Abstract
Capping protein (CP) is a ubiquitously expressed, 62-kDa heterodimer that binds the barbed end of the actin filament with ∼0.1 nm affinity to prevent further monomer addition. CARMIL is a multidomain protein, present from protozoa to mammals, that binds CP and is important for normal actin dynamics in vivo. The CARMIL CP binding site resides in its CAH3 domain (CARMIL homology domain 3) located at or near the protein's C terminus. CAH3 binds CP with ∼1 nm affinity, resulting in a complex with weak capping activity (30–200 nm). Solution assays and single-molecule imaging show that CAH3 binds CP already present on the barbed end, causing a 300-fold increase in the dissociation rate of CP from the end (i.e. uncapping). Here we used nuclear magnetic resonance (NMR) to define the molecular interaction between the minimal CAH3 domain (CAH3a/b) of mouse CARMIL-1 and CP. Specifically, we show that the highly basic CAH3a subdomain is required for the high affinity interaction of CAH3 with a complementary “acidic groove” on CP opposite its actin-binding surface. This CAH3a-CP interaction orients the CAH3b subdomain, which we show is also required for potent anti-CP activity, directly adjacent to the basic patch of CP, shown previously to be required for CP association to and high affinity interaction with the barbed end. The importance of specific residue interactions between CP and CAH3a/b was confirmed by site-directed mutagenesis of both proteins. Together, these results offer a mechanistic explanation for the barbed end uncapping activity of CARMIL, and they identify the basic patch on CP as a crucial regulatory site.
Keywords: Actin, Biophysics, Cytoskeleton, NMR, Protein-Protein Interactions, CARMIL, Barbed Ends, Capping Protein
Introduction
Capping protein (CP)4 is a ubiquitously expressed, 62-kDa α/β heterodimer that binds the barbed end of the actin filament with high affinity (Kd = 0.1 nm) to prevent further actin monomer association and dissociation, thereby limiting the extent of filament elongation in vivo (1, 2). Consistent with such a central role in actin filament assembly, CP is one of only five proteins required for the reconstitution of actin-based motility in vitro (3–5), and cells lacking CP have profound deficiencies in actin cytoskeleton assembly (6–10).
Determination of the CP crystal structure led to the “tentacles” model of barbed end capping by CP (11). The two structurally homologous CP subunits form a central β-sheet, which comprises the bulk of the protein core, above which there are two antiparallel α-helices, one belonging to each subunit (11). At the end of these helices, each subunit contains a C-terminal “tentacle,” which, on CPα, is composed of an unstructured region punctuated in the middle by a short, 4-residue helix and, on CPβ, is composed of a longer amphipathic helix that protrudes from the protein core. Based on crystallographic evidence, it was proposed that these C-terminal tentacles are flexible in solution, allowing them to bind and cap the barbed end. Extensive mutational studies in yeast (12) and vertebrate (13) CP that focused on the tentacles provided strong support for the tentacles model of capping. Specifically, deletion of the α tentacle decreased the affinity of CP for the barbed end by 6000-fold and its on-rate by 20-fold, whereas deletion of the β tentacle decreased the CP affinity by 400-fold, with no effect on its on-rate (12, 13). Moreover, deletion of both tentacles rendered CP unable to cap filaments (12). These results indicated that both the α and β tentacles are important for capping, and they emphasized the greater importance of the α tentacle for fast association with the barbed end and for overall capping.5
Subsequently, the structure of CP bound to the barbed end was determined at 23 Å resolution by cryoelectron microscopy and was then fitted with the crystal structure of CP and the proposed F-actin structure (14–16). The resulting structure identified additional residues in CP involved in barbed end capping and led to the proposal of a two-step capping mechanism. First, conserved basic residues in the α tentacle, together with additional nearby basic residues from the core of CPβ, present a basic patch that interacts coordinately with a complementary acidic cluster on the barbed end. This electrostatic interaction would drive the initial association of CP with the barbed end as well as much of the overall binding strength. Second, with CP electrostatically bound to the filament end, the β tentacle undergoes conformational sampling in order to contact the hydrophobic cleft between subdomains 1 and 3 on the outside of the terminal actin subunit (14). This model requires that the β-tentacle be flexible, although this was not directly observed. Overall, this two-step capping mechanism incorporates all available structural and biochemical data to date.
Because actin monomers add mainly onto the barbed end of the filament, the rate of actin-based motility is thought to be largely controlled by the availability of free barbed ends (17). Because the primary barbed end capping protein in cells, CP is probably a major focal point for regulation. Indeed, the huge discrepancy between the half-life of CP bound to the barbed end in vitro (∼30 min) (18, 19) and in vivo (∼1 s) (20) suggests that CP activity is significantly controlled by regulatory molecules in vivo. In fact, several proteins have been found to bind CP to alter its activity (1, 2, 21–24). One possible cellular regulator of CP is V-1 (myotrophin), which binds CP in vitro with high affinity (Kd ∼40 nm) in a 1:1 complex that has no affinity for the barbed end (i.e. V-1 sequesters CP in a totally inactive complex) (23, 25). Moreover, V-1 is unable to accelerate the dissociation of CP already present on the barbed end (23, 25). These interactions represent a model for CP “sequestering” in which V-1 sequesters CP in an inactive complex and is completely incapable of uncapping CP-capped barbed ends.
Another likely cellular regulator of CP is CARMIL. CARMIL proteins comprise a recently identified family of molecules whose two most conspicuous features are a central, leucine-rich repeat domain and a C-terminal proline-rich domain (24, 26). Genes encoding CARMIL proteins have been identified in Acanthamoeba, Dictyostelium, Caenorhabditis elegans, Drosophila, mice, and humans (24, 26–30). The first CARMIL protein to be identified was Acan125 (31). This 125-kDa Acanthamoeba protein was discovered based on its ability to bind to the isolated Src homology 3 domain of Acanthamoeba myosin-IC, a monomeric unconventional myosin. Using a similar approach, Jung et al. (24) subsequently identified p116, the Dictyostelium homologue of Acan125. Importantly, the eluates of their myosin I Src homology 3 domain affinity columns contained not only p116, but also CP and the Arp2/3 complex. Immunoprecipitation reactions and other experiments provided evidence that p116 forms a complex with CP, Arp2/3, and myosin I in vivo and that p116 serves as the scaffold for assembly of this complex by binding CP, Arp2/3, and myosin I at independent sites. Given its central role in forming the complex, Jung et al. (24) gave p116 the name CARMIL for capping protein, Arp2/3, myosin I linker. Subsequent efforts to purify Acanthamoeba CARMIL provided evidence that CARMIL interacts very tightly with CP (32). Consistently, a GST fusion protein containing the C-terminal ∼200 residues of Acanthamoeba CARMIL was found to bind CP with an affinity of ∼10 nm (33).
Sequence alignments identify three regions of strong similarity within CARMIL family proteins that we refer to as CAH1–CAH3 (CARMIL homology domains 1–3) (33). Recent studies of both Acanthamoeba CARMIL (33) and mouse CARMIL-1 (mCARMIL-1; the product of one of three mouse CARMIL genes) (30) have established that the CAH3 domain forms the core of the CARMIL CP binding site. In the case of Acanthamoeba CARMIL, the CAH3 domain falls very near the protein's C terminus, and the isolated C-terminal 51-residue region of Acanthamoeba CARMIL, which contains the protein's entire CAH3 domain sequence, binds CP very tightly (Kd ∼10 nm by pull-down assay) (33). In the case of mCARMIL-1, mapping studies localized its CP binding site to an internal 123-residue fragment that encompasses the protein's CAH3 domain (30). This fragment, referred to as C-1, was estimated by kinetic modeling to bind CP with an affinity of ∼1.5 nm (30). Importantly, C-1 exhibits essentially the same affinity for CP that lacks both of its tentacles, indicating that the tentacles do not play a significant role in CAH3·CP complex formation. For all of these CAH3 domain-containing fragments, conversion of a highly conserved arginine residue (Arg993 in mCARMIL-1) to either an alanine or glutamate residue essentially abrogates their interaction with CP (30, 33). In addition, the double mutation R1104E/R1105E in the CAH3 domain of Acanthamoeba CARMIL essentially ablates its affinity for CP, whereas other mutations, such as H1093L/D, L1091E, and R1098E, affect affinity to a significant but lesser extent (33).
Bulk solution assays performed using CAH3 domain-containing fragments from both Acanthamoeba CARMIL (33) and mCARMIL-1 (30) argue strongly that this domain potently antagonizes CP function in two ways. First, assays in which CAH3 and CP are mixed together and then added to actin assembly assays indicate that the binding of CAH3 to CP dramatically reduces the affinity of CP for the barbed end. For the Acanthamoeba CARMIL CAH3 domain, the complex of CAH3 and CP was estimated to bind the barbed end with an affinity of ∼150 nm, down ∼1500-fold relative to CP alone (33). For the mCARMIL-1 CAH3 domain, the complex of C-1 and CP was estimated to bind the barbed end with an affinity of ∼30 nm, down approximately ∼300-fold relative to CP alone (30). Second, assays in which the CAH3 domain is added to actin filaments already capped with CP are consistent with the CAH3 domain-catalyzed removal of CP from the barbed end (i.e. with CAH3-driven uncapping). Importantly, the rapidity of this effect argues strongly that the CAH3-dependent acceleration of CP dissociation from the barbed end is due to the binding of CAH3 to CP at the barbed end, which then results in a rapid and large decrease in the affinity of CP for the filament end (as opposed to a mechanism in which free CAH3 serves merely as a sink for the slow, spontaneous dissociation of CP from the barbed end) (30, 33). This type of uncapping mechanism is completely consistent with the fact that the preformed complex of CP and CAH3 still has weak barbed end capping activity. Indeed, just as sequestering and the inability to uncap CP-capped barbed ends are probably mechanistically coupled in V-1, weak capping and barbed end uncapping are probably mechanistically coupled in CARMIL (34). Importantly, recent single-molecule imaging of CP-capped actin filaments following the addition of the CAH3 domain-containing fragment of mCARMIL-1 used here (CAH3a/b) has confirmed both the uncapping activity of the CAH3 domain and the ability of the CAH3-CP complex to cap the barbed end weakly (34).
Given the high affinity of CARMIL for CP, its potent anti-CP activities, and its cellular concentration (0.5 μm in both Acanthamoeba (32, 35) and fibroblasts5 versus ∼1 μm for CP (36), one would predict that the loss of CARMIL would significantly alter actin assembly-based processes in cells. Consistently, Dictyostelium cells in which the CARMIL gene has been rendered nonfunctional by homologous recombination exhibit striking defects in several actin-based processes, including chemotactic migration and macropinocytosis (24). Similarly, cultured vertebrate cells in which mCARMIL-1 has been knocked down using RNAi exhibit dramatic defects in cell migration and leading edge dynamics (30, 37).
The purpose of the work presented here was to use structural biology to provide a mechanistic explanation for the two signature biochemical activities of the CARMIL CAH3 domain: uncapping of CP-capped filaments and weak barbed end capping when CAH3 is in a complex with CP. Specifically, we used NMR chemical shift mapping and intermolecular paramagnetic relaxation enhancement experiments to determine the interaction surface between CP and a minimal CAH3 domain fragment from mCARMIL-1 (CAH3a/b). Our results reveal the overall binding topology of CAH3a/b on CP and offer a likely mechanism for the two anti-CP activities of CAH3a/b.
EXPERIMENTAL PROCEDURES
Protein Preparation
Mouse CPα1 and CPβ2 were co-expressed on a pET3d plasmid and purified as described previously (33, 38).
The mouse CAH3a/b domain, which had been previously isolated (33), was purified as before, with significant alterations. Escherichia coli BL21 DE3 cells were transformed with the pGEX-2T vector expressing the GST-CAH3a/b fusion protein and were grown to A600 of 2–3 and were then induced by adding isopropyl-β-d-thiogalactopyranoside to a final concentration of 1 mm and continued protein expression at 37 °C for 4–6 h. Cells were collected by centrifugation at 7,000 × g and resuspended in 3 ml/g 1× PBS (pH 7.4) supplemented with 1 mm DTT and complete protease inhibitor mixture (Roche Applied Science). Cells were lysed by passing through a high pressure cell homogenizer (Emulsi-Flex®-C3, Avestin, Inc., Ottawa, Canada) three times at 1500 p.s.i. Cell membranes and insoluble material were pelleted by centrifugation at 22,000 × g for 45 min at 4 °C, after which the supernatant containing CAH3a/b was removed for further purification. The soluble cell lysate was applied to column packed with glutathione-Sepharose (5 ml of resin/1 liter of culture) pre-equilibrated in 1× PBS. The resin was washed at 4 °C with 30 column volumes of 1× PBS. The column was equilibrated with 2 column volumes of TEV protease buffer (50 mm Tris-HCl, pH 8.0, 0.5 mm EDTA, 150 mm NaCl, 0.5 mm DTT), after which the resin was suspended in 1 column volume of protease buffer and 200 μl of TEV protease (generously supplied by Dr. Grace Liou) (A280 = 1.48) per 20 ml of resin. Digestion was carried out at room temperature with shaking for 4 h. The cleaved CAH3 was eluted with 1× PBS, and fractions containing the protein were pooled and applied to a reversed-phase HPLC column packed with C4 resin and eluted with increasing concentrations of acetonitrile. The fractions containing pure CAH3a/b were lyophilized, resuspended, and washed into NMR buffer (see below).
Actin was purified from rabbit skeletal muscle as described previously (39, 40). Monomeric actin was purified by gel filtration chromatography using S-300 resin to remove oligomers. G-actin was stored in G-buffer (0.2 mm ATP, 1 mm NaN3, 0.1 mm CaCl2, 0.5 mm DTT, and 2 mm Tris-HCl (pH 8.0)). Mg-actin in KMEI buffer (50 mm KCl, 1 mm MgCl2, 1 mm EGTA, 10 mm imidazole (pH7.0)) was labeled with pyrene (Invitrogen) and purified by centrifugation, dialysis, and gel filtration with G-25 resin. The concentrations of actin and pyrene were estimated using the extinction coefficients A290 = 26,600 m−1 cm−1 and A344 = 22,000 m−1 cm−1 (40). The concentration of labeled actin was determined by subtracting 0.127 times the A344 value from the A290 value.
NMR Samples
In general, NMR samples were prepared at ∼0.2–0.3 mm in 20 mm sodium acetate, pH 6.45, 100 mm KCl, 1 mm DTT, 0.5 mm EDTA, and 8% D2O. All NMR experiments were performed at 32 °C. The same conditions were used for CAH3, and sample homogeneity was confirmed by comparing the two-dimensional 1H-15N HSQC spectrum with a reference spectrum.
NMR Spectroscopy and Chemical Shift Indexing
All NMR experiments involving CP used TROSY selection and were performed on a Bruker 800-MHz spectrometer equipped with a pulsed-field gradient cryoprobe. CP backbone and 13Cβ assignments, as described elsewhere,6 were obtained using a combination of four-dimensional HNCACO and HNCOCA and three-dimensional HNCACB and HN(CO)CACB experiments. Of 548 expected backbone resonance peaks, 435 (79%) were observed, and of these, 91% were assigned, with unassigned residues mainly in the hydrophobic core of the protein (56).6 For the CARMIL CAH3 domain, experiments were recorded with a Bruker 600-MHz spectrometer equipped with a pulsed-field gradient cryoprobe. Sequential connectivities and amino acid residue identities were determined using three-dimensional HN(CO)CACBi - 1 and HNCACBi recorded with (1024 × 72 × 114) complex data points in the F1 (1H), F2 (15N), and F1 (CACB) dimensions, having spectral widths of 7184, 1582, and 8446 Hz. Despite its length (78 amino acids), the unstructured CAH3 domain H-N NMR spectrum contained significant spectral overlaps, and these were resolved with a set of high resolution three-dimensional HNCAi and HNCOi − 1 experiments recorded using (1024 × 62 × 140 (Cα) or 128 (C′)) complex data points, respectively, with spectral widths of 7184, 1582, and 4528 Hz (Cα) or 2500 (C′).
Secondary structure of CAH3a/b was inferred by comparing its secondary 13Cα and 13Cβ chemical shifts with a reference for random coil amino acids, and secondary structures were determined as described previously (41, 42). For CP, all experiments were recorded on a Bruker 800-MHz spectrometer equipped with a pulsed field gradient cryoprobe. All pulse sequences used for CP analysis contained TROSY selection, as described previously (43, 44), and gradient selection (45, 46) during the TROSY component.
Chemical Shift Mapping
Because binding between CP and CAH3 is tight (Kd = 1 nm), and the system was in slow chemical exchange on the NMR time scale, free CP and CAH3a/b-bound CP were assigned independently. CP resonances whose chemical shift changed by ≥0.1 or ≥0.015 ppm for CAH3a/b resonances (calculated as described previously (47) were considered significant and were used to determine the chemical shift maps.
Paramagnetic Relaxation Enhancement
CAH3a/b was paramagnetically labeled with 3-(2-iodoacetamido)-2,2,5,5-tetramethyl-1-pyrolidinyloxy, free radical (Toronto Research Chemicals Inc., catalog number I684000), which contained a nitroxide free radical. Five spin-labeled CAH3a/b samples were prepared. Wild-type CAH3a/b contained a single cysteine (Cys1008), which was labeled for one series of experiments, and in other samples, the native cysteine was mutated to serine, and a cysteine was introduced at the N terminus, Ser980, Val1026, or the C terminus. For the N-terminal label, a short extension of GSWGC replaced Ser964 for labeling at Cys963, and for the C-terminal label, a short TGC extension was added C-terminal to Val1038). Cysteine mutants were confirmed for binding CP by covalently labeling them with a fluorophore at their cysteine and measuring their Kd for CP binding by fluorescence anisotropy (data not shown). All CAH3 samples produced CD spectra characteristic of unfolded proteins, as did wild-type CAH3a/b. In order to produce a sufficient quantity of 2H-labeled CAH3a/b, a culture was grown in 2 liters of M9 minimal medium in D2O, and the mutant protein was purified in the same way as wild-type CAH3a/b. Prior to labeling, the purified protein was soaked in buffer containing 20 mm DTT for 30 min at room temperature in order to reduce disulfide bonds, which formed readily, and then exchanged into buffer lacking DTT. Spin labeling was carried out as described previously.7 The reaction was confirmed by LC-MS (data not shown), and no unlabeled protein was observed. The excess spin label was removed by reversed-phase HPLC using C4 resin and a 10–95% acetonitrile gradient. The spin-labeled CAH3a/b was added to 15N-, 2H-labeled CP at a 1.1:1.0 molar ratio. The diamagnetic sample was made by soaking labeled 2H-CAH3a/b with 20 mm ascorbic acid for 6 h at room temperature prior to binding with CP. The buffer was exchanged to remove ascorbic acid, and CP was added to the sample. A 15NH,1HN HSQC spectrum of each sample was recorded, and peak intensities were directly compared.
Structure Calculation
Chemical shift perturbations and PRE intensities were translated into distance restraints in XPLOR-NIH (48) calculations to determine the structure of the complex of CP and CAH3a/b. A total of 27 ambiguous restraints from CAH3a/b, consisting of residues experiencing chemical shift changes upon CP binding as well as residues within the CP-binding consensus motif were obtained. From CP there were 76 (20 from α, 56 from β) ambiguous restraints. Although 246 residues in CP experienced significant chemical shift changes, most were eliminated as restraints because they were far from the trace of CAH3a/b highlighted by PRE experiments. Additionally, the remaining ambiguous restraints were informed by PRE label sites on CAH3a/b. For example, residues on CPβ experiencing chemical shift changes were given ambiguous restraints to only residues between Ser980 and Cys1008 on CAH3 because this region of CPβ was between those that were affected by the two labels on CAH3a/b. PREs were translated into distances between atoms in CP experiencing an effect and each labeled side chain in CAH3a/b and were empirically grouped into bins of increasing target distances. Distance restraints from both data sets were enforced using a soft square potential with a switching value of 2.0 or 0.5 Å for chemical shift and PRE data, respectively. The structures were calculated using the simulated annealing protocol within NIH-XPLOR (48, 49) with both ambiguous and PRE restraints included and randomized starting positions for CP and CAH3a/b. CP was treated as a rigid body in structure refinement, except that the side chains were allowed to be flexible during dynamics calculations, whereas only experimentally obtained restraints were placed on CAH3a/b. Of 200 total structures calculated, the 10 lowest energy structures were chosen having no distance violations greater than 1.0 Å from either ambiguous or PRE distance restraints.
Biochemical Assays
Pyrene-actin polymerization assays were preformed as described previously (19) but using preformed actin filaments as seeds, supplied to the assay at 1 μm final concentration.
Capping protein and CAH3 variants used in titration calorimetry experiments were dialyzed overnight in the same buffer (30 mm potassium phosphate, pH 7.2, 90 mm KCl, 1 mm EDTA, 0.5 mm DTT). ITC titrations were performed at 37 °C using a VP-ITC calorimeter (MicroCal, Northampton, MA). CAH3 protein variants at the concentration of ∼70 μm were loaded into the syringe and injected in 3–5-μl aliquots at 400-s intervals into a 1.3-ml cell containing 4 μm CP protein. Data collection and analysis were performed with Origin software (OriginLab, Northampton, MA).
Fluorescence spectra were obtained using the Quantamaster spectrofluorometer (PTI, London, Canada). All measurements were performed at 20 °C and 285-nm excitation wavelength, with Glan polarizers at magic angle condition. Concentration of both CP and CAH3 constructs was 8 μm.
RESULTS
The Minimal CAH3 Domain of mCARMIL-1
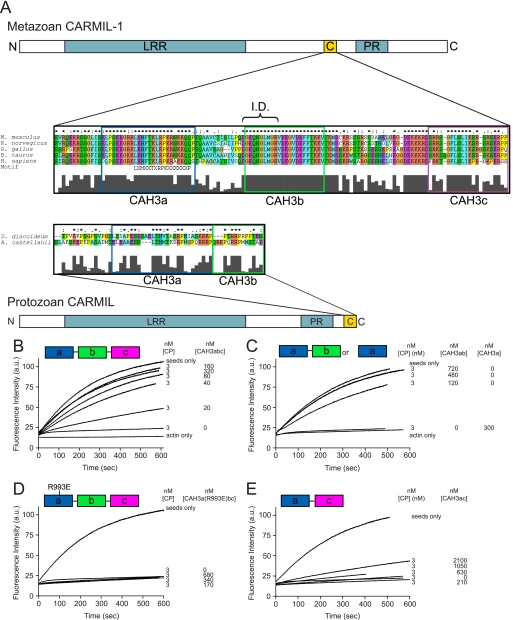
Based on sequence comparisons among and between protozoan and metazoan CARMIL proteins, we subdivide the CAH3 domain of protozoan CARMIL into two subdomains (CAH3a and CAH3b) and the CAH3 domain of metazoan CARMIL into three subdomains (CAH3a, CAH3b, and CAH3c) (Fig. 1A). The CAH3a subdomain is highly conserved across all phyla and contains the consensus sequence LXHXTKXRPKX6P that also is present in other proteins that bind CP, such as CD2AP/CIN85 and CKIP-1 (21, 22, 50, 51). In contrast, the CAH3b subdomain differs significantly between protozoan and metazoan CARMILs, with protozoan CAH3b being rich in basic residues and prolines, whereas metazoan CAH3b is rich in acidic and hydrophobic residues. Finally, the additional CAH3 subdomain specific to metazoan CARMILs, CAH3c, is rich in basic residues (Fig. 1A). The CAH3 domain of mCARMIL-1 used by Yang et al. (30) (which they referred to as C-1) corresponds essentially to CAH3a/b/c.
FIGURE 1.
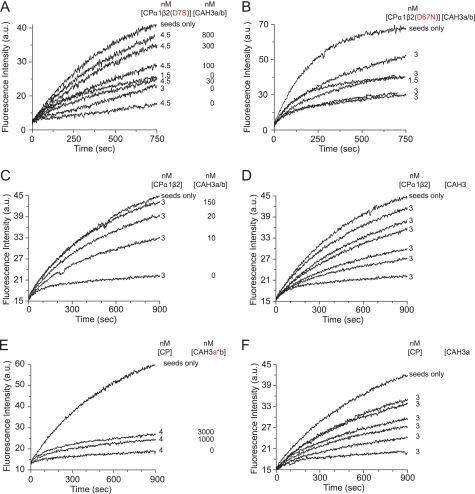
The organization of CARMIL proteins and the conserved CAH3 domain and the identification of CAH3a/b as the minimal CAH3 domain-containing fragment of mCARMIL-1. A, domain organization of protozoan and metazoan CARMIL proteins. Both contain a leucine-rich repeat domain (blue), a proline-rich (PR) domain (blue), and a CAH3 domain. The CAH3 domain (gold), which is necessary and sufficient for the anti-CP activities of CARMIL, is N-terminal to the PR domain in metazoan CARMIL proteins and C-terminal to the PR domain in protozoan CARMIL proteins. Sequence alignments of the CAH3 domains from several species are also shown. These alignments reveal that metazoan CARMIL CAH3 domains can be subdivided into three conserved subdomains, CAH3a, CAH3b, and CAH3c, whereas protozoan CARMIL CAH3 domains contain only the CAH3a and CAH3b subdomains. The consensus CP-binding motif LXHXTXXRPKX6P (21) is indicated, as is the capping “interference domain” (I.D.) defined in this work. B–E, shown are actin polymerization assays using 10% pyrene-labeled G-actin, actin seeds, and the indicated concentrations of CP and the following CAH3 domain fragments of mCARMIL-1: CAH3a/b/c (B), CAH3a/b or CAH3a only (C), CAH3a(R993E)/b/c (D), or CAH3a/c (E). Actin polymerization was monitored by pyrene fluorescence. The CAH3a, CAH3b, and CAH3c subdomains are shaded green, pink, and blue, respectively.
To determine the minimal portion of the mCARMIL-1 CAH3 domain (CAH3a/b/c) that is required for potent anti-CP activity, we generated recombinant fragments containing the CAH3a, -b, and -c subdomains in various combinations and tested their ability to antagonize CP function in solution-based polymerization assays. In our standard assay, 3 nm CP was sufficient to totally inhibit the elongation of actin filament seeds (Fig. 1, B–E). The addition of CAH3a/b/c (Ser964–Pro1086) inhibited CP activity in a dose-dependent manner, with 160 nm CAH3a/b/c sufficient to yield elongation rates similar to those of seeds alone (Fig. 1B). This result is consistent with previous studies using C-1 from mCARMIL-1 (30) as well as the CAH3 domain of Acanthamoeba CARMIL (33). Like CAH3a/b/c, CAH3a/b (Ser964–Val1038) was also sufficient for potent anti-CP activity, with ∼480 nm CAH3a/b sufficient to return actin polymerization to the seed-only rate (Fig. 1C). In contrast, CAH3a alone (Ser964–Gln1019) could not effectively antagonize CP because assays containing 300 nm CAH3a showed no restoration of polymerization (Fig. 1C). Previously, mutation of R993E in the CAH3a subdomain of both protozoan and metazoan CAH3 domains was shown to result in an almost complete ablation of their affinity for CP and their anti-CP activities (30, 33), and this mutation has been used as a simple way to block CAH3 domain function. Consistently, CAH3a(R993E)/b/c exhibited no anti-CP activity even at concentrations approaching 1 μm (Fig. 1D). This result also argues that CAH3b/c is not sufficient for potent anti-CP activity. Moreover, a chimeric fragment containing the CAH3a subdomain fused directly to the CAH3c subdomain (CAH3a/c; Ser964–Gln1019 plus Thr1039–Pro1086) exhibited minimal anti-CP activity, requiring a concentration of ∼2 μm to see even minimal recovery of actin polymerization (Fig. 1E). Based on these results, we define the minimal, fully functional CAH3 domain of mCARMIL-1 as containing only its CAH3a and CAH3b subdomains, and we performed all of our structural and functional studies in this work using this fragment. We thus refer to this fragment simply as CAH3a/b.
NMR Resonance Assignment and Chemical Shift Mapping of CAH3a/b Show That It Is Unstructured and Experiences Changes in Multiple Regions upon CP Binding
To characterize the structure of CAH3a/b, NMR backbone resonances were assigned. The 1H,15N HSQC spectra of both free and CP-bound CAH3a/b were both indicative of a random coil protein, having 1HN resonances largely between 7.7 and 8.5ppm (supplemental Fig. S1). Because CAH3a/b binds CP tightly and is in slow exchange with its free form, NMR resonances of CAH3a/b in its CP-bound form were independently assigned. Comparison of 13Cα and 13Cβ secondary shifts (41, 42) showed that CAH3a/b is unstructured in solution in both its free and CP-bound forms (supplemental Fig. S2).
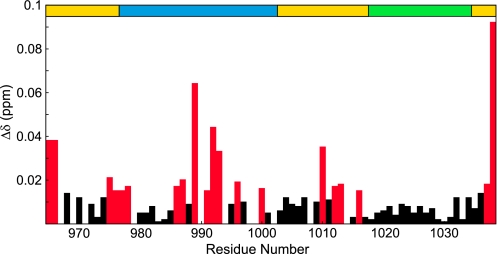
To determine the regions of CAH3a/b involved in binding CP, chemical shift mapping experiments were used to identify residues experiencing changes in local chemical environment upon binding. The chemical shift map of CP binding on CAH3 revealed chemical shift changes in multiple, discrete areas spanning the entire length of CAH3 (Fig. 2). Significant changes (Δδ ≥ 0.015 ppm) were observed at residues Val965 and Arg966 of CAH3a/b, and smaller, but still significant changes were observed at Ile975–Leu978. These residues are primarily N-terminal to the CAH3a subdomain, which begins at residue Glu977. The CAH3a subdomain, which contains the CP-binding motif, as well as the basic cluster previously shown to be involved in binding CP (33), experienced the largest chemical shift changes upon CP binding, with average Δδ = 0.016 ppm. For the conserved CP-binding motif (Leu986–Pro1002) within the CAH3a domain, the average chemical shift change was larger (0.022 ppm), indicating that this motif indeed is involved in CP binding. Other significant chemical shift changes were observed from Ile1010 to Asp1016, a sequence that lies between the CAH3a and CAH3b subdomains. Finally, the only significant chemical shift changes observed in the CAH3b subdomain were at the two C-terminal residues of CAH3a/b, Lys1037 and Val1038. This suggests that the CAH3b subdomain does not make the extensive contacts required for the tight binding of the CAH3a/b domain to CP.
FIGURE 2.
Chemical shift map of CP binding on CAH3a/b. Shown are the chemical shift change magnitudes of residues in CAH3a/b, plotted versus residue number. Significant values (Δδ > 0.015 ppm) are highlighted red. The domain organization of CAH3a/b is shown at the top (gold), with the CAH3a subdomain (blue) and the CAH3b subdomain (green) highlighted.
NMR Chemical Shift Mapping Reveals Extensive Changes in CP upon CAH3a/b Binding
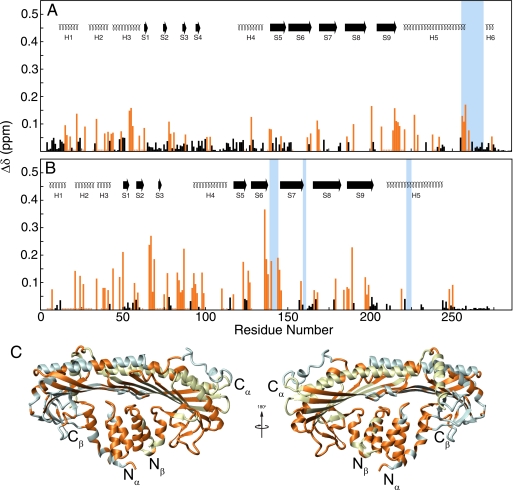
The chemical shift changes in CP upon CAH3a/b binding were extensive (supplemental Fig. S3) (significant changes for 111 residues in CPα and 109 residues in CPβ) and involved almost all of CP, although changes on CPβ (Fig. 3B) were in general larger in magnitude than those in CPα (Fig. 3A). In CPα, the edges of the peripheral loop domain (Lys66–Thr77, Leu82–Phe87, Gln93–Asp99, and Glu104–Lys118) and in helix 4 (Ala130–Ser137) were unaffected by CAH3a/b binding (Fig. 3A), whereas in CPβ, almost all of the residues in the corresponding regions were affected by CAH3a/b binding (Fig. 3B). Surprisingly, the only other extensive regions not experiencing chemical shift changes upon CAH3a/b binding were the C-terminal regions of both subunits, from Thr265 to the C terminus in CPα and from Asp251 to the C terminus in CPβ. These regions contain the C-terminal tentacles shown previously to be necessary for barbed end capping (12, 13). The surface area on CP experiencing significant chemical shift perturbations was larger than the total surface of CAH3a/b, suggesting that some chemical shift changes were probably due to a conformational change in CP. Despite these extensive changes in 1HN,15NH chemical shift, secondary structure was conserved in CP upon CAH3a/b binding (data not shown), indicating that the conformational changes may involve a subtle rearrangement of the structure of CP rather than a dramatic allosteric effect.
FIGURE 3.
Chemical shift map of CAH3a/b binding on CP. Shown are the chemical shift change magnitudes of residues in CPα (A) and CPβ (B), plotted versus residue number. Significant values (Δδ > 0.10 ppm) are highlighted orange. The secondary structures for each subunit are shown at the top.7 Residues experiencing chemical shift changes are highlighted on the crystal structure of CP (C), including residues qualitatively assigned as being affected by CAH3a/b binding.
Residues within the basic patch on CP, shown previously to be critical for CP association with and overall affinity for the barbed end (13, 14), experienced large chemical shift changes upon CAH3a/b binding (Fig. 3). In CPα, one cluster that showed large chemical shift changes was the “joint” of the α tentacle (for Lys256–Gln260, average Δδ = 0.120 ppm). In CPβ, residues comprising the loop between strands 6 and 7 of the central β-sheet (Lys136–Val146) experienced the largest chemical shift changes of all of those observed in CP (average Δδ = 0.174 ppm). This loop contains two residues that contribute to the basic patch, Lys142 and Lys143. Finally, the majority of chemical shift changes in CP occurred opposite its actin binding surface, composed of the two long antiparallel α-helices (H5 in CPα and CPβ) and the N-terminal region of each subunit (Fig. 3C). This indicates that CAH3a/b interacts primarily with the N-terminal domains of CP, although it also affects the basic patch.
Multiple Paramagnetic Labels Identify the CAH3 Binding Site on CP
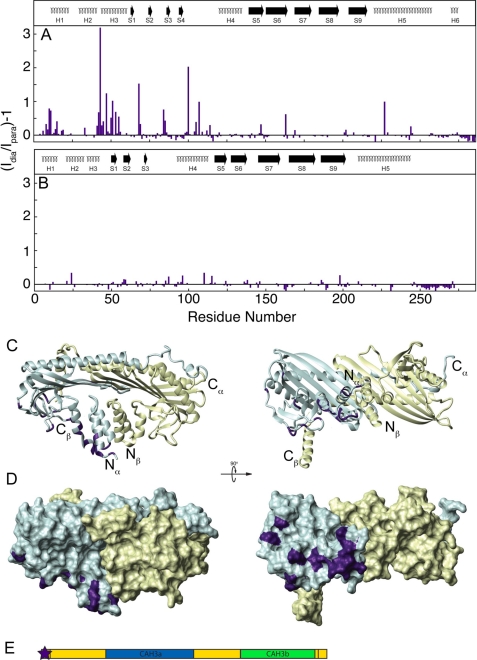
To obtain a clearer picture of the binding interface between CAH3a/b and CP as well as to determine the relative orientation of the proteins, intermolecular paramagnetic relaxation enhancement experiments were performed using CAH3a/b containing a paramagnetic spin label. This technique involves the introduction at a specific site in a protein of a paramagnetic label, which increases the magnetic relaxation of nearby (≤∼25 Å) atoms in a distance-dependent manner. The increase in relaxation rate of each resonance peak can be translated into its distance from the paramagnetic site and used as a discrete distance restraint in structure calculations (52, 53). Because CAH3a/b is unstructured and affects a large area on CP, the information provided by a single spin label would give little information about distant sites on CAH3a/b. Therefore, spin labels were introduced via native or added cysteine residues at five positions throughout the CAH3a/b sequence in order to probe the entire CAH3a/b binding interface on CP: N terminus to Ser964, C terminus to Val1038, and at internal positions S980C, C1008, and V1026C (see “Experimental Procedures”). Together, labeling at these sites provided a map of the trace of CAH3a/b binding on CP. For example, CAH3a/b containing a paramagnetic label introduced at its N terminus produced changes only in CPα, centered between its N-terminal helix bundle (at residues Val8, Glu12–Gly27, Glu31–Asn44, Leu46, Gly49–Ala51, and Phe54-Ala55) and its peripheral loop region (at residues Glu68-Gly69, Asn84-Ser85, His100, Ala105, Asp107, Asp114, Thr147, and Phe163) (Fig. 4, A–D). The CAH3a/b samples labeled at other sites produced effects in other regions of CP (supplemental Figs. S4–S7). Collectively, the results of these intermolecular PRE experiments showed that residues Ser964–Cys1008 of CAH3a/b make a loop from the N-terminal helix bundle on CPα, around the N-terminal helix bundle and peripheral loop domain of CPβ. CAH3a/b residues Thr1009–Val1038 then extend across the central β-sheet loops present in both CP subunits. CP resonance peak intensity changes resulting from PRE in the presence of each labeled CAH3a/b protein were translated into distance restraints, which were then used to determine the model of the CP·CAH3a/b complex.
FIGURE 4.
Intermolecular paramagnetic relaxation enhancement from N-terminally labeled CAH3a/b to CP. For this sample, a short extension of GSWGC replaced the CARMIL CAH3a/b N terminus (Ser964) for labeling through an iodoacetamido-reactive spin label to the -SH group of Cys963. The intensity ratio of each CP resonance peak in its diamagnetic form compared with its paramagnetic form is plotted versus residue number for CPα (A) and CPβ (B). The secondary structures for each subunit are shown at the top, as in Fig. 2. The ribbon diagram in C, where the N and C termini of each subunit are indicated, shows the orientation of the surface diagram in D and of those in supplemental Figs. S4-S7. CP subunits are shaded the same as in Fig. 2. Residues experiencing significant PRE (Id/Ip > 1.7) are highlighted on CP in D in purple. The location of the paramagnetic label is shown as a star on the domain diagram of CAH3a/b (E), where the coloring is the same as in Fig. 2.
Interestingly, in spectra obtained using CAH3a/b labeled at position V1026C and bound to CP, some resonance peak frequencies were shifted compared with those from wild-type CAH3a/b bound to CP. This indicates that introduction of the spin label at V1026C interferes with this portion of the CAH3a/b CP binding site without significantly affecting the affinity. When the chemical shifts of CP bound to this mutant were compared with those of CP bound to wild-type CAH3a/b, differences localized to two main regions (supplemental Fig. S8). The first was the loop between CPβ β-strands 6 and 7 in the central β-sheet. Interestingly, this loop contributes two residues, Lys142 and Lys143, to the basic patch on CP, which is responsible for much of the binding affinity of CP for the barbed end. The second region was the loop between β-strand 9 and α-helix 5 in CPα, which coincides with the region experiencing PRE (supplemental Fig. S8). Because this region corresponds to regions experiencing PRE, these changes were probably due to direct interactions with the paramagnetic label, resulting in an exchange of free and bound conformations of these residues, which were reflected in the trend of the observed chemical shift changes toward those of free CP. Thus, because PRE effects observed using CAH3a/b labeled at this site contained contributions from bound and unbound exchanges, distance restraints were effectively larger than those obtained from labeling at other sites in CAH3a/b. However, because labeling within the CAH3b subdomain affects residues in the basic patch on CP, which is important for barbed end capping, we take this as strong evidence that binding by the CAH3b subdomain is involved in anti-CP activity.
Structure of the CP·CAH3a/b Complex
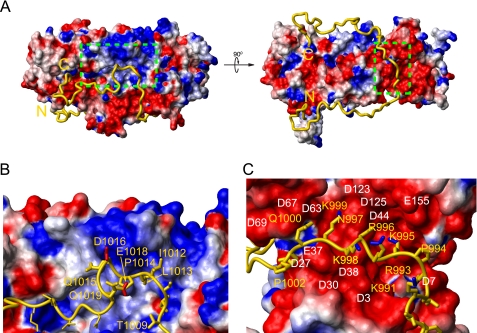
Using ambiguous restraints generated from chemical shift mapping as well as distance restraints from PRE experiments, 200 structures of CAH3a/b bound to CP were calculated. No residual dipolar coupling values were obtained for restraints during structure calculations. The presence of CP in an NMR sample composed of aligned phage Pf1 prevented deuterium splitting in the sample, indicative of CP destroying the integrity of the phage alignment (data not shown). Based on its propensity for interacting with both protein and lipid samples, CP could not be observed in the presence of alignment media for measuring residual dipolar coupling values. We present the 10 lowest energy structures (supplemental Fig. S9) and their statistics (Table 1). Because CAH3a/b was known to be unstructured in solution, and no secondary structure was induced upon CP binding, CAH3a/b was unrestrained during structure calculations, whereas the CP backbone was treated as a rigid body. Overall, the model of the CP·CAH3a/b complex shows that CAH3a/b makes extensive contacts with both CP subunits and forms a “U” shape by wrapping around the N-terminal helix bundles of CP (Fig. 5A). The CP-CAH3a/b interaction is dominated by two striking features. First, the CAH3a subdomain binds across a large acidic groove between the CPβ N-terminal helix bundle and the peripheral loop, composed of residues Asp2, Asp7, Glu37, Asp38, Asp44, Asp63, Asp67, Glu84, Asp85, Asp123, Asp125, Glu155, and Glu198 (Fig. 5C). Specifically, residues Arg984-Pro1002 from CAH3a, containing the consensus CP-binding motif (LXHXTXXRPKX6P), lie extended across this groove on the surface of CPβ. This binding site is characterized by extensive electrostatic complementarity between the largely basic CAH3a subdomain (+7 net charge) and CPβ. Within this basic region of the CAH3a subdomain, the side chain of the critical CP-binding residue Arg993 extends into a pocket in the acidic groove composed of Asp7, Asp11, Glu22, Asp38, and Asp44 from CPβ (supplemental Fig. S10). In our model of this interaction, the side chain of Arg993 most likely forms a salt bridge with the side chain from Asp7 of CPβ. Notably, the CP binding motif contains a proline residue (Pro1002), which, in our structure, forms a turn, orienting CAH3b toward the actin binding surface on CP. C-terminal to the CAH3a subdomain, several residues between the CAH3a and CAH3b subdomains (Ile1010–Pro1014) align near a narrow hydrophobic region on the surface of CPβ composed of β-sheet 1, the loop formed by residues Glu113-Val116, and β-strand 5, between the acidic groove and the basic patch (Fig. 5B).
TABLE 1.
Structure statistics of the 10 lowest energy CP·CAH3a/b complex NMR structures
Shown are structure statistics of the 10 lowest energy structures of the CP-V-1 complex calculated using XPLOR-NIH using rigid body refinement of CP with ambiguous and PRE restraints only.
| Parameters | Values |
|---|---|
| Backbone r.m.s. deviation (Å) of CAH3 with CP coordinates fixed | |
| All backbonea | 8.76 ± 1.94 |
| Backbone r.m.s. deviation (Å) of CAH3 R993-S1011 with CP coordinates fixed | |
| All backbone | 3.42 ± 0.89 |
| Backbone r.m.s. deviation (Å) of complex with respect to the mean | |
| Interfaceb backbone | 1.07 ± 0.39 |
| All backbone | 4.05 ± 1.02 |
| No. of restraints | |
| Ambiguous from CPα | 19 |
| Ambiguous from CPβ | 61 |
| Ambiguous from CAH3a/b | 27 |
| PRE (N terminus) | 18 |
| PRE (S980C) | 20 |
| PRE (C1008) | 24 |
| PRE (V1026C) | 6 |
| PRE (C terminus) | 33 |
| Interaction surface area (Å2)c | 4451 ± 510 |
a Backbone residues were defined as NH, Cα, and C′.
b Interface residues were defined as those experiencing significant chemical shift perturbations upon binding, as defined under “Experimental Procedures.” Interface backbone residues included, from CPα, residues 7, 8, 12–27, 31–44, 46, 49–51, 54–65, 78–81, 88, 92, 100–103, 113, 114,119–123, 128, 129, 138–143, 148, 151–153, 162–164, 168, 169, 172, 182, 186, 187, 189, 190, 202, 208–221, 228, 229, 238–242, 255–262; from CPβ, 4–7, 12–31, 34, 38–50, 55–59, 62–99, 110, 111, 113, 116, 117, 123–128, 136–145, 158–159, 166–168, 178, 179, 184–190, 195–201, 221, 244, 248, 250, 251; and all residues from CAH3a/b.
c Interaction area calculated using a sphere of radius 1.4 Å.
FIGURE 5.
The structure of the CP·CAH3a/b complex. CP is displayed as a surface charge diagram (A) in the same orientation as in Fig. 3. CAH3a/b (gold) is shown as a ribbon with its N and C termini indicated. The green boxed area on the left in A indicates the basic patch on CP and is shown enlarged in C. The boxed area on the right in A represents the acidic groove on CP and is shown enlarged in B. Selected residues from CAH3a/b (yellow) and CPβ (white) are indicated in B and C, and the side chains of CAH3a/b residues are shown as gold sticks with side chain C–NH bonds highlighted blue in B and side chain C–O bonds highlighted red in C.
The second striking feature of our CP·CAH3a/b structure is that C-terminal to the turn at Pro1002, the N-terminal part of the CAH3b lies directly adjacent to the basic patch on CP (Fig. 5B). Specifically, the region of CAH3a/b from Thr1009 to Gln1019, including part of the interdomain region and CAH3b, is situated just below the two loops between strands 6 and 7 and between strands 8 and 9 of the central β-sheet of CP. This region of CAH3a/b contains several hydrophobic residues that interact with a hydrophobic surface on CP adjacent to its basic patch. Additionally, the side chain of Asp1016 within this part of CAH3a/b is oriented toward the basic patch. The loops between strands 6 and 7 (Gly138–Lys143) and between strands 8 and 9 (Lys181–Ser184) in the central β-sheet of CPβ contribute three important residues (Lys142, Lys143, and Lys181) to the basic patch, and mutation of these residues significantly decreases the ability of CP to cap barbed ends.7 The interaction of CAH3a/b with this region probably confers its potent anti-CP activity (see “Discussion”). C-terminal to this interaction site, the CAH3b subdomain is additionally aligned between the central β-sheet and the N-terminal helix bundle of CPα, forming several hydrophobic contacts with CP.
Site-directed Mutagenesis Confirms the Mechanism of Weak Capping/Uncapping
Based on the above structure, we sought to identify specific residues in both CP and CAH3a/b whose interactions are required for weak capping by the complex and uncapping by CAH3a/b. Two mutations were introduced into CP to attenuate its interaction with CAH3a/b. Because the CAH3a/b binding interface on CP was large, we focused our site-directed mutagenesis on the acidic groove of CP determined here to bind the CAH3a subdomain, which is necessary for tight CP binding. CP containing the mutation D7S in its β-subunit showed a significant decrease in sensitivity to inhibition by CAH3a/b because actin polymerization was not restored even at a concentration of 800 nm CAH3a/b (Fig. 6A) (compared with wild-type CP, where 150 nm CAH3a/b was sufficient to return polymerization to the seed-only rate (Fig. 6C)). Mutation of the two other acidic residues (D3S and D11S) that are close to Asp7 in the same helix of CP had no effect on CAH3a/b binding, indicating that among these three aspartates, Asp7 is the important one for CP·CAH3a/b complex formation (data not shown). A second CP mutant containing the mutation D67N in CPβ was also resistant to antagonism by CAH3a/b because a concentration of 800 nm CAH3a/b resulted in only a partial restoration of the rate of actin polymerization (Fig. 6B). Together, these two CP mutants show that residues in the acidic groove are important for the tight binding of CP to CAH3a/b, consistent with our structure of the complex.
FIGURE 6.
Site-directed mutagenesis of CP and CAH3a/b. Pyrene-actin polymerization assays were performed as in Fig. 1. The concentrations of CP and CAH3a/b are indicated for each experiment. Two CP mutants were tested for their responsiveness to CAH3a/b (A and B). CAH3a/b mutants were tested for their ability to rescue actin polymerization (C–F). CAH3a/b (A–C) corresponds to residues Ser964–Val1038 of mCARMIL-1, CAH3a (D) corresponds to residues Ser964–Gln1019, CAH3a*b (E) contains the R993E mutation, and CAH3ab* (F) contains Val1026, Val1030, Phe1033, and Phe1034 mutations. a.u., absorbance units.
Our structure as well as biochemical assays performed in Fig. 1 argue that although the CAH3a subdomain is primarily responsible for the high affinity binding of CAH3a/b to CP, it possesses only weak anti-CP activity on its own. Our data also argue that although the CAH3b subdomain is required for the potent anti-CP activity of CAH3a/b, it has little impact on the overall binding affinity of CAH3a/b for CP. To test these ideas, we measured binding affinities by isothermal titration calorimetry (supplemental Fig. S11 and Table 2) and anti-CP activities by actin-polymerization assays (Fig. 6) for the following four recombinant proteins: wild-type CAH3a/b, CAH3a subdomain only, CAH3a(R993E)/b, and CAH3a/b(V1026H/V1030H/F1033A/F1034A) (Fig. 6, C–F). Wild-type CAH3a/b bound CP with Kd = 1.25 nm, and a concentration of 150 nm of this protein was sufficient to restore actin polymerization to the seed-only rate (Fig. 6C). The CAH3a subdomain bound CP with Kd = 27 nm, or 22-fold weaker than wild-type CAH3a/b, and exhibited at least a 20-fold decrease in anti-CP activity relative to wild-type CAH3a/b (Fig. 6D). CAH3a/b containing the critical R993E mutation bound CP with Kd = 1423 nm, or 1100-fold weaker than wild-type CAH3a/b, and exhibited no anti-CP activity, consistent with its minimal interaction with CP (Fig. 6E). This result also indicates that the CAH3b subdomain is itself insufficient for binding CP and for potent anti-CP activity. Finally, CAH3a/b(V1026H/V1030H/F1033A/F1034A) bound CP with Kd = 30 nm and exhibited a 30-fold decrease in anti-CP activity (Fig. 6F). These values for CP affinity and anti-CP activity are very similar to those obtained for the CAH3a subdomain only, indicating that one or more of these four residues in the CAH3b subdomain are necessary for the potent anti-CP activity of CAH3a/b. In total, these experiments show that residues in both the CAH3a and CAH3b subdomains are important for CP binding and anti-CP activity, although they also indicate that each subdomain makes somewhat different contributions to CP binding and anti-CP activity.
TABLE 2.
Equilibrium dissociation constants for CP binding to CAH3a/b mutants determined using isothermal titration calorimetry
| Proteins | Kda |
|---|---|
| nm | |
| WT CP + CAH3a/b | 1.3 |
| WT CP + CAH3a | 27 |
| WT CP + CAH3ab(V1026H/V1030H/F1033A/F1034A) | 30 |
| WT CP + CAH3▵R993E)/b | 1423 |
a Equilibrium dissociation constants were measured by isothermal titration calorimetry, and data were fit with standard binding equations using Origin® software.
Upon CAH3a/b binding, CP showed a large increase in intrinsic tryptophan fluorescence at 330 nm (supplemental Fig. S12), indicative of a change in the local environment of a native tryptophan residue in CP. This result was unexpected because no tryptophans are located in the interface between CP and CAH3a/b. Both the CAH3a and CAH3b subdomains are required for this increase in tryptophan fluorescence intensity (supplemental Fig. S12A). To determine which tryptophan causes this effect, those tryptophans that are close to CAH3a/b in the structure (CPβ Trp122, Trp75, and Trp176) were mutated to phenylalanine. All of these mutant CPs bound CAH3a/b with an affinity similar to that of wild-type CP (data not shown), indicating that the intensity change was not caused by direct interaction with CAH3. Importantly, mutation of Trp176 to Phe or Ser, and only Trp176 to Phe or Ser, blocked the enhancement of tryptophan fluorescence upon CAH3a/b binding (supplemental Fig. S12B). The side chain of Trp176 is located in strand 8 of the central β-sheet and is oriented directly above the 3-helix bundle. We conclude, therefore, that CAH3a/b binding causes a change in the chemical environment of CPβ Trp176, consistent with complex formation leading to a conformational change in CP.
DISCUSSION
Shortly after its discovery, CARMIL was shown to bind CP very tightly both in vitro and in vivo (24, 26, 31, 32). Two biochemical studies then pinpointed the CAH3 domain of CARMIL as containing its CP binding site (30, 33), demonstrated the two anti-CP activities exhibited by the CAH3 domain, and identified several residues that are important for CAH3 domain function. However, the molecular mechanism by which the CAH3 domain antagonizes CP function has remained unknown.
Based on previous studies of CP-CARMIL interaction (30, 32–34), there are three main requirements that must be satisfied for CAH3 to effectively uncap CP-capped barbed ends. First, CAH3a/b must be able to bind CP already present on the barbed end, implying that the binding site on CP for CAH3a/b must be available when CP is on the barbed end. Second, the fact that the binding of CAH3a/b to CP on the barbed end causes a ∼1000-fold decrease in the affinity of CP for the barbed end (33) necessitates that CAH3a/b binding alters the actin binding surface on CP. Third, CP must retain weak capping activity when complexed with CAH3a/b. Based on the structure of the CP·CAH3a/b complex determined here, we propose a three-step mechanism for uncapping by CAH3a/b that satisfies these three requirements.
First, the CAH3a subdomain (Ser964–Pro1002) binds with high affinity to an acidic groove on CP opposite its actin-binding surface. This interaction is probably well defined, as suggested by its overall lower r.m.s. deviation than that for the rest of CAH3a/b (3.42 Å compared with 8.7 Å). This interaction satisfies the first requirement of uncapping (i.e. that CAH3a/b can bind CP already present on the barbed end). Specifically, residues in the conserved CP-binding motif (21), which are mostly basic, make extensive electrostatic contacts with the acidic groove of CP. By mutating residues within this contact region in both CP (D7S and D67S) and CAH3a/b (R993E), we confirmed that this interaction is largely responsible for the affinity of CAH3a/b for CP. This interaction then orients the interdomain linker and the CAH3b subdomain on the appropriate side of CP and in roughly the appropriate orientation to interfere with the interaction of CP with the barbed end via the CP basic patch.
Second, two distinct, independent interactions between CAH3a/b and CP serve to anchor what we call the CAH3a/b “interference domain” (Gln1015-Arg1025) on CP (Fig. 1). The first of these anchor sites is composed of hydrophobic interactions between residues Ile1010, Ser1011, Ile1012, and Leu1013 in the interdomain linker of CAH3a/b and residues from β-strand 1 of CPβ (Leu47, Ile49, and Ala50). The second anchor is composed of interactions between the two C-terminal residues of the CAH3b subdomain (Lys1037 and Val1038) and the pocket on CPα between its central β-sheet and N-terminal helix bundle. These two anchoring interactions serve to maintain a high local concentration of the interference domain near the major actin-binding surface on CP (i.e. its basic patch on and around its α-tentacle).
Third, the interference domain, thus anchored, intercalates between the barbed end and a portion of the basic patch on CP. In so doing, the interference domain decreases the affinity of CP for the barbed end, which satisfies the second requirement for uncapping. Importantly, the length of the interference domain peptide backbone (∼96 Å), when compared with the distance on CP that it spans (∼38 Å), is long enough to reach the CP-barbed end interface. Also of importance, the absence of significant chemical shift changes in the interference domain upon CP binding indicates that it does not interact strongly with CP and thus should be free to reach the major actin-binding surface on CP. In addition to this steric mechanism, the interference domain may also weaken the affinity of CP for the barbed end allosterically by causing a transient local conformational change in CP at its basic patch. This idea is based on two observations. First, we observe significant chemical shift changes in the basic patch on CP that could not be accounted for by direct interaction with CAH3a/b. Second, upon CAH3a/b binding, CP undergoes an increase in tryptophan fluorescence due to a change in the local chemical environment of Trp176 in CPβ, which is structurally linked to the basic patch. Importantly, this increase in fluorescence required the integrity of both the CAH3a and CAH3b subdomains. Also of importance, the chemical shift changes in the CP basic patch upon CAH3a/b binding were the largest of all of those occurring on CP and were localized to only part of the basic patch (Lys142 and Lys143 from CPβ and Lys256, Arg259, and Arg260 from CPα). Because the interference domain only partially interferes with this major barbed end binding site on CP, the CP·CAH3a/b complex functions as a weak capper, which satisfies the third requirement for uncapping. In summary, this three-step mechanism of uncapping is dominated by the direct interference of CP-barbed end interaction by the interference domain of CAH3a/b and involves a contribution from a local conformational change in the CP basic patch.
During preparation of this manuscript, the crystal structure of the CAH3a/b subdomain bound to CP was published (54). Overall, the NMR structure of the CP·CAH3a/b complex determined here is consistent with the crystal structure. In addition to agreeing generally with the crystal structure, we determined the binding sites on CP for the N-terminal part of CAH3a/b (Ser964–Leu974) and for residues Thr1009–Val1026, all of which were not observed in the crystal. We also identify residues Thr1009–Val1026 as being important for CAH3a/b activity. Additionally, we observe that paramagnetic labeling of Val1026 in CAH3a/b affects the loop between strands 6 and 7 of CPβ, which lends support to the idea that this region is required for the local conformational change at the basic patch occurring upon CAH3a/b binding. Interestingly, no dramatic conformational differences were found between free CP (11) and CP bound to the CPI motif (Ser972–Cys1008) or the CBR115 fragment (Glu968–Ser1082) of CARMIL (54). One possible explanation for this result is that residues Thr1009–Val1026, which were not observed in the crystal, are involved in driving the conformational change in CP and that this region does not form a stable (i.e. crystallizable) structure, accounting for the lack of electron density for this region in the crystal (54). Based on the crystal structure, two likely mechanisms for the anti-CP activities of CAH3a/b were proposed: 1) part of the CAH3a subdomain sterically prevents CP from binding the barbed end (the direct competition model), and 2) CAH3a/b binding causes an allosteric change in CP that decreases its affinity for the barbed end (the allosteric model) (54). Our data argue that the mechanism of uncapping by CAH3a/b is driven largely by direct interference of CP-barbed end interaction by a portion of the CAH3b subdomain (Gln1015–Arg1025), which we name the interference domain. That said, our results are also consistent with allosteric changes in CP induced by CAH3a/b binding as contributing to uncapping. Importantly, most of this interference domain is not visible in the crystal structure (54).
Our conclusion that CAH3-driven uncapping involves interference in the function of the CP basic patch on and around its α tentacle provides further evidence that this region of CP, which is known to drive the association of CP with the barbed end and much of its overall binding strength, is the crucial site for CP regulation, because it is also the binding site for V-17 and most likely the binding site for the anionic phospholipid phosphatidylinositol 4,5-bisphosphate (19, 55). We note, however, that CAH3a/b is the first molecule identified whose effect on the function of the CP basic patch results from binding elsewhere on CP. Importantly, the mechanism of uncapping by CAH3a/b proposed here is completely consistent with the two-step model of barbed end capping described by Narita et al. (14). Equally interesting is the fact that the CAH3-driven uncapping mechanism described here and the mechanism of CP sequestration by V-1, which we recently determined,7 can be fully integrated with each other. Specifically, because V-1 binding occludes almost the entire basic patch on CP, the first step in barbed end capping is completely blocked. This effect, coupled with the likelihood that the CP β-tentacle cannot find its binding site on the barbed end when CP is complexed with V-1, explains the sequestering activity of V-1. Moreover, this mode of interaction between V-1 and CP also explains why V-1 cannot uncap CP-capped filaments, the second of the two signature biochemical activities of V-1. In contrast, CAH3a/b couples tight binding at the acidic groove of CP with uncapping activity driven by steric inhibition of CP-barbed end interaction via its interference domain, together with local conformational changes induced in the basic patch of CP. This binding mechanism confers upon CAH3a/b the ability to bind to CP on the barbed end and to then “uncap” the end, and it also explains why the complex of CP·CAH3a/b retains weak barbed end capping activity. Because the basic patch on CP is responsible for the overall affinity of CP for the barbed end as well as the on-rate of capping, our results show that, within the context of the CP barbed end capping activity, the basic patch represents an important regulatory node on CP.
Supplementary Material
Acknowledgments
We thank Marie-Paule Strub for generating the CAH3a/b cysteine mutants. We thank Dr. Duck-Yeon Lee (Protein Analysis Core Facility, NHLBI, National Institutes of Health) for expertise and advice regarding the mass spectrometry-related experiments performed in this paper.
This work was supported, in whole or in part, by National Institutes of Health Intramural Research Program, NHLBI (to J. A. H. and N. T.).
The atomic coordinates and structure factors (code 2KZ7) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S12.
J. A. Hammer, unpublished observation.
In a parallel study (A. Zwolak, T. Uruno, G. Piszczek, J. A. Hammer III, and N. Tjandra, submitted for publication), we measured the effective T2 relaxation times for CP to determine the picosecond-nanosecond time scale flexibility of its C-terminal “tentacles.” We defined the CP tentacles as only those C-terminal residues that are flexible on this time scale. By this definition, we determined that the β tentacle is composed of residues Phe249–Cys272, essentially the same as previously suggested. However, the α tentacle includes only the C-terminal 12 residues (Leu275–Ala286). We refer to the remaining residues historically included as part of the α tentacle (residues Arg259–Ile274) as the “joint” of the α tentacle.
- CP
- capping protein
- PRE
- paramagnetic relaxation enhancement
- mCARMIL-1
- mouse CARMIL-1
- r.m.s.
- root mean square.
REFERENCES
- 1.Wear M. A., Cooper J. A. (2004) Trends Biochem. Sci. 29, 418–428 [DOI] [PubMed] [Google Scholar]
- 2.Cooper J. A., Sept D. (2008) Int. Rev. Cell Mol. Biol. 267, 183–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loisel T. P., Boujemaa R., Pantaloni D., Carlier M. F. (1999) Nature 401, 613–616 [DOI] [PubMed] [Google Scholar]
- 4.Iwasa J. H., Mullins R. D. (2007) Curr. Biol. 17, 395–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akin O., Mullins R. D. (2008) Cell 133, 841–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amatruda J. F., Cannon J. F., Tatchell K., Hug C., Cooper J. A. (1990) Nature 344, 352–354 [DOI] [PubMed] [Google Scholar]
- 7.Amatruda J. F., Gattermeir D. J., Karpova T. S., Cooper J. A. (1992) J. Cell Biol. 119, 1151–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sizonenko G. I., Karpova T. S., Gattermeir D. J., Cooper J. A. (1996) Mol. Biol. Cell 7, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mejillano M. R., Kojima S., Applewhite D. A., Gertler F. B., Svitkina T. M., Borisy G. G. (2004) Cell 118, 363–373 [DOI] [PubMed] [Google Scholar]
- 10.Hug C., Jay P. Y., Reddy I., McNally J. G., Bridgman P. C., Elson E. L., Cooper J. A. (1995) Cell 81, 591–600 [DOI] [PubMed] [Google Scholar]
- 11.Yamashita A., Maeda K., Maéda Y. (2003) EMBO J. 22, 1529–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim K., Yamashita A., Wear M. A., Maéda Y., Cooper J. A. (2004) J. Cell Biol. 164, 567–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wear M. A., Yamashita A., Kim K., Maéda Y., Cooper J. A. (2003) Curr. Biol. 13, 1531–1537 [DOI] [PubMed] [Google Scholar]
- 14.Narita A., Takeda S., Yamashita A., Maéda Y. (2006) EMBO J. 25, 5626–5633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Narita A., Maéda Y. (2007) J. Mol. Biol. 365, 480–501 [DOI] [PubMed] [Google Scholar]
- 16.Lorenz M., Popp D., Holmes K. C. (1993) J. Mol. Biol. 234, 826–836 [DOI] [PubMed] [Google Scholar]
- 17.Pollard T. D., Borisy G. G. (2003) Cell 112, 453–465 [DOI] [PubMed] [Google Scholar]
- 18.Schafer D. A., Jennings P. B., Cooper J. A. (1996) J. Cell Biol. 135, 169–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuhn J. R., Pollard T. D. (2007) J. Biol. Chem. 282, 28014–28024 [DOI] [PubMed] [Google Scholar]
- 20.Miyoshi T., Tsuji T., Higashida C., Hertzog M., Fujita A., Narumiya S., Scita G., Watanabe N. (2006) J. Cell Biol. 175, 947–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bruck S., Huber T. B., Ingham R. J., Kim K., Niederstrasser H., Allen P. M., Pawson T., Cooper J. A., Shaw A. S. (2006) J. Biol. Chem. 281, 19196–19203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Canton D. A., Olsten M. E., Kim K., Doherty-Kirby A., Lajoie G., Cooper J. A., Litchfield D. W. (2005) Mol. Cell. Biol. 25, 3519–3534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taoka M., Ichimura T., Wakamiya-Tsuruta A., Kubota Y., Araki T., Obinata T., Isobe T. (2003) J. Biol. Chem. 278, 5864–5870 [DOI] [PubMed] [Google Scholar]
- 24.Jung G., Remmert K., Wu X., Volosky J. M., Hammer J. A., 3rd (2001) J. Cell Biol. 153, 1479–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhattacharya N., Ghosh S., Sept D., Cooper J. A. (2006) J. Biol. Chem. 281, 31021–31030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu P., Mitchelhill K. I., Kobe B., Kemp B. E., Zot H. G. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 3685–3690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vanderzalm P. J., Pandey A., Hurwitz M. E., Bloom L., Horvitz H. R., Garriga G. (2009) Development 136, 1201–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rogers S. L., Wiedemann U., Stuurman N., Vale R. D. (2003) J. Cell Biol. 162, 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuzaka Y., Okamoto K., Mabuchi T., Iizuka M., Ozawa A., Oka A., Tamiya G., Kulski J. K., Inoko H. (2004) Gene 343, 291–304 [DOI] [PubMed] [Google Scholar]
- 30.Yang C., Pring M., Wear M. A., Huang M., Cooper J. A., Svitkina T. M., Zigmond S. H. (2005) Dev. Cell 9, 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu P., Zot A. S., Zot H. G. (1995) J. Biol. Chem. 270, 25316–25319 [DOI] [PubMed] [Google Scholar]
- 32.Remmert K., Olszewski T. E., Bowers M. B., Dimitrova M., Ginsburg A., Hammer J. A., 3rd (2004) J. Biol. Chem. 279, 3068–3077 [DOI] [PubMed] [Google Scholar]
- 33.Uruno T., Remmert K., Hammer J. A., 3rd (2006) J. Biol. Chem. 281, 10635–10650 [DOI] [PubMed] [Google Scholar]
- 34.Fujiwara I., Remmert K., Hammer J. A., 3rd (2010) J. Biol. Chem. 285, 2707–2720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee W. L., Ostap E. M., Zot H. G., Pollard T. D. (1999) J. Biol. Chem. 274, 35159–35171 [DOI] [PubMed] [Google Scholar]
- 36.Cooper J. A., Blum J. D., Pollard T. D. (1984) J. Cell Biol. 99, 217–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang Y., Niederstrasser H., Edwards M., Jackson C. E., Cooper J. A. (2009) Mol. Biol. Cell 20, 5290–5305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Remmert K., Uruno T., Hammer J. A., 3rd (2009) Protein Expr. Purif. 67, 113–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuhn J. R., Pollard T. D. (2005) Biophys. J. 88, 1387–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kouyama T., Mihashi K. (1980) Eur. J. Biochem. 105, 279–287 [DOI] [PubMed] [Google Scholar]
- 41.Wishart D. S., Bigam C. G., Holm A., Hodges R. S., Sykes B. D. (1995) J. Biomol. NMR 5, 67–81 [DOI] [PubMed] [Google Scholar]
- 42.Spera S., Bax A. (1991) J. Am. Chem. Soc. 113, 5490–5492 [Google Scholar]
- 43.Pervushin K., Riek R., Wider G., Wüthrich K. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 12366–12371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pervushin K. V., Wider G., Wuthrich K. (1998) J. Biomol. NMR 12, 345–348 [DOI] [PubMed] [Google Scholar]
- 45.Kay L., Keifer P., Saarinen T. (1992) J. Am. Chem. Soc. 114, 10663–10665 [Google Scholar]
- 46.Schleucher J., Schwendinger M., Sattler M., Schmidt P., Schedletzky O., Glaser S. J., Sørensen O. W., Griesinger C. (1994) J. Biomol. NMR 4, 301–306 [DOI] [PubMed] [Google Scholar]
- 47.Garrett D. S., Seok Y. J., Liao D. I., Peterkofsky A., Gronenborn A. M., Clore G. M. (1997) Biochemistry 36, 2517–2530 [DOI] [PubMed] [Google Scholar]
- 48.Schwieters C. D., Kuszewski J. J., Tjandra N., Clore G. M. (2003) J. Magn. Reson. 160, 65–73 [DOI] [PubMed] [Google Scholar]
- 49.Clore G. M., Schwieters C. D. (2003) J. Am. Chem. Soc. 125, 2902–2912 [DOI] [PubMed] [Google Scholar]
- 50.Canton D. A., Olsten M. E., Niederstrasser H., Cooper J. A., Litchfield D. W. (2006) J. Biol. Chem. 281, 36347–36359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hutchings N. J., Clarkson N., Chalkley R., Barclay A. N., Brown M. H. (2003) J. Biol. Chem. 278, 22396–22403 [DOI] [PubMed] [Google Scholar]
- 52.Battiste J. L., Wagner G. (2000) Biochemistry 39, 5355–5365 [DOI] [PubMed] [Google Scholar]
- 53.Iwahara J., Tang C., Marius Clore G. (2007) J. Magn. Reson. 184, 185–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hernandez-Valladares M., Kim T., Kannan B., Tung A., Aguda A. H., Larsson M., Cooper J. A., Robinson R. C. (2010) Nat. Struct. Mol. Biol. 17, 497–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim K., McCully M. E., Bhattacharya N., Butler B., Sept D., Cooper J. A. (2007) J. Biol. Chem. 282, 5871–5879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zwolak A., Fujiwara I., Hammer J. A., III, Tjandra N. (2010) J. Biol. Chem. 285, 25767–25781 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.