Abstract
The mammalian target of rapamycin (mTOR) kinase acts as a cellular rheostat that integrates signals from a variety of cellular signal transduction pathways that sense growth factor and nutrient availability as well as intracellular energy status. It was previously reported that the human papillomavirus type 16 (HPV16) E6 oncoprotein may activate the S6 protein kinase (S6K) through binding and E6AP-mediated degradation of the mTOR inhibitor tuberous sclerosis complex 2 (TSC2) (Z. Lu, X. Hu, Y. Li, L. Zheng, Y. Zhou, H. Jiang, T. Ning, Z. Basang, C. Zhang, and Y. Ke, J. Biol. Chem. 279:35664-35670, 2004; L. Zheng, H. Ding, Z. Lu, Y. Li, Y. Pan, T. Ning, and Y. Ke, Genes Cells 13:285-294, 2008). Our results confirmed that HPV16 E6 expression causes an increase in mTORC1 activity through enhanced phosphorylation of mTOR and activation of downstream signaling pathways S6K and eukaryotic initiation factor binding protein 1 (4E-BP1). However, we did not detect a decrease in TSC2 levels in HPV16 E6-expressing cells. We discovered, however, that HPV16 E6 expression causes AKT activation through the upstream kinases PDK1 and mTORC2 under conditions of nutrient deprivation. We show that HPV16 E6 expression causes an increase in protein synthesis by enhancing translation initiation complex assembly at the 5′ mRNA cap and an increase in cap-dependent translation. The increase in cap-dependent translation likely results from HPV16 E6-induced AKT/mTORC1 activation, as the assembly of the translation initiation complex and cap-dependent translation are rapamycin sensitive. Lastly, coexpression of the HPV16 E6 and E7 oncoproteins does not affect HPV16 E6-induced activation of mTORC1 and cap-dependent translation. HPV16 E6-mediated activation of mTORC1 signaling and cap-dependent translation may be a mechanism to promote viral replication under conditions of limited nutrient supply in differentiated, HPV oncoprotein-expressing proliferating cells.
Human papillomaviruses (HPVs) are small DNA viruses with a pronounced tropism for epithelial cells. Of the greater than 200 HPV types that have been identified, a subgroup specifically infects mucosal epithelia. These mucosal HPVs are classified as “low-risk” and “high-risk,” depending on the propensity for malignant progression of the lesions that they cause. Low-risk HPVs, such as HPV type 6 (HPV6) and HPV11, cause genital warts, whereas high-risk HPVs, such as HPV16 and HPV18, cause squamous intraepithelial lesions, which can undergo malignant progression. High-risk HPVs are associated with greater than 99% of cervical carcinomas and are also associated with other anogenital tumors and approximately 25% of oral cancers (reviewed in references 43 and 58). During carcinogenic progression the HPV genome frequently integrates into a host cell chromosome, resulting in persistent and dysregulated expression of the HPV E6 and E7 proteins. High-risk HPV E6 and E7 proteins have oncogenic activities and are necessary for the induction and maintenance of the transformed phenotype of cervical cancer cells (reviewed in reference 43). The best described cellular targets of the HPV E6 and E7 oncoproteins are the tumor suppressors p53 and retinoblastoma (pRB), respectively (reviewed in references 25 and 42). High-risk HPV E6 proteins cause proteasomal degradation of p53 through association with the cellular ubiquitin ligase E6AP (57). In addition to p53, other high-risk HPV E6-associated cellular proteins, including PDZ proteins, such as hDlg, hScribble, MUPP1, and MAGI 1 that bind to E6 through a carboxyl-terminal PDZ binding domain, may also be degraded by the E6/E6AP complex (19, 21, 29, 32, 33, 47, 61). High-risk HPV E6 proteins also transcriptionally activate hTERT, the catalytic protein subunit of human telomerase (30).
HPVs initially infect basal epithelial cells, where the viral episome is maintained extrachromosomally at a low copy number. High-level viral genome replication and production of progeny virus, however, is confined to the outer, terminally differentiated layers of the infected squamous epithelium, where metabolic activity of the host cells and nutrient availability are presumably more limited. Moreover, HPV16 E7 expression may induce metabolic stress by inducing the “Warburg effect,” a switch from an oxidative phosphorylation-based mode to a glycolytic mode of glucose metabolism (67). Consistent with this notion, we reported that HPV16 E7 expression in human keratinocytes triggers an autophagy-like process (65), which may serve to generate metabolites that can be used for energy-consuming processes, including viral replication.
The mammalian target of the rapamycin complex 1 (mTORC1) signaling cascade serves as a metabolic sensor, integrating a diverse array of signals, including nutrient and growth factor availability. mTORC1 signaling regulates a variety of cellular processes, including cell growth, viability, and proliferation, at least in part through the activation of protein translation (reviewed in reference 38). mTORC1 kinase activity is negatively regulated through TSC1/TSC2-mediated inhibition of the Ras homologue and the mTOR activator Rheb (63). TSC2 itself is regulated through phosphorylation at multiple sites by a diverse set of kinases, including AKT (reviewed in reference 38). TSC2 phosphorylation by AKT inhibits TSC2, releasing mTOR from repression for subsequent activation of downstream signaling cascades that regulate protein translation—the ribosomal S6 protein kinase (S6K) and the eukaryotic initiation factor binding protein 1 (4E-BP1) pathways (reviewed in references 38 and 39).
Previous studies have suggested that HPV16 E6 may activate mTORC1 signaling (37). A yeast two-hybrid screen with E6 as the bait identified peptides corresponding to TSC2 and the E6-associated GTPase activating protein (GAP) E6TP1 (12, 17). Subsequent studies suggested that HPV16 E6 may bind and degrade TSC2 through an E6AP-dependent mechanism, thereby activating mTORC1 (64). These latter studies, however, were performed by transient transfection, and it remained unclear in cells with stable expression of E6 whether TSC2 levels are decreased, whether increased mTORC1 activity can be detected, whether this is relevant in the context of a nutrient-deprived state, and if the increase in mTORC1 signaling in E6-expressing cells results in a corresponding increase in cap-dependent translation.
Here we report that HPV16 E6 expression does not cause a reduction in the steady-state levels of TSC2, but instead HPV16 E6 activates mTORC1 as a result of increased AKT activity through the PDK1 and mTORC2 pathways. Moreover, mTORC1 activity is sustained in HPV16 E6-expressing primary human foreskin keratinocyte (HFK) populations under conditions of nutrient deprivation. Furthermore, HPV16 E6 expression causes activation of the S6K and 4E-BP1 translation regulatory pathways, causes enhanced binding of translation initiation factors to a synthetic cap structure, and increases cap-dependent translation as measured by luciferase reporter assays. The HPV16 E6-mediated increases in binding of translation initiation factors to the cap and cap-dependent translation are rapamycin sensitive, suggesting a connection between the HPV16 E6-mediated increase in mTORC1 activation and enhanced cap-dependent translation. Lastly, coexpression of the HPV E7 oncoprotein does not affect these processes, suggesting that the ability of E6 to activate mTORC1 signaling and cap-dependent translation may be relevant in the context of an HPV infection.
MATERIALS AND METHODS
Plasmids.
Plasmids used in this study include the retroviral vectors pLXSN (control), pLXSN HPV16 E6, pLXSN HPV16 E7, and pLXSN HPV16 E6/E7 (22); a set of human β-actin-promoter-driven expression vectors, p1318 (control), p1435 (HPV16 E7), p1436 (HPV16 E6), p1321 (HPV16 E6/E7), and p1319 (HPV16 early coding region) (46); a pCMV Bam Neo-based vector (2), pCMV HPV16 NE6 (with Flag-hemagglutinin [HA] epitope tags fused to the amino terminus of HPV16 E6); and the pCMV Bam Neo N control plasmid (2, 46). The pFR_CrPV_xb bicistronic firefly/Renilla luciferase reporter plasmid (50) was used for translation reporter assays and was obtained from Phil Sharp through Addgene (plasmid 11509).
Cell lines and culture.
293, 293T, and U2OS cells (ATCC) were maintained in Dulbecco's modified Eagle medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum (FBS), 50 U/ml penicillin, and 50 μg/ml streptomycin. RKO pC (pCMV control cells) and RKO 10.2 (HPV16 E6-expressing) cells (28) were generously provided by Kathleen Cho (University of Michigan, Ann Arbor, MI) and maintained in modified McCoy's medium (Invitrogen) supplemented with 10% newborn calf serum (NCS), 50 U/ml penicillin, 50 μg/ml streptomycin, and 500 μg/ml G418.
Primary human foreskin keratinocytes (HFKs) were isolated from anonymous newborn circumcisions as previously described (41) and maintained in keratinocyte serum-free medium (KSFM) supplemented with human recombinant epidermal growth factor 1-53, bovine pituitary extract (Invitrogen), 50 U/ml penicillin, 50 μg/ml streptomycin, 20 μg/ml gentamicin, and 1 μg/ml amphotericin B. HPV oncogene-expressing HFK populations were generated by infection with the corresponding LXSN-based retroviral vectors and selected and maintained as previously described (52). All experiments were performed with HFKs passaged less than 10 times. For growth factor withdrawal experiments, HFKs were seeded onto poly-d-lysine-coated plates (BD). For nutrient deprivation assays, 90% confluent HFKs were washed twice with phosphate-buffered saline (PBS), followed by incubation in PBS for 15 to 30 min prior to lysis.
Western blotting and antibodies.
Cell lysates were prepared by incubating the cells in ML buffer (300 mM NaCl, 0.5% Nonidet P-40 [NP-40], 20 mM Tris-HCl [pH 8.0], 1 mM EDTA supplemented with one complete EDTA-free protease inhibitor cocktail tablet [Roche] per 25 ml lysis buffer, and one PhosSTOP phosphatase inhibitor cocktail tablet [Roche] per 5 ml lysis buffer) (41). Cells were then scraped and lysates cleared by centrifugation at 16,110 × g for 10 min at 4°C. Protein concentrations were determined using the Bradford method (Bio-Rad). Proteins were separated by SDS-PAGE and transferred onto polyvinylidene difluoride membranes (Immobilon-P; Millipore). The membranes were blocked in 5% nonfat dry milk in TBST (137 mM NaCl, 2.7 mM KCl, 25 mM Tris [pH 7.4], 0.1% Tween 20) and probed with the appropriate antibody. The following primary antibodies were used at a 1:1,000 dilution unless otherwise specified: β-actin (1501; Chemicon), p53 (Ab-6; Calbiochem), SGK1 (ab43606; Abcam), Firefly luciferase (ab498; Abcam), Renilla luciferase (PM047; MBL), SGK1 S422 (1:500 dilution; sc-16745-R; Santa Cruz), Flag (3165; Sigma), mTOR (2972), mTOR S2448 (2971), S6K (9202), S6K T389 (9206), S6 (2317), S6 S235/36 (4858), S6 240/44 (4838), TSC2 (3635), 4E-BP1 (9644), 4E-BP1 T37/46 (2855), 4E-BP1 S65 (9451), 4E-BP1 T70 (9455), Akt (9272), Akt S473 (4060), Akt T308 (9275), eIF4G (2498), and SGK1 T256 (2939), all from Cell Signaling Technology. Secondary anti-mouse and anti-rabbit antibodies conjugated to horseradish peroxidase (Amersham) were used at dilutions of 1:10,000 and 1:15,000, respectively. Proteins were visualized by enhanced chemiluminescence (PerkinElmer) and exposed on Kodak BioMax XAR film or electronically acquired and quantified with a Kodak Image Station 4000R equipped with Kodak Imaging Software, version 4.0.
7-Methyl-GTP binding assays.
Proteins that interact with a synthetic 7-methyl-GTP RNA cap structure were purified as previously described (31). In brief, 250-μg aliquots of cell lysates were precleared with 25 μl Sepharose prewashed in buffer D (50 mM HEPES, pH 7.4, 40 mM NaCl, 2 mM EDTA, 0.1% Triton X-100) for 1 h and combined with 30 μl of a 50% slurry of 7-methyl-GTP-Sepharose (GE Healthcare, United Kingdom) prewashed in buffer D and incubated for 1 h at 4°C. After washing the resin three times with buffer D, samples were analyzed by SDS-PAGE and immunoblotting.
Transfections and luciferase assays.
U2OS cells were transfected in six-well plates in triplicate for luciferase reporter assays using FuGene6 reagent (Roche). One microgram of pFR_CrPV_xb was cotransfected with 2 μg p1318, p1435, p1436, p1319, or p1321. HFKs were transfected in six-well plates in triplicate (seeded at 300,000 cells/well) using FuGene6 reagent (Roche) with 0.5 μg pFR_CrPV_xb and 1.5 μg of the previously described plasmids. Both U2OS cells and HFKs were lysed 48 h posttransfection in 450 μl passive lysis buffer (dual luciferase reporter kit; Promega) per well. The supernatants were subjected to the dual luciferase reporter assay. The fold change in activity was determined by calculating the ratio of firefly luciferase activity to Renilla luciferase activity compared to control vector-transfected cells. At least three independent experiments were performed.
Real-time qRT-PCR.
Total RNA was extracted from U2OS cells cotransfected as described above with pFR_CrPV_xb and either p1318 or p1436 using the total RNA isolation mini kit (Agilent). Quantitative reverse transcription-PCR (qRT-PCR) was performed using a 7300 real-time PCR system (Applied Biosystems) and SYBR green fluorescence. Primers for firefly luciferase and Renilla luciferase were as follows: firefly luciferase Fwd, 5′-CCTCTGGATCTACTGGGTTACCTAAG-3′; firefly luciferase Rev, 5′-TCTGGCATGCGAGAATCTGA-3′; Renilla luciferase Fwd, 5′-GAATTTGCAGCATATCTTGAACCAT-3′; Renilla luciferase Rev, 5′-GGATTTCACGAGGCCATGATAA-3′. For cDNA synthesis and quantitative PCR, the QuantiTect SYBR green RT-PCR kit (Qiagen) was used. Cycling parameters were as previously described (41). Dissociation curve analysis (95°C for 15 s, 60°C for 15 s, and 95°C for 15 s) was performed at the end of 40 cycles to verify PCR product identity. qRT-PCR was performed for each sample in triplicate, and data were analyzed using the threshold cycle (2−ΔΔCT) method (35).
RESULTS
mTORC1 signaling is increased in HPV16 E6-expressing cells.
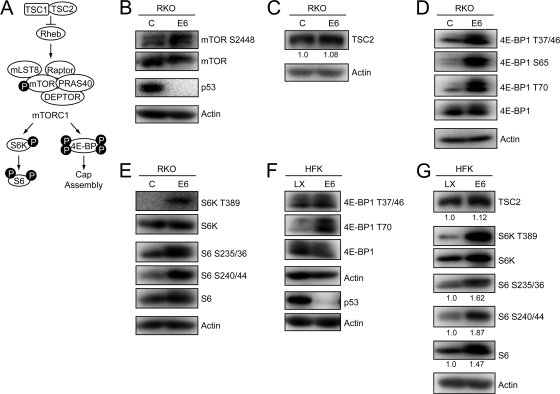
The tuberous sclerosis tumor suppressor 2 (TSC2), sometimes also referred to as tuberin, is a negative regulator of mTORC1 activity (Fig. 1 A). Previous studies suggested that the HPV16 E6 oncoprotein may be able to associate with TSC2 (12, 37). Moreover, transient expression studies in HEK 293 cells suggested that HPV16 E6 not only binds to but can also enhance E6AP-mediated TSC2 degradation, thereby activating mTORC1 signaling (64). Based on these findings, we evaluated mTORC1 signaling in RKO human colon cancer cells with stable expression of HPV16 E6 (RKO E6) (28) as well as control RKO cells. RKO cells have intact p53 and pRB tumor suppressor pathways, and previous work has shown that p53 activities are lost upon E6 expression (23, 28). Consistent with the published results (37), RKO E6 cells showed evidence of increased mTORC1 activity as evaluated by phosphorylation of the mTOR kinase at serine residue 2448 (S2448) (Fig. 1B). TSC2 steady-state levels, however, were not decreased in RKO E6 cells compared to those in control cells (Fig. 1C). In contrast, p53 tumor suppressor levels were dramatically decreased in RKO E6 cells, indicating that there are no defects in E6/E6AP-induced proteasomal degradation in these cells (Fig. 1B).
FIG. 1.
HPV16 E6 expression activates mTOR1, 4E-BP1, S6K, and S6 phosphorylation through a TSC2-independent mechanism. (A) Schematic diagram of mTORC1 signaling. See text for details. (B to E) Western blot analysis of mTOR phosphorylation (B), TSC2 expression (with quantifications shown below) (C), 4E-BP1 phosphorylation (D), and S6K and S6 phosphorylation (E) in HPV16 E6-expressing and control RKO cells. A p53 blot is shown in panel B to document HPV16 E6 expression, and actin blots are shown as loading controls. Also shown are results from Western blot analysis of 4E-BP1 (F) and TSC2 expression and S6K and S6 phosphorylation (G) (with quantifications shown below) in HPV16 E6-expressing and control (LX) primary human foreskin keratinocyte cultures (HFKs). A p53 blot is shown in panel F to document HPV16 E6 expression, and actin blots are shown as loading controls.
To determine whether the observed increased mTOR S2448 phosphorylation causes increased mTORC1 activity, we evaluated the phosphorylation status of downstream mTORC1 phosphorylation targets in RKO E6 and control RKO cells. The eukaryotic translation initiation factor 4E binding protein 1 (4E-BP1) regulates formation of the mRNA cap structure. Hypophosphorylated 4E-BP1 inhibits functional interaction of eukaryotic translation initiation factor 4E (eIF4E) with the 5′ mRNA cap structure (reviewed in reference 26). Upon mTORC1 activation, 4E-BP1 is sequentially phosphorylated by mTOR at at least four residues. 4E-BP1 phosphorylation at threonine 37 (T37) and T46 serve as priming phosphorylation events that are required for subsequent phosphorylation and activation at T70 and S65. Hyperphosphorylated 4E-BP1 is released from the cap, allowing for recruitment of eIF4E and other translation initiation factors to the 5′ mRNA cap (20). Consistent with increased mTORC1 activity in RKO E6 cells, phosphorylation of 4E-BP1 at T37/46, S65, and T70 was strikingly increased in these cells (Fig. 1D).
The S6 kinase (S6K) is another well-established mTORC1 substrate. Once phosphorylated at T389 by mTORC1, S6K activates and phosphorylates the ribosomal subunit 6 (S6), an important factor in ribosome biogenesis and a component of the 40S ribosome, at serines 235, 236, 240, and 244 (14). Phosphorylated S6 is incorporated into the 40S ribosome at the mRNA binding site and has been correlated with an increase in protein synthesis (reviewed in reference 27). Phosphorylation of S6K at T389 and its substrate S6 at S235/236 and S240/244 was markedly increased in RKO E6 cells compared to that in control RKO cells (Fig. 1E). These results further support the notion that HPV16 E6 expression in RKO cells causes increased mTORC1 signaling.
To ensure that the observed activation of mTORC1 by HPV16 E6 is not specific to the RKO cell line, we performed similar experiments in HPV16 E6-expressing primary HFK populations. Compared to control vector-transfected HFKs, HFK E6 cells showed increased phosphorylation of 4E-BP1 (Fig. 1F) as well as S6K and its substrate S6, while TSC2 steady-state levels were unchanged (Fig. 1G). Similar results were obtained with a second independently derived set of HFK E6 and HFK control populations (data not shown). Of note, in some experiments we also found evidence for increased S6K and S6 steady-state levels in HKF E6 populations (Fig. 1G), which in combination with mTORC1 activation, as evidenced by increased 4E-BP1 phosphorylation (Fig. 1F), may contribute to the increased detection of S6 phosphorylation at S235/36 and S240/44.
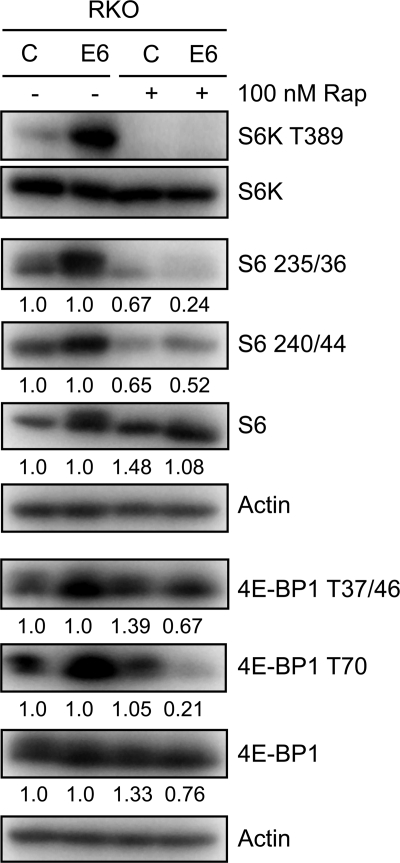
In order to confirm that the observed effects on 4E-BP1, S6K, and S6 phosphorylation are a result of mTORC1 activation, we treated RKO E6 and RKO control cells with 100 nM of the mTORC1 inhibitor rapamycin for 1 h. Phosphorylation of S6K, S6, and 4E-BP1 was decreased in RKO E6 cells, but to a lesser extent in RKO cells. These results are consistent with our model that HPV16 E6 expression in RKO cells causes mTORC1 activation (Fig. 2).
FIG. 2.
HPV16 E6 expression causes increased S6K, S6, and 4E-BP1 phosphorylation through mTORC1 activation. Western blot analysis of mTORC1 downstream signaling components in RKO control and HPV16 E6-expressing RKO cells, treated with dimethyl sulfoxide (DMSO) or 100 nM rapamycin (Rap) for 1 h prior to lysis. Relative levels of unphosphorylated and phosphorylated species of S6 and 4E-BP1 are indicated. Actin blots are shown as loading controls.
In combination, these results show that HPV16 E6 expression causes increases mTORC1 activity through a mechanism that does not appear to involve TSC2 degradation.
HPV16 E6-mediated mTORC1 activation is mediated by PDK1 and mTORC2 activation.
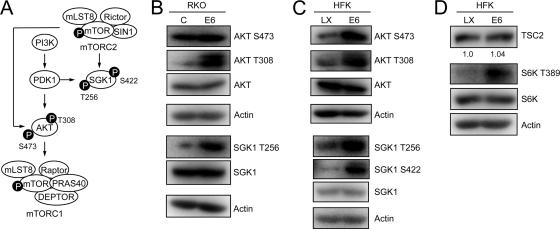
Since we found no evidence for decreases in TSC2 steady-state levels in HPV16 E6-expressing cells (Fig. 1C and G and Fig. 3 D) and we did not detect an association of HPV16 E6 with TSC2 by immunoprecipitation experiments (data not shown), we evaluated alternative signaling events upstream of mTORC1 activation.
FIG. 3.
HPV16 E6 expression causes AKT activation. (A) Schematic diagram of AKT phosphorylation through PDK1 and mTORC2 pathways. (B) Western blot analysis of AKT phosphorylation in control and HPV16 E6-expressing RKO cells. SGK1 is phosphorylated by PDK1 at T256 and is included as a control for PDK1 activation in HPV16 E6-expressing RKO cells. Actin blots are shown as loading controls. (C) Sustained AKT activation in control (LX) and HPV16 E6-expressing HFK populations under conditions of nutrient deprivation. SGK1 is phosphorylated by PDK1 at T256 and by mTORC2 at S422 and is included as a control for PDK1 and mTORC2 activation in HPV16 E6-expressing HFKs. Actin blots are shown as loading controls. (D) Sustained S6K activation in control (LX) and HPV16 E6-expressing HFK populations under conditions of nutrient deprivation. A TSC2 blot with quantification is shown to document similar expression in the two cell populations after nutrient deprivation; an actin blot is shown as a loading control.
Members of the AKT serine/threonine kinase family are important activators of mTORC1 signaling (reviewed in reference 49). 3-Phosphoinositide-dependent kinase 1 (PDK1) is downstream of phosphoinositide 3-kinase (PI3K) and activates AKT by T308 phosphorylation, which in turn causes mTORC1 activation (1), and the mTORC2 kinase complex activates AKT by S473 phosphorylation (Fig. 3A) (56). Hence, we assessed AKT T308 and S422 phosphorylation in RKO E6 and RKO control cells. RKO E6 cells showed increased AKT T308 phosphorylation compared to control RKO cells. In contrast, RKO E6 and RKO control cells each showed high levels of AKT S473 phosphorylation, which may explain why there is only a modest increase in mTOR S2448 phosphorylation in RKO E6 cells (Fig. 1B). To confirm that PDK1 activity is increased in RKO E6 cells, we also evaluated T256 phosphorylation of the PDK1 substrate serum- and glucocorticoid-inducible kinase 1 (SGK1). Consistent with increased PDK1 activity, SGK1 T256 phosphorylation was increased in RKO E6 compared to that in RKO control cells (Fig. 3B).
Given that RKO cells are a colon cancer-derived line that may harbor mutations that may cause aberrant AKT phosphorylation, we next evaluated AKT T308 and S473 phosphorylation in HPV16 E6-expressing primary HFK and control HFK populations. When grown in growth factor containing keratinocyte serum-free medium, AKT was phosphorylated at T308 and S473 in HPV16 E6-expressing HFKs as well as control HFKs (data not shown). To assess AKT phosphorylation in these cells under conditions of nutrient deprivation, we incubated HPV16 E6-expressing and control HFKs in phosphate-buffered saline (PBS) for 15 or 30 min. Under these conditions, we detected increased AKT S473 and T308 phosphorylation in HFK E6 compared to that in control HFKs (Fig. 3C). Similar results were obtained when these cells were treated with Earle's balanced salt solution containing 1 mg/ml glucose (data not shown). These results suggest that AKT S473 and T308 phosphorylation is maintained in HPV16 E6-expressing HFKs under conditions of limited growth factor availability.
To assess whether sustained AKT S473 and T308 phosphorylation in HPV16 E6-expressing HFKs is a result of sustained PDK1 and mTORC2 activity, respectively, we evaluated SGK1 phosphorylation. PDK1 phosphorylates SGK1 at T256 (3), whereas mTORC2 phosphorylates SGK1 at S422 (18). Consistent with our model, SGK1 S256 and S422 phosphorylation was sustained in HFK E6 cells under conditions of nutrient deprivation (Fig. 3C). Moreover, S6K T389 phosphorylation was detected in HFK E6 cells but not in HFK control cells under conditions of nutrient deprivation. Of note, TSC2 levels were not decreased in HFK E6 cells undergoing growth factor restriction as a result of PBS treatment (Fig. 3D).
These results suggest that HPV16 E6 expression activates mTORC1 at least in part through PDK1- and mTORC2-mediated AKT phosphorylation and that this activation is sustained during conditions of nutrient deprivation.
HPV16 E6 expression increases the assembly of the translation initiation complex at the mRNA cap.
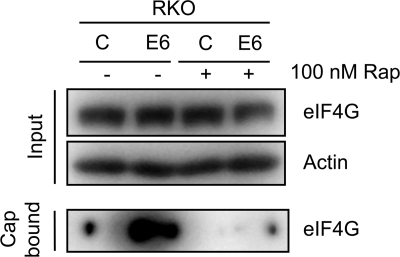
4E-BP1 phosphorylation by mTORC1 allows association of translation initiation factors to the 5′ mRNA cap structure, thereby activating cap-dependent translation (reviewed in reference 26). To evaluate whether HPV16 E6 expression enhances the assembly of the translation initiation complex at the mRNA cap, we performed in vitro cap-binding assays. Lysates from RKO E6 and RKO control cells were incubated with 7-methyl-GTP-Sepharose, and association of initiation factor eIF4G was evaluated by Western blotting. As expected, we detected increased eIF4G binding to the synthetic cap structure with lysates from RKO E6 cells compared to those from RKO control cells (Fig. 4). To determine whether the increase in eIF4G binding observed with RKO E6 cell lysates is caused by increased mTORC1 activity, we also performed experiments with cell lysates prepared from RKO E6 and RKO control cells that were treated with the mTORC1 inhibitor rapamycin for 1 h prior to harvesting. Inhibition of mTORC1 abrogated eIF4G binding to the cap structure in RKO E6 cells (Fig. 4).
FIG. 4.
Increased binding of the translation initiation factor eIF4G to a synthetic 7-methyl-GTP (7MeGTP) mRNA cap structure in HPV16 E6-expressing RKO cell lysates, which is sensitive to rapamycin treatment. Control and HPV16 E6-expressing RKO cells were treated with dimethyl sulfoxide (DMSO) or 100 nM rapamycin (Rap) for 1 h prior to lysis. Cap binding assays were performed as described in Materials and Methods. Levels of eIF4G in a 50-μg sample, representing 25% of the cap-binding reaction, together with an actin blot, are shown in the top panel (Input). Blot results for cap-bound eIF4G are shown in the bottom panel.
These results show that binding of translation initiation factors to the 5′ mRNA cap is increased in HPV16 E6-expressing cells and that this most likely represents a consequence of mTORC1 activation.
HPV16 E6 expression causes increased translation of capped mRNA.
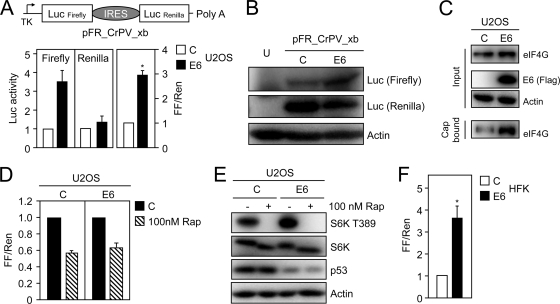
Given the observed increased binding of eIF4G to a synthetic cap in vitro with RKO E6 cells, we next determined if HPV16 E6 expression might increase cap-dependent translation. The U2OS human osteosarcoma line was used for the initial experiments because it contains wild-type p53 and is highly transfectable. We performed dual luciferase reporter assays utilizing a bicistronic reporter vector, pFR_CrPV_xb (50), which drives expression of the firefly and Renilla luciferase genes from a minimal thymidine kinase promoter. Firefly luciferase is translated by a cap-dependent mechanism, whereas translation of Renilla luciferase is through a cap-independent mechanism from a cricket paralysis virus (CrPV) internal ribosomal entry site (IRES) (Fig. 5 A, top). Coexpression of HPV16 E6 caused a 3.56-fold ± 0.68-fold increase in firefly luciferase activity compared to control vector cotransfection. In contrast, Renilla luciferase activity was increased only 1.22-fold ± 0.24-fold compared to vector cotransfection. When normalized to Renilla luciferase activity, HPV16 E6 cotransfection caused a statistically significant 2.92-fold ± 0.33-fold (P < 0.0001) increase in firefly luciferase activity (Fig. 5A, bottom) compared to vector-transfected cells. To confirm that the HPV16 E6-mediated increase in cap-dependent translation is not a result of transcriptional regulation or aberrant splicing of the bicistronic mRNA, we performed quantitative real-time reverse transcription-PCR for firefly and Renilla luciferases. These experiments showed that the mRNA levels of firefly and Renilla luciferase were unchanged (data not shown). Moreover, we also directly evaluated steady-state levels of firefly and Renilla luciferase proteins by Western blotting in U2OS cells that were transiently cotransfected with the reporter plasmid and HPV16 E6 or the control vector. Consistent with the enzyme activity results, expression of HPV16 E6 caused an increase in steady-state levels of firefly but not Renilla luciferase levels (Fig. 5B). We also performed cap-binding experiments, and similar to what we observed in RKO cells with stable expression of HPV16 E6 (Fig. 4), transient expression of HPV16 E6 in U2OS cells caused increased association of eIF4G with a synthetic mRNA cap structure (Fig. 5C). To confirm that mTORC1 signaling is necessary for the HPV16 E6-mediated increase in cap-dependent translation, dual luciferase reporter assays were performed with cells that were treated with 100 nM rapamycin for 18 h prior to harvesting. Results from these experiments show that cap-dependent translation is reduced in rapamycin-treated HPV16 E6 cells as well as in control vector-transfected U2OS cells (Fig. 5D). To further confirm that HPV16 E6 expression increases mTORC1 activity in U2OS cells and that this is inhibited by rapamycin treatment, we also evaluated mTORC1-dependent S6K phosphorylation at T389. As expected, transient expression of HPV16 E6 caused S6K T389 phosphorylation that was reduced upon treatment with rapamycin (Fig. 5E).
FIG. 5.
HPV16 E6 expression causes as increase in cap-dependent translation, which is sensitive to rapamycin treatment. (A) Diagram of bicistronic firefly Renilla reporter plasmid, pFR_CrPV_xb, used for these experiments. Firefly luciferase is translated through a cap-dependent mechanism, whereas Renilla luciferase is expressed from an internal ribosomal entry site (IRES) through a cap-independent mechanism (top). HPV16 E6 expression causes an increase in firefly but not Renilla luciferase activity (bottom). U2OS cells were transfected with control or HPV16 E6 expression vector, and lysates were processed for Renilla and firefly luciferase assays at 48 h posttransfection. The data are presented as the change of firefly and Renilla luciferase activities normalized to control vector-transfected cells (left and middle) and the fold change of normalized firefly compared to normalized Renilla luciferase activity (FF/Ren) (right). The bar graphs represent averages and standard deviations of four experiments, each performed in triplicate. The asterisk denotes statistical significance (P < 0.0001). (B) Western blot analysis of firefly and Renilla luciferase expression in U2OS cells transiently transfected with the indicated plasmids. U, untransfected cells. (C) Western blot analysis of eIF4G binding to a synthetic 7-methyl-GTP (7MeGTP) mRNA cap upon transient transfection of HPV16 E6 or control vector in U2OS cells. (D) HPV16 E6-mediated increase in cap-dependent translation is rapamycin sensitive. U2OS cells were transfected with pFR_CrPV_xb and the indicated plasmids; 18 h prior to lysis, cells were treated with dimethyl sulfoxide (DMSO) or 100 nM rapamycin (Rap). The graph represents averages and standard deviations of four experiments, each performed in triplicate. (E) Western blot analysis of S6K phosphorylation in U2OS cells transiently transfected with HPV16 E6 or control vector. One hour prior to lysis, cells were treated with DMSO or 100 nM rapamycin (Rap). Decreases in p53 levels are shown to document HPV16 E6 expression, and an actin blot is included as a loading control. (F) Transient transfection of HPV16 E6 activates cap-dependent translation in primary HFKs. Cells were transfected with pFR_CrPV_xb and the indicated plasmids and processed for Renilla and firefly luciferase assays at 48 h posttransfection. Firefly and Renilla luciferase activities were normalized to control vector-transfected cells and are presented as fold changes of normalized firefly relative to normalized Renilla luciferase activity. The bar graph represents the average and standard deviation of four experiments, each performed in triplicate; asterisks indicate statistical significance (P = 0.0001).
To assess whether HPV16 E6 expression can cause increased cap-dependent translation in biologically relevant cells, we performed dual luciferase reporter assays in primary HFKs. Similar to what we observed with U2OS cells, cotransfection of HPV16 E6 caused a statistically significant 3.49-fold ± 0.56-fold (P = 0.0001) increase in firefly luciferase compared to that in control vector-transfected cells (Fig. 5F).
Hence, HPV16 E6 expression can increase cap-dependent translation and mTORC1 signaling is necessary for HPV16 E6 to modulate this process.
HPV16 E7 coexpression does not affect E6-induced activation of mTORC1 and cap-dependent translation.
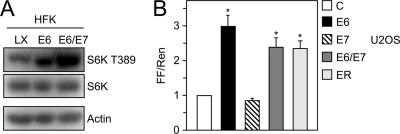
Since HPV E6 and E7 oncoproteins are coexpressed in high-risk HPV-associated lesions and cancers, we also evaluated mTORC1 signaling and cap-dependent translation in HPV16 E6/E7-coexpressing cells. Phosphorylation of S6K at T389 by mTORC1 was increased in HFK populations with coexpression of HPV16 E6/E7, similar to that in HPV16 E6-expressing HFKs (Fig. 6 A). While expression of HPV16 E7 alone did not affect cap-dependent translation, expression of HPV16 E6/E7 or the entire HPV16 early coding region in U2OS cells caused statistically significant (2.40-fold ± 0.30-fold [P = 0.0013] and 2.36-fold ± 0.23-fold [P = 0.0005], respectively) increases in firefly luciferase activity, similar to that of E6 cotransfection (2.97-fold ± 0.31-fold [P < 0.0001]) (Fig. 6B).
FIG. 6.
HPV16 E7 coexpression does not affect E6-induced S6K T389 phosphorylation or cap-dependent translation. (A) Western blot analysis of S6K T389 phosphorylation in HFK populations with stable expression of HPV16 E6 or HPV16 E6/E7 or control vector (LX)-transduced HFKs. An actin blot is shown as a loading control. (B) U2OS cells were transiently transfected with pFR_CrPV_xb and human β-actin-promoter-driven expression vectors for HPV16 E6, E7, E6/E7, the entire HPV16 early coding region (ER), or empty vector as a control and processed for Renilla and firefly luciferase assays at 48 h posttransfection. Firefly and Renilla luciferase activities were normalized to control vector-transfected cells and are presented as fold changes of normalized firefly relative to normalized Renilla luciferase activity. The bar represents the average and standard deviation of four experiments, each performed in triplicate; asterisks indicate statistical significance (P ≤ 0.0013).
Hence, HPV16 E7 coexpression does not markedly affect the ability of HPV16 E6 to activate mTORC1 activity and to augment cap-dependent translation.
DISCUSSION
Previous reports have suggested that mTOR is activated in cells transiently expressing HPV16 E6, as indicated by an increase in S6K phosphorylation (37). This activity was attributed to the ability of HPV16 E6 to interact with and accelerate TSC2 degradation through an E6AP-dependent pathway (64). In our experiments, TSC2 steady-state levels were unaltered in HPV16 E6-expressing RKO cells and HFKs relative to that in control cells (Fig. 1C and G and Fig. 3D) and upon transient transfection of HPV16 E6 in HEK293 or U2OS cells (data not shown). Moreover, we did not detect association of HPV16 E6 with TSC2 by immunoprecipitation experiments (data not shown). Hence, the reported E6AP-mediated TSC2 degradation by HPV16 E6 is not a rate-limiting mechanism by which HPV16 E6 expression causes mTORC1 activation in our experimental systems.
Here we report that cells with stable HPV16 E6 expression show evidence of active mTORC1 signaling, as evidenced by activation of the S6K and 4E-BP1 downstream cascades (Fig. 1). Most importantly, mTORC1 activity is sustained in HPV16 E6-expressing HFKs under conditions of nutrient deprivation (Fig. 3). In contrast to the previously published studies, we did not find any evidence for HPV16 E6 binding to TSC2 and/or lowering its steady-state levels in the cells that we studied (Fig. 1C and G and Fig. 3D). Our results, however, suggest that HPV16 E6 expression causes mTORC1 activation, at least in part, through an AKT-dependent mechanism. HPV16 E6 expression in primary human epithelial cells caused AKT activation through at least two distinct pathways, PDK1 and mTORC2 (Fig. 3C). As with mTORC1, our results show that AKT remains active in HPV16 E6-expressing HFKs under conditions of nutrient deprivation. HPV16 E6 expression also caused an increase in cap-dependent translation (Fig. 5 and 6). This effect correlated with increased binding of translation initiation factors to a synthetic cap (Fig. 4 and 5C) and was inhibited by the mTORC1 inhibitor rapamycin (Fig. 4 and Fig. 5D and E), suggesting that HPV16 E6-mediated activation of translation may represent a consequence of mTORC1 activation.
The HPV16 E6 and E7 oncoproteins play important functions during the viral life cycle (15, 60). Whereas HPVs initially infect proliferative basal epithelial cells, high-level viral genome replication and synthesis of viral progeny is restricted to terminally differentiated epithelial cells. The HPV E6 and E7 proteins contribute to the viral life cycle by uncoupling the process of epithelial cell differentiation from cell cycle withdrawal. The HPV E7 protein, in particular, through degradation of the retinoblastoma tumor suppressor pRB and the related family members p107 and p130, causes increased transcription of E2F-responsive genes, many of which encode enzymes that are rate limiting for cellular DNA synthesis (reviewed in references 24, 36, and 43). Since HPV genome replication is acutely dependent on expression of host cellular replication proteins, one might envision that the ability of HPV16 E6 to activate translation of capped mRNAs represents an additional facet of this strategy in order to ensure adequate expression of cellular proteins that are necessary for viral genome replication. In addition, or alternatively, the ability of HPV16 E6 to activate protein synthesis may also contribute to high-level synthesis of viral proteins, particularly the L1 and L2 capsid proteins that need to be abundantly expressed during productive viral replication. While there is no direct evidence for such a mechanism, translational control of the L1 capsid protein synthesis has been suggested by results from experiments where HPV31 episome-containing human epithelial cells were induced to undergo differentiation by suspension in methylcellulose-containing medium. Under these conditions, the authors observed abundant expression of L1-encoding mRNAs; however, there was no evidence for L1 protein synthesis (55). There is also evidence for translational regulation of early protein synthesis during epithelial cell differentiation in HPV-positive cells. When HPV16-positive CaSki cervical carcinoma cells were cultured in methylcellulose- or CaCl2-containing medium to induce differentiation, increased expression of the E7 oncoprotein was observed. This increase was not at the level of transcription or protein stability, but rather the authors observed an increase in association of E7-encoding mRNAs to polysomes. These authors also observed sustained phosphorylation of 4E-BP1 upon differentiation of CaSki cells but not with HPV-negative HaCaT cells or primary HFKs. Moreover, mTORC1 inhibition by rapamycin treatment reduced 4E-BP1 phosphorylation and HPV16 E7 oncoprotein expression in these cells (48).
Increased mTOR S2448 and S6K T389 phosphorylation was also observed in HPV-positive high-grade cervical squamous intraepithelial lesions (13), and there is also evidence for increased AKT phosphorylation in HPV-positive high-grade cervical squamous intraepithelial lesions (44). Given our results, it is tempting to speculate that these effects may at least in part represent a consequence of HPV16 E6 expression.
There are several reports that have shown that HPV16 E7 expression may also cause AKT activation (44, 53). Several mechanisms have been proposed. HPV16 E7 may activate AKT by a pRB-dependent process, causing p27kip1 cytoplasmic accumulation and induction of cellular migration (5, 44). It has also been reported that HPV16 E7 can activate AKT independently of the pRB pathway through binding and inhibition of protein phosphatase 2A (53). In another study, however, cells that ectopically expressed HPV16 E7 and activated AKT showed a significantly higher rate of cellular proliferation and migration than either AKT or HPV16 E7-expressing cells (8). These results would suggest that HPV16 E7 expression is not sufficient to fully activate AKT. While our experiments did not directly address the possible contribution of HPV16 E7 in AKT phosphorylation, there was no evidence that coexpression of HPV16 E6 and E7 caused an increase in mTORC1 signaling compared to HPV16 E6-expressing cells (Fig. 6A). Moreover, HPV16 E7 expression did not increase cap-dependent translation in our reporter assays (Fig. 6B).
Aberrant activation of AKT and mTORC1/2 is frequently observed in human cancers, and mTORC1 inhibitors have been evaluated as antineoplastic agents (11, 45, 59). As the regulation of mTORC2 and its downstream signaling pathways are increasingly understood, it is becoming apparent that the development of mTORC2-specific rictor inhibitors may also limit aberrant cellular growth and proliferation associated with human cancers. Hence it is conceivable that HPV16 E6-mediated AKT and mTORC1 and mTORC2 activation may also contribute to the transforming activities of HPV16 E6. If that was the case, inhibition of AKT and mTORC1 and/or mTORC2 should be evaluated as a therapeutic modality for HPV-associated lesions and cancers.
Our studies presented here were focused on AKT, but they do not exclude the possibility that HPV16 E6 expression may also affect mTORC1 activity through other pathways. Activation of the p53 tumor suppressor inhibits mTORC1 activity through sestrin 1 and sestrin 2. These two proteins are transcriptional targets of p53 and activate the AMP-responsive protein kinase (AMPK). AMPK phosphorylates and activates the mTOR inhibitor TSC2, thereby inhibiting mTOR (4). E6/E6AP-mediated p53 degradation may therefore be predicted to short-circuit this regulatory loop and may contribute to sustained mTORC1 activity.
In addition, several PDZ proteins have been implicated in mTOR signaling. Inactivation of hScribble, which is targeted for degradation by HPV16 E6 (47), was shown to dysregulate MAP kinase signaling (10), which is predicted to activate mTORC1. More recently, Sabatini's group identified a novel mTORC1/mTORC2-associated inhibitor, DEPTOR, which contains a PDZ domain (51) and thus may be a potential candidate for HPV16 E6 association and degradation.
Our results show that HPV16 E6 expression in primary epithelial cells activates AKT through at least two pathways, PDK1 and mTORC2, but the exact mechanism remains unknown. PDK1 is downstream of PI3K signaling. Several transforming viral proteins have been reported to activate PI3K, including SV40 small tumor antigen and the mouse polyomavirus middle tumor antigen (reviewed in reference 7). Our future experiments will explore whether PI3K is activated by HPV16 E6 expression. A number of scenarios are possible, including activation of upstream signaling events, direct activation of PI3K, or inhibition of the phosphatase and tumor suppressor PTEN. Alternatively, HPV16 E6 may activate PDK1 by a PI3K-independent mechanism. Importantly, PDK1 also activates kinases other than AKT, including SGK1 (Fig. 3) and the Rho/Rac effector target PKN (9), a serine/threonine protein kinase, with a catalytic domain that is similar to that of protein kinase C. Interestingly, PKN has been reported to associate with high-risk HPV E6 proteins (16). It will be interesting to determine the biological consequences of PDK1-mediated activation of kinases other than AKT.
Our results also suggest that HPV16 E6 activates mTORC2 signaling. Recent reports suggest that rictor expression is critical to the activation of mTORC2, with rictor overexpression activating the kinase complex and resulting in increased cell growth and motility in gliomas, and rictor short hairpin RNA (shRNA) knockdown inhibiting cellular proliferation in colon cancer cell lines (40, 54). Interestingly, the FOXO1 transcription factor regulates rictor transcription, which is in turn regulated by AKT (6). These authors suggest that FOXO1 balances mTORC1 inhibition and mTORC2 activation through two separable transcriptional activities of FOXO1: direct inhibition of mTORC1 through sestrin-3 gene transcription and activation of mTORC2 through rictor gene transcription as a coactivator of a distinct transcriptional activating complex. Collectively this results in the maintenance of cellular energy homoeostasis even under conditions of nutrient stress. It is possible that the HPV16 E6 oncoprotein expression uncouples these processes through independent activation of mTORC1 and mTORC2. Alternatively, the PDZ protein and mTOR inhibitor DEPTOR described above inhibits both mTORC1 and mTORC2 and thus should be evaluated as a potential candidate for HPV16 E6-mediated mTORC2 regulation.
We initiated these studies after we discovered that HPV16 E7 expression in normal human epithelial cells triggers an autophagy-like response (65). Autophagy is a survival pathway that allows survival of cells under conditions of metabolic stress (reviewed in reference 34). While we do not know the exact mechanism by which E7 expression may trigger such a response, it has been reported that HPV16 E7 expression causes the “Warburg effect,” a metabolic switch from an oxidative phosphorylation-based pathway to a glycolytic pathway (67). While such a switch may offer a number of advantages for a rapidly proliferating cell, including efficient growth under conditions of lower oxygen concentrations and increased synthesis of metabolic precursors (62), conversion of glucose to lactate generates far less energy in the form of ATP than conversion to CO2 through oxidative phosphorylation. Particularly under conditions of limiting supply of nutrients, as may be the case in terminally differentiated cells in a squamous epithelium, autophagy may eventually lead to the demise of the cell. It is thus tempting to speculate that the ability of HPV16 E6 to activate mTORC1 signaling, a major regulator of autophagy, may function to dampen the autophagy response to HPV16 E7 expression and limited availability to nutrients. In such a model, expression of the HPV16 E6 protein would induce a cellular state of “blissful ignorance” and allow metabolically stressed, HPV-infected cells to survive long enough to support synthesis of viral progeny (66).
Acknowledgments
This work was supported by NIH grants R01CA081135 (K.M.) and T32CA009031 (J.M.S.). J.M.S. is a Ryan Fellow.
We thank John Blenis, Nick Dyson, Tom Roberts, Sheila Thomas, and Fred Wang for helpful discussions and technical advice, Kathleen Cho for generously providing the RKO 16E6 cell line, and Margaret McLaughlin-Drubin for critical review of the manuscript.
Footnotes
Published ahead of print on 14 July 2010.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Alessi, D. R., S. R. James, C. P. Downes, A. B. Holmes, P. R. Gaffney, C. B. Reese, and P. Cohen. 1997. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Bα. Curr. Biol. 7:261-269. [DOI] [PubMed] [Google Scholar]
- 2.Baker, S. J., S. Markowitz, E. R. Fearon, J. K. Willson, and B. Vogelstein. 1990. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science 249:912-915. [DOI] [PubMed] [Google Scholar]
- 3.Biondi, R. M., A. Kieloch, R. A. Currie, M. Deak, and D. R. Alessi. 2001. The PIF-binding pocket in PDK1 is essential for activation of S6K and SGK, but not PKB. EMBO J. 20:4380-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Budanov, A. V., and M. Karin. 2008. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell 134:451-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charette, S. T., and D. J. McCance. 2007. The E7 protein from human papillomavirus type 16 enhances keratinocyte migration in an Akt-dependent manner. Oncogene 26:7386-7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, C. C., S. M. Jeon, P. T. Bhaskar, V. Nogueira, D. Sundararajan, I. Tonic, Y. Park, and N. Hay. 2010. FoxOs inhibit mTORC1 and activate Akt by inducing the expression of Sestrin3 and Rictor. Dev. Cell 18:592-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng, J., J. A. DeCaprio, M. M. Fluck, and B. S. Schaffhausen. 2009. Cellular transformation by Simian Virus 40 and Murine Polyoma Virus T antigens. Semin. Cancer Biol. 19:218-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Debnath, J., S. J. Walker, and J. S. Brugge. 2003. Akt activation disrupts mammary acinar architecture and enhances proliferation in an mTOR-dependent manner. J. Cell Biol. 163:315-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong, L. Q., L. R. Landa, M. J. Wick, L. Zhu, H. Mukai, Y. Ono, and F. Liu. 2000. Phosphorylation of protein kinase N by phosphoinositide-dependent protein kinase-1 mediates insulin signals to the actin cytoskeleton. Proc. Natl. Acad. Sci. U. S. A. 97:5089-5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dow, L. E., I. A. Elsum, C. L. King, K. M. Kinross, H. E. Richardson, and P. O. Humbert. 2008. Loss of human Scribble cooperates with H-Ras to promote cell invasion through deregulation of MAPK signalling. Oncogene 27:5988-6001. [DOI] [PubMed] [Google Scholar]
- 11.Dowling, R. J., I. Topisirovic, B. D. Fonseca, and N. Sonenberg. 2010. Dissecting the role of mTOR: lessons from mTOR inhibitors. Biochim. Biophys. Acta 1804:433-439. [DOI] [PubMed] [Google Scholar]
- 12.Elston, R. C., S. Napthine, and J. Doorbar. 1998. The identification of a conserved binding motif within human papillomavirus type 16 E6 binding peptides, E6AP and E6BP. J. Gen. Virol. 79:371-374. [DOI] [PubMed] [Google Scholar]
- 13.Feng, W., X. Duan, J. Liu, J. Xiao, and R. E. Brown. 2009. Morphoproteomic evidence of constitutively activated and overexpressed mTOR pathway in cervical squamous carcinoma and high grade squamous intraepithelial lesions. Int. J. Clin. Exp. Pathol. 2:249-260. [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrari, S., H. R. Bandi, J. Hofsteenge, B. M. Bussian, and G. Thomas. 1991. Mitogen-activated 70K S6 kinase. Identification of in vitro 40 S ribosomal S6 phosphorylation sites. J. Biol. Chem. 266:22770-22775. [PubMed] [Google Scholar]
- 15.Flores, E. R., B. L. Allen-Hoffmann, D. Lee, and P. F. Lambert. 2000. The human papillomavirus type 16 E7 oncogene is required for the productive stage of the viral life cycle. J. Virol. 74:6622-6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao, Q., A. Kumar, S. Srinivasan, L. Singh, H. Mukai, Y. Ono, D. E. Wazer, and V. Band. 2000. PKN binds and phosphorylates human papillomavirus E6 oncoprotein. J. Biol. Chem. 275:14824-14830. [DOI] [PubMed] [Google Scholar]
- 17.Gao, Q., S. Srinivasan, S. N. Boyer, D. E. Wazer, and V. Band. 1999. The E6 oncoproteins of high-risk papillomaviruses bind to a novel putative GAP protein, E6TP1, and target it for degradation. Mol. Cell. Biol. 19:733-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.García-Martínez, J. M., and D. R. Alessi. 2008. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1). Biochem. J. 416:375-385. [DOI] [PubMed] [Google Scholar]
- 19.Gardiol, D., C. Kuhne, B. Glaunsinger, S. S. Lee, R. Javier, and L. Banks. 1999. Oncogenic human papillomavirus E6 proteins target the discs large tumour suppressor for proteasome-mediated degradation. Oncogene 18:5487-5496. [DOI] [PubMed] [Google Scholar]
- 20.Gingras, A. C., B. Raught, S. P. Gygi, A. Niedzwiecka, M. Miron, S. K. Burley, R. D. Polakiewicz, A. Wyslouch-Cieszynska, R. Aebersold, and N. Sonenberg. 2001. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 15:2852-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glaunsinger, B. A., S. S. Lee, M. Thomas, L. Banks, and R. Javier. 2000. Interactions of the PDZ-protein MAGI-1 with adenovirus E4-ORF1 and high-risk papillomavirus E6 oncoproteins. Oncogene 19:5270-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halbert, C. L., G. W. Demers, and D. A. Galloway. 1992. The E6 and E7 genes of human papillomavirus type 6 have weak immortalizing activity in human epithelial cells. J. Virol. 66:2125-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Havre, P. A., J. Yuan, L. Hedrick, K. R. Cho, and P. M. Glazer. 1995. p53 inactivation by HPV16 E6 results in increased mutagenesis in human cells. Cancer Res. 55:4420-4424. [PubMed] [Google Scholar]
- 24.Helt, A. M., and D. A. Galloway. 2003. Mechanisms by which DNA tumor virus oncoproteins target the Rb family of pocket proteins. Carcinogenesis 24:159-169. [DOI] [PubMed] [Google Scholar]
- 25.Howie, H. L., R. A. Katzenellenbogen, and D. A. Galloway. 2009. Papillomavirus E6 proteins. Virology 384:324-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson, R. J., C. U. Hellen, and T. V. Pestova. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11:113-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jastrzebski, K., K. M. Hannan, E. B. Tchoubrieva, R. D. Hannan, and R. B. Pearson. 2007. Coordinate regulation of ribosome biogenesis and function by the ribosomal protein S6 kinase, a key mediator of mTOR function. Growth Factors 25:209-226. [DOI] [PubMed] [Google Scholar]
- 28.Kessis, T. D., R. J. Slebos, W. G. Nelson, M. B. Kastan, B. S. Plunkett, S. M. Han, A. T. Lorincz, L. Hedrick, and K. R. Cho. 1993. Human papillomavirus 16 E6 expression disrupts the p53-mediated cellular response to DNA damage. Proc. Natl. Acad. Sci. U. S. A. 90:3988-3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kiyono, T., A. Hiraiwa, M. Fujita, Y. Hayashi, T. Akiyama, and M. Ishibashi. 1997. Binding of high-risk human papillomavirus E6 oncoproteins to the human homologue of the Drosophila discs large tumor suppressor protein. Proc. Natl. Acad. Sci. U. S. A. 94:11612-11616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klingelhutz, A. J., S. A. Foster, and J. K. McDougall. 1996. Telomerase activation by the E6 gene product of human papillomavirus type 16. Nature 380:79-82. [DOI] [PubMed] [Google Scholar]
- 31.Kumar, V., D. Sabatini, P. Pandey, A. C. Gingras, P. K. Majumder, M. Kumar, Z. M. Yuan, G. Carmichael, R. Weichselbaum, N. Sonenberg, D. Kufe, and S. Kharbanda. 2000. Regulation of the rapamycin and FKBP-target 1/mammalian target of rapamycin and cap-dependent initiation of translation by the c-Abl protein-tyrosine kinase. J. Biol. Chem. 275:10779-10787. [DOI] [PubMed] [Google Scholar]
- 32.Lee, S. S., B. Glaunsinger, F. Mantovani, L. Banks, and R. T. Javier. 2000. Multi-PDZ domain protein MUPP1 is a cellular target for both adenovirus E4-ORF1 and high-risk papillomavirus type 18 E6 oncoproteins. J. Virol. 74:9680-9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, S. S., R. S. Weiss, and R. T. Javier. 1997. Binding of human virus oncoproteins to hDlg/SAP97, a mammalian homolog of the Drosophila discs large tumor suppressor protein. Proc. Natl. Acad. Sci. U. S. A. 94:6670-6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levine, B., and G. Kroemer. 2008. Autophagy in the pathogenesis of disease. Cell 132:27-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 36.Longworth, M. S., and L. A. Laimins. 2004. Pathogenesis of human papillomaviruses in differentiating epithelia. Microbiol. Mol. Biol. Rev. 68:362-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu, Z., X. Hu, Y. Li, L. Zheng, Y. Zhou, H. Jiang, T. Ning, Z. Basang, C. Zhang, and Y. Ke. 2004. Human papillomavirus 16 E6 oncoprotein interferences with insulin signaling pathway by binding to tuberin. J. Biol. Chem. 279:35664-35670. [DOI] [PubMed] [Google Scholar]
- 38.Ma, X. M., and J. Blenis. 2009. Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 10:307-318. [DOI] [PubMed] [Google Scholar]
- 39.Mamane, Y., E. Petroulakis, O. LeBacquer, and N. Sonenberg. 2006. mTOR, translation initiation and cancer. Oncogene 25:6416-6422. [DOI] [PubMed] [Google Scholar]
- 40.Masri, J., A. Bernath, J. Martin, O. D. Jo, R. Vartanian, A. Funk, and J. Gera. 2007. mTORC2 activity is elevated in gliomas and promotes growth and cell motility via overexpression of rictor. Cancer Res. 67:11712-11720. [DOI] [PubMed] [Google Scholar]
- 41.McLaughlin-Drubin, M. E., K. W. Huh, and K. Munger. 2008. Human papillomavirus type 16 E7 oncoprotein associates with E2F6. J. Virol. 82:8695-8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McLaughlin-Drubin, M. E., and K. Munger. 2009. The human papillomavirus E7 oncoprotein. Virology 384:335-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McLaughlin-Drubin, M. E., and K. Munger. 2009. Oncogenic activities of human papillomaviruses. Virus Res. 143:195-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Menges, C. W., L. A. Baglia, R. Lapoint, and D. J. McCance. 2006. Human papillomavirus type 16 E7 up-regulates AKT activity through the retinoblastoma protein. Cancer Res. 66:5555-5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Menon, S., and B. D. Manning. 2008. Common corruption of the mTOR signaling network in human tumors. Oncogene 27(Suppl. 2):S43-S51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Münger, K., W. C. Phelps, V. Bubb, P. M. Howley, and R. Schlegel. 1989. The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J. Virol. 63:4417-4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakagawa, S., and J. M. Huibregtse. 2000. Human scribble (Vartul) is targeted for ubiquitin-mediated degradation by the high-risk papillomavirus E6 proteins and the E6AP ubiquitin-protein ligase. Mol. Cell. Biol. 20:8244-8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oh, K. J., A. Kalinina, N. H. Park, and S. Bagchi. 2006. Deregulation of eIF4E: 4E-BP1 in differentiated human papillomavirus-containing cells leads to high levels of expression of the E7 oncoprotein. J. Virol. 80:7079-7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pearce, L. R., D. Komander, and D. R. Alessi. 2010. The nuts and bolts of AGC protein kinases. Nat. Rev. Mol. Cell Biol. 11:9-22. [DOI] [PubMed] [Google Scholar]
- 50.Petersen, C. P., M. E. Bordeleau, J. Pelletier, and P. A. Sharp. 2006. Short RNAs repress translation after initiation in mammalian cells. Mol. Cell 21:533-542. [DOI] [PubMed] [Google Scholar]
- 51.Peterson, T. R., M. Laplante, C. C. Thoreen, Y. Sancak, S. A. Kang, W. M. Kuehl, N. S. Gray, and D. M. Sabatini. 2009. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell 137:873-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Piboonniyom, S. O., S. Duensing, N. W. Swilling, J. Hasskarl, P. W. Hinds, and K. Munger. 2003. Abrogation of the retinoblastoma tumor suppressor checkpoint during keratinocyte immortalization is not sufficient for induction of centrosome-mediated genomic instability. Cancer Res. 63:476-483. [PubMed] [Google Scholar]
- 53.Pim, D., P. Massimi, S. M. Dilworth, and L. Banks. 2005. Activation of the protein kinase B pathway by the HPV-16 E7 oncoprotein occurs through a mechanism involving interaction with PP2A. Oncogene 24:7830-7838. [DOI] [PubMed] [Google Scholar]
- 54.Roulin, D., Y. Cerantola, A. Dormond-Meuwly, N. Demartines, and O. Dormond. 2010. Targeting mTORC2 inhibits colon cancer cell proliferation in vitro and tumor formation in vivo. Mol. Cancer. 9:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ruesch, M. N., F. Stubenrauch, and L. A. Laimins. 1998. Activation of papillomavirus late gene transcription and genome amplification upon differentiation in semisolid medium is coincident with expression of involucrin and transglutaminase but not keratin-10. J. Virol. 72:5016-5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sarbassov, D. D., D. A. Guertin, S. M. Ali, and D. M. Sabatini. 2005. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307:1098-1101. [DOI] [PubMed] [Google Scholar]
- 57.Scheffner, M., J. M. Huibregtse, R. D. Vierstra, and P. M. Howley. 1993. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75:495-505. [DOI] [PubMed] [Google Scholar]
- 58.Schiffman, M., P. E. Castle, J. Jeronimo, A. C. Rodriguez, and S. Wacholder. 2007. Human papillomavirus and cervical cancer. Lancet 370:890-907. [DOI] [PubMed] [Google Scholar]
- 59.Shor, B., J. J. Gibbons, R. T. Abraham, and K. Yu. 2009. Targeting mTOR globally in cancer: thinking beyond rapamycin. Cell Cycle 8:3831-3837. [DOI] [PubMed] [Google Scholar]
- 60.Thomas, J. T., W. G. Hubert, M. N. Ruesch, and L. A. Laimins. 1999. Human papillomavirus type 31 oncoproteins E6 and E7 are required for the maintenance of episomes during the viral life cycle in normal human keratinocytes. Proc. Natl. Acad. Sci. U. S. A. 96:8449-8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thomas, M., R. Laura, K. Hepner, E. Guccione, C. Sawyers, L. Lasky, and L. Banks. 2002. Oncogenic human papillomavirus E6 proteins target the MAGI-2 and MAGI-3 proteins for degradation. Oncogene 21:5088-5096. [DOI] [PubMed] [Google Scholar]
- 62.Vander Heiden, M. G., L. C. Cantley, and C. B. Thompson. 2009. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324:1029-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang, Y., X. Gao, L. J. Saucedo, B. Ru, B. A. Edgar, and D. Pan. 2003. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat. Cell Biol. 5:578-581. [DOI] [PubMed] [Google Scholar]
- 64.Zheng, L., H. Ding, Z. Lu, Y. Li, Y. Pan, T. Ning, and Y. Ke. 2008. E3 ubiquitin ligase E6AP-mediated TSC2 turnover in the presence and absence of HPV16 E6. Genes Cells 13:285-294. [DOI] [PubMed] [Google Scholar]
- 65.Zhou, X., and K. Munger. 2009. Expression of the human papillomavirus type 16 E7 oncoprotein induces an autophagy-related process and sensitizes normal human keratinocytes to cell death in response to growth factor deprivation. Virology 385:192-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou, X., J. M. Spangle, and K. Munger. 2009. Expression of a viral oncoprotein in normal human epithelial cells triggers an autophagy-related process: is autophagy an “Achilles' heel” of human cancers? Autophagy 5:578-579. [DOI] [PubMed] [Google Scholar]
- 67.Zwerschke, W., S. Mazurek, P. Massimi, L. Banks, E. Eigenbrodt, and P. Jansen-Durr. 1999. Modulation of type M2 pyruvate kinase activity by the human papillomavirus type 16 E7 oncoprotein. Proc. Natl. Acad. Sci. U. S. A. 96:1291-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]