Abstract
Full-genome sequencing of 11 Australian and 1 New Zealand avian influenza A virus isolate (all subtype H7) has enabled comparison of the sequences of each of the genome segments to those of other subtype H7 avian influenza A viruses. The inference of phylogenetic relationships for each segment has been used to develop a model of the natural history of these viruses in Australia. Phylogenetic analysis of the hemagglutinin segment indicates that the Australian H7 isolates form a monophyletic clade. This pattern is consistent with the long-term, independent evolution that is, in this instance, associated with geographic regions. On the basis of the analysis of the other H7 hemagglutinin sequences, three other geographic regions for which similar monophyletic clades have been observed were confirmed. These regions are Eurasia plus Africa, North America, and South America. Analysis of the neuraminidase sequences from the H7N1, H7N3, and H7N7 genomes revealed the same region-based relationships. This pattern of independent evolution of Australian isolates is supported by the results of analysis of each of the six remaining genomic segments. These results, in conjunction with the occurrence of five different combinations of neuraminidase subtypes (H7N2, H7N3, H7N4, H7N6, H7N7) among the 11 Australian isolates, suggest that the maintenance host(s) is nearly exclusively associated with Australia. The single lineage of Australian H7 hemagglutinin sequences, despite the occurrence of multiple neuraminidase types, suggests the existence of a genetic pool from which a variety of reassortants arise rather than the presence of a small number of stable viral clones. This pattern of evolution is likely to occur in each of the regions mentioned above.
The emergence of highly pathogenic avian influenza viruses of subtype H5N1 as a potential human pandemic disease threat has focused attention on the roles that wild birds play in the maintenance and distribution of avian influenza viruses (18, 22). Moreover, the H5 and H7 subtypes of avian influenza A virus are major causes of economic loss in poultry production through disease. In Australia, there have been five documented outbreaks of H7 subtype avian influenza A virus disease, with evidence of adaptation to the poultry host being provided by sequence data supporting the presence of high-pathogenicity avian influenza virus (HPAI) isolates in poultry. Waterfowl (Anseriformes order, particularly ducks, geese, and swans) and the waders and gulls (Charadriiformes order, particularly gulls, terns, and waders) have been found to be the major global natural reservoirs of influenza A viruses. Transmission of avian influenza viruses from wild birds to production poultry and geographic spread are dependent upon the migratory behavior of the wild bird reservoir hosts. Members of the Anseriformes and Charadriiformes orders undertake both irregular and regular transcontinental and intercontinental migrations. During these migrations, large numbers of birds congregate at aquatic feeding locations, providing ideal sites for cross-species transmission of avian influenza viruses. A variety of mechanisms have been observed whereby influenza A viruses adapt rapidly. These include genetic shifts facilitated through genome segment reassortment, as well as genetic drift through the insertion, deletion, and substitution of nucleotides. The error-prone RNA replication and a lack of error correction are the causes of drift. In vivo, this results in viral genetic diversity within any viral sample, or a quasispecies, thus providing a pool of closely related variant viruses from enabling events, such as viral adaptation to new hosts (25). Long-term sampling of water birds in North America and Europe has started to elucidate the ecology and biology of the avian influenza A virus types in the natural reservoirs in these regions (8, 18, 22). There is a suggestion that two superfamilies, the Eurasia (which in the context of this paper includes Europe, Asia, and Africa) and the Americas superfamilies, exist; however, the extent of overlap and the rate of transfer of influenza viruses between these two regions are not well-defined. Recent studies suggest that intercontinental virus exchange is slow and limited (17), while a detailed analysis of the differences between H7 hemagglutinin (HA) segments circulating in Europe and China showed that the H7 hemagglutinin segments shared a recent common ancestor and limited sequence divergence on a background of multiple reassortant virus genotypes between 1999 and 2005 (7).
Avian influenza A viruses of the Oceania region (Australia, New Zealand, and southwest Pacific) have been far less well studied (3). Australia and New Zealand are at the southern extremity of a number of major bird migration pathways. Waders in the Charadriidae family migrate to south and southeastern Australia and New Zealand from their summer breeding grounds in Arctic regions of Siberia and Alaska, where they freely mix with the same or other species which migrate into the shared breeding grounds of Eurasia and the Americas (30). Pelagic seabirds of the Procellariformes order breed on and around Australian and New Zealand coasts during the southern hemisphere summer and migrate to maritime regions of the northern Pacific associated with Japan, Russia, and Alaska. Some move as far as the west coasts of North and South America (28). Unlike North and South America and Europe, where regular migrations of ducks, geese, swans, etc., are established, the members of the Anatidae family (ducks, etc.) in Australia and New Zealand are mainly endemic residents (30). However, within Australia, ducks undertake long-distance movements in response to water availability. Movements of waterfowl from northern Australia to nearby areas of Southeast Asia are believed to occur but are limited, as suggested by Wallace's Line (19). Generally, these waterfowl movements have not been well studied (30). The risks to Australian poultry production systems by movement of H5N1 via migratory shorebirds and nomadic wildfowl have been assessed to be low using risk-based analysis techniques (9, 10).
Regular and extensive surveillance sampling of migratory birds has been undertaken in North America and northern Europe (17, 18). The findings have shed significant insights into the ecology of the viruses and their hosts (8, 17). In contrast, surveillance sampling of wild birds in Asia and Oceania has been spasmodic and sparse, until the recent emergence of H5N1 highly pathogenic avian influenza virus as a poultry and human disease threat. Spasmodic and small-scale outbreaks of highly pathogenic avian influenza virus have occurred in Australian poultry production flocks located in the southeastern region of the continent. These poultry production areas are concentrated close to large human population centers (26, 33, 34). Each of the Australian outbreaks has been rapidly controlled by slaughter of infected flocks. All have been caused by avian influenza viruses of the H7 subtype, which appear to have entered production poultry from water birds, possibly wild ducks, via contaminated water supplies used on the poultry farms. Disease has occurred on five occasions: 1976 (H7N7), 1985 (H7N7), 1992 (H7N3), 1994 (H7N3), and 1997 (H7N4) (13, 14, 26, 27, 31, 34). National on-farm biosecurity measures have been focused on reducing the likelihood of future outbreaks. The availability of avian influenza virus isolates from poultry and wild birds associated with these outbreaks, along with a small number of subtype H7 avian influenza viruses isolated from wild ducks during recent national surveillance programs in Australia and New Zealand, provided the opportunity to explore the relationships of Australian and New Zealand subtype H7 avian influenza virus isolates with viruses circulating elsewhere in the world.
MATERIALS AND METHODS
Subtype H7 Australian and New Zealand avian influenza virus isolates.
The subtype H7 avian influenza viruses subjected to full-genome sequencing in this study are listed in Table 1. The avian influenza viruses analyzed in this study were collected over a 31-year period (1976 to 2007). Virus isolates associated with disease outbreaks in 1976 (H7N7), 1985 (H7N7), 1992 (H7N3), 1994 (H7N3), and 1997 (H7N4) were sequenced along with three isolates from wild birds (two Australian and one New Zealand) obtained during wild bird surveillance programs from 2005 to 2007. The wild bird isolates were derived from cloacal swab samples, and virus isolation was by inoculation of embryonated eggs.
TABLE 1.
Australian and New Zealand avian influenza virus subtype H7 isolates used in this study
| Virus | Subtype | Species of origina | Collection date | Location | GenBank entries | Pathogenicity | Reference(s) |
|---|---|---|---|---|---|---|---|
| A/Chicken/Victoria/76 | H7N7 | Chicken (D, F) | January 1976 | Keysborough, Victoria | CY024786-CY024793 | Pathogenic | 2, 6, 31, 33, 35 |
| A/Duck/Victoria/76 | H7N7 | Duck (D, F) | January 1976 | Keysborough, Victoria | CY061602-CY061609 | Nonpathogenic | 2, 6, 31, 33, 35 |
| A/Starling/Victoria/85 | H7N7 | European starling (I, W) | May 1985 | Bendigo, Victoria | CY024778-CY024785 | Pathogenic | 5, 13, 20, 21, 33 |
| A/Chicken/Victoria/1/85 | H7N7 | Chicken (D, F) | May 1985 | Bendigo, Victoria | CY025069-CY025076 | Pathogenic | 5, 13, 20, 21, 33 |
| A/Chicken/Victoria/224/92 | H7N3 | Chicken (D, F) | July 1992 | Bendigo, Victoria | CY025077-CY025084 | Pathogenic | 14, 26 |
| A/Chicken/Queensland/94 | H7N3 | Chicken (D, F) | December 1994 | Brisbane, Queensland | CY022685-CY022692 | Pathogenic | 34 |
| A/Chicken/New South Wales/2/97 | H7N4 | Chicken (D, F) | November 1997 | Tamworth, NSW | CY022693-CY022700 | Pathogenic | 26 |
| A/Chicken/New South Wales/327/97 | H7N4 | Chicken (D, F) | November 1997 | Tamworth, NSW | CY022701-CY022708 | Pathogenic | 26 |
| A/Emu/New South Wales/775/97 | H7N4 | Emu (E, F) | November 1997 | Tamworth, NSW | CY022709-CY022716 | Pathogenic | 26 |
| A/Mallard/NZ/1365-355/05 | H7N7 | Mallard duck (I, W) | 2005 | Piako River, New Zealand | CY061618-CY061625 | Nonpathogenic | Unpublishedb |
| A/Gray Teal/Victoria/07 | H7N6 | Gray teal duck (E, W) | 2007 | Victoria | CY061610-CY061617 | Nonpathogenic | Unpublishedc |
| A/duck/Tasmania/277/2007 | H7N2 | Duck (E, W) | February 2007 | Tasmania | CY033161-CY033168 | Nonpathogenic | Unpublishedd |
Classifications: D, domestic; I, introduced, nonendemic; E, endemic; W, wild; F, farmed.
Wlodek Stanislawek, Ministry of Agriculture and Forestry, New Zealand.
Simone Warner, Department of Primary Industries, Victoria, Australia.
Stephen Pyecroft, Department of Primary Industries and Water, Tasmania, Australia.
On each occasion, the outbreaks were of limited scale and duration and were rapidly controlled by slaughter of infected flocks. In addition to viruses isolated from domestic chickens with disease, H7 avian influenza A viruses isolated from healthy birds associated with the outbreaks are also available; these include isolates from a domestic duck (1976), a feral European starling (1985), and a farmed emu (1997). Attempts during these outbreaks to isolate H7 avian influenza viruses from wild waterfowl were unsuccessful, although on each occasion, the introduction of H7 virus into domestic chickens was most likely caused by water supplies contaminated by wild waterfowl.
In response to the emergence of H5N1 in Asia, renewed surveillance activities in Australia and New Zealand were successful in isolating avian influenza viruses from wild and feral waterfowl. Among these viruses were three H7 subtype isolates: two from southeastern Australia (Victoria H7N6 and Tasmania H7N2, 2007) and one from New Zealand H7N7 (2005). These isolates were from wild indigenous ducks in the case of Australia and wild feral mallard ducks in the case of New Zealand (Table 1).
Cultivation of influenza isolates.
The viruses were cultivated in the allantoic cavity of 7- to 10-day-old embryonated eggs. Virus isolates were recovered from the lowest passage available in the collection held at the Australian Animal Health Laboratory. Virus stocks were prepared after a single round of limiting-dilution purification by inoculation of embryonated eggs with 10-fold dilutions of virus (three to five eggs per dilution). Eggs with the highest dilution positive for virus growth by death (highly pathogenic strains) and/or eggs that were hemagglutinin positive (low pathogenic strains) were used for the preparation of virus seed stocks, which were then passaged once more for the preparation of allantoic fluids for RNA extraction. Allantoic fluids were harvested 3 to 5 days after inoculation of the eggs.
Preparation of RNA for sequencing.
RNA was extracted directly from allantoic fluids, which had been clarified by centrifugation at 1,500 × g for 10 to 15 min, using an RNeasy minikit (Qiagen). Alternatively, the virus in the allantoic fluid was concentrated by ultracentrifugation at 100,000 × g for 1.5 h prior to extraction using the RNeasy minikit (Qiagen). After spectrophotometric determination of the concentration, the RNA was precipitated with ethanol and shipped to the J. Craig Venter Institute for sequencing.
Confirmation of avian influenza virus.
The presence of avian influenza virus RNA in each of the samples was confirmed by a reverse transcription-PCR (RT-PCR) TaqMan assay targeted at the matrix (M) gene; this assay is capable of detecting all influenza A virus types (16).
Nucleotide sequencing.
Each of the viruses was subjected to full-genome sequencing from the purified RNA using the pipeline developed at the J. Craig Venter Institute. Briefly, the RNA samples were reverse transcribed and amplified with a OneStep RT-PCR kit (Qiagen), using primers designed for a universal high-throughput sequencing pipeline. An M13 sequencing tag was added to the 5′ end of each primer to facilitate sequencing. Sequencing reactions were conducted with BigDye Terminator chemistry (Applied Biosystems). An ABI 3730 sequencer was used for sequence determination, and assembly was undertaken with a software pipeline developed specifically for the large-scale sequencing project (8). The primers used in this study are listed in Table S1 in the supplemental material.
Sequence quality assurance and editing were undertaken as described previously before the finished annotated full-genome sequences were submitted to GenBank (15). GenBank access numbers are listed in Table 1.
Phylogenetic and bioinformatics analysis.
Details for the subtype H7 influenza A virus isolates from Australia and New Zealand are listed in Table 1. The details for the isolates and the sequence accession numbers of the other subtype H7 influenza A sequences used in this study are listed in Table S2 and Table S3 in the supplemental material. The sequences were downloaded from the NCBI Influenza Virus Resource Center (http://www.ncbi.nlm.nih.gov/genomes/FLU/FLU.html) in April 2009, using the database search facility available at this site (4).
Multiple-sequence alignments of nucleic acid or protein sequences were constructed using the MUSCLE program (11). Phylogenetic analysis was performed using features of the MEGA4 suite of programs (29). In most instances, trees were inferred using the minimum-evolution method (24), and the bootstrap method was used to evaluate statistically the robustness of the trees presented with bootstrap values above 70%, considered to show a statistically supported node (12).
RESULTS
Subtype H7 hemagglutinin sequence relationships.
The hemagglutinin segment of each of the Australian and New Zealand isolates encodes a predicted HA0 protein of between 560 and 563 amino acid residues, with the variability being associated with the number of basic residues at the HA1-HA2 cleavage site (Fig. 1). The isolates recovered from wild ducks in 2007, the domestic duck in 1976, and the New Zealand feral duck in 2005 have three basic residues associated with the cleavage site, typical of low pathogenic isolates, while the remaining highly pathogenic isolates have either two (1992 and 1994 isolates) or three (1976 and 1985 isolates) additional residues. Outbreaks for which multiple isolates are available (1976, 1985, and 1997) have HA0 proteins with greater than 99% identity. In addition, the recent wild duck isolates from 2007 (Victoria and Tasmania) have greater than 98% sequence identity. The most distantly related Australian isolates are the 1976 and 2007 isolates, with 92% identity. The most related Eurasian isolate has 90% identity with an Australian isolate. The New Zealand isolate is most related to several Eurasian isolates, with the identities being up to 94.1%. It also shares up to 89.6% identity with the Australian isolates. The minimum HA0 sequence identity between two H7 isolates is 76.4% (determined from a total of 371 full-length sequences; see Table S2 in the supplemental material). The differences between the Australian and New Zealand HA0 sequences and the HA0 sequence of the A/turkey/Italy/8535/2002 (H7N3) isolate used in structural comparisons (23) reveal uniform divergence patterns across the protein, with the exception of the leader sequence and the cleavage site, where lower levels of sequence conservation were observed, thus discounting the possibility of intrasegment recombination. Approximately 93% sequence similarity was observed across the protein, with minimal deviation from this similarity being detected in any of the major regions of HA1 (E, F, F′, and R) and HA2. In addition, N-glycosylation sites were found to be conserved, including the group-specific (H7, H10, and H15) glycosylation site at Asn 82 of HA2 (data not shown).
FIG. 1.
Alignment of the protein sequence near the HA cleavage site for the Australian and New Zealand isolates. The top line shows the protein from which the sequence is derived. The cleavage site region is indicated by §.
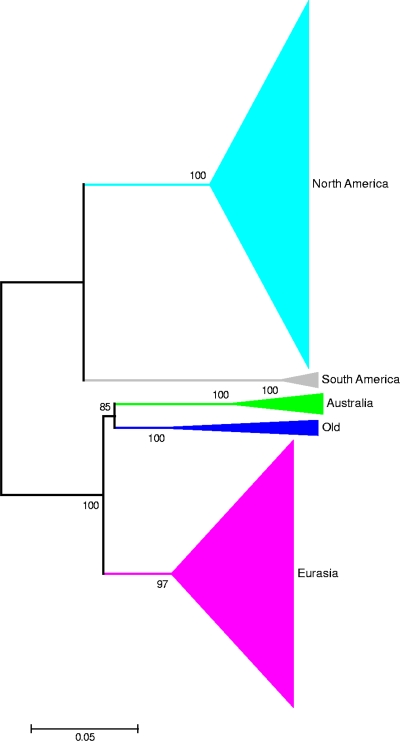
The nucleotide sequences of the HA0-coding region were aligned using the MUSCLE program, and the minimum-evolution method was used to infer relationships between the 371 full-length sequences (see Table S2 in the supplemental material). The inferred trees show strongly supported geographically based clades (Fig. 2; see also Fig. S1 in the supplemental material), with the exception of a small number of isolates from about 1930 which form a separate, statistically well supported clade (labeled “Old clade”). Concurring with the amino acid sequence similarity analysis presented above, the Australian isolates form a deep branching clade most related to the Eurasia and Old clades, and the New Zealand isolate is not closely related to any other isolate but is present as a deep branching taxon associated with the Eurasia clade (Fig. 2).
FIG. 2.
Inferred relationships for 371 subtype H7 protein-coding sequences for HA0. Clades based on geographic regions are shown. Individual taxa are not shown on the tree; the size of the shaded area is proportional to the number of taxa. The evolutionary history was inferred using the minimum-evolution method. The optimal tree with the sum of branch length of 3.05693659 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (100 replicates) is shown next to the branches. The tree is drawn to scale, with branch lengths being in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the maximum-composite-likelihood method and are in units of the number of base substitutions per site. The minimum-evolution tree was searched using the close-neighbor-interchange algorithm at a search level of 1. The neighbor-joining algorithm was used to generate the initial tree. All positions containing gaps and missing data were eliminated from the data set (complete deletion option). There were a total of 1,557 positions in the final data set. Phylogenetic analyses were conducted in the MEGA4 program (29).
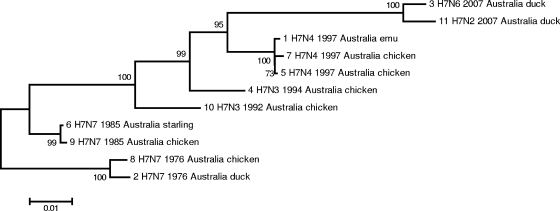
The relationship between the HA nucleotide sequences of the Australian isolates was inferred using the minimum-evolution method. In the tree shown in Fig. 3, the 1976 isolates have been used as the outgroup, and bootstrap values indicate that the inferred relationships are statistically supported; these relationships are related to the time of isolation and therefore indicate that the sequences are diverging from a single genetic pool.
FIG. 3.
Inferred relationships for the 11 subtype H7 protein-coding sequences for HA0 showing the chronological relationship between the Australian isolates. The evolutionary history was inferred using the minimum-evolution method. The optimal tree with the sum of branch length of 0.19747488 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) is shown next to the branches. The tree is drawn to scale, with branch lengths being in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the maximum-composite-likelihood method and are in units of the number of base substitutions per site. The minimum-evolution tree was searched using the close-neighbor-interchange algorithm at a search level of 1. The neighbor-joining algorithm was used to generate the initial tree. The codon positions included were 1st + 2nd + 3rd + noncoding. All positions containing gaps and missing data were eliminated from the data set (complete deletion option). There were a total of 1,674 positions in the final data set. Phylogenetic analyses were conducted in the MEGA4 program (29).
As indicated by the tree (Fig. 3), the HA0 sequences of intraoutbreak isolates are highly related, although the cleavage sites of the 1976 isolates differ, with the duck isolate being low pathogenic and the chicken isolate being highly pathogenic (Fig. 1). Modification to the cleavage site resulting in the HPAI polybasic cleavage site is a common adaptation to passage in chickens (1).
Neuraminidase sequences of subtype H7 isolates.
Isolates of the subtype H7 hemagglutinin have been found in combination with each of the nine neuraminidase (N) subtypes. Remarkably, five out of the nine combinations have been observed among the 11 Australian isolates (H7N2, H7N3, H7N4, H7N6, and H7N7), while the New Zealand isolate is H7N7. Among the 285 subtype H7 isolates for which full neuraminidase sequences are available (see Table S2 in the supplemental material), the distribution of subtypes is as follows: N1 (49 sequences), N2 (121 sequences), N3 (79 sequences), N4 (3 sequences), N5 (1 sequences), N6 (2 sequences), N7 (23 sequences), N8 (4 sequences), and N9 (3 sequences). Examination of the relationship between the sequences and the geographic location from which isolates were obtained is, in this instance, limited to where there are sufficient sequences for comparison and there is a geographic distribution of isolates; for this reason the analysis of neuraminidase sequences is limited to H7N1, H7N2, H7N3, and H7N7. In overview, it is noted that H7N1 isolates are predominantly from Eurasia and that H7N2 isolates are predominantly from North America; the small sample size means that it is unclear if there is a geographic bias for particular neuraminidase subtypes or if the distribution pattern is merely a sampling artifact.
Relationships between neuraminidase sequences from H7N3 isolates.
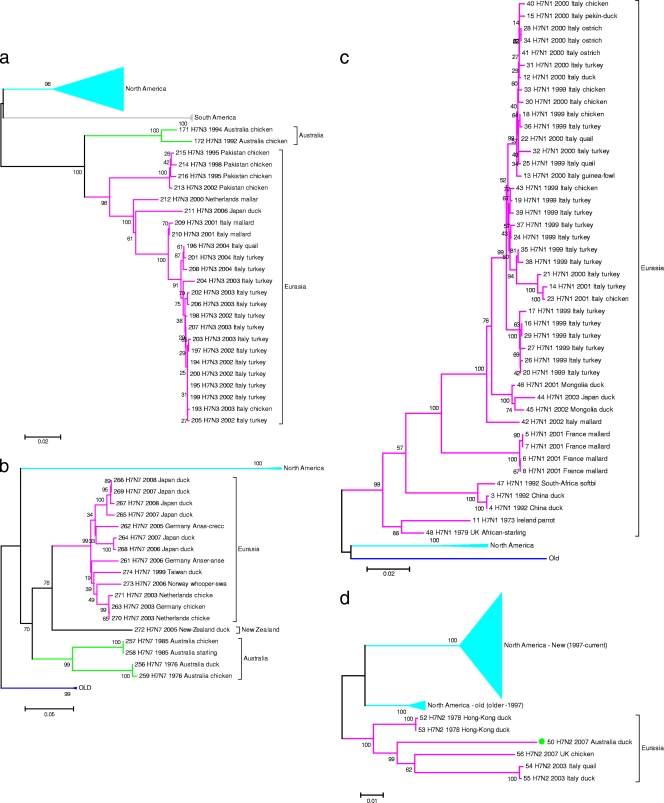
Seventy-nine full-length H7N3 neuraminidase nucleotide sequences, representing the coding regions of the gene, were aligned using the MUSCLE program, and the relationships between these sequences were inferred using the minimum-evolution method. While substantially fewer sequences were examined in this analysis, with only two sequences from Australian isolates being examined, the overall relationships were similar to the relationship found for the HA nucleotide sequences. There were four statistically supported clades associated with the geographic origins of the isolates. Again, the North America clade is most related to the South America clade and the Australia clade is most related to the Eurasia clade (Fig. 4a; see also Fig. S2a in the supplemental material); the only exception was a gull isolate from 1980. Expanding the analysis, all subtype N3 neuraminidase sequences revealed that the 1980 gull isolate is part of a deep branching clade derived from isolates collected in both Eurasia and North America. For this clade, the hosts were exclusively shorebirds (see Fig. S3 in the supplemental material). Another notable feature of the relationships inferred from the expanded set of N3 sequences is the loss of separate Eurasia and Australia clades.
FIG. 4.
(a) Inferred relationships for 79 H7N3 neuraminidase protein-coding regions. The tree shows the relationships between the major geographic clades of the H7N3 neuraminidase protein-coding region. The optimal tree with the sum of branch length of 0.81060971 is shown. There were a total of 1,247 positions in the final data set. (b) Inferred relationships for 23 H7N7 neuraminidase protein-coding regions. The tree shows the relationships between the major geographic clades of the H7N7 neuraminidase protein-coding region. The optimal tree with the sum of branch length of 0.88321959 is shown. There were a total of 527 positions in the final data set. (c) Inferred relationships for 48 H7N1 neuraminidase protein-coding regions. The tree shows the relationships between the major geographic clades of the H7N1 neuraminidase protein-coding region. The optimal tree with the sum of branch length of 0.52780887 is shown. There were a total of 1,271 positions in the final data set. (d) Inferred relationships for 121 H7N2 neuraminidase protein-coding regions. The tree shows the relationships between the major geographic clades of the H7N2 neuraminidase protein-coding region. The optimal tree with the sum of branch length of 0.64898981 is shown. There were a total of 1,312 positions in the final data set. (a to d) The evolutionary histories were inferred using the minimum-evolution method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) is shown next to the branches. The trees are drawn to scale, with the branch lengths being in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the maximum-composite-likelihood method and are in units of the number of base substitutions per site. The trees were searched using the close-neighbor-interchange algorithm at a search level of 1. The neighbor-joining algorithm was used to generate the initial trees. All positions containing gaps and missing data were eliminated from the data set (complete deletion option). Phylogenetic analyses were conducted in the MEGA4 program (29). Virus name and sequence number for each taxon are listed in Table S2 in the supplemental material.
Relationships between neuraminidase sequences from H7N7, H7N1, and H7N2 isolates.
The relationships inferred between the H7N7 neuraminidases again show an exclusive correlation between the geographic region of isolation and the clades. Despite the availability of only 23 taxa for analysis, there are four statistically supported clades, exclusively representing North America, Eurasia, Australia, and Old isolates, respectively (Fig. 4b; see also Fig. S2b in the supplemental material). The New Zealand isolate is the deepest taxon in the Eurasia clade. This topography shows considerable congruency with the relationships inferred for HA0 and the H7N3 neuraminidase protein-coding sequences. The 49 H7N1 nucleotide sequences are derived from isolates with a narrower geographic base, and in this instance, only three statistically supported clades (North America, Eurasia, and Old) were identified (Fig. 4c; see also Fig. S2c in the supplemental material), with no isolates being from Australia or New Zealand. Again, these clades are associated with geographic regions. There are incongruencies between the H7N2 tree (inferred using 121 nucleotide sequences using the minimum-evolution method [Fig. 4d; see also Fig. S2d in the supplemental material]) and the trees presented in Fig. 2 and Fig. 4. The correlation between region of isolation and genetic grouping is not exclusive. In this case, three statistically supported clades were observed (Fig. 4d): two North American clades and a Eurasian clade that included an Australian isolate. Despite being part of the Eurasia clade, the long branch length indicates that the Australian isolate is distantly related to other taxa in the clade. The dearth of Eurasia taxa may mean that sublineages, such as those apparent for the North America taxa, are not obvious. The two North America clades indicated in Fig. 4d represent two distinct sublineages of the N2 subtype. The North America taxa, in this instance, show temporal as well as geographic relationships.
Relationships between NS1 gene sequences from subtype H7.
Two distinct subtypes of the nonstructural (NS1) gene are known and are distributed globally. The relationship between the subtype B NS1 nucleotide sequences (258 NS1 sequences, inferred using the minimum-evolution method; see Fig. S4 in the supplemental material) shows an association between geographic region of isolation and genetic grouping similar to that observed with the neuraminidase and hemagglutinin sequences, with statistically supported North America, South America, and Eurasia clades being identified. A statistically supported and geographically exclusive Australia clade was not identified for the four subtype B Australian taxa. However, the tree indicates that these taxa, while not closely related to any other taxa, are most related to the Eurasia taxa.
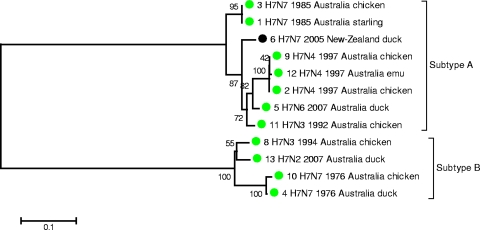
The inferred relationship between the subtype A NS1 nucleotide sequences again shows the grouping of taxa into geographically based clades of North America and Eurasia as well as the Old isolate clade. The Eurasia clade includes some Australian taxa as well as the New Zealand taxon; again, the isolates in these Oceania taxa are distinct from other Eurasia isolates, despite being part of the Eurasia clade. Among the Australian taxa, there are clearly multiple sublineages. It seems that the multiple sublineages are being maintained exclusively within Australian host populations, as indicated by the close relationship between the 1994 chicken and the 2007 duck isolate sequences (Fig. 5). Also striking is that while both the 1992 and 1994 Australian isolates are H7N3, they contain different NS1 subtypes, further reinforcing the notion of a pool of influenza A virus segments from which reassortant viruses are frequently arising and circulating in populations of Australian water birds.
FIG. 5.
Inferred relationships for the NS1 protein-coding regions of the 12 subtype H7 isolates from Australia and New Zealand. Subtypes A and B are indicated. The evolutionary history was inferred using the minimum-evolution method. The optimal tree with the sum of branch length of 1.16593153 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) is shown next to the branches. The tree is drawn to scale, with the branch lengths being in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the maximum-composite-likelihood method and are in units of the number of base substitutions per site. The minimum-evolution tree was searched using the close-neighbor-interchange algorithm at a search level of 1. The neighbor-joining algorithm was used to generate the initial tree. The codon positions included were the 1st + 2nd + 3rd + noncoding. All positions containing gaps and missing data were eliminated from the data set (complete deletion option). There were a total of 693 positions in the final data set. Phylogenetic analyses were conducted in the MEGA4 program (29).
Phylogenetic relationships of other genes from subtype H7 isolates.
The relationships between the polymerase PA subunit (252 sequences), M protein (222 sequences), polymerase PB1 subunit (229 sequences), polymerase PB2 subunit (229 sequences), and nucleoprotein (NP) (225 sequences) were inferred from the nucleotide sequence of the protein-coding regions of these segments using the minimum-evolution method. Constraints on sequence divergence are higher for these nonstructural proteins than for the neuraminidase and hemagglutinin, and as a result, the phylogenetic signals are lower. Despite this, the same exclusive, geographically based clades are apparent, with the Eurasia, North America, and South America clades, as well as the Old isolate clade, being statistically supported for each of the five genes. An Australia clade was statistically supported in the PB1 tree (see Fig. S5 to S9 in the supplemental material). The long branch lengths associated with the Australian and New Zealand taxa in the trees for these nonstructural genes may indicate independent evolutions for the lineages that are present in Australia, despite their relationship to other Eurasian taxa.
DISCUSSION
Evidence of genetic isolation for Australian and New Zealand subtype H7 avian influenza viruses.
Previous studies investigating the relationships between HA0 sequences from subtype H7 influenza A virus isolates have demonstrated vast geographic regions, such as Eurasia (Europe, Asia, and Africa), across which subtype H7 avian influenza A viruses rapidly traverse. There are published examples of nearly identical viruses being isolated as far apart as Italy and China (7). Despite this viral mobility, there are apparent barriers across which there is no or very limited virus movement, for example, the Bering Straits (32), that are likely to related to the migratory patterns of the host. The analysis presented in this paper indicates that Australia and New Zealand may represent two additional regions for which there is evidence that influenza viruses are genetically isolated with respect to subtype H7 isolates. The tree in Fig. 2, representing the inferred relationships between the HA0 nucleotide sequences from the available subtype H7 isolates from around the world, shows that the Australian isolates form a geographically exclusive clade, within which the Australian subtype H7 isolates have undergone a period of independent evolution. In addition, the New Zealand isolate, which is included as a deep branching member of the Eurasia clade in the HA0 tree (Fig. 2), has undergone a period of genetic isolation, based on the long branch length to its nearest relative. The relationships inferred for other segments shows a similar pattern of genetic isolation.
Biosecurity and role of host migration in pattern of avian influenza viruses in Australia and New Zealand.
From a biosecurity perspective, the circulating subtype H7 avian influenza viruses in Australia and New Zealand appear to have undergone an extended period of genetic isolation, with the most recent genetic contact coming from Eurasia. This overview of influenza virus evolution provides insight into the role of various avian virus species in the movement of avian influenza virus in Australia and New Zealand. Previous studies have suggested that members of the Anseriformes order, particularly those in the Anatidae family, generally play a central role as the natural reservoirs for avian influenza A virus. The patterns of localized migration of these birds in Australia or New Zealand are consistent with the patterns of genetic isolation of subtype H7 avian influenza virus observed not only for the hemagglutinin- and neuraminidase-encoding segments but also for each of the viral segments. We speculate that the pelagic seabirds of the Procellariformes order and the Charadriidae family that often migrate into North America do not, in fact, play a role in the movement of influenza viruses to Australia and New Zealand. This is based on the closer relationship of the Australian and New Zealand isolates to Eurasia avian influenza virus isolates than to North America isolates, despite the flyways that link the Australia, New Zealand, and North America regions. The limited exchange of genetic material between Australia/New Zealand and Eurasia, as indicated by the distant relationship of the viral sequences between these regions, is more likely to occur as a result of the occasional interaction of endemic Australian Anatidae species with the Asian Anatidae species and is supported by Wallace's Line (19). We emphasize that these overviews are based on a small sample of virus isolates; it will be of interest to see if the relationships hold in a broader survey of Australian and New Zealand influenza virus isolates that is under way.
Epidemiology of subtype H7 avian influenza virus circulating in Australia.
Despite the limited sampling of the Australian subtype H7 viral genetic pool (11 isolates, equivalent to seven sampling events [five poultry outbreaks and two independent wild bird isolations]), there is extraordinary variety in the associated neuraminidase subtypes. Consider that the seven isolation events have yielded five of the nine known neuraminidase subtypes (H7N2, H7N3, H7N4, H7N6, and H7N7). We have also demonstrated that the inferred relationship between the Australian HA0 sequences (Fig. 3) indicates a relationship consistent with hemagglutinin sequences evolving from a single source. In combination with the isolation of multiple neuraminidase subtypes in Australia, this suggests that stable influenza virus clones (as is seen with seasonal human influenza viruses) are not likely to exist in the Australian waterfowl populations. Furthermore, analysis of each of the remaining six segments (see Fig. S4 to S9 in the supplemental material) reveals lineages (summarized in Table 2) that demonstrate that the virus from each of the seven isolation events is a unique reassortant. These data support a model whereby reassortants are arising regularly. This pattern of continuous reassortment is more obvious in the small, isolated waterfowl populations in Australia, although there is evidence that similar patterns of regular reassortment occur in the more expansive and complex Eurasia and North America avian influenza virus regions. This is based on the fact that multiple neuraminidase subtypes are maintained in these populations, and there is no evidence of H7 sublineages being associated with a particular neuraminidase subtype. The absence of this pattern argues strongly for continuous reassortment in the North America and Eurasia clades.
TABLE 2.
Relationships between gene segments among the Australian isolates
| Isolate namea | HAb | NAb | NS1c | PAd | Md | PB1d | PB2d | NPd |
|---|---|---|---|---|---|---|---|---|
| A/duck/Victoria/76 (H7N7) | H7 | N7 | NS1.B.3 | PA.3 | M.2 | PB1.2 | PB2.1 | NP.3 |
| A/chicken/Victoria/1976 (H7N7) | H7 | N7 | NS1.B.3 | PA.3 | M.2 | PB1.2 | PB2.1 | NP.3 |
| A/chicken/Victoria/1/1985 (H7N7) | H7 | N7 | NS1.A.1 | PA.5 | M.4 | PB1.3 | PB2.1 | NP.2 |
| A/starling/Victoria/1985 (H7N7) | H7 | N7 | NS1.A.1 | PA.5 | M.4 | PB1.3 | PB2.1 | NP.2 |
| A/chicken/Victoria/224/1992 (H7N3) | H7 | N3 | NS1.A.2 | PA.4 | M.1 | PB1.1 | PB2.1 | NP.2 |
| A/chicken/Queensland/1994 (H7N3) | H7 | N3 | NS1.B.4 | PA.4 | M.1 | PB1.1 | PB2.1 | NP.1 |
| A/emu/New South Wales/775/97 (H7N4) | H7 | N4 | NS1.A.2 | PA.1 | M.1 | PB1.1 | PB2.1 | NP.1 |
| A/chicken/New South Wales/327/1997 (H7N4) | H7 | N4 | NS1.A.2 | PA.1 | M.1 | PB1.1 | PB2.1 | NP.1 |
| A/chicken/New South Wales/2/1997 (H7N4) | H7 | N4 | NS1.A.2 | PA.1 | M.1 | PB1.1 | PB2.1 | NP.1 |
| A/duck/Victoria/512/2007 (H7N6) | H7 | N6 | NS1.A.2 | PA.2 | M.3 | PB1.1 | PB2.1 | NP.1 |
| A/duck/Tasmania/277/2007 (H7N2) | H7 | N2 | NS1.B.4 | PA.2 | M.3 | PB1.1 | PB2.1 | NP.2 |
The seven outbreaks/isolation events are highlighted by alternate boldface type.
Hemagglutinin and neuraminidase subtypes.
NS1 subtypes (subtype A or B), followed by further classification into four phylogenetic lineages.
Phylogenetic lineages assigned to each of the remaining segments. Lineage numbers are arbitrary. Lineages were assigned on the basis of the relationships shown in Fig. S4 to S9 in the supplemental material.
Conclusions.
Despite the limited sample of subtype H7 avian influenza A virus isolates in Australia and New Zealand, the relationship between the sequences of these viruses, when they are considered in conjunction with the sequences of other related viruses from around the world, provides a relatively clear picture of the dynamics of the circulation of this virus in Australia and New Zealand. This understanding of viral epizootiology is critical for managing biosecurity risks given that it is now clear that, first, there is circulating endemic subtype H7 influenza virus in Australia and New Zealand and, second, that the risk of incursion of exotic avian influenza virus is less from the migratory birds that traverse the Australia/New Zealand to North America flyway than from bird migration between Australia and Southeast Asia. The observation that subtype H7 avian influenza virus in Australia has undergone an extended period of genetic isolation has provided a unique insight into the dynamics of the subtype H7 influenza virus in its natural host. The fact that there is a single, time-related lineage of Australian H7 sequences, despite the multiple neuraminidase subtypes, suggests that virus is not circulating in the natural host as a series of stable viral clones but, rather, that virus diversity is maintained through frequent reassortment.
Supplementary Material
Acknowledgments
We acknowledge the technical support of Rachel Amos-Ritchie, Danielle Anderson, and Lee Trinidad at the Australian Animal Health Laboratory (AAHL). Paul Selleck of the CSIRO Australian Animal Health Laboratory and Wlodek Stanislawek MAF Biosecurity New Zealand provided assistance with accessing the relevant Australian and New Zealand avian influenza virus collections. Access to recent subtype H7 isolates from wild birds in Australia was provided by Simone Warner and Stephen Pyecroft.
Australian funding support was provided by the Australian Biosecurity Co-Operative Research Centre for Emerging Infectious Disease. This project was funded in whole or part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract N01-AI-30071.
Footnotes
Published ahead of print on 28 July 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Alexander, D. J. 2003. Should we change the definition of avian influenza for eradication purposes? Avian Dis. 47:976-981. [DOI] [PubMed] [Google Scholar]
- 2.Alexander, D. J., W. H. Allan, D. G. Parsons, and G. Parsons. 1978. Pathogenicity of 4 avian influenza-viruses for fowls, turkeys and ducks. Res. Vet. Sci. 24:242-247. [PubMed] [Google Scholar]
- 3.Banks, J., E. C. Speidel, J. W. McCauley, and D. J. Alexander. 2000. Phylogenetic analysis of H7 haemagglutinin subtype influenza A viruses. Arch. Virol. 145:1047-1058. [DOI] [PubMed] [Google Scholar]
- 4.Bao, Y. M., P. Bolotov, D. Dernovoy, B. Kiryutin, L. Zaslavsky, T. Tatusova, J. Ostell, and D. Lipman. 2008. The influenza virus resource at the National Center for Biotechnology Information. J. Virol. 82:596-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barr, D. A., A. P. Kelly, R. T. Badman, A. R. Campey, M. D. Orourke, D. C. Grix, and R. L. Reece. 1986. Avian influenza on a multi-age chicken farm. Aust. Vet. J. 63:195-196. [DOI] [PubMed] [Google Scholar]
- 6.Bashiruddin, J. B., A. R. Gould, and H. A. Westbury. 1992. Molecular pathotyping of 2 avian influenza-viruses isolated during the Victoria 1976 outbreak. Aust. Vet. J. 69:140-142. [DOI] [PubMed] [Google Scholar]
- 7.Campitelli, L., A. Di Martino, D. Spagnolo, J. Gavin, D. Smith, L. Di Trani, M. Facchini, M. A. De Marco, E. Foni, C. Chiapponi, A. M. Martin, H. Chen, Y. Guan, M. Delogu, and I. Donatelli. 2008. Molecular analysis of avian H7 influenza viruses circulating in Eurasia in 1999-2005: detection of multiple reassortant virus genotypes. J. Gen. Virol. 89:48-59. [DOI] [PubMed] [Google Scholar]
- 8.Dugan, V. G., R. Chen, D. J. Spiro, N. Sengamalay, J. Zaborsky, E. Ghedin, J. Nolting, D. E. Swayne, J. A. Runstadler, G. M. Happ, D. A. Senne, R. X. Wang, R. D. Slemons, E. C. Holmes, and J. K. Taubenberger. 2008. The evolutionary genetics and emergence of avian influenza viruses in wild birds. Plos Pathog. 4:e1000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.East, I. J., S. Hamilton, and G. Garner. 2008. Identifying areas of Australia at risk of H5N1 avian influenza infection from exposure to migratory birds: a spatial analysis. Geospatial Health 2:203-213. [DOI] [PubMed] [Google Scholar]
- 10.East, I. J., S. A. Hamilton, L. A. Sharp, and M. G. Garner. 2008. Identifying areas of Australia at risk for H5N1 avian influenza infection from exposure to nomadic waterfowl moving throughout the Australo-Papuan region. Geospatial Health 3:17-27. [DOI] [PubMed] [Google Scholar]
- 11.Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felsenstein, J. 1985. Confidence-limits on phylogenies—an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 13.Forman, A. J., I. M. Parsonson, and W. J. Doughty. 1986. The pathogenicity of an avian influenza-virus isolated in Victoria. Aust. Vet. J. 63:294-296. [DOI] [PubMed] [Google Scholar]
- 14.Forsyth, W. M., D. C. Grix, and C. A. Gibson. 1993. Diagnosis of highly pathogenic avian influenza in chickens—Bendigo 1992. Aust. Vet. J. 70:118-119. [DOI] [PubMed] [Google Scholar]
- 15.Ghedin, E., N. A. Sengamalay, M. Shumway, J. Zaborsky, T. Feldblyum, V. Subbu, D. J. Spiro, J. Sitz, H. Koo, P. Bolotov, D. Dernovoy, T. Tatusova, Y. M. Bao, K. St. George, J. Taylor, D. J. Lipman, C. M. Fraser, J. K. Taubenberger, and S. L. Salzberg. 2005. Large-scale sequencing of human influenza reveals the dynamic nature of viral genome evolution. Nature 437:1162-1166. [DOI] [PubMed] [Google Scholar]
- 16.Heine, H. G., L. Trinidad, P. Selleck, and S. Lowther. 2007. Rapid detection of highly pathogenic avian influenza H5N1 virus by TaqMan reverse transcriptase-polymerase chain reaction. Avian Dis. 51:370-372. [DOI] [PubMed] [Google Scholar]
- 17.Krauss, S., C. A. Obert, J. Franks, D. Walker, K. Jones, P. Seiler, L. Niles, S. P. Pryor, J. C. Obenauer, C. W. Naeve, L. Widjaja, R. J. Webby, and R. G. Webster. 2007. Influenza in migratory birds and evidence of limited intercontinental virus exchange. Plos Pathog. 3:1684-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krauss, S., D. Walker, S. P. Pryor, L. Niles, C. H. Li, V. S. Hinshaw, and R. G. Webster. 2004. Influenza A viruses of migrating wild aquatic birds in North America. Vector Borne Zoonotic Dis. 4:177-189. [DOI] [PubMed] [Google Scholar]
- 19.McCallum, H. I., D. A. Roshier, J. P. Tracey, L. Joseph, and R. Heinsohn. 2008. Will Wallace's Line save Australia from avian influenza? Ecol. Soc. 13:41. [Google Scholar]
- 20.Morgan, I. R., and A. P. Kelly. 1990. Epidemiology of an avian influenza outbreak in Victoria in 1985. Aust. Vet. J. 67:125-128. [DOI] [PubMed] [Google Scholar]
- 21.Nestorowicz, A., Y. Kawaoka, W. J. Bean, and R. G. Webster. 1987. Molecular analysis of the hemagglutinin genes of Australian H7n7 influenza viruses: role of passerine birds in maintenance or transmission? Virology 160:411-418. [DOI] [PubMed] [Google Scholar]
- 22.Olsen, B., V. J. Munster, A. Wallensten, J. Waldenstrom, A. D. M. E. Osterhaus, and R. A. M. Fouchier. 2006. Global patterns of influenza A virus in wild birds. Science 312:384-388. [DOI] [PubMed] [Google Scholar]
- 23.Russell, R. J., S. J. Gamblin, L. F. Haire, D. J. Stevens, B. Xiao, Y. Ha, and J. J. Skehel. 2004. HI and H7 influenza haemagglutinin structures extend a structural classification of haemagglutinin subtypes. Virology 325:287-296. [DOI] [PubMed] [Google Scholar]
- 24.Rzhetsky, A., and M. Nei. 1992. A simple method for estimating and testing minimum-evolution trees. Mol. Biol. Evol. 9:945-967. [Google Scholar]
- 25.Scholtissek, C. 1995. Molecular evolution of influenza viruses. Virus Genes 11:209-215. [DOI] [PubMed] [Google Scholar]
- 26.Selleck, P. W., G. Arzey, P. D. Kirkland, R. L. Reece, A. R. Gould, P. W. Daniels, and H. A. Westbury. 2003. An outbreak of highly pathogenic avian influenza in Australia in 1997 caused by an H7N4 virus. Avian Dis. 47:806-811. [DOI] [PubMed] [Google Scholar]
- 27.Selleck, P. W., L. J. Gleeson, P. T. Hooper, H. A. Westbury, and E. Hansson. 1997. Identification and characterisation of an H7N3 influenza A virus from an outbreak of virulent avian influenza in Victoria. Aust. Vet. J. 75:289-292. [DOI] [PubMed] [Google Scholar]
- 28.Shaffer, S. A., Y. Tremblay, H. Weimerskirch, D. Scott, D. R. Thompson, P. M. Sagar, H. Moller, G. A. Taylor, D. G. Foley, B. A. Block, and D. P. Costa. 2006. Migratory shearwaters integrate oceanic resources across the Pacific Ocean in an endless summer. Proc. Natl. Acad. Sci. U. S. A. 103:12799-12802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 30.Tracey, J. P., R. Woods, D. Roshier, P. West, and G. R. Saunders. 2004. The role of wild birds in the transmission of avian influenza for Australia: an ecological perspective. Emu 104:109-124. [Google Scholar]
- 31.Turner, A. J. 1976. Isolation of fowl plague virus in Victoria. Aust. Vet. J. 52:384. [DOI] [PubMed] [Google Scholar]
- 32.Wahlgren, J., J. Waldenstrom, S. Sahlin, P. D. Haemig, R. A. M. Fouchier, A. D. M. E. Osterhaus, J. Pinhassi, J. Bonnedahl, M. Pisareva, M. Grudinin, O. Kiselev, J. Hernandez, K. I. Falk, A. Lundkvist, and B. Olsen. 2008. Gene segment reassortment between American and Asian lineages of avian influenza virus from waterfowl in the Beringia area. Vector Borne Zoonotic Dis. 8:783-790. [DOI] [PubMed] [Google Scholar]
- 33.Westbury, H. A. 1989. Avian influenza. Aust. Vet. J. 66:427-428. [DOI] [PubMed] [Google Scholar]
- 34.Westbury, H. A. 1997. History of highly pathogenic avian influenza in Australia, p. 23-30. Proc. Fourth Int. Symp. Avian Influenza.
- 35.Westbury, H. A., A. J. Turner, and L. Kovesdy. 1979. Pathogenicity of 3 Australian fowl plague viruses for chickens, turkeys and ducks. Vet. Microbiol. 4:223-234. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.