Abstract
Dyshomeostasis of transition metals iron and copper as well as accumulation of oxidative DNA damage have been implicated in multitude of human neurodegenerative diseases, including Alzheimer disease and Parkinson disease. These metals oxidize DNA bases by generating reactive oxygen species. Most oxidized bases in mammalian genomes are repaired via the base excision repair pathway, initiated with one of four major DNA glycosylases: NTH1 or OGG1 (of the Nth family) or NEIL1 or NEIL2 (of the Nei family). Here we show that Fe(II/III) and Cu(II) at physiological levels bind to NEIL1 and NEIL2 to alter their secondary structure and strongly inhibit repair of mutagenic 5-hydroxyuracil, a common cytosine oxidation product, both in vitro and in neuroblastoma (SH-SY5Y) cell extract by affecting the base excision and AP lyase activities of NEILs. The specificity of iron/copper inhibition of NEILs is indicated by a lack of similar inhibition of OGG1, which also indicated that the inhibition is due to metal binding to the enzymes and not DNA. Fluorescence and surface plasmon resonance studies show submicromolar binding of copper/iron to NEILs but not OGG1. Furthermore, Fe(II) inhibits the interaction of NEIL1 with downstream base excision repair proteins DNA polymerase β and flap endonuclease-1 by 4–6-fold. These results indicate that iron/copper overload in the neurodegenerative diseases could act as a double-edged sword by both increasing oxidative genome damage and preventing their repair. Interestingly, specific chelators, including the natural chemopreventive compound curcumin, reverse the inhibition of NEILs both in vitro and in cells, suggesting their therapeutic potential.
Keywords: Copper, DNA Damage, DNA Repair, Iron, Neurodegeneration, Base Excision Repair, DNA Glycosylase, NEIL1, NEIL2, Oxidative DNA Damage, Curcumin, Dyshomeostasis, Neuroprotection
Introduction
Excessive accumulation of transition metals, particularly copper and iron, have been implicated in various human neurodegenerative diseases and aging (1–4). In the brain, dysregulation of metal homeostasis may occur in stroke and other neurodegenerative disorders, including Alzheimer disease (AD),2 Parkinson disease (PD), Huntington disease (HD), hereditary ferritinopathy, and Wilson disease (2, 3). Toxicity due to transition metals has a complex basis because of their essentiality as micronutrients under normal physiological conditions and tight regulation of their levels. These metals are typically sequestered in catalytically inactive form by metal storage proteins in vivo, such as ferritin, transferrin, or cerruloplasmin (5). Their bioavailable forms bound to low affinity ligands are chelatable. According to one estimate, the chelatable iron is present at 1–10 μm in normal mammalian cells (6). In PD brains, increased iron is often accompanied with decreased ferritin synthesis, resulting in free iron overload (7). Several studies have implicated copper, aluminum, and iron in AD as well (3, 8). Further, Lewy bodies in PD and amyloid deposits or neurofibrillary tangles in AD are enriched in transition metals (9, 10).
Metal accumulation has also been implicated in cancer; copper and iron along with other heavy metals are 5–10 times more abundant in human lung, breast, and prostate tumors than the respective normal tissues (11, 12). However, the levels of these metals were shown to be lower in liver tumors than in the normal tissue (11). Thus, an imbalance in metal homeostasis is associated with various cancers, although the causal link has not been established.
Accumulation of oxidative genome damage is a unifying feature of most pathologies associated with metal toxicity (13–15). Redox-cycling iron and copper could generate O2 free radicals (ROS) via a Fenton reaction, which oxidizes cellular components, including DNA. Furthermore, accumulation of oxidative DNA damage may be accompanied by a decrease in the DNA repair capacity for many neurodegenerative disorders, including PD (16). However, the precise mechanism of how metals affect DNA repair is not understood. Few studies have been reported on the genotoxicity of these metals on DNA repair, particularly BER, the major pathway for repairing oxidized bases in mammalian neuronal cells (17–19). Repair of oxidized bases in mammalian genomes is initiated with one of four DNA glycosylases that excise the base lesion (19). These glycosylases with overlapping substrate range belong to two families, OGG1 and NTH1, in the Nth family and NEIL1 and NEIL2, more recently discovered by us and others, in the Nei family (20–24). All of these glycosylases have intrinsic base excision and AP lyase activities, such that after removing the base, these enzymes cleave the DNA strand at the AP site, resulting in a single-stranded break with blocked ends (19). The NEILs, unlike OGG1/NTH1, excise base lesion from single-stranded (ss) or bubble DNA substrates, and hence their activity could be selectively analyzed (25). Furthermore, OGG1 and NTH1 carry out β-elimination, generating 3′-deoxyribose phosphate, which is subsequently removed by AP-endonuclease (APE1), whereas NEIL1 and NEIL2 with βδ-elimination lyase activity generate 3′-P, which is then removed by polynucleotide kinase (PNK). The resulting 1-nucleotide gap in the damaged strand is then filled by a DNA polymerase, typically DNA polymerase β (Polβ) in the nucleus, and the final nick sealing is carried out by DNA ligase IIIα (LigIIIα) for single-nucleotide repair (19). In addition, Polβ could collaborate with FEN-1 to carry out long-patch repair synthesis (26).
The genomes of neurons and their progenitor cells are particularly susceptible to oxidative damage because of the abundance of ROS generated due to high O2 consumption. Whereas the levels of a subset of BER enzymes decrease during brain ontogeny (proliferative to postmitotic transition), the levels of NEIL1 and NEIL2 were shown to be higher during brain development (27), suggesting a role for the NEIL glycosylases in maintaining genome integrity in brain cells. Recently, few studies have documented inhibition of BER enzymes by heavy metals. Cadmium was shown to irreversibly inhibit OGG1 activity and its substrate affinity (18). More recently, iron was found to inhibit FEN-1 and DNA ligase (28). Inhibition of Fpg (formamidopyrimidine-DNA glycosylase) family DNA glycosylases, including NEIL1, by heavy metals has been recently reported (29). However, most of these studies attributed the inhibition to the metal binding of DNA. In contrast, we show in this study for the first time that subtoxic levels of iron and copper directly bind to NEIL1 and NEIL2 with high affinity and abrogate their activities both in vitro and in cells. We also show further inhibition of overall repair due to inhibition of the interaction of NEIL1 in the presence of Fe(II) with downstream BER proteins. Interestingly, chelators, including the natural compound curcumin, restored NEIL activities, suggesting their therapeutic potential in reversing metal toxicity.
EXPERIMENTAL PROCEDURES
Chemicals and Antibodies
NaEDTA, CaEDTA, desferrioxamine, FeSO4, FeCl3, CuCl2, AlCl3, ZnCl2, tris-(2-carboxyethyl)-phosphine (TCEP), sodium borohydride (NaBH4), and curcumin were purchased from Sigma. Freshly dissolved metal solutions were used for each experiment. NEIL1 antibody was described earlier (21). Anti-His antibody was obtained from Sigma.
Preparation of Single-oxidized Base Lesion-containing Oligonucleotide and Plasmid Substrates
A 51-mer oligonucleotide containing 5-OHU or 8-oxoG at position 26 and undamaged complementary oligonucleotides containing G or C opposite the lesion, respectively, and other oligonucleotides for ligase or polymerase assays (supplemental Fig. S1) were purchased from Midlands Inc. Duplex oligonucleotides were prepared by annealing complementary oligonucleotides after heating at 94 °C for 3–5 min, followed by slow cooling to room temperature. To produce 5′-32P-labeled substrates, the ss oligonucleotides were labeled at the 5′ terminus with [γ-32P]ATP and T4 PNK before annealing. The unincorporated label was removed by gel filtration on a Sephadex G25 column (30).
The repair substrate pUC19CPD plasmid containing a single 5-OHU was generated as described previously (31, 32). Briefly, the parent plasmid containing two ss nicking sites on the same strand, 32 nt apart, was digested with N.BstNB1 (New England Biolabs) and heated at 65 °C for 10 min to dissociate the 32-nt ss oligonucleotide (5′-GCG GAT ATT AAT GTG ACG GTA GCG AGT CGC TC-3′), which was then annealed with excess biotinylated complementary 32-nt oligonucleotide and removed from the solution by binding to streptavidin-agarose Dynabeads (Sigma). The gapped plasmid was extracted with phenol/chloroform followed by ethanol precipitation and then annealed with 5-OHU-containing 5′-phosphorylated 32-nt oligonucleotide (5′-pGCG GAT ATT AAT GTG ACG G 5-OHU A GCG AGT CGC TC-3′) at 45 °C in TE buffer containing 50 mm NaCl. 5-OHU-containing form I plasmid generated by sealing the nick with T4 DNA ligase was purified by equilibrium ultracentrifugation in CsCl/ethidium bromide. The presence of 5-OHU was verified by treating the plasmid with NEIL1, which converted the form I plasmid into nicked form II.
Expression and Purification of Proteins
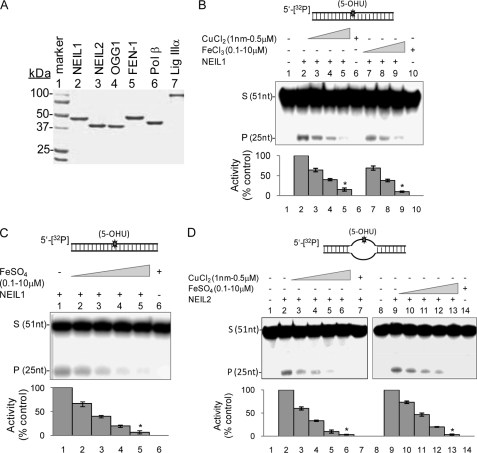
The human recombinant enzymes, including NEIL1, NEIL2, OGG1, PNK, Polβ, FEN-1, and LigIIIα, were purified as described previously (20, 21, 30, 33, 34). Specifically, untagged NEIL1 and NEIL2 cDNA were subcloned in pET 22(b) vector with NdeI and XhoI restriction sites and purified from the extract of plasmid-bearing Escherichia coli by sequential anion exchange and cation exchange chromatography. The purity of these enzymes was confirmed by SDS-PAGE analysis (Fig. 1A).
FIGURE 1.
Inhibition of NEILs by iron and copper salts. A, Coomassie-stained 10% SDS-PAGE of the BER protein preparations used in this study. B–D, inhibition of NEIL1 glycosylase activity by iron/copper. B, NEIL1 activity with 5-OHU-containing duplex (upper panel), in the absence (lane 2) or presence of 1, 100, or 500 nm CuCl2 (lanes 3–5) or 0.1, 1, or 10 μm FeCl3 (lanes 7 and 8). Lane 1, substrate alone control; lane 6, substrate plus copper (500 nm); lane 10, substrate plus iron (10 μm). Quantitation of activity (% control) is represented in the histogram (lower panel). *, statistical significance at p < 0.01. C, effect of FeSO4 (0.1, 1, and 10 μm; lanes 2–5) on NEIL1 activity with 5-OHU-duplex substrate. Lane 1, substrate alone; lane 6, substrate plus 10 μm FeSO4. D, effect of CuCl2 (1, 10, 100, or 500 nm; lanes 3–6) or FeSO4 (0.1, 0.5, 1, or 10 μm; lanes 10–13) on NEIL2 activity (lane 2 or 9) with 5-OHU-containing bubble substrate (upper panel) (*, p < 0.01). S, substrate; P, product; Error bars, S.E.
Cell Culture and Treatments
The human neuroblastoma SH-SY5Y cells were grown in 1:1 DMEM and F-12 medium containing 10% FBS, 4 mm l-glutamate. The undifferentiated cells (40% confluence) were treated twice in succession with 1–100 μm FeSO4 or CuCl2 for 24 h. At 12 h after the first treatment, the medium was replaced with a freshly prepared metal solution-containing medium for another 12 h. In some cases, the cells were treated with 1–10 μm curcumin (Sigma) at 1 h after metal addition. The curcumin stock solution (0.5 m) in DMSO was serially diluted in PBS so that the final DMSO concentration in the culture medium was less than 0.5%. The cells were harvested, and nuclear extracts (NE) were prepared as described previously (33). Briefly, cells were lysed in EB buffer (20 mm Tris-HCl, pH 8.0, 1.5 mm MgCl2, 10 mm KCl, 10% glycerol, and protease inhibitors), centrifuged at 13,000 × g for 1 min. The nuclear pellet was then lysed in NE buffer (20 mm Tris-HCl, pH 8.0, 1.5 mm MgCl2, 420 mm NaCl, and 20% glycerol) and centrifuged at 14,000 × g for 5 min. The supernatant was diluted 4 times with EB buffer to reduce the NaCl concentration to 100 mm before using in repair assays. EDTA and DTT were deliberately excluded from the extraction buffers.
Analysis of DNA Glycosylase Activity
DNA strand scission activity of NEIL1, NEIL2, or OGG1 after lesion base excision was analyzed using the 51-mer duplex oligonucleotide containing 5-OHU or 8-oxoG, as described previously (30). The 5′-32P-labeled oligonucleotide (2 pmol) was incubated with the enzymes (0.2 pmol) alone or in the presence of indicated amount of metals, at 37 °C for 15 min in a 10-μl reaction mixture containing BER buffer (40 mm Hepes, pH 7.5, 50 mm KCl, 100 μg/ml bovine serum albumin (BSA), and 5% glycerol). Linear reaction rate was maintained during incubation. After stopping the reaction with formamide dye (80% formamide, 20 mm NaOH, 20 mm EDTA, 0.05% bromphenol blue, and 0.05% xylene cyanol), the products were separated by 20% polyacrylamide gel containing 8 m urea in Tris borate-EDTA buffer, pH 8.4 (35), and the radioactivity in the substrate and product was quantitated in a PhosphorImager (Amersham Biosciences) with ImageQuant software.
Measurement of AP Lyase Activity
We generated an AP site-containing duplex oligonucleotide substrate starting with the 51-nt oligonucleotide as described earlier, except for incorporation of U at position 26. After annealing with the complementary strand, the duplex was treated with 100 units of uracil-DNA glycosylase (New England Biolabs) at 37 °C for 30 min to generate an AP site by excising U. Subsequent incubation with NEIL1, NEIL2, or OGG1 was carried out in the presence or absence of metals at 37 °C for 8 min, the reaction was stopped with 80% formamide dye (without NaOH), and the products were analyzed as before.
DNA Trapping Assay
Oxidized base-specific DNA glycosylases form covalent adducts, named trapped complexes, with the oligonucleotide substrate at the AP site after reduction of the transient Schiff base intermediate (36). We carried out the trapping reactions under glycosylase assay conditions with 5′-32P-labeled substrate oligonucleotide, in the presence of 100 mm freshly prepared NaBH4, and incubated for 30 min at 37 °C. The trapped complexes were separated in 12% SDS-PAGE, and the radioactivity was analyzed as before.
Affinity Measurements of Metal Binding to DNA Glycosylases Using Fluorescence and Surface Plasmon Resonance (SPR)
Interaction of NEIL1 and NEIL2 with metals was monitored by the increase in the intrinsic tyrosine/tryptophan fluorescence of NEILs (λex = 280 nm, λem = 290–400 nm) upon titration with increasing amounts of FeSO4 and CuCl2 (1 nm-10 μm), in a FluoroMax, SPEX spectrofluorometer (Horiba Scientific). For binding studies, the enzymes were incubated with iron and copper salts in 25 mm Tris buffer, pH 7.4, containing 50 mm NaCl, at 25 °C for 5 min. The binding constant, KD, was calculated by plotting ΔF (change in fluorescence maximum) versus ligand (metal) concentrations according to the equation ΔF = ΔFmax × [ligand]/KD + [ligand], as described previously (30).
Interaction between NEILs and CuCl2 was also analyzed by SPR using Biacore T100 (GE Healthcare). Full-length NEIL1 (ligand, 20 μg/ml) was immobilized to a CM5 sensor chip via amine coupling (30). CuCl2 (1 nm to 10 μm in 25 mm Tris, pH 7.5, 0.1 m NaCl, 1 mm MgCl2) in analyte was injected at 30 μl/min for 2 min. The response units were corrected for the blank obtained from the reference flow cell, and the data were analyzed using a 1:1 (Langmuir) binding model (BIAevaluation Software, GE Healthcare).
Isothermal Titration Calorimetry (ITC)
The enthalpy of binding of Cu(II) with NEIL1 was quantitated by ITC in a VP-ITC (MicroCal, Amherst, MA) as described elsewhere (37). The ITC cell was filled with 0.5 μm NEIL1 in 20 mm HEPES, pH 7.4, 50 mm NaCl, and the syringe was filled with CuCl2 solution in the same buffer. The HEPES was shown to have negligible interference with Cu(II) (37). In a control experiment, the cell was filled with NEIL1, and the syringe was filled with buffer without Cu(II). The titration involved 30 serial injections of 10 μl with an initial delay of 60 s, duration of 20 s, and spacing of 180 s. Background titration consisting of identical copper amounts in the syringe with blank buffer in the cell was subtracted from each titration to account for the heat of dilution (38). From the non-constrained curve fitting to plot the heat evolved/mol of Cu(II) ions versus the molar ratio of copper to NEIL1, the binding stoichiometry (N), binding affinity (KD), enthalpy (ΔH), and entropy (ΔS) were determined using MicroCal ORIGIN 6.0 software.
Secondary Structure Analysis Using Circular Dichroism (CD) Spectroscopy
The CD spectra (195–260 nm) were recorded for NEIL1 or NEIL2 (2 μm) in the presence/absence of FeSO4 or CuCl2 in 25 mm Tris buffer (pH 7.4), 50 mm NaCl in a JASCO J-720 spectropolarimeter (39), using a 1-mm cuvette, and four repetitions of the CD spectra were used for averaging and then corrected for the contribution from the metal alone. The secondary structures of the enzyme were calculated from the CD spectra using K2d software.
Blue Native PAGE Analysis
Protein multimer formation was analyzed by blue native PAGE (40). Briefly, NEIL1, NEIL2, or OGG1 (100 pmol each) in the presence or absence of 1 μm CuCl2 or 10 μm FeSO4 was separated in a 4–16% polyacrylamide gel under non-reducing/non-denaturing conditions, using a blue native PAGE kit (Invitrogen) and the manufacturer's protocol, and then stained with Coomassie Brilliant Blue.
His Affinity Pull-down Assay
Histidine affinity pull-down assays were carried out as described previously (30, 41). NEIL1 (20 pmol) in the presence or absence of FeSO4 or CuCl2 (0.5 and 1 μm) was mixed with His-tagged Polβ, FEN-1, or LigIIIα (40 pmol each), prebound to His-select magnetic nickel beads (Ni2+-NTA; Invitrogen), and incubated for 1 h at 4 °C with constant rotation in a buffer containing TBS (20 mm Tris, pH 7.4, 100 mm NaCl), 5% BSA, 10% glycerol. After washing the beads with TBS containing 0.1% Triton X-100 and 400 mm NaCl, the bound proteins were eluted with SDS sample dye mix and tested for the presence of NEIL1 and -2 by immunoblotting.
Complete Repair Assay
NEIL1-initiated repair of a plasmid or oligonucleotide substrate was carried out in a reconstituted system containing recombinant proteins (PNK, Polβ, and LigIIIα, 50 fmol each) in the presence or absence of metals and chelators as described previously (30). Briefly, the enzymes and transition metals were incubated with 2 pmol of damage-containing duplex oligonucleotide in a 20-μl reaction mixture containing 1 mmol of ATP, 25 μmol of dNTPs, and one 10-μmol [α-32P]dNTP (the concentration of the corresponding unlabeled dNTP was lowered to 5 μm unless otherwise specified) in BER buffer and incubated for 30 min at 37 °C. Appropriate controls minus the enzymes were simultaneously analyzed, and the products were analyzed as described before. OGG1-initiated repair was similarly performed using 8-oxoG-containing substrate, and APE1 was substituted with PNK.
DNA Polymerase Assay
A partial duplex oligonucleotide substrate with 3′-OH-containing primer for DNA polymerase assay was generated by annealing the 25-nt complementary oligonucleotide (pol 25; supplemental Fig. S1) with the 51-nt strand of the same sequence as used for repair studies but lacking the oxidized base lesion. The assay was based on a single nt (position 26, [α-32P]dTMP) incorporation reaction by Polβ. The reaction was carried out by incubating 100 fmol of Polβ with 2 pmol of substrate and 1 mCi of [α-32P]dTTP in the presence or absence of either iron or copper. After incubation for 15 min at 37 °C and stopping the reaction with an equal volume of formamide/dye, the products were separated on 20% polyacrylamide, urea gel as described before.
DNA Ligase Assay
The DNA substrate for the LigIIIα reaction was prepared by 5′-labeling a 26-nt oligonucleotide (lig 26) with [γ-32P]ATP and annealed with the 51-nt complementary oligonucleotide and pol 25 oligonucleotide, the same sequences as used for repair studies (supplemental Fig. S1). This oligonucleotide has a ligatable nick between nt 26 and 27 nucleotide, and the ligation produces a 51-nt product from the 26-nt labeled substrate. The ligation reaction (substrate, 2 pmol; LigIIIα, 200 fmol) in the absence or presence of iron/copper, as indicated, was carried out at 37 °C for 30 min, and the products were analyzed as before.
Statistical Analysis
Data shown in the figures represent mean values ± S.E. from at least three independent experiments unless otherwise stated. Statistical analysis was performed using either Microsoft Excel or Sigmaplot software. Statistical significance has been set at p < 0.01 (*) or p < 0.05 (**).
RESULTS
Inhibition of NEILs but Not OGG1 by Iron and Copper
We examined the effect of CuCl2 (1 nm to 500 nm), FeCl3, and FeSO4 (100 nm to 10 μm) on base excision and AP lyase activities of NEIL1 (20 fmol), using 5-OHU-containing 5′-32P-labeled oligonucleotide duplex (200 fmol) as described under “Experimental Procedures” (Fig. 1B). CuCl2 strongly inhibited the strand scission activity of NEIL1, with 30 and 70% inhibition at 1 and 100 nm CuCl2, respectively (Fig. 1B), whereas 500 nm CuCl2 abolished the activity nearly completely (>90% inhibition; Fig. 1B, lanes 4 and 5). As expected, CuCl2 (500 nm) by itself did not cleave the substrate (Fig. 1B, lane 6). Similarly, both FeCl3 (Fig. 1B, lanes 7–10) and FeSO4 (Fig. 1C) strongly inhibited NEIL1, albeit at a higher concentration than copper but still at physiologically relevant levels. 10 μm iron completely abolished the activity of NEIL1 without direct effect on the DNA (Fig. 1, B and C).
We then showed that CuCl2 and FeSO4 reduced NEIL2 activity with 5-OHU-containing bubble substrate to a similar extent for NEIL1 (Fig. 1D). FeCl3 (0.1–10 μm) also inhibited NEIL2 (data not shown). Because both NEIL1 and NEIL2 have higher activity with ss DNA than with duplex substrates (25), we tested the effect of Cu(II) and Fe(III) on NEIL1 with 5-OHU bubble substrate and observed similar inhibition (supplemental Fig. S2).
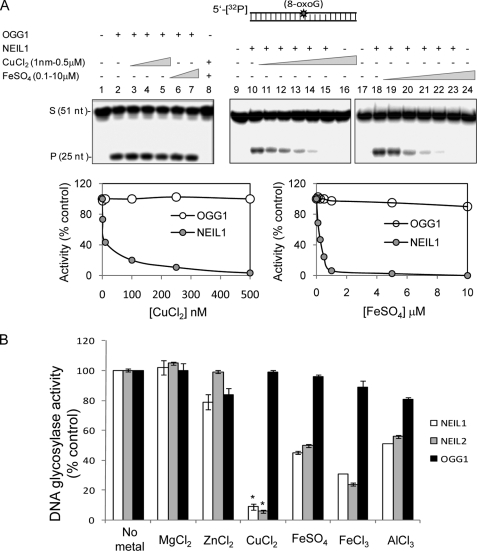
To investigate whether the metal inhibition of NEILs is specific, we carried out similar experiments with OGG1 using 8-oxoG-containing duplex substrate of the same sequence (Fig. 2A). Whereas 8-oxoG in a duplex is the preferred substrate of OGG1, it is also a weak substrate for NEIL1 (21, 25). It is clear that CuCl2 (1–500 nm) or FeSO4 (0.1–10 μm) did not affect OGG1 (20 fmol) activity (Fig. 2A, lanes 1–8), whereas NEIL1 inhibition was confirmed (lanes 9–24). The histogram shows comparative inhibition of OGG1 and NEIL1 by Cu(II) and Fe(II). FeCl3 also showed similar inhibition of NEILs but not of OGG1 (data not shown). These results strongly suggest that inhibition of NEILs could involve their direct binding to the metals rather than to the DNA.
FIGURE 2.
Lack of OGG1 inhibition by iron/copper. A, effect of CuCl2 and FeSO4 on OGG1 (25 fmol) and NEIL1 (50 fmol) activity with 8-oxoG-containing duplex substrate (40 fmol). B, effect of 500 nm each of Mg(II), Zn(II), Cu(II), Fe(II), Fe(III), and Al(III) on the activities of NEIL1, NEIL2, or OGG1. 5-OHU-containing bubble substrate for NEILs and 8-oxoG-containing duplex for OGG1 were used. The activity in the absence of metals was set at 100 to compare relative activity in the presence of metals. *, p < 0.01. S, substrate; P, product; Error bars, S.E.
To further address the issue of electrostatic effects via metal binding to DNA, we tested the effect of MgCl2 (1, 2, and 5 mm), which did not affect NEIL1 activity (supplemental Fig S3A). We also show similar iron/copper-mediated inhibition of NEIL1 in the background of 1 mm MgCl2 as in its absence (supplemental Fig S3B).
We also tested the effect of other metals, namely, MgCl2, ZnCl2, and AlCl3, on the activities of NEILs and OGG1 (Fig. 2B). These metals are essential for humans and other mammals (3, 42). AlCl3 (500 nm) also inhibited NEIL1 and NEIL2, in the order, CuCl2 > FeCl3 > FeSO4 > AlCl3, at equimolar level, whereas MgCl2 and ZnCl2 did not affect the activity. At the same time, OGG1 was not significantly affected by any of these metals at similar concentrations. Taken together, these studies showed selective inhibition of NEIL1 and NEIL1 but not of OGG1 by physiological levels of iron, copper, and aluminum.
Copper and Iron Affect both Base Excision and AP Lyase Activities of NEILs
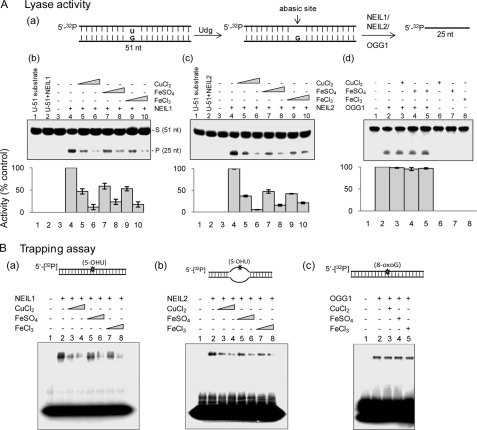
Base excision and AP lyase activities are tightly coupled for NEILs (33, 43), whereas for some DNA glycosylases, particularly OGG1, these activities could be uncoupled (35, 43). To test whether base excision and AP lyase activity are similarly inhibited by copper and iron, we examined the effect of these metals on the AP lyase activity using an AP site-containing oligonucleotide (Fig. 3A, a). CuCl2 (0.1–0.5 μm; Fig. 1A, b, lanes 5 and 6), FeSO4 (1–10 μm; lanes 7 and 8), and FeCl3 (1–10 μm; lanes 9 and 10) strongly inhibited the AP lyase activity of NEIL1, whereas NEIL1 by itself did not cleave the U-containing substrate, as expected (lanes 1 and 2). AP lyase function of NEIL2 was likewise affected by these metals (Fig. 3A, c). In contrast, OGG1 activity was unaffected (Fig. 3A, d).
FIGURE 3.
Iron/copper-induced inhibition of both base excision and AP lyase activities for NEILs. A, AP lyase activity in the presence of iron and copper. a, the AP site substrate was generated by treating U-containing 51-nt duplex with Udg (E. coli). The effect of CuCl2 (0.1 or 0.5 μm), FeSO4 (1 or 10 μm) and FeCl3 (1 or 10 μm) on the AP lyase activity of NEIL1 (b), NEIL2 (c), and OGG1 (d) was analyzed as described under “Experimental Procedures.” Lower panels, relative activity as in Fig. 1. B, analysis of trapped complexes. 50 fmol each of NEIL1 (a), NEIL2 (b), or OGG1 (c) was incubated with the substrate (upper panel) and metals (copper, 0.1 or 0.5 μm; iron, 1 or 10 μm) in the presence of 100 mm NaBH4 at 37 °C for 45 min and analyzed by 12% SDS-PAGE. Lane 1, substrate alone. S, substrate; P, product; Error bars, S.E.
We also examined iron/copper inhibition of trapped complex formation as a measure of AP lyase activity. CuCl2 (100–500 nm), FeSO4 (1–10 μm), and FeCl3 (1–10 μm) reduced trapped complex formation for both NEIL1 and NEIL2 up to 6–8-fold in a dose-dependent fashion (Fig. 3B, a and b) but not for OGG1 (Fig. 3B, c). These results further confirm that both the base excision and AP lyase activities of NEILs but not of OGG1 are inhibited by Cu(II) and Fe(II/III).
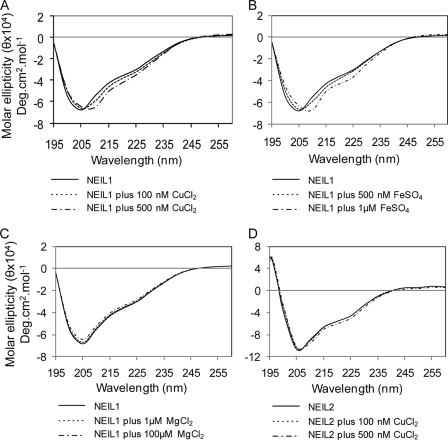
Fluorescence Analysis of Cu(II) and Fe(II) Binding to NEILs
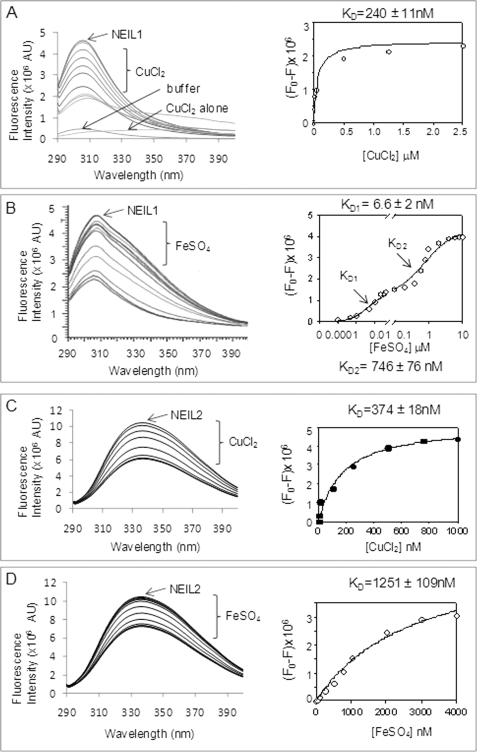
We tested the effect of copper and iron binding on the secondary structures of NEILs by measuring their intrinsic fluorescence. The protein in 25 mm Tris buffer, pH 7.4, containing 50 mm NaCl was excited at 280 nm, and the emission was measured at 290–400 nm. The emission maximum of NEIL1 at 307 nm was observed previously (30, 44), whereas the emission maximum of NEIL2 is at 340 nm. Titration with CuCl2 (1 nm-2.5 μm) or FeSO4 (1 nm-4 μm) decreased the fluorescence of NEIL1 and NEIL2 (Fig. 4). Cu(II) showed monophasic binding with KD of 40 nm (Fig. 4A), whereas Fe(II) showed biphasic binding to NEIL1 with KD1 of 6.6 nm and KD2 of 746 nm (Fig. 4B). The KD values calculated for Cu(II) and Fe(II) binding with NEIL2 were 374 nm and 1.2 μm, respectively (Fig. 4, C and D). We also tested the effect of iron/copper on OGG1 fluorescence and did not observe specific interaction at a similar metal concentration (data not shown).
FIGURE 4.
Fluorescence analysis of Cu(II) and Fe(II) binding with NEILs. NEIL1 or NEIL2 (2 μm) in 25 mm Tris buffer, pH 7.4, 50 mm NaCl, were titrated with CuCl2 or FeSO4 (1 nm to 10 μm), the solution was excited at 280 nm, and emission of intrinsic Tyr/Trp fluorescence was measured at 290–400 nm. A, NEIL1 plus Cu(II); B, NEIL1 plus Fe(II); C, NEIL2 plus Cu(II); D, NEIL2 plus Fe(II). AU, arbitrary units.
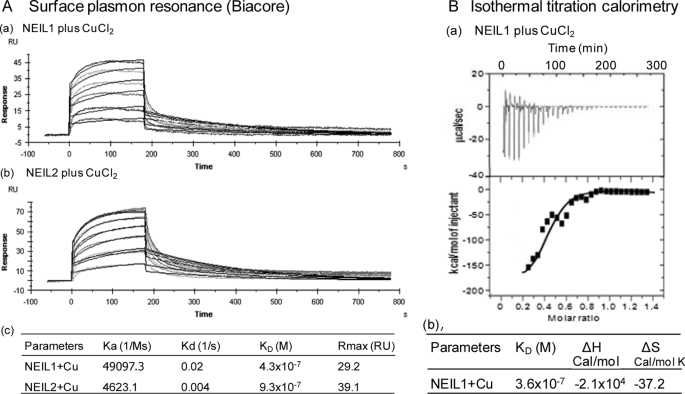
Surface Plasmon Resonance Analysis
Using CuCl2 in the analyte with NEIL1 or NEIL2 immobilized on a CM5 sensorchip, we measured the kinetics of copper binding to NEIL1 and NEIL2 in Biacore sensorgrams. Using a 1:1 (Langmuir) binding model, we calculated the binding parameters of NEIL1 and NEIL2 with copper (Fig. 5A). The KD values for copper binding for NEIL1 and NEIL2 were 430 and 930 nm, respectively. The small discrepancy in the affinity values from those measured by fluorescence could be ascribed to difference in the buffer conditions used in the two methods. In any case, both methods showed strong affinity of NEILs for Cu(II).
FIGURE 5.
Kinetic and thermodynamic parameters of copper binding to NEILs. A, SPR analysis of NEIL1 (a) and NEIL2 (b) binding to CuCl2 (1 nm to 10 μm) and calculated binding parameters (c). B, ITC analysis of NEIL1 binding to CuCl2. The binding isotherms (a) indicate ITC measurements for Cu(II) binding to NEIL1 (upper panel). The binding constant (KD), enthalpy (ΔH), and entropy (ΔS) were calculated from the plot of heat evolved/mol of Cu(II) versus the molar ratio of Cu(II) to NEIL1 using MicroCal ORIGIN 6.0 software (b).
Thermodynamic Parameters of NEIL1 Binding to Cu(II)
ITC studies with 10 μm NEIL1 bound CuCl2 at 25 °C showed stable binding, as indicated by the enthalpy change as a function of molar ratios of Cu(II) to NEIL1 (Fig. 5B). The binding constant, KD, suggested one-site binding (KD = 360 nm). The calculated stoichiometry (N) of 0.84. This indicates 1:1 binding, assuming that 84% of NEIL1 was active and competent for binding. The binding enthalpy (ΔH) and entropy (ΔS) were calculated to be −2.1 × 104 cal/mol and −37.2 cal/mol K, respectively (Fig. 5B, b). The large negative enthalpy indicates a strong exothermic process (45). Taken together, these data indicate stable, enthalpically driven Cu(II) binding to NEIL1.
Cu(II) and Fe(II) Alter Secondary Structure of NEIL1
We analyzed CD spectra to assess copper/iron-induced change in the NEIL1 secondary structure. A small but clear change in the secondary conformation of NEIL1 was observed in the presence of CuCl2 or FeSO4 (Fig. 6, A and B). A deep negative CD signal of NEIL1 at 205 nm and a smaller negative signal around 220 nm indicate mixed secondary conformation with about 39% helix, 33% sheet, and 19–25% random coil conformation. Both Cu(II) and Fe(II) shifted the negative signal at 205 nm and increased the 220 nm signal, indicating an increase in folding. On the other hand, MgCl2 used as control did not affect NEIL1 activity or NEIL1 conformation (Fig. 6C).
FIGURE 6.
Effect of copper/iron on the secondary structure of NEILs. A, NEIL1 plus CuCl2 (0.1 or 0.5 μm). B, NEIL1 plus FeSO4 (0.5 or 1 μm). C, NEIL1 plus MgCl2 (0.1 or 1 μm). D, NEIL2 plus CuCl2 (0.1 or 0.5 μm). The CD spectra were recorded at 195–260 nm, and CD contribution from metal alone was subtracted from metal-protein complex spectra.
Interestingly, unlike for NEIL1, the secondary structure of NEIL2 was not affected by copper (Fig. 6D) and iron (data not shown). It is possible that zinc in the zinc finger motif present in NEIL2 but absent in NEIL1 (46), could be replaced with Cu(II) or Fe(II) (17) without a major change in its conformation.
Finally, copper and iron binding did not induce oligomerization of NEIL1 and OGG1, as analyzed by blue native PAGE at the levels of CuCl2 and FeSO4 used in earlier studies (supplemental Fig. S4). However, a small amount of NEIL2 dimer was observed in the presence of both Cu(II) and Fe(II).
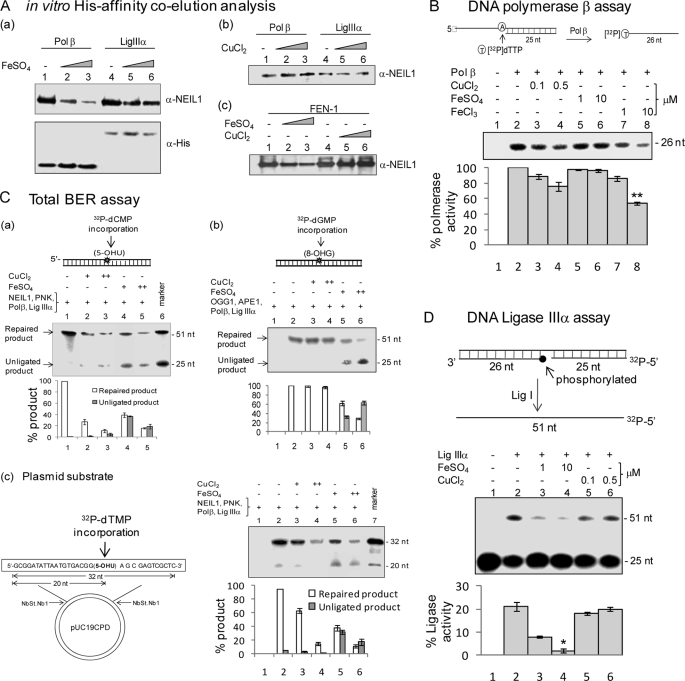
Cu(II) Disrupts the Interaction of NEIL1 with Polβ and FEN-1
We have previously shown that NEIL1 stably interacts with the downstream BER proteins Polβ, FEN-1, and LigIIIα and that these interactions are required for efficient, overall repair (30, 33). We used affinity co-elution analysis to test whether iron and copper affect the interaction of NEIL1 with these proteins. His-tagged Polβ, LigIIIα, or FEN-1 (50 pmol) was bound to Ni2+-NTA beads and then incubated with untagged NEIL1 (25 pmol), following its interaction with metals. After extensive washing, the bound NEIL1 was analyzed by 10% SDS-PAGE and immunoblotting. We observed ∼6-fold reduction in the interaction of NEIL1 with Polβ in the presence of 10 μm Fe(II) (Fig. 7A, a). Fe(II) also reduced the interaction of NEIL1 with FEN-1 (about 2-fold) but did not affect interaction with LigIIIα (Fig. 7A, b and c). Cu(II) did not affect any of these interactions. Western blotting with anti-His antibody showed a similar level of His-tagged proteins in the reactions (Fig. 6A, a, bottom).
FIGURE 7.
A, effect of Fe(II)/Cu(II) on NEIL1 interactions with Polβ (a and b) and FEN-1 (c). Affinity of NEIL1 for His-tagged Polβ, LigIIIα, or FEN-1 was analyzed in the presence of Fe(II) or Cu(II). After appropriate washing, the bound proteins were eluted for Western analysis with NEIL1 antibody (upper panel) or anti-His antibody (lower panel). B, effect of Cu(II) and Fe(II/III) on Polβ activity with a primer-template substrate (upper panel). The single nucleotide incorporation by Polβ was measured by the incorporation of [32P]dTMP in the absence or presence of iron/copper. **, p < 0.05. C, effect of Cu(II)/Fe(II) on NEIL1-initiated total BER with 5-OHU-containing oligonucleotide duplex (a) or plasmid substrate (c) and OGG1-initiated BER with 8-oxoG-containing duplex substrate (b). The 20-μl reaction contained 50 fmol of the indicated proteins, 2 pmol of substrate, 1 mmol of ATP, 25 μmol of unlabeled dNTPs, and 10 μmol of [α-32P]dNTP and the indicated amounts of CuCl2 (+, 0.1 μm; ++, 0.5 μm) or FeSO4 (+, 1 μm; ++, 10 μm). Product quantitation is shown in a histogram (lower panel). D, effect of Cu(II)/Fe(II) on DNA LigIIIα activity. The 5′-32P-labeled nicked duplex with 3′-OH and 5′-P ends at the nick was used for LigIIIα assay (upper panel). Quantitation is shown in the lower panel; *, p < 0.01. Error bars, S.E.
We next tested whether copper and iron affect Polβ activity using a primer-template substrate and measuring single-nucleotide ([32P]dTMP) incorporation (Fig. 7B). FeCl3 (10 μm) decreased the polymerase activity about 3-fold, whereas Cu(II) and Fe(II) did not have any significant effect. This further suggested that the mechanism of inhibition of NEIL1-Polβ interaction at least by Fe(II) involves Fe(II) binding to NEIL1 rather than its binding to Polβ.
Effect of Cu(II) and Fe(II) on NEIL1-initiated Total Repair
We recently characterized point mutants of NEIL1 and FEN-1 that disrupt their interaction without affecting individual activity but leading to significant reduction in total repair (30). Here we show that both Cu(II) and Fe(II) inhibited total repair of 5-OHU-containing duplex oligonucleotide by up to 80% at 10 μm FeSO4 and 0.5 μm CuCl2 (Fig. 7C, a). Interestingly, an increase in the fraction of unligated repair intermediate in the presence of FeSO4 (Fig. 7C, a, lanes 4 and 5), suggests that Fe(II) could independently affect LigIIIα activity. We similarly examined the effect of these metals on OGG1-initiated total repair using the same proteins except for substituting PNK with APE1. Cu(II) did not affect OGG1-initiated total repair, which was expected because OGG1 itself is not inhibited by Cu(II). However, as with NEIL1, Fe(II) increased the formation of unligated intermediate in OGG1-initiated repair (Fig. 7C, b).
Because metal ions could interfere with protein binding particularly to short oligonucleotides, we used plasmid substrate containing 5-OHU for repair, as described under “Experimental Procedures” (Fig. 7C, c, left). We used the reconstituted system with NEIL1, PNK, Polβ, and LigIIIα for repair, and incorporation of [32P]dTMP at the site of 5-OHU was monitored. Similar to their effect with oligonucleotide substrate, 0.5 μm Cu(II) and 10 μm Fe(II) reduced total BER by about 80%, and furthermore, Fe(II) affected the ligation step (Fig. 7C, c, right).
In view of the previous observation suggesting inhibition of LigIIIα by iron, we directly tested this with a 5′-32P-labeled nicked duplex substrate, where ligation converts the 26-nt substrate to a 51-nt product (Fig. 7D, top). FeSO4 inhibited LigIIIα activity by 60% at 1 μm and 85% at 10 μm, whereas copper (0.1–0.5 μm) had no effect (Fig. 7D, bottom). A recent report indicating that ZnCl2 strongly inhibited ligase activity (28) is consistent with our finding that accumulation of unligated repair products results from LigIIIα inhibition by Fe(II).
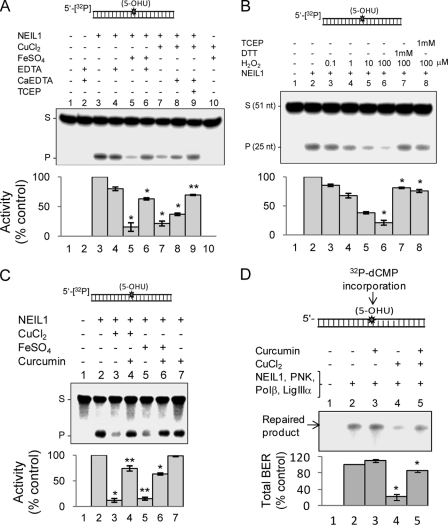
Metal Chelators and Curcumin Reverse Repair Inhibition by Copper/Iron
We then investigated the ability of various metal chelators to reverse the inhibition of NEIL1 due to iron/copper. NaEDTA restored repair activity by ∼75–85% to reverse Fe(II)-induced inhibition (Fig. 8A, lanes 2–6, p < 0.01). CaEDTA, a strong chelator of Cu(II), only partially reversed NEIL1 inhibition due to 0.5 mm copper (Fig. 8A, lane 8, about 30–40% at p < 0.01). However, the addition of both CaEDTA and TCEP, a reducing agent, restored NEIL1 activity to about 80% (Fig. 8A, lane 9). DTT was as effective as TCEP for reversing inhibition by cadmium as well as copper (data not shown). These results suggest that Cu(II)-induced inhibition partly results from oxidation of Cys residues. Previous studies have shown the ability of copper and cadmium to oxidize Cys residues in proteins (47, 48). To further test whether oxidation inhibits the activity of NEIL1, we treated NEIL1 with hydrogen peroxide (H2O2, 0.1–100 μm). Dose-dependent reduction in the glycosylase activity of NEIL1 was reversed by DTT or TCEP, indicating reversibility of Cys oxidation in NEIL1 (Fig. 8B).
FIGURE 8.
Prevention of iron/copper-induced inhibition of NEIL1 by chelators and curcumin. A, NEIL1 (50 fmol) was mixed with copper (0.5 μm) or iron (10 μm) and/or EDTA, CaEDTA, or TCEP (1 mm), and the NEIL1 activity was measured with 5-OHU duplex substrate as before. Constituents in each reaction are indicated. Lane 1, substrate alone; lane 2, substrate plus EDTA/CaEDTA; lane 10, substrate plus Fe(II)/Cu(II). B, effect of H2O2 (0.1–100 μm) on NEIL1 activity and reversal of inhibition by DTT or TCEP (1 mm) with 5′-32P-labeled 5-OHU duplex substrate. C, prevention of CuCl2 (0.5 μm)-induced and FeSO4 (1 μm)-induced inhibition of NEIL1 by curcumin (1 μm). Lane 1, substrate alone; lane 2, NEIL1 alone; lane 7, NEIL1 plus curcumin alone. D, prevention of CuCl2 (0.5 μm)-mediated inhibition of NEIL1-initiated total repair by curcumin (1 μm). *, p < 0.01; **, p < 0.05. S, substrate; P, product; Error bars, S.E.
We also tested the ability of curcumin to reverse iron/copper inhibition of NEIL1. Curcumin, a spice predominantly used in South Asia, has multiple beneficial effects (49, 50). Recently, curcumin was shown to have a strong metal chelating ability in addition to its antioxidant properties, as when two curcumin molecules bind to one Cu(II) (8, 51). We observed prevention of Cu(II)- and Fe(II)-mediated inhibition of NEIL1 in vitro by curcumin (Fig. 8C), which by itself did not affect the activity of NEIL1 (Fig. 8C, lane 7). Furthermore, inhibition of NEIL1-initiated total BER was reversed by 10 μm curcumin (Fig. 8D). Curcumin, in the absence of metals, slightly stimulated overall repair (Fig. 8D, lane 2).
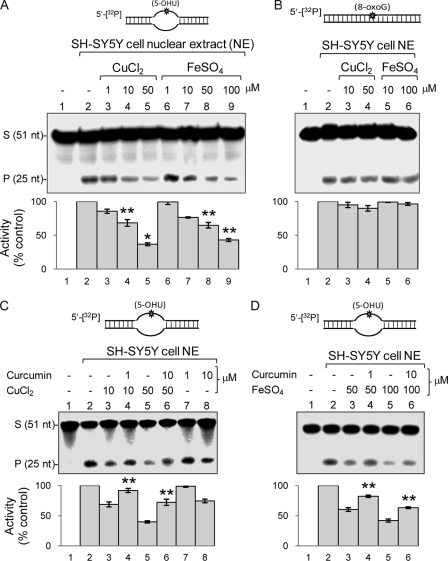
Inhibition of NEIL-specific Repair in SH-SY5Y Cells by Iron/Copper
We tested whether copper/iron inhibition of NEIL-mediated in vitro repair could be recapitulated in cells. Undifferentiated human neuroblastoma SH-SY5Y cells were treated twice with CuCl2 (1–50 μm) or FeSO4 (1–100 μm) for 24 h, as described under “Experimental Procedures.” The cells were then harvested, and NE was tested for DNA strand scission activity with a 5-OHU-containing bubble oligonucleotide to selectively measure total NEIL1 and NEIL2 activities because OGG1 and NTH1 are inactive with the ss substrate (25). Fig. 9A shows that NE from both copper-treated (50 μm) and iron-treated (100 μm) cells had significantly lower glycosylase/AP lyase activity than untreated cells (50–60%, p < 0.05). A single dose of copper (50 μm) or iron (100 μm) for 12 h also reduced the activity by about 30% (data not shown); however, about 60% reduction occurred with double dose for 24 h, which presumably overcame the threshold for cellular transition metal load, with accumulation of free metal ions. The ability of NE to excise 8-oxoG from duplex substrates, predominantly catalyzed by OGG1, was not affected by copper/iron (Fig. 9B), confirming that these metal ions inhibit NEILs exclusively. To test the presence of metals in the NE from SH-SY5Y cells (treated with FeSO4), we treated the NE with metal chelators EDTA or desferrioxamine (1 mm) and then measured strand cleavage with 5′-labeled 5-OHU-containing bubble substrate (supplemental Fig. S5). Both EDTA and desferrioxamine increased glycosylase activity by 2–3-fold, suggesting reversal of metal-mediated inhibition by chelation. This further confirmed that the decreased base excision activity of NE was primarily due to reversible inhibition of NEILs by iron/copper ions.
FIGURE 9.
Interference of Fe(II)/Cu(II) with NEIL-initiated repair in SH-SY5Y cells. A, after exposing cells to CuCl2 (1–50 μm) or FeSO4 (1–100 μm) as described under “Experimental Procedures,” repair was measured from the NE with 5′-32P-labeled 5-OHU-bubble (A) or 5′-32P-labeled 8-oxoG duplex substrate (B), which predominantly represent activity of NEILs and OGG1, respectively. C and D, prevention of copper/iron-mediated inhibition of repair by curcumin. Curcumin was added to cells after 1 h of treatment with CuCl2 (C) or FeSO4 (D). *, p < 0.01; **, p < 0.05. S, substrate; P, product; Error bars, S.E.
Curcumin Protects SH-SY5Y Cells from Iron/Copper-induced Inhibition of BER
We finally tested the protective effect of curcumin against copper/iron-mediated inhibition of NEIL activities in cells. Curcumin is non-toxic even at 50 μm, and its neuroprotective properties are well documented (52). After treating the SH-SY5Y cells with Cu(II) or Fe(II) for 1 h, curcumin (1–10 μm) was added to the medium. The NEIL activity was measured in NE isolated from these cells with 5-OHU-containing bubble substrate oligonucleotide as before. Curcumin (1 μm) completely protected (p < 0.05) the cells from the inhibitory effect of 10 μm copper (Fig. 9C, lanes 2–4), and 10 μm curcumin partially protected the cells even at 50 μm copper (Fig. 9C, lanes 5 and 6). Curcumin alone did not affect the 5-OHU repair activity in the NE (Fig. 9C, lanes 7 and 8). Similarly, curcumin was also able to protect the cells from the inhibitory effect of FeSO4 (Fig. 9D, p < 0.05).
DISCUSSION
The mechanisms involving neurotoxicity of essential transition metals are complex. In the normal tissue, the metal content is tightly regulated and released from the metal-storing proteins in response to metabolic need (53). For example, in the brain, the majority of iron (∼70%) is sequestered in ferritin and copper in ceruloplasmin (54). However, aging-related neurodegenerative disorders often involve gradual, clinically silent accumulation of free copper and iron, followed by sudden onset of toxicity when the metal load exceeds a threshold, leading to dysregulation in metal homeostasis and neuronal dysfunction (55). MRI imaging studies as well as post-mortem evaluations of brain samples of patients affected with neurodegenerative disorders confirmed the presence of elevated iron (2). Close association between brain metal dyshomeostasis and the onset and/or progression of AD and PD has been established in a number of studies, although the underlying biochemical mechanisms remain obscure (56). Recent studies have strongly implicated physiologically essential transition metals, such as copper, iron, and zinc and nonessential elements, such as aluminum, as key factors in the pathophysiology of AD and PD (3, 57). Indeed, very high levels of copper (400 μm) and zinc (1 mm) were found in amyloid plaques and AD neuropil regions in comparison with healthy brain (70 μm copper and 350 μm zinc) (58, 59). Progressive accumulation of metals in AD brain during disease progression from moderate to severe AD was also observed (3). Remarkably, the levels of Cu(II), Fe(II), and Zn(II) were shown to increase in the early phase of AD, whereas the increased levels of trivalent Fe(III) and Al(III) occurred in the later phase of AD, mainly in the frontal cortex and hippocampus. In general, a common trait of neurodegeneration appears to be (direct or indirect) perturbation in the homeostasis of copper, zinc, iron, aluminum, etc., leading to an increase in free metal ions in the affected cells. Moreover, increased iron level in the PD brain was not found to be associated with an increase in ferritin synthesis. On the contrary, decreased ferritin levels may lead to accumulation of excess free iron in PD (7).
The metal toxicity disorders could severely affect genomic integrity in brain cells with accumulation of oxidative DNA damage (60–62). Redox metals have been implicated in ROS generation and other free radicals, causing DNA damage (63). Although few earlier studies suggested inhibition of DNA repair activities by heavy metals (18, 64), the effect of physiologically normal or pathologically relevant levels of copper and iron ions on the repair of oxidized DNA bases in mammalian cells has not been characterized. Some recent studies showed that these metals could bind to DNA and interfere with damage scanning and detection by BER proteins and consequently reduce the efficiency of repair (28, 29). In the present study, we investigated the effect of copper and iron on the human DNA glycosylases, NEIL1, NEIL2, and OGG1. We showed that Fe(II/III) in micromolar and Cu(II) in submicromolar concentrations specifically inhibit the NEILs and not OGG1. We also showed for the first time that Cu(II) and Fe(II) stably bind to NEILs with high affinity. Earlier studies have shown that Cu(II) binding to proteins probably occurs through hydrophobic interactions, electrostatic interactions, or both (65). Our ITC data showed that the formation of Cu(II)-NEIL1 complex is enthalpically driven, which is unlikely to be due to hydrophobic interactions. Metal binding caused the conformational change of NEIL1, as indicated by CD and fluorescence analysis. In contrast, NEIL2 conformation was not significantly affected, although the intrinsic Trp fluorescence of NEIL2 was quenched. Additionally, partial reversal of copper-induced inhibition of NEIL1 by reducing agents suggested that the mechanism could involve oxidation of cysteine residues by copper. Previous studies have shown oxidation of cysteines in proteins by copper and cadmium (48).
We made an interesting observation that Fe(II) also inhibited the interaction of NEIL1 with Polβ and FEN-1. Further, both Cu(II) and Fe(II) inhibited NEIL1-initiated total repair but not of OGG1-initiated repair. We confirmed earlier reports (18, 66) that Cu(II) and Cd(II) at high concentration (>5 mm) inhibited OGG1 activity (data not shown). However, we have clearly shown that at physiologically relevant concentration, OGG1 is not affected by copper/iron.
Although both NEIL1 and NEIL2 were affected to similar extents by iron and copper, the mechanism(s) involved could be somewhat different. Inhibition of NEIL2 could at least partially involve displacement of the intrinsic Zn2+ in the zinc finger with Fe2+ or Cu2+. Earlier studies have shown the sensitivity of zinc finger proteins, such as bacterial Fpg and the mammalian XPA protein, to heavy metals that could compete for the zinc finger (67–69). However, NEIL1 lacks the zinc finger motif in its sequence, although it contains a motif named the zincless finger (70). This could explain why copper/iron-induced conformational change in NEIL1 is more pronounced than in NEIL2. It was previously shown that N-methylpurine-DNA glycosylase is inhibited by heavy metals, cadmium, nickel, and zinc at 50–1000 μm (71). Using a molecular dynamics simulation, the authors suggested that these metals bind to the N-methylpurine-DNA glycosylase active site, which has a potential zinc-binding site.
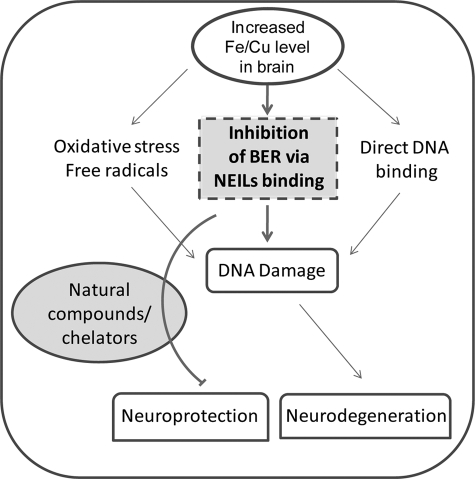
Our results showing that the NEIL-initiated BER is preferentially compromised in neuronal precursor SH-SY5Y cells by Fe(II) and Cu(II) raises the possibility that a similar situation could occur in the early phase of neurodegenerative disorders, with increased levels of divalent metals in the brain (3, 8). The higher metal concentration required to cause a similar level of inhibition in cell versus in vitro may be due to metal absorption efficiency of the cells as well as their sequestering by cellular proteins. We showed earlier that NEIL1 down-regulation in mammalian cells results in elevated oxidative damage in the genome and increased spontaneous mutations (72). Further, NEILs play a vital role in repair of oxidized bases in brain cells (27). Thus, if accumulation of metals is responsible for neuronal death, it is possible that oxidative genome damage is a proximal cause. Our results show that the excessive iron/copper not only cause DNA damage but could also inhibit repair of such damage in early neurodegenerative conditions (Fig. 10).
FIGURE 10.
Genome damage in neurodegenerative diseases due to elevated iron/copper level, a case of double jeopardy. Iron/copper cause oxidative genome damage due to ROS production. Iron/copper also inhibit NEIL-mediated repair of oxidized base damage. Reversal of inhibition by chelating agents, including curcumin, could provide neuroprotection.
Our observation that metal chelators, including curcumin, could reverse the iron/copper-mediated inhibition of NEIL-initiated repair both in vitro and in cells suggested the potential of a chelation approach for restoring repair after metal toxicity. Curcumin (1,7-bis(4-hydroxy 3-methoxyphenyl)-1,6-heptadiene-3,5-dione) is a common dietary pigment abundantly used in cuisines of several Asian countries and in traditional Indian medicine (49, 50). Curcumin is one of the three curcuminoids in turmeric (8). The phenolic and the enolic hydroxyls in curcumin could form metal complexes (51). Curcumin protects PC12 cells against apoptosis induced by a lethal dose of H2O2 and 1-methyl-4-phenylpiriinium ion (MPP+)-induced apoptosis (73). Recent studies have suggested that curcumin significantly reduces metal-induced neurotoxicity in rat hippocampal neurons (74, 75). It was proposed that the ability of curcumin to bind toxic metals and to form tight and inactive complexes could be a plausible pathway by which curcumin offers protection to the brain (76). In fact, the ability of curcumin to bind to several metals (51, 74, 77) and thereby potentially reduce their toxicity is well documented. The interaction of curcumin with copper and iron reaches half-maximum at ∼3–12 and 2.5–5 μm levels, respectively (77). Additionally, the antioxidant and anti-inflammatory properties of curcumin suggest its potential neuroprotective role (78).
In conclusion, prevailing studies show that elevated transition metals in neurodegenerative brain could cause genomic instability and DNA damage by direct metal toxicity on DNA or via oxidative DNA damage induced by metal-mediated excess free radical generation. These multiple toxic mechanisms of metal-mediated DNA damage have been schematically illustrated in Fig. 10. Our studies showed a new mode of transition metal toxicity of inhibition of NEIL1 and NEIL2 and other BER proteins via formation of metal-protein complexes that could play an important role in the accumulation of DNA damage in the early neurodegenerative conditions and highlight the potential for metal-targeted therapeutic approaches.
Supplementary Material
Acknowledgments
We thank Dr. Vince Hilser for the Biophysical Core Facility at UTMB. We acknowledge the help of Dr. T. Wood of the NIEHS Center for CsCl purification of pUC19CPD plasmid. We thank Dr. R. W. Bolen (Department of Biochemistry and Molecular Biology), Dr. Istvan Boldogh (Department of Microbiology and Immunology), and Mitra laboratory members, including Drs. A. K. Mantha and K. K. Bhakat at the University of Texas Medical Branch for valuable discussions. We also thank Wanda Smith for secretarial help.
This work was supported, in whole or in part, by National Institutes of Health, USPHS, Grants CA81063 and P01 CA092584 (to S. M.) and CA102271 (to T. K. H.). This research was also supported by an American Parkinson's Disease Association postdoctoral fellowship award (to M. L. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- AD
- Alzheimer disease
- BER
- base excision repair
- PD
- Parkinson disease
- Polβ
- DNA polymerase β
- FEN-1
- flap endonuclease 1
- LigIIIα
- DNA ligase IIIα
- ss
- single-stranded
- ITC
- isothermal titration calorimetry
- SPR
- surface plasmon resonance
- 5-OHU
- 5-hydroxyuracil
- HD
- Huntington disease
- PNK
- polynucleotide kinase
- TCEP
- tris-(2-carboxyethyl)-phosphine
- NE
- nuclear extract(s)
- nt
- nucleotide(s)
- 8-oxoG
- 8-oxoguanine.
REFERENCES
- 1.Lipscomb D. C., Gorman L. G., Traystman R. J., Hurn P. D. (1998) Stroke 29, 487–492; discussion 493 [DOI] [PubMed] [Google Scholar]
- 2.Zecca L., Youdim M. B., Riederer P., Connor J. R., Crichton R. R. (2004) Nat. Rev. Neurosci. 5, 863–873 [DOI] [PubMed] [Google Scholar]
- 3.Rao K. S. J., Rao R. V., Shanmugavelu P., Menon R. B. (1999) Alz. Rep. 2, 241–246 [Google Scholar]
- 4.Zatta P., Lucchini R., van Rensburg S. J., Taylor A. (2003) Brain Res. Bull. 62, 15–28 [DOI] [PubMed] [Google Scholar]
- 5.Koeppen A. H. (2003) J. Neurol. Sci. 207, 95–97 [DOI] [PubMed] [Google Scholar]
- 6.Petrat F., de Groot H., Sustmann R., Rauen U. (2002) Biol. Chem. 383, 489–502 [DOI] [PubMed] [Google Scholar]
- 7.Dexter D. T., Carayon A., Vidailhet M., Ruberg M., Agid F., Agid Y., Lees A. J., Wells F. R., Jenner P., Marsden C. D. (1990) J. Neurochem. 55, 16–20 [DOI] [PubMed] [Google Scholar]
- 8.Hegde M. L., Bharathi P., Suram A., Venugopal C., Jagannathan R., Poddar P., Srinivas P., Sambamurti K., Rao K. J., Scancar J., Messori L., Zecca L., Zatta P. (2009) J. Alzheimers Dis. 17, 457–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirsch E. C., Brandel J. P., Galle P., Javoy-Agid F., Agid Y. (1991) J. Neurochem. 56, 446–451 [DOI] [PubMed] [Google Scholar]
- 10.Dexter D. T., Carayon A., Javoy-Agid F., Agid Y., Wells F. R., Daniel S. E., Lees A. J., Jenner P., Marsden C. D. (1991) Brain 114, 1953–1975 [DOI] [PubMed] [Google Scholar]
- 11.Farah I. O., Trimble Q., Ndebele K., Mawson A. (2010) Biomed. Sci. Instrum. 46, 404–409 [PMC free article] [PubMed] [Google Scholar]
- 12.Banas A., Kwiatek W. M., Banas K., Gajda M., Pawlicki B., Cichocki T. (2010) J. Biol. Inorg. Chem., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yasuhara T., Hara K., Sethi K. D., Morgan J. C., Borlongan C. V. (2007) Brain Res. 1133, 49–52 [DOI] [PubMed] [Google Scholar]
- 14.Arai T., Fukae J., Hatano T., Kubo S., Ohtsubo T., Nakabeppu Y., Mori H., Mizuno Y., Hattori N. (2006) Acta Neuropathol. 112, 139–145 [DOI] [PubMed] [Google Scholar]
- 15.Rao K. S. (2007) Neuroscience 145, 1330–1340 [DOI] [PubMed] [Google Scholar]
- 16.Hart R. W., Setlow R. B. (1974) Proc. Natl. Acad. Sci. U.S.A. 71, 2169–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beyersmann D., Hartwig A. (2008) Arch. Toxicol. 82, 493–512 [DOI] [PubMed] [Google Scholar]
- 18.Zharkov D. O., Rosenquist T. A. (2002) DNA Repair 1, 661–670 [DOI] [PubMed] [Google Scholar]
- 19.Hegde M. L., Hazra T. K., Mitra S. (2008) Cell Res. 18, 27–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hazra T. K., Kow Y. W., Hatahet Z., Imhoff B., Boldogh I., Mokkapati S. K., Mitra S., Izumi T. (2002) J. Biol. Chem. 277, 30417–30420 [DOI] [PubMed] [Google Scholar]
- 21.Hazra T. K., Izumi T., Boldogh I., Imhoff B., Kow Y. W., Jaruga P., Dizdaroglu M., Mitra S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 3523–3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bandaru V., Sunkara S., Wallace S. S., Bond J. P. (2002) DNA Repair 1, 517–529 [DOI] [PubMed] [Google Scholar]
- 23.Morland I., Rolseth V., Luna L., Rognes T., Bjørås M., Seeberg E. (2002) Nucleic Acids Res. 30, 4926–4936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takao M., Kanno S., Shiromoto T., Hasegawa R., Ide H., Ikeda S., Sarker A. H., Seki S., Xing J. Z., Le X. C., Weinfeld M., Kobayashi K., Miyazaki J., Muijtjens M., Hoeijmakers J. H., van der Horst G., Yasui A., Sarker A. H. (2002) EMBO J. 21, 3486–3493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dou H., Mitra S., Hazra T. K. (2003) J. Biol. Chem. 278, 49679–49684 [DOI] [PubMed] [Google Scholar]
- 26.Podlutsky A. J., Dianova, Podust V. N., Bohr V. A., Dianov G. L. (2001) EMBO J. 20, 1477–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Englander E. W., Ma H. (2006) Mech. Ageing Dev. 127, 64–69 [DOI] [PubMed] [Google Scholar]
- 28.Li H., Swiercz R., Englander E. W. (2009) J. Neurochem. 110, 1774–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grin I. R., Konorovsky P. G., Nevinsky G. A., Zharkov D. O. (2009) Biochemistry 74, 1253–1259 [DOI] [PubMed] [Google Scholar]
- 30.Hegde M. L., Theriot C. A., Das A., Hegde P. M., Guo Z., Gary R. K., Hazra T. K., Shen B., Mitra S. (2008) J. Biol. Chem. 283, 27028–27037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Das A., Wiederhold L., Leppard J. B., Kedar P., Prasad R., Wang H., Boldogh I., Karimi-Busheri F., Weinfeld M., Tomkinson A. E., Wilson S. H., Mitra S., Hazra T. K. (2006) DNA Repair 5, 1439–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H., Hays J. B. (2003) J. Biol. Chem. 278, 28686–28693 [DOI] [PubMed] [Google Scholar]
- 33.Wiederhold L., Leppard J. B., Kedar P., Karimi-Busheri F., Rasouli-Nia A., Weinfeld M., Tomkinson A. E., Izumi T., Prasad R., Wilson S. H., Mitra S., Hazra T. K. (2004) Mol. Cell 15, 209–220 [DOI] [PubMed] [Google Scholar]
- 34.Prasad R., Kumar A., Widen S. G., Casas-Finet J. R., Wilson S. H. (1993) J. Biol. Chem. 268, 22746–22755 [PubMed] [Google Scholar]
- 35.Hill J. W., Hazra T. K., Izumi T., Mitra S. (2001) Nucleic Acids Res. 29, 430–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hazra T. K., Izumi T., Maidt L., Floyd R. A., Mitra S. (1998) Nucleic Acids Res. 26, 5116–5122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bharathi, Rao K. S. (2007) Biochem. Biophys. Res. Commun. 359, 115–120 [DOI] [PubMed] [Google Scholar]
- 38.Wiseman T., Williston S., Brandts J. F., Lin L. N. (1989) Anal. Biochem. 179, 131–137 [DOI] [PubMed] [Google Scholar]
- 39.Hegde M. L., Rao K. S. (2007) Arch. Biochem. Biophys. 464, 57–69 [DOI] [PubMed] [Google Scholar]
- 40.Niepmann M., Zheng J. (2006) Electrophoresis 27, 3949–3951 [DOI] [PubMed] [Google Scholar]
- 41.Dou H., Theriot C. A., Das A., Hegde M. L., Matsumoto Y., Boldogh I., Hazra T. K., Bhakat K. K., Mitra S. (2008) J. Biol. Chem. 283, 3130–3140 [DOI] [PubMed] [Google Scholar]
- 42.Hegde M. L., Shanmugavelu P., Vengamma B., Rao T. S., Menon R. B., Rao R. V., Rao K. S. (2004) J. Trace Elem. Med. Biol. 18, 163–171 [DOI] [PubMed] [Google Scholar]
- 43.Zharkov D. O., Rosenquist T. A., Gerchman S. E., Grollman A. P. (2000) J. Biol. Chem. 275, 28607–28617 [DOI] [PubMed] [Google Scholar]
- 44.Das A., Boldogh I., Lee J. W., Harrigan J. A., Hegde M. L., Piotrowski J., de Souza Pinto N., Ramos W., Greenberg M. M., Hazra T. K., Mitra S., Bohr V. A. (2007) J. Biol. Chem. 282, 26591–26602 [DOI] [PubMed] [Google Scholar]
- 45.Grossoehme N. E., Mulrooney S. B., Hausinger R. P., Wilcox D. E. (2007) Biochemistry 46, 10506–10516 [DOI] [PubMed] [Google Scholar]
- 46.Das A., Rajagopalan L., Mathura V. S., Rigby S. J., Mitra S., Hazra T. K. (2004) J. Biol. Chem. 279, 47132–47138 [DOI] [PubMed] [Google Scholar]
- 47.Bravard A., Campalans A., Vacher M., Gouget B., Levalois C., Chevillard S., Radicella J. P. (2010) Mutat. Res. 685, 61–69 [DOI] [PubMed] [Google Scholar]
- 48.Jalilehvand F., Mah V., Leung B. O., Mink J., Bernard G. M., Hajba L. (2009) Inorg. Chem. 48, 4219–4230 [DOI] [PubMed] [Google Scholar]
- 49.Aggarwal B. B., Sung B. (2009) Trends Pharmacol. Sci. 30, 85–94 [DOI] [PubMed] [Google Scholar]
- 50.Sharma R. A., Gescher A. J., Steward W. P. (2005) Eur. J. Cancer 41, 1955–1968 [DOI] [PubMed] [Google Scholar]
- 51.Barik A., Mishra B., Shen L., Mohan H., Kadam R. M., Dutta S., Zhang H. Y., Priyadarsini K. I. (2005) Free Radic. Biol. Med. 39, 811–822 [DOI] [PubMed] [Google Scholar]
- 52.Mendonça L. M., Dos Santos G. C., Antonucci G. A., Dos Santos A. C., Bianchi Mde L., Antunes L. M. (2009) Mutat. Res. 675, 29–34 [DOI] [PubMed] [Google Scholar]
- 53.Gaasch J. A., Lockman P. R., Geldenhuys W. J., Allen D. D., Van der Schyf C. J. (2007) Neurochem. Res. 32, 1196–1208 [DOI] [PubMed] [Google Scholar]
- 54.Texel S. J., Xu X., Harris Z. L. (2008) Biochem. Soc. Trans. 36, 1277–1281 [DOI] [PubMed] [Google Scholar]
- 55.Llanos R. M., Mercer J. F. (2002) DNA Cell Biol. 21, 259–270 [DOI] [PubMed] [Google Scholar]
- 56.Cuajungco M. P., Frederickson C. J., Bush A. I. (2005) Subcell. Biochem. 38, 235–254 [DOI] [PubMed] [Google Scholar]
- 57.Molina-Holgado F., Hider R. C., Gaeta A., Williams R., Francis P. (2007) Biometals 20, 639–654 [DOI] [PubMed] [Google Scholar]
- 58.Lovell M. A., Robertson J. D., Teesdale W. J., Campbell J. L., Markesbery W. R. (1998) J. Neurol. Sci. 158, 47–52 [DOI] [PubMed] [Google Scholar]
- 59.Huang X., Atwood C. S., Moir R. D., Hartshorn M. A., Vonsattel J. P., Tanzi R. E., Bush A. I. (1997) J. Biol. Chem. 272, 26464–26470 [DOI] [PubMed] [Google Scholar]
- 60.Hegde M. L., Gupta V. B., Anitha M., Harikrishna T., Shankar S. K., Muthane U., Subba Rao K., Jagannatha Rao K. S. (2006) Arch. Biochem. Biophys. 449, 143–156 [DOI] [PubMed] [Google Scholar]
- 61.Gupta V. B., Hegde M. L., Rao K. S. (2006) Curr. Alzheimer Res. 3, 297–309 [DOI] [PubMed] [Google Scholar]
- 62.Hartwig A., Schwerdtle T. (2002) Toxicol. Lett. 127, 47–54 [DOI] [PubMed] [Google Scholar]
- 63.Berg D., Youdim M. B. (2006) Top. Magn. Reson. Imaging 17, 5–17 [DOI] [PubMed] [Google Scholar]
- 64.McNeill D. R., Narayana A., Wong H. K., Wilson D. M., 3rd (2004) Environ. Health Perspect. 112, 799–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rasia R. M., Bertoncini C. W., Marsh D., Hoyer W., Cherny D., Zweckstetter M., Griesinger C., Jovin T. M., Fernández C. O. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4294–4299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hamann I., Schwerdtle T., Hartwig A. (2009) Mutat Res. 669, 122–130 [DOI] [PubMed] [Google Scholar]
- 67.Hartwig A., Asmuss M., Blessing H., Hoffmann S., Jahnke G., Khandelwal S., Pelzer A., Bürkle A. (2002) Food Chem. Toxicol. 40, 1179–1184 [DOI] [PubMed] [Google Scholar]
- 68.Hartwig A., Asmuss M., Ehleben I., Herzer U., Kostelac D., Pelzer A., Schwerdtle T., Bürkle A. (2002) Environ. Health Perspect. 110, Suppl. 5, 797–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Asmuss M., Mullenders L. H., Eker A., Hartwig A. (2000) Carcinogenesis 21, 2097–2104 [DOI] [PubMed] [Google Scholar]
- 70.Doublié S., Bandaru V., Bond J. P., Wallace S. S. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10284–10289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang P., Guliaev A. B., Hang B. (2006) Toxicol. Lett. 166, 237–247 [DOI] [PubMed] [Google Scholar]
- 72.Maiti A. K., Boldogh I., Spratt H., Mitra S., Hazra T. K. (2008) DNA Repair 7, 1213–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen J., Tang X. Q., Zhi J. L., Cui Y., Yu H. M., Tang E. H., Sun S. N., Feng J. Q., Chen P. X. (2006) Apoptosis 11, 943–953 [DOI] [PubMed] [Google Scholar]
- 74.Daniel S., Limson J. L., Dairam A., Watkins G. M., Daya S. (2004) J. Inorg. Biochem. 98, 266–275 [DOI] [PubMed] [Google Scholar]
- 75.Dairam A., Limson J. L., Watkins G. M., Antunes E., Daya S. (2007) J. Agric. Food Chem. 55, 1039–1044 [DOI] [PubMed] [Google Scholar]
- 76.Baum L., Ng A. (2004) J. Alzheimers Dis. 6, 367–377; discussion 443–449 [DOI] [PubMed] [Google Scholar]
- 77.Barreto R., Kawakita S., Tsuchiya J., Minelli E., Pavasuthipaisit K., Helmy A., Marotta F. (2005) Chin. J. Dig. Dis. 6, 31–36 [DOI] [PubMed] [Google Scholar]
- 78.Cole G. M., Teter B., Frautschy S. A. (2007) Adv. Exp. Med. Biol. 595, 197–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.