutagenesis kit (Stratagene). Xic1WT-NPIP1, Xic1I174A-NPIP1, and Xic1N160-NPIP1 were generated by PCR mutagenesis, using pCS2+/Xic1 as the template plasmid, followed by subcloning into the BamHI/EcoRI sites of pCS2+ and pGEX-4T. For the generation of Xic1WT-NPIP2, Xic1I174A-NPIP2, and Xic1N160-NPIP2, pGEM2/p21Δ156-161 was used as the template for PCR, and the products were cloned into the BamHI/EcoRI sites of pCS2+ and pGEX-4T. Subsequently, the PCR products of Xic1WT(2-210), Xic1I174A(2-210), and Xic1N160(2-160) were cloned into the EcoRI and XhoI sites of pCS2+/p21135-164(Δ156-161) or pGEX-4T/p21135-164(Δ156-161). All mutations were verified by DNA sequencing. The primer sequences are available upon request.
Preparation of Xenopus interphase egg extracts and demembranated sperm chromatin.
Xenopus interphase egg extracts (low-speed supernatant [LSS]), membrane-free high-speed supernatant (HSS), and demembranated Xenopus sperm chromatin (XSC) were prepared as described previously (6, 7, 21).
Analysis of proteins by mass spectrometry.
The GST-Xic1 fusion proteins were expressed in BL21(pLysS)DE3, as described previously (21). The GST and GST-Xic1 proteins (5 μg) were coupled to glutathione-Sepharose 4B (GE Healthcare) and incubated in 250 μl of LSS for 1 h at 4°C. The beads were washed extensively with NETN buffer (50 mM Tris at pH 8, 250 mM NaCl, 5 mM EDTA, and 0.5% Nonidet P-40). The GST or GST-Xic1 binding proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the corresponding acrylamide gel lanes were sliced into 8 pieces and digested with 10 ng/μl trypsin (Promega) at 37°C for 18 h. The peptides were extracted in 5% formic acid and 50% acetonitrile and dried in a SpeedVac (Savant). The tryptic peptides were separated by UltiMate Nano liquid chromatography (LC) systems (LC Packings) and sequenced using a QStar mass spectrometer (Applied Biosystems) as described previously (23). Acquired mass data were searched against the NCBI protein database using the MASCOT software package (Matrix Sciences). Only sequences exhibiting P values of <0.05 were considered. Notably, XCdt2 was identified by mass spectrometry as a Xic1-interacting protein, but the P value was above the 0.05 cutoff point. This may be due to a difficulty in eluting the Cdt2 peptides from the gel, a failure to recover negatively charged peptides of Cdt2, or posttranslational modification of Cdt2, resulting in phosphorylated Cdt2-derived peptides.
In vitro transcription and translation and antibody preparation.
Wild-type and mutant Xic1 and XCdt2 were in vitro transcribed and translated from the SP6 promoter in pCS2+ using the TNT-coupled reticulocyte lysate system (Promega) and [35S]methionine (PerkinElmer Life Sciences). The generation of antibodies to XPCNA, XCdt2, and XDDB1 was described previously (2, 6, 15).
Xic1 degradation assay and p21 ubiquitination assay.
Xic1 degradation assays were conducted as described previously (6, 7). To examine the ability of exogenously added XCdt2 to promote Xic1 degradation, 2.4 μl in vitro-translated XCdt2WT, XCdt2R247A, XCdt21-400, XCdt2401-710, unprogrammed rabbit reticulocyte lysate (RRL), or Xenopus extracts supplemented with buffer (XB−; 100 mM KCl, 0.1 mM CaCl2, 1 mM MgCl2, 10 mM HEPES at pH 7.7) was added to HSS (12 μl). The samples were incubated with a 0.1 volume of [35S]methionine-labeled Xic1 in the presence or absence of 15 ng/μl ΦX174 single-stranded DNA (New England BioLabs) for 0, 45, 90, 135, or 180 min at 23°C. The samples were then analyzed by SDS-PAGE, followed by PhosphorImager analysis (Molecular Dynamics), and quantitation was performed using ImageQuant software (GE Healthcare). In general, the addition of RRL alone inhibited Xic1 turnover by between ∼20 to 25% compared to that of XB−. p21 ubiquitination assays were performed as described above for the Xic1 degradation assay, except that in vitro-translated human Cdt2 (hCdt2) was added. We performed a statistical analysis of the percentage of Xic1 remaining at the 1.5- and 3-h time points (LSS) or at the 1- and 2-h time points (HSS) for the Xic1-K5R mutant compared to that of the Xic1 wild type (WT), K6R, K8R, or K13R using a dependent sample or “paired” t test analysis, where the P values for the two time points were averaged.
Xic1 ubiquitination assay.
In vitro-translated [35S]methionine- and [35S]cysteine-labeled wild-type Xic1 and mutants (K2R, K3R, K5R, K6R, K8R, and K13R) (0.5 μl) were incubated in 3 μl HSS in the presence of 15 ng/μl ΦX174 single-stranded DNA (New England BioLabs) and 3 μg/μl methyl-ubiquitin (Boston Biochem, Inc.) for 120 min at 23°C, followed by SDS-PAGE (4 to 20% precise protein gel; Pierce) and PhosphorImager analysis.
Recombinant protein expression and purification.
The MBP-Xic1 fusion proteins and His6-XPCNA cloned into pET28a were expressed in BL21(pLysS)DE3 and purified with amylose agarose (NEB) or nickel-nitrilotriacetic acid-Sepharose (Qiagen), respectively (6). Recombinant proteins were dialyzed into buffer containing 50 mM Tris, 150 mM NaCl, and 5 mM EDTA (pH 7.5) and concentrated before use.
In vitro binding assays.
For glutathione S-transferase (GST) pulldown assays, GST-Xic1, GST-hp21, and GST-hp27 fusion proteins (5 μg) were bound to glutathione-Sepharose 4B (GE Healthcare) and incubated with [35S]methionine-labeled XCdt2 (4 μl) for 1.5 h at 23°C. For GST-XCdt21-400, GST-XCdt2401-710, and GST-XPCNA, beads were incubated with purified MBP-Xic1 or XPCNA (5, 15, 25, or 50 μg). The beads were washed with NETN buffer (50 mM Tris, 250 mM NaCl, 5 mM EDTA at pH 7.5, and 0.1% NP-40) and subjected to SDS-PAGE and phosphorimager analysis or Coomassie blue staining. Protein bands from Coomassie blue staining were analyzed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (Voyager-DE Pro; Applied Biosystems) as described previously (18). To examine the binding between p21 and XPCNA, 2.5 μg GST or GST-p21 wild-type or mutant proteins immobilized on glutathione-Sepharose beads was incubated with 10 μl of HSS in NETN buffer, followed by immunoblotting with anti-hPCNA antibody (PC-10; Santa Cruz). To examine the binding between XPCNA and Xic1, GST-PCNA fusion proteins (5 μg) were bound to glutathione-Sepharose 4B and incubated with [35S]methionine-labeled Xic1-NPIP mutants (4 μl) for 1.5 h at 23°C. The beads were washed with NETN buffer and subjected to SDS-PAGE and PhosphorImager analysis. For the coimmunoprecipitation assay, antibody to XPCNA, XCdt2, or XDDB1 was incubated with 15 μl of protein A-Sepharose (GE Healthcare) for 1.5 h at 23°C. The beads were washed with NETN and incubated with [35S]methionine-labeled Xic1 or XCdt2 for 1.5 h at 23°C in NETN, followed by SDS-PAGE and PhosphorImager analysis.
Immunodepletion and rescue assays.
Specific antibody to XDDB1 or XCdt2 was covalently coupled to protein A-Sepharose and then incubated in the egg extract for 1 h at 4°C. The depleted extracts were separated from the beads by centrifugation at 4°C, and the immunodepletion protocol was repeated. Control depletions were performed using normal rabbit serum (NRS). To restore XCdt2 activity to the immunodepleted extract, in vitro-translated wild-type or mutant XCdt2 or unprogrammed RRL was added.
Sequence alignment analysis.
AlignX of Vector NTI (Invitrogen) software was used for sequence alignment analysis. The Xenopus laevis hypothetical protein MGC115611 (gi 71679818) was compared to human Cul4a, and MGC114697 was compared to human Cdt2.
RESULTS
Identification of Xic1-associated proteins from the Xenopus interphase egg extract.
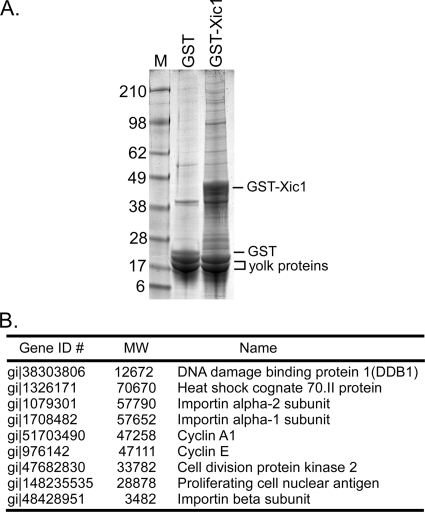
In an effort to identify E3 of Xic1 in the egg, we purified Xic1-associated proteins from the egg extract using recombinant GST-Xic1 and identified the proteins by mass spectrometry (Fig. 1). The proteins identified as Xic1-associating proteins included known Xic1 partners, such as cyclins A and B, CDK2, and PCNA. In addition, Hsp70 and importin family members were found to be associated with Xic1. Our previous studies have indicated that the nuclear localization of Xic1 requires both CDK2-cyclin binding and the three basic nuclear localization sequences within the Xic1 C terminus (7). We had also previously noted an interaction between Xic1 and importin α. Together, these findings suggest that importin family members may play an important role in Xic1 nuclear localization. Interestingly, Xic1 was also found to be associated with DDB1, a component of the Cul4 ubiquitin ligase (CRL4), implying that Xic1 might be a substrate of Cul4-DDB1. Our previous studies demonstrated that Xic1 turnover in the egg extract was absolutely dependent upon its binding to PCNA (6). Recent studies indicated that the replication licensing factor Cdt1 was targeted for degradation in the Xenopus egg and mammalian cells in a PCNA- and CRL4Cdt2-dependent manner (2, 15). In fact, the molecular mechanism of Cdt1 turnover in the egg appeared remarkably similar to the mechanism of Xic1 turnover in the egg. Based on these findings, we tested whether CRL4Cdt2 mediated the turnover of Xic1 in the egg extract.
FIG. 1.
DDB1 associates with Xic1 in the egg extract. (A) GST or GST-Xic1 binding proteins purified from the Xenopus interphase egg extract (LSS) and stained with Coomassie blue. GST-Xic1, GST, and yolk proteins are indicated, along with molecular mass markers in kilodaltons (M). (B) Xic1-interacting proteins identified by LC-tandem mass spectrometry (MS-MS). Proteins are listed according to their molecular weights (MW). Each protein is also identified by its gene identification number and common name.
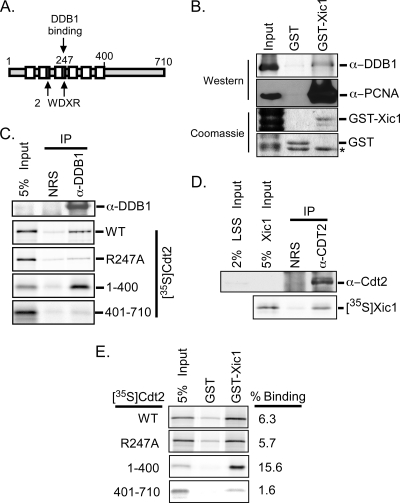
Xic1 interacts with DDB1 and the N-terminal domain of XCdt2.
Based on studies of Cdt1 in the Xenopus extract, we wondered whether XCdt2 might function as the substrate-binding component to recruit Xic1 to the CRL4 ubiquitin ligase. To examine this possibility, we performed a BLAST search of the NCBI database using the human Cdt2 amino acid sequence and identified a related Xenopus cDNA sequence encoding hypothetical protein MGC114697 (gi 66911503). The Xenopus MGC114697 and human Cdt2 proteins exhibited 55% amino acid identity and 65% amino acid similarity, suggesting that MGC114697 is the Xenopus paralog of human Cdt2 (see Fig. 7A). Further sequence analysis of the putative XCdt2 coding sequence indicated that it was organized into 6 WD40 domains and 2 WDXR motifs, similar to its human counterpart (Fig. 2 A). Studies of human Cdt2 suggested that the WDXR motifs and, in particular, residue R246 are essential for binding to DDB1 (15). We generated the conservative mutation in the putative XCdt2 protein at residue R247 and tested the ability of the wild-type XCdt2 (XCdt2WT) and R247A mutant (XCdt2R247A) to bind to DDB1 from the Xenopus egg extract. Immunopurified DDB1 from the egg extract was incubated with 35S-labeled XCdt2WT, XCdt2R247A, XCdt21-400 (N-terminal amino acids [aa] 1 to 400), or XCdt2401-710 (C-terminal aa 401 to 710). Consistent with studies of human Cdt2, DDB1 associated with XCdt2WT and XCdt21-400 but did not appreciably associate with XCdt2R247A or XCdt2401-710 (Fig. 2C). These studies strongly suggest that the hypothetical MGC114697 gene encodes the Xenopus Cdt2 protein.
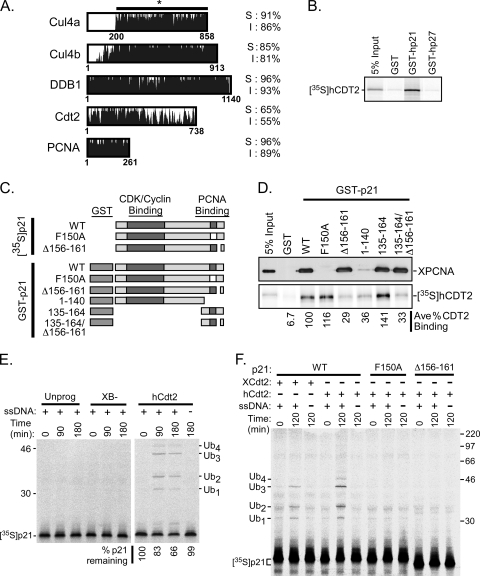
FIG. 7.
p21 is ubiquitinated during the events of DNA polymerase switching/elongation in the Xenopus egg extract. (A) Amino acid sequence similarity between Xenopus and human Cul4a, Cul4b, DDB1, Cdt2, and PCNA. Xenopus residue numbers are indicated at the bottom of the sequence alignments, and the percentages of similarity (S) and identity (I) between the Xenopus and human proteins are shown on the right. Xenopus Cul4a, the MGC115611 protein (gi 71679818), contains 200 additional residues in the N terminus compared to human Cul4a, so only residues 200 to 858 of Xenopus Cul4a were compared in the alignment. (B) GST pulldown assay. GST, GST-p21, or GST-p27 proteins were immobilized on glutathione-Sepharose beads and incubated with 35S-hCDT2. A total of 5% of the input hCdt2 is shown (5% input). (C) Schematic representation of p21 mutants. CDK-cyclin and PCNA binding domains for untagged and GST-tagged p21 mutants are indicated. In the p21 point mutant F150A, phenylalanine is replaced by alanine at residue 150. Mutant Δ156-161 contains a deletion of residues 156 to 161, while other deletion mutants are named by the remaining residues of p21. (D) GST pulldown assay. (Top) GST or GST-p21 wild-type or mutant proteins were bound to glutathione-Sepharose beads, followed by incubation with 10 μl of HSS in NETN buffer. The bead fraction was analyzed by immunoblotting with anti-hPCNA antibody (Santa Cruz), and 0.5 μl HSS was included as an input control (5% input). (Bottom) GST or GST-p21 wild-type or mutant proteins were immobilized onto glutathione-Sepharose beads, followed by incubation with 35S-hCDT2 and analysis by SDS-PAGE and phosphorimaging. The average percentage of hCdt2 bound (ave % CDT2 binding) was calculated using results from 2 independent experiments and was normalized to the level of hCdt2 binding to wild-type p21, which was set at 100%. (E) p21 ubiquitination and degradation assay. 35S-labeled wild-type p21 was incubated in HSS supplemented with 2.5 μl XB− buffer, unprogrammed reticulocyte lysate (unprog; lysate programmed with vector DNA), or in vitro-translated hCdt2 with (+) or without (−) single-stranded DNA (ssDNA). Samples were analyzed at time points between 0 and 180 min as indicated. Ubiquitinated p21 species (Ubn) are shown on the right, and molecular mass markers are shown in kilodaltons on the left. The percentage of p21 remaining at each time point was calculated as a percentage of the amount of p21 at the zero time point, which was normalized to 100%. (F) p21 ubiquitination assay. 35S-labeled wild-type p21 (WT), the p21F150A point mutant (F150A), or the p21Δ156-161 deletion mutant (Δ156-161) was incubated in HSS supplemented with 2.5 μl in vitro-translated Xenopus Cdt2 (XCdt2) or human Cdt2 (hCdt2) as indicated in the presence (+) or absence (−) of single-stranded DNA (ssDNA), followed by analysis at 0 and 120 min. Ubiquitinated p21 species (Ubn) are shown on the left, and molecular mass markers are shown in kilodaltons on the right.
FIG. 2.
Xic1 interacts with DDB1 and XCdt2. (A) Schematic representation of XCdt2. XCdt2 contains two WDXR motifs (gray boxes), six WD40 domains (white boxes), and a conserved arginine residue (R247) essential for DDB1 binding. (B) GST pulldown assay. GST or GST-Xic1 immobilized on glutathione-Sepharose beads was incubated with Xenopus interphase egg extract and immunoblotted with antibody against Xenopus DDB1 and PCNA (Western blot). GST and GST-Xic1 proteins (20% of Western blot reaction) were stained with Coomassie brilliant blue. The input (4%) is shown in lane 1. α, anti; *, nonspecific bacterial protein. (C) Coimmunoprecipitation assay. Immunoprecipitated DDB1 (IP) from the egg extract was bound to protein A beads and incubated with 35S-labeled wild-type XCdt2 (WT), XCdt2R247A (R247A), XCdt21-400 (1-400), or XCdt2401-710 (401-710). As a control, nonspecific normal rabbit serum (NRS) was used in the place of DDB1 antiserum. Efficient immunoprecipitation of XDDB1 was confirmed by immunoblotting with anti-DDB1 antibody (top). Binding of XCdt2 proteins (35S-Cdt2) was analyzed by SDS-PAGE and phosphorimaging, and 5% of the input proteins is shown (5% input). (D) Coimmunoprecipitation assay. Immunoprecipitated XCdt2 (anti-CDT2, IP) from the egg extract was incubated with 35S-labeled Xic1 and subjected to SDS-PAGE and phosphorimager analysis. Efficient immunoprecipitation of XCdt2 was confirmed by immunoblotting with anti-Cdt2 antibody (top). Immunoprecipitation with normal rabbit serum (NRS) was included as a control, and input samples are indicated. (E) GST pulldown assay. Bacterially expressed GST or GST-Xic1 (5 μg) was immobilized on glutathione-Sepharose beads and incubated with 35S-labeled XCdt2 proteins, as indicated. A total of 5% of the input reaction is shown. The percentage of Cdt2 bound by all GST-Xic1 proteins (% binding) is an average value obtained from 2 independent experiments.
To investigate a role for DDB1 and XCdt2 in the turnover of Xic1, we first studied whether Xic1 could associate with DDB1, and in particular with XCdt2, as would be expected if XCdt2 functioned as the substrate-binding domain of Cul4-DDB1. For these studies, we added to the egg extract either GST or GST-tagged Xic1 bound to glutathione-Sepharose and immunoblotted the bound fraction with anti-DDB1 antibody. The immunoblot revealed weak binding between Xic1 and DDB1 from the extract, despite Xic1's ability to efficiently bind PCNA (Fig. 2B). We next immunopurified XCdt2 from the egg extract and examined its ability to associate with 35S-labeled Xic1 (Fig. 2D). Our results suggested that Xic1 readily associated with XCdt2, and to further study the nature of this interaction, we studied the binding of XCdt2WT, XCdt2R247A, XCdt21-400, and XCdt2401-710 to GST-Xic1 (Fig. 2E). Our studies indicated that Xic1 efficiently associated with the XCdt2 amino-terminal domain (15.6% binding) and much less efficiently bound to the XCdt2 C-terminal domain (1.6% binding) (Fig. 2E). Nevertheless, Xic1 still retained some specific binding to the C terminus compared to background binding (Fig. 2E). These studies suggest that both Xic1 and DDB1 associate with the N-terminal residues of XCdt2 and that Xic1 might be a substrate of CRL4XCdt2.
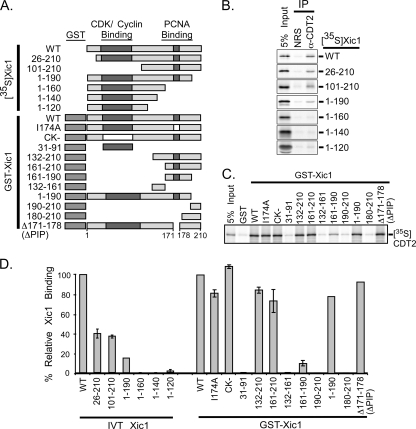
Xic1 residues 161 to 170 and 179 to 190 are important for XCdt2 binding.
To better understand the requirements for XCdt2 binding to Xic1, we generated and tested a number of Xic1 mutants in order to identify the cis-acting sequences of Xic1 that were necessary and sufficient for XCdt2 binding (Fig. 3 A). Using XCdt2 immunopurified from the egg extract and 35S-labeled Xic1 N- and C-terminal truncation mutants, we found that the amino-terminal half of Xic1 was not absolutely required, nor was it sufficient for XCdt2 binding (Fig. 3A, B, and D, left). Upon further examination, we found that C-terminal sequences within Xic1 residues 161 to 190 were important for XCdt2 binding. Analysis of the ability of GST-Xic1 mutants to bind XCdt2 revealed that point mutants CK− (defective for CDK2-cyclin binding) and I174A (defective for PCNA binding) were able to associate with XCdt2 as well as wild-type Xic1 (Fig. 3C and D, right). Moreover, while Xic1 residues 161 to 190 appeared to represent the minimal domain of Xic1 that could bind to XCdt2, a deletion of the residues comprising the PIP box (PCNA binding motif, aa 171 to 178) in the Xic1 mutant, Δ171 to 178 (ΔPIP), indicated that the PIP box was not required for XCdt2 binding since this mutant bound Cdt2 similar to wild-type Xic1 (Fig. 3C and D). These results suggested that Xic1 residues 161 to 170 and 179 to 190 were the amino acids that were necessary and sufficient for XCdt2 binding. However, residues upstream and downstream of amino acids 161 and 190 appeared to contribute toward establishing an efficient interaction between Xic1 and XCdt2, as evidenced by the robust binding of Xic1161-210 and Xic11-190 to XCdt2 compared to the ∼7-fold-lower binding of Xic1161-190 to XCdt2 (Fig. 3D). Consistent with this finding, our previous studies indicated that although Xic1161-190 could be modestly ubiquitinated, it was not efficiently degraded (8). These studies suggest that the XCdt2 and PCNA binding regions of Xic1 are immediately adjacent to one another, perhaps suggesting that some type of cooperation may occur between PCNA and XCdt2 to mediate Xic1 ubiquitination and degradation.
FIG. 3.
Xic1 residues immediately upstream and downstream of its PCNA binding domain are important for Cdt2 binding. (A) Schematic representation of full-length Xic1 and Xic1 deletion mutants, with CDK/cyclin and PCNA binding domains indicated. Amino- or carboxy-terminal serial deletion mutants of Xic1 were in vitro-translated (35S-Xic1) or bacterially expressed as GST-Xic1 fusion proteins (GST-Xic1). The Xic1 wild type (WT), point mutant I174A deficient for PCNA binding (I174A), CK− mutant deficient for CDK2-cyclin binding (CK−), or Xic1 deletion mutants indicated by the residues contained within the mutant or deleted (Δ) within the mutant are shown. (B) Coimmunoprecipitation assay. Immunoprecipitated XCdt2 (anti-CDT2, IP) from the egg extract was incubated with the 35S-Xic1 wild type (WT) or mutants as indicated. Equivalent immunoprecipitation of XCdt2 for each sample was confirmed by immunoblotting with anti-Cdt2 antibody (data not shown). Immunoprecipitation with normal rabbit serum (NRS) was conducted as a control, and 5% of the input 35S-Xic1 is shown (5% input). (C) GST pulldown assay. GST or GST-Xic1 wild-type or mutant proteins as indicated were immobilized on glutathione-Sepharose beads and incubated with 35S-CDT2. A total of 5% of the input XCdt2 for each reaction is shown (5% input). (D) Quantitation of the results shown in panels B and C. The relative XCdt2 binding value (% relative Cdt2 binding) for each Xic1 mutant is shown, where wild-type Xic1 (WT) binding was normalized to 100% for each experiment. Each sample was tested at least 2 or 3 times, and the standard error of the mean (SEM) is shown as an error bar for samples tested at least three times. IVT, in vitro transcribed.
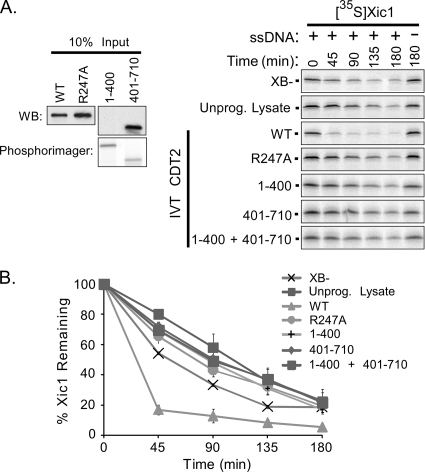
XCdt2 promotes DNA-dependent Xic1 proteolysis in the Xenopus interphase egg extract.
The interaction between Xic1 and DDB1-Cdt2 suggested that Xic1 might be a substrate of the CRL4XCdt2 ubiquitin ligase and that XCdt2 might function as the Xic1 substrate-binding component of Cul4-DDB1. If XCdt2 functions as an important mediator of Xic1 turnover in the egg extract, then it is predicted that if XCdt2 is a limiting component in the extract, the addition of XCdt2 will promote the degradation of Xic1. To examine this possibility, we added in vitro-translated XCdt2WT, XCdt2R247A, XCdt21-400, or XCdt2401-710 (Fig. 4 A, left) to the interphase extract in the presence or absence of DNA and analyzed the degradation of 35S-labeled Xic1 over time. Our results indicated that while the half-life of Xic1 in the control sample supplemented with unprogrammed reticulocyte lysate was ∼105 min, when the extract was supplemented with lysate programmed with the XCdt2WT protein, the half-life of Xic1 was substantially reduced to ∼30 min. Further, the addition of XCdt21-400 and XCdt2401-710 did not promote Xic1 turnover, even though the XCdt21-400 polypeptide was sufficient to bind both Xic1 and DDB1 (Fig. 4A and B). Similarly, the DDB1 binding-deficient mutant XCdt2R247A and the XCdt21-400 and XCdt2401-710 polypeptides added together did not promote Xic1 turnover (Fig. 4A and B). Taken together, these results suggest, first, that XCdt2 promotes Xic1 degradation through Cul4-DDB1-mediated ubiquitination and, second, that XCdt2 binding to Xic1 and DDB1 is not sufficient to support efficient Xic1 turnover. One possibility is that in addition to Xic1 binding and recruitment to Cul4A-DDB1, XCdt2 may mediate an additional as-yet-uncharacterized function encoded within the XCdt2 C terminus that is required for efficient Xic1 turnover. This possibility is examined below (see Fig. 9).
FIG. 4.
XCdt2 promotes the turnover of Xic1 in the egg extract. (A, right) Xic1 degradation assay. 35S-Xic1 was incubated in the HSS supplemented with XB− buffer, unprogrammed reticulocyte lysate (Unprog. Lysate), XCdt2WT (WT), XCdt2R247A (R247A), XCdt21-400 (1-400), XCdt2401-710 (401-710), or both XCdt21-400 and XCdt2401-710 with (+) or without (−) single-stranded DNA (ssDNA), and samples were analyzed at time points between 0 and 180 min as indicated. (Left) Input amounts of unlabeled in vitro-translated XCdt2 proteins added were quantitated by Western blotting (WB) using anti-Cdt2 antibody. 35S-labeled Cdt2 (1-400) was also quantitated by phosphorimager analysis since it is not recognized by the anti-Cdt2 antibody which was generated against the C-terminal fragment of Cdt2. (B) Quantitation of Xic1 turnover. The mean percentage of Xic1 remaining from two or three independent experiments as described in the legend to panel A is shown, where the 0 h time point was normalized to 100% of Xic1 remaining for each sample. SEMs are shown as error bars.
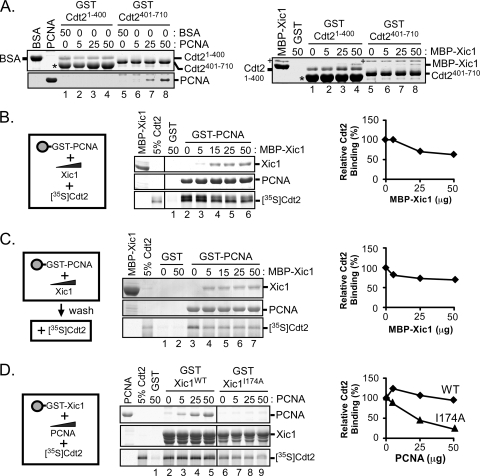
FIG. 9.
PCNA directly interacts with the C-terminal domain of XCdt2. (A) GST pulldown assay. Bacterially expressed GST, GST-XCdt21-400, or GST-XCdt2401-710 was bound to glutathione-Sepharose and incubated with purified XPCNA (0, 5, 25, and 50 μg) or bovine serum albumin (BSA; 0 and 50 μg) (left) as indicated and MBP-Xic1 (0, 5, 25, and 50 μg) (right), followed by staining with Coomassie blue. Protein bands were identified by mass spectrometry and are labeled accordingly. Several bacterial contaminants were identified. “+” was identified as the bacterial DnaK protein, and “*” was identified as the bacterial GroEL protein. (B) GST pulldown and competitive binding assay. Bacterially expressed GST or GST-PCNA (5 μg) was bound to glutathione-Sepharose beads and incubated with 0 to 50 μg of purified MBP-Xic1 or GST as indicated and 35S-labeled wild-type XCdt2. (C) GST pulldown and competition study. GST or GST-PCNA was bound to glutathione-Sepharose beads and incubated with 0 to 50 μg of purified MBP-Xic1 as indicated. Following a washing step, samples were incubated with 35S-labeled XCdt2. (D) GST pulldown assay and competitive binding assay. GST, GST-Xic1WT, or GST-Xic1I174A bound to glutathione-Sepharose beads was incubated with 0 to 50 μg of purified XPCNA and 35S-labeled wild-type XCdt2. (B to D) Samples were analyzed by Coomassie blue staining and phosphorimaging. (Left) Schematic representation of proteins analyzed in binding assays. (Right) The average relative Cdt2 binding values [relative Cdt2 binding (%)] of results from at least 2 independent experiments are shown, where the “zero competitor” value was normalized to 100%.
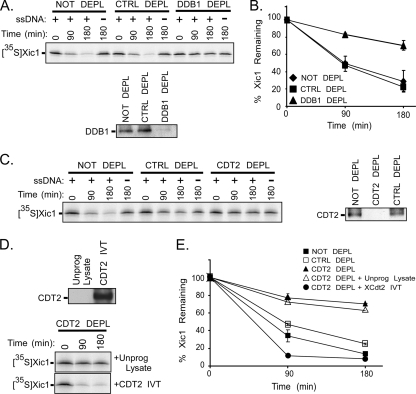
DDB1 and XCdt2 are required for Xic1 degradation during DNA polymerase switching.
Our studies suggest that XCdt2 is a limiting factor in the interphase egg extract for Xic1 ubiquitination, and the addition of XCdt2 to the extract promotes Xic1 turnover. To more directly study a requirement for CRL4XCdt2 in Xic1 turnover, we performed immunodepletion studies of the Xenopus interphase extract called the high-speed supernatant (HSS), an extract that fully supports Xic1 ubiquitination and degradation as well as DNA polymerase switching, the step of replication initiation required for Xic1 turnover (6). The interphase extract was either not depleted or immunodepleted with control antibody or DDB1 antibody coupled to beads and was then analyzed for the ability to support Xic1 degradation (Fig. 5 A and B). The results indicated that upon the depletion of DDB1, Xic1 was significantly stabilized in the extract. Similarly, upon the immunodepletion of XCdt2 from the egg extract, Xic1 turnover was inhibited (Fig. 5C and E). Importantly, when extract depleted of XCdt2 protein was subsequently supplemented with the in vitro-translated XCdt2 protein, Xic1 turnover was completely restored compared to that with the addition of unprogrammed reticulocyte lysate (Fig. 5D and E). This indicated that although the depletion of XCdt2 might have codepleted another regulator of Xic1 turnover, the addition of XCdt2 to the extract was sufficient to fully restore Xic1 turnover. These studies demonstrate the requirement for DDB1 and XCdt2 in Xic1 turnover in the egg extract and strongly suggest that CRL4XCdt2 is the ubiquitin ligase for Xic1 in the replicating Xenopus interphase egg extract.
FIG. 5.
Both DDB1 and XCdt2 are required for Xic1 turnover in the interphase egg extract. (A, top) Xic1 degradation assay. HSS that was not depleted (NOT DEPL), control depleted using nonspecific rabbit serum (CTRL DEPL), or immunodepleted (DDB1 DEPL) of DDB1 was supplemented with (+) or without (−) single-stranded DNA, 35S-Xic1, and assayed at the time points indicated. (Bottom) Samples of the treated extracts were immunoblotted with anti-DDB1 antibody. (B) Quantitation of Xic1 turnover. The mean percentage of Xic1 remaining from two or three independent experiments described in the legend to panel A is shown, where the 0-h time point was normalized to 100% of Xic1 remaining. SEMs are shown as error bars. (C, left) Xic1 degradation assay. HSS that was not depleted (NOT DEPL), control depleted (CTRL DEPL), or immunodepleted (CDT2 DEPL) of XCdt2 was supplemented with (+) or without (−) single-stranded DNA, 35S-Xic1, and assayed at the time points indicated. (Right) Samples of the treated extracts were immunoblotted with anti-XCdt2 antibody. (D, top) Unprogrammed reticulocyte lysate (Unprog. Lysate) or lysate programmed with XCdt2 was immunoblotted with anti-XCdt2 antibody. (Bottom) HSS immunodepleted of XCdt2 as described in the legend to panel C was supplemented with 35S-Xic1, single-stranded DNA, and unprogrammed reticulocyte lysate (+Unprog. Lysate) or lysate programmed with XCdt2 (+CDT2 IVT) as indicated. Samples were analyzed at the indicated time points. (E) Quantitation of Xic1 turnover. The mean percentage of Xic1 remaining from two or three independent experiments as described in the legends to panels C and D is shown, where the 0-h time point was normalized to 100% of Xic1 remaining. SEMs are shown as error bars for the sample time points shown in panel C.
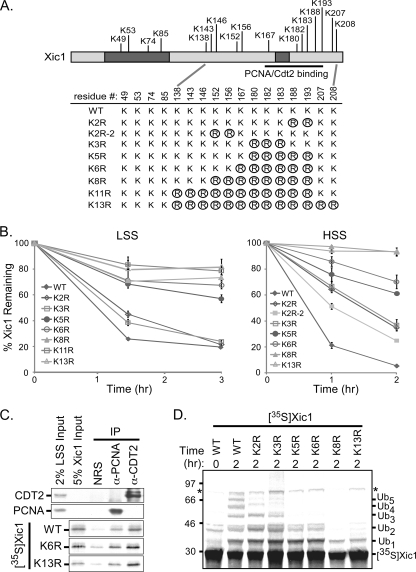
Five lysine residues of Xic1 directly downstream of the PCNA binding domain and within the XCdt2 binding domain are critical for efficient Xic1 turnover.
Our studies suggest that both PCNA and XCdt2 bind to Xic1 residues within the Xic1 C terminus and perhaps cooperate to target Xic1 for ubiquitination. To further examine the molecular mechanism of Xic1 ubiquitination by XCdt2, we wanted to identify the lysine residues of Xic1 that are critical for efficient Xic1 degradation. Xic1 contains a total of 17 lysine residues spanning the entire length of the Xic1 polypeptide, although the majority of the Xic1 lysine residues are clustered within the C terminus of Xic1 (Fig. 6 A). Our past studies suggested that Xic1 was ubiquitinated on ∼3 to 4 lysine residues within the C-terminal domain of Xic1 (8). In order to identify the precise lysine residues of Xic1 essential for efficient Xic1 turnover, lysine residues were mutated to arginine to prevent possible ubiquitination at that site but to preserve the overall charge of Xic1 (Fig. 6A). The stability of Xic1 mutants was then examined in both the membrane-containing LSS and membrane-free HSS extracts. We found that mutation of three lysine residues (K3R) or two lysine residues (K2R, or in the HSS only, K2R-2) resulted in only subtle stabilizations of Xic1 in the LSS while roughly doubling the half-life of Xic1 in the HSS (Fig. 6B). In contrast, upon the mutation of five of these lysine residues (K5R; K180, K182, K183, K188, and K193), Xic1 turnover was substantially inhibited in both extracts (Fig. 6B). Additional mutation of Xic1 lysine residues to include a total of six lysine residues (K6R) or eight lysine residues (K8R) had only a modest effect in the LSS (K5R versus K8R; P = 0.13), while Xic1-K8R was statistically more stabilized in the HSS (K5R versus K8R; P = 0.04). Mutation of all 13 lysine residues within the Xic1 C-terminal domain (K13R) also further stabilized Xic1 beyond the mutation of five lysine residues in both extracts (K5R versus K13R; P = 0.04) (Fig. 6A and B). Studies to examine Xic1 ubiquitination confirmed that the K5R, K6R, K8R, and K13R Xic1 mutants were less efficiently ubiquitinated in the egg extract, as expected (Fig. 6D). To ensure that the lysine mutants of Xic1 were not grossly misfolded, we confirmed that Xic1WT, Xic1K6R, and Xic1K13R were able to comparably associate with XCdt2 and PCNA from the egg extract (Fig. 6C). These studies suggest that CRL4XCdt2 mediates the ubiquitination of at least five critical lysine residues of Xic1 and that ubiquitination of these sites is required for the efficient degradation of Xic1. Interestingly, the critical lysine residues for Xic1 turnover are located directly downstream of the PCNA binding domain and within the second XCdt2 binding domain of Xic1, suggesting that this might be a general feature of ubiquitination by CRL4Cdt2. We postulate that this organization of PCNA and XCdt2 binding to Xic1 helps to link Xic1 ubiquitination to PCNA binding and DNA polymerase switching.
FIG. 6.
Xic1 lysine residues adjacent to the PCNA binding domain are critical for efficient Xic1 turnover. (A) Schematic representation of Xic1, the PCNA and Cdt2 binding regions of Xic1, all the lysine residues of Xic1, and the targeted lysine-to-arginine mutations generated within Xic1. The shaded portions indicate the CDK2-cyclin binding domain (left) and the PCNA binding domain (right). Wild-type Xic1 and seven Xic1 mutants were generated containing lysine (K)-to-arginine (R, circled) mutations at specific residues as indicated. (B) Quantitation of Xic1 degradation assays. The 35S-labeled Xic1 wild type and mutants were incubated in LSS supplemented with Xenopus sperm chromatin or HSS supplemented with ΦX174 single-stranded DNA and analyzed at the time points indicated. The mean percentage of Xic1 remaining from at least three independent experiments is shown, where the 0-h time point was normalized to 100% of Xic1 remaining. SEMs are shown as error bars for each sample. Dependent sample or paired t test analysis between K5R and WT, K6R, K8R, or K13R was performed. K5R versus WT, P values of 0.01 (LSS) and 0.02 (HSS); K5R versus K6R, P value of 0.2 (LSS and HSS); K5R versus K8R, P values of 0.13 (LSS) and 0.04 (HSS); K5R versus K13R, P value of 0.04 (LSS and HSS). (C) PCNA and Cdt2 coimmunoprecipitation assay. Immunoaffinity-purified PCNA (anti-PCNA) and XCdt2 (anti-CDT2) from the egg extract were incubated with 35S-labeled WT Xic1, K6R, and K13R. Normal rabbit serum was used as a negative control for the immunoprecipitations, and samples were immunoblotted with antibody to Cdt2 or PCNA to confirm efficient immunoprecipitation of these proteins. Input samples for the Cdt2 and PCNA immunoblots and for the 35S-labeled Xic1 are shown. (D) Xic1 ubiquitination assay. 35S-labeled WT Xic1 or mutants (K2R, K3R, K5R, K6R, K8R, K13R) were incubated in the HSS supplemented with 3 μg/μl methyl ubiquitin at 0 and 120 min. Monoubiquitinated Xic1 species (Ubn) are shown on the right, and molecular mass markers are shown in kilodaltons on the left. The asterisk indicates a nonspecific in vitro translation product.
p21 is ubiquitinated in a DNA-, PCNA-, and Cdt2-dependent manner during the events of DNA polymerase switching/elongation in the Xenopus egg extract.
Our previous studies had indicated that human p21 and p27 were not readily ubiquitinated or degraded in the Xenopus interphase egg extract in the presence or absence of DNA (our unpublished results). This led us to speculate that the ubiquitination machinery in the Xenopus egg extract might not support mammalian CDK inhibitor turnover or that the mechanisms regulating frog and mammalian CDK inhibitor turnover were different. In light of our current studies and recent studies (1, 17, 24), we now postulate that the mechanisms regulating Cip/Kip-type CDK inhibitor turnover in vertebrates are actually highly conserved, while the substrate-binding domains of CRL-type E3s are more divergent. Examination of the amino acid sequences of human and frog Cul4A, DDB1, and PCNA demonstrate that all share >90% amino acid similarity. In comparison, the conservation between human and Xenopus Cdt2 is significantly lower (∼65% similarity) (Fig. 7 A). Because our previous and current studies indicate that Xic1 is targeted for ubiquitination and proteolysis during DNA polymerase switching by CRL4XCdt2, we wanted to examine whether human p21 was also targeted for proteolysis in a DNA-dependent manner during DNA polymerase switching using the Xenopus extract system. Consistent with recent studies, we found that in an in vitro binding assay, p21 readily associated with human Cdt2 (hCdt2), while human p27 did not (Fig. 7B) (1, 17, 24). Our studies also suggested that while the PIP box of p21 (residues 144 to 151) and residue F150, both being regions of p21 shown to be essential for PCNA binding, were not required for Cdt2 binding, residues 156 to 161 of p21 directly downstream of the PIP box were critical for human Cdt2 binding (Fig. 7C and D). Additionally, residues 135 to 164 were sufficient for wild-type levels of binding to both PCNA and human Cdt2. Because our previous studies indicated that the Xenopus extract could not support p21 ubiquitination or proteolysis, we decided to “humanize” the Xenopus extract by supplementing human Cdt2 under the assumption that Xenopus Cdt2 did not readily support p21 ubiquitination. As predicted, Xenopus extracts supplemented with buffer (XB−) or unprogrammed rabbit reticulocyte lysate did not support p21 ubiquitination, while extracts supplemented with in vitro-translated human Cdt2 exhibited DNA-dependent ubiquitination and modest proteolysis of p21 (Fig. 7E). Further studies indicated that the DNA-dependent ubiquitination of p21 observed in the Xenopus extract supplemented with human Cdt2 required both PCNA and Cdt2 binding since the p21 mutants, F150A and Δ156 to 161, defective for PCNA and Cdt2 binding, respectively, were not ubiquitinated (Fig. 7F). When the extract was supplemented with additional Xenopus Cdt2, p21 ubiquitination was observed, suggesting that at higher concentrations, Xenopus Cdt2 can support p21 ubiquitination (Fig. 7F). This study suggests that the ubiquitination of Xenopus Xic1 and that of human p21 are remarkably similar, both requiring DNA, Cdt2, and binding to PCNA. This study also suggests that like Xic1, the timing of p21 ubiquitination may also be during DNA polymerase switching.
Adjacent localization of Cdt2 and PCNA binding domains on Xic1 is not required for efficient Xic1 turnover.
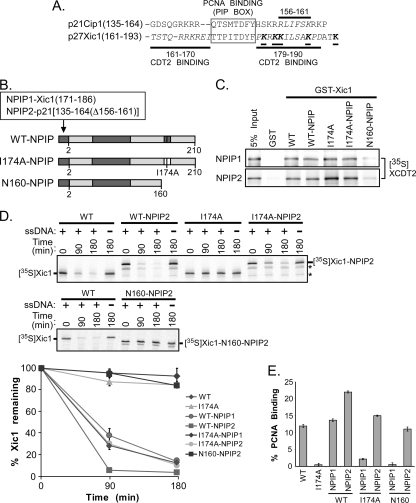
Our studies suggest that within the Xic1 C-terminal domain, the Cdt2 binding region directly flanks the PIP box (PCNA binding motif) on either side. This places the Cdt2 binding domain, the PCNA binding domain, and the critical lysine residues of Xic1 within a small 33-amino-acid region (Xic1 aa 161 to 193) we call the “Xic1 ubiquitination domain” (Fig. 8 A). Similarly, our results for p21 also indicate that the Cdt2 and PCNA binding domains are directly adjacent to each other. These studies suggest that perhaps the organization of the Xic1 ubiquitination domain may be important to mediate some type of cooperation between PCNA and Cdt2 in the ubiquitination of Xic1. Our studies suggest that the minimal region of Xic1 for wild-type binding to PCNA includes residues 161 to 190, while the minimal region of Xic1 for wild-type binding to Cdt2 includes residues 161 to 190 as well as additional N-terminal or C-terminal residues of Xic1. Additionally, based on the crystal structure of the C-terminal domain of p21 with PCNA, a conserved isoleucine residue within Xic1 (I184) predicted to mediate efficient binding to PCNA lies within the second Cdt2 binding domain of Xic1 (11). This implies that the binding regions for PCNA and Cdt2 are overlapping and suggests either that this particular arrangement may be important for cooperation between PCNA and Cdt2 or that the binding of these two Xic1 regulators may be mutually exclusive. To study the organization of the Xic1 33-amino-acid ubiquitination domain, we first asked a simple question of whether it was critical for the PCNA and Cdt2 binding regions to be directly adjacent to one another by separating the two domains. We reengineered Xic1 so that the C-terminal PCNA binding region of Xic1 was eliminated through a point mutation (I174A) while leaving the C-terminal Cdt2 binding region of Xic1 intact (Fig. 8B, C, and E). We next added an efficient PCNA binding site to the N terminus of Xic1, which in the end we could more easily achieve by using residues from p21 than by using residues from Xic1 (Fig. 8B, C, and E). Initially, we utilized Xic1 residues 171 to 186 for fusion to the Xic1 N terminus (NPIP1), reasoning that this region included all the sequences predicted to mediate PCNA binding (Fig. 8A and B). However, when fused to Xic1-I174A or Xic1-N160 (containing the N-terminal 160 amino acids of Xic1 and used as a control), this Xic1 mutant was not able to efficiently bind to PCNA, and as expected, it was not degraded (Fig. 8C to E). We then proceeded to fuse p21 residues 135 to 164/Δ156 to 161 to the N terminus of Xic1 (NPIP2) and finally obtained a Xic1 mutant which could efficiently bind to both Cdt2 within its C-terminal domain and PCNA within its N-terminal domain (Fig. 8B, C, and E). Contrary to our expectations, when the PCNA binding domain was physically separated from the Cdt2 binding domain in the mutant I174A-NPIP2, Xic1 was still efficiently degraded (Fig. 8D). The Xic1 mutant N160-NPIP2, expressing the PCNA binding domain from p21 at the Xic1 N terminus but missing the Xic1 Cdt2 binding domain, was stabilized as expected (Fig. 8D). These studies suggest that the specific organization of adjacent or overlapping PCNA and Cdt2 binding domains within Xic1 is not essential for Xic1 turnover during DNA polymerase switching. However, because Cip/Kip-type CDK inhibitors are thought to be generally unstructured (26) and that it is possible that the Xic1 protein could bend to bring the amino and carboxyl termini into close proximity and because trimeric PCNA has 3 binding domains for Xic1, our studies cannot completely rule out some type of cooperation between PCNA and Cdt2 in mediating Xic1 turnover.
FIG. 8.
Xic1 turnover does not require the tandem arrangement of PCNA and Cdt2 binding domains. (A) Amino acid sequence alignment of p21 (p21Cip1) and Xic1 (p27Xic1). Cdt2 binding regions indicated by italicized amino acid residues and bold lines, the PCNA binding element (PIP box) indicated by gray box, and critical lysine residues of Xic1 indicated by underlining, italicizing, and boldfacing of amino acid residues. (B) Schematic representation of mutant Xic1 proteins. CDK2-cyclin and wild-type PCNA binding domains are indicated by dark gray shading, while the I174A PCNA binding mutant is indicated by a white box. Xic1 residue numbers are indicated below each schematic. The NPIP1 and NPIP2 domains are fused to the N terminus of wild-type Xic1 (WT-NPIP), Xic1-I174A (I174A-NPIP), or amino acids 1 to 160 of Xic1 (N160-NPIP) as indicated and includes Xic1 amino acids 171 to 186 (TTPITDYFPKRKKILS) for NPIP1 and p21 residues 135 to 164 with an internal deletion of residues 156 to 161 for NPIP2. The NPIP2 domain serves solely as a PCNA binding domain and does not retain the ability to efficiently bind Cdt2. (C) GST pulldown assay. GST or GST-Xic1 wild-type and mutant proteins (top, NPIP1; bottom, NPIP2) were immobilized on glutathione-Sepharose beads and incubated with 35S-labeled Xenopus Cdt2 (35S-XCDT2). The 35S-XCdt2 input control (5% input) is shown in lane 1. (D) Xic1 degradation assay. (Top and middle) 35S-labeled Xic1 wild-type (WT) and mutant proteins (WT-NPIP2, I174A, I174A-NPIP2, and N160-NPIP2) as indicated were incubated in HSS with (+) or without (−) single-stranded DNA for the indicated times, followed by SDS-PAGE and phosphorimager analysis. Asterisks indicate internal initiation translation products. (Bottom) Quantitation of Xic1 degradation. The mean percentage of Xic1 remaining from two (WT, WT-NPIP1, I174A-NPIP1, and N160-NPIP1) or three (WT-NPIP2, I174A, I174A-NPIP2, and N160-NPIP2) independent experiments as described above is shown, where the 0-h time point was normalized to 100% of Xic1 remaining for each sample. SEMs are shown as error bars. (E) Quantitation of Xic1 binding to PCNA. GST or GST-PCNA proteins were immobilized on glutathione-Sepharose beads and incubated with 35S-labeled Xic1 wild-type (WT) or mutant proteins (I174A, WT-NPIP1, I174A-NPIP1, N160-NPIP1, WT-NPIP2, I174A-NPIP2, and N160-NPIP2). The average percentage of Xic1 bound by GST-PCNA (% PCNA binding) is shown, where values for WT Xic1 and I174A are averages of results from 4 independent experiments, and the values of the NPIP mutants (WT-NPIP1, I174A-NPIP1, N160-NPIP1, WT-NPIP2, I174A-NPIP2, and N160-NPIP2) are averages of results from 2 independent experiments. SEMs are shown as error bars.
The C-terminal domain of Cdt2 binds directly to PCNA.
While the studies shown in Fig. 8 suggested that on the Xic1 molecule, the binding domains for PCNA and Cdt2 need not exist adjacent to one another, we wanted to better understand the binding dynamics between Xic1 and PCNA or Cdt2 by performing competitive binding studies with PCNA, Cdt2, and Xic1. From these studies, we made the surprising discovery that in the absence of Xic1, in vitro-translated Cdt2 can bind to PCNA (Fig. 9 B, middle, lane 2). Using bacterially expressed and purified proteins, we show that PCNA binds directly to the C terminus of Cdt2 (residues 401 to 710) (Fig. 9A, left, lanes 7 and 8). In contrast, Xic1 appears to bind preferentially to the Cdt2 N-terminal domain, when normalized to the Cdt2 input (Fig. 4 and 9A, right). The studies shown in Fig. 4 suggested that while the N-terminal domain of Cdt2 was sufficient for both Xic1 and DDB1 binding, it could not promote Xic1 turnover when added to the egg extract, suggesting that another function of Cdt2 was required to mediate Xic1 ubiquitination and degradation. We postulate that this other function of Cdt2 required for Xic1 turnover is direct binding to PCNA. Our further studies indicate that Xic1 can compete with Cdt2 for binding to PCNA (Fig. 9B and C) and that PCNA can compete with Xic1 for binding to Cdt2 (Fig. 9D). Additionally, the studies of the Xic1-I174A mutant, defective for PCNA binding, suggest that Cdt2 bound to PCNA may not be able to bind to Xic1 (Fig. 9D) since the addition of PCNA could reduce Cdt2 binding to Xic1, despite our previous finding that Xic1-I174A binds to Cdt2 like WT Xic1 (Fig. 3C and D). Our studies suggest not only that Xic1 binds to both PCNA and Cdt2 directly but also that there is a previously undescribed direct association between PCNA and the C terminus of Cdt2. Additionally, the competitive binding studies suggest that certain interactions among Xic1, Cdt2, and PCNA are mutually exclusive.
DISCUSSION
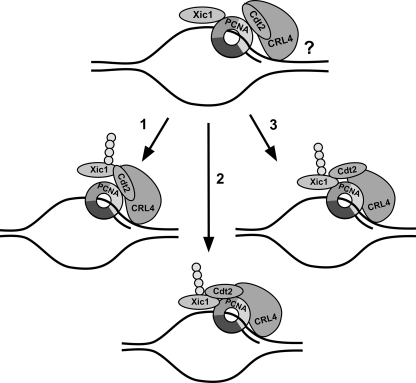
Based on our studies, we postulate that the ubiquitination and degradation of Xic1 requires the following steps: (i) PCNA binding to the Xic1 PIP box, (ii) PCNA binding to the Cdt2 C-terminal domain, (iii) Cdt2 binding to Xic1 through two elements located directly upstream and downstream of the Xic1 PIP box, and (iv) ubiquitination of Xic1 by CRL4XCdt2 at five critical lysine residues within the second Cdt2 binding domain. In our working model, we hypothesize that Xic1 and Cdt2 initially associate with different subunits of the PCNA trimer in solution and on DNA (Fig. 10). It is unclear whether CRL4 components can themselves associate with DNA in the absence of PCNA. Our studies have shown that Xic1 can freely associate with soluble PCNA and Cdt2 in the absence of DNA, but the presence of DNA is essential to trigger Xic1 proteolysis. In our model, PCNA serves as a platform to nucleate both the substrate and CRL4Cdt2, while we postulate that DNA may serve to trigger a conformational shift in PCNA. We hypothesize that chromatin-bound PCNA is conformationally distinct and provides the structural constraints necessary to bring a PIP box-containing substrate like Xic1 into functional proximity with the CRL4Cdt2 ubiquitin ligase.
FIG. 10.
Model for PCNA-mediated Xic1 ubiquitination by CRL4Cdt2. In our working model, we hypothesize that Xic1 and Cdt2 can associate to different subunits of the PCNA trimer in the absence of DNA. We postulate that in the absence of DNA, the binding of PCNA and Cdt2 to Xic1 and the binding of Xic1 and Cdt2 to monomeric PCNA may be mutually exclusive. It is unclear whether the CRL4 ubiquitin ligase can associate with DNA independently or what role this might play in CRL4 substrate ubiquitination. We propose the following three possible scenarios for Xic1 ubiquitination following the chromatin recruitment of PCNA in association with Xic1 and Cdt2 (top) and a proposed PCNA conformational shift: (1) Xic1 dissociates from PCNA and binds to the N-terminal domain of PCNA-bound Cdt2, resulting in Xic1 ubiquitination; (2) chromatin-bound PCNA mediates a stable interaction among Xic1, Cdt2, and PCNA on DNA, resulting in Xic1 ubiquitination; and (3) Cdt2 dissociates from PCNA and binds to Xic1, resulting in Xic1 ubiquitination.
Our past and current studies provide support for this model. Our previous studies demonstrated that the proteolysis of Xic1 required DNA and binding to trimeric PCNA (6). Although Xic1 could bind to monomeric PCNA, this did not support Xic1 proteolysis in the Xenopus extract (6). Importantly, we identified a direct interaction between PCNA and the C-terminal domain of Cdt2. This finding is significant because previous models of PCNA- and CRL4Cdt2-mediated ubiquitination of PIP box-containing substrates indicated that it was the substrate, bound to PCNA on chromatin, that was required for recruitment of the E3 (12, 15), while our studies suggest that PCNA serves to recruit both substrate and CRL4Cdt2 directly to DNA. Additionally, studies have demonstrated that Xic1 can associate directly with both PCNA and Cdt2 in the absence of DNA, yet Xic1 is ubiquitinated and degraded only when bound to chromatin (6, 7, 10). This suggests that perhaps Xic1 cannot associate with both PCNA and Cdt2 simultaneously in solution. Our studies identified overlapping binding domains for PCNA and Cdt2, and our competitive binding studies suggested that the binding dynamics among Xic1, PCNA, and Cdt2 are complex and that certain associations may be mutually exclusive. While the role of DNA in the molecular mechanism of Xic1 turnover is still unclear, one possibility is that DNA binding alters the conformation of PCNA, CRL4, or both of these. Another possibility is that a chromatin-bound factor mediates a posttranslational modification of one of these components, thereby triggering a conformational change and favoring a functional association between the substrate and CRL4Cdt2. Further studies are necessary to fully understand the molecular dynamics of substrate ubiquitination by PCNA and CRL4Cdt2 during DNA replication.
Subsequent to our finding that Xic1 binding to PCNA was required for its proteolysis during DNA replication initiation (6), the turnover of other proteins, including the prereplication complex protein Cdt1 and the mammalian Cip-type CDK inhibitor p21, was shown to require PCNA binding during UV damage and DNA replication (1, 2, 17, 24). Several of these findings also indicated that these proteins were targeted for ubiquitination by the CRL4Cdt2 ubiquitin ligase (1, 15, 17). Initially, our finding that Xic1 proteolysis strictly required PCNA binding suggested that the mechanism of Xic1 turnover was different than the mechanism of mammalian CDK inhibitors such as p21. However, our studies and the recent studies on p21 ubiquitination by CRL4Cdt2 clearly demonstrate that the turnover of Cip-type CDK inhibitors is remarkably conserved among higher eukaryotes (1, 17, 24).
In Xenopus, studies have shown that both Xic1 and Cdt1 are targeted for proteolysis by the same PCNA- and DNA-dependent mechanism during DNA replication initiation (2, 6). However, recent studies suggest that there are notable differences between the ubiquitination of Xenopus Cdt1 (12) and Xic1 by CRL4Cdt2. First, Cdt1 does not appreciably bind to free soluble PCNA or Cdt2, while as mentioned above, Xic1 does bind efficiently to both soluble PCNA and Cdt2 (Fig. 2, 3, 7, and 9) (6). Second, efficient binding of Xic1 to Cdt2 does not require a PIP box but does require sequences both upstream and downstream of the PIP box (Fig. 3 and 7), while Cdt1 binding to Cdt2 requires a specialized PIP box “degron” (PIP degron), containing a specialized PIP box and a basic amino acid 4 residues downstream of the PIP box. Additionally, while the PIP degron was sufficient for ubiquitination and degradation of Cdt1 by CRL4Cdt2, our studies have shown that this minimal PIP degron is not sufficient for Xic1 turnover (Fig. 8, see the studies of Xic1-I174A-NPIP1 and Xic1-N160-NPIP2). Thus, our data are more consistent with those studies that demonstrate that Cdt1 and p21 can bind to free soluble PCNA in the absence of DNA and that the binding of Cdt1 and p21 to Cdt2 does not require a PIP box (1, 13, 14, 17, 24).
During DNA replication, it appears that PCNA serves as the hub for proteolysis of CRL4Cdt2 substrates, but how the order of substrate ubiquitination is determined is not known. Moreover, many more substrates have been shown to be targeted for proteolysis by CRL4Cdt2 in response to UV damage or during a checkpoint, including p21, Cdt1, PCNA, fly E2F, worm DNA polymerase η, and possibly Chk1 (1, 15-17, 20, 24, 29, 34). Again, how the ubiquitination of these various CRL4Cdt2 substrates is orchestrated by PCNA during DNA repair is unclear. Because the ubiquitination of a CRL4Cdt2 substrate has not been reconstituted from purified components, it is not known whether other essential regulators may be required. However, based on our studies, it is now clear that PCNA plays a direct central role in mediating the ubiquitination of substrates by CRL4Cdt2.
Acknowledgments
We thank Johannes Walter and Emily Arias (Harvard Medical School) for providing the XCdt2 and DDB1 antibodies, J. Wade Harper and Jianpin Jin (Harvard Medical School) for providing the Homo sapiens Cdt2 cDNA, Martin J. Allday (Ludwig Institute for Cancer Research, Imperial College of Science) for providing the p21Δ156 to 161 cDNA, Guem Hee Baek for the generation of the Xic1K11R and Xic1K13R mutants, Angelica Hernandez for the generation of the XCdt2R247A mutant, and Ethan Lee (Vanderbilt University) for providing FA cloning plasmids. We also thank Ikjin Kim and Nam Hee Kim (UTHSCSA) for helpful comments and advice and Michael J. Parker (UTHSCSA) for excellent technical support.
This work was supported by a Career Development Award to P.R.Y. (award DAMD17-02-1-0589) from the U.S. Army Department of Defense and by the National Institute of Health (grant RO1-GM066226 to P.R.Y.).
Footnotes
Published ahead of print on 6 July 2010.
REFERENCES
- 1.Abbas, T., U. Sivaprasad, K. Terai, V. Amador, M. Pagano, and A. Dutta. 2008. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 22:2496-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arias, E. E., and J. C. Walter. 2006. PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat. Cell Biol. 8:84-90. [DOI] [PubMed] [Google Scholar]
- 3.Blow, J. J., and R. A. Laskey. 1986. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell 47:577-587. [DOI] [PubMed] [Google Scholar]
- 4.Bornstein, G., J. Bloom, D. Sitry-Shevah, K. Nakayama, M. Pagano, and A. Hershko. 2003. Role of the SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 in S phase. J. Biol. Chem. 278:25752-25757. [DOI] [PubMed] [Google Scholar]
- 5.Carrano, A. C., E. Eytan, A. Hershko, and M. Pagano. 1999. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat. Cell Biol. 1:193-199. [DOI] [PubMed] [Google Scholar]
- 6.Chuang, L. C., and P. R. Yew. 2005. Proliferating cell nuclear antigen recruits cyclin-dependent kinase inhibitor Xic1 to DNA and couples its proteolysis to DNA polymerase switching. J. Biol. Chem. 280:35299-35309. [DOI] [PubMed] [Google Scholar]
- 7.Chuang, L. C., and P. R. Yew. 2001. Regulation of nuclear transport and degradation of the Xenopus cyclin-dependent kinase inhibitor, p27Xic1. J. Biol. Chem. 276:1610-1617. [DOI] [PubMed] [Google Scholar]
- 8.Chuang, L. C., X. N. Zhu, C. R. Herrera, H. M. Tseng, C. M. Pfleger, K. Block, and P. R. Yew. 2005. The C-terminal domain of the Xenopus cyclin-dependent kinase inhibitor, p27Xic1, is both necessary and sufficient for phosphorylation-independent proteolysis. J. Biol. Chem. 280:35290-35298. [DOI] [PubMed] [Google Scholar]
- 9.Daniels, M., V. Dhokia, L. Richard-Parpaillon, and S. Ohnuma. 2004. Identification of Xenopus cyclin-dependent kinase inhibitors, p16Xic2 and p17Xic3. Gene 342:41-47. [DOI] [PubMed] [Google Scholar]
- 10.Furstenthal, L., C. Swanson, B. K. Kaiser, A. G. Eldridge, and P. K. Jackson. 2001. Triggering ubiquitination of a CDK inhibitor at origins of DNA replication. Nat. Cell Biol. 3:715-722. [DOI] [PubMed] [Google Scholar]
- 11.Gulbis, J. M., Z. Kelman, J. Hurwitz, M. O'Donnell, and J. Kuriyan. 1996. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell 87:297-306. [DOI] [PubMed] [Google Scholar]
- 12.Havens, C. G., and J. C. Walter. 2009. Docking of a specialized PIP box onto chromatin-bound PCNA creates a degron for the ubiquitin ligase CRL4Cdt2. Mol. Cell 35:93-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higa, L. A., I. S. Mihaylov, D. P. Banks, J. Zheng, and H. Zhang. 2003. Radiation-mediated proteolysis of CDT1 by CUL4-ROC1 and CSN complexes constitutes a new checkpoint. Nat. Cell Biol. 5:1008-1015. [DOI] [PubMed] [Google Scholar]
- 14.Hu, J., and Y. Xiong. 2006. An evolutionarily conserved function of proliferating cell nuclear antigen for Cdt1 degradation by the Cul4-Ddb1 ubiquitin ligase in response to DNA damage. J. Biol. Chem. 281:3753-3756. [DOI] [PubMed] [Google Scholar]
- 15.Jin, J., E. E. Arias, J. Chen, J. W. Harper, and J. C. Walter. 2006. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol. Cell 23:709-721. [DOI] [PubMed] [Google Scholar]
- 16.Kim, S. H., and W. M. Michael. 2008. Regulated proteolysis of DNA polymerase eta during the DNA-damage response in C. elegans. Mol. Cell 32:757-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, Y., N. G. Starostina, and E. T. Kipreos. 2008. The CRL4Cdt2 ubiquitin ligase targets the degradation of p21Cip1 to control replication licensing. Genes Dev. 22:2507-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwon, C., and I. K. Chung. 2004. Interaction of an Arabidopsis RNA-binding protein with plant single-stranded telomeric DNA modulates telomerase activity. J. Biol. Chem. 279:12812-12818. [DOI] [PubMed] [Google Scholar]
- 19.Lengronne, A., and E. Schwob. 2002. The yeast CDK inhibitor Sic1 prevents genomic instability by promoting replication origin licensing in late G(1). Mol. Cell 9:1067-1078. [DOI] [PubMed] [Google Scholar]
- 20.Leung-Pineda, V., J. Huh, and H. Piwnica-Worms. 2009. DDB1 targets Chk1 to the Cul4 E3 ligase complex in normal cycling cells and in cells experiencing replication stress. Cancer Res. 69:2630-2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin, H. R., L. C. Chuang, H. Boix-Perales, A. Philpott, and P. R. Yew. 2006. Ubiquitination of cyclin-dependent kinase inhibitor, Xic1, is mediated by the Xenopus F-box protein xSkp2. Cell Cycle 5:304-314. [DOI] [PubMed] [Google Scholar]
- 22.Morgan, D. O. 1995. Principles of CDK regulation. Nature 374:131-134. [DOI] [PubMed] [Google Scholar]
- 23.Nam, H. W., R. Simpson, and Y. S. Kim. 2005. N-terminal isotope tagging with propionic anhydride: proteomic analysis of myogenic differentiation of C2C12 cells. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 826:91-107. [DOI] [PubMed] [Google Scholar]
- 24.Nishitani, H., Y. Shiomi, H. Iida, M. Michishita, T. Takami, and T. Tsurimoto. 2008. CDK inhibitor p21 is degraded by a proliferating cell nuclear antigen-coupled Cul4-DDB1Cdt2 pathway during S phase and after UV irradiation. J. Biol. Chem. 283:29045-29052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts, B. T., C. Y. Ying, J. Gautier, and J. L. Maller. 1999. DNA replication in vertebrates requires a homolog of the Cdc7 protein kinase. Proc. Natl. Acad. Sci. U. S. A. 96:2800-2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Russo, A. A., P. D. Jeffrey, A. K. Patten, J. Massague, and N. P. Pavletich. 1996. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature 382:325-331. [DOI] [PubMed] [Google Scholar]
- 27.Sherr, C. J. 1994. G1 phase progression: cycling on cue. Cell 79:551-555. [DOI] [PubMed] [Google Scholar]
- 28.Sherr, C. J., and J. M. Roberts. 1999. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 13:1501-1512. [DOI] [PubMed] [Google Scholar]
- 29.Shibutani, S. T., A. F. de la Cruz, V. Tran, W. J. Turbyfill III, T. Reis, B. A. Edgar, and R. J. Duronio. 2008. Intrinsic negative cell cycle regulation provided by PIP box- and Cul4Cdt2-mediated destruction of E2f1 during S phase. Dev. Cell 15:890-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shou, W., and W. G. Dunphy. 1996. Cell cycle control by Xenopus p28Kix1, a developmentally regulated inhibitor of cyclin-dependent kinases. Mol. Biol. Cell 7:457-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spruck, C. H., K. A. Won, and S. I. Reed. 1999. Deregulated cyclin E induces chromosome instability. Nature 401:297-300. [DOI] [PubMed] [Google Scholar]
- 32.Su, J. Y., R. E. Rempel, E. Erikson, and J. L. Maller. 1995. Cloning and characterization of the Xenopus cyclin-dependent kinase inhibitor p27XIC1. Proc. Natl. Acad. Sci. U. S. A. 92:10187-10191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sutterluty, H., E. Chatelain, A. Marti, C. Wirbelauer, M. Senften, U. Muller, and W. Krek. 1999. p45SKP2 promotes p27Kip1 degradation and induces S phase in quiescent cells. Nat. Cell Biol. 1:207-214. [DOI] [PubMed] [Google Scholar]
- 34.Terai, K., T. Abbas, A. A. Jazaeri, and A. Dutta. 2010. CRL4(Cdt2) E3 ubiquitin ligase monoubiquitinates PCNA to promote translesion DNA synthesis. Mol. Cell 37:143-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsvetkov, L. M., K. H. Yeh, S. J. Lee, H. Sun, and H. Zhang. 1999. p27(Kip1) ubiquitination and degradation is regulated by the SCF(Skp2) complex through phosphorylated Thr187 in p27. Curr. Biol. 9:661-664. [DOI] [PubMed] [Google Scholar]
- 36.Yew, P. R., and M. W. Kirschner. 1997. Proteolysis and DNA replication: the CDC34 requirement in the Xenopus egg cell cycle. Science 277:1672-1676. [DOI] [PubMed] [Google Scholar]