Abstract
Smith-Lemli-Opitz syndrome (SLOS) and lathosterolosis are malformation syndromes with cognitive deficits caused by mutations of 7-dehydrocholesterol reductase (DHCR7) and lathosterol 5-desaturase (SC5D), respectively. DHCR7 encodes the last enzyme in the Kandutsch-Russel cholesterol biosynthetic pathway, and impaired DHCR7 activity leads to a deficiency of cholesterol and an accumulation of 7-dehydrocholesterol. SC5D catalyzes the synthesis of 7-dehydrocholesterol from lathosterol. Impaired SC5D activity leads to a similar deficiency of cholesterol but an accumulation of lathosterol. Although the genetic and biochemical causes underlying both syndromes are known, the pathophysiological processes leading to the developmental defects remain unclear. To study the pathophysiological mechanisms underlying SLOS and lathosterolosis neurological symptoms, we performed quantitative proteomics analysis of SLOS and lathosterolosis mouse brain tissue and identified multiple biological pathways affected in Dhcr7Δ3–5/Δ3–5 and Sc5d−/− E18.5 embryos. These include alterations in mevalonate metabolism, apoptosis, glycolysis, oxidative stress, protein biosynthesis, intracellular trafficking, and cytoskeleton. Comparison of proteome alterations in both Dhcr7Δ3–5/Δ3–5 and Sc5d−/− brain tissues helps elucidate whether perturbed protein expression was due to decreased cholesterol or a toxic effect of sterol precursors. Validation of the proteomics results confirmed increased expression of isoprenoid and cholesterol synthetic enzymes. This alteration of isoprenoid synthesis may underlie the altered posttranslational modification of Rab7, a small GTPase that is functionally dependent on prenylation with geranylgeranyl, that we identified and validated in this study. These data suggested that although cholesterol synthesis is impaired in both Dhcr7Δ3–5/Δ3–5 and Sc5d−/− embryonic brain tissues the synthesis of nonsterol isoprenoids may be increased and thus contribute to SLOS and lathosterolosis pathology. This proteomics study has provided insight into the pathophysiological mechanisms of SLOS and lathosterolosis, and understanding these pathophysiological changes will help guide clinical therapy for SLOS and lathosterolosis.
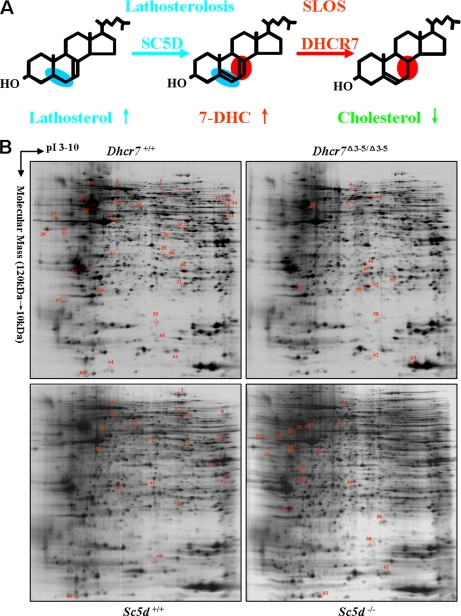
Smith-Lemli-Opitz syndrome (SLOS1; Online Mendelian Inheritance in Man 270400) is a multiple malformation syndrome with cognitive and behavioral deficiencies due to an inborn error of cholesterol synthesis. Typical findings in SLOS include dysmorphic facial features, limb defects, genital anomalies, growth retardation, cognitive disabilities, behavioral problems, and autistic features (for a review, see Ref. 1). The incidence of SLOS has been estimated to be on the order of 1/20,000–1/70,000 (1). SLOS is an autosomal recessive disorder caused by mutation of the 7-dehydrocholesterol reductase gene (DHCR7) (2–4). DHCR7 catalyzes the final step in the Kandutsch-Russel cholesterol biosynthetic pathway. Impaired DHCR7 activity results in increased 7-dehydrocholesterol (7DHC) and decreased cholesterol levels (Fig. 1A). Lathosterolosis is a rare “SLOS-like” malformation syndrome due to mutations of lathosterol 5-desaturase (SC5D) (5–7). SC5D catalyzes the conversion of lathosterol to 7DHC. Thus, in lathosterolosis, like SLOS, there is a deficiency of cholesterol. However, the accumulating precursor sterol is lathosterol rather than 7DHC (Fig. 1A). Because of its rarity and the fact that all known cases of lathosterolosis were ascertained due to similarity with SLOS, the phenotypic spectrum of lathosterolosis has not been defined.
Fig. 1.
Representative 2-DE maps of SLOS and lathosterolosis mouse brain proteins. A, SLOS and lathosterolosis are inborn errors of cholesterol synthesis. SLOS is caused by mutations in the DHCR7 gene. DHCR7 catalyzes the final step in cholesterol synthesis. Lathosterolosis is caused by mutations of the SC5D gene. Cholesterol levels are decreased in both SLOS and lathosterolosis, but the accumulating precursor sterol differs. In SLOS, 7DHC accumulates, whereas in lathosterolosis, the accumulating sterol is lathosterol. B, representative 2-DE maps of control (Dhcr7+/+ and Sc5d+/+), Dhcr7Δ3–5/Δ3–5, and Sc5d−/− mouse brain proteins. Eighty micrograms of the pooled protein sample from Dhcr7+/+, Dhcr7Δ3–5/Δ3–5, Sc5d+/+, and Sc5d−/− embryonic mouse brain tissues were separated on a pH 3–10 nonlinear IPG strip followed by electrophoretic separation on a 12% SDS-polyacrylamide gel. Acidic pH is to the left, and increased molecular mass is at the top. Compared with Dhcr7+/+ mouse brains, the protein spots with significantly decreased or increased expression in Dhcr7Δ3–5/Δ3–5 mouse brains are marked in Dhcr7+/+ and Dhcr7Δ3–5/Δ3–5 mouse brain 2-DE maps, respectively. Compared with Sc5d+/+ mouse brains, the protein spots with significantly decreased or increased expression in Sc5d−/− mouse brains are marked in Sc5d+/+ and Sc5d−/− mouse brain 2-DE maps, respectively. Supplemental Table 2 provides detailed information on the differentially expressed protein spots.
Although the genetic and biochemical causes of SLOS are defined, the pathophysiological mechanisms contributing to specific malformations have not been delineated. The classic paradigm for the pathogenesis of an inborn error of metabolism includes the accumulation of a toxic precursor and/or deficiency of an essential product. In the case of SLOS, the observed defects are postulated to be caused, either singly or in combination, by cholesterol deficiency or the accumulation of 7DHC (8, 9).
Cholesterol is an essential lipid with multiple critical functions. In addition to being a structural lipid in membranes and myelin, cholesterol is the precursor for bile acid, steroid hormone, neuroactive steroid, and oxysterol synthesis. In cellular membranes, cholesterol rafts are microdomains that function in receptor-mediated signal transduction. Functional defects in IgE receptor-mediated mast cell degranulation and cytokine production (10), N-methyl-d-aspartate receptor function (11), and serotonin 1A receptor ligand binding (12, 13) have been reported in SLOS. The altered sterol composition in SLOS affects the physiochemical properties and function of lipid rafts. Membrane domains incorporating 7DHC differ from those containing only cholesterol in protein composition (14), packing (15), and stability (16–18). Substitution of 7DHC for cholesterol also decreases membrane bending rigidity (19). In addition, model membranes mimicking SLOS membranes have been reported to exhibit atypical membrane organization (20) and curvature (19). These alterations may have functional consequences. Depletion of cholesterol from hippocampal membranes and replenishment with 7-dehydrocholesterol does not restore ligand binding activity of the serotonin 1A receptor despite the recovery of the overall membrane order (12). Cholesterol is also necessary for maturation and function of the hedgehog family of morphogens during embryonic development, and several mechanisms by which sonic hedgehog signaling might be impaired in SLOS have been proposed (21–23).
To understand the pathophysiological processes underlying cognitive defects found in SLOS, we need to consider the potential detrimental effects of decreased cholesterol/functional sterol levels versus the potential toxic effects of increased 7DHC. To give insight into pathological effects due to cholesterol deficiency and precursor accumulation, we have produced mouse models deficient in either 7-dehydrocholesterol reductase (11) or lathosterol reductase (6) activity (Dhcr7Δ3–5/Δ3–5 and Sc5d−/−, respectively). Although the two models are similar in many respects, significant differences exist. Dhcr7 pups have relatively few physical malformations other than a low frequency of cleft palate but die during the 1st day of life due to failure to feed (11). In contrast Sc5d mutant embryos are stillborn and have multiple developmental malformations (6). In addition, although secretory granule formation is altered in both models, consistent with differing physiochemical properties of the two precursor sterols, the specific changes differ between the two models (19). For these reasons, a comparison of the two models will provide insight into common mechanisms that are likely due to cholesterol/sterol deficiency and syndrome-specific mechanisms that are due to specific effects of one of the two precursors.
We now report the use of two-dimensional electrophoresis (2-DE) mass spectrometry proteomics analysis to identify proteins with altered expression in brain tissue from both Dhcr7 and Sc5d mutants with the goal of identifying novel pathophysiological mechanisms contributing to the neurological deficits in these two inborn errors of cholesterol synthesis. Because our focus was on identifying processes that could contribute to abnormal neurological development, our analysis was focused on brain tissue from E18.5 embryos. This embryonic age was selected because the biochemical defect increases with embryonic age (6, 11), and it is the latest time point for which we could obtain viable tissue for both mutants. Western blot analysis was used to validate selected individual proteins and pathways. Functional annotation suggested that alterations in mevalonate metabolism, glycolysis, oxidative stress, apoptosis, protein biosynthesis, intracellular trafficking, and cytoskeleton may contribute to the pathology of inborn errors of cholesterol synthesis. In addition, our data are consistent with the hypothesis that both cholesterol deficiency and increased precursor sterol levels contribute to SLOS and lathosterolosis pathology.
EXPERIMENTAL PROCEDURES
Materials
Analytical reagent grade chemicals were used unless stated otherwise. Distilled-deionized water was used throughout the experiments. Chemicals used for gel electrophoresis were purchased from Bio-Rad. Sodium orthovanadate (Na3VO4) and sodium fluoride (NaF) were obtained from Sigma. Protease inhibitor mixture was purchased from GE Healthcare. High performance LC grade acetonitrile was obtained from Fisher. Sequencing grade trypsin was obtained from Roche Applied Science.
Animal Care and Husbandry
Animal work was performed under an NICHD Animal Care and Use Committee-approved animal study protocol. For timed matings, identification of a copulatory plug was used to define E0.5. After euthanasia of the dam, E18.5 embryos were rapidly dissected from the uterus. Embryonic age was confirmed by inspection, and embryos were genotyped as described previously (6, 11). E18.5 tissue was examined because Dhcr7 mutant pups die soon after birth and Sc5d mutant pups are stillborn. E18.5 is thus the optional gestational age to allow for both central nervous system maturation and development of the biochemical defect. All mice were on a mixed 129/B6 background. To ameliorate variability resulting from a non-isogenic background, mutants were matched with littermate controls, two independent control sets were used for the Dhcr7 and the Sc5d experiments, and seven to eight individual brains corresponding to each genotype were pooled for analysis.
Protein Sample Preparation
Brain tissue was rapidly dissected from E18.5 embryos and immediately washed with ice-cold 1× PBS. Each brain was then dissolved in 500 μl of lysis buffer (7 m urea, 2 m thiourea, 4% CHAPS, 65 mm DTT, 40 mm Tris, protease inhibitor mixture, and phosphoprotease inhibitor (0.2 mm Na3VO4 and 1 mm NaF)). After homogenization, the suspension was sonicated at 100 watts for 30 s and centrifuged at 25,000 × g for 1 h. The supernatant contained proteins solubilized in an IEF-compatible buffer. Protein samples from seven or eight individual E18.5 embryos corresponding to each genotype (Dhcr7+/+, Dhcr7Δ3–5/Δ3–5, Sc5d+/+, and Sc5d−/−) were pooled and used for both 2-DE analysis and initial Western blot validation. Protein concentration was determined using a Bradford assay. Samples were stored at −80 °C until analysis.
Two-dimensional Electrophoresis
2-DE was performed as described previously (24). Briefly, IPG-IEF was performed on an Ettan IPGphor 3 IEF System (GE Healthcare). Eighty micrograms of protein were mixed with DeStreakTM Rehydration Solution (GE Healthcare) containing 1% IPG buffer, pH 3–10 nonlinear, 0.2% DTT, and a trace of bromphenol blue to a total volume of 250 μl. The protein preparations were pipetted into an IPG strip holder. Precast IPG dry strips, pH 3–10 nonlinear (130 × 3 × 0.5 mm) (GE Healthcare), were applied and overlaid with mineral oil. The IPG strip holders were then placed onto the electrode plate of the IPGphor platform. After rehydration at 30 V for 12 h, IEF was conducted at 500 V for 1 h, 1000 V for 1 h, and then 8000 V for 8 h to obtain a total of 50–60 kV-h. Temperature was maintained at 20 °C for the duration of the isoelectric focusing.
Following IEF separation, the gel strips were equilibrated twice for 15 min each in buffer containing 50 mm Tris-HCl, pH 8.8, 6 m urea, 30% glycerol, 2% SDS, and a trace of bromphenol blue. One percent DTT was added to the first equilibration buffer, and in the second equilibration buffer, DTT was replaced with 4.5% iodoacetamide. The equilibrated gel strips were then applied onto 1-mm-thick 12% polyacrylamide vertical slab gels and sealed with 0.5% agarose. SDS-PAGE was performed in a Hoefer SE600 Ruby Electrophoresis Unit (GE Healthcare) for 15 min at a constant current of 10 mA/gel and then switched to 20 mA/gel until the bromphenol blue reached the bottom of the gels. Gels corresponding to each of the four genotypes were run in triplicate. Preparative 2-DE gels were loaded with 2 mg of protein.
Gel Staining
After 2-DE, the analytic gels were stained with ammoniacal silver nitrate as described previously (24). Briefly, the analytic gels were washed for 5 min in water and soaked for 1 h in ethanol:acetic acid:water (40:10:50) followed by overnight immersion in ethanol:acetic acid:water (5:5:90). Gels were then washed twice for 20 min each in water, fixed with 5% glutaraldehyde for 1 h, and then washed four times for 30 min each in water. The gels were stained for 45 min in 23.5 mm silver nitrate, 1% ammonia, and 20 mm NaOH. After staining, the gels were washed three times for 3 min each with water and developed using 0.005% citric acid and 0.0185% formaldehyde for 5–10 min. Development was terminated with 5% acetic acid. Preparative gels were stained using Coomassie Brilliant Blue R-250 (Bio-Rad).
Image and Statistical Analysis
Silver-stained gels were scanned at an optical resolution of 42.3 × 42.3 μm using a GS-800 calibrated densitometer (Bio-Rad) in transmissive mode. Spot detection, matching, quantification, normalization, and statistical analysis for the triplicate silver-stained analysis gels were performed in one match set using PDQuest 8.0 software (Bio-Rad) as described previously (24). The PDQuest software models protein spots mathematically as a three-dimensional Gaussian distribution and determines maximum absorption after raw image correction and background subtraction. A total of 1036 protein spots were matched among the four groups of gels. The positional deviation of the matched spots among the 12 130 × 150-mm 2-DE gels is 0.92 ± 0.57 mm in the IEF direction and 0.82 ± 0.56 mm in the SDS-PAGE direction. Spot intensities were then obtained by integration of the Gaussian function with unit of intensity calculated as “Intensity × Area as parts per million.” The intensity of each protein spot was normalized using the “total intensity of all the spots detected” method to compensate for non-expression-related variations in spot quantities between gels. Normalized spot intensity values for the three replicated gels corresponding to each genotype were analyzed by applying an unpaired Student's t test (p < 0.05) comparing control with mutant. We experimentally derived a predicted false positive rate by comparing control embryos from both Dhcr7 and Sc5d litters using intensity differences of 1.2-, 1.5-, and 2.0-fold (supplemental Table 1). An intensity difference of >1.5-fold change optimized protein identification and minimized the predicted false positive rate (6.5–8.0%).
Protein Identification by In-gel Digestion, Mass Spectrometry, and Database Searching
Protein spots were excised from preparative gels and “in-gel” digested with trypsin as described previously (25) with the following modifications. Briefly, gel pieces were dried in a vacuum centrifuge, and 200 ng of modified trypsin (Promega) in 100 mm ammonium bicarbonate were added followed by incubation overnight at 37 °C. The supernatant was then collected, and the peptides were extracted from the gel pieces using 5% formic acid and ACN (1:1, v/v) followed by a second extraction with 5% formic acid and ACN (5:95, v/v). The peptide extracts were vacuum-dried, brought up in 0.1% TFA, and cleaned using C18 ZipTips (Millipore). The resulting peptides were extracted and analyzed both by MALDI-TOF/TOF using an ABI 4800 Proteomics Analyzer (Applied Biosystems) and by combined LC/ESI/MS/MS using an LCQ Deca ion trap mass spectrometer (Thermo Fischer).
In the MALDI-TOF/TOF work, tryptic peptides were applied as unseparated mixtures to sample plate wells. Typically, 0.5 μl of extracted peptide, 5% of total sample in acetonitrile and 0.1% TFA (1:1), was applied with an equal volume of cyano-4-hydroxycinnamic acid matrix (5 mg/ml in the same solvent as the sample); two internal standards were added to the matrix and along with tryptic autolysis peptides were used for mass scale calibration. The typical operating procedure is to acquire a 400-laser shot spectrum of the full peptide mixture in reflector mode and then to fragment four to six peptides that are both intense and well separated from others within the limits of the timed ion selector operated at a resolution of 300; fragmentation spectra were acquired as unimolecular decompositions (collision gas off) using 1000 laser shots. There is no evidence of sample depletion from this approach.
For the LC/ESI/MS/MS measurements, tryptic peptides were separated by reversed phase chromatography and electrosprayed directly into the sampling orifice of the mass spectrometer. MS/MS spectra were collected in a data-dependent manner with up to three of the most intense ions in each full MS scan subjected to isolation and fragmentation. BioWorks v2.0 (Thermo Fisher) was used to extract the MS/MS spectra as dta files using the default parameters, and the dta files for each LC/MS/MS run were merged into one file for database analysis.
MS/MS spectra from both the MALDI and LC/ESI measurements were then analyzed, and both b and y ion series were used to match to protein sequences present in the Mus musculus subset of the UniProtKB/Swiss-Prot/TrEMBL database (database version 56.0; 392,667 sequence entries, July 22, 2008) using the Mascot v2.2 (Matrix Science) search program. Parameters for the database search with Mascot MS/MS Ions Search software were set as follows: enzyme, trypsin; one missed tryptic cleavage site permitted; fixed modification, carbamidomethylation of cysteine; variable modification, methionine oxidation; mass tolerance for precursor ions, ±1.2 (Deca) and ±0.15 Da (4800 Proteomics Analyzer); mass tolerance for fragment ions, ±0.6 (Deca) and ±0.06 Da (4800 Proteomics Analyzer). Only peptides with individual ions Mowse scores ≥32 indicating significant identity or extensive homology (p < 0.05) were used for protein identification. Scaffold v2.2 (Proteome Software, Portland, OR) was used to validate peptide and protein identifications. In Scaffold, the peptide identifications from Mascot were verified using the X! Tandem (v2007.01.0.2) database search program (26). Probabilistic validations of the peptide identifications were performed using Peptide Prophet (27), and corresponding protein probabilities were calculated by Protein Prophet (28). The cutoff for peptide identification was set at greater than 95.0% probability, and the cutoff for protein identification was set at greater than 99% probability with two or more identified peptides. In the case of peptides matching to multiple members of a protein family, the presented protein was selected based on the highest score and the highest number of matching peptides.
Preparation of Detergent-resistant Membrane of Mouse Brain Tissue
The detergent-soluble fraction (DSF) and detergent-resistant membrane (DRM) of mouse brain tissue were prepared with the ReadyPrepTM Protein Extraction kit (Bio-Rad) according to the manufacturer's protocol. Briefly, brains were isolated from E18.5 embryos and lysed using the provided detergent solution. After centrifugation at 16,000 × g for 20 min at 4 °C, the supernatant was collected as the DSF, and the DRM pellet was dissolved in the provided protein solubilization buffer. Protein samples from six individual E18.5 embryos corresponding to each genotype (Dhcr7+/+ and Dhcr7Δ3–5/Δ3–5) were pooled and used for Western blot analysis. Protein concentration was determined using the Bio-Rad RC DC Protein Assay. Western blot analysis of clathrin heavy chain and transferrin receptor was used to validate the preparation efficiency as marker proteins for DRM and DSF, respectively.
Validation of Candidate Proteins
For Western blot analysis, 20 μg of individual or pooled E18.5 Dhcr7+/+, Dhcr7Δ3–5/Δ3–5, Sc5d+/+, or Sc5d−/− mouse brain protein preparations were separated by 4–12% NuPAGE Tris acetate gels (Invitrogen) according to the manufacturer's protocol. Proteins were then transferred to a nitrocellulose membrane using iBlot (Invitrogen) according to the manufacturer's protocol, and chemoluminescence detection was performed using WesternBreeze kits from Invitrogen. The following primary antibodies were used for Western blot analysis: rabbit polyclonal anti-cytoplasmic hydroxymethylglutaryl-CoA synthase (HMGCS1) (1:1000; Santa Cruz Biotechnology), rabbit polyclonal anti-geranylgeranyl-diphosphate synthase (GGPS) (1:1000; Abgent), rabbit polyclonal anti-sterol regulatory element-binding protein 1 (SREBP-1) (1:500; Santa Cruz Biotechnology), rabbit polyclonal anti-SREBP-2 (1:500; Santa Cruz Biotechnology), mouse monoclonal anti-Rab7 (1:100,000; Sigma), mouse monoclonal anti-Rab5 (1:100,000; Sigma), rabbit polyclonal anti-cleaved caspase-3 (1:500; Cell Signaling Technology), mouse monoclonal anti-clathrin heavy chain (1:10,000; Sigma), mouse monoclonal anti-transferrin receptor (1:2,000; Invitrogen), rabbit polyclonal anti-cofilin-1 (1:2000; Cell Signaling Technology), rabbit monoclonal anti-phosphocofilin (Ser-3) (1:200; Cell Signaling Technology), rabbit polyclonal anti-vacuolar proton pump F subunit (1:500; Santa Cruz Biotechnology), rabbit polyclonal anti-pyruvate kinase isozyme M2 (1:1000; Cell Signaling Technology), rabbit polyclonal anti-dihydropyrimidinase-related protein 2 (1:1000; Santa Cruz Biotechnology), and mouse monoclonal anti-actin (1:5,000; Sigma). The appropriate secondary antibody (rabbit or mouse) was applied, and the resulting chemiluminescence signal was detected. Band intensity was quantified with the Quantity One software (Bio-Rad) and normalized to actin.
RESULTS
Differential Protein Expression in Dhcr7Δ3–5/Δ3–5 and Sc5d−/− Mouse Brain Tissues
A procedural schematic for 2-DE proteomics analysis is presented in supplemental Fig. 1. To minimize effects of individual variation, whole brain protein extracts prepared from seven or eight E18.5 embryos corresponding to each genotype (Dhcr7+/+, Dhcr7Δ3–5/Δ3–5, Sc5d+/+ or Sc5d−/−) were pooled. For each genotype, triplicate 2-DE silver-stained gels were prepared for image analysis. Representative silver-stained gels are shown in Fig. 1B. A total of 1036 protein spots were matched among the four groups of gels.
After gel-to-gel matching and normalization, statistics analyses were performed to identify potential proteins with differential expression. To experimentally determine a predicted false positive rate, we compared control Dhcr7 and Sc5d brain tissues. Predicted false positive rates are shown in supplemental Table 1. Using criteria of p < 0.05 and intensity differences of either 1.2-, 1.5-, or 2.0-fold, we found predicted false positive rates of 33.9–36.5, 6.5–7.9, and 5.3–10.0%, respectively. Thus, to optimize candidate protein identification and minimize false positives, we selected criteria of r > 1.5 and p < 0.05 to identify proteins with altered expression. Using these criteria, 46 (46 of 1036; 4.4%) protein spots showed altered intensity in gels from Dhcr7Δ3–5/Δ3–5 embryonic brain tissue compared with control brain tissue. Of these protein spots, 33 were decreased (r < 0.67, p < 0.05), and 13 were increased (r > 1.50, p < 0.05) in mutant tissue. Similarly, 38 (38 of 1036; 3.5%) protein spots were differentially expressed in Sc5d−/− embryonic brain tissue. Relative to controls, 19 protein spots showed decreased expression (r < 0.67, p < 0.05), and 19 protein spots showed increased expression (r > 1.50, p < 0.05) in mutant tissue. In total, 66 protein spots showed significant expression changes of more than 1.5-fold in Dhcr7Δ3–5/Δ3–5 or Sc5d−/− mouse brains (Fig. 1B and supplemental Table 2). Of these 66 protein spots, 14 showed concordant changes in both mutants, four were discordant, 28 were only observed in the Dhcr7 comparison, and 20 were only observed in the Sc5d comparison (Fig. 1B and supplemental Table 2).
Protein Identification and Validation
These 66 differentially expressed protein spots were identified and excised from Coomassie Brilliant Blue R-250-stained preparative gels. After in-gel digestion with trypsin, the resulting tryptic peptides were extracted and analyzed by both MALDI-tandem TOF mass spectrometry and LC/ESI/MS/MS. MS/MS spectra were searched using Mascot. Identified proteins are listed in Table II, and the peptide list used for identification of each protein spot is provided in supplemental Table 3. The peptide and protein identifications were validated using Scaffold (supplemental Table 4). Fifty-two proteins (52 of 66; 78.8%) were identified based on at least two unique peptides. Thirty-six (36 of 46; 78.3%) and 30 (30 of 38; 78.9%) were identified in the Dhcr7 and Sc5d experiments, respectively (Table I and Table II).
Table II. Identified proteins that exhibited significant differential expression (>1.5-fold) in Dhcr7Δ3–5/Δ3–5 or Sc5d−/− embryonic brain tissue.
| Spot no.a | Accession no.b | Protein name | Ratio Dhcr7 (Δ3–5/Δ3–5/+/+) | Ratio Sc5d (−/−/+/+) | Theoretical molecular mass (Da)/pI | Experimental molecular mass (kDa)/pI | No. of unique peptides (Mascot)c | No. of unique peptides (Scaffold)d | Sequence coveragee |
|---|---|---|---|---|---|---|---|---|---|
| % | |||||||||
| Mevalonate metabolism | |||||||||
| 17 | Q8JZK9 | Hydroxymethylglutaryl-CoA synthase, cytoplasmic | 1.55 | 1.75 | 58,160/5.65 | 58.2/5.65 | 17 | 19 | 36 |
| 49 | P58044 | Isopentenyl-diphosphate Δ-isomerase 1 | 1.61 | 1.44 | 26,615/5.79 | 26.0/5.90 | 3 | 8 | 19 |
| Apoptosis-related | |||||||||
| 39 | P70677 | Caspase-3 precursor | 0.56 | 0.72 | 31,911/6.45 | 34.0/6.08 | 5 | 7 | 22 |
| 47 | P61982 | 14-3-3 protein γf | 0.48 | 1.18g | 28,456/4.80 | 28.5/4.71 | 8 | 12 | 51 |
| 53 | P63028 | Translationally controlled tumor protein | 2.02 | 1.85 | 19,564/4.76 | 21.5/4.85 | 4 | 4 | 30 |
| Intracellular trafficking | |||||||||
| 6 | O08599 | Syntaxin-binding protein 1 | 0.62 | 0.56 | 67,925/6.49 | 66.1/6.94 | 13 | 26 | 28 |
| 11 | Q62188 | Dihydropyrimidinase-related protein 3 | 1.56 | 0.85g | 62,296/6.04 | 59.5/5.96 | 20 | 17 | 51 |
| 50 | P51150 | Ras-related protein Rab7a | 2.06 | 1.43 | 23,760/6.40 | 22.7/6.90 | 7 | 8 | 40 |
| 51 | P51150 | Ras-related protein Rab7a | 0.26 | 0.34 | 23,760/6.40 | 22.6/6.97 | 9 | 10 | 54 |
| 56 | Q9QZ88 | Vacuolar protein sorting-associated protein 29 | 1.10g | 1.68 | 20,654/6.29 | 20.2/6.64 | 5 | 6 | 31 |
| 59 | P84078 | ADP-ribosylation factor 1 | 0.57 | 0.97g | 20,741/6.32 | 17.1/6.02 | 4 | 5 | 33 |
| 64 | Q9D1K2 | Vacuolar proton pump F subunit | 0.65 | 1.34 | 13,362/5.52 | 13.4/5.42 | 4 | 6 | 41 |
| Antioxidant | |||||||||
| 40 | Q99J99 | 3-Mercaptopyruvate sulfurtransferase | 0.11 | 0.76g | 33,231/6.11 | 33.2/6.11 | 6 | 9 | 27 |
| 54 | Q9CPU0 | Lactoylglutathione lyase | 0.60 | 1.07g | 20,967/5.24 | 21.2/5.11 | 7 | 11 | 39 |
| 60 | P08228 | Superoxide dismutase (Cu,Zn) | 0.51 | 0.62 | 16,104/6.02 | 16.1/6.02 | 6 | 7 | 46 |
| Glycolysis | |||||||||
| 12 | P52480 | Pyruvate kinase isozymes M1/M2 | 0.32 | 0.84g | 58,378/7.18 | 59.8/8.99 | 8 | 15 | 37 |
| 13 | P52480 | Pyruvate kinase isozymes M1/M2 | 0.29 | 0.72 | 58,378/7.18 | 59.9/9.01 | 6 | 11 | 26 |
| 36 | Q93092 | Transaldolase | 0.67 | 1.07g | 37,534/6.57 | 38.7/6.04 | 13 | 13 | 34 |
| 48 | Q9DBJ1 | Phosphoglycerate mutase 1 | 0.65 | 1.11g | 28,928/6.67 | 28.2/6.96 | 12 | 14 | 68 |
| Protein biosynthesis | |||||||||
| 1 | P58252 | Elongation factor 2 (EF-2) | 1.13g | 0.59 | 96,222/6.41 | 91.0/6.88 | 26 | 40 | 42 |
| 24 | P60843 | Eukaryotic initiation factor 4A-I | 0.36 | 0.15 | 46,353/5.32 | 46.4/5.32 | 5 | 13 | 14 |
| 45 | P63073 | Eukaryotic translation initiation factor 4E | 0.78g | 0.62 | 25,266/5.79 | 26.0/5.48 | 4 | 4 | 20 |
| 61 | P63323 | 40 S ribosomal protein S12 | 0.67 | 1.71 | 14,858/6.82 | 14.9/6.82 | 3 | 5 | 22 |
| Cytoskeleton-associated | |||||||||
| 4 | O08553 | Dihydropyrimidinase-related protein 2 | 1.69 | 2.13 | 62,638/5.95 | 71.6/5.65 | 9 | 22 | 24 |
| 7 | O08553 | Dihydropyrimidinase-related protein 2 | 0.57 | 1.01g | 62,638/5.95 | 64.2/6.08 | 6 | 13 | 13 |
| 15 | P99024 | Tubulin β-5 chainf | 0.65 | 1.08g | 50,095/4.78 | 58.6/4.72 | 15 | 17 | 46 |
| 19 | P99024 | Tubulin β-5 chainf | 0.54 | 0.86g | 50,095/4.78 | 47.2/4.96 | 21 | 26 | 62 |
| 22 | P20152 | Vimentin | 0.91g | 1.93 | 53,712/5.06 | 46.6/4.90 | 7 | 9 | 15 |
| 58 | P18760 | Cofilin-1 | 3.81 | 2.61 | 18,776/8.22 | 17.8/6.05 | 4 | 7 | 33 |
| Molecular chaperone | |||||||||
| 3 | P20029 | 78-kDa glucose-regulated protein | 0.66 | 1.15g | 72,492/5.07 | 72.5/5.07 | 14 | 18 | 26 |
| 10 | P80317 | T-complex protein 1 subunit ζ | 0.96g | 0.54 | 58,424/6.63 | 60.5/6.96 | 21 | 32 | 48 |
| 16 | P80314 | T-complex protein 1 subunit β | 0.65 | 0.66 | 57,783/5.97 | 58.8/5.56 | 25 | 28 | 63 |
| 62 | Q9CWM4 | Prefoldin subunit 1 | 1.71 | 1.04g | 14,246/7.93 | 14.4/5.96 | 4 | 5 | 29 |
| mRNA processing | |||||||||
| 29 | Q9Z204 | Heterogeneous nuclear ribonucleoproteins C1/C2 | 0.81g | 1.57 | 34,421/4.92 | 41.0/5.02 | 8 | 11 | 29 |
| 32 | Q9Z204 | Heterogeneous nuclear ribonucleoproteins C1/C2 | 0.95g | 1.54 | 34,421/4.92 | 39.4/5.01 | 4 | 7 | 21 |
| 34 | P60335 | Poly(rC)-binding protein 1 | 0.60 | 0.58 | 37,987/6.66 | 38.7/6.04 | 8 | 14 | 49 |
| 37 | O88569 | Heterogeneous nuclear ribonucleoproteins A2/B1 | 1.11g | 0.52 | 37,437/8.97 | 37.4/8.97 | 8 | 17 | 26 |
| Transcription regulation | |||||||||
| 8 | Q8C854 | Myelin expression factor 2 | 1.04g | 0.44 | 63,482/8.96 | 61.0/9.00 | 6 | 14 | 13 |
| 9 | Q8C854 | Myelin expression factor 2 | 0.29 | 0.80 | 63,482/8.96 | 60.7/9.12 | 9 | 11 | 17 |
| 14 | Q99K48 | Non-POU domain-containing octamer-binding protein | 0.18 | 0.89g | 54,620/9.01 | 59.4/9.01 | 2 | 6 | 4 |
| 27 | Q921F2 | TAR DNA-binding protein 43 | 1.16g | 0.36 | 44,918/6.26 | 43.6/5.87 | 9 | 13 | 30 |
| 46 | Q8CCT4 | Transcription elongation factor A protein-like 5 | 1.39g | 1.94 | 22,082/5.95 | 29.1/5.74 | 3 | 5 | 16 |
| 65 | P84089 | Enhancer of rudimentary homolog | 0.92g | 1.51 | 12,422/5.63 | 12.4/5.46 | 2 | 3 | 26 |
| Tricarboxylic acid cycle | |||||||||
| 28 | P54071 | Isocitrate dehydrogenase (NADP), mitochondrial | 1.08g | 0.41 | 51,330/5.88 | 44.4/8.99 | 11 | 19 | 27 |
| 35 | Q9D6R2 | Isocitrate dehydrogenase (NAD) subunit α | 0.89g | 1.54 | 40,069/6.27 | 39.0/5.47 | 6 | 8 | 18 |
| Other | |||||||||
| 25 | Q9EQU5 | Phosphatase 2A inhibitor I2PP2A | 0.43 | 1.82 | 33,358/4.22 | 45.0/4.31 | 4 | 9 | 12 |
| 26 | Q05186 | Reticulocalbin-1 precursor | 1.38g | 2.01 | 38,090/4.70 | 45.4/4.62 | 4 | 6 | 15 |
| 30 | Q9EQU5 | Phosphatase 2A inhibitor I2PP2A | 0.25 | 1.07g | 33,358/4.22 | 39.5/4.20 | 4 | 4 | 16 |
| 31 | Q9EQU5 | Phosphatase 2A inhibitor I2PP2A | 0.61 | 1.78 | 33,358/4.22 | 39.4/4.41 | 4 | 6 | 16 |
| 42 | Q80XN0 | d-β-Hydroxybutyrate dehydrogenase | 0.92g | 0.65 | 38,603/9.14 | 32.9/9.14 | 7 | 11 | 23 |
| 44 | P70195 | Proteasome subunit β type-7 precursor | 1.54 | 0.64 | 30,214/8.14 | 29.6/5.88 | 8 | 14 | 38 |
| 63 | P02088 | Hemoglobin subunit β-1f | 1.58 | 1.47 | 15,944/7.12 | 13.7/7.96 | 12 | 15 | 95 |
a The gel locations for the numbered spots are indicated in Fig. 1B.
b Swiss-Prot/TrEMBL primary accession number.
c Unique peptides matched using Mascot with scores greater than identity cutoff score (ion score ≥32 indicates identity or close homology, p < 0.05).
d Unique peptides matched using Scaffold (Mascot and X! Tandem combined searches) with probability score ≥95% as specified by Peptide Prophet.
e Percentage of amino acids in protein matched to peptides identified using Mascot.
f A mixture of protein isozymes were detected in gel spot, but only the predominant form is listed here (see supplemental Table 3 for a complete list of isozyme peptides).
g p > 0.05; no statistical significance.
Table I. Protein identification.
| Ratio (mutant/control) | Student's t test p < 0.05 |
||
|---|---|---|---|
| r < 0.67 | r > 1.50 | Total | |
| Dhcr7 (identified/total) | 26/33 | 10/13 | 36/46 (78%) |
| Sc5d (identified/total) | 15/19 | 15/19 | 30/38 (79%) |
To validate the differentially expressed proteins identified by 2-DE mass spectrometry proteomics analysis, Western blot analysis was performed for seven proteins, including HMGCS1, Rab7, caspase-3, vacuolar proton pump F subunit, cofilin-1, pyruvate kinase isozymes M1/M2, and dihydropyrimidinase-related protein 2. Altered expression consistent with the above proteomics analysis was confirmed for five (71%) of these proteins. The two proteins that failed validation were pyruvate kinase isozymes M1/M2 and dihydropyrimidinase-related protein 2. It should be noted that multiple issues, other than the identified protein being a false positive, could contribute to failure to validate a given protein. These include posttranslational modifications or alteration of a specific isoform with the total level of all isoforms being unaltered. These types of changes could be appreciated on a 2-DE gel but not detected on a one-dimensional gel. An example of the former situation is that although total cofilin-1 levels were similar, phosphocofilin levels were significantly increased in mutant embryos (29). Thus, this 71% validation rate is a minimal estimate.
Functional Annotation
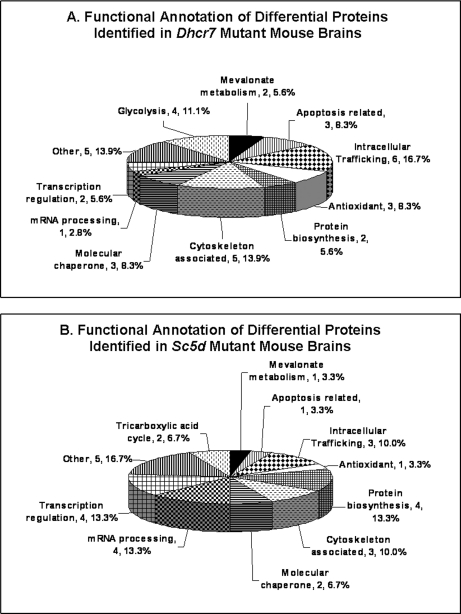
Functional annotation of the identified proteins suggests that multiple biological pathways are perturbed in embryonic Dhcr7 and Sc5d mutant brain tissues (Fig. 2 and Table II). These include mevalonate metabolism, glycolysis and tricarboxylic acid cycle, oxidative stress, apoptosis, protein biosynthesis, intracellular trafficking, cytoskeleton, transcriptional regulation, and mRNA processing. Comparison of the two data sets showed similar perturbation of mevalonate metabolism, protein biosynthesis, intracellular trafficking, and cytoskeleton in the two mutants, whereas glycolysis appeared to be more affected in Dhcr7 mutants, and the tricarboxylic acid cycle appeared to be more affected in Sc5d mutants (Fig. 2 and Table II). In the subsequent sections, we confirm the identification of altered mevalonate metabolism, intracellular trafficking, and apoptosis as functional groupings with altered expression. In addition, we have recently completed a functional analysis of cofilin-1 confirming alteration in cytoskeletal proteins in both Dhcr7 and Sc5d mutant brain tissues (29).
Fig. 2.
Functional annotation of differentially expressed proteins that were identified in Dhcr7Δ3–5/Δ3–5 and Sc5d−/− embryonic brain tissues. A, functional annotation of 36 differentially expressed (r > 1.5, p < 0.05) proteins in Dhcr7Δ3–5/Δ3–5 embryonic brain tissue. B, functional annotation of 30 differentially expressed (r > 1.5, p < 0.05) proteins in Sc5d−/− embryonic brain tissue.
Activation of Presqualene Cholesterol Biosynthetic Pathway
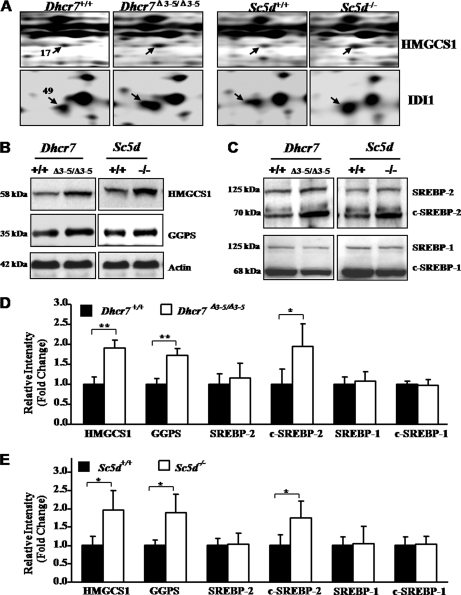
Cholesterol is synthesized in the endoplasmic reticulum of mammalian cells, and the first committed step of this pathway involves the synthesis of mevalonate by hydroxymethylglutaryl-CoA reductase (30–32) (supplemental Fig. 2). The presqualene portion of the cholesterol biosynthetic pathway generates isoprenoid precursor molecules for the synthesis of nonsterol isoprenoids, such as dolichol, ubiquinone, and geranylgeranyl diphosphate. Farnesyl and geranylgeranyl are involved in posttranslational modification of specific proteins, such as small GTPases (supplemental Fig. 2) (30–32). Two enzymes in the mevalonate pathway, HMGCS1 and isopentenyl-diphosphate Δ-isomerase 1 (IDI1), were identified in differentially expressed spots based on 18 and three unique peptides, respectively (Fig. 3A, Table II, and supplemental Table 3). HMGCS1 showed 1.55- and 1.75-fold increased expression in Dhcr7 and Sc5d mutant brain tissues compared with controls, respectively. Similarly, IDI1 showed 1.61- and 1.44-fold increased expression in Dhcr7 and Sc5d mutant brain tissues compared with controls, respectively (Fig. 3A and Table II). Increased expression of HMGCS1 in Dhcr7Δ3–5/Δ3–5 and Sc5d−/− brain tissues was validated by Western blot (Fig. 3, B and D). Confirming the extrapolation of individual protein identification to the identification of altered pathways by functional annotation, Western blot analysis also demonstrated increased expression of GGPS, another enzyme in the mevalonate pathway, in both Dhcr7Δ3–5/Δ3–5 and Sc5d−/− (Fig. 3, B and D).
Fig. 3.
Activation of mevalonate metabolism pathway in Dhcr7Δ3–5/Δ3–5 and Sc5d−/− embryonic brain tissues. A, representative regions from the 2-DE gels demonstrating differential expression of protein spots 17 and 49, which were subsequently identified as HMGCS1 and IDI1, respectively. B, Western blot analysis of pooled protein samples (20 μg) with anti-HMGCS1 and anti-GGPS validated altered expression of these two proteins. C, Western blot analysis of the pooled protein samples (20 μg) with SREBP-2 and SREBP-1 antibodies showed increased activation of SREBP-2 but no change in SREBP1. D, Western blot analysis of HMGCS1, GGPS, c-SREBP-2, and c-SREBP-1 from five independent Dhcr7+/+ or Dhcr7Δ3–5/Δ3–5 brain tissue protein samples (20 μg) confirmed increased protein expression for HMGCS1, GGPS, and c-SREBP2 (mean ± S.D., n = 5, *, p < 0.05; **, p < 0.01). E, Western blot analysis of HMGCS1, GGPS, c-SREBP-2, and c-SREBP-1 from five independent Sc5d+/+ or Sc5d−/− brain tissue protein samples (20 μg) confirmed increased protein expression for HMGCS1, GGPS, and c-SREBP2 (mean ± S.D.; n = 5; *, p < 0.05). Band intensity was normalized to that of actin. Error bars indicate the S.D. calculated from five individual samples.
SREBP-2 regulates the expression of cholesterol biosynthetic and lipid synthetic enzymes in response to intracellular cholesterol levels. In response to decreased cholesterol, SREBP-2 undergoes a series of proteolytic cleavages to yield a transcriptionally active protein (c-SREBP-2) (33–38). Consistent with increased protein expression of enzymes in the mevalonate pathway (Fig. 3, A, B, and D), Western blot analyses demonstrated increased c-SREBP-2 in Dhcr7Δ3–5/Δ3–5 (Fig. 3, C and D) and Sc5d−/− (Fig. 3, C and E) mouse brains. This appears to be specific because a corresponding activation of SREBP-1, a homologous regulator of lipid synthetic genes, was not observed (Fig. 3, C and D) in either of the two mutants. It should be noted that altered expression of GGPS and SREBP-2 was not detected in our proteomics screen. Rather, the potential of these changes was inferred from the functional annotation of the identified proteins (Fig. 2).
Altered Expression of Rab7 and Rab5 in Dhcr7Δ3–5/Δ3–5 and Sc5d−/− Brain Tissues
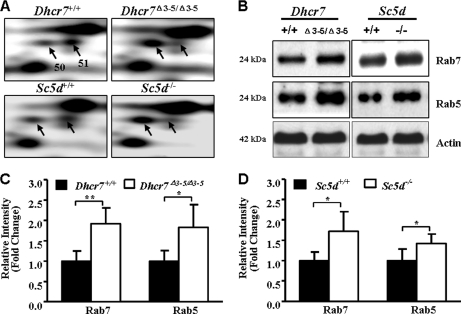
The Rab family of small GTPases functions to regulate intracellular vesicular transport. Over 60 mammalian Rab proteins have been identified, and each Rab protein regulates a distinct step in intracellular vesicular transport (39, 40). In this study, we observed differential expression of Rab7 in two independent protein spots (spots 50 and 51), which migrated with distinct isoelectric points. The identification of Rab7 was based on seven and nine peptides, respectively, for the two spots (Fig. 4A, Table II, and supplemental Table 3). The identification of Rab7 in two spots suggested the possibility of altered posttranslational modification. Rab proteins are known to be modified by prenylation, methylation, and phosphorylation. In the more acidic spot (spot 50), expression was increased 2.06- and 1.43-fold relative to control animals in Dhcr7Δ3–5/Δ3–5 and Sc5d−/− embryonic brain tissues, respectively. However, in the second, more basic Rab7 spot (spot 51), expression was decreased 3.85- and 2.94-fold in Dhcr7Δ3–5/Δ3–5 and Sc5d−/−, respectively (Fig. 4A and Table II). Western blot analyses confirmed increased expression of total Rab7 in Dhcr7Δ3–5/Δ3–5 (Fig. 4, B and C) and Sc5d−/− (Fig. 4, B and D) mouse brains.
Fig. 4.
Differential expression of Rab7 and Rab5 in Dhcr7Δ3–5/Δ3–5 and Sc5d−/− embryonic brain tissues. A, representative 2-DE gel sections of protein spots 50 and 51 identified as Rab7. B, Western blot analysis of pooled protein samples (20 μg) with anti-Rab7 and anti-Rab5 antibodies. C, increased expression of both Rab7 and Rab5 was confirmed in five independent Dhcr7+/+ or Dhcr7Δ3–5/Δ3–5 brain tissue samples (mean ± S.D.; n = 5; *, p < 0.05; **, p < 0.01). D, increased expression of both Rab7 and Rab5 was confirmed in five independent Sc5d+/+ or Sc5d−/− brain tissue samples (mean ± S.D.; n = 5; *, p < 0.05). Band intensity on Western blots was quantified and normalized to that of actin. Error bars indicate the S.D. calculated from five individual samples.
Rab7 functions subsequent to Rab5, and sequential action of Rab5 and Rab7 regulates early and late endosomal trafficking, respectively (40–44). Thus, we investigated whether expression of Rab5 could be altered in either Dhcr7 or Sc5d mutant brain tissue. Total Rab5 was increased in both Dhcr7Δ3–5/Δ3–5 (Fig. 4, B and C) and in Sc5d−/− (Fig. 4, B and D) mouse brains, indicating that endocytosis regulated by Rab5 and Rab7 might be affected in Dhcr7Δ3–5/Δ3–5 and Sc5d−/− mouse brains. Identification of altered Rab5 expression further validates the use of functional annotation to infer disturbances in biological pathways.
Activation of Caspase-3 in Dhcr7Δ3–5/Δ3–5 and Sc5d−/− Brain Tissue
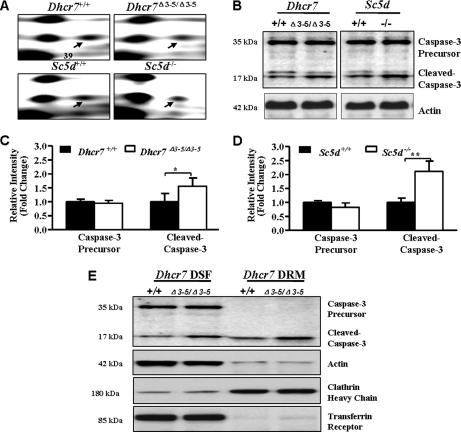
Caspase-3 is a critical protein that promotes apoptosis by catalyzing the proteolysis of members of the Bcl-2 family of apoptosis-related proteins. In response to various death signals, the caspase-3 proenzyme is cleaved by initiator caspases at Asp-28 and Asp-175 to generate the active large (p17) and small (p12) subunits, forming an active heterotetramer (45). In this study, we identified caspase-3 precursor based on five unique peptides from the differential protein spot 39, which showed 1.79- and 1.39-fold decreased expression in Dhcr7 and Sc5d mutant brain tissues compared with controls, respectively (Fig. 5A, Table II, and supplemental Table 3). To validate the identification of caspase-3, we performed Western blot analysis with an antibody that detects both precursor and activated caspase-3. Although no significant decrease of caspase-3 precursor was observed by Western blot analysis, the cleaved caspase-3 (p17) was significantly increased in Dhcr7Δ3–5/Δ3–5 and Sc5d−/− brain tissues (Fig. 5, B, C, and D). Functional characterization further indicated that cleaved caspase-3 was increased in both detergent-soluble fraction and detergent-resistant membrane of Dhcr7Δ3–5/Δ3–5 brain tissue (Fig. 5E). However, caspase-3 precursor was not detected in detergent-resistant membrane of control and Dhcr7Δ3–5/Δ3–5 brain tissues (Fig. 5E).
Fig. 5.
Activation of caspase-3 in Dhcr7Δ3–5/Δ3–5 and Sc5d−/− embryonic brain tissues. A, representative regions from the 2-DE gels demonstrating differential expression of protein spot 39, which was subsequently identified as caspase-3 precursor. B, Western blot analysis of pooled protein samples (20 μg) with anti-caspase-3 showed increased cleaved caspase-3 in Dhcr7Δ3–5/Δ3–5 and Sc5d−/− brain tissues. C, Western blot analysis of caspase-3 from five independent Dhcr7+/+ or Dhcr7Δ3–5/Δ3–5 brain tissue protein samples (20 μg) confirmed increased cleaved caspase-3 (mean ± S.D.; n = 5; *, p < 0.05). D, Western blot analysis of caspase-3 from five independent Sc5d+/+ or Sc5d−/− brain tissue protein samples (20 μg) confirmed increased cleaved caspase-3 (mean ± S.D.; n = 5; **, p < 0.01). Band intensity was normalized to that of actin. Error bars indicate the S.D. calculated from five individual samples. E, Western blot analysis showed that cleaved caspase-3 increased both in DSF and in DRM of Dhcr7Δ3–5/Δ3–5 brain tissue. Transferrin receptor and clathrin heavy chain were used as marker proteins for DSF and DRM, respectively.
DISCUSSION
Although the primary molecular and enzymatic defects that cause SLOS and lathosterolosis are known, the pathophysiological processes that underlie the congenital malformations and cognitive impairment are not defined. It is unlikely that a single pathogenic mechanism can explain the pleiotropic phenotypic findings and symptoms observed in SLOS and lathosterolosis (1, 46). To identify biological pathways that potentially contribute to the SLOS and lathosterolosis phenotypes, we performed comparative proteomics analysis of control, Dhcr7Δ3–5/Δ3–5, and Sc5d−/− embryonic mouse brains. Comparison of the proteomic changes in Dhcr7Δ3–5/Δ3–5 and Sc5d−/− E18.5 embryonic brain tissue may also help elucidate whether protein expression was altered due to decreased cholesterol or a toxic effect of sterol precursors.
Both Cholesterol Deficiency and Sterol Precursor Accumulation May Contribute to Pathology of SLOS and Lathosterolosis
Cholesterol levels are decreased in brain tissue from both Dhcr7Δ3–5/Δ3–5 and Sc5d−/− mice; however, the accumulating precursor sterol is 7DHC and lathosterol, respectively (6, 11). Sterol precursors, such as 7DHC and lathosterol, could be bioactive themselves or give rise to bioactive products (8, 9). Because the cholesterol deficiency is similar in both mouse models, our comparison of proteomic changes in Dhcr7Δ3–5/Δ3–5 and Sc5d−/− mice allows for identification of changes that might be specifically due to or potentiated by a toxic effect of the accumulating precursor sterol. Of the 66 protein spots with differential expression >1.5-fold (Table II, supplemental Table 2), 14 showed concordant changes in both mutants, suggesting that altered expression was a consequence of decreased cholesterol. In contrast, 42 protein spots showed discordant changes, suggesting that altered expression may be due to a specific effect of either 7DHC or lathosterol. Toxic effects of either 7DHC or lathosterol could either be a direct consequence of the sterol precursor or a metabolite of the precursor sterol. These toxic effects could be potentiated by the concurrent cholesterol deficiency. Functional annotation of the differentially expressed proteins showed concordant changes in mevalonate metabolism, protein biosynthesis, apoptosis, intracellular trafficking, and cytoskeleton in both Dhcr7 and Sc5d mutants. In contrast, the tricarboxylic acid cycle appeared to be specifically affected in Sc5d mutants, whereas glycolysis appeared to be more affected in Dhcr7 mutants (Fig. 2 and Table II). Although future studies will need to explore this in detail for all of the identified proteins and pathways, both identification of specific proteins and functional annotation support the hypothesis that both decreased cholesterol/functional sterol and toxic effects of elevated concentrations of sterol precursors contribute to the phenotypic findings and symptoms observed in SLOS and lathosterolosis. This is consistent with the different but related phenotypes observed for SLOS and lathosterolosis mouse models and consistent with the fact that the syndromic phenotypes associated with mutations of NSHDL (congenital hemidysplasia with ichthyosiform erythroderma and limb defects (CHILD) syndrome) and EBP (Conradi-Hünermann-Happle syndrome), two genes that encode postsqualene cholesterol synthetic enzymes, are distinct from SLOS (47). The concordant changes observed for multiple proteins and biological pathways likely result from sterol deficiency that is common between the two disorders.
Multiple Biological Pathways, Including Isoprenoid Synthesis, Are Affected in Developing Dhcr7Δ3–5/Δ3–5 or Sc5d−/− Mouse Brain Tissue
Compared with control, we identified 46 and 38 differentially expressed protein spots in Dhcr7Δ3–5/Δ3–5 and Sc5d−/− embryonic mouse brains, respectively. Mass spectrometric analysis identified 36 (36 of 46; 78%) and 30 (30 of 38; 79%) of these spots, respectively (Tables I and II). An experimentally determined false positive rate of 6.5–7.9% and a minimal validation rate of 71% strongly support the use of the reported protein identifications to guide development of future experiments investigating pathological processes contributing to the development of the SLOS and lathosterolosis phenotypes. Consistent with pleiotropic phenotypic findings and symptoms observed in SLOS and lathosterolosis, functional annotation of the differentially expressed proteins identified multiple biological pathways that may be affected in Dhcr7Δ3–5/Δ3–5 or Sc5d−/− brain tissue. These include the mevalonate metabolism, glycolysis, antioxidant system, apoptosis, protein biosynthesis, intracellular trafficking, and cytoskeleton system (Fig. 2 and Table II).
Oxidative stress results from an imbalance between biochemical processes, leading to the production of reactive oxygen species (ROS) and those responsible for the removal of ROS, the so-called antioxidant cascade. Because of high oxygen consumption and relatively poor antioxidant defense, the central nervous system is susceptible to ROS-mediated toxicity. There is growing evidence that oxidative stress contributes to the pathology of neurological disorders, such as Alzheimer disease, Parkinson disease, amyotrophic lateral sclerosis, multiple sclerosis, and Huntington disease (48–51), and in inborn errors of metabolism with neurological symptoms, such as phenylketonuria (52–54). Previous work suggests that increased oxidative stress is present in SLOS. Using a pharmacological model of SLOS, Richards et al. (55) found increased lipid hydroperoxides and retinal degeneration in AY9944-treated rats in response to intense light exposure. Treatment with dimethylthiourea, a synthetic antioxidant, was effective in preventing retinal degeneration in this model system (56). Data from this study further support the hypothesis that increased oxidative stress may contribute to SLOS neuropathology. In this study, we found decreased expression of Cu,Zn-superoxide dismutase, which catalyzes the metabolism of superoxide and plays a central role in reactive oxidative stress (48, 49), in both Dhcr7Δ3–5/Δ3–5 and Sc5d−/− brain tissues (Table II). Decreased expression of two other enzymes involved in the cellular response to oxidative stress, 3-mercaptopyruvate sulfurtransferase and lactoylglutathione lyase, was also observed in Dhcr7Δ3–5/Δ3–5 embryonic mouse brain tissue (Table II). Although increased oxidative stress cannot be excluded in lathosterolosis, in this study, we only identified a change in a single protein. Increased oxidative stress is a biochemical process that can be targeted with various therapeutic agents.
Aberrant neuronal death occurs in various neurological diseases, such as Down syndrome, Alzheimer disease, Parkinson disease, and Huntington diseases (57, 58). Caspase-3 is the major effecter in neuronal apoptosis triggered by various stimuli (59). In our study, we found that the cleaved caspase-3 (p17) was significantly increased in Dhcr7Δ3–5/Δ3–5 and Sc5d−/− brain tissues (Fig. 5, B, C, D, and E). It has been reported that cleaved caspase-3 shows co-localization with detergent-resistant lipid rafts (60), and caspase-3 is a component of Fas death-inducing signaling complex in lipid rafts (61). In our study, we also found that cleaved caspase-3 was increased in both the detergent-soluble fraction and detergent-resistant membrane of Dhcr7Δ3–5/Δ3–5 brain tissue, whereas caspase-3 precursor was not detected in detergent-resistant membrane of control and Dhcr7Δ3–5/Δ3–5 brain tissues (Fig. 5E). These data indicate that apoptosis may contribute to the neuropathology found in SLOS and lathosterolosis.
Altered glucose metabolism has been implicated as a pathological mechanism contributing to pathophysiology in neurodegenerative disorders, including neuronal ceroid lipofuscinosis (62, 63), chromosomal disorders, such as Down syndrome (64), and inborn errors of metabolism, such as phenylketonuria (65–68). In the current study, we identified decreased expression of pyruvate kinase isozymes M1/M2, transaldolase, and phosphoglycerate mutase 1 in DhcrΔ3–5/Δ3–5 brain tissue (Table II). Decreased expression of these three glycolytic enzymes suggests that there may be a functional disturbance of glucose metabolism in SLOS. The brain is dependent upon either glucose or ketone body metabolism for energy; thus, it is plausible that impaired glucose utilization may contribute to the neurological problems found in SLOS. We did not identify decreased expression of glycolytic enzymes in Sc5d−/− brain tissue. Further work will need to be done to confirm a functional deficit of glycolysis in SLOS and to determine whether this is due to a toxic effect of 7DHC. Identification of a functional disturbance of glucose metabolism has potential therapeutic implications. In contrast to fixed developmental malformations, biochemical alterations resulting in functional impairment, such as decreased glucose utilization, may be amendable to therapeutic intervention.
Mevalonate metabolism encompasses multiple presqualene and postsqualene enzymatic steps that contribute to the synthesis of cholesterol. Cellular cholesterol levels are tightly regulated by a feedback regulation mechanism in which sterol regulatory element-binding proteins coordinately regulate expression of the cholesterol biosynthetic enzymes (30–35). The data presented in this study show increased expression of HMGCS1, IDI1, and GGPS as well as increased activation of SREBP-2 in both Dhcr7Δ3–5/Δ3–5 and Sc5d−/− embryonic mouse brain tissues (Fig. 3). Consistent with our results, microarray analysis showed increased mRNA expression of mevalonate pathway enzymes in E14 Dhcr7Δ3–5/Δ3–5 hindbrain tissue (69).
In addition to the synthesis of cholesterol, presqualene cholesterol synthetic enzymes contribute to the synthesis of nonsterol isoprenoids such as dolichol, ubiquinone, farnesyl, and geranylgeranyl diphosphate (supplemental Fig. 2). Thus, due to the coordinate regulation of the expression of cholesterol synthetic genes by SREBP-2, altered regulation of cholesterol synthesis may also affect synthesis of nonsterol isoprenoids. Nonsterol isoprenoids play critical roles in multiple cellular processes. Clinical studies support the possibility of increased synthesis of presqualene isoprenoids in SLOS. Steiner et al. (70) have shown that total sterol synthesis is reduced to 60% of normal in SLOS patients. However, under the same conditions, urinary mevalonate excretion is normal (71). Normal mevalonate excretion in the context of decreased sterol synthesis could be explained by diversion of mevalonate into the synthesis of nonsterol isoprenoids or metabolism by the mitochondrial mevalonate shunt pathway. Evidence for both processes is provided by prior clinical studies. Kelley and Kratz (72) reported increased mevalonate shunt activity in SLOS children, and Pappu et al. (46) reported increased urinary excretion of ubiquinone and dolichol. Previous work by Dallner and co-workers (31, 73, 74) has shown that the first committed enzymes of dolichol and ubiquinone biosynthesis have higher affinities for the branch point intermediate farnesyl pyrophosphate than the first committed enzyme of sterol biosynthesis, thus favoring nonsterol isoprenoid synthesis. Increased activation of SREBP-2 and increased expression of presqualene isoprenoid synthetic enzymes as reported in this study combined with increased affinities of the enzymes involved in nonsterol isoprenoid synthesis are consistent with increased synthesis of nonsterol isoprenoids in both SLOS and lathosterolosis.
Increased synthesis of biologically active nonsterol isoprenoids may contribute to the SLOS and lathosterolosis phenotype. Specifically altered nonsterol isoprenoid synthesis may contribute to defects in intracellular trafficking and cytoskeletal function, two functional domains identified in this study. Geranylgeranyl pyrophosphate is a nonsterol isoprenoid that is utilized in the posttranslational modification of Rho and Rab GTPases. Prenylation of Rho and Rab GTPases is essential for membrane attachment and cycling between active GTP-bound and inactive GDP-bound states (39, 40). Increased prenylation due to increased production of geranylgeranyl in mutant tissue might lead to increased activation of Rho GTPases and then induce phosphorylation of cofilin in Dhcr7 and Sc5d mutant brain tissues (29). In this study, we identified altered expression of Rab7 in both Dhcr7 and Sc5d mutants (Fig. 4 and Table II). Rab7 was identified in two protein spots, suggesting altered posttranslational modification. Extending this primary observation, we were able to demonstrate increased expression of Rab5 in both Dhcr7 and Sc5d mutants (Fig. 4, B, C, and D); Rab5 is a protein that acts in concert with Rab7 to regulate endosomal vesicular transport (40–44). Along with the identification of altered Rab5 and Rab7 expression, functional annotation supports the hypothesis of altered intracellular trafficking (Fig. 2 and Table II). In addition to Rab7, identified proteins include syntaxin-binding protein 1, dihydropyrimidinase-related protein 3, vacuolar proton pump F subunit, ADP-ribosylation factor 1, and vacuolar protein sorting-associated protein 29 (Table II). This observation is consistent with previous work showing defects in intracellular trafficking for both SLOS and lathosterolosis. Our initial lathosterolosis patient was initially described as a severely affected SLOS patient with mucolipidosis (75). Gondré-Lewis et al. (19) reported a decreased number of secretory granules in the pancreas and pituitary and adrenal glands of Dhcr7Δ3–5/Δ3–5 and Sc5d−/− mice and abnormal morphology of secretory granules in Dhcr7 mutant exocrine pancreatic acinar cells. Wassif et al. (76) reported a secondary defect in intracellular cholesterol transport in SLOS fibroblasts resembling that seen in Niemann-Pick disease, type C. The degree to which alterations of intracellular cholesterol transport contribute to the pathology of SLOS and lathosterolosis still remains to be defined.
The cytoskeleton is a network of elongated protein polymer fibers that support cell shape, compartmentalization, and intracellular trafficking or even whole-cell movement. The cytoskeleton also plays a major role in development of neuronal processes. The three key filament systems of the cytoskeleton are microfilaments, microtubules, and intermediate filaments (77). In our study, multiple cytoskeleton-associated proteins and regulators were found to be differentially expressed in Dhcr7 and Sc5d mutant brain tissues. These included dihydropyrimidinase-related protein 2, tubulin β-5 chain, vimentin, and cofilin-1 (Table II). Although we were unable to directly validate dihydropyrimidinase-related protein 2 by Western blot, its identification in two independent spots with discordant changes in the SLOS mouse model suggest that posttranslational modifications or isoforms may underlie this observation (78). Cofilin-1 is an actin-depolymerizing factor that regulates neuronal dendrite and axon formation. We were able to validate altered cofilin-1 phosphorylation, and further functional analysis identified alteration of the Rho-Rock-Limk-cofilin pathway in both SLOS and lathosterolosis (29). This is of major interest because Rho/Rac signaling impairs normal dendritic and axonal formation, and mutations in genes encoding regulators and effectors of the Rho GTPases underlie other human mental retardation syndromes (79, 80). Developmental abnormalities of neuronal process formation may contribute to the neurocognitive deficits found in SLOS and may represent a potential target for therapeutic intervention.
In conclusion, this study reports the first quantitative proteomics analysis of SLOS and lathosterolosis tissues. In addition to identifying multiple biological pathways where altered function could contribute to the pathophysiological processes underlying SLOS and lathosterolosis and separating the effects of decreased cholesterol from increased sterol precursors, this work also suggests potential novel therapeutic interventions that can be tested in subsequent studies.
Supplementary Material
Acknowledgments
The technical assistance of Kirstyn Brownson, Meghan Lyman, and Erin Merkel was greatly appreciated.
* This work was supported, in whole or in part, by the National Institutes of Health intramural program of the NICHD.
 This article contains supplemental Figs. 1 and 2 and Tables 1–4.
This article contains supplemental Figs. 1 and 2 and Tables 1–4.
1 The abbreviations used are:
- SLOS
- Smith-Lemli-Opitz syndrome
- 2-DE
- two-dimensional electrophoresis
- 7DHC
- 7-dehydrocholesterol
- DHCR7
- 7-dehydrocholesterol reductase
- GGPS
- geranylgeranyl-diphosphate synthase
- HMGCS1
- cytoplasmic hydroxymethylglutaryl-CoA synthase
- IDI1
- isopentenyl-diphosphate Δ-isomerase 1
- ROS
- reactive oxygen species
- SC5D
- lathosterol 5-desaturase
- SREBP
- sterol regulatory element-binding protein
- c-SREBP
- cleaved sterol regulatory element-binding protein
- DSF
- detergent-soluble fraction
- DRM
- detergent-resistant membrane.
REFERENCES
- 1.Porter F. D. (2008) Smith-Lemli-Opitz syndrome: pathogenesis, diagnosis and management. Eur. J. Hum. Genet. 16, 535–541 [DOI] [PubMed] [Google Scholar]
- 2.Fitzky B. U., Witsch-Baumgartner M., Erdel M., Lee J. N., Paik Y. K., Glossmann H., Utermann G., Moebius F. F. (1998) Mutations in the Delta7-sterol reductase gene in patients with the Smith-Lemli-Opitz syndrome. Proc. Natl. Acad. Sci. U.S.A. 95, 8181–8186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wassif C. A., Maslen C., Kachilele-Linjewile S., Lin D., Linck L. M., Connor W. E., Steiner R. D., Porter F. D. (1998) Mutations in the human sterol delta7-reductase gene at 11q12–13 cause Smith-Lemli-Opitz syndrome. Am. J. Hum. Genet. 63, 55–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waterham H. R., Wijburg F. A., Hennekam R. C., Vreken P., Poll-The B. T., Dorland L., Duran M., Jira P. E., Smeitink J. A., Wevers R. A., Wanders R. J. (1998) Smith-Lemli-Opitz syndrome is caused by mutations in the 7-dehydrocholesterol reductase gene. Am. J. Hum. Genet. 63, 329–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunetti-Pierri N., Corso G., Rossi M., Ferrari P., Balli F., Rivasi F., Annunziata I., Ballabio A., Russo A. D., Andria G., Parenti G. (2002) Lathosterolosis, a novel multiple-malformation/mental retardation syndrome due to deficiency of 3beta-hydroxysteroid-delta5-desaturase. Am. J. Hum. Genet. 71, 952–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krakowiak P. A., Wassif C. A., Kratz L., Cozma D., Kovárová M., Harris G., Grinberg A., Yang Y., Hunter A. G., Tsokos M., Kelley R. I., Porter F. D. (2003) Lathosterolosis: an inborn error of human and murine cholesterol synthesis due to lathosterol 5-desaturase deficiency. Hum. Mol. Genet. 12, 1631–1641 [DOI] [PubMed] [Google Scholar]
- 7.Rossi M., D'Armiento M., Parisi I., Ferrari P., Hall C. M., Cervasio M., Rivasi F., Balli F., Vecchione R., Corso G., Andria G., Parenti G. (2007) Clinical phenotype of lathosterolosis. Am. J. Med. Genet. A 143A, 2371–2381 [DOI] [PubMed] [Google Scholar]
- 8.Porter F. D. (2003) Human malformation syndromes due to inborn errors of cholesterol synthesis. Curr. Opin. Pediatr. 15, 607–613 [DOI] [PubMed] [Google Scholar]
- 9.Waterham H. R. (2006) Defects of cholesterol biosynthesis. FEBS Lett. 580, 5442–5449 [DOI] [PubMed] [Google Scholar]
- 10.Kovarova M., Wassif C. A., Odom S., Liao K., Porter F. D., Rivera J. (2006) Cholesterol deficiency in a mouse model of Smith-Lemli-Opitz syndrome reveals increased mast cell responsiveness. J. Exp. Med. 203, 1161–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wassif C. A., Zhu P., Kratz L., Krakowiak P. A., Battaile K. P., Weight F. F., Grinberg A., Steiner R. D., Nwokoro N. A., Kelley R. I., Stewart R. R., Porter F. D. (2001) Biochemical, phenotypic and neurophysiological characterization of a genetic mouse model of RSH/Smith-Lemli-Opitz syndrome. Hum. Mol. Genet. 10, 555–564 [DOI] [PubMed] [Google Scholar]
- 12.Singh P., Paila Y. D., Chattopadhyay A. (2007) Differential effects of cholesterol and 7-dehydrocholesterol on the ligand binding activity of the hippocampal serotonin(1A) receptor: implications in SLOS. Biochem. Biophys. Res. Commun. 358, 495–499 [DOI] [PubMed] [Google Scholar]
- 13.Paila Y. D., Murty M. R., Vairamani M., Chattopadhyay A. (2008) Signaling by the human serotonin(1A) receptor is impaired in cellular model of Smith-Lemli-Opitz Syndrome. Biochim. Biophys. Acta 1778, 1508–1516 [DOI] [PubMed] [Google Scholar]
- 14.Keller R. K., Arnold T. P., Fliesler S. J. (2004) Formation of 7-dehydrocholesterol-containing membrane rafts in vitro and in vivo, with relevance to the Smith-Lemli-Opitz syndrome. J. Lipid Res. 45, 347–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berring E. E., Borrenpohl K., Fliesler S. J., Serfis A. B. (2005) A comparison of the behavior of cholesterol and selected derivatives in mixed sterol-phospholipid Langmuir monolayers: a fluorescence microscopy study. Chem. Phys. Lipids 136, 1–12 [DOI] [PubMed] [Google Scholar]
- 16.Wolf C., Chachaty C. (2000) Compared effects of cholesterol and 7-dehydrocholesterol on sphingomyelin-glycerophospholipid bilayers studied by ESR. Biophys. Chem. 84, 269–279 [DOI] [PubMed] [Google Scholar]
- 17.Wolf C., Koumanov K., Tenchov B., Quinn P. J. (2001) Cholesterol favors phase separation of sphingomyelin. Biophys. Chem. 89, 163–172 [DOI] [PubMed] [Google Scholar]
- 18.Megha, Bakht O., London E. (2006) Cholesterol precursors stabilize ordinary and ceramide-rich ordered lipid domains (lipid rafts) to different degrees. Implications for the Bloch hypothesis and sterol biosynthesis disorders. J. Biol. Chem. 281, 21903–21913 [DOI] [PubMed] [Google Scholar]
- 19.Gondré-Lewis M. C., Petrache H. I., Wassif C. A., Harries D., Parsegian A., Porter F. D., Loh Y. P. (2006) Abnormal sterols in cholesterol-deficiency diseases cause secretory granule malformation and decreased membrane curvature. J. Cell Sci. 119, 1876–1885 [DOI] [PubMed] [Google Scholar]
- 20.Tulenko T. N., Boeze-Battaglia K., Mason R. P., Tint G. S., Steiner R. D., Connor W. E., Labelle E. F. (2006) A membrane defect in the pathogenesis of the Smith-Lemli-Opitz syndrome. J. Lipid Res. 47, 134–143 [DOI] [PubMed] [Google Scholar]
- 21.Cooper M. K., Wassif C. A., Krakowiak P. A., Taipale J., Gong R., Kelley R. I., Porter F. D., Beachy P. A. (2003) A defective response to Hedgehog signaling in disorders of cholesterol biosynthesis. Nat. Genet. 33, 508–513 [DOI] [PubMed] [Google Scholar]
- 22.Koide T., Hayata T., Cho K. W. (2006) Negative regulation of Hedgehog signaling by the cholesterogenic enzyme 7-dehydrocholesterol reductase. Development 133, 2395–2405 [DOI] [PubMed] [Google Scholar]
- 23.Bijlsma M. F., Spek C. A., Zivkovic D., van de Water S., Rezaee F., Peppelenbosch M. P. (2006) Repression of smoothened by patched-dependent (pro-) vitamin D3 secretion. PLoS Biol. 4, e232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang X. S., Tang L. Y., Cao X. J., Zhou H., Xia Q. C., Wu J. R., Zeng R. (2005) Two-dimensional gel electrophoresis maps of the proteome and phosphoproteome of primitively cultured rat mesangial cells. Electrophoresis 26, 4540–4562 [DOI] [PubMed] [Google Scholar]
- 25.Hellman U., Wernstedt C., Góñez J., Heldin C. H. (1995) Improvement of an “In-Gel” digestion procedure for the micropreparation of internal protein fragments for amino acid sequencing. Anal. Biochem. 224, 451–455 [DOI] [PubMed] [Google Scholar]
- 26.Craig R., Beavis R. C. (2003) A method for reducing the time required to match protein sequences with tandem mass spectra. Rapid Commun. Mass Spectrom. 17, 2310–2316 [DOI] [PubMed] [Google Scholar]
- 27.Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 28.Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 29.Jiang X. S., Wassif C. A., Backlund P. S., Song L., Holtzclaw L. A., Li Z., Yergey A. L., Porter F. D. (2010) Activation of Rho GTPases in Smith-Lemli-Opitz syndrome: pathophysiological and clinical implications. Hum. Mol. Genet. 19, 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buhaescu I., Izzedine H. (2007) Mevalonate pathway: a review of clinical and therapeutical implications. Clin. Biochem. 40, 575–584 [DOI] [PubMed] [Google Scholar]
- 31.Grünler J., Ericsson J., Dallner G. (1994) Branch-point reactions in the biosynthesis of cholesterol, dolichol, ubiquinone and prenylated proteins. Biochim. Biophys. Acta 1212, 259–277 [DOI] [PubMed] [Google Scholar]
- 32.Dallner G., Sindelar P. J. (2000) Regulation of ubiquinone metabolism. Free Radic. Biol. Med. 29, 285–294 [DOI] [PubMed] [Google Scholar]
- 33.Sato R. (2009) SREBPs: protein interaction and SREBPs. FEBS J. 276, 622–627 [DOI] [PubMed] [Google Scholar]
- 34.Espenshade P. J., Hughes A. L. (2007) Regulation of sterol synthesis in eukaryotes. Annu. Rev. Genet. 41, 401–427 [DOI] [PubMed] [Google Scholar]
- 35.McPherson R., Gauthier A. (2004) Molecular regulation of SREBP function: the Insig-SCAP connection and isoform-specific modulation of lipid synthesis. Biochem. Cell Biol. 82, 201–211 [DOI] [PubMed] [Google Scholar]
- 36.Sakakura Y., Shimano H., Sone H., Takahashi A., Inoue N., Toyoshima H., Suzuki S., Yamada N., Inoue K. (2001) Sterol regulatory element-binding proteins induce an entire pathway of cholesterol synthesis. Biochem. Biophys. Res. Commun. 286, 176–183 [DOI] [PubMed] [Google Scholar]
- 37.Horton J. D., Goldstein J. L., Brown M. S. (2002) SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Investig. 109, 1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horton J. D., Shah N. A., Warrington J. A., Anderson N. N., Park S. W., Brown M. S., Goldstein J. L. (2003) Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. U.S.A. 100, 12027–12032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jordens I., Marsman M., Kuijl C., Neefjes J. (2005) Rab proteins, connecting transport and vesicle fusion. Traffic 6, 1070–1077 [DOI] [PubMed] [Google Scholar]
- 40.Markgraf D. F., Peplowska K., Ungermann C. (2007) Rab cascades and tethering factors in the endomembrane system. FEBS Lett. 581, 2125–2130 [DOI] [PubMed] [Google Scholar]
- 41.Konstantinopoulos P. A., Karamouzis M. V., Papavassiliou A. G. (2007) Post-translational modifications and regulation of the RAS superfamily of GTPases as anticancer targets. Nat. Rev. Drug Discov. 6, 541–555 [DOI] [PubMed] [Google Scholar]
- 42.Rojas R., van Vlijmen T., Mardones G. A., Prabhu Y., Rojas A. L., Mohammed S., Heck A. J., Raposo G., van der Sluijs P., Bonifacino J. S. (2008) Regulation of retromer recruitment to endosomes by sequential action of Rab5 and Rab7. J. Cell Biol. 183, 513–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Croizet-Berger K., Daumerie C., Couvreur M., Courtoy P. J., van den Hove M. F. (2002) The endocytic catalysts, Rab5a and Rab7, are tandem regulators of thyroid hormone production. Proc. Natl. Acad. Sci. U.S.A. 99, 8277–82782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mukhopadhyay A., Barbieri A. M., Funato K., Roberts R., Stahl P. D. (1997) Sequential actions of Rab5 and Rab7 regulate endocytosis in the Xenopus oocyte. J. Cell Biol. 136, 1227–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nicholson D. W., Ali A., Thornberry N. A., Vaillancourt J. P., Ding C. K., Gallant M., Gareau Y., Griffin P. R., Labelle M., Lazebnik Y. A., Mungay N. A., Raju S. M., Smulson M. E., Yamin T. T., Yu Y. L., Miller D. K. (1995) Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature 376, 37–43 [DOI] [PubMed] [Google Scholar]
- 46.Pappu A. S., Connor W. E., Merkens L. S., Jordan J. M., Penfield J. A., Illingworth D. R., Steiner R. D. (2006) Increased nonsterol isoprenoids, dolichol and ubiquinone, in the Smith-Lemli-Opitz syndrome: effects of dietary cholesterol. J. Lipid Res. 47, 2789–2798 [DOI] [PubMed] [Google Scholar]
- 47.Nwokoro N. A., Wassif C. A., Porter F. D. (2001) Genetic disorders of cholesterol biosynthesis in mice and humans. Mol. Genet. Metab. 74, 105–119 [DOI] [PubMed] [Google Scholar]
- 48.Sayre L. M., Perry G., Smith M. A. (2008) Oxidative stress and neurotoxicity. Chem. Res. Toxicol. 21, 172–188 [DOI] [PubMed] [Google Scholar]
- 49.Pong K. (2003) Oxidative stress in neurodegenerative diseases: therapeutic implications for superoxide dismutase mimetics. Expert Opin. Biol. Ther. 3, 127–139 [DOI] [PubMed] [Google Scholar]
- 50.Méndez-Alvarez E., Soto-Otero R., Hermida-Ameijeiras A., López-Real A. M., Labandeira-García J. L. (2002) Effects of aluminum and zinc on the oxidative stress caused by 6-hydroxydopamine autoxidation: relevance for the pathogenesis of Parkinson's disease. Biochim. Biophys. Acta 1586, 155–168 [DOI] [PubMed] [Google Scholar]
- 51.Karelson E., Bogdanovic N., Garlind A., Winblad B., Zilmer K., Kullisaar T., Vihalemm T., Kairane C., Zilmer M. (2001) The cerebrocortical areas in normal brain aging and in Alzheimer's disease: noticeable differences in the lipid peroxidation level and in antioxidant defense. Neurochem. Res. 26, 353–361 [DOI] [PubMed] [Google Scholar]
- 52.Sirtori L. R., Dutra-Filho C. S., Fitarelli D., Sitta A., Haeser A., Barschak A. G., Wajner M., Coelho D. M., Llesuy S., Belló-Klein A., Giugliani R., Deon M., Vargas C. R. (2005) Oxidative stress in patients with phenylketonuria. Biochim. Biophys. Acta 1740, 68–73 [DOI] [PubMed] [Google Scholar]
- 53.Martinez-Cruz F., Pozo D., Osuna C., Espinar A., Marchante C., Guerrero J. M. (2002) Oxidative stress induced by phenylketonuria in the rat: prevent by melatonin, vitamin E and vitamin C. J. Neurosci. Res. 69, 550–558 [DOI] [PubMed] [Google Scholar]
- 54.Kienzle Hagen M. E., Pederzolli C. D., Sgaravatti A. M., Bridi R., Wajner M., Wannmacher C. M., Wyse A. T., Dutra-Filho C. S. (2002) Experimental hyperphenylalaninemia provokes oxidative stress in rat brain. Biochim. Biophys. Acta 1586, 344–352 [DOI] [PubMed] [Google Scholar]
- 55.Richards M. J., Nagel B. A., Fliesler S. J. (2006) Lipid hydroperoxide formation in the retina: correlation with retinal degeneration and light damage in a rat model of Smith-Lemli-Opitz syndrome. Exp. Eye Res. 82, 538–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vaughan D. K., Peachey N. S., Richards M. J., Buchan B., Fliesler S. J. (2006) Light-induced exacerbation of retinal degeneration in a rat model of Smith-Lemli-Opitz syndrome. Exp. Eye Res. 82, 496–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sawa A. (1999) Neuronal cell death in Down's syndrome. J. Neural Transm. Suppl. 57, 87–97 [DOI] [PubMed] [Google Scholar]
- 58.Gorman A. M. (2008) Neuronal cell death in neurodegenerative diseases: recurring themes around protein handling. J. Cell. Mol. Med. 12, 2263–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yakovlev A. G., Faden A. I. (2001) Caspase-dependent apoptotic pathways in CNS injury. Mol. Neurobiol. 24, 131–144 [DOI] [PubMed] [Google Scholar]
- 60.Szymczyk K. H., Freeman T. A., Adams C. S., Srinivas V., Steinbeck M. J. (2006) Active caspase-3 is required for osteoclast differentiation. J. Cell. Physiol. 209, 836–844 [DOI] [PubMed] [Google Scholar]
- 61.Aouad S. M., Cohen L. Y., Sharif-Askari E., Haddad E. K., Alam A., Sekaly R. P. (2004) Caspase-3 is a component of Fas death-inducing signaling complex in lipid rafts and its activity is required for complete caspase-8 activation during Fas-mediated cell death. J. Immunol. 172, 2316–2323 [DOI] [PubMed] [Google Scholar]
- 62.Ristow M. (2004) Neurodegenerative disorders associated with diabetes mellitus. J. Mol. Med. 82, 510–529 [DOI] [PubMed] [Google Scholar]
- 63.Pietrini P., Dani A., Furey M. L., Alexander G. E., Freo U., Grady C. L., Mentis M. J., Mangot D., Simon E. W., Horwitz B., Haxby J. V., Schapiro M. B. (1997) Low glucose metabolism during brain stimulation in older Down's syndrome subjects at risk for Alzheimer's disease prior to dementia. Am. J. Psychiatry 154, 1063–1069 [DOI] [PubMed] [Google Scholar]
- 64.Kitani K., Senda M., Toyama H., Miyasaka K., Kanai S., Ohta M., Ivy G. O., Koppang N. (1995) Decline in glucose metabolism in the brain in neuronal ceroid lipofuscinosis (NCL) in English setter—evidence by positron emission tomography (PET). Gerontology 41, Suppl. 2, 249–257 [DOI] [PubMed] [Google Scholar]
- 65.Wasserstein M. P., Snyderman S. E., Sansaricq C., Buchsbaum M. S. (2006) Cerebral glucose metabolism in adults with early treated classic phenylketonuria. Mol. Genet. Metab. 87, 272–277 [DOI] [PubMed] [Google Scholar]
- 66.Weber G. (1969) Inhibition of human brain pyruvate kinase and hexokinase by phenylalanine and phenylpyruvate: possible relevance to phenylketonuric brain damage. Proc. Natl. Acad. Sci. U.S.A. 63, 1365–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Glazer R. I., Weber G. (1971) The effects of L-phenylalanine and phenylpyruvate on glycolysis in rat cerebral cortex. Brain Res. 33, 439–450 [DOI] [PubMed] [Google Scholar]
- 68.Pietz J., Rupp A., Ebinger F., Rating D., Mayatepek E., Boesch C., Kreis R. (2003) Cerebral energy metabolism in phenylketonuria: findings by quantitative in vivo 31P MR spectroscopy. Pediatr. Res. 53, 654–662 [DOI] [PubMed] [Google Scholar]
- 69.Waage-Baudet H., Dunty W. C., Jr., Dehart D. B., Hiller S., Sulik K. K. (2005) Immunohistochemical and microarray analyses of a mouse model for the Smith-Lemli-Opitz syndrome. Dev. Neurosci. 27, 378–396 [DOI] [PubMed] [Google Scholar]
- 70.Steiner R. D., Linck L. M., Flavell D. P., Lin D. S., Connor W. E. (2000) Sterol balance in the Smith-Lemli-Opitz syndrome. Reduction in whole body cholesterol synthesis and normal bile acid production. J. Lipid Res. 41, 1437–1447 [PubMed] [Google Scholar]
- 71.Pappu A. S., Steiner R. D., Connor S. L., Flavell D. P., Lin D. S., Hatcher L., Illingworth D. R., Connor W. E. (2002) Feedback inhibition of the cholesterol biosynthetic pathway in patients with Smith-Lemli-Opitz syndrome as demonstrated by urinary mevalonate excretion. J. Lipid Res. 43, 1661–1669 [DOI] [PubMed] [Google Scholar]
- 72.Kelley R. I., Kratz L. (1995) 3-Methylglutaconic acidemia in Smith-Lemli-Opitz syndrome. Pediatr. Res. 37, 671–674 [DOI] [PubMed] [Google Scholar]
- 73.Thelin A., Runquist M., Ericsson J., Swiezewska E., Dallner G. (1994) Age-dependent changes in rat liver prenyltransferases. Mech. Ageing Dev. 76, 165–176 [DOI] [PubMed] [Google Scholar]
- 74.Ericsson J., Thelin A., Chojnacki T., Dallner G. (1992) Substrate specificity of cis-prenyltransferase in rat liver microsomes. J. Biol. Chem. 267, 19730–19735 [PubMed] [Google Scholar]
- 75.Parnes S., Hunter A. G., Jimenez C., Carpenter B. F., MacDonald I. (1990) Apparent Smith-Lemli-Opitz syndrome in a child with a previously undescribed form of mucolipidosis not involving the neurons. Am. J. Med. Genet. 35, 397–405 [DOI] [PubMed] [Google Scholar]
- 76.Wassif C. A., Vied D., Tsokos M., Connor W. E., Steiner R. D., Porter F. D. (2002) Cholesterol storage defect in RSH/Smith-Lemli-Opitz syndrome fibroblasts. Mol. Genet. Metab. 75, 325–334 [DOI] [PubMed] [Google Scholar]
- 77.Pollard T. D. (2003) The cytoskeleton, cellular motility and the reductionist agenda. Nature 422, 741–745 [DOI] [PubMed] [Google Scholar]
- 78.Petratos S., Li Q. X., George A. J., Hou X., Kerr M. L., Unabia S. E., Hatzinisiriou I., Maksel D., Aguilar M. I., Small D. H. (2008) The beta-amyloid protein of Alzheimer's disease increases neuronal CRMP-2 phosphorylation by a Rho-GTP mechanism. Brain 131, 90–108 [DOI] [PubMed] [Google Scholar]
- 79.van Galen E. J., Ramakers G. J. (2005) Rho proteins, mental retardation and the neurobiological basis of intelligence. Prog. Brain Res. 147, 295–317 [DOI] [PubMed] [Google Scholar]
- 80.Linseman D. A., Loucks F. A. (2008) Diverse roles of Rho family GTPases in neuronal development, survival, and death. Front. Biosci. 13, 657–676 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.