Abstract
DNA microarray technology has already revolutionized basic research in infectious diseases, and whole-genome sequencing efforts have allowed for the fabrication of tailor-made spotted microarrays for an increasing number of bacterial pathogens. However, the application of microarrays in diagnostic microbiology is currently hampered by the high costs associated with microarray experiments and the specialized equipment needed. Here, we show that a thorough bioinformatic postprocessing of the microarray design to reduce the amount of unspecific noise also allows the reliable use of spotted gene expression microarrays for gene content analyses. We further demonstrate that the use of only single-color labeling to halve the costs for dye-labeled nucleotides results in only a moderate decrease in overall specificity and sensitivity. Therefore, gene expression microarrays using only single-color labeling can also reliably be used for gene content analyses, thus reducing the costs for potential routine applications such as genome-based pathogen detection or strain typing.
In recent years, molecular applications in the diagnosis of infectious diseases have become commonplace in academic medical centers and tertiary-care facilities and are becoming also more tangible in community-based settings. However, to be implemented in clinical microbiology laboratories with ease and accuracy, the further advancement of molecular infectious disease diagnostics is dependent on the ability of multiplexing technologies or the ability to detect and identify more than one pathogen simultaneously from the same specimen (18).
One approach to multiplex detection and characterization is microarray analysis which, since its first description in the 1990s, has already revolutionized basic research in infectious diseases (reviewed in references 7 and 18). Accordingly, microbial diagnostic microarrays (MDMs) have also been used in a number of research applications in clinical microbiology (18). For example, an oligonucleotide microarray targeting the 16S rRNA gene was recently developed for the detection of a panel of 40 predominant human intestinal bacterial pathogens in human fecal samples (35), and assays using broad-range PCR along with microarrays have been shown to allow rapid bacterial detection and identification with positive blood culture (2). Another promising application of microarray techniques in clinical microbiology is the determination of antimicrobial resistance by simultaneously detecting a panel of drug resistance-related mutations in microbial genomes, and oligonucleotide microarrays were developed to analyze and identify drug-resistant Mycobacterium tuberculosis strains with results that were comparable to those of standard antimicrobial susceptibility testing but obtained in less than 24 h (12, 17). Likewise, an oligonucleotide microarray outperforming the standard procedures in terms of assay time and the depth of information provided was designed for the rapid identification of extended-spectrum beta-lactamases in Gram-negative bacteria by simultaneously genotyping blaTEM, blaSHV, and blaCTX-M (14). The accurate identification and prompt typing of pathogens is finally another important area where MDMs have numerous potential applications, and microarray-based approaches that can be used to support or replace the classical serotyping methods for several conventional diarrhea bacterial pathogens, including pathogenic Escherichia coli (16), as well as Salmonella (34) and Campylobacter (33) species, have already been proposed. As a consequence, this multitude of possible clinical applications, as well as their recent technical evaluation by the MicroArray Quality Control (MAQC) consortium (27), resulted in the approval of microarray technology by the U.S. Food and Drug Administration (26), and recent advancements in whole-genome sequencing technologies will allow for the fabrication of tailor-made spotted microarrays for an ever-increasing number of bacterial pathogens.
Unfortunately, one of the biggest challenges for the use of MDMs in routine microbiological diagnostic laboratories is still the high price not only for the design and manufacturing of microarrays but also for the downstream experimental steps (19). Moreover, since transcription profiling is still the most widespread application of microarrays and the design of oligonucleotides for gene expression arrays differs from the design of oligonucleotides used, e.g., for strain identification and typing or the detection of antibiotic resistance genes by comparative genome hybridization (aCGH) (4), tailor-made microarrays are required for each of these applications. Since the costs for dye-labeled deoxynucleoside triphosphates (dNTPs) are also still considerable, possibilities to reduce the costs would therefore include the dual use of gene expression microarrays also for aCGH experiments and/or the use of only a single fluorescent dye for DNA labeling. However, the difference in quality between one- and two-color designs have only been assessed for measuring gene expression differences (21), as well as with respect to tissue classification tasks (3), and thus far no systematic comparisons have been made in the context of aCGH studies. Likewise, a systematic comparison of the effect of spotted array design on aCGH performance is also still missing.
Since quality and reproducibility are critical issues in microarray experiments (6), we compared here computationally unprocessed and postprocessed spotted oligonucleotide microarrays originally designed for transcriptional profiling in Neisseria meningitidis as a test case in a one-color and a two-color aCGH setup with respect to specificity, sensitivity, and prediction variability using the results from computational genome comparisons as a reference.
MATERIALS AND METHODS
Microarray design and fabrication.
There are already two PCR-product-based spotted microarrays available for N. meningitidis which were both termed pan-Neisseria microarray (30, 31) and representing open reading frames (ORFs) from the genomes of the invasive N. meningitidis strains Z2491 (20), MC58 (32), and FAM18 (5) among others. However, oligonucleotide-based microarrays were shown to offer important advantages over PCR product-based microarrays, including a reduction in cross-hybridization and an increase in the differentiation of overlapping genes or highly homologous regions (reviewed in reference 10). Therefore, to also include the recently sequenced genome of the meningococcal carriage strain α14 (25), an oligonucleotide-based microarray was constructed in collaboration with Eurofins MWG Operon (Ebersberg, Germany) containing 2,872 oligonucleotides representing 2,098 open reading frame (ORFs) from N. meningitidis MC58 (NCBI Ref_Seq NC_003112), 2,119 ORFs from N. meningitidis Z2491 (NCBI Ref_Seq NC_003116), 2,131 ORFs from N. meningitidis FAM18 (NCBI Ref_Seq NC_008767), and 2,067 ORFs from N. meningitidis α14 (GenBank accession no. AM889136), respectively. The oligonucleotides were designed according to the method described by Li and Stormo (15). Accordingly, 2,078 oligonucleotides were directly designed from the primary source N. meningitidis MC58, but the probes were selected such that a large number of N. meningitidis Z2491, N. meningitidis FAM18, and N. meningitidis α14 ORFs were also represented. An ORF was said to be represented by an oligonucleotide if the oligonucleotide had a greater than 93% identity over the entire length of the oligonucleotide in the corresponding ORF. The cross-hybridization percent identity was calculated from the number of matched bases divided by oligonucleotide length times 100 using ungapped BLASTN (1) comparisons against the top non-self-hit gene for that oligonucleotide. All oligonucleotides were first designed for each ORF in the primary source N. meningitidis MC58 minimized for cross-hybridization identity to other non-self ORFs in strain MC58 and to maximize cross-hybridization identity to all other ORFs of the three other genomes. Since N. meningitidis MC58 has a number of duplicate ORFs with almost identical nucleotide sequences, only one of these duplicate ORFs was used for oligonucleotide design. Accordingly, 296 oligonucleotides were directly designed from N. meningitidis Z2491, 102 oligonucleotides were directly designed from N. meningitidis FAM18, and 396 oligonucleotides were directly designed from N. meningitidis α14, respectively. All oligonucleotides had ≤70% cross-hybridization identity to all other non-self ORFs and were designed to be 70mers with a Tm of 75 ± 5°C except for a few cases with too-high or too-low GC content. In addition, 14 oligonucleotides that represent different regions of the luciferase gene (from pGL3-Basic Vector; Promega, Manheim, Germany) were spotted, and 100 ng of a PCR product (using the primer pair LucF [5-CTAGCAAAATAGGCTGTCCC-3′] and LucR [5′-GACGATAGTCATGCCCCGCG-3′]) was used as a spike in controls in each slide to assess the quality of hybridization. The oligonucleotides were resuspended in spotting buffer (3× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 1.5 M betaine) to a final concentration of 25 μM and were spotted onto Schott Nexterion E epoxy-coated slides (Schott AG, Mainz, Germany) using an OmniGrid spotter (Genomic Solutions, Ann Arbor, MI) according to the manufacturer's protocol. Test scans of slides hybridized with Cy3-labeled nonamer probe [4× SSC, 1 mg of poly(dA)/ml, 50 mM HEPES, 0.2% sodium dodecyl sulfate, 7.5 μM Cy3 random nonamer (Qiagen, Hilden, Germany)] were performed to assess the quality of the spotted slides with respect to spot morphology and missing spots.
The layout of the spotted microarray slides and the data associated with the present study have been deposited in NCBI's Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through the GEO series accession number GSE18158 (two-color data set) and GSE18159 (one-color data set), respectively.
Microarray hybridization.
Chromosomal DNA from the meningococcal strains were isolated by using Qiagen Genomic-Tip 20/G (Qiagen, Hilden, Germany), and the microarray slides were prehybridized according to the manufacturer's protocols (Schott AG, Germany).
In a low-cost setting using only one fluorescently labeled dNTP, 4 μg of chromosomal DNA was labeled with Cy3 dCTP (GE Healthcare, Munich, Germany) and Klenow enzyme. Briefly, 4 μg of meningococcal DNA (denatured at 95°C for 10 min) was mixed with 100 μM dATP, dTTP, and dGTP and 50 μM dCTP; 10 μg of random nonamers (Sigma Aldrich, Heidenheim, Germany); 1 nmol of Cy3dCTP; and 2 U of Klenow enzyme (Roche Diagnostics, Mannheim, Germany) with a 1× reaction buffer (50 mM Tris-HCl, 10 mM MgCl2, 100 μM dithiothreitol, 2 μg of bovine serum albumin). The labeling reaction was performed at 37°C overnight. The reaction was stopped by using 200 μM EDTA (pH 8.0) and 4 M sodium acetate (pH 4.5), and the labeled DNA was precipitated using absolute ethanol. The labeled DNA pellet was resuspended in distilled water after washing with 70% ethanol and drying. Labeling efficiencies were measured by using Nanodrop 1000 (PeqLab Biotechnologie GmbH, Erlangen, Germany) to assess the quantity of labeled DNA and also the rate of incorporation of the labeled dCTPs (Cy3/Cy5). Standard rates of incorporation of labeled nucleotides were adapted from reference 24 and, accordingly, a probe specific activity as defined by (pmol of dye per μl)/(μg of DNA per μl) > 25 pmol/μg and a yield of labeled DNA > 6 μg were used for hybridizations. The DNA probe for hybridization onto the microarray slides was prepared by mixing the labeled DNA with 3 μg of salmon sperm DNA (Sigma Aldrich) and two volumes of hybridization buffer (Eurogentec, Cologne, Germany). The probes were denatured at 95°C for 10 min, manually hybridized onto prehybridized microarray slides using a coverslip, and incubated overnight at 50°C. After hybridization, the slides were washed according to the manufacturer's protocols and scanned using ScanArray HT (Perkin-Elmer, Jügesheim, Germany), and the resulting images were analyzed using Imagene 4.0 (BioDiscovery, El Segundo, CA) to generate the raw files for further analyses.
In the high-cost setting using two fluorescently labeled dNTPs, an aliquot of all DNAs to be tested was pooled together to form the common reference. Then, 4 μg of test DNA was labeled with Cy3 dCTP, and 4 μg of reference DNA was labeled with Cy5 dCTP using the protocol described above. Hybridization in this case was carried out on a Tecan 4800 Pro hybridization station (Tecan Trading AG, Switzerland). The slides were scanned by using GenePix 4200, and the raw data files were extracted by using GenePix Pro 4.0. Spots were flagged in obvious instances of high background or stray fluorescent signals in both experiments. In both cases (i.e., the low-cost and high-cost settings), three microarrays were performed for each probe.
Prediction of gene presence or absence from the genomic data.
For computational gene presence or absence predictions, we performed gapped BLASTN searches (word size, 11) of all 2,870 oligonucleotides against both strands of the genomes of N. meningitidis MC58, N. meningitidis Z2491, N. meningitidis FAM18, N. meningitidis α14, N. meningitidis α153 (GenBank accession no. AM889137), and N. meningitidis α275 (GenBank accession no. AM889138). Hits below an E-value cutoff of 0.001 were considered significant and predicted the presence of the oligonucleotide in the genome and therefore of a hybridization signal.
Postprocessing of the microarray design for aCGH.
To minimize the chance for cross-hybridizations with noncoding regions, we used the available genome annotations to distinguish true gene hits from hits in intergenic regions. Oligonucleotides that resulted in a significant BLAST hit in an intergenic region within one or more genomes were therefore removed from downstream analyses.
Prediction of gene absence or presence from aCGH data.
Microarrays were analyzed using Limma (29) implemented in the R language (23). All processing and normalization steps were performed identically on both one- and two-color arrays. Normalization was carried out using variance stabilization (13) and, after fitting to the reference channel, the two-color arrays were transformed into single-channel intensities using Limma's intraspot correlation routine. The normalized single-channel intensities were used for the absence or presence prediction of individual genes. To determine the intensity threshold above which a gene was predicted to be present, we fitted receiver operating characteristic (ROC) curves to the linear predictor and used the threshold minimizing the misclassification error, thus treating specificity and sensitivity as equal (28). To assess robustness of the threshold estimator, we generated 1,000 bootstrap samples by drawing with replacement from the original data set and repeated the threshold estimation for each bootstrap sample to create ca. 95% bootstrapped percentile confidence bounds for the threshold parameter. All normalization and bootstrapping steps were performed using R.
RESULTS AND DISCUSSION
Statistically rigid validation of the hybridization results with respect to prediction accuracy has seldom been performed thus far in aCGH studies. Therefore, we used available whole-genome data as a “gold standard” against which we individually compared the results of aCGH experiments obtained under two different test conditions representing different complexity and cost scales: a simple one-color manually hybridized intensity array versus the log fold changes from a two-color study against a pooled common reference probeset, hybridized with specialized hybridization station equipment. Microarray experiments for both one- and two-color arrays were carried out using identical arrays originally designed for transcriptional profiling studies. Although there are steps in the experimental design in both settings that are not identical, the robust postprocessing of the obtained data in both cases and their comparison to the available genome data allowed for a robust and meaningful comparison between the one-color and two-color arrays. Prediction accuracy was measured both with and without computational postprocessing of the arrays to cope with the danger of cross-hybridization to intergenic regions.
Effect of array design.
Typically, aCGH microarrays are specifically designed for the task, and oligonucleotides to be used in expression analyses are usually not suited for aCGH studies since they are designed to minimize cross-hybridization with other coding regions but not with genomic regions that are not expressed (15). To cope with possible cross-hybridizations from intergenic regions, we performed a filtering step as described above that excludes these oligonucleotides from the downstream analyses. Since most laboratories use microarrays for gene expression analyses this filtering provides a cost-efficient way to exploit a specific microarray to its fullest by using the same array design for both expression and aCGH studies.
By excluding oligonucleotides with possible cross-hybridizations in intergenic regions 1,174 oligonucleotides were removed, thus reducing the number of oligonucleotides available for aCGH to 1696. However, for one- and two-color arrays, comparison of the array performance before and after postprocessing showed that false-positive rates dramatically decreased in the filtered setting (Table 1). For example, in α14 the false-positive rate decreased from 31% without filtering to only 1.3%, and this rather simple computational postprocessing therefore results in a drastically improved performance of gene expression arrays in aCGH studies.
TABLE 1.
Comparison of array performancea
| Result | Relative error (±SD) |
|||
|---|---|---|---|---|
| Two-color array |
One-color array |
|||
| Filtered | Unfiltered | Filtered | Unfiltered | |
| True positive | 0.99 (±0.00) | 0.99 (±0.00) | 0.98 (±0.01) | 0.97 (±0.01) |
| False negative | 0.01 (±0.00) | 0.01 (±0.00) | 0.02 (±0.01) | 0.03 (±0.01) |
| True negative | 0.95 (±0.05) | 0.58 (±0.08) | 0.81 (±0.04) | 0.52 (±0.06) |
| False positive | 0.05 (±0.05) | 0.42 (±0.08) | 0.19 (±0.04) | 0.48 (±0.06) |
| Total correct | 0.98 (±0.01) | 0.88 (±0.04) | 0.94 (±0.01) | 0.85 (±0.03) |
| Total wrong | 0.02 (±0.01) | 0.12 (±0.04) | 0.06 (±0.01) | 0.15 (±0.03) |
Relative error and correct classification rates for gene absence or presence predictions were averaged over all six strains comparing one- and two-color arrays before and after postprocessing.
Prediction accuracy for one- and two-color arrays.
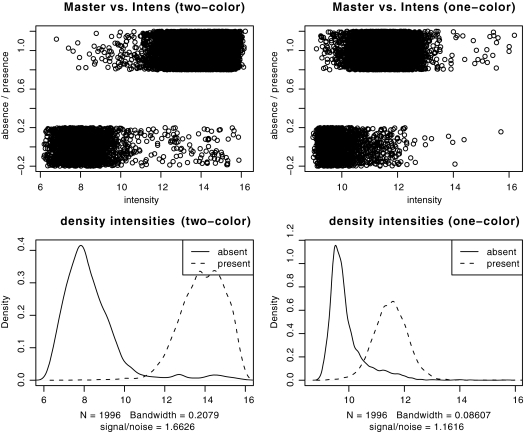
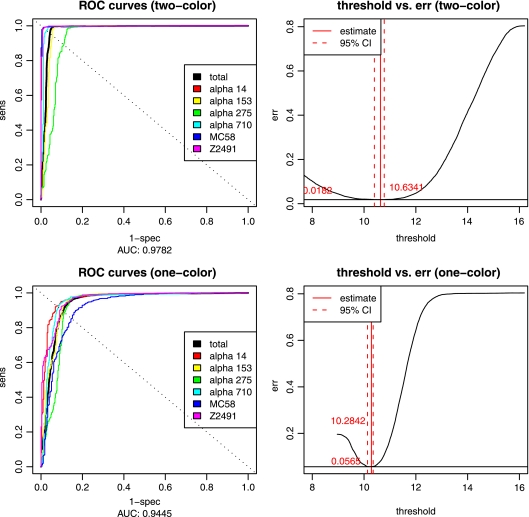
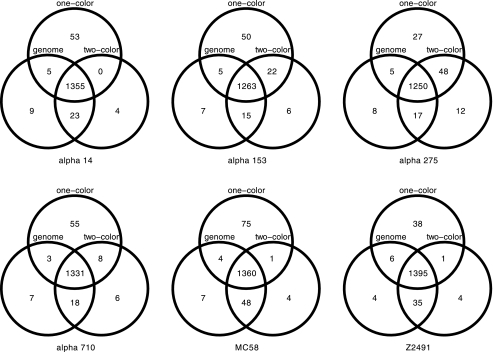
Explorative analysis of the experimental signal intensities with respect to the results from computational genome comparisons further revealed that the cheaper one-color arrays had lower signal-to-noise ratios (1.17 compared to 1.66) and greater overlap between genes predicted as absent and present by computational genome comparisons (Fig. 1, right panel) than the two-color arrays (Fig. 1, left panel). This was also confirmed by analysis of the ROC of the linear classifiers, where the area under the curve (AUC) indicated a higher accuracy of the two-color experiments (AUC = 0.98) compared to the one-color design (AUC = 0.94). The optimality criterion for choosing the intensity threshold was to minimize the combined specificity and sensitivity error (left panel of Fig. 2) and was comparable for both data sets (10.6743 in the one-color case versus 10.6341 in the two-color case), but the curvature of the error function (right panel of Fig. 2) clearly indicated lower confidence in the one-color case. However, individual comparisons of the results of genome analysis with these gene absence or presence classifications further showed that both one- and two-color studies were indeed able to reproduce the results from the computational genome analyses for the most part (Fig. 3), and this could be done largely independent of whether the analyzed genome was spotted on the chip (misclassification rates of 2 and 6%, respectively, for one-color and two-color studies). The difference in quality between one- and two-color experiments is only apparent with 76 genes that the one-color arrays failed to classify correctly compared to the two-color experiments. Investigation of the misclassified genes showed that their intensities lay close to the decision boundary, and misclassification could therefore be due to the lower signal-to-noise ratio of the one-color arrays. The two-color arrays thus outperformed the results from the one-color studies by about a factor of 3, lowering the total misclassification rates from 5.68 to 1.86%. However, even a combination of state-of-the-art hybridization technology with an increased experimental effort requiring an automated hybridization station and twice the amount of fluorescently labeled dCTP only led to an overall increase in prediction accuracy from 94 to 98% compared to manual hybridization and a one-color setup, provided a careful selection of oligonucleotide probes.
FIG. 1.
Comparison of spot intensities (upper panels) and the corresponding densities (lower panels) between one- and two-color arrays. The two-color arrays (left panels) have clearly a wider spread, lower overlap, and therefore better differentiation between absent or present genes. Note that the absence or presence data in the first plots (upper panels) are binary values that have been jittered solely for visualization purposes.
FIG. 2.
ROC curves for one- and two-color arrays. ROC curves are shown for both one-color (top left) and two-color (bottom left) arrays for all six strains (colored) and the complete data set (black), which was used for determination of the optimal threshold (right). The threshold fitted on the original data set had its optimum at 10.6341 at an error rate of 0.0182 in the two-color case and 10.2842 at 0.0565 in the one-color case. err, error.
FIG. 3.
Gene absence and presence comparisons for six meningococcal strains. Venn diagrams of gene presence comparing genome sequencing with one-color and two-color arrays over all six test strains are shown. For the vast majority of the genes (92.94, 92.26, 91.61, 93.15, 91.14, and 94.09% from left to right, top to bottom) for genome sequencing, the one- and two-color arrays were in agreement (total intersection set and universe). Prediction errors accumulate in the intersections, where one-color predictions contradict both two-color and genome analyses (66 and 19 in α14, 52 and 13 in α153, 30 and 13 in α275, 56 and 17 in α710, 79 and 39 in MC58, and 48 and 24 in Z2491).
Relative cost assessment and potential cost savings.
As shown in this work, a DNA microarray experiment in its most reduced version requires in addition to custom-made slides only a microarray scanner and one fluorescently labeled dNTP for labeling. Therefore, for institutions where microarrays are already used for genome-wide gene expression analyses, the dual use of the same microarray also for aCGH obviates the need for extra array design and fabrication. Further, since manual hybridization did not drastically impair the overall performance of the aCGH, the acquirement of a costly hybridization station is also not required for laboratories with only a low throughput of clinical specimens to be analyzed by aCGH. Finally, cost differences between one- and two-color arrays are mainly due to the twice-as-high consumption of the fluorescent dCTP, Klenow enzyme, and dNTP in the two-color case.
Conclusion and outlook.
Based on a careful selection of oligonucleotide probes, our results show that the accuracy of gene absence/presence prediction of manually hybridized one-color microarrays in aCGH studies remains remarkable, with an overall of 94% of genes correctly annotated compared to two-color hybridization using specialized equipment. At least for institutions where microarrays are already used for genome-wide gene expression analyses, the dual use of the same microarray also for aCGH thus obviates the need for an extra array design and, in particular, the one-color aCGHs could be applied in diagnostic microbiology laboratories complementing more traditional cultural or molecular approaches. Since the computational postprocessing of the gene expression microarray design for aCGH application presented here is based on the availability of whole-genome sequences, the clinical applicability of MDMs might indeed be fostered by the increasing number of whole-genome sequences currently generated by numerous genome sequencing efforts using so-called next-generation sequencing technologies (9). The considerable number of oligonucleotides that had be excluded from further analyses is usually not a major drawback if the aCGH outcome is used for simple pathogen identification by simultaneous detection of pathogen-specific genes (8), for strain typing for epidemiologic studies based on gene content (11), or for the detection of multiple antibiotic resistance genes (22).
Acknowledgments
This study was supported by the PathoGenoMik-Plus funding initiative grant 0313801A from the Bundesministerium für Bildung und Forschung.
Footnotes
Published ahead of print on 30 June 2010.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anthony, R. M., T. J. Brown, and G. L. French. 2000. Rapid diagnosis of bacteremia by universal amplification of 23S ribosomal DNA followed by hybridization to an oligonucleotide array. J. Clin. Microbiol. 38:781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Attoor, S., E. R. Dougherty, Y. Chen, M. L. Bittner, and J. M. Trent. 2004. Which is better for cDNA-microarray-based classification: ratios or direct intensities. Bioinformatics 20:2513-2520. [DOI] [PubMed] [Google Scholar]
- 4.Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. [DOI] [PubMed] [Google Scholar]
- 5.Bentley, S. D., G. S. Vernikos, L. A. Snyder, C. Churcher, C. Arrowsmith, T. Chillingworth, A. Cronin, P. H. Davis, N. E. Holroyd, K. Jagels, M. Maddison, S. Moule, E. Rabbinowitsch, S. Sharp, L. Unwin, S. Whitehead, M. A. Quail, M. Achtman, B. Barrell, N. J. Saunders, and J. Parkhill. 2007. Meningococcal genetic variation mechanisms viewed through comparative analysis of serogroup C strain FAM18. PLoS Genet. 3:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brazma, A., P. Hingamp, J. Quackenbush, G. Sherlock, P. Spellman, C. Stoeckert, J. Aach, W. Ansorge, C. A. Ball, H. C. Causton, T. Gaasterland, P. Glenisson, F. C. Holstege, I. F. Kim, V. Markowitz, J. C. Matese, H. Parkinson, A. Robinson, U. Sarkans, S. Schulze-Kremer, J. Stewart, R. Taylor, J. Vilo, and M. Vingron. 2001. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat. Genet. 29:365-371. [DOI] [PubMed] [Google Scholar]
- 7.Bryant, P. A., D. Venter, R. Robins-Browne, and N. Curtis. 2004. Chips with everything: DNA microarrays in infectious diseases. Lancet Infect. Dis. 4:100-111. [DOI] [PubMed] [Google Scholar]
- 8.Cleven, B. E., M. Palka-Santini, J. Gielen, S. Meembor, M. Krönke, and O. Krut. 2006. Identification and characterization of bacterial pathogens causing bloodstream infections by DNA microarray. J. Clin. Microbiol. 44:2389-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coppée, J.-Y. 2008. Do DNA microarrays have their future behind them? Microbes Infect. 10:1067-1071. [DOI] [PubMed] [Google Scholar]
- 10.Dorrell, N., S. J. Hinchliffe, and B. W. Wren. 2005. Comparative phylogenomics of pathogenic bacteria by microarray analysis. Curr. Opin. Microbiol. 8:620-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorrell, N., J. A. Mangan, K. G. Laing, J. Hinds, D. Linton, H. Al-Ghusein, B. G. Barrell, J. Parkhill, N. G. Stoker, A. V. Karlyshev, P. D. Butcher, and B. W. Wren. 2001. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 11:1706-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gryadunov, D., V. Mikhailovich, S. Lapa, N. Roudinskii, M. Donnikov, S. Pan'kov, O. Markova, A. Kuz'min, L. Chernousova, O. Skotnikova, A. Moroz, A. Zasedatelev, and A. Mirzabekov. 2005. Evaluation of hybridisation on oligonucleotide microarrays for analysis of drug-resistant Mycobacterium tuberculosis. Clin. Microbiol. Infect. 11:531-539. [DOI] [PubMed] [Google Scholar]
- 13.Huber, W., A. von Heydebreck, H. Sueltmann, A. Poustka, and M. Vingron. 2002. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18(Suppl. 1):S96-S104. [DOI] [PubMed] [Google Scholar]
- 14.Leinberger, D. M., V. Grimm, M. Rubtsova, J. Weile, K. Schroppel, T. A. Wichelhaus, C. Knabbe, R. D. Schmid, and T. T. Bachmann. 2010. Integrated detection of extended-spectrum beta-lactam resistance by DNA microarray-based genotyping of TEM, SHV, and CTX-M genes. J. Clin. Microbiol. 48:460-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, F., and G. D. Stormo. 2001. Selection of optimal DNA oligonucleotides for gene expression arrays. Bioinformatics 17:1067-1076. [DOI] [PubMed] [Google Scholar]
- 16.McNamara, S. E., U. Srinivasan, L. Zhang, T. S. Whittam, C. F. Marrs, and B. Foxman. 2009. Comparison of probe hybridization array typing to multilocus sequence typing for pathogenic Escherichia coli. J. Clin. Microbiol. 47:596-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mikhailovich, V., S. Lapa, D. Gryadunov, A. Sobolev, B. Strizhkov, N. Chernyh, O. Skotnikova, O. Irtuganova, A. Moroz, V. Litvinov, M. Vladimirskii, M. Perelman, L. Chernousova, V. Erokhin, A. Zasedatelev, and A. Mirzabekov. 2001. Identification of rifampin-resistant Mycobacterium tuberculosis strains by hybridization, PCR, and ligase detection reaction on oligonucleotide microchips. J. Clin. Microbiol. 39:2531-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller, M. B., and Y. W. Tang. 2009. Basic concepts of microarrays and potential applications in clinical microbiology. Clin. Microbiol. Rev. 22:611-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nolte, F. S., and A. M. Caliendo. 2007. Molecular detection and identification of microorganisms, p. 218-244. In P. Murray, E. J. Baron, J. H. Jorgensen, M. L. Landry, and M. A. Pfaller (ed.), Manual of clinical microbiology. ASM Press, Washington, DC.
- 20.Parkhill, J., M. Achtman, K. D. James, S. D. Bentley, C. Churcher, S. R. Klee, G. Morelli, D. Basham, D. Brown, T. Chillingworth, R. M. Davies, P. Davis, K. Devlin, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Leather, S. Moule, K. Mungall, M. A. Quail, M. A. Rajandream, K. M. Rutherford, M. Simmonds, J. Skelton, S. Whitehead, B. G. Spratt, and B. G. Barrell. 2000. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404:502-506. [DOI] [PubMed] [Google Scholar]
- 21.Patterson, T. A., E. K. Lobenhofer, S. B. Fulmer-Smentek, P. J. Collins, T. M. Chu, W. Bao, H. Fang, E. S. Kawasaki, J. Hager, I. R. Tikhonova, S. J. Walker, L. Zhang, P. Hurban, F. de Longueville, J. C. Fuscoe, W. Tong, L. Shi, and R. D. Wolfinger. 2006. Performance comparison of one-color and two-color platforms within the MicroArray Quality Control (MAQC) project. Nat. Biotechnol. 24:1140-1150. [DOI] [PubMed] [Google Scholar]
- 22.Perreten, V., L. Vorlet-Fawer, P. Slickers, R. Ehricht, P. Kuhnert, and J. Frey. 2005. Microarray-based detection of 90 antibiotic resistance genes of gram-positive bacteria. J. Clin. Microbiol. 43:2291-2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.R Development Core Team. 2008. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
- 24.Redon, R., D. Rigler, and N. P. Carter. 2009. Comparative genomic hybridization: DNA preparation for microarray fabrication. Methods Mol. Biol. 529:259-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schoen, C., J. Blom, H. Claus, A. Schramm-Gluck, P. Brandt, T. Muller, A. Goesmann, B. Joseph, S. Konietzny, O. Kurzai, C. Schmitt, T. Friedrich, B. Linke, U. Vogel, and M. Frosch. 2008. Whole-genome comparison of disease and carriage strains provides insights into virulence evolution in Neisseria meningitidis. Proc. Natl. Acad. Sci. U. S. A. 105:3473-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi, L., R. G. Perkins, H. Fang, and W. Tong. 2008. Reproducible and reliable microarray results through quality control: good laboratory proficiency and appropriate data analysis practices are essential. Curr. Opin. Biotechnol. 19:10-18. [DOI] [PubMed] [Google Scholar]
- 27.Shi, L., L. H. Reid, W. D. Jones, R. Shippy, J. A. Warrington, S. C. Baker, P. J. Collins, F. de Longueville, E. S. Kawasaki, K. Y. Lee, Y. Luo, Y. A. Sun, J. C. Willey, R. A. Setterquist, G. M. Fischer, W. Tong, Y. P. Dragan, D. J. Dix, F. W. Frueh, F. M. Goodsaid, D. Herman, R. V. Jensen, C. D. Johnson, E. K. Lobenhofer, R. K. Puri, U. Schrf, J. Thierry-Mieg, C. Wang, M. Wilson, P. K. Wolber, L. Zhang, S. Amur, W. Bao, C. C. Barbacioru, A. B. Lucas, V. Bertholet, C. Boysen, B. Bromley, D. Brown, A. Brunner, R. Canales, X. M. Cao, T. A. Cebula, J. J. Chen, J. Cheng, T. M. Chu, E. Chudin, J. Corson, J. C. Corton, L. J. Croner, C. Davies, T. S. Davison, G. Delenstarr, X. Deng, D. Dorris, A. C. Eklund, X. H. Fan, H. Fang, S. Fulmer-Smentek, J. C. Fuscoe, K. Gallagher, W. Ge, L. Guo, X. Guo, J. Hager, P. K. Haje, J. Han, T. Han, H. C. Harbottle, S. C. Harris, E. Hatchwell, C. A. Hauser, S. Hester, H. Hong, P. Hurban, S. A. Jackson, H. Ji, C. R. Knight, W. P. Kuo, J. E. LeClerc, S. Levy, Q. Z. Li, C. Liu, Y. Liu, M. J. Lombardi, Y. Ma, S. R. Magnuson, B. Maqsodi, T. McDaniel, N. Mei, O. Myklebost, B. Ning, N. Novoradovskaya, M. S. Orr, T. W. Osborn, A. Papallo, T. A. Patterson, R. G. Perkins, E. H. Peters, R. Peterson, et al. 2006. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat. Biotechnol. 24:1151-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sing, T., O. Sander, N. Beerenwinkel, and T. Lengauer. 2007. ROCR: visualizing the performance of scoring classifiers. [DOI] [PubMed]
- 29.Smyth, G. K. 2005. Limma: linear models for microarray data, p. 397-420. In R. Gentleman, V. Carey, S. R. Dudoit, R. Irizarry, and W. Huber (ed.), Bioinformatics and computational biology solutions using R and Bioconductor. Springer, New York, NY.
- 30.Snyder, L. A. S., J. K. Davies, and N. J. Saunders. 2004. Microarray genomotyping of key experimental strains of Neisseria gonorrhoeae reveals gene complement diversity and five new neisserial genes associated with minimal mobile elements. BMC Genomics 5:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stabler, R. A., G. L. Marsden, A. A. Witney, Y. Li, S. D. Bentley, C. M. Tang, and J. Hinds. 2005. Identification of pathogen-specific genes through microarray analysis of pathogenic and commensal Neisseria species. Microbiology 151:2907-2922. [DOI] [PubMed] [Google Scholar]
- 32.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 33.Volokhov, D., V. Chizhikov, K. Chumakov, and A. Rasooly. 2003. Microarray-based identification of thermophilic Campylobacter jejuni, C. coli, C. lari, and C. upsaliensis. J. Clin. Microbiol. 41:4071-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Willse, A., T. M. Straub, S. C. Wunschel, J. A. Small, D. R. Call, D. S. Daly, and D. P. Chandler. 2004. Quantitative oligonucleotide microarray fingerprinting of Salmonella enterica isolates. Nucleic Acids Res. 32:1848-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.You, Y., C. Fu, X. Zeng, D. Fang, X. Yan, B. Sun, D. Xiao, and J. Zhang. 2008. A novel DNA microarray for rapid diagnosis of enteropathogenic bacteria in stool specimens of patients with diarrhea. J. Microbiol. Methods 75:566-571. [DOI] [PubMed] [Google Scholar]