Abstract
The identification of surface proteins on the plasma membrane of pathogens is of fundamental importance in understanding host-pathogen interactions. Surface proteins of the extracellular parasite Trichomonas are implicated in the initial adherence to mucosal tissue and are likely to play a critical role in the long term survival of this pathogen in the urogenital tract. In this study, we used cell surface biotinylation and multidimensional protein identification technology to identify the surface proteome of six strains of Trichomonas vaginalis with differing adherence capacities to vaginal epithelial cells. A combined total of 411 proteins were identified, and of these, 11 were found to be more abundant in adherent strains relative to less adherent parasites. The mRNA levels of five differentially expressed proteins selected for quantitative RT-PCR analysis mirrored their observed protein levels, confirming their up-regulation in highly adherent strains. As proof of principle and to investigate a possible role in pathogenesis for differentially expressed proteins, gain of function experiments were performed using two novel proteins that were among the most highly expressed surface proteins in adherent strains. Overexpression of either of these proteins, TVAG_244130 or TVAG_166850, in a relatively non-adherent strain increased attachment of transfected parasites to vaginal epithelial cells ∼2.2-fold. These data support a role in adhesion for these abundant surface proteins. Our analyses demonstrate that comprehensive profiling of the cell surface proteome of different parasite strains is an effective approach to identify potential new adhesion factors as well as other surface molecules that may participate in establishing and maintaining infection by this extracellular pathogen.
The flagellated protozoan parasite Trichomonas vaginalis is the etiologic agent of trichomoniasis, the most common non-viral sexually transmitted infection worldwide with an estimated 174 million new cases annually (1). Although asymptomatic infection by T. vaginalis is common, multiple symptoms and pathologies can arise in both men and women, including vaginitis, urethritis, prostatitis, low birth weight infants and preterm delivery, premature rupture of membranes, and infertility (2–5). T. vaginalis has also emerged as an important cofactor in amplifying human immunodeficiency virus spread (6) as individuals infected with T. vaginalis have a significantly increased incidence of human immunodeficiency virus transmission (7, 8). T. vaginalis infection likewise increases the risk of cervical and aggressive prostate cancers (9–11).
Despite the serious consequences that can arise from trichomoniasis, the underlying biochemical processes that lead to T. vaginalis pathogenesis are not well defined. Because T. vaginalis is an obligate extracellular pathogen, adherence to epithelial cells is critical for parasite survival within the human host (12). Several in vitro studies indicate that adhesion of the parasite to target mucosal epithelial cells is essential for the maintenance of infection and for cytopathogenicity (13, 14). T. vaginalis adherence to host cells is mediated, in part, by a lipophosphoglycan (LPG)1 that coats the surface of the parasite, and altering the sugar content of this LPG reduces both adherence and cytotoxicity (15). Moreover, the mammalian protein galectin-1 binds to T. vaginalis in a carbohydrate-dependent manner via a direct interaction with parasite LPG (16). Knockdown of galectin-1 in mammalian cells, however, reduces parasite binding only by ∼17% (16). Although galectin-1-mediated interactions between T. vaginalis LPG and host cell glycoconjugates may be central in establishing infection, it is clear that parasite adhesion factors in addition to LPG are likely to be involved in host-parasite interaction. Surface proteins are likely to play important roles in the initial adherence to mucosal tissue as well as the long term survival of the pathogen on mucosal surfaces.
The outcome of infection with T. vaginalis is highly variable. Possible explanations for this phenomenon include host immunity, host nutritional status, and the vaginal microbiota. Additionally, genetic differences between T. vaginalis isolates leading to differences in adherence and cytotoxicity capacities are likely to result in differences in disease progression. Recently, geographically diverse T. vaginalis strains that are significantly more cytotoxic to host cells than laboratory-adapted strains have become available (17, 18), paving the way toward comparative studies aimed at identifying proteins that correlate with virulent phenotypes.
Despite the importance of T. vaginalis surface proteins as a critical interface for pathogen-host interactions, there has been no systematic investigation of the surface proteins of this parasite. The T. vaginalis genome is large and encodes a massive proteome with a considerable and diverse repertoire of candidate surface proteins (19). For example, sequence analysis programs that predict transmembrane protein topology identified over 5100 T. vaginalis proteins with one or more transmembrane domains (20). Furthermore, over 300 annotated proteins with predicted transmembrane domains also contain protein motifs common to surface proteins from other pathogens known to contribute to mucosal colonization and other pathogenic processes (20). The vast number and diversity of possible surface proteins necessitates a multitiered approach using complementary genomics and proteomics analyses to identify candidates for focused functional studies.
Biotinylation of proteins at the cell surface with an impermeable reagent followed by specific purification of these proteins using streptavidin has successfully been used for the enrichment and identification of surface proteins (21–24). The high avidity binding of biotin to streptavidin greatly enhances membrane protein purification, a challenging feat because of the low abundance of membrane proteins in total cellular extracts. Here, we used this approach to profile the surface plasma membrane proteome of T. vaginalis and to identify proteins that are differentially expressed in adherent relative to less adherent strains of the parasite. To the best of our knowledge, this is the first study to systematically identify and characterize proteins at the surface of Trichomonas parasites. Defining the parasite cell surface proteome is a critical step toward understanding the relative abundance of surface proteins in strains with varying virulence properties. This information will be critical for defining the role surface proteins play in mediating contact between the parasite and host cells as well as the resulting intracellular and extracellular signals that contribute to establishing and maintaining infection. Additionally, conserved surface molecules unique to T. vaginalis that might serve as specific vaccine candidates can be revealed using this approach. The prevalence of trichomoniasis among women of reproductive age (25) and its correlation with AIDS transmission and cervical and prostate cancers (6, 8–11) provide strong arguments for the need to develop vaccines against this human pathogen.
EXPERIMENTAL PROCEDURES
Parasites, Cell cultures, and Media
T. vaginalis strains B7RC2 (PA strain, ATCC 50 167), T1 (J. H. Tai, Institute of Biomedical Sciences, Taipei, Taiwan), B7268 (18), G3 (ATCC PRA-98; Kent, UK), SD7, and SD10 were cultured in trypticase, yeast extract, maltose medium supplemented with 10% horse serum, penicillin and streptomycin (Invitrogen), and iron (26). Parasites were grown at 37 °C and passaged daily. The human ectocervical epithelial cell lines Ect1 E6/E7 (ectocervical, ATCC CRL-2614), also known as vaginal epithelial cells (VECs), were grown as described previously (27) in defined keratinocyte serum-free medium complemented with the provided recombinant protein supplements, penicillin, and streptomycin (Invitrogen) and cultured at 37 °C in 5% CO2.
Biotinylation of Cell Surface Proteins
Parasites (5 × 107 cells) were washed twice with prewarmed PBS-S (PBS, 5% sucrose) and then incubated with 0.5 mg/ml EZ-Link sulfo-NHS-SS-biotin (Pierce) in PBS-S for 45 min on ice. Excess biotin was quenched with 50 mm Tris-HCl, pH 7.5 to terminate the reaction, and cells were washed three times with PBS-S.
Assessing Permeabilization after Biotinylation
The degree of permeabilization after biotinylation was assessed using an immunofluorescence assay (IFA) with anti-biotin antibody (Jackson Immuno Research Laboratories). Cells were washed in PBS-S and fixed in 4% formaldehyde for 20 min. After three washes, cells were permeabilized with 0.2% Triton X-100 in PBS for 15 min, blocked with 3% BSA in PBS for 30 min, incubated with anti-biotin antibody diluted 1:1000 in 3% BSA in PBS, washed, and then incubated with a 1:5000 dilution of Alexa Fluor®-conjugated secondary antibody (Molecular Probes). The coverslips were mounted onto microscope slips using Mowiol (Calbiochem). Stained parasites were examined using an Axioskop 2 epifluorescence microscope (Zeiss), and images were recorded with an AxioCam camera and processed with the AxioVision 3.2 program (Zeiss).
Preparation of Membrane Fractions and Purification of Biotinylated Proteins
Biotinylated cells were resuspended in PBS and subjected to freezing-thawing. The homogenate was clarified by centrifugation (13,000 × g for 30 min at 4 °C) to yield a membrane-enriched pellet. The membrane-enriched pellet was solubilized in 2 ml of lysis buffer (0.1% Nonidet P-40, 0.5% deoxycholate, 2% SDS, 50 mm Tris, pH 8, 5 mm EDTA, 150 mm NaCl). Streptavidin-Sepharose slurry (150 μl/mg of total proteins; Amersham Biosciences) was equilibrated by two washes in lysis buffer, and binding of biotinylated membrane proteins was allowed overnight in a rotating mixer at 4 °C. The resin was then washed once with each of the following buffers: A (8 m urea, 200 mm NaCl, 2% SDS, 100 mm Tris, pH 8), B (8 m urea, 1.2 m NaCl, 0.2% SDS, 100 mm Tris, pH 8, 10% ethanol, 10% isopropanol), and C (8 m urea, 200 mm NaCl, 0.2% SDS, 100 mm Tris, pH 8, 10% ethanol, 10% isopropanol). The resin was then washed twice with buffer D (8 m urea, 100 mm Tris, pH 8). Binding of biotinylated proteins to the resin and washing efficiencies were checked by SDS-PAGE and blot analysis using an anti-biotin antibody (Jackson Immuno Research Laboratories).
On-bead Digestion and Shotgun Proteomics Analysis of Biotinylated Proteins
After the final wash, the resin was resuspended in a minimal volume of digestion buffer (100 mm Tris-HCl, pH 8, 8 m urea). Biotinylated proteins were reduced and alkylated by the sequential addition of 5 mm tris(2-carboxyethyl)phosphine and 10 mm iodoacetamide as described previously (28). The samples were then digested by Lys-C (Princeton Separations) and trypsin proteases (Promega) (28). First, Lys-C protease (∼1:50 (w/w) ratio of enzyme:substrate) was added to each sample and incubated for 4 h at 37 °C with gentle shaking. The digests were then diluted to 2 m urea by the addition of digestion buffer lacking urea, and trypsin was added to a final enzyme:substrate ratio of 1:20 (w/w) and incubated for 8 h at 37 °C with gentle shaking. Digests were stopped by the addition of formic acid to a final concentration of 5%. Supernatants were carefully removed from the resin and analyzed further by proteomics mass spectrometry.
Digested samples were then analyzed using a shotgun proteomics platform comprising on-line two-dimensional liquid chromatography separation coupled to tandem mass spectrometric analysis of the fractionated peptide mixture as described previously (29, 30). Briefly, digested samples were loaded onto a triphasic fused silica capillary column with a 5-μm electrospray tip and packed sequentially in house with 3 cm of Luna C18 resin (Phenomenex), 3 cm of Luna strong cation exchange resin, and 9 cm of Luna C18 resin (Phenomenex). The column was then placed in line with an LTQ-Orbitrap mass spectrometer (Thermo Fisher), and peptides were fractionated using a seven-step separation strategy in which alternating pulses of increasing salt concentration and gradients of increasing acetonitrile were used to separate peptides on adjacent strong cation exchange and C18 reversed phased resins. Peptides were eluted directly into the LTQ-Orbitrap mass spectrometer where MS/MS spectra were collected. The data-dependent spectral acquisition strategy consisted of a repeating cycle of one full MS spectrum using the Orbitrap mass analyzer (resolution, 60,000) followed by MS/MS in the linear ion trap of the eight most intense precursor ions from the full MS scan.
Raw binary files containing MS/MS spectra were converted to MS2 files using RawExtractor v.1.8 (31). MS2 files were then analyzed using the SEQUEST algorithm (v.27) (32) and searching against a fasta protein database consisting of all predicted open reading frames downloaded from TrichDB on April 14, 2005 (33) concatenated to a decoy database in which the amino acid sequence of each entry was reversed. The following search parameters were used: 1) precursor ion tolerance was 20 ppm, 2) fragment ion tolerance was 0.4 Da, 3) no modifications were allowed, 4) peptides must be fully tryptic, and 5) no consideration was made for missed cleavages. False positive rates for peptide identifications were estimated using a decoy database approach and then filtered using the DTASelect algorithm (v.2.0.6) (34–36). XCorr and ΔCn cutoffs were identified dynamically using a linear discriminant analysis (36). Proteins identified by at least two fully tryptic unique peptides, each with a false positive rate of less than 5%, were considered to be present in the sample. Differential protein expression analysis was performed using the ACfold algorithm as implemented in the PatternLab software suite (37). Spectral counts were normalized using the Row Sigma method of PatternLab (37). Proteins were considered differentially expressed if they met the following criteria: 1) a -fold change cutoff of 2.5, 2) an estimated -fold change false discovery rate of less than 0.1, and 3) an ACtest p value of less than 0.01.
Adherence to Ectocervical Epithelial Cells
A modified version of an in vitro assay to qualify the binding of T. vaginalis to host cell monolayers (15) was performed. Briefly, ectocervical epithelial cells were seeded on 12-mm coverslips in 24-well plates at 3 × 105 cells/well in culture medium and grown to confluence at 37 °C in 5% CO2 for 2 days. Cell monolayers were washed before the addition of parasites. T. vaginalis was labeled with 10 mm CellTracker Blue CMAC (7-amino-4-chloromethylcoumarin) (Invitrogen), and ∼105 labeled parasites in 0.5 ml of keratinocyte medium (Invitrogen) were added (1:3 parasite:host cell ratio) in triplicate. Plates were incubated at 37 °C in 5% CO2 for 30 min. Coverslips were subsequently washed in PBS, fixed with 4% paraformaldehyde, and mounted on slides with Mowiol (Calbiochem). Ten 20× magnification fields were analyzed per coverslip with three coverslips per treatment. Fluorescent parasites adhered to host cells were counted using Scion Image for Windows, v.Beta 4.0.2 (Scion Corp.). All experiments were repeated at least three independent times.
Immunolocalization Experiments
Parasites were incubated at 37 °C to let them attach to glass coverslips. All further incubations were carried out at room temperature. Cells were washed in PBS-S and fixed in 4% formaldehyde for 20 min. After three washes, cells were permeabilized with 0.2% Triton X-100 in PBS for 15 min, blocked with 3% BSA in PBS (PBS-BSA) for 30 min, incubated with primary antibody diluted in PBS-BSA, washed, and then incubated with a 1:5000 dilution of Alexa Fluor-conjugated secondary antibody (Molecular Probes). The coverslips were mounted onto microscope slips using ProLong Gold antifade reagent with 4′,6′-diamidino-2-phenylindole (Invitrogen). Stained parasites were examined using an Axioskop 2 epifluorescence microscope (Zeiss), and images were recorded with an AxioCam camera and processed with the AxioVision 3.2 program (Zeiss).
Quantitative PCR (qPCR)
Total RNA was extracted from ∼4 × 106 T. vaginalis using TRIzol (Invitrogen) following the manufacturer's instructions. RNA was treated with amplification grade DNase I (Invitrogen) and reverse transcribed using SuperScript III reverse transcriptase and oligo(dT) primers (Invitrogen). Real time PCRs were performed using Brilliant SYBR Green qPCR Master Mix (Stratagene), a 150–450 nm concentration of each primer, and 200–500 ng of cDNA in a 20-μl reaction volume using an Eppendorf Mastercycler and realplex v.1.5 (Eppendorf). Parallel reactions performed without reverse transcriptase were included as negative controls. During the exponential phase of the qPCR, a threshold cycle (CT) and base line were set according to Eppendorf Mastercycler protocols. Data from different samples were interpolated on a line created by running standard curves for each primer set and then normalized against the tubulin housekeeping gene. Every sample and points of the standard curve were done in duplicates in three independent experiments. qPCR primer pair sequences were as follows: TUB-F, GTCTCGGCACACTCCTTCTC and TUB-R, AGACGTGGGAATGGAACAAG; TVAG_020780-F, TGCTGTTTCTGTTTCACCTTC and TVAG_020780-R, TCATCATGATATCCGCGTAA; TVAG_147050-F, TGTTTATCCAGAATGCGACA and TVAG_147050-R, CCTGCAATGGCATATGTTTT; TVAG_244130-F, AAGGCAATTACGCCATCATA and TVAG_244130-R, TGTTGGAACAGGGTGTTTCT; TVAG_166850-F, GAAGAACCCAGCTGTTTTGA and TVAG_166850-R, TTCAGGACCACCATTGAACT; and TVAG_110300-F, GGAATACCCATCCACAATGA and TVAG_110300-R, TGCGTAATCTTCGACTGTGA.
Plasmid Construction and Expression in T. vaginalis
To corroborate localization of five detected putative surface proteins, we expressed the corresponding proteins in T. vaginalis under the control of their own promoter. The respective genes were PCR-amplified using the following primer pairs: TVAG_258230F, GAGCTCCCTTTGGAACGATATTAACCA and TVAG_258230R, GGTACCAGCATTTAATGGTTCATCAG; TVAG_230400F, GAGCTCCTAAAACTCGGTAAACAACTCC and TVAG_230400R, GGTACCAACGGCATCAGCGCCATAAAT; TVAG_020780F, GAGCTCTAAAATAATAAATAAAAGTGA and TVAG_020780R, GGTACCGTCACCTCTGTTTGAATAAG; and TVAG_019180F, GAGCTCACCTTGCTAATTTCTTTCCAGT and TVAG_019180R, GGTACCAACGTATGTGATGCCTTCCTT. SacI and KpnI restriction sites were engineered into the 5′- and 3′-primers, respectively, and the PCR fragments were then cloned into the master-Neo-(HA)2 plasmid (38). To generate the N-terminal hemagglutinin (HA)-tagged version of TVAG_166850 and TVAG_244130-HA, each ORF was amplified using the following primers: TVAG_166850, 5′-AAACCGCGGATGTTACCACTATTTTACACA-3′ and 5′-AAAGGATCCAGCTGGGAAGATTCCTTCGAC-3′; and TVAG_244130, 5′-AAACCGCGGATGTTTTGCTCATTACTTATC-3′ and 5′-AAAGGATCCATCGACATCTTCGAATTCAT-3′. SacII and BamHI restriction sites were engineered into the 5′- and 3′-primers, respectively. The PCR fragments were then cloned into the Nt-(HA)-master-Neo plasmid to generate constructs to transfect T. vaginalis. Electroporation of T. vaginalis strain T1 was carried out as described previously (39) with 50 μg of circular plasmid DNA. Transfectants were selected with 100 μg/ml G418 (Sigma).
RESULTS
Preparation of T. vaginalis Surface Protein-enriched Fractions
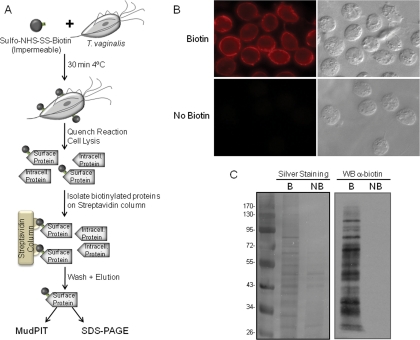
To identify surface proteins of T. vaginalis, we developed a method to label intact parasites with a non-permeable biotin reagent, sulfo-NHS-SS-biotin, which binds specifically to exposed primary amines (Fig. 1A). Labeled cells were assessed by IFAs using an anti-biotin antibody. The pattern of staining was confined to the surface with little visible penetration inside the cells (Fig. 1B, top panel). Cells that had been washed to remove unbound biotin were lysed, and membrane proteins were solubilized and loaded onto a streptavidin column. Following high salt and high urea washes to remove contaminating proteins, biotin-labeled proteins retained on the column were eluted with SDS-PAGE sample buffer. Bound and eluted proteins from biotinylated and control non-biotinylated samples were compared by SDS-PAGE and silver staining (Fig. 1C, left panel). Although very little nonspecific binding to the streptavidin column was observed, a few non-biotinylated proteins were shown to bind nonspecifically to the column. To verify that the proteins recovered from the beads were biotinylated, an aliquot was separated by SDS-PAGE, blotted onto a PVDF membrane, and probed with anti-biotin antibody (Fig. 1C, right panel). A range of proteins, varying in molecular mass from >100 to < 30 kDa were observed, demonstrating the ability to label many proteins using this approach (Fig. 1C, right panel). Non-biotinylated samples were included on the blot to further test for endogenous or nonspecific binding of streptavidin-conjugated proteins, and no signal was detected (Fig. 1B, right panel). Similar results were also obtained using IFA (Fig. 1B, bottom panel). These data demonstrate that the T. vaginalis biotin-labeled fractions are highly enriched with surface proteins.
Fig. 1.
Biotinylation of T. vaginalis surface proteins using sulfo-NHS-SS-biotin. A, scheme of the protocol for biotinylating and purifying surface proteins. B, immunofluorescence assay with anti-biotin antibody. Top panel, biotinylated parasites; bottom panel, non-biotinylated parasites. Data for only one strain (T1) are shown; however, all had comparable staining patterns. C, SDS-PAGE analysis of proteins purified by streptavidin. Biotin-labeled (B) and unlabeled proteins (NB) were recovered by affinity purification, separated by SDS-PAGE, and silver-stained (left panel) or detected by Western blot (WB) with anti-biotin antibody (right panel). The molecular weight marker is shown on the far left. Intracell., intracellular.
Shotgun Proteomics Analysis of Surface Protein-enriched Fractions
The identity of proteins in surface-enriched fractions obtained from six different parasites strains was determined using multidimensional protein identification technology (MudPIT). To assess the binding specificity of the streptavidin column, non-biotinylated cells were processed identically to the biotinylated samples except for the omission of biotin in the reactions and subsequently analyzed by MudPIT. Only 17 proteins were identified in non-biotinylated samples, and these are most likely abundant, “sticky” proteins that bind nonspecifically to the streptavidin beads. Compiling protein identifications across all six biotinylated samples, 411 total parasite proteins were identified (supplemental Table 1). In addition to three independent experiments comparing these six strains, multiple experiments were also done comparing two of the six strains, and with the exception of varying degrees of contamination, a core set of putative membrane proteins was reproducibly identified.
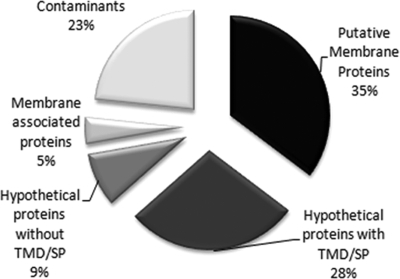
Of the 411 proteins identified, 35% are proteins previously predicted to be membrane proteins based on Trichomonas genome analyses (19) or proteins with domains found in membrane proteins of other organisms. Another group of proteins, which comprises 28% of the total, are hypothetical proteins that are unique to T. vaginalis and are predicted to have transmembrane domains and/or signal peptides (Fig. 2). Likewise, we identified a group of novel, hypothetical proteins (9% of the total) without a transmembrane domain and/or signal peptide. Analyses of known T. vaginalis membrane proteins indicate that many may not have conventional N-terminal signal peptides or transmembrane domains identifiable by bioinformatics analyses2; hence, it is possible that these are transmembrane proteins. Alternatively, they may be directly anchored to the plasma membrane. A small minority (5%; membrane-associated proteins) of identified proteins are cytoskeletal and most likely reflect a tight apposition of the plasma membrane to the underlying cytoskeleton. Additionally, 23% of the proteins were labeled as contaminants as many of these are known to reside in other subcellular compartments. These include abundant hydrogenosomal, ribosomal, and rab proteins that are common contaminants of subcellular fractions and immunoprecipitation experiments because of their abundance (40). The labeling of these proteins may result from leaky or dying cells. It is also possible that these proteins are labeled by virtue of a close association between a subset of the subcellular compartments in which they reside and the inner leaflet of the plasma membrane.
Fig. 2.
Proteins identified by MudPIT analysis. A total of 411 proteins were identified in the six examined strains. Putative membrane proteins (35%) have conserved domains with known or predicted membrane proteins in other organisms, 28% are hypothetical proteins with transmembrane domain (TMD) and/or signal peptide (SP), 9% represent hypothetical proteins without transmembrane domain or signal peptide, 23% represent suspected protein contaminants, and 5% were classified as membrane-associated proteins and are largely cytoskeleton proteins.
Predicted and identified integral membrane proteins were sorted into functional groups according to BLAST analysis and genome annotation (Table I). The largest group (116 of 271) of predicted coding sequences is hypothetical proteins, representing about 44% of all integral membrane proteins (supplemental Table 1). Among the predicted membrane proteins with identifiable domains that allow the assignment of a predicted function, ∼18% are proteins predicted to be involved in host-parasite interaction, ∼8% are transporters, ∼8% are cyclases, and ∼5% are peptidases; the remaining ∼60% include phosphatases and proteins involved in vesicle transport, ATPase activity, or hydrolase activity (Table I). Of the proteins previously predicted to be membrane-associated and possibly involved in host-parasite interaction by the genome analyses (19, 20), chlamydial polymorphic outer membrane proteins, GP63-like proteins, surface antigen BspA-like proteins, P270-related proteins, and immunodominant variable surface antigen-like proteins (41) are present in our membrane proteome. Interestingly, a number of novel proteins putatively related to host-parasite interaction, like the tetraspanin family protein and biofilm-associated protein-like proteins, are also present.
Table I. Putative membrane proteins sorted into functional groups according to BLAST analysis and genome annotation.
TMD, transmembrane domain; SP, signal peptide; SNARE, soluble N-ethylmaleimide-sensitive factor attachment protein receptor; v-SNARE, vesicle SNARE; PMCA, plasma membrane Ca2+-ATPase.
| No. identified here | No. genes in genome | |
|---|---|---|
| Total putative membrane proteins | 271 | |
| Hypothetical proteins with TMD/SP | 116 | |
| Host-cell interaction | ||
| GP63-like | ≥16 | 77 |
| Surface antigen BspA-like | 11 | 658 |
| Chlamydial polymorphic outer membrane protein | 5 | 27 |
| Tetraspanin family protein | 3 | 9 |
| P270-related protein | ≥9 | 25 |
| Immunodominant variable surface antigen-like | 2 | 15 |
| Transporters | ||
| ABC transporter family protein | 6 | 104 |
| Ctl transporter, putative | 5 | 9 |
| Major facilitator superfamily protein | 2 | 20 |
| Nucleoside transporter family protein | 2 | 10 |
| Auxin efflux carrier family protein | 1 | 31 |
| Transmembrane amino acid transporter protein | 1 | 32 |
| Na+-driven multidrug efflux pump | 1 | 60 |
| Sodium-sulfate symporter transmembrane region | 1 | 13 |
| Cyclases | ||
| Adenylate and guanylate cyclase catalytic domain | 20 | 167 |
| Peptidases | ||
| Subtilase family protein | 7 | 27 |
| Clan CA, family C2, calpain-like cysteine peptidase | 2 | 10 |
| Clan S, family S54, rhomboid-like serine peptidase | 1 | 9 |
| Nicastrin precursor, putative | 1 | 2 |
| Vesicle transport | ||
| Synaptobrevin family protein | 3 | 16 |
| Vesicle transport v-SNARE protein | 2 | 22 |
| Novel plant SNARE | 1 | 4 |
| Secretory carrier membrane protein | 1 | 6 |
| Phosphatases | ||
| Ser/Thr protein phosphatase family protein | 5 | 20 |
| Nucleoside phosphatase family protein | 1 | 8 |
| Adenosine diphosphatase | 1 | 6 |
| ATPase activity | ||
| Phospholipid-transporting ATPase | 1 | 34 |
| Calcium-translocating P-type ATPase, PMCA-type | ≥4 | 28 |
| V-type ATPase, C subunit family protein | 1 | 5 |
| V-type ATPase 116-kDa subunit family protein | 1 | 7 |
| Hydrolase activity | ||
| Class I nuclease-related | ≥1 | 16 |
| Glycosyl hydrolase family 14 protein | 1 | 4 |
| Other | 36 |
The recently released genome of T. vaginalis revealed multiple paralogs of many putative plasma membrane proteins (19). Consistent with genome analysis, our membrane proteome contains multiple representatives of many protein families (Table I and supplemental Table 1). However, only a specific subset of paralogs within any protein family was reproducibly identified in multiple, independent experiments. For example, 104 ABC transporter genes were predicted in the genome; however, only six members of this family were systematically found in the proteome (Table I), indicating that these are the most abundant and/or biotin-accessible members of the family.
Localization of Candidate Surface Proteins by Immunofluorescence Microscopy
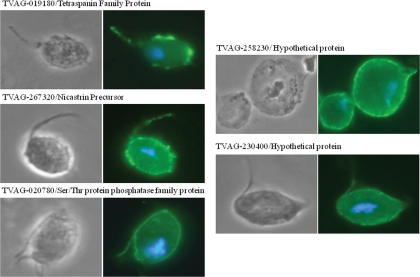
After excluding predicted contaminating proteins, 271 proteins identified by multiple peptides were predicted to be putative plasma membrane proteins (Fig. 2 and supplemental Table 1). To validate this prediction, we selected five proteins that have predicted transmembrane domains but were not previously predicted to be membrane proteins in T. vaginalis to determine their subcellular localization. Two were hypothetical proteins (TVAG_258230 and TVAG_230400), and three proteins had conserved domains found in proteins in other organisms (TVAG_020780/Ser/Thr protein phosphatase family protein, TVAG_019180/tetraspanin family protein, and TVAG_267320/nicastrin precursor) (supplemental Table 1). The genes encoding these proteins were cloned and expressed under control of their own promoters as C-terminal HA-tagged fusion proteins in strain T1 transgenic parasites. We then determined the localization in transgenic cells by IFA. All selected proteins localized to the surface of the parasite (Fig. 3), validating our experimental design as a reliable approach for surface protein identification.
Fig. 3.
Subcellular localization of five randomly selected putative membrane proteins in T. vaginalis transfectants. Cells expressing C-terminal HA-tagged versions of the indicated selected proteins were stained for immunofluorescence microscopy using a mouse HA-tagged antibody. The nucleus (blue) was also stained with 4′,6′-diamidino-2-phenylindole.
Identification of Membrane Proteins That Are Differentially Expressed in Parasites That Differ in Their Capacity to Adhere to Host Cells
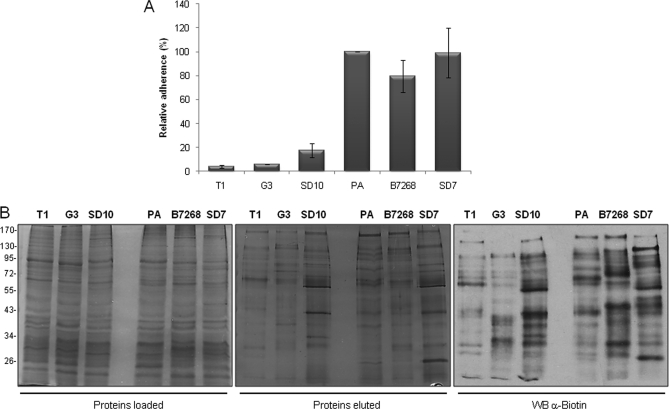
T. vaginalis is an extracellular parasite that adheres to vaginal epithelial cells to colonize its human host. To identify surface proteins that are differentially expressed in adherent and less adherent parasites, we first examined a number of strains to determine their relative adherence to monolayers of VECs using previously described assays (15, 16) (Fig. 4A). Six strains were selected for comparison of their surface proteins; three (T1, G3, and SD10) were only ∼5–17% as adherent as our standard laboratory adherent strain PA (15, 16). In addition to PA, two other strains with comparable adherence properties (SD7 and B7268) were selected for analyses. The three adherent strains (PA, SD7, and B7268) (Fig. 4A) were also significantly more cytotoxic to VECs relative to less adherent T1, G3, and SD10 strains (data not shown).
Fig. 4.
Analyses of six parasite strains for adherence and biotinylation. A, binding to ectocervical cells. Parasites from six different strains were fluorescently labeled and incubated with ectocervical cell monolayers at 37 °C. Coverslips were washed to remove non-adherent parasites, mounted, and visualized and quantified by fluorescence microscopy. Data are expressed as percentage of adhesion related to the PA strain ± the standard deviation of the mean. A representative experiment of three independent experiments is shown. B, surface protein biotinylation of strains with different adherence capacities. A comparable amount of protein extract of three less adherent (T1, G3, and SD10) and three highly adherent strains (PA, B7268, and SD7) was loaded onto a streptavidin column (left panel). Biotinylated labeled proteins were recovered, separated by SDS-PAGE, and silver-stained (middle panel) or detected by Western blot (WB) with an anti-biotin antibody (right panel). The molecular weight marker is shown on the far left.
To identify proteins that are differentially expressed in the highly adherent and less adherent strains, comparative analyses of their biotinylated plasma membrane proteins were performed. A comparison of the protein profiles of fractions loaded on the streptavidin columns and the eluted proteins showed that the biotinylated proteins are not among the more abundant cellular proteins, indicating the presence of low abundance surface molecules (Fig. 4B). The overall protein profiles of the six strains were highly similar (left panel); however, biotinylated proteins eluted from the streptavidin column varied notably (middle and right panels). These data indicate that there is a substantial variation in relatively low abundance membrane proteins between strains, consistent with data shown in Fig. 1C.
To search for proteins that are over-represented in highly adherent relative to less adherent strains, spectral counts were normalized to minimize run to run variation and compared between strains using the ACfold algorithm (37). Proteins that exhibited changes in normalized spectral counts that were greater than 2.5-fold and possessed p values from the ACtest of less than 0.01 were considered differentially expressed. Additionally, we required that these proteins contain either a putative signal peptide or transmembrane domain and that the spectral counts of these proteins taken from previous analyses exhibited the same quantitative trends. Using this approach, 16 proteins were determined to be differentially expressed between highly adherent and less adherent strains, 11 of which were up-regulated in more adherent strains (Table II). Two related hypothetical proteins were either ∼21- or ∼43-fold more abundant in adherent strains, and a chlamydial polymorphic outer membrane-like protein was observed to be ∼27-fold more abundant. Other proteins with similarity to known proteins that were significantly more abundant in highly adherent strains include a serine/threonine protein phosphatase (18-fold), an adenylate/guanylate cyclase (15-fold), a subtilisin-like serine protease (∼6-fold), and an additional chlamydial polymorphic outer membrane-like protein (∼5-fold). Three additional hypothetical proteins were found to be ∼3-, 7-, and 9-fold more abundant on the surface of adherent parasite strains. We also identified five proteins that were expressed at higher levels in the less adherent strains: an ABC transporter, an auxin efflux carrier, and three hypothetical proteins (Table II). All of these proteins have at least one transmembrane domain, but all but one lack a detectable signal peptide. This indicates that signal peptides for T. vaginalis surface proteins are atypical and thus not readily detected with standard search algorithms.
Table II. Differentially expressed proteins in strains with differing adherent capacities.
| Locus | -Fold change | p value | Description |
|---|---|---|---|
| More abundant in highly adherent strains | |||
| TVAG_166850 | 42.7 | 1e−13 | Hypothetical protein |
| TVAG_140850 | 26.5 | 2e−08 | Chlamydial polymorphic outer membrane protein |
| TVAG_244130 | 20.7 | 1e−06 | Hypothetical protein |
| TVAG_020780 | 18.0 | 8e−06 | Ser/Thr protein phosphatase family protein |
| TVAG_299950 | 15.0 | 6e−05 | Adenylate and guanylate cyclase catalytic domain |
| TVAG_147050 | 9.0 | 4e−03 | Hypothetical protein |
| TVAG_239650 | 6.8 | 2e−30 | Hypothetical protein |
| TVAG_142720 | 5.8 | 3e−03 | Subtilase family protein |
| TVAG_110300 | 5.2 | 2E−13 | Hypothetical protein |
| TVAG_212570 | 5.1 | 9e−04 | Chlamydial polymorphic outer membrane protein |
| TVAG_494220 | 2.6 | 1e−02 | Hypothetical protein |
| More abundant in less adherent strains | |||
| TVAG_190320 | 13.0 | 9e−04 | ABC transporter family protein |
| TVAG_384150 | 4.8 | 8e−03 | Hypothetical protein |
| TVAG_108920 | 4.6 | 1e−19 | Hypothetical protein |
| TVAG_411910 | 3.8 | 5e−03 | Auxin efflux carrier family protein |
| TVAG_307990 | 3.7 | 7e−03 | Hypothetical protein |
Validation of Differential Protein Expression by Examining Relative mRNA Levels
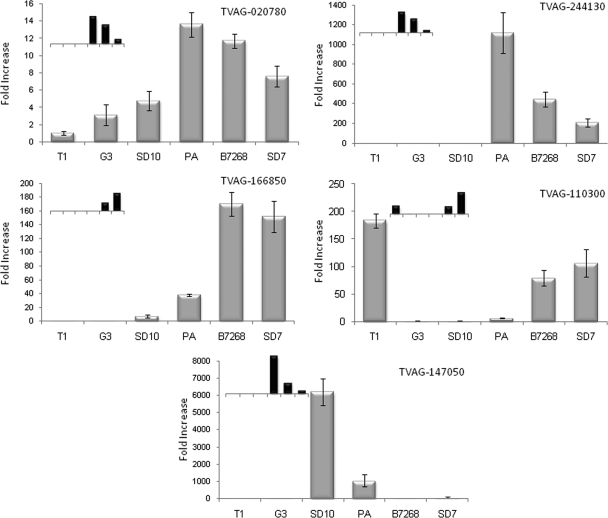
Using an independent method to validate the MudPIT data and to assess the relative expression of genes encoding membrane proteins in adherent and less adherent T. vaginalis strains, we utilized qPCR to compare mRNA levels of five of the 11 genes encoding proteins found to be highly expressed in adherent parasites (Fig. 5). These data indicate that protein levels measured by MudPIT analyses correlate well with mRNA levels determined by qPCR. A strict relative correlation between protein and mRNA abundance was observed for all six strains for four of these genes (TVAG_166850, TVAG_244130, TVAG_020780, and TVAG_110300) and for five of the six strains for the other gene (TVAG_147050). In addition to validating the differential protein expression levels, the qPCR analyses also revealed that the mRNA encoding TVAG_244130 is 850- and 350-fold more highly expressed in adherent strains PA and B7268, respectively, relative to the less adherent G3 strain (Fig. 5); this is the greatest difference in mRNA levels observed for the tested genes. The mRNA encoding TVAG_166850 was similarly shown to be induced 150- and 135-fold in the B7268 and SD7 adherent strains, respectively, compared with the G3 strain, and this transcript was undetectable in non-adherent T1 cells (Fig. 5). Interestingly, these two genes (TVAG_244130 and TVAG_166850) belong to the same family and have no predicted known domain identified by bioinformatics analysis. Consistent with our qPCR data, the proteins encoded by these hypothetical genes are two of the three proteins with the greatest -fold change in abundance between highly adherent and less adherent strains (Table II).
Fig. 5.
qPCR analysis validates MudPIT differential expression. mRNA expression levels of five transcripts identified as differentially expressed by MudPIT analysis were examined by qPCR. Data are expressed as -fold increase compared with the G3 strain ± the standard deviation of the mean. Inset graphs represent MudPIT values for the corresponding gene and share the same x axis with the qPCR data. TVAG_166850, hypothetical protein; TVAG_244130, hypothetical protein; TVAG_020780, Ser/Thr protein phosphatase family protein; TVAG_110300, hypothetical protein; TVAG_147050, hypothetical protein.
Two Highly Expressed Proteins in Adherent Strains Significantly Increase Adherence of a Less Adherent Strain When Expressed Exogenously
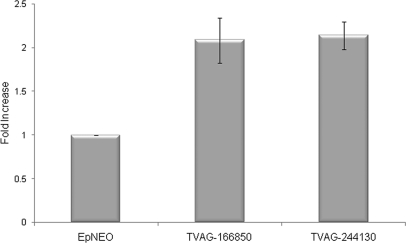
The striking difference in expression between adherent and less adherent strains of mRNAs encoding TVAG_166850 and TVAG_244130 led us to determine whether exogenous expression of these proteins in the poorly adherent T1 strain would confer an adherent phenotype. Both genes were cloned into episomal expression vectors under the control of a strong promoter (39) and transfected independently into T1 parasites. Adherence assays were then performed to evaluate whether the sole gain of expression of either protein influences adherence capacity. Expression of either N-terminal HA-tagged TVAG_166850 or TVAG_244130 in transfected parasites was capable of increasing attachment to VECs ∼2.1- and ∼2.2-fold, respectively, compared with control parasites transfected with an empty plasmid (Fig. 6). Similar results were also observed using protein HA-tagged at the C terminus (data not shown). No effect on cytotoxicity of VECs in vitro was observed using T1 parasites overexpressing either protein (data not shown). This indicates a specific role in adherence for these proteins.
Fig. 6.
Overexpression of TVAG_244130 and TVAG_166850 enhances adherence of T. vaginalis to ectocervical cells. Exogenous expression of an N-terminal HA-tagged TVAG_244130 or TVAG_166850 protein in the less adherent T1 strain was analyzed by introducing the gene on an episome under the control of a strong promoter. Attachment of parasites expressing the protein to VECs was assessed and compared with parasites transfected with empty plasmid (EpNEO). Data are expressed as -fold increase compared with empty plasmid control ± the standard deviation of the mean. A representative experiment of four independent experiments is shown.
DISCUSSION
We developed a straightforward approach for the isolation of surface proteins and combined this with high throughput proteomics to identify proteins on the surface of the parasite T. vaginalis. This constitutes the first large scale description of surface proteins on this extracellular human pathogen. Utilizing a non-membrane-permeable biotin reagent, we preferentially labeled plasma membrane and plasma membrane-associated proteins while largely excluding intracellular proteins. Combined with MudPIT analyses, 411 proteins were identified when examining six strains of T. vaginalis. The majority (63%) possess a predicted transmembrane domain and/or signal peptide sequence. Of these, ∼56% (35% of total) are predicted to be membrane proteins based on similarity to known membrane proteins in other organisms. Within this group, members of seven of 10 protein families that were predicted by in silico analyses of the T. vaginalis genome to be membrane proteins with possible roles in Trichomonas pathogenesis (19, 20) are present (see Table I). These include proteins with similarity to BspA proteins of mucosal bacteria known to mediate adherence to host cells (20, 42); three types of proteinases, a metalloproteinase (GP63) implicated in Leishmania virulence (20, 43), a subtilisin-like serine peptidase implicated in egress of apicomplexan parasites (44, 45), and calpain-like cysteine proteases (46); Chlamydia polymorphic membrane-like proteins (47); immunodominant variable surface antigen (48, 49); and the P270 surface immunogen (50, 51). The presence of these predicted membrane proteins in our proteome further validates our approach.
It is notable that relatively few members of the unusually large protein families comprising previously predicted surface proteins (17, 18) were found in our membrane proteome (Table I). However, these data do not preclude the expression of many additional family members that may not be as abundant or as accessible to labeling as the ones identified. Additional family members might also be found if more strains were surveyed, consistent with the proposal that subsets of these protein families may be differentially expressed in different strains or during different stages of infection (20). Differential expression may also explain why only a limited number of proteins from other large gene families in our membrane proteome were detected. For example, only six ABC transporters were found in the proteome of 104 genes encoded in the genome (19). Nevertheless, these data underscore the value of a proteomics approach in an organism such as T. vaginalis where multiple, large gene families are common to identify specific proteins for further functional studies. Proteomics is particularly useful as high density microarrays for T. vaginalis are not yet available because of the complexity and repetition of its genome.
Interestingly, 37% of proteins found in the membrane proteome are hypothetical proteins that are unique to T. vaginalis. The ability to identify these novel surface proteins provides a foundation for future studies with the potential of uncovering unpredicted host-parasite interactions that may be important for pathogenesis. The majority (77%) of these newly identified novel hypothetical proteins have predicted signal peptides and/or transmembrane domains; however, 23% do not. These proteins may be directly anchored to the membrane or contain unidentified membrane-targeting/assembly domains. Of all putative surface proteins identified, 63% have predicted signal peptides and/or transmembrane domains. The remaining 37% include membrane-associated cytoskeletal proteins (5%) and proteins predicted to be intracellular (23%). The majority of the proteins in the latter category are highly abundant soluble proteins that are common contaminants observed in subcellular localization experiments (20, 40).2 However, we cannot strictly preclude the possibility that some of these previously defined intracellular proteins have an additional uncharacterized external localization, contributing an unknown function. Alternatively, such cytosolic proteins, for example enolase, glycerol-3-phosphate dehydrogenase, and HSP70, may be located immediately beneath the plasma membrane.
Several new protein families that have yet to be examined in T. vaginalis and that may play an important role in parasite biology were revealed by our analyses. These include the tetraspanin and β-lactamase families of proteins, the nicastrin precursor, and rhomboid-like and subtilisin-like serine peptidases. Based on functional analyses of these proteins in other organisms, a role in T. vaginalis pathogenesis for these surface molecules is predicted. For example, tetraspanin family proteins have been shown to play a role in adhesion, migration, and tissue invasion in mammalian cells (52, 53); these are processes required for T. vaginalis to colonize its human host (12, 54). Moreover, tetraspanins on the surface of Schistosoma mansoni are protective antigens against schistosomiasis (55, 56). Evidence for multiple roles for rhomboid intramembrane and subtilisin-like serine proteases in the pathogenesis of divergent protozoan parasites has emerged in recent years. An Entamoeba histolytica rhomboid protease has been demonstrated to cleave a surface lectin involved in phagocytosis and immune evasion (57). Moreover, multiple rhomboid proteases and subtilisin-like proteases are known to play critical roles in adherence, invasion, and survival of Toxoplasma and malaria parasites (58–60). The work reported here sets the stage for detailed studies to address the roles of these newly identified surface proteins in Trichomonas biology.
As an obligate extracellular parasite, the surface of T. vaginalis is likely to function in multiple ways to establish, maintain, and modulate infection. Prior to this study, only two proteins have been shown to be surface-localized (20, 61), and neither have been demonstrated to modulate adherence of T. vaginalis to host cells. As a first step toward identifying proteins that may be involved in adherence, numerous T. vaginalis strains were examined to determine their capacity to adhere to vaginal epithelial cells. From these data, six strains were selected for proteomics analyses: three of these were ≥5 times more adherent to vaginal epithelial cells than the three others. Comparison of the surface proteomes of the six strains revealed only 16 proteins that were detectably differentially expressed (Table II). Eleven of these were more abundant in adherent parasites, whereas five were less abundant. Two Chlamydia polymorphic membrane-like proteins, a serine/threonine protein phosphatase, an adenylate/guanylate cyclase, a subtilase-like serine protease, and six hypothetical proteins were 2.5–43-fold more abundant in adherent strains. It is notable that related proteins are implicated in the pathogenesis of other pathogens, meriting a closer examination of a possible function in T. vaginalis pathogenesis. Chlamydia polymorphic membrane protein PmpD is an autotransporter component of the chlamydial outer membrane thought to be involved in bacterial invasion and host inflammation (47). Serine peptidases are known to participate in invasion of malaria and Toxoplasma (58–60). Phosphatases and adenylate/guanylate cyclases play critical roles in cell signaling, a mechanism by which cells sense their environment. Further studies are necessary to examine whether these proteins are directly or indirectly involved in T. vaginalis adherence to host cells.
Two of six novel, hypothetical proteins that were more abundant in highly adherent strains, TVAG_244130 and TVAG_166850, share 33% similarity on the amino acid level and belong to a family encoded by ∼100 genes in the T. vaginalis genome. When either of these proteins was expressed upon transfection of the less adherent strain, T1 attachment to VECs was increased ∼2-fold. Several other members of this family were detected in the membrane proteome; however, only these two exhibited differential expression. Whether other members of this protein family are also involved in adherence or other pathogenic processes remains to be determined. The T. vaginalis genome has a multitude of unique, hypothetical genes, and 152 were found in our surface proteome. In addition to revealing possible novel functions for these proteins, further analyses are warranted as parasite-specific surface proteins could serve as good vaccine candidates.
Comprehensive profiling of the cell surface proteome of different parasite strains provides an effective approach for the identification of potential new surface adhesion factors. The data described here will facilitate studies aimed toward a better understanding of host-pathogen interactions as well as genetic and strain differences that may account for the variation in symptoms and disease progression observed in women infected by this pathogen.
Supplementary Material
Acknowledgments
We thank Dr. Jacqui Upcroft and Professor Lynette Corbeil for parasite strains, Dr. Jennifer Gordon for critical comments on the manuscript, and our colleagues in the laboratory for helpful discussions.
* This work was supported, in whole or in part, by National Institutes of Health Grant R01 AI069058.
 This article contains supplemental Table 1.
This article contains supplemental Table 1.
2 R. Schneider, M. T. Brown, and P. J. Johnson, unpublished data.
1 The abbreviations used are:
- LPG
- lipophosphoglycan
- MudPIT
- multidimensional protein identification technology
- BLAST
- basic local alignment search tool
- VEC
- vaginal epithelial cell
- ABC
- ATP-binding cassette
- BspA
- basic surface-exposed protein, leucine-rich repeat
- IFA
- immunofluorescence assay
- HA
- hemagglutinin
- qPCR
- quantitative PCR
- sulfo-NHS-SS-biotin
- sulfosuccinimidyl 2-(biotinamido)-ethyl-1,3-dithiopropionate.
REFERENCES
- 1.Gerbase A. C., Rowley J. T., Heymann D. H., Berkley S. F., Piot P. (1998) Global prevalence and incidence estimates of selected curable STDs. Sex. Transm. Infect. 74, S12–S16 [PubMed] [Google Scholar]
- 2.Gardner W. A., Jr., Culberson D. E., Bennett B. D. (1986) Trichomonas vaginalis in the prostate gland. Arch. Pathol. Lab. Med. 110, 430–432 [PubMed] [Google Scholar]
- 3.Minkoff H., Grunebaum A. N., Schwarz R. H., Feldman J., Cummings M., Crombleholme W., Clark L., Pringle G., McCormack W. M. (1984) Risk factors for prematurity and premature rupture of membranes: a prospective study of the vaginal flora in pregnancy. Am. J. Obstet. Gynecol. 150, 965–972 [DOI] [PubMed] [Google Scholar]
- 4.Cotch M. F., Pastorek J. G., 2nd, Nugent R. P., Hillier S. L., Gibbs R. S., Martin D. H., Eschenbach D. A., Edelman R., Carey J. C., Regan J. A., Krohn M. A., Klebanoff M. A., Rao A. V., Rhoads G. G., Yaffe S. J., Catz C. S., McNellis D., Berendes H. W., Blackwelder W. C., Kaslow R. A., Reed G. F., Greenberg E. M., Williams S., Rettig P. J. (1997) Trichomonas vaginalis associated with low birth weight and preterm delivery. Sex. Transm. Dis. 24, 353–360 [DOI] [PubMed] [Google Scholar]
- 5.Reimer T., Ulfig N., Friese K. (1999) Antibiotics: treatment of preterm labor. J. Perinat. Med. 27, 35–40 [DOI] [PubMed] [Google Scholar]
- 6.Sorvillo F., Kerndt P. (1998) Trichomonas vaginalis and amplification of HIV-1 transmission. Lancet 351, 213–214 [DOI] [PubMed] [Google Scholar]
- 7.Miller M., Liao Y., Gomez A. M., Gaydos C. A., D'Mellow D. (2008) Factors associated with the prevalence and incidence of Trichomonas vaginalis infection among African American women in New York city who use drugs. J. Infect. Dis. 197, 503–509 [DOI] [PubMed] [Google Scholar]
- 8.Van Der Pol B., Kwok C., Pierre-Louis B., Rinaldi A., Salata R. A., Chen P. L., van de Wijgert J., Mmiro F., Mugerwa R., Chipato T., Morrison C. S. (2008) Trichomonas vaginalis infection and human immunodeficiency virus acquisition in African women. J. Infect. Dis. 197, 548–554 [DOI] [PubMed] [Google Scholar]
- 9.Yap E. H., Ho T. H., Chan Y. C., Thong T. W., Ng G. C., Ho L. C., Singh M. (1995) Serum antibodies to Trichomonas vaginalis in invasive cervical cancer patients. Genitourin. Med. 71, 402–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Z. F., Begg C. B. (1994) Is Trichomonas vaginalis a cause of cervical neoplasia: results from a combined analysis of 24 studies. Int. J. Epidemiol. 23, 682–690 [DOI] [PubMed] [Google Scholar]
- 11.Sutcliffe S., Giovannucci E., Alderete J. F., Chang T. H., Gaydos C. A., Zenilman J. M., De Marzo A. M., Willett W. C., Platz E. A. (2006) Plasma antibodies against Trichomonas vaginalis and subsequent risk of prostate cancer. Cancer Epidemiol. Biomarkers Prev. 15, 939–945 [DOI] [PubMed] [Google Scholar]
- 12.Petrin D., Delgaty K., Bhatt R., Garber G. (1998) Clinical and microbiological aspects of Trichomonas vaginalis. Clin. Microbiol. Rev. 11, 300–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alderete J. F., Garza G. E. (1988) Identification and properties of Trichomonas vaginalis proteins involved in cytadherence. Infect. Immun. 56, 28–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hernández-Gutiérrez R., Avila-González L., Ortega-López J., Cruz-Talonia F., Gómez-Gutierrez G., Arroyo R. (2004) Trichomonas vaginalis: characterization of a 39-kDa cysteine proteinase found in patient vaginal secretions. Exp. Parasitol. 107, 125–135 [DOI] [PubMed] [Google Scholar]
- 15.Bastida-Corcuera F. D., Okumura C. Y., Colocoussi A., Johnson P. J. (2005) Trichomonas vaginalis lipophosphoglycan mutants have reduced adherence and cytotoxicity to human ectocervical cells. Eukaryot. Cell 4, 1951–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okumura C. Y., Baum L. G., Johnson P. J. (2008) Galectin-1 on cervical epithelial cells is a receptor for the sexually transmitted human parasite Trichomonas vaginalis. Cell. Microbiol. 10, 2078–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Upcroft J. A., Delgadillo-Correa M. G., Dunne R. L., Sturm A. W., Johnson P. J., Upcroft P. (2006) Genotyping Trichomonas vaginalis. Int. J. Parasitol. 36, 821–828 [DOI] [PubMed] [Google Scholar]
- 18.Upcroft J. A., Upcroft P. (2001) Drug susceptibility testing of anaerobic protozoa. Antimicrob. Agents Chemother. 45, 1810–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carlton J. M., Hirt R. P., Silva J. C., Delcher A. L., Schatz M., Zhao Q., Wortman J. R., Bidwell S. L., Alsmark U. C., Besteiro S., Sicheritz-Ponten T., Noel C. J., Dacks J. B., Foster P. G., Simillion C., Van de Peer Y., Miranda-Saavedra D., Barton G. J., Westrop G. D., Müller S., Dessi D., Fiori P. L., Ren Q., Paulsen I., Zhang H., Bastida-Corcuera F. D., Simoes-Barbosa A., Brown M. T., Hayes R. D., Mukherjee M., Okumura C. Y., Schneider R., Smith A. J., Vanacova S., Villalvazo M., Haas B. J., Pertea M., Feldblyum T. V., Utterback T. R., Shu C. L., Osoegawa K., de Jong P. J., Hrdy I., Horvathova L., Zubacova Z., Dolezal P., Malik S. B., Logsdon J. M., Jr., Henze K., Gupta A., Wang C. C., Dunne R. L., Upcroft J. A., Upcroft P., White O., Salzberg S. L., Tang P., Chiu C. H., Lee Y. S., Embley T. M., Coombs G. H., Mottram J. C., Tachezy J., Fraser-Liggett C. M., Johnson P. J. (2007) Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science 315, 207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirt R. P., Noel C. J., Sicheritz-Ponten T., Tachezy J., Fiori P. L. (2007) Trichomonas vaginalis surface proteins: a view from the genome. Trends Parasitol. 23, 540–547 [DOI] [PubMed] [Google Scholar]
- 21.Jang J. H., Hanash S. (2003) Profiling of the cell surface proteome. Proteomics 3, 1947–1954 [DOI] [PubMed] [Google Scholar]
- 22.Shin B. K., Wang H., Yim A. M., Le Naour F., Brichory F., Jang J. H., Zhao R., Puravs E., Tra J., Michael C. W., Misek D. E., Hanash S. M. (2003) Global profiling of the cell surface proteome of cancer cells uncovers an abundance of proteins with chaperone function. J. Biol. Chem. 278, 7607–7616 [DOI] [PubMed] [Google Scholar]
- 23.Zhao Y., Zhang W., Kho Y., Zhao Y. (2004) Proteomic analysis of integral plasma membrane proteins. Anal. Chem. 76, 1817–1823 [DOI] [PubMed] [Google Scholar]
- 24.Sostaric E., Georgiou A. S., Wong C. H., Watson P. F., Holt W. V., Fazeli A. (2006) Global profiling of surface plasma membrane proteome of oviductal epithelial cells. J. Proteome Res. 5, 3029–3037 [DOI] [PubMed] [Google Scholar]
- 25.Sutton M., Sternberg M., Koumans E. H., McQuillan G., Berman S., Markowitz L. (2007) The prevalence of Trichomonas vaginalis infection among reproductive-age women in the United States, 2001–2004. Clin. Infect. Dis. 45, 1319–1326 [DOI] [PubMed] [Google Scholar]
- 26.Clark C. G., Diamond L. S. (2002) Methods for cultivation of luminal parasitic protists of clinical importance. Clin. Microbiol. Rev. 15, 329–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fichorova R. N., Rheinwald J. G., Anderson D. J. (1997) Generation of papillomavirus-immortalized cell lines from normal human ectocervical, endocervical, and vaginal epithelium that maintain expression of tissue-specific differentiation proteins. Biol. Reprod. 57, 847–855 [DOI] [PubMed] [Google Scholar]
- 28.Florens L., Carozza M. J., Swanson S. K., Fournier M., Coleman M. K., Workman J. L., Washburn M. P. (2006) Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods 40, 303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Washburn M. P., Wolters D., Yates J. R., 3rd (2001) Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–247 [DOI] [PubMed] [Google Scholar]
- 30.Wohlschlegel J. A. (2009) Identification of SUMO-conjugated proteins and their SUMO attachment sites using proteomic mass spectrometry. Methods Mol. Biol. 497, 33–49 [DOI] [PubMed] [Google Scholar]
- 31.McDonald W. H., Tabb D. L., Sadygov R. G., MacCoss M. J., Venable J., Graumann J., Johnson J. R., Cociorva D., Yates J. R., 3rd (2004) MS1, MS2, and SQT: three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid Commun. Mass Spectrom. 18, 2162–2168 [DOI] [PubMed] [Google Scholar]
- 32.Eng J. K., Mccormack A. L., Yates J. R. (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 33.Aurrecoechea C., Brestelli J., Brunk B. P., Carlton J. M., Dommer J., Fischer S., Gajria B., Gao X., Gingle A., Grant G., Harb O. S., Heiges M., Innamorato F., Iodice J., Kissinger J. C., Kraemer E., Li W., Miller J. A., Morrison H. G., Nayak V., Pennington C., Pinney D. F., Roos D. S., Ross C., Stoeckert C. J., Jr., Sullivan S., Treatman C., Wang H. M. (2009) GiardiaDB and TrichDB: integrated genomic resources for the eukaryotic protist pathogens Giardia lamblia and Trichomonas vaginalis. Nucleic Acids Res. 37, D526–D530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tabb D. L., McDonald W. H., Yates J. R., 3rd (2002) DTASelect and contrast: Tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1, 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 36.Cociorva D., Tabb D. L., Yates J. R. (2007) Validation of tandem mass spectrometry database search results using DTASelect, in Current Protocols in Bioinformatics (Baxevanis A. D. ed) Unit 13.4, John Wiley and Sons, New York: [DOI] [PubMed] [Google Scholar]
- 37.Carvalho P. C., Fischer J. S., Chen E. I., Yates J. R., 3rd, Barbosa V. C. (2008) PatternLab for proteomics: a tool for differential shotgun proteomics. BMC Bioinformatics 9, 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dyall S. D., Koehler C. M., Delgadillo-Correa M. G., Bradley P. J., Plümper E., Leuenberger D., Turck C. W., Johnson P. J. (2000) Presence of a member of the mitochondrial carrier family in hydrogenosomes: conservation of membrane-targeting pathways between hydrogenosomes and mitochondria. Mol. Cell. Biol. 20, 2488–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Delgadillo M. G., Liston D. R., Niazi K., Johnson P. J. (1997) Transient and selectable transformation of the parasitic protist Trichomonas vaginalis. Proc. Natl. Acad. Sci. U.S.A. 94, 4716–4720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Addis M. F., Rappelli P., Fiori P. L. (2000) Host and tissue specificity of Trichomonas vaginalis is not mediated by its known adhesion proteins. Infect. Immun. 68, 4358–4360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lehker M. W., Alderete J. F. (2000) Biology of trichomonosis. Curr. Opin. Infect. Dis. 13, 37–45 [DOI] [PubMed] [Google Scholar]
- 42.Hirt R. P., Harriman N., Kajava A. V., Embley T. M. (2002) A novel potential surface protein in Trichomonas vaginalis contains a leucine-rich repeat shared by micro-organisms from all three domains of life. Mol. Biochem. Parasitol. 125, 195–199 [DOI] [PubMed] [Google Scholar]
- 43.Yao C., Donelson J. E., Wilson M. E. (2003) The major surface protease (MSP or GP63) of Leishmania sp. Biosynthesis, regulation of expression, and function. Mol. Biochem. Parasitol. 132, 1–16 [DOI] [PubMed] [Google Scholar]
- 44.Blackman M. J. (2008) Malarial proteases and host cell egress: an ‘emerging’ cascade. Cell. Microbiol. 10, 1925–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roiko M. S., Carruthers V. B. (2009) New roles for perforins and proteases in apicomplexan egress. Cell. Microbiol. 11, 1444–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosenthal P. J. (2004) Cysteine proteases of malaria parasites. Int. J. Parasitol. 34, 1489–1499 [DOI] [PubMed] [Google Scholar]
- 47.Wehrl W., Brinkmann V., Jungblut P. R., Meyer T. F., Szczepek A. J. (2004) From the inside out: processing of the chlamydial autotransporter PmpD and its role in bacterial adhesion and activation of human host cells. Mol. Microbiol. 51, 319–334 [DOI] [PubMed] [Google Scholar]
- 48.Edman U., Meraz M. A., Rausser S., Agabian N., Meza I. (1990) Characterization of an immunodominant variable surface antigen from pathogenic and nonpathogenic Entamoeba histolytica. J. Exp. Med. 172, 879–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marion S., Guillén N. (2006) Genomic and proteomic approaches highlight phagocytosis of living and apoptotic human cells by the parasite Entamoeba histolytica. Int. J. Parasitol. 36, 131–139 [DOI] [PubMed] [Google Scholar]
- 50.Musatovova O., Alderete J. F. (1999) The Trichomonas vaginalis phenotypically varying P270 immunogen is highly conserved except for numbers of repeated elements. Microb. Pathog. 27, 93–104 [DOI] [PubMed] [Google Scholar]
- 51.Musatovova O., Alderete J. F. (1998) Molecular analysis of the gene encoding the immunodominant phenotypically varying P270 protein of Trichomonas vaginalis. Microb. Pathog. 24, 223–239 [DOI] [PubMed] [Google Scholar]
- 52.Hemler M. E. (2005) Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 6, 801–811 [DOI] [PubMed] [Google Scholar]
- 53.Hemler M. E. (2003) Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu. Rev. Cell Dev. Biol. 19, 397–422 [DOI] [PubMed] [Google Scholar]
- 54.Fiori P. L., Rappelli P., Addis M. F. (1999) The flagellated parasite Trichomonas vaginalis: new insights into cytopathogenicity mechanisms. Microbes Infect. 1, 149–156 [DOI] [PubMed] [Google Scholar]
- 55.Loukas A., Tran M., Pearson M. S. (2007) Schistosome membrane proteins as vaccines. Int. J. Parasitol. 37, 257–263 [DOI] [PubMed] [Google Scholar]
- 56.Tran M. H., Pearson M. S., Bethony J. M., Smyth D. J., Jones M. K., Duke M., Don T. A., McManus D. P., Correa-Oliveira R., Loukas A. (2006) Tetraspanins on the surface of Schistosoma mansoni are protective antigens against schistosomiasis. Nat. Med. 12, 835–840 [DOI] [PubMed] [Google Scholar]
- 57.Baxt L. A., Baker R. P., Singh U., Urban S. (2008) An Entamoeba histolytica rhomboid protease with atypical specificity cleaves a surface lectin involved in phagocytosis and immune evasion. Genes Dev. 22, 1636–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Srinivasan P., Coppens I., Jacobs-Lorena M. (2009) Distinct roles of Plasmodium Rhomboid 1 in parasite development and malaria pathogenesis. PLoS Pathogens 5, e1000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim K. (2004) Role of proteases in host cell invasion by Toxoplasma gondii and other apicomplexa. Acta Trop. 91, 69–81 [DOI] [PubMed] [Google Scholar]
- 60.Miller S. A., Blackman M., Carruthers V. B., Kim K. (2000) The role of subtilisin proteases in invasion by Toxoplasma gondii tachyzoites. Mol. Biol. Cell 11, 291a [Google Scholar]
- 61.Alderete J. F. (1999) Iron modulates phenotypic variation and phosphorylation of P270 in double-stranded RNA virus-infected Trichomonas vaginalis. Infect. Immun. 67, 4298–4302 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.