Abstract
The phosphatidylinositol 3-kinase-mammalian target of rapamycin (PI3K-mTOR) pathway plays pivotal roles in cell survival, growth, and proliferation downstream of growth factors. Its perturbations are associated with cancer progression, type 2 diabetes, and neurological disorders. To better understand the mechanisms of action and regulation of this pathway, we initiated a large scale yeast two-hybrid screen for 33 components of the PI3K-mTOR pathway. Identification of 67 new interactions was followed by validation by co-affinity purification and exhaustive literature curation of existing information. We provide a nearly complete, functionally annotated interactome of 802 interactions for the PI3K-mTOR pathway. Our screen revealed a predominant place for glycogen synthase kinase-3 (GSK3) A and B and the AMP-activated protein kinase. In particular, we identified the deformed epidermal autoregulatory factor-1 (DEAF1) transcription factor as an interactor and in vitro substrate of GSK3A and GSK3B. Moreover, GSK3 inhibitors increased DEAF1 transcriptional activity on the 5-HT1A serotonin receptor promoter. We propose that DEAF1 may represent a therapeutic target of lithium and other GSK3 inhibitors used in bipolar disease and depression.
The phosphatidylinositol 3-kinases (PI3Ks)1 are a conserved family of lipid kinases that phosphorylate phosphatidylinositol on phosphoinositide 3′-hydroxyl group, forming second messengers. Class IA PI3K (hereafter “PI3K”) is activated by growth factor receptor tyrosine kinases, and upon stimulation by insulin or IGF1, PI3K triggers the formation of phosphatidylinositol 3,4,5-trisphosphate, which recruits and activates kinases such as AKT and PDK1, mediating most effects of insulin and IGF1 on cell metabolism, growth, proliferation, and differentiation (1, 2). Downstream of AKT, the mammalian target of rapamycin (mTOR) kinase is an essential activator of protein synthesis, promoting cell growth and proliferation (1, 3, 4). mTOR is regulated by growth factors through AKT, by energy availability through the AMP-activated kinase (AMPK), and by amino acid content through class III PI3K. Glycogen synthase kinase-3 (GSK3) is another major target of the PI3K pathway, and its inhibitory phosphorylation by AKT relieves its negative impact on cell cycle progression and cell growth (5). The PI3K-mTOR pathway is central for cell metabolism and proliferation, and its perturbation is implicated in many human diseases (1, 3, 4, 6). Mutations leading to PI3K-mTOR pathway activation are important steps in the initiation and progression of tumors and are frequently encountered in human cancers. In contrast, down-regulation of the PI3K pathway impairs cell responses to insulin, leading to type 2 diabetes. Perturbations of the PI3K-mTOR pathway are also linked to muscle atrophy and autoimmune and cardiovascular diseases. Moreover, GSK3 dysregulation is associated with mood disorders and Alzheimer disease (7). Several components of the PI3K-mTOR pathway are promising targets for antitumoral, metabolic, and neurological therapies (3, 4, 7).
The complexity of the PI3K-mTOR pathway necessitates innovative strategies to identify its exact involvement in physiology and pathology and to predict the consequences of its manipulation in therapy. A better comprehension of cell responses to PI3K-mTOR pathway activation may come from the identification of new regulators or effectors of this pathway, and this goal can now be reached via high throughput approaches (8–11). We conducted a large scale yeast two-hybrid screen of 33 components of the PI3K-mTOR pathway. The resulting interactions were supplemented with a manually curated set of literature interactions, providing a comprehensive and annotated interactome for the PI3K-mTOR pathway. Our screen revealed a predominant place for GSK3A, GSK3B, and AMPK and highlights their role in cancer, metabolic diseases, immune response, and neurological disorders. In particular, we characterized a functional interaction of GSK3A and GSK3B with deformed epidermal autoregulatory factor-1 (DEAF1) transcription factor in the serotonergic pathway.
EXPERIMENTAL PROCEDURES
Cloning, Yeast Two-hybrid (Y2H) Screens, and Co-affinity Purification (Co-AP) Experiments
Detailed descriptions of cloning, Y2H screens, and co-AP experiments are available in the supplemental material and methods. Briefly, full-length ORFs for our baits were cloned, using the Gateway technology, as DNA binding domain (DB) and activation domain (AD) expression vectors and transformed in MaV203 and MaV103 yeast strains. DB expression vectors were used for screening with an AD-cDNA library from E10.5 mouse embryo, and both DB and AD expression vectors were used for screening with the hORFeome1.1 library as described previously (9, 12, 13). The activation of three reporter genes (HIS3, URA3, and lacZ) was assessed in comparison with controls, and an interaction was considered positive if at least two reporter genes were activated. For the cDNA screen, after selection of in-frame cDNAs, interactions were retested using the gap repair technique (14). The gap repair technique was also used to test interactions with GSK3A/GSK3B and RHEB/RHEBL1 constructs. A subset of 40 interactions was further tested in HEK293T cells using full-length ORFs cloned as GST-bait and Myc-prey vectors using the Gateway technology. Experiments were essentially performed as described (9).
Literature-completed Interactome
For each selected PI3K-mTOR component, the Biomolecular Interaction Network Database (BIND) (15), Molecular INTeraction database (MINT) (16), human protein reference database (HPRD) (17), Agile Protein Interaction DataAnalyzer (APID) (18), and PubMed databases were investigated for protein-protein interactions (PPIs). Each PPI was verified in the corresponding study to check the exact reference of the protein and the technique. Only binary interactions were retained, corresponding to the followings techniques: yeast or mammalian two-hybrid experiments, in vitro kinase or other enzymatic assays with relevant controls, in vitro binding of recombinant proteins purified from mammalian cell-free systems, binding of a protein purified from mammalian cell-free systems to a membrane-immobilized protein, crystallography, and surface plasmon resonance analysis. Careful attention was paid to check that the interaction could not be indirect due to a third component. Co-immunoprecipitation and GST pulldown performed in vivo were not retained. However, in vitro kinase assays involving mTOR or STK11/LKB1 kinases were often performed with kinases isolated from cells as a complex (mammalian target of rapamycin complexes 1 and 2 for mTOR and in complex with MO25 and STE20-related adapter protein for STK11). If controls for these specific kinase activities were suitable, interactions were included with the corresponding indication. Interactions involving a purified protein for which the exact isoform could not be determined were not retained. Described interactions mostly involve human proteins but sometimes involve proteins from mouse or other mammals. For each interaction, the PMID reference referring to the study describing the binary interaction is indicated.
Functional annotation of each interactor for its molecular and subcellular functions and its pathway involvement was deduced from NCBI-Gene and PubMed data banks. Proteins were classified according to a limited number of chosen terms rather than to gene ontology annotations that were either much too detailed or not accurate enough according to the considered protein. As a protein can be engaged in several PPIs with different components of the PI3K-mTOR pathway, interactors may be redundant, whereas interactions are not. Therefore, the annotation of interactors is formally referred to as the annotation of interactions. Figs. 2, 3, and 4 were realized using Cytoscape software (19).
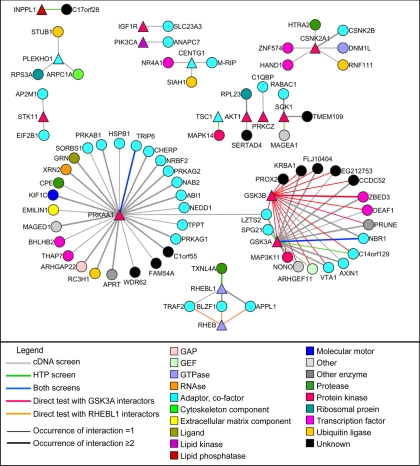
Fig. 2.
Yeast-two hybrid interaction network for PI3K-mTOR pathway. Y2H baits are depicted are triangles, and the discovered interactors are depicted as circles. Baits for which no interaction was found are not represented. The color of the links is indicative of the Y2H method that led to its discovery. The thickness of the links is indicative of the number of times an interaction was found in the Y2H cDNA or high throughput (HTP) screen. The molecular function of each protein is described by a color code. GAP, GTPase-activating protein.
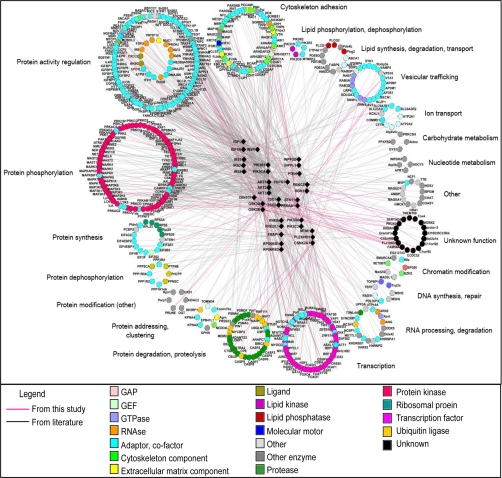
Fig. 3.
Functional annotation of literature-completed interactome of PI3K-mTOR pathway. PI3K-mTOR pathway components are depicted are triangles, and interactors are depicted as circles. Pink links represent interactions found in this study, and gray links are manually curated interactions from the literature. The molecular function of each protein is described by a color code, and interactors are grouped according to their subcellular functions. GAP, GTPase-activating protein.
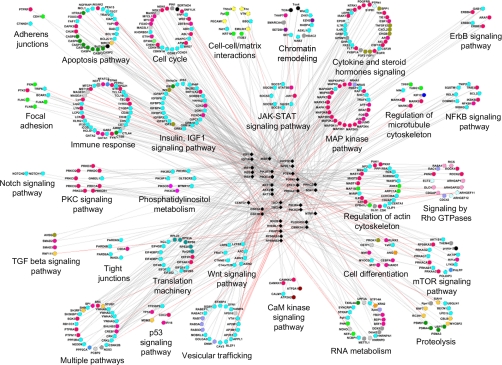
Fig. 4.
Connections of literature-completed interactome of PI3K-mTOR pathway with other signaling pathways. PI3K-mTOR pathway components as depicted as triangles, and interactors are depicted as circles. Pink links represent interactions found in this study, and gray links are manually curated interactions from the literature. The molecular function of each protein is described by a color code, and interactors are grouped according to their belonging to specific signaling pathways. GAP, GTPase-activating protein; TGF, transforming growth factor; PKC, protein kinase C; MAP, mitogen-activated protein; NFKB, nuclear factor κB; JAK, Janus kinase; STAT, signal transducer and activator of transcription; CaM, calmodulin.
Kinase Assay with GSK3, MLK3, and DEAF1 Constructs
Protein Purification
GST-GSK3A, GST-GSK3B, GST-GSK3A-KA, and GST-GSK3B-KA Gateway vectors were transfected in HEK293T cells as described for co-AP experiments. Cells were lysed with co-AP lysis buffer for 30 min on ice. Lysates were sonicated and precleared by centrifugation for 10 min at 14,000 rpm at 4 °C. Precleared lysates were incubated with immobilized glutathione beads (Perbio) for 1 h at 4 °C. Beads were washed extensively three times with lysis buffer. GST fusion proteins were eluted with PBS, 50 mm glutathione (Perbio) for 15 min at room temperature. Aliquots of eluates were run on a polyacrylamide-SDS gel in parallel to a BSA scale. The gel was stained with a Coomassie solution, and the quantity of GST fusion proteins was estimated. Adjusted amounts of proteins were then analyzed by Western blot using anti-GST antibody (see Fig. 6B, left gel). Myc-DEAF1, Myc-DEAF1-1, Myc-DEAF1-2, Myc-DEAF1-3 Gateway and pCDN3-M2-MLK3 (kindly provided by K. A. Gallo) vectors were transfected in HEK293T cells as described for co-AP experiments. Each well of 6-well plates was lysed with 250 μl of IP buffer 1 (300 mm NaCl, 50 mm Tris, pH 7.4, 0.5 mm EDTA, 1% Triton X-100, Complete protease inhibitor mixture (Roche Applied Science)) for 30 min on ice. Lysates were sonicated and precleared by centrifugation for 10 min at 14,000 rpm at 4 °C. 250 μl of IP buffer 2 (50 mm Tris, pH 7.4, 0.5 mm EDTA, 1% Triton X-100, Complete protease inhibitor mixture (Roche Applied Science)) and mouse anti-Myc 9E10 antibody (Covance) or anti-M2 FLAG (Sigma) were added to the lysates for overnight agitation at 4 °C. Protein A- and G-Sepharose beads were added for 1 h at 4 °C and then extensively washed with IP buffer 3 (150 mm NaCl, 50 mm Tris, pH 7.4, 0.5 mm EDTA, 1% Triton X-100, Complete protease inhibitor mixture (Roche Applied Science)). All three IP buffers were supplemented with phosphatase inhibitors (200 μm NaF, 1 mm sodium orthovanadate, 100 μm β-glycerophosphate). Finally, beads were resuspended in 1× kinase buffer (25 mm Hepes, pH 7.4, 10 mm MgCl2), and 10 μl of beads suspensions were run on a polyacrylamide-SDS gel, transferred to PVDF membranes, and analyzed by Western blotting with anti-Myc (see Fig. 6C) and -M2 (see Fig. 6B, right gel) antibodies to estimate the quantity of Myc-DEAF1 and M2-MLK3 proteins.
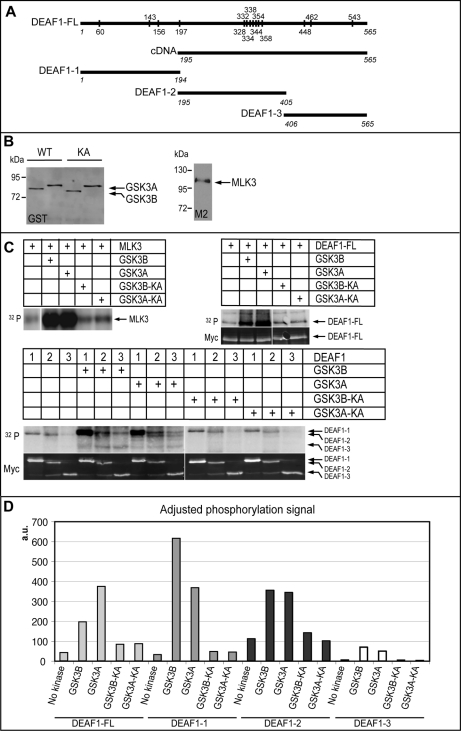
Fig. 6.
DEAF1 in vitro phosphorylation by GSK3A and GSK3B kinases. A, DEAF1-FL, DEAF1-1, DEAF1-2, and DEAF1-3 constructs used for kinase assay are represented in parallel to the cDNA clone that was isolated from the Y2H screen. Numbers in italic indicate the first and last amino acids of the constructs in comparison with DEAF1-FL. Conserved residues between human and mouse that are potential sites of phosphorylation by GSK3A and GSK3B are shown on the DEAF1-FL construct. B, quantification of purified wild-type (WT) and kinase-dead (KA) GST-GSK3A and GST-GSK3B as well as M2-MLK3 by Western blot with anti-GST and -M2 antibodies, respectively. C, upper parts, 32P radioactive signal from kinase assay reveals MLK3 (positive control), DEAF1-FL, DEAF1-1, DEAF1-2, and DEAF1-3 protein phosphorylation by wild-type GSK3A and GSK3B kinases. Lower parts, quantitative fluorescent Western blot for Myc on the kinase assay membranes reveals the respective amount of Myc-DEAF1 proteins. D, adjusted quantification of DEAF1 construct phosphorylation by GSK3A and GSK3B was calculated as the ratio (32P radioactive signal/Myc signal quantification) for each DEAF1 construct. a.u., arbitrary units.
Kinase Assay
Similar amounts of GST-GSK3 proteins (around 70 ng) were added to Myc-DEAF1 or M2-MLK3 recombinant proteins (around 50 ng). The mixture was adjusted to final concentrations of 25 mm Hepes, pH 7.4 and 10 mm MgCl2. 30μCi of [γ-32P]ATP were added to the mixture. After incubation for 30 min at 30 °C, the reaction was stopped by addition of 5× loading buffer and denaturation at 95 °C for 5 min. Aliquots run on polyacrylamide-SDS gels and transferred to PVDF membranes were revealed using a phosphorimaging system (Fuji) for radioactivity measures.
Quantification of Myc-DEAF1 Constructs
The membranes were then subjected to Western blotting with mouse anti-Myc (9E10) and goat anti-mouse IRDye 800 secondary antibody (LI-COR) to reveal and quantify Myc-DEAF1 proteins on an Odyssey LI-COR scanner (ScienceTec). Adjusted radioactive signal from Myc-DEAF1 proteins was calculated by the ratio radioactive signal/protein quantity.
RT-PCRs for 5-HT1A and DEAF1
mRNAs were extracted from RN46A and HEK293T cells using a NucleoSpin RNA II kit (Macherey-Nagel). 500 ng of gel-verified RNA were retrotranscribed using RevertAid H Minus Moloney murine leukemia virus reverse transcriptase (Fermentas). DNA from RN46A cells was extracted by lysis in 5 mm EDTA, 5 mm Tris, pH 8, 100 mm NaCl, 0.2% SDS, 0.3 μg/μl proteinase K followed by precipitation. PCR were performed using Qiagen Taq polymerase on the retrotranscribed products to check the expression of actin, 5-HT1A, and Deaf1 in RN46A cells and of actin and DEAF1 in HEK293T cells. Pairs of oligos used for 5-HT1A amplification were the following: sense and antisense 5-HT1A oligos (20), sense 5-HT1A (20) plus 5-HT1A new antisense oligo (GCGGTGCCGACGAAGTTC), and 5-HT1A-FOR (CCGCACGCTTCCGAATC) plus 5-HT1A-REV (ACCTGGCTGTCCGTTCAG) oligos. Oligos used for Deaf1 amplification were the following: rDeaf1-FOR (TGCACCTGTGCTGCCTGTTG) and rDeaf1-REV (CGATCTGGCAGCTGTCCTGAT) (on rat RN46A products) and hDEAF1-FOR (GAACGCGGCATCCATCTCAG) and hDEAF1-REV (CTTGCGTTGGCAGAAGGTGG) (on human HEK293T products). PCR for actin and Deaf1 were positive on all tested RT products as well as PCR for 5-HT1A on RN46A cells DNA (data not shown). The identity of RT-PCR products for Deaf1 from RN46A and 293T cells was assessed by sequencing.
Plasmids and Luciferase Assays
Plasmids
The luciferase plasmid 5-HT1A(C) was kindly provided by Dr. Paul R. Albert's laboratory and is described in Lemonde et al. (20). GST, DEAF1-Myc, and Myc Gateway vectors were described above. pCMV-SPORT-βgal (Invitrogen) and pGFP-S6 (21) plasmids were used as reporter plasmids for normalization.
Transfections and Drug Treatment
HEK293T cells were grown at 37 °C in 5% CO2 in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Cells were seeded in 12-well plates and transfected using calcium chloride. Each well was transfected with 200 ng of 5-HT1A(C) vector; 50 ng of pCMV-SPORT-βgal; 150 ng of GST, Deaf1-Myc, or Myc vector; and 400 ng of pcDNA3 to complete the DNA mixture. Drugs were added at transfection. Lithium and sodium chloride were added at a final concentration of 5 mm. Azakenpaullone (Calbiochem) was diluted in DMSO and added at a final concentration of 1 μm or 500 nm. An equivalent volume of DMSO (0.25 μl) was added in control wells. Medium was changed 12 h after transfection, and drugs were replaced at the same concentrations.
RN46A cells (kindly provided by Dr. Scott Whittemore's laboratory) were grown at 33 °C in 5% CO2 in Neurobasal medium (Invitrogen) supplemented with 10% fetal bovine serum and glutamine. Cells were seeded in 12-well plates. The transfection mixture was composed of 100 μl of Opti-MEM (Invitrogen); 4 μl of FuGENE HD (Roche Applied Science); 200 ng of 5-HT1A(C) vector; 50 ng of CMV-GFP vector; 150 ng of GST, Deaf1-Myc, or Myc vector; and 400 ng of pcDNA3. Drugs were added in the same conditions as for HEK293T cells except that that the medium was not changed after transfection.
Luciferase Assay
Cells were harvested 36 h after transfection. HEK293T cells were lysed with 250 μl of Passive Lysis Buffer (Promega). 10 μl of lysate were used for luciferase activity detection using the Luciferase Assay System (Promega). 50 μl of lysate were used for β-galactosidase activity detection and were added to 50 μl of detection buffer containing 1 mm MgCl2, 50 μm β-mercaptoethanol, and 40 μg of ortho-nitrophenyl-β-galactoside. After incubation at 37 °C, the plate was read at 405 nm.
RN46A cells were dissociated with Versene solution (Invitrogen) and resuspended in 1 ml of PBS. 200 μl were used for fluorescence-activated cell sorting analysis (BD Biosciences) of GFP-transfected cells. The remaining cells were lysed with 100 μl of Reporter Lysis Buffer (Promega). 10 μl of lysate were used for luciferase activity detection using the Luciferase Assay System (Promega). 10 μl of lysate were used for the protein assay using Bio-Rad Protein Assay.
Statistical Analysis
Independent experiments were performed at least six times. Adjusted luciferase activity was corrected for transfection efficiency and cell density by calculating the ratio of luciferase activity/β-galactosidase activity for HEK293T cells. As β-galactosidase activity detection was not sensitive enough for normalization in RN46A cells (data not shown), adjusted luciferase activity was calculated as the ratio of luciferase activity/GFP transfection rate/protein concentration. Statistical analysis was performed using Student's t test.
RESULTS
Cloning of PI3K-mTOR Pathway Components
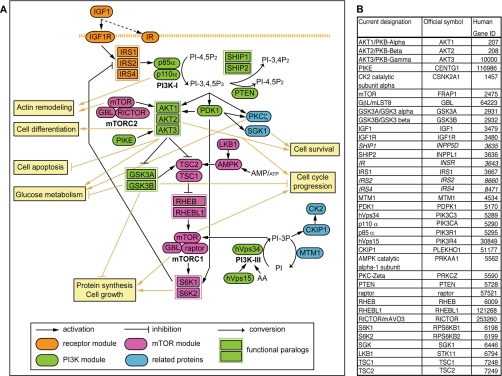
The PI3K-mTOR pathway is multimodular, each functional module integrating various inputs for downstream effectors (1, 2). Thirty-seven components, considered as essential mediators involved in IGF1 signaling, were selected (Fig. 1). They are schematized as part of a receptor module (IGF1, IGF1 receptor, insulin receptor, IRS1, IRS2, and IRS4), a PI3K module (p85α, p110α, hVps34, hVps15, SHIP1, SHIP2, PTEN (phosphatase and tensin homolog), PDK1, AKT1, AKT2, AKT3, phosphoinositide 3-kinase enhancer, GSK3A, and GSK3B), an mTOR module (mTOR, G protein β subunit-like, RICTOR (RPTOR-independent companion of mTOR), raptor, TSC1, TSC2, RHEB, RHEBL1, S6K1, S6K2, LKB1, and AMPK), or a related protein module (protein kinase C ζ, SGK1, CK2, CKIP1, and MTM1). The functional paralogy for RHEB and RHEBL1 or GSK3A and GSK3B is indicated. A description of all components is provided in supplemental Data 1.
Fig. 1.
Description of PI3K-mTOR pathway. A, functional relationships between the proteins of the PI3K-mTOR pathway and major outputs of the pathway activation. Modules are color-coded: orange, green, pink, and blue correspond to the receptor, the PI3K, the mTOR, and the related protein modules, respectively. Functional paralogs are depicted as joined rectangles. PI, phosphatidylinositol; PIP, phosphatidylinositide phosphate. B, current designations of the PI3K-mTOR pathway components are presented in parallel to their official symbols and to their gene ID. Genes indicated in italic could not be included in our yeast-two hybrid screen but were integrated in the literature-completed interactome. GβL, G protein β subunit-like; IR, insulin receptor; PIKE, phosphoinositide 3-kinase enhancer; PKB, protein kinase B; PTEN, phosphatase and tensin homolog; IGF1R, insulin-like growth factor 1 receptor.
Selected components were cloned by Gateway recombinational cloning into Y2H destination vectors to generate GAL4 DB or AD fusion proteins (9, 22). Insulin receptor, IRS2, IRS4, and SHIP1 failed to clone, leaving 33 constructs for Y2H screening.
Yeast Two-hybrid Screens and Co-affinity Purification Experiments
As the PI3K-mTOR pathway is conserved and ubiquitously expressed, it was important to screen using the broadest possible libraries. Two stringent Y2H screens were carried out, one with an E10.5 mouse embryo library and the other with the human ORFeome v1.1 ORF library (9, 22). Both screens followed optimized protocols in which high quality parameters are assessed (14, 23, 24).
In total, the cDNA and the ORFeome screens identified 68 and eight PPIs, respectively, involving 15 PI3K-mTOR pathway components (Fig. 2 and supplemental Table I). Two PPIs were found in both screens, showing the complementarity of these screens. Three PPIs have been described to occur in the literature, and four other interactions found with GSK3A had been previously characterized with its close paralog GSK3B, giving 67 of 74 (91%) new potential interactions (supplemental Table II). There were 35 PPIs (47%) found at least twice independently (Fig. 2 and supplemental Table I). GSK3A and PRKAA1, the AMPK α1 catalytic subunit, were the most connected proteins with 17 and 27 PPIs, respectively.
Paralogs in the screens had sharply different numbers of PPIs. We found four PPIs for RHEBL1 but none for RHEB and 17 PPIs for GSK3A but two for GSK3B. Using the gap repair technique (14), we tested the interaction between RHEBL1 interactors and RHEB. Y2H interactions were retrieved with RHEB and RHEBL1 only when their last amino acids that constitute a prenylation motif (25) were removed (supplemental Fig. 1). In this case, RHEB interacted with three of four interactors of RHEBL1 (supplemental Fig. 1, Fig. 2, and supplemental Table I).
To validate Y2H interactions, we tested half (n = 40) of the PPIs by co-AP assays in human HEK293T cells (9). Transfected proteins were expressed in 33 co-APs, and 19 co-APs (58%) tested positive (supplemental Fig. 2 and supplemental Table I), which is in line with expectation from previous such validation efforts (26, 27). The protein interactions reported here have been submitted to the International Molecular Exchange (IMEx) Consortium (http://imex.sf.net) through IntAct (28) (PMID 17145710) and assigned the identifier IM-11703.
Literature-completed Interactome
We manually curated from the literature all binary interactions for each PI3K-mTOR pathway component. Verified interactions were added to our new interactions in a resulting “literature-completed interactome” containing 802 distinct interactions (supplemental Table II). The interactors were annotated for their molecular function and their subcellular functions based on validated data from the literature (supplemental Table II and Fig. 3). Accordingly, 648 interactors were annotated for their participation in particular signaling pathways (supplemental Table II and Fig. 4).
GSK3A and GSK3B Protein-Protein Interactions
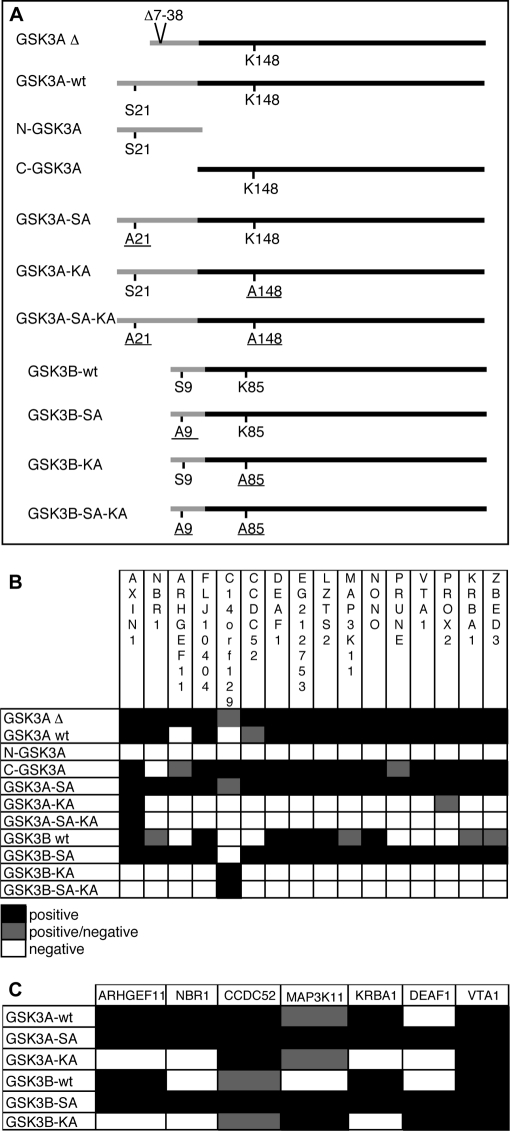
The GSK3A and GSK3B paralogs have high sequence similarity and almost identical kinase domains (29) but have divergent N-terminal regions, spanning from amino acids (aa) 1 to 90 for GSK3A and from 1 to 27 for GSK3B (Fig. 5A). To determine whether GSK3A PPIs involved its specific N-terminal domain or the homologous part shared with GSK3B, we made the following constructs: full-length GSK3A (GSK3A-wt) and GSK3B (GSK3B-wt), N-GSK3A (aa 1–90), and C-GSK3A (aa 91–483) (Fig. 5A). Two additional constructs, GSK3A-SA and GSK3B-SA, had Ser-21 and Ser-9, respectively, mutated to alanine to render them constitutionally active kinases (30, 31). These constructs can be compared with the initial GSK3A construct used in our screens (GSK3AΔ), which has an in-frame deletion of 31 amino acids from aa 7 to 38, removing the negative Ser-21 regulatory site (29) (Fig. 5A). These six constructs were screened against 16 Y2H interactors of GSK3AΔ (Fig. 5B). GSK3A-SA behaved as GSK3AΔ, interacting with all tested interactors, and GSK3A-wt exhibited 15 interactions. N-GSK3A did not interact with any GSK3AΔ interactor, indicating that these PPIs did not involve the GSK3A-specific N-terminal domain. In contrast, C-GSK3A interacted with 14 GSK3AΔ interactors. GSK3B-SA interacted with 15 of the 16 interactors, whereas GSK3B-wt exhibited 10 interactions. In conclusion, all tested interactions involved the homologous parts of GSK3A and GSK3B. Constitutive activation of these kinases facilitated Y2H interaction, especially for GSK3B.
Fig. 5.
Interactions for GSK3A and GSK3B variant constructs. A, the different constructs used for Y2H and co-AP experiments are presented. The black lines represent the homologous parts of the proteins, whereas the divergent parts are in gray. Amino acids Ser-21 and Ser-9 are inhibitory sites, and Lys-148 and Lys-85 are essential residues for the catalytic sites in GSK3A and GSK3B, respectively. Mutated residues are underlined. B, the table recapitulates results obtained for Y2H experiments with the different GSK3A and GSK3B constructs. The experiments were performed twice except for GSK3A-SA-KA and GSK3B-SA-KA constructs that were tested once. A positive/negative result reflects a discrepancy between both experiments except for GSK3AΔ-C14orf129 interaction that was twice at the positive threshold. C, co-AP results for a set of interactors are presented. Experiments were done once, and a positive/negative result reflects an ambiguous profile for Myc detection in eluates.
To test whether the kinase activity of GSK3A and GSK3B was necessary for these Y2H interactions, we cloned the kinase-dead forms, GSK3A-KA and GSK3B-KA, in which Lys-148 and Lys-85, respectively, were replaced by an alanine (32, 33). To prevent a deleterious phosphorylation on Ser-21 and Ser-9, we also constructed GSK3A-SA-KA and GSK3B-SA-KA forms, respectively, combining both described mutations. The KA mutations abolished most interactions for both kinases independently of the presence of the inactivation site. There was only one interactor for GSK3B-KA and two for GSK3A-KA (Fig. 5B).
We tested seven of these interactions by co-AP in HEK293T cells (Fig. 5C and supplemental Table I). GSK3A-SA and GSK3B-SA interacted with all tested GSK3AΔ interactors; GSK3A-wt and GSK3B-wt interacted with six and four GSK3AΔ interactors, respectively; and GSK3A-KA and GSK3B-KA interacted with three and four GSK3AΔ interactors, respectively. Seemingly, the co-AP assay was less sensitive to GSK3 kinase activity than Y2H.
In conclusion, all GSK3AΔ tested interactors were shared by GSK3B and were favored by constitutive activation of the kinases, whereas kinase inactivation abolished most interactions. Different sets of interactors were observed for wild-type, constitutively active, and inactive forms of GSK3A and GSK3B (Fig. 5, B and C), suggesting specificities for these two kinases.
DEAF1 Is a New GSK3 Substrate
Our Y2H screens identified the DEAF1 (also called NUDR) transcription factor as a new interactor for GSK3A and GSK3B, and these interactions were confirmed by co-AP assay in human cells (Figs. 2 and 5, supplemental Fig. 2, and supplemental Table I).
We tested whether DEAF1 was a substrate for GSK3A and GSK3B kinase activity. GSK3A or GSK3B mutated for kinase activity showed impaired Y2H interactions with DEAF1 (Fig. 5B). DEAF1 has 14 putative conserved sites for GSK3 phosphorylation ((S/T)XXX(S/T) sequences where S/T should be primed by another kinase (34)) with a stretch of seven sites from aa 328 to 358, a region also present in the mouse DEAF1 cDNA isolated in the cDNA Y2H screen (aa 195–565) (Fig. 6A). Several Myc-DEAF1 fusion proteins were tested for their capacities to be phosphorylated by GSK3A and GSK3B: full-length DEAF1 (DEAF1-FL, aa 1–565), DEAF1-1 (aa 1–194), DEAF1-2 (aa 195–405), and DEAF1-3 (aa 406–565) (Fig. 6A). Wild-type and kinase-dead GSK3A and GSK3B kinases were purified and used in in vitro kinase assays (Fig. 6, B and C). MLK3, a recently identified GSK3 substrate (Ref. 35 and our screen) was used as positive control (Fig. 6, B and C). DEAF1-FL, DEAF1-1, and DEAF1-2 proteins were phosphorylated by wild-type but not kinase-dead forms of GSK3A and GSK3B (Fig. 6C). The DEAF1-FL and DEAF1-2 proteins were phosphorylated to a similar extent, whereas DEAF1-1 showed a little more phosphorylation (Fig. 6, C and D). DEAF1-3 phosphorylation levels remained at background in all conditions (Fig. 6, C and D). DEAF1 likely represents a substrate for GSK3A and GSK3B kinases with several phosphorylation sites distributed from aa 1 to 405.
DEAF1 Transcriptional Activity upon 5-HT1A Promoter Is Increased by GSK3 Inhibition
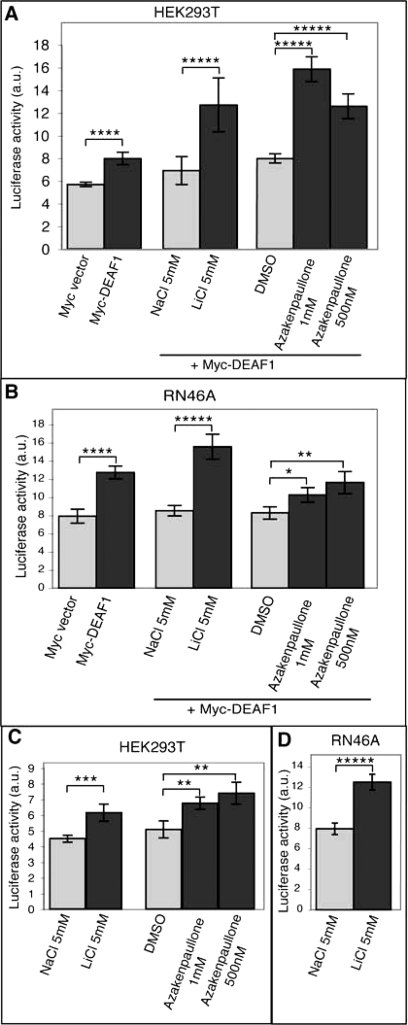
We next tested whether GSK3 kinase activity affected DEAF1 transcriptional activity. A major transcriptional target for DEAF1 is the 5-HT1A serotonin receptor gene (20, 36). We examined DEAF1 transcriptional activity upon GSK3 inhibition with a luciferase reporter under the control of 5-HT1A promoter (20). Transfection of the full-length Myc-DEAF1 construct into HEK293T cells caused luciferase activity to increase (Fig. 7A; p < 0.005). Both lithium and azakenpaullone, two inhibitors of GSK3 acting through different mechanisms (37, 38), significantly increased Myc-DEAF1 activity as reflected by increased luciferase activity in HEK293T cells (Fig. 7A; p < 0.001). Similarly, Myc-DEAF1 transfection into rat RN46A serotonergic cells markedly increased luciferase activity (Fig. 7B; p < 0.005), and treatment by lithium or azakenpaullone stimulated Myc-DEAF1 activation of luciferase activity. Whereas lithium stimulated luciferase activity in RN46A cells as strongly as in HEK293T cells (p < 0.001), azakenpaullone only moderately stimulated luciferase activity (p < 0.05 at 1 μm and p < 0.025 at 500 nm).
Fig. 7.
Activation of 5-HT1A promoter by DEAF1 and GSK3 inhibitors. A and B, results of luciferase assays on HEK293T cells (A) and RN46A cells (B) show the influence of DEAF1 on 5-HT1A promoter activity alone or in addition to GSK3 inhibitors, i. e. lithium chloride and azakenpaullone, in comparison with control conditions (Myc vector, sodium chloride, and dimethyl sulfoxide, respectively). C and D, results of luciferase assays on HEK293T cells (C) and RN46A cells (D) show the influence of GSK3 inhibitors on 5-HT1A promoter activity in comparison with control conditions. Independent experiments were performed at least six times. The luciferase level was adjusted to the transfection conditions (see “Experimental Procedures”). The histograms show means ± S.E. Statistical analysis was performed using Student's t test. *, p < 0.05; **, p < 0.025; ***, p < 0.01; ****, p < 0.005; *****, p < 0.001. a.u., arbitrary units.
DEAF1 is broadly expressed (39), and RT-PCR experiments confirmed that DEAF1 was expressed in HEK293T and RN46A cells (data not shown; see “Experimental Procedures”). We tested the effect of GSK3 inhibition upon endogenous DEAF1 activity. In both cell lines, luciferase activity under the control of 5-HT1A promoter was significantly increased by GSK3 inhibitors with the strongest stimulation by lithium in RN46A cells (Fig. 7, C and D; p < 0.001). In summary, DEAF1 activation of 5-HT1A promoter activity increased upon GSK3 inhibition in HEK293T and RN46A cells, suggesting an inhibitory role of GSK3 phosphorylation on DEAF1.
DISCUSSION
Protein-Protein Interaction Mapping for PI3K-mTOR Pathway
Y2H mapping for 33 components of the PI3K-mTOR pathway against two different libraries identified 74 interactions, 67 of which have not been described previously. This screen revealed two highly connected proteins: PRKAA1, the AMPK α1 catalytic subunit (27 interactors) and GSK3A (17 interactors). Literature curation provided a set of 802 functionally annotated PPIs for the PI3K-mTOR pathway (literature-completed interactome) that offers a comprehensive picture of the connectivity and the predominant cellular processes involving the PI3K-mTOR pathway (supplemental Data 2). The low overlap between our screen and the interactions from the literature (seven of 74) may be explained by two parameters: sampling sensitivity and assay sensitivity. Sampling sensitivity refers to the interactions that can be identified in a single trial of an assay. Sampling sensitivity was estimated at 45% in a comparable Y2H screen (24), and six screens were needed to reach 90% saturation. Successive iterations of our screen would likely achieve a higher degree of saturation. In parallel, assay sensitivity designates the fraction of all interactions that can be identified by an assay with its specific experimental conditions. In parallel to the high stringency of the Y2H criteria used in this study (low copy plasmids and two reporters activated), its intrinsic limitations may include the cDNA and ORF isoforms and the level of representation in the libraries used, the activation status, and the post-translational modifications of the baits in yeast that may be different from the ones encountered in previous screens. Moreover, full-length baits, instead of domains, may have prevented some PPIs (27), and lipid-interacting domains present in several components of the PI3K-mTOR pathway may have impaired the nuclear translocation required in Y2H screening. Interestingly, assay sensitivity for different methods of PPI identification was estimated to a similar value around 20–35%, and our screen probably lies in the same range (24). Altogether, sampling and assay sensitivities may explain the complementarities of the different approaches used in PPI identification for PI3K-mTOR components. The newly discovered interactions from this study provide insights on GSK3A and GSK3B specificities and suggest new paths for the comprehension of PI3K-mTOR pathway involvement in cancer, metabolism, diabetes, immune response, and neurobiology.
Implications for GSK3A and GSK3B Specificities
GSK3A and GSK3B kinases are highly related paralog kinases that share a large redundancy, although recent studies revealed specific functions for each of them (29, 32, 40–43). Understanding isoform redundancies and specificities represents an important stake for cell biology and drug development (44). However, GSK3A received little attention compared with GSK3B. Our screens identified 17 common interactors for GSK3A and GSK3B, reinforcing a functional redundancy between them. Y2H interactions were nearly abrogated when kinase-dead forms of GSK3A and GSK3B were used, indicating that these interactors may be substrates for both kinases.
However, all interactors did not interact with the same strength and with the equivalent constructs for each kinase. For example, C14orf129 interacted with C-GSK3A and constitutively active GSK3A but with kinase-dead GSK3B in Y2H. These different affinities for GSK3A and GSK3B, depending on their activation status, may lead in vivo to a preferential interaction with one of these two kinases or to a sequestration by one of them. Beyond an apparent redundancy, the status of GSK3A and GSK3B for their kinase activity may represent a determinant element of their specificities. Characterization of these respective interactions could help understanding the coordinated activities of these kinases and developing focused strategies for targeting specific interactions.
PI3K-mTOR Pathway and Cancer
Many components of the PI3K-mTOR pathway act as oncogenes or tumor suppressor genes, and mutations leading to an activation of the PI3K-mTOR pathway may account for 30% of human cancers (45). Many drugs targeting PI3K, AKT, or mTOR are currently tested for cancer therapy (44). Several interactions discovered in our Y2H may participate in specific aspects of tumor initiation or progression.
Our screen identified a new interaction between TSC1 and MAPK14 (p38α), one of the four members of the p38 mitogen-activated protein kinase family. p38 mitogen-activated protein kinases regulate inflammatory and stress responses and behave as tumor suppressors (46). The TSC1-TSC2 complex inhibits mTOR, a translational activator promoting cell growth and proliferation, and mutations in TSC1 or TSC2 lead to a tumor syndrome (47). p38 may regulate mTOR activation through TSC1 phosphorylation.
New interactions may link the PI3K-mTOR pathway to the Wnt-β-catenin pathway, which is also involved in tumorigenesis (48). GSK3 is at the crossroad of these two signaling pathways. Upon activation of Wnt signaling pathway, GSK3, in complex with Axin, phosphorylates β-catenin and targets it to degradation (34). In our Y2H screens, GSK3A and GSK3B interacted with LZTS2 and ZBED3. LZTS2 tumor suppressor promotes β-catenin nuclear export, impairing its transcriptional activity (49). ZBED3, an Axin-binding protein, inhibits β-catenin phosphorylation by GSK3B (50). The interactions among GSK3, LZTS2, and ZBED3 suggest new coordination for β-catenin inhibition, ensuring a precise control of this potent oncogene.
AMPK, the “cell energy sensor,” inhibits mTOR through the activation of the TSC1-TSC2 complex (51), and AMPK-activating drugs are candidate for cancer therapies (52). Interestingly, LZTS2 was also found as a new interactor for PRKAA1, suggesting a putative coordination between GSK3 and AMPK in β-catenin regulation. A similar link between these two kinases already exists in sequential phosphorylation and activation of TSC2 (53). This interaction between AMPK and LZTS2 may reveal another role for AMPK in cancer.
Moreover, AMPK is involved in the control of epithelial polarity, actin organization, and cell proliferation downstream of LKB1 (54–56). We found a new interaction between PRKAA1 and Abelson interactor protein-1 (ABI1). ABI1 regulates actin polymerization, and its expression correlates with cell migration and invasiveness of cancer cell lines (57, 58). ABI1 may thus mediate part of AMPK control of cell polarity and tumor progression.
Finally, we describe a new interaction between PRKAA1 and KIF1C, a plus-end microtubule motor that promotes podosome dynamics (59). KIF1C expression is highly predictive of brain metastasis in lung cancer (60), and immunotherapy against KIF1C appears promising in glioma (61). We suggest that KIF1C interaction with AMPK may modulate its activity in cancer progression.
PI3K-mTOR Pathway, Metabolism, and Diabetes
AMPK is activated upon a high AMP/ATP ratio and turns on ATP-generating processes while switching off ATP-consuming activities (62). AMPK-activating drugs, such as aminoimidazole carboxamide ribonucleotide and metformin, are used in type 2 diabetes treatment. It was recently shown that AMPK regulates the circadian clock of peripheral organs (63). We describe a new interaction between PRKAA1 and BHLHB2 (or DEC1), a transcription factor regulating the expression of metabolic and clock genes that are required for circadian rhythm (64, 65). This interaction may participate in the control of circadian rhythm by AMPK, likely adapting metabolism to animal activity.
Carboxypeptidase E (CPE) was another new interactor of PRKAA1. Mutation of CPE is associated with type 2 diabetes (66–68). The regulation of CPE activity by AMPK could contribute to the therapeutic benefits of AMPK activation in type 2 diabetes.
GSK3 is another prime target for type 2 diabetes management (48). In our screen, GSK3A and GSK3B interacted with ARHGEF11 (or PDZ-RhoGEF), a small GTPase activator. ARHGEF11 polymorphism is associated with insulin resistance and type 2 diabetes (69–71). Several small GTPases are implicated in the trafficking of glucose transporter 4 (GLUT4) following insulin stimulation (72). We propose that GSK3 interaction with ARHGEF11 may regulate glucose uptake in response to insulin through the modulation of GLUT4 transport.
Finally, we describe a new interaction between RHEB/RHEBL1 and APPL1, an adaptor protein that mediates adiponectin-induced sensitization of the insulin pathway (73). APPL1 activates AMPK, leading to S6K inhibition and subsequent release of IRS1 inhibition (see Fig. 1) (74). This likely occurs through RHEB inhibition as RHEB overexpression impairs the APPL1 effect (74). APPL1 direct interaction with RHEB and RHEBL1 may thus reinforce APPL1-mediated sensitization of insulin signaling.
PI3K-mTOR Pathway and Immune Response
The PI3K-mTOR pathway participates in many aspects of the immune response, and rapamycin, an mTOR inhibitor, is used as an immunosuppressive drug (75, 76). We found a new interaction between RHEB/RHEBL1, two mTOR activators, and TRAF2, an adaptor protein that lies downstream of TNFR1 and CD40 receptors and mediates the activation of the MAPK and nuclear factor κB pathways (77). TRAF2 participates in various aspects of the immune response (78, 79), and RHEB negatively regulates MAPK activation through B-Raf inhibition (80). TRAF2 interaction with RHEB and RHEBL1 may represent another possible level of control of immune responses by PI3K downstream of tumor necrosis factor α and CD40 ligand.
In addition, we identified an interaction between PRKAA1 and RC3H1 (or Roquin). In mouse, Roquin mutation impairs the degradation of particular mRNAs, leading to the development of severe autoimmunity (81, 82). AMPK may influence Roquin activity and modulate certain aspects of autoimmunity.
PI3K-mTOR Pathway and Neurobiology
During development, GSK3 regulates neurite extension and neuronal architecture (5). In the adult, GSK3 has been implicated in specific behavioral responses (83). In our Y2H screens, GSK3A and GSK3B interacted with ARHGEF11, which increases EAAT4 glutamate transporter activity in brain and participates in neuronal morphogenesis (84, 85). GSK3 interaction with ARHGEF11 may explain part of GSK3 involvement in synaptic plasticity. More broadly, we suggest that this interaction may participate in regulated trafficking in different contexts, such as the insulin pathway (see above) and neurogenesis.
Our Y2H screens also found SPG21 (or Maspardin) interacting with GSK3A. SPG21 mutation leads to hereditary spastic paraplegia, a neurodegenerative disease associated with dementia (86). Spastic disorders are also encountered in hereditary cases of Alzheimer disease (87). Importantly, GSK3 is involved in the progression of Alzheimer disease, and chemical inhibition of GSK3 alleviates β-amyloid accumulation (7). The interaction between GSK3A and SPG21 may reveal a mechanistic proximity between hereditary spastic paraplegia and Alzheimer disease.
Possible Involvement of DEAF1-GSK3 Interaction in Mood Disorders
Finally, we identified DEAF1 transcription factor as an interactor and an in vitro substrate for GSK3A and GSK3B. GSK3 inhibitors increased DEAF1 transcriptional activity on the 5-HT1A promoter in both HEK293T and RN46A cells, suggesting an inhibitory regulation of DEAF1 by GSK3.
Variations in 5-hydroxytryptamine (serotonin) (5-HT) availability and signaling cascade are associated with psychiatric disorders such as major depression disease. Among several receptors, the 5-HT1A receptor for serotonin is a major target of antidepressant treatments (88–91). Presynaptic 5-HT1A receptors act as inhibitory autoreceptors in serotonergic neurons located in midbrain raphe nuclei. On the contrary, postsynaptic 5-HT1A receptors are found in 5-HT-responsive areas that are limbic regions and specific cortical layers.
In previous reports, DEAF1 repressed 5-HT1A expression in HEK293T cells and in the serotonergic RN46A cells that express 5-HT1A but enhanced 5-HT1A expression in non-serotonergic 5-HT1A-expressing cells such as SN48, NG108-15, and SKN-SH cells (20, 36). However, DEAF1 enhanced 5-HT1A expression in several hippocampal and septal cells (20). These data likely reflect an opposite regulation on presynaptic versus postsynaptic 5-HT1A expression (36) and perhaps a phenotypic derivation of the HEK293T cells and RN46A cells we used. The RN46A cells we used did not express 5-HT1A (data not shown) and may behave as non-serotonergic cells. The stimulation of 5-HT1A expression by DEAF1 that we observed in both cell lines is in accordance with neither of them expressing 5-HT1A.
A reduction of 5-HT1A expression is observed in postsynaptic areas (88, 92–95) and in raphe nuclei (88, 92, 96, 97) during depression or bipolar disease (91). Accordingly, a blunted response to 5-HT1A activation has been associated with depression. Most antidepressants increase serotonin transmission and lead to improved 5-HT1A function in postsynaptic areas (88, 89, 91, 92). These data suggest that increasing 5-HT1A expression should alleviate depression symptoms.
In parallel, lithium has long been used in the treatment of bipolar disease and as an adjuvant to antidepressant drugs (38, 83, 88). The preeminent mechanism of action of lithium in psychiatric disorders is likely represented by GSK3 inhibition (7, 38). Accordingly, lithium behavioral responses are increased by GSK3B haploinsufficiency and reproduced by another GSK3 inhibitor (98), whereas GSK3B haploinsufficiency and GSK3B inhibition alleviates aberrant behaviors due to 5-HT deficiency (99). Other mood-stabilizing agents such as clozapine and valproate also inhibit GSK3 activity (83, 100). Conversely, an increased GSK3B activity is observed in the prefrontal cortex of major depression disease subjects (101), whereas both 5-HT1A and DEAF1 were decreased in the prefrontal cortex of depressed women, suggesting a positive regulation of 5-HT1A transcription by DEAF1 in these areas (95). Importantly, treatment with lithium or divalproex increased 5-HT1A expression in bipolar disorder patients (102). Our finding that GSK3 phosphorylates DEAF1 and impairs DEAF1-driven expression of 5-HT1A suggests that abnormal GSK3B activation reduces 5-HT1A expression, contributing to depressive symptoms, whereas GSK3 inhibition by lithium or another drug would increase DEAF1-driven 5-HT1A expression in 5-HT-responsive areas and restore 5-HT transmission. GSK3 and DEAF1 interaction might explain part of GSK3 involvement in bipolar disease and depression, and DEAF1 may represent a therapeutic target of lithium and other GSK3 inhibitors used in these disorders.
Acknowledgments
We are grateful to J. Woodgett, P. R. Albert, S. Whittemore, K. A. Gallo, J. J. Zhao, T. M. Roberts, M. Billaud, C. Erneux, G. Thomas, and K. Salehi-Ashtiani for sharing reagents and to P. Lamesch, T. Hao, and K. Gauthier for technical advice.
* This work was supported in part by Association de Recherche sur le Cancer Grant 3853.
 This article contains supplemental Figs. 1 and 2, Data 1 and 2, Tables I and II, and materials and methods.
This article contains supplemental Figs. 1 and 2, Data 1 and 2, Tables I and II, and materials and methods.
1 The abbreviations used are:
- PI3K
- phosphatidylinositol 3-kinase
- ABI1
- Abelson interactor protein-1
- AD
- activation domain
- AKT
- protein kinase B
- AMPK
- AMP-activated kinase
- APPL1
- adaptor protein-containing pleckstrin homology domain, phosphotyrosine binding domain, and leucine zipper motif
- ARHGEF11
- Rho guanine nucleotide exchange factor (GEF) 11
- BHLHB2/DEC1
- basic helix-loop-helix domain-containing, class B, 2
- CK2
- casein kinase 2
- CKIP1/PLEKHO1
- pleckstrin homology domain-containing, family O member 1
- co-AP
- co-affinity purification
- CPE
- carboxypeptidase E
- DB
- DNA binding domain
- DEAF1/NUDR
- deformed epidermal autoregulatory factor-1
- EAAT4
- excitatory amino acid transporter 4
- GLUT4
- glucose transporter 4
- GSK3
- glycogen synthase kinase 3
- HIS3
- imidazoleglycerol-phosphate dehydratase
- 5-HT
- 5-hydroxytryptamine (serotonin)
- 5-HT1A
- serotonin receptor 1A
- hVps15
- human vacuolar protein sorting 15 or phosphoinositide 3-kinase, class 3, regulatory subunit
- hVps34
- human vacuolar protein sorting 34 or phosphoinositide 3-kinase, class 3, catalytic subunit
- IGF1
- insulin-like growth factor 1
- IP
- immunoprecipitation
- IRS
- insulin receptor substrate
- KIF1C
- kinesin family member 1C
- lacZ
- β-d-galactosidase LacZ
- LZTS2
- leucine zipper, putative tumor suppressor 2
- MAPK
- mitogen-activated protein kinase
- MO25
- calcium-binding protein 39
- MTM1
- myotubularin 1
- mTOR
- mammalian target of rapamycin, subunit 1
- PDK1
- 3-phosphoinositide-dependent protein kinase
- PPI
- protein-protein interaction
- PRKAA1
- 5′-AMP-activated protein kinase, catalytic α-1 chain
- RC3H1/Roquin
- ring finger and CCCH-type zinc finger domains 1
- RHEB
- Ras homolog enriched in brain
- RHEBL1
- Ras homolog enriched in brainlike 1
- S6K
- ribosomal protein S6 kinase
- SGK1
- serum/glucocorticoid-regulated kinase 1
- SHIP
- SH2-containing inositol phosphatase
- SPG21/Maspardin
- spastic paraplegia 21
- STK11/LKB1
- serine/threonine kinase 11
- TNFR1
- tumor necrosis factor receptor superfamily, member 1
- TRAF2
- tumor necrosis factor receptor-associated factor 2
- TSC
- tuberous sclerosis
- URA3
- orotidine-5′-phosphate decarboxylase
- Y2H
- yeast two-hybrid
- ZBED3
- zinc finger, BED domain-containing 3
- PMID
- PubMed Unique Identifier
- GFP
- green fluorescent protein
- aa
- amino acids.
REFERENCES
- 1.Engelman J. A., Luo J., Cantley L. C. (2006) The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat. Rev. Genet. 7, 606–619 [DOI] [PubMed] [Google Scholar]
- 2.Taniguchi C. M., Emanuelli B., Kahn C. R. (2006) Critical nodes in signalling pathways: insights into insulin action. Nat. Rev. Mol. Cell Biol. 7, 85–96 [DOI] [PubMed] [Google Scholar]
- 3.Wullschleger S., Loewith R., Hall M. N. (2006) TOR signaling in growth and metabolism. Cell 124, 471–484 [DOI] [PubMed] [Google Scholar]
- 4.Shaw R. J., Cantley L. C. (2006) Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature 441, 424–430 [DOI] [PubMed] [Google Scholar]
- 5.Jope R. S., Johnson G. V. (2004) The glamour and gloom of glycogen synthase kinase-3. Trends Biochem. Sci. 29, 95–102 [DOI] [PubMed] [Google Scholar]
- 6.Wymann M. P., Marone R. (2005) Phosphoinositide 3-kinase in disease: timing, location, and scaffolding. Curr. Opin. Cell Biol. 17, 141–149 [DOI] [PubMed] [Google Scholar]
- 7.Cohen P., Goedert M. (2004) GSK3 inhibitors: development and therapeutic potential. Nat. Rev. Drug Discov. 3, 479–487 [DOI] [PubMed] [Google Scholar]
- 8.Stelzl U., Worm U., Lalowski M., Haenig C., Brembeck F. H., Goehler H., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S., Timm J., Mintzlaff S., Abraham C., Bock N., Kietzmann S., Goedde A., Toksöz E., Droege A., Krobitsch S., Korn B., Birchmeier W., Lehrach H., Wanker E. E. (2005) A human protein-protein interaction network: a resource for annotating the proteome. Cell 122, 957–968 [DOI] [PubMed] [Google Scholar]
- 9.Rual J. F., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., Li N., Berriz G. F., Gibbons F. D., Dreze M., Ayivi-Guedehoussou N., Klitgord N., Simon C., Boxem M., Milstein S., Rosenberg J., Goldberg D. S., Zhang L. V., Wong S. L., Franklin G., Li S., Albala J. S., Lim J., Fraughton C., Llamosas E., Cevik S., Bex C., Lamesch P., Sikorski R. S., Vandenhaute J., Zoghbi H. Y., Smolyar A., Bosak S., Sequerra R., Doucette-Stamm L., Cusick M. E., Hill D. E., Roth F. P., Vidal M. (2005) Towards a proteome-scale map of the human protein-protein interaction network. Nature 437, 1173–1178 [DOI] [PubMed] [Google Scholar]
- 10.Colland F., Jacq X., Trouplin V., Mougin C., Groizeleau C., Hamburger A., Meil A., Wojcik J., Legrain P., Gauthier J. M. (2004) Functional proteomics mapping of a human signaling pathway. Genome Res. 14, 1324–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tewari M., Hu P. J., Ahn J. S., Ayivi-Guedehoussou N., Vidalain P. O., Li S., Milstein S., Armstrong C. M., Boxem M., Butler M. D., Busiguina S., Rual J. F., Ibarrola N., Chaklos S. T., Bertin N., Vaglio P., Edgley M. L., King K. V., Albert P. S., Vandenhaute J., Pandey A., Riddle D. L., Ruvkun G., Vidal M. (2004) Systematic interactome mapping and genetic perturbation analysis of a C. elegans TGF-beta signaling network. Mol. Cell 13, 469–482 [DOI] [PubMed] [Google Scholar]
- 12.Li S., Armstrong C. M., Bertin N., Ge H., Milstein S., Boxem M., Vidalain P. O., Han J. D., Chesneau A., Hao T., Goldberg D. S., Li N., Martinez M., Rual J. F., Lamesch P., Xu L., Tewari M., Wong S. L., Zhang L. V., Berriz G. F., Jacotot L., Vaglio P., Reboul J., Hirozane-Kishikawa T., Li Q., Gabel H. W., Elewa A., Baumgartner B., Rose D. J., Yu H., Bosak S., Sequerra R., Fraser A., Mango S. E., Saxton W. M., Strome S., Van Den Heuvel S., Piano F., Vandenhaute J., Sardet C., Gerstein M., Doucette-Stamm L., Gunsalus K. C., Harper J. W., Cusick M. E., Roth F. P., Hill D. E., Vidal M. (2004) A map of the interactome network of the metazoan C. elegans. Science 303, 540–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim J., Hao T., Shaw C., Patel A. J., Szabó G., Rual J. F., Fisk C. J., Li N., Smolyar A., Hill D. E., Barabási A. L., Vidal M., Zoghbi H. Y. (2006) A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell 125, 801–814 [DOI] [PubMed] [Google Scholar]
- 14.Walhout A. J., Vidal M. (2001) High-throughput yeast two-hybrid assays for large-scale protein interaction mapping. Methods 24, 297–306 [DOI] [PubMed] [Google Scholar]
- 15.Bader G. D., Donaldson I., Wolting C., Ouellette B. F., Pawson T., Hogue C. W. (2001) BIND—the Biomolecular Interaction Network Database. Nucleic Acids Res. 29, 242–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zanzoni A., Montecchi-Palazzi L., Quondam M., Ausiello G., Helmer-Citterich M., Cesareni G. (2002) MINT: a Molecular INTeraction database. FEBS Lett. 513, 135–140 [DOI] [PubMed] [Google Scholar]
- 17.Mishra G. R., Suresh M., Kumaran K., Kannabiran N., Suresh S., Bala P., Shivakumar K., Anuradha N., Reddy R., Raghavan T. M., Menon S., Hanumanthu G., Gupta M., Upendran S., Gupta S., Mahesh M., Jacob B., Mathew P., Chatterjee P., Arun K. S., Sharma S., Chandrika K. N., Deshpande N., Palvankar K., Raghavnath R., Krishnakanth R., Karathia H., Rekha B., Nayak R., Vishnupriya G., Kumar H. G., Nagini M., Kumar G. S., Jose R., Deepthi P., Mohan S. S., Gandhi T. K., Harsha H. C., Deshpande K. S., Sarker M., Prasad T. S., Pandey A. (2006) Human protein reference database—2006 update. Nucleic Acids Res. 34, D411–D414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prieto C., De Las Rivas J. (2006) APID: Agile Protein Interaction DataAnalyzer. Nucleic Acids Res. 34, W298–W302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemonde S., Turecki G., Bakish D., Du L., Hrdina P. D., Bown C. D., Sequeira A., Kushwaha N., Morris S. J., Basak A., Ou X. M., Albert P. R. (2003) Impaired repression at a 5-hydroxytryptamine 1A receptor gene polymorphism associated with major depression and suicide. J. Neurosci. 23, 8788–8799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones G., Moore C., Hashemolhosseini S., Brenner H. R. (1999) Constitutively active MuSK is clustered in the absence of agrin and induces ectopic postsynaptic-like membranes in skeletal muscle fibers. J. Neurosci. 19, 3376–3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rual J. F., Hirozane-Kishikawa T., Hao T., Bertin N., Li S., Dricot A., Li N., Rosenberg J., Lamesch P., Vidalain P. O., Clingingsmith T. R., Hartley J. L., Esposito D., Cheo D., Moore T., Simmons B., Sequerra R., Bosak S., Doucette-Stamm L., Le Peuch C., Vandenhaute J., Cusick M. E., Albala J. S., Hill D. E., Vidal M. (2004) Human ORFeome version 1.1: a platform for reverse proteomics. Genome Res. 14, 2128–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vidalain P. O., Boxem M., Ge H., Li S., Vidal M. (2004) Increasing specificity in high-throughput yeast two-hybrid experiments. Methods 32, 363–370 [DOI] [PubMed] [Google Scholar]
- 24.Venkatesan K., Rual J. F., Vazquez A., Stelzl U., Lemmens I., Hirozane-Kishikawa T., Hao T., Zenkner M., Xin X., Goh K. I., Yildirim M. A., Simonis N., Heinzmann K., Gebreab F., Sahalie J. M., Cevik S., Simon C., de Smet A. S., Dann E., Smolyar A., Vinayagam A., Yu H., Szeto D., Borick H., Dricot A., Klitgord N., Murray R. R., Lin C., Lalowski M., Timm J., Rau K., Boone C., Braun P., Cusick M. E., Roth F. P., Hill D. E., Tavernier J., Wanker E. E., Barabási A. L., Vidal M. (2009) An empirical framework for binary interactome mapping. Nat. Methods 6, 83–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakai K., Horton P. (1999) PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24, 34–36 [DOI] [PubMed] [Google Scholar]
- 26.Braun P., Tasan M., Dreze M., Barrios-Rodiles M., Lemmens I., Yu H., Sahalie J. M., Murray R. R., Roncari L., de Smet A. S., Venkatesan K., Rual J. F., Vandenhaute J., Cusick M. E., Pawson T., Hill D. E., Tavernier J., Wrana J. L., Roth F. P., Vidal M. (2009) An experimentally derived confidence score for binary protein-protein interactions. Nat. Methods 6, 91–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boxem M., Maliga Z., Klitgord N., Li N., Lemmens I., Mana M., de Lichtervelde L., Mul J. D., van de Peut D., Devos M., Simonis N., Yildirim M. A., Cokol M., Kao H. L., de Smet A. S., Wang H., Schlaitz A. L., Hao T., Milstein S., Fan C., Tipsword M., Drew K., Galli M., Rhrissorrakrai K., Drechsel D., Koller D., Roth F. P., Iakoucheva L. M., Dunker A. K., Bonneau R., Gunsalus K. C., Hill D. E., Piano F., Tavernier J., van den Heuvel S., Hyman A. A., Vidal M. (2008) A protein domain-based interactome network for C. elegans early embryogenesis. Cell 134, 534–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kerrien S., Alam-Faruque Y., Aranda B., Bancarz I., Bridge A., Derow C., Dimmer E., Feuermann M., Friedrichsen A., Huntley R., Kohler C., Khadake J., Leroy C., Liban A., Lieftink C., Montecchi-Palazzi L., Orchard S., Risse J., Robbe K., Roechert B., Thorneycroft D., Zhang Y., Apweiler R., Hermjakob H. (2007) IntAct—open source resource for molecular interaction data. Nucleic Acids Res. 35, D561–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ali A., Hoeflich K. P., Woodgett J. R. (2001) Glycogen synthase kinase-3: properties, functions, and regulation. Chem. Rev. 101, 2527–2540 [DOI] [PubMed] [Google Scholar]
- 30.Ohteki T., Parsons M., Zakarian A., Jones R. G., Nguyen L. T., Woodgett J. R., Ohashi P. S. (2000) Negative regulation of T cell proliferation and interleukin 2 production by the serine threonine kinase GSK-3. J. Exp. Med. 192, 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McManus E. J., Sakamoto K., Armit L. J., Ronaldson L., Shpiro N., Marquez R., Alessi D. R. (2005) Role that phosphorylation of GSK3 plays in insulin and Wnt signalling defined by knockin analysis. EMBO J. 24, 1571–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doble B. W., Patel S., Wood G. A., Kockeritz L. K., Woodgett J. R. (2007) Functional redundancy of GSK-3alpha and GSK-3beta in Wnt/beta-catenin signaling shown by using an allelic series of embryonic stem cell lines. Dev. Cell 12, 957–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He X., Saint-Jeannet J. P., Woodgett J. R., Varmus H. E., Dawid I. B. (1995) Glycogen synthase kinase-3 and dorsoventral patterning in Xenopus embryos. Nature 374, 617–622 [DOI] [PubMed] [Google Scholar]
- 34.Doble B. W., Woodgett J. R. (2003) GSK-3: tricks of the trade for a multi-tasking kinase. J. Cell Sci. 116, 1175–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mishra R., Barthwal M. K., Sondarva G., Rana B., Wong L., Chatterjee M., Woodgett J. R., Rana A. (2007) Glycogen synthase kinase-3beta induces neuronal cell death via direct phosphorylation of mixed lineage kinase 3. J. Biol. Chem. 282, 30393–30405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Czesak M., Lemonde S., Peterson E. A., Rogaeva A., Albert P. R. (2006) Cell-specific repressor or enhancer activities of Deaf-1 at a serotonin 1A receptor gene polymorphism. J. Neurosci. 26, 1864–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kunick C., Lauenroth K., Leost M., Meijer L., Lemcke T. (2004) 1-Azakenpaullone is a selective inhibitor of glycogen synthase kinase-3 beta. Bioorg. Med. Chem. Lett. 14, 413–416 [DOI] [PubMed] [Google Scholar]
- 38.Phiel C. J., Klein P. S. (2001) Molecular targets of lithium action. Annu. Rev. Pharmacol. Toxicol. 41, 789–813 [DOI] [PubMed] [Google Scholar]
- 39.Huggenvik J. I., Michelson R. J., Collard M. W., Ziemba A. J., Gurley P., Mowen K. A. (1998) Characterization of a nuclear deformed epidermal autoregulatory factor-1 (DEAF-1)-related (NUDR) transcriptional regulator protein. Mol. Endocrinol. 12, 1619–1639 [DOI] [PubMed] [Google Scholar]
- 40.MacAulay K., Doble B. W., Patel S., Hansotia T., Sinclair E. M., Drucker D. J., Nagy A., Woodgett J. R. (2007) Glycogen synthase kinase 3alpha-specific regulation of murine hepatic glycogen metabolism. Cell Metab. 6, 329–337 [DOI] [PubMed] [Google Scholar]
- 41.Hoeflich K. P., Luo J., Rubie E. A., Tsao M. S., Jin O., Woodgett J. R. (2000) Requirement for glycogen synthase kinase-3beta in cell survival and NF-kappaB activation. Nature 406, 86–90 [DOI] [PubMed] [Google Scholar]
- 42.Phiel C. J., Wilson C. A., Lee V. M., Klein P. S. (2003) GSK-3alpha regulates production of Alzheimer's disease amyloid-beta peptides. Nature 423, 435–439 [DOI] [PubMed] [Google Scholar]
- 43.Patel S., Doble B. W., MacAulay K., Sinclair E. M., Drucker D. J., Woodgett J. R. (2008) Tissue-specific role of glycogen synthase kinase 3beta in glucose homeostasis and insulin action. Mol. Cell. Biol. 28, 6314–6328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu P., Cheng H., Roberts T. M., Zhao J. J. (2009) Targeting the phosphoinositide 3-kinase pathway in cancer. Nat. Rev. Drug Discov. 8, 627–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samuels Y., Ericson K. (2006) Oncogenic PI3K and its role in cancer. Curr. Opin. Oncol. 18, 77–82 [DOI] [PubMed] [Google Scholar]
- 46.Cuenda A., Rousseau S. (2007) p38 MAP-kinases pathway regulation, function and role in human diseases. Biochim. Biophys. Acta 1773, 1358–1375 [DOI] [PubMed] [Google Scholar]
- 47.Rosner M., Hanneder M., Siegel N., Valli A., Hengstschläger M. (2008) The tuberous sclerosis gene products hamartin and tuberin are multifunctional proteins with a wide spectrum of interacting partners. Mutat. Res. 658, 234–246 [DOI] [PubMed] [Google Scholar]
- 48.Patel S., Doble B., Woodgett J. R. (2004) Glycogen synthase kinase-3 in insulin and Wnt signalling: a double-edged sword? Biochem. Soc. Trans. 32, 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thyssen G., Li T. H., Lehmann L., Zhuo M., Sharma M., Sun Z. (2006) LZTS2 is a novel beta-catenin-interacting protein and regulates the nuclear export of beta-catenin. Mol. Cell. Biol. 26, 8857–8867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen T., Li M., Ding Y., Zhang L. S., Xi Y., Pan W. J., Tao D. L., Wang J. Y., Li L. (2009) Identification of zinc-finger BED domain-containing 3 (Zbed3) as a novel Axin-interacting protein that activates Wnt/beta-catenin signaling. J. Biol. Chem. 284, 6683–6689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krymskaya V. P. (2003) Tumour suppressors hamartin and tuberin: intracellular signalling. Cell. Signal. 15, 729–739 [DOI] [PubMed] [Google Scholar]
- 52.Hadad S. M., Fleming S., Thompson A. M. (2008) Targeting AMPK: a new therapeutic opportunity in breast cancer. Crit. Rev. Oncol. Hematol. 67, 1–7 [DOI] [PubMed] [Google Scholar]
- 53.Inoki K., Ouyang H., Zhu T., Lindvall C., Wang Y., Zhang X., Yang Q., Bennett C., Harada Y., Stankunas K., Wang C. Y., He X., MacDougald O. A., You M., Williams B. O., Guan K. L. (2006) TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell 126, 955–968 [DOI] [PubMed] [Google Scholar]
- 54.Lee J. H., Koh H., Kim M., Kim Y., Lee S. Y., Karess R. E., Lee S. H., Shong M., Kim J. M., Kim J., Chung J. (2007) Energy-dependent regulation of cell structure by AMP-activated protein kinase. Nature 447, 1017–1020 [DOI] [PubMed] [Google Scholar]
- 55.Mirouse V., Swick L. L., Kazgan N., St Johnston D., Brenman J. E. (2007) LKB1 and AMPK maintain epithelial cell polarity under energetic stress. J. Cell Biol. 177, 387–392 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 56.Zheng B., Cantley L. C. (2007) Regulation of epithelial tight junction assembly and disassembly by AMP-activated protein kinase. Proc. Natl. Acad. Sci. U.S.A. 104, 819–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang C., Navab R., Iakovlev V., Leng Y., Zhang J., Tsao M. S., Siminovitch K., McCready D. R., Done S. J. (2007) Abelson interactor protein-1 positively regulates breast cancer cell proliferation, migration, and invasion. Mol. Cancer Res. 5, 1031–1039 [DOI] [PubMed] [Google Scholar]
- 58.Yu W., Sun X., Clough N., Cobos E., Tao Y., Dai Z. (2008) Abi1 gene silencing by short hairpin RNA impairs Bcr-Abl-induced cell adhesion and migration in vitro and leukemogenesis in vivo. Carcinogenesis 29, 1717–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kopp P., Lammers R., Aepfelbacher M., Woehlke G., Rudel T., Machuy N., Steffen W., Linder S. (2006) The kinesin KIF1C and microtubule plus ends regulate podosome dynamics in macrophages. Mol. Biol. Cell 17, 2811–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grinberg-Rashi H., Ofek E., Perelman M., Skarda J., Yaron P., Hajdúch M., Jacob-Hirsch J., Amariglio N., Krupsky M., Simansky D. A., Ram Z., Pfeffer R., Galernter I., Steinberg D. M., Ben-Dov I., Rechavi G., Izraeli S. (2009) The expression of three genes in primary non-small cell lung cancer is associated with metastatic spread to the brain. Clin. Cancer Res. 15, 1755–1761 [DOI] [PubMed] [Google Scholar]
- 61.Harada M., Ishihara Y., Itoh K., Yamanaka R. (2007) Kinesin superfamily protein-derived peptides with the ability to induce glioma-reactive cytotoxic T lymphocytes in human leukocyte antigen-A24+ glioma patients. Oncol. Rep. 17, 629–636 [PubMed] [Google Scholar]
- 62.Hardie D. G. (2005) New roles for the LKB1–>AMPK pathway. Curr. Opin. Cell Biol. 17, 167–173 [DOI] [PubMed] [Google Scholar]
- 63.Lamia K. A., Sachdeva U. M., DiTacchio L., Williams E. C., Alvarez J. G., Egan D. F., Vasquez D. S., Juguilon H., Panda S., Shaw R. J., Thompson C. B., Evans R. M. (2009) AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science 326, 437–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nakashima A., Kawamoto T., Honda K. K., Ueshima T., Noshiro M., Iwata T., Fujimoto K., Kubo H., Honma S., Yorioka N., Kohno N., Kato Y. (2008) DEC1 modulates the circadian phase of clock gene expression. Mol. Cell. Biol. 28, 4080–4092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Iizuka K., Horikawa Y. (2008) Regulation of lipogenesis via BHLHB2/DEC1 and ChREBP feedback looping. Biochem. Biophys. Res. Commun. 374, 95–100 [DOI] [PubMed] [Google Scholar]
- 66.Cawley N. X., Zhou J., Hill J. M., Abebe D., Romboz S., Yanik T., Rodriguiz R. M., Wetsel W. C., Loh Y. P. (2004) The carboxypeptidase E knockout mouse exhibits endocrinological and behavioral deficits. Endocrinology 145, 5807–5819 [DOI] [PubMed] [Google Scholar]
- 67.Varlamov O., Fricker L. D., Furukawa H., Steiner D. F., Langley S. H., Leiter E. H. (1997) Beta-cell lines derived from transgenic Cpe(fat)/Cpe(fat) mice are defective in carboxypeptidase E and proinsulin processing. Endocrinology 138, 4883–4892 [DOI] [PubMed] [Google Scholar]
- 68.Chen H., Jawahar S., Qian Y., Duong Q., Chan G., Parker A., Meyer J. M., Moore K. J., Chayen S., Gross D. J., Glaser B., Permutt M. A., Fricker L. D. (2001) Missense polymorphism in the human carboxypeptidase E gene alters enzymatic activity. Hum. Mutat. 18, 120–131 [DOI] [PubMed] [Google Scholar]
- 69.Ma L., Hanson R. L., Que L. N., Cali A. M., Fu M., Mack J. L., Infante A. M., Kobes S., Bogardus C., Shuldiner A. R., Baier L. J. (2007) Variants in ARHGEF11, a candidate gene for the linkage to type 2 diabetes on chromosome 1q, are nominally associated with insulin resistance and type 2 diabetes in Pima Indians. Diabetes 56, 1454–1459 [DOI] [PubMed] [Google Scholar]
- 70.Fu M., Sabra M. M., Damcott C., Pollin T. I., Ma L., Ott S., Shelton J. C., Shi X., Reinhart L., O'Connell J., Mitchell B. D., Baier L. J., Shuldiner A. R. (2007) Evidence that Rho guanine nucleotide exchange factor 11 (ARHGEF11) on 1q21 is a type 2 diabetes susceptibility gene in the Old Order Amish. Diabetes 56, 1363–1368 [DOI] [PubMed] [Google Scholar]
- 71.Böttcher Y., Schleinitz D., Tönjes A., Blüher M., Stumvoll M., Kovacs P. (2008) R1467H variant in the rho guanine nucleotide exchange factor 11 (ARHGEF11) is associated with impaired glucose tolerance and type 2 diabetes in German Caucasians. J. Hum. Genet. 53, 365–367 [DOI] [PubMed] [Google Scholar]
- 72.Ishikura S., Koshkina A., Klip A. (2008) Small G proteins in insulin action: Rab and Rho families at the crossroads of signal transduction and GLUT4 vesicle traffic. Acta Physiol. 192, 61–74 [DOI] [PubMed] [Google Scholar]
- 73.Deepa S. S., Dong L. Q. (2009) APPL1: role in adiponectin signaling and beyond. Am. J. Physiol. Endocrinol. Metab. 296, E22–E36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang C., Mao X., Wang L., Liu M., Wetzel M. D., Guan K. L., Dong L. Q., Liu F. (2007) Adiponectin sensitizes insulin signaling by reducing p70 S6 kinase-mediated serine phosphorylation of IRS-1. J. Biol. Chem. 282, 7991–7996 [DOI] [PubMed] [Google Scholar]
- 75.Fruman D. A., Bismuth G. (2009) Fine tuning the immune response with PI3K. Immunol. Rev. 228, 253–272 [DOI] [PubMed] [Google Scholar]
- 76.Weichhart T., Säemann M. D. (2009) The multiple facets of mTOR in immunity. Trends Immunol. 30, 218–226 [DOI] [PubMed] [Google Scholar]
- 77.Karin M., Gallagher E. (2009) TNFR signaling: ubiquitin-conjugated TRAFfic signals control stop-and-go for MAPK signaling complexes. Immunol. Rev. 228, 225–240 [DOI] [PubMed] [Google Scholar]
- 78.Jabara H. H., Weng Y., Sannikova T., Geha R. S. (2009) TRAF2 and TRAF3 independently mediate Ig class switching driven by CD40. Int. Immunol. 21, 477–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dupoux A., Cartier J., Cathelin S., Filomenko R., Solary E., Dubrez-Daloz L. (2009) cIAP1-dependent TRAF2 degradation regulates the differentiation of monocytes into macrophages and their response to CD40 ligand. Blood 113, 175–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Karbowniczek M., Cash T., Cheung M., Robertson G. P., Astrinidis A., Henske E. P. (2004) Regulation of B-Raf kinase activity by tuberin and Rheb is mammalian target of rapamycin (mTOR)-independent. J. Biol. Chem. 279, 29930–29937 [DOI] [PubMed] [Google Scholar]
- 81.Vinuesa C. G., Cook M. C., Angelucci C., Athanasopoulos V., Rui L., Hill K. M., Yu D., Domaschenz H., Whittle B., Lambe T., Roberts I. S., Copley R. R., Bell J. I., Cornall R. J., Goodnow C. C. (2005) A RING-type ubiquitin ligase family member required to repress follicular helper T cells and autoimmunity. Nature 435, 452–458 [DOI] [PubMed] [Google Scholar]
- 82.Yu D., Tan A. H., Hu X., Athanasopoulos V., Simpson N., Silva D. G., Hutloff A., Giles K. M., Leedman P. J., Lam K. P., Goodnow C. C., Vinuesa C. G. (2007) Roquin represses autoimmunity by limiting inducible T-cell co-stimulator messenger RNA. Nature 450, 299–303 [DOI] [PubMed] [Google Scholar]
- 83.Beaulieu J. M., Gainetdinov R. R., Caron M. G. (2009) Akt/GSK3 signaling in the action of psychotropic drugs. Annu. Rev. Pharmacol. Toxicol. 49, 327–347 [DOI] [PubMed] [Google Scholar]
- 84.Jackson M., Song W., Liu M. Y., Jin L., Dykes-Hoberg M., Lin C. I., Bowers W. J., Federoff H. J., Sternweis P. C., Rothstein J. D. (2001) Modulation of the neuronal glutamate transporter EAAT4 by two interacting proteins. Nature 410, 89–93 [DOI] [PubMed] [Google Scholar]
- 85.Perrot V., Vazquez-Prado J., Gutkind J. S. (2002) Plexin B regulates Rho through the guanine nucleotide exchange factors leukemia-associated Rho GEF (LARG) and PDZ-RhoGEF. J. Biol. Chem. 277, 43115–43120 [DOI] [PubMed] [Google Scholar]
- 86.Simpson M. A., Cross H., Proukakis C., Pryde A., Hershberger R., Chatonnet A., Patton M. A., Crosby A. H. (2003) Maspardin is mutated in mast syndrome, a complicated form of hereditary spastic paraplegia associated with dementia. Am. J. Hum. Genet. 73, 1147–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Karlstrom H., Brooks W. S., Kwok J. B., Broe G. A., Kril J. J., McCann H., Halliday G. M., Schofield P. R. (2008) Variable phenotype of Alzheimer's disease with spastic paraparesis. J. Neurochem. 104, 573–583 [DOI] [PubMed] [Google Scholar]
- 88.Drevets W. C., Thase M. E., Moses-Kolko E. L., Price J., Frank E., Kupfer D. J., Mathis C. (2007) Serotonin-1A receptor imaging in recurrent depression: replication and literature review. Nucl. Med. Biol. 34, 865–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Celada P., Puig M., Amargós-Bosch M., Adell A., Artigas F. (2004) The therapeutic role of 5-HT1A and 5-HT2A receptors in depression. J. Psychiatry Neurosci. 29, 252–265 [PMC free article] [PubMed] [Google Scholar]
- 90.Sharp T., Boothman L., Raley J., Quérée P. (2007) Important messages in the ‘post’: recent discoveries in 5-HT neurone feedback control. Trends Pharmacol. Sci. 28, 629–636 [DOI] [PubMed] [Google Scholar]
- 91.Savitz J., Lucki I., Drevets W. C. (2009) 5-HT(1A) receptor function in major depressive disorder. Prog. Neurobiol. 88, 17–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Drevets W. C., Frank E., Price J. C., Kupfer D. J., Holt D., Greer P. J., Huang Y., Gautier C., Mathis C. (1999) PET imaging of serotonin 1A receptor binding in depression. Biol. Psychiatry 46, 1375–1387 [DOI] [PubMed] [Google Scholar]
- 93.Moses-Kolko E. L., Wisner K. L., Price J. C., Berga S. L., Drevets W. C., Hanusa B. H., Loucks T. L., Meltzer C. C. (2008) Serotonin 1A receptor reductions in postpartum depression: a positron emission tomography study. Fertil. Steril. 89, 685–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bain E. E., Nugent A. C., Carson R. E., Luckenbaugh D., Lang L., Eckekman W. C., Neumeister A., Bonne O., Williams J., Gordon J., Charney D. S., Drevets W. C. (2004) Decreased 5-HT1A receptor binding in bipolar depression. Biol. Psychiatry 55, 178S [Google Scholar]
- 95.Szewczyk B., Albert P. R., Burns A. M., Czesak M., Overholser J. C., Jurjus G. J., Meltzer H. Y., Konick L. C., Dieter L., Herbst N., May W., Rajkowska G., Stockmeier C. A., Austin M. C. (2009) Gender-specific decrease in NUDR and 5-HT1A receptor proteins in the prefrontal cortex of subjects with major depressive disorder. Int. J. Neuropsychopharmacol. 12, 155–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sargent P. A., Kjaer K. H., Bench C. J., Rabiner E. A., Messa C., Meyer J., Gunn R. N., Grasby P. M., Cowen P. J. (2000) Brain serotonin1A receptor binding measured by positron emission tomography with [11C]WAY-100635: effects of depression and antidepressant treatment. Arch. Gen. Psychiatry 57, 174–180 [DOI] [PubMed] [Google Scholar]
- 97.Meltzer C. C., Price J. C., Mathis C. A., Butters M. A., Ziolko S. K., Moses-Kolko E., Mazumdar S., Mulsant B. H., Houck P. R., Lopresti B. J., Weissfeld L. A., Reynolds C. F. (2004) Serotonin 1A receptor binding and treatment response in late-life depression. Neuropsychopharmacology 29, 2258–2265 [DOI] [PubMed] [Google Scholar]
- 98.Beaulieu J. M., Marion S., Rodriguiz R. M., Medvedev I. O., Sotnikova T. D., Ghisi V., Wetsel W. C., Lefkowitz R. J., Gainetdinov R. R., Caron M. G. (2008) A beta-arrestin 2 signaling complex mediates lithium action on behavior. Cell 132, 125–136 [DOI] [PubMed] [Google Scholar]
- 99.Beaulieu J. M., Zhang X., Rodriguiz R. M., Sotnikova T. D., Cools M. J., Wetsel W. C., Gainetdinov R. R., Caron M. G. (2008) Role of GSK3 beta in behavioral abnormalities induced by serotonin deficiency. Proc. Natl. Acad. Sci. U.S.A. 105, 1333–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kang U. G., Seo M. S., Roh M. S., Kim Y., Yoon S. C., Kim Y. S. (2004) The effects of clozapine on the GSK-3-mediated signaling pathway. FEBS Lett. 560, 115–119 [DOI] [PubMed] [Google Scholar]
- 101.Karege F., Perroud N., Burkhardt S., Schwald M., Ballmann E., La Harpe R., Malafosse A. (2007) Alteration in kinase activity but not in protein levels of protein kinase B and glycogen synthase kinase-3beta in ventral prefrontal cortex of depressed suicide victims. Biol. Psychiatry 61, 240–245 [DOI] [PubMed] [Google Scholar]
- 102.Carlson P. J., Bain E., Tinsley R., Luckenbaugh D., Manji H. K., Drevets W. C. (2007) Serotonin-1A receptor binding in bipolar depression before and after mood stabilizer treatment. Biol. Psychiatry 61, 57S [Google Scholar]