Abstract
In this study, we used imaging and proteomics to identify the presence of virus-associated cellular proteins that may play a role in respiratory syncytial virus (RSV) maturation. Fluorescence microscopy of virus-infected cells revealed the presence of virus-induced cytoplasmic inclusion bodies and mature virus particles, the latter appearing as virus filaments. In situ electron tomography suggested that the virus filaments were complex structures that were able to package multiple copies of the virus genome. The virus particles were purified, and the protein content was analyzed by one-dimensional nano-LC MS/MS. In addition to all the major virus structural proteins, 25 cellular proteins were also detected, including proteins associated with the cortical actin network, energy pathways, and heat shock proteins (HSP70, HSC70, and HSP90). Representative actin-associated proteins, HSC70, and HSP90 were selected for further biological validation. The presence of β-actin, filamin-1, cofilin-1, HSC70, and HSP90 in the virus preparation was confirmed by immunoblotting using relevant antibodies. Immunofluorescence microscopy of infected cells stained with antibodies against relevant virus and cellular proteins confirmed the presence of these cellular proteins in the virus filaments and inclusion bodies. The relevance of HSP90 to virus infection was examined using the specific inhibitors 17-N-Allylamino-17-demethoxygeldanamycin. Although virus protein expression was largely unaffected by these drugs, we noted that the formation of virus particles was inhibited, and virus transmission was impaired, suggesting an important role for HSP90 in virus maturation. This study highlights the utility of proteomics in facilitating both our understanding of the role that cellular proteins play during RSV maturation and, by extrapolation, the identification of new potential targets for antiviral therapy.
Respiratory syncytial virus (RSV)1 belongs to the paramyxovirus group of viruses, and it is the most important respiratory virus causing lower respiratory tract infection in young children and neonates. The mature RSV particle comprises a ribonucleoparticle (RNP) core formed by the interaction between the viral genomic RNA (vRNA), the nucleocapsid (N) protein (42 kDa), the phospho (P) protein (35 kDa), and the large (L) protein (250 kDa). The RNP core is visualized by electron microscopy as a strand of repeating N protein subunits that form a herringbone-like structure of ∼10–20 nm in diameter (1). Although the minimal functional polymerase activity requires an association between the N, P, and L proteins and the virus genome vRNA (2–4), additional viral proteins called the M2-1 protein (22 kDa), M2-2 protein, and M protein (28 kDa) regulate the activity of the polymerase (5–8). The virus is surrounded by a lipid envelope that is formed from the host cell during the budding process in which the three virus membrane proteins are inserted. The G protein (90 kDa) mediates attachment of the virus to the cell during virus entry (9), and the fusion (F) protein (10) mediates the fusion of the virus and host cell membranes during virus entry, whereas the role of the SH protein is currently unknown. In addition, two non-structural proteins called NS1 and NS2, which are thought not to be present in the virus particle but play a role in countering the host innate immune response (11), are expressed.
During virus infection two distinct virus structures are formed, virus filaments and inclusion bodies. The virus filaments are membrane-bound structures that are ∼150–200 nm thick and can be up to 6 μm in length (1, 12–16); they form at the sites of virus assembly and are the progeny viruses. The inclusion bodies form in the cytoplasm and can be several μm in diameter, consisting of accumulations of RNP cores (17–19). Inclusion bodies are found in all RSV-infected tissue culture cells, and they have also been observed in biopsy material isolated from RSV-infected patients (20) suggesting a clinical relevance. Although the cellular processes that lead to assembly of the mature virus filaments are still poorly understood, the involvement of lipid raft microdomains and the cortical cytoskeleton network appear to play an important role in this process (16, 21–25). For example, rhoA kinase is a raft-associated signaling molecule that is involved in regulating actin structure (26), and it has been implicated in virus filament formation (27, 28). Virus filament formation also requires phosphoinositide 3-kinase (PI3K) activity (25, 29, 30); PI3K is a raft-associated kinase activated by rhoA kinase (31). The identification of cellular proteins that interact with the virus particles should further facilitate the identification of the cellular pathways that are involved in RSV maturation. In this study, we examined virus-host cell interactions during RSV assembly using a combination of advanced imaging techniques and analyzed the protein content of purified virus particles by proteomics technology. Our analysis provides evidence that cellular proteins that regulate actin structures in the cell may also play an important role in formation of infectious RSV particles, and that the HSP90 protein plays an important role in the virus assembly process.
EXPERIMENTAL PROCEDURES
The RSV A2 strain and the human respiratory airway cell line HEp2 were used throughout this study. Cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS) and antibiotics, and infections were carried out in DMEM with 2% FCS. Infected cells were incubated at 33 °C in 5% CO2. Unless otherwise stated, the cells were infected with a multiplicity of infection (m.o.i.) of 2.
Antibodies and Specific Reagents
Anti-RSV (RCL3) was purchased from Novacastra Laboratories, and MAb19 (anti-F) and MAb30 (anti-G) were obtained from Geraldine Taylor (Institute for Animal Health, Compton, Berks, UK). Anti-N and anti-M2-1 have been described previously (32). The RSV anti-F protein rabbit polyclonal antibody was a gift from Jose Melero. Anti-β-actin was purchased from Sigma-Aldrich; anti-filamin-1, anti-HSC70, anti-HSP70, anti-actin, anti-caveolin-1, and anti-HSP90A were obtained from Santa Cruz Laboratories; and anti-cofilin-1 was purchased from Cytoskeleton. Geldanamycin (GA) and 17AAG were purchased from Calbiochem and reconstituted in DMSO at 10 mm. This was diluted into tissue culture medium prior to use, giving a final concentration of 2 μm (GA and 17AAG).
Purification of RSV Particles
This was based on the method of Ueba (33). Cells grown to 90% confluence were infected with RSV using an m.o.i. of 0.1. At 48 h post-infection, the infected cells were harvested, and the virus was released at 4 °C by gentle agitation using acid-washed glass beads. The virus suspension was clarified by centrifugation (5000 × g for 20 min) at 4 °C, and the virus in the clarified supernatant was precipitated by the addition of PEG 6000 to a final concentration of 10% (w/v) with stirring and incubated at 4 °C for 1 h. The virus was pelleted by centrifugation (5000 × g for 20 min at 4 °C) after which the virus pellet was resuspended in 20% sucrose in Hanks' buffered saline solution (HBSS). This was overlaid on a 30% sucrose cushion and pelleted again by centrifugation 51,000 × g for 1 h. The pellet was resuspended in 20% sucrose and overlaid on a discontinuous gradient consisting of 35, 45, and 60% sucrose prepared in HBSS and centrifuged for 1 h at 165,000 × g at 4 °C. The opalescent band at the interface of 35 and 45% sucrose was harvested, diluted with HBSS, and overlaid on a 30–60% sucrose gradient prepared using HBSS. This was centrifuged for 3 h at 165,000 × g at 4 °C, and the virus band (located at ∼45% sucrose) was harvested. The virus suspension was diluted with HBSS, and the virus was recovered as a pellet after centrifugation at 284,000 × g for 90 min at 4 °C.
SDS-PAGE and Western Blotting
Protein samples were made up to 1× boiling mixture (1% SDS, 15% glycerol, 1% β-mercaptoethanol, 60 mm sodium phosphate, pH 6.8) and heated at 100 °C for 2 min. The protein samples were analyzed by SDS-PAGE using a Mini-PROTEAN 3 (Bio-Rad). The proteins were visualized by either (a) staining the polyacrylamide gels with Coomassie Brilliant Blue (G-250) and imaging the protein bands using a Bio-Rad Gel Doc system or (b) staining the polyacrylamide gels with SYPRO Ruby Red (Molecular Probes) and imaging the protein bands using a Typhoon Trio scanner. In all cases, the apparent molecular masses were estimated using Precision Plus protein standards (Bio-Rad). Relative quantification of protein bands detected in SYPRO Ruby-stained gels was performed using ImageQuantTM TL.
In Western blotting, the proteins were transferred onto PVDF membranes using a mini blotting apparatus (Bio-Rad) after which the membranes were washed with PBS and blocked for 18 h at 4 °C in PBSA containing 1% Marvel milk powder and 0.05% Tween 20. The membrane was incubated with the specific primary antibody followed by the appropriate anti-mouse or anti-rabbit IgG (whole molecule)-peroxidase conjugate (Sigma). The protein bands were visualized using the ECL protein detection system (Amersham Biosciences), and the apparent molecular masses were estimated using Kaleidoscope protein standards (Bio-Rad).
Microplaque Titration of Purified Virus Preparations
The viral infectivity was assessed using a microplaque assay in 96-well tissue culture plates as described previously (34). Viral preparations were serially diluted using PBS, and 50 μl of each dilution was added to a confluent cell monolayer in each well and allowed to absorb for 2 h at 33 °C. After adsorption, the virus dilutions were replaced with DMEM + 2% FCS, and the plates were incubated at 33 °C for 48 h after which the monolayers were washed with PBS and fixed with methanol for 20 min. The monolayers were washed with PBS and then incubated with anti-RSV MAb (Novacastra Laboratories) for 60 min after which the monolayers were then washed and incubated for a further 60 min using a rabbit anti-mouse IgG MAb conjugated to HRP. The microplaques were visualized using staining solution (200 μg/ml 3-amino-9-ethylcarbazole, 0.05% hydrogen peroxide, 20 mm sodium acetate, pH 5.5) and counted using a low magnification inverted microscope.
One-dimensional LC-MS/MS Analysis
Sample Preparation
The purified virus preparation was treated with trypsin using the in-gel digestion procedure described recently (35, 36). Briefly, the virus pellet (5 μg) was dissolved in 25 mm ammonium bicarbonate (ABC) and 2% (w/v) SDS and then incorporated into a 40% acrylamide/bisacrylamide (29:1) gel matrix. The gel was cut into small pieces, washed with 25 mm ABC containing 50% acetonitrile (ACN), and vacuum-dried. The dried gel was treated with 10 mm DTT in ABC at 60 °C for 30 min after which it was then treated with 55 mm iodoacetamide in ABC at 25 °C in the dark for 30 min. The gel pieces were washed with 25 mm ABC, dehydrated with 100% ACN, and vacuum-dried. The proteolytic digestion was then carried out with trypsin in ABC (protein/trypsin = 10:1, g/g) at 37 °C for 16 h. The peptides were sequentially extracted from the gel using 25 mm ABC, 5% formic acid (FA) in water, 5% FA in ACN, and 100% ACN, and the pooled peptide extracts were vacuum-dried.
MS Analysis
The peptide mixture was reconstituted in buffer A (0.1% FA, 2% ACN), and aliquots were injected for LC separation. An autosampler (Famos, Dionex, San Francisco, CA) was used for injection of peptides. A nanoflow liquid chromatography system (UltiMate Plus, Dionex) with a nano-LC column (C18 PepMap100, 3 μm, 0.075 × 150 mm, Dionex) was used for peptide separation. A solvent gradient with buffer B (0.1% FA, 90% ACN) increasing from 0 to 45% in 45 min was used to elute the peptides followed by a 10-min wash with 80% buffer B and a 25-min equilibration with 100% buffer A. The flow rate was kept at 300 nl/min throughout. The column was coupled to an LTQ Orbitrap (Thermo, Bremen, Germany) with a T-union and a nanospray tip (20-μm-inner diameter tubing, 10-μm-inner diameter tip). The spray voltage was 1.8 kV applied through the T-union. The mass spectrometer was operated in data-dependent mode to switch between MS and MS/MS. Full-scan MS spectra were acquired from m/z 350 to 1800 Da in the Orbitrap with a resolution of 60,000 at m/z 400. The five most intense precursor ions were selected for MS/MS scans with collision-induced dissociation (CID) in the linear ion trap LTQ.
Database Search
The acquired spectra in the .raw file format were converted to the standard mzXML input file format in the Trans Proteomic Pipeline (TPP; v.4.2 JETSTREAM revision 0 Build 200902190939) and searched using the Sequest database search engine (Bioworks, v.3.3, Thermo Electron Co.) The database was constructed by extracting the Protein Knowledgebase (UniProtKB) with taxonomy “Homo sapiens (9606)” or “human respiratory syncytial virus A (strain A2) (11259)” and concatenated with their reverse sequences (generated in TPP). The database contained 95,673 entries for human proteins, 11 entries for RSV (strain A2), and the same numbers of reverse entries. The search settings were as follows: mass values, monoisotopic; precursor mass tolerance, ±20 ppm; fragment mass tolerance, ±0.5 Da; enzyme, trypsin; maximum missed cleavages allowed, 1; fixed modification, carbamidomethyl-Cys; variable modifications, oxidation of Met and phosphorylation of Ser, Thr, and Tyr. The search results were converted to the pepXML format, and peptide identifications were validated with PeptideProphet and InterProphet and protein identifications were validated with ProteinProphet in TPP with default settings (filter out results below probability 0.05 and minimum peptide length 7). The statistical calculation predicted sensitivity and error rate versus the minimum protein probability. The final list of protein hits was obtained by setting the minimum protein probability to 0.95 and the minimum peptide probability to 0.95.
Electron Microscopy
Negative Staining
A droplet of the virus suspension was placed on a Formvar-coated grid for 10 min and subsequently stained with phosphotungstic acid, pH 7.5. Specimens were viewed at 80 kV in a JEOL 1200 EX electron microscope.
Immunolabeled Ultrathin Sections
Cell monolayers were pelleted in BEEM capsules (TAAB Laboratories) and fixed for 5 h at 4 °C using 0.5% glutaraldehyde in PBS after which the fixed pellet was washed with PBS. The pellet was dehydrated through a gradient of increasing ethanol concentrations. The cell pellet was subsequently infiltrated with Unicryl (TAAB Laboratories), and the resin was polymerized by UV irradiation at −15 °C for 48 h. Ultrathin sections prepared using a Leica UC6 Ultratome were placed on uncoated nickel grids and incubated with the appropriate monoclonal antibody for 4 h at 25 °C, washed with PBS, and incubated for 4 h in anti-mouse IgG (whole molecule)-10-nm colloidal gold conjugate (Sigma). The grids were washed in PBS and fixed in osmium tetroxide vapor for 2 h. The specimens were then stained using uranyl acetate (saturated in 50:50 ethanol/water), washed in distilled water, and counterstained with lead citrate. Specimens were then viewed at 80 kV in a JEOL 100S electron microscope.
Whole Cell Tomography
Cell monolayers were pelleted in BEEM capsules (TAAB Laboratories) and fixed with 3% glutaraldehyde, 1% tannic acid, 100 mm sodium cacodylate, pH 7.2. The cell pellet was washed with 100 mm sodium cacodylate, pH 7.2 and postfixed with 1% osmium tetroxide solution (TAAB Laboratories). The cell pellet was then dehydrated through a gradient of increasing ethanol concentrations. The pellet was infiltrated with Epon812 (TAAB Laboratories), and the resin was polymerized at 65 °C for 24 h. Thin sections (300 nm thick) were cut on a Leica UC6 Ultratome and collected on uncoated copper 200 mesh (hexagonal) grids (TAAB Laboratories). Dual axis tomography was done in a JEOL JEM-2100 electron microscope operating at 200 kV. The sections were mounted in a Fischione 2040 dual axis holder. Dual axis tilt series were acquired between ±64° at 1.5° increments using SerialEM (37, 38). Processing of the dual axis data sets was done using IMOD (39). The rotation between the two tilt series making up the dual axis data sets was determined as 89.6°. Denoising of the final tomogram was done using the IMOD implementation of the nonlinear anisotropic diffusion algorithm according to Frangakis and Hegerl (40).
Immunofluorescence Microscopy
Cells were labeled as described previously (16, 21). Briefly, cells on 13-mm glass cover slips were generally fixed with 3% paraformaldehyde in PBS for 30 min at 4 °C, permeabilized using 0.1% saponin, and then labeled with the relevant primary antibody and appropriate secondary antibodies (conjugated to either FITC or Cy5). In the case of anti-β-actin staining, the cells were fixed using methanol for 5 min at 4 °C prior to antibody labeling. The stained cells were mounted on slides using Citifluor and visualized using either a Nikon ECLIPSE 80i fluorescence microscope or a Zeiss Axioplan 2 confocal microscope using appropriate machine settings. Confocal microscopy images were processed using LSM510 software. For three-dimensional reconstruction, 50–80 slices were gathered in a Z-stack series using a 1:1:1 setting. Autodeblur and Autovisualize software (Media Cybernetics Inc.) were used to generate three-dimensional reconstruction of the area of interest. Stacks were processed by three-dimensional blind deconvolution (10 iterations) and then imported into Autovisualize for imaging in the 5D Viewer.
Transfection of siRNA Molecules
Cells were transfected with siGFP, siHSC70, or siHSP90 (Dharmacon) at 100 nm using Lipofectamine. At 36 h post-transfection, the cells were infected with RSV, and at 18 h postinfection, the cells were examined by confocal microscopy. The transfection efficiency of each experiment was determined using siGlow (Dharmacon).
Radioimmunoprecipitation
Mock- and virus-infected cells were incubated at 33 °C for 8 h after which the medium was removed and replaced with DMEM minus methionine (Invitrogen) for 1 h prior to radiolabeling with 100 μCi/ml [35S]methionine (GE Healthcare). At 24 h postinfection, the monolayers (on 60-mm dishes) were extracted at 4 °C for 20 min with 500 μl of lysis buffer (1% Nonidet P-40, 150 mm NaCl, 1 mm EDTA, 2 mm PMSF, 20 mm Tris-HCl, pH 7.5) and then clarified by centrifugation (12,000 × g for 10 min). Aliquots of the clarified lysate (100 μl) were added to 600 μl of binding buffer (0.5% Nonidet P-40, 150 mm NaCl, 1 mm EDTA, 0.25% BSA, 20 mm Tris-HCl, pH 8) together with the appropriate antibody and incubated for 18 h at 4 °C. The immune complexes were isolated by adding protein A-Sepharose for 2 h at 4 °C. The protein A-Sepharose was washed three times with wash buffer (1% Triton X-100, 150 mm NaCl, 1 mm EDTA, 10 mm sodium phosphate, pH 7.0). The protein A-Sepharose was resuspended in 40 μl of boiling mixture and incubated at 100 °C for 2 min. The proteins were separated using SDS-PAGE, and the protein bands were detected in the dried polyacrylamide gels using a Bio-Rad personal Fx phosphorimaging system using Quantity One software (Bio-Rad, version 4).
RESULTS
In Situ Characterization of RSV-infected Cells by Three-dimensional Imaging
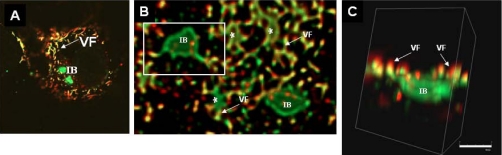
To assist in the proteomics analysis of virus particles we first characterized RSV-infected cells using three-dimensional imaging, which allowed us to examine the distribution of the virus filaments and inclusion bodies in infected cells (Fig. 1) Cells were infected with RSV, and at 20 h postinfection (hpi), the cells were fixed and stained using MAb19 and anti-P, antibodies that recognize the F and P proteins respectively (16, 21, 41). The virus glycoproteins can only be detected in the virus filaments, whereas the polymerase-associated proteins are present in both the virus filaments and inclusions bodies (41). Therefore, as expected, anti-P stained both the virus filaments and inclusion bodies, whereas MAb19 only stained the virus filaments (Fig. 1A), and co-localization of both antibodies was restricted to the virus filaments. Most infected cells contained several inclusion bodies, and these were of varying sizes and up to several μm in diameter. Confocal microscopy was used to obtain a series of images from individual infected cells in the Z-dimension, and these data were processed as described under “Experimental Procedures” to allow visualization of the infected cells as three-dimensional images. Whereas the larger inclusion bodies were visible in the two-dimensional image, the distribution of the smaller inclusion bodies together with the virus filaments was clearly defined in the three-dimensional images (Fig. 1B, highlighted by *). Furthermore, this analysis showed that the inclusion bodies appeared to be irregular in shape, suggesting that they form complex structures, consistent with recent ultrastructural studies (25). Connectivity between the inclusion bodies and virus filaments was observed (Fig. 1C), supporting recent observations that virus proteins can be transported from these structures into maturing virus filaments (42).
Fig. 1.
Association of virus filaments and inclusion bodies in virus-infected cells. Virus-infected cells were labeled using antibodies to P protein (green) and F protein (red). A series of images was obtained from the same cell at different focal planes in the Z-axis by confocal microscopy. The Z-stacks were then processed and visualized in three-dimensional as described under “Experimental Procedures.” A is a single optical slice visualized in parallel to the substrate and viewed two-dimensionally. B is a low magnification deconvolved image showing a region near the surface of the cell, again parallel to the substrate. The inclusion body (IB) can be seen to be irregular in shape and labeled only by anti-P, whereas the virus filaments (VF) are labeled by both anti-P and anti-F. C is a higher magnification image within the cell showing an individual inclusion body and its associated virus filaments. The three-dimensional projection has now been rotated through 90° and tilted slightly, showing that the filaments are predominantly above the inclusion body and in a vertical orientation. * highlights the smaller sized inclusion body.
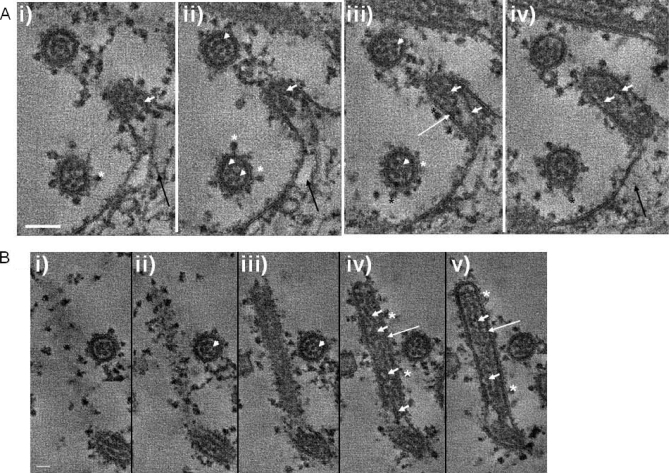
Because RSV is largely cell-associated and filamentous, ultrastructural analysis of the virus particles is therefore difficult using conventional methods such as single particle reconstruction that requires virus particles to be uniform and symmetrical (43). An ultrastructural analysis on the virus filaments was performed on the cell-associated virus using a combination of transmission electron microscopy (TEM) and in situ tomography (IST) of infected cells (Fig. 2). The virus filaments imaged in this way resembled the structures reported previously in electron microscopy studies (13, 14, 22), appearing as finger-like projections of ∼150 nm in diameter extending from the surface of infected cells. IST allowed a series of consecutive high resolution images through individual virus filaments at approximately 11-nm increments, and a comparison of several sections allowed a more detailed characterization of the virus filaments than has been observed previously. Structures on the surface of the virus filaments were observed (Fig. 2, A and B, highlighted by *), and on the basis of previous observations, these were interpreted as the virus envelope proteins (22). An electron-dense layer corresponding to the expected location of M protein (44) was also observed beneath the surface of the virus filaments (Fig. 2, A (ii) and B (ii and iii), highlighted by long white arrows). RNP structures were observed as strands along the length of the virus filaments, and more than one of these strands appeared to be present in a virus filament (Fig. 2B (ii and iii), highlighted by short white arrows). Examination of the virus filaments in cross-section (Fig. 2, A (ii and iii) and B (i)), highlighted by white arrowheads) showed a dotted pattern within the circular structure similar to that observed previously (12). The distribution pattern presumably reflects the packing of multiple RNP strands in the virus filaments. Because there appear to be several RNP strands in the virus filament, this observation also suggests that each virus filament may be able to package multiple copies of the virus genome.
Fig. 2.
TEM tomography of virus-infected cells. Infected cells were embedded in Epon and processed as described under “Experimental Procedures.” In each case, consecutive images from two tomograms at different planes through the section are shown, providing an image series at 11-nm intervals through the infected cells. In each case, plate i is a view closer to the top of the section, and subsequent plates ii–iv (A) and ii–v (B) are views progressing through the section. A shows the relationship between the newly assembled virus filaments and the cell surface, in particular the packaging of the virus RNP into a virus filament. B is a longitudinal cross-section through a virus filament showing the distribution of the RNPs in the virus filament. Also seen in both A and B is a virus filament viewed in radial cross-section where the RNPs are highlighted by white arrowheads. The virus envelope proteins (*), electron-dense M protein layer beneath the surface of the filaments (long white arrows), RNPs within the virus filaments (short white arrows), and cortical cytoskeleton (black arrows) are highlighted. Scale bars, 100 (A) and 50 nm (B).
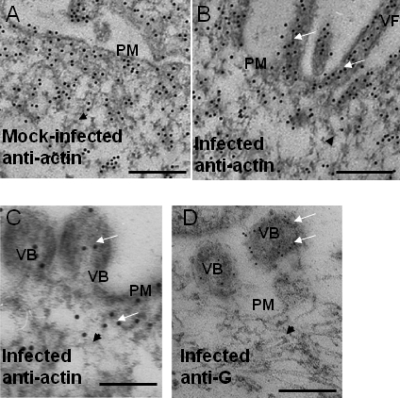
IST allowed us to visualize the process of RNP packaging into the virus filament (Fig. 2A). At the base of the filaments, structures that resembled the actin cytoskeleton was observed (Fig. 2A, indicated by a black arrow), but these were distinguishable from the significantly larger diameter and more electron-dense virus RNPs (Fig. 2A, indicated by a short white arrow). A role for the cortical actin network in virus filament formation has been suggested (24, 25, 42), and distribution of the cortical actin network in relation to the virus filaments was examined using immuno-TEM. Ultrathin sections were prepared from similar mock-infected and infected cells and labeled with anti-actin or anti-G, and the presence of the bound primary antibody was detected with a second antibody conjugated to either 5- or 10-nm colloidal gold (Fig. 3). In mock-infected and infected cells, actin-like structures near the plasma membrane were clearly visible (Fig. 3, highlighted by black arrowheads) that stained with anti-actin, indicating the presence of the cortical actin network (Fig. 3A). In virus-infected cells, similar structures were also seen under the plasma membrane that were clearly labeled with the actin antibody. In addition, we noted the presence of structures that resembled virus filaments (Fig. 3B, highlighted by white arrows) and that were labeled with the actin antibody. Similarly, we observed the presence of budding viruses that were labeled with anti-actin and, as expected, anti-G (Fig. 3, C and D, highlighted by white arrows), the latter recognizing the virus G protein.
Fig. 3.
Immuno-TEM of infected cells. Mock-infected and infected cells were embedded in Unicryl, and thin sections were prepared. These labeled with either anti-actin (A–C) or anti-G (D), and the presence of bound antibody was visualized using anti-mouse IgG conjugated to either 10- or 5-nm colloidal gold, respectively. The virus filaments (VF), plasma membrane (PM), and budding virus (VB) are highlighted. The cortical actin network (black arrowheads) and colloidal gold particles (white arrows) are indicated. Scale bar, 400 (A and B) and 200 nm (C and D).
Proteomics Analysis of RSV Particles
Although during virus infection RSV remains mainly cell-associated, upon cell detachment these particles change into their characteristic pleiomorphic morphology (1, 12). However, these cell-free particles retain their infectivity, and both the terms virus filaments and virus particles can be regarded as synonymous. To obtain a clearer description of the role that the host plays in the virus replication process, the virus particles were purified, and the protein content was examined. The rationale for this experimental approach was that host cell proteins that play a direct role in virus assembly may also co-purify with the virus particles, either by remaining attached to the virus filaments upon release from the infected cell or because they become incorporated into the virus particle, e.g. by inserting into the virus envelope. Recent proteomics analysis of purified influenza virus particles revealed the presence of low levels of contaminating host cell proteins (45) primarily due to the presence of exosomes released by the cell (46, 47). Although low levels of host cell protein contaminants may also be present in the RSV preparation, this strategy served as an initial screen to identify relevant cellular proteins that would then be further examined to confirm their virus association.
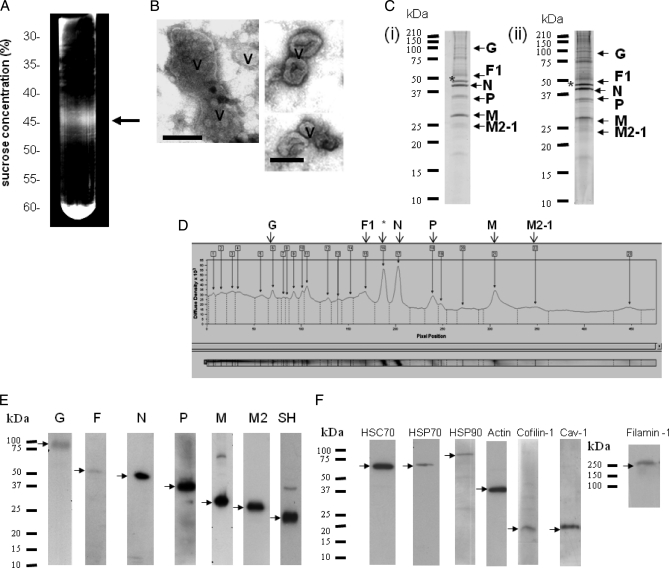
Purification of the virus particles was based on the method described previously (33) that has been used to obtain purified RSV suitable for a variety of purposes, e.g. examining the host response to RSV infection (e.g. Ref. 48). We modified the procedure by using buffers that contain relatively high levels of magnesium sulfate to stabilize the virus particles (49). In the final stage of the purification methodology, the visible opalescent virus band was harvested from the sucrose gradient (Fig. 4A) and was typically recovered in 2–3 ml with an average infectivity of 7 × 107 plaque-forming units/ml. Each purified virus preparation obtained from 1 × 108 cells produced an average yield of ∼320 μg of protein. Examination of the virus preparation by TEM revealed the presence of pleiomorphic virus particles that were between 100 and 300 nm in diameter (Fig. 4B), a size range and morphology similar to those reported previously (12). Although it is difficult to assess the degree of purity on the virus preparation using TEM, we failed to detect the presence of cellular debris, and the TEM analysis confirmed the presence of intact virus particles in the virus preparation. The peak fractions identified in the preparative sucrose gradient were pooled, and the recovered virus was examined by 15% SDS-PAGE to assess the purity of the preparation. Polyacrylamide gels stained using both Coomassie Brilliant Blue and SYPRO Ruby Red (SRR) revealed prominent protein bands at 50, 42, 35, and 28 kDa (Fig. 4C (i and ii)), which are the expected sizes for the F1, N, P, and M proteins. An additional 45-kDa protein band was also observed that is the expected size for actin and whose appearance has been reported previously (50). Densitometry analysis of the SRR-stained gel revealed that the N protein accounts for ∼23% of the protein detected and was the most abundant virus protein detected (Fig. 4D). The M and P proteins accounted for 13 and 10% of the total protein respectively. This analysis also indicated that the 45-kDa protein represented the most abundant cellular protein in the virus preparation, accounting for 20% of the total protein detected. As expected, because of the increased sensitivity of SRR staining, the presence of several minor protein species was also revealed. We hypothesized that some of these proteins may represent co-purifying virus-associated cellular proteins, and the virus preparation was examined further by using nano-LC MS/MS to identify significant proteins. The analysis was performed using two individual virus preparations, referred to as Samples 1 and 2. The peptides identified by the MS/MS analysis in each preparation were matched against the virus and human database as described under “Experimental Procedures.” After statistical analysis using PeptideProphet, InterProphet, and ProteinProphet (51, 52), a plot of sensitivity and error rate versus minimum protein probability was generated for each data set (supplemental Fig. S1, A and B). At a minimum protein probability of 0.95, the statistical analysis predicted 35 correct hits (85% of the predicted total number of correct hits) and no incorrect hit (0.1% error rate) for Sample 1, and 49 correct hits (83% of the predicted total number of correct hits) and no incorrect hits (0.2% error rate) for Sample 2. The decoy sequence hits did not appear in either of these data sets if the minimum protein probability was set at ≥0.1 or 0.3, respectively. The two lists are presented (supplemental Table S1, A and B) for evaluation of data reproducibility. MS/MS spectra for protein identifications based on a single peptide sequence (which may be observed multiple times and with different charge states) are shown (supplemental Fig. S2, A and B). The virus and human proteins or protein groups identified in both virus preparations are summarized in Table I.
Fig. 4.
Purification of virus particles by sucrose gradient centrifugation. A, RSV was purified as described under “Experimental Procedures.” At the final stage, the virus was applied to a 30–60% continuous sucrose gradient, the virus band was detected using a focused light, and the band was harvested (indicated by a black arrow). B, the material in the virus band was viewed by negative staining using an electron microscope (bar, 100 nm), and the presence of virus particles (V) was demonstrated. C, the proteins in the pooled virus fraction were separated by SDS-PAGE, and proteins were visualized in the polyacrylamide gel viewed after Coomassie Brilliant Blue (i) or SYPRO Ruby Red (ii) staining. Protein bands whose sizes correspond to known virus bands are labeled. *, this protein was identified as β-actin by MALDI-TOF/TOF analysis. D, the SYPRO Ruby Red-stained polyacrylamide gel was scanned using a densitometer to determine the relative abundance of proteins detected. Confirmation of the presence of either virus (E) or specific host cell (F) proteins in the virus preparation by immunoblotting is shown. Protein bands corresponding in size to the respective virus or host cell proteins are indicated (black arrows).
Table I. Protein analysis of purified virus particles.
In-gel digestion of purified viruses was performed, and the peptides were analyzed using LC MS/MS. Virus (A) and host (B) proteins identified in two virus preparations are shown. Homologous proteins are not listed when there is a specific protein identified in the same group. Protein groups are listed when there are no unique peptides identified to infer a specific protein. Numbers in parentheses are the number of unique peptides identified. Statistics collected from Sample 1 and Sample 2 are separated by “/.” When peptides identified can only infer a group of proteins, these entries are labeled with “group” in the “Accession number” column, and other statistics referring to a single protein are labeled with “NA.” For a complete list of proteins identified from Sample 1 or 2, refer to supplemental Table S1, A or B, respectively. sp, Swiss-Prot.
| Protein name | Accession number | Protein probability | Peptides total (unique) | Sequence coverage | Group probability |
|---|---|---|---|---|---|
| % | |||||
| A | |||||
| Nucleoprotein (protein N) (nucleocapsid protein) | sp|P03418|NCAP_HRSVA | 1/1 | 218 (11)/165 (12) | 51.4/51.2 | 1/1 |
| Matrix protein (M) | sp|P03419|MATRX_HRSVA | 1/1 | 239 (12)/192 (12) | 69.5/69.5 | 1/1 |
| Fusion glycoprotein | sp|P03420|FUS_HRSVA | 1/1 | 144 (14)/135 (13) | 39.9/44.4 | 1/1 |
| Phosphoprotein (protein P) | sp|P03421|PHOSP_HRSVA | 1/1 | 97 (6)/48 (6) | 51.9/44 | 1/1 |
| Matrix M2 (envelope-associated 22-kDa protein) | sp|P04545|M21_HRSVA | 1/1 | 90 (9)/94 (7) | 60.8/59.8 | 1/1 |
| Polymerase L | sp|P28887|L_HRSVA | 1/1 | 31 (8)/37 (10) | 7.2/8.8 | 1/1 |
| Major surface glycoprotein G | sp|P03423|GLYC_HRSVA | 1/0.9904 | 5 (1)/1 (1) | 5.4/5.4 | 1/0.9904 |
| B | |||||
| Chaperones | |||||
| Heat shock protein HSP90α | sp|P07900|HS90A_HUMAN/group | 0.9937/NA | 4 (2)/NA | 4.4/NA | 0.9999/1 |
| Heat shock protein HSP90β | sp|P08238|HS90B_HUMAN | 0.9937/1 | 4 (2)/14 (3) | 4.4/14.9 | 0.9999/1 |
| Heat shock cognate 71-kDa protein | Group/sp|P11142|HSP7C_HUMAN | NA/1 | NA/11 (5) | NA/21.1 | 1/1 |
| Heat shock 70-kDa protein 6 | Group/sp|P17066|HSP76_HUMAN | NA/0.9584 | NA/1 (1) | NA/5.1 | 1/1 |
| Cytoskeleton | |||||
| β-Actin-like protein 2 | Group/sp|Q562R1|ACTBL_HUMAN | NA/1 | NA/22 (3) | NA/14.1 | 1/1 |
| Cofilin-1 | sp|P23528|COF1_HUMAN | 1/1 | 23 (3)/7 (2) | 31.3/27.7 | 1/1 |
| Keratin, type II cytoskeletal 1 | sp|P04264|K2C1_HUMAN | 1/1 | 9 (2)/13 (6) | 6.1/16.3 | 1/1 |
| Moesin | sp|P26038|MOES_HUMAN | 0.9979/1 | 5 (1)/39 (5) | 6.4/23.3 | 0.9984/1 |
| Filamin A | sp|P21333|FLNA_HUMAN | 0.9989/1 | 2 (2)/22 (7) | 1/4.2 | 0.9989/1 |
| Tubulin, β | Group/group | NA/NA | NA/NA | NA/NA | 1/1 |
| Tubulin, α-1B | sp|P68363|TBA1B_HUMAN/group | 1/NA | 6 (2)/NA | 11.8/NA | 1/1 |
| Putative tubulin-like protein α-4B | Group/sp|Q9H853|TBA4B_HUMAN | NA/0.9999 | NA/6 (2) | NA/10.8 | 1/1 |
| Energy pathways | |||||
| Alkaline phosphatase | sp|P05187|PPB1_HUMAN | 1/1 | 50 (9)/52 (11) | 35.7/43.7 | 1/1 |
| Intestinal type alkaline phosphatase | sp|P09923|PPBI_HUMAN | 0.9999/1 | 4 (1)/20 (5) | 12.1/24.1 | 0.9999/1 |
| Pyruvate kinase | sp|P14618|KPYM_HUMAN | 0.9997/1 | 3 (1)/22 (7) | 5.8/26.4 | 0.9998/1 |
| Glyceraldehyde-3-phosphate dehydrogenase | sp|P04406|G3P_HUMAN | 0.9996/1 | 6 (2)/9 (2) | 25.9/16.1 | 0.9998/1 |
| Host immune response-related | |||||
| Putative annexin A2-like protein | sp|A6NMY6|AXA2L_HUMAN/group | 1/NA | 25 (6)/NA | 23.3/NA | 1/1 |
| Annexin A1 | sp|P04083|ANXA1_HUMAN | 0.9953/1 | 1 (1)/9 (5) | 13/23.1 | 0.9953/1 |
| Complement decay acceleration factor (CD55) | sp|P08174|DAF_HUMAN | 0.9978/1 | 17 (2)/37 (2) | 13.7/23.9 | 1/1 |
| Ubiquitination | |||||
| Ubiquitin | sp|P62988|UBIQ_HUMAN | 1/1 | 12 (3)/15 (3) | 44.7/56.6 | 1/1 |
| Transcription/translation | |||||
| Elongation factor 1α | Group/group | NA/NA | NA/NA | NA/NA | 1/1 |
| Membrane transport | |||||
| Monocarboxylate transporter 1 | sp|P53985|MOT1_HUMAN | 1/0.9983 | 2 (2)/2 (1) | 7.9/2.8 | 1/0.9983 |
| Monocarboxylate transporter 4 | sp|O15427|MOT4_HUMAN | 0.9953/1 | 4 (1)/6 (3) | 2.4/7.5 | 0.9953/1 |
| 4F2 cell surface antigen heavy chain | sp|P08195|4F2_HUMAN | 1/1 | 13 (5)/13 (5) | 11.7/15.2 | 1/1 |
| Others | |||||
| Brain acid-soluble protein 1 | sp|P80723|BASP1_HUMAN | 1/1 | 18 (6)/30 (7) | 42.3/61.2 | 1/1 |
Virus Proteins
Proteins associated with the virus polymerase (N, P, M2, and L proteins), the M protein, and the two main virus glycoproteins (F and G proteins) were identified (Table IA). The virus proteins identified by MS/MS showed high sequence coverage of between 40 and 70% except for L and G proteins. The G protein is extensively modified by O- and N-linked glycosylation, accounting for up to 50% of its mass in SDS-PAGE (53). This makes assigning of the G protein peptides difficult and presumably accounts for its relative poor sequence coverage. The presence of most of these virus proteins was confirmed by Western blotting (Fig. 4E). Although the MS/MS analysis revealed only a single G protein peptide, the high quality fragmentation spectra for this single peptide in both samples (supplemental Fig. S2, A and B) gave a good indication of its presence in the virus preparation. Furthermore, immunoblotting with a G protein-specific antibody revealed a single protein species of 90 kDa, the expected size for the fully mature form of the G protein. We identified 8 and 11 unique peptides for the L protein in Samples 1 and 2 respectively, corresponding to 7.2 and 8.8% sequence coverage respectively. The large size of the L protein presumably accounts for the relatively low sequence coverage. Although an L protein antibody has been described (19), such reagents are generally not available in the scientific community, and we were unable to confirm its presence by immunoblotting. The two virus-encoded non-structural proteins were not detected either in the MS/MS analysis or by immunoblotting (data not shown), consistent with their absence in the mature virus particles.
Host Cell Proteins
The host cell proteins identified in the MS/MS analysis were grouped into categories based on their major functional activities or cellular location. Cytoskeleton or cytoskeleton-associated proteins were most represented in this analysis, and most of these were actin-related (Table IB). We identified several actin isoforms in the virus preparations, including a recently described novel actin isoform called β-actin-like protein 2 (κ-actin) (54). In Table IB, although only κ-actin was identified with a non-degenerate peptide sequence, peptides corresponding to other actin isoforms, including α-actin and β-actin, were present in the sample (supplemental Table 1, A and B). Presumably, because of a high degree of sequence conservation among different actin isoforms (55), many of the actin-related peptides identified in the virus preparation are present in these different isoforms, accounting for these degenerate peptide identifications. Therefore, the actin isoforms identified in our analysis are listed within an actin group of proteins. Most of the actin-binding proteins identified play a role in regulating the structure of the cortical actin network, for example filamin-1 (56) and cofilin-1 (57). Furthermore, there are many more known actin-binding proteins than were observed in our proteomics analysis, suggesting that this small subset of actin-binding proteins represents a specific set of actin-binding proteins that are relevant to the virus particles. The next largest group was those proteins involved in energy pathways, accounting for 20% of the host cell proteins detected. Three chaperones, HSP70, HSC70, and HSP90, were also detected, and a role for chaperones in the replication cycle of several viruses has been described (58).
There are two major families of HSP90 proteins, which are referred to as HSP90A (located in the cytosol) and HSP90B (located in the endoplasmic reticulum), but only the human HSP90A protein was detected in the virus preparation. Two closely related HSP90A isoforms exist, HSP90α and HSP90β, which have 85% sequence identity. Peptides present in both HSP90A isoforms were identified in the virus preparation (supplemental Table 1, A and B), and this apparent degeneracy of the HSP90 peptides identified presumably reflects the high degree of sequence conservation between both HSP90A isoforms (59). We are currently unable to identify whether one or both isoforms were present in the virus preparation, and as a consequence, in this study, the term HSP90 refers to both HSP90A isoforms.
Validation of Proteomics Analysis
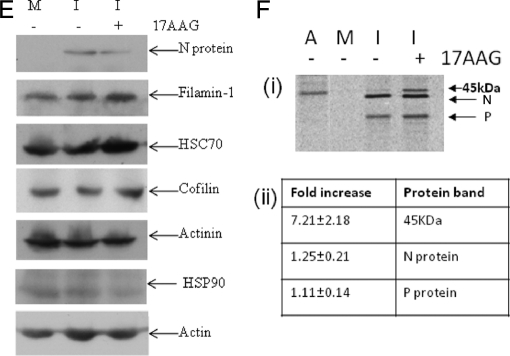
Virus-associated Host Cell Proteins
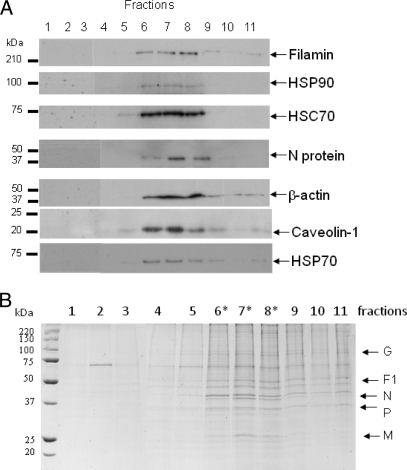
Although the analysis identified several host cell proteins that were confirmed previously as being RSV-associated, e.g. CD55, HSP70, and actin (41, 50, 60, 61), most of the cellular proteins identified in the pick list had no previous documented association with RSV. Because actin and HSP70 have both been previously implicated in RSV replication and shown to interact with the virus polymerase complex (16, 50, 61), the biological validation studies focused mainly on some of the new actin-associated proteins and chaperones identified in this proteomics analysis. The presence of specific cellular proteins in the virus preparation was first validated by immunoblotting using antibodies that recognize HSC70, HSP70, HSP90, filamin-1, β-actin, caveolin-1, and cofilin-1, and protein bands corresponding in size to these proteins were detected (Fig. 4F). We have previously demonstrated the presence of caveolin-1 in virus filaments (16). Although we failed to observe peptides corresponding to caveolin-1 in the pick list (Table IB), its presence was confirmed by immunoblotting. This observation is similar to our previous analysis on lipid raft membranes where the presence of this lipid raft-abundant protein was detected by immunoblotting but not by MS/MS (32). It is likely that the extremely hydrophobic nature of caveolin-1 accounts for the apparent inability to detect it by MS/MS in the virus preparations, and in our previous study. The sedimentation profile of filamin-1, HSP90, HSC70, HSP70, caveolin-1, and β-actin was compared with that of the virus N protein (the major virus structural protein) in the sucrose gradient by immunoblotting (Fig. 5A). This analysis revealed co-migration of all proteins, suggesting co-migration of the host cell proteins and virus particles. The peak fractions (Fractions 6–8) corresponded to the fractions in which the virus band was visualized (Fig. 5A) and that contained the highest level of protein in the gradient (Fig. 5B).
Fig. 5.
Co-migration of host cell proteins identified by MS/MS analysis and RSV N protein. The 30–60% continuous sucrose gradient was fractionated, the proteins in the individual gradient fractions were separated by SDS-PAGE, and either the presence of specific proteins was detected by immunoblotting using antibodies against filamin-1, HSP90, HSC70, β-actin, HSP70, caveolin-1, and the RSV N protein (Fraction 1, top; Fraction 11, bottom) (A) or the total proteins were visualized in the SYPRO Ruby Red-stained polyacrylamide gel (B). * indicates the peak virus fractions.
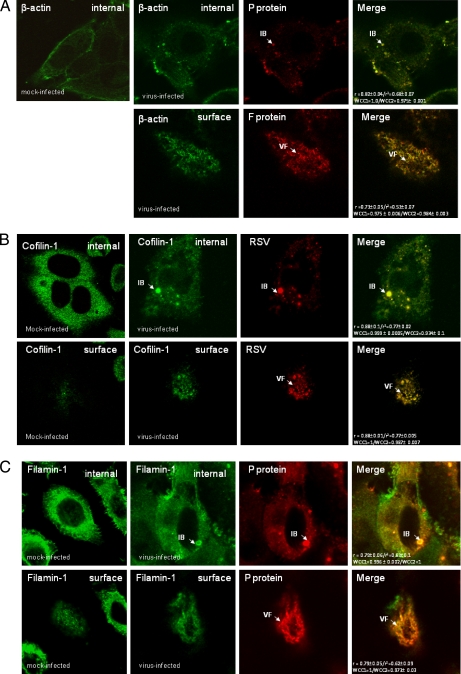
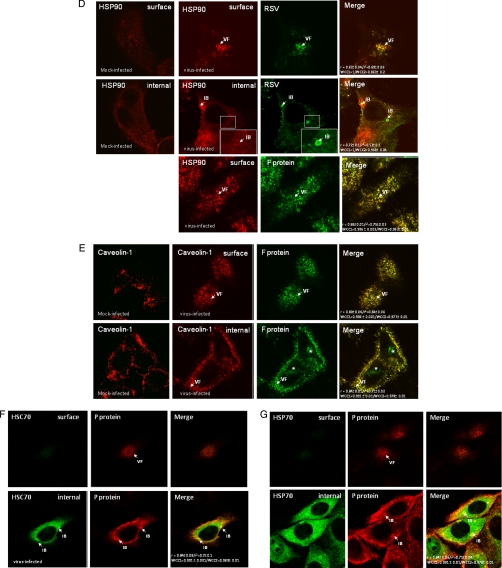
Because the virus-induced structures can be visualized using fluorescence microscopy (Fig. 1), this technique was used to confirm the virus association of these cellular protein. Cells were either mock-infected or infected with RSV, and at 24 hpi, the cells were fixed and stained using appropriate antibodies to visualize the virus filaments and inclusion bodies together with antibodies against either β-actin, filamin-1, cofilin-1, HSC70, caveolin-1, HSP70, or HSP90 (Fig. 6). The cells were examined at focal planes that allowed mainly visualization of the inclusion bodies (internal) or the virus filaments (surface). Staining of infected cells with either MAb19 or anti-P and anti-β-actin showed good levels of co-localization within the virus filaments and inclusion bodies (Fig. 6A), suggesting the presence of actin within these virus structures. Similarly, cells co-stained with antibodies against cofilin-1 (Fig. 6B), filamin-1 (Fig. 6C), and HSP90 (Fig. 6D) showed a significant level of co-localization within virus filaments and inclusion bodies. Significant levels of caveolin-1 were only detected in the virus filaments (Fig. 6E), supporting previous observations that RSV assembly occurs in lipid raft microdomains (16). Although anti-HSC70 failed to stain the virus filaments, a significant level of co-staining with anti-P protein was observed in the inclusion bodies (Fig. 6F). As expected, staining infected cells with anti-HSP70 and anti-P showed staining of HSP70 within the inclusion bodies, but no significant HSP70 staining within the virus filaments was observed (Fig. 6G), confirming previous observations (61). In staining combinations that exhibited co-localization within the virus filaments or inclusion bodies, high correlation coefficients (>0.9) and Pearson's coefficients (>0.8) were obtained in the merged images.
Fig. 6.
Validation of association with virus structures in infected cells. Cells were either mock- or virus-infected, and at 24 hpi, the cells were fixed and stained using β-actin and either anti-P or anti-F (A), anti-cofilin-1 and anti-RSV (B), anti-filamin-1 and anti-P (C), anti-HSP90 and either anti-RSV or anti-F (D), anti-caveolin-1 and anti-F (E), anti-HSC70 and anti-P (F), and anti-HSP70 and anti-P (G). The merged images showing both distributions in the same image are shown, and co-localization in the merged image is indicated by the yellow staining pattern and quantified using the correlation coefficient and Pearson's coefficients. The cells were viewed in a confocal microscope at optical planes representing predominantly the cell interior (internal) or surface. Golgi staining of the F protein (*), the inclusion bodies (IB), and virus filaments (VF) is highlighted.
Role for HSP90 in Virus Filament Formation
The above data suggested that actin and the actin-associated proteins filamin-1 and cofilin are readily detected in both the virus inclusion bodies and virus filaments. This suggests that that they may play an important role in virus assembly process and directly contribute to the architecture of the virus particle. In contrast, although HSC70 was detected in the inclusion bodies and the virus preparation, we could not detect its presence in virus filaments by fluorescence microscopy. We have reported similar findings for RSV-associated HSP70 (61) where a possible role in virus polymerase activity was suggested. This suggests that both HSP70 and HSC70 may be present in the virus particles but at relatively low levels. Therefore, the primary function of HSC70 may be related to the activity of the virus polymerase complex rather than playing a direct role in virus assembly, and its presence in the virus particle may therefore be due to packaging of the virus polymerase complex into the virus particles.
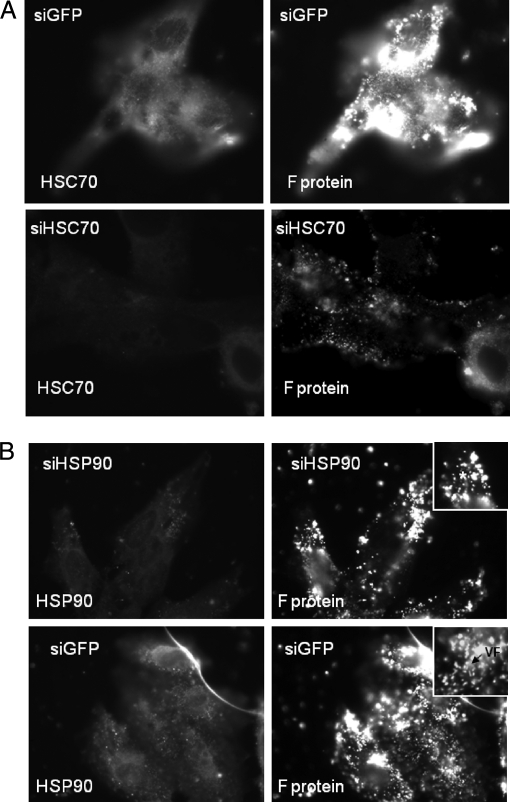
Previous studies have suggested a temporal increase in the expression of several HSPs during the early stages of RSV infection, including HSC70 and HSP90 (62), and we have confirmed this finding.2 However, although the changes in expression of these host cell proteins were suggestive of a role for HSC70 and HSP90 in the RSV replication cycle, no functional significance of either protein to virus replication was demonstrated in this earlier study. We therefore examined the effect of HSC70 on RSV replication using a specific siRNA molecule to inhibit the transient increase in HSC70 gene expression that occurs during the early phase of RSV infection. Infected cells were transfected either with siGFP or siHSC70, and at 20 hpi, the transfected cells were fixed and stained with anti-HSC70 and anti-F protein (Fig. 7A). The siGFP is targeted against GFP and was used as a control transfection to determine any effect of the transfection procedure on virus infection. In siGFP-transfected cells, staining of virus antigen revealed the presence of HSC70 and F protein staining at levels similar to that observed in non-transfected cells (Fig. 6). In contrast, in siHSC70-transfected cells, a significant reduction in HSC70 staining, which coincided with a reduction in the level of F protein staining, was seen, consistent with an important role of HSC70 in virus gene expression.
Fig. 7.
The effect of HSC70 and HSP90 expression on RSV replication is shown. Cells were transfected with siGFP or siHSC70 (A) or siGFP or siHSP90 (B). At 36 h post-transfection, the cells were infected with RSV using an m.o.i. of 1, and at 20 hpi, the cells were fixed and either stained using anti-F and anti-HSC70 (A) or stained with anti-F (MAb19) and anti-HSP90 (B). Insets, enlarged images showing the F protein staining pattern in siGFP- and siHSP90-tranfected cells. In all cases, the stained cells were imaged at ×100 (oil immersion) with a Nikon ECLIPSE TE2000-U using appropriate machine settings and identical exposure times. VF, virus filaments; * highlights the punctate F protein staining pattern in siHSP90 treated cells.
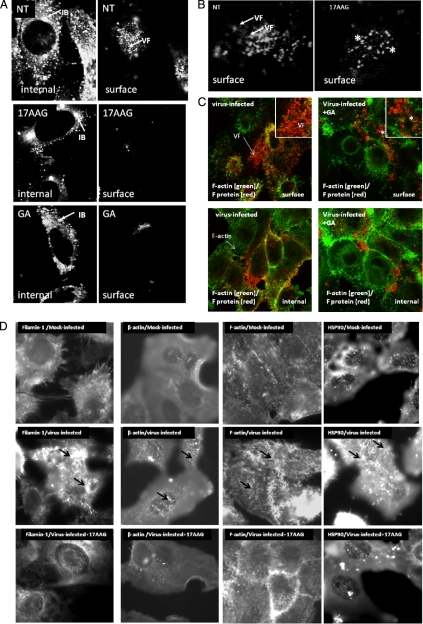
In contrast, the HSP90 protein was readily detected within the virus filaments, suggesting a role in virus assembly. The significance of the association of HSP90 and RSV was evaluated using GA and 17AAG, two well established specific small molecular weight inhibitors of HSP90 (63). Cells were infected with RSV, and at 8 hpi, the cells were either left non-treated or alternatively treated with either 2 μm GA or 2 μm 17AAG. At 18 hpi, the cells were fixed, labeled with anti-RSV or MAb19, and then examined by confocal microscopy (Fig. 8). As expected, in anti-RSV-labeled non-treated cells, the presence of inclusion bodies and numerous virus filaments was observed (Fig. 8A). We failed to detect the presence of the virus filaments in anti-RSV-labeled GA- or 17AAG-treated cells, although inclusion bodies were observed. Staining with MAb19 revealed a changed staining pattern for the F protein upon drug treatment (Fig. 8B); the filamentous staining pattern was replaced with a punctate staining pattern on the surface of drug-treated infected cells. Furthermore, co-staining of infected cells with MAb19 and phalloidin-FITC allowed both the virus filaments and F-actin network to be clearly distinguished in non-treated and GA-treated cells (Fig. 8C). In addition, the staining pattern of anti-β-actin, F-actin, anti-filamin-1, and HSP90 was examined in mock-infected, virus-infected, and 17AAG-treated virus-infected cells using fluorescence microscopy (Fig. 8D). We noted that although mock-infected cells stained with these antibodies showed a filamentous staining pattern this filamentous staining pattern was enhanced on the surface of infected cells. However, treatment of virus-infected cells with 17AAG greatly diminished this filamentous staining pattern (Fig. 8D). This is consistent with localized virus-induced changes in the structure of the F-actin network together with changes in the distribution of F-actin-associated proteins. However, these changes were not observed in virus-infected cells treated with 17AAG where the reduction in the filamentous staining pattern resembled that observed in mock-infected cells. Examination of actin and several actin-associated proteins in mock-infected, virus-infected, and virus-infected 17AAG-treated cells by immunoblotting with the relevant antibodies revealed similar levels of protein detection (Fig. 8E). This suggested that although an altered staining was observed following drug treatment this did not arise due to drug-induced degradation of these proteins.
Fig. 8.
Effect of HSP90 on progeny virus formation. Infected cells were either non-treated (NT) or treated with 17AAG or GA, and at 24 hpi, the cells were fixed and stained with either anti-RSV (A) or MAb19 (B). C, infected cells were either non-treated or GA-treated and stained using phalloidin-FITC (to visualize the F-actin network) and MAb19. The cells were then viewed in a confocal microscope (×64; oil immersion) at optical planes representing predominantly either the cell interior (internal) or surface. Insets, enlarged images showing the F protein staining pattern in non-treated and drug-treated cells. D, mock-infected, virus-infected, and virus-infected cells treated with 17AAG were stained using anti-β-actin, phalloidin-FITC, anti-filamin, and anti-HSP90, and the cells were imaged at ×100 (oil immersion) using a Nikon ECLIPSE TE2000-U. E, mock-infected and virus-infected cells (I), either non-treated (−) or treated (+) with 17-AAG, were harvested at 18 hpi and examined by immunoblot using relevant antibodies. The respective proteins are highlighted. F, mock-infected and virus-infected cells (I), either non-treated (−) or treated (+) with 17-AAG, were radiolabeled with [35S]methionine between 8 and 18 hpi. Lysates were prepared and immunoprecipitated using either anti-RSV or anti-β-actin and then examined by SDS-PAGE (i). The presence of the N and P proteins are indicated, and the presence of an additional band corresponding in size to actin was also found in the treated cells. Immunoprecipitation of mock-infected cells (M) using anti-RSV or actin (A) is also shown. ii, the radiolabeled bands in the non-treated and 17AAG-treated virus-infected cells were quantified by densitometry, which allowed determination of the -fold increase of the relevant protein bands in the 17AAG-treated cells compared with the non-treated cells.
The effect of HSC90 expression on RSV replication was further examined using siRNA targeted against HSP90 gene expression. HEp2 cells were transfected with either siHSC90 or siGFP, and at 20 hpi, the infected cells were fixed, stained using anti-HSP90 and MAb13 (F protein), and examined by fluorescence microscopy (Fig. 7B). In siGFP-transfected cells, staining using MAb13 revealed the presence of HSC70 and virus filaments. In contrast, in siHSC90-transfected cells, a significant reduction in HSP90 gene expression was observed that coincided with a prominent punctate F protein staining pattern, consistent with the F protein staining pattern on drug-treated cells.
The microscopic and Western blot analysis suggested that similar levels of virus protein were expressed in the absence and presence of the drug, and this was confirmed by examining the effect of 17AAG on the expression levels of the N protein in [35S]methionine-labeled cells (Fig. 8F). Virus-infected cells were radiolabeled with [35S]methionine in the presence and absence of drug treatment, and virus N protein was immunoprecipitated using anti-N (Fig. 8F (i)). In the absence of drug, a protein band corresponding in size to the N protein was observed, and the level of this protein remained unchanged following drug treatment. The P protein is known to interact with the N protein within the RNP, and in virus-infected cells, it is usually co-precipitated with the N protein (17). Similarly, previous studies have also demonstrated that the N protein interacts with actin and that actin can be co-precipitated with the N protein (50). Although we observed similar levels of co-precipitated P protein in the absence or presence of drug, in drug-treated cells there was an approximate 7-fold increase in the co-precipitation of a band corresponding in size to actin (Fig. 8F (ii)), suggesting that inhibiting HSP90 may alter the association between actin and the N protein and, by extrapolation, between actin and the virus RNP.
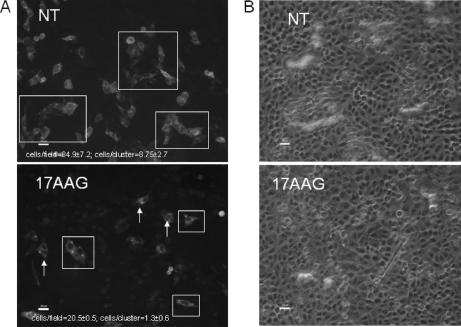
RSV remains largely cell-associated, and infection occurs via the virus filaments by direct cell to cell contact. In a final analysis, we examined the effect of HSP90 on virus infectivity and transmission (Fig. 9). Cell monolayers were infected with RSV using an m.o.i. of 0.1, and at 8 hpi, the cells were either mock-treated or treated with 17AAG. At 30 hpi, the cells were fixed, labeled using anti-RSV, and examined at low magnification using bright field and fluorescence microscopy (Nikon ECLIPSE TE2000-U). Several individual images from non-treated and drug-treated (17AAG) infected cells were obtained, and the average number of stained (i.e. infected) cells was estimated in each image. In the non-treated cells, an average of 84.9 ± 7.2 cells/image field of view was observed compared with 20.5 ± 0.5 observed in the drug-treated cells. In addition, we noted in the absence of drug clusters of labeled cells within the monolayers that consisted of 8.75 ± 2.7 cells/cluster compared with 1.3 ± 0.60 cells/cluster in drug-treated cells. This observation is consistent with the efficient cell to cell spread of virus from the initial point of infection in non-treated cells and a reduced efficiency of cell to cell transmission in the presence of 17AAG, observations that were consistent with impaired virus maturation in the presence of 17AAG.
Fig. 9.
Inhibiting HSP90 activity impairs cell transmission of RSV. Cells were infected using an m.o.i. of 0.1, and at 8 hpi, the cells were treated with 17AAG. At 30 hpi, the cells were fixed and stained using anti-RSV. The same cell area was viewed at ×20 magnification using fluorescence (A) and bright field (B) microscopy. The presence of clusters of labeled cells (white boxes) or individual labeled cells (white arrows) are highlighted. Scale bar, 20 μm. The average numbers of stained cells per image and the average numbers of stained cells per cluster (highlighted by white boxes) were estimated in both non-treated and 17AAG-treated cells. NT, non-treated.
DISCUSSION
Structure of Virus Filaments and Inclusion Bodies
The increasing body of experimental evidence suggests that at various stages of the RSV replication cycle there are a complex series of interactions between components of the virus and the host cell (for a review, see Ref. 64). For example, the functional association of the cortical actin network and lipid raft microdomains has been proposed (65–67), and both these cell structures are implicated in virus filament formation (16, 21–25). At least 25 host cell proteins that co-purified with the virus particles were identified, and we can speculate about their possible role during virus morphogenesis based on our understanding of their cellular functions. Actin plays a role in the morphogenesis of several viruses (68), and recent studies have identified an association between actin and both inclusion bodies and virus filaments in virus-infected cells (24, 25). In our current analysis, actin was detected in these virus structures, supporting earlier studies showing that the actin network plays an important role in RSV maturation (50). A role for actin as a cofactor for the virus polymerase complex has also been demonstrated in virus-infected cells, and its association with partially purified RSV preparations has been demonstrated (60). Several early studies have identified actin in purified preparations of other related paramyxovirus preparations (69, 70), suggesting that its presence is a common feature of this virus family. It was also interesting that several actin-binding proteins were also detected in the proteomics analysis, and the implications of these are discussed.
RSV-infected cells were examined in unprecedented detail using three-dimensional imaging, which suggests that virus filaments appear to package several RNPs, consistent with a recent observation (71). This suggests that the RSV filament structure would be an efficient way for a single virus particle to transfer multiple copies of the virus genome into an adjacent non-infected cell. The proteomics analysis also suggested that the virus filaments are complex structures consisting of both virus structural proteins and a small number of cellular proteins. Our imaging analysis suggested a physical association between the virus filaments and inclusion bodies, consistent with recent observations (25), and it was interesting to note that most of the cellular proteins were present in both the virus filaments and inclusion bodies. This provides supporting evidence that inclusion bodies may play a role in the recruitment of host and virus factors prior to packaging into newly assembled virus, consistent with recent reports that virus proteins can be transported from these inclusion bodies into maturing virus filaments (42).
The presence of virus-associated cofilin-1, caveolin-1, and filamin-1 in the mature virus was also detected in this analysis, and the presence of filamin-1 together with actin in the virus particles may indicate an important role in maintaining the virus architecture. For example, filamin-1 is a 280-kDa protein that cross-links actin filaments into orthogonal networks within the cortical actin network. It has been implicated in the formation of membrane protrusions, presumably enhancing the stability of these structures (72). Cofilin-1 was also detected in this analysis. It is able to promote the extension of F-actin strands, and its presence is consistent with a role for the actin-binding protein profilin in virus RSV morphogenesis (73). Although we were able to demonstrate the presence of actin in the virus filaments by antibody labeling, we have previously been unable to stain these virus structures using phalloidin (25), suggesting that actin may be present either as actin monomers (G-actin) or in a form of F-actin that cannot bind phalloidin.
RSV assembly occurs in lipid raft microdomains (21, 23), and in addition to the actin-binding proteins described above, several raft-associated proteins were also detected, including caveolin-1, placental alkaline phosphatase, and CD55. The majority of cellular proteins identified in our previous studies on lipid raft membranes isolated from infected HEp2 cells (32) were different from those identified in the virus preparation. However, alkaline phosphatase has been detected with the most number (11) of unique peptides of all host cell proteins in both current and previous studies, presumably reflecting its relative abundance in lipid raft membranes and hence its presence in the purified virus preparation. The biological consequence of the presence of these raft-associated proteins in the virus envelope is currently unclear, and this will require further examination. The presence of CD55 in the envelope of virus filaments has been demonstrated previously using immuno-TEM (41). It is envisaged that the presence of host cell proteins that can prevent complement-mediated virus inactivation would have advantages for the virus. The presence of CD55 in the envelope of HIV-1 has been shown to increase its resistance to complement-mediated inactivation (74, 75). It is unclear whether the RSV-associated CD55 can also perform a similar role, and this will require further examination.
The presence of caveolin-1 in virus filaments has been reported previously (16), and it is also interesting to note that filamin-1 has been shown to bind to caveolin-1 (76), an interaction that creates a physical connection between actin and raft membranes. Although it is not clear whether a similar interaction also occurs in the virus filaments, it may suggest that these proteins mediate the interaction of actin with the virus envelope in the virus structures. The presence of caveolin-1 and several lipid raft-associated proteins also suggests that the virus envelope may have properties that are akin to those in cellular lipid raft membranes and supports previous observations for the assembly of virus filaments within lipid raft microdomains (21). Recent studies have suggested that lipid raft membranes may require the cell cytoskeleton for their maintenance and stability, observations that emphasize the possible functional interaction between the cortical actin network and these membrane structures (77).
HSP90 Plays a Role in Formation of Virus Filaments
Chaperones play important roles in the replication cycle of several viruses (58), and previous studies have demonstrated temporal changes in the expression levels of several chaperones during RSV infection (62). Our studies presented here demonstrate an association between RSV and at least three cellular chaperones, namely HSP70, HSC70, and HSP90. An interaction between HSP70 and the RSV polymerase complex has been demonstrated in virus-infected cells where HSP70 enhanced the enzyme activity of the virus polymerase complex (61) in a manner similar to that reported for measles virus (78). Similarly, an association between the virus and HSC70 was also demonstrated. Our data provided evidence for the association of HSP90 with virus inclusion bodies and virus filaments, consistent with its increased expression in RSV-infected cells (62). Geldanamycin and 17AAG are established specific inhibitors of HSP90, and although drug treatment did not appear to inhibit the production of virus proteins or the formation of inclusion bodies, virus filament formation and virus cell to cell transmission were impaired. The interaction between actin and the virus polymerase complex in RSV-infected cells has been documented, and this appears to have important consequences for the activity of the polymerase (60). It is interesting to note that inhibiting HSP90 activity correlated with an increased co-precipitation of actin with the N protein, suggesting that HSP90 may play a role in the interaction of actin with the N protein and, by extrapolation, the virus RNP. The increased association of actin with the N protein may be related to the impaired transport of the N protein into the virus filaments that we observed.
A number of specific functions have been ascribed to HSP90 that range from protein folding, intracellular transport, proteasome degradation of proteins, and mediating the interaction of specific proteins with actin (79), and several client proteins of HSP90, for example N-WASP (80), are known. N-WASP is activated by HSP90 and mediates actin reorganization by the arp2/3 complex (80–84), and N-WASP has been implicated in virus maturation (85–88). N-WASP was not detected in the RSV preparation, and it is currently unclear whether the changes observed during RSV assembly involve either N-WASP or the arp2/3 complex. In addition, the actin cytoskeleton regulatory protein rhoA and its down effectors have been implicated in RSV assembly (28–30), and the activation of rhoA via HSP90 has been suggested (89, 90). It is therefore possible that HSP90 may play an early role in the activation of cellular pathways that lead to virus filament formation. Other virus-specific activities have also been associated with HSP90, such as the stimulation of the influenza virus polymerase complex (91), the stabilization of virus polymerases (92), and the formation of the mature influenza virus polymerase complex (93). Although HSP90 appears to be important for RSV assembly, the precise mechanism involved will require further examination and will form the basis of future studies on RSV maturation.
Exosomes represent a source of minor contaminants in the virus preparation, and exosome-associated proteins have been reported in purified virus preparations in other studies (45). We did not validate the virus association of some proteins identified in our proteomics analysis, and it is possible that they may be derived from similar cell structures. Although several cellular proteins identified in our analysis have not been reported previously in exosomes (e.g. filamin-1 and cofilin-1), several, including HSP90 and HSC70 (47), have been reported. Our analysis showed clear evidence of HSC70 and HSP90 virus association and, in the case of HSP90, a role in virus filaments formation. Exosomes form in cellular structures called multivesicular bodies (MVBs), and the fusion of MVBs with the cell plasma membrane releases the exosomes from the cell. Several viruses have been shown to use MVB in the mechanism of cell release (94), and the presence of several exosome-associated proteins in the virus filaments may indicate a role for MVB in RSV assembly, although this possibility will require further examination.
The virus association of several cellular proteins identified in the proteomics analysis was not validated, and their relevance to what we currently know about RSV assembly is unclear. For example, β-enolase and α-enolase were specifically identified in Samples 1 and 2, respectively (supplemental Tables S1, A and B). Enolase is an enzyme that is involved in the glycolytic pathway and has been detected in the purified preparations of other viruses. It is interesting to note that some biological activities have been ascribed to enolase in other related viruses, for example in the transcription of Sendai virus vRNA (95). Its presence in the RSV preparation may indicate a similar function during RSV replication and may provide the basis for future investigations in new areas of RSV research. The identification of these cellular proteins in our study is a first step toward improving our understanding of the RSV maturation process, and future studies will focus on understanding the molecular basis for the role that they play in virus particle formation. This increased understanding may also facilitate the development of novel strategies for the prevention of RSV infection. For example, effective drugs have already been developed that target the cellular chaperones identified in our analysis (96–99). In addition, understanding the molecular basis of the interaction between lipid raft membranes and the cortical actin network during virus assembly may serve as a useful model system with which to facilitate our understanding of the general molecular processes involved in the formation of cell surface structures. Finally, it is also worth pointing out that although RSV has been the focus of this study several other negative strand RNA viruses mature as filamentous particles on the surface of infected cells. It will be interesting to determine whether host cell proteins similar to those identified in this study are also involved in the assembly and maturation of these other viruses.
Supplementary Material
* This work was supported by the Medical Research Council (UK), Defense Science Technology Agency Singapore, and Singapore-Massachusetts Institute of Technology Alliance for Research and Technology.
 This article contains supplemental Figs. S1 and S2 and Table S1.
This article contains supplemental Figs. S1 and S2 and Table S1.
2 D. Yeo, B. H. Tan, and R. J. Sugrue, unpublished observations.
1 The abbreviations used are:
- RSV
- respiratory syncytial virus
- 17AAG
- 17-allylaminogeldanamycin
- RNP
- ribonucleoparticle
- vRNA
- viral genomic RNA
- N
- nucleocapsid
- P
- phospho
- L
- large
- F
- fusion
- m.o.i.
- multiplicity of infection
- GA
- geldanamycin
- HBSS
- Hanks' buffered saline solution
- MAb
- monoclonal antibody
- ABC
- ammonium bicarbonate
- FA
- formic acid
- TPP
- Trans Proteomic Pipeline
- hpi
- h postinfection
- TEM
- transmission electron microscopy
- IST
- in situ tomography
- SRR
- SYPRO Ruby Red
- MVB
- multivesicular body.
REFERENCES
- 1.Norrby E., Marusyk H., Orvell C. (1970) Morphogenesis of respiratory syncytial virus in a green monkey kidney cell line (Vero). J. Virol. 6, 237–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grosfeld H., Hill M. G., Collins P. L. (1995) RNA replication by respiratory syncytial virus (RSV) is directed by the N, P, and L proteins; transcription also occurs under these conditions but requires RSV superinfection for efficient synthesis of full-length mRNA. J. Virol. 69, 5677–5686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu Q., Hardy R. W., Wertz G. W. (1995) Functional cDNA clones of the human respiratory syncytial (RS) virus N, P, and L proteins support replication of RS virus genomic RNA analogs and define minimal trans-acting requirements for RNA replication. J. Virol. 69, 2412–2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins P. L., Hill M. G., Cristina J., Grosfeld H. (1996) Transcription elongation factor of respiratory syncytial virus, a nonsegmented negative-strand RNA virus. Proc. Natl. Acad. Sci. U.S.A. 93, 81–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hardy R. W., Wertz G. W. (1998) The product of the respiratory syncytial virus M2 gene ORF1 enhances readthrough of intergenic junctions during viral transcription. J. Virol. 72, 520–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bermingham A., Collins P. L. (1999) The M2–2 protein of human respiratory syncytial virus is a regulatory factor involved in the balance between RNA replication and transcription. Proc. Natl. Acad. Sci. U.S.A. 96, 11259–11264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fearns R., Collins P. L. (1999) Role of the M2–1 transcription antitermination protein of respiratory syncytial virus in sequential transcription. J. Virol. 73, 5852–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghildyal R., Mills J., Murray M., Vardaxis N., Meanger J. (2002) Respiratory syncytial virus matrix protein associates with nucleocapsids in infected cells. J. Gen. Virol. 83, 753–757 [DOI] [PubMed] [Google Scholar]
- 9.Levine S., Klaiber-Franco R., Paradiso P. R. (1987) Demonstration that glycoprotein G is the attachment protein of respiratory syncytial virus. J. Gen. Virol. 68, 2521–2524 [DOI] [PubMed] [Google Scholar]
- 10.Scheid A., Choppin P. W. (1977) Two disulfide-linked polypeptide chains constitute the active F protein of paramyxoviruses. Virology 80, 54–66 [DOI] [PubMed] [Google Scholar]
- 11.Schlender J., Bossert B., Buchholz U., Conzelmann K. K. (2000) Bovine respiratory syncytial virus nonstructural proteins NS1 and NS2 cooperatively antagonize alpha/beta interferon-induced antiviral response. J. Virol. 74, 8234–8242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bächi T., Howe C. (1973) Morphogenesis and ultrastructure of respiratory syncytial virus. J. Virol. 12, 1173–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parry J. E., Shirodaria P. V., Pringle C. R. (1979) Pneumoviruses: the cell surface of lytically and persistently infected cells. J. Gen. Virol. 44, 479–491 [DOI] [PubMed] [Google Scholar]
- 14.Roberts S. R., Compans R. W., Wertz G. W. (1995) Respiratory syncytial virus matures at the apical surfaces of polarized epithelial cells. J. Virol. 69, 2667–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arslañagic E., Matsumoto M., Suzuki K., Nerome K., Tsutsumi H., Hung T. (1996) Maturation of respiratory syncytial virus within HEp-2 cell cytoplasm. Acta Virol. 40, 209–214 [PubMed] [Google Scholar]
- 16.Brown G., Aitken J., Rixon H. W., Sugrue R. J. (2002) Caveolin-1 is incorporated into mature respiratory syncytial virus particles during virus assembly on the surface of virus-infected cells. J. Gen. Virol. 83, 611–621 [DOI] [PubMed] [Google Scholar]
- 17.García J., García-Barreno B., Vivo A., Melero J. A. (1993) Cytoplasmic inclusions of respiratory syncytial virus-infected cells: formation of inclusion bodies in transfected cells that coexpress the nucleoprotein, the phosphoprotein, and the 22K protein. Virology 195, 243–247 [DOI] [PubMed] [Google Scholar]
- 18.Santangelo P., Nitin N., LaConte L., Woolums A., Bao G. (2006) Live-cell characterization and analysis of a clinical isolate of bovine respiratory syncytial virus, using molecular beacons. J. Virol. 80, 682–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carromeu C., Simabuco F. M., Tamura R. E., Farinha Arcieri L. E., Ventura A. M. (2007) Intracellular localization of human respiratory syncytial virus L protein. Arch. Virol. 152, 2259–2263 [DOI] [PubMed] [Google Scholar]
- 20.Parham D. M., Bozeman P., Killian C., Murti G., Brenner M., Hanif I. (1993) Cytologic diagnosis of respiratory syncytial virus infection in a bronchoalveolar lavage specimen from a bone marrow transplant recipient. Am. J. Clin. Pathol. 99, 588–592 [DOI] [PubMed] [Google Scholar]
- 21.Brown G., Rixon H. W., Sugrue R. J. (2002) Respiratory syncytial virus assembly occurs in GM1-rich regions of the host-cell membrane and alters the cellular distribution of tyrosine phosphorylated caveolin-1. J. Gen. Virol. 83, 1841–1850 [DOI] [PubMed] [Google Scholar]
- 22.Jeffree C. E., Rixon H. W., Brown G., Aitken J., Sugrue R. J. (2003) Distribution of the attachment (G) glycoprotein and GM1 within the envelope of mature respiratory syncytial virus filaments revealed using field emission scanning electron microscopy. Virology 306, 254–267 [DOI] [PubMed] [Google Scholar]
- 23.McCurdy L. H., Graham B. S. (2003) Role of plasma membrane lipid microdomains in respiratory syncytial virus filament formation. J. Virol. 77, 1747–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kallewaard N. L., Bowen A. L., Crowe J. E., Jr. (2005) Cooperativity of actin and microtubule elements during replication of respiratory syncytial virus. Virology 331, 73–81 [DOI] [PubMed] [Google Scholar]
- 25.Jeffree C. E., Brown G., Aitken J., Su-Yin D. Y., Tan B. H., Sugrue R. J. (2007) Ultrastructural analysis of the interaction between F-actin and respiratory syncytial virus during virus assembly. Virology 369, 309–323 [DOI] [PubMed] [Google Scholar]
- 26.Brown J. H., Del Re D. P., Sussman M. A. (2006) The Rac and Rho hall of fame: a decade of hypertrophic signaling hits. Circ. Res. 98, 730–742 [DOI] [PubMed] [Google Scholar]
- 27.Gower T. L., Graham B. S. (2001) Antiviral activity of lovastatin against respiratory syncytial virus in vivo and in vitro. Antimicrob. Agents Chemother. 45, 1231–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gower T. L., Pastey M. K., Peeples M. E., Collins P. L., McCurdy L. H., Hart T. K., Guth A., Johnson T. R., Graham B. S. (2005) RhoA signaling is required for respiratory syncytial virus-induced syncytium formation and filamentous virion morphology. J. Virol. 79, 5326–5336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas K. W., Monick M. M., Staber J. M., Yarovinsky T., Carter A. B., Hunninghake G. W. (2002) Respiratory syncytial virus inhibits apoptosis and induces NF-kappa B activity through a phosphatidylinositol 3-kinase-dependent pathway. J. Biol. Chem. 277, 492–501 [DOI] [PubMed] [Google Scholar]
- 30.Yeo D. S., Chan R., Brown G., Ying L., Sutejo R., Aitken J., Tan B. H., Wenk M. R., Sugrue R. J. (2009) Evidence that selective changes in the lipid composition of raft-membranes occur during respiratory syncytial virus infection. Virology 386, 168–182 [DOI] [PubMed] [Google Scholar]
- 31.Arcaro A., Aubert M., Espinosa del Hierro M. E., Khanzada U. K., Angelidou S., Tetley T. D., Bittermann A. G., Frame M. C., Seckl M. J. (2007) Critical role for lipid raft-associated Src kinases in activation of PI3K-Akt signalling. Cell. Signal. 19, 1081–1092 [DOI] [PubMed] [Google Scholar]
- 32.McDonald T. P., Pitt A. R., Brown G., Rixon H. W., Sugrue R. J. (2004) Evidence that the respiratory syncytial virus polymerase complex associates with lipid rafts in virus-infected cells: a proteomic analysis. Virology 330, 147–157 [DOI] [PubMed] [Google Scholar]
- 33.Ueba O. (1978) Respiratory syncytial virus. I. Concentration and purification of the infectious virus. Acta Med. Okayama 32, 265–272 [PubMed] [Google Scholar]
- 34.Sugrue R. J., Brown C., Brown G., Aitken J., McL Rixon H. W. (2001) Furin cleavage of the respiratory syncytial virus fusion protein is not a requirement for its transport to the surface of virus-infected cells. J. Gen. Virol. 82, 1375–1386 [DOI] [PubMed] [Google Scholar]
- 35.Han C. L., Chien C. W., Chen W. C., Chen Y. R., Wu C. P., Li H., Chen Y. J. (2008) A multiplexed quantitative strategy for membrane proteomics: opportunities for mining therapeutic targets for autosomal dominant polycystic kidney disease. Mol. Cell. Proteomics 7, 1983–1997 [DOI] [PubMed] [Google Scholar]
- 36.Lu X., Zhu H. (2005) Tube-gel digestion: a novel proteomic approach for high throughput analysis of membrane proteins. Mol. Cell. Proteomics 4, 1948–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mastronarde D. (2003) SerialEM: a program for automated tilt series acquisition on Tecnai microscopes using prediction of specimen position. Microsc. Microanal. 9, 1182–1183 [Google Scholar]
- 38.Mastronarde D. N. (2005) Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 [DOI] [PubMed] [Google Scholar]
- 39.Kremer J. R., Mastronarde D. N., McIntosh J. R. (1996) Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 [DOI] [PubMed] [Google Scholar]
- 40.Frangakis A. S., Hegerl R. (2001) Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J. Struct. Biol. 135, 239–250 [DOI] [PubMed] [Google Scholar]
- 41.Brown G., Jeffree C. E., McDonald T., Rixon H. W., Aitken J. D., Sugrue R. J. (2004) Analysis of the interaction between respiratory syncytial virus and lipid-rafts in Hep2 cells during infection. Virology 327, 175–185 [DOI] [PubMed] [Google Scholar]
- 42.Santangelo P. J., Bao G. (2007) Dynamics of filamentous viral RNPs prior to egress. Nucleic Acids Res. 35, 3602–3611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang W., Ludtke S. J. (2005) Electron cryomicroscopy of single particles at subnanometer resolution. Curr. Opin. Struct. Biol. 15, 571–577 [DOI] [PubMed] [Google Scholar]
- 44.Rixon H. W., Brown G., Aitken J., McDonald T., Graham S., Sugrue R. J. (2004) The small hydrophobic (SH) protein accumulates within lipid-raft structures of the Golgi complex during respiratory syncytial virus infection. J. Gen. Virol. 85, 1153–1165 [DOI] [PubMed] [Google Scholar]
- 45.Shaw M. L., Stone K. L., Colangelo C. M., Gulcicek E. E., Palese P. (2008) Cellular proteins in influenza virus particles. PLoS Pathog. 4, e1000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Denzer K., Kleijmeer M. J., Heijnen H. F., Stoorvogel W., Geuze H. J. (2000) Exosome: from internal vesicle of the multivesicular body to intercellular signaling device. J. Cell Sci. 113, 3365–3374 [DOI] [PubMed] [Google Scholar]
- 47.Hegmans J. P., Bard M. P., Hemmes A., Luider T. M., Kleijmeer M. J., Prins J. B., Zitvogel L., Burgers S. A., Hoogsteden H. C., Lambrecht B. N. (2004) Proteomic analysis of exosomes secreted by human mesothelioma cells. Am. J. Pathol. 164, 1807–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu P., Li K., Garofalo R. P., Brasier A. R. (2008) Respiratory syncytial virus induces RelA release from cytoplasmic 100-kDa NF-kappa B2 complexes via a novel retinoic acid-inducible gene-I{middle dot}NF-kappa B-inducing kinase signaling pathway. J. Biol. Chem. 283, 23169–23178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fernie B. F., Gerin J. L. (1980) The stabilization and purification of respiratory syncytial virus using MgSO4. Virology 106, 141–144 [DOI] [PubMed] [Google Scholar]
- 50.Ulloa L., Serra R., Asenjo A., Villanueva N. (1998) Interactions between cellular actin and human respiratory syncytial virus (HRSV). Virus Res. 53, 13–25 [DOI] [PubMed] [Google Scholar]
- 51.Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 52.Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 53.Collins P. L., Mottet G. (1992) Oligomerization and post-translational processing of glycoprotein G of human respiratory syncytial virus: altered O-glycosylation in the presence of brefeldin A. J. Gen. Virol. 73, 849–863 [DOI] [PubMed] [Google Scholar]
- 54.Chang K. W., Yang P. Y., Lai H. Y., Yeh T. S., Chen T. C., Yeh C. T. (2006) Identification of a novel actin isoform in hepatocellular carcinoma. Hepatol. Res. 36, 33–39 [DOI] [PubMed] [Google Scholar]
- 55.Hennessey E. S., Drummond D. R., Sparrow J. C. (1993) Molecular genetics of actin function. Biochem. J. 291, 657–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gardel M. L., Nakamura F., Hartwig J. H., Crocker J. C., Stossel T. P., Weitz D. A. (2006) Prestressed F-actin networks cross-linked by hinged filamins replicate mechanical properties of cells. Proc. Natl. Acad. Sci. U.S.A. 103, 1762–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bamburg J. R., Wiggan O. P. (2002) ADF/cofilin and actin dynamics in disease. Trends Cell Biol. 12, 598–605 [DOI] [PubMed] [Google Scholar]
- 58.Mayer M. P. (2005) Recruitment of Hsp70 chaperones: a crucial part of viral survival strategies. Rev. Physiol. Biochem. Pharmacol. 153, 1–46 [DOI] [PubMed] [Google Scholar]
- 59.Chen B., Piel W. H., Gui L., Bruford E., Monteiro A. (2005) The HSP90 family of genes in the human genome: insights into their divergence and evolution. Genomics 86, 627–637 [DOI] [PubMed] [Google Scholar]
- 60.Burke E., Dupuy L., Wall C., Barik S. (1998) Role of cellular actin in the gene expression and morphogenesis of human respiratory syncytial virus. Virology 252, 137–148 [DOI] [PubMed] [Google Scholar]
- 61.Brown G., Rixon H. W., Steel J., McDonald T. P., Pitt A. R., Graham S., Sugrue R. J. (2005) Evidence for an association between heat shock protein 70 and the respiratory syncytial virus polymerase complex within lipid-raft membranes during virus infection. Virology 338, 69–80 [DOI] [PubMed] [Google Scholar]
- 62.Brasier A. R., Spratt H., Wu Z., Boldogh I., Zhang Y., Garofalo R. P., Casola A., Pashmi J., Haag A., Luxon B., Kurosky A. (2004) Nuclear heat shock response and novel nuclear domain 10 reorganization in respiratory syncytial virus-infected a549 cells identified by high-resolution two-dimensional gel electrophoresis. J. Virol. 78, 11461–11476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schulte T. W., Neckers L. M. (1998) The benzoquinone ansamycin 17-allylamino-17-demethoxygeldanamycin binds to HSP90 and shares important biologic activities with geldanamycin. Cancer Chemother. Pharmacol. 42, 273–279 [DOI] [PubMed] [Google Scholar]
- 64.Sugrue R. J. (2006) Interactions between respiratory syncytial virus and the host cell: opportunities for antivirus strategies? Expert Rev. Mol. Med. 8, 1–17 [DOI] [PubMed] [Google Scholar]
- 65.Caroni P. (2001) New EMBO members' review: actin cytoskeleton regulation through modulation of PI(4,5)P(2) rafts. EMBO J. 20, 4332–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Villalba M., Bi K., Rodriguez F., Tanaka Y., Schoenberger S., Altman A. (2001) Vav1/Rac-dependent actin cytoskeleton reorganization is required for lipid raft clustering in T cells. J. Cell Biol. 155, 331–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chadda R., Howes M. T., Plowman S. J., Hancock J. F., Parton R. G., Mayor S. (2007) Cholesterol-sensitive Cdc42 activation regulates actin polymerization for endocytosis via the GEEC pathway. Traffic 8, 702–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Döhner K., Sodeik B. (2005) The role of the cytoskeleton during viral infection. Curr. Top. Microbiol. Immunol. 285, 67–108 [DOI] [PubMed] [Google Scholar]
- 69.Tyrrell D. L., Norrby E. (1978) Structural polypeptides of measles virus. J. Gen. Virol. 39, 219–229 [DOI] [PubMed] [Google Scholar]
- 70.Giuffre R. M., Tovell D. R., Kay C. M., Tyrrell D. L. (1982) Evidence for an interaction between the membrane protein of a paramyxovirus and actin. J. Virol. 42, 963–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Loney C., Mottet-Osman G., Roux L., Bhella D. (2009) Paramyxovirus ultrastructure and genome packaging: cryo-electron tomography of sendai virus. J. Virol. 83, 8191–8197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stossel T. P., Condeelis J., Cooley L., Hartwig J. H., Noegel A., Schleicher M., Shapiro S. S. (2001) Filamins as integrators of cell mechanics and signalling. Nat. Rev. Mol. Cell Biol. 2, 138–145 [DOI] [PubMed] [Google Scholar]
- 73.Burke E., Mahoney N. M., Almo S. C., Barik S. (2000) Profilin is required for optimal actin-dependent transcription of respiratory syncytial virus genome RNA. J. Virol. 74, 669–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stoiber H., Pintér C., Siccardi A. G., Clivio A., Dierich M. P. (1996) Efficient destruction of human immunodeficiency virus in human serum by inhibiting the protective action of complement factor H and decay accelerating factor (DAF, CD55). J. Exp. Med. 183, 307–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Saifuddin M., Hedayati T., Atkinson J. P., Holguin M. H., Parker C. J., Spear G. T. (1997) Human immunodeficiency virus type 1 incorporates both glycosyl phosphatidylinositol-anchored CD55 and CD59 and integral membrane CD46 at levels that protect from complement-mediated destruction. J. Gen. Virol. 78, 1907–1911 [DOI] [PubMed] [Google Scholar]
- 76.Stahlhut M., van Deurs B. (2000) Identification of filamin as a novel ligand for caveolin-1: evidence for the organization of caveolin-1-associated membrane domains by the actin cytoskeleton. Mol. Biol. Cell 11, 325–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lillemeier B. F., Pfeiffer J. R., Surviladze Z., Wilson B. S., Davis M. M. (2006) Plasma membrane-associated proteins are clustered into islands attached to the cytoskeleton. Proc. Natl. Acad. Sci. U.S.A. 103, 18992–18997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang X., Glendening C., Linke H., Parks C. L., Brooks C., Udem S. A., Oglesbee M. (2002) Identification and characterization of a regulatory domain on the carboxyl terminus of the measles virus nucleocapsid protein. J. Virol. 76, 8737–8746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wandinger S. K., Richter K., Buchner J. (2008) The Hsp90 chaperone machinery. J. Biol. Chem. 283, 18473–18477 [DOI] [PubMed] [Google Scholar]
- 80.Park S. J., Suetsugu S., Takenawa T. (2005) Interaction of HSP90 to N-WASP leads to activation and protection from proteasome-dependent degradation. EMBO J. 24, 1557–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Machesky L. M., Mullins R. D., Higgs H. N., Kaiser D. A., Blanchoin L., May R. C., Hall M. E., Pollard T. D. (1999) Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc. Natl. Acad. Sci. U.S.A. 96, 3739–3744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rohatgi R., Ma L., Miki H., Lopez M., Kirchhausen T., Takenawa T., Kirschner M. W. (1999) The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell 97, 221–231 [DOI] [PubMed] [Google Scholar]
- 83.Rohatgi R., Nollau P., Ho H. Y., Kirschner M. W., Mayer B. J. (2001) Nck and phosphatidylinositol 4,5-bisphosphate synergistically activate actin polymerization through the N-WASP-Arp2/3 pathway. J. Biol. Chem. 276, 26448–26452 [DOI] [PubMed] [Google Scholar]
- 84.Park S. J., Suetsugu S., Sagara H., Takenawa T. (2007) HSP90 cross-links branched actin filaments induced by N-WASP and the Arp2/3 complex. Genes Cells 12, 611–622 [DOI] [PubMed] [Google Scholar]
- 85.Cudmore S., Cossart P., Griffiths G., Way M. (1995) Actin-based motility of vaccinia virus. Nature 378, 636–638 [DOI] [PubMed] [Google Scholar]
- 86.Frischknecht F., Moreau V., Röttger S., Gonfloni S., Reckmann I., Superti-Furga G., Way M. (1999) Actin-based motility of vaccinia virus mimics receptor tyrosine kinase signalling. Nature 401, 926–929 [DOI] [PubMed] [Google Scholar]
- 87.Moreau V., Frischknecht F., Reckmann I., Vincentelli R., Rabut G., Stewart D., Way M. (2000) A complex of N-WASP and WIP integrates signalling cascades that lead to actin polymerization. Nat. Cell Biol. 2, 441–448 [DOI] [PubMed] [Google Scholar]
- 88.Scaplehorn N., Holmström A., Moreau V., Frischknecht F., Reckmann I., Way M. (2002) Grb2 and Nck act cooperatively to promote actin-based motility of vaccinia virus. Curr. Biol. 12, 740–745 [DOI] [PubMed] [Google Scholar]
- 89.Pai K. S., Mahajan V. B., Lau A., Cunningham D. D. (2001) Thrombin receptor signaling to cytoskeleton requires Hsp90. J. Biol. Chem. 276, 32642–32647 [DOI] [PubMed] [Google Scholar]
- 90.Beckers C. M., van Hinsbergh V. W., van Nieuw Amerongen G. P. (2010) Driving Rho GTPase activity in endothelial cells regulates barrier integrity. Thromb. Haemost. 103, 40–55 [DOI] [PubMed] [Google Scholar]
- 91.Momose F., Naito T., Yano K., Sugimoto S., Morikawa Y., Nagata K. (2002) Identification of Hsp90 as a stimulatory host factor involved in influenza virus RNA synthesis. J. Biol. Chem. 277, 45306–45314 [DOI] [PubMed] [Google Scholar]
- 92.Connor J. H., McKenzie M. O., Parks G. D., Lyles D. S. (2007) Antiviral activity and RNA polymerase degradation following Hsp90 inhibition in a range of negative strand viruses. Virology 362, 109–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Naito T., Momose F., Kawaguchi A., Nagata K. (2007) Involvement of Hsp90 in assembly and nuclear import of influenza virus RNA polymerase subunits. J. Virol. 81, 1339–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Calistri A., Salata C., Parolin C., Paù G. (2009) Role of multivesicular bodies and their components in the egress of enveloped RNA viruses. Rev. Med. Virol. 19, 31–45 [DOI] [PubMed] [Google Scholar]
- 95.Ogino T., Yamadera T., Nonaka T., Imajoh-Ohmi S., Mizumoto K. (2001) Enolase, a cellular glycolytic enzyme, is required for efficient transcription of Sendai virus genome. Biochem. Biophys. Res. Commun. 285, 447–455 [DOI] [PubMed] [Google Scholar]
- 96.Brodsky J. L., Chiosis G. (2006) Hsp70 molecular chaperones: emerging roles in human disease and identification of small molecule modulators. Curr. Top. Med. Chem. 6, 1215–1225 [DOI] [PubMed] [Google Scholar]
- 97.Pearl L. H., Prodromou C., Workman P. (2008) The Hsp90 molecular chaperone: an open and shut case for treatment. Biochem. J. 410, 439–453 [DOI] [PubMed] [Google Scholar]
- 98.Verkhivker G. M., Dixit A., Morra G., Colombo G. (2009) Structural and computational biology of the molecular chaperone Hsp90: from understanding molecular mechanisms to computer-based inhibitor design. Curr. Top. Med. Chem. 9, 1369–1385 [DOI] [PubMed] [Google Scholar]
- 99.Solit D. B., Chiosis G. (2008) Development and application of Hsp90 inhibitors. Drug Discov. Today 13, 38–43 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.