Abstract
Poxviruses produce complement regulatory proteins to subvert the host's immune response. Similar to the human pathogen variola virus, ectromelia virus has a limited host range and provides a mouse model where the virus and the host's immune response have coevolved. We previously demonstrated that multiple components (C3, C4, and factor B) of the classical and alternative pathways are required to survive ectromelia virus infection. Complement's role in the innate and adaptive immune responses likely drove the evolution of a virus-encoded virulence factor that regulates complement activation. In this study, we characterized the ectromelia virus inhibitor of complement enzymes (EMICE). Recombinant EMICE regulated complement activation on the surface of CHO cells, and it protected complement-sensitive intracellular mature virions (IMV) from neutralization in vitro. It accomplished this by serving as a cofactor for the inactivation of C3b and C4b and by dissociating the catalytic domain of the classical pathway C3 convertase. Infected murine cells initiated synthesis of EMICE within 4 to 6 h postinoculation. The levels were sufficient in the supernatant to protect the IMV, upon release, from complement-mediated neutralization. EMICE on the surface of infected murine cells also reduced complement activation by the alternative pathway. In contrast, classical pathway activation by high-titer antibody overwhelmed EMICE's regulatory capacity. These results suggest that EMICE's role is early during infection when it counteracts the innate immune response. In summary, ectromelia virus produced EMICE within a few hours of an infection, and EMICE in turn decreased complement activation on IMV and infected cells.
Poxviruses encode in their large double-stranded DNA genomes many factors that modify the immune system (30, 56). The analysis of these molecules has revealed a delicate balance between viral pathogenesis and the host's immune response (2, 21, 31, 61). Variola, vaccinia, monkeypox, cowpox, and ectromelia (ECTV) viruses each produce an orthologous complement regulatory protein (poxviral inhibitor of complement enzymes [PICE]) that has structural and functional homology to host proteins (14, 29, 34, 38, 41, 45, 54). The loss of the regulatory protein resulted in smaller local lesions with vaccinia virus lacking the vaccinia virus complement control protein (VCP) (29) and in a greater local inflammatory response in the case of cowpox lacking the inflammation-modulatory protein (IMP; the cowpox virus PICE) (35, 45, 46). Additionally, the complete loss of the monkeypox virus inhibitor of complement enzymes (MOPICE) may account for part of the reduced mortality observed in the West African compared to Congo basin strains of monkeypox virus (12).
The complement system consists of proteins on the cell surface and in blood that recognize and destroy invading pathogens and infected host cells (36, 52). Viruses protect themselves from the antiviral effects of complement activation in a variety of ways, including hijacking the host's complement regulatory proteins or producing their own inhibitors (7, 8, 15, 20, 23). Another effective strategy is to incorporate the host's complement regulators in the outermost viral membrane, which then protects the virus from complement attack (62). The extracellular enveloped virus (EEV) produced by poxviruses acquires a unique outer membrane derived from the Golgi complex or early endosomes that contain the protective host complement regulators (58, 62). Poxviruses have multiple infectious forms, and the most abundant, intracellular mature virions (IMV), are released when infected cells lyse (58). The IMV lacks the outermost membrane found on EEV and is sensitive to complement-mediated neutralization. The multiple strategies viruses have evolved to evade the complement system underscore its importance to innate and adaptive immunity (15, 36).
The most well-characterized PICE is VCP (24-29, 34, 49, 50, 53, 55, 59, 60). Originally described as a secreted complement inhibitor (34), VCP also attaches to the surface of infected cells through an interaction with the viral membrane protein A56 that requires an unpaired N-terminal cysteine (26). This extra cysteine also adds to the potency of the inhibitor by forming function-enhancing dimers (41). VCP and the smallpox virus inhibitor of complement enzymes (SPICE) bind heparin in vitro, and this may facilitate cell surface interactions (24, 38, 50, 59). The coevolution of variola virus with its only natural host, humans, likely explains the enhanced activity against human complement observed with SPICE compared to the other PICEs (54, 64).
Our recent work with ECTV, the causative agent of mousepox infection, demonstrated that the classical and alternative pathways of the complement system are required for host survival (48). The mouse-specific pathogen ECTV causes severe disease in most strains and has coevolved with its natural host, analogous to variola virus in humans (9). This close host-virus relationship is particularly important for evaluating the role of the complement system, given the species specificity of many complement proteins, receptors, and regulators (10, 47, 62). Additionally, the availability of complement-deficient mice permits dissection of the complement activation pathways involved. Naïve C57BL/6 mouse serum neutralizes the IMV of ECTV in vitro, predominately through opsonization (48). Maximal neutralization requires natural antibody, classical-pathway activation, and amplification by the alternative pathway. C3 deficiency in the normally resistant C57BL/6 strain results in acute mortality, similar to immunodeficiencies in important elements of the antiviral immune response, including CD8+ T cells (19, 32), natural killer cells (18, 51), and gamma interferon (33). During ECTV infection, the complement system acts in the first few hours and days to delay the spread of infection, resulting in lower levels of viremia and viral burden in tissues (48).
This study characterized the PICE produced by ECTV, ectromelia virus inhibitor of complement enzymes (EMICE), and assessed its complement regulatory activity. Recombinant EMICE (rEMICE) decreased activation of both human and mouse complement. Murine cells produced EMICE at 4 to 6 h postinfection prior to the release of the majority of the complement-sensitive IMV from infected cells. rEMICE protected ECTV IMV from complement-mediated neutralization. Further, EMICE produced during natural infection inhibited complement deposition on infected cells by the alternative pathway. ECTV likely produces this abundance of EMICE to protect both the IMV and infected cells.
MATERIALS AND METHODS
Cell lines and culture.
Chinese hamster ovary cells (CHO; American Type Culture Collection CHO-K1 line) were cultured in Ham's F12 medium supplemented with 10% heat-inactivated (HI) fetal calf serum (FCS), 2 mM l-glutamine, 100 U/ml penicillin G, and 100 μg/ml streptomycin sulfate. A murine fibroblast cell line (L929) and African green monkey kidney cell lines (BS-C-1 and CV-1) were cultured in Dulbecco's modified Eagle's medium (DMEM; BioWhittaker) supplemented with 10% HI-FCS (HyClone). L929 cells were passaged in bacterial petri dishes (Falcon) in normal medium to generate a suspension cell line.
Mouse strains and sera.
The following strains of C57BL/6 mice were acquired: factor B (FB) deficient (43, 63) from H. Molina, Washington University Medical School; C4 deficient (22) from M. Carroll, Harvard Medical School; and wild type (WT) from Jackson Laboratories. For the CHO cell experiments, mouse serum was collected in Eppendorf tubes and clotted on ice for 30 min, and the supernatant was stored at 4°C until use. Sera collected on ice in microtainer tubes (BD) were pooled, aliquoted, and frozen at −70°C until used in IMV, EEV, and infected cell assays. Anti-ECTV sera were collected in microtainer tubes from wild-type mice 4 weeks after intranasal infection with 100 PFU, pooled, heat-inactivated, and frozen in aliquots at −70°C.
rEMICE production.
Using EMICE cDNA (in pSG5) as a template, the coding sequence of EMICE was generated by PCR using the following primers: 5′-CCGGAATTCGGAATGTGCTGTACTATTCCGTCACG-3′ and 5′-ATAAGAATGCGGCCGCTTATTCGCGTACACATTTTGG-3′. The resulting PCR fragment was ligated into the EcoRI and NotI sites of pET28a-2 (38), a derivative of pET28a (EMD/Novagen) generated in our laboratory. Recombinant protein was produced using previously described methods (38).
EMICE quantification and detection.
rEMICE was quantified using carbonic anhydrase (C5024; Sigma) as a standard on a Coomassie-stained sodium dodecyl sulfate-polyacrylamide gel (SDS-PAG). EMICE in tissue culture samples was compared to rEMICE via Western blotting (41). Nonreduced samples were electrophoresed in a 12% SDS-PAG (Invitrogen), transferred to nitrocellulose membranes, and probed with a 1:5,000 dilution of a previously described rabbit anti-VCP antibody (41), followed by horseradish peroxidase (HRP)-goat anti-rabbit IgG (GE Healthcare). rEMICE and rSPICE were detected via flow cytometry (38). Cells were incubated with a 1:2,000 dilution of rabbit anti-VCP for 30 min at 4°C followed by fluorescein isothiocyanate (FITC)-donkey anti-rabbit IgG secondary antibody (Sigma) diluted 1:100 for 30 min at 4°C. After a washing step, the cells were resuspended in 0.5% paraformaldehyde and analyzed with a BD Biosciences FACSCalibur system.
In vitro complement regulatory assays.
Binding to C3b and C4b was detected using a previously described enzyme-linked immunosorbent assay (ELISA) format (39, 41) in at least three separate experiments. Decay-accelerating assays for the classical pathway C3 convertase were performed as previously described (41) four times under each condition in duplicate. The cofactor assays were performed a minimum of three times under each condition in duplicate as previously described (40, 41).
Complement challenge assay.
The standard procedure for initiating the complement pathways (4) was modified to use mouse serum as the source of complement. CHO cells (about 70% confluence) were freed using cell detachment buffer (4 mM EDTA [Sigma] and 10% HI-FCS in phosphate-buffered saline [PBS]), collected, and washed in PBS-1% HI-FCS. CHO cells (1 × 106/well) in a V-bottom 96-well plate were mixed with rEMICE or rSPICE diluted in PBS-1% HI-FCS to the concentrations indicated in the figure legends (100 μl final volume) and incubated at 30°C for 30 min (300 rpm). An Eppendorf Thermomixer was used for all incubations. Following incubation, cells were placed on ice and washed twice. Unless otherwise indicated, cells were washed with PBS-1% HI-FCS and centrifuged at 466 × g for 5 min. The sensitizing antibody (38), IgG from rabbits injected with whole CHO cells (Harlan Bioproducts for Science), was diluted in PBS-1% HI-FCS and added to the cells at 1 mg/ml, and the mixture was incubated for 20 min at 4°C (600 rpm).
For complement activation, the cells were washed twice in 100 μl and resuspended in 50 μl of gelatin veronal buffer with Ca++/Mg++ ([GVB++] G-6514; Sigma). An equal volume of freshly isolated mouse serum, diluted to a 20% concentration in the same buffer, was added and mixed thoroughly. After a 45-min incubation at 37°C (300 rpm), the samples were washed twice. Cells were stained in 100 μl of FITC-conjugated goat F(ab′)2 fragment to mouse C3 (Cappel 55510) diluted 1:200 in PBS-1% HI-FCS for 30 min at 4°C (600 rpm), washed twice, and resuspended in 0.5% paraformaldehyde in PBS. C3 deposition was detected by flow cytometry, and the geometric mean was used for calculations. The dose-response experiment used wild-type serum diluted to a 20% final concentration in GVB without Ca++/Mg++ ([GVB°] B103; CompTech) with added 7 mM MgCl2 and 10 mM EGTA, which limits activation to the alternative pathway. rEMICE and rSPICE experiments used C4-deficient serum at a final concentration of 10 to 40% and varied the sensitizing antibody from 1 to 4 mg/ml.
Virus production and culture.
Plaque-purified Moscow strain ECTV was used to generate the EMICE-deficient (ΔEMICE) virus. The left- and right-flanking segments of the EMICE gene (017) were selected to produce a central 600-bp deletion (11), where 017 designates the open reading frame of EMICE in the ECTV genome (11). The segments were amplified by PCR using EVM017 LF-5′ (GCGGGCGCCGTGGAGTTTATACCACGTATGAG) with EVM017 LF-3′ (GCGACGCATTGCGTCGACGCTAGCGGACGTGACGGAATAGTACAG) and EVM017 RF-5′ (GCGACCGTACTCGAGGCGGCCGCAAGCTTGATCATACTCATACAAGCACAATG) with EVM017 RF-3′ (GCGGAATTCCGTATCTCCGACAAGCACGTAG) and then ligated into pUCP7.5-gpt-1 to yield pNCEV017. This plasmid was recombined into ECTV as previously described (17). Briefly, six-well plates of CV-1 cells were infected with ECTV (5 × 104 PFU/well) and then transfected with 2 μg of pNCEV017 using Lipofectamine 2000 (Invitrogen). After 48 h, cell lysates were collected, and isolates were subjected to two rounds of plaque purification in the presence of mycophenolic acid, followed by three rounds without mycophenolic acid.
The EMICE gene was reintroduced to ΔEMICE using a similar protocol and a 10-kb PCR product from genomic DNA (bp 23179 to 33190). Crude ΔEMICE stock (4 × 106 PFU) was combined with 2 μg of psoralen, 120 μg of bovine serum albumin, and DMEM to a total volume of 1 ml. After a 10-min incubation, the mixture was exposed to a UV lamp in a 12-well tissue culture plate and applied to a BS-C-1 monolayer. The 10-kb PCR product containing the wild-type EMICE gene and the ΔEMICE viral DNA were transfected into the cells with Lipofectamine 2000 at a 40:1 molar ratio (4 ng total). The resulting virus was collected and subjected to four rounds of plaque purification on BS-C-1 cells. Multiple plaques were isolated at each round and screened for the restoration of EMICE by PCR using the primers EVM 017 LF-5′ and EVM 017 RF-3′. Viral DNA for PCR was isolated from infected BS-C-1 cells using a DNeasy blood and tissue kit (cultured cell protocol; Qiagen). Western blotting confirmed that EMICE was produced by the rescue virus (+EMICE ECTV).
Plaque-purified ECTV strains were propagated in murine L929 cells. IMV stocks were purified through a sucrose cushion as described previously (16) and titrated on BS-C-1 cells (13). A single stock of each virus was aliquoted, titrated, and used for all experiments.
In the EMICE production studies, 24-well plates of L929 cell cultures (106 cells/well) were infected at a multiplicity of infection (MOI) of 1 with ΔEMICE or +EMICE ECTV for 1 h. Each well was washed with 1 ml of 37°C DMEM two times. Finally, 1,000 μl of DMEM was added to each well, and this marked the 0-h time point. At each time point, 800 μl of supernatant was collected for analysis, and the remaining supernatant was discarded. The cell layer was washed two times with 1 ml of DMEM; 100 μl of DMEM was added, and the cells were scraped off, resuspended, and collected. The viral titer was determined in 200 μl of supernatant. At early time points, 500 μl of supernatant was concentrated 10-fold in a Millipore concentrator unit (UFV5BCC25). All samples were stored at −70°C. Frozen samples were thawed, mixed 1:1 with Laemmli sample buffer (Bio-Rad), boiled for 10 min, and analyzed by Western blotting.
IMV neutralization assay.
The previously used procedure (48) with the following modifications was used to evaluate the effect of rEMICE on IMV neutralization by mouse serum. rEMICE or a control protein was serially diluted 1:10 in GVB° (B103; CompTech), producing a final volume of 20 μl. Wild-type mouse serum was thawed and diluted to 60% in GVB°, and 10 μl of this was added to the protein-containing tubes. Purified wild-type ECTV IMV was prepared (48), and an equal volume of virus (30 μl, or ≈150 PFU) was added rapidly using an Eppendorf Repeater Plus to the diluted complement/protein at room temperature. Samples were mixed and incubated at 37°C, and infectious virus was detected as plaques on a BS-C-1 monolayer as previously described (48). The data were normalized by setting the HI-serum control as 0% neutralization and 0 plaques as 100% neutralization.
EEV neutralization assay.
Fresh EEVs were produced prior to each experiment by infecting murine L929 cells with ΔEMICE and +EMICE ECTV at an MOI of 0.1 to 0.2 for at least 1 hour. Cells were washed twice with warm PBS (37°C), covered with warm DMEM without added FCS, and incubated at 37°C for 2 days. Harvested culture supernatants were centrifuged at 400 × g for 10 min to remove detached cells and debris. Anti-L1R monoclonal antibody (NR-417 [BEI Resources], similar to VMC-2 [3]) was diluted in the culture supernatant (1:20) to neutralize the IMV. Prior to neutralization, the titer was on average 4 × 104 PFU/ml with about 20% EEVs. The samples were kept on ice until needed.
Thawed mouse serum with or without heat inactivation (5 μl) and heat-inactivated anti-ΔEMICE ECTV serum (1 μl) were diluted in GVB°, producing a final volume of 30 μl on ice, and then vortexed and centrifuged. Culture supernatant with anti-L1R antibody (20 μl) was added rapidly to the diluted complement using an Eppendorf Repeater Plus. Final concentration for serum was 10% and for anti-L1R antibody was 20 μg/ml, which neutralizes >99% of purified IMV. Unlike the IMV assay, samples were not vortexed before or after the incubation at 37°C. Otherwise, the samples were incubated, diluted with medium, applied to monolayer, and stained to detect plaques as described above. The data were normalized to the sample without serum added, which had on average 160 plaques per well.
Infected cell complement challenge assay.
Suspension L929 cells were passed through a 40-μm-pore-size strainer, centrifuged (500 × g for 5 min was used throughout experiment), and resuspended in DMEM-10% HI-FCS. Cells were infected with ΔEMICE or +EMICE ECTV at an MOI of 5 and aliquoted into a V-bottom 96-well plate (1 × 105 to 2 × 105 cells/well). At 10 h postinfection, supernatant was collected after centrifugation for a few samples, and the corresponding cells were transferred to Eppendorf tubes, washed with 1 ml of PBS, and resuspended in a small volume of PBS for Western blotting. Otherwise, the cells were washed with 200 μl of PBS-1% HI-FCS, centrifuged again, and resuspended in PBS-1% HI-FCS (50 μl/well) with or without a 1:10 dilution of anti-ΔEMICE ECTV serum. Samples were incubated on ice at 100 rpm (Innova 2000 Platform Shaker) for 20 min in the dark. Afterwards, 100 μl of PBS-1% HI-FCS was added prior to centrifugation, and this was followed by a second wash with 200 μl of PBS-1% HI-FCS. After centrifugation, cells were resuspended in 50 μl of mouse serum diluted 1:10 in GVB++ on ice. The plate was incubated in a 37°C water bath for 2 min and then transferred to a rotating platform (Nutator) for 30 min at 37°C. The cells were washed as before and resuspended in 50 μl of a 1:200 dilution of FITC-conjugated goat F(ab′)2 fragment to mouse C3 (Cappel 55510) in PBS-1% HI-FCS. After incubation on ice at 100 rpm for 20 min in the dark, the cells were washed as before and resuspended in PBS-0.5% paraformaldehyde. C3 deposition was measured using a FACSCalibur system and analyzed with FlowJo software, version 7.5, for Windows.
Statistical analysis.
All statistical analysis was performed using GraphPad Prism software, version 5.01 (GraphPad Software). The CHO complement deposition and EEV neutralization were analyzed with an unpaired t test (two-tailed). The 50% effective concentration (EC50) was determined using nonlinear regression [log(agonist) versus normalized response (variable slope)]. Complement deposition on the infected cells was analyzed with one-way analysis of variance (ANOVA), followed by the Tukey multiple comparison test.
RESULTS
Production of rEMICE.
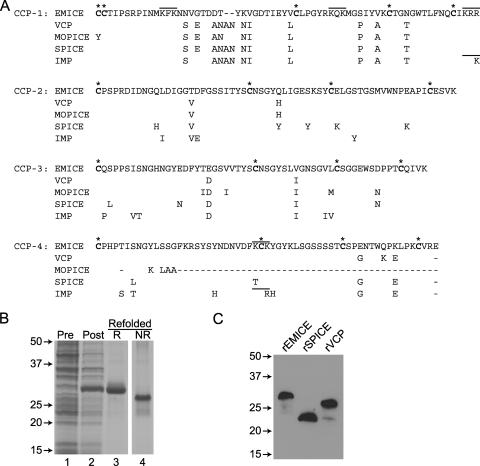
EMICE has approximately 90% sequence identity to the other PICEs at the amino acid level, and all possess a similar structure consisting of four complement control protein modules (CCP-1 to CCP-4) (Fig. 1 A). EMICE most closely resembles VCP (difference of 18 amino acids [aa] and a 2-aa deletion) and MOPICE (difference of 19 aa prior to the frameshift mutation and a 1-aa deletion), then SPICE (difference of 26 aa and a 2-aa deletion), and finally IMP (difference of 29 aa and a 2-aa deletion).
FIG. 1.
Comparison of the PICEs. (A) Protein sequence of EMICE was compared to sequences of PICEs from vaccinia (VCP), monkeypox (MOPICE), variola (SPICE), and cowpox (IMP) viruses. The differences are noted below the EMICE sequence. Each line represents a single CCP module, which contains four conserved cysteines that pair Cys1 with Cys3 and Cys2 with Cys4 (27). Asterisks and boldface mark the cysteines. The bars denote heparin binding sites (K/R-X-K/R) below. A single base pair deletion in CCP-4 of MOPICE produces a truncated protein. Sequence does not include the signal peptide. (B) rEMICE produced in an E. coli expression system. Bacterial samples preinduction (lane 1) and postinduction with IPTG (lane 2) along with rEMICE refolded from the inclusion bodies (lanes 3 and 4) were electrophoresed on a 12% SDS-PAG and stained with Coomassie blue. Samples in lanes 1 to 3 were reduced (R), while lane 4 contained nonreduced (NR) rEMICE. Numbers to the left indicate the relative electrophoretic mobility (Mr × 10−3). (C) Western blotting of rPICEs. Samples (0.1 ng) were electrophoresed on 12% SDS-PAG under nonreducing conditions. After transfer to nitrocellulose, blots were developed using a polyclonal rabbit anti-VCP antiserum. Numbers to the left indicate the relative electrophoretic mobility (Mr × 10−3).
Of these PICEs, EMICE has the greatest number of unique residues, with two-thirds (10 of 15) occurring in CCP-1 where the other PICEs are nearly identical. The majority (five of nine) of unique residues in SPICE are located in CCP-2, and some of them mediate SPICE's enhanced activity against human complement compared to VCP's activity (25, 42, 64). IMP has a similar number of unique residues compared to EMICE, but they are concentrated in CCP-3 and CCP-4 (10 of 14). In contrast, only one amino acid in VCP is not found in the other PICEs, and only a few residues are unique to MOPICE prior to the frameshift mutation.
Similar to our previous studies with recombinant PICEs (rVCP, rMOPICE, and rSPICE) (38, 41, 42), we synthesized rEMICE in Escherichia coli (Fig. 1B). Isopropyl-β-d-thiogalactopyranoside (IPTG) induction produced a single band, rEMICE (Fig. 1B, lane 2). The protein was refolded from purified inclusion bodies. The reduced protein migrated more slowly than the nonreduced form (Fig. 1B, lane 3 versus lane 4) as each CCP is held together by two disulfide bonds (27). rEMICE was compared to rSPICE and rVCP by Western blotting (Fig. 1C). rSPICE migrated faster than rVCP, as previously noted (41, 42, 54). A single residue in CCP-3 produces this difference, and when this residue is mutated to match the rest of the PICEs (L131S), rSPICE migrates in the same manner as rVCP (42). rVCP migrated slightly faster than rEMICE.
Characterization of rEMICE's complement regulatory activity.
We analyzed rEMICE's regulatory activity in assays previously used to evaluate the activity of rVCP, rMOPICE, and rSPICE against human complement components (41). Throughout these experiments, rEMICE was compared to rSPICE, a potent regulator of the human complement system, as a positive control.
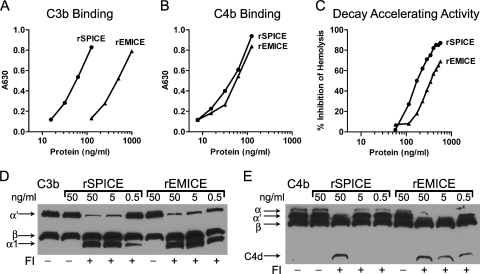
The PICEs must first bind C3b and C4b, components of the C3 convertases, to regulate complement activation. rEMICE's interaction with these two opsonic cleavage fragments, which are also part of the convertases, was assessed using an established ELISA protocol (39, 41). In the case of human C3b (Fig. 2 A), rEMICE bound approximately 10-fold less than rSPICE (EC50 of 45 ng/ml [1.8 nM] versus 5.4 ng/ml [0.2 nM]), whereas they bound C4b similarly (Fig. 2B) (EC50 of 5.2 ng/ml [0.21 nM] versus 3.9 ng/ml [0.14 nM]). rSPICE has the highest affinity for C3b of the PICEs tested (41) and binds C3b and C4b approximately equally (Fig. 2A and B). The higher affinity for C4b than for C3b found with rEMICE was also observed for rMOPICE and rVCP (about 5- and 30-fold, respectively) (41). Upon comparison to published data (41), rEMICE's binding ability most closely resembled that of rMOPICE and was greater than that of rVCP, which is about 100-fold less than that of rSPICE.
FIG. 2.
rEMICE bound to and regulated C3 convertase components C3b and C4b. (A and B) rEMICE's ability to interact with human C3b (A) or C4b (B) was measured using an ELISA format. (C) The classical pathway C3 convertase was assembled on the surface of antibody-coated erythrocytes using purified human complement components. The PICEs' ability to dissociate the convertase was detected as decreased hemolysis once the cells were exposed to the terminal pathway complement components. (D and E) Comparison of rEMICE and rSPICE cofactor activities against C3b and C4b. Factor I (FI) inactivates C3b by cleaving the α′ chain into two fragments, α1 and α2. Similarly, factor I cleaves the α′ chain of C4b twice to release C4d from C4c. The β chain remains intact.
Given the efficient binding to C4b and previous reports of decay-accelerating activity being mediated by the PICEs (41, 49, 64), we determined whether rEMICE dissociates (decays) the subunits that compose the human classical pathway C3 convertase (Fig. 2C). In this experimental method, the convertase (C4bC2a) was assembled on the surface of antibody-coated erythrocytes using purified human complement components. These cells were then incubated with the regulatory protein, followed by C3 and the late components of the complement system. If the regulatory protein efficiently dissociates the convertases, then there is less hemolysis of the cells. rEMICE decayed the human classical-pathway C3 convertase, with about 50% of the activity of rSPICE in this regard.
In contrast to decay-accelerating activity, cofactor activity irreversibly inactivates the convertases. Factor I, in the presence of a cofactor like the PICEs, cleaves the α′ chain of C3b or C4b, and the resulting cleavage fragments no longer can form a convertase. rEMICE's ability to serve as a cofactor protein was assessed by combining it with the protease factor I (human) and biotinylated human C3b or C4b (Fig. 2D or E, respectively). rEMICE serves as a cofactor protein for the cleavage of C3b and C4b at amounts as low as 0.5 ng/ml. No cleavage occurred in the absence of factor I.
rEMICE bound CHO cells and protected against mouse complement.
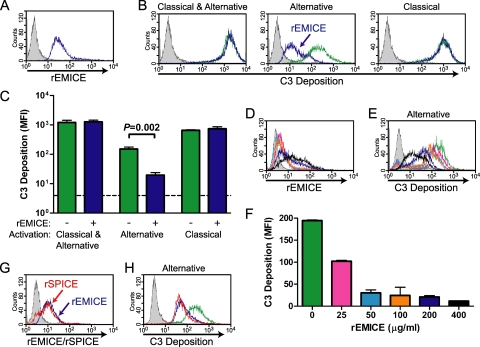
The three heparin binding sites that mediate SPICE's interaction with the surface of cells and an additional site in CCP-4 of VCP are conserved in EMICE (24, 38, 41, 50, 59) (Fig. 1A). We evaluated whether secreted EMICE could limit complement deposition on infected cells in a CHO cell model. rEMICE bound to the cell surface (Fig. 3 A) similarly to rSPICE (38) and the human regulator factor H (44). This property enabled us to assess how well rEMICE regulated mouse complement on a cell surface.
FIG. 3.
rEMICE bound to cells and regulated the mouse alternative complement pathway. (A) rEMICE bound to CHO cells. CHO cells were mixed with rEMICE (200 μg/ml), and rEMICE was detected with a polyclonal rabbit anti-VCP antibody (blue). The gray peak represents cells not exposed to rEMICE. (B and C) rEMICE significantly reduced C3 deposition by the alternative pathway. CHO cells (green) or CHO cells with bound rEMICE (blue) were sensitized with anti-CHO antibody and then exposed to sera from wild-type, C4-deficient, or FB-deficient mouse strains. In wild-type serum, both the classical and alternative pathways were active. When activation was limited to only the alternative pathway by using C4-deficient serum, rEMICE decreased C3 deposition, as indicated by the arrow. To limit activation to the classical pathway, FB-deficient serum was used. The gray peak represents cells not exposed to serum. C3 deposition was measured as mean fluorescence intensity (MFI). The bar graph in panel C displays the mean ± standard error of the mean from two experiments performed in duplicate, with representative images from one experiment shown in panel B. (D to F) rEMICE regulated the alternative pathway in a dose-dependent manner. CHO cells were mixed with increasing concentrations (μg/ml) of rEMICE (25, pink; 50, light blue; 100, orange; 200, dark blue; 400, black) or not exposed to rEMICE (green). (D) rEMICE binding as in panel A. (E and F) Cells were exposed to wild-type mouse sera in the presence of Mg++ EGTA, which limits activation to the alternative pathway. C3 deposition was measured, displayed, and quantitated as described above for panels B and C. (G and H) rEMICE and rSPICE regulated mouse complement similarly. rEMICE (blue) and rSPICE (red) were mixed with CHO cells at 50 μg/ml, and the level of binding was analyzed as described in panel A. C3 deposition on the cells was measured after exposure to sera from C4-deficient mice as described in panel B. The data shown are representative of five separate experiments in which the concentration of sensitizing antibody (4-fold range) and of serum (10 to 40%) and the quantity of rEMICE (4-fold range) were varied. In each case, the degree of inhibition by rEMICE or rSPICE was similar.
CHO cells, with or without prior exposure to rEMICE, were mixed with an anti-CHO antibody, and then wild-type, C4-deficient, and FB-deficient mouse sera were added. Flow cytometry was employed to measure the quantity of C3 fragments deposited (Fig. 3B and C). Classical pathway activation (wild-type or FB−/− serum) by the antibody in this assay most closely mimics an infection after the production of specific antiviral IgG antibody. In this setting, rEMICE did not impact C3 deposition. When only the alternative pathway was activated (C4−/− serum), however, rEMICE significantly reduced C3 deposition by about 90% (P = 0.002) (Fig. 3C). Varying the concentration of rEMICE (Fig. 3D) demonstrated that rEMICE had dose-dependent activity against the alternative pathway (Fig. 3E and F). Comparing rSPICE to rEMICE across a range of conditions showed that both proteins had similar abilities to regulate activation of the alternative pathway in mouse serum (Fig. 3G and H).
Generation of EMICE-deficient virus.
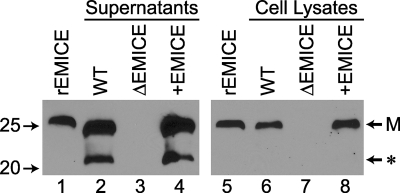
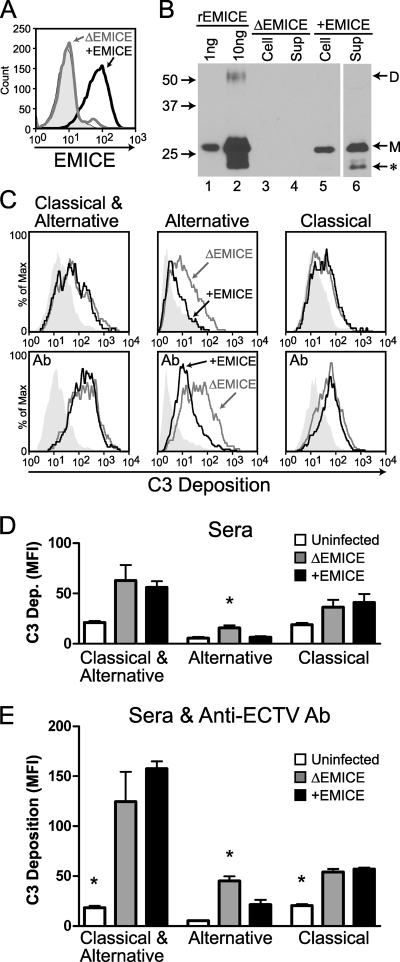
To address the role of EMICE during viral infection, an EMICE-deficient (ΔEMICE) ECTV was produced by deleting about 600 bp from the middle of the EMICE gene in the wild-type (WT) Moscow strain of ECTV. The functional EMICE gene was restored in the ΔEMICE strain to produce the rescue strain (+EMICE). Clones lacking or containing the wild-type EMICE gene were selected by PCR, and EMICE production was confirmed by Western blotting (Fig. 4).
FIG. 4.
Generation of EMICE-deficient (ΔEMICE) ECTV. Supernatants (lanes 2 to 4) and cell lysates (lanes 6 to 8) were collected from L929 cells infected with the three viral strains listed above the gel at an MOI of 1 and analyzed by Western blotting. EMICE-deficient ECTV (ΔEMICE) was generated from wild-type ECTV (WT) using a construct with a 600-bp deletion in the middle of the EMICE gene. The wild-type genomic sequence was added back to ΔEMICE to create the rescue strain (+EMICE). EMICE-producing clones were selected by PCR and confirmed by Western blotting. rEMICE (1 ng) in lanes 1 and 5 served as a positive control. The EMICE monomer (M) and a smaller antigenically related protein of unknown significance (*) are indicated. Numbers to the left indicate the relative electrophoretic mobility (Mr × 10−3).
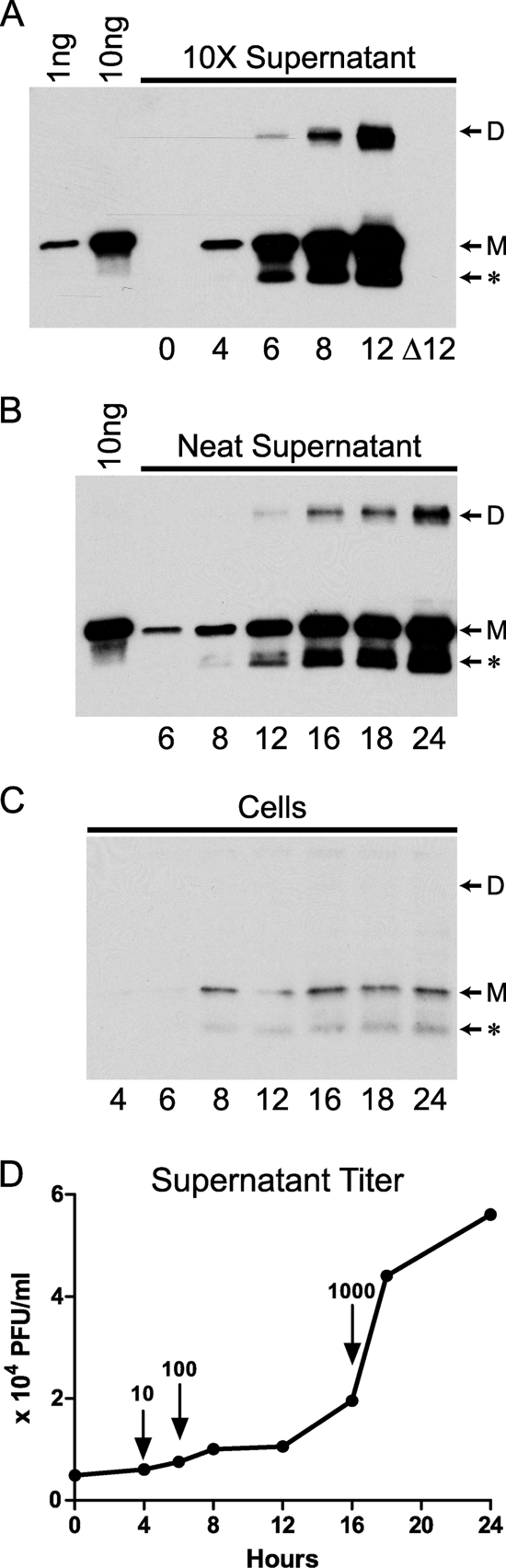
High levels of secreted EMICE preceded the release of IMV from infected cells.
To understand when an infected cell synthesizes EMICE, murine L929 cells were infected in vitro with +EMICE ECTV at an MOI of 1, washed, and then cultured in serum-free medium. The kinetics of EMICE production was assessed in the supernatants and cells via Western blotting. Given the low levels of EMICE observed at the earliest time points in prior experiments, the supernatants at initial time points were concentrated 10-fold (Fig. 5 A). The blots of concentrated or neat supernatant show the EMICE contained in 10% or 1% of the total supernatant, respectively (Fig. 5A and B). To enable comparison, 10% of the total cells were also analyzed (Fig. 5C). The quantity of EMICE produced was approximated using rEMICE as a standard. The effect of Ara-C on EMICE production was also evaluated to determine whether EMICE is an early or late protein. Ara-C (25 μg/ml) inhibited EMICE production at 8 h postinfection, which indicates that EMICE is a late protein (data not shown).
FIG. 5.
EMICE synthesis by infected cells began 4 to 6 h postinfection and reached high levels prior to the release of IMV. The production of EMICE was followed over time in murine L929 cells infected at an MOI of 1. After infection for 1 h, the cells were rinsed to remove free virus and then cultured in 1,000 μl of medium. The EMICE produced was assessed via Western blotting in 10-fold-concentrated supernatants (10% of total) (A), unconcentrated supernatants (1% of total) (B), or washed cells (10% of total) (C). The blots shown were developed on the same film to facilitate comparison. rEMICE is labeled with the amount loaded (1 or 10 ng). The numbers (0 to 24) below indicate the number of hours postinfection when the samples were obtained. The delta (Δ) indicates ECTV with a deletion of EMICE. The forms of EMICE are indicated: dimer (D), monomer (M), and smaller antigenically related protein (*), which was previously described in vaccinia virus culture supernatants (41). (D) The supernatant was titrated on BS-C-1 cells at the time of collection. The concentration of EMICE in the supernatant (A and B) was based on comparison to rEMICE in multiple exposures and appears above the arrows (ng/ml).
EMICE was undetectable in the supernatant (Fig. 5A) and cells (data not shown) at 0 h and in the samples from the ΔEMICE virus (Fig. 5A), which served as a control for potential cross-reaction of the antibody with other viral proteins. In the concentrated supernatants at 4 h, greater than 1 ng of EMICE was detected, indicating that the concentration was at least 10 ng/ml. Over the next 2 h, the concentration of EMICE increased 10-fold to 100 ng/ml, as indicated by the 10 ng in the concentrated sample (Fig. 5A) and about 1 ng in the unconcentrated supernatant (Fig. 5B). An additional 10-fold increase in EMICE concentration to 1,000 ng/ml occurred between 12 and 16 h since there was greater than 10 ng of EMICE in the unconcentrated supernatant at 16 h (Fig. 5B). On average, each cell infected at an MOI of 1 had produced 1 pg of EMICE by 16 h. In contrast, minimal quantities of EMICE were detected in or on the cells at all time points (Fig. 5C), which indicates that in this system EMICE is predominately a secreted protein.
The largest increase in infectious virus in the supernatant occurred between 16 and 18 h postinfection (Fig. 5D). In other experiments evaluating EEV production, the majority of the virus detected at 18 and 24 h postinfection was IMV (data not shown). At 16 h, the EMICE level was 1,000 ng/ml (Fig. 5E), and rEMICE's complement regulatory activity occurred in vitro at lower concentrations than this (Fig. 2). The large quantity of EMICE secreted from infected cells prior to the release of IMV could provide needed protection from complement-mediated destruction for the virus.
Soluble rEMICE protected ECTV IMV.
Mouse complement neutralizes the IMV form of ECTV effectively in vitro, even at serum concentrations as low as 10% (48). Sera from mice genetically deficient in antibody or in either classical- or alternative-pathway components have decreased neutralizing activity (48). Our prior data indicate that natural antibody initiates the classical complement cascade, and the subsequent engagement of the alternative pathway amplifies opsonization, the predominant process of neutralization in wild-type serum (48).
EMICE's ability to protect IMV from complement-mediated neutralization was tested using a plaque reduction assay (Fig. 6). rEMICE was mixed with wild-type mouse serum just prior to the addition of purified IMV, and infectious IMV was detected as plaques. The interleukin-18 (IL-18) binding protein of ECTV was used as a control protein because it was also produced in E. coli and is similar in size. Mouse serum at a final concentration of 10% neutralized 87% of the virus in the absence of recombinant protein. The IL-18 binding protein did not affect neutralization, except slightly at the highest concentration when the IL-18 binding protein and its buffer were 40% of the reaction volume (≤4% under all other conditions).
FIG. 6.
Soluble rEMICE protected IMV from complement-mediated neutralization. Prior to the addition of purified IMV, naïve wild-type C57BL/6 mouse serum was mixed with rEMICE or recombinant ECTV IL-18 binding protein (rIL-18 BP) as a control. Following incubation at a final concentration of 10% serum, the samples were applied to BS-C-1 monolayers. Percent neutralization was calculated by setting zero plaques to 100% neutralization and the average number of plaques in heat-inactivated sera (HI-Sera) to 0% neutralization. The dashed line indicates the level of neutralization by the wild-type serum alone (87%). The rEMICE line represents the mean ± standard error of the mean from two independent experiments, with each condition performed in duplicate.
rEMICE provided greater protection than the control protein with 1,000-fold less protein. At the highest concentration, rEMICE reduced the neutralizing activity of the serum by 86%, or greater than 7-fold (from 87 to 12%). However, protection of 50% of the virus occurred at a lower concentration, an EC50 of 283 ng/ml, as determined by nonlinear regression. This level was about 4-fold lower than the 1 μg/ml concentration of EMICE observed at 16 h in vitro (Fig. 5D). Therefore, the microenvironment of an infected cell likely contains sufficient EMICE to significantly inhibit complement activation.
EMICE not required for EEV resistance to complement-mediated neutralization.
Poxviruses produce a second form of the virus, the EEV, which mediates dissemination over long distances (58). In the case of vaccinia virus, the EEV avoids complement-mediated neutralization by incorporating the host regulators (62) and has greater resistance to antibody-mediated neutralization than IMV (37). To evaluate the role of EMICE on the EEV form, we established a homologous system using only mouse reagents to mimic a natural infection (Fig. 7).
FIG. 7.
EMICE did not impact EEV's resistance to neutralization by complement and antibody. EEV was produced by infecting murine L929 cells at a low MOI with ΔEMICE (gray) or +EMICE (black) ECTV for 2 days. Contaminating IMV in the supernatant was eliminated using a neutralizing anti-L1R monoclonal antibody (3). The EEV was added to heat-inactivated or active mouse serum (10%). Active sera were supplemented with convalescent-phase sera (Ab) from mice infected with ΔEMICE ECTV. Infectious virus was detected as plaques on a BS-C-1 monolayer. The number of plaques was normalized to the samples not exposed to serum. No difference was detected between the ΔEMICE and +EMICE EEV under any condition (P = 0.24, HI-Sera; P = 0.77, Sera; 0.37, P = Sera & Ab). The data shown are means standard errors of the means of four experiments, with each condition performed in triplicate.
EEV from ΔEMICE or +EMICE ECTV was produced in murine L929 cells. The culture supernatant contains EEV as well as “contaminating” IMV, which was neutralized by a monoclonal antibody to the IMV protein L1R (3). At the concentration used (10 μg/ml), this antibody neutralized greater than 99% of purified IMV (data not shown). Convalescent-phase sera from wild-type mice infected with ΔEMICE ECTV were used as a source of anti-ECTV antibody to avoid anti-EMICE antibodies. VCP is immunogenic following vaccination (1), and anti-VCP antibodies block complement regulatory function (28). Finally, sera from the same strain of mice were used as the source of complement.
The ΔEMICE EEV behaved similarly to +EMICE EEV under all conditions tested (Fig. 7). Serum alone or heat-inactivated serum produced a modest reduction viral titer compared to the samples not exposed to serum. Serum supplemented with anti-ECTV antibody neutralized about half of the virions, and this neutralization was independent of EMICE.
EMICE reduced complement activation on infected cells.
To evaluate the importance of EMICE on the surface of infected cells, we again used this homologous system consisting only of mouse reagents. The ability of EMICE to protect the surface of infected cells from complement deposition was evaluated by comparing murine L929 cells infected with ΔEMICE or +EMICE ECTV (Fig. 8 ). Most assays to evaluate complement regulatory proteins on the cell surface, like the experiments employing CHO cells (Fig. 3B and C), use antibody to initiate the complement cascade (38). The same convalescent-phase sera were used as a source of anti-ECTV antibody to enhance complement activation. Finally, sera from wild-type or complement-deficient mouse strains were used as the source of complement. The complement-deficient mouse sera enabled selective activation of the alternative or classical pathways.
FIG. 8.
EMICE protected infected murine cells from alternative-pathway activation in mouse serum. Murine L929 cells were infected at an MOI of 5 with ΔEMICE (gray) or +EMICE (black) ECTV for 10 h. (A) EMICE expression on the cell surface was evaluated by flow cytometry. The shaded peak represents +EMICE virus-infected cells exposed to secondary antibody only. (B) EMICE associated with infected cells was also evaluated by Western blotting. Lanes 1 and 2 contain 1 and 10 ng of rEMICE, respectively, as standards. Lanes 3 and 5 contain 105 washed cells infected with the indicated virus. Lanes 4 and 6 contain the supernatant (Sup) from 104 cells. Numbers to the left indicate the relative electrophoretic mobility (Mr × 10−3), and dimer (D), monomer (M), and smaller antigenically related protein (*) are indicated to the right. (C) Cells infected with ΔEMICE (gray line) or +EMICE (black line) ECTV or left uninfected (shaded peak) were divided, and a subset was incubated with convalescent-phase sera (Ab) from mice infected with ΔEMICE ECTV. The samples were exposed to sera from wild-type, C4-deficient, or FB-deficient strains of mice to evaluate EMICE's ability to regulate complement activation by both pathways, the alternative pathway, or the classical pathway, respectively. C3 deposition was measured using flow cytometry. Representative images from the experiment analyzed in the following panels are shown. EMICE significantly reduced C3 deposition by the alternative pathway in the absence (D) and presence (E) of anti-ECTV antibody. Uninfected (white), ΔEMICE virus-infected (gray) or +EMICE virus-infected (black) cells were treated as described in panel C. An asterisk indicates a statistically significant difference between that cell type and the other two types (P > 0.05, using one-way ANOVA followed by Tukey's multiple comparison test). The data shown are mean ± standard error of the mean from one experiment performed in triplicate and are representative of three independent experiments.
EMICE was detected on the surface of +EMICE ECTV-infected cells by flow cytometry (Fig. 8A) and on or in these cells by Western blotting (Fig. 8B). The cell lysate (Fig. 8B, lane 5) has protein from 105 cells, whereas the supernatant (Fig. 8B, lane 6) has protein from only 104 cells, indicating that only a minority of EMICE was cell associated.
Upon exposure to mouse serum, more C3 was deposited on the cells infected with either ΔEMICE or +EMICE virus than on uninfected cells (Fig. 8C and D). The addition of convalescent-phase serum amplified the difference between the infected and uninfected cells (Fig. 8C and E). Activation of both pathways (wild-type serum) or selective activation of the classical pathway (FB-deficient serum) produced similar levels of C3 deposition on the ΔEMICE and +EMICE ECTV-infected cells (Fig. 8D and E) although more C3 deposition occurred if both pathways were active. Significantly less C3 was deposited on +EMICE virus-infected cells than on ΔEMICE virus-infected cells (80% [P = 0.006] and 57% [P = 0.04] with anti-ECTV antibody) when only the alternative pathway was active (C4-deficient serum) (Fig. 8D and E).
DISCUSSION
In this study, we characterized EMICE and its ability to regulate complement activation in the ECTV model. The ECTV model system is arguably the best poxvirus animal model for studying the role of complement in pathogenesis as ECTV is a natural mouse pathogen, and there is an abundance of reagents and mouse strains available for dissection of the complement activation pathway.
EMICE shares high homology with the other PICEs, with the greatest divergence occurring in CCP-1, which is nearly identical in the PICEs of variola, vaccinia, cowpox, and monkeypox viruses. The first CCP domain of VCP is required for binding to C3b and C4b (49, 53). The concentration of differences (10 of 15 unique residues) in CCP-1 of EMICE likely results from the coevolution of ECTV with its mouse host and improves viral fitness. SPICE and EMICE had similar abilities to regulate complement activation in mouse sera, suggesting that the changes in CCP-1 may influence EMICE's ability to interact with other virus- or host-specific proteins in vivo. Alternatively, natural ECTV infection is not subject to the selective pressures that have preserved CCP-1 in VCP, MOPICE, IMP, and SPICE.
Despite the structural variation, rEMICE's affinity for C3b mirrored what was previously observed with rMOPICE in vitro (41). The two proteins differ by 18 residues, with the majority being in CCP-1, and by the truncation of CCP-4 in MOPICE. Based on this line of reasoning, CCP-2/CCP-3 are the most important for ligand binding, which is consistent with electrostatic modeling (57) as well as a report of function-blocking antibodies that recognize either CCP-2 or CCP-3/CCP-4 (28). Moreover, over half (five of nine) of the unique residues in SPICE are located in CCP-2, and these differences mediate SPICE's enhanced activity for human complement compared to VCP (42, 57, 64).
While PICEs bind both C3b and C4b, they bind C4b with higher affinity (41, 49). Additionally, they have decay-accelerating activity for the C4b-containing convertases generated by the classical or lectin pathways. The pressures that have selected for this enhanced activity against C4 suggest that C4 has an important role in the immune response to a poxviral infection. Since complement activation has antiviral effects that enhance host survival (48), the ability to inactivate C4b would be advantageous for the virus if the classical or lectin pathway initiates complement activation.
Prior in vitro work showed that natural antibody triggers the complement-mediated neutralization of IMV (48). rEMICE effectively prevented this neutralization, which demonstrates EMICE's capacity to inhibit the classical pathway activated by natural antibody. Serum deficient in either classical- or alternative-pathway components retains partial neutralizing capacity (48). The deficient serum neutralizes more IMV than wild-type serum did in the presence of rEMICE, which indicates that EMICE also decreases activation of the alternative pathway. rEMICE's protection of IMV from the complement-mediated neutralization demonstrates EMICE's potential to regulate mouse complement and influence events early during infection.
EMICE is secreted in vitro prior to the release of the majority of IMV, allowing it to protect IMV as they are released. Additionally, EMICE's ability to dimerize to produce a more efficient complement inhibitor (versus the monomer) and to bind to cell surfaces may further enhance its regulatory activity in the microenvironment of the infected cell. These experiments model what may occur in vivo; they demonstrated that infected cells produce large quantities of EMICE, which impedes neutralization of ECTV IMV by the host's innate immune system.
In contrast, the EEV form does not require EMICE to resist complement-mediated neutralization, likely due to protective host regulators in its outermost envelope (62). The modest decrease in titer with the heat-inactivated serum was also previously observed with vaccinia virus (6). The relatively equivalent results after exposing EEV to either active or heat-inactivated serum establishes the resistance of the EEV form to complement-mediated neutralization in a second virus. The prior study that used vaccinia virus found similar resistance to complement when a homologous system (rabbit cells and complement or human cells and complement) was used; conversely, the EEV produced in rabbit cells was readily neutralized (over 90%) by human complement (62). The ability of serum when combined with anti-ECTV antibody to neutralize about half of the EEV demonstrates the activity of the serum. Consistent with published findings (5, 6, 62), the addition of antiviral antibody can overcome the EEV's normal resistance to complement. Taken together, these results highlight the importance of employing a homologous system, preferably from the natural host, in analyzing the importance of complement inhibitors to virulence.
EMICE also reduced complement activation on the cell surface. Similar to rSPICE (38), rEMICE bound to the surface of CHO cells. There, rEMICE protected CHO cells against complement activation by the alternative pathway in an in situ model system employing antibody as the activator. In a logical extension of these studies, we evaluated EMICE's ability to protect infected mouse cells from mouse immune serum and complement. Similarly, L929 cells infected with the virus containing EMICE were protected from complement activation by the alternative pathway. Cofactor activity likely mediates this protection since the other PICEs have limited or absent decay-accelerating activity for the alternative pathway convertases in vitro (41, 49). EMICE's regulation of the alternative pathway and the classical pathway when activated in a less robust manner by natural antibody may help ECTV evade the innate and early adaptive immune responses. Prior data from in vivo experiments using PICE-deficient poxviruses also supports a role for PICEs early during infection. The loss of IMP from cowpox virus (35, 45, 46) or VCP from vaccinia virus (29) affects local inflammation by 3 to 5 days postinfection, prior to the induction of a robust adaptive immune response.
Rabbit IgG directed against CHO cells efficiently engages the classical pathway, and it overwhelmed rEMICE's regulatory capacity. These conditions mimic an infectious state after the adaptive immune response produces abundant antibody. Prior reports using similar methodology showed that rSPICE decreased C3b deposition by the classical pathway by about 50% (38); however, this decrease would likely be insufficient to protect the cells from the detrimental effects of complement activation. Classical pathway activation on ΔEMICE and +EMICE virus-infected cells produced equivalent levels of complement deposition. The impact of the PICEs during infection likely occurs prior to the induction of a strong humoral immune response.
Interestingly, complement deposition occurred on the surface of all infected cells but not on uninfected cells, even before anti-ECTV serum was added. The increased complement activation could result from increased expression of viral antigens that are recognized by natural antibody or from decreased expression of the normal cell surface regulatory proteins. Hypothetically, host cells may decrease production of their regulators during infection as a defense mechanism to increase their susceptibility to complement, or decreased expression may result from the viral takeover of the cells' translational machinery. These interesting hypotheses are currently being explored. Independent of the mechanism, the increased susceptibility of infected cells to complement activation demonstrates the need for viral complement inhibitors.
ECTV infection produces multiple forms of EMICE. The unpaired N-terminal cysteine of VCP economically enables both the formation of dimers with enhanced regulatory activity (41) and an interaction with A56 that tethers the complement inhibitor to the surface of infected cells (26) and possibly to the EEV form as well. The multiple forms of the PICEs appear to be advantageous to the virus as the only PICE that lacks a free N-terminal cysteine, MOPICE, has a free cysteine in the truncated CCP-4 that mediates dimerization (41). The generation of ECTV mutants carrying EMICE lacking the N-terminal cysteine could address the importance of the multiple PICE forms in vivo; however, differentiating between the effects of dimerization versus A56-mediated expression could be difficult. This potential interaction between EMICE and A56 and its contribution to complement resistance as well as virulence in vivo require future investigation.
Vaccination-induced anti-PICE antibodies neutralize these virulence factors (1, 28); however, alone they provide insufficient protection in mice challenged with a lethal dose of vaccinia virus. Passive administration of high-affinity function-blocking antibodies should facilitate viral clearance and may aid recovery. Such antibodies could enhance complement activity by disabling the inhibitor while simultaneously tagging infected cells that express PICEs on their surfaces for destruction by complement or other immune-mediated mechanisms.
In summary, we have characterized the complement-inhibitory profile of EMICE produced recombinantly or by infected cells. In standard complement assays, rEMICE served as cofactor for the cleavage and, thereby, inactivation of C3b and C4b and dissociated the classical pathway C3 convertase. When bound to cells, EMICE protected them from complement attack, especially by the alternative pathway. Immune serum largely overcame this activity. EMICE was produced by infected cells at 4 to 6 h postinfection and in large amounts by 16 h, which would protect IMV from neutralization by complement. These results represent an informative example of the workings of a single virulence factor that inhibits a major player in the innate immune system's response.
Acknowledgments
We thank R. J. Eisenberg and G. H. Cohen for contributing the NR-417 antibody to BEI Resources, Chung Lee for the generous gift of recombinant IL-18 binding protein, Xiaobo Wu for providing fresh mouse sera, Ed Hembrador for growing the viral stocks, Jill Schriewer for generating the suspension L929 cells and for technical assistance producing and analyzing the EMICE rescue virus, M. Kathryn Liszewski for assistance with the Western blotting, and Joy Eslick and Sherri Koehm for help performing and analyzing the flow cytometry data. We additionally thank Scott Parker, Herbert Virgin IV, Michael S. Diamond, Wayne Yokoyama, Hector Molina, and Ted Hansen for helpful discussions, Andrew Zimolzak for reading the manuscript, and Madonna Bogacki for secretarial and administrative assistance.
This work was supported by NIAID grant NOI-AI-15436 and contracts U54-AI-057169 and U54-AI-057160 from the NIAID to the Midwestern Regional Center of Excellence for Biodefense and Emerging Infectious Diseases. Additional support was also provided by NIH T32 HL07317, Principles in Pulmonary Research, and T32 GM07200, National Research Service Award, Medical Scientist.
Footnotes
Published ahead of print on 7 July 2010.
REFERENCES
- 1.Adamo, J. E., C. A. Meseda, J. P. Weir, and M. J. Merchlinsky. 2009. Smallpox vaccines induce antibodies to the immunomodulatory, secreted vaccinia virus complement control protein. J. Gen. Virol. 90:2604-2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alcami, A. 2007. New insights into the subversion of the chemokine system by poxviruses. Eur. J. Immunol. 37:880-883. [DOI] [PubMed] [Google Scholar]
- 3.Aldaz-Carroll, L., J. C. Whitbeck, M. Ponce de Leon, H. Lou, L. K. Pannell, J. Lebowitz, C. Fogg, C. L. White, B. Moss, G. H. Cohen, and R. J. Eisenberg. 2005. Physical and immunological characterization of a recombinant secreted form of the membrane protein encoded by the vaccinia virus L1R gene. Virology 341:59-71. [DOI] [PubMed] [Google Scholar]
- 4.Barilla-LaBarca, M. L., M. K. Liszewski, J. D. Lambris, D. Hourcade, and J. P. Atkinson. 2002. Role of membrane cofactor protein (CD46) in regulation of C4b and C3b deposited on cells. J. Immunol. 168:6298-6304. [DOI] [PubMed] [Google Scholar]
- 5.Benhnia, M. R., M. M. McCausland, J. Laudenslager, S. W. Granger, S. Rickert, L. Koriazova, T. Tahara, R. T. Kubo, S. Kato, and S. Crotty. 2009. Heavily isotype-dependent protective activities of human antibodies against vaccinia virus extracellular virion antigen B5. J. Virol. 83:12355-12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benhnia, M. R., M. M. McCausland, J. Moyron, J. Laudenslager, S. Granger, S. Rickert, L. Koriazova, R. Kubo, S. Kato, and S. Crotty. 2009. Vaccinia virus extracellular enveloped virion neutralization in vitro and protection in vivo depend on complement. J. Virol. 83:1201-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernet, J., J. Mullick, A. K. Singh, and A. Sahu. 2003. Viral mimicry of the complement system. J. Biosci. 28:249-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blue, C. E., O. B. Spiller, and D. J. Blackbourn. 2004. The relevance of complement to virus biology. Virology 319:176-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buller, R. M., and F. Fenner. 2007. Mousepox, p. 67-72. In J. G. Fox, M. T. Davisson, F. W. Quimby, S. W. Barthold, C. E. Newcomer, and A. L. Smith (ed.), The mouse in biomedical research, 2nd ed., vol. 2. Elsevier, New York, NY. [Google Scholar]
- 10.Chaudhri, G., V. Panchanathan, H. Bluethmann, and G. Karupiah. 2006. Obligatory requirement for antibody in recovery from a primary poxvirus infection. J. Virol. 80:6339-6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, N., M. I. Danila, Z. Feng, R. M. Buller, C. Wang, X. Han, E. J. Lefkowitz, and C. Upton. 2003. The genomic sequence of ectromelia virus, the causative agent of mousepox. Virology 317:165-186. [DOI] [PubMed] [Google Scholar]
- 12.Chen, N., G. Li, M. K. Liszewski, J. P. Atkinson, P. B. Jahrling, Z. Feng, J. Schriewer, C. Buck, C. Wang, E. J. Lefkowitz, J. J. Esposito, T. Harms, I. K. Damon, R. L. Roper, C. Upton, and R. M. Buller. 2005. Virulence differences between monkeypox virus isolates from West Africa and the Congo basin. Virology 340:46-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, W., R. Drillien, D. Spehner, and R. M. Buller. 1992. Restricted replication of ectromelia virus in cell culture correlates with mutations in virus-encoded host range gene. Virology 187:433-442. [DOI] [PubMed] [Google Scholar]
- 14.Ciulla, E., A. Emery, D. Konz, and J. Krushkal. 2005. Evolutionary history of orthopoxvirus proteins similar to human complement regulators. Gene 355:40-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cummings, K. L., S. N. Waggoner, R. Tacke, and Y. S. Hahn. 2007. Role of complement in immune regulation and its exploitation by virus. Viral Immunol. 20:505-524. [DOI] [PubMed] [Google Scholar]
- 16.Earl, P. L., B. Moss, L. S. Wyatt, and M. W. Carroll. 2001. Generation of recombinant vaccinia viruses. Curr. Protoc. Mol. Biol. 43:16.17.1-16.17.19. [DOI] [PubMed] [Google Scholar]
- 17.Falkner, F. G., and B. Moss. 1990. Transient dominant selection of recombinant vaccinia viruses. J. Virol. 64:3108-3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang, M., L. L. Lanier, and L. J. Sigal. 2008. A role for NKG2D in NK cell-mediated resistance to poxvirus disease. PLoS Pathog. 4:e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang, M., and L. J. Sigal. 2005. Antibodies and CD8+ T Cells are complementary and essential for natural resistance to a highly lethal cytopathic virus. J. Immunol. 175:6829-6836. [DOI] [PubMed] [Google Scholar]
- 20.Favoreel, H. W., G. R. Van de Walle, H. J. Nauwynck, and M. B. Pensaert. 2003. Virus complement evasion strategies. J. Gen. Virol. 84:1-15. [DOI] [PubMed] [Google Scholar]
- 21.Finlay, B. B., and G. McFadden. 2006. Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell 124:767-782. [DOI] [PubMed] [Google Scholar]
- 22.Fischer, M., M. Ma, S. Goerg, X. Zhou, J. Xia, O. Finco, S. Han, G. Kelsoe, R. Howard, T. Rothstein, E. Kremmer, F. Rosen, and M. Carroll. 1996. Regulation of the B cell response to T-dependent antigens by classical pathway complement. J. Immunol. 157:549-556. [PubMed] [Google Scholar]
- 23.Fries, L. F., H. M. Friedman, G. H. Cohen, R. J. Eisenberg, C. H. Hammer, and M. M. Frank. 1986. Glycoprotein C of herpes simplex virus 1 is an inhibitor of the complement cascade. J. Immunol. 137:1636-1641. [PubMed] [Google Scholar]
- 24.Ganesh, V. K., S. K. Muthuvel, S. A. Smith, G. J. Kotwal, and K. H. Murthy. 2005. Structural basis for antagonism by suramin of heparin binding to vaccinia complement protein. Biochemistry 44:10757-10765. [DOI] [PubMed] [Google Scholar]
- 25.Ghebremariam, Y. T., O. O. Odunuga, K. Janse, and G. J. Kotwal. 2005. Humanized recombinant vaccinia virus complement control protein (hrVCP) with three amino acid changes, H98Y, E102K, and E120K creating an additional putative heparin binding site, is 100-fold more active than rVCP in blocking both classical and alternative complement pathways. Ann. N. Y. Acad. Sci. 1056:113-122. [DOI] [PubMed] [Google Scholar]
- 26.Girgis, N. M., B. C. Dehaven, X. Fan, K. M. Viner, M. Shamim, and S. N. Isaacs. 2008. Cell surface expression of the vaccinia virus complement control protein is mediated by interaction with the viral A56 protein and protects infected cells from complement attack. J. Virol. 82:4205-4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henderson, C. E., K. Bromek, N. P. Mullin, B. O. Smith, D. Uhrin, and P. N. Barlow. 2001. Solution structure and dynamics of the central CCP module pair of a poxvirus complement control protein. J. Mol. Biol. 307:323-339. [DOI] [PubMed] [Google Scholar]
- 28.Isaacs, S. N., E. Argyropoulos, G. Sfyroera, S. Mohammad, and J. D. Lambris. 2003. Restoration of complement-enhanced neutralization of vaccinia virus virions by novel monoclonal antibodies raised against the vaccinia virus complement control protein. J. Virol. 77:8256-8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Isaacs, S. N., G. J. Kotwal, and B. Moss. 1992. Vaccinia virus complement-control protein prevents antibody-dependent complement-enhanced neutralization of infectivity and contributes to virulence. Proc. Natl. Acad. Sci. U. S. A. 89:628-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnston, J. B., and G. McFadden. 2003. Poxvirus immunomodulatory strategies: current perspectives. J. Virol. 77:6093-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnston, J. B., and G. McFadden. 2004. Technical knockout: understanding poxvirus pathogenesis by selectively deleting viral immunomodulatory genes. Cell Microbiol. 6:695-705. [DOI] [PubMed] [Google Scholar]
- 32.Karupiah, G., R. M. Buller, N. Van Rooijen, C. J. Duarte, and J. Chen. 1996. Different roles for CD4+ and CD8+ T lymphocytes and macrophage subsets in the control of a generalized virus infection. J. Virol. 70:8301-8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karupiah, G., T. N. Fredrickson, K. L. Holmes, L. H. Khairallah, and R. M. Buller. 1993. Importance of interferons in recovery from mousepox. J. Virol. 67:4214-4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kotwal, G. J., S. N. Isaacs, R. McKenzie, M. M. Frank, and B. Moss. 1990. Inhibition of the complement cascade by the major secretory protein of vaccinia virus. Science 250:827-830. [DOI] [PubMed] [Google Scholar]
- 35.Kotwal, G. J., C. G. Miller, and D. E. Justus. 1998. The inflammation modulatory protein (IMP) of cowpox virus drastically diminishes the tissue damage by down-regulating cellular infiltration resulting from complement activation. Mol. Cell. Biochem. 185:39-46. [DOI] [PubMed] [Google Scholar]
- 36.Lambris, J. D., D. Ricklin, and B. V. Geisbrecht. 2008. Complement evasion by human pathogens. Nat. Rev. Microbiol. 6:132-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Law, M., and G. L. Smith. 2001. Antibody neutralization of the extracellular enveloped form of vaccinia virus. Virology 280:132-142. [DOI] [PubMed] [Google Scholar]
- 38.Liszewski, M. K., P. Bertram, M. K. Leung, R. Hauhart, L. Zhang, and J. P. Atkinson. 2008. Smallpox inhibitor of complement enzymes (SPICE): regulation of complement activation on cells and mechanism of its cellular attachment. J. Immunol. 181:4199-4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liszewski, M. K., M. Leung, W. Cui, V. B. Subramanian, J. Parkinson, P. N. Barlow, M. Manchester, and J. P. Atkinson. 2000. Dissecting sites important for complement regulatory activity in membrane cofactor protein (MCP; CD46). J. Biol. Chem. 275:37692-37701. [DOI] [PubMed] [Google Scholar]
- 40.Liszewski, M. K., M. K. Leung, and J. P. Atkinson. 1998. Membrane cofactor protein: importance of N- and O-glycosylation for complement regulatory function. J. Immunol. 161:3711-3718. [PubMed] [Google Scholar]
- 41.Liszewski, M. K., M. K. Leung, R. Hauhart, R. M. Buller, P. Bertram, X. Wang, A. M. Rosengard, G. J. Kotwal, and J. P. Atkinson. 2006. Structure and regulatory profile of the monkeypox inhibitor of complement: comparison to homologs in vaccinia and variola and evidence for dimer formation. J. Immunol. 176:3725-3734. [DOI] [PubMed] [Google Scholar]
- 42.Liszewski, M. K., M. K. Leung, R. Hauhart, C. J. Fang, P. Bertram, and J. P. Atkinson. 2009. Smallpox inhibitor of complement enzymes (SPICE): dissecting functional sites and abrogating activity. J. Immunol. 183:3150-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsumoto, M., W. Fukuda, A. Circolo, J. Goellner, J. Strauss-Schoenberger, X. Wang, S. Fujita, T. Hidvegi, D. D. Chaplin, and H. R. Colten. 1997. Abrogation of the alternative complement pathway by targeted deletion of murine factor B. Proc. Natl. Acad. Sci. U. S. A. 94:8720-8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meri, S., and M. K. Pangburn. 1994. Regulation of alternative pathway complement activation by glycosaminoglycans: specificity of the polyanion binding site on factor H. Biochem. Biophys. Res. Commun. 198:52-59. [DOI] [PubMed] [Google Scholar]
- 45.Miller, C. G., D. E. Justus, S. Jayaraman, and G. J. Kotwal. 1995. Severe and prolonged inflammatory response to localized cowpox virus infection in footpads of C5-deficient mice: investigation of the role of host complement in poxvirus pathogenesis. Cell Immunol. 162:326-332. [DOI] [PubMed] [Google Scholar]
- 46.Miller, C. G., S. N. Shchelkunov, and G. J. Kotwal. 1997. The cowpox virus-encoded homolog of the vaccinia virus complement control protein is an inflammation modulatory protein. Virology 229:126-133. [DOI] [PubMed] [Google Scholar]
- 47.Morgan, B. P., C. W. Berg, and C. L. Harris. 2005. “Homologous restriction” in complement lysis: roles of membrane complement regulators. Xenotransplantation 12:258-265. [DOI] [PubMed] [Google Scholar]
- 48.Moulton, E. A., J. P. Atkinson, and R. M. L. Buller. 2008. Surviving mousepox infection requires the complement system. PLoS Pathog. 4:e1000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mullick, J., J. Bernet, Y. Panse, S. Hallihosur, A. K. Singh, and A. Sahu. 2005. Identification of complement regulatory domains in vaccinia virus complement control protein. J. Virol. 79:12382-12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murthy, K. H., S. A. Smith, V. K. Ganesh, K. W. Judge, N. Mullin, P. N. Barlow, C. M. Ogata, and G. J. Kotwal. 2001. Crystal structure of a complement control protein that regulates both pathways of complement activation and binds heparan sulfate proteoglycans. Cell 104:301-311. [DOI] [PubMed] [Google Scholar]
- 51.Parker, A. K., S. Parker, W. M. Yokoyama, J. A. Corbett, and R. M. Buller. 2007. Induction of natural killer cell responses by ectromelia virus controls infection. J. Virol. 81:4070-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roozendaal, R., and M. C. Carroll. 2006. Emerging patterns in complement-mediated pathogen recognition. Cell 125:29-32. [DOI] [PubMed] [Google Scholar]
- 53.Rosengard, A. M., L. C. Alonso, L. C. Korb, W. M. Baldwin III, F. Sanfilippo, L. A. Turka, and J. M. Ahearn. 1999. Functional characterization of soluble and membrane-bound forms of vaccinia virus complement control protein (VCP). Mol. Immunol. 36:685-697. [DOI] [PubMed] [Google Scholar]
- 54.Rosengard, A. M., Y. Liu, Z. Nie, and R. Jimenez. 2002. Variola virus immune evasion design: expression of a highly efficient inhibitor of human complement. Proc. Natl. Acad. Sci. U. S. A. 99:8808-8813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sahu, A., S. N. Isaacs, A. M. Soulika, and J. D. Lambris. 1998. Interaction of vaccinia virus complement control protein with human complement proteins: factor I-mediated degradation of C3b to iC3b1 inactivates the alternative complement pathway. J. Immunol. 160:5596-5604. [PubMed] [Google Scholar]
- 56.Seet, B. T., J. B. Johnston, C. R. Brunetti, J. W. Barrett, H. Everett, C. Cameron, J. Sypula, S. H. Nazarian, A. Lucas, and G. McFadden. 2003. Poxviruses and immune evasion. Annu. Rev. Immunol. 21:377-423. [DOI] [PubMed] [Google Scholar]
- 57.Sfyroera, G., M. Katragadda, D. Morikis, S. N. Isaacs, and J. D. Lambris. 2005. Electrostatic modeling predicts the activities of orthopoxvirus complement control proteins. J. Immunol. 174:2143-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith, G. L., A. Vanderplasschen, and M. Law. 2002. The formation and function of extracellular enveloped vaccinia virus. J. Gen. Virol. 83:2915-2931. [DOI] [PubMed] [Google Scholar]
- 59.Smith, S. A., N. P. Mullin, J. Parkinson, S. N. Shchelkunov, A. V. Totmenin, V. N. Loparev, R. Srisatjaluk, D. N. Reynolds, K. L. Keeling, D. E. Justus, P. N. Barlow, and G. J. Kotwal. 2000. Conserved surface-exposed K/R-X-K/R motifs and net positive charge on poxvirus complement control proteins serve as putative heparin binding sites and contribute to inhibition of molecular interactions with human endothelial cells: a novel mechanism for evasion of host defense. J. Virol. 74:5659-5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith, S. A., R. Sreenivasan, G. Krishnasamy, K. W. Judge, K. H. Murthy, S. J. Arjunwadkar, D. R. Pugh, and G. J. Kotwal. 2003. Mapping of regions within the vaccinia virus complement control protein involved in dose-dependent binding to key complement components and heparin using surface plasmon resonance. Biochim. Biophys. Acta 1650:30-39. [DOI] [PubMed] [Google Scholar]
- 61.Turner, P. C., and R. W. Moyer. 2002. Poxvirus immune modulators: functional insights from animal models. Virus Res. 88:35-53. [DOI] [PubMed] [Google Scholar]
- 62.Vanderplasschen, A., E. Mathew, M. Hollinshead, R. B. Sim, and G. L. Smith. 1998. Extracellular enveloped vaccinia virus is resistant to complement because of incorporation of host complement control proteins into its envelope. Proc. Natl. Acad. Sci. U. S. A. 95:7544-7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu, X., D. Spitzer, D. Mao, S. L. Peng, H. Molina, and J. P. Atkinson. 2008. Membrane protein Crry maintains homeostasis of the complement system. J. Immunol. 181:2732-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yadav, V. N., K. Pyaram, J. Mullick, and A. Sahu. 2008. Identification of hot spots in the variola virus complement inhibitor (SPICE) for human complement regulation. J. Virol. 82:3283-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]