Abstract
Reactive oxygen species (ROS) are both physiological intermediates in cellular signaling and mediators of oxidative stress. The cysteine-specific redox-sensitivity of proteins can shed light on how ROS are regulated and function, but low sensitivity has limited quantification of the redox state of many fundamental cellular regulators in a cellular context. Here we describe a highly sensitive and reproducible oxidation analysis approach (OxMRM) that combines protein purification, differential alkylation with stable isotopes, and multiple reaction monitoring mass spectrometry that can be applied in a targeted manner to virtually any cysteine or protein. Using this approach, we quantified the site-specific cysteine oxidation status of endogenous p53 for the first time and found that Cys182 at the dimerization interface of the DNA binding domain is particularly susceptible to diamide oxidation intracellularly. OxMRM enables analysis of sulfinic and sulfonic acid oxidation levels, which we validate by assessing the oxidation of the catalytic Cys215 of protein tyrosine phosphatase-1B under numerous oxidant conditions. OxMRM also complements unbiased redox proteomics discovery studies as a verification tool through its high sensitivity, accuracy, precision, and throughput.
Oxidation of cysteine residues plays a critical role in modifying the structure and function of many proteins. Although cysteine oxidation is a tightly regulated biological process, nonenzymatic processes can contribute substantially to its levels, such as during oxidative stress. Regulatory oxidation states such as disulfide bonding and S-nitrosylation are readily modulated (1) and play an essential role in many physiological processes, including cell cycle, growth, death, and differentiation (2). In contrast, prolonged accumulation of reactive oxygen species is associated with many pathological conditions and leads to stable overoxidized states (sulfinic and sulfonic acid) that may disrupt redox regulation and protein function (3) and, in most cases, are thought to be nonregenerative.
Assays capable of comprehensively assessing the dynamic changes in site-specific oxidation states are especially critical to understanding the contribution of redox status to many diseases. Numerous redox-sensitive proteins, including essential cellular regulators such as p53, have been described previously (for review, see ref. 4). However, technical factors have hampered the identification of specific site(s) of modification and characterization of their redox status in cells. Site-directed mutagenesis is often employed to determine whether specific cysteines have redox-regulated functional roles (1), but this approach provides no information on the oxidation status of the endogenous protein. In addition, cysteine oxidation is dynamically dependent on the concentration, location, and specificity of small-molecule oxidants (5) and regulators of various antioxidant enzymes (6). Thiol pKa (7), solvent accessibility, and subcellular compartment (8, 9) also contribute to the dynamics of cysteine oxidation. Because the interface between chronic oxidative stress and disruption of essential cellular signaling has substantial biological relevance to disease and age-related pathological conditions (10–13), there is a strong need to develop sensitive and flexible assays capable of quantifying dynamic changes in the redox status of specific endogenous proteins.
Direct analysis of most regulatory cysteine modifications is not suitable for robust quantitation because the modifications tend to be labile and susceptible to artifactual changes. Accurate preservation of the thiol oxidation state is commonly achieved with a three-step differential alkylation labeling strategy in which nonoxidized cysteines are 1) labeled with a tag, 2) chemically reduced, and 3) labeled with a distinguishing tag. The value of this process is that it replaces the labile oxidation-modified cysteines with highly stable alkylated forms (1). Differential alkylation specifically targets cysteine oxidation species that are susceptible to reaction with chemical reductants (DTT or TCEP1) because higher oxidation states such as sulfinic and sulfonic acid are chemically irreversible. Fluorescent or epitope tags have been employed to evaluate redox sensitivity at the protein level (14, 15); however, combining differential alkylation using stable isotope-labeled reagents with mass spectrometry simultaneously identifies the specifically oxidized site and quantifies its reversible oxidation status. Although labeled iodoacetic acid (16) and N-ethylmaleimide (17) (NEM) have often been used, commercial ICAT reagents have become the preferred stable isotope label for redox analysis (18–21). A recent ICAT-based study identified and quantified the reversible oxidation of 120 redox-sensitive cysteines in Escherichia coli in an unbiased manner (18). However, because of the limited sensitivity, dynamic range, number of testable conditions, and stochastic sampling inherent in unbiased proteomics experiments, this approach has limited utility for interrogating targeted moderate- to low-abundance proteins or comprehensively characterizing multiple cysteines within a single protein. The use of an ICAT-based approach for targeted analysis of specific proteins is significantly limited by both the difficulty in scaling down the ICAT protocol and the disruption of protein structure that occurs after alkylation with numerous ICAT adducts, each over a kilodalton in size, which may occlude antibody epitopes useful for immunoaffinity enrichment.
To overcome these limitations, we developed a highly sensitive method, OxMRM, that integrates protein purification, differential alkylation using a generic d5 stable isotope-labeled NEM, and multiple reaction monitoring (MRM). OxMRM can quantify the oxidation status, both reversible and irreversible, of virtually any targeted cysteine or protein, even if in low abundance. We validate the OxMRM approach using the low-abundance tumor suppressor protein p53 and an established overoxidizable signaling regulator, protein tyrosine phosphatase-1B (PTP1B) (22, 23). These proteins serve as benchmark examples of nuclear and cytoplasmic redox-regulated proteins bearing reversibly and irreversibly oxidized cysteines. The flexibility of the OxMRM method allows it to be applied to essentially any protein or cysteine of interest with equal ease, allowing in-depth, site-specific analysis of putative and established redox-sensitive proteins. This makes OxMRM an ideal complement to large-scale redox proteomics studies as a verification tool with high sensitivity, precision, accuracy, and capacity to assess numerous oxidation conditions.
EXPERIMENTAL PROCEDURES
Materials
All chemicals and HPLC-grade solvents were purchased from Sigma, except for those listed below, and were of the highest grade available. d5 NEM was from Cambridge Isotope Laboratories (Andover, MA); anti-p53 antibodies FL-393 and DO-1 were from Santa Cruz Biotechnology (Santa Cruz, CA); anti-PTP1B (FG-6) antibody was from Abcam (Cambridge, MA); anti-PTP Cys215 sulfonic acid oxidized antibody was from R&D Systems (Minneapolis, MN); protein G-Sepharose was from GE Healthcare (Waukesha, WI); Econopak 10DG (packed with 10 ml of Bio-Gel P-6) columns were from Bio-Rad Laboratories; and sequencing-grade modified trypsin was from Promega (Madison, WI).
Cell Lysis, Acid Trapping, and Differential Alkylation
Both MCF7 and HCA2 cells were passaged in 20% O2 conditions (10% CO2, 70% N2) and transferred to 3% O2 (10% CO2, 87% N2) conditions for 3–4 days before treatment or lysis. Cells were not serum-starved. Once the 15-cm plates of cells reached 80–90% confluence, the media were removed and cells were quickly washed once with room-temperature PBS. One 15-cm plate of MCF7 cells typically had 6 × 107 cells and one plate of HCA2 cells had 6 × 106 cells. To culture biological replicates, several T75 flasks of cells were grown, split into separate plates, and treated independently from then on. 1.5 ml of ice-cold 20% (v/v) TCA was added to the cells and quickly spread to cover the entire plate followed by incubation for at least 20 min at 4 °C. Cells were scraped into a microcentrifuge tube and washed with 10 and 5% ice-cold TCA one time, and care was taken to break up the pellet each time. Samples were resuspended in 900 μl of d0 TUNES buffer (200 mm Tris pH 7.0, 8 m urea, 100 mm d0 NEM, 10 mm EDTA, and 2% SDS). Samples were sonicated on ice to resuspend the protein pellet and incubated at 50 °C for 30 min at 1000 rpm on a shaker/incubator. The reaction was stopped and proteins were reprecipitated by bringing the sample to 20% TCA and then washing once with 10% TCA and twice with 5% TCA to remove the d0 NEM. Samples were then reduced by resuspension in TUNES buffer without NEM but with 5.5 mm TCEP, sonicated, and incubated at 50 °C for 30 min while shaking at 1200 rpm. To complete the differential alkylation, 15 μl of 100 mm d5 NEM was added and samples were incubated at 50 °C for 30 min at 1200 rpm. At this point samples proceeded to immunoaffinity purification or were frozen at −80 °C.
Protein Immunoaffinity Purification and Proteolysis
Before immunoaffinity purification, samples were brought to 3 ml with water and desalted using a Bio-Gel P-6 column according to manufacturer's instructions and eluted with 4 ml of H2O. For the immunoaffinity purification, samples from a 15-cm plate were diluted into 12 ml of 0.1% SDS, 0.5% (w/v) sodium deoxycholic acid, 20 mm Tris, pH 7.0, and 100 ml of NaCl. Samples were precleared overnight with 15 μl of protein G-Sepharose. For p53, 15 μl each of 200 ng/μl FL-393 and DO-1 antibodies were incubated for >4 h, followed by addition of 10 μl of protein G-Sepharose overnight. For PTP1B, 30 μl of 100 μg/ml FG-6 antibody was added as above.
After purification, samples were placed into a siliconized tube and washed twice with ice-cold PBS + 1% (v/v) Nonidet P-40. Samples were then washed once with ice-cold 100 mm Tris, pH 7.0, and three times with ice-cold 50 mm ammonium bicarbonate. For trypsin proteolysis, 320 μl of 12.5% (v/v) acetonitrile with 150 ng of trypsin were added to each sample and incubated overnight at 37 °C, 1200 rpm. Samples were removed, concentrated, and used for LC-MRM/MS analysis.
LC-MRM/MS
Samples were analyzed by nano-LC-MRM/MS on a 4000 QTRAP hybrid triple quadrupole/linear ion trap mass spectrometer (Applied Biosystems, Foster City, CA). Chromatography was performed using a NanoLC-2D LC system (Eksigent, Dublin, CA) with buffer A (0.1% (v/v) formic acid) and buffer B (90% (v/v) acetonitrile in 0.1% formic acid). Digests were loaded at 20 μl/min (0.1% formic acid) onto a 5 mm × 300 μm reversed-phase C18 trap column (5 μm, 100 Å; Dionex, Sunnyvale, CA) and eluted at 300 nL/min with a 75-μm inner diameter Integrafrit analytical column (New Objective, Woburn, MA) packed in-house with 10–12 cm of ReproSil-Pur C18-AQ 3 μm reversed-phase resin (Dr. Maisch GmbH, Ammerbuch-Entringen, Germany) with a gradient of 2–70% buffer B over 32 min. To detect the hydrophilic p53 peptide CPHHER corresponding to Cys176, the trap column was bypassed, and the sample was directly loaded onto the analytical column. Peptides were ionized using a PicoTip emitter (75 μm, 15 μm tip; New Objective). Data acquisition was performed using Analyst 1.5 with an ion spray voltage of 2450 V, curtain gas of 10 psi, nebulizer gas of 20 p.s.i., and an interface heater temperature of 150°C. Collision energy, declustering potential, and collision cell exit potential were optimized using recombinant p53 and PTP1B for maximum sensitivity and listed in supplemental Table 1. MRM transitions were monitored and acquired at unit resolution in both the first and third quadrupoles (Q1 and Q3). Multiquant version 1.1 (Applied Biosystems/MDS Analytical Technologies) was used to process all MRM data. Each transition was individually integrated to generate peak areas. If a peak was not detected, the level of background was integrated.
LC-MS/MS Identification of p53 and PTP1B Peptides and Post-translational Modifications
Samples were analyzed by nano-LC-MS/MS on a 4000 QTRAP hybrid triple quadrupole/linear ion trap mass spectrometer (AB Sciex) as described under “LC-MRM/MS.” MS/MS spectra were collected in the linear ion trap mode with a mass range of 100–1400 using Q0 trapping. Often, as in Fig. 4, experiments were designed in which MRM transitions were used to trigger MS/MS data acquisition to detect specific oxidized peptides at low levels. Identification of peptides and post-translational modifications was also performed on samples using a nano-HPLC-ESI Q-TOF mass spectrometer (QSTAR Elite; AB Sciex). Chromatographic separation of peptides was carried out using an Eksigent nano-LC 2D HPLC system that was directly connected to the QSTAR Elite mass spectrometer. Peptide mixtures were loaded onto a guard column (C18 Acclaim PepMap100, 300 μm inner diameter × 5 mm, 5-μm particle size, 100-Å pore size; Dionex) and washed with the loading solvent (0.1% formic acid, 98% H2O, and 2% acetonitrile; flow rate, 20 μl/min) for 10 min. Subsequently, samples were transferred onto the analytical C18-nanocapillary HPLC column (C18 Acclaim PepMap100; 300-μm inner diameter × 15 cm, 3-μm particle size, 100-Å pore size; Dionex) and eluted at a flow rate of 300 nL/min using a 60-min gradient. Mass spectra (ESI-MS) and tandem mass spectra (ESI-MS/MS) were recorded in positive-ion mode with a resolution of 12,000–15,000 full-width half-maximum. The peaklist generating software was Analyst Mascot.dll v1.6b23 (Applied Biosystems), and the search engine used was Mascot Version 2.1.0 (Matrix Science Inc., Boston, MA). The database searched was SwissProt 57.12 (513877 entries). The search parameters were as follows. The enzyme specificity was trypsin with one missed cleavage permitted; variable modifications included oxidation of methionine as well as NEM, dioxidation, or trioxidation of cysteine. The mass tolerance for data for precursor ions for the QTRAP was set at 0.6 Da, and the mass tolerance for fragment ions was 0.4 Da. For the QSTAR, both the precursor and fragment ion mass tolerance was set at 0.4 Da. The maximum expectation value for accepting individual MS/MS spectra was 0.05 when searched against the SwissProt database (Table I), except for the peptide CPHHER, which is hand-annotated in supplemental Fig. 1 with detail assignment criteria addressed.
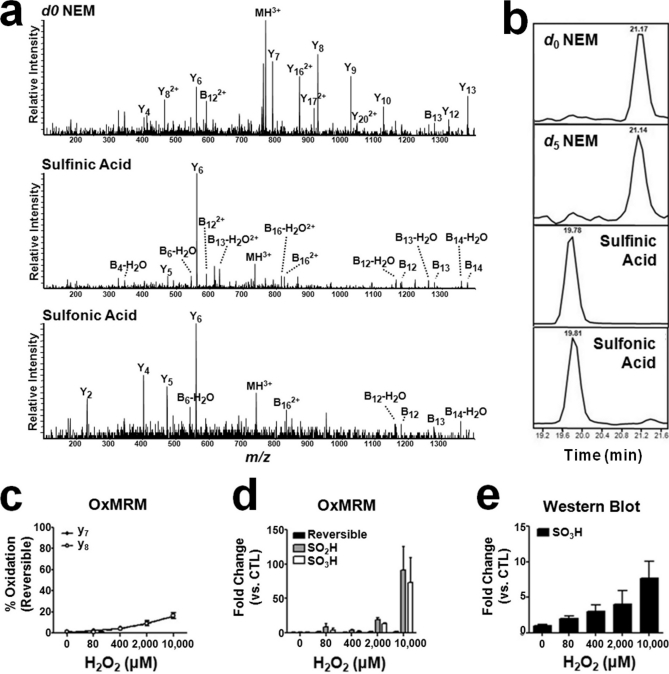
Fig. 4.
Quantitation of irreversible oxidation of endogenous PTP1B Cys215 by H2O2. a, MS/MS spectra of the MH33+ (m/z 767.4) d0 NEM-alkylated peptide ESGSLSPEHGPVVVHCSAGIGR, which includes PTP1B Cys215 (in bold). The MS/MS spectra of the MH33+ precursor ions for sulfinic acid (m/z 736.4) and sulfonic acid (m/z 741.7)-modified peptides, which were generated from tryptic digestion of PTP1B isolated from MCF7 cells, are also shown. b, the relative retention of each species in LC-MS/MRM analysis. c, OxMRM analysis of the reversible oxidation of PTP1B Cys215 in response to exogenous H2O2 oxidation. d, H2O2 increases the level of sulfinic and sulfonic acid-modified PTP1B Cys215 in a dose-dependent manner when normalized to the level of nonoxidized (d0 NEM) peptide. e, the relative increase of PTP1B Cys215 sulfonic acid oxidation (normalized to total PTP1B levels) after H2O2 treatment using an established antibody. In all cases, error bars represent the S.D. of three biological replicates.
Table I. OxMRM analysis of 15 total cysteines from p53 and PTP1B.
Table of the peptides and cysteines in endogenous p53 and PTP1B analyzed by OxMRM. The charge state and MRM transitions measured for each peptide are listed. Oxidation of specific cysteines in p53 and PTP1B that have been previously characterized by mass spectrometry are summarized, with the source being recombinant protein unless indicated by an #. For MH+, cysteines alkylated with d0 NEM are shown.
| Protein | Peptide | Cys | MH+ (mono) | Charge | Ion type | Oxidation | Reference |
|---|---|---|---|---|---|---|---|
| p53 | SVTCTYSPALNK | 124 | 1408.7 | 2 | y8,y9 | GSH adduct | (42) |
| p53 | MFCQLAK | 135 | 965.5 | 2 | y6,y5 | ||
| p53 | TCPVQLWVDSTPPPGTR | 141 | 1979.0 | 2 | y6,y9 | GSH adduct, DTT reversible oxidation | (40, 42) |
| p53 | CPHHER | 176 | 903.4 | 2 | y4,y5 | ||
| p53 | CSDSDGLAPPQHLIR | 182 | 1733.8 | 2 | y7,y8 | GSH adduct | (42) |
| p53 | VCACPGR | 275/277 | 955.4 | 2 | y4,y5 | ||
| p53 | HEASDFPCR | 32 | 1186.5 | 3 | b5,y3 | SNO adduct | (29) |
| PTP1B | SYILTQGPLPNTCGHFWEMVWEQK | 91 | 2012.0 | 3 | y152+,y172+ | SNO adduct | (29) |
| PTP1B | CAQYWPQK | 121 | 1148.5 | 2 | b5,y5 | ||
| PTP1B | CAQYWPQKEEK | 121 | 1534.7 | 2 | y7,y6 | ||
| PTP1B | ESGSLSPEHGPVVVHCSAGIGR | 215 | 2300.1 | 3 | y8,y9 | SNO/GSH Adduct, Sulfinic#/sulfonic# acid | (29, 48, 49, 50) (35)# |
| PTP1B | SGTFCLADTCLLLMDKR | 226/231 | 2137.0 | 3 | y11,y10 | ||
| PTP1B | CREFFPNHQWVK | 324 | 1715.8 | 3 | b5,b7 | ||
| PTP1B | DCPIKEEK | 343 | 1086.5 | 3 | y62+,y4 |
Pro-oxidant Treatment Conditions
MCF7 cells were grown in 15-cm plates at 3% O2 without serum starvation. Treatments were as follows: N,N′-dimethyl-4,4′-bipyridinium dichloride (Paraquat): 1.2 mm, 20 h; N-acetylcysteine: 5 mm, 30 min; rotenone: 32 μm, 2 h; TNFα: 10 ng/ml, 15 min; glutathione ethyl ester: 100 μm, 30 min; pyrrolidine dithiocarbamate: 75 μm, 2 h; and S-nitroso-N-acetyl-penicillamine: 500 μm, 1 h. All hydrogen peroxide (H2O2) treatments were for 10 min, and diamide treatments were for 15 min at the concentrations indicated in Figure 5. DNA damage was induced by 500 ng/ml doxorubicin for 24 h. Plates were washed with PBS and lysed as above.
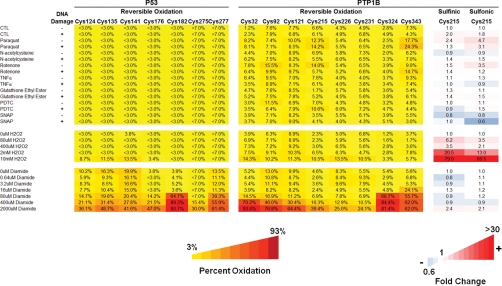
Fig. 5.
Quantitation of p53 and PTP1B oxidation in response to varied pro-oxidants. OxMRM characterization of reversible and irreversible oxidation of seven cysteines in p53 and eight cysteines in PTP1B in MCF7 cells after the pro-oxidant treatments listed and detailed under Materials and Methods. The conditions are grouped into three batches: 1) treatments with or without DNA damage, 2) H2O2 concentration curve, and 3) diamide concentration curve. Only PTP1B Cys215 was detected as an irreversibly oxidized species. Reversible oxidation is measured as percentage oxidation and irreversible oxidation as the fold change of the sulfinic and sulfonic acid-modified peptide relative to the corresponding control condition. When the oxidized peptide was not detected, the upper limit of oxidation that may potentially be present, but not detected, was determined by ratio of the unmodified peptide signal to the baseline level of the oxidized peptide transition and is represented in the heat map with a less than sign. Data shown are from a single transition and biological replicate. The full transition list is detailed in supplemental Table 1.
Oxidation of Recombinant p53
Recombinant p53 protein was diluted in Tris buffer (10 mm, pH 7.0) and 10 mm EDTA. 10 μm TCEP was added to reduce the level of background oxidation that occurs during purification and storage. Oxidants were added as described in supplementary Fig. 8. For differential alkylation, d5 NEM was added at final concentration of 5 mm, quenched by adding cysteine to 40 mm, reduced with 25 mm TCEP, and brought to 50 mm d0 NEM, with each step incubated for 30 min at room temperature. d0 NEM was quenched with excess cysteine before in-solution trypsin digestion overnight at 37 °C, and proteolysis was stopped by addition of formic acid.
Immunoblot Analysis of Sulfonic Acid Oxidized PTP1B Cys215
Cells were grown in 3% O2 as above, washed once with room temperature PBS, and lysed in TUNES buffer plus 5.5 mm TCEP. Samples were sonicated twice and incubated at 50 °C for 30 min during shaking at 1200 rpm. The total lysate was immunoblotted for sulfonic acid-oxidized PTP1B (1:200; anti-PTP active site sulfonic acid-oxidized antibody) or total PTP1B levels (1:1000; anti-PTP1B).
RESULTS
Method Overview
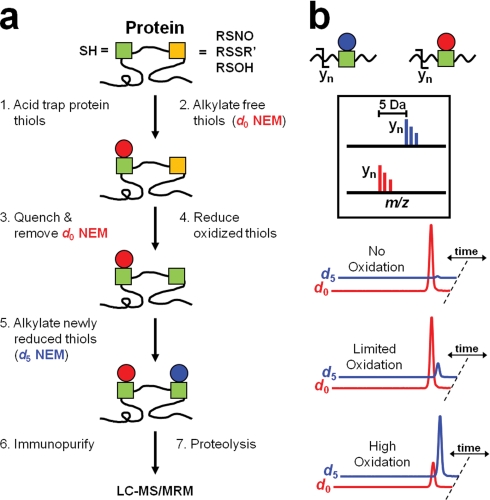
Cysteine oxidation analysis is susceptible to artifacts during sample preparation (24). To minimize such artifacts, cells were lysed in TCA, the most rapid method to quench cellular redox reactions (rate constant = 109 s−1 m−1) (25). To maximize the kinetics of alkylation and reduce artifacts during resuspension at neutral pH for the initial alkylation of free thiols, we used unlabeled (d0) NEM, a faster and more cysteine-specific reagent than iodoacetamide-based reagents such as ICAT (25). Alkylation was performed in highly denaturing conditions (8 m urea, 2% (w/v) SDS) to maximize solvent accessibility and labeling of all cysteines. Subsequently, differential alkylation was carried out with a chemically equivalent, stable isotope version of NEM with 5 deuteriums (d5), which is widely commercially available and allows calculation of the absolute ratio of the reduced to oxidized forms of each cysteine within a single sample (Fig. 1a). Thus, the percentage reversible oxidation, rather than fold change, can be calculated (18). However, very high levels of irreversible oxidation of the cysteine may lead to an overestimation of this percentage because a substantial amount of the cysteine will not be differentially alkylated.
Fig. 1.
Summary of the OxMRM method that integrates differential thiol alkylation, protein purification, and MRM analysis. a, cellular redox reactions are first quenched by TCA lysis, which protonates the highly reactive thiolate anions. Nonoxidized (green) cysteines are differentiated from reducibly oxidized (yellow) cysteines by differential alkylation using an unlabeled (d0, red) and stable isotope-labeled version (d5, blue) of NEM. The protein of interest is then affinity-purified and proteolyzed before MRM analysis. b, a triple-quadrupole mass spectrometer can isolate a narrow m/z region in Q1 corresponding to a precursor ion of interest, fragment the peptide using collisionally activated dissociation (CAD) in Q2, and transfer ions within a different m/z region corresponding to a known fragment ion in Q3. MRM rapidly scans through unique Q1/Q3 transitions for each peptide serially and can differentiate the d0 and d5 NEM-alkylated peptides. Representative chromatograms depicting the relative levels of d0 and d5 NEM-alkylated peptide under no, limited, and high oxidation states are shown.
Quantitation of cysteine oxidation in low-abundance proteins pose both sensitivity and dynamic range challenges that can be addressed by MRM. MRM analysis has unparalleled sensitivity for targeted analysis, offering quantitation with a dynamic range of 5–6 orders of magnitude and the capability to detect proteins with as few as 50 copies per cell (26). To analyze a low-abundance target protein such as p53, which has a half-life of 6–30 min (27), protein purification (via immunoaffinity purification, epitope tagging, or other means) allows for maximum sensitivity (Fig. 1a). After the differentially alkylated protein is purified, the protein is enzymatically proteolyzed and the resulting peptides are subjected to LC-MS/MRM analysis. We used direct proteolysis of the target protein while attached to protein G-Sepharose to limit the loss of sample and minimize oxidation artifacts that can occur during gel electrophoresis. MRM can differentially quantitate d0 and d5 NEM-alkylated peptides (Fig. 1b) by integrating the area of each chromatographic peak and calculating the percentage oxidation. Example chromatograms for no, limited, and high oxidation are shown in Fig. 1b. The overlap of the chromatograms for the d0 and d5 NEM-alkylated peptides is greater than 95%, arguing against any substantial matrix effects (supplemental Fig. 2). Tryptic digestion of p53 and PTP1B both generate a peptide containing two cysteines (Table I). OxMRM is unique in that it can differentiate the oxidation status of individual cysteines when two are present in the same peptide (supplemental Fig. 3).
We were able to monitor the intracellular oxidation status of 7 of 10 cysteines in p53 and 8 of 10 cysteines in PTP1B after tryptic digestion (Table I). Although almost all of the previous mass spectrometry studies on these proteins have been performed on recombinant (18, 19, 28–30) or overexpressed (21) proteins (Table I), the improved sensitivity of OxMRM allows for a greater depth of coverage and the ability to analyze even low-abundance endogenous proteins such as these at their native concentration levels. However, 3 of 10 cysteines in p53 and 2 of 10 cysteines in PTP1B were not covered in this analysis. This lack of coverage occurred because trypsin produced cysteine-containing peptides in both p53 and PTP1B that were not readily amenable to MS analysis as a result of their large size (>30 amino acids).
Sensitivity of Oxidation Analysis by OxMRM
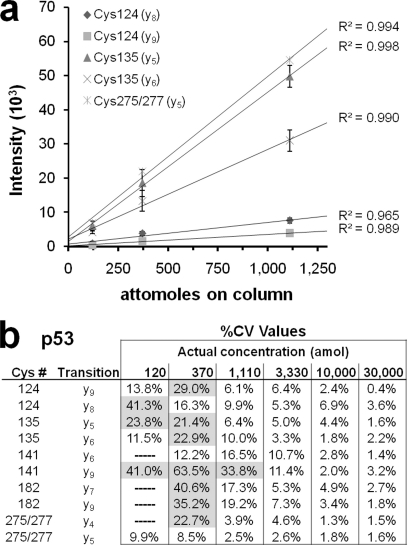
To estimate OxMRM detection limits for each p53 peptide, we assessed a dilution series using known quantities of recombinant protein. The starting amount of protein was verified by comparing the one-dimensional-SDS-PAGE gel-staining intensity of the p53 band with that of standard proteins loaded at known concentration (supplemental Fig. 4). Each concentration in the standard curve was coinjected with 500 ng of tryptic peptides from yeast, which lacks p53, as a matrix. The results demonstrate that MRM analysis has a linear response into the attomole range with high correlation coefficients (Fig. 2a and supplemental Fig. 5), which is typical for peptide MRM assays (26, 31). To assess technical reproducibility and percentage coefficient of variation, samples were analyzed in triplicate. The results showed that acceptable coefficients of variation under 20% (32) can be achieved at the level of ∼1 fmol for all peptides but one (Fig. 2b).
Fig. 2.
Sensitivity and reproducibility of OxMRM analysis of p53. MS/MS spectra were collected for each NEM-alkylated peptide and the optimal precursor charge state, fragment ions, collision energy, and declustering potential were determined for MRM analysis. Dwell times for each transition were adjusted to have an equal detection limit for all peptides or scheduled based on retention time. a, known quantities of trypsin-digested recombinant p53 (d0 NEM-alkylated) were used to generate a calibration curve from 120 amol to 30 fmol. The regression line for several individual MRM transitions are shown and demonstrate that the linear range extends to the attomole level. Correlation coefficients (R2 value) for each line are listed. Error bars represent the S.D. from three technical replicates. “Cys275/277” represents the single peptide that includes both Cys275 and 277. b, percentage coefficient of variation (%CV) for all p53 transitions in the calibration curve. %CVs above 20% are shaded. A dashed line indicates that no peak was detected for the transition. The full transition list is detailed in supplemental Table 1.
Oxidation Analysis of Endogenous p53 and PTP1B
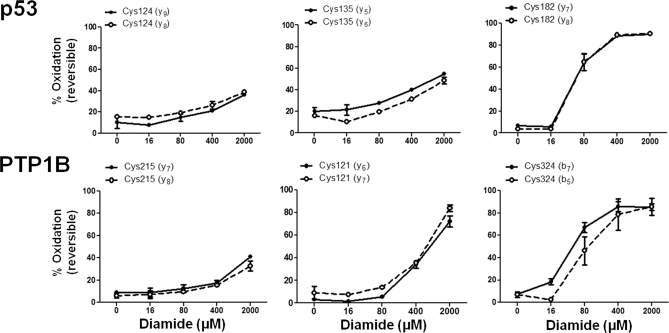
To assess the ability of OxMRM to measure intracellular oxidation of p53 and PTP1B, MCF7 (human breast cancer) cells (one 15-cm plate per condition) were treated with the thiol-specific oxidant diamide and differentially alkylated with d0 and d5 NEM. p53 and PTP1B were then serially immunoaffinity-purified and analyzed by MRM. Representative examples of p53 and PTP1B cysteines with low, moderate, and high susceptibility to diamide oxidation are shown in Fig. 3; data for all cysteines is shown in supplemental Fig. 6. For each peptide, similar levels of oxidation were determined by distinct MRM transitions, demonstrating the capacity of OxMRM to measure even low-abundance endogenous proteins with precision. p53 is even less abundant in primary human fibroblasts such as HCA2 cells. In these cells, the OxMRM approach also quantified oxidation of seven cysteines in p53 (using two 15-cm plates per condition), revealing a very similar overall diamide oxidation susceptibility profile (supplemental Fig. 7). Within cells, Cys182 is most susceptible to diamide oxidation of all p53 cysteines tested. Previous studies have hypothesized that, because Cys182 lies at the dimer-dimer interface of the p53 tetramer (33), oxidation of this residue may affect DNA binding (34).
Fig. 3.
Intracellular oxidation of endogenous p53 and PTP1B. To determine the susceptibility of p53 and PTP1B cysteines to intracellular oxidation, MCF7 cells cultured under normoxic conditions (3% O2) were treated with varying doses of diamide for 15 min. The percentage oxidation of three p53 and PTP1B cysteines are shown that represent slight, moderate, and high diamide sensitivity. The error bars represent the S.D. of three biological replicates (a single 15-cm plate of cells each), and the percentage oxidation is shown for two MRM transitions per peptide. The full transition list is detailed in supplemental Table 1.
OxMRM Analysis of Irreversible Oxidation
OxMRM allows quantitation of irreversible oxidation, because sulfinic and sulfonic acid-modified cysteines are stable during sample preparation and can be directly detected by MRM. Most differential alkylation approaches, such as ICAT, use an epitope tag to enrich for alkylated cysteines and cannot measure irreversible oxidation. With OxMRM, affinity purification of the target protein does not require cysteine alkylation and therefore has the unique potential to detect both essential, regulated oxidation and chronic oxidation in disease processes. To explore the well documented irreversible oxidation of PTP1B Cys215 (22, 29, 35) using OxMRM, we treated cells with hydrogen peroxide (H2O2) and collected MS/MS data for the sulfinic and sulfonic acid-modified peptide (Fig. 4a and Table II). These spectra showed the characteristic increase in intensity for the y fragment ion (y6) C-terminal to the sulfinic and sulfonic acid modified cysteine (36). This is especially true for the sulfinic acid-modified peptide, as has been observed, and the limited number of fragment ions produced can lead to decreased expectation scores when searching these modified peptides using search algorithms. MRM transitions to the y6 ion determined that the sulfonic and sulfinic acid-modified peptides elute ∼1.5 min earlier than the NEM-alkylated species (Fig. 4b). Using OxMRM, we quantified PTP1B Cys215 reversible oxidation, as well as both sulfinic and sulfonic acid modifications, of intracellular PTP1B Cys215 in MCF7 cells across a range of H2O2 concentrations. As with diamide oxidation, reversible oxidation of Cys215 increased only slightly (Fig. 4c). Because the sulfinic and sulfonic acid-modified Cys215 peptides are not chemically equivalent to the NEM-modified forms, it is not possible to compare the absolute levels of irreversible and reversible oxidation. However, comparing the relative levels of irreversible oxidation and the nonoxidized peptide determines the fold change under different conditions. The induction of PTP1B Cys215 sulfinic and sulfonic acid across H2O2 concentrations is shown in Fig. 4d. Both are markedly increased relative to control, as reported previously using a mass spectrometry based approach (29). The fold increase in PTP1B Cys215 sulfonic acid was determined by Western blot by normalizing the signal intensity from a commercially available antibody (29) generated against sulfonic acid modified Cys215 to total PTP1B protein levels (Fig. 4e). The fold increase determined by OxMRM was larger than that seen on the immunoblot, possibly because of OxMRM normalization to the unmodified PTP1B Cys215-containing peptide. If a large percentage of the cysteine becomes oxidized, ratio expansion can possibly result. Alternatively, the improved signal to noise of the OxMRM data may more accurately reflect the increase. Unlike specific antibodies, however, OxMRM is capable of multiplexed quantitation of essentially any cysteine.
Table II. Modified peptides identified from tryptic digestion of PTP1B representing each post-translational modification detected to Cys215.
C(O2) represents the sulfinic acid-modified cysteine and C(O3) represents the sulfonic acid-modified peptide.
| Peptide | m/z observed | Change in mass | Peptide score | Expectation value |
|---|---|---|---|---|
| Da | ||||
| ESGSLSPEHGPVVVHC(O2)SAGIGR | 736.33 | −0.08 | 28 | 0.048 |
| ESGSLSPEHGPVVVHC(O3)SAGIGR | 741.62 | −0.40 | 47 | 0.0026 |
Multiplexed Quantitation of Redox Regulation As a Result of Diverse Pro-oxidant Conditions
We expanded our redox analysis of p53 and PTP1B by OxMRM beyond diamide and H2O2 to assess how a range of physiological and exogenous oxidants (DNA damage, paraquat, rotenone, and others) affect oxidation of these two proteins in MCF7 cells. The modified heat map in Fig. 5 shows the multiplexed analysis of cysteines under a variety of experimental conditions (see “Materials and Methods”). When only a small amount of the oxidized protein was present and not detected, the theoretical maximum percentage oxidation was estimated by the ratio of the area of the unmodified peptide to the baseline level of the reversibly oxidized transition. This determines the maximum threshold level of oxidized peptide that could be present but not detected and is represented by a > sign.
To generate and optimize MRM transitions to the irreversible oxidation of all the cysteines in p53 and PTP1B for Fig. 5, we treated recombinant p53 and PTP1B with H2O2 and collected MS/MS spectra for peptides containing irreversibly oxidized cysteines. These modified peptides generally elute slightly earlier than the NEM-alkylated forms as seen for PTP1B Cys215 (Fig. 4b), and their MS/MS spectra typically have similar relative fragment ion abundance ratios with an enhancement of the y ion C-terminal to the cysteine (Fig. 4a). To obtain a comprehensive set of MRM transitions to irreversibly oxidized cysteine peptides that were not detected, we constructed hypothetical MRM transitions based on predicted abundant fragment ions from the unmodified peptide MS/MS spectra. The full transitions list is detailed in supplemental Table 1. However, other than the irreversible oxidation of Cys215, we did not detect intracellular overoxidation of any additional cysteines in the samples analyzed, including those oxidized by 10 mm H2O2.
We demonstrate that endogenous oxidation from Adriamycin-induced DNA damage is sufficient to increase both sulfinic and sulfonic acid oxidation of PTP1B Cys215 by 2.0- and 1.8-fold, respectively. We also show that nitrosylation (generated by S-nitroso-N-acetyl-penicillamine) abrogates the irreversible oxidation, supporting previous results (29). However, in contrast to the previous study, which examined pretreatment of PTP1B Cys215 with NO before H2O2 treatment, we find that a 1-h pulse of nitrosylation may abrogate irreversible oxidation of PTP1B Cys215 generated during a long period of Adriamycin-induced DNA damage.
DISCUSSION
Quantitative oxidation analysis of specific cysteines in targeted, moderate- to low-abundance proteins in cells poses a significant analytical challenge. Unbiased proteomics-based approaches such as ICAT have identified and quantified numerous oxidized cysteines (18–20, 28, 37), but the limited dynamic range, sensitivity, and difficulty in scaling down the protocol make it suitable for targeted analysis of only the most abundant proteins. As such, all studies of which we are aware that apply ICAT analysis to distinct cellular proteins have relied on either recombinant proteins (18, 19, 28–30) or protein overexpression systems (21). Both approaches require labor-intensive preparations and may not recapitulate the status of the protein in cells under physiological conditions. The capability to measure proteins at endogenous levels eliminates a significant barrier to the characterization of novel or putative redox-sensitive proteins. The increased sensitivity of MRM analysis also permits comprehensive evaluation of as many cysteines as possible within a targeted protein, which depends on the reproducible detection of peptides that do not ionize with high efficiency.
Comprehensive quantitation of sulfinic acid, sulfonic acid, and reversible oxidation of 8 cysteines in endogenous PTP1B (Fig. 5) allowed us to compare the effects of various conditions that create oxidative stress, such as DNA damage and exposure to paraquat, rotenone, TNFα and others. Physiologically relevant responses to these manipulations can be assessed only by examining the effect on a protein of interest within living cells. Most treatments caused little change, other than irreversible oxidation of Cys215. However, distinct pro-oxidants affected the relative levels of Cys215 sulfinic versus sulfonic acid differentially and may play an important role in PTP1B regulation (Fig. 5). A recent study of PTP1B found that pretreatment with an NO donor dramatically decreased irreversible oxidation of Cys215 by H2O2, providing a key insight into the chemical balance between reversible and irreversible cysteine oxidation (29). Using OxMRM, we show that this holds true for intracellular DNA damage-induced irreversible Cys215 oxidation. However, our data suggest that subsequent NO treatment may be sufficient to abrogate irreversible oxidation levels as well though further investigation is warranted (Fig. 5).
More than 15 years ago, p53 was suggested to be a redox-sensitive protein (38). Subsequently, oxidant-dependent changes in sequence-specific DNA binding (39, 40) and protein-protein interactions (41) were reported. Studies examining the nature of the oxidative modifications however, were confined to recombinant protein tested in vitro (40, 42). Little is known about the oxidation of intracellular p53, including which cysteines are affected and the conditions under which they are oxidized. This gap in our knowledge is due in large measure to the fact that endogenous levels of p53 are below the detection limits of current unbiased quantitative mass spectrometry-based assays. The use of site-directed mutagenesis to examine redox regulation of p53 is confounded by the fact that most cysteines in p53 are required for maintaining the essential folded structure of the protein (43), imposing a serious limitation to interpreting these studies. We demonstrate the sensitivity of the OxMRM approach by reliably detecting 7 of the 10 cysteines of p53 from both primary fibroblasts (supplemental Fig. 6) and an epithelial cancer cell line (Fig. 3) at endogenous levels. Furthermore, OxMRM quantified the oxidation of 8 of 10 cysteines in PTP1B in the cancer cell line, whereas a recent study using the ICAT reagent to quantify recombinant PTP1B reported oxidation of only three cysteines (29). In our study, the low variance between biological replicates in the diamide and H2O2 data sets (Figs. 3 and 4) demonstrates the reproducibility achievable throughout the entire OxMRM procedure. However, although within-batch oxidation is closely correlated, variation in oxidation levels between batches can occur (see Fig. 5 controls) and is probably due to variability in cell density or passage number.
Although oxidant treatment of isolated p53 can lead to dramatic reversible (supplemental Fig. 8) and irreversible oxidation, only diamide leads to a robust increase in oxidation of the measurable cysteines in endogenous p53 (Fig. 5). It is noteworthy that diamide oxidizes via a nonradical mechanism. Pyrrolidine dithiocarbamate was reported to oxidize a small fraction of p53 (44), which we did not observe. Moreover, no irreversibly oxidized endogenous p53 cysteines were detected even after 10 mm H2O2 treatment. These observations suggest that rapid protein synthesis and degradation, intracellular compartmentalization (45), interaction with other proteins, and/or antioxidant activities may work in concert to protect p53 and minimize specific types of oxidation. Diamide oxidation of endogenous p53 was highly selective for Cys182 (Fig. 3), but its effects on recombinant p53 were relatively nonspecific (supplemental Fig. 8). This finding emphasizes the importance of intracellular factors in regulating oxidation of endogenous proteins, an important consideration for future studies into the redox sensitivity of any isolated protein. Our finding that intracellular p53 is insensitive to a wide variety of oxidant conditions in cells contrasts the conclusion that p53 is redox-sensitive, which was based on evidence from the use of isolated protein under in vitro conditions. The increased PTP1B oxidation observed under identical cell conditions confirm that the treatments did increase intracellular pro-oxidant levels (Fig. 5); in addition, the robust increase in diamide oxidation supports the conclusion that reversible intracellular oxidation is preserved during sample preparation. Moreover, to verify that even potentially labile S-nitrosylation oxidation (46) was stable throughout our procedure, samples were spiked with nitrosylated recombinant p53 at different points during the protocol, from the initial TCA lysis through the incubation with d0 NEM. Because no decrease in the spiked levels of p53-S-nitrosylation was seen throughout the procedure (supplemental Fig. 9), we are confident that OxMRM measurements accurately reflect the redox state of intracellular p53. However, we cannot rule out the possibility that different oxidants or exposure intervals might lead to different degrees of intracellular p53 oxidization. Because we assessed total intracellular p53, it is also possible that p53 restricted to a subcellular compartment is much more susceptible to oxidation. A very small proportion of total intracellular p53, at maximum 2% (47), is thought to localize in or around mitochondria. Even if this mitochondrial p53 were extensively oxidized in response to our pro-oxidant treatments, this small change to the total pool of intracellular p53 would probably not be detectable by the current OxMRM procedure.
It is important to note that although we found OxMRM to provide quantitative data on the redox status of most cysteine residues in p53 and PTP1B, a small number of cysteines (i.e. 3 and 2 of 10 cysteines, respectively) were not covered in our analysis. As discussed earlier, the lack of obtaining a complete OxMRM coverage map for these two proteins was due largely to the lack of appropriate trypsin cleavage sites surrounding these five residues. This limitation is likely to be general because this method does require that the cysteine-containing peptides chosen as targeted analytes for OxMRM be amenable to HPLC separation and MS detection, conditions that are influenced by their size, charge state, chromatographic properties, and ionization efficiencies. Although we used trypsin exclusively in our studies here, other proteases with well defined cleavage specificities can also be used (e.g. Glu-C and Asp-N), either in combination or alone, to obtain the desired coverage, requiring some level of optimization for each protein to be analyzed. This, however, is a problem common to all peptide-based proteomic strategies that attempt a comprehensive coverage of posttranslational modifications.
OxMRM addresses several unmet needs in the analysis of cysteine oxidation and is applicable to any cellular or in vivo model that is compatible with acidic conditions. First, OxMRM offers robust quantitation and replicate validation for large unbiased proteomic screens designed to uncover redox-sensitive cysteines in susceptible proteins. It is noteworthy that development of an antibody to NEM could allow the OxMRM approach to be used as a discovery tool as well, with the benefits of improved dynamic range and quantitation at the MS/MS level as a result of MRM analysis. Second, this new method yields in-depth analysis and coverage of putative redox-sensitive proteins that are undetectable by less sensitive methods and circumvents the need for antibodies or site-directed mutants. Third, OxMRM provides a highly sensitive and robust assay to explore endogenous regulation of established redox-regulated cysteines. The technology and reagents needed to perform OxMRM are readily available, triple quadrupole mass spectrometers are widely used and the only required biochemical reagent is an antibody or epitope-tagged protein for affinity purification. OxMRM can be applied to essentially any protein, with the specific set of cysteines that can be quantified dependent to some degree on the primary sequence and proteolytic digestion conditions employed. In-depth oxidation analysis of targeted cellular regulators of oncogenesis, apoptosis, or signaling networks under a variety of oxidant conditions, including reversible and irreversible oxidation, will greatly aid our understanding of oxidation in health and disease.
Supplementary Material
* This article contains supplemental Tables 1 and 2 and supplemental Figures 1–8.
1 The abbreviations used are:
- TCEP
- tris(2-carboxyethyl)phosphine
- NEM
- N-ethylmaleimide
- ICAT
- isotope coded affinity tag
- MRM
- multiple reaction monitoring
- PTP1B
- protein tyrosine phosphatase-1B.
REFERENCES
- 1.Janssen-Heininger Y. M., Mossman B. T., Heintz N. H., Forman H. J., Kalyanaraman B., Finkel T., Stamler J. S., Rhee S. G., van der Vliet A. (2008) Redox-based regulation of signal transduction: principles, pitfalls, and promises. Free Radic. Biol. Med. 45, 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rhee S. G. (2006) Cell signaling. H2O2, a necessary evil for cell signaling. Science 312, 1882–1883 [DOI] [PubMed] [Google Scholar]
- 3.Tonks N. K. (2005) Redox redux: revisiting PTPs and the control of cell signaling. Cell 121, 667–670 [DOI] [PubMed] [Google Scholar]
- 4.Hess D. T., Matsumoto A., Kim S. O., Marshall H. E., Stamler J. S. (2005) Protein S-nitrosylation: purview and parameters. Nat. Rev. Mol. Cell Biol. 6, 150–166 [DOI] [PubMed] [Google Scholar]
- 5.Winterbourn C. C. (2008) Reconciling the chemistry and biology of reactive oxygen species. Nat. Chem. Biol. 4, 278–286 [DOI] [PubMed] [Google Scholar]
- 6.Mitchell D. A., Marletta M. A. (2005) Thioredoxin catalyzes the S-nitrosation of the caspase-3 active site cysteine. Nat. Chem. Biol. 1, 154–158 [DOI] [PubMed] [Google Scholar]
- 7.Kallis G. B., Holmgren A. (1980) Differential reactivity of the functional sulfhydryl groups of cysteine-32 and cysteine-35 present in the reduced form of thioredoxin from Escherichia coli. J. Biol. Chem. 255, 10261–10265 [PubMed] [Google Scholar]
- 8.Riemer J., Bulleid N., Herrmann J. M. (2009) Disulfide formation in the ER and mitochondria: two solutions to a common process. Science 324, 1284–1287 [DOI] [PubMed] [Google Scholar]
- 9.Sevier C. S., Qu H., Heldman N., Gross E., Fass D., Kaiser C. A. (2007) Modulation of cellular disulfide-bond formation and the ER redox environment by feedback regulation of Ero1. Cell 129, 333–344 [DOI] [PubMed] [Google Scholar]
- 10.Uehara T., Nakamura T., Yao D., Shi Z. Q., Gu Z., Ma Y., Masliah E., Nomura Y., Lipton S. A. (2006) S-nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature 441, 513–517 [DOI] [PubMed] [Google Scholar]
- 11.Giles G. I. (2006) The redox regulation of thiol dependent signaling pathways in cancer. Curr. Pharm. Des. 12, 4427–4443 [DOI] [PubMed] [Google Scholar]
- 12.Gu Z., Kaul M., Yan B., Kridel S. J., Cui J., Strongin A., Smith J. W., Liddington R. C., Lipton S. A. (2002) S-nitrosylation of matrix metalloproteinases: signaling pathway to neuronal cell death. Science 297, 1186–1190 [DOI] [PubMed] [Google Scholar]
- 13.Ago T., Liu T., Zhai P., Chen W., Li H., Molkentin J. D., Vatner S. F., Sadoshima J. (2008) A redox-dependent pathway for regulating class II HDACs and cardiac hypertrophy. Cell 133, 978–993 [DOI] [PubMed] [Google Scholar]
- 14.Hurd T. R., Prime T. A., Harbour M. E., Lilley K. S., Murphy M. P. (2007) Detection of reactive oxygen species-sensitive thiol proteins by redox difference gel electrophoresis: implications for mitochondrial redox signaling. J. Biol. Chem. 282, 22040–22051 [DOI] [PubMed] [Google Scholar]
- 15.Jaffrey S. R., Snyder S. H. (2001) The biotin switch method for the detection of S-nitrosylated proteins. Sci. STKE 2001, pl1. [DOI] [PubMed] [Google Scholar]
- 16.Meza J. E., Scott G. K., Benz C. C., Baldwin M. A. (2003) Essential cysteine-alkylation strategies to monitor structurally altered estrogen receptor as found in oxidant-stressed breast cancers. Anal. Biochem. 320, 21–31 [DOI] [PubMed] [Google Scholar]
- 17.Schilling B., Yoo C. B., Collins C. J., Gibson B. W. (2004) Determining cysteine oxidation status using differential alkylation. Int. J. Mass Spectrom. 236, 117–127 [Google Scholar]
- 18.Leichert L. I., Gehrke F., Gudiseva H. V., Blackwell T., Ilbert M., Walker A. K., Strahler J. R., Andrews P. C., Jakob U. (2008) Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc. Natl. Acad. Sci. U.S.A. 105, 8197–8202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sethuraman M., Clavreul N., Huang H., McComb M. E., Costello C. E., Cohen R. A. (2007) Quantification of oxidative posttranslational modifications of cysteine thiols of p21ras associated with redox modulation of activity using isotope-coded affinity tags and mass spectrometry. Free Radic. Biol. Med. 42, 823–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sethuraman M., McComb M. E., Heibeck T., Costello C. E., Cohen R. A. (2004) Isotope-coded affinity tag approach to identify and quantify oxidant-sensitive protein thiols. Mol. Cell. Proteomics 3, 273–278 [DOI] [PubMed] [Google Scholar]
- 21.Yi L., Jenkins P. M., Leichert L. I., Jakob U., Martens J. R., Ragsdale S. W. (2009) Heme regulatory motifs in heme oxygenase-2 form a thiol/disulfide redox switch that responds to the cellular redox state. J. Biol. Chem. 284, 20556–20561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Montfort R. L., Congreve M., Tisi D., Carr R., Jhoti H. (2003) Oxidation state of the active-site cysteine in protein tyrosine phosphatase 1B. Nature 423, 773–777 [DOI] [PubMed] [Google Scholar]
- 23.Salmeen A., Andersen J. N., Myers M. P., Meng T. C., Hinks J. A., Tonks N. K., Barford D. (2003) Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature 423, 769–773 [DOI] [PubMed] [Google Scholar]
- 24.Leichert L. I., Jakob U. (2006) Global methods to monitor the thiol-disulfide state of proteins in vivo. Antioxid. Redox Signal. 8, 763–772 [DOI] [PubMed] [Google Scholar]
- 25.Zander T., Phadke N. D., Bardwell J. C. (1998) Disulfide bond catalysts in Escherichia coli. Methods Enzymol. 290, 59–74 [DOI] [PubMed] [Google Scholar]
- 26.Picotti P., Bodenmiller B., Mueller L. N., Domon B., Aebersold R. (2009) Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 138, 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riley T., Sontag E., Chen P., Levine A. (2008) Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 9, 402–412 [DOI] [PubMed] [Google Scholar]
- 28.Winter J., Ilbert M., Graf P. C., Ozcelik D., Jakob U. (2008) Bleach activates a redox-regulated chaperone by oxidative protein unfolding. Cell 135, 691–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y. Y., Chu H. M., Pan K. T., Teng C. H., Wang D. L., Wang A. H., Khoo K. H., Meng T. C. (2008) Cysteine S-nitrosylation protects protein-tyrosine phosphatase 1B against oxidation-induced permanent inactivation. J. Biol. Chem. 283, 35265–35272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozarova A., Sliskovic I., Mutus B., Simon E. S., Andrews P. C., Vacratsis P. O. (2007) Identification of redox sensitive thiols of protein disulfide isomerase using isotope coded affinity technology and mass spectrometry. J. Am. Soc. Mass Spectrom. 18, 260–269 [DOI] [PubMed] [Google Scholar]
- 31.Kuzyk M. A., Smith D., Yang J., Cross T. J., Jackson A. M., Hardie D. B., Anderson N. L., Borchers C. H. (2009) Multiple reaction monitoring-based, multiplexed, absolute quantitation of 45 proteins in human plasma. Mol. Cell. Proteomics 8, 1860–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderson L., Hunter C. L. (2006) Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol. Cell. Proteomics 5, 573–588 [DOI] [PubMed] [Google Scholar]
- 33.Cho Y., Gorina S., Jeffrey P. D., Pavletich N. P. (1994) Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science 265, 346–355 [DOI] [PubMed] [Google Scholar]
- 34.Sun X. Z., Vinci C., Makmura L., Han S., Tran D., Nguyen J., Hamann M., Grazziani S., Sheppard S., Gutova M., Zhou F., Thomas J., Momand J. (2003) Formation of disulfide bond in p53 correlates with inhibition of DNA binding and tetramerization. Antioxid. Redox Signal. 5, 655–665 [DOI] [PubMed] [Google Scholar]
- 35.Lou Y. W., Chen Y. Y., Hsu S. F., Chen R. K., Lee C. L., Khoo K. H., Tonks N. K., Meng T. C. (2008) Redox regulation of the protein tyrosine phosphatase PTP1B in cancer cells. FEBS J. 275, 69–88 [DOI] [PubMed] [Google Scholar]
- 36.Men L., Wang Y. (2005) Further studies on the fragmentation of protonated ions of peptides containing aspartic acid, glutamic acid, cysteine sulfinic acid, and cysteine sulfonic acid. Rapid Commun. Mass Spectrom. 19, 23–30 [DOI] [PubMed] [Google Scholar]
- 37.Sethuraman M., McComb M. E., Huang H., Huang S., Heibeck T., Costello C. E., Cohen R. A. (2004) Isotope-coded affinity tag (ICAT) approach to redox proteomics: identification and quantitation of oxidant-sensitive cysteine thiols in complex protein mixtures. J. Proteome Res. 3, 1228–1233 [DOI] [PubMed] [Google Scholar]
- 38.Hainaut P., Milner J. (1993) Redox modulation of p53 conformation and sequence-specific DNA binding in vitro. Cancer Res. 53, 4469–4473 [PubMed] [Google Scholar]
- 39.Sablina A. A., Budanov A. V., Ilyinskaya G. V., Agapova L. S., Kravchenko J. E., Chumakov P. M. (2005) The antioxidant function of the p53 tumor suppressor. Nat. Med. 11, 1306–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Augustyn K. E., Merino E. J., Barton J. K. (2007) A role for DNA-mediated charge transport in regulating p53: Oxidation of the DNA-bound protein from a distance. Proc. Natl. Acad. Sci. U.S.A. 104, 18907–18912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheffner M., Takahashi T., Huibregtse J. M., Minna J. D., Howley P. M. (1992) Interaction of the human papillomavirus type 16 E6 oncoprotein with wild-type and mutant human p53 proteins. J. Virol. 66, 5100–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Velu C. S., Niture S. K., Doneanu C. E., Pattabiraman N., Srivenugopal K. S. (2007) Human p53 is inhibited by glutathionylation of cysteines present in the proximal DNA-binding domain during oxidative stress. Biochemistry 46, 7765–7780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rainwater R., Parks D., Anderson M. E., Tegtmeyer P., Mann K. (1995) Role of cysteine residues in regulation of p53 function. Mol. Cell. Biol. 15, 3892–3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu H. H., Thomas J. A., Momand J. (2000) p53 protein oxidation in cultured cells in response to pyrrolidine dithiocarbamate: a novel method for relating the amount of p53 oxidation in vivo to the regulation of p53-responsive genes. Biochem. J. 351, 87–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hansen J. M., Go Y. M., Jones D. P. (2006) Nuclear and mitochondrial compartmentation of oxidative stress and redox signaling. Annu. Rev. Pharmacol. Toxicol. 46, 215–234 [DOI] [PubMed] [Google Scholar]
- 46.Mannick J. B., Schonhoff C. M. (2008) Measurement of protein S-nitrosylation during cell signaling. Methods Enzymol. 440, 231–242 [DOI] [PubMed] [Google Scholar]
- 47.Marchenko N. D., Zaika A., Moll U. M. (2000) Death signal-induced localization of p53 protein to mitochondria. A potential role in apoptotic signaling. J. Biol. Chem. 275, 16202–16212 [DOI] [PubMed] [Google Scholar]
- 48.von Montfort C., Sharov V. S., Metzger S., Schoneich C., Sies H., Klotz L. O. (2006) Singlet oxygen inactivates protein tyrosine phosphatase-1B by oxidation of the active site cysteine. Biol. Chem. 387, 1399–1404 [DOI] [PubMed] [Google Scholar]
- 49.Barrett W. C., DeGnore J. P., König S., Fales H. M., Keng Y. F., Zhang Z. Y., Yim M. B., Chock P. B. (1999) Regulation of PTP1B via glutathionylation of the active site cysteine 215. Biochemistry 38, 6699–6705 [DOI] [PubMed] [Google Scholar]
- 50.Chen Y. Y., Huang Y. F., Khoo K. H., Meng T. C. (2007) Mass spectrometry-based analyses for identifying and characterizing S-nitrosylation of protein tyrosine phosphatases. Methods 42, 243–249 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.