Abstract
Many diseases with unknown etiology may be caused by unidentified viruses. Sequence-independent amplification revealed a new astrovirus, similar to VA1, in a 4-year-old male diagnosed with celiac disease. This expands the geographic range of this virus to include Europe and may associate astrovirus infection with the onset of celiac disease.
Infectious diseases pose a continuous disease burden on humans, and many diseases with unknown etiology may be caused by unidentified viruses. Key to effectively counter the potential public health threat caused by emerging infectious diseases requires a systematic exploration of infectious viruses. Diarrhea is a common cause of morbidity and mortality. In developing countries, the mortality associated with gastroenteritis has been estimated at 3 to 5 million cases per year (24). In industrialized countries, although typically self-limited, diarrheal diseases are a significant cause of morbidity among all age groups. Most gastroenteritis cases in industrialized countries are caused by viruses (24). However, the etiological cause of a large proportion of diarrhea cases (up to ∼40%) remains unresolved. The implementation of new technologies for virus discovery, based on microarrays or sequence-independent amplification of nucleic acids, already resulted in identification of many previously unknown viruses, among which are several new human astroviruses in human stool samples (1, 2, 5-9, 11, 14-16, 19-21, 23).
From the Diagnostic Unit, Department of Virology, Erasmus Medical Center, Rotterdam, Netherlands, we received 84 diarrhea samples of unknown etiology for further analysis on the potential presence of viral pathogens. Random sequence-independent amplification of RNA on an initial subset of these samples revealed the presence of new picobirnaviruses and a rhinovirus (22) and was now also performed on diarrhea from patient VS34. A total of 138 clones were sequenced, and most of the sequences were unclassified or of bacterial origin. Three out of 138 clones contained sequences most closely related to mink and ovine astroviruses by BLASTn similarity searches (3).
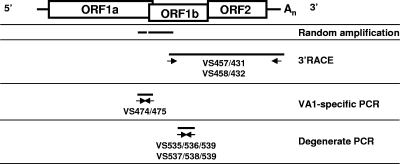
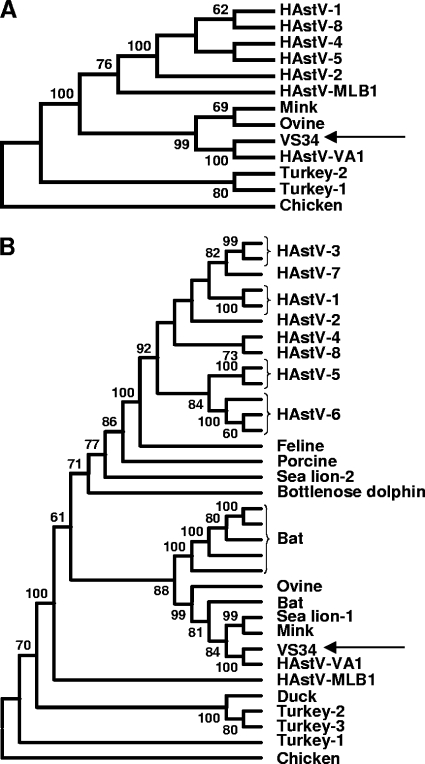
To acquire the 3′ ∼4,400 bp of the astrovirus genome, the astrovirus sequence reads from a stool sample from patient VS34 were assembled in different contigs and the nucleic acid between the contigs was obtained by reverse transcriptase-PCR (RT-PCR) with 400 nM primers VS474 and VS475 (Fig. 1; Table 1), 1× AmpliTaq Gold reaction buffer, 2.5 mM MgCl2, 200 μM each deoxynucleoside triphosphates (dNTPs), and 2.5 U AmpliTaq Gold with Geneamp (Roche). The conditions for amplification were 95°C for 5 min, followed by 40 cycles of 95°C for 30 s, 50°C for 1 min, and 72°C for 1 min, followed by a single incubation at 72°C for 10 min. In addition, 3′ rapid amplification of cDNA ends (RACE) PCR was employed, using primers VS430, VS431, VS457, VS432, and VS458 (Fig. 1; Table 1), as described previously (22). Amino acid sequences from open reading frame 1b (ORF1b) and ORF2 of the astrovirus from sample VS34 were aligned to the respective complete ORFs of other astroviruses in GenBank with ClustalX 2.0. Maximum parsimony trees were generated using MEGA version 4.1beta (18) in a manner similar to that used in previous astrovirus studies (5-8). This analysis demonstrated that the astrovirus from sample VS34 was closely related (>99% identical) to the recently described astrovirus variant VA1, which is most closely related to mink and ovine astroviruses in ORF1b and to mink and California sea lion astroviruses in ORF2 (Fig. 2) (8, 14).
FIG. 1.
Schematic outlines of the strategies used for PCR amplification. The upper panel shows a schematic representation of the astrovirus genome. The boxes represent the ORFs encoding the astrovirus proteins. Indicated are the 5′ end and the poly(A) tail (An). The lower panels show a schematic outline of the RT-PCR assays employed to amplify astrovirus sequences, using random amplification, specific amplification, degenerate PCR, and 3′ RACE PCR. The orientations and positions of the oligonucleotides on the astrovirus genome are shown.
TABLE 1.
Oligonucleotide primers used in this study
| Primer | Nucleotide sequence (5′ → 3′) | Assay |
|---|---|---|
| VS430 | GCGAGCACAGAATTAATACGACTCACTATAGGT12VN | 3′ RACE |
| VS431 | GCTGATGGCGATGAATGAACACTG | 3′ RACE |
| VS432 | CGCGGATCCGAATTAATACGACTCACTATAGG | 3′ RACE |
| VS457 | GAATGCTGACTCCACACCTG | 3′ RACE |
| VS458 | CCACACCTGCTTATCCAAAGAC | 3′ RACE |
| VS474 | CCCAGAAGACTCTGAGGAG | Astrovirus VA1 |
| VS475 | GTGGACCCATGGCATAGTC | Astrovirus VA1 |
| VS535 | GARTTYGATTGGRCKCGKTAYGA | Degenerate astrovirus PCR |
| VS536 | GARTTYGATTGGRCKAGGTAYGA | Degenerate astrovirus PCR |
| VS537 | CGKTAYGATGGKACKATICC | Degenerate astrovirus PCR |
| VS538 | AGGTAYGATGGKACKATICC | Degenerate astrovirus PCR |
| VS539 | GGYTTKACCCACATICCRAA | Degenerate astrovirus PCR |
FIG. 2.
Phylogenetic analysis of astroviruses. Panel A shows the phylogenetic tree of the ORF1b-encoded amino acids generated using MEGA 4.1, using the maximum parsimony method with 1,000 bootstrap replicates. Significant bootstrap values are shown. Panel B depicts the phylogenetic tree of the ORF2-encoded amino acids generated using MEGA 4.1, using the maximum parsimony method with 1,000 bootstrap replicates. Significant bootstrap values are shown. HAstV, human astrovirus. The other astroviruses are named by the host species in which they were identified. GenBank accession numbers are available on request.
To determine whether astrovirus VA1 is prevalent in gastroenteritis cases in the Netherlands, the other 83 diarrhea samples were screened with an astrovirus VA1-specific PCR, as described above, and a degenerate astrovirus PCR (14) (Fig. 1; Table 1). This sample set was skewed for young age as 40 of these patients were children <5 years of age (22). No evidence for infection with astrovirus VA1 was obtained. Subsequently, 245 diarrhea samples from Dutch patients with gastroenteritis in 2009, not prescreened for known pathogens, were screened negative for the presence of astrovirus VA1 with the astrovirus VA1-specific PCR. These data suggest that the prevalence of astrovirus VA1 in patients with gastroenteritis in the Netherlands is low.
Patient VS34 was a 4-year-old male with a history of malaise and abdominal pain for over 6 weeks. A gastroduodenoscopy was performed, and histological examination of duodenal biopsies showed evidence for celiac disease, characterized by total villous atrophy and lesions of type Marsh IIIc. To evaluate whether astrovirus infection is associated with onset of celiac disease, diarrhea samples from four additional patients with onset of celiac disease (<4 years old) were analyzed for known pathogens using routine diagnostic assays and/or for the presence of astroviruses (Table 2). Standard diagnostic real-time PCR revealed that a 1-year-old female was positive for enterovirus, and a 3-year-old male was positive for astrovirus using a diagnostic enzyme immunoassay (ProSpecT Astrovirus test; Oxoid). None of the patients, however, was positive for astrovirus VA1.
TABLE 2.
Viruses detected in patients with onset of celiac disease
| Patient | Age (yr) | Sex | Symptoms | Virus detected |
|---|---|---|---|---|
| VS170 | 0 | Female | Diarrhea | |
| VS171 | 1 | Male | Vomiting, diarrhea, weight loss | |
| VS34 | 4 | Male | Diarrhea, weight loss | Astrovirus VA1 |
| VS169 | 3 | Male | Vomiting, diarrhea | Astrovirus |
| VS167 | 1 | Female | Diarrhea | Enterovirus |
In this study, we obtained evidence for the presence of a new astrovirus that resembled the recently described astrovirus variant VA1 (5, 8). This astrovirus variant is highly divergent from all previously described astroviruses, explaining how this virus escaped detection during routine astrovirus diagnostic assays in the Diagnostic Unit, Department of Virology, Erasmus Medical Center, Rotterdam, Netherlands. The detection of astrovirus VA1 in a Dutch diarrhea sample expands the known geographic range of this virus beyond North America and Nepal to include Europe (8, 14). A total of 328 patients with diarrhea were screened negative for the presence of astrovirus VA1, suggesting a low prevalence of astrovirus VA1 in patients with gastroenteritis in the Netherlands, which is in line with previous observations (5, 14). Future studies are needed to define a causative role of this virus in gastroenteritis. In addition, the previously unrecognized high diversity of astroviruses (5-8, 14) warrants the development of new diagnostic assays.
It remains interesting that the new astrovirus variant VA1 was detected in a patient with new-onset celiac disease. Celiac disease is precipitated by the ingestion of gluten in genetically susceptible individuals and can cause a broad range of signs and symptoms, among which are constipation, fatigue, headaches, mild gastrointestinal complaints, diarrhea, and weight loss (10). In two out of five patients with onset of celiac disease, astroviruses were detected, and one out of five patients was enterovirus positive. Infectious agents have been implicated in the pathogenesis of celiac disease, among which are adenovirus, hepatitis C virus, and rotavirus (4, 12, 13, 17). Whether entero- and/or astroviruses are associated with development of celiac disease remains to be determined.
Acknowledgments
We thank J. Kruining and G. I. Arron from the Department of Virology, Erasmus Medical Center, Rotterdam, Netherlands, for providing diarrhea samples.
This work was funded by EU Framework project-7 grant 223498 and Nobilon International B.V.
A. D. M. E. Osterhaus is part-time chief scientific officer of ViroClinics Biosciences B.V.
Footnotes
Published ahead of print on 23 June 2010.
REFERENCES
- 1.Allander, T., S. U. Emerson, R. E. Engle, R. H. Purcell, and J. Bukh. 2001. A virus discovery method incorporating DNase treatment and its application to the identification of two bovine parvovirus species. Proc. Natl. Acad. Sci. U. S. A. 98:11609-11614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allander, T., M. T. Tammi, M. Eriksson, A. Bjerkner, A. Tiveljung-Lindell, and B. Andersson. 2005. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. U. S. A. 102:12891-12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fine, K. D., F. Ogunji, Y. Saloum, S. Beharry, J. Crippin, and J. Weinstein. 2001. Celiac sprue: another autoimmune syndrome associated with hepatitis C. Am. J. Gastroenterol. 96:138-145. [DOI] [PubMed] [Google Scholar]
- 5.Finkbeiner, S. R., L. R. Holtz, Y. Jiang, P. Rajendran, C. J. Franz, G. Zhao, G. Kang, and D. Wang. 2009. Human stool contains a previously unrecognized diversity of novel astroviruses. Virol. J. 6:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finkbeiner, S. R., C. D. Kirkwood, and D. Wang. 2008. Complete genome sequence of a highly divergent astrovirus isolated from a child with acute diarrhea. Virol. J. 5:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finkbeiner, S. R., B. M. Le, L. R. Holtz, G. A. Storch, and D. Wang. 2009. Detection of newly described astrovirus MLB1 in stool samples from children. Emerg. Infect. Dis. 15:441-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finkbeiner, S. R., Y. Li, S. Ruone, C. Conrardy, N. Gregoricus, D. Toney, H. W. Virgin, L. J. Anderson, J. Vinje, D. Wang, and S. Tong. 2009. Identification of a novel astrovirus (astrovirus VA1) associated with an outbreak of acute gastroenteritis. J. Virol. 83:10836-10839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fouchier, R. A., N. G. Hartwig, T. M. Bestebroer, B. Niemeyer, J. C. de Jong, J. H. Simon, and A. D. Osterhaus. 2004. A previously undescribed coronavirus associated with respiratory disease in humans. Proc. Natl. Acad. Sci. U. S. A. 101:6212-6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Green, P. H., and C. Cellier. 2007. Celiac disease. N. Engl. J. Med. 357:1731-1743. [DOI] [PubMed] [Google Scholar]
- 11.Jones, M. S., A. Kapoor, V. V. Lukashov, P. Simmonds, F. Hecht, and E. Delwart. 2005. New DNA viruses identified in patients with acute viral infection syndrome. J. Virol. 79:8230-8236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kagnoff, M. F., R. K. Austin, J. J. Hubert, J. E. Bernardin, and D. D. Kasarda. 1984. Possible role for a human adenovirus in the pathogenesis of celiac disease. J. Exp. Med. 160:1544-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kagnoff, M. F., Y. J. Paterson, P. J. Kumar, D. D. Kasarda, F. R. Carbone, D. J. Unsworth, and R. K. Austin. 1987. Evidence for the role of a human intestinal adenovirus in the pathogenesis of coeliac disease. Gut 28:995-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kapoor, A., L. Li, J. Victoria, B. Oderinde, C. Mason, P. Pandey, S. Z. Zaidi, and E. Delwart. 2009. Multiple novel astrovirus species in human stool. J. Gen. Virol. 90:2965-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kistler, A., P. C. Avila, S. Rouskin, D. Wang, T. Ward, S. Yagi, D. Schnurr, D. Ganem, J. L. DeRisi, and H. A. Boushey. 2007. Pan-viral screening of respiratory tract infections in adults with and without asthma reveals unexpected human coronavirus and human rhinovirus diversity. J. Infect. Dis. 196:817-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mihindukulasuriya, K. A., G. Wu, J. St. Leger, R. W. Nordhausen, and D. Wang. 2008. Identification of a novel coronavirus from a beluga whale by using a panviral microarray. J. Virol. 82:5084-5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stene, L. C., M. C. Honeyman, E. J. Hoffenberg, J. E. Haas, R. J. Sokol, L. Emery, I. Taki, J. M. Norris, H. A. Erlich, G. S. Eisenbarth, and M. Rewers. 2006. Rotavirus infection frequency and risk of celiac disease autoimmunity in early childhood: a longitudinal study. Am. J. Gastroenterol. 101:2333-2340. [DOI] [PubMed] [Google Scholar]
- 18.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 19.Urisman, A., R. J. Molinaro, N. Fischer, S. J. Plummer, G. Casey, E. A. Klein, K. Malathi, C. Magi-Galluzzi, R. R. Tubbs, D. Ganem, R. H. Silverman, and J. L. DeRisi. 2006. Identification of a novel Gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2:e25. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.van den Hoogen, B. G., J. C. de Jong, J. Groen, T. Kuiken, R. de Groot, R. A. Fouchier, and A. D. Osterhaus. 2001. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat. Med. 7:719-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Hoek, L., K. Pyrc, M. F. Jebbink, W. Vermeulen-Oost, R. J. Berkhout, K. C. Wolthers, P. M. Wertheim-van Dillen, J. Kaandorp, J. Spaargaren, and B. Berkhout. 2004. Identification of a new human coronavirus. Nat. Med. 10:368-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Leeuwen, M., M. M. Williams, P. Koraka, J. H. Simon, S. L. Smits, and A. D. Osterhaus. 2010. Human picobirnaviruses identified by molecular screening of diarrhea samples. J. Clin. Microbiol. 48:1787-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Victoria, J. G., A. Kapoor, K. Dupuis, D. P. Schnurr, and E. L. Delwart. 2008. Rapid identification of known and new RNA viruses from animal tissues. PLoS Pathog. 4:e1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilhelmi, I., E. Roman, and A. Sanchez-Fauquier. 2003. Viruses causing gastroenteritis. Clin. Microbiol. Infect. 9:247-262. [DOI] [PMC free article] [PubMed] [Google Scholar]