Abstract
The cell surface glycoprotein γ-glutamyl transpeptidase (GGT) was isolated from healthy human kidney and liver to characterize its glycosylation in normal human tissue in vivo. GGT is expressed by a single cell type in the kidney. The spectrum of N-glycans released from kidney GGT constituted a subset of the N-glycans identified from renal membrane glycoproteins. Recent advances in mass spectrometry enabled us to identify the microheterogeneity and relative abundance of glycans on specific glycopeptides and revealed a broader spectrum of glycans than was observed among glycans enzymatically released from isolated GGT. A total of 36 glycan compositions, with 40 unique structures, were identified by site-specific glycan analysis. Up to 15 different glycans were observed at a single site, with site-specific variation in glycan composition. N-Glycans released from liver membrane glycoproteins included many glycans also identified in the kidney. However, analysis of hepatic GGT glycopeptides revealed 11 glycan compositions, with 12 unique structures, none of which were observed on kidney GGT. No variation in glycosylation was observed among multiple kidney and liver donors. Two glycosylation sites on renal GGT were modified exclusively by neutral glycans. In silico modeling of GGT predicts that these two glycans are located in clefts on the surface of the protein facing the cell membrane, and their synthesis may be subject to steric constraints. This is the first analysis at the level of individual glycopeptides of a human glycoprotein produced by two different tissues in vivo and provides novel insights into tissue-specific and site-specific glycosylation in normal human tissues.
Keywords: Glycoprotein, Glycosylation, Kidney, Liver, Post-translational Modification, GGT, Glycopeptides, gamma-Glutamyltranspeptidase
Introduction
In mammalian systems, the carbohydrate structures present on glycoproteins, glycolipids, and proteoglycans play pivotal roles in the regulation of protein folding and sorting as well as cell-cell recognition, communication, adhesion, and migration (1–3). The array of glycans isolated from human tissues reveals tissue-specific patterns, with some glycans common to multiple tissues. Studies in mouse model systems have correlated the general structural features of glycans on glycoproteins with the tissue-specific expression pattern of glycosylation enzymes (4–6). Many additional factors, including compartmentalization of glycosyltransferases and the supply and transport of sugars and sugar nucleotides, have also been shown to influence glycan synthesis (7, 8). This multifaceted process is altered in cancer and other diseases (9, 10). Thus, disease-specific modifications to glycan structures provide potential biomarkers. The cell surface protein γ-glutamyl transpeptidase (GGT)2 is shed into the urine and serum and may be useful as a biomarker of kidney and liver disease. To identify the glycan profile and site-specific glycosylation of GGT in normal human tissue, we isolated and characterized GGT from healthy human kidney and liver. Unlike profiles of tissue glycans, which are a mixture of glycans produced by the many cell types within the tissue, GGT is expressed only in the proximal tubule cells in the kidney and is expressed on the bile canaliculi of the hepatocytes (11). Therefore, the glycans on isolated GGT reveal the glycosylation of this protein by a single cell type in each tissue. Recent advances in mass spectrometry (MS) enabled us to identify the microheterogeneity and relative abundance of glycans on specific glycopeptides within the GGT isolated from each tissue. This is the first comparative analysis at the level of individual glycopeptides from a human glycoprotein produced by two different tissues in vivo. GGT was isolated independently from multiple healthy kidneys and livers to assess variations in glycosylation from donor to donor.
GGT (EC 2.3.2.2) is a type II cell surface membrane glycoprotein, which plays a critical role in glutathione and cysteine homeostasis by catalyzing the first enzymatic step in the catabolism of extracellular glutathione and glutathione conjugates (11–15). GGT is expressed as a single polypeptide that undergoes an intramolecular autocatalytic cleavage that resolves the protein into a large (amino acids 1–380) and a small (amino acids 381–569) subunit (16–19). Both subunits are required for enzymatic activity (20). The protein is anchored in the plasma membrane by a transmembrane domain at the N terminus of the large subunit (amino acids 5–26). The heterodimer faces the extracellular environment. Human GGT has seven potential N-glycosylation sites (sequon, NX(S/T), X ≠ P) as follows: six on the large subunit and one on the small subunit. Early studies of the glycans on human GGT identified tissue-specific patterns of glycosylation based on differential lectin affinities, distinct electrophoretic mobility patterns, and exoglycosidase cleavage patterns (21–29). Further progress was hampered by technical limitations in the sensitivity of the methods used to characterize glycoforms, site differences in multiply glycosylated proteins, and the challenge posed by glycan microheterogeneity. Based on these early studies of GGT from normal human kidney and liver tissue, we hypothesized that tissue-specific glycopeptides could be identified. To test this hypothesis, a combination of matrix-assisted laser desorption/ionization-time-of-flight (MALDI-TOF)/MS-MS and high resolution liquid chromatography (LC)-MS-MS analyses were used to analyze immunopurified human renal and hepatic GGT.
EXPERIMENTAL PROCEDURES
Isolation of Membrane Proteins
Normal human kidneys were obtained from the National Disease Research Interchange (Philadelphia, PA). Three perfused kidneys (prepared but not used for transplant) were acquired on ice from two Caucasian females (46 and 75 years old) and one Caucasian male (51 years old). The kidney cortex was frozen at −80 °C until use. Two livers, one from a 90-year-old Caucasian female and one from an 80-year-old Caucasian male, were received frozen and stored at −80 °C until use. The exclusion criteria for the liver tissue were as follows: no alcoholism, no HIV, no hepatitis, no cancer or other liver disease, and no sepsis. Both livers were frozen within 12 h post-mortem. Membrane fractions were prepared as described previously (30). A 0.5% (v/v) Triton X-100 extract of the membrane fraction was used for SDS-PAGE and Western blot analysis, and a 0.5% (w/v) CHAPS extract was used for immunopurification of GGT and mass spectrometric analysis.
Electrophoresis on Nondenaturing (Native) Gels
Triton X-100-solubilized membranes were incubated for 18 h at 37 °C with papain (1 μg of papain/4 of μg protein; Sigma) to cleave GGT from its hydrophobic anchor. Papain cleaves human GGT at Ser-30 (31, 32). Samples were incubated as indicated in the presence or absence of Clostridium perfringens neuraminidase (New England Biolabs, Ipswich, MA). Samples were separated on 10% polyacrylamide Tris-glycine gels, pH 8.3, containing 0.1% Triton X-100 and resolved at 100 V for 4.5 h in 25 mm Tris, 100 mm glycine buffer, pH 8.3. Using a modification of the method described by Rutenberg et al. (33), GGT activity was localized in the gels by the formation of a red precipitate during incubation at 37 °C in a solution containing 400 μm l-glutamic acid γ-(4-methoxy-β-naphthylamide) (Sigma), 30 mm glycyl-glycine, 1.2 mm Fast Blue, 100 mm NaCl, and 25 mm Tris-HCl, pH 7.5.
SDS-PAGE and Western Blotting
Triton X-100-solubilized membranes were incubated at 95 °C for 10 min in 25 mm Tris-HCl, pH 7.4, 25 mm β-mercaptoethanol, 0.1% (w/v) SDS, and 0.5% Triton X-100. The solution was cooled to room temperature, supplemented with leupeptin (1 μm) and aprotinin (1 μg/ml), and incubated in the presence or absence of peptide-N-glycosidase F (PNGase F, EC 3.5.1.52; New England Biolabs, Ipswich, MA) for 18 h at 37 °C. The reactions were stopped by boiling in Laemmli sample buffer (2% SDS, 5% glycerol, 5% 2-mercaptoethanol, 0.002% bromphenol blue, 62.5 mm Tris-HCl, pH 6.8), subjected to electrophoresis on 8 or 10% SDS-polyacrylamide gels, and electroblotted to nitrocellulose. The nitrocellulose blots were blocked in Tris-buffered saline with 0.1% Tween 20 (TBST) containing 3% dry milk followed by incubation in TBST buffer containing primary antibodies directed against either the large (GGT129 (11)) or small (GGT1, Abnova, Taipei City, Taiwan) subunits of GGT. The blots were washed extensively in TBST, incubated with HRP-conjugated secondary antibodies, and visualized by chemiluminescence according to the manufacturer's protocol (ECL Plus, GE Healthcare).
Immunopurification of GGT
The GGT129 antibody, directed against a 19-amino acid peptide sequence at the C terminus of the large subunit of GGT, was used to immunopurify GGT (11). The peptide sequence was not predicted to undergo post-translational modification. Affinity-purified GGT129 antibody was bound to a protein A affinity matrix (ImmunoPure Protein A IgG Plus, Pierce) and covalently linked using the NHS-ester cross-linking agent disuccinimidyl suberate. CHAPS-solubilized membranes were heated at 85 °C in 1% SDS for 15 min, diluted 50-fold in binding buffer (25 mm Tris-HCl, pH 7.4, 100 mm NaCl, 0.5% CHAPS, 1 μm leupeptin, 1 μg/ml aprotinin), and incubated with the immunoaffinity matrix (with constant mixing) for 2 h at 22 °C. The matrix was poured into a 10-ml column, washed extensively with binding buffer (20× bed volume), and eluted with low pH buffer (0.1 m glycine-HCl, pH 2.8, 0.2 m NaCl, 0.1% CHAPS) into tubes containing 1.0 m Na2HPO4, pH 8.5. GGT-containing fractions were pooled and concentrated in Ultracel YM-10 micro-concentrators (Millipore, Bedford, MA) followed by vacuum centrifugation of the retentates to dryness.
Trypsin Digestion
Immunopurified GGT was resuspended in 10 mm ammonium bicarbonate, reduced (in the absence of urea), alkylated, and digested with proteomic grade trypsin (Sigma) as described previously (34).
Release of N-Glycans
To release N-glycans from GGT, trypsin-digested GGT was treated with 5 milliunits of PNGase F for 18 h at 37 °C. To release N-glycans from total membrane glycoproteins, CHAPS-solubilized membranes (400 μg of protein) were incubated at 95 °C for 10 min in 25 mm Tris-HCl, pH 7.4, 25 mm β-mercaptoethanol, 0.1% SDS, and 0.5% CHAPS, cooled for 10 min at 22 °C, and incubated with 17 units of PNGase F for 18 h at 37 °C. The released N-glycans (50-μl aliquots diluted with 350 μl of water) were adsorbed to activated charcoal micro spin columns (Harvard Apparatus, Holliston, MA), preconditioned by rinsing three times with 400-μl aliquots of 85% acetonitrile/water (v/v) containing 0.1% (v/v) trifluoroacetic acid, and twice with 400-μl aliquots of 5% acetonitrile/water, 0.1% trifluoroacetic acid, eluted with 300 μl of 40% acetonitrile/water with 0.1% trifluoroacetic acid, and dried by vacuum centrifugation.
Solid-phase Spin Column Permethylation
NaOH beads (Aldrich) were suspended in acetonitrile, packed in microcolumns (Harvard Apparatus, Holliston, MA), and preconditioned with dimethyl sulfoxide (35, 36). Dried N-glycans were resuspended in a mixture of 60 μl of dimethyl sulfoxide, 44.8 μl of iodomethane, and 2.4 μl of water and recycled through the spin columns three times by centrifugation at 800 rpm for 30 s. A second 44.8-μl aliquot of iodomethane was added to the sample, which was again recycled through the column three times. Permethylated glycans were extracted by the addition of 1 ml of 500 mm NaCl and 400 μl of CHCl3 (EM Science, Gibbstown, NJ). The aqueous layer was discarded, and the CHCl3 layer was washed repeatedly with water by shaking and dried by vacuum centrifugation.
Mass Spectrometric Glycomic Analysis
The N-glycans released from purified GGT were characterized by MALDI-TOF/TOF mass spectrometry. Dried permethylated samples were resuspended in 1:1 (v/v) MeOH/H2O containing 0.5 mm sodium acetate, mixed 1:1 (v/v) on the MALDI plate with 10 mg/ml 2,5-dihydroxybenzoic acid in 1:1 (v/v) MeOH/H2O, and dried by vacuum. Samples were analyzed with an Applied Biosystems 4800 Proteomic Analyzer (Applied Biosystems, Framingham, MA) equipped with an Nd:YAG laser (355-nm wavelength) operated in the positive-ion mode or on an Ultraflex II (Bruker Daltonics, Billerica, MA) with a SMARTbeam laser operated in reflectron positive mode. MALDI collision-induced dissociation spectra were obtained at collision energy set to 1 kV (defined by the potential difference between source and collision cell). Cross-ring fragmentation was enhanced using air as the collision gas at 8.0 × 10−6 torr. MS and tandem MS data were processed using DataExplorer 4.0 (Applied Biosystems, Framingham, MA).
Permethylated N-glycans (∼300 pmol) from kidney GGT samples were also analyzed by ESI by resuspending in 30 μl of 50% methanol and diluted 1:100 or 1:20 into 50% methanol and 1 mm NaOH. The glycans were analyzed on a linear ion trap mass spectrometer (LTQ, Thermo Fisher Scientific, Waltham, MA). Each glycan mixture was infused directly into the LTQ at a flow rate of 0.1–0.3 ml/min and electrosprayed through a silica capillary (New Objective, Woburn, MA). MSn experiments in the LTQ were carried out in positive ion mode using a collision energy of 25–35%. Structures of the PNGase F-released and permethylated glycans were initially assigned on the basis of exact mass match of the singly charged sodiated ions and biosynthetic rules of N-glycosylation (37).
Glycopeptide Analyses
Tryptic digests of immunopurified GGT were analyzed on a Dionex 3000 Ultimate nano-LC system (Dionex, Sunnyvale, CA) interfaced to a linear trap quadrupole-Fourier transform (LTQ-FT) or LTQ Orbitrap XL mass spectrometer (Thermo Fisher, San Jose, CA). A 5-μl aliquot (2 μg of protein equivalent) was loaded onto a PepMap300 C18 cartridge (5 μm, 300 Å, Dionex) and eluted through a pulled tip capillary column (150 mm × 75 μm inner diameter) packed with 90-Å Jupiter Proteo 90 packing materials (Phenomenex, Torrance, CA), using a reversed-phase gradient from 10 to 55% solvent B (solvent A: 3% acetonitrile, 0.1% formic acid; solvent B: 97% acetonitrile, 0.1% formic acid) over 50 min at 300 nl/min (total run time 85 min). Alternative methods included a reversed-phase gradient from 0 to 35% solvent B (solvent A: 0.1% formic acid; solvent B: 97% acetonitrile, 0.1% formic acid) over 150 min at 300 nl/min or a 20-min wash interval with 3% acetonitrile prior to initiating the gradient. The LTQ-FT mass spectrometer was operated in an automated data-dependent mode that performed seven scans per interval (an initial MS scan, then skimmer in source decay-MS, then MS/MS scans of the five most abundant ions). The total cycle (seven scans) took less than 1 s and was continuously repeated for the entire LC-MS run under data-dependent conditions with a dynamic exclusion window of 60 s. In-source fragmentation was attained by applying a maximum voltage between the transfer tube and the skimmer. This promotes ion acceleration, which in the presence of air molecules induces fragmentation. The Orbitrap was also operated in a similar data-dependent scanning mode by performing an initial MS scan (300–2000 m/z) and then selecting the five most abundant ions for MS/MS fragmentation.
Candidate glycopeptides were identified by the program GlycID that analyzes LC-MS/MS data based on selection criteria, including the presence of glycan oxonium ions (singly protonated HexNAc and HexNAc + galactose at m/z 204.08 and 366.14, respectively) in the skimmer ISD spectrum co-eluting with the glycopeptide, a high resolution mass match to a predicted tryptic peptide from GGT with a predicted glycoform attached, and diagnostic ions present in the MS/MS spectra. Potential glycopeptides identified by the algorithm were then manually verified for the presence of glycan oxonium ions and diagnostic peptide fragment ions (b- and y-type ions). The search was then expanded by manually probing the entire LC/MS-MS run for mass matches against predicted glycopeptides and expanded by the inclusion of glycans of related compositions, including those predicted by the glycans identified in the glycomics data. Assuming equal ionization efficiency of each glycoform at a particular glycosylation site, the relative abundance of each glycoform on a single glycosylation site was determined by summing the total ion intensities for all of the glycoforms within a glycopeptide family over the entire elution window. Each of the total ion intensities was then calculated as a percentile relative to the other family members. The reported percentiles represent the average of at least three separate LC/MS-MS experiments.
N-Glycosylation of the sites was independently confirmed by removing buffer salts and trypsin by C18 SepPak of a tryptic GGT digest and incubating with PNGase F in a 1:1 mixture of 16O-water and 18O-water (97% isotope purity, Cambridge Isotope Laboratories, Andover, MA) at 37 °C for 3 h. Deglycosylated peptides were analyzed by MALDI-TOF/MS on an Ultraflex II (Bruker Daltonics, Billerica, MA) with a SMARTbeam laser operated in reflectron-positive mode. Enzymatic cleavage of N-linked glycans by PNGase F converts glycosylated asparagine residues into aspartates and incorporates either an 16O or 18O atom, which results in a 1- or 3-Da increase in mass of the residue from the unmodified asparagines. By performing the deglycosylation reaction in a 1:1 ratio of heavy and light water, peptides bearing N-glycosylated sites were observed as doublet offset by two mass units.
Homology Model
The human GGT homology model was constructed using MODELLER version 9 software (38). The large subunit of GGT was generated using Escherichia coli GGT (Protein Data Bank code 2DBW), which exhibits 29% sequence identity, as a template (39). The small subunit of GGT was modeled using E. coli GGT (Protein Data Bank code 2DG5) with 38% sequence identity (39). Structural models of N-glycans were added according to GLYCAM04 force field protocols (40). Specific structural linkages were informed by the exoglycosidase experiments conducted by Yamashita et al. (28). The heterodimer model was energy-minimized within the MODELLER protocol.
RESULTS
Electrophoretic Behavior of GGT from Human Kidney and Liver
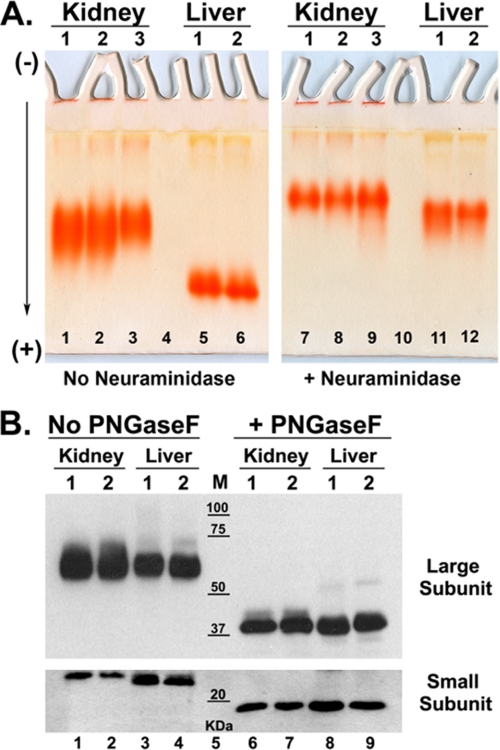
GGT from human kidney and human liver differed in their migration in native (nondenaturing) gels, indicating tissue-specific differences in size and/or inherent charge. Detergent-solubilized GGT was treated with papain, which cleaves GGT at amino acid 30, releasing the enzyme from its N-terminal transmembrane domain (32). The papain-cleaved GGT readily migrates in nondenaturing gels. GGT from three human kidneys had a similar migration pattern, consisting of a single broad band, which indicates microheterogeneity in the inherent charge of the glycoprotein (Fig. 1A, lanes 1–3). GGT from two human livers migrated more rapidly than kidney-derived GGT, suggesting the liver protein was smaller or had a higher inherent negative charge (Fig. 1A, lanes 1–6). The migration of liver GGT from two donors was similar (Fig. 1A, lanes 5 and 6). The kidney and liver samples were treated with recombinant nonspecific clostridial neuraminidase, releasing the negatively charged sialic acid residues. GGT from both tissues migrated more slowly after neuraminidase treatment consistent with a reduction in negative charge (Fig. 1A, lanes 7–12). A greater reduction in mobility was observed for the liver GGT than kidney GGT, indicating that GGT is more heavily sialylated in the liver than in the kidney. Treatment with neuraminidase also sharpened the kidney GGT band, which suggests heterogeneous sialylation.
FIGURE 1.
Electrophoresis of GGT from human kidney and liver microsomes. A, native gel analysis of GGT. Triton X-100-solubilized microsomes from normal human kidney (lanes 1–3 and 7–9) or liver (lanes 5 and 6 and 11 and 12) tissue were incubated at 37 °C with papain alone (lanes 1–6) or with papain and clostridial neuraminidase (lanes 7–12) and resolved in parallel on 10% nondenaturing polyacrylamide gels. GGT was localized in situ with the histochemical stain for GGT activity (red precipitate). B, SDS-PAGE analysis of intact and deglycosylated GGT. Non-papain-treated samples from human kidney (lanes 1 and 2 and 6 and 7) and liver (lanes 3 and 4 and 8 and 9) were denatured and incubated in the absence (lanes 1–4) or presence (lanes 6–9) of PNGase F and resolved on an 8% (upper panel) or 10% (lower panel) SDS-polyacrylamide gel. Resolved proteins were electroblotted onto nitrocellulose and Western blotted using antibodies against the large and small subunits of GGT. Positions of molecular mass (M) markers are indicated.
Analysis of GGT by SDS-PAGE indicated that N-linked glycosylation is a major post-translational modification. Intact, detergent-solubilized human kidney and liver GGT (not treated with papain) were resolved on reducing SDS-polyacrylamide gels. The large and small subunits of GGT migrated as discrete bands with apparent molecular masses of 62 and 22 kDa, respectively (Fig. 1B, lanes 1–4). The large subunit from the kidney migrated more diffusely than the large subunit from liver GGT, suggesting greater heterogeneity in size or charge. The small subunit from kidney GGT migrated slightly slower than the liver small subunit, suggesting a larger size or less negative charge. Pretreatment with PNGase F, to remove the N-glycans, increased the electrophoretic mobility of both kidney and liver GGT and also reduced the heterogeneity in the migration of the large subunit (Fig. 1B, lanes 6–9). The apparent molecular masses of the deglycosylated large and small subunits were 42 and 19 kDa, respectively, consistent with the predicted molecular masses of the unmodified large (41.4 kDa) and small (20.0 kDa) subunits. The shifts in apparent molecular masses following deglycosylation (a reduction of 20 kDa for the large subunit and 3 kDa for the small subunit) indicate extensive N-glycosylation at the six potential glycosylation sites in the large subunit and the single site in the small subunit.
N-Glycomic Analysis of Kidney Membrane Glycoproteins and Kidney GGT
To investigate the N-glycans produced in the kidney and the N-glycans on renal GGT, we analyzed the N-glycans released from renal membranes and from isolated kidney GGT. We began our analysis by investigating N-glycosylation in the kidney. The renal membranes include glycoproteins expressed by many cell types within the kidney, and the N-glycan profile revealed a diverse array of glycan ions (Fig. 2A). High mannose type and GlcNAc-bisected, complex type N-glycans were prevalent. Only a subset of the complex type N-glycan structures in the kidney profile were sialylated. Relative levels of specific glycans were inferred from the ion abundance (41, 42). The structures for most of the high abundance glycoforms observed by MS, including the bisecting GlcNAc modification, core and peripheral fucose, and sialylation, were confirmed by tandem MS (data not shown). Virtually identical profiles were obtained for the kidney glycans from three different donors (see supplemental Fig. 1).
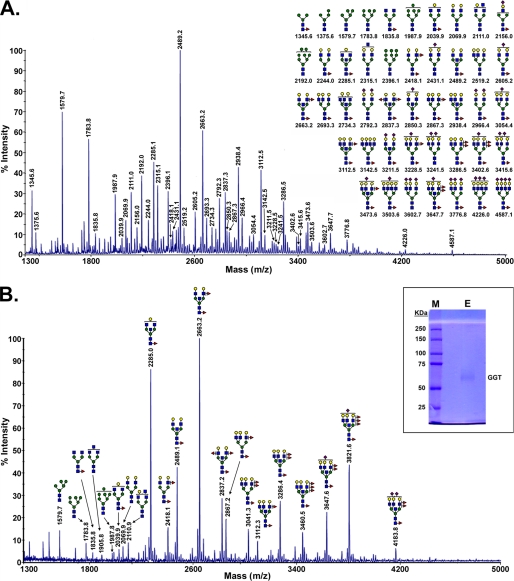
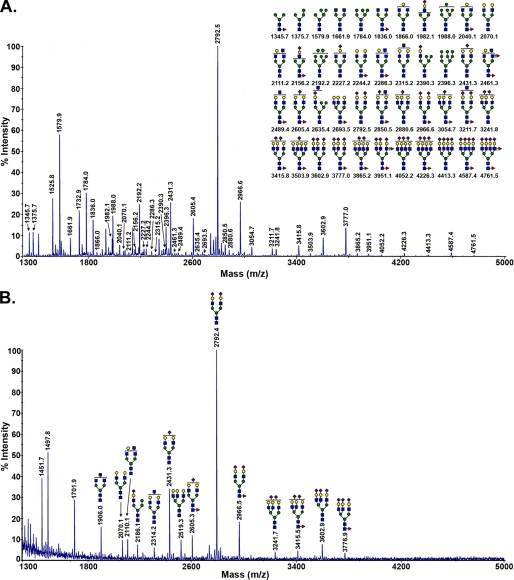
FIGURE 2.
MALDI-TOF-MS glycomic profile of permethylated N-glycans derived by PNGase F digestion of a soluble detergent extract of membranes from the cortex of normal human kidney 1 (A) and GGT purified from human kidney 1 (B) are shown. B, inset, the immunopurified large subunit of kidney GGT used for glycomic analysis was analyzed by SDS-PAGE and stained with Coomassie Blue (E, sample; M, molecular mass markers). Symbols used are as follows: blue box, N-acetylglucosamine; green circle, mannose; yellow circle, galactose; half-green and half-yellow circle, hexose (galactose or mannose); red triangle, fucose; purple diamond, N-acetylneuraminic acid.
The N-glycans on renal GGT were characterized and compared with the total profile of N-glycans on membrane glycoproteins in the kidney. Immunopurified large subunit of GGT used for glycan analysis is shown in Fig. 2B, inset. N-Glycans released from purified renal GGT consisted primarily of complex type N-glycans dominated by bisected bi- and triantennary structures with a high degree of fucosylation on both core and peripheral GlcNAc residues and a low degree of sialylation (Fig. 2A). The most abundant structures, at m/z 2285.0 and 2663.2, corresponded to neutral, core-fucosylated biantennary complex type N-glycans modified with bisecting GlcNAc residues, with the m/z 2663.2 ion exhibiting both core and peripheral fucosylation. These structures were also prominent among the N-glycans observed in the renal membrane glycoprotein pool. In both profiles, relatively low sialylation was observed.
There were also marked differences between the two profiles. Three of the 10 most abundant glycans in the kidney N-glycomic profile were high mannose type structures. In contrast, these structures constituted only a minor portion of the GGT N-glycans. The bisected biantennary structure at m/z 2489.2 dominated the kidney glycans (Fig. 2A), but it was less prominent among the GGT glycans (Fig. 2B). The total kidney glycans exhibited less fucosylation than the N-glycans conjugated to GGT. The prominent tetra-fucosylated, bisected triantennary structure observed on kidney GGT (m/z 3821.6) was below the limit of detection in the total kidney N-glycan pool. The difference in the relative abundance of specific glycans between the two profiles may reflect cell type-specific glycosylation, as GGT is only expressed in the proximal tubule cells of the kidney.
Site-specific Glycosylation of Renal GGT
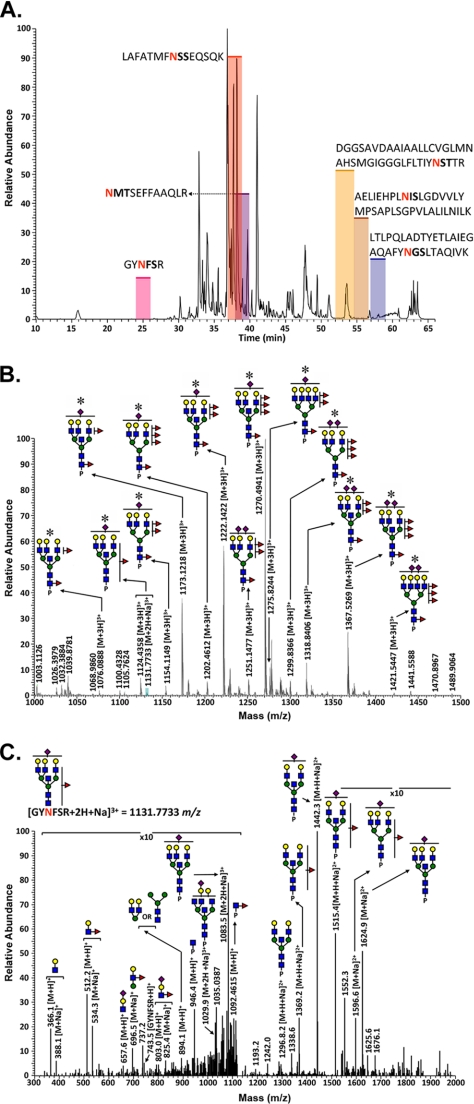
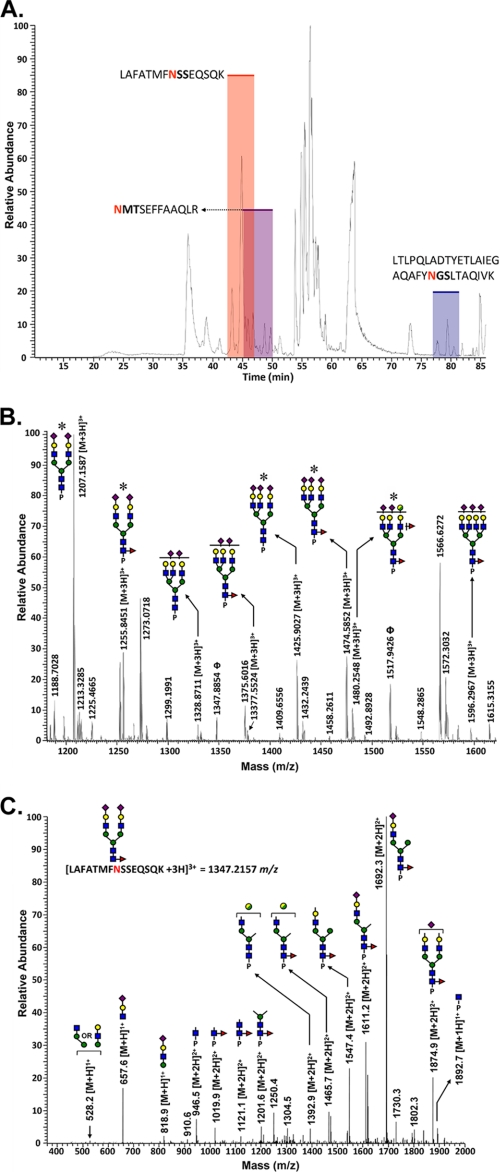
Analysis of the renal GGT glycopeptides provided specific, detailed information regarding the structural compositions and relative abundance of the N-glycans at each potential glycosylation site. Each glycosylation site on the large subunit of GGT is contained within a unique tryptic fragment (Fig. 3). A LC-MS/MS base peak intensity chromatogram of the tryptic peptides is shown in Fig. 4A. The elution intervals during which the individual glycopeptide families eluted are highlighted. Individual glycopeptide families eluted over wider (2–3-fold) chromatographic intervals than peptides that were not glycosylated, indicating that the elution time is influenced primarily by the peptide and modulated by the glycoform type (Fig. 4A). The glycans on the GYNFSR (Asn-297) glycopeptide are shown in Fig. 4B. The composition of each glycan was associated with a mass accuracy of 5 ppm or less with respect to the theoretical m/z value for each glycopeptide. The structural models within each glycopeptide family were identified by data-dependent collision-induced dissociation MS/MS. An example is shown in Fig. 4C for the low abundance GYNFSR glycopeptide ion at m/z = 1131.8, which is the monosodiated form of the m/z = 1124.4 glycopeptide ion (Table 1). The presence of diagnostic oxonium ions at m/z = 366.1, 512.2, and 657.6 are readily discernible within the fragmentation pattern. Fragments matching the m/z values for the core GlcNAc-modified peptide (m/z = 946.4), the fucosylated core GlcNAc-modified peptide (m/z = 1092.5), and the peptide backbone (m/z = 743.5) confirm assignment of the glycosylation site to the GYNFSR tryptic peptide fragment. Fragment ions corresponding to Lewis-type fucosylation on antennary GlcNAc residues (m/z = 696.5 and 803.0) were also observed in this spectrum, indicative of a mixture of isoforms containing a single fucose on either the core-GlcNAc residue or on a distal antennary GlcNAc.
FIGURE 3.
Tryptic glycopeptide sequence map for the large subunit of human GGT. Predicted tryptic glycopeptides are underlined. Arrowheads denote the positions of putative N-glycosylation sites.
FIGURE 4.
Glycopeptide analysis of GGT isolated from normal human kidney cortex. A, base peak intensity chromatogram of the tryptic digest of GGT immunopurified from normal human kidney. The shaded regions depict the retention times of the different glycopeptides associated with this glycoprotein, and Table 1 summarizes the microheterogeneity at each site. B, averaged LC/Orbitrap-FT MS spectrum for the GGT Asn-297 (GYNFSR) family of glycopeptides from kidney tissue, showing the identified glycoconjugates within the elution interval. Asterisks denote glycoconjugates that were confirmed by tandem MS analysis. P represents the GYNFSR tryptic peptide to which the N-glycans are attached. Unannotated peaks represent nonglycosylated peptides that co-eluted with this glycopeptide family. C, MS/MS spectrum showing the fragmentation pattern of the monosodiated glycopeptide at m/z = 1131.7 identified on the Asn-297 glycosylation site. For symbol definitions, see Fig. 2.
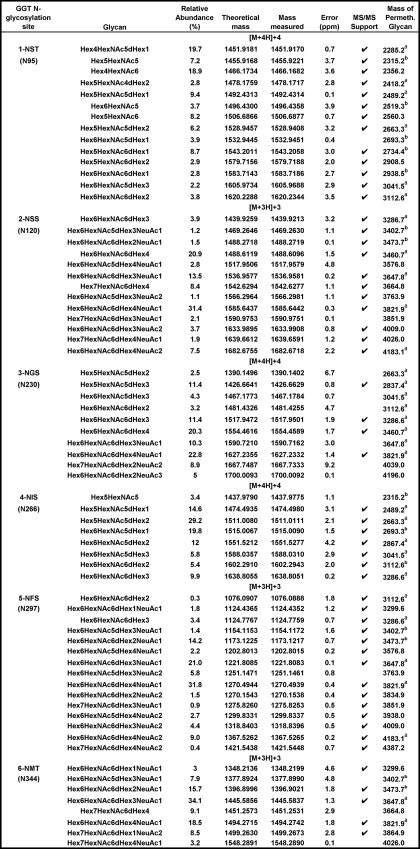
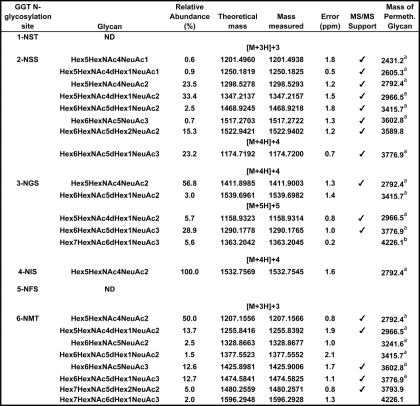
TABLE 1.
N-Glycan microheterogeneity of renal GGT as determined by LC/MS-MS
Relative abundance values were deduced from comparative ion intensities among glycopeptide family members. The tryptic peptides in which each glycosylation site is contained are depicted in Fig. 3. Calculated m/z values for the corresponding permethylated N-glycans enzymatically released from either renal GGT (a) or from the pool of kidney membrane glycoproteins (b) in Fig. 2 are included for reference. Abbreviations used are as follows: Hex, hexose (mannose and galactose); HexNac, N-acetylglucosamine; dHex, deoxyhexose (fucose); NeuAc, neuraminic (sialic) acid; ND, not detected.
Table 1 shows the composition and relative abundance of the glycans identified on the renal GGT glycopeptides. The data reveal 8–15 glycans at each of the six N-glycosylation sites. A total of 36 glycan compositions were observed. The Hex6HexNAc6dHex1NeuAc1 glycan composition, which was observed on two of the sites, was identified as a mixture of both core and antennary mono-fucosylated structures as described above for the analysis of the monosodiated form of the 1124.4 glycopeptide ion. The array of N-glycans differed among the glycopeptides, highlighting site-specific glycosylation within a single protein. Notably, the N-glycans identified at the NST (Asn-95) and NIS (Asn-266) sites were limited to neutral carbohydrates, although sialylated glycans were observed at each of the other four sites. The NST and NIS sites were also unique in having nonfucosylated glycans. On the NST site, nonfucosylated glycans constituted 38% of the N-glycans. At the NIS site, the nonfucosylated glycans were of low abundance, constituting only 3.4% of the glycans. Of the 36 glycan compositions observed, 17 were sialylated and 19 were not. All of the glycans at the NSS (Asn-120), NFS (Asn-297), and NMT (Asn-344) sites were tri- and tetra-antennary glycans, although the other three sites included less processed glycans. The variation among the glycans at a single site was greatest on the NGS (Asn-230) glycopeptide, where both neutral and sialylated, small and large glycans were observed. All glycopeptides were identified with a mass accuracy of <5 ppm with the exception of two of the glycans at the NGS site. Independent confirmation of site occupancy was obtained for all but the NGS peptide based on mass match coupled with enzymatic incorporation of the 18O stable isotope when glycosylated asparagine residues were converted to aspartate in the presence of PNGase F and H218O (data not shown). No peptides corresponding to unmodified sites were observed within the MALDI-TOF/MS data. All six potential glycosylation sites appear to be fully occupied.
A comparison of the information obtained from the analysis of the kidney GGT glycopeptides to the analysis of the glycans released from kidney GGT shows that the data from each approach confirm and further expand the insight into glycosylation of renal GGT. There was general agreement between the analysis of the GGT glycopeptides and the N-glycans released from GGT. On the NIS (Asn-N266) site, all of the glycans identified in the glycopeptide analysis were observed among the glycans released from GGT or the kidney glycans. The most abundant glycan composition at this site (Hex5HexNAc5dHex2) was also the most abundant glycan released from GGT (m/z 2663.3). The two most abundant glycan compositions on the NSS (Asn-120) site (Hex6HexNAc6dHex4) and Hex6HexNAc6dHex4 NeuAc1) are also among the most abundant released from GGT (m/z 3460.7, 3821.9).
Among the 36 unique glycan compositions identified in the glycopeptide analysis, 17 were sialylated (Table 1). Only three (m/z 3647.8, 3821.9, and 4183.1) of the sialylated glycans had been detected among the glycans enzymatically released from GGT, and two additional glycans (m/z 3402.7, 3473.7) had been identified in the total kidney glycans pool (Fig. 2). The identification of 12 novel sialylated glycans by glycopeptide analysis indicates the enhanced sensitivity of this approach. Of the 19 nonsialylated glycans identified in the glycopeptide analysis, 11 were detected among the glycans released from GGT, and 4 additional glycans were detected in the total glycan pool. Four novel nonsialylated glycans were identified by glycopeptide analysis. Of note is the Hex4HexNAc6 glycan identified as the second-most abundant at the NST site, which was not identified among the released glycans (predicted m/z 2356.2 for permethylated N-glycan). Data from the N-glycans released from GGT provided novel information not present in the glycopeptide analysis. All 13 N-glycans with m/z >2200 identified among the N-glycans released from kidney GGT (Fig. 2B) were observed in the kidney GGT glycopeptide analysis (Table 1). However, none of the eight N-glycans with m/z <2200, released from GGT were observed in the glycopeptide analysis.
N-Glycomic Analysis of Liver Membrane Glycoproteins and Liver GGT
The profile of N-glycans from liver membrane glycoproteins reveals a diverse mixture of high mannose and complex type structures, which is dominated by a biantennary, di-sialylated N-glycan (m/z 2792.5, see Fig. 5A). There is considerable overlap between the glycan ions identified among those released from the total liver and kidney membranes. Of the 44 glycan ions identified in the liver, 30 were also present in the kidney (Fig. 2A). The structures that were unique in the liver were primarily complex type N-glycans, including the tri- and tetra-antennary, di-fucosylated structures. Profiles of the N-glycans from the membrane glycoproteins of two liver donors were indistinguishable (see supplemental Fig. 2).
FIGURE 5.
MALDI-TOF-MS glycomic profile of permethylated N-glycans derived by PNGase F digestion of a soluble detergent extract of membranes from normal human liver 1 (A) and GGT isolated from normal human liver 1 (B). For symbol definitions, see Fig. 2.
The N-glycans released from hepatic GGT are shown in Fig. 5B and constitute a subset of the glycans from the total liver membrane glycoproteins. The hepatic GGT N-glycans were dominated by the same biantennary, di-sialylated structure (m/z 2792.4) that dominated the liver membrane glycan profile. The pattern of complex type N-glycans on liver GGT mirrors the relative abundance pattern of these glycans in the liver membranes. However, the high mannose structures identified in the liver membrane pool (Fig. 5A) were not observed on purified liver GGT (Fig. 5B), similar to the pattern observed in the kidney (Fig. 2, A and B).
The N-glycan profile from liver GGT (acidic, unbisected, and rare antennary fucosylation) contrasts markedly with that of renal GGT (low sialylation, bisected, and abundant antennary fucosylation, see Fig. 2B). Only three low abundance glycoforms were identified on both the kidney and liver GGT (m/z 1906, 2070, and 2110). The N-glycans identified on liver and kidney GGT confirm the variations in the molecular composition of hepatic and renal glycosylation proposed by the electrophoretic mobility differences (Fig. 1). The N-glycans on liver GGT exhibited a higher degree of sialylation consistent with the increased mobility of liver GGT on native gels and its more pronounced migrational shift following neuraminidase treatment relative to the kidney enzyme (Fig. 1A). In addition, there is less microheterogeneity among the liver GGT glycans compared with the kidney GGT glycans, which correlates with the more compact migration pattern observed for the liver GGT compared with the broad band observed for kidney GGT in native gels (Fig. 1A).
Site-specific Glycosylation of Liver GGT
Tryptic peptides from liver GGT were analyzed for their site-specific glycosylation patterns, and the data were compared with those observed for kidney GGT. The time intervals during which the liver GGT glycopeptides eluted are indicated on the LC-MS/MS base peak chromatogram (Fig. 6A). The liver glycopeptides eluted as families. The averaged MS spectrum for the liver NMT (Asn-344) glycopeptide family shows eight distinct glycans present at this site (Fig. 6B), six of which were observed among the glycans released from hepatic GGT (Fig. 5B). The low abundance composition at m/z = 1480.3 (Fig. 6B) was unique to the NMT site (among the glycopeptides detected on liver GGT), but its structure is not clearly defined.
FIGURE 6.
Glycopeptide analysis of GGT isolated from normal human liver tissue. A, base peak intensity chromatogram of GGT tryptic digests immunopurified from liver tissue. The shaded regions depict the retention times of the different glycopeptides associated with this glycoprotein. Table 2 summarizes the microheterogeneity of each site. B, averaged LC/Orbitrap-FT MS spectrum for the liver GGT Asn-344 (NMTSEFFAAQLR) glycopeptide, showing the associated glycoconjugates within the elution interval. Asterisks denote glycoconjugates that were confirmed by MS/MS. Peaks annotated by Φ represent members of the Asn-120 (N-sequon = NSS) family of glycopeptides that co-eluted over this same interval. P represents the NMTSEFFAAQLR tryptic peptide to which the N-glycans are attached. Unannotated peaks represent unmodified peptides that co-eluted over this interval. C, MS/MS spectrum showing the fragmentation pattern of the Asn-120 (NSS) glycopeptide at m/z = 1347.2. For symbol definitions, see Fig. 2.
The fragmentation pattern for the NSS (Asn-120) liver GGT glycopeptide bearing a biantennary, di-sialylated, core-fucosylated complex type N-glycan (m/z = 1347.2) is shown in Fig. 6C. The collision-induced dissociation fragmentation pattern contains the diagnostic sialylated oxonium ion at m/z = 657.9 that assisted in its preliminary assignment as a glycopeptide. Additional oxonium ions at m/z = 528.2 and 818.9 were also evident. Two fragment ions at different charge states confirm the assignment of the modified peptide backbone (m/z = 946.5 and 1892.7). Core fucosylation is evidenced by the detection of a series of fragment ions bearing a single fucose residue on substructural elements composed solely of proximal GlcNAc residues attached to the peptide backbone (Fig. 6C, m/z = 1019.9, 1121.1, and 1201.6).
The results from the liver GGT glycopeptide analyses are presented in Table 2. The glycopeptide analysis yielded evidence for glycosylation at four of the six sites. Of the 11 unique glycan compositions identified in the glycopeptide analysis, all were sialylated, although seven were also fucosylated. The nonfucosylated, biantennary, di-sialylated N-glycan (m/z 2792.5), which dominated the liver glycomics profile from both the total membrane fraction and the GGT-specific pool of N-glycans (Fig. 5), was also the most prominent glycan on the glycopeptides. It was the only glycan composition detected for the NIS site by mass match and constituted greater than 50% of the N-glycans on the NGS (Asn-230) and NMT (Asn-344) sites (Table 2). This glycan constituted 23.5% of the glycans on the NSS site, although its fucosylated derivative (m/z 2966.5) accounted for 33.4% of the glycans on this site. As a result, the NSS site was the only site at which fucosylated glycans were more abundant than nonfucosylated glycans. Eight of the 11 glycan compositions identified in situ by the glycopeptide analysis were also observed among the glycans released from liver GGT, and an additional glycan Hex7HexNAc6dHex1NeuAc3 (m/z 4226.1) was observed within the total liver membrane N-glycan pool (Fig. 5A). Both of the two di-fucosylated glycans that were identified by the glycopeptide analysis, Hex6HexNAc5dHex2NeuAc2 and Hex7HexNAc5dHex2NeuAc2, were not observed among the glycans released from GGT or liver membranes (predicted m/z 3589.8 and 3793.9 for permethylated N-glycans). A comparison of the N-glycans released from hepatic GGT to the liver glycopeptides shows that the low molecular weight glycans (m/z <2450) identified in the GGT glycomic analysis were not observed on the glycopeptides, although all of the larger glycans (m/z >2450) identified in the glycomic analysis were observed on the glycopeptides.
TABLE 2.
N-Glycan microheterogeneity of hepatic GGT as determined by LC/MS-MS
Relative abundance values were deduced from comparative ion intensities among glycopeptide family members. The tryptic peptides in which each glycosylation site is contained are depicted in Fig. 3. Calculated m/z values for the corresponding permethylated N-glycans identified on either hepatic GGT (a) or from the pool of liver membrane glycoproteins (b) in Fig. 5 are included for reference. For abbreviations, see Table 1.
As shown in Table 2, the analysis of liver GGT glycopeptides yielded evidence for glycosylation at four (Asn-120, Asn-230, Asn-266, and Asn-344) of the six sites in the large subunit. Neither modified nor unmodified tryptic peptides containing the NFS (Asn-297) or NST (Asn-95) sites were detected over the course of multiple experiments, using GGT immunopurified from two independent normal human livers. The corresponding unglycosylated peptides for these sites were also absent in LC-MS/MS analyses. Multiple attempts were made to address this discrepancy instrumentally, including increasing the sample load, altering the elution parameters, using an alternative ion source, and re-probing the MS/MS data for evidence of incomplete tryptic digestion, for tyrosine-sulfated peptides, for the presence of O-glycans, or for the most common natural sequence variants and single nucleotide polymorphisms in human GGT (10). This discrepancy may reflect a lack of glycosylation at these sites in the liver, a pervasive and unanticipated form of post-translational modification, or sequence variation within these two tryptic peptides.
Unique Glycosylation of GGT in Human Liver Versus Human Kidney
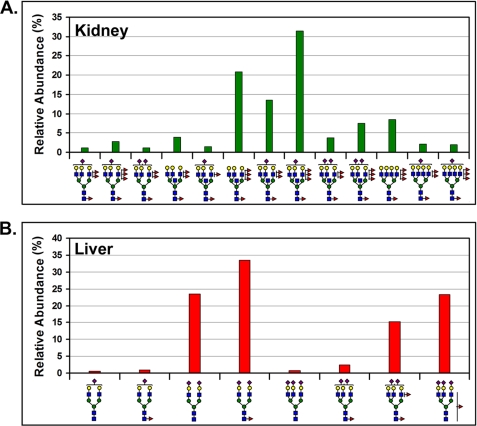
A comparison of the glycopeptides identified in liver GGT shows that all of these glycopeptides are distinct from the glycopeptides identified in kidney GGT. In addition, none of the 11 glycan compositions identified on liver GGT glycopeptides were in common with any of the 36 glycan compositions identified on kidney GGT glycopeptides (Tables 1 and 2). N-Glycans identified on the NSS (Asn-120) site are illustrated in Fig. 7 with their corresponding relative abundances. Thirteen glycan compositions were found on the kidney GGT at this site and eight on the liver GGT. Tandem MS analysis of the tri-antennary, tri-sialylated, and mono-fucosylated glycan identified on the liver NSS site revealed a mixture of both core and antennary-fucosylated structures for this glycan as indicated in Fig. 7B. A distinct pattern of tissue-specific glycans can be illustrated for all GGT glycosylation sites analyzed. These data demonstrate that investigation of tissue-specific glycosylation of proteins cannot rely upon analysis of total glycans released from a tissue. Analysis of glycans released from an isolated protein produced by a single cell type within a tissue provides more refined information. However, as observed in the N-glycan characterization of both liver and kidney GGT, glycopeptide analysis reveals a more complete spectrum of the glycans that modify this protein and eliminates the possibility of detecting glycans on a minor contaminating protein that co-purified with the glycoprotein. The exquisite sensitivity of the glycopeptide analysis has revealed that the liver synthesizes an array of N-glycans on the GGT polypeptide that are completely distinct from those that are synthesized by the kidney.
FIGURE 7.
Tissue-specific N-glycosylation patterns on the Asn-120 site of GGT. The relative abundances of distinct N-glycan structures on the Asn-120 (NSS) glycosylation site, as inferred from glycopeptide ion intensities, are graphically depicted for GGT immunopurified from either kidney (A) or liver (B) tissue. For symbol definitions, see Fig. 2.
DISCUSSION
Previous studies have reported differences in the N-glycans produced by different tissues. However, in most studies there has been little appreciation of the many cell types within tissues. This study of the glycosylation of GGT in human kidney and liver highlights the importance of understanding the composition of a tissue and protein expression within the tissue in studies of glycosylation. Analysis of the total N-glycans released from membrane glycoproteins revealed that more than 65% of the glycan compositions were common between kidney and liver tissue. A major finding of this study was that, despite the overlap in composition among the N-glycans released from the two tissues, there was no overlap among the glycans identified in the glycopeptide analysis of human kidney and liver GGT. Despite distinct glycosylation, GGT from both tissues retains full enzymatic activity as shown on native gels (Fig. 1A). Additional insights into site-specific glycosylation were also gained from the glycopeptide analysis of GGT isolated from both tissues.
The kidney consists of many different cells, including endothelial cells, podocytes, mesangial cells, epithelial cells of the proximal tubules, distal tubules and collecting ducts, fibroblasts, smooth muscle, and nerves. A kidney that has not been perfused also contains circulating blood cells and plasma. We focused on the glycosylation of GGT, which, in the kidney, is expressed only by the proximal tubule cells (11). We compared the glycosylation of GGT isolated from human kidney to GGT isolated from human liver. The liver differs from the kidney in that 95% of the total mass of the liver is composed of a single cell type, the hepatocyte (43). GGT is expressed by hepatocytes and is localized to their bile canalicular surface. GGT is also expressed by bile duct cells in the liver, but these cells contribute only a minor fraction to the GGT in the liver (11). Therefore, the GGT isolated from kidney and from liver provides insight into the biosynthesis of N-glycans within a single cell type in each tissue.
We have identified 40 distinct N-glycan structures among the glycopeptides from human kidney GGT with the potential for additional structural and linkage-specific isomers. The N-glycans identified were of the complex type and ranged from bi- to tetra-antennary structures, with a predominance of bisecting N-acetylglucosamine residues. These multiantennary structures exhibited a high degree of fucosylation, with some structures containing both core fucosylation and up to three Lewis-type peripheral fucose residues. Variable degrees of sialylation (0–3) were also observed on the tri- and tetra-antennary structures. These results confirm and extend those reported by Yamashita et al. (28) who, based on exoglycosidase and paper electrophoresis analysis of human kidney GGT N-glycans released by hydrazinolysis, concluded that human kidney GGT is modified with bisected bi- and triantennary N-glycans with variable degrees of core and LewisX peripheral fucosylation as well as sialylation. The sensitivity of the mass spectrometric approach in this study revealed site specificity and a higher degree of compositional microheterogeneity than previously known.
Our data are the first to demonstrate that each of the six putative N-glycosylation sites on the large subunit of the kidney enzyme is glycosylated. In addition, Western analysis (Fig. 1B) demonstrates that the single putative N-glycosylation site in the small subunit is also quantitatively occupied in the kidney enzyme. The microheterogeneity of the GGT glycans at each site in the kidney indicates all are accessible to the glycosylation machinery as the heterodimer undergoes remodeling and maturation in the endoplasmic reticulum and Golgi network. However, the smaller sizes and uniform neutrality of the N-glycans on the NST (Asn-95) and NIS (Asn-266) glycopeptides relative to the other glycosylation sites and the high relative abundance of nonfucosylated glycans at the NST site suggested that steric hindrance at these sites may restrict processing of the glycans by the resident glycosyltransferases.
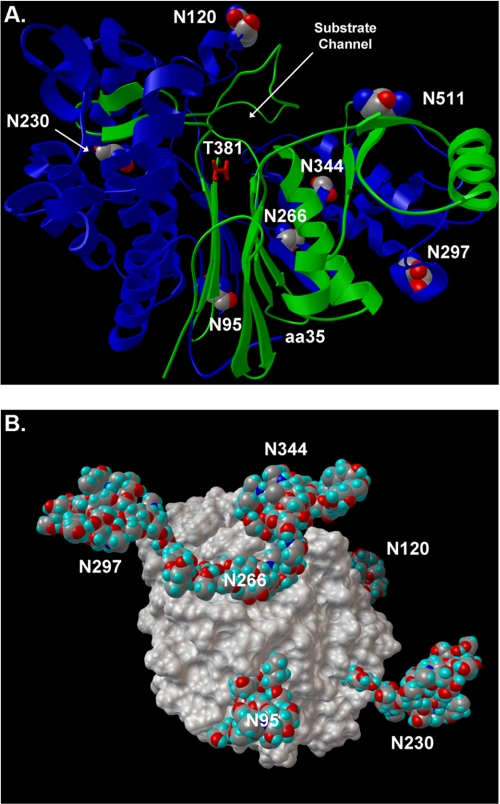
A three-dimensional structural model of human GGT was developed. The model was based on the crystal structure of GGT from E. coli, which shares 29% identity in the large subunit and 38% identity in the small subunit with human GGT (39). Crystal structures have only been reported for bacterial GGT. None of the eukaryotic GGT enzymes have been crystallized. The ribbon model of human GGT reveals two parallel β-sheets that form a hairpin and open into a channel through which the substrate gains access to threonine 381 in the active site of the enzyme (Fig. 8A). All seven of the N-glycosylation sites are on the surface of the molecule. NST (Asn-95) and NIS (Asn-266), the two sites bearing small neutral glycans in the renal enzyme, are localized on the hairpin of the β-sheets. They are on the surface of the protein from which the N terminus of the large subunit extends. GGT is a type II membrane protein, and the N terminus of the large subunit contains a single transmembrane domain (amino acids 5–26). The proximity of the Asn-95 and Asn-266 sites to the N terminus of the large subunit indicates that the glycans modifying these sites are on the surface of the protein facing the plasma membrane. Analysis of a space-filling model that includes the most common glycan at each site reveals that the glycans at these two sites are located in small clefts on the surface of the protein (Fig. 8B). These two glycans are locked in a more rigid conformation than the five other glycans, each of which arises from N-glycosylation sites located on ridges facing the extracellular environment. The localization of the glycans on Asn-95 and Asn-266 to small clefts facing the membrane strongly supports the contention that the smaller sizes and uniform neutrality of the N-glycans at these two sites result from steric hindrance. Further studies of GGT glycosylation may provide additional insights into the temporal and spatial processing of the protein, particularly with regard to folding, autocleavage, and glycosylation as it traverses the endoplasmic reticulum and Golgi. The space-filling model has been rotated to show the maximal number of glycans (Fig. 8B). Six of the seven are observed, including Asn-95 and Asn-266.
FIGURE 8.
Human GGT homology model. A, ribbon diagram of human GGT (large subunit, blue; small subunit, green). Glycosylated asparagine residues are highlighted with CPK atom coloring, and their positions in the amino acid sequence are indicated with white numbers. The position of the substrate channel and catalytic threonine (T381, red sticks) are shown. The homology model is based on the crystal structure of soluble GGT from E. coli. The first 34 amino acids of human GGT contain the transmembrane domain and do not have homology to E. coli GGT. The position of amino acid 35 (aa35) is shown, which defines the orientation of the enzyme on the cell surface. B, space-filling model of human renal GGT with the most abundant N-glycans identified at each site (Table 1). Rotational view of the renal heterodimeric enzyme, featuring a view from the intracellular perspective (i.e. base of transmembrane stalk). The Asn-511 glycosylation site is located on the distal surface of GGT and is obscured by the N-glycan at position Asn-344.
We have identified 12 N-glycan structures among four glycopeptides within the large subunit of human liver GGT. Western analysis (Fig. 1B) demonstrates that the putative N-glycosylation site in the small subunit is also quantitatively occupied on the liver enzyme. The glycosylation pattern in liver is strikingly different from that of the kidney enzyme, with none of the glycan compositions found in the liver identified on kidney GGT glycopeptides. The relative abundance data reveal liver GGT glycopeptides are dominated by a di-sialylated, nonbisected biantennary complex type N-glycan. This structure was previously inferred from lectin studies and identified by Yamashita and co-workers on human liver GGT (24, 27, 29). Our data identified a more extensive array of N-glycans on human liver GGT than recognized previously. Although a larger number of glycoforms were detected by MS, these could be related in a homologous series of incremental additions of monosaccharides, as shown in the example of glycoforms observed on the NSS site (Fig. 7B). Analysis of the glycans on liver GGT in the context of the GGT structural model (Fig. 8) reveals that the glycans on Asn-120, Asn-230, and Asn-344, which are predicted to be located on ridges readily exposed to the extracellular environment, are complex and charged glycans. This is similar to what was observed on the kidney enzyme. We were unable to identify the Asn-95 liver glycopeptide and found only a single mass match (not confirmed by MS/MS) for the Asn-266 liver glycopeptide; thus, we were unable to determine whether the glycans on these sites were smaller and less charged, as was observed on renal GGT.
Our data are in agreement with recent studies from other investigators on large pools of tryptic protein fragments from a variety of human tissues that provided preliminary evidence for GGT glycosylation at sites Asn-120 (in liver and T-lymphocytes), Asn-230 (liver), and Asn-511 (liver and bile) (44–46). These studies employed general glycopeptide-enrichment strategies (lectin or hydrazide chemistry) and tandem MS identification of the tryptic peptides released from the immobilized N-glycans. No information was reported with respect to the composition of the N-glycans attached to these sites.
The organ- and GGT-specific glycosylation patterns observed during the course of our investigation were conserved among tissue donors. In addition, the N-glycans identified in our MS characterization of GGT isolated from kidney and liver tissues and those documented by Yamashita et al. (28, 29) on samples derived from geographically distinct donors further document tissue-specific, gender-independent glycosylation patterns within the human population. The N-glycoforms that we identified on liver GGT isolated from two distinct donors are similar in the composition and abundance of glycans identified by others on liver-derived serum glycoproteins (47–49). These data suggest that GGT is glycosylated in a manner analogous to other liver glycoproteins and that the glycosylation pattern is highly conserved among individuals. Babu et al. (50) reported a similar degree of conservation in the glycosylation patterns acquired from human neutrophils originating from geographically isolated samples. These findings indicate that factors such as age, gender, and environment have negligible effects on N-glycosylation patterns in human tissues. This is in contrast to data derived from total N-glycan profiling of human serum, which is variable from person to person (51).
Analysis of glycopeptides from human tissues is the optimal approach for identifying the products of in vivo glycan biosynthesis. Cell lines and primary cell cultures are problematic as glycosylation patterns are altered when cells are cultured in vitro. Animal model systems also have limitations. Although bisecting GlcNAc and peripheral fucosylation are commonly observed among both human and mouse kidney N-glycans, a biantennary, bisected, and nonsialylated core-fucosylated structure was the predominant species in our glycomic analysis of the human kidney tissue, and a tri-fucosylated variant of this structure is predominant in mouse kidney (4–6). In addition, the N-glycomic analysis from human kidney tissue revealed a greater degree of peripheral sialylation than previously detected in the mouse. The human and mouse liver profiles also differ in that the oxygen-substituted form of sialic acid, N-glycolylneuraminic acid, which is abundant in the mouse, was not detected in the human samples as expected, because CMP-Neu5Ac hydroxylase is inactive in human tissues (52).
In the past, characterization of human glycoproteins produced by a single cell type within a tissue has been hindered by the amount of purified protein required. Glycan modifications at individual sites on glycoproteins present additional challenges because glycopeptide ionization is often suppressed by the presence of unmodified peptides, and glycan heterogeneity in each glycopeptide reduces the abundance of individual species, and some structures share the same mass requiring MS-MS or orthogonal methods to differentiate them. These obstacles have been overcome in this study by the use of nano-reverse-phase LC to enrich for glycopeptide families over an elution interval, the implementation of sensitive strategies to monitor these elution intervals, marked by the co-elution of sugar oxonium ions, on-line screening for candidate glycopeptides based on a mass list of potential peptides and glycans from the glycomics analysis, which is benefited by the 5 ppm mass accuracy of the FT detector, and selected data-dependent MS-MS to obtain confirmatory fragment ion support for the preliminary assignments.
Glycans are receiving increased scrutiny as potential biomarkers of cancer and other diseases. Membrane-bound glycoproteins are often shed into the serum as a by-product of the disease process. Our overall goal is to define disease-specific alterations in the glycosylation of GGT and develop the disease-specific glycopeptides as biomarkers that can be detected on GGT released into the urine and serum. To identify and develop glycoproteins as biomarkers requires sensitive methodology to define the glycopeptides within the proteins expressed in normal tissue and the alteration in the glycopeptides produced in the diseased tissue. The data presented in this study are the first to provide an in-depth characterization of the glycopeptides and site-specific microheterogeneity of the glycans from human kidney and liver GGT produced in vivo.
Supplementary Material
Acknowledgments
We gratefully acknowledge the assistance of Dr. Michael Kinter, Oklahoma Medical Research Foundation, for assistance with the MS3 analysis on kidney N-glycans.
This work was supported, in whole or in part, by National Institutes of Health Grant F32CA128338 (to M. B. W.), Grants RO1CA57530 and R56CA057530 (to M. H. H.), and Grant RR018942 from the NCRR for the National Center for Glycomics and Glycoproteomics at Indiana University (to M. V. N. and Y. M.). This work was also supported by the Indiana Metabolomics and Cytomics Initiative (METACyt) funded by a grant from Eli Lilly Endowment (to Y. M.). The Oklahoma Center for Medical Glycobiology is supported by the Office of the Vice President for Research and the Department of Biochemistry and Molecular Biology at University of Oklahoma Health Sciences Center.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- GGT
- γ-glutamyl transpeptidase
- PNGase F
- peptide:N-glycosidase F
- LTQ-FT
- linear trap quadrupole-Fourier transform
- Neu5Gc
- N-glycolylneuraminic acid.
REFERENCES
- 1.Varki A. (1993) Glycobiology 3, 97–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Helenius A., Aebi M. (2001) Science 291, 2364–2369 [DOI] [PubMed] [Google Scholar]
- 3.Haltiwanger R. S., Lowe J. B. (2004) Annu. Rev. Biochem. 73, 491–537 [DOI] [PubMed] [Google Scholar]
- 4.Haslam S. M., North S. J., Dell A. (2006) Curr. Opin. Struct. Biol. 16, 584–591 [DOI] [PubMed] [Google Scholar]
- 5.Comelli E. M., Head S. R., Gilmartin T., Whisenant T., Haslam S. M., North S. J., Wong N. K., Kudo T., Narimatsu H., Esko J. D., Drickamer K., Dell A., Paulson J. C. (2006) Glycobiology 16, 117–131 [DOI] [PubMed] [Google Scholar]
- 6.Nairn A. V., York W. S., Harris K., Hall E. M., Pierce J. M., Moremen K. W. (2008) J. Biol. Chem. 283, 17298–17313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schachter H. (2000) Glycoconj. J. 17, 465–483 [DOI] [PubMed] [Google Scholar]
- 8.Czlapinski J. L., Bertozzi C. R. (2006) Curr. Opin. Chem. Biol. 10, 645–651 [DOI] [PubMed] [Google Scholar]
- 9.Dube D. H., Bertozzi C. R. (2005) Nat. Rev. Drug Discov. 4, 477–488 [DOI] [PubMed] [Google Scholar]
- 10.Peracaula R., Barrabés S., Sarrats A., Rudd P. M., de Llorens R. (2008) Dis. Markers 25, 207–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanigan M. H., Frierson H. F., Jr. (1996) J. Histochem. Cytochem. 44, 1101–1108 [DOI] [PubMed] [Google Scholar]
- 12.Hughey R. P., Rankin B. B., Elce J. S., Curthoys N. P. (1978) Arch. Biochem. Biophys. 186, 211–217 [DOI] [PubMed] [Google Scholar]
- 13.Hanigan M. H., Ricketts W. A. (1993) Biochemistry 32, 6302–6306 [DOI] [PubMed] [Google Scholar]
- 14.Lieberman M. W., Wiseman A. L., Shi Z. Z., Carter B. Z., Barrios R., Ou C. N., Chévez-Barrios P., Wang Y., Habib G. M., Goodman J. C., Huang S. L., Lebovitz R. M., Matzuk M. M. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 7923–7926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ikeda Y., Taniguchi N. (2005) Methods Enzymol. 401, 408–425 [DOI] [PubMed] [Google Scholar]
- 16.Inoue M., Hiratake J., Suzuki H., Kumagai H., Sakata K. (2000) Biochemistry 39, 7764–7771 [DOI] [PubMed] [Google Scholar]
- 17.Suzuki H., Kumagai H. (2002) J. Biol. Chem. 277, 43536–43543 [DOI] [PubMed] [Google Scholar]
- 18.Kinlough C. L., Poland P. A., Bruns J. B., Hughey R. P. (2005) Methods Enzymol. 401, 426–449 [DOI] [PubMed] [Google Scholar]
- 19.Boanca G., Sand A., Okada T., Suzuki H., Kumagai H., Fukuyama K., Barycki J. J. (2007) J. Biol. Chem. 282, 534–541 [DOI] [PubMed] [Google Scholar]
- 20.Gardell S. J., Tate S. S. (1981) J. Biol. Chem. 256, 4799–4804 [PubMed] [Google Scholar]
- 21.Shaw L. M., London J. W., Petersen L. E. (1978) Clin. Chem. 24, 905–915 [PubMed] [Google Scholar]
- 22.Shaw L. M., Petersen-Archer L. (1979) Clin. Biochem. 12, 256–260 [DOI] [PubMed] [Google Scholar]
- 23.Huseby N. E. (1981) Clin. Chim. Acta 111, 39–45 [DOI] [PubMed] [Google Scholar]
- 24.Tsuchida S., Yamazaki T., Camba E. M., Morita T., Matsue H., Yoshida Y., Sato K. (1985) Clin. Chim. Acta 152, 17–26 [DOI] [PubMed] [Google Scholar]
- 25.Arai K., Yoshida K., Komoda T., Kobayashi N., Saitoh H., Sakagishi Y. (1989) Clin. Chim. Acta 184, 75–84 [DOI] [PubMed] [Google Scholar]
- 26.Arai K., Yoshida K., Komoda T., Kobayashi N., Sakagishi Y. (1992) Clin. Chim. Acta 210, 35–46 [DOI] [PubMed] [Google Scholar]
- 27.Evjen G., Huseby N. E. (1992) Clin. Chim. Acta 209, 27–34 [DOI] [PubMed] [Google Scholar]
- 28.Yamashita K., Hitoi A., Matsuda Y., Miura T., Katunuma N., Kobata A. (1986) J. Biochem. 99, 55–62 [DOI] [PubMed] [Google Scholar]
- 29.Yamashita K., Totani K., Iwaki Y., Takamisawa I., Tateishi N., Higashi T., Sakamoto Y., Kobata A. (1989) J. Biochem. 105, 728–735 [DOI] [PubMed] [Google Scholar]
- 30.King J. B., West M. B., Cook P. F., Hanigan M. H. (2009) J. Biol. Chem. 284, 9059–9065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tate S. S., Khadse V., Wellner D. (1988) Arch. Biochem. Biophys. 262, 397–408 [DOI] [PubMed] [Google Scholar]
- 32.Thioudellet C., Oster T., Wellman M., Siest G. (1994) Eur. J. Biochem. 222, 1009–1016 [DOI] [PubMed] [Google Scholar]
- 33.Rutenberg H. L., Pamintuan J. C., Soloff L. A. (1969) Lancet 2, 559–564 [DOI] [PubMed] [Google Scholar]
- 34.Alley W. R., Jr., Mechref Y., Novotny M. V. (2009) Rapid Commun. Mass Spectrom. 23, 495–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang P., Mechref Y., Klouckova I., Novotny M. V. (2005) Rapid Commun. Mass Spectrom. 19, 3421–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang P., Mechref Y., Novotny M. V. (2008) Rapid Commun. Mass Spectrom. 22, 721–734 [DOI] [PubMed] [Google Scholar]
- 37.Goldberg D., Bern M., North S. J., Haslam S. M., Dell A. (2009) Bioinformatics 25, 365–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eswar N., Marti-Renom M. A., Webb B., Madhusudhan M. S., Eramian D., Shen M., Pieper U., Sali A. (2006) in Current Protocols in Bioinformatics, Supplement 15, 5.6.1–5.6.30, p. 200, John Wiley & Sons, Inc., New York: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okada T., Suzuki H., Wada K., Kumagai H., Fukuyama K. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 6471–6476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kirschner K. N., Woods R. J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10541–10545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jang-Lee J., North S. J., Sutton-Smith M., Goldberg D., Panico M., Morris H., Haslam S., Dell A. (2006) Methods Enzymol. 415, 59–86 [DOI] [PubMed] [Google Scholar]
- 42.Aoki K., Perlman M., Lim J. M., Cantu R., Wells L., Tiemeyer M. (2007) J. Biol. Chem. 282, 9127–9142 [DOI] [PubMed] [Google Scholar]
- 43.Miki K., Kubota K., Inoue Y., Vera D. R., Makuuchi M. (2001) J. Nucl. Med. 42, 733–737 [PubMed] [Google Scholar]
- 44.Kristiansen T. Z., Bunkenborg J., Gronborg M., Molina H., Thuluvath P. J., Argani P., Goggins M. G., Maitra A., Pandey A. (2004) Mol. Cell. Proteomics 3, 715–728 [DOI] [PubMed] [Google Scholar]
- 45.Chen R., Jiang X., Sun D., Han G., Wang F., Ye M., Wang L., Zou H. (2009) J. Proteome Res. 8, 651–661 [DOI] [PubMed] [Google Scholar]
- 46.Wollscheid B., Bausch-Fluck D., Henderson C., O'Brien R., Bibel M., Schiess R., Aebersold R., Watts J. D. (2009) Nat. Biotechnol. 27, 378–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wada Y., Azadi P., Costello C. E., Dell A., Dwek R. A., Geyer H., Geyer R., Kakehi K., Karlsson N. G., Kato K., Kawasaki N., Khoo K. H., Kim S., Kondo A., Lattova E., Mechref Y., Miyoshi E., Nakamura K., Narimatsu H., Novotny M. V., Packer N. H., Perreault H., Peter-Katalinic J., Pohlentz G., Reinhold V. N., Rudd P. M., Suzuki A., Taniguchi N. (2007) Glycobiology 17, 411–422 [DOI] [PubMed] [Google Scholar]
- 48.Fenaille F., Le Mignon M., Groseil C., Ramon C., Riandé S., Siret L., Bihoreau N. (2007) Glycobiology 17, 932–944 [DOI] [PubMed] [Google Scholar]
- 49.Gil G. C., Velander W. H., Van Cott K. E. (2009) Proteomics 9, 2555–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Babu P., North S. J., Jang-Lee J., Chalabi S., Mackerness K., Stowell S. R., Cummings R. D., Rankin S., Dell A., Haslam S. M. (2009) Glycoconj. J. 26, 975–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Knezeviæ A., Polasek O., Gornik O., Rudan I., Campbell H., Hayward C., Wright A., Kolcic I., O'Donoghue N., Bones J., Rudd P. M., Lauc G. (2009) J. Proteome Res. 8, 694–701 [DOI] [PubMed] [Google Scholar]
- 52.Chou H. H., Takematsu H., Diaz S., Iber J., Nickerson E., Wright K. L., Muchmore E. A., Nelson D. L., Warren S. T., Varki A. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 11751–11756 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.