Abstract
The obligate intracellular parasite, Toxoplasma gondii, modulates host immunity in a variety of highly specific ways. Previous work revealed a polymorphic, injected parasite factor, ROP16, to be a key virulence determinant and regulator of host cell transcription. These properties were shown to be partially mediated by dysregulation of the host transcription factors STAT3 and STAT6, but the molecular mechanisms underlying this phenotype were unclear. Here, we use a Type I Toxoplasma strain deficient in ROP16 to show that ROP16 induces not only sustained activation but also an extremely rapid (within 1 min) initial activation of STAT6. Using recombinant wild-type and kinase-deficient ROP16, we demonstrate in vitro that ROP16 has intrinsic tyrosine kinase activity and is capable of directly phosphorylating the key tyrosine residue for STAT6 activation, Tyr641. Furthermore, ROP16 co-immunoprecipitates with STAT6 from infected cells. Taken together, these data strongly suggest that STAT6 is a direct substrate for ROP16 in vivo.
Keywords: Immunology, Parasitology, Signal Transduction, STAT Transcription Factor, Tyrosine Protein Kinase (Tyrosine Kinase), STAT6, Toxoplasma gondii
Introduction
Toxoplasma gondii, a ubiquitous, intracellular parasite of the phylum Apicomplexa, infects an estimated one-third of the human population as well as a broad range of warm-blooded animals. Infection is typically asymptomatic and chronic; however, severe and even fatal disease can result from acute infections of congenitally infected or immunocompromised hosts. Strikingly, Toxoplasma-induced inflammatory pathology in adult immunocompetent patients has also been reported (1, 2).
Understanding the host/parasite interactions that underpin these various disease manifestations has become an area of active investigation, and recent work has implicated as key players a set of polymorphic proteins that are secreted from the apical organelles known as rhoptries (3). One of these, a putative serine-threonine kinase known as ROP16, was revealed as a parasite factor responsible for many changes in host gene expression (4). Much of the effect of ROP16 on transcription was found to depend on sustained activation of STAT3 and STAT6, two host transcription factors that can negatively regulate Th1 inflammatory responses. ROP16 is thus poised to mediate the inflammatory status of the host, an intriguing role for a parasite effector because Th1-driven immunopathology, not just uncontrolled parasite growth and dissemination, has been proposed to be a significant contributor to the disease outcome (5).
Although the potential significance of the ROP16 effect on STATs is clear, the molecular mechanism by which it accomplishes this sustained activation was not apparent from the initial studies. STAT activation requires phosphorylation at specific, conserved tyrosine residues; this phosphorylation, which induces dimerization and nuclear translocation of the STATs, is canonically initiated and maintained in the cytosol by either receptor-associated tyrosine kinases (such as the JAKs) or non-receptor tyrosine kinases (e.g. Src family kinases) (6). The duration of STAT activation is usually limited by negative feedback because STATs up-regulate transcription of many negative regulators, such as suppressors of cytokine signaling (SOCS).2 ROP16-induced STAT activation, however, can persist despite transcriptional up-regulation of these negative regulators. As a predicted serine-threonine kinase, we hypothesized that ROP16 might regulate STAT activation and bypass conventional host negative feedback by phosphorylation of one or more host substrates.
To define the molecular function of ROP16, we generated parasites deficient in ROP16. We show that ROP16 is required for not only sustained activation of STAT3 and 6 but also initial activation upon parasite invasion. We further show that ROP16 is able to interact with STAT6 in infected cells and, surprisingly, that it can directly phosphorylate the critical tyrosine for STAT6 activation in vitro.
EXPERIMENTAL PROCEDURES
Mice, Host Cell Culture, and Parasites
2fTGH (human fibrosarcoma) cells, 2fTGH-derived mutant cells deficient in JAK1 (U4A), JAK2 (γ2A), Tyk2 (U1A) (a gift from G. R. Stark, The Cleveland Clinic Foundation, OH), and primary human foreskin fibroblasts (HFFs) were maintained in complete DMEM, consisting of Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% heat-inactivated fetal calf serum (FCS; Hyclone, Logan, UT), 2 mm l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin.
JAK3 knock-out (B6;129S4-JAK3tm1Ljb/J) and B6129SF2/J mice were purchased from the Jackson Laboratory (Bar Harbor, ME). Mouse bone marrow-derived macrophages were generated as described previously (7). Briefly, femurs were harvested from 10–20-week-old mice. Bone marrow was flushed from the femurs, and red blood cells were lysed with ACK lysis buffer (Invitrogen). Cells were then cultured in complete DMEM supplemented with macrophage colony-stimulating factor for 3 days. Half the medium was then replaced with fresh medium, and the cells were cultured for an additional 3–4 days before use in infection assays. T. gondii tachyzoites were maintained in vitro by serial passage on confluent monolayers of HFFs grown in complete DMEM at 37 °C with 5% CO2.
Generation of ROP16 Knock-out Parasites
A targeting construct was engineered using the pTKO vector (a gift from Gus Zeiner, Stanford University, Stanford, CA) essentially as described elsewhere (8). Briefly, the construct flanks a hypoxanthine-xanthine-guanine phosphoribosyltransferase-selectable marker 5′ with the 2144 bp immediately upstream of the ROP16 start codon and 3′ with the 3363 bp of ROP16 Type I (RH strain) genomic sequence immediately downstream of the ROP16 stop codon. A GFP marker upstream of the 5′-flanking sequence facilitated negative selection against single recombination events. The genomic regions flanking the ROP16 open reading frame (ORF) in the RH strain were amplified by PCR using the following primers: 5′-ATTGAGGTCGTCCGTGTATTTG-3′ and 5′-TGCTTCGTTTCCCATTTTAGTTG-3′ (5′-targeting sequence) and 5′-CGAGGATGTTACCGCTTTTG-3′ and 5′-CGTTTGGAGGGGAGTGTCTT-3′ (3′-targeting sequence). The 5′-targeting sequence was cloned into the KpnI and EcoRV restriction sites of the pTKO vector, and the 3′-targeting sequence was cloned into the NheI and HpaI restriction sites of pTKO to generate the targeting plasmid. The plasmid was linearized by restriction digest with NotI, and 25 μg of plasmid was transfected into the RHΔhpt strain of parasites by electroporation, as described previously (9). Stable integrants were selected with mycophenolic acid (MPA)/xanthine essentially as described elsewhere (10) and cloned by limiting dilution.
MPA/xanthine-resistant, GFP-negative clones were selected for screening. Disruption of the ROP16 locus was confirmed by PCR of parasite genomic DNA using the following primers: 5′-TTTTAGGTGTCGGCTTTGCTG-3′ and 5′-ACGAGATGTTCCGCGACTTC-3′ (integration of 5′ targeting sequence); 5′-TGTGGATGTCATACGCCAAC-3′ and 5′-TAGGTTCTGGCGTTTGGAGG-3′ (integration of 3′ targeting sequence); 5′-GCAACTGCACGATACATGTCG-3′ and 5′-CATCCGATGTGAAGAAAGTTC-3′ (presence of ROP16 ORF). Initial characterization of the knock-out was performed using two clones derived from independent populations to verify phenotypes; all results shown in this paper utilize one clone, B23.
Generation of Transgenic Parasites Ectopically Expressing ROP16 Variants
Transgenic parasites expressing engineered forms of ROP16 were generated essentially as described previously (4). Briefly, RHΔhpt parasites were transfected by electroporation with a plasmid expressing HA-tagged Type I ROP16-K404N (pTKO-mutK404N), followed by selection in MPA/xanthine and single-cell cloning by limiting dilution as described above. Site-directed mutagenesis to generate ROP16-K404N was carried out using splice overlap extension PCR as described elsewhere (11). The Type I ROP16 point mutant, from 2144 bp upstream of the predicted ATG start codon to the base pair immediately preceding the stop codon that was replaced with a single HA tag, was cloned into the KpnI and EcoRV sites of the pTKO vector, which provided the GRA2 3′-UTR from Type I parasites and the HXGPRT gene. The following primers were used: 5′-TGCCTCAACTGTAAACGTCTT-3′ and 5′-TTACGCGTAGTCCGGGACGT-3′. Ectopic expression of HA-tagged ROP16-K404N in MPA/xanthine-resistant clones was confirmed by immunofluorescence and immunoblot for the HA tag.
ROP16 knock-out parasites were complemented with Type I ROP16-HA essentially as described elsewhere (12). Briefly, Δrop16 parasites were transfected by electroporation with a plasmid expressing Type I ROP16-HA and the Tn5 bleomycin resistance gene flanked by the SAG1 5′- and 3′-UTRs, followed by selection in bleomycin and single-cell cloning by limited dilution as described above. As above, the ROP16-HA gene was flanked by 2144 bp of upstream sequence and the GRA2 3′-UTR. Ectopic expression of Type I ROP16-HA was confirmed by immunofluorescence and immunoblot for the HA tag.
Expression of ROP16 in E. coli
The coding sequences for C-terminally HA-tagged ROP16-WT or ROP16-K404N, excluding the predicted signal peptide, were cloned into the expression vector pET28a (Novagen, Madison, WI) using the following primers: 5′-CCATGGCTCGATACATGTCGTTTGAGGAA-3′ and 5′-TCCGTCGACCGCGTAGTCCGGGACGTC-3′.
The resulting constructs were used to transform chemically competent E. coli strain BL21 (DE3) cells. Overnight cultures were diluted 1:1000 into 1.5-liter 2X YT cultures and grown shaking at 37 °C. When an A600 of 0.6 was reached, protein expression was induced by the addition of isopropyl 1-thio-β-d-galactopyranoside (to a final concentration of 300 μm), and cultures were grown shaking at 18 °C for 16 h. Polyhistidine-tagged recombinant protein was purified from soluble lysates of induced bacterial pellets using standard nickel resin affinity chromatography (Qiagen, Valencia, CA). Enrichment for full-length recombinant ROP16 was accomplished by adding ammonium sulfate to a final concentration of 1.5 m and reserving the soluble fraction. Purified fractions were dialyzed against 10 mm Tris (pH 7.5) overnight at 4 °C.
In Vitro Kinase Assays on Immunocomplexes
Parasites expressing HA-tagged ROP16-WT or ROP16-K404N were harvested by syringe lysis. Pellets of 107 parasites were lysed in 1 ml of buffer containing 50 mm Tris-HCl (pH 7.5), 5 mm EDTA, 150 mm NaCl, 0.1% Nonidet P-40, and EDTA-free complete protease inhibitor mixture (Roche Applied Science) and immunoprecipitated using anti-HA 3F10 affinity matrix (Roche Applied Science) for 12 h at 4 °C with gentle mixing. The beads were washed three times with lysis buffer and then once with kinase assay buffer containing 20 mm HEPES (pH 7.5), 10 mm MnCl2, 1 mm DTT, and 1× Halt phosphatase inhibitor (Pierce). The immunoprecipitates were then incubated with 30 μl of kinase buffer and 10 μCi of [γ-32P]ATP (MP Biomedicals, Solon, OH) at 30 °C for 30 min with shaking. Kinase reactions were stopped by the addition of SDS sample buffer and boiled to elute the immunocomplexes. Samples were separated by electrophoresis on polyacrylamide gels and blotted to PVDF membranes (Millipore Corp., Billerica, MA). To assess ROP16-HA levels, membranes were immunoblotted using 3F10 anti-HA HRP-conjugated antibodies (Roche Applied Science). To assess 32P incorporation, the membrane was dried and exposed to a PhosphorImager screen overnight and then analyzed using the STORM PhosphorImager system (Amersham Biosciences).
In Vitro Kinase Assays with Recombinant ROP16
All assays were performed using a kinase assay buffer containing 50 mm Tris-HCl (pH 7.5), 10 mm MgCl2, 1 mm EGTA, 0.01% Brij 35, and 2 mm DTT. For autophosphorylation assays, reaction volumes of 30 μl were incubated with 10 μCi of [γ-32P]ATP at 30 °C for 30 min with shaking and analyzed as described above for immunocomplex kinase assays. Tyrosine kinase activity assays were performed using the Antibody Beacon tyrosine kinase assay kit from Molecular Probes, Inc. (Eugene, OR). Recombinant JAK3-GST was obtained from Active Motif (Carlsbad, CA). Pan-JAK inhibitor (JAK Inhibitor I) was obtained from Calbiochem. 50-μl kinase reactions composed of recombinant kinase in kinase assay buffer, poly(Glu-Tyr) peptide substrate, and 1 mm ATP were incubated at 30 °C; fluorescence was read using excitation/emission of 492/517 nm on a Flexstation II-384 (Amersham Biosciences). Phosphorylation assays on STAT6 were performed under similar conditions, using 0.5 μg of recombinant STAT6-GST as substrate and 1 mm ATP (Abnova, Taipei, Taiwan). Reactions were incubated at 30 °C for 30 min, stopped by the addition of SDS sample buffer, and boiled. Samples were analyzed by SDS-PAGE and immunoblot using the primary antibodies: a rat monoclonal anti-HA antibody (3F10) that has been directly conjugated to HRP (Roche Applied Science) and rabbit polyclonal anti-STAT6 antibodies (Cell Signaling Technology, Danvers, MA). For STAT6 immunoblots, a goat anti-rabbit-IgG secondary antibody conjugated to HRP (Bio-Rad) was used. HRP activity was detected using the SuperSignal West Pico chemiluminescent system (Pierce).
Microarray Analysis
Parasites were harvested by syringe lysis and extensively washed (three washes in 50 ml of complete DMEM). Confluent monolayers of serum-starved HFFs in T25 flasks were infected at multiplicity of infection (MOI) 5, and total RNA was extracted at 1 and 5 h postinfection using TRIzol (Invitrogen). Two biological replicates were performed for each strain. Total RNA from each sample was labeled using either the Affymetrix one-cycle labeling kit or the Affymetrix 3′ IVT express kit as indicated (Affymetrix, Santa Clara, CA). 20 μg of the resulting cRNA was hybridized onto Affymetrix Human Genome U133 Plus 2.0 gene chips. Gene expression values were computed by implementing the robust multichip average procedure for normalization (13). Data were subjected to Significance Analysis of Microarrays (SAM 2.0) analysis (14). Genes meeting the threshold of a <5% false discovery rate and absolute expression -fold change greater than 2 were considered as significantly differentially expressed.
Immunofluorescence Assays
Monolayers of host cells (HFFs, bone marrow-derived macrophages, or 2fTGH-derived) were grown on 12-mm glass coverslips. To investigate the kinetics of STAT6 activation, a potassium buffer shift was used to synchronize invasion as described elsewhere (15, 16). Briefly, tachyzoites were harvested, washed, and resuspended in Endo buffer (44.7 mm K2SO4, 106 mm sucrose, 10 mm Mg2SO4, 20 mm Tris (pH 8.2), 5 mm glucose, and 3.5 mg/ml BSA). Parasites were added to cells at MOI ∼1 and allowed to settle at 37 °C for 15 min. Endo buffer was exchanged for prewarmed, serum-free DMEM (Invitrogen) to initiate parasite invasion. To assess the kinetics of IL-4 activation, uninfected cells were treated with serum-free DMEM containing either DMSO or recombinant human IL-4 at 50 ng/ml (R&D Systems, Minneapolis, MN). Cells were fixed at defined time points in ice-cold 100% methanol for 10 min.
For other immunofluorescence assays, parasites were washed, resuspended, and allowed to settle on host cells in serum-free DMEM for 10–20 min at room temperature. Parasites and cells were then shifted to a 37 °C incubator. To assess IL-4 activation, uninfected host cells were treated with serum-free DMEM containing either DMSO, recombinant human IL-4 (R&D Systems, Minneapolis, MN), or murine IL-4 (BioLegend, San Diego, CA) at 100 ng/ml. For experiments with chemical inhibitors, host cells were pretreated for 1 h at 37 °C. Chemical inhibitors (PP2 and PP3 for Src family kinases; JAK inhibitor I for JAKs) were obtained from Calbiochem. Cells were fixed at defined time points after temperature shift or cytokine addition as described above.
For all immunofluorescence assays, coverslips were blocked in PBS supplemented with 3% BSA and permeabilized with 0.2% Triton X-100 for 1 h at room temperature and then incubated in block/permeabilization buffer with mouse monoclonal anti-ROP2/4 antibody (T34A7; a gift from J. F. Dubremetz) and antibodies specific for phospho-STAT6 (Tyr641) (Cell Signaling Technology, Danvers, MA) overnight at 4 °C. Fluorescent antibodies (Invitrogen/Molecular Probes) were used for antigen visualization. Coverslips were mounted using Vectashield mounting medium containing 4′,6-diamidino-2-phenylindole (Vector Laboratories, Burlingame, CA) to stain DNA and visualized with an Olympus BX60 microscope with a ×100 oil objective.
Phosphoamino Acid Analysis
In vitro kinase assays containing recombinant WT ROP16 and 20 μCi of [γ-32P]ATP were performed as described above. Samples were separated by electrophoresis on polyacrylamide gels and blotted to PVDF membranes (Millipore Corp.). The membrane was dried, and following exposure to film (Eastman Kodak Co. Biomax MR), bands labeled with 32P were excised and subjected to acid hydrolysis and phosphoamino acid analysis as described elsewhere (18).
Co-immunoprecipitation
Cells were infected at MOI ∼10 for 2 h with RH:ROP16-WT or RH:ROP16-K404N parasites. Cells were lysed in lysis buffer containing 20 mm Tris-HCl (pH 7.5), 1 mm EDTA, 150 mm NaCl, 0.1% Nonidet P-40, and EDTA-free complete protease inhibitor mixture (Roche Applied Science). Clarified lysates were incubated with agarose-conjugated rabbit polyclonal M-20 (anti-STAT6) antibodies (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) or an isotype control, agarose-conjugated rabbit IgG (Santa Cruz Biotechnology, Inc.) for 12 h at 4 °C with gentle mixing. Beads were washed three times with lysis buffer, and immunocomplexes were eluted by boiling with SDS sample buffer. Samples were separated by SDS-PAGE and blotted onto PVDF membrane. Immunoblots were carried out using the following primary antibodies: mouse monoclonal anti-STAT6 (BD Biosciences), rat monoclonal anti-HA (3F10, Roche Applied Science), mouse monoclonal anti-ROP1 (Tg49) (19), or mouse polyclonal anti-toxofilin antiserum (20).
RESULTS
ROP16 Has Kinase Activity and Autophosphorylates in Vitro
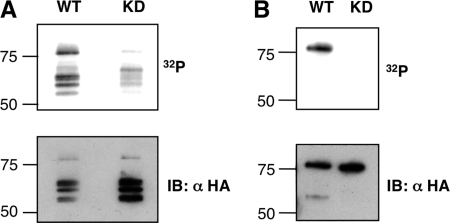
ROP16 is a ∼79-kDa bipartite protein; its N-terminal domain lacks homology to known proteins, but its C-terminal domain has homology to serine-threonine protein kinase domains, and the residues generally required for catalytic activity are conserved (21, 22). Many protein kinases show marked autophosphorylation activity. As a first step to determining if ROP16 is an active kinase, therefore, we performed in vitro assays on Type I ROP16 (ROP16-WT) immunoprecipitated using an anti-HA monoclonal antibody from parasites expressing an HA-tagged form of the protein (previously described in Ref. 4). Phosphorylation assays were carried out in the presence of [γ-32P]ATP; reaction mixtures were resolved by SDS-PAGE and analyzed by autoradiography and immunoblot (Fig. 1A). 32P labeling of bands co-migrating with the various forms of ROP16-HA was strongly suggestive of autophosphorylation (ROP16 appears highly susceptible to proteolysis and is routinely obtained from parasite lysates as a reproducible collection of four or five bands in the 55–80 kDa size range) (4). To confirm that the kinase activity in the immunocomplexes was due to ROP16 catalytic activity, phosphorylation assays were carried out using parasites expressing a kinase-deficient (KD) Type I ROP16 (ROP16-K404N) where the critical residue Lys404 (necessary for ATP binding) was mutated to Asn (Fig. 1A). The results showed that in reactions containing comparable amounts of the ROP16-WT and ROP16-K404N immunoprecipitates (as revealed by HA immunoblot), 32P labeling of the ROP16-comigrating bands was eliminated. Low amounts of 32P labeling were still detectable using the ROP16-K404N immunoprecipitate; this seems most likely to be due to activity of a small amount of a parasite kinase that associates with the epitope-tagged ROP16, possibly endogenous wild-type, non-epitope-tagged ROP16. Regardless, these results strongly suggest that ROP16 is indeed an active protein kinase.
FIGURE 1.
ROP16 is an active kinase. In vitro kinase assays were performed using WT or kinase-deficient (KD) K404N point mutants of ROP16-HA, obtained either by immunoprecipitation from Type I parasites ectopically expressing these proteins (A) or by recombinant expression in E. coli and purification (B). Kinase reactions including [γ-32P]ATP were performed directly on the immunocomplexes (A) or with the recombinant proteins (B) for 30 min at 30 °C. Following SDS-PAGE and transfer to PVDF membranes, 32P incorporation was measured by PhosphorImager, and membranes were immunoblotted (IB) for ROP16-HA. Molecular masses are noted in kDa on the left.
To facilitate biochemical characterization of ROP16 kinase activity, full-length WT and K404N ROP16 (C-terminally His6- and HA-tagged but lacking the predicted signal peptide) were recombinantly expressed in E. coli and purified by nickel-agarose chromatography followed by ammonium sulfate precipitation. In vitro kinase assays using equivalent amounts of WT and K404N recombinant ROP16 were carried out and analyzed as described for native ROP16. As expected, 32P labeling consistent with ROP16 autophosphorylation was only observed in reactions containing WT recombinant ROP16 and not K404N recombinant ROP16 (Fig. 1B). These results confirm that ROP16 has catalytic kinase activity.
Loss of ROP16 Leads to Loss of STAT6 Activation
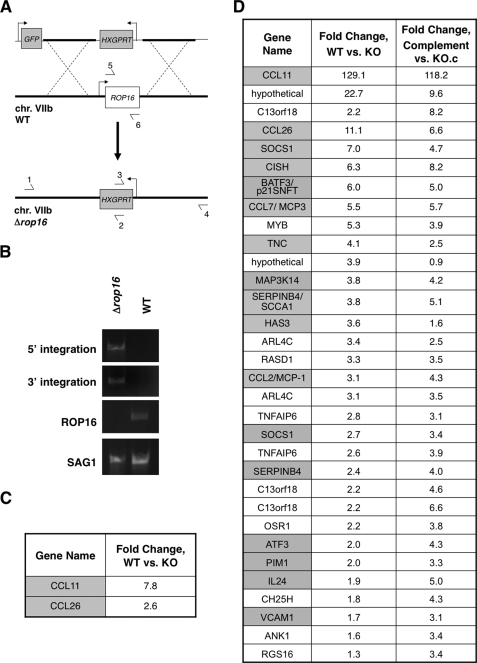
Given the role of ROP16 in virulence and its demonstrated catalytic activity, we hypothesized that it might regulate host signaling pathways by phosphorylating one or more host substrates. To find candidate substrates, we first sought to identify signaling pathways that were altered in a ROP16-dependent manner. For this, we needed a Type I (RH) parasite line deficient for ROP16 (Δrop16), which we generated by homologous recombination. The targeting construct contained the selectable marker HXGPRT flanked by ∼2 kb of the ROP16 upstream and downstream genomic regions as well as a GFP marker to allow for negative selection against heterologous recombinants (Fig. 2A). Disruption of the ROP16 locus in Δrop16 parasites was confirmed by showing a failure to amplify by PCR using primers for the ROP16 ORF (Fig. 2B). Replacement of the complete ROP16 ORF with the selectable marker HXGPRT was also confirmed by PCR (Fig. 2B).
FIGURE 2.
Deletion of the ROP16 locus identifies STAT6 as a potential target. A, schematic of Δrop16 generation (not to scale). The top line shows the construct used with the Toxoplasma sequences derived from the ROP16 flanks shown as a thick line. Bent arrows indicate promoters driving the negative (GFP) and positive (HXGPRT) selectable markers. The middle line represents the region of chromosome VIIb containing ROP16. The bottom line shows the resulting deletion of the ROP16 locus. Primers used for PCR detection of homologous integration of the targeting construct are indicated by horizontal arrows. B, confirmation of replacement of the ROP16 locus with HXGPRT by PCR. Correct 5′- and 3′-integration was demonstrated with primer pairs 1/2 and 3/4 of Fig. 2A, respectively. Primers 5 and 6 were used to demonstrate the loss of the ROP16 coding region in the Δrop16 mutant. Toxoplasma surface antigen 1 (SAG1) primers were used as a positive control for the PCRs. C and D, microarray analysis of HFFs infected with either WT or Δrop16 (KO) parasites at 1 h (C) and 5 h (D) postinfection. Expression data for HFFs 5 h post-infection with ROP16-complemented Δrop16 parasites or control Δrop16 parasites that underwent transfection and selection alongside the complemented parasites (KO.c) are shown in D. Two biological replicates of each infection per time point were analyzed. Genes shown are those identified as significantly up-regulated by SAM analysis in cells infected with WT versus Δrop16 parasites and >2-fold change in expression (C) or >3-fold change in expression in cells infected with either WT or complemented parasites versus Δrop16 parasites (D). Genes that have previously been characterized as STAT6-regulated are highlighted in gray; genes not known to be regulated by STATs are in white.
We reasoned that signaling pathways directly modulated by ROP16 might be revealed as a ROP16-dependent signature at the level of host cell transcription. To test this, we used Affymetrix microarrays to compare host transcriptional response to infection by parental (WT) parasites and Δrop16 parasites. Parasites were harvested by syringe lysis from HFFs and extensively washed to minimize cytokine carry-over; serum-starved HFFs were then infected at MOI ∼5, and total RNA was harvested at 1 or 5 h postinfection. These conditions were chosen to ensure that the vast majority of host cells would be infected and that there would be time for transcriptional up-regulation in response to signaling perturbations while minimizing secondary or tertiary transcriptional effects that might be dominant at later time points. Inspection of host genes significantly up-regulated by infection with parental versus Δrop16 parasites revealed a striking enrichment for genes characterized as STAT6-regulated (Fig. 2, C and D) (23–30).
To control for the possibility that the observed differences in host gene expression were due to some unrelated defect in the Δrop16 parasite line, comparative host gene expression profiling at 5 h postinfection was also conducted using Δrop16 parasites that had been complemented with HA-tagged Type I ROP16. Analysis of the host genes revealed to be up-regulated in the complemented parasites versus control ROP16-deficient parasites (KO.c) recapitulated the pattern observed for parental versus ROP16-deficient parasites, confirming that the differences observed are ROP16-dependent. Although STAT6 activation was by no means sufficient to explain all of the genes that were up-regulated, this well characterized transcription factor appeared to be one of the most dominant signatures among this set of host genes altered in a ROP16-dependent manner.
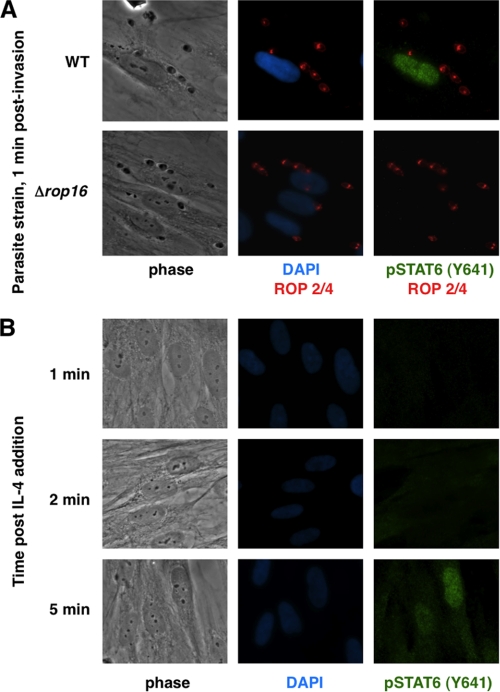
The results shown in Fig. 2 indicate that, even 1 h after infection, the STAT6-dependent genes CCL11 and CCL26 are markedly more expressed in cells infected with WT versus Δrop16 parasites. These results strongly suggested that initial ROP16-dependent STAT6 activation is an extremely rapid event. To investigate this further, preliminary experiments were performed using immunofluorescence analysis of phospho-STAT6 (Tyr641) in HFFs 20 min postinfection; phosphorylated STAT6 was detected in cell nuclei upon infection with WT or ROP16-complemented parasites but not in cells infected with either of two independently generated Δrop16 parasites, confirming that STAT6 activation is rapid and dependent on the ROP16 locus (data not shown). To examine the kinetics of this initial activation more closely, HFFs were infected with Δrop16 or WT parasites using a potassium buffer shift (15, 31) to allow more precise control of invasion. Strikingly, phosphorylated STAT6 was detected in host cell nuclei within 1 min of parasite invasion upon infection with the control parasites (the earliest time point that was experimentally tractable; Fig. 3A). This effect appeared to be ROP16-dependent in that infection with Δrop16 parasites resulted in no activation of STAT6 at this or any other time point (up to 24 h postinfection; Fig. 3A and data not shown). In contrast, and as previously reported by others (32), stimulation of HFFs with IL-4 did not induce levels of STAT6 activation similar to that seen in infected cells until ∼5 min poststimulation (Fig. 3B). These results suggest that ROP16-dependent activation of STAT6 is somehow circumventing the normal signal transduction pathways.
FIGURE 3.
STAT6 activation is ROP16-dependent and much faster than IL-4-induced activation. A, parasite infection was synchronized by potassium shift as described under “Experimental Procedures.” Infected HFF cells were fixed 1 min postinvasion with methanol and stained with DAPI and the indicated antibodies. B, HFF cells were stimulated with IL-4 (50 ng/ml), fixed at the indicated time points, and stained with DAPI and the indicated antibodies.
ROP16 Directly Phosphorylates STAT6 on Tyr641 in Vitro
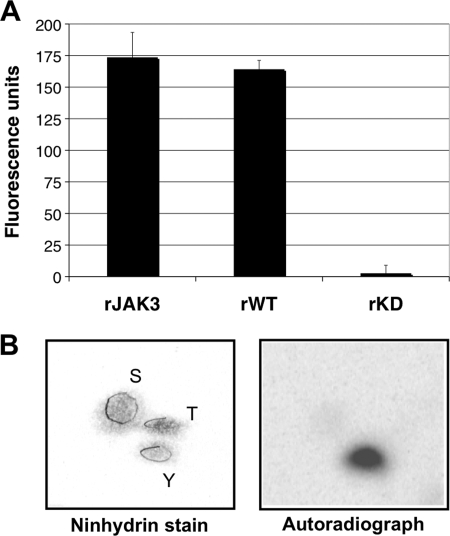
We next sought to dissect the mechanism underlying this unconventional activation. Given the speed with which parasites containing a functional ROP16 induce STAT6 activation, we hypothesized that ROP16 might directly phosphorylate STAT6 or some host tyrosine kinase immediately upstream of STAT6. Because most tyrosine kinases are activated by tyrosine phosphorylation, both models invoke such an activity as a key step. This raised the question of whether ROP16, contrary to what was predicted by its amino acid sequence (4), might also possess tyrosine kinase activity. To address this, an in vitro tyrosine kinase assay was performed using recombinant ROP16. The assay makes use of a small molecule fluorescent ligand that is quenched when bound to anti-phosphotyrosine antibodies. Tyrosine phosphorylation of substrate peptides results in competition for antibody binding and liberation of the fluorescent ligand, providing a quantitative measure of the extent of tyrosine kinase activity in a reaction mixture. Incubation of recombinant ROP16-WT with the detection complex and ATP resulted in a marked increase in fluorescence, comparable with fluorescence levels from reactions containing the known tyrosine kinase JAK3, whereas incubation of an equivalent amount of recombinant ROP16-K404N did not (Fig. 4A). To confirm tyrosine kinase activity by an independent approach, phosphoamino acid analysis of autophosphorylated ROP16 was performed. This revealed the presence of phosphotyrosine residues only (Fig. 4B). These results together demonstrate that, contrary to original predictions, ROP16 has intrinsic tyrosine kinase activity.
FIGURE 4.
ROP16 has tyrosine kinase activity. A, Antibody Beacon tyrosine kinase detection complex was incubated with either equivalent amounts of recombinant ROP16-WT (rWT) and kinase-deficient ROP16-K404N (rKD) or 0.6 μg of JAK3 (rJAK3) as described under “Experimental Procedures”; tyrosine kinase peptide substrate (poly(Glu-Tyr), 4:1) and ATP were then added to each well, and the reactions were incubated at 30 °C for 30 min. Fluorescence was measured in a fluorescence microplate reader using excitation at 492 nm and emission at 517 nm. Background fluorescence, or fluorescence from reaction mixtures lacking kinase, was subtracted to normalize samples. Each sample was analyzed in triplicate; data shown are from one assay representative of at least three independent replicates. B, phosphoamino acid analysis of 32P-labeled ROP16 by semidry cellulose thin layer electrophoresis. Left, photograph of the TLC plate stained with ninhydrin; positions of phosphoserine (S), phosphothreonine (T), and phosphotyrosine (Y) standards are circled and labeled. Right, autoradiograph of the TLC plate.
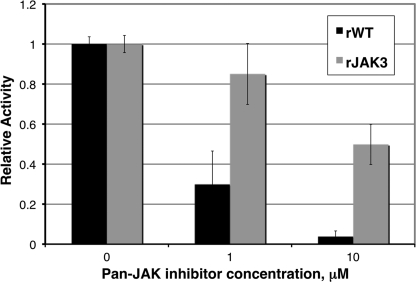
To distinguish between the two models of direct and indirect STAT6 activation proposed above, we next investigated whether ROP16 might act via a tyrosine kinase known to be upstream of STAT6. To do this, we examined the role of the Src family kinases and the Janus kinase family (JAK1, JAK2, JAK3, and TYK2) by employing chemical inhibitors that are reported to be largely specific for these kinases. HFFs were pretreated with the inhibitors and infected with Δrop16 or WT parental parasites, and STAT6 activation was assessed by immunofluorescence analysis as described above. Pretreatment with the Src family kinase inhibitor PP2 had no effect on parasite-induced STAT6 activation compared with pretreatment with the control compound PP3 (data not shown). Pretreatment with the pan-JAK inhibitor at 10 μm (the concentration preliminary experiments showed was necessary to diminish IL-4 stimulation of STAT6 activation), however, diminished WT parasite-induced STAT6 activation relative to pretreatment with DMSO (data not shown). Because chemical inhibitors can often have “off-target” effects, we asked whether the pan-JAK inhibitor might also affect the ability of ROP16 to phosphorylate substrates. To test this, we used the inhibitor in in vitro tyrosine kinase assays on ROP16 as described above. Strong (greater than 90%) inhibition of ROP16 tyrosine kinase activity was indeed observed in the presence of the pan-JAK inhibitor at the concentration used for the cell-based assay (p < 0.05; Fig. 5). Inhibition of ROP16 activity was significantly greater than inhibition of JAK3, one of the specifically intended targets of the inhibitor (p < 0.05; Fig. 5). As such, inhibition of either ROP16 or JAK activity might account for the observed effects of the pan-JAK inhibitor on STAT6 activation. As an independent approach to testing whether JAKs might contribute to ROP16-induced STAT6 activation, we obtained a panel of singly JAK-deficient cells that have been described elsewhere: JAK1-deficient U4A, JAK2-deficient γ2A, and TYK2-deficient U1A fibrosarcoma cells (33, 34). JAK3-deficient cells were obtained by generating bone marrow-derived macrophages from JAK3-deficient mice (35). By immunofluorescence analysis of phospho-STAT6 localization, similar levels of STAT6 activation upon parasite infection were observed in all singly JAK-deficient and parental cell lines (data not shown). Although these results do not completely exclude a role for intermediary kinases (e.g. multiple JAKs operating in a redundant manner or some other upstream tyrosine kinase), they provide further support for a model where ROP16 acts directly upon STAT6.
FIGURE 5.
A pan-JAK inhibitor inhibits ROP16 in vitro. ROP16 tyrosine kinase activity was assessed by fluorescence essentially as described in the legend to Fig. 4. Recombinant ROP16-WT (rWT) or recombinant JAK3 (rJAK3) were incubated with detection complex and either DMSO or pan-JAK inhibitor at the indicated concentrations for 15 min at room temperature; ATP was then added to each well, and reactions were incubated at 30 °C for 30 min. Fluorescence (excitation at 492 nm and emission at 517 nm) was measured at 1 and 30 min post-ATP addition. Background fluorescence at 1 min was subtracted; changes in fluorescence for recombinant ROP16-WT (black bars) and recombinant JAK3 (gray bars) reactions are shown normalized to DMSO-treated reactions.
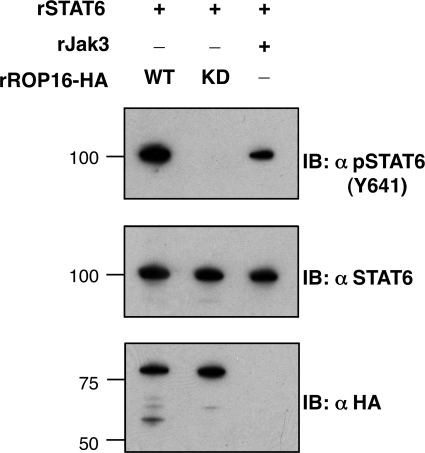
To ask whether ROP16 could directly phosphorylate STAT6 on the critical Tyr641 residue required for activation, in vitro kinase assays with recombinant ROP16 were carried out using full-length recombinant STAT6-GST as a substrate. Efficient phosphorylation of Tyr641 on STAT6 was indeed readily detected by immunoblot in reactions containing WT but not K404N recombinant ROP16 (Fig. 6). These results demonstrate that ROP16 can recognize this specific tyrosine residue of STAT6 as a phosphorylation substrate.
FIGURE 6.
Tyrosine 641 of STAT6 is directly phosphorylated by ROP16. Recombinant STAT6 (rSTAT6) was incubated with recombinant JAK3 (rJAK3) or recombinant (E. coli-produced) ROP16-HA (WT) or the kinase-deficient K404N (KD) in kinase assay conditions for 30 min at 30 °C, followed by SDS-PAGE and immunoblotting (IB) using antibodies specific for STAT6 phosphorylated on the key activation residue for STAT6, tyrosine 641 (Y641; top), antibodies that recognize STAT6 regardless of activation state (middle), or antibodies for HA-tagged ROP16 (bottom). Molecular masses are noted in kDa on the left.
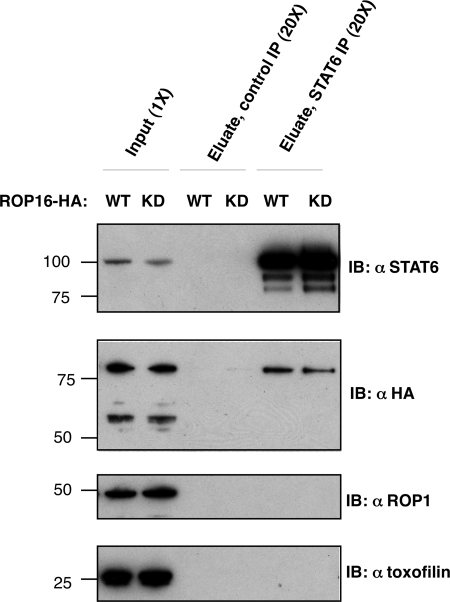
Because not all in vitro kinase substrates are bona fide physiological substrates, we sought to determine whether ROP16 and STAT6 interact in their native environments. To this end, we analyzed whether ROP16 could be co-immunoprecipitated with endogenous STAT6 from infected cells. HFFs were infected at MOI ∼10 with parasites ectopically expressing either HA-tagged ROP16-WT or ROP16-K404N. To ensure a high rate of invasion and secretion, parasites were not synchronized by potassium buffer shift but allowed to settle and invade for 2 h, at which point lysates were collected for immunoprecipitation. Both WT and K404N HA-tagged ROP16 were detected in STAT6 immunoprecipitates but not in immunoprecipitates using an isotype-matched control antibody (Fig. 7). This interaction was specific for ROP16 because two other rhoptry-secreted, soluble proteins, toxofilin (8) and ROP1 (36), were not co-precipitated with STAT6 (Fig. 7). These results suggest that the co-immunoprecipitation of ROP16 and STAT6 represents a genuine, specific interaction.
FIGURE 7.
Co-immunoprecipitation of ROP16 and STAT6 in infected cells. HFFs were infected at MOI ∼10 with Type I parasites ectopically expressing HA-tagged wild type ROP16 (WT) or ROP16-K404N (KD). 2 h postinfection, cells were washed with cold PBS and lysed in situ. Immunoprecipitation was carried out on clarified lysate (input) for 12 h at 4 °C using either nonspecific rabbit IgG-conjugated to agarose beads (Control IP) or anti-STAT6 polyclonal rabbit IgG conjugated to agarose beads (STAT6 IP). Immunocomplexes were washed in lysis buffer at 4 °C and then eluted in SDS sample buffer. Input and eluates were analyzed by SDS-PAGE and immunoblotting (IB) using antibodies specific for STAT6 (top panel), HA epitope of ROP16 (second panel), and parasite-secreted proteins ROP1 (third panel) and toxofilin (bottom panel). Cell equivalents are noted (1× or 20×) at the top; molecular masses are noted in kDa on the left.
DISCUSSION
We have used a reverse genetic approach to identify ROP16-dependent host responses and show that immediate as well as sustained STAT6 activation by Toxoplasma is dependent on ROP16. We further demonstrate that a STAT6/ROP16 interaction can be detected from infected cells and that recombinant, purified ROP16 has intrinsic tyrosine kinase activity in vitro and can directly phosphorylate STAT6 on the crucial activation residue Tyr641. Together, these results indicate that ROP16 may directly activate STAT6 upon secretion into host cells; they also provide a molecular basis for observations by Ahn et al. (37, 38) and Saeij et al. (4) that Toxoplasma induces rapid STAT6 activation in an invasion-dependent manner.
While this manuscript was in preparation, Yamamoto et al. (39) described similar results for host STAT3. Although their elegant work clearly demonstrates that STAT3 phosphorylation is dependent on ROP16 kinase activity, their use of immunoprecipitated ROP16 from overexpressing host cells for their in vitro STAT3 phosphorylation assay did not allow them to exclude the possibility that a co-precipitating host cell tyrosine kinase dependent upon ROP16 for activation (e.g. a JAK) might be directly responsible for the observed STAT3 phosphorylation. Although our data do not exclude the possibility that ROP16 might also target intermediary tyrosine kinases for phosphorylation and thereby amplify STAT6 activation, our results do clearly indicate that ROP16 in the absence of host tyrosine kinases is competent to directly phosphorylate STAT6.
Our results further indicate that there are probably additional targets of ROP16 beyond the STAT6 shown here and STAT3 indicated by Yamamoto et al. (39). This is indicated by the fact that many of the genes that we observe to be up-regulated at 5 h postinfection with wild type and ROP16-complemented parasites relative to infection with ROP16-deficient parasites are not known to be under the control of either of these STATs or factors downstream of them. Of the 11 unique genes shown in Fig. 2D that have not been previously characterized as STAT6-regulated, only one (RGS16) has been characterized as STAT3-regulated (40); of the remaining 10 genes, only TNFAIP6 has an in silico predicted STAT binding site (STAT6) in its promoter region. Yamamoto et al. (39) suggested based on in silico modeling that ROP16 may possess dual specificity kinase activity. Although we have only found evidence for tyrosine kinase activity to date, it is possible that under the right conditions, ROP16 may indeed possess dual specificity kinase activity. Hence, the other genes that appear dependent on ROP16 for their activation could be controlled by a factor that is regulated by ROP16-driven Ser/Thr phosphorylation.
In our hands, phosphorylated STAT6 was observed by immunofluorescence analysis in infected cell nuclei within 1 min of a single parasite invasion event, which was considerably more rapid than IL-4 stimulation of STAT6 activation that we and others have observed (41). This extreme speed further argues that ROP16 directly activates STAT6 rather than acting through stimulation of signaling cascades involving receptor cross-linking and JAKs. Recent studies have shown that unphosphorylated STATs may freely cycle between the cytosol and the nucleus (42, 43). Because ROP16 also traffics to the host cell nucleus, it is possible that it is phosphorylating STAT6 there as well as or even instead of in the cytosol. Arguing against this possibility, we do not see ROP16 in the nucleus 1 min post-invasion by immunofluorescence, but detectable levels have been seen at 10 min postinvasion (4), and we cannot exclude the possibility that the failure to see it at 1 min is a matter of the sensitivity of the reagents used. Finally, it should be noted that detecting phosphorylated STAT6 in the nucleus this rapidly does not require that it start in this compartment because this is sufficient time for proteins to translocate from the cytoplasm (44). Resolution of this particular issue will require generation and analysis of parasites exclusively expressing a mutated ROP16 that lacks a nuclear localization signal.
In contrast to the results described here, Yamamoto et al. (39) reported that ROP16-dependent STAT3 phosphorylation was slower than IL-6- or IL-10-induced activation and suggested that this could be due to a lag in injection of ROP16 or lower activity of ROP16 toward STAT3 compared with cellular tyrosine kinases, such as the JAKs. One possible explanation for the discrepancy between their results and ours is that STAT6 may be more rapidly phosphorylated by ROP16 than is STAT3. Some support for this comes from our analysis of host transcriptional responses at 1 and 5 h postinfection, which reveals a STAT6-dominated rather than STAT3-dominated signature at these early time points. Arguing against this explanation, however, is the fact that, as with STAT6, we also observe phosphorylated STAT3 in infected cell nuclei within 1 min of parasite invasion (data not shown), but this level of activation may not be sufficient for turning on STAT3-dependent genes, and/or another factor may interfere with its activity. Another possible explanation for the discrepancy between our results and those of Yamamoto et al. (39) is that the latter group based their conclusions on immunoblot analysis of total phospho-STAT3 from populations of non-synchronously infected cells rather than examining STAT activation at a single-cell level in cultures where invasion was synchronized. As such, the lag of several h in maximal STAT3 activation might simply reflect a lag in parasite attachment and invasion, which we have also observed using the method employed by Yamamoto et al. (39), where we find the bulk of the invasion events are occurring 1 h or longer after the addition of the parasites.
The results presented here also suggest a molecular model for how Toxoplasma might induce sustained STAT6 activation despite negative feedback induction. Our microarray results show that the suppressors of cytokine signaling (SOCS1, -2, and -3) as well as CISH (cytokine-inducible SH2-containing protein) are all up-regulated by 5 h postinfection in a ROP16-dependent manner. These proteins down-regulate STAT activation by inhibition of JAK kinase activity or competition for key receptor residues that are required for STAT recruitment to their cognate receptors (45). Our results indicate that JAK kinases do not play a direct role in ROP16-dependent STAT6 activation, so the up-regulation of SOCS may have little to no effect on the activation of this STAT. Another mode by which cells normally exert feedback control on STAT activation is dephosphorylation of JAKs or direct dephosphorylation of either cytoplasmic or nuclear STATs (46). Direct phosphorylation of STAT6 by Type I ROP16 would allow ROP16 to bypass at least the mechanisms involving JAKs. This is similar to the mechanisms operating with some oncogenic cellular tyrosine kinases, such as Src family kinases (47). Collectively, although we cannot exclude the possibility that ROP16 might also have activity on JAKS, the data strongly support the more parsimonious model in which all of the parasite-mediated early activation of STAT6 is a result of the direct phosphorylation of STAT6 by ROP16. The ability of Toxoplasma to then sustain the STAT6 phosphorylation may also indicate an inhibition of the regulating phosphatases and/or simply being more efficient at adding the key phosphate than the cell is at removing it.
Direct activation of STAT6 by Toxoplasma could have many advantages for the parasite. Toxoplasma has been observed to subvert host response by dysregulation of apoptosis (48) and IFN-γ-induced STAT1 activation (49) as well as inhibition of dendritic cell maturation (50). All of these host responses have been shown to be influenced by STAT6 activation (17, 24, 51). The evolution of such a process by an obligate intracellular parasite represents an important and potent addition to the means by which such organisms successfully occupy their unusual intracellular niche.
Acknowledgments
We thank all members of the Boothroyd laboratory for helpful discussion, in particular Dr. Gusti Zeiner for providing the pTKO plasmid, Dr. Jon Boyle for help with the microarray analysis, Dr. Sandeep Ravindran for discussions on the synchronized invasion assays, and Dr. Melissa Lodoen for help with macrophage work. We also thank Dr. James Ferrell for providing equipment and reagents for the phosphoamino acid analysis, Dr. Aimee Shen for help generating recombinant ROP16, and Dr. Jeroen Saeij (MIT) for discussion on ROP16 and for sharing unpublished data.
This work was supported, in whole or in part, by National Institutes of Health Grants AI21423 and AI73756 (to J. C. B.). This work was also supported by a National Science Foundation Graduate Research Fellowship and Smith Stanford Graduate Fellowship (to Y.-C. O.) and an American Cancer Society fellowship (to M. L. R.).
- SOCS
- suppressors of cytokine signaling
- HFF
- human foreskin fibroblast
- MOI
- multiplicity of infection
- HXGPRT or HPT
- hypoxanthine-xanthine-guanine phosphoribosyltransferase
- KD
- kinase-deficient
- ROP
- rhoptry protein.
REFERENCES
- 1.Carme B., Demar M., Ajzenberg D., Dardé M. L. (2009) Emerging Infect. Dis. 15, 656–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grigg M. E., Ganatra J., Boothroyd J. C., Margolis T. P. (2001) J. Infect. Dis. 184, 633–639 [DOI] [PubMed] [Google Scholar]
- 3.Boothroyd J. C., Dubremetz J. F. (2008) Nat. Rev. Microbiol. 6, 79–88 [DOI] [PubMed] [Google Scholar]
- 4.Saeij J. P., Coller S., Boyle J. P., Jerome M. E., White M. W., Boothroyd J. C. (2007) Nature 445, 324–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaddi P. J., Yap G. S. (2007) Immunol. Cell Biol. 85, 155–159 [DOI] [PubMed] [Google Scholar]
- 6.O'Shea J. J., Murray P. J. (2008) Immunity 28, 477–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamerman J. A., Tchao N. K., Lowell C. A., Lanier L. L. (2005) Nat. Immunol. 6, 579–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lodoen M. B., Gerke C., Boothroyd J. C. (2010) Cell Microbiol. 12, 55–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soldati D., Boothroyd J. C. (1993) Science 260, 349–352 [DOI] [PubMed] [Google Scholar]
- 10.Donald R. G., Carter D., Ullman B., Roos D. S. (1996) J. Biol. Chem. 271, 14010–14019 [DOI] [PubMed] [Google Scholar]
- 11.Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. (1989) Gene 77, 51–59 [DOI] [PubMed] [Google Scholar]
- 12.Soldati D., Kim K., Kampmeier J., Dubremetz J. F., Boothroyd J. C. (1995) Mol. Biochem. Parasitol. 74, 87–97 [DOI] [PubMed] [Google Scholar]
- 13.Irizarry R. A., Bolstad B. M., Collin F., Cope L. M., Hobbs B., Speed T. P. (2003) Nucleic Acids Res. 31, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tusher V. G., Tibshirani R., Chu G. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kafsack B. F., Beckers C., Carruthers V. B. (2004) Mol. Biochem. Parasitol. 136, 309–311 [DOI] [PubMed] [Google Scholar]
- 16.Endo T., Yagita K. (1990) J. Protozool. 37, 133–138 [DOI] [PubMed] [Google Scholar]
- 17.Jackson S. H., Yu C. R., Mahdi R. M., Ebong S., Egwuagu C. E. (2004) J. Immunol. 172, 2307–2315 [DOI] [PubMed] [Google Scholar]
- 18.Boyle W. J., van der Geer P., Hunter T. (1991) Methods Enzymol. 201, 110–149 [DOI] [PubMed] [Google Scholar]
- 19.Ossorio P. N., Schwartzman J. D., Boothroyd J. C. (1992) Mol. Biochem. Parasitol. 50, 1–15 [DOI] [PubMed] [Google Scholar]
- 20.Bradley P. J., Ward C., Cheng S. J., Alexander D. L., Coller S., Coombs G. H., Dunn J. D., Ferguson D. J., Sanderson S. J., Wastling J. M., Boothroyd J. C. (2005) J. Biol. Chem. 280, 34245–34258 [DOI] [PubMed] [Google Scholar]
- 21.Qiu W., Wernimont A., Tang K., Taylor S., Lunin V., Schapira M., Fentress S., Hui R., Sibley L. D. (2009) EMBO J. 28, 969–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanks S. K., Quinn A. M., Hunter T. (1988) Science 241, 42–52 [DOI] [PubMed] [Google Scholar]
- 23.Hoeck J., Woisetschläger M. (2001) J. Immunol. 167, 3216–3222 [DOI] [PubMed] [Google Scholar]
- 24.Zhang W. J., Li B. H., Yang X. Z., Li P. D., Yuan Q., Liu X. H., Xu S. B., Zhang Y., Yuan J., Gerhard G. S., Masker K. K., Dong C., Koltun W. A., Chorney M. J. (2008) Cytokine 42, 39–47 [DOI] [PubMed] [Google Scholar]
- 25.Hebenstreit D., Wirnsberger G., Horejs-Hoeck J., Duschl A. (2006) Cytokine Growth Factor Rev. 17, 173–188 [DOI] [PubMed] [Google Scholar]
- 26.Zimmermann N., Mishra A., King N. E., Fulkerson P. C., Doepker M. P., Nikolaidis N. M., Kindinger L. E., Moulton E. A., Aronow B. J., Rothenberg M. E. (2004) J. Immunol. 172, 1815–1824 [DOI] [PubMed] [Google Scholar]
- 27.Chen Z., Lund R., Aittokallio T., Kosonen M., Nevalainen O., Lahesmaa R. (2003) J. Immunol. 171, 3627–3635 [DOI] [PubMed] [Google Scholar]
- 28.Tomkinson A., Kanehiro A., Rabinovitch N., Joetham A., Cieslewicz G., Gelfand E. W. (1999) Am. J. Respir. Crit. Care Med. 160, 1283–1291 [DOI] [PubMed] [Google Scholar]
- 29.Schnyder B., Schnyder-Candrian S., Panski A., Bömmel H., Heim M., Duschl A., Moser R. (2002) Biochem. Biophys. Res. Commun. 292, 841–847 [DOI] [PubMed] [Google Scholar]
- 30.Wei L., Vahedi G., Sun H. W., Watford W. T., Takatori H., Ramos H. L., Takahashi H., Liang J., Gutierrez-Cruz G., Zang C., Peng W., O'Shea J. J., Kanno Y. (2010) Immunity 32, 840–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsiao W. L., Mendosa G., Kothari N. H., Fan H. (1996) Carcinogenesis 17, 2771–2777 [DOI] [PubMed] [Google Scholar]
- 32.Daines M. O., Andrews R. P., Chen W., El-Zayaty S. A., Hershey G. K. (2003) J. Biol. Chem. 278, 30971–30974 [DOI] [PubMed] [Google Scholar]
- 33.McKendry R., John J., Flavell D., Müller M., Kerr I. M., Stark G. R. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 11455–11459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kohlhuber F., Rogers N. C., Watling D., Feng J., Guschin D., Briscoe J., Witthuhn B. A., Kotenko S. V., Pestka S., Stark G. R., Ihle J. N., Kerr I. M. (1997) Mol. Cell. Biol. 17, 695–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomis D. C., Gurniak C. B., Tivol E., Sharpe A. H., Berg L. J. (1995) Science 270, 794–797 [DOI] [PubMed] [Google Scholar]
- 36.Bradley P. J., Hsieh C. L., Boothroyd J. C. (2002) Mol. Biochem. Parasitol. 125, 189–193 [DOI] [PubMed] [Google Scholar]
- 37.Ahn H. J., Kim J. Y., Nam H. W. (2009) Korean J. Parasitol. 47, 117–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahn H. J., Kim J. Y., Ryu K. J., Nam H. W. (2009) Parasitol. Res. 105, 1445–1453 [DOI] [PubMed] [Google Scholar]
- 39.Yamamoto M., Standley D., Takashima S., Saiga H., Okuyama M., Kayama H., Kubo E., Ito H., Takaura M., Matsuda T., Soldati-Favre D., Takeda K. (2009) J. Exp. Med. 206, 2747–2760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bourillot P. Y., Aksoy I., Schreiber V., Wianny F., Schulz H., Hummel O., Hubner N., Savatier P. (2009) Stem Cells 27, 1760–1771 [DOI] [PubMed] [Google Scholar]
- 41.Montag D. T., Lotze M. T. (2006) Clin. Immunol. 121, 215–226 [DOI] [PubMed] [Google Scholar]
- 42.Meyer T., Vinkemeier U. (2004) Eur. J. Biochem. 271, 4606–4612 [DOI] [PubMed] [Google Scholar]
- 43.Chen H. C., Reich N. C. (2010) J. Immunol. 185, 64–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ribbeck K., Görlich D. (2001) EMBO J. 20, 1320–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Croker B. A., Kiu H., Nicholson S. E. (2008) Semin. Cell Dev. Biol. 19, 414–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu D., Qu C. K. (2008) Front. Biosci. 13, 4925–4932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hayakawa F., Naoe T. (2006) Ann. N.Y. Acad. Sci. 1086, 213–222 [DOI] [PubMed] [Google Scholar]
- 48.Carmen J. C., Sinai A. P. (2007) Mol. Microbiol. 64, 904–916 [DOI] [PubMed] [Google Scholar]
- 49.Kim S. K., Fouts A. E., Boothroyd J. C. (2007) J. Immunol. 178, 5154–5165 [DOI] [PubMed] [Google Scholar]
- 50.McKee A. S., Dzierszinski F., Boes M., Roos D. S., Pearce E. J. (2004) J. Immunol. 173, 2632–2640 [DOI] [PubMed] [Google Scholar]
- 51.Ohmori Y., Hamilton T. A. (2000) J. Biol. Chem. 275, 38095–38103 [DOI] [PubMed] [Google Scholar]