Abstract
Using a gas chromatography-mass spectrometry-time of flight technique, we determined major metabolite changes during induction of the carbon-concentrating mechanism in the unicellular green alga Chlamydomonas reinhardtii. In total, 128 metabolites with significant differences between high- and low-CO2-grown cells were detected, of which 82 were wholly or partially identified, including amino acids, lipids, and carbohydrates. In a 24-h time course experiment, we show that the amino acids serine and phenylalanine increase transiently while aspartate and glutamate decrease after transfer to low CO2. The biggest differences were typically observed 3 h after transfer to low-CO2 conditions. Therefore, we made a careful metabolomic examination at the 3-h time point, comparing low-CO2 treatment to high-CO2 control. Five metabolites involved in photorespiration, 11 amino acids, and one lipid were increased, while six amino acids and, interestingly, 21 lipids were significantly lower. Our conclusion is that the metabolic pattern during early induction of the carbon-concentrating mechanism fit a model where photorespiration is increasing.
Most microalgae express a carbon-concentrating mechanism (CCM; Raven et al., 2005). The green alga Chlamydomonas reinhardtii is one of the species with a CCM (Giordano et al., 2005). The CCM in C. reinhardtii is typically induced when the concentration of CO2 in the air bubbled through the cultures is decreased to around 0.5% or lower (Vance and Spalding, 2005). Within the first few hours after starting the induction, numerous genes are either up- or down-regulated (Miura et al., 2004; Yamano et al., 2008; Yamano and Fukuzawa, 2009). However, the change in gene expression is only manifested as rather limited detectable changes in the abundance of proteins (Manuel and Moroney, 1988; Spalding and Jeffrey, 1989). Even though the response to decreased concentrations of inorganic carbon (Ci) is fast (Eriksson et al., 1998), the algal cells go through a transient phase before the CCM is fully operational.
Many genes coding for enzymes of the photorespiratory pathway are up-regulated (Marek and Spalding, 1991; Miura et al., 2002, 2004) within 20 min and show a transient expression pattern (Tural and Moroney, 2005; Yamano et al., 2008). A decline in starch content is also detectable within 30 min after transfer to low CO2 (Kuchitsu et al., 1988). This decline in starch is followed, after about 2 h, by a net synthesis of starch (Thyssen et al., 2001) that is mainly deposited around the pyrenoid, rather than as the starch grains distributed in the stroma normally found in high-CO2-grown cells. When the CCM is fully induced, the alga can concentrate Ci inside the cell/chloroplast against a free-energy gradient. Accumulation of Ci increases the CO2-oxygen ratio at the site of Rubisco (Giordano et al., 2005), with a corresponding increase in photosynthesis, a decrease in photorespiration, and a greater capacity for net organic carbon production at low external Ci (Giordano et al., 2003).
Induction of the CCM is thus known to affect genes of many different pathways, and especially the photorespiratory pathway has been studied extensively (Moroney et al., 1986; Tural and Moroney, 2005). The aim of this work was to screen for metabolic changes in order to find key metabolites that could trigger the expression of genes that regulate the CCM. We have extended the analyses by using metabolomics to detect changes in major metabolites, particularly in the beginning of the induction period but also in preliminary experiments, over a 24-h time period. The resulting metabolic changes have enabled us to propose a working model for the coordinated regulation of cellular metabolism during the induction of the CCM in C. reinhardtii.
RESULTS
Experiments were designed to obtain data on major changes of metabolites during acclimation to air concentrations of CO2 in C. reinhardtii cells.
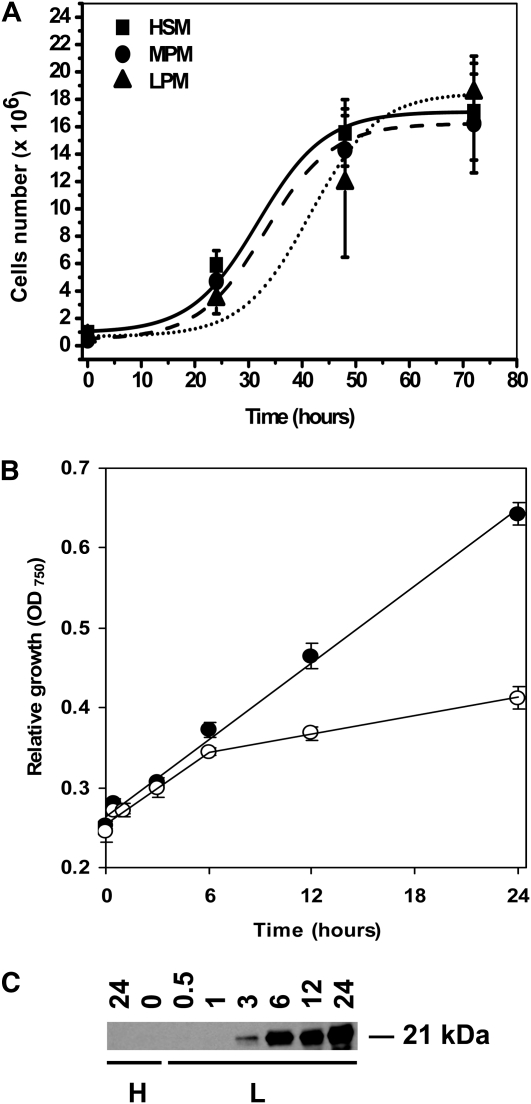
In order to characterize our experimental material, three control experiments were done. First, the population growth was examined for cultures in high CO2 (5% CO2-enriched air) at three different phosphate concentrations (for details, see “Materials and Methods”; Fig. 1A). The population growth was identical when cells were grown in full Sueoka high-salt medium (HSM) and in a medium where the concentration of phosphate was decreased by 25 times (MPM). A somewhat prolonged lag phase in growth was found in cultures grown at a 50 times less phosphate concentration (LPM; Fig. 1A). After the prolonged lag phase, the population growth was resumed and similar to populations grown at the higher phosphate concentrations. Relative population growth was also compared between cultures switched to low CO2 at time zero (ambient air, 0.04% CO2; MPM) and cultures grown in high CO2. It is evident from Figure 1B that cultures switched to low CO2 bubbling at time zero grew more slowly compared with the high-CO2 controls, and the differences became visible approximately 6 h after transfer to low CO2. After 24 h, the number of cells in low CO2 was 40% lower than in the corresponding high-CO2 control.
Figure 1.
Culture growth and CCM induction control. A, The growth of high-CO2 cultures in different types of media. All points represent the average ± sd of four individual replicates. B, Relative growth of the low-CO2 (white circles) and high-CO2 (black circles) cultures in MPM. All points represent the average ± sd of four individual replicates. C, A 24-h time course of the mtCA protein expression. H, High CO2; L, low CO2.
To be sure that the cultures induced CCM after transfer to low CO2, two separate control experiments were done. A western-blot analysis was done to detect the time course of induction of the low-CO2-induced mitochondrial carbonic anhydrase protein (mtCA), and also the increased affinity for CO2 in cultures switched to low-CO2 conditions for 3 h was determined (Fig. 1C; Table I). The mtCA gene is silent under high-CO2 conditions, and no transcript can be detected under noninducing conditions, although under CCM-inducing conditions this transcript is easily detected (Eriksson et al., 1998). The mtCA in this setup was first detected 3 h after changing to low CO2 bubbling and showed a gradual increase with time in low CO2 (Fig. 1C). The mtCA could not, even under prolonged exposure of the western-blot filter, be detected in cultures grown under high-CO2 conditions. As an additional indication that CCM is induced under our growth conditions, we determined the Km value for Ci in both high- and low-CO2-grown cells 3 h after transfer to CCM-inducing conditions. As is visible in Table I, the affinity for Ci had already increased after 3 h in all of the three phosphate concentrations. The maximal rate of oxygen evolution was lower in low-CO2-grown compared with high-CO2-grown cells, but there was no difference between cultures with different concentrations of phosphate, indicating that none of the concentrations of phosphate were limiting (Kozlowska-Szerenos et al., 2004; Beardall et al., 2005a). In summary, both the induction of mtCA as well as the decreased Km values indicate that CCM was at least partly induced already after 3 h in low CO2.
Table I. The photosynthetic oxygen evolution parameters Vmax and Km of cells grown at high (5%) CO2 and adapted for 3 h to low CO2.
The cells were grown in three media types: HSM, MPM, and LPM. The data (an average from three individual replicates) were analyzed using a Lineweaver-Burk plot.
| CO2 Level | Parameter | HSM | MPM | LPM |
| High | Km | 20.8 | 16.4 | 12.0 |
| Vmax | 117.6 | 112.4 | 112.4 | |
| Low | Km | 5.9 | 5.3 | 4.3 |
| Vmax | 58.8 | 55.6 | 58.8 |
Only small changes in total chlorophyll, chlorophyll a/b ratios, and protein-to-chlorophyll ratios occurred during the course of the experiment (Supplemental Fig. S1).
Lowering of the CO2 Concentration Causes a Transient Metabolic Change
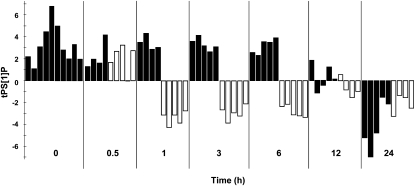
To find out how rapidly metabolic changes occur in cells transferred to low CO2, a preliminary metabolomics study was performed, analyzing cells sampled at different time points after lowering the CO2. The data obtained were analyzed by calculating orthogonal projections to latent structures (OPLS) models for the high-CO2 samples and predict the low-CO2 samples into the models (for details, see “Materials and Methods”). The transient behavior of cells transferred to low CO2 is shown in Figure 2. As expected, the samples at 0 min were similar according to the criteria used in the model. The low-CO2 samples at 0.5 h were also predicted to be similar to the high-CO2 control, indicating a delay in the metabolic response. However, low-CO2-grown samples between 1 and 6 h show a clear difference in predictive values as compared with the high-CO2-grown cells, showing a change to the entire metabolic profile over 5 h. The samples at 12 and 24 h indicate that metabolic differences decreased with time, so major differences in the metabolic profiles of control and low-CO2-grown cells could not be detected 12 h after transfer to CCM-inducing conditions.
Figure 2.
Differences over 24 h in the metabolic profile of control (high-CO2) and low-CO2-grown cells. Predictive score values are from the four-component (one predictive + three orthogonal; R2X = 0.604, R2Y = 0.967, Q2Y = 0.632) OPLS-DA model, fitted to separate low CO2 and control samples collected at 1, 3, and 6 h. Each bar represents the metabolic profile of an individual replicate at each time point (control, black bars; low CO2, white bars). Each time point is highlighted on the x axis as white and black fields marking the samples taken at that time. Predictive score values for low-CO2 and control samples, collected at 0, 0.5, 12, and 24 h, were predicted by the model. As expected, the samples at 0 h were all predicted as control samples. The low-CO2 samples at 0.5 h were also predicted as control samples, indicating a delay in the metabolic response. Low-CO2-grown samples between 1 and 6 h show a clear difference in predictive values as compared with the cells grown under control conditions. The samples at 12 and 24 h indicate that metabolic differences were reduced over time. tPS[1]P, Predicted score values for low CO2 and control samples.
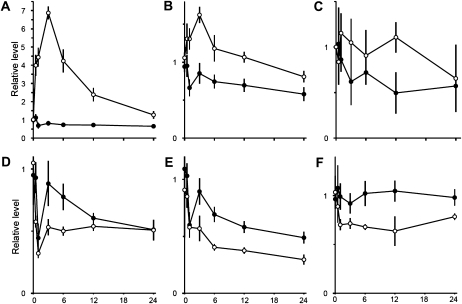
The time course of some metabolites showing differences between the high-CO2 control and the low-CO2-induced cells are presented in Figure 3. At 0.5 h after transfer to low CO2, there is a marked increase of Ser (Fig. 3A). Malic acid (Fig. 3B) also increases, but not as rapidly as Ser. The increased levels of Ser and malic acid are transient, and both metabolites peak 3 h after transfer, while the change in Phe is slower (Fig. 3C). Ser is an essential amino acid and a metabolite in the photorespiratory pathway, which is known to be up-regulated upon induction of the CCM (Marek and Spalding, 1991; Tural and Moroney, 2005). Malate is a central metabolite in many different metabolic pathways (e.g. the tricarboxylic acid [TCA] cycle) and shuttle mechanisms for redox transfer between compartments, such as the malate valve exporting reducing equivalents from the chloroplast (Scheibe, 2004; Foyer et al., 2009).
Figure 3.
Time course of single metabolites. Selection of metabolites showing significantly different levels in high-CO2 control (black circles) and under low-CO2 conditions (white circles). All points represent the average ± sd of at least four biological replicates. Data have been standardized to the average of all samples collected at time zero. All graphs center around 1 at time zero. The y axis shows the level of up- and/or down-regulation, with ticks every 0.5-level shift. The x axis shows the 24-h time frame, with ticks every 6 h. A, Ser. B, Malic acid. C, Phe. D, Asp. E, Glu. F, α-Linolenic acid.
Decreased levels of Asp, Glu, and α-linolenic acid are observed 1 to 3 h after transfer to low-Ci conditions (Fig. 3, D–F). The level of Asp is already similar to the high-CO2 control after 12 h (Fig. 3D), while Glu decreases in both high- and low-CO2-grown cells between 3 and 24 h after transfer to low-CO2 conditions, with a greater decrease in cells inducing the CCM (Fig. 3E). α-Linolenic acid shows a somewhat similar pattern to Ser, although the levels are not identical in cells from the two growth conditions at the end of the 24-h experiment (Fig. 3F).
It is striking that several of the metabolites with rather rapid changes immediately after transfer to low-Ci conditions are close to, or at, the initial level within 24 h. This transient behavior, with an initial divergence of the metabolic levels and a final convergence between cultures grown with high or low CO2, was also observed in additional experiments (data not shown).
Focus on Differences 3 h after CCM Induction
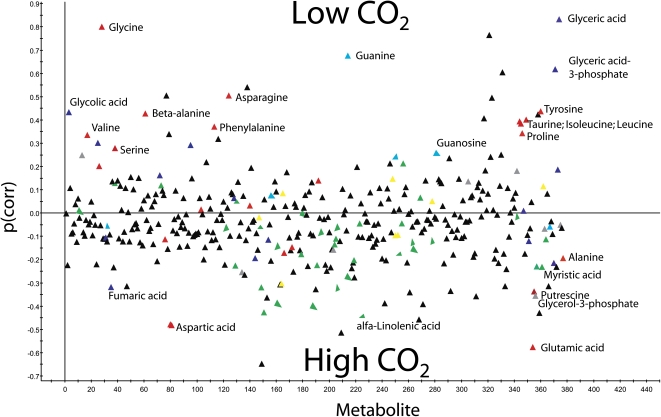
Since the biggest difference in metabolic profile as well as in single metabolites between the low-CO2-induced culture and the control culture was observed approximately 3 h after transfer to low CO2 (Figs. 2 and 3), we conducted another experiment to increase the resolution at this time point. To verify that there were no differences between the flasks at time zero, a OPLS-discriminant analysis (DA) model was calculated, showing no significant differences between the two classes (predictive ability of variation of Y less than 0%; Q2Y = −0.013). The difference between high and low CO2 was determined by calculating an OPLS-DA model with the 3-h samples (one predictive and three orthogonal components; R2X = 0.43, R2Y = 0.96, Q2Y = 0.90). Statistical tests involve a 95% confidence interval and are expressed as loadings ± sd of the OPLS-DA analysis. All significant detectable changes in metabolite levels at 3 h after transfer to CCM-inducing conditions are listed in Supplemental Table S1. A total of 377 putative metabolites were detected in the samples (for overview, see Fig. 4; Supplemental Fig. S2), and 82 of those were either identified or classified to compound class. A total of 128 metabolites were significantly changed between high- and low-CO2-grown cultures at 3 h after transfer to CCM-inducing conditions. Of those, 65 were identified; 27 had increased levels in low CO2, while 38 had decreased levels. Of the compounds with a clear change, many were amino acids, carbohydrates, or lipids. Seventeen amino acids were identified; 11 increased and six decreased in the low-CO2-grown cultures compared with the control. A different pattern was observed for the 22 compounds classified as lipids, where only one showed an increased level while 20 had decreased levels compared with the control cells in high CO2. Among the carbohydrate group, the distribution between those that had increased and decreased was about equal. In agreement with the time course results (Fig. 3), the increase in Ser levels was significant, as was the decrease in Glu and Asp content. Other amino acids with increased levels were Gly, which was dramatically increased after 3 h, β-Ala, Thr, Pro, and Tyr.
Figure 4.
Predictive p-loadings (correlation scaled) for 377 metabolites. The 1+3 OPLS-DA model was calculated in order to optimize the separation between low-CO2 and high-CO2 samples at 3 h. Positive p(corr) values indicate higher concentrations for low-CO2 samples, and negative p(corr) values indicate higher concentrations for high-CO2 samples. Red, Amino acids and amines; green, lipids; blue, organic acids; turquoise, nucleosides; yellow, carbohydrates; gray, other classes; black, unknown compounds
DISCUSSION
C. reinhardtii is the major eukaryotic algal model system for studies of cellular functions, a result of its well-defined classical genetics and early development and/or adaptation of molecular tools to fit to Chlamydomonas (Rochaix, 1995) and the sequencing of its nuclear genome (Merchant et al., 2007). Recent work has emphasized the regulation of gene expression, signaling pathways, and the use of transcriptome analysis.
The response of growth when Chlamydomonas cells are acclimated to a lowered CO2 concentration in the medium is clearly visible, with decreased relative growth after about 6 h (Fig. 1B). Despite different population growth, the total protein content as well as the chlorophyll a/b ratios are relatively similar in high- and low-CO2-grown cells up to 24 h (Supplemental Fig. S1, A and B). The chlorophyll/protein values increase more in high-CO2 cells, particularly at the end of the experimental period (Supplemental Fig. S1C). The induction was performed in a medium with a decreased phosphate concentration, because in a full phosphate medium algal cells store phosphate as polyphosphate granules that mask metabolite detection in the mass spectrometric analyses. It can be seen (Fig. 1; Table I) that the decrease in phosphate concentration during the relatively short experimental time period does not influence growth or affinity for CO2.
Ultrastructural changes (Ramazanov et al., 1994; Geraghty and Spalding, 1996) as well as induction of additional proteins (Manuel and Moroney, 1988; Spalding and Jeffrey, 1989) occur after transfer to low-CO2 conditions, leading to acclimation of the photosynthetic machinery (Spalding et al., 1984), possibly maximizing photosynthesis through adjusting the supply of ATP for the transport of Ci into the cells. Therefore, it is a reasonable conclusion that CCM is a function of several relatively small but orchestrated changes that are taking place in the cells during the first 3 to 12 h of induction.
The reason that major differences in metabolites between high- and low-CO2 cells disappear after 12 h of induction may be that the CCM is already functional after that time (Matsuda and Colman, 1995a, 1995b; Bozzo and Colman, 2000). It should be pointed out that although the model predicts no major changes between high- and low-CO2-grown cells, it is still possible that there are differences in individual metabolites not detected in our experiments. The major advantage with a metabolomics approach is that it gives nonbiased information about pool sizes of a large number of substances under specific conditions. This can also give useful insights into unexpected changes that could easily be missed in targeted approaches where only a few specific compounds are analyzed. However, the interpretation of metabolomics data is complicated for several reasons. Many metabolites are not end products but intermediates of metabolic pathways. For such compounds, the steady-state concentration is determined by the relative rates of its formation and removal. Thus, the concentration is not directly related to the metabolic flux through the compound. Complications for the interpretation also arise when a metabolite is involved in several pathways and/or is present in separate subcellular pools. A technical limitation is also that not all metabolites can be detected and that not all detected metabolites can be identified. In addition, it is not obvious what should be the appropriate control, since there may also be changes in the high-CO2-grown cultures that are independent of the concentration of Ci. It cannot be excluded that a partial synchronization of the cultures occurs with cell populations that progress differently during the cell cycle. In addition, high-CO2-grown cultures grow faster with more self shading and a faster depletion of mineral nutrients. However, we have chosen to sample cells grown in high CO2 under identical conditions and at the same time points as when low-CO2 samples were harvested.
Since several of the metabolites most specific for low-CO2 conditions are intermediates of the photorespiratory glycolate pathway (Fig. 4; Supplemental Table S1), we present an attempt to interpret our results in relation to an increased flux through the photorespiratory pathway and closely related primary metabolic pathways. By combining transcriptomic data (Yamano et al., 2008) and physiological/biochemical data (Marek and Spalding, 1991; Bozzo and Colman, 2000) on CCM-inducing cells from the literature and metabolomic data in this work, we propose a model that fits both data sets and therefore may have some relevance (Fig. 5).
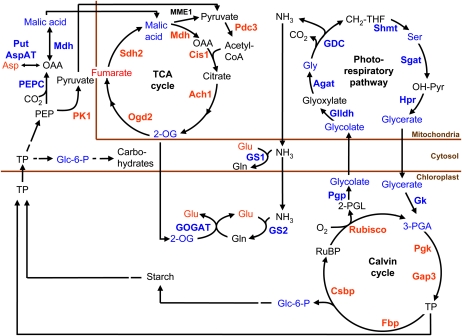
Figure 5.
A model of the metabolic changes after 3 h of CCM induction, including photorespiration and closely related metabolic pathways. The metabolomics data come from this paper, and the transcriptomics data indicated in the pathways have been obtained from Yamano et al. (2008). Blue indicates an increased metabolite content/expression level, and red indicates a decreased metabolite content/expression level as compared with high-CO2-grown control cells.
When the cells are transferred from high to low CO2, the increase in the oxygenation reaction of Rubisco is presumably larger than in the steady state in low CO2, because the CCM has not yet been expressed and the CO2-oxygen ratio around Rubisco is low, resulting in the production of 2-phosphoglycolate. Initially, this results in the excretion of glycolate (Merrett and Lord, 1973), but very soon genes for enzymes of the photorespiratory pathway are transcribed (Tural and Moroney, 2005). When the CCM is expressed and when corresponding enzymes are produced in sufficient amounts, glycolate can be metabolized within the cell and excretion stops. Over a similar time, genes of the Calvin cycle are down-regulated (Yamano et al., 2008).
Localization of enzymes of the photorespiratory pathway (photorespiratory carbon oxidation cycle [Tural and Moroney, 2005]) to mitochondria has been shown for Chlamydomonas (Atteia et al., 2009). These and other results on the Chlorophyceae agree with data on Eremosphaera viridis from the sister class Trebouxiophyceae (Stabenau and Winkler, 2005).
After 3 h in low CO2, Gly and glyceric acid are the two identified metabolites most specific for this condition, but also glycolate, Ser, and 3-phosphoglyceraldehyde increase. These metabolites are all intermediates of the photorespiratory glycolate pathway. The increased expression of all genes in this pathway, except for Glu:glyoxylate aminotransferase, has been shown (Tural and Moroney, 2005; Yamano et al., 2008). There could also be a role for another low CO2-inducible gene, Ala:2-oxoglutarate aminotransferase, in the photorespiratory carbon oxidation cycle (Chen et al., 1996; see Fig. 1 in Tural and Moroney, 2005).
As photorespiration is activated and flux through the Gly decarboxylate complex increases, there is a release of NH3 in the mitochondria. This ammonia can be refixed by glutamine synthetase (GS), and both the cytosolic GS1 and the chloroplastic GS2 gene expression increase (Yamano et al., 2008). Glu is a central metabolite in primary nitrogen metabolism and is directly involved in recycling of NH3 during photorespiration. The observed decrease in Glu can be linked to increased demand for refixation of NH3. Furthermore, Asp is a major source for transaminations, and its decrease can be related to the decrease in Glu, since the two amino acids are linked via Asp aminotransferase. Asp is also involved in the synthesis of Lys and Thr that increases at 3 h, but a direct link is not obvious and more studies will be needed to clarify this aspect. It is interesting that a large number of amino acids increase in low-CO2 conditions (Supplemental Table S1). These amino acids represent several different biosynthetic classes, making a biosynthetic link difficult to see. Instead, it could be possible that the general accumulation of amino acids is linked to cellular rearrangements, where some proteins are degraded while others are synthesized. The same argument may be used to explain the increase in free bases (A,U, and G) and guanosine/uridine by RNA degradation. There are very significant interactions between CO2 availability and nitrogen availability on the expression of the CCM in C. reinhardtii (Giordano et al., 2003), and interactions between carbon and nitrogen metabolism with changing CO2 supply are to be expected (Thyssen et al., 2001). There seems to be no information on the effects of growth at different CO2 concentrations on the carbon-nitrogen ratio of C. reinhardtii, and data for other algae show that an increased carbon-nitrogen ratio in microalgal cells grown at high CO2 does occur but is by no means universal (Beardall et al., 2005b; Finkel et al., 2010).
Rearrangement of starch from granules to the pyrenoid sheath is characteristic for the early part of the low-CO2 acclimation response (Thyssen et al., 2001). The absolute level of starch synthesis is lower in low-CO2 cells, although it represents a larger fraction of the total carbon fixed. Thus, less carbon will be available for synthesis of other carbohydrates, and this may be the explanation for the observed decrease in some carbohydrates (Supplemental Table S1).
An increase in transcripts for phosphoenolpyruvate carboxylase (PEPC) and cytosolic NAD-dependent malate dehydrogenase was reported (Yamano et al., 2008) at 0.3 h after transfer to CCM-inducing conditions. Interestingly, the transcript for cytosolic pyruvate kinase (PK1) declined steeply around 1 h after transfer to low CO2 (Yamano et al., 2008), suggesting a shift at the end of glycolysis from PK to PEPC as the final step. In C. reinhardtii, there are five isoforms of PK, all thought to be cytosolic. The expression of PK3 and PK5 is largely unchanged and may complicate the picture. In potato (Solanum tuberosum) tubers, however, it has been shown that gene silencing by RNA interference of the most highly expressed cytosolic PK resulted in a decreased pyruvate synthesis, even though another four isoforms of cytosolic PK were present (Oliver et al., 2008). In C. reinhardtii, the isoform PK1 has about three times more supporting EST sequences in the National Center for Biotechnology Information database than the other isoforms, giving good support to this being the most abundant cytosolic PK. It is plausible, therefore, that the down-regulated cytosolic PK1 affects the synthesis of pyruvate. Within roughly the same time interval, we observed an increase in malate (Fig. 3) that can be formed from PEP via oxaloacetate by the action of PEPC and cytosolic NAD-dependent malate dehydrogenase. Malate is also a central metabolite for redox communication between compartments such as the “malate valve” (Scheibe, 2004). Transfer to low CO2 will lead to decreased consumption of reductants for CO2 fixation in the Calvin cycle. This would possibly lead to a more reduced redox state and shift the oxaloacetate to malate equilibrium toward malate. Malate can subsequently enter the TCA cycle, in which the transiently down-regulated genes are back to, or above, the high-CO2 levels again after 6 h in low CO2 (Yamano et al., 2008). Other metabolites of the TCA cycle show different patterns: fumarate is higher in high CO2, while 2-oxoglutarate is high in low CO2. The reactivation of the TCA cycle, we suggest, will produce 2-oxoglutarate, which in turn can fill up the Glu pool by the action of GOGAT in the chloroplast. A sign of an increased metabolite transport between compartments is the up-regulation of genes encoding two different translocators. One of these is Lci19 (for low CO2 induced protein 19), which is believed to encode a Glu/malate translocator. The Lci19 gene is immediately up-regulated upon induction of the CCM. The other gene encodes a putative 2-oxoglutarate/malate translocator (Yamano et al., 2008).
A further possibility, granted the up-regulation of PEPC and cytosolic malate dehydrogenase and the increase in malate upon transfer to low Ci, is that the CCM resulting in the C4-like physiology induced by acclimation to decreased Ci has at least a component of C4 photosynthetic biochemistry. This possibility has been suggested for certain diatoms but is still a matter of debate (Kroth et al., 2008; Raven, 2010). Some of the evidence used to support a C4 contribution to a diatom CCM has been shown not to apply to (a marine) Chlamydomonas (Reinfelder et al., 2004), and work on C. reinhardtii also agrees with the absence of C4-like photosynthesis (Harris, 1989; Raven, 2010). The absence of C4 photosynthesis is in agreement with the location of a potential C4 photosynthesis decarboxylase, PEPCK, in mitochondria (Fig. 5; Kroth et al., 2008; Atteia et al., 2009). Furthermore, Giordano et al. (2003) showed that the activity of PEPC in C. reinhardtii is influenced by nitrogen availability to a greater extent than carbon availability, which is consistent with a predominantly anaplerotic role for this enzyme.
It has been known for a long time from biochemical evidence that, despite the induction of a CCM, photorespiration increases after transfer of cells to air CO2 levels; this is also supported by transcriptomic data (Yamano et al., 2008) and now also by metabolomic data (this work). Not all changes in metabolites can be fitted to changes in photorespiration, and most interesting is the decrease in 21 out of 22 analyzed lipid compounds. At this point, we can only speculate about why most of the detected lipids decrease during CCM induction. One possibility is that cells are light stressed directly after the transfer because of less CO2 as e- acceptor and that this causes oxidation of lipids by free radicals, a possibility supported by transcriptome data from C. reinhardtii (Im et al., 2003) and the glaucocystophyte alga Cyanophora paradoxa (Burey et al., 2007; Raven, 2010).
Our results indicate that there exists a good relation between changes in gene expression and metabolic activities in CCM-inducing Chlamydomonas cells. However, significant changes in fluxes are not necessarily manifested as changes in metabolite pools; therefore, in order to get a detailed picture of the changes in metabolism during induction, labeling experiments and determination of fluxes of metabolites would be required. The rationale, strategies, and design of such experiments were thoroughly described (Ratcliffe and Shachar-Hill, 2006, and refs. therein). Such experiments are planned with the aim to identify key metabolites that are controlling the induction of CCM in C. reinhardtii.
MATERIALS AND METHODS
Algal Strain and Culture Conditions
The Chlamydomonas reinhardtii cell-wall-less mutant cw92 was obtained from the Chlamydomonas Culture Collection at Duke University. The cells were precultured in TAP medium (Harris, 1989) at 20°C, 60 rpm, with 25 μmol m−2 s−1 continuous irradiation in an incubator (Innova 4340; New Brunswick Scientific). For autotrophic growth, the cells were grown in full HSM (Suoeka, 1960) or HSM modified to facilitate metabolite identification: MPM, with 25 times reduced phosphate concentration, or LPM, with 50 times reduced phosphate concentration. To retain the buffering capacity, 20 mm HEPES, pH 7.5, was added to both MPM and LPM media. The culture grown on HSM, MPM, and LPM was diluted on a daily basis to 0.5 × 106 cells mL−1 each morning for 3 d before each experiment. Temperature was kept at 23°C with continuous irradiation from cool-white fluorescent lamps (Philips Master TLD 36W/830) at 200 ± 10 μmol m−2 s−1. Cultures were bubbled either with air enriched with 5% CO2 (high CO2) or with 0.04% CO2 (low CO2) for induction of the CCM.
Experimental Procedures and Sample Collection
To compare algae growth in full and modified media, HSM, MPM, and LPM were inoculated with cells to a final density of 0.5 × 106 cells mL−1, and the culture density was determined microscopically at 24, 48, and 72 h using the standard procedure.
At the start of CCM induction experiments, all cultures (optical density at 750 nm [OD750] = 0.25 ± 0.05) were pooled and poured through a mesh, to remove big aggregates, and redistributed into 500-mL flasks under controlled conditions. To maximize acclimation of the cells to the experimental conditions, a 2-h interlude at 5% CO2 bubbling and a light intensity of 200 ± 10 μmol m−2 s1 (Philips Master TLD 36W/830) was inserted before time zero, when the cultures were subjected to bubbling with ambient air to start the CCM induction. At time zero, the first set of samples were collected from all 10 cultures. After all samples at time zero had been collected, half of the cultures were switched to air bubbling (five cultures) and the remaining half were retained in high CO2. Samples were also taken at 0.5, 1, 3, 6, 12, and 24 h from each bottle for metabolomics, protein content, chlorophyll content, and OD measurements. Samples for protein and chlorophyll contents were immediately pelleted and frozen at −20°C.
Samples used for metabolomics analyses were harvested from culture flasks by a pipette and immediately quenched according to Bölling and Fiehn (2005). A quenching solution composed of 32.5% methanol in water with 300 μm CaCl2, 400 μm MgCl2, and 7 mm KCl was prepared. The quenching solution was precooled to −25°C in an ethanol/dry ice bath. Three milliliters of cells was pipetted into 12 mL of the quenching solution and kept at −20°C. Immediately after sampling, 10 samples for each time point were centrifuged for 5 min at 2,500g using a Hermle ZK380 centrifuge precooled to −20°C. The supernatants were removed, and the pellets were directly frozen in liquid nitrogen and stored at −80°C.
Chlorophyll and Protein Measurements
Chlorophyll concentrations were determined spectroscopically after extraction in 80% acetone according to Methods in Enzymology (Yocum, 1988). Total protein was extracted with 0.5 n NaOH and incubated at 80°C for 30 min (Teoh et al., 2004).
Protein concentrations were measured using Bio-Rad protein assay solution based on the Bradford method (Bradford, 1976). Cell-free extracts of protein for immunodetection were prepared by resolving the pelleted cells in Tris, pH 6.8, followed by three consecutive cycles of freeze and thaw. The proteins were then extracted with loading buffer containing mercaptoethanol and incubated at 90°C for 5 min. As a measure of relative growth rates, OD750 was used.
Metabolomics
Pellets collected for metabolomics analysis were freeze dried at −40°C to remove any residual supernatant. The samples were extracted and analyzed according to the methods described by Gullberg et al. (2004) with minor changes. Briefly, stable isotope reference compounds (5 ng μL−1 each [13C3]myristic acid, [13C4]hexadecanoic acid, [2H4]succinic acid, [13C5,15N]Glu, [2H7]cholesterol, [13C5]Pro, [13C4]disodium 2-oxoglutarate, [13C12]Suc, [2H4]putrescine, [2H6]salicylic acid, and [13C6]Glc) were added to an extraction mixture consisting of chloroform:methanol:water (1:3:1). The samples (pellet from 3 mL of liquid culture each) were then extracted in standardizing volumes (according to OD750) of the extraction mixture by vortexing for 30 min at 5°C. The extracts were centrifuged for 10 min at 14,000 rpm before 275 μL of the supernatant was transferred to a gas chromatograph vial and evaporated to dryness. The samples were then derivatized by shaking them with 10 μL of methoxyamine hydrochloride (15 mg mL−1) in pyridine for 10 min at 5°C prior to incubation for 16 h at room temperature. The samples were trimethylsilylated by adding 10 μL of N-methyl-N-(trimethylsilyl)trifluoroacetamide with 1% trimethylchlorosilane and incubating them for 1 h at room temperature. After silylation, 10 μL of heptane including 15 ng of methylstearate was added.
Samples were analyzed, according to Gullberg et al. (2004), using gas chromatography-mass spectrometry-time of flight (GC-MS-TOF) together with blank control samples and a series of n-alkanes (C12–C40), which allowed retention indices to be calculated (Schauer et al., 2005). One microliter of each derivatized sample was injected without splitting into a gas chromatograph equipped with a 10-m × 0.18-mm i.d. fused silica capillary column with a chemically bonded 0.18-μm DB 5-MS stationary phase. The injector temperature was 270°C, the septum purge flow rate was 20 mL min−1, and the purge was turned on after 60 s. The gas flow rate through the column was 1 mL min−1; the column temperature was held at 70°C for 2 min, then increased by 40°C min−1 to 320°C, and held there for 2 min. The column effluent was introduced into the ion source of a Pegasus III GC-MS-TOF apparatus (Leco). The transfer line and the ion source temperatures were 250°C and 200°C, respectively. Ions were generated by a 70-eV electron beam, at an ionization current of 2.0 mA, and 30 spectra s−1 were recorded in the mass range 50 to 800 mass-to-charge ratio. The acceleration voltage was turned on after a solvent delay of 150 s. The detector voltage was 1,660 V.
All nonprocessed MS files from the metabolic analysis were exported into MATLAB, in which all data pretreatment procedures, such as baseline correction, chromatogram alignment, and hierarchical multivariate curve resolution, were performed using custom scripts (Jonsson et al., 2005). All manual integrations were performed using ChromaTOF 2.32 (Leco) software or custom scripts.
Multivariate Analysis
Multivariate analyses such as principal component analysis (PCA; Wold et al., 1987) and OPLS (Trygg and Wold, 2002) were performed using SIMCA-P+ 12.0 (Umetrics). To provide an overview of the data, a PCA model was initially calculated on the X-matrix (i.e. the metabolite data). In PCA, a small number of latent variables (principal components) are calculated; these latent variables describe the largest variation in the X-matrix (transcript or metabolite data), reflecting the largest systematic variations. Consequently, the influence of noise is reduced and the dimensionality of the data is greatly reduced, which simplifies interpretation. R2X values vary between 0 and 1 (i.e. describe 0%–100% of the variation in the data) without losing predictability. The coordinates of the samples in the reduced space (the principal component space) are called scores. The relation between the original variables and the principal components are given by the loadings. The predictive ability of the model according to cross-validation is the Q2X value (0–1). Score plots can then be used to find trends and outliers and to reveal clustering of samples. To find variations in the data related to different prior information on the samples, a regression model is useful. PLS (Wold et al., 2001) bears some relation to PCA and is used to find relations between descriptors (X) and responses (Y). However, if systematic variation unrelated to Y is present in X, the model will be difficult to interpret. In OPLS, the systematic variation in X is divided into two parts: one that is linearly related to Y (predictive; RP2X) and one that is unrelated to Y (orthogonal; Trygg and Wold, 2002; Bylesjö et al., 2006; Wiklund et al., 2008). Thus, the Y-related variation in X is readily described by the loadings along the predictive component, which greatly simplifies the interpretation of the model. The total explained variance in Y is R2Y (0–1), and the cross-validated predictive ability is Q2Y. To perform OPLS, a DA model was performed to account for the variation in X related to classes of samples (e.g. high CO2 versus low CO2).
CCM Induction
SDS-PAGE and Western Blot
Western blots using mtCA antibodies were used to determine the time course of CCM induction during the experiments (Eriksson et al., 1996). Cell-free extracts prepared as described above were run on 12% SDS-PAGE gels (Laemmli, 1970). The proteins were then blotted onto a nitrocellulose membrane, and immunodetection of proteins was done using anti-mtCA IgGs raised against the protein in C. reinhardtii. Secondary antibodies labeled with horseradish peroxidase were hybridized to the IgG, and enhanced chemiluminescence was used to detect the antibody-antigen conjugate (Amersham ECL Western Blotting Detection Reagents; GE Healthcare).
Measurement of Cell Affinity for Dissolved Ci
An estimation of Ci affinity of high- and low-CO2-growing cells was done according to Kozłowska-Szerenos et al. (2000). Cells were harvested by centrifugation (1,200g, 10 min), resuspended in 25 mm HEPES-KOH to a constant OD750 of 0.35 ± 0.05, and transferred into the electrode chamber. All experiments were carried out at a temperature of 25°C with an irradiation (400–700 nm) of 1,000 μmol m−2 s−1 at pH 8.2. Following consumption of all accessible Ci by the cells, the bicarbonate was added in the concentration range of 5 to 200 μm, and the oxygen evolution rate was measured with a Clark-type oxygen electrode (Hansatech). Km and Vmax values were estimated using a Lineweaver-Burk plot.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Changes in protein and chlorophyll content during CCM induction.
Supplemental Figure S2. Significantly differing metabolites during CCM induction.
Supplemental Table S1. List of metabolites showing significantly changed levels at 3 h after transfer to CCM-inducing conditions.
Supplementary Material
References
- Atteia A, Adrait A, Brugiere S, Tardif M, van Lis R, Deusch O, Dagan T, Kuhn L, Gontero B, Martin W, et al. (2009) A proteomic survey of Chlamydomonas reinhardtii mitochondria sheds new light on the metabolic plasticity of the organelle and on the nature of the a-proteobacterial mitochondrial ancestor. Mol Biol Evol 26: 1533–1548 [DOI] [PubMed] [Google Scholar]
- Beardall J, Griffiths H, Raven JA. (2005a) Carbon isotope discrimination and the CO2 accumulating mechanism in Chlorella emersonii. J Exp Bot 33: 729–737 [Google Scholar]
- Beardall J, Roberts S, Raven JA. (2005b) Regulation of inorganic carbon acquisition by phosphorous limitation in the green alga Chlorella emersonii. Can J Bot 83: 859–864 [Google Scholar]
- Bölling C, Fiehn O. (2005) Metabolite profiling of Chlamydomonas reinhardtii under nutrient deprivation. Plant Physiol 139: 1995–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozzo GG, Colman B. (2000) The induction of inorganic carbon transport and external carbonic anhydrase in Chlamydomonas reinhardtii is regulated by external CO2 concentration. Plant Cell Environ 23: 1137–1144 [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Burey SC, Poroyko V, Ergen ZN, Fathi-Nejad S, Schuller C, Ohnishi N, Fukuzawa H, Bohnert HJ, Loffelhardt W. (2007) Acclimation to low [CO2] by an inorganic carbon-concentrating mechanism in Cyanophora paradoxa. Plant Cell Environ 30: 1422–1435 [DOI] [PubMed] [Google Scholar]
- Bylesjö M, Rantalainen M, Cloarec O, Nicholson JK, Holmes E, Trygg J. (2006) OPLS discriminant analysis: combining the strengths of PLS-DA and SIMCA classification. J Chemometr 20: 341–351 [Google Scholar]
- Chen ZY, Burow MD, Mason CB, Moroney JV. (1996) A low-CO2-inducible gene encoding an alanine-α-ketoglutarate aminotransferase in Chlamydomonas reinhardtii. Plant Physiol 112: 677–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson M, Karlsson J, Ramazanov Z, Gardestrom P, Samuelsson G. (1996) Discovery of an algal mitochondrial carbonic anhydrase: molecular cloning and characterization of a low-CO2-induced polypeptide in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 93: 12031–12034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson M, Villand P, Gardestrom P, Samuelsson G. (1998) Induction and regulation of expression of a low-CO2-induced mitochondrial carbonic anhydrase in Chlamydomonas reinhardtii. Plant Physiol 116: 637–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel ZV, Beardall J, Flynn KJ, Quigg A, Rees TAV, Raven JA. (2010) Phytoplankton in a changing world: cell size and elemental stoichiometry. J Plankton Res 32: 119–137 [Google Scholar]
- Foyer CH, Bloom AJ, Queval G, Noctor G. (2009) Photorespiratory metabolism: genes, mutants, energetics, and redox signaling. Annu Rev Plant Biol 60: 455–484 [DOI] [PubMed] [Google Scholar]
- Geraghty AM, Spalding MH. (1996) Molecular and structural changes in Chlamydomonas under limiting CO2: a possible mitochondrial role in adaptation. Plant Physiol 111: 1339–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano M, Beardall J, Raven JA. (2005) CO2 concentrating mechanisms in algae: mechanisms, environmental modulation, and evolution. Annu Rev Plant Biol 56: 99–131 [DOI] [PubMed] [Google Scholar]
- Giordano M, Norici A, Forssen M, Eriksson M, Raven JA. (2003) An anaplerotic role for mitochondrial carbonic anhydrase in Chlamydomonas reinhardtii. Plant Physiol 132: 2126–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullberg J, Jonsson P, Nordstrom A, Sjostrom M, Moritz T. (2004) Design of experiments: an efficient strategy to identify factors influencing extraction and derivatization of Arabidopsis thaliana samples in metabolomic studies with gas chromatography/mass spectrometry. Anal Biochem 331: 283–295 [DOI] [PubMed] [Google Scholar]
- Harris E. (1989) The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. Academic Press, San Diego: [DOI] [PubMed] [Google Scholar]
- Im CS, Zhang ZD, Shrager J, Chang CW, Grossman AR. (2003) Analysis of light and CO2 regulation in Chlamydomonas reinhardtii using genome-wide approaches. Photosynth Res 75: 111–125 [DOI] [PubMed] [Google Scholar]
- Jonsson P, Johansson AI, Gullberg J, Trygg J, A J, Grung B, Marklund S, Sjöström M, Antti H, Moritz T. (2005) High-throughput data analysis for detecting and identifying differences between samples in GC/MS-based metabolomic analyses. Anal Chem 77: 5635–5642 [DOI] [PubMed] [Google Scholar]
- Kozlowska-Szerenos B, Bialuk I, Maleszewski S. (2004) Enhanced photosynthetic O2 evolution in Chlorella vulgaris under high light and increased CO2 concentration as a sign of acclimation to phosphate deficiency. Plant Physiol Biochem 42: 403–409 [DOI] [PubMed] [Google Scholar]
- Kozłowska-Szerenos B, Zieliński P, Maleszewski S. (2000) Involvement of glycolate metabolism in acclimation of Chlorella vulgaris cultures to low phosphate supply. Plant Physiol Biochem 38: 727–734 [Google Scholar]
- Kroth PG, Chiovitti A, Gruber A, Martin-Jezequel V, Mock T, Schnitzler-Parker M, Stanley MS, Kaplan A, Caron L, Weber T, et al. (2008) A model for carbohydrate metabolism in the diatom Phaeodactylum tricornutum deduced from comparative whole genome analysis. PLoS ONE 3: e1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchitsu K, Tsuzuki M, Miyachi S. (1988) Changes of starch localization within the chloroplast induced by changes in CO2 concentration during growth of Chlamydomonas reinhardtii: independent regulation of pyrenoid starch and stroma starch. Plant Cell Physiol 29: 1269–1278 [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Manuel LJ, Moroney JV. (1988) Inorganic carbon accumulation by Chlamydomonas reinhardtii: new proteins are made during adaptation to low CO2. Plant Physiol 88: 491–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek LF, Spalding MH. (1991) Changes in photorespiratory enzyme-activity in response to limiting CO2 in Chlamydomonas reinhardtii. Plant Physiol 97: 420–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda Y, Colman B. (1995a) Induction of CO2 and bicarbonate transport in the green alga Chlorella ellipsoidea. I. Time course of induction of the two systems. Plant Physiol 108: 247–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda Y, Colman B. (1995b) Induction of CO2 and bicarbonate transport in the green alga Chlorella ellipsoidea. II. Evidence for induction in response to external CO2 concentration. Plant Physiol 108: 253–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Marechal-Drouard L, et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrett MJ, Lord JM. (1973) Glycollate formation and metabolism by algae. New Phytol 72: 751–767 [Google Scholar]
- Miura K, Kohinata T, Yoshioka S, Ohyama K, Fukuzawa H. (2002) Regulation of a carbon concentrating mechanism through CCM1 in Chlamydomonas reinhardtii. Funct Plant Biol 29: 211–219 [DOI] [PubMed] [Google Scholar]
- Miura K, Yamano T, Yoshioka S, Kohinata T, Inoue Y, Taniguchi F, Asamizu E, Nakamura Y, Tabata S, Yamato KT, et al. (2004) Expression profiling-based identification of CO2-responsive genes regulated by CCM1 controlling a carbon-concentrating mechanism in Chlamydomonas reinhardtii. Plant Physiol 135: 1595–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroney JV, Wilson BJ, Tolbert NE. (1986) Glycolate metabolism and excretion by Chlamydomonas reinhardtii. Plant Physiol 82: 821–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver SN, Lunn JE, Urbanczyk-Wochniak E, Lytovchenko A, van Dongen JT, Faix B, Schmalzlin E, Fernie AR, Geigenberger P. (2008) Decreased expression of cytosolic pyruvate kinase in potato tubers leads to a decline in pyruvate resulting in an in vivo repression of the alternative oxidase. Plant Physiol 148: 1640–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramazanov Z, Rawat M, Henk MC, Mason CB, Matthews SW, Moroney JV. (1994) The induction of the CO2-concentrating mechanism is correlated with the formation of the starch sheath around the pyrenoid of Chlamydomonas reinhardtii. Planta 195: 210–216 [Google Scholar]
- Ratcliffe RG, Shachar-Hill Y. (2006) Measuring multiple fluxes through plant metabolic networks. Plant J 45: 490–511 [DOI] [PubMed] [Google Scholar]
- Raven JA. (2010) Inorganic carbon acquisition by eukaryotic algae: four current questions. Photosynth Res (in press) [DOI] [PubMed] [Google Scholar]
- Raven JA, Ball LA, Beardall J, Giordano M, Maberly SC. (2005) Algae lacking carbon-concentrating mechanisms. Can J Bot 83: 879–890 [Google Scholar]
- Reinfelder JR, Milligan AJ, Morel FMM. (2004) The role of the C-4 pathway in carbon accumulation and fixation in a marine diatom. Plant Physiol 135: 2106–2111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix J. (1995) Chlamydomonas reinhardtii as the photosynthetic yeast. Annu Rev Genet 29: 209–230 [DOI] [PubMed] [Google Scholar]
- Schauer N, Steinhauser D, Strelkov S, Schomburg D, Allison G, Moritz T, Lundgren K, Roessner-Tunali U, Forbes MG, Willmitzer L, et al. (2005) GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett 579: 1332–1337 [DOI] [PubMed] [Google Scholar]
- Scheibe R. (2004) Malate valves to balance cellular energy supply. Physiol Plant 120: 21–26 [DOI] [PubMed] [Google Scholar]
- Spalding MH, Critchley C, Govindjee, Orgren WL. (1984) Influence of carbon-dioxide concentration during growth on fluorescence induction characteristics of the green-alga Chlamydomonas reinhardtii. Photosynth Res 5: 169–176 [DOI] [PubMed] [Google Scholar]
- Spalding MH, Jeffrey M. (1989) Membrane-associated polypeptides induced in Chlamydomonas by limiting CO2 concentrations. Plant Physiol 89: 133–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabenau H, Winkler U. (2005) Glycolate metabolism in green algae. Physiol Plant 123: 235–245 [Google Scholar]
- Suoeka N. (1960) Mitotic replication of deoxyribonucleic acid in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 46: 83–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teoh ML, Chu WL, Marchant H, Phang SM. (2004) Influence of culture temperature on the growth, biochemical composition and fatty acid profiles of six Antarctic microalgae. J Appl Phycol 16: 421–430 [Google Scholar]
- Thyssen C, Schlichting R, Giersch C. (2001) The CO2-concentrating mechanism in the physiological context: lowering the CO2 supply diminishes culture growth and economises starch utilisation in Chlamydomonas reinhardtii. Planta 213: 629–639 [DOI] [PubMed] [Google Scholar]
- Trygg J, Wold S. (2002) Orthogonal projections to latent structures (O-PLS). J Chemometr 16: 119–128 [Google Scholar]
- Tural B, Moroney JV. (2005) Regulation of the expression of photorespiratory genes in Chlamydomonas reinhardtii. Can J Bot 83: 810–819 [Google Scholar]
- Vance P, Spalding MH. (2005) Growth, photosynthesis, and gene expression in Chlamydomonas over a range of CO2 concentrations and CO2/O2 ratios: CO2 regulates multiple acclimation states. Can J Bot 83: 796–809 [Google Scholar]
- Wiklund S, Johansson E, Sjöström L, Mellerowicz EJ, Edlund U, Schockcor JP, Gottfries J, Moritz T, Trygg J. (2008) Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal Chem 80: 115–122 [DOI] [PubMed] [Google Scholar]
- Wold S, Esbensen K, Geladi P. (1987) Principal component analysis. Chemom Intell Lab Syst 2: 37–52 [Google Scholar]
- Wold S, Sjöström M, Eriksson L. (2001) PLS-regression: a basic tool of chemometrics. Chemom Intell Lab Syst 58: 109–130 [Google Scholar]
- Yamano T, Fukuzawa H. (2009) Carbon-concentrating mechanism in a green alga, Chlamydomonas reinhardtii, revealed by transcriptome analyses. J Basic Microbiol 49: 42–51 [DOI] [PubMed] [Google Scholar]
- Yamano T, Miura K, Fukuzawa H. (2008) Expression analysis of genes associated with the induction of the carbon-concentrating mechanism in Chlamydomonas reinhardtii. Plant Physiol 147: 340–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yocum CF, editor (1988) Methods in Enzymology, Vol 148, Plant Cell Membranes. Academic Press, Orlando, FL [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.