Abstract
Acetylenic specialized metabolites containing one or more carbon-carbon triple bonds are widespread, being found in fungi, vascular and lower plants, marine sponges and algae, and insects. Plants, moss, and most recently, insects, have been shown to employ an energetically difficult, sequential dehydrogenation mechanism for acetylenic bond formation. Here, we describe the cloning and heterologous expression in yeast of a linoleoyl 12-desaturase (acetylenase) and a bifunctional desaturase with Δ12-/Δ14-regiospecificity from the Pacific golden chanterelle. The acetylenase gene, which is the first identified from a fungus, is phylogenetically distinct from known plant and fungal desaturases. Together, the bifunctional desaturase and the acetylenase provide the enzymatic activities required to drive oleate through linoleate to crepenynate and the conjugated enyne (14Z)-dehydrocrepenynate, the branchpoint precursors to a major class of acetylenic natural products.
Keywords: Enzymes, Fatty Acid, Fatty Acid Metabolism, Fungi, Lipid Synthesis, Acetylenase, Basidiomycetes, Desaturase, Polyacetylene, Secondary Metabolism
Introduction
The dehydrogenation of oleoyl (18:1 Δ9c) phosphatidylcholine (PC) to linoleoyl (18:2 Δ9c,12c) PC in plant and fungal primary metabolism is catalyzed in the endoplasmic reticulum (ER)2 by the Δ12 desaturase, FAD2, and is central to the functionality of biological membrane systems, cellular signaling, thermal adaptation, and energy storage (1, 2). The oxidative potential of the FAD2 architecture is apparent in the diversification of this non-heme diiron enzyme to a panorama of regio- and chemoselective microsomal desaturases that provide most of the functional group diversity in natural acyl lipids, including acetylenic fatty acids (3–5). Bioengineering of seed oil in agronomically important oil-producing plants by controlling flux through seed-specific diverged FAD2s (6, 7) and the manipulation of variant desaturase/acetylenase enzymes may diversify biobased feedstocks. Although acetylenic bond formation is a fundamental transformation, the origin of such highly unsaturated natural products is not well established. In this report we reveal two cooperative multifunctional FAD2 enzymes that produce cis-enyne and trans-alkene building blocks of the naturally occurring acetylenes.
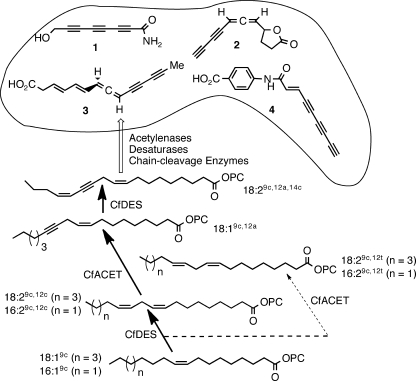
Of the acetylenic natural products, the majority are found in plants, moss, and fungi (8). Agrocybin (Fig. 1, 1), the likely origin of fairy rings in grass turf, may promote cell death via apoptosis in trypanosomes (9). Nemotin (2), mycomycin (3), and aryl amide (4) have potent antibiotic activities against nematodes, bacilli (e.g. tuberculosis), and drug-resistant strains of Staphylococcus aureus, respectively (10–12). (14Z)-Dehydrocrepenynic acid (18:2 Δ9c,12a,14c) was found to be an inhibitor of bacterial conjugation, suggesting a target for reducing the horizontal gene transfer involved in multi-drug resistance (13). During recent years, a small number of FAD2-diverged genes from plants, moss, and insects have been functionally demonstrated to encode acetylene-forming enzymes or acetylenases (14–17). Basidiomycete (club) fungi are some of the most prolific producers of acetylenic natural products, yet the genes responsible for the critical C C bond-forming reactions have yet to be described.
C bond-forming reactions have yet to be described.
FIGURE 1.
Biosynthesis of polyacetylenic metabolites in Basidiomycetes. The metabolic progression is supported by the feeding studies leading to highly unsaturated, bioactive metabolites, exemplified by compounds 1-4. Biotransformations functionally characterized in this paper produce lipids accumulated by (bold lines) or absent from (dashed lines) C. formosus.
Mushrooms of the genera Cantharellus (chanterelles), Craterellus and Hydnum accumulate up to 62% of their total acyl lipids as 18:2 Δ9c,12a,14c (18). Chanterelles are valuable, widely distributed, nutritious mushrooms whose ectomycorrhizae are symbiotic with oak, beech, and conifers. Chanterelles appear to be resistant to insect and snail damage, and although the mechanism of this defense is unproven, acetylenic lipids or oxylipins are potential repellant or insecticidal natural products (19).
Radiochemical tracer experiments support acyl procession from 18:2 Δ9c,12c via dehydrocrepenynate, which rarely accumulates, with an exception being in Cantharellaceae family members, to more highly unsaturated specialized products in both plants and fungi (20–22). To synthesize 18:2 Δ9c,12a,14c, we hypothesized that a set of active FAD2 desaturases and an acetylenase must be expressed in the chanterelle family. In this paper we describe the cloning and functional characterization of the first acetylenase and a FAD2 desaturase from a chanterelle, Cantharellus formosus. Heterologous expression of these genes demonstrated sequential substrate utilization, sensitivity to epitope tagging, and by reconstitution of the Basidiomycetes pathway in yeast, allowed the first definition of the enzymatic transformations leading to dehydrocrepenynic acid.
EXPERIMENTAL PROCEDURES
Strains and Growth Conditions
The untransformed yeast strain, InvSc1 (MATa/α his3Δ1 leu2 trp1-289 ura3-52; Invitrogen) was cultivated on rich medium (1% Bacto yeast extract, 2% Bacto peptone (Difco), 2% dextrose) at 30 °C. Transformed yeast strains were cultured in complete minimal media lacking uracil or lacking uracil and histidine (CM−ura or CM−ura−his) supplemented with dextrose (for propagation) or galactose (2 g/100 ml, for expression experiments) either without fatty acid supplementation or with linoleic acid (Sigma) or crepenynic acid (prepared from Crepis alpina seeds as described (23)) supplied at 0.5 mm in 1 mg/ml Nonidet P-40 (Sigma). Unless otherwise specified, for the heterologous expression of Cf0745 and Cf0807, cultures were grown at 15 °C with shaking at 250 rpm. Escherichia coli strains XL1-Blue and XL-1 MRF′ (Stratagene) and JM109 (Promega) were used for DNA cloning and amplification of plasmids. Plasmids carrying Aleurites fordii Myc-tagged ω-3 desaturase (24), Euphorbia FAD2 (25), and soybean GmFAD2–1a and GmFAD2–1b (26) desaturase genes were transformed into InvSc1 and expressed at 15–22 °C as described below. Chanterelle fruiting bodies were acquired from Ken Ottoboni Mushrooms (Ridgefield, WA).
Cloning and Sequencing of C. formosus Desaturase Genes
mRNA was prepared from C. formosus fruiting bodies using Trizol (Invitrogen) and poly(dT) beads (Dynal), and a Uni-ZAP XR cDNA library was prepared according to the manufacturer's protocol (Stratagene). Excised phagemids were sequenced with a T3 primer, and the resulting sequences were searched for homologs of PFAD2 and other known desaturases by BlastX (27, 28). Selected clones were resequenced with T7 primer.
In an effort to obtain the 5′ ends of two Cantharellus cibarius PFAD2 homologs identified by shotgun sequencing (0745 and 0807), the phage library was screened with DNA probes (∼600 base pairs) prepared by PCR-amplification (Vent DNA polymerase; New England Biolabs) from excised phagemids with primers specific for the FAD2 homologs (supplemental Table S1). The probes were radiolabeled with dCTP* (PerkinElmer Life Sciences; 185 MBq, 50 mCi) by a random-primed reaction using the Ladderman labeling kit (TaKaRa, Shiga, Japan). Plaque lifts, hybridization, and screening were carried out by standard methods (29, 30). Positive phage clones from a two-step screen were recovered, and phagemids were excised with ExAssist helper phage in the SOLR E. coli strain (Stratagene). Excised plasmids were sequenced by in-house automated DNA sequencing (Dyenamic ET Terminator Cycle Sequencing kit; Amersham Biosciences) with a T7 promoter primer and compared with the initially sequenced EST clones. A cDNA that was slightly shorter than the original 0807 shot gun sequenced phagemid was obtained from this screen. Comparison of the sequences with each other and other diverged desaturases indicated that the initial phagemid clone was full-length, which was later confirmed by expression in yeast. Despite a second round of library screening for 0745 using a probe amplified with primers closer to the 5′ end of the initial EST clone (0745s@100, 0745s@600), we were unable to obtain the entire 5′ end of the gene.
A 5′ rapid amplification of cDNA ends (5′-RACE) approach was used to obtain the remainder of the 0745 PFAD2 homolog from poly(A)+ RNA isolated from C. formosus fruiting bodies with the BD SMARTRACE cDNA amplification kit (BD Biosciences/Clontech). First-strand cDNA synthesis was accomplished with a poly(dT) primer and BD Power Script reverse transcriptase (BD Biosciences/Clontech), the C. formosus 0745 desaturase gene was amplified from the 5′-RACE-ready cDNA using a universal primer mix and a gene-specific primer (0745s@190), and the product was isolated by agarose gel electrophoresis gel extraction (BD Biosciences Nucleotrap Gel Extraction kit). 5′-RACE products were then subcloned into pGEM-T by T/A cloning (Promega pGEM-T Easy Vector System II) and transformed into ultra-competent JM109 E. coli (Promega), and insert-containing plasmids were sequenced as described. All oligonucleotide primers used are shown in supplemental Table S1.
Construction of Yeast Expression Vectors and Cell Lines
The full-length 0745 and 0807 open reading frames were amplified with primers containing BamHI (sense) and EcoRI (antisense) restriction sites for insertion into yeast expression vectors (supplemental Table S1). PCR products and DNA fragments were purified by Gene Clean (Bio 101), and purified, digested (BamHI/EcoRI; Promega) C. formosus desaturase inserts were ligated into pYES2 or pESc-His (Stratagene) with or without an N-terminal His6GlySer fusion tag with T4 DNA ligase (Invitrogen) and transformed into competent XL-1 Blue cells by standard techniques (30). Sequence-validated plasmids were transformed into competent InvSc1 by a standard lithium acetate method (31). Strains for co-expression were transformed simultaneously with dual selection.
Desaturase Expression and GC/MS Analysis of Fungal Lipids
Heterologous expression of C. formosus-diverged desaturase genes was induced by cultivation of transformed Saccharomyces cerevisiae in 5 ml of CM−ura or CM−ura−his with galactose (2 g/100 ml) at the indicated temperatures with shaking at 250 rpm. Fatty acid methyl esters (FAMEs) of yeast lipids were prepared and analyzed by gas chromatography-mass spectrometry (GC-MS, HP 5890-HP 5972 MS; DB-23 column) as previously described (28) and identified by comparison of retention time and parent ions to authentic standards. Cells supplemented with exogenous fatty acids were washed twice with 10 mg/ml Nonidet P-40 and once with H2O to remove fatty acids not incorporated into cellular lipids. Dimethyloxazolines were prepared according to the method of Christie (32). Synthesis and characterization of trans fatty acid authentic standards are provided in the supplemental data.
FAME stereoisomers were additionally separated with an HP 5890 gas chromatograph using a SP-2380 capillary column (Aldrich, 30 m × 0.25 mm). The temperature program was 150 °C for 10 min, ramped to 180 °C at 2 °C/min, then ramped at 20 °C/min to 250 °C and cooled by 20 °C/min back to the initial temperature. The injector and detector were set to 250 °C, and the gas flow rate was 0.9 ml/min. Dimethyloxazoline derivatives were separated using the temperature program 100 °C or 3 min, ramped 5 °C/min to 250 °C, 250 °C for 3 min, and cooled 50 °C/min to 100 °C; inlet and detector temperatures, 250 °C.
RESULTS
Cloning of C. formosus Diverged Desaturases
We initiated our search for diverged desaturases in the C. formosus, recently distinguished from C. cibarius as a unique species (33), genome through a cDNA sequencing approach. Phagemids (1037) were sequenced, and the resulting sequences were screened using a BLAST algorithm for homologs of known plant and fungal desaturases allowing us to identify two putative diverged Δ12 desaturases, Cf0745 and Cf0807 (5 representatives), and a Δ9 desaturase, Cf0820. The initial sequences were used in hybridization screening of the cDNA phage library for full-length clones. Of 1 × 105 plaque-forming units screened, 22 were identified as positives for Cf0807. A secondary screen provided three clones, and sequencing demonstrated that the full-length cDNA was represented in the Cf0807 initial clone. The predicted Cf0807 fatty acid desaturase cDNA contained 1248 base pairs encoding a 415-amino acid protein (GenBankTM accession number HM036207).
Consistent with the initial shotgun sequencing, the predicted Cf0745 Δ12 desaturase was less well represented in the fruiting body cDNA library; of 1.5 × 105 plaque-forming units screened, 7 were identified as positive. A secondary screen resulted in 3 positive clones; however, a 5′-RACE strategy was necessary to obtain the 5′ end of the Cf0745 cDNA, affording a predicted 1389-bp FAD2 homolog cDNA encoding a 462-amino acid protein (accession number HM036206). Full-length nucleotide and amino acid sequences for both Cf0807 and Cf0745 are found in supplemental Fig. S2 and have been submitted to GenBankTM.
Predicted Protein Structure Analysis of Cf0745 and Cf0807
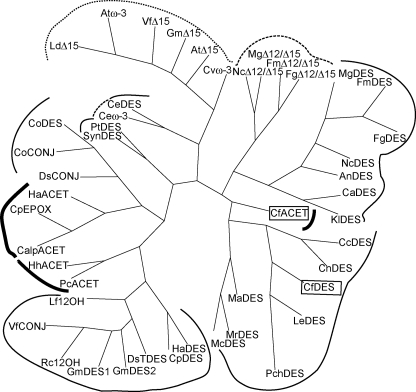
An unrooted phylogenetic tree demonstrated the relationships of the chanterelle proteins to recognized desaturases and was suggestive of biochemical function (Fig. 2). Cf0745 (CfDES) was found to segregate with fungal proteins known only to carry out the prototypical oleate desaturase reaction. Cf0807 (CfACET), however, was not imbedded deeply within the mapped fungal clades but was, rather, lying on a discrete spur.
FIGURE 2.
Cladogram relating the chanterelle enzymes to representative FAD2, FAD3, and bifunctional desaturases. The graph shows the phylogenetic relationships between non-heme diiron enzymes diverged in chemo- and regioselectivity, assembled by a neighbor-joining algorithm from boot-strapped (n = 100) sequence alignments, plotted using DRAWGRAM (Phylip 3.65) (51). Families of desaturases and hydroxylases are bracketed by normal-weight lines, acetylenases by bold lines, enzymes with activity regioselective to the Δ15/ω-3 position by the dotted line, and enzymes with multiple locations of reactivity by the dashed lines. Chemoselectivity of the associated sequences are: DES, Δ12 desaturase; Δ12/Δ15, bifunctional Δ12/Δ15 desaturase; ACET, Δ12 acetylenase; CONJ, Δ12 conjugase; ω-3, ω-3 desaturase; ω-6, microsomal ω-6 desaturase; EPOX, 12 epoxygenase; 12-OH, 12-hydroxylase; TDES, trans- Δ12 desaturase; Δ15, Δ15 linoleoyl desaturase. The GenBankTM accession numbers for the displayed sequences are: AnDES, AAG36933; AtD15 (ER), P48623; Atw-3 chloroplast, P46310; CaDES, XP_722258; CalpACET, CAA76158; CcDES, AAU12575; CeDES, NP_502560; Cew-3, NP_001023560; CnDES, AAW42920; CoDES, AAG42259; CoCONJ, AAG42260; CpDES, CAA76157; CpEPOX, CAA76156; Cvw-3, BAB78717; DsCONJ, AAS72901; DsTDES, AAS72902; FgDES, EAA75859; FgD12/D15, ABB88516; FmDES, ABB88515; FmD12/D15, ABB88516; GmDES1, AAB00859; GmDES2, AAB00860; GmD15 (ER), P48625; HaACET, AAO38032; HaDES, AAL68982; HhACET, AAO38031; KlDES, XP_455402; LdD15, AAA86690; LeDES, BAD51484; Lf12OH, AAC32755; MaDES, BAA81754; McDES, BAB69056; MgDES, XP_365283; MgD12/D15, XP_362963; MrDES, AAM97924; NcDES, XP_959528; NcD12/D15, XP_329856; PcACET, AAB80697; PchDES, ACL13289; PtDES, AAO23564; Rc12OH, AAC49010; SynDES, NP_896789; VfCONJ, AAN87574; VfD15, AAC98967. An, Aspergillus nidulans; At, Arabidopsis thaliana; Ca, Candida albicans SC5314; Calp, C. alpina; Cc, Cryptococcus curvatus; Ce, Caenorhabditis elegans; Cn, Cryptococcus neoformans var. neoformans JEC21; Co, Calendula officinalis; Cp, Crepis palaestina; Cv, Chlorella vulgaris; Ds, Dimorphotheca sinuata; Fg, Gibberella zeae PH-1; Fm, Fusarium moniliforme; Gm, Glycine max; Ha, Helianthus annuus; Hh, Hedera helix; Kl, Kluyveromyces lactis; Ld, Limnanthes douglasii; Le, Lentinula edodes; Lf, Lesquerella fendleri; Ma, Mortierella alpina; Mc, Mucor circinelloides; Mg, Magnaporthe grisea 70-15; Mr, Mucor rouxii; Nc, Neurospora crassa OR74A; Pc, Petroselinum crispum; Pch, Phanerochaete chrysosporium; Rhizopus oryzae Pt, Phaeodactylum tricornutum; Rc, Ricinus communis; RoDES; Syn, Synechococcus sp. WH 8102; Vf, Vernicia fordii.
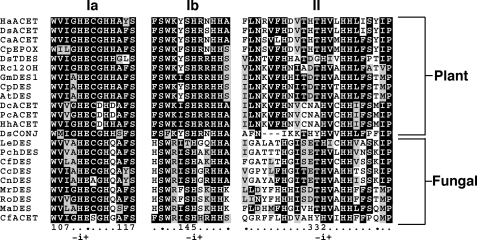
Local sequence alignments showed several distinguishing features between the plant and fungal kingdoms. Histidine box motifs that are believed to bind two active-site iron atoms and are critical to enzyme function (34, 35) are present and are positionally conserved in translations of both Cf0745 (CfDES) and Cf0807 (CfACET) (Fig. 3). The i-1 position of motif Ia in most diverged desaturases (i.e. residue 108 in Cf0807) is Gly rather than Ala in stricto sensu desaturases (i.e. Cf0745). In His box motif Ib, the i-3 residue to the initial histidine is Arg in all fungal proteins but Lys in plants. Among the three His boxes, the only apparent structural correlation to acetylenase activity is in the i-5 to i-2 positions in box II, where (D/N)VX(H/N) occurs in all plant acetylenases and Cf0807 but not in other desaturase-like sequences. Hydropathy predictions strongly indicted that Cf0745 and Cf0807 have four potential helical transmembrane domains and putative KXKXX and KKXX ER retention sequences (36–38). Based on this model, the His boxes are found near the predicted cytosolic face of the ER bilayer.
FIGURE 3.
Local sequence alignments comparing the three iron-binding His box domains in FAD2-diverged desaturases from plants and fungi. The first His residue in each box is labeled i, and features are indexed negatively amino-proximate and positively to the carboxyl side of i. The sequence abbreviations are described in the Fig. 2 legend except: CaACET (=CalpACET), CAA76158 (14); AtDES, AAA32782 (1); DcACET, AAO38033 (15); DsACET, AAO38036 (15); RoDES, AAV52631.
Expression of C. formosus Diverged Desaturases
We have explored the catalytic properties of the C. formosus predicted diverged desaturases through heterologous expression in S. cerevisiae. Our experience with Basidiomycete diverged desaturases indicates that an N-terminal hexahistidine tag results in increased levels of in vivo desaturation activity when compared with the untagged protein (28). We expressed both the untagged and the His6-tagged predicted diverged desaturases cloned from C. formosus and determined the temperature dependence of desaturase-product accumulation. Acid-catalyzed methanolysis of whole yeast preparations produced FAMEs that were analyzed by GC-MS. Expression of each untagged Cf0745 and Cf0807 and the His6 fusion proteins H6Cf0745 and H6Cf0807 resulted in the production of new fatty acids not present in the empty vector control (Table 1 and supplemental Table S2, A and B). Expression of H6CfDES and H6CfACET increased the doubling time compared with the empty vector control by 13 and 34%, respectively, at 15 °C. However, the fusion proteins resulted in significantly higher product accumulation (up to 4-fold greater for the dominant acyl products) than their untagged counterparts. Therefore, to more easily follow in vivo activity of Cf0745 and Cf0807, we conducted subsequent heterologous expression studies with the His6 fusion proteins.
TABLE 1.
Effect of temperature on fatty acid desaturation activity of Cf0745 and Cf0807 on yeast total fatty acid composition in the heterologous expression of C. cibarius enzymes with no added fatty acid was determined by GC-MS analysis of FAMEs
Results are expressed as the mean ± S.D., n = 3–6.
| Temperature | Construct | Fatty acids |
|||||
|---|---|---|---|---|---|---|---|
| 16:0 | 16:1 | 16:2 | 18:0 | 18:1 | 18:2 | ||
| °C | |||||||
| 15 | pYES2 | 12.2 ± 0.9 | 57.7 ± 1.8 | 0 | 3.51 ± 0.20 | 26.6 ± 0.8 | 0 |
| 22 | 19.8 ± 0.5 | 52.9 ± 1.4 | 0 | 3.22 ± 0.14 | 24.0 ± 0.8 | 0 | |
| 30 | 20.6 ± 0.8 | 44.3 ± 4.1 | 0 | 7.26 ± 1.90 | 27.9 ± 1.4 | 0 | |
| 15 | H6Cf0745a | 13.8 ± 1.5 | 45.9 ± 1.3 | 7.57 ± 0.73 | 4.32 ± 0.15 | 9.58 ± 0.2 | 18.9 ± 1.7 |
| 22 | 21.2 ± 0.9 | 49.4 ± 1.5 | 3.31 ± 0.40 | 4.01 ± 0.26 | 10.6 ± 0.5 | 11.4 ± 0.9 | |
| 30 | 18.0 ± 1.0 | 49.0 ± 1.3 | 0.61 ± 0.29 | 5.37 ± 0.06 | 25.0 ± 1.0 | 1.90 ± 1.99 | |
| 15 | H6Cf0807b | 11.8 ± 0.3 | 59.6 ± 0.6 | 3.02 ± 0.21 | 2.74 ± 0.16 | 21.9 ± 0.2 | 0.90 ± 0.07 |
| 22 | 19.9 ± 0.9 | 53.8 ± 2.0 | 0.64 ± 0.03 | 2.75 ± 0.29 | 22.6 ± 0.8 | 0.30 ± 0.04 | |
| 30 | 17.6 ± 1.0 | 51.3 ± 0.6 | 0 | 4.56 ± 0.23 | 26.5 ± 0.6 | 0 | |
a For diunsaturated lipids, Δ9c,12c.
b For diunsaturated lipids, Δ9c,12t.
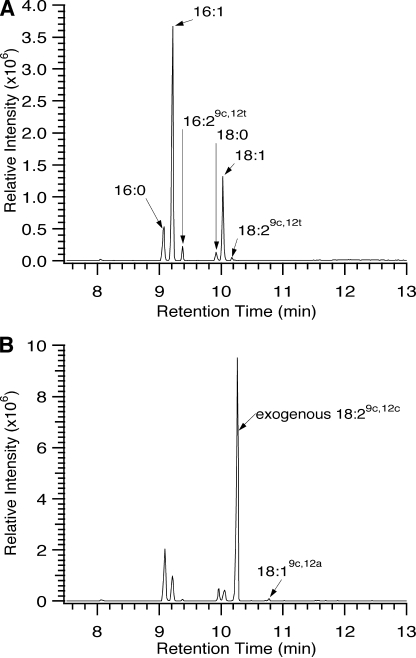
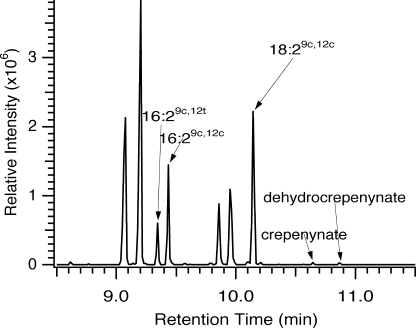
Total ion chromatograms and mass spectra of the new fatty acids produced by Cf0745 and Cf0807 were consistent with 16:2 Δ9,12 and 18:2 Δ9,12, demonstrating that both of the predicted enzymes have Δ12 desaturation activity and are functionally related to the FAD2 family of desaturases, as suggested by the gene and protein alignments. H6Cf0745 was an extremely active desaturase with a preference for the C18 substrate producing almost 20% of the total fatty acids as 18:2 Δ9,12 and more than 7% 16:2 Δ9,12 at 15 °C. Close inspection of the retention times of the C16- and C18-diunsaturated products of H6Cf0807 (Fig. 4A) suggested that they were not identical to the H6Cf0745 products. Rather, the 16:2 and 18:2 products of H6Cf0807 were demonstrated to have exclusively the trans double bond configuration at the C12 position by separation of the stereoisomers with a specialized SP-2380 capillary GC column and co-elution of the H6Cf0807 16:2 and 18:2 products with authentic Δ9c,12t FAMEs prepared in our laboratory (data not shown). H6Cf0807 demonstrated a preference for the C16 substrate producing ∼3% 16:2 Δ9c,12t and 0.9% 18:2 Δ9c,12t at 15 °C (Table 1).
FIGURE 4.
GC-MS analysis of the novel acyl modifications resulting from the recombinant chanterelle desaturase and acetylenase. FAME total ion chromatograms collected for total acyl lipids of Cf0807 are expressed in yeast at 15 °C in the absence (A) and presence (B) of 0.5-mm exogenous linoleic acid.
Growth of the expression cultures at 15, 22, and 30 °C resulted in dramatic differences in the level of diunsaturated fatty acids for both H6Cf0745 and H6Cf0807 fusions (Table 1) and the untagged proteins (supplemental Table S2) with each showing substantially more activity at lower temperatures. H6Cf0745 desaturation products made up more than 26% of the total FAMEs at 15 °C compared with 2.5% at 30 °C. Similarly, H6Cf0807 products were almost 4% that of the total FAMEs at 15 °C and were not detectable at 30 °C (Table 1). This temperature dependence is consistent with plant FAD2 homologs when heterologously expressed in yeast (39); however, it is opposite that of another fungal desaturase that we recently described (PchFAD2) that demonstrated higher activity at 30 °C compared with 15 °C (28).
To produce the accumulated natural product dehydrocrepenynic acid, chanterelle must have both FAD2/3 desaturase and acetylenase activities. The yeast expression of H6Cf0745 and H6Cf0807 establishes that H6Cf0745 is a classical FAD2 desaturase. The production of the trans Δ12 double bond by H6Cf0807 suggested that this enzyme is an acetylenase, as this activity has also been observed in the C. alpina acetylenase CREP1 (40). To test for acetylenase activity in the C. formosus FAD2 homologs, we supplemented yeast expressing H6Cf0745 or H6Cf0807 with 18:2 Δ9c,12c and analyzed FAMEs. In all cell lines the supplemented 18:2Δ9,12 composed >75% of the total FAMEs (Table 2). H6Cf0745 expression resulted in the production of only 16:2 Δ9,12 and 18:2 Δ9,12 (indicated by increased levels above the empty vector control) as seen in the non-supplemented cultures. However, expression of H6Cf0807 resulted in the production of crepenynic acid (18:1 Δ9c,12a) to >0.5% of the total FAMEs as determined by comparison of retention time and mass spectra with dimethyloxazoline and FAME authentic standards and literature data (Figs. 4B and supplemental Fig. S3) (15, 32). No C16 acetylenic fatty acid was detected. Supplementation with 18:2 Δ9c,12t, α-linolenic acid (18:3 Δ9,12,15), or γ-linolenic acid (18:3 Δ6,9,12) did not result in the production of new fatty acids (data not shown). Crepenynate supplementation of cultures at low concentrations to avoid its toxicity resulted in the production of another new fatty acid upon heterologous expression of H6Cf0745 (>3.2% of total FAMEs; Table 2). This fatty acid was determined by mass spectrometry of authentic dimethyloxazoline and FAME standards to be dehydrocrepenynic acid (supplemental Fig. S3) (15).
TABLE 2.
Substrate specificity of C. formosus diverged desaturases
Shown are fatty acids as % of yeast total fatty acid composition, as determined by GC-MS analysis of FAMEs, in the heterologous expression of Cf0745 and Cf0807 supplemented with linoleic (+18:2 Δ9c,12c, 0.5 mm) or crepenynic acid (+18:1 Δ9c,12a, 0.5 mm). Cultures were grown at 15 °C, 250 rpm in media containing NP-40 (1 mg/ml). Results are expressed as the mean ± sd, n = 3–6. ND, not detected.
| Constructs | Fatty acids |
|||||||
|---|---|---|---|---|---|---|---|---|
| 16:0 | 16:1 | 16:2 Δ9,12 | 18:0 | 18:1 | 18:2 Δ9,12 | 18:1 Δ9c,12a | 18:2 Δ9c,12a,14c | |
| +18:2Δ9,12a | ||||||||
| pYES2 | 13.6 ± 3.1 | 4.00 ± 0.12 | ND | 2.47 ± 0.62 | 2.06 ± 0.16 | 77.9 ± 3.8 | ND | ND |
| H6Cf0745 | 10.3 ± 0.1 | 2.52 ± 0.17 | 0.40 ± 0.02b | 1.81 ± 0.06 | 0.63 ± 0.05 | 84.4 ± 0.2b | ND | ND |
| H6Cf0807 | 12.8 ± 1.4 | 3.75 ± 0.40 | 0.17 ± 0.02c | 2.43 ± 0.38 | 2.11 ± 0.27 | 78.2 ± 2.5c | 0.52 ± 0.08 | ND |
| +18:1Δ9,12aa | ||||||||
| pYES2 | 17.0 ± 0.7 | 21.3 ± 1.5 | ND | 4.98 ± 0.10 | 18.1 ± 1.2 | ND | 38.6 ± 3.3 | ND |
| vH6Cf0745 | 21.1 ± 0.6 | 20.6 ± 1.3 | 1.48 ± 0.29b | 5.75 ± 0.70 | 7.7 ± 1.0 | 8.20 ± 0.30b | 31.9 ± 1.4 | 3.26 ± 0.19 |
| H6Cf0807 | 21.6 ± 0.2 | 21.7 ± 3.6 | 0.96 ± 0.26c | 6.45 ± 0.66 | 15.0 ± 2.0 | 0.43 ± 0.19c | 33.9 ± 4.9 | ND |
| PchFAD2d | 34.8 ± 1.3 | 18.2 ± 1.4 | 1.40 ± 0.22b | 10.7 ± 0.5 | 7.99 ± 0.48 | 4.68 ± 0.15b | 18.7 ± 3.6 | 3.46 ± 0.26 |
a Fatty acid supplemented to expression culture.
b Δ9c,12c.
c Δ9c,12t.
d Expression temperature of 30 °C.
Metabolic channeling in dehydrocrepenynate biosynthesis between the desaturase and acetylenase enzymes was demonstrated by the co-expression of Cf0745 and Cf0807 in S. cerevisiae. When H6Cf0745 and H6Cf0807 were co-expressed from independent plasmids, the production of 18:2 Δ9c,12c, 18:1 Δ9c,12a, and 18:2 Δ9c,12a,14c from endogenous oleic acid was observed (Fig. 5). Channeling through the biosynthetic pathway from oleic to dehydrocrepenynic acid was more efficient than when each Cf0745 and Cf0807 were supplied substrates through media supplementation as demonstrated by the % conversion of 18:2 Δ9c,12c into 18:1 Δ9c,12a (0.67% for supplementation versus 1.51% conversion of enzymatically produced 18:2 Δ9c,12c) and the subsequent conversion of 18:1 Δ9c,12a to 18:2 Δ9c,12a,14c (20.9% versus 43%, respectively) (Table 3). The Δ12c desaturation efficiency ratio for C18 versus C16 substrates, and acetylenation/trans-desaturation ratio were 3.43 and 1.37, respectively, upon co-expression.
FIGURE 5.
Total ion chromatogram of yeast total FAMEs demonstrating desaturase-acetylenase co-expression.
TABLE 3.
Coexpression of C. formosus diverged desaturases
Shown are fatty acids as % of yeast total fatty acid composition, as determined by GC-MS analysis of FAMEs, in the simultaneous heterologous expression of Cf0745 and Cf0807. Cultures were grown at 22 °C for 1 day then 15 °C for 3 days at 250 rpm. Results are expressed as the mean ± S.D., n = 6; ND, not detected.
| Constructs | Fatty acids |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 16:0 | 16:1 | 16:2 Δ9c,12t | 16:2 Δ9c,12c | 18:0 | 18:1 | 18:2 Δ9c,12c | 18:1 Δ9c,12a | 18:2 Δ9c,12a,14c | |
| pYES2/pESc | 18.3 ± 1.4 | 43.0 ± 2.4 | ND | ND | 4.00 ± 0.29 | 34.8 ± 2.3 | ND | ND | ND |
| H6Cf0745/ H6Cf0807 | 20.1 ± 1.0 | 32.0 ± 1.4 | 3.56 ± 0.13 | 7.87 ± 1.3 | 6.1 ± 0.28 | 11.0 ± 0.78 | 18.9 ± 0.87 | 0.29 ± 0.06 | 0.22 ± 0.07 |
DISCUSSION
In this report we describe the cloning and functional characterization of two diverged desaturases from the Basidiomycete C. formosus and show that the production of dehydrocrepenynic acid from oleic acid in fungi may be accomplished by the activity of just two FAD2-like activities.
Simple acetylenic fatty acids are known from a small number of plants and fungi, but unfortunately, yeast expression of plant FAD2 acetylenases and epoxidases, a straightforward system for functional characterization of both plant and fungal desaturases, often results in little or no detectable activity (14, 15). A similar result was observed during the current studies of Cf0807; however, His6 fusion-tagging serendipitously amplified the heterologous acetylenase activity and allowed us to redesignate the gene CfACET.
A number of basidiomycotal Δ12 desaturases have been characterized by our group and others (28, 41–43). In this study, Cf0745, an enzyme similar to known fungal desaturases, showed high desaturation activity for both C16 and C18 substrates and was consequently renamed CfDES. Although fungal desaturases have been observed not to be transcriptionally regulated by unsaturated fatty acid levels, decreasing temperature normally boosts activity (43). The temperature dependence of heterologously expressed CfDES and CfACET activity was similar to plants but dissimilar to the Phanerochaete FAD2 (28) and is consistent with ectomycorrhizal C. formosus thriving in a temperate plant-like environment.
Although the expected spacing and organization of the three His box domains for a fungal Δ12 or Δ15/ω-3 desaturase is present, a pairwise comparison with CfACET revealed a maximum sequence identity of 48% to fungal Δ12 desaturases and a meager 25% to plant FAD2-like acetylenases. Local analysis of the histidine box motifs established clear attributes of both plant acetylenases and fungal desaturases in CfACET. Acetylenases may have diverged from a single ancient Δ12 or ω-3 desaturase paralog. Subsequent functional diversification (44) resulted in relatively homogeneous subfamilies of the fungal FAD2, plant acetylenases, and mixed regioselectivity desaturases; CfACET appears to be the first member of a new acetylenase subfamily. Given the large evolutionary distance between plants and fungi, resolving whether the ability of fungi to produce polyacetylenes is a gain or loss of function phenotype requires substantially more data.
The multifunctional nature of the diverged Δ12-desaturases demonstrated by both of the C. formosus enzymes is apparently of broad occurrence. Although the details of the hydrogen abstraction are sparse due to the difficulty in gaining structural and spectroscopic data into these integral membrane proteins, oxygen activation by a reduced Fe(II)-Fe(II) core and stepwise fission of the carboxyl-proximate C-H bond followed by chemoselective loss of the second hydrogen atom is the generally held model for both C C and C
C and C C bonds (45–47). Nevertheless, the origins of selectivity leading to acetylenases and desaturases are not currently distinguishable de rigueur by comparisons at the primary sequence level, and residues promoting acetylenase activity have yet to be located.
C bonds (45–47). Nevertheless, the origins of selectivity leading to acetylenases and desaturases are not currently distinguishable de rigueur by comparisons at the primary sequence level, and residues promoting acetylenase activity have yet to be located.
The observation that trans- and acetylenic fatty acid production is intertwined in the Cantharellus enzyme deserves comment. The 9c,12t products of CfACET trans-desaturase activity are apparent metabolic deadends as the trans double bond appears to prevent the correct positioning for hydrogen abstraction leading to the acetylenic unit (16, 40). The absence of 18:2 Δ9c,12t in chanterelle lipids is consistent with relatively low oleate concentrations in the mushroom. In contrast, C. alpina accumulates 3.3% 18:2 Δ9c12t produced by its 12-acetylenase CREP1 (40). Striking is the stereochemical continuum of the desaturase activities from purely 18:2 Δ9c,12c-producing FAD2s through the cis/trans mixtures of CREP1 and noted in insect pheromone biosynthesis and plant sphingolipid Δ8 desaturation (48) to pure 18:2 Δ9c,12t in CfACET. Efficient streaming in chanterelle of oleate through linoleate to the acetylenase, selectivity of lipid storage pathway enzymes, or modifications to the structure or environment of CfACET in fungal membranes may suppress in vivo trans-desaturation.
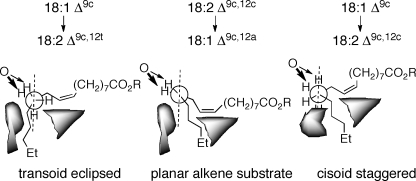
Although in the absence of high resolution structural data, a precise structural determinant of trans-desaturation cannot be unequivocably defined, this activity does seem to be an inherent property of the Δ12-acetylenases and other diverged FAD2 enzymes. Stymne and co-workers (40) showed that 18:29c,12c is produced by abstraction of the pro-(R) hydrogens at C12 and C13, whereas 18:29c,12t is produced by scission of the (12R,13S) hydrogens; similar selectivity of prochiral hydrogens has been noted for Δ8-sphinolipid desaturases (48). Diverged FAD2 desaturases exhibit a primary kinetic isotope effect of 4–15 at C12 and near unity at C13 revealing sequential C-H abstraction (45, 49). The short lifetime anticipated for the putative alkyl radical at C12 makes improbable a 180° cis-eclipsed to anti-staggered rotation before excision of the activated hydrogen at C13. Although evidence points to a transversely flexible binding cleft in the microsomal desaturases rather than the constrained tunnel of the soluble desaturases (50), to avoid electrophilic attack of activated oxygen on the π-system, acetylenase binding pockets must precisely position the rigid linoleate substrate while templating oleate in a range of conformations capable of producing one or both alkene stereochemistries (supplemental Fig. S4). The combined data suggest that stereochemical control in desaturases and acetylenases may be better described by a window model rather than the cis-desaturase eclipsed binding paradigm (Fig. 6). In a 12-trans-yielding eclipsed transoid conformation, the presentation of the reactive hydrogens to the activated oxygen is expected to only deviate slightly from linoleate. A less restrictive binding pocket could allow a small rotation to a torsionally strained cisoid conformation and permit the formation of an E/Z mixture. Normal Δ12-desaturases may have substrate channels or pockets within the active site fully capitulated toward a cis-eclipsed conformation. In this model, CfACET has a less flexible active site than CREP1. Variations in diverged desaturase activity may additionally lie in the precise positioning of the iron atoms or perturbations of the protein shells surrounding the active site.
FIGURE 6.
Consolidated binding pocket model for cis- and trans-desaturases and acetylenases. To accommodate the regiospecific production of (12E)-alkenes in all of the methyl-proximal acetylenases, the alkyl chain at C12-C13 is preorganized within a 90–120° dihedral angle window. Within these bounds, the projection of C13 normal to the linoleate-defined plane for a cis-desaturation is minimized, whereas the vinyl C-H bond orientation is constrained. Torsion about the C12-C13 bond coordinated with H13 abstraction is postulated to lead to cis- and trans-alkenes.
Expression of plant acetylenases in somatic soybean embryos resulted in the accumulation of crepenynic and dehydrocrepenynic acids (15); however, the metabolic origin of the conjugated Δ14 double bond was not clear at the time. A possibility was that crepenynic acid had an altered substrate conformation in the active site of an endogenous ω-3/Δ15 desaturase (FAD3), resulting in the desaturation at the Δ14 position. Our studies argue against this, as S. cerevisiae does not have FAD3 activity. Additionally, we found that a Myc-tagged ω-3-desaturase from tung was not active with crepenynate. Another possibility was that the acetylenases were able to introduce a triple bond into both 18:2 Δ9,12 as well as 18:3 Δ9,12,15, both of which are present in high levels in soybean. For 18:3 Δ9,12,15, this would occur with concomitant migration of the (15Z)-double bond to the 14-position. We found that CfACET is inactive with α-linolenic acid (18:3 Δ9,12,15), an observation that could not be easily made in the soybean system and is strong evidence against this possibility. Here we showed that only when S. cerevisiae cells were supplemented with crepenynic acid and expressing CfDES or co-expressing CfDES and CfACET was dehydrocrepenynic acid produced. This clearly demonstrates that Δ14-desaturating activity lies within the catalytic abilities of the canonical CfDES and may result from altered substrate geometries in the FAD2 active site. Furthermore, we observed the general ability to form a (Z)-enyne with PchFAD2 (Table 2) and the oleate desaturases from Euphorbia and soybean, which are not known to produce acetylenic compounds. Together, and bolstered by the observation that the Tpi-PGFAD enzyme responsible for the production of a moth sex phermone is capable of successive Δ11-desaturation, Δ11-acetylenation, and Δ13-desaturation reactions to produce 16:2 Δ11a,13 (17), these results are consistent with a generally open active site in the Δ12- but not ω-3 desaturases. A shortened acetylenic chain sets up a ν+2 desaturation reaction despite the limited conformational possibilities for C11-C14 in 18:19c,12a. This facility to use a collection of acyl substrates to produce multiple products implies a flexible binding pocket, a hallmark of secondary metabolic processes.
As the pathway to dehydrocrepenynic acid was reconstituted in our S. cerevisiae expression system, the compositional importance of this lipid in the chanterelle, and the fact that no other FAD2 diverged desaturase was found in our shotgun sequencing, CfDES and CfACET fulfill all of the FAD2-like activities required to produce dehydrocrepenynic acid in fungi and demonstrate that the two FAD2 homologs are necessary and sufficient for the production of dehydrocrepenynic acid from oleic acid. Additionally, because the conversion of enzymatically derived intermediates of the three-step pathway in the co-expression experiments was more efficient than conversion of supplemented substrates, it appears that the flux of desaturation and acetylenation products produced in situ in the ER is more directed than that of exogenously supplied fatty acids. Many complexities of polyacetylenic natural product biosynthesis remain to be resolved; however, the current work clearly indicates that the number of required diverged desaturases may be lower than the number of dehydrogenation steps due to strong multifunctional activities.
In conclusion, enzymatic participants necessary for the initial three polyunsaturation reactions in C. formosus were discovered that give underlying molecular support for the crepenynate pathway. Significant variation between fungal and plant acetylenases and desaturases provide a wealth of diversity in dehydrogenative lipid specialized metabolism. Future correlation of entrained structure/function relationships will yield an improved microsomal desaturase structural model aiding in the engineering of biobased lipid products.
Supplementary Material
Acknowledgments
NMR and MS data were collected using instruments acquired through the support of National Science Foundation Grants CHE 0619254 and DBI 0821661. We acknowledge John Dyer and Ed Cahoon for helpful discussions. Plasmids for Δ12 and Myc-tagged Δ15 desaturases were appreciated gifts from Ed Cahoon, Ralph Dewey, and John Dyer. C. alpina seed oil was provided by Richard Adlof (United States Department of Agriculture, Peoria, IL). We greatly appreciate the efforts of many undergraduate researchers and Errol Huffman, who earlier tested necessary but unfruitful approaches to the isolation of a fungal acetylenase.
This work was supported, in whole or in part, by National Institutes of Health Grant R15 GM06493-02. This work was also supported by a grant from the American Cancer Society, Ohio Division and startup support from Miami University (Ohio) and the School of Science, Indiana University-Purdue University Indianapolis.

The on-line version of this article (available at http://www.jbc.org) contains supplemental data, Tables S1 and S2, and Figs. S1–S4.
- ER
- endoplasmic reticulum
- CM
- complete minimal media
- FAME
- fatty acid methyl esters
- 5′-RACE
- 5′ rapid amplification of cDNA ends.
REFERENCES
- 1.Okuley J., Lightner J., Feldmann K., Yadav N., Lark E., Browse J. (1994) Plant Cell 6, 147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vance J. E., Vance D. E. (eds) (2008) Biochemistry of Lipids, Lipoproteins, and Membranes, Elsevier Science, Boston [Google Scholar]
- 3.Millar A. A., Smith M. A., Kunst L. (2000) Trends Plant. Sci. 5, 95–101 [DOI] [PubMed] [Google Scholar]
- 4.Sperling P., Ternes P., Zank T. K., Heinz E. (2003) Prostaglandins Leukot. Essent. Fatty Acids 68, 73–95 [DOI] [PubMed] [Google Scholar]
- 5.Badami R. C., Patil K. B. (1980) Prog. Lipid Res. 19, 119–153 [DOI] [PubMed] [Google Scholar]
- 6.Jaworski J., Cahoon E. B. (2003) Curr. Opin. Plant Biol. 6, 178–184 [DOI] [PubMed] [Google Scholar]
- 7.Singh S. P., Zhou X. R., Liu Q., Stymne S., Green A. G. (2005) Curr. Opin. Plant Biol. 8, 197–203 [DOI] [PubMed] [Google Scholar]
- 8.Minto R. E., Blacklock B. J. (2008) Prog. Lipid Res. 47, 233–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosa L. H., Souza-Fagundes E. M., Machado K. M. G., Alves T. M. A., Martins-Filho O. A., Romanha A. J., Oliveira R. C., Rosa C. A., Zani C. L. (2006) World J. Microbiol. Biotechnol. 22, 539–545 [Google Scholar]
- 10.Bu'Lock J. D., Jones E. H. R., Leeming P. R. (1955) J. Chem. Soc. 4270–4276 [Google Scholar]
- 11.Celmer W. D., Solomons I. A. (1952) J. Am. Chem. Soc. 74, 1870–1871 [Google Scholar]
- 12.Parish C. A., Huber J., Baxter J., González A., Collado J., Platas G., Diez M. T., Vicente F., Dorso K., Abruzzo G., Wilson K. (2004) J. Nat. Prod. 67, 1900–1902 [DOI] [PubMed] [Google Scholar]
- 13.Fernandez-Lopez R., Machón C., Longshaw C. M., Martin S., Molin S., Zechner E. L., Espinosa M., Lanka E., de la Cruz F. (2005) Microbiology 151, 3517–3526 [DOI] [PubMed] [Google Scholar]
- 14.Lee M., Lenman M., Banaś A., Bafor M., Singh S., Schweizer M., Nilsson R., Liljenberg C., Dahlqvist A., Gummeson P. O., Sjödahl S., Green A., Stymne S. (1998) Science 280, 915–918 [DOI] [PubMed] [Google Scholar]
- 15.Cahoon E. B., Schnurr J. A., Huffman E. A., Minto R. E. (2003) Plant J. 34, 671–683 [DOI] [PubMed] [Google Scholar]
- 16.Sperling P., Lee M., Girke T., Zähringer U., Stymne S., Heinz E. (2000) Eur. J. Biochem. 267, 3801–3811 [DOI] [PubMed] [Google Scholar]
- 17.Serra M., Piña B., Abad J. L., Camps F., Fabriàs G. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 16444–16449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hiroi M., Tsuyuki H. (1992) Trans. Mycol. Soc. Japan 33, 517–525 [Google Scholar]
- 19.Pang Z., Sterner O. (1991) J. Org. Chem. 56, 1233–1235 [Google Scholar]
- 20.Bohlmann F., Burkhardt T., Zdero C. (1973) Naturally Occurring Acetylenes, Academic Press, Inc., London [Google Scholar]
- 21.Barley G. C., Graf U., Higham C. A., Jarrah M. Y., Jones E. R. H., O'Neill I., Tachikawa R., Thaller V., Turner J. L., Hodge A. V. (1987) J. Chem. Res. (S) 232–233 [Google Scholar]
- 22.Farrell I. W., Higham C. A., Jones E. R. H., Thaller V. (1987) J. Chem. Res. (S) 234–235 [Google Scholar]
- 23.Ford G. L. (1985) J. Chromatogr. 346, 431–434 [DOI] [PubMed] [Google Scholar]
- 24.Dyer J. M., Chapital D. C., Kuan J.-C. W., Shepherd H. S., Tang F., Peppeman A. B. (2004) J. Am. Oil Chem. Soc. 81, 647–651 [Google Scholar]
- 25.Cahoon E. B., Kinney A. J. (2004) J. Biol. Chem. 279, 12495–12502 [DOI] [PubMed] [Google Scholar]
- 26.Tang G. Q., Novitzky W. P., Carol Griffin H., Huber S. C., Dewey R. E. (2005) Plant J. 44, 433–446 [DOI] [PubMed] [Google Scholar]
- 27.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Nucl. Acid Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minto R. E., Blacklock B. J., Younus H., Pratt A. C. (2009) Appl. Environ. Microbiol. 75, 1156–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. (1997) Current Protocols in Molecular Biology, Wiley-Interscience, New York [Google Scholar]
- 30.Sambrook J. F. T., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 31.Gietz R. D., Woods R. A. (2002) Methods Enzymol. 350, 87–96 [DOI] [PubMed] [Google Scholar]
- 32.Christie W. W. (1998) Chem. Phys. Lipids 94, 35–41 [DOI] [PubMed] [Google Scholar]
- 33.Redhead S. A., Norvell L., Danell E. (1997) Mycotaxon 65, 285–322 [Google Scholar]
- 34.Shanklin J., Whittle E., Fox B. G. (1994) Biochemistry 33, 12787–12794 [DOI] [PubMed] [Google Scholar]
- 35.Avelange-Macherel M. H., Macherel D., Wada H., Murata N. (1995) FEBS Lett. 361, 111–114 [DOI] [PubMed] [Google Scholar]
- 36.Rost B., Casadio R., Fariselli P., Sander C. (1995) Protein Sci. 4, 521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jackson M. R., Nilsson T., Peterson P. A. (1990) EMBO J. 9, 3153–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson M. R., Nilsson T., Peterson P. A. (1993) J. Cell Biol. 121, 317–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Covello P. S., Reed D. W. (1996) Plant Physiol. 111, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carlsson A. S., Thomaeus S., Hamberg M., Stymne S. (2004) Eur. J. Biochem. 271, 2991–2997 [DOI] [PubMed] [Google Scholar]
- 41.Zhang S., Sakuradani E., Ito K., Shimizu S. (2007) FEBS Lett. 581, 315–319 [DOI] [PubMed] [Google Scholar]
- 42.Sakai H., Kajiwara S. (2005) Mol. Genet. Genomics 273, 336–341 [DOI] [PubMed] [Google Scholar]
- 43.Watanabe K., Oura T., Sakai H., Kajiwara S. (2004) Biosci. Biotechnol. Biochem. 68, 721–727 [DOI] [PubMed] [Google Scholar]
- 44.Hashimoto K., Yoshizawa A. C., Okuda S., Kuma K., Goto S., Kanehisa M. (2008) J. Lipid Res. 49, 183–191 [DOI] [PubMed] [Google Scholar]
- 45.Buist P. H. (2004) Nat. Prod. Rep. 21, 249–262 [DOI] [PubMed] [Google Scholar]
- 46.Broadwater J. A., Whittle E., Shanklin J. (2002) J. Biol. Chem. 277, 15613–15620 [DOI] [PubMed] [Google Scholar]
- 47.Shanklin J., Whittle E. (2003) FEBS Lett. 545, 188–192 [DOI] [PubMed] [Google Scholar]
- 48.Beckmann C., Rattke J., Oldham N. J., Sperling P., Heinz E., Boland W. (2002) Angew. Chem. Int. Ed. Engl. 41, 2298–2300 [DOI] [PubMed] [Google Scholar]
- 49.Reed D. W., Polichuk D. R., Buist P. H., Ambrose S. J., Sasata R. J., Savile C. K., Ross A. R., Covello P. S. (2003) J. Am. Chem. Soc. 125, 10635–10640 [DOI] [PubMed] [Google Scholar]
- 50.Shanklin J., Guy J. E., Mishra G., Lindqvist Y. (2009) J. Biol. Chem. 284, 18559–18563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Felsenstein J. (1989) Cladistics 5, 164–166 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.