Abstract
A plastid-derived signal plays an important role in the coordinated expression of both nuclear- and chloroplast-localized genes that encode photosynthesis-related proteins. Arabidopsis GUN (genomes uncoupled) loci have been identified as components of plastid-to-nucleus signal transduction. Unlike wild-type plants, gun mutants have nuclear Lhcb1 expression in the absence of chloroplast development. We observed a synergistic phenotype in some gun double-mutant combinations, suggesting there are at least two independent pathways in plastid-to-nucleus signal transduction. There is a reduction of chlorophyll accumulation in gun4 and gun5 mutant plants, and a gun4gun5 double mutant shows an albino phenotype. We cloned the GUN5 gene, which encodes the ChlH subunit of Mg-chelatase. We also show that gun2 and gun3 are alleles of the known photomorphogenic mutants, hy1 and hy2, which are required for phytochromobilin synthesis from heme. These findings suggest that certain perturbations of the tetrapyrrole biosynthetic pathway generate a signal from chloroplasts that causes transcriptional repression of nuclear genes encoding plastid-localized proteins. The comparison of mutant phenotypes of gun5 and another Mg-chelatase subunit (ChlI) mutant suggests a specific function for ChlH protein in the plastid-signaling pathway.
A number of components required for plastid structure and development are encoded by the nuclear genome. There is a considerable body of evidence that suggests that the proper and timely expression of these genes depends in part on the functional state of the chloroplast. For example, nuclear mutations that result in developmentally arrested chloroplasts also result in the reduced expression of nuclear-localized photosynthetic genes (1). These results tie the functional state of the chloroplast to nuclear function and suggest that the chloroplast signals the nucleus in a retrograde fashion (2).
Such retrograde signaling between chloroplast and nucleus has been studied primarily by using carotenoid-deficient plants induced either by mutations or by carotenoid biosynthesis inhibitors such as Norflurazon (Nf; refs. 3 and 4). Carotenoids prevent the production of reactive oxygen species by excited triplet states of chlorophyll. Carotenoid-deficient plants thus suffer a rapid photooxidation of most chloroplast components under intense light. Although most nuclear-encoded cytoplasmic enzymes are present at normal levels in photooxidatively damaged plants, a small set of nuclear-encoded chloroplast enzymes is absent (3, 4). Accordingly, it was hypothesized that plastids send an unknown signal(s) to the nucleus that regulates the expression of a small subset of nuclear-localized photosynthetic genes (2, 5).
The molecular nature of the plastid signal(s) and the mechanism by which it is relayed to the nucleus remain ambiguous. Although both plastid transcription and translation are necessary for the production of the plastid signal (4, 6), the plastid signal is not a direct translational product of a plastid gene. Light does not seem necessary for the activity of the plastid signal. Dark-grown pea lip1 and Arabidopsis cop1-4 mutants contain plastids that do not arrest as etioplasts but continue to differentiate into prechloroplasts. Nuclear Lhcb1 is derepressed in these dark-grown mutants, implying that the plastid signal does not depend on photosynthesis (7). The plastid signal observed in these mutants grown in the dark still depends on early plastid translation, because Lhcb1 expression is sensitive to lincomycin (7).
Early studies using chlorophyll-deficient mutants led to the proposal that the plastid signal does not depend on chlorophyll biosynthesis (8). However, Johanningmeier and Howell (9) reported an inhibition of light-induced Lhcb mRNA accumulation in Chlamydomonas treated with 2, 2′-dipyridyl (DP), which is presumed to cause Mg-protoporphyrin IX (MgProto) and Mg-protoporphyrin IX monomethyl ester (MgProtoME) accumulation [MgProto(ME)]. They proposed that chlorophyll precursors are negative regulators of Lhcb mRNA accumulation. Kropat et al. (10, 49) recently reported that the addition of MgProto or Mg-protoporphyrin IX dimethyl ester (MgProtoME2) in the dark can substitute for light signals in the induction of nuclear HSP70 genes in Chlamydomonas. Together, this evidence suggests that chlorophyll precursors can act as plastid-derived signals that influence nuclear gene expression. However, the mechanism by which these putative signals are relayed to the nucleus is still unclear.
We have taken a genetic approach to understanding the mechanisms of the plastid-to-nucleus signal transduction pathway(s). We have reported previously the isolation of Arabidopsis mutants, referred to as gun (genomes uncoupled) mutants, that express nuclear-encoded Lhcb and RbcS transcripts in the absence of chloroplast development (11). We described three nonallelic loci (GUN1, GUN2, and an unspecified locus) that are involved in the plastid-dependent regulation of the Lhcb1 promoter.
In this paper, we report the identification of previously unidentified GUN loci (GUN3, GUN4, and GUN5) and the genetic interactions between GUN1, GUN4, and GUN5 genes. We cloned the GUN5 gene and show that it encodes the ChlH subunit of Mg-chelatase. A comparison of Mg-chelatase activity and the gun phenotypes in gun5 and the cs/ch42 mutants, which have defects in the ChlI subunit of Mg-chelatase, shows that the ChlH subunit may have a distinct function in plastid signaling in addition to acting as a subunit of the Mg-chelatase enzyme.
Materials and Methods
Strains and Media.
The transgenic line, pOCA107–2 (12), contains a pOCA18-based Lhcb1*2-hpt/Lhcb1*2-β-glucuronidase (GUS) transgene integrated near GPA1 and was used as the wild type in all the experiments described here. gun mutants were isolated by their elevated expression of Lhcb1*2-GUS in the presence of Nf, as described (11). Because a gun3 allele was not specified in the previous study, we assigned the 11th isolate of 12 gun mutants as gun3-1. gun4-1 and gun5-1 were designated originally as gun1-4 and gun0-6, respectively (11). Unless specified, gun1, gun2, gun3, gun4, and gun5 represent gun1-1, gun2-1, gun3-1, gun4-1, and gun5-1, respectively. cch (conditional chlorina) was isolated from ethyl-methanesulfonate-treated Columbia for its light-sensitive reduction of chlorophyll accumulation (ref. 13 and J.A.B., unpublished data). ch42 and cs are Mg-chelatase chl I mutants isolated by x-ray irradiation (14) and transferred-DNA insertion (15), respectively. Plants were grown on soil or on Murashige–Skoog (Wako-Jyunyaku, Japan) agar medium containing 2% (wt/vol) sucrose. Seedlings were photobleached by including 5 μM Nf (provided by Sandoz Pharmaceutical) in the growth media and exposing the seedlings to continuous white light (100 μmol/m2/sec).
GUS Assay.
GUS activity was measured according to the methods of Jefferson (16). For quantitative assays, plant extracts were prepared from 5- to 6-day-old seedlings, and protein concentrations were determined by Protein Assay reagent (Bio-Rad). One unit of GUS activity was defined as the amount of activity that could produce 1 pmol of 4-methylumbelliferone/minute/mg protein. For a semiquantitative assay, a piece of cotyledon was excised from a 5- to 6-day-old plant and placed into 100 μl of GUS-assay mixture (16). After a 12-h incubation at 37°C, the reaction was terminated by adding 100 μl of 1 M sodium bicarbonate. The fluorescence (excitation wavelength, 365 nm; emission wavelength, 455 nm) was measured with a Perkin–Elmer Luminescence Spectrophotometer LS50B. For histochemical staining, harvested tissues were incubated for 12 h at 37°C in 2 mg/ml 5-bromo-4-chloro-3-indolyl β-d-glucuronide (X-Gluc) in 10 mM sodium phosphate, pH 7.0 (16).
Chlorophyll Measurements.
Chlorophyll was extracted from approximately 0.1 g of fresh 1-week-old seedlings with N,N′-dimethylformamide for 12 h at 4°C in complete darkness. The extract was subjected to spectrophotometric measurements at 603, 647, and 664 nm. Specific chlorophyll content was calculated by using the equations of Moran (17) and normalized to the total fresh weight for each sample.
Porphyrin Measurements.
Porphyrins were extracted on the basis of the method of Rebeiz et al. (18). Approximately 0.1 g of tissue was homogenized with a microtube pestle in ammonium-acetone (0.1 M NH4OH/acetone 1:9) and centrifuged for 15 min at 0°C. The supernatant was extracted with hexane three times. The excitation (Ex) and emission (Em) wavelength for each porphyrin was: Proto, Ex 400 nm and Em 632 nm; MgProto(ME), Ex 420 nm and Em 595 nm (19).
5-Aminolevulinate (ALA)–DP Feeding.
ALA–DP feeding experiments were performed on the basis of the methods of Falbel and Staehelin (20). Seedlings were grown on Murashige–Skoog agar plates containing 50 μM gibberellin A3 in the dark for 4 days at 24°C. Then, 3 ml of an ALA–DP feeding solution (10 mM ALA/10 mM DP/5 mM MgCl2/10 mM KPO4, pH 7.0) was added to each plate. After a 12-h incubation at 24°C in the dark, porphyrin levels were determined as described above. Fluorescence intensity of the extract was normalized to the total fresh weight for each sample.
Northern Analysis.
Total RNA was purified by using the TRIzol reagent (GIBCO/BRL). Then, 3 μg of total RNA was separated on a denaturing agarose gel and blotted onto a nylon membrane. Hybridization was carried out as described (21). Lhcb1 DNA probes were generated from a plasmid containing the Lhcb1*2 genomic DNA (22), and an 18S rRNA genomic sequence was cloned by PCR and used as a probe.
DNA Markers and Clones for GUN5 Cloning.
Molecular markers (23–25) were used to map the GUN5 locus. Yeast-artificial chromosome DNA (CIC7F6, CIC5H3) and a full-length cDNA clone of CHLH from the Columbia ecotype were provided generously by M. Seki and K. Shinozaki (The Institute of Physical and Chemical Research, Tsukuba, Japan). P1 clones were from S. Tabata (Kazusa DNA Research Institute, Chiba, Japan). The plant transformation vector, pPZP221 (26), was from P. Maliga (Rutgers University, Piscataway, NJ). We noticed that the published cDNA sequences of CHLH from C24 ecotype (27) have an insertion and a deletion of a T-residue in the third exon, which results in three amino acid differences from 987–989, compared with the amino acid sequences predicted from the cDNA sequence of Columbia CHLH. We sequenced the corresponding region of genomic DNA from C24, Columbia, and Landsberg erecta. All sequences were identical to the Columbia CHLH in this region. In addition, the Columbia amino acid sequence is conserved perfectly between known BchH/CHLH genes. Therefore, the Columbia sequence is used as the wild-type CHLH sequence in this paper.
Results
Genetic Characterization of gun Mutants.
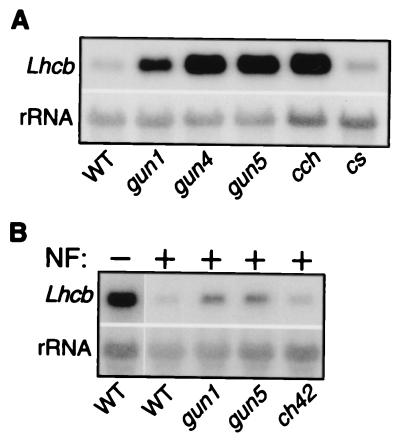
In a previous study, we reported the isolation and characterization of 12 gun-mutant candidates (11). To understand the genetic relationships between the mutants better, they were backcrossed to the wild-type parent (pOCA107–2), and complementation tests were done. The progeny were assayed for the mutant phenotype in the F1 and F2 generations and a total of five GUN loci (GUN1-GUN5) were identified (data not shown). gun2 and gun3 have long hypocotyl phenotypes under white light, and they were found to be alleles of hy1 and hy2, respectively (data not shown). We examined hy1-1 and hy2-1 (28) for the gun phenotype and found that they had elevated Lhcb expression in the presence of Nf (N.M., unpublished data). We roughly mapped gun5 to approximately 29.5 centimorgans on chromosome 5, and we noticed that cch, which affects chlorophyll accumulation, was located near GUN5 (J.A.B., unpublished data). Complementation analysis and a comparison of Lhcb1 mRNA levels in photobleached seedlings indicated that cch and gun5 are allelic (N.M., unpublished data, and Fig. 6A). Although cch is paler than gun5 (Figs. 2 and 3), Lhcb1 mRNA levels were equivalent in photobleached seedlings (Fig. 6A).
Figure 6.
Lhcb1 mRNA accumulation in photobleached pOCA107–2 (WT), gun, cch, cs, and ch42 mutants. (A) cs is not a gun mutant. Wild-type and mutant plants were photobleached for 6 days as described in Materials and Methods, and Lhcb1 mRNA levels were determined as described in Fig. 1B. (B) ch42 is not a gun mutant. Seedlings were grown on Murashige–Skoog media for 3 days. After 3 days, albino ch42 homozygotes, wild-type, and gun mutant seedlings were transferred to media containing Nf and grown for 4 days under strong continuous light. Lhcb1 mRNA levels were determined as described in Materials and Methods.
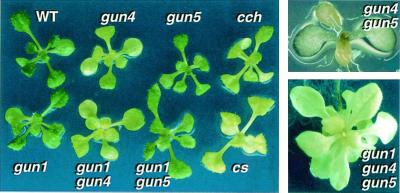
Figure 2.
Comparison of the leaf color of wild-type, gun, cch, and cs plants. Two-week-old wild-type and mutant plants (Left ), a 1-week-old putative gun4gun5 double mutant (Upper Right), and a 1-month-old putative gun1gun4gun5 triple mutant (Bottom Right). Mutants were grown in long-day conditions at fluence rates of 50 μmol/m2/sec.
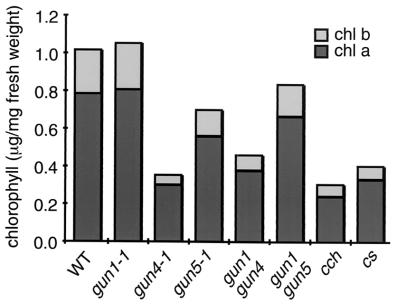
Figure 3.
Comparison of chlorophyll levels in wild-type, gun, cch, and cs mutants. Average chlorophyll content of wild-type and mutant plants. Chlorophyll “a” is presented as a black bar and Chlorophyll “b” as a gray bar. n = 4.
Genetic Interactions Between gun Mutants.
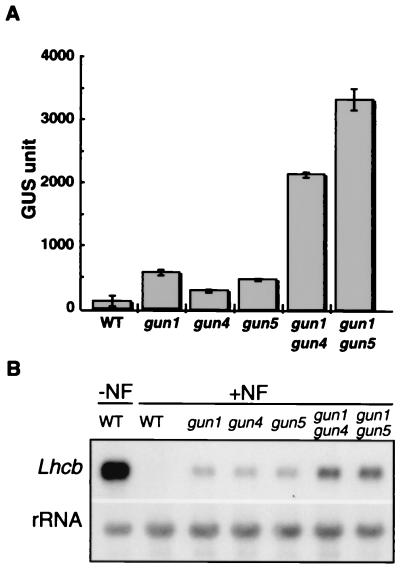
During complementation analysis, we noticed that approximately 1/16 of the F2 individuals from the gun1 × gun4 and the gun1 × gun5 crosses expressed extremely high levels of Lhcb1*2-GUS in the presence of Nf (data not shown). Thus, we hypothesized that they were gun1gun4 and gun1gun5 double mutants. We backcrossed self-fertilized F3 progeny of the putative double mutants with pOCA107–2. As expected for a double mutant, the phenotype segregated in an approximately 9:7 (wild-type/mutant) ratio (data not shown). Lhcb1*2-GUS expression in the gun1gun4 and gun1gun5 double mutants was 4.8- and 6.3-fold higher than the single mutants, respectively, and was about 11–17% of that observed in nonphotobleached wild-type plants (Fig. 1A). The endogenous Lhcb1 mRNA levels were also derepressed synergistically in these double mutants (Fig. 1B).
Figure 1.
Lhcb1*2-GUS and Lhcb1 mRNA accumulation in wild-type and mutant seedlings. (A) Lhcb1*2-GUS activity in photobleached wild-type (WT), gun single, and gun double mutants. Seedlings were photobleached for 5 days, and GUS was extracted and assayed as described in Materials and Methods. In the absence of Nf, wild type expressed 20,120 ± 2,197 GUS units. (B) Lhcb1 mRNA accumulation in photobleached wild-type, gun single, and gun double mutants. Total RNA was analyzed as described in Materials and Methods from seedlings that were grown as described in A.
Individuals with extremely high levels of Lhcb1*2-GUS expression were not observed among gun4 × gun5 F2 progeny, but we found approximately 1/16 of gun4 × gun5 F2 progeny were extremely pale under normal growth conditions (Fig. 2). We also crossed gun1gun4 with gun1gun5 to obtain a gun1gun4gun5 triple mutant, and albino seedlings appeared in a ratio of approximately 1/16 in the F2 generation (data not shown). We were unable to backcross the putative gun4gun5 double mutants and the putative triple mutants because of low fertility. gun4 and gun5 have lower chlorophyll levels than wild type, but gun1 mutants accumulate normal levels of chlorophyll (Figs. 2 and 3). Additionally, gun1gun4 and gun1gun5 accumulate the same levels of chlorophyll as gun4 and gun5 single mutants, respectively (Figs. 2 and 3). Thus, it is most likely that the extreme chlorophyll phenotype of the gun4gun5 double mutant is because of the additive effect of these two mutations. Although we were able to extract low levels of chlorophyll “a” from the putative gun4gun5 double mutants, we could not extract measurable levels of chlorophyll from the putative triple mutants (N.M., unpublished results). It is not surprising to see a more severe chlorophyll phenotype in the triple mutant, because the gun1 mutation can impair chloroplast development under some conditions (29). These results suggest that there are at least two separate but partially redundant pathways for plastid-to-nucleus signal transduction and that GUN1 affects a separate pathway from GUN4 and GUN5. Also, the GUN4/GUN5 pathway is required for proper chlorophyll accumulation.
gun5-1 and cch Are Defective in Mg-Protoporphyrin IX Synthesis.
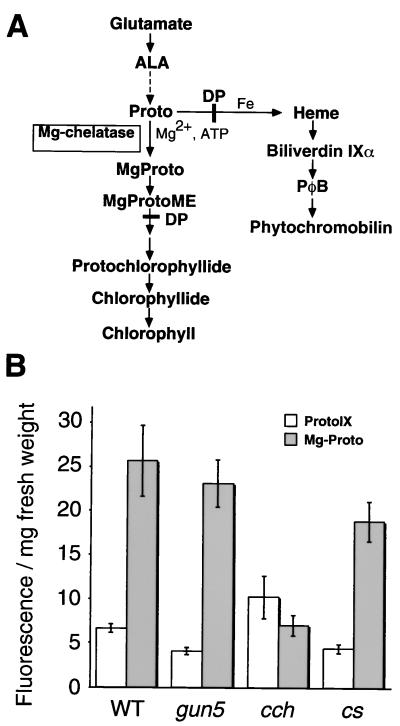
Because gun5 and cch have pale phenotypes, we decided to search for defects in the chlorophyll biosynthetic pathway. Potential bottleneck steps in chlorophyll biosynthesis were determined by measuring chlorophyll precursor levels after feeding dark-grown seedlings with ALA and DP (ref. 20 and Fig. 4A). ALA is an early precursor, and DP inhibits ferrochelatase isocyclic ring formation and causes MgProto(ME) accumulation in dark-grown plants (30). The cs mutant, which has a lesion in the ChlI subunit of Mg-chelatase, produced approximately 74% of the MgProto(ME) levels as wild type (cf. ref. 20 and Fig. 4B). cch made only approximately 28% of the MgProto(ME) that was synthesized by wild type, and cch accumulated more Proto than wild type, which suggests that there is a defect in Mg-chelatase in this mutant. MgProto(ME) levels were not reduced significantly in gun5, and both gun5 and cs produced less Proto than wild type (Fig. 4B). gun5 has a weak chlorophyll phenotype (Figs. 2 and 3), which may explain why a significant difference in MgProto(ME) levels was not detected between gun5 and wild type with this assay.
Figure 4.
Proto and MgProto levels in ALA–DP-fed plants. (A) Schematic diagram of chlorophyll biosynthetic pathway. Dashed arrows represent multiple steps, and the steps that are inhibited by DP are indicated by solid lines across the arrows. (B) Proto and MgProto accumulation in seedlings fed with ALA–DP. Etiolated seedlings (4 days old) were fed with ALA and DP, and porphyrin levels were measured as described in Materials and Methods.
Cloning of GUN5/CCH.
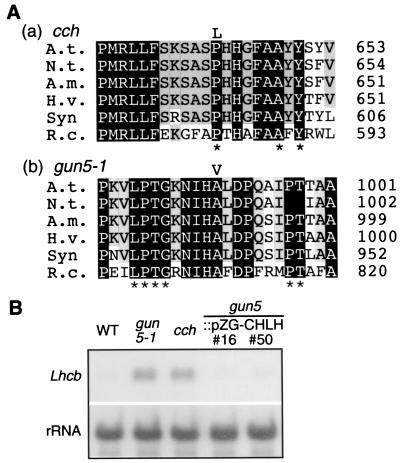
We crossed gun5 to L. erecta to create an F2 mapping population. F2 plants were grown on Nf under continuous bright light, and gun5 mutants were selected by the semiquantitative GUS assay. Then gun5 F2 recombinants were subjected to genetic-linkage analysis by using PCR-based markers as described in Materials and Methods. In the initial mapping effort, GUN5 was localized to the interval between mi174 and CHS (<0.1 centimorgans) on chromosome 5. Previous work by Falbel and Staehelin had suggested that mutants in several crop species with phenotypes similar to cch might have reduced Mg-chelatase activity (20). On the basis of this work, we decided to focus our cloning efforts on the genes that encode the subunits of Mg-chelatase. Mg-chelatase is composed of three subunits, which are commonly referred to as ChlD, ChlH, and ChlI. Because the sequence information was available only for CHLI and CHLH when we mapped GUN5, and because CHLI was mapped on chromosome 4, we tested chromosome 5 yeast-artificial chromosomes and P1 clones from the interval for the presence of the CHLH gene by using PCR. We obtained a specific amplification of the CHLH sequence when CIC7F6, CIC5H3 and MSH12 were used as PCR templates (data not shown). We then amplified the CHLH region (8.5 kb) containing 5′-upstream sequence (2 kb), all the exons and introns, and 3′-sequence (1.9 kb) from Columbia, gun5, and cch and sequenced the products. Both gun5 and the cch mutants have nucleotide substitutions in the third exon. In gun5, the change is from a C to a T, resulting in an A990V mutation. In cch, another C to T substitution results in a P642L mutation (Fig. 5A). Both amino acid substitutions reside in the region conserved among all of the reported ChlH subunits, and P642 is also conserved in Co-chelatase CobN from P. denitrificans (Fig. 5A).
Figure 5.
Cloning of GUN5/CCH. (A) Partial amino acid sequence alignment of ChlH around the mutation site of (a) cch and (b) gun5. The substituted residues in the mutants are indicated above the aligned sequences. The derived amino acid sequences of Arabidopsis thaliana ChlH, Nicotiana tabacum ChlH, Antirrhinum majus ChlH, Hordeum vulgare ChlH, Synechocystis sp. PCC6803 ChlH, and Rhodobacter capsulatus BchH are shown. Conserved residues in six ChlH/BchH and in the ChlH counterparts of Co-chelatase CobN from Paracoccus denitrificans, putative Co-chelatase from Methanococcus jannaschii, and Ni-chelatase from Synechocystis PCC 6803 are marked by asterisks. (B) The wild-type CHLH gene rescues gun5. Lhcb mRNA levels were measured as described in Fig. 1B from two representative gun5 lines (#16 and #50) that were transformed with pZG-CHLH.
The gun5 phenotype was rescued by introducing the wild-type CHLH gene into the gun5 mutant, as described below. We subcloned a 9.1-kb BamHI–XbaI fragment from MSH12 that contains the genomic sequence of CHLH into pPZP221. The resulting construct, pZG–CHLH, was introduced into gun5-1 by Agrobacterium-mediated vacuum infiltration (31). Gentamicin-resistant plants (T1 generation) were self-fertilized, and their progeny (T2 generation) were tested for the zygosity of transferred-DNA (T-DNA) insertion. T2 and T3 generations that were homozygous for T-DNA insertion (five independent lines) were subsequently tested for the gun phenotype. These plant lines showed reduced (i.e., wild-type) levels of Lhcb1 mRNA accumulation in the presence of Nf (Fig. 5B). Therefore, we conclude that the gun5/cch phenotype is caused by defects in the CHLH gene.
chlI Subunit Mutants Are Not gun Mutants.
Our analysis of gun2, -3, -4, and -5 indicates that perturbations in tetrapyrrole metabolism uncouple Lhcb expression from chloroplast development and suggest that Proto and/or MgProto(ME) levels might be responsible for the gun phenotype. To test this hypothesis, we measured Lhcb1 mRNA levels in photobleached cs and ch42 mutants. cs is an insertional mutant in which the last four residues of ChlI are replaced with 11 new residues (15), and the chlorophyll phenotype of cs is comparable to cch (Figs. 2 and 3). ch42 is an x-ray generated allele of chlI that is completely albino (14). Because ch42 homozygotes were unable to grow without sucrose, we used albino progeny derived from CH42/ch42 heterozygous parents for this analysis. Unlike cch and the gun mutants, cs and ch42 Lhcb1 mRNA levels were comparable to wild type in the presence of Nf (Fig. 6 A and B). Thus, plastid-to-nucleus signaling is fully functional in cs and ch42, even though cs has less Mg-chelatase activity than gun5 (Fig. 4B), and the chlorophyll phenotype of ch42 is severe. These data strongly suggest that modulation of Proto and/or MgProto(ME) levels is not sufficient to derepress Lhcb1 genes in photobleached seedlings and that ChlH is an interorganelle signaling molecule.
Discussion
Here we show that the GUN5/CCH gene encodes the ChlH subunit of Mg-chelatase, which is a key enzyme in the chlorophyll branch of the tetrapyrrole biosynthetic pathway. We also found that ChlI subunit of Mg-chelatase is not necessary for plastid signal transduction, because the chlI (cs and ch42) mutants do not exhibit a gun phenotype. Thus, our results provide strong genetic evidence in support of earlier studies that suggest that chlorophyll precursors may function as signal molecules in the plastid-to-nucleus signal transduction pathway; however, they also point to a specific role for the ChlH protein in this signaling pathway.
Multiple Signaling Pathways from Plastids to the Nucleus.
At present, we have identified five loci that uncouple Lhcb1 transcription from chloroplast development. Double-mutant studies suggest that GUN1 might function in a pathway that is separate from but partially redundant with the GUN4/GUN5 pathway. Strong enhancements of the gun phenotype were observed in gun1gun4 and gun1gun5 double mutants. The putative gun4gun5 double mutant did not exhibit such an enhanced gun phenotype despite a drastic reduction in chlorophyll accumulation. There is another difference between the mutants: gun1 is defective in greening after a prolonged period of dark growth, whereas neither gun4 nor gun5 has such a defect (refs. 11 and 29, and N.M., unpublished data). Therefore, it is likely that there are at least two independent GUN pathways, one that is composed of GUN1 and the other composed of GUN4 and GUN5. However, it is possible that GUN1 and GUN2, -3, -4, and -5 function in the same pathway as our results, which might be due to the synthetic enhancement of leaky mutant combinations (32). Lastly, it should be noted that chloroplasts also regulate Lhcb genes with a light-intensity signal that is mediated by the redox status of plastoquinone (33), thereby suggesting yet a third pathway of signaling from plastids to the nucleus.
Tetrapyrroles as Signals from the Plastid.
We have found that gun2 and gun3 are alleles of hy1 and hy2, respectively, and that hy1-1 and hy2-1 (28) are gun mutants. hy1-6.2, a null allele (34), is also a gun mutant (48). HY1 and HY2 are required for the synthesis of phytochromobilin (PΦB) from heme. HY1 encodes a heme oxygenase that converts heme to biliverdin IXα (BV; refs. 35 and 36), and HY2 is thought to encode a PΦB synthase that converts BV to 3(Z)-PΦB (37). Chlorophyll synthesis is repressed in gun2 (R.L., unpublished results), hy1, hy2, the corresponding mutants of tomato and pea (ref. 38 and M. Terry, personal communication), and in transgenic plants that attenuate PΦB synthesis by targeting mammalian biliverdin IXα reductase to the plastids (39). Heme, PΦB, and chlorophyll are derived from a group of common precursors in the chloroplast. Repression of chlorophyll synthesis in these mutants is thought to be mediated by repressive effects of heme (ref. 38 and M. Terry, personal communication) or other linear tetrapyrroles (39) on the synthesis of early tetrapyrrole precursors. hy1gun4 and hy1gun5 double mutants have albino phenotypes that resemble the gun4gun5 double mutant (48). Because gun2 is an allele of hy1, and because gun2 and gun3 affect chlorophyll accumulation like gun4 and gun5, it is likely that gun2–gun5 mutations affect the same plastid-to-nucleus signaling pathway by perturbing tetrapyrrole synthesis.
One technical concern is that gun2–gun5 are not completely photobleached by Nf. Nf inhibits chloroplast development by blocking carotenoid synthesis. Carotenoids quench excited triple states of chlorophyll, which can interact with molecular oxygen and produce reactive oxygen species. It is possible that there is less photooxidative damage in the photobleached plastids of chlorophyll-deficient mutants, but two observations indicate that inefficient photobleaching is not the cause of the gun phenotype in gun2–gun5. First, Lhcb1 expression is derepressed in these mutants when chloroplast development is blocked with chloramphenicol (N.M., unpublished results), which inhibits chloroplast translation. Second, the Arabidopsis chlorophyll-deficient mutants cs, ch42, ch1-2, and ch5-1 are not gun mutants (Fig. 6 and N.M., unpublished results).
The ChlH Subunit in Chlorophyll Biosynthesis and Plastid Signaling.
We found that the GUN5/CCH gene encodes the BchH/ChlH subunit of Mg-chelatase. The subunit varies in size (123–154 kDa among different species), binds to Proto, and is thought to be largely responsible for catalysis. The two smaller subunits of Mg-chelatase, ChlI and ChlD, form an ATP-dependent complex that associates with ChlH–Proto and stimulates this reaction. After Mg insertion, the complex is thought to dissociate into ChlH–MgProto and ChlI-ChlD–MgADP (40). The residues that are altered in gun5 and cch (A990 and P642, respectively) are in highly conserved regions, and P642 is even conserved in Co-chelatase and Ni-chelatase (Fig. 5A and ref. 40). Although these substitutions have dramatically different effects on chlorophyll synthesis, the gun phenotypes of these alleles are very similar. The functional topography of ChlH has not been determined, but our results indicate that catalytic and signaling functions of ChlH can be uncoupled.
Plastid Signal Transduction: A Model.
Feeding and inhibitor experiments have implicated intermediates in the chlorophyll biosynthetic pathway, especially Proto(ME), in plastid-mediated repression of Lhcb genes in Chlamydomonas and cress seedlings (9, 41, 42), and Proto(ME) feeding activates nuclear heat-shock genes through a light-responsive promoter element in dark-grown Chlamydomonas cultures (10, 49). MgProto(ME) levels should be reduced in gun2–gun5 and cch, which is consistent with MgProto(ME) acting as a plastid signal in Arabidopsis. However, MgProto(ME) levels are reduced also in the ChlI subunit mutants cs and ch42, which are not gun mutants. Also, Proto and MgProto(ME) are below the limits of detection in Arabidopsis seedlings that have been treated with Nf for 6 days (N.M., unpublished results). Also, over- or underexpressing the ChlH and the ChlI subunit in tobacco has pleiotropic effects on tetrapyrrole metabolism, including inhibition of Mg-chelatase activity and suppression of early tetrapyrrole biosynthetic enzyme levels (43, 44). However, ChlH subunit levels were not affected by altered ChlI levels in these studies, and therefore we expect that cs and ch42 mutants might contain wild-type levels of the ChlH subunit.
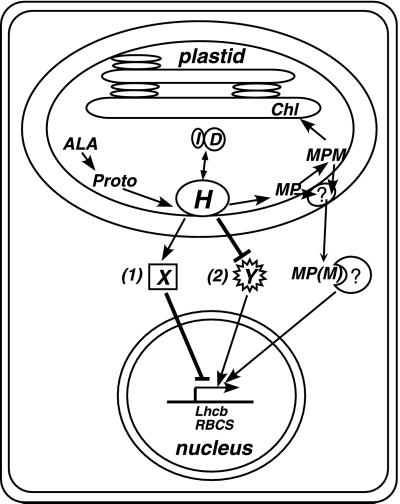
Together, these results suggest a model (Fig. 7). We propose that ChlH measures the flux at the beginning of the chlorophyll branch of the plastid tetrapyrrole biosynthetic pathway and sends information about the rate of chlorophyll synthesis to the nucleus. ChlH might exist in free, Proto-bound, or MgProto-bound states, and ChlI and ChlD might interact with each of these ChlH forms. Each of these ligands might affect the signaling activity of ChlH. gun2, -3, and -4 might affect the signaling activity of ChlH indirectly by modulating the levels of one or more of these ligands or by altering ChlH levels. In contrast, cs and ch42 mutations may not reduce the levels of MgProto(ME), ChlD, or ChlH below a threshold that is required to block the signaling activity of ChlH and uncouple Lhcb1 expression from chloroplast development.
Figure 7.
Model for plastid signaling mediated by the ChlH subunit of Mg-chelatase. The details of the model are explained in the text. “X” and “Y” represent a putative repressor and an activator of Lhcb1 transcription, respectively. MP, MgProto; MPM, MgProtoMe.
Mg-chelatase has been found in the inner envelope of chloroplasts in the presence of 5 μM Mg2+, which is within the estimated range of chloroplast Mg2+ concentrations (45), although the enzyme is also localized in the stroma at lower Mg2+ concentrations (27, 46). Some steps in chlorophyll biosynthesis including MgProto(ME) production are presumed to occur on the inner envelope. Thus, the chloroplast inner envelope seems like a reasonable location for a receptor or a sensor of chlorophyll precursors. ChlH might monitor porphyrin levels by binding excess Proto and/or MgProto and (i) send a negative signal or (ii) inhibit a positive signal to the nucleus via a hypothetical downstream factor(s) (Fig. 7). Proto and/or MgProto(ME) synthesis might be out of balance with the plastid's needs when the plastid is damaged by reactive oxygen species, as described in these experiments, or when the levels of chlorophyll precursors fluctuate in response to internal and external stimuli (47). If MgProto(ME) is exported from the chloroplast as proposed by Kropat et al. (10, 49), another factor(s) probably mediates the transduction of this porphyrin signal from the chloroplast inner envelope to the cytosol (Fig. 7). It is possible also that very small amounts of ChlH are transported across the chloroplast envelope and interact with porphyrins that are released from the chloroplast.
Acknowledgments
We thank Dr. Satoshi Tabata for P1 clones, Drs. Motoaki Seki and Kazuo Shinozaki for the CHLH cDNA clone, Dr. Pal Maliga for pPZP221, and Dr. Marci Surpin for critically discussing the manuscript. N.M. is grateful to Dr. Masanobu Nakamura and other laboratory members for helpful discussions. This work was supported by Grants-in-Aid for Scientific Research in Priority Areas (No. 11151219) to N.M. from the Ministry of Education, Science, and Culture of Japan and by a grant from the Department of Energy to J.C. (ER13993). R.L. was supported by a U.S. Department of Agriculture fellowship. J.C. is an Associate Investigator of the Howard Hughes Medical Institute.
Abbreviations
- Nf
Norflurazon
- DP
2, 2′-dipyridyl
- MgProto
Mg-protoporphyrin IX
- MgProtoME
Mg-protoporphyrin IX monomethyl ester
- MgProto(ME)
MgProto and MgProtoME
- MgProtoME2
Mg-protoporphyrin IX dimethyl ester
- GUS
β-glucuronidase
- ALA
5-aminolevulinate
- PΦB
phytochromobilin
References
- 1.Bradbeer J W, Atkinson Y E, Borner T, Hagemann R. Nature (London) 1979;279:816–817. [Google Scholar]
- 2.Oelmuller R. Photochem Photobiol. 1989;49:229–239. [Google Scholar]
- 3.Oelmuller R, Mohr H. Planta. 1986;167:106–113. doi: 10.1007/BF00446376. [DOI] [PubMed] [Google Scholar]
- 4.Oelmuller R, Levitant I, Bergfeld R, Rajasekhar V K, Mohr H. Planta. 1986;168:482–492. doi: 10.1007/BF00392267. [DOI] [PubMed] [Google Scholar]
- 5.Taylor W C. Annu Rev Plant Pysiol Plant Mol Biol. 1989;40:211–233. [Google Scholar]
- 6.Lukens J H, Mathews D E, Durbin R D. Plant Physiol. 1987;84:808–813. doi: 10.1104/pp.84.3.808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sullivan J A, Gray J C. Plant Cell. 1999;11:901–910. doi: 10.1105/tpc.11.5.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mayfield S P, Taylor W C. Eur J Biochem. 1984;144:79–84. doi: 10.1111/j.1432-1033.1984.tb08433.x. [DOI] [PubMed] [Google Scholar]
- 9.Johanningmeier U, Howell S H. J Biol Chem. 1984;259:13541–13549. [PubMed] [Google Scholar]
- 10.Kropat J, Oster U, Rudiger W, Beck C F. Proc Natl Acad Sci USA. 1997;94:14168–14172. doi: 10.1073/pnas.94.25.14168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Susek R E, Ausubel F M, Chory J. Cell. 1993;74:787–799. doi: 10.1016/0092-8674(93)90459-4. [DOI] [PubMed] [Google Scholar]
- 12.Chory J, Altschmied L, Cabrera H, Li H M, Susek R. In: Cellular Communication in Plants. Amashino R M, editor. New York: Plenum; 1993. pp. 57–62. [Google Scholar]
- 13.Espineda C E, Linford A S, Devine D, Brusslan J A. Proc Natl Acad Sci USA. 1999;96:10507–10511. doi: 10.1073/pnas.96.18.10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischerova H. Biol Plant. 1975;17:182–188. [Google Scholar]
- 15.Koncz C, Mayerhofer R, Koncz-Kalman Z, Nawrath C, Reiss B, Redei G P, Schell J. EMBO J. 1990;9:1337–1346. doi: 10.1002/j.1460-2075.1990.tb08248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jefferson R A. Plant Mol Biol Rep. 1987;5:387–405. [Google Scholar]
- 17.Moran R. Plant Physiol. 1982;69:1376–1381. doi: 10.1104/pp.69.6.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rebeiz C A, Mattheis J R, Smith B B, Rebeiz C C, Dayton D F. Arch Biochem Biophys. 1975;171:549–567. doi: 10.1016/0003-9861(75)90065-x. [DOI] [PubMed] [Google Scholar]
- 19.Tripathy B C, Rebeiz C A. Anal Biochem. 1985;149:43–61. doi: 10.1016/0003-2697(85)90475-0. [DOI] [PubMed] [Google Scholar]
- 20.Falbel T G, Staehelin L A. Physiol Plant. 1996;97:311–320. [Google Scholar]
- 21.Chory J, Nagpal P, Peto C A. Plant Cell. 1991;3:445–459. doi: 10.1105/tpc.3.5.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leutwiler L S, Meyerowitz E M, Tobin E M. Nucleic Acids Res. 1986;14:4051–4064. doi: 10.1093/nar/14.10.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konieczny A, Ausubel F M. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- 24.Bell C, Ecker J. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- 25.Liu Y G, Mitsukawa N, Lister C, Dean C, Whittier R F. Plant J. 1996;10:733–736. doi: 10.1046/j.1365-313x.1996.10040733.x. [DOI] [PubMed] [Google Scholar]
- 26.Hajdukiewicz P, Svab Z, Maliga P. Plant Mol Biol. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- 27.Gibson L C, Marrison J L, Leech R M, Jensen P E, Bassham D C, Gibson M, Hunter C N. Plant Physiol. 1996;111:61–71. doi: 10.1104/pp.111.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koornneef M, Rolffe E, Spruit C J P. Z Pflanzenphysiol. 1980;100:147–160. [Google Scholar]
- 29.Mochizuki N, Susek R, Chory J. Plant Physiol. 1996;112:1465–1469. doi: 10.1104/pp.112.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duggan J, Gassman M. Plant Physiol. 1974;53:206–215. doi: 10.1104/pp.53.2.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bechtold, N. & Pelletier, G. (1998). eds. Martinez-Zapater, J. M. & Salinas, J. (Humana, Totowa, NJ), pp. 259–266.
- 32.Guarente L. Trends Genet. 1993;9:362–366. doi: 10.1016/0168-9525(93)90042-g. [DOI] [PubMed] [Google Scholar]
- 33.Escoubas J M, Lomas M, LaRoche J, Falkowski P G. Proc Natl Acad Sci USA. 1995;92:10237–10241. doi: 10.1073/pnas.92.22.10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lopez-Juez E, Paul Jarvis R, Takeuchi A, Page A M, Chory J. Plant Physiol. 1998;118:803–815. doi: 10.1104/pp.118.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muramoto T, Kohchi T, Yokota A, Hwang I, Goodman H M. Plant Cell. 1999;11:335–348. doi: 10.1105/tpc.11.3.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davis S J, Kurepa J, Vierstra R D. Proc Natl Acad Sci USA. 1999;96:6541–6546. doi: 10.1073/pnas.96.11.6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parks B M, Quail P H. Plant Cell. 1991;3:1177–1186. doi: 10.1105/tpc.3.11.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Terry M J, Kendrick R E. Plant Physiol. 1999;119:143–152. doi: 10.1104/pp.119.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Montgomery B L, Yeh K C, Crepeau M W, Lagarias J C. Plant Physiol. 1999;121:629–639. doi: 10.1104/pp.121.2.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walker C J, Willows R D. Biochem J. 1997;327:321–333. doi: 10.1042/bj3270321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kittsteiner U, Brunner H, Rüdiger W. Physiol Plant. 1991;81:190–196. [Google Scholar]
- 42.Oster U, Brunner H, Rüdiger W. J Photochem Photobiol B. 1996;36:255–261. [Google Scholar]
- 43.Papenbrock J, Pfundel E, Mock H P, Grimm B. Plant J. 2000;22:155–164. doi: 10.1046/j.1365-313x.2000.00724.x. [DOI] [PubMed] [Google Scholar]
- 44.Papenbrock J, Mock H P, Tanaka R, Kruse E, Grimm B. Plant Physiol. 2000;122:1161–1169. doi: 10.1104/pp.122.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leegood R C, Walker D A, Foyer C H. In: Photosynthetic Mechanisms and the Environment. Barber J, editor. Amsterdam: Elsevier Science; 1985. pp. 190–258. [Google Scholar]
- 46.Walker C J, Weinstein J D. Proc Natl Acad Sci USA. 1991;88:5789–5793. doi: 10.1073/pnas.88.13.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Papenbrock J, Mock H P, Kruse E, Grimm B. Planta. 1999;208:264–273. [Google Scholar]
- 48.Vinti G, Hills A, Campbell S, Bowyer J R, Mochizuki N, Chory J, López-Juez E. Plant J. 2000;24:883–894. doi: 10.1046/j.1365-313x.2000.00936.x. [DOI] [PubMed] [Google Scholar]
- 49.Kropat J, Oster U, Rüdiger W, Beck C F. Plant J. 2000;24:523–531. doi: 10.1046/j.1365-313x.2000.00898.x. [DOI] [PubMed] [Google Scholar]