Abstract
We have investigated the origin of the Pto disease resistance (R) gene that was previously identified in the wild tomato species Lycopersicon pimpinellifolium and isolated by map-based cloning. Pto encodes a serine-threonine protein kinase that specifically recognizes strains of Pseudomonas syringae pv. tomato (Pst) that express the avirulence gene avrPto. We examined an accession of the distantly related wild species Lycopersicon hirsutum var. glabratum that exhibits avrPto-specific resistance to Pst. The Pst resistance of L. hirsutum was introgressed into a susceptible Lycopersicon esculentum background to create the near-isogenic line 96T133-3. Resistance to Pst(avrPto) in 96T133-3 was inherited as a single dominant locus and cosegregated with a restriction fragment length polymorphism detected by the Pto gene. This observation suggested that a member of the Pto gene family confers Pst(avrPto) resistance in this L. hirsutum line. Here we report the cloning and characterization of four members of the Pto family from 96T133-3. One gene (LhirPto) is 97% identical to Pto and encodes a catalytically active protein kinase that elicits a hypersensitive response when coexpressed with avrPto in leaves of Nicotiana benthamiana. In common with the Pto kinase, the LhirPto protein physically interacts with AvrPto and downstream members of the Pto signaling pathway. Our studies indicate that R genes of the protein kinase class may not evolve rapidly in response to pathogen pressure and rather that their ability to recognize specific Avr proteins can be highly conserved.
The Pto gene in tomato encodes a serine-threonine protein kinase and confers resistance to isolates of Pseudomonas syringae that express the avirulence gene avrPto (1, 2). The AvrPto protein is believed to be secreted from Pseudomonas into the plant cell by a type III secretion system where its interaction with the Pto kinase initiates responses leading to disease resistance (3–6). Only Pto alleles that encode proteins which interact with AvrPto in a yeast two-hybrid system can elicit avrPto-dependent resistance response (4, 5). Similarly, only AvrPto variants that interact with the Pto kinase possess avirulence activity (7). We have recently demonstrated that the specificity of the Pto–AvrPto interaction is determined by a threonine residue located within the activation loop of the Pto kinase (8), a domain that participates in substrate binding and catalytic regulation in other protein kinases (9).
In nature, it is likely that mechanisms such as horizontal gene transfer and mutation change the types and specificity of avr genes present in plant pathogens (10). Consequently it is thought that the host responds by evolving new disease resistance (R) genes that recognize new pathogen avirulence specificities. In contrast to Pto, the majority of R genes cloned to date encode proteins with a region of leucine-rich repeats (LRRs) and a putative nucleotide-binding site (NBS). In these proteins, the LRR domain is hypothesized to mediate recognition specificity through a direct or indirect interaction with the Avr protein (11–14). Recent reports indicate that the sequences of some LRR-type R genes are particularly subject to rapid evolution (15–17) and that regions of the LRR proteins implicated in recognition specificity are affected by diversifying selection (16, 17). Furthermore, the organization of many R genes in clustered families appears to make them amenable to unequal crossing over and gene conversion events. These rearrangements give rise to duplications and deletions of LRR regions and to recombination between R genes, all of which can alter recognition specificities (16–19).
Despite the apparent ability to evolve rapidly, recent data suggest that some recognition specificities are ancient and that hosts and pathogens may have coexisted for millions of years. One example of an ancient R gene specificity occurs in Arabidopsis, where a homolog of the RPM1 gene was cloned from an isolate of Arabidopsis lyrata that is resistant to Pseudomonas syringae(avrRpm1) (20). Although it was not confirmed that the A. lyrata RPM1 gene actually conferred Pseudomonas resistance, this report suggests that recognition of avrRpm1 predates the divergence of Arabidopsis thaliana from A. lyrata (20). In fact, avrRpm1 recognition may have arisen even earlier, as avrRpm1 (and avrB) recognition also has been reported in soybean (21).
We reported previously that an accession of Lycopersicon hirsutum var. glabratum, PI134418, is resistant to strains of P. syringae pv. tomato (Pst) that express avrPto but is susceptible to strains that do not express this avirulence gene (22). Both restriction fragment length polymorphisms (RFLPs) and morphological features indicate that L. hirsutum is a distant relative of Lycopersicon pimpinellifolium, from which the Pto gene was originally isolated (23). In earlier work the Pst(avrPto) resistance of PI134418 was introgressed into a susceptible Lycopersicon esculentum variety to create the near-isogenic line 96T133-3 (22). In progeny derived from a cross with this line Pst(avrPto) resistance cosegregates with an RFLP detected by the Pto resistance gene and maps to the same location on chromosome 5 as the Pto gene in L. pimpinellifolium (22). These observations raised the possibility that a member of the Pto gene family in L. hirsutum is responsible for conferring resistance to Pst(avrPto).
We now report the cloning and molecular characterization of four Pto gene family members from accession PI134418 and demonstrate that one of them encodes a protein conferring avrPto-specific disease resistance. Our results indicate that the evolution of Pto recognition specificity for the Pseudomonas AvrPto protein predates the divergence of L. pimpinellifolium and L. hirsutum.
Experimental Procedures
Standard Methods and Plant Treatments.
Standard methods were used for tomato genomic DNA isolation, restriction enzyme digestion, DNA blotting, and DNA gel blot hybridization (24). Bacterial growth measurements were performed as described previously (1). Fenthion sensitivity assays were performed by vacuum infiltrating 4-week-old tomato plants with a solution containing 0.05% fenthion (Chem Service, West Chester, PA), 0.15% isopropyl alcohol, and 0.004% Silwet (OSI Specialties, South Charleston, WV) for 2 min.
Plant Materials.
Seeds of 96T133-3, TA537, and TA209 were a gift of Steven Tanksley (Cornell Univ.). Line 96T133-3 was developed by introgressing Pst (avrPto) resistance into cultivar TA209 as described in ref. 22. TA537 was developed by a similar process, which introgressed the Pto locus from an Israeli processing line, H14-Pto, into TA209 (25).
cDNA Library Construction and cDNA Cloning.
mRNA was isolated from 96T133-3 leaves 6 h after infiltration with 107 colony-forming units/ml Pst (avrPto) and used to construct a cDNA library in a ZAP Express cDNA kit (Stratagene). Plaque lifts were performed on >106 plaques according to the Stratagene protocol. Filters were hybridized with radiolabeled Pto probe and washed to a stringency of 0.5× SSC (1× SSC is 0.15 M NaCl/0.015 M sodium citrate, pH 7.0). megalign 4.02, part of the DNASTAR package (DNASTAR, Madison, WI) was used to create sequence alignments and dendrograms.
Yeast Two-Hybrid Analysis.
Primers which introduced a BamHI or a SalI site to the 5′ and 3′ ends, respectively, were designed for the LhirPto open reading frames. The LhirPto inserts were PCR amplified by using Pwo high-fidelity polymerase (Boehringer Mannheim), cloned into pBluescript (Stratagene) and confirmed by sequencing. LhirPto genes were cloned into the pEG202 bait vector and transformed into yeast strain EGY48 by using the lithium acetate method (24). Western blots and two-hybrid assays were done as previously described (26–28).
Glutathione S-Transferase (GST) Fusion Protein Expression and Kinase Assays.
Primers that introduced a BamHI or a SalI site to the 5′ and 3′ ends, respectively, were designed for each LhirPto open reading frame. The cDNA insert was PCR amplified by using Pwo high-fidelity DNA polymerase (Boehringer Mannheim), cloned into pBluescript (Stratagene), and confirmed by sequencing. The insert was removed from pBluescript, ligated into pGEX-KG, and electroporated into Escherichia coli strain DH5α. GST-fusion protein purification and phosphorylation assays were performed as previously described (27, 29, 30). Autoradiography was performed with a Storm System Imager (Molecular Dynamics). Purified Pti1 and Pti4 were gifts from Yong Gu (Boyce Thompson Institute).
Transient Assays.
LhirPto genes were removed from pBluescript (see above), ligated onto the cauliflower mosaic virus 35S promoter, and cloned into pBTEX or pBTEX∷35S∷avrPto. These constructs were electroporated into Agrobacterium tumefaciens strain EHA105. Transient assays were performed as described previously (8).
Results
Tomato Line 96T133-3 Contains the Pto Locus from L. hirsutum Accession PI134418.
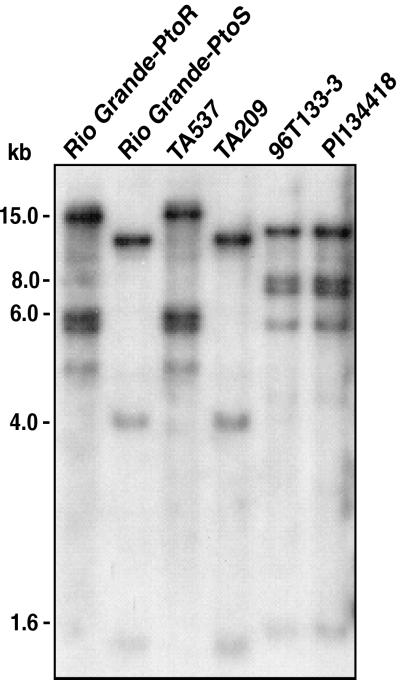
In common with L. pimpinellifolium, the original source of the Pto resistance gene, L. hirsutum var. glabratum (accession PI134418) is resistant to strains of Pst that express the avirulence gene avrPto and is susceptible to strains of Pst that do not express avrPto (22). Hybridization of a genomic DNA blot with the Pto gene revealed that PI134418 contains a Pto gene family that is polymorphic with respect to either Rio Grande-PtoR (RG-PtoR) or Rio Grande-PtoS (RG-PtoS) (Fig. 1). These near isogenic lines contain the Pto locus from L. pimpinellifolium and L. esculentum, respectively. Resistance to Pst(avrPto) was introgressed from PI134418 into the susceptible L. esculentum cultivar TA209 (22). The introgression involved six backcrosses to the L. esculentum parent with selection in each generation for avrPto-mediated resistance, and a final selfing to create the homozygous resistant line 96T133-3. All RFLPs detected by Pto in PI134418 were also detected in 96T133-3 (Fig. 1) and cosegregated with Pst(avrPto) resistance. Furthermore, the Pto RFLP and Pst(avrPto) resistance mapped to the same location on chromosome 5 as the Pto gene found in L. pimpinellifolium (22). These data suggested that a member of the clustered Pto gene family in this L. hirsutum line is responsible for conferring resistance to Pst(avrPto).
Figure 1.
RFLP patterns detected by the Pto gene in wild L. hirsutum, L. esculentum, and near-isogenic L. esculentum lines. Genomic DNA (3 μg) isolated from each of the indicated lines was digested with EcoRI, separated on a 1% agarose gel, and transferred onto Hybond N+ membrane. The membrane was hybridized to a 32P-labeled PCR product of the Pto open reading frame and washed to a stringency of 0.5× SSC at 65°C. Rio Grande-PtoR and TA537 contain the Pto locus from L. pimpinellifolium, Rio Grande-PtoS and TA209 contain the Pto locus from L. esculentum, and 96T133-3 contains the Pto locus from the L. hirsutum tomato accession PI134418.
The L. hirsutum and L. pimpinellifolium Pto Loci Confer Identical Levels of Resistance to P. syringae.
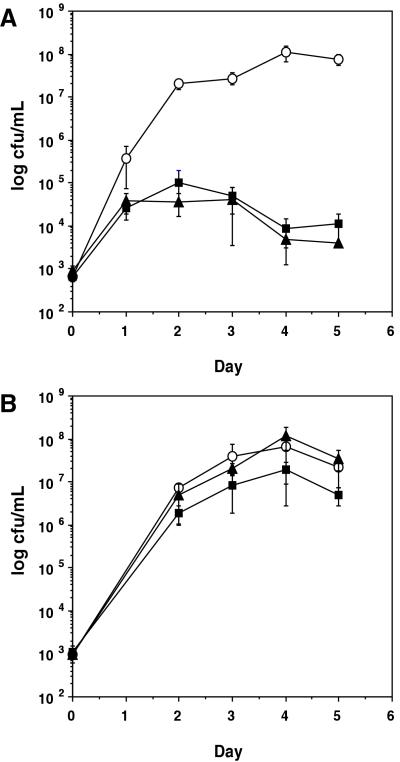
To compare the resistance conferred by the L. hirsutum and L. pimpinellifolium Pto loci, we studied their abilities to suppress the growth of Pst in near-isogenic tomato lines. The L. pimpinellifolium Pto locus was introgressed into the susceptible TA209 L. esculentum background to create the near-isogenic line TA537 (Fig. 1). Five days after inoculation, Pst(avrPto) populations in leaves of 96T133-3 were equivalent to those in TA537 and were 104 times lower than in susceptible TA209 leaves (Fig. 2A). Neither 96T133-3 nor TA537 showed disease symptoms 1 week after being inoculated with Pst(avrPto). However, TA209 developed bacterial speck symptoms 3 days after inoculation. All three lines were susceptible to Pst lacking the avrPto gene (Fig. 2B). These data demonstrate that the L. hirsutum Pto locus confers a level of avrPto-specific resistance indistinguishable from that conferred by the L. pimpinellifolium Pto locus.
Figure 2.
Growth of Pst(avrPto) (A) or Pst (B) in leaves of near-isogenic lines containing different Pto loci. Four-week-old plants of TA209 (L. esculentum Pto locus, ○), TA537 (L. pimpinellifolium Pto locus, ■), and 96T133-3 (L. hirsutum Pto locus, ▴) were vacuum-infiltrated with 105 colony-forming units per milliliter (cfu/ml) of Pseudomonas, and bacterial growth was measured as cfu/ml at the time points specified. Each time point represents the average of three samples. Each sample contains three 1-cm2 leaf discs in 1 ml of 10 mM MgCl2. The error bars represent the standard error.
The L. hirsutum Pto Locus Does Not Confer Fenthion Sensitivity.
In L. pimpinellifolium Pto is closely linked to the Fen gene, which confers sensitivity to the insecticide fenthion (31). Given that the LhirPto locus confers avrPto-specific resistance, we tested whether 96T133-3 might also exhibit fenthion sensitivity. TA209, TA537, and 96T133-3 leaves were infiltrated with a solution of 0.05% fenthion and observed over a 4-day period. Only TA537, which contains the Pto locus from L. pimpinellifolium, developed necrotic specks typical of the fenthion response (data not shown). Neither TA209 nor 96T133-3 exhibited symptoms of fenthion sensitivity, indicating that, like L. esculentum, 96T133-3 does not contain a functional Fen gene. Previous tests for fenthion sensitivity of PI134418 and other L. hirsutum accessions have yielded similar results (22, 32).
96T133-3 Expresses Genes That Are Orthologous with Pto Gene Family Members from Rio Grande-PtoR and VFNT Cherry.
The linkage analysis described above suggested that a member of the Pto gene family in 96T133-3 is responsible for conferring resistance to Pst(avrPto). To isolate members of the Pto gene family from 96T133-3, we hybridized a 96T133-3 cDNA library with the Pto gene. Restriction fragment analysis of 46 independent cDNAs indicated that there were four different classes of cDNAs (data not shown). We sequenced representatives from each class and found that each class represented a different LhirPto gene that shared between 83% and 97% nucleotide identity with Pto. Each gene contained an open reading frame encoding a protein that is 314–323 amino acids in length (Fig. 5, which is published as supplemental data on the PNAS web site, www.pnas.org).
The entire Pto locus has been sequenced from the tomato lines Rio Grande-PtoR (RG-PtoR) and VFNT Cherry (VFNT) which are resistant and susceptible to Pst(avrPto), respectively (GenBank accession nos. AF220602 and AF220603). An analysis of these data indicated that the RG-PtoR and VFNT genes present at corresponding locations within the Pto gene cluster (orthologs) are more similar to each other than they are to other genes within their respective haplotypes (paralogs) (18). On the basis of these data the Pto family members were named by assigning each gene a prefix to denote the species from which it was identified, and a suffix that arbitrarily numbers each family member. Orthologs in VFNT and RG-PtoR were given the same number but were not numbered in any particular order (GenBank accession nos. AF220602 and AF220603). To simplify future comparisons of the Pto loci from various tomato species we propose a new nomenclature for the Pto locus. With this system a prefix will identify the wild species from which the gene was isolated, and a letter suffix will denote the location of the gene along tomato chromosome 5, based on its orthology to the RG-PtoR Pto genes. A summary of our proposed nomenclature is presented in Table 1.
Table 1.
Proposed nomenclature for the Pto gene family
| Rio Grande-PtoR
ortholog
|
VFNT ortholog
|
L.
hirsutum ortholog
|
||
|---|---|---|---|---|
| Current* | Proposed† | Current* | Proposed† | Proposed† |
| LpimPth4* | LpimPtoA | LescPth4 | LescPtoA | — |
| Fen | LpimPtoB/Fen | fen | LescPtoB/fen | LhirPtoB/Lhirfen |
| LpimPth3 | LpimPtoC | LescPth3 | LescPtoC | — |
| LpimPth2 | LpimPtoD | LescPth2 | LescPtoD | LhirPtoD |
| Pto | LpimPtoE/Pto | Not present | Not present | LhirPtoE/LhirPto |
| LpimPth5 | LpimPtoF | LescPth5 | LescPtoF | LhirPtoF |
Nomenclature for Pto orthologs and paralogs is based on genomic sequences of the Pto loci from Rio Grande-PtoR and VFNT Cherry (GenBank accession nos. AF220602 and AF220603, respectively).
Proposed gene names are based on their species of origin and designated in alphabetical order according to their position within the Pto loci of L. pimpinellifolium and L. esculentum. A — denotes that cDNAs corresponding to these orthologs were not found in this study.
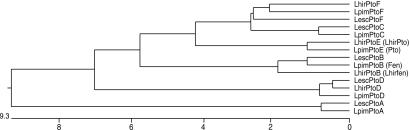
We compared the nucleotide sequences of the LhirPto genes with the Pto gene families from RG-PtoR (LpimPto) and VFNT Cherry (LescPto). A dendrogram based on nucleotide identity placed each LhirPto gene with the orthologous genes from RG-PtoR and VFNT (Fig. 3). We named each L. hirsutum Pto gene according to its putative ortholog from RG-PtoR and VFNT by using the nomenclature presented in Table 1. LhirPtoE is most closely related to the Pto gene from L. pimpinellifolium (RG-PtoR), with 97% nucleotide identity. Because of its close sequence identity (and functional identity, see below) with Pto we refer to LhirPtoE as simply LhirPto. Interestingly, the Pto and LhirPto genes do not appear to have an ortholog in the susceptible VFNT haplotype (GenBank accession no. AF220603). LhirPtoB is most closely related to the genes Fen (LpimPtoB) and fen (LescPtoB) and shares 95% and 96% nucleotide identity with them, respectively (26, 31). Because of its close sequence identify to Fen and fen we refer to LhirPtoB as Lhirfen. LhirPtoD is nearly identical to the genes LescPtoD (99% nucleotide identity) and LpimPtoD (98% nucleotide identity) genes from VFNT and RG-PtoR haplotypes, respectively. LhirPtoF is 93% identical to both LescPtoF and LpimPtoF. LescPtoF was previously isolated from L. esculentum line Ailsa Craig and reported to be the ortholog (susceptible allele) of Pto (26). Recent data, however, show the L. esculentum line VFNT Cherry does not contain a Pto ortholog (GenBank accession no. AF220603). It is possible, therefore, that in Ailsa Craig LescPtoF is actually a Pto paralog that is directly adjacent to the location where a Pto ortholog would be if it were present in the VFNT haplotype Pto gene cluster.
Figure 3.
Relationships among the Pto gene family. Phylogenetic tree based on nucleotide sequence identity comparing LhirPto genes with Pto gene family members from Rio Grande-PtoR (LpimPto, GenBank accession no. AF220602) and VFNT Cherry (LescPto, GenBank accession no. AF220603). Scale units represent the number of substitution events.
We did not find L. hirsutum cDNA clones corresponding to two predicted open reading frames (PtoA and PtoC) that are present in both RG-PtoR and VFNT. It is possible that the Pto locus has undergone rearrangements since the L. hirsutum species diverged, resulting in loss of these Pto paralogs. Previous studies have suggested that such rearrangements have taken place within the L. esculentum Pto locus, resulting in the deletion of a Pto family member (18). However, it is more likely that these genes are simply not expressed in leaf tissue, from which our cDNA library was derived.
LhirPto Encodes an Active Kinase That Elicits an avrPto-Dependent Hypersensitive Response (HR).
Gene-for-gene resistance often manifests as rapid, localized cell death, termed the HR, that occurs at the site of attempted pathogen ingress. Both tomato and tobacco cells undergo an HR when the Pto resistance gene and the avrPto avirulence gene are expressed within the same cell. Previous studies also demonstrated that for an avrPto-dependent HR to occur, the Pto kinase must possess autophosphorylation activity (4, 5).
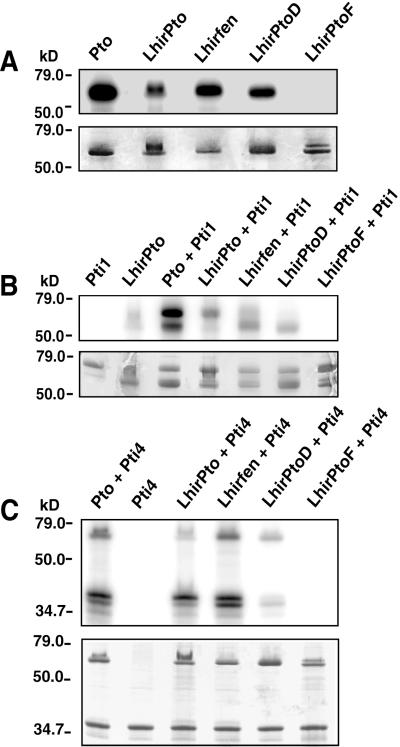
To determine whether the LhirPto proteins have autophosphorylation activity we expressed each one as a GST fusion and performed in vitro kinase assays. Autoradiography revealed that LhirPto, Lhirfen, and LhirPtoD each autophosphorylated (Fig. 4A). We were unable, however, to detect autophosphorylation by LhirPtoF. Comparing LhirPtoF with other functional protein kinases revealed that LhirPtoF contains a serine residue (Ser-45) in a position normally occupied by an invariant glycine in the nucleotide-binding motif (33). This substitution might be responsible for the observed lack of autophosphorylation activity.
Figure 4.
Phosphorylation assays of the LhirPto proteins. (A) Pto and the LhirPto proteins were expressed in bacteria as GST fusions and purified on glutathione-agarose. Each protein (2 μg) was incubated in a kinase reaction with [γ-32P]ATP, separated by SDS/PAGE, and analyzed by autoradiography. (Upper) Autoradiogram of autophosphorylated LhirPto proteins. (Lower) Coomassie blue-stained gel. The locations of protein standards used to estimate molecular masses are indicated in kilodaltons (kD). (B) Assay to test phosphorylation of Pti1 by LhirPto proteins. GST-LhirPto proteins (2 μg) were incubated with 2 μg of purified kinase-deficient GST-Pti1-(K69N) in the presence of [γ-32P]ATP, separated by SDS/PAGE, and analyzed by autoradiography. (Upper) Autoradiogram of phosphorylated proteins. (Lower) Coomassie blue-stained gel. (C) Assay to test phosphorylation of Pti4 by LhirPto proteins. GST-LhirPto proteins (2 μg) were incubated with His-tagged Pti4 (2 μg) in the presence of [γ-32P]ATP, separated by SDS/PAGE, and analyzed by autoradiography. (Upper) Autoradiogram of phosphorylated proteins. (Lower) Coomassie blue-stained gel.
Pto recognition specificity for AvrPto is highly specific as Fen, which shares 87% amino acid similarity with Pto, does not elicit an avrPto-dependent HR. A threonine residue (Thr-204) located within the catalytic domain of Pto determines recognition specificity of the kinase for AvrPto (8). Mutation of Thr-204 in Pto abolishes its ability to elicit an HR when it is coexpressed with avrPto. Interestingly, LhirPto is the only active LhirPto kinase to contain a threonine in the position corresponding to Thr-204, suggesting this protein may elicit an avrPto-dependent HR. To test this possibility, we placed each L. hirsutum Pto gene under control of the cauliflower mosaic virus 35S promoter and used Agrobacterium to transiently express them with avrPto in leaves of Nicotiana benthamiana. Indeed, expression of LhirPto with avrPto in N. benthamiana induced an HR 3–5 days after infiltration identical to coexpression of Pto and avrPto (Table 2; Fig. 6, which is published as supplemental data on the PNAS web site at www.pnas.org). Expression of either LhirPto or avrPto alone did not elicit the HR. Lhirfen, LhirPtoD, and LhirPtoF were unable to elicit an HR when expressed either with or without avrPto. These results indicate that LhirPto is the functional Pto gene from L. hirsutum var. glabratum and that Lhirfen, LhirPtoD, and LhirPtoF do not mediate avrPto-specific resistance.
Table 2.
Agrobacterium-mediated transient expression assays of the LhirPto genes
| Gene | HR |
|---|---|
| LhirPto | − |
| avrPto | − |
| Pto/avrPto | + |
| LhirPto/avrPto | + |
| Lhirfen/avrPto | − |
| LhirPtoD/avrPto | − |
| LhirPtoF/avrPto | − |
The indicated genes were placed under control of the cauliflower mosaic virus 35S promoter and cloned into the pBTEX plasmid. These constructs were transformed into Agrobacterium tumefaciens strain EHA105. A. tumefaciens strains were induced with acetosyringone and syringe-infiltrated into mature Nicotiana benthamiana leaves. Leaves were scored for HR 4 days after infiltration. + indicates that total tissue collapse occurred in the infiltrated tissue; − indicates that no tissue collapse was observed.
LhirPto Physically Interacts with AvrPto and Pti Proteins.
Only Pto alleles which interact with AvrPto in a yeast two-hybrid system can mediate avrPto-specific resistance to P. syringae (4, 5, 34). We tested each LhirPto gene in the yeast two-hybrid system to determine whether the proteins encoded by them could interact with AvrPto. We cloned each LhirPto gene in a bait plasmid and performed Western blotting to confirm that each LhirPto-LexA fusion protein was expressed in yeast (data not shown). After confirming that none of the bait constructs activated the lacZ reporter gene on their own, we transformed each yeast strain with an AvrPto prey construct and tested for lacZ activation (5). Only LhirPto activated the lacZ reporter gene when expressed with the AvrPto prey, indicating that LhirPto specifically interacts with AvrPto (Table 3).
Table 3.
Physical interactions of the LhirPto proteins with AvrPto and Pto-interacting proteins
| AvrPto | Pti1 | Pti4 | Pti5 | Pti6 | |
|---|---|---|---|---|---|
| Pto | ++++ | +++ | ++ | ++ | +++ |
| LhirPto | ++++ | − | ++ | +++ | +++ |
| Lhirfen | − | − | − | − | − |
| LhirPtoD | − | − | − | ++++ | ++++ |
| LhirPtoF | − | − | − | − | − |
| Bicoid | − | − | − | − | − |
EGY48 yeast cells expressing the LhirPto, Pto, or Bicoid proteins from the bait plasmid pEG202 were transformed with AvrPto, Pti1, Pti4, Pti5, or Pti6 in the prey plasmid pJG4-5. Ten independent transformants for each bait–prey combination were grown on galactose medium containing X-gal and lacking uracil, histidine, and tryptophan to test for lacZ reporter gene activation. Representative colonies were scored on the following scale: −, completely white; ++++, dark blue.
Pti1, Pti4, Pti5, and Pti6 encode putative downstream members of the Pto signaling pathway (27, 28). We next tested whether the products of the LhirPto genes could interact with these signaling molecules. LhirPto activated lacZ expression when it was coexpressed with Pti4, Pti5, or Pti6, indicating that LhirPto physically interacts with these transcription factors (Table 3). Surprisingly, LhirPto did not appear to interact with the Pti1 kinase. Neither Lhirfen nor LhirPtoF interacted with any of the Pti proteins tested. LhirPtoD was unable to interact with Pti1 or Pti4 but did interact with Pti5 and Pti6, suggesting that the residues of Pto (and LhirPto) that are required for interaction with these proteins are conserved in this kinase.
LhirPto Phosphorylates Pti1 and Pti4.
We have reported previously that the Pto kinase phosphorylates both Pti1 and Pti4 in vitro (27, 29). Phosphorylation of Pti1 occurs on residue Thr-233, which is present in the activation domain of Pti1 (34). Thr-233 is also required for the interaction of Pto and Pti1 in a yeast two-hybrid assay, and phosphorylation of Pti1 by Pto may be required for downstream signaling mediated by Pti1. Phosphorylation of Pti4 by Pto enhances the affinity of Pti4 for PR gene promoter elements in mobility-shift assays (29). We performed in vitro kinase assays to determine whether the LhirPto proteins could phosphorylate kinase-deficient Pti1(K69N) or Pti4. Although we were unable to detect interaction of LhirPto with Pti1 in a yeast two-hybrid assay, LhirPto did phosphorylate Pti1 in vitro (Fig. 4B). The ability to phosphorylate Pti1 was specific to LhirPto, as none of the other LhirPto proteins phosphorylated Pti1. LhirPto also phosphorylated Pti4 in vitro (Fig. 4C). However, unlike the phosphorylation of Pti1, the ability to phosphorylate Pti4 was not specific to LhirPto as the Lhirfen protein also strongly phosphorylated Pti4.
Discussion
We have presented evidence that, in common with the product of the Pto gene isolated from L. pimpinellifolium, the L. hirsutum Pto kinase interacts specifically with the Pseudomonas AvrPto protein and mediates an avrPto-specific HR in an N. benthamiana transient expression assay. We have further demonstrated that the LhirPto kinase interacts with downstream members of the Pto pathway, suggesting that these signaling components are also conserved among different tomato species. These results indicate that the specificity of the Pto gene for the Pseudomonas AvrPto protein evolved before the divergence of L. hirsutum and L. pimpinellifolium.
Evidence for an Ancient Origin of Pto Kinase Recognition Specificity.
The Pto locus itself appears to be of ancient origin. Pto homologs map to syntenic regions in tomato, potato, and pepper, and the genome organization of the Pto locus is mostly conserved between L. esculentum and L. pimpinellifolium, indicating that the locus predates the divergence of these species (35, 36). Despite many decades of disease resistance breeding, however, R genes have yet to be identified that map to the Pto locus in potato or pepper. This observation indicates that a role in pathogen recognition and disease resistance for the Pto gene family might have evolved subsequent to the divergence of the Lycopersicon lineage or, alternatively, has since been lost from potato and pepper.
We earlier confirmed that the L. hirsutum accession PI134418 expresses avrPto-specific resistance and mapped that resistance to the Pto locus (22). We have now demonstrated that a member of the Pto gene family in PI134418, LhirPto, encodes a protein that bears remarkable sequence identity and functional similarity to the L. pimpinellifolium Pto kinase. The presence of functional Pto orthologs in two distantly related tomato species suggests that Pto evolved the capability to recognize the AvrPto protein before the divergence of L. pimpinellifolium and L. hirsutum. We cannot, of course, exclude the possibility that Pto may possess an additional function that predates AvrPto recognition and is the driving force behind the conservation of Pto. However, the absence of Pto in L. esculentum and other Lycopersicon species (see below) demonstrates that any putative secondary function is dispensable. It seems more likely, therefore, that Pto and AvrPto coevolved and that AvrPto recognition provided the impetus to conserve the resistance function of Pto.
In contrast to AvrPto recognition specificity, sensitivity to fenthion, which has been identified only in L. pimpinellifolium, appears to be more recent. Fen orthologs have been identified in both L. esculentum and L. hirsutum and share 97% and 95% nucleotide identity with Fen, respectively. However only the Fen kinase is capable of recognizing fenthion. Thus relatively few (and perhaps recent) changes in the Fen amino acid sequence appear to underlie the specificity involved in this recognition mechanism. Similarly, single amino acid substitutions within the Pto and Fen kinases can confer or eliminate recognition of AvrPto (8, 34, 37). Despite the apparent delicate nature of recognition specificity in the kinase class of R gene, 17 amino acid differences that have arisen to distinguish Pto and LhirPto do not disrupt Pto disease resistance function. This further suggests that there has been selective pressure to maintain AvrPto recognition specificity.
It is interesting to note that Pst resistance originating in the cultivated tomato, L. esculentum, has yet to be identified. Although some L. esculentum varieties express avrPto-specific Pto-mediated resistance, RFLP analyses show that this resistance likely originated in L. pimpinellifolium (ref. 38; G.B.M., unpublished data). Furthermore, it has been reported recently that Pto orthologs are entirely absent from two different L. esculentum haplotypes (18). Why would a gene that has retained its recognition specificity over much of Lycopersicon evolution be absent from an entire species? A simple explanation is that during the course of domestication of wild L. esculentum a genetic bottleneck occurred that resulted in the selection of L. esculentum lines that, by chance alone, lacked the Pto gene. An alternative possibility is that in the absence of Pst expressing the avrPto gene there is a fitness cost associated with the Pto locus in L. esculentum that has resulted in selection against lines containing Pto either in nature or in cultivated fields (39). Tanksley and colleagues (25) compared 17 processing traits between L. esculentum near-isogenic lines with and without the introgressed Pto locus. They concluded that with respect to these 17 traits, Pto/Pto plants were not significantly different from pto/pto plants. Therefore it seems unlikely that the Pto locus exacts a fitness cost that might result in negative selection. It remains a possibility that the Pto locus confers deleterious effects on fitness that might be difficult to detect experimentally.
LhirPto and Pto Mediate Resistance Through a Conserved Signaling Pathway.
We have demonstrated that in an L. esculentum background LhirPto mediates avrPto-specific disease resistance that is indistinguishable from that conferred by the Pto gene. We infer from this observation that Pto and LhirPto use the same signaling components to effect Pseudomonas resistance in L. esculentum. Our present study demonstrates that Pto and LhirPto do indeed interact with a similar array of downstream components. Previous investigations have identified L. esculentum signaling molecules with which Pto interacts both physically and biochemically (27–29). In common with the Pto kinase, LhirPto interacts with the transcription factors Pti4, Pti5, and Pti6 in the yeast two-hybrid system and phosphorylates Pti4 in vitro. On the basis of these results we speculate that, similar to Pto, the interaction and phosphorylation of Pti4 by LhirPto enhances the binding of Pti4 to the GCC box cis element to activate PR gene expression (29). Although we were unable to detect an interaction between the LhirPto and Pti1 proteins in the yeast two-hybrid system, LhirPto retains the ability to specifically phosphorylate Pti1 in an in vitro kinase assay. The structural and functional similarities that exist between LhirPto and Pto and the phosphorylation of Pti1 by LhirPto in vitro suggest that LhirPto does use the Pti1 kinase in defense signaling. Taken together, our data suggest that L. esculentum, L. pimpinellifolium, and the more distantly related L. hirsutum share a common signaling pathway that mediates resistance to bacterial speck disease. Considering that the Pto and LhirPto genes also function in N. benthamiana, this pathway likely predates speciation within the Solanaceae.
An Ancient Origin of AvrPto Recognition Specificity Supports the Trench Warfare Hypothesis.
Several recent reports have presented evidence that the LRR class of R genes evolve rapidly to develop new recognition specificities (16–19, 40). These reports indicate that the LRR domain is under diversifying selection to create novel recognition capabilities in response to pathogen pressure and thus support an “arms-race” model for R–avr gene evolution (41). In contrast, a comparison of nucleotide sequences from the RPM1 locus of different Arabidopsis ecotypes suggested that some R–avr gene pairs have coexisted for millions of years (20). According to the “trench warfare” hypothesis proposed in that study, gene-for-gene interactions are ancient and polymorphism at R gene loci is governed by frequency-dependent selection as a function of both disease frequency and a “cost of resistance” associated with the R gene.
We have reported genetic and molecular analyses demonstrating that Pto orthologs from L. hirsutum and L. pimpinellifolium mediate identical recognition specificity for AvrPto. These findings indicate that AvrPto recognition arose before these two species diverged and suggest that Pto and AvrPto have coexisted for millions of years. Surveys of wild tomato germ plasm have previously shown that not all Lycopersicon accessions are resistant to Pst, and recent sequencing of the Pto locus in VFNT Cherry found that a Pto ortholog is not even present in that L. esculentum haplotype (18, 42, 43). Thus significant functional polymorphism exists at the Pto locus in Lycopersicon. In conjunction with the ancient origins of the Pto–AvrPto interaction, the polymorphic nature of the Pto locus supports the trench-warfare hypothesis in which the “rise and fall” of a specific R gene maintains variation for disease resistance in a plant population (20).
Supplementary Material
Acknowledgments
We thank Steve Tanksley for providing seeds of 96T133-3, TA537, and TA209, Richard Michelmore for sharing unpublished data, and Yong Gu for sharing Pti1 and Pti4 protein and for assisting with the phosphorylation assays. We also thank Alan Collmer, Molly Jahn, and Todd Vision for their critical reading of the manuscript. This work was supported in part by National Research Initiative Competitive Grants Program/U.S. Department of Agriculture Award 99-35301-7973, National Science Foundation Grant MCB-9896308, and a David and Lucile Packard Foundation Fellowship (to G.B.M.). B.K.R. was supported, in part, by a National Science Foundation Training Grant at Purdue University.
Abbreviations
- LRR
leucine-rich repeat
- RFLP
restriction fragment length polymorphism
- Pst, Pseudomonas syringae pv. tomato
GST, glutathione S-transferase
- HR
hypersensitive response
Footnotes
References
- 1.Martin G B, Brommonschenkel S H, Chunwongse J, Frary A, Ganal M W, Spivey R, Wu T, Earle E D, Tanksley S D. Science. 1993;262:1432–1436. doi: 10.1126/science.7902614. [DOI] [PubMed] [Google Scholar]
- 2.Thilmony R T, Chen Z, Bressan R A, Martin G B. Plant Cell. 1995;7:1529–1536. doi: 10.1105/tpc.7.10.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galan J E, Collmer A. Science. 1999;284:1322–1328. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- 4.Scofield S R, Tobias C M, Rathjen J P, Chang J H, Lavelle D T, Michelmore R W, Staskawicz B J. Science. 1996;274:2063–2065. doi: 10.1126/science.274.5295.2063. [DOI] [PubMed] [Google Scholar]
- 5.Tang X, Frederick R D, Zhou J, Halterman D A, Jia Y, Martin G B. Science. 1996;274:2060–2063. doi: 10.1126/science.274.5295.2060. [DOI] [PubMed] [Google Scholar]
- 6.van Dijk K, Fouts D E, Rehm A H, Hill A R, Collmer A, Alfano J R. J Bacteriol. 1999;181:4790–4797. doi: 10.1128/jb.181.16.4790-4797.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shan L, Thara V K, Martin G B, Zhou J-M, Tang X. Plant Cell. 2000;12:2323–2337. doi: 10.1105/tpc.12.12.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frederick R D, Thilmony R L, Sessa G, Martin G B. Mol Cell. 1998;2:241–245. doi: 10.1016/s1097-2765(00)80134-3. [DOI] [PubMed] [Google Scholar]
- 9.Johnson L N, Noble M E M, Owen D J. Cell. 1996;85:149–158. doi: 10.1016/s0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]
- 10.Leach J E, White F F. Annu Rev Phytopathol. 1996;34:153–179. doi: 10.1146/annurev.phyto.34.1.153. [DOI] [PubMed] [Google Scholar]
- 11.Ellis J G, Lawrence G J, Luck J E, Dodds P N. Plant Cell. 1999;11:495–506. doi: 10.1105/tpc.11.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jia Y, McAdams S A, Bryan G J, Hershey H P, Valent B. EMBO J. 2000;29:4004–4014. doi: 10.1093/emboj/19.15.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leister R T, Katagiri F. Plant J. 2000;22:345–354. doi: 10.1046/j.1365-313x.2000.00744.x. [DOI] [PubMed] [Google Scholar]
- 14.Thomas C, Jones D, Parniske M, Harrison K, Balint-Kurti P J, Hatzixanthis K, Jones J. Plant Cell. 1997;9:2209–2224. doi: 10.1105/tpc.9.12.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caicedo A L, Schaal B A, Kunkel B N. Proc Natl Acad Sci USA. 1999;96:302–306. doi: 10.1073/pnas.96.1.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDowell J M, Dhandaydham M, Long T A, Aarts M G, Goff S, Holub E B, Dangl J L. Plant Cell. 1998;10:1861–1874. doi: 10.1105/tpc.10.11.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parniske M, Hammond-Kosack K E, Golstein C, Thomas C M, Jones D A, Harrison K, Wulff B B, Jones J D. Cell. 1997;91:821–832. doi: 10.1016/s0092-8674(00)80470-5. [DOI] [PubMed] [Google Scholar]
- 18.Michelmore R W, Meyers B C. Genome Res. 1998;8:1113–1130. doi: 10.1101/gr.8.11.1113. [DOI] [PubMed] [Google Scholar]
- 19.Simons G, Groenendijk J, Wijbrandi J, Reijans M, Groenen J, Diergaarde P, Van der Lee T, Bleeker M, Onstenk J, de Both M, et al. Plant Cell. 1998;10:1055–1068. doi: 10.1105/tpc.10.6.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stahl E A, Dwyer G, Mauricio R, Kreitman M, Bergelson J. Nature (London) 1999;400:667–671. doi: 10.1038/23260. [DOI] [PubMed] [Google Scholar]
- 21.Ashfield T, Danzer J R, Held D, Clayton D, Keim P, Maroof M A S, Webb D M, Innes R W. Theor Appl Genet. 1998;96:1013–1021. [Google Scholar]
- 22.Tanksley S, Brommonschenkel S, Martin G. Rep Tomato Genet Coop. 1996;46:28–29. [Google Scholar]
- 23.Miller J C, Tanksley S D. Theor Appl Genet. 1990;80:437–448. doi: 10.1007/BF00226743. [DOI] [PubMed] [Google Scholar]
- 24.Ausubel F, Brent R, Kingston R E, Moore D, Smith J A, Struhl K. Protocols in Molecular Biology. New York: Wiley; 1987. [Google Scholar]
- 25.Tanksley S, Bernacchi D, Fulton T M, Beck-Bunn T, Emmatty D, Eshed Y, Inai S, Lopez J, Petiard V, Sayama H, et al. Rep Tomato Genet Coop. 1997;47:40–42. [Google Scholar]
- 26.Jia Y, Loh Y-T, Zhou J, Martin G B. Plant Cell. 1997;9:61–73. doi: 10.1105/tpc.9.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou J, Loh Y-T, Bressan R A, Martin G B. Cell. 1995;83:925–935. doi: 10.1016/0092-8674(95)90208-2. [DOI] [PubMed] [Google Scholar]
- 28.Zhou J, Tang X, Martin G B. EMBO J. 1997;16:3207–3218. doi: 10.1093/emboj/16.11.3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu Y, Yang C-H, Venkatappa T K, Zhou J, Martin G B. Plant Cell. 2000;12:771–785. doi: 10.1105/tpc.12.5.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loh Y-T, Martin G B. Proc Natl Acad Sci USA. 1995;92:4181–4184. doi: 10.1073/pnas.92.10.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin G B, Frary A, Wu T, Brommonschenkel S, Chunwongse J, Earle E D, Tanksley S D. Plant Cell. 1994;6:1543–1552. doi: 10.1105/tpc.6.11.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laterrot H, Moretti A. Rep Tomato Genet Coop. 1989;39:21–22. [Google Scholar]
- 33.Hanks S, Hunter T. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- 34.Sessa G, D'Ascenzo M, Martin G B. EMBO J. 2000;19:2257–2269. doi: 10.1093/emboj/19.10.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grube R C, Radwanski E R, Jahn M. Genetics. 2000;155:873–877. doi: 10.1093/genetics/155.2.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leister D, Ballvora A, Salamini F, Gebhardt C. Nat Genet. 1996;14:421–429. doi: 10.1038/ng1296-421. [DOI] [PubMed] [Google Scholar]
- 37.Rathjen J P, Chang J H, Staskawicz B J, Michelmore R W. EMBO J. 1999;18:3232–3240. doi: 10.1093/emboj/18.12.3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pitblado R E, MacNeill B H. Can J Plant Pathol. 1983;5:251–255. [Google Scholar]
- 39.Simms E L. Bioscience. 1996;46:136–145. [Google Scholar]
- 40.Noël L, Moores T L, van der Biezen E A, Parniske M, Daniels M, Parker J, Jones J. Plant Cell. 1999;11:2099–2111. [PMC free article] [PubMed] [Google Scholar]
- 41.Dawkins R, Krebs J R. Proc R Soc London. 1979;205:489–511. doi: 10.1098/rspb.1979.0081. [DOI] [PubMed] [Google Scholar]
- 42.Lawson V F, Summers W L. Plant Dis. 1984;68:139–141. [Google Scholar]
- 43.Pilowsky M. Plant Dis. 1982;66:46–47. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.