Abstract
The transcription of tissue-specific and inducible genes is usually subject to the dynamic control of multiple activators. Dedifferentiated hepatic cell lines lose the expression of tissue-specific activators and many characteristic hepatic genes, such as drug-metabolizing cytochrome P450. Here we demonstrate that by combining adenoviral vectors for CCAAT/enhancer-binding protein α (C/EBPα), hepatocyte nuclear factor 4α (HNF4α), and constitutive androstane receptor, the CYP2B6 expression and inducibility by CITCO are restored in human hepatoma HepG2 cells at levels similar to those in cultured human hepatocytes. Moreover, several other phase I and II genes are simultaneously activated, which suggests that this is an effective approach to endow dedifferentiated human hepatoma cells with a particular metabolic competence and response to inducers. In order to gain insight into the molecular mechanism, we examined the cooperation of these three transcription factors on the CYP2B6 5′-flanking region. We show new CYP2B6-responsive sequences for C/EBPα and HNF4α and a novel synergistic regulatory mechanism whereby C/EBPα, HNF4α, and constitutive androstane receptor bind and cooperate through proximal and distal response elements to confer a maximal level of expression. The results obtained from human liver also suggest that important differences in the expression and binding of C/EBPα and HNF4α could account for the large interindividual variability of the hepatic CYP2B6 enzyme, which metabolizes commonly used drugs.
Keywords: C/EBP transcription factor, Cytochrome P450, Drug metabolism, Hepatocyte, Transcription regulation, CYP2B6, Constitutive androstane receptor, Hepatocyte nuclear factor 4 alpha, gene-induction, human hepatocellular model
Introduction
Cytochromes P450 (CYPs)3 are involved in the oxidative metabolism of drugs, carcinogens, and environmental pollutants to more polar metabolites, thereby facilitating their excretion and preventing these potentially harmful compounds from accumulating. In addition, many CYPs participate in the metabolism and conversion of a diverse range of endogenous compounds, including steroid hormones, bile acids, fatty acids, and prostaglandins (1, 2).
CYP2B6 accounts for 2–10% of the total CYP content in the human liver, where it plays an important role in the metabolism of an increasing number of clinically important drugs (3). These include anti-neoplastics like cyclophosphamide, ifosfamide, and tamoxifen (4, 5); the anti-malarial artemisinin (6); the anti-retrovirals nevirapine (7) and efavirenz (8); anesthetics like propofol (9) and ketamine (10); the anti-Parkinsonian selegiline (11); the anti-epileptic mephobarbital (12); and the anti-depressant bupropion, which is now the most commonly used probe drug for CYP2B6 (13). Moreover, CYP2B6 plays a key role in the metabolism of many environmental pollutants, which can serve as substrates, inhibitors, and/or inducers of CYP2B6 and can cause metabolic interactions between environmental chemicals and clinical drugs (14).
A 20–250-fold interindividual variation in CYP2B6 expression has been demonstrated, which is presumably due to polymorphisms and the induction of transcription by xenobiotics. These interindividual differences may result in variable systemic exposure to drugs metabolized by CYP2B6 and may also affect drug efficacy and cause drug-drug interactions (3).
The accurate prediction of drug metabolism and toxicity and drug-mediated induction is a key issue in drug development that greatly influences the success of therapeutics in the market. Prediction studies are further challenged by the lack of suitable models capable of mimicking the human liver. Human hepatic cell lines could be a suitable model for screening strategies. Unfortunately, commonly used hepatic cell lines have consistently demonstrated a very low or marginal CYP expression, which hinders their use as alternative models for drug metabolism and induction studies (15). A feasible explanation for this lack of CYP expression is that the hepatic-specific transcription factors controlling CYP genes are down-regulated in these cell systems. Indeed, several recent studies have demonstrated that transfection of individual liver-enriched transcription factors or coactivators in HepG2 cells up-regulates particular CYPs but to a limited extent (16–21). The reactivation of the metabolic competence in human hepatoma cells by the simultaneous transfection of several of these key factors still remains to be tested.
The transcriptional regulation of many CYP genes in hepatocytes requires both tissue-specific activators (e.g. liver-enriched transcription factors) and ligand-activated nuclear receptors. The transcriptional activation of CYP2B6 by xenobiotics is mediated by the interaction of the nuclear receptors constitutive androstane receptor (CAR) and pregnane X receptor (PXR), with two regulatory sequences that lie upstream of the CYP2B6 gene promoter: the phenobarbital-responsive enhancer module (PBREM, from −1683 to −1733 bp) (22) and the xenobiotic-responsive enhancer module (XREM, from −8495 to −8597 bp) (23). Both regions respond to CAR and PXR and act coordinately to mediate the optimal drug-induced expression of CYP2B6.
More recently, the activation of PBREM by xenobiotics has been seen to be synergistically increased by co-treatment with the protein phosphatase inhibitor okadaic acid (OA) (24). Deletion assays delineate the OA-responsive activity to a proximal 50 bp (−217 to −268 bp) sequence in the CYP2B6 promoter (24, 25). In this OA-responsive module, EGR1 (early growth response 1) and HNF4α (hepatocyte nuclear factor 4α) bind and determine the CAR-mediated induction of the human CYP2B6 gene. Two additional EGR1 binding sites have been found downstream of PBREM (26). Both the distal and proximal EGR1 sites are probably essential to loop the distal PBREM toward the proximal OA-responsive module, thus enabling the xenobiotic sensor, CAR, to communicate with HNF4α to synergistically activate the CYP2B6 promoter.
In this study, we investigated in detail the role of the liver-enriched transcription factors CCAAT/enhancer-binding protein α (C/EBPα) and HNF4α in CYP2B6 transactivation and in the cooperation of these factors with CAR during CYP2B6 induction. By combining the adenoviral expression vectors for C/EBPα, HNF4α and CAR in HepG2 cells, we restored the expression and inducibility of human CYP2B6 at levels similar to those in cultured human hepatocytes. Moreover, several other phase I and II genes were simultaneously activated, thus suggesting that this is an effective approach to endow partially dedifferentiated hepatoma cells with metabolic competence and to generate a human cell system for particular drug induction and metabolism studies. In order to gain insight into the molecular mechanism of CYP2B6 transactivation, we examined the cross-talk of these transcription factors in the CYP2B6 5′-flanking region. We show herein new responsive sequences for C/EBPα and HNF4α and a novel synergistic regulatory mechanism whereby C/EBPα, HNF4α, and CAR bind and cooperate through proximal and distal response elements to confer a maximal CYP2B6 expression level in the human liver.
EXPERIMENTAL PROCEDURES
Cell Culture and Human Liver Samples
Human hepatoma HepG2 cells were plated in Ham's F-12/Leibovitz L-15 (1:1, v/v), supplemented with 5% fetal bovine serum and cultured to 30–40% confluence. Cultures were routinely supplemented with 50 units/ml penicillin and 50 μg/ml streptomycin. Liver samples (1–4 g) from healthy areas of cadaveric grafts of donors were obtained in compliance with the rules of the hospital ethics committee. Donors regularly consumed neither alcohol nor other drugs, nor were they suspected of harboring infectious diseases. They tested negative for human immunodeficiency virus and hepatitis. Liver samples were processed for RNA isolation, ChIP assay, or hepatocyte isolation by using a two-step perfusion technique (27). Isolated hepatocytes were seeded on fibronectin-coated plastic dishes (3.5 μg/cm2) at a density of 8 × 104 viable cells/cm2. Hepatocytes were cultured at 37 °C with Ham's F-12/Williams' E medium (1:1) supplemented with 2% newborn calf serum, 50 milliunits/ml penicillin, 50 μg/ml streptomycin, 0.1% BSA, 10 nm insulin, 25 μg/ml transferrin, 0.1 μm sodium selenite, 65.5 μm ethanolamine, 7.2 μm linoleic acid, 17.5 mm glucose, 6.14 mm ascorbic acid, and 0.64 mm N-ω-nitro-l-arginine methyl ester. The medium was changed 1 h later to remove unattached hepatocytes. By 24 h, the cells were shifted to serum-free hormone-supplemented medium (100 nm dexamethasone and 10 nm insulin). Human hepatocytes in culture show phenotypic instability as a result of tissue disorganization. In our culture conditions, CYP mRNA levels decrease during the early culture and stabilize by 24–48 h at around 10–20% of liver level. Enzyme activities decline at a much lower rate and are better preserved during the culture life span.
Plasmid Constructs
Chimeric luciferase reporter constructs with different lengths of the 5′-flanking region of the human CYP2B6 gene were prepared as follows.
pGL3-B−2654, −1788, −800, −481, and −347
These plasmids were obtained by cloning CYP2B6 5′-flanking sequences into the enhancerless, promoterless pGL3-Basic vector (Promega). Five DNA fragments spanning nucleotides +31 to −2654 bp, −1788 bp, −800 bp, −481 bp, and −347 bp of the CYP2B6 5′-flanking region were PCR-cloned from human genomic DNA (Roche Applied Science) using the Expand 20kbPLUS PCR System (Roche Applied Science). The PCR primers used are described in supplemental Table 1. In order to facilitate cloning, the PCR fragments were first ligated into pCR-XL TOPO (Invitrogen). The resulting vectors were digested with XhoI and HindIII, and the released DNA inserts were ligated into the pGL3-Basic vector previously digested with the same enzymes. All of the pGL3-B-CYP2B6 constructs were confirmed by restriction digestion and double-stranded DNA sequencing.
pGL3-B−1614, −1441, and −218
These plasmids were prepared from pGL3-B−1788, −2654, and −347, respectively, following standard techniques using restriction enzymes.
pGL3-B(−10,311/−8097)+(−2654)
This plasmid was constructed to analyze the influence of a 2200-bp enhancer region located far upstream of the CYP2B6 start site containing the XREM region. A fragment spanning nucleotides from −10,918 to −7428 bp was PCR-amplified as described above, digested with KpnI + XbaI, and ligated into the pGL3-B−2654.
pGL3-B(−1788/−1386)+(−347), (−1788/−1386)+(−218), and (−1788/−1614)+(−218)
These plasmids were constructed to analyze in more detail the responsive sequences within the −1.8 kb CYP2B6 promoter. A fragment spanning bases −1788 to −1386 bp was released from plasmid pGL3-B−2654 with SacI and ligated into the pGL3-B−347 or pGL3-B−218 vectors. A fragment spanning bases −1788 to −1433 bp was PCR-cloned from the plasmid pGL3-B−2654 using the Expand high fidelity PCR system (Roche Applied Science). The amplified fragment was digested with BamHI (at −1614 bp), blunted, and ligated into the pGL3-B−218.
Mutants for C/EBPα and HNF4α
The mutants for C/EBPα and HNF4α were synthesized by GenScript Technologies (Piscataway, NJ). CYP2B6 5′-flanking sequences (from +31 to −347 and from −1386 to −1788) with base substitutions in the predicted C/EBPα binding sites (located at −184/−121 bp, −1510/−1502 bp, and −1597/−1589 bp) and in the predicted HNF4α binding site (at −1642/−1630 bp) were cloned in the pUC57 plasmid and excised by restriction enzymes to be subcloned in pGL3-Basic, pGL3-B−218 or pGL3-B−347.
Adenoviral Vectors
The recombinant adenoviruses encoding CAR, C/EBPα, and HNF4α were developed in our laboratory as described elsewhere (CAR (16), C/EBPα (28), and HNF4α (17)). The resulting viruses (called Ad-CAR, Ad-C/EBPα, and Ad-HNF4α) were plaque-cloned, amplified by standard protocols, and purified and concentrated with the Vivapure AdenoPACK TM100 kit (Sartorius). Titration was performed by a plaque-forming assay as described previously (29). HepG2 cells were infected with recombinant adenoviruses for 18 h at a multiplicity of infection (MOI) ranging from 15 to 65 plaque-forming units/cell. Then cells were washed, and fresh medium was added. At 48 h post-transfection, cells were harvested and analyzed or frozen in liquid N2.
Transfection and Reporter Gene Assays
Plasmid DNAs were purified with Qiagen Maxiprep kit columns (Qiagen) and quantified by A260. The day before infection, cells were plated in 12-well plates to reach 50% confluence. Cells were infected with recombinant adenovirus and were transfected the next day with 0.5 μg of firefly luciferase expression constructs (pGL3-Basic and pGL3-CYP2B6-LUC) and Fugene HD (Roche Applied Science) as transfection reagent. In parallel, 0.08 μg of pRL-SV40 (a plasmid expressing Renilla reniformis luciferase) was cotransfected to correct the variations in transfection efficiency. Prior to measuring luciferase activities using the Dual-Luciferase® reporter kit (Promega), cells were incubated for 48 h more. CITCO (500 nm) was present in the medium for the last 24 h.
Quantification of mRNA Levels
Total cellular RNA was extracted with the RNeasy Plus minikit (Qiagen), which removes contaminating genomic DNA. Total RNA (1 μg) was reverse transcribed as described (30, 31). Diluted cDNA (3 μl) was amplified with a rapid thermal cycler (LightCycler Instrument LC480, Roche Applied Science) in 15 μl of LightCycler DNA Master SYBR Green I (Roche Applied Science) and a 0.3 μm concentration of each primer. We designed specific primers for 10 different cDNAs, including CYPs and transcription factors (supplemental Table 1). In parallel, we always analyzed the mRNA concentration of the human housekeeping porphobilinogen deaminase or glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as internal controls for normalization (supplemental Table 1). PCR amplicons were confirmed to be specific by both size (agarose gel electrophoresis) and melting curve analyses. After denaturing for 30 s at 95 °C, amplification was performed in 40 cycles of 1 s at 94 °C, 10 s at 60 or 62 °C, and 15–20 s at 72 °C. Relative expression levels were calculated with the LightCycler relative quantification analysis software. Briefly, the efficiency of each PCR was estimated from a serially diluted human liver cDNA standard curve. Based on the PCR efficiencies, the software calculates the relative concentration of target and reference (porphobilinogen deaminase or GAPDH) cDNAs and their ratio. Moreover, a positive sample with a stable target/reference ratio (a calibrator) was included in each PCR run to normalize all of the samples in one run and to provide a constant calibration point among several amplification runs.
Preparation of Microsomes and Immunoblotting Analysis
To prepare liver microsomes, liver samples were immediately dissected into small pieces and homogenized in 50 mm Tris HCl, pH 7.4, containing 150 mm KCl and 1 mm EDTA. Homogenates were centrifuged at 9000 × g for 30 min at 4 °C, and the supernatant obtained (S9 fraction) was subsequently centrifuged at 100,000 × g for 1 h at 4 °C. The microsomal pellet was resuspended in 100 mm phosphate buffer, pH 7.4, containing 20% glycerol, quickly frozen in liquid nitrogen, and stored in aliquots at −80 °C. Protein content was determined by using a protein assay kit (Bio-Rad). Microsomal proteins were resolved by 7.5% SDS-PAGE (15 μg of protein/lane), transferred to Immobilion membranes (Millipore), and incubated with anti-CYP2B6 (1:300, sc-67224, Santa Cruz Biotechnology, Inc. (Santa Cruz, CA)). After washing, blots were developed with horseradish peroxidase-labeled IgG using an ECL kit (Amersham Biosciences). Equal loading was verified by Coomassie Brilliant Blue staining of the membrane blots.
Evaluation of CYP Activities
Activity assays were performed by directly incubating cell monolayers with a mixture of five substrates, as described previously (32, 33). The substrate mixture stock solutions were prepared in DMSO to be then conveniently diluted in the incubation medium to obtain the following final concentrations: 10 μm bupropion (CYP2B6), 10 μm diclofenac (CYP2C9), 10 μm bufuralol (CYP2D6), 50 μm chlorzoxazone (CYP2E1), and 5 μm midazolam (CYP3A4). The final DMSO concentration during incubation was 0.5% (v/v). After a 2-h incubation period with the mixture of substrates, aliquots of the medium (300 μl) were collected and stored at −80 °C until analysis. Subsequently, samples were extracted twice with ethyl acetate (1:1, v/v), and dextrometorphan was used as a recovery standard. The organic phase was transferred to a clean tube, evaporated under vacuum, dissolved in 100 μl of HEPES (5% acetonitrile), and analyzed. The metabolites formed and released into the culture medium were quantified by high performance liquid chromatography tandem mass spectrometry (LC/MS/MS). The system comprised a Micromass Quattro Micro (Waters) triple quadrupole mass spectrometer in the electrospray ionization mode, interfaced with an Alliance 2795 HPLC (Waters). After chromatography, the column eluent was directed to an electrospray ionization interface without splitting, operating at 320 °C and using nitrogen as cone gas (50 liters/h). The MS/MS experiments were carried out with a triple quadrupole as analyzer operating in multiple-reaction monitoring mode (32, 33). Enzymatic activities were expressed as pmol of metabolites formed/h/mg of total protein.
Chromatin Immunoprecipitation Assay
Cells were infected with Ad-CAR, Ad-C/EBPα, and Ad-HNF4α. The next day, the medium was changed, and cells were incubated with 500 nm CITCO for another 24-h period. Then cells were treated with 1% formaldehyde in PBS buffer by gentle agitation for 10 min at room temperature to cross-link proteins to DNA. Next, cells were collected by centrifugation, washed, resuspended in lysis buffer, and sonicated on ice for eight steps of 15 s at a 20% output in a Branson Sonicator. Cross-linking and sonication of chromatin from human livers (≤1 g) was carried out following a slightly different protocol (34). Sonicated samples were centrifuged to clear supernatants. DNA content was quantified and properly diluted to maintain an equivalent amount of DNA in all of the samples (input DNA). For the immunoprecipitation of C/EBPα-DNA and HNF4α-DNA complexes, 4 μg of specific antibodies (sc-61 and sc-6556, Santa Cruz Biotechnology, Inc.) were added. Samples were incubated overnight at 4 °C on a 360° rotator (antibody-bound DNA fraction). For each cell preparation, an additional mock immunoprecipitation with rabbit/goat preimmune IgG (sc-2027 and sc-2028, Santa Cruz Biotechnology, Inc.) was performed in parallel (background DNA fraction). Immunocomplexes were affinity-absorbed with 60 mg of protein G-agarose/salmon sperm DNA (Millipore) (prewashed with lysis buffer for 2 h at 4 °C by gentle rotation) and collected by centrifugation (6500 × g, 1 min). The antibody-bound and background DNA fractions were washed as described (34). Cross-links were reversed using 100 μl of 10% Chelex (Bio-Rad), which were added directly to the washed protein G beads and vortexed. After 10 min of boiling, the Chelex/protein G bead suspensions were allowed to cool to room temperature. Proteinase K (20 mg/ml) was then added, and beads were incubated for 30 min at 55 °C while shaking, followed by another 10-min boiling. Suspensions were centrifuged, and supernatants were collected. Chelex/protein G bead pellets were resuspended with 100 μl of water, vortexed, and centrifuged again. The first and second eluates were combined and used directly as a template for quantitative PCR with a LightCycler 480 instrument. Amplification was real time-monitored, stopped in the exponential phase of amplification, and analyzed by agarose gel electrophoresis. Amplifications of the CYP2B6 gene sequences (5′-flanking) among the pull of DNA were performed with specific primers flanking these regions (supplemental Table 1).
Microarray Expression Analysis
Total RNA was purified from HepG2 infected with C/EBPα, HNF4α, and CAR or from HepG2 infected with a control adenovirus (insertless Ad-pACC). Their expression profiles were analyzed with the GeneChip® Human Gene 1.0 ST Array (Affymetrix, Santa Clara, CA). The amount and integrity of purified RNA were estimated by microcapillary electrophoresis (2100 Bioanalyzer, Agilent Technologies), whereas purity was assessed by the A260/A280 ratio.
Amplified and biotinylated sense strand DNA was generated from total RNA according to the Affymetrix whole transcript sense target labeling assay. Labeled single-stranded DNA (5.5 μg) was combined with hybridization and spike controls and hybridized with a pre-equilibrated Affymetrix chip for 16–18 h. Following hybridization, arrays were washed and stained with a streptavidin phycoerythrin conjugate using an automated GeneChip® Fluidics Station 450. They were then scanned with a GeneChip® Scanner 3000 using a 570-nm excitation wavelength laser. Sample preparation and microarray hybridization and scanning were performed at the Gene Analysis Service (Central Research Unit (UCIM), Faculty of Medicine, University of Valencia). Microarray quality control assessment and data acquisition were performed with the GeneChip® Operating Software (Affymetrix). Normalization among the different microarray data files was performed by robust multiarray analysis. Next, we applied a conservative probe filtering (log2 expression value ≥ 5 in at least one sample), which resulted in the selection of a total of 17,095 probes of the original 32,320. Unsupervised hierarchical clustering analysis and a principal component analysis were performed with the DChip software to establish non-forced groups of samples. Differential expression was determined by using linear models and empirical Bayes paired moderated t statistics. False discovery rates were determined by following the Benjamini-Hochberg procedure.
Statistical Analysis
Data were expressed as mean ± S.D. Student's t test was done to determine the statistical significance of the pairwise comparisons.
RESULTS
Transfection of C/EBPα, HNF4α, and CAR Restores Functional CYP2B6 Expression and Response to Inducers in Human Hepatoma HepG2 Cells
Three adenoviral vectors were used to investigate the role of C/EBPα, HNF4α, and CAR in the constitutive and inducible expression of CYP2B6. A preliminary series of experiments was performed to determine the effective doses for each adenovirus, which were set at 20 MOI for Ad-C/EBPα, 65 MOI for Ad-HNF4α, and 15 MOI for Ad-CAR. Each vector, at the selected dose, up-regulated well characterized direct target genes to a similar extent (data not shown). We also determined the level of expression of the transfected transcription factors and found that C/EBPα and CAR increased in HepG2 cells to average liver levels, whereas HNF4α was overexpressed (5–6-fold over the liver level).
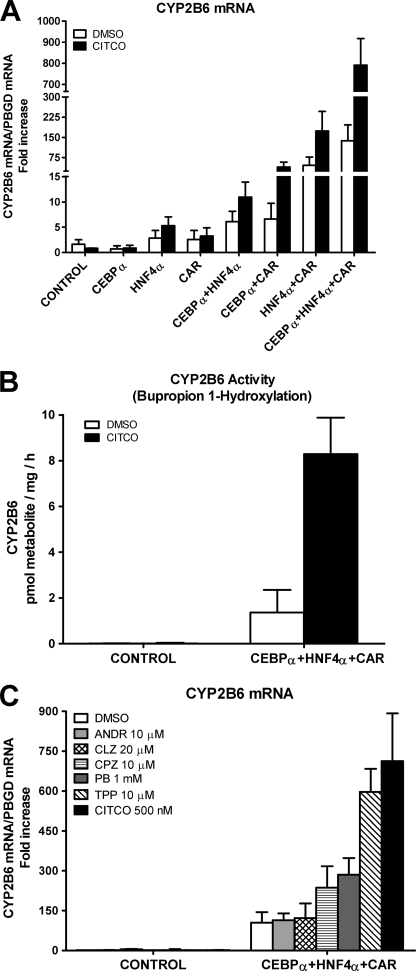
Control HepG2 cells had a very low level of CYP2B6 mRNA (crossing point > cycle 32), and no induction was observed after 24-h exposure to 500 nm CITCO (Fig. 1A). The individual transfection of each factor did not significantly improve the basal expression CYP2B6 level or its inducibility by CITCO. However, combining these factors in pairs had a more important effect; C/EBPα and HNF4α caused a significant increase in the basal and inducible CYP2B6 mRNA of 6- and 11-fold, respectively. Transfection of C/EBPα or HNF4α with CAR had a greater effect, and, as expected, induction by CITCO was much larger (Fig. 1A). These results indicate cooperations between C/EBPα and HNF4α, C/EBPα and CAR, and HNF4α and CAR during CYP2B6 transcription activation. We next examined the effect of these three factors together and found a surprising synergistic up-regulation of CYP2B6 of 138- and 791-fold over the control HepG2 in the non-induced and CITCO-induced cells, respectively (5.7-fold induction) (Fig. 1A). This result demonstrates that C/EBPα, HNF4α, and CAR cooperate synergistically to sustain a high transcription level of human CYP2B6. Our results also reveal that the induction of CYP2B6 by CITCO is not only dependent on CAR but also requires C/EBPα and HNF4α.
FIGURE 1.
C/EBPα, HNF4α, and CAR cooperate to synergistically activate human CYP2B6. HepG2 cells were infected with Ad-C/EBPα, Ad-HNF4α, and Ad-CAR either individually or combined. Twenty-four hours later, 500 nm CITCO or solvent (DMSO) was incorporated, and at 48 h postinfection, cells were harvested and processed. A, CYP2B6 mRNA concentration was measured by quantitative RT-PCR. In parallel, the mRNA concentration of the housekeeping porphobilinogen deaminase was also analyzed for normalization. Data represent the mean ± S.D. of 4–5 independent experiments, expressed as the -fold increase over the control HepG2 cells. B, CYP2B6 activity was measured by determining the bupropion hydroxylation rate in cultured cells by LC/MS/MS. Enzymatic activity was expressed as pmol of metabolite formed/h/mg of total protein. Each bar represents mean ± S.D.; n = 4. C, adenovirus-transduced and control HepG2 cells were incubated with prototypical CYP2B6 inducers (chlorpromazine (CPZ) and PB) and with CAR agonists (triphenyl phosphate (TPP) and CITCO) and inverse agonists (androstanol (ANDR) and clotrimazole (CLZ)). CYP2B6 mRNA levels were measured 24 h later. Data represent the mean ± S.D. of 3–4 independent experiments, expressed as the -fold increase over control cells.
We next sought to determine whether the up-regulation of CYP2B6 mRNA by the co-transfection of C/EBPα, HNF4α, and CAR was followed by a concomitant increase in CYP2B6 activity. Analysis of bupropion hydroxylation shows that the control HepG2 cells have negligible or null CYP2B6 activity (less than the detection limit, 0.01 pmol/mg/h). Co-transfection with the three adenoviral vectors considerably increased CYP2B6 activity to accurately measurable levels (Fig. 1B). More importantly, the incubation of the transfected HepG2 cells with CITCO led to a 6.1-fold induction in the bupropion hydroxylation rate (Fig. 1B). These results show a good correlation between mRNA and activity levels. Our experimental approach upgrades HepG2 cells to a cell model with both significant CYP2B6 activity and induction response.
We next investigated whether the upgraded HepG2 cells respond to CYP2B6 inducers and to human CAR activators and deactivators. HepG2 cells were transfected with the three transcription factors for 24 h to be then exposed to selected concentrations of the different compounds for another 24-h period. We investigated the induction potential of two human CAR agonists, CITCO (35) and triphenyl phosphate (36), and the well known CYP2B6 inducer phenobarbital (PB), which does not bind to CAR but stimulates the translocation of the receptors to the nucleus. We also investigated the response to chlorpromazine, which is an activator of mouse (22, 37) and human (38) CAR. Moreover, we assayed the effect of two CAR inverse agonists or deactivators: clotrimazole (35, 39, 40) and androstanol) (39–41) (they are also PXR activators) (23, 39). Fig. 1C shows that the control HepG2 cells did not respond to any of the assayed CYP2B6 inducers. However, the HepG2 cells transfected with C/EBPα, HNF4α, and CAR responded significantly to the CAR activators (CITCO 6.8-fold > triphenyl phosphate 5.7-fold > chlorpromazine 2.3-fold). We also observed a significant induction of CYP2B6 by PB (2.7-fold) despite the cytoplasmic retention of CAR being impaired in control HepG2 cells (42). Conversely, the two CAR deactivators (clotrimazole and androstanol) did not change the CYP2B6 expression level in the upgraded HepG2 cells (Fig. 1C). These data indicate that the upgraded HepG2 cells recovered a significant response to human CAR activators, thus rendering this engineered cell line a potential tool for screening inducers acting via CAR.
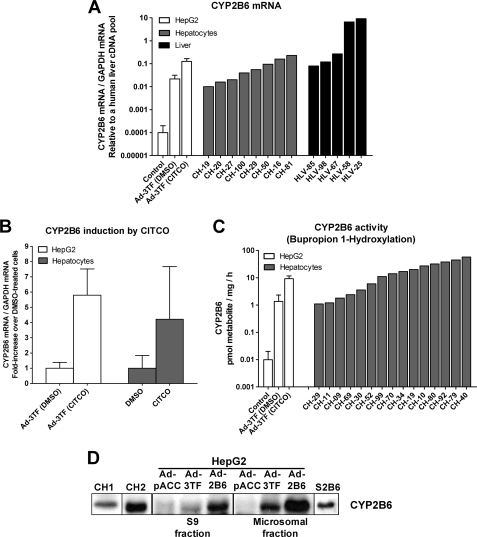
One important question is whether the activation of CYP2B6 in HepG2 cells leads to physiological levels as in cultured hepatocytes or human liver. To answer this question, we compared the CYP2B6 levels in both the modified HepG2 cells and several cultures of human hepatocytes and human livers. The results in Fig. 2, A and C, depict how the CYP2B6 expression achieved in the upgraded HepG2 cells (mRNA and activity) fell within the range of that observed in cultured hepatocytes. The data also confirm previous findings showing the great interindividual variability of this CYP in the population. Immunoblotting analysis (Fig. 2D) shows a lack of CYP2B6 protein in control HepG2 cells and the presence of an immunoreactive CYP2B6 band in the upgraded hepatoma. This band has the same mobility as those derived from either CYP2B6 supersomes (Gentest) or microsomes from HepG2 cells overexpressing CYP2B6 (25 MOI of Ad-CYP2B6). Finally, we compared the extent of CYP2B6 induction by CITCO (Fig. 2B) and found an induction in C/EBPα/HNF4α/CAR-transfected HepG2 cells (5.7-fold increase) similar to that in cultured human hepatocytes (4.2-fold increase). This -fold induction in cultured human hepatocytes coincides with the literature (35, 43).
FIGURE 2.
Comparative analysis of CYP2B6 expression in C/EBPα/HNF4α/CAR-transduced HepG2 cells, cultured hepatocytes, and human livers. HepG2 cells were transduced with the combined adenoviral vectors for 24 h and then exposed to 500 nm CITCO or DMSO for an additional 24-h period. Total RNA was purified from HepG2, cultured human hepatocytes (48 h), or human liver samples (50 mg). A, CYP2B6 mRNA levels were determined by quantitative RT-PCR, normalized with the housekeeping GAPDH, and expressed as relative to a human liver cDNA pool. B, the induction of CYP2B6 mRNA by CITCO was determined by quantitative RT-PCR. Data were expressed as the -fold increase over solvent-treated cells and represent the mean ± S.D. of 4–6 independent experiments. C, CYP2B6 activity was assayed using bupropion as specific substrate. Enzymatic activity was expressed as pmol of metabolite formed/h/mg of total protein. D, CYP2B6 protein in S9 and microsomal fractions was analyzed by immunoblotting. Proteins were electrophoresed on 7.5% SDS-PAGE, transferred to Immobilon membranes, and immunoblotted using a rabbit anti-human CYP2B6 antibody. CH, cultured hepatocytes; HLV, human liver; S2B6, CYP2B6 supersomes (Gentest); Ad-2B6, adenoviral vector encoding CYP2B6; Ad-3TF, adenoviral vectors for the three transcription factors: CEBPα, HNF4α, and CAR.
Cooperation among C/EBPα, HNF4α, and CAR Depends on the Responsive Sequences Located within 1.8 kb Upstream of the CYP2B6 Transcription Start Site
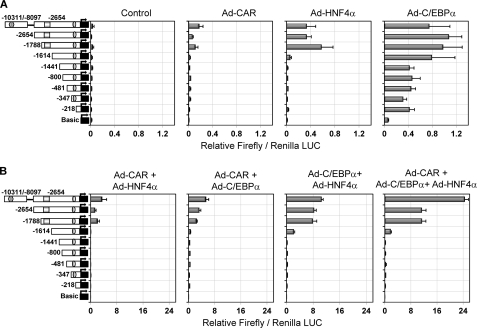
To delineate the cis-acting regions of the CYP2B6 gene that are directly responsible for the synergistic cooperation of C/EBPα, HNF4α, and CAR, a series of reporter assays was performed. Initially, we cloned 2654 bp of the CYP2B6 5′-flanking region in the pGL3-Basic reporter vector. This region contains the PBREM regulatory module (from −1733 to −1683 bp) with the CAR functional elements (NR1 and -2), and the OA-responsive module (from −268 to −217 bp) with the EGR1- and HNF4α-binding elements. We generated seven deletion constructs from this pGL3-B−2654 vector (Fig. 3). Moreover, a distal 2.2-kb CYP2B6 fragment (from −10,311 to −8097 bp) was subcloned into the pGL3-B−2654 vector to create a longer construct containing the CYP2B6 promoter region (2654 bp) plus a distal region containing the CAR-responsive XREM module (from −8597 to −8495 bp). The resulting construct was called pGL3-B(−10311/−8097)+(−2654) vector. All of these reporter constructs show negligible basal reporter activity in the control HepG2 cells (Fig. 3A), which is in agreement with the very low CYP2B6 gene expression level. The individual transfection of each transcription factor had a small effect on the reporter activity (see the LUC activity scale in Fig. 3A). Transactivation by HNF4α or CAR was consistently detected only when the fragment from −1614 to −1788 bp containing the PBREM was present. Regarding C/EBPα, two regions from +1 to −218 bp and from −1441 to −1614 bp appeared important for the basal activity of this factor on CYP2B6 (Fig. 3A). The transfection of the three factors together or in pairs led to much greater responses (see the LUC activity scale in Fig. 3B), suggesting cooperation among these factors in CYP2B6 transactivation. These results also demonstrate that the region from −1614 to −1788 bp containing the PBREM was required for the cooperative effect of any tested combination (Fig. 3B). The region from −1441 to −1614 bp also played a role in the cooperation between C/EBPα and HNF4α. Finally, the presence of the distal region containing the XREM only favored the combined response to the three factors and was not critical for the synergistic effect (Fig. 3B).
FIGURE 3.
Transactivation of CYP2B6 5′-flanking reporter constructs by C/EBPα, HNF4α, and CAR. Sequential deletion fragments of the 5′-flanking region of the CYP2B6 promoter were cloned into the firefly luciferase pGL3-Basic reporter vector. HepG2 cells were infected with the three adenoviral vectors individually (A) or combined (B). Twenty-four hours later, cells were transfected with the different CYP2B6 promoter constructs (0.5 μg) and the normalization plasmid pRL-SV40 (0.08 μg). At 48 h postinfection, cells were treated with 500 nm CITCO, and at 72 h postinfection, cells were lysed, and both firefly and Renilla reniformis luciferase activities were determined. Bars, mean ± S.D. of five independent experiments, expressed as firefly/Renilla luciferase activity ratios. The symbols in the left-hand diagrams denote OA-responsive module (ellipse), PBREM (square), and XREM (hexagon).
Transactivation by C/EBPα, HNF4α, and CAR Requires a Proximal Promoter Sequence (−347 bp) and a Distal Region (from −1386 to −1788 bp) of the CYP2B6
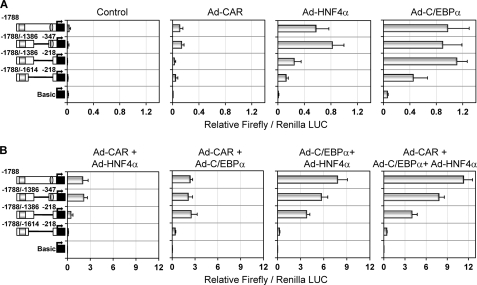
We next dissected the −1.8kb-2B6-Luc vector, which encompasses all of the sequences needed for most of the synergistic response. Deletion of 1 kb from −347 to −1386 bp (the pGL3-B(−1788/−1386)+(−347) vector) did not significantly change the response to C/EBPα, HNF4α, or CAR (Fig. 4A). Further deletion of the OA-responsive module (the pGL3-B(−1788/−1386)+(−218) vector) led to a significant decrease in the response to HNF4α (p < 0.001) and CAR (p < 0.001). In order to reduce the response to C/EBPα, a 225-bp fragment in the distal region (from −1386 to −1614) also had to be deleted (the pGL3-B(−1788/−1614)+(−218) vector) (Fig. 4A). These results, along with the results in Fig. 3, suggest that the C/EBPα-responsive sequences are located in two regions: +1 to −218 bp (near the OA-responsive module) and −1386 to −1614 (near the PBREM). Moreover, our results also suggest that two HNF4α-responsive sequences are involved: the previously described one (26) located in the OA-responsive module (from −217 to −229 bp) and the other in the PBREM region (from −1614 to −1788 bp).
FIGURE 4.
Synergistic CYP2B6 transactivation by C/EBPα, HNF4α, and CAR requires both proximal and distal promoter sequences. The −1.8 kb CYP2B6 promoter region was dissected into several chimeric reporter constructs. HepG2 cells were infected with the recombinant adenoviral vectors individually (A) or combined (B) for 24 h. Then cells were transfected with the CYP2B6 promoter constructs. At 48 h postinfection, cells were treated with 500 nm CITCO, and at 72 h postinfection, cells were lysed, and both firefly and R. reniformis luciferase activities were determined. Bars, mean ± S.D. of six independent experiments, expressed as firefly/Renilla luciferase activity ratios. The symbols in the left-hand diagrams denote OA-responsive module (ellipse) and PBREM (square).
Regarding the combined effect of the three transcription factors, the results in Fig. 4B demonstrate that the cooperation between CAR and HNF4α requires the PBREM and OA-responsive regions. However, the cooperation between C/EBPα and CAR principally requires the PBREM and a distal region from −1386 to −1614 bp. Similarly, that between C/EBPα and HNF4α apparently also requires this C/EBPα-responsive distal region (Fig. 4B).
New Proximal C/EBPα-responsive Sequences Are Essential for the Constitutive CYP2B6 Expression, whereas the New Distal C/EBPα and HNF4α Elements Play a Significant Role in the Synergistic CYP2B6 Induction by CAR
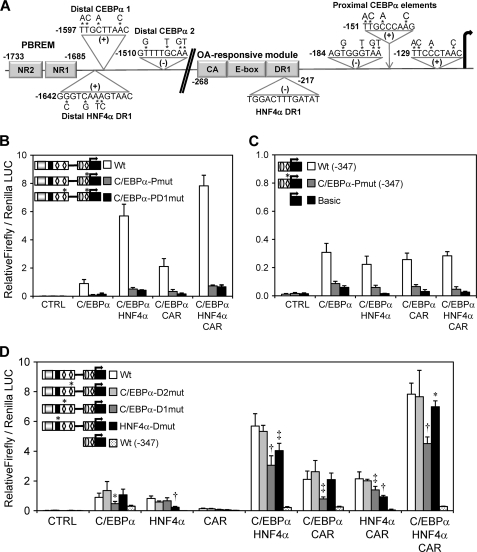
We used the MatInspector 8.0 software (44) to analyze the newly identified regions for potential C/EBPα and HNF4α binding sites. Three consensus-adjacent C/EBPα response elements (45) were identified in a short 67-bp region located between −121 and −184 bp. Moreover, two additional C/EBPα response elements were found at −1510 bp and at −1597 bp in the distal C/EBPα-responsive region near the PBREM (Fig. 5A). Regarding HNF4α, a new consensus DR1 response element was identified at −1642 bp, which is at only 91 bp from the CAR/PXR NR2 site in the PBREM region. Another HNF4α DR1 binding site at −229 bp in the OA-responsive module was also described by Inoue and Negishi (26) (Fig. 5A).
FIGURE 5.
Proximal C/EBPα-responsive elements are essential for the constitutive CYP2B6 transcription, whereas new HNF4α and C/EBPα distal elements are required to achieve a maximal synergistic response along with CAR. A, schematic diagram showing the putative binding sites for C/EBPα and HNF4α in the CYP2B6 promoter region identified by in silico analysis. The positions relate to the transcriptional start site +1. A new distal HNF4α binding site was located at −1642 bp, just 43 bp from the CAR NR sites in the PBREM. The putative distal C/EBPα sites were at −1597 and −1510 bp, whereas the proximal C/EBPα sites were within a short region located between −184 and −121 bp. Base substitutions in the mutated reporter constructs are indicated by asterisks. B–D, HepG2 cells were infected with the three adenoviral vectors for 24 h. Then they were transfected with the different wild-type and mutated CYP2B6 reporter constructs. At 48 h postinfection, cells were treated with CITCO, and at 72 h postinfection, cells were lysed, and both firefly and R. reniformis luciferase activities were determined. Bars, mean ± S.D. of four independent experiments, expressed as luciferase ratios. The relevance of the three proximal C/EBPα-responsive elements was investigated with (B) or without (C) the influence of the −1788/−1386 bp (PBREM) region. The relevance of the distal C/EBPα- and HNF4α-response elements is shown in D. The symbols in the left-hand diagrams denote OA-responsive module (gray ellipse), PBREM (gray rectangle), C/EBPα-binding elements (white diamonds), and the HNF4α distal element (black rectangle). *, p < 0.05; ‡, p < 0.01; †, p < 0.001.
We generated a series of reporter constructs with point mutations to determine the relevance of these putative C/EBPα and HNF4α binding sites. The simultaneous mutation of the three proximal C/EBPα sites in the pGL3-B(−1788/−386)+(−347) vector abolished most of the transactivation by C/EBPα either alone or in combination with HNF4α or/and CAR (Fig. 5B). The same mutations in the pGL3-B−347 vector lowered the promoter activity up to marginal background Luc levels, which were comparable only with those of the promoterless pGL-3-Basic (Fig. 5C). These results suggest that activation by C/EBPα, through the new functional C/EBPα binding sites in the proximal promoter, is critical for the constitutive expression of CYP2B6 in the liver. Moreover, the requirement of the proximal C/EBPα-responsive sequences for cooperation with HNF4α and CAR cannot be ruled out.
The mutation of the two distal C/EBPα elements reveals that only one of them, C/EBPα-mt-1 at −1598 bp, was important for the pGL3-B(−1788/−386)+(−347) reporter activity. The mutation of this element was sufficient to reduce the response to C/EBPα by 53% and to significantly decrease the cooperation of this factor with HNF4α and with CAR by 45–60% (Fig. 5D). The mutation of the new distal DR1 HNF4α element at −1642 bp significantly lowered the transactivation by HNF4α and its cooperation with C/EBPα and CAR. However, its potential relevance was less evident when the three transcription factors were coexpressed (Fig. 5D). These results suggest that the new C/EBPα and HNF4α elements in the distal CYP2B6 promoter, which are located close to the CAR elements in the PBREM, are required to cooperate and build up effective CYP2B6 activation.
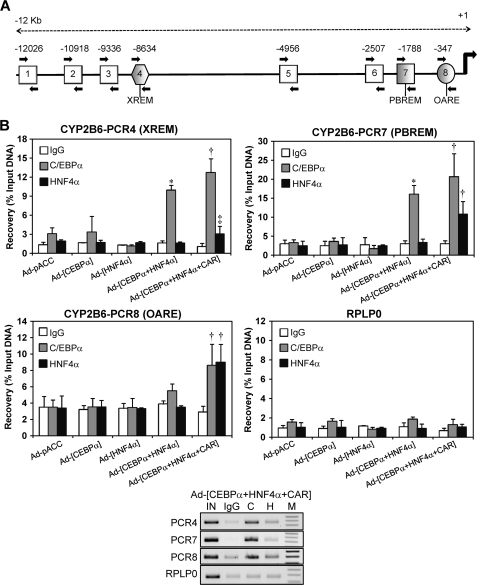
Chromatin Immunoprecipitation Demonstrates the Binding of C/EBPα and HNF4α to Active Promoter and Enhancer Modules of the CYP2B6 in Both Transfected HepG2 Cells and Human Liver
Formaldehyde cross-linked chromatin from HepG2 cells was immunoprecipitated with antibodies against C/EBPα and HNF4α. We designed primers to assess the binding of C/EBPα and HNF4α to eight sequences of the CYP2B6 5′-flanking region, which not only covered both the PBREM and the OA-responsive module but also more distal regions, including the XREM and beyond (Fig. 6A). Control HepG2 cells (transduced with insertless adenoviral vector) showed no binding of C/EBPα or HNF4α to the promoter and enhancer CYP2B6 sequences (Fig. 6B). Similarly, HepG2 cells transduced with only Ad-C/EBPα or Ad-HNF4α did not exhibit a significant binding of the encoded transcription factors. However, the coexpression of C/EBPα and HNF4α substantially enhanced C/EBPα binding to both the PBREM and XREM, suggesting that the binding of this factor depends on HNF4α. Finally, when HepG2 cells were transfected with the three transcription factors, a significant binding of both C/EBPα and HNF4α was observed in the OA-responsive module in the PBREM and XREM regions (Fig. 6B) but not in the other five 5′-flanking regions of the CYP2B6, which were interrogated (data not shown). Because HNF4α binding was only robustly detected with all three factors together, HNF4α binding should then be dependent on CAR.
FIGURE 6.
ChIP assays demonstrate the binding of C/EBPα and HNF4α to both the proximal and distal CYP2B6 promoter sequences and to the far enhancer region containing the XREM. A, schematic diagram showing the positions of the eight PCR primer sets designed to assess C/EBPα and HNF4α binding through 12 kb of the CYP2B6 5′-flanking region. PCRs 4, 7, and 8 encompass the XREM, PBREM, and OA-responsive modules, respectively. Moreover, PCR7 includes the new identified HNF4α and C/EBPα elements, whereas PCR8 comprises very proximal C/EBPα sites. B, formaldehyde cross-linked chromatin from adenovirus-infected HepG2 cells was incubated with antibodies against C/EBPα and HNF4α or with preimmune IgG. To assess the binding to the different regions, quantitative PCR was performed on immunoprecipitated, purified DNA. A genomic region covering exons 3 and 4 of human RPLP0 (ribosomal protein large P0) was also amplified as a negative binding control. Recovery was calculated as the percentage of input DNA (100%). Bars, mean ± S.D., n = 4. *, p < 0.05; ‡, p < 0.01. Lower panel, aliquots (10 μl) of PCR amplifications from a representative experiment were subjected to electrophoresis on 1.5% agarose gel and stained with ethidium bromide. IN, input; C, anti-C/EBPα; H, anti-HNF4α; M, 100-bp DNA ladder.
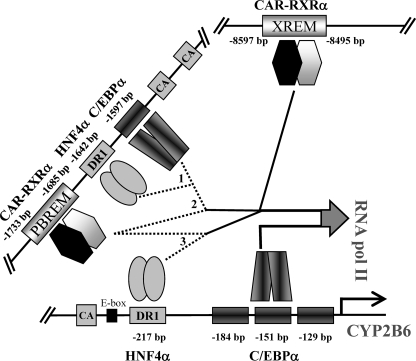
These results support the notion that C/EBPα and HNF4α bind to the previously identified sequences in the proximal (PCR8) and distal (PCR7) CYP2B6 promoter. Moreover, additional binding sites for these factors may be present in the XREM region (PCR4), although the binding to the XREM could also be explained by a looping mechanism, whereby CAR and additional factors close the XREM to the promoter region (PBREM and OA-responsive module) and enable CAR to cross-talk with HNF4α and C/EBPα to synergistically transactivate CYP2B6 (see Fig. 8).
FIGURE 8.
Schematic representation of C/EBPα, HNF4α, and CAR binding to the different regulatory CYP2B6 sequences, and their potential cooperation mechanism by looping the DNA to achieve maximal transactivation. Major cooperative activity was noted between the distal HNF4α and C/EBPα elements (1); CAR in the PBREM and the distal C/EBPα element (2), and CAR in the PBREM and the proximal HNF4α element (3). Moreover, indirect evidence suggests that unidentified protein-protein interactions involving other transcription factors and coregulators probably close the XREM and PBREM regions to the proximal promoter region by looping the DNA. Proximal C/EBPα elements could also play a role in cooperation with HNF4α and CAR, but they seem to be more critical for the constitutive CYP2B6 transcription in hepatocytes.
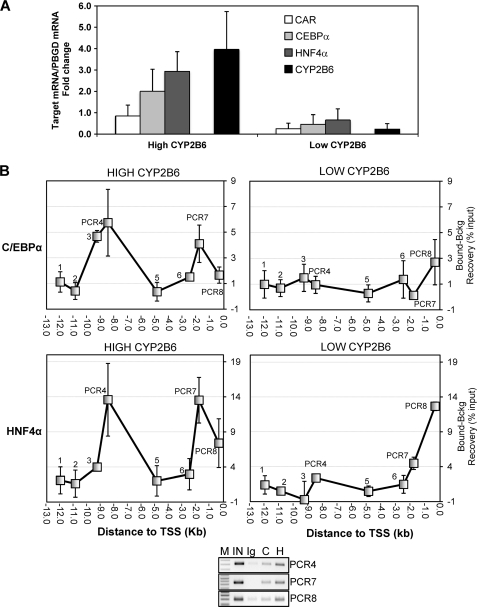
In order to confirm and reinforce the relevance of these results, we performed ChIP assays in human liver samples from the Hospital La Fe-CIBERehd Human Liver Bank. We selected livers from six donors showing high (n = 3) and low (n = 3) CYP2B6 expressions (activity and mRNA). We found that in agreement with the different CYP2B6 level, these livers also presented important differences in the expression of C/EBPα, HNF4α, and CAR (Fig. 7A). ChIP assays also reveal striking differences in the binding of C/EBPα and HNF4α between these two groups. The analysis of the livers with a high CYP2B6 expression demonstrated important binding for C/EBPα and HNF4α to the distal XREM, the proximal PBREM, and the OA-responsive regions (Fig. 7B). However, much less binding of C/EBPα and HNF4α was observed in the livers with a low CYP2B6 expression level (Fig. 7B). These results extrapolate our findings in HepG2 cells to human beings and indicate a potential relevant role of HNF4α and C/EBPα, along with CAR, in sustaining the coordinated transcription of CYP2B6 in the human liver and in determining the high interindividual variability in the CYP2B6 expression.
FIGURE 7.
Analysis of C/EBPα and HNF4α binding to the CYP2B6 5′-flanking region in human livers. A, total RNA from six liver donors showing high (n = 3) and low (n = 3) CYP2B6 expression was purified, and the mRNA levels of CYP2B6, C/EBPα, HNF4α, and CAR were determined by real-time quantitative RT-PCR analysis. In parallel, we also analyzed the mRNA concentration of the housekeeping porphobilinogen deaminase for normalization. Data represent the mean ± S.D. (error bars) expressed as -fold increase/decrease compared with a reference human liver cDNA pool (n = 12). B, formaldehyde cross-linked chromatin from human liver tissues was immunoprecipitated with antibodies against C/EBPα and HNF4α or with preimmune IgG. Quantitative PCR was performed on immunoprecipitated, purified DNA (bound DNA fraction) using primers specific to eight CYP2B6 5′-flanking regions (as depicted in Fig. 6A). Parallel PCRs were performed with both input and IgG-immunoprecipitated DNA (background DNA fraction). Data represent the mean ± S.D. of three different livers and are expressed as the difference between the recoveries in the bound and in the background fractions. Lower panel, PCR aliquots (10 μl) from a representative amplification were subjected to electrophoresis on 1.5% agarose gel and stained with ethidium bromide. IN, input; C, anti-C/EBPα; H, anti-HNF4α; M, 100-bp DNA ladder.
Transfection of C/EBPα, HNF4α, and CAR in Human HepG2 Cells Up-regulates Phase I and Phase II Drug-metabolizing Enzymes, Enhances Drug Detoxification Pathways, and Improves the Hepatic Phenotype
A large number of target genes should be activated after increasing the expression of three key hepatic transcription factors. We first investigated the impact of this combination of factors on other phase I drug-metabolizing CYPs and found that CYP3A4 and CYP2C9 were also up-regulated (supplemental Fig. 1A). However, the relative -fold increase in mRNA (10–15-fold) was much lower than for CYP2B6. A slight induction by CITCO was only observed for CYP2C9 mRNA in the transfected HepG2 cells.
CYP-associated activities were also determined. Diclofenac 4′-hydroxylation by CYP2C9 increased 13-fold in the upgraded HepG2 cells, and incubation with CITCO further induced this activity by up to 23-fold over the control HepG2 cells (supplemental Fig. 1A). However, midazolam 1′-hydroxylation activity only increased ∼2.5-fold in the upgraded HepG2, which did not correlate with the more significant increase in CYP3A4 mRNA (supplemental Fig. 1A). The other CYP activities simultaneously measured by LC/MS/MS with a substrate-mixture approach demonstrated a slight 1.8-fold increase in CYP2D6 bufarolol hydroxylation, and no significant increase in CYP2E1 chlorzoxazone 6-hydroxylation was observed (data not shown).
We next determined the effect of C/EBPα, HNF4α, and CAR cotransfection on the mRNA concentration of several phase II genes (supplemental Fig. 1B). Our results show that our experimental approach also leads to an important up-regulation of GSTA1 and UDP glucuronosyltransferases 1A1 and 2B4. Moreover, the bile acid transporter OSTβ (organic solute transporter β) mRNA was also robustly induced in the upgraded HepG2 cells (supplemental Fig. 1B). Induction by CITCO was only observed in UDP glucuronosyltransferase 1A1 mRNA. Our results demonstrate that the upgraded HepG2 cells not only show a high basal and inducible CYP2B6 expression but also the up-regulation of several other key drug-metabolizing CYPs, phase II conjugating enzymes, and transporters.
To further characterize the impact of the three transcription factors in the transcriptome, we did a microarray expression analysis using the GeneChip® human gene 1.0 ST array of Affymetrix, which offers whole-transcript coverage of 28,869 human genes. We performed triplicate experiments to compare the control HepG2 (control adenovirus-transfected and CITCO-induced) versus the upgraded HepG2 cells (C/EBPα-, HNF4α-, and CAR-transfected and CITCO-induced). Unsupervised principal component analysis of the microarray data indicates that experimental samples cluster into two groups: control and upgraded HepG2 (supplemental Fig. 2A). We set the cut-off for the differentially expressed genes at a q value (false discovery rate) of <0.05 and a ±5/3-fold change. The results reveal 415 differentially expressed genes, of which 282 were up-regulated by the cotransfection of C/EBPα, HNF4α and CAR, whereas only 133 were down-regulated. A heat map of the top 50 differentially expressed genes demonstrates that up-regulation was much more significant in the transfected HepG2 cells than repression (supplemental Fig. 2B). We next analyzed the overrepresented biochemical pathways in our up-regulated gene list by using the ConsensusPathDB tool (Max Planck Institute for Molecular Genetics), which was set at a next neighbor radius of 1, an overlap with input list of ≥2, and a p value cut-off of ≤0.001. Forty-three pathway-based sets were enriched. Supplemental Table 2 shows some of the most representative overrepresented pathways, of which it is noteworthy to remark the up-regulation of biological oxidations, the functionalization of compounds and xenobiotics, and the induction of several hepatic functions, such as glycogen and urea synthesis and the metabolism of steroids and bile acids. The PPAR signaling pathway and a number of genes related to lipid and fatty acid metabolism (ACSL1, ACSL5, ELOVL4, ELOVL3, SLC27A2, CD36, APOL2, APOL1, APOL4, and APOM) were also significantly induced. Moreover, the rate-limiting gluconeogenic enzyme, phosphoenol-pyruvate carboxykinase (PCK1), was up-regulated 6.9-fold. A more detailed analysis of the overrepresented pathways by means of the ConsensusPathDB with the pharmacogenomics knowledge data base (PharmGKB) shows how, in the upgraded HepG2 cells, there is a significant up-regulation of the genes involved in the metabolism and disposition of many important clinically used drugs (e.g. clopidogrel, tamoxifen, ifosfamide, cyclophosphamide, erlotinib, fluoropyrimidine, and several statins) (supplemental Table 3).
Collectively, our results indicate that the upgraded HepG2 cells, with an improved hepatic phenotype, provide a better in vitro human liver model, which could be suitable for particular drug metabolism and toxicity studies or for drug-mediated CYP induction analyses during early drug development.
DISCUSSION
The transcription of tissue-specific and inducible genes is generally subject to the dynamic control of multiple activators and coregulators working in concert to confer the maximal level of expression. The aim of the present study was to gain insight into the molecular mechanism by which C/EBPα, HNF4α, and CAR cooperate in the transcription of CYP2B6, an important drug-metabolizing human liver gene. Despite the complexity of such a study, we offer novel results, which demonstrate that 1) for the efficient transcriptional activation of CYP2B6, the nuclear receptor CAR requires the cooperation of two specific liver-enriched transcription factors, HNF4α and C/EBPα; 2) both C/EBPα and HNF4α regulate CYP2B6 through several recognition sequences in its regulatory 5′ region; 3) the three factors cooperate with one another, CAR with C/EBPα, CAR with HNF4α, and C/EBPα with HNF4α; 4) cooperation between each pair of factors does not equally involve all of their binding sites but depends on specific response elements of each factor. Defining the functional cooperation of C/EBPα, HNF4α, and CAR has led to an increased understanding of the mechanisms by which the CYP2B6 gene is regulated and how these factors collaborate to mediate transcriptional activation.
The overall impact of HNF4α on the control of human CYP genes has been assessed by gain- and loss-of-function strategies in cultured cells. HNF4α has been seen to be a critical factor for the expression of many major drug-metabolizing CYP genes, including CYP2A6, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP3A4, and CYP3A5 (46–48). Other studies have also demonstrated that the cross-talk of HNF4α with CAR or PXR is critical for the appropriate drug-mediated induction of major human CYP genes, such as CYP3A4 or CYP2C9 (49–52). More recently, the important role of the proximal HNF4α response element in the cooperation between EGR1 and CAR for CYP2B6 induction has also been demonstrated (25). All of these results, along with those of the present study, suggest that the cooperation of HNF4α and CAR is a common feature in the regulation of drug-metabolizing CYPs and that clusters of CAR and HNF4α response elements are likely a common signature in the regulatory modules of these genes.
Although HNF4α are well expressed in HepG2 cells, we had to overexpress HNF4α to observe an activating effect on CYP2B6. The poor activity of endogenous HNF4α on the CYP2B6 promoter could be due to several reasons, including an increased level of transcription repressors/corepressors (e.g. COUP-TFs (53)) or an altered HNF4α isoform pattern (e.g. increased expression of HNF4α7) in hepatoma cells. We also know that HNF4α is enriched in the liver but is not as abundantly expressed as other transactivators, such as ATF5, C/EBPα, or HNF3γ (16). This probably indicates that HNF4α at its physiological concentration may not fully activate all its target genes.
We have previously shown that the re-expression of the coactivator PGC1α in HepG2 cells enhances endogenous HNF4α activity on particular target genes (17). Consequently, we investigated whether PGC1α, by coactivating endogenous HNF4α, is able to improve CYP2B6 expression in cooperation with CAR and C/EBPα. First, we investigated the role of PGC1α alone and found that Ad-PGC1α is able to up-regulate HNF4α target genes, such as CYP7A1 or ApoCIII, but has no effect on CYP2B6 levels. Surprisingly, we also found that the cotransfection of PGC1α with C/EBPα and CAR has a negative effect on CYP7A1 and ApoCIII and abolishes the synergistic activation of CYP2B6 (data not shown). These results are in agreement with studies showing that CAR can inhibit HNF4α activity in particular promoters (e.g. CYP7A1) by competing for binding to common coactivators, such as PGC1α (54). Therefore, it is feasible that the transfection of PGC1α along with CAR leads to nonfunctional PGC1α-CAR complexes, which disrupt the synergistic interaction among CAR, C/EBPα, and HNF4α in the CYP2B6 promoter. The interaction of transcription factors with coactivators depends on the promoter context. It is feasible that PGC1α does not cooperate with HNF4α in all of the HNF4α-activated promoters, which could be the case in the CYP2B6 gene.
The regulation of CYP genes by C/EBPα has been much less explored. We have previously shown that adenovirus-mediated overexpression of C/EBPs reactivates the expression of CYP3A4, CYP2B6, CYP2C9, and CYP2A6 (19, 21, 28) to anticipate that the overall relevance of C/EBPα on CYP gene regulation could also be significant. However, as far as we know, this is the first work showing how C/EBPα cooperates with CAR in drug-mediated induction of CYP genes.
We have demonstrated that C/EBPα elements in the CYP2B6 proximal promoter are critical for the constitutive CYP2B6 expression. However, for C/EBPα to cooperate with HNF4α or CAR, a distal C/EBPα element near the PBREM seems preferable. Therefore, we hypothesize that the different elements of C/EBPα are involved in different scenarios for CYP2B6 transcription. A similar conclusion may be drawn for the HNF4α response elements, where the proximal promoter DR1 element is preferred for cooperation with CAR, whereas the new distal DR1 element is apparently favored for cooperation with C/EBPα (Fig. 8). The molecular mechanism by which these factors cooperate is not yet understood.
Our reporter assay data suggest that the distal XREM enhancer does not play a major role in either activation by C/EBPα and HNF4α or the cross-talk of these factors with CAR. However, ChIP assays demonstrate significant binding of C/EBPα and HNF4α to the distal XREM region. One possible explanation for this apparent discrepancy is that the distal promoter and enhancer regulatory DNA elements have to loop to come closer to the transcription start site in order to constitute a cooperative assembly of activators into a enhanceosome, which will synergistically favor transcription initiation by RNApol II (55, 56). The cross-linking of such a multiprotein complex will result in the immunoprecipitation of clogged DNA sequences, some of which are indirectly bound to the target protein. Along these lines, a recent study has shown that an antibody against coactivator NCOA6 precipitates both the distal CAR and the proximal HNF4α-binding elements of the CYP2C9 promoter (57), thus demonstrating the cross-link and co-immunoprecipitation of the distal and proximal sequences in the ChIP assay after the cooperative assembly of transcription factors via intermediate coactivators. The looping hypothesis is further reinforced by the recent findings of Inoue and Negishi (26), who demonstrate that EGR1 loops the CYP2B6 promoter region to achieve synergistic activation by CAR and HNF4α. Nevertheless, this mechanism is not proven in the present study, and we cannot rule out that HNF4α and C/EBPα also bind directly to the XREM region. Multiple protein-DNA and protein-protein interaction steps (involving other transcription factors and coregulators) expected to occur during this process will be subject to intense future research.
Notable differences in the expression level and binding of HNF4α and C/EBPα were found in the livers of different donors. These differences correlate with significant differences in CYP2B6 expression and activity, thus supporting the notion that regulatory variability could account for CYP2B6 interindividual variability to a great extent. The correlation between CYP2B6 and CAR in population-based studies has been demonstrated previously (58). A 278-fold variability was found in human CYP2B6 mRNA, whereas a 240-fold variability was seen in CAR mRNA. In a more recent study, CAR and HNF4α were identified as the nuclear receptors whose expression levels most strongly associated with the expression of xenobiotic metabolism genes, including CYP2B6 (59). This study, with 20 human liver samples, shows a more than 7-fold variability for HNF4α expression (59). In another study, HNF4α expression was highly variable in the fetal liver and fluctuated in the postnatal liver by up to 16-fold (60). Thus, variations in these transacting regulators may account for the differential expression of a large amount of CYP2B6 and also determine both complex traits and the individual's clearance rate of a wide range of prescribed drugs. A potential association between C/EBPα and CYP2B6 expression levels in the population still remains to be investigated.
Given the intrinsic variability and limited accessibility of primary cultured human hepatocytes, researchers have attempted to discover or develop suitable alternative human hepatic cell models for drug metabolism studies. Unfortunately, the human hepatic cell lines available (e.g. HepG2) are not a real alternative because they express only marginal levels of drug-metabolizing CYPs and do not respond to CYP inducers (15). This could result from an altered (mostly repressed) expression of liver-enriched transcription factors and co-regulators. We have previously shown that this could be the case for C/EBPα, HNF4α, and CAR, three key transcription factors for the hepatic phenotype, which are down-regulated or dysfunctional in human HepG2 cells (16, 17, 21, 27). The functional re-expression of these three factors in HepG2 cells has not only improved the critical pathways associated with a differentiated hepatic phenotype but also activated a number of drug metabolism and disposition enzymes. CYP2B6 was so robustly reactivated in HepG2 cells that its mRNA and activity reached levels comparable with those found in cultured human hepatocytes. Moreover, the induction of CYP2B6 by CAR ligands, such as CITCO, was similar in this upgraded cell model to that in cultured human hepatocytes. In contrast, induction by PB (ligand-independent mechanism) was lower in upgraded HepG2 cells than in cultured human hepatocytes. We observed a 2.7-fold increase in C/EBPα/HNF4α/CAR-transfected HepG2 cells, whereas the induction for PB reported in the literature in cultured hepatocytes ranges from 6.5- to 9-fold (61, 62). We conclude that this cell system may be suitable for screening compounds with CAR-agonist properties but may underestimate PB-like inducers.
Despite our results being proof of concept in demonstrating that this strategy is suitable for achieving metabolically competent cell lines, several limitations still need to be addressed: 1) not all of the important drug-metabolizing CYPs are reactivated at functional levels; 2) the profile of the various drug metabolism enzymes re-expressed in the upgraded HepG2 cells may not be similar to that in the human liver; and 3) the expression level of PXR, a major transcription factor in CYP2B6 regulation, is low in HepG2 cells; therefore, this upgraded cell system may only be suitable for particular drug metabolism studies or for screening drugs with an induction potential for specific CAR-dependent phase I and II enzymes.
The restitution of a significant induction of CYP2B6 by PB in upgraded HepG2 cells is intriguing. CAR is sequestered primarily in the cytoplasm of non-induced hepatocytes and undergoes a two-step activation process after exposure to activators. The first step is the translocation of CAR from the cytoplasm to the nucleus. PB is not a direct ligand of human CAR (39), but it stimulates the translocation process (42, 63). However, CAR accumulates spontaneously in the nuclei of immortalized cell lines, such as HepG2, in the absence of xenobiotic activators (42, 63). Consequently, we did not expect a PB-mediated induction of CYP2B6 in the upgraded HepG2 cells. However, we observed a statistically significant 2.7-fold increase in CYP2B6 mRNA in response to PB (p < 0.01), which may indicate that the nuclear translocation of CAR by PB is restored in the presence of expressed C/EBPα and/or HNF4α in HepG2 cells. Nevertheless, a significant induction of CYP2B6 mRNA by PB was also observed in HepG2 cells transfected only with mCAR and pretreated with 3α-androstenol (22).
Transfection of C/EBPα, HNF4α, and CAR also enhances the expression of other CYPs and phase II conjugating enzymes. Moreover, the microarray analysis also suggests that drug metabolism pathways are activated in upgraded HepG2 cells. However, whether C/EBPα, HNF4α, and CAR display cooperative or synergistic cross-talk in the transactivation of these other genes and pathways or only one or two of these factors are responsible for the observed up-regulation remains to be investigated. Several of these genes (e.g. CYP3A4 or GSTA1) are not sensitive to CITCO, suggesting that CAR may not play a determining role in the up-regulation of these genes. Indeed, a weak effect of activated CAR on human CYP3A4 has been seen (43). Similarly, GSTA1 seems to be strongly induced by activated CAR in the mouse liver (64). However, a much weaker induction of GSTA1 has been reported in PB-induced human hepatocytes (65).
As expected, the transfection of the three transcription factors has a considerable impact on the transcriptome. Many up-regulated genes belong to pathways associated with the hepatic phenotype (glycogenesis, ureagenesis, lipid metabolism, etc.). Nevertheless (and as stated above), the activations of some of these pathways may not be CAR-dependent; they are more likely to be associated with C/EBPα and/or HNF4α. For instance, gluconeogenesis and glycogen storage are severely impaired in mice with a targeted deletion of the C/EBPα gene (66, 67), and those mice lacking hepatic HNF4α expression exhibited increased serum ammonia and lower ureagenesis (68).
Supplementary Material
This work was supported in part by Fondo de Investigación Sanitaria (Instituto de Salud Carlos III) Grant PI 070550; by the EC Integrated Project, CARCINOGENOMICS, Grant PL037712; and by Spanish Ministry of Science and Innovation/Instituto de Salud Carlos III Miguel Servet Contract CP08/00125 (to A. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–3 and Figs. 1 and 2.
- CYP
- cytochrome P450
- PBREM
- phenobarbital-responsive enhancer module
- XREM
- xenobiotic-responsive enhancer module
- OA
- okadaic acid
- C/EBPα
- CCAAT/enhancer-binding protein α
- CAR
- constitutive androstane receptor
- PXR
- pregnane X receptor
- MOI
- multiplicity of infection
- CITCO
- 6-(4-chlorophenyl)imidazo[2,1-b][1,3]thiazole-5-carbaldehyde-O-(3,4-dichlorobenzyl)oxime
- PB
- phenobarbital.
REFERENCES
- 1.Ioannides C., Lewis D. F. (2004) Curr. Top. Med. Chem. 4, 1767–1788 [DOI] [PubMed] [Google Scholar]
- 2.Nelson D. R., Koymans L., Kamataki T., Stegeman J. J., Feyereisen R., Waxman D. J., Waterman M. R., Gotoh O., Coon M. J., Estabrook R. W., Gunsalus I. C., Nebert D. W. (1996) Pharmacogenetics 6, 1–42 [DOI] [PubMed] [Google Scholar]
- 3.Wang H., Tompkins L. M. (2008) Curr. Drug Metab. 9, 598–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roy P., Yu L. J., Crespi C. L., Waxman D. J. (1999) Drug Metab. Dispos. 27, 655–666 [PubMed] [Google Scholar]
- 5.Coller J. K., Krebsfaenger N., Klein K., Endrizzi K., Wolbold R., Lang T., Nüssler A., Neuhaus P., Zanger U. M., Eichelbaum M., Mürdter T. E. (2002) Br. J. Clin. Pharmacol. 54, 157–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Svensson U. S., Ashton M. (1999) Br. J. Clin. Pharmacol. 48, 528–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erickson D. A., Mather G., Trager W. F., Levy R. H., Keirns J. J. (1999) Drug Metab. Dispos. 27, 1488–1495 [PubMed] [Google Scholar]
- 8.Desta Z., Saussele T., Ward B., Blievernicht J., Li L., Klein K., Flockhart D. A., Zanger U. M. (2007) Pharmacogenomics 8, 547–558 [DOI] [PubMed] [Google Scholar]
- 9.Court M. H., Duan S. X., Hesse L. M., Venkatakrishnan K., Greenblatt D. J. (2001) Anesthesiology 94, 110–119 [DOI] [PubMed] [Google Scholar]
- 10.Yanagihara Y., Kariya S., Ohtani M., Uchino K., Aoyama T., Yamamura Y., Iga T. (2001) Drug Metab. Dispos. 29, 887–890 [PubMed] [Google Scholar]
- 11.Hidestrand M., Oscarson M., Salonen J. S., Nyman L., Pelkonen O., Turpeinen M., Ingelman-Sundberg M. (2001) Drug Metab. Dispos. 29, 1480–1484 [PubMed] [Google Scholar]
- 12.Kobayashi K., Abe S., Nakajima M., Shimada N., Tani M., Chiba K., Yamamoto T. (1999) Drug Metab. Dispos. 27, 1429–1433 [PubMed] [Google Scholar]
- 13.Faucette S. R., Hawke R. L., Lecluyse E. L., Shord S. S., Yan B., Laethem R. M., Lindley C. M. (2000) Drug Metab. Dispos. 28, 1222–1230 [PubMed] [Google Scholar]
- 14.Hodgson E., Rose R. L. (2007) Pharmacol. Ther. 113, 420–428 [DOI] [PubMed] [Google Scholar]
- 15.Castell J. V., Jover R., Martínez-Jiménez C. P., Gómez-Lechón M. J. (2006) Expert Opin. Drug Metab. Toxicol. 2, 183–212 [DOI] [PubMed] [Google Scholar]
- 16.Pascual M., Gómez-Lechón M. J., Castell J. V., Jover R. (2008) Drug Metab. Dispos. 36, 1063–1072 [DOI] [PubMed] [Google Scholar]
- 17.Martínez-Jiménez C. P., Gómez-Lechón M. J., Castell J. V., Jover R. (2006) J. Biol. Chem. 281, 29840–29849 [DOI] [PubMed] [Google Scholar]
- 18.Martínez-Jiménez C. P., Castell J. V., Gómez-Lechón M. J., Jover R. (2006) Mol. Pharmacol. 70, 1681–1692 [DOI] [PubMed] [Google Scholar]
- 19.Martínez-Jiménez C. P., Gómez-Lechón M. J., Castell J. V., Jover R. (2005) Mol. Pharmacol. 67, 2088–2101 [DOI] [PubMed] [Google Scholar]
- 20.Bort R., Gómez-Lechón M. J., Castell J. V., Jover R. (2004) Arch. Biochem. Biophys. 426, 63–72 [DOI] [PubMed] [Google Scholar]
- 21.Jover R., Bort R., Gómez-Lechón M. J., Castell J. V. (1998) FEBS Lett. 431, 227–230 [DOI] [PubMed] [Google Scholar]
- 22.Sueyoshi T., Kawamoto T., Zelko I., Honkakoski P., Negishi M. (1999) J. Biol. Chem. 274, 6043–6046 [DOI] [PubMed] [Google Scholar]
- 23.Wang H., Faucette S., Sueyoshi T., Moore R., Ferguson S., Negishi M., LeCluyse E. L. (2003) J. Biol. Chem. 278, 14146–14152 [DOI] [PubMed] [Google Scholar]
- 24.Swales K., Kakizaki S., Yamamoto Y., Inoue K., Kobayashi K., Negishi M. (2005) J. Biol. Chem. 280, 3458–3466 [DOI] [PubMed] [Google Scholar]
- 25.Inoue K., Negishi M. (2008) J. Biol. Chem. 283, 10425–10432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inoue K., Negishi M. (2009) FEBS Lett. 583, 2126–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moya M., Jose Gómez-Lechón M., Castell J. V., Jover R. (2010) Chem. Biol. Interact. 184, 376–387 [DOI] [PubMed] [Google Scholar]
- 28.Rodríguez-Antona C., Bort R., Jover R., Tindberg N., Ingelman-Sundberg M., Gómez-Lechón M. J., Castell J. V. (2003) Mol. Pharmacol. 63, 1180–1189 [DOI] [PubMed] [Google Scholar]
- 29.Castell J. V., Hernández D., Gómez-Foix A. M., Guillén I., Donato T., Gómez-Lechón M. J. (1997) Gene Ther. 4, 455–464 [DOI] [PubMed] [Google Scholar]
- 30.Pérez G., Tabares B., Jover R., Gómez-Lechón M. J., Castell J. V. (2003) Toxicol. In Vitro 17, 643–649 [DOI] [PubMed] [Google Scholar]
- 31.Rodríguez-Antona C., Jover R., Gómez-Lechón M. J., Castell J. V. (2000) Arch. Biochem. Biophys. 376, 109–116 [DOI] [PubMed] [Google Scholar]
- 32.Lahoz A., Donato M. T., Castell J. V., Gómez-Lechón M. J. (2008) Curr. Drug Metab. 9, 12–19 [DOI] [PubMed] [Google Scholar]
- 33.Lahoz A., Donato M. T., Picazo L., Castell J. V., Gómez-Lechón M. J. (2008) Drug Metab. Lett. 2, 205–209 [DOI] [PubMed] [Google Scholar]
- 34.Sandoval J., Rodríguez J. L., Tur G., Serviddio G., Pereda J., Boukaba A., Sastre J., Torres L., Franco L., López-Rodas G. (2004) Nucleic Acids Res. 32, e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maglich J. M., Parks D. J., Moore L. B., Collins J. L., Goodwin B., Billin A. N., Stoltz C. A., Kliewer S. A., Lambert M. H., Willson T. M., Moore J. T. (2003) J. Biol. Chem. 278, 17277–17283 [DOI] [PubMed] [Google Scholar]
- 36.Honkakoski P., Palvimo J. J., Penttilä L., Vepsäläinen J., Auriola S. (2004) Biochem. Pharmacol. 67, 97–106 [DOI] [PubMed] [Google Scholar]
- 37.Wei P., Zhang J., Dowhan D. H., Han Y., Moore D. D. (2002) Pharmacogenomics J. 2, 117–126 [DOI] [PubMed] [Google Scholar]
- 38.Faucette S. R., Zhang T. C., Moore R., Sueyoshi T., Omiecinski C. J., LeCluyse E. L., Negishi M., Wang H. (2007) J. Pharmacol. Exp. Ther. 320, 72–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore L. B., Parks D. J., Jones S. A., Bledsoe R. K., Consler T. G., Stimmel J. B., Goodwin B., Liddle C., Blanchard S. G., Willson T. M., Collins J. L., Kliewer S. A. (2000) J. Biol. Chem. 275, 15122–15127 [DOI] [PubMed] [Google Scholar]
- 40.Auerbach S. S., Dekeyser J. G., Stoner M. A., Omiecinski C. J. (2007) Drug Metab. Dispos. 35, 428–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Forman B. M., Tzameli I., Choi H. S., Chen J., Simha D., Seol W., Evans R. M., Moore D. D. (1998) Nature 395, 612–615 [DOI] [PubMed] [Google Scholar]
- 42.Kawamoto T., Sueyoshi T., Zelko I., Moore R., Washburn K., Negishi M. (1999) Mol. Cell. Biol. 19, 6318–6322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Faucette S. R., Sueyoshi T., Smith C. M., Negishi M., Lecluyse E. L., Wang H. (2006) J. Pharmacol. Exp. Ther. 317, 1200–1209 [DOI] [PubMed] [Google Scholar]
- 44.Quandt K., Frech K., Karas H., Wingender E., Werner T. (1995) Nucleic Acids Res. 23, 4878–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lefterova M. I., Zhang Y., Steger D. J., Schupp M., Schug J., Cristancho A., Feng D., Zhuo D., Stoeckert C. J., Jr., Liu X. S., Lazar M. A. (2008) Genes Dev. 22, 2941–2952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Naiki T., Nagaki M., Shidoji Y., Kojima H., Moriwaki H. (2004) Cell Transplant. 13, 393–403 [DOI] [PubMed] [Google Scholar]
- 47.Kamiyama Y., Matsubara T., Yoshinari K., Nagata K., Kamimura H., Yamazoe Y. (2007) Drug Metab. Pharmacokinet. 22, 287–298 [DOI] [PubMed] [Google Scholar]
- 48.Jover R., Bort R., Gómez-Lechón M. J., Castell J. V. (2001) Hepatology 33, 668–675 [DOI] [PubMed] [Google Scholar]
- 49.Chen Y., Kissling G., Negishi M., Goldstein J. A. (2005) J. Pharmacol. Exp. Ther. 314, 1125–1133 [DOI] [PubMed] [Google Scholar]
- 50.Tirona R. G., Lee W., Leake B. F., Lan L. B., Cline C. B., Lamba V., Parviz F., Duncan S. A., Inoue Y., Gonzalez F. J., Schuetz E. G., Kim R. B. (2003) Nat. Med. 9, 220–224 [DOI] [PubMed] [Google Scholar]
- 51.Rana R., Chen Y., Ferguson S., Kissling G. E., Surapureddi S., Goldstein J. A. (2010) Drug Metab. Dispos. 38, 591–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jover R., Moya M., Gómez-Lechón M. J. (2009) Curr. Drug Metab. 10, 508–519 [DOI] [PubMed] [Google Scholar]
- 53.Kimura A., Nishiyori A., Murakami T., Tsukamoto T., Hata S., Osumi T., Okamura R., Mori M., Takiguchi M. (1993) J. Biol. Chem. 268, 11125–11133 [PubMed] [Google Scholar]
- 54.Miao J., Fang S., Bae Y., Kemper J. K. (2006) J. Biol. Chem. 281, 14537–14546 [DOI] [PubMed] [Google Scholar]
- 55.Carey M. (1998) Cell 92, 5–8 [DOI] [PubMed] [Google Scholar]
- 56.Martinez E. (2002) Plant Mol. Biol. 50, 925–947 [DOI] [PubMed] [Google Scholar]
- 57.Surapureddi S., Rana R., Reddy J. K., Goldstein J. A. (2008) Mol. Pharmacol. 74, 913–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chang T. K., Bandiera S. M., Chen J. (2003) Drug Metab. Dispos. 31, 7–10 [DOI] [PubMed] [Google Scholar]
- 59.Wortham M., Czerwinski M., He L., Parkinson A., Wan Y. J. (2007) Drug Metab. Dispos. 35, 1700–1710 [DOI] [PubMed] [Google Scholar]
- 60.Vyhlidal C. A., Gaedigk R., Leeder J. S. (2006) Drug Metab. Dispos. 34, 131–137 [DOI] [PubMed] [Google Scholar]
- 61.Madan A., Graham R. A., Carroll K. M., Mudra D. R., Burton L. A., Krueger L. A., Downey A. D., Czerwinski M., Forster J., Ribadeneira M. D., Gan L. S., LeCluyse E. L., Zech K., Robertson P., Jr., Koch P., Antonian L., Wagner G., Yu L., Parkinson A. (2003) Drug Metab. Dispos. 31, 421–431 [DOI] [PubMed] [Google Scholar]
- 62.Gerbal-Chaloin S., Pascussi J. M., Pichard-Garcia L., Daujat M., Waechter F., Fabre J. M., Carrère N., Maurel P. (2001) Drug Metab. Dispos. 29, 242–251 [PubMed] [Google Scholar]
- 63.Li H., Chen T., Cottrell J., Wang H. (2009) Drug Metab. Dispos. 37, 1098–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maglich J. M., Stoltz C. M., Goodwin B., Hawkins-Brown D., Moore J. T., Kliewer S. A. (2002) Mol. Pharmacol. 62, 638–646 [DOI] [PubMed] [Google Scholar]
- 65.Duret C., Daujat-Chavanieu M., Pascussi J. M., Pichard-Garcia L., Balaguer P., Fabre J. M., Vilarem M. J., Maurel P., Gerbal-Chaloin S. (2006) Mol. Pharmacol. 70, 329–339 [DOI] [PubMed] [Google Scholar]
- 66.Wang N. D., Finegold M. J., Bradley A., Ou C. N., Abdelsayed S. V., Wilde M. D., Taylor L. R., Wilson D. R., Darlington G. J. (1995) Science 269, 1108–1112 [DOI] [PubMed] [Google Scholar]
- 67.Lee Y. H., Sauer B., Johnson P. F., Gonzalez F. J. (1997) Mol. Cell Biol. 17, 6014–6022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Inoue Y., Hayhurst G. P., Inoue J., Mori M., Gonzalez F. J. (2002) J. Biol. Chem. 277, 25257–25265 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.