Abstract
Members of the plant type I MADS domain subfamily have been reported to be involved in reproductive development in Arabidopsis (Arabidopsis thaliana). However, from the 61 type I genes in the Arabidopsis genome, only PHERES1, AGAMOUS-LIKE80 (AGL80), DIANA, AGL62, and AGL23 have been functionally characterized, which revealed important roles for these genes during female gametophyte and early seed development. The functions of the other genes are still unknown, despite the fact that the available single T-DNA insertion mutants have been largely investigated. The lack of mutant phenotypes is likely due to a considerable number of recent intrachromosomal duplications in the type I subfamily, resulting in nonfunctional genes in addition to a high level of redundancy. To enable a breakthrough in type I MADS box gene characterization, a framework needs to be established that allows the prediction of the functionality and redundancy of the type I genes. Here, we present a complete atlas of their expression patterns during female gametophyte and seed development in Arabidopsis, deduced from reporter lines containing translational fusions of the genes to green fluorescent protein and β-glucuronidase. All the expressed genes were revealed to be active in the female gametophyte or developing seed, indicating that the entire type I subfamily is involved in reproductive development in Arabidopsis. Interestingly, expression was predominantly observed in the central cell, antipodal cells, and chalazal endosperm. The combination of our expression results with phylogenetic and protein interaction data allows a better identification of putative redundantly acting genes and provides a useful tool for the functional characterization of the type I MADS box genes in Arabidopsis.
The MADS box family of transcription factors in plants can be subdivided into MIKC-type and type I genes. While the MIKC-type genes are famous for their role in floral organ development, the type I genes were never found in forward genetic studies and were only discovered after the completion of the Arabidopsis (Arabidopsis thaliana) genome sequence (Alvarez-Buylla et al., 2000). The type I genes, which outnumber the MIKC-type genes in the Arabidopsis genome (61 versus 46), can be further subdivided into Mα, Mβ, and Mγ classes based on the sequence of the MADS box and the presence of additional motifs (De Bodt et al., 2003a; Pařenicová et al., 2003). Although the different subclasses may not be monophyletic (De Bodt et al., 2003b), the type I MADS box genes have several features in common. Most of the genes contain no introns (De Bodt et al., 2003a), seem to be weakly expressed (Kofuji et al., 2003; Pařenicová et al., 2003), and were duplicated after the divergence of monocots and dicots via intrachromosomal duplications (De Bodt et al., 2003b). Many recent duplications have occurred in the type I subfamily, and the genes are subject to high birth and death rates, suggesting that several type I genes may undergo nonfunctionalization (De Bodt et al., 2003b; Nam et al., 2004; Bemer et al., 2010).
Recently, several type I MADS box transcription factors have been reported to play important roles in reproductive development in Arabidopsis. Like most angiosperms, Arabidopsis has a polygonum-type embryo sac, which consists of seven cells: three antipodal cells that degenerate before fertilization, two synergid cells, which play a role in the attraction and guidance of the pollen tube, and two gametic cells, the egg cell and the central cell (Christensen et al., 1997). Reproduction occurs via a double fertilization process, in which two haploid male gametes are delivered into the female gametophyte by the pollen tube, where one fuses with the haploid egg cell and the other with the diploid central cell. The fertilized egg cell subsequently forms the diploid embryo, and the fertilized central cell develops into the triploid endosperm, a tissue that provides the nutrients for the growing embryo. While the zygote develops into a globular embryo, the endosperm nuclei undergo several rounds of mitosis without cytokinesis, resulting in a syncytium that contains three specific domains: the micropylar endosperm that surrounds the embryo, the peripheral endosperm, and the chalazal endosperm. The micropylar and peripheral endosperm become cellularized when the embryo reaches heart stage, whereas the chalazal endosperm remains a syncytium (Brown et al., 1999).
The type I MADS domain proteins AGAMOUS-LIKE80 (AGL80) and DIANA (DIA) are supposed to form a complex in planta to specify the formation of the central cell in the embryo sac. In both agl80 and dia mutants, central cell development is impaired, which results in a maternal-lethal phenotype (Portereiko et al., 2006a; Bemer et al., 2008; Steffen et al., 2008). Two other type I genes, PHERES1 (PHE1) and AGL62, were found to be involved in endosperm development. PHE1 is regulated by the Polycomb group gene MEDEA (MEA) via imprinting and is expressed only from the paternal allele shortly after fertilization. Although phe1 mutants show a wild-type phenotype, reduced expression levels of PHE1 in mea seed development mutants partially restored the mutant phenotype (Köhler et al., 2003b). AGL62 regulates the timing of endosperm cellularization: in agl62 mutants, the endosperm undergoes precocious cellularization, resulting in the arrest of embryo growth (Kang et al., 2008). Finally, AGL23 was reported to play a role in both female gametophyte and seed development. agl23 mutant ovules are partially arrested after megasporogenesis, suggesting a role for AGL23 in early embryo sac development. In addition, homozygous mutant seeds show defects in chloroplast formation during embryogenesis (Colombo et al., 2008).
In the last decade, a few other transcription factors were found to function in the Arabidopsis fertilization process. The synergid-expressed gene MYB98 was revealed to be essential for pollen tube guidance (Kasahara et al., 2005), while the homeodomain factor OESTRE/BLH1 (for BEL1-like homeodomain 1) is involved in the determination of cell fates in the mature embryo sac (Pagnussat et al., 2007). In addition, the Polycomb group proteins FERTILIZATION INDEPENDENT ENDOSPERM (FIE), FERTILIZATION INDEPENDENT SEED2 (FIS2), MULTICOPY SUPPRESSOR OF IRA1 (MSI1), and MEA also play important roles in the fertilization process. In the absence of fertilization, fie, fis2, msi1, and mea mutants develop seed-like structures, while the fertilized mutant seeds abort due to overproliferation of the embryo and endosperm (Ohad et al., 1996; Chaudhury et al., 1997; Grossniklaus et al., 1998; Köhler et al., 2003a). However, most of the genes identified in screens for embryo sac mutants are housekeeping genes, often involved in cell cycle regulation (Springer et al., 2000; Kwee and Sundaresan, 2003; Pagnussat et al., 2005; Portereiko et al., 2006b). The transcription factors that function specifically in female gametophyte and early seed development remain largely unknown. This is at least partly due to the fact that forward genetic studies for a distorted segregation often yield housekeeping genes, due to the (partial) lethality of the mutation to the haploid cell (Dresselhaus, 2006).
Therefore, studies on female gametophyte and early seed development have recently focused on transcriptome analysis, which resulted in the identification of a list of genes that are predominantly expressed before or after double fertilization (Yu et al., 2005; Johnston et al., 2007; Steffen et al., 2007; Day et al., 2008; Wuest et al., 2010). The list with expressed genes can subsequently be used for reverse genetic mutagenesis. However, also in these studies, cell cycle and signaling genes are overrepresented and the usually lower expressed transcription factors remain largely unidentified.
The functional characterization of the five type I MADS box transcription factors suggests that the type I subfamily plays an important role in female gametophyte and early seed development in Arabidopsis. Unraveling the functions of the other 56 Arabidopsis type I genes, therefore, will likely contribute to a better understanding of the molecular and genetic aspects that underlie the double fertilization process and subsequent seed development. However, despite considerable efforts by several research groups, the functional characterization of the remaining 56 type I genes has not been successful so far. This appears to be mainly caused by the high level of duplications that occurred in this subclass of MADS box genes, resulting in many paralogous genes that either function in a redundant manner or are subjected to nonfunctionalization (De Bodt et al., 2003b; Pařenicová et al., 2003; Bemer et al., 2010). To tackle this problem and enable a breakthrough in type I MADS box gene characterization, a framework needs to be established that allows the prediction of the functionality and redundancy of the type I proteins. Here, we present such a framework, in which highly detailed expression data are integrated with phylogenetic and protein-protein interaction data.
We generated plants transformed with translational fusions of the genes to GFP and GUS, which yielded expression profiles for 38 genes, while 20 genes appeared not expressed or expressed at a too low level. All the expressed genes are active in the female gametophyte and developing seed, indicating that the entire type I subfamily is involved in reproductive development in Arabidopsis. Interestingly, expression was predominantly observed in the central cell, antipodal cells, and chalazal endosperm, suggesting that the type I MADS domain transcription factors function mainly in these three cell types. The combination of our expression results with phylogenetic and protein interaction data allows a better identification of putative redundantly acting genes and provides a useful tool for the functional characterization of the type I MADS box genes in Arabidopsis.
RESULTS
Generation of Constructs to Study MADS Box Type I Gene Expression
The few characterized type I MADS box genes from Arabidopsis (PHE1, AGL80, DIA, AGL62, and AGL23) are all expressed in the female gametophyte or developing seed. Therefore, we expected that several of the uncharacterized type I genes would also be expressed in these structures. Both the female gametophyte and the endosperm have a syncytial phase during their development, which complicates the interpretation of expression patterns derived from promoter::reporter constructs. To obtain a detailed overview of the expression patterns of the Arabidopsis type I MADS box genes, therefore, we generated translational fusions of the genes with GFP and GUS (pAGLx::AGLx-GFP-GUS). Because MADS box genes belong to a family of transcription factors, the fusion proteins are expected to be translocated to the nucleus, which facilitates the precise determination of spatial expression in the syncytial stages.
We generated fusion constructs for all Arabidopsis type I MADS box genes identified by Pařenicová et al. (2003), except for AGL88, which has been annotated as a pseudogene (The Arabidopsis Information Resource), and AGL105, which is inserted in the coding region of another gene and therefore probably is a pseudogene as well. In addition, we also investigated the expression of the Mβ-type gene AGL101, which for unclear reasons is not present in the study of Pařenicová et al. (2003). In total, we studied the expression of 60 type I MADS box genes from Arabidopsis, 24 Mα-type genes, 16 Mγ-type genes, and 20 Mβ-type genes. For each construct, we amplified the genomic region of the gene from 2,000 bp upstream of the ATG until just before the stop codon, including any predicted intronic regions. However, if the next upstream locus was present within the 2-kb upstream region, only the part up to that locus was used in the construct. All constructs were successfully introduced to Arabidopsis by floral dip transformation, except for the constructs for AGL76 and AGL87, which repeatedly failed to yield transgenic seedlings.
Overview of the Expression Profiles of the Arabidopsis Type I MADS Box Genes
To determine the expression patterns of the Arabidopsis type I MADS box genes, tissues from plants transformed with pAGLx::AGLx-GFP-GUS constructs were incubated in GUS buffer for 1 to 3 d. GFP signal could only be observed if the GUS signal was already visible after one night of staining. However, in many cases, the expression level of the genes is so low that GUS expression could only be detected after 3 d of staining, and no GFP profile was obtained. The expression patterns for every gene are presented in Supplemental Figure S1, where information about the number of analyzed lines is also included.
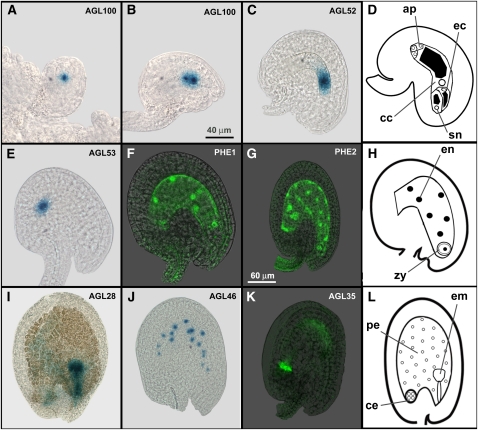
We obtained expression profiles for 38 of the 58 genes for which transgenic pAGLx::AGLx-GFP-GUS lines were studied. No GUS signal was observed in the transgenic lines of the remaining 20 genes. All expressed genes were found to be predominantly active in the female gametophyte or the developing seed. The observed expression profiles in both structures are shown in Figure 1. For each cell type in which GUS or GFP signal was observed, a representative image from one of the expressed genes is displayed. A complete overview of the expression patterns of the Arabidopsis type I MADS box genes is presented in Figure 2. The genes in the expression table are arranged according to their phylogeny, to facilitate the comparison of the expression patterns of paralogous genes.
Figure 1.
Cell types in which GUS/GFP expression was observed in the embryo sac and developing seed. A, AGL100 in the functional megaspore (FG1; Christensen et al., 1997). B, AGL100 in the two-nucleate embryo sac (FG2-3). C, AGL52 in the central cell (FG6). D, Schematic representation of the embryo sac in stage FG6. E, AGL53 in the antipodal cells. F, PHE1 in the endosperm nuclei approximately 16 h after pollination (hap). G, PHE2 in the peripheral and chalazal endosperm nuclei approximately 42 hap. H, Schematic representation of the developing seed approximately 16 hap. I, AGL28 in the globular embryo approximately 60 hap. J, AGL46 in the peripheral endosperm nuclei approximately 48 hap. K, AGL35 in the chalazal endosperm approximately 60 hap. L, Schematic representation of the developing seed approximately 60 hap. ap, Antipodal cells; cc, central cell; ce, chalazal endosperm; ec, egg cell; em, embryo; en, endosperm nuclei; pe, peripheral endosperm; sn, synergids; zy, zygote. Images A to F have the same scale (bar in B), and images G to K have the same scale (bar in G).
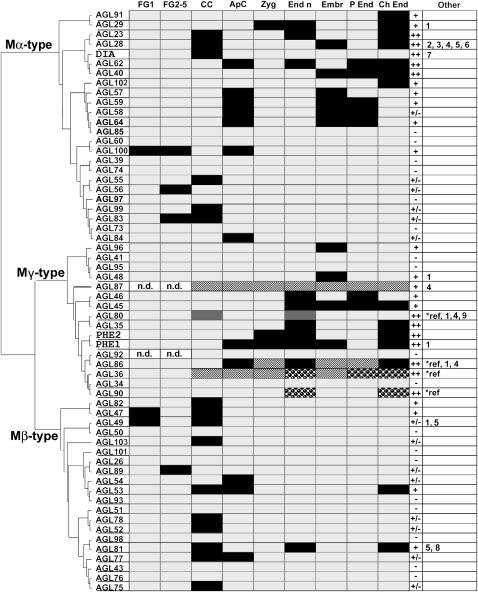
Figure 2.
Overview of the expression patterns of the type I MADS box genes. The genes are arranged according to their relationships in a neighbor-joining tree. The first nine columns represent different cell types as follows: FG1, embryo sac with megaspore mother cell; FG2-5, two- to eight-nucleate embryo sac; CC, central cell in stage FG6-7; ApC, antipodal cells in stage FG6; Zyg, zygote (approximately 12 h after pollination [hap]); End n, endosperm nuclei from 6 to 36 hap; Embr, globular/heart-stage embryo; P End, peripheral endosperm after cellularization; Ch End, chalazal endosperm (embryo sac stages after Christensen et al., 1997). Black boxes indicate GUS/GFP signal observed; light gray boxes indicate no signal observed; dark gray boxes indicate expression detected in other studies (references are given below); white/black dashed boxes indicate expression detected with q-RT-PCR in tissue including the cell type; spotted boxes indicate expression detected with q-RT-PCR and in other studies. n.d., Not determined. Column 10 indicates the expression levels as follows: ++, strong, signal visible after overnight staining in the majority of the transgenic lines; +, intermediate, signal visible after one night of staining in the minority of the lines or visible in the majority of the lines after 2 to 3 d of staining; ±, weak, signal visible in the minority of the lines after 2 to 3 d of staining; −, no signal observed. The last column indicates if the genes are also expressed in other tissues as follows: 1, pollen/anthers; 2, leaf primordia; 3, hypocotyls; 4, cotyledon/leaf; 5, pedicel; 6, roots; 7, filament; 8, silique; 9, flowers at stages 1 to 10. References are as follows: AGL80, Portereiko et al. (2006a); AGL36/AGL90/AGL86, Day et al. (2008); AGL36/AGL90, Walia et al. (2009).
In the embryo sac, expression was predominantly found in the central cell and antipodal cells in the final developmental stages (FG6–FG8; Christensen et al., 1997). Although a few genes are also expressed at earlier stages of megagametogenesis, expression was never observed in the egg cell or synergids, suggesting that type I MADS box genes specifically play a role in the determination and functioning of the chalazal cell types. In the developing seed, expression was often detected shortly after fertilization in the endosperm and sometimes in the embryo. During seed development, the expression of many type I genes became restricted to the chalazal endosperm, which remains a syncytium.
The overview in Figure 2 focuses on the expression in the female gametophyte and developing seed. However, several genes were found to be expressed in other tissues as well. This expression is summarized in the last column of the table but is presented in more detail in Supplemental Figure S1. We relatively often observed blue staining in mature pollen of the different transgenic lines. However, pollen of wild-type Arabidopsis plants also appears bluish after GUS staining. Therefore, we only assigned a signal to “pollen” in the expression data if it was clearly higher than background levels.
Twenty Arabidopsis Type I MADS Box Genes May Not Be Expressed
We did not observe GUS signal in any of the transgenic lines from 20 of the 58 studied genes. From these 20 genes, six belong to the Mα class (AGL39, AGL60, AGL73, AGL74, AGL85, AGL97), eight to the Mβ class (AGL26, AGL43, AGL50, AGL51, AGL78, AGL93, AGL98, AGL101), and six are Mγ-type genes (AGL34, AGL36, AGL41, AGL90, AGL92, AGL95). The absence of GUS expression in these transgenic lines probably has different causes.
First, it is possible that we did not detect the GUS signal, because the gene is only active under certain environmental conditions or because the expression is too weak or very transient. We primarily focused on the female gametophyte and developing seed and may have overlooked specific expression in other tissue types. A drawback of promoter::coding sequence-GFP-GUS constructs is that the signal of the chimeric protein is usually lower than that of GFP-GUS under the control of the promoter alone. This difference in signal strength was obvious for DIA (AGL61) and AGL29, for which we studied both promoter::reporter and promoter::coding sequence-reporter lines (Supplemental Fig. S1; Bemer et al., 2008). The levels of GUS and GFP expression and the number of expressing transgenic lines were considerably higher in the plants transformed with the promoter::reporter construct. Therefore, we cannot exclude that we did not observe GUS expression for very weakly expressed genes, which we would have observed if a promoter::reporter construct had been used. In addition, it is possible that the gene is expressed in a certain tissue but that the (chimeric) protein is absent because it is unstable or actively degraded.
A second explanation is that the upstream regulatory region used in our constructs did not cover all cis-regulatory elements required for the expression of the gene. In a few cases, the 2-kb promoter region that we used for the constructs may not have been sufficient, because cis-elements farther upstream or 3′ from the coding sequence are required for the correct expression of the gene. This may very well be the case for the Mγ-type genes AGL36 and AGL90, which do not appear to be expressed in our studies but have recently been found to play a role in endosperm development (Walia et al., 2009). The intergenic region upstream of both genes is approximately 10 kb, and important cis-elements may thus be located beyond the 2-kb fragment used in the construct. The fragment used for AGL86, a close homolog of AGL36 and AGL90, probably also does not contain all elements important for correct expression. Although we found expression for AGL86 in the anthers and the antipodal cells (Fig. 2), Day et al. (2008) reported the presence of AGL86 transcripts in the endosperm at 4 d after pollination. Similarly, the other genes in the same clade, AGL92 and AGL34, may also be expressed in the endosperm, despite the fact that we did not observe any GUS expression in the corresponding transgenic lines.
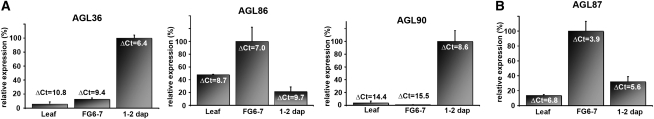
To test the expression of these five genes in the endosperm, we performed quantitative reverse transcription (q-RT)-PCR experiments with cDNA from siliques 1 to 2 d after pollination, pistils with stage FG6-7 embryo sacs, and leaves. We did not detect any significant transcript for AGL34 and AGL92 in any of the three tissues, suggesting that both genes are really not expressed in the female gametophyte or developing seed. The q-RT-PCR results for the other genes are presented in Figure 3A. The graphs show the relative expression of the genes in the different tissues. The graphs confirm the expression for AGL36 and AGL90 in developing seeds shortly after fertilization and show that both genes are not evidently expressed in the embryo sac or the leaf. AGL86 appears to be more ubiquitously expressed, since distinct expression can be found in all three tissues. In addition to the observed expression in the antipodal cells and anthers, AGL86 is probably also active in leaves and developing seeds.
Figure 3.
q-RT-PCR experiments for AGL36, AGL86, AGL90, and AGL87. Relative transcript levels are given for AGL36, AGL86, and AGL90 (A) and AGL87 (B) in three different tissues as follows: leaf, rosette and cauline leaves; FG6-7, pistils harvested just before pollination, containing embryo sacs in stage FG6-7 (Christensen et al., 1997); 1–2 dap, siliques harvested 1 to 2 d after pollination. The error bars depict the se of two biological replicates. To indicate the expression level, ΔCt values (CtAGLx – Ctref) are shown near the top of every bar (a low ΔCt value signifies a high expression level).
To investigate if a larger promoter fragment of AGL86 would indeed result in an extended activity, we generated a promoter::reporter construct with the complete 3.6-kb upstream region of AGL86. Several pAGL86(3.6kb)::GFP-GUS lines showed expression in the developing seed, although the expression appeared relatively weak. The expression profile resembles that of PHE1, with expression in the early endosperm nuclei, embryo, and chalazal endosperm (Supplemental Fig. S1). These results reveal, in combination with the q-RT-PCR results, that the promoter fragments that we used to generate the fusion constructs for AGL36, AGL86, and AGL90 were most likely not sufficient and lack certain essential cis-regulatory elements.
A third possibility is that the genes are really not expressed, because they lost essential regulatory elements during evolution. A high number of type I MADS box genes originated recently via intrachromosomal duplications (De Bodt et al., 2003b; Bemer et al., 2010). This occurs often via unequal crossing over, where one of the chromosomes acquires a duplicated region while that region is deleted from the other chromosome. If a gene is duplicated via this mode, its regulatory region may be disrupted, resulting in an altered expression or silencing of the gene. In Arabidopsis, these recent duplication events can be found in particular within the subclass of Mβ-type genes, where clusters of duplicated genes were identified on chromosomes I and V (De Bodt et al., 2003a). To investigate if some type I MADS box genes may have lost (part of) their regulatory region due to intrachromosomal duplication, we analyzed the sizes of the putative promoters (upstream region up to the next gene; Supplemental Table S2). This revealed that the promoters of the nonexpressed genes AGL74, AGL26, AGL43, and AGL51 are less than 500 bp, suggesting that these genes may have been immediately silenced after duplication by the loss of their regulatory region. However, whether these genes are really pseudogenes cannot be conclusively determined based on their promoter size. Also, the weakly expressed genes AGL52 and AGL75 have a promoter region that is shorter than 500 bp.
Expression of AGL76 and AGL87
The constructs for the Mβ-type gene AGL76 and the Mγ-type gene AGL87 were not successfully transformed to Arabidopsis; therefore, detailed expression data for these genes are not presented. To obtain some information about their activity, q-RT-PCR was performed on the same tissues as for AGL34, AGL36, AGL86, AGL90, and AGL92. Expression for AGL76 was not detected, but AGL87 transcript levels were high in leaves, pistils, and siliques (low ΔCt [threshold cycle] values for all three tissues; Fig. 3B), and it may thus be worthwhile to characterize this gene further in the future.
Analysis of the Expression Patterns of the Mα, Mβ, and Mγ Subclasses
The type I MADS box genes in plants are further subdivided into Mα-, Mβ-, and Mγ-type genes. Phylogenetic studies revealed that the Mβ- and Mγ-type genes from Arabidopsis share a common ancestor, while the Mα-type genes may not be monophyletic with the Mβ/Mγ clade (De Bodt et al., 2003b; Leseberg et al., 2006). Although all duplications within the three type I subclasses presumably occurred after the divergence of monocots and dicots (Arora et al., 2007), comparison of the Arabidopsis and poplar (Populus trichocarpa) type I MADS box genes suggests that the expansion and subsequent divergence of genes occurred earlier in the Mα-type subclass than in the Mβ- and Mγ-type subclasses (Leseberg et al., 2006). In agreement with this observation, we find that the expression patterns of the Arabidopsis Mα-type genes have diverged more than those of the Mβ- and Mγ-type genes.
The Mα-Type MADS Box Genes
The Mα-type genes show a wide range of different expression patterns in the female gametophyte and developing seed and can be divided into two groups based on the levels of their expression (Fig. 2). The first group contains genes that are distinctly expressed (the GUS signal is visible after 1 d of staining); the second group contains only weakly expressed genes. Within the first group, two distinct clades can be recognized, in which the genes share more or less the same expression patterns.
The first clade contains the genes AGL23, AGL28, AGL40, DIA, and AGL62. AGL23, DIA, and AGL62 have already been functionally characterized and revealed to play a role in central cell development (DIA [Bemer et al., 2008]), endosperm development (AGL62 [Kang et al., 2008]), and early embryo sac and seed development (AGL23 [Colombo et al., 2008]). We confirmed the expression patterns of DIA and AGL62 and the expression of AGL23 in the developing seed. However, GUS signal for AGL23 during early female gametophyte development, as reported by Colombo et al. (2008), was not detected in our studies, although we used a comparable upstream region for the analysis. We detected the earliest nucleus-localized GUS signal for AGL23 in the central cell of stage FG6 ovules. The functions of the other two genes in the clade, AGL28 and AGL40, are still unknown, but their expression in the embryo, endosperm nuclei, and chalazal endosperm of developing seeds suggests that both genes may have overlapping functions with AGL62 and AGL23. AGL28 is a remarkable type I MADS box gene, because it is expressed in a wide range of tissues in addition to its expression in the embryo sac and developing seed. We observed expression in hypocotyl, cotyledon, root, and leaf tips, in the stem and inflorescence, and in young leaves. These data are in conformance with the study of Yoo et al. (2006), who found that AGL28 is expressed in vegetative tissues. Whether AGL28 really plays a role in flowering, as suggested by the AGL28 overexpression phenotype (Yoo et al., 2006), remains to be investigated.
The second clade contains the paralogs AGL57, AGL58, AGL59, and AGL64. These genes are all distinctly expressed in the embryo and the peripheral endosperm. The fact that no mutant phenotypes have been found for any of these genes (Pařenicová et al., 2003) suggests that they function in a redundant manner. This may also be the case for the paralogous genes AGL29 and AGL91, which have overlapping expression patterns in the chalazal endosperm.
Finally, we also observed a distinct and interesting expression pattern for AGL100, which is highly homologous to AGL60. The gene is already expressed during the early stages of megagametogenesis, from the one-nucleate stage onward (FG1). The GUS signal can be detected in the course of embryo sac development in several nuclei, until it becomes restricted to the antipodal nuclei in stage FG5-6. Although we did not precisely determine the spatial and temporal expression of AGL100, its activity in certain nuclei during megagametogenesis may indicate a role in cell fate specification. We did not observe GUS signal in the pAGL60::AGL60-GFP-GUS transgenic lines, suggesting that AGL60 does not function in a redundant manner with AGL100.
The group of weakly expressed genes constitutes one large clade, containing AGL39, AGL74, AGL55, AGL56, AGL97, AGL99, AGL83, AGL73, and AGL84. We did not observe any GUS expression for AGL39, AGL74, AGL97, and AGL73, while the other five genes are specifically expressed in the female gametophyte. Occasionally, GUS signal for AGL56 and AGL83 was observed in unspecified nuclei of stage FG2-5 embryo sacs, but the clearest signal was found for AGL83, AGL73, and AGL99 in the central cell and for AGL84 in the antipodal cells of stage FG6-7 female gametophytes.
The Mγ-Type MADS Box Genes
The Mγ-type genes for which expression was observed are all relatively strongly expressed. Except for AGL80, which is predominantly expressed in the central cell (Portereiko et al., 2006a), all Mγ-type genes were revealed to be most strongly expressed in the developing seed. We did not detect GUS signal for AGL36, AGL86, and AGL90, but these genes are active in the developing seed, as discussed above. The gene clade consisting of AGL34, AGL35, AGL36, PHE1, PHE2, AGL80, AGL86, AGL90, and AGL92 contains interesting genes that have been shown to be involved in endosperm proliferation. PHE1 is an imprinted gene that is paternally expressed in the developing seed (Köhler et al., 2003b). Although the phe1 single mutant does not show an aberrant phenotype, down-regulation of PHE1 in a mea mutant does partially rescue the embryo and endosperm overproliferation phenotype. Recently, Walia et al. (2009) found that, in addition to PHE1, also PHE2, AGL35, AGL36, and AGL90 are important for endosperm development. In developing seeds from incompatible interhybrid crosses between Arabidopsis and Arabidopsis arenosa, the expression of the five genes is increased compared with seeds from compatible crosses (Walia et al., 2009). In correspondence with the fact that we did not observe expression for AGL34 and AGL92, both genes were not identified in the study of Walia et al. (2009), suggesting that they are actually silenced and may be considered as pseudogenes. In support of this idea, the AGL34 coding sequence lacks the Mγ motif, which is hypothesized to be required for heterodimerization (Bemer et al., 2010). Several single mutants of genes in the PHE1 cluster have been studied, but none has been found to exhibit a mutant phenotype (Köhler et al., 2003b; Pařenicová et al., 2003; Walia et al., 2009), suggesting that the genes function in a redundant manner. Whether AGL86 also contributes to endosperm development is still unclear, but the broad expression profile of this gene suggests that its function has at least partly diverged.
AGL48 is expressed in the globular embryo and probably in the embryo surrounding endosperm (Supplemental Fig. S1). The protein was found to interact with the Mα-type protein AGL64 (De Folter et al., 2005), which is colocalized in the embryo. The gene clade to which AGL48 belongs also contains AGL41 and AGL95, for which no expression was observed, and AGL96, which is also embryo expressed. AGL41 is most likely a silenced pseudogene, because its coding sequence contains an early stop codon just downstream from the MADS box. The activity of both AGL48 and AGL96 in the embryo suggests that both genes function in a redundant manner in embryo development, probably in a complex with the embryo-expressed Mα-type paralogs AGL64, AGL57, AGL58, and AGL59. However, evidence for the interaction between the members of both clades is confined to the observation by De Folter et al. (2005) that AGL48 and AGL64 form a heterodimer in yeast.
Finally, the closely related genes AGL45 and AGL46 are both expressed in the endosperm. AGL46 is an interesting gene, because it is the only type I MADS box gene that is specifically expressed in the peripheral endosperm and not in the chalazal endosperm. The gene is highly homologous to AGL45, for which we did not manage to generate a pAGL45::AGL45-GFP-GUS construct. Although we produced several entry clones of the genomic AGL45 sequence, these clones repeatedly failed to recombine with the destination vector. Therefore, we analyzed pAGL45::GFP-GUS plants, which revealed that AGL45 is expressed in the peripheral and chalazal endosperm of developing seeds. Possibly, AGL45 functions redundantly with AGL46 in the peripheral endosperm.
The Mβ-Type MADS Box Genes
In contrast to the Mγ-type genes, the Mβ-type MADS box genes are mainly expressed in the female gametophyte. Their expression is in general weak, comparable to the levels of the Mα-type genes AGL55, AGL56, AGL73, AGL83, AGL84, and AGL99, the proteins of which interact with the Mβ-type proteins (De Folter et al., 2005). Clades with different expression patterns cannot be distinguished within the Mβ subclass, and it appears that all genes belong to one large cluster, which is active in the central cell (Fig. 2). This suggests a high redundancy and would explain why no mutant of an Mβ-type gene has been identified so far. However, within the large Mβ-clade, several individual genes evolved additional expression patterns and may have acquired other functions. Especially AGL53 and AGL81, which are more strongly expressed than the other Mβ-type genes and also active in other tissues, are interesting in this respect. Other interesting genes are AGL47, AGL49, and AGL89, which are active in the central cell and during early megagametogenesis, and AGL54 and AGL77, which are expressed in the antipodal cells.
Type I MADS Domain Complexes in the Embryo Sac and Developing Seed
MADS domain transcription factors act predominantly as heterodimers or higher order complexes to regulate the expression of downstream targets. De Folter et al. (2005) published an interaction map of the Arabidopsis MADS domain proteins based on yeast two-hybrid experiments. This study revealed that type I MADS domain proteins interact predominantly with other type I proteins and that Mα-type proteins preferably heterodimerize with Mβ- or Mγ-type proteins, whereas interactions within one subclass are rare. Interestingly, the Mα-type genes that are distinctly expressed and belong to the AGL62 clade encode proteins that only form heterodimers with Mγ-type proteins, whereas the genes that are only weakly expressed in the female gametophyte (the AGL83 clade) encode proteins that interact with Mβ-type proteins. This suggests that both gene clades not only diverged in their expression patterns but that the proteins also evolved different interaction specificities. This protein divergence also occurred to a certain extent within the different clades, resulting in less redundancy than was inferred on the basis of the expression profiles and phylogeny. Therefore, integration of our expression data with the interaction data of De Folter et al. (2005) allows a more reliable prediction of which type I MADS domain complexes actually play a role in planta and provides more insight into the possible redundancy among the type I genes.
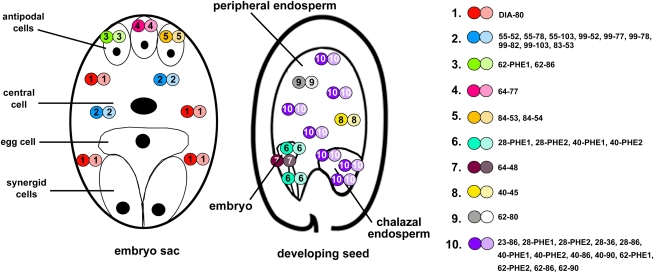
De Folter et al. (2005) reported that three type I MADS box proteins, AGL39, AGL74, and AGL97, form interactions with many other type I and MIKC-type proteins. However, our study shows that these genes are not expressed and that the heterodimers are thus not formed in planta. The majority of the other type I heterodimers identified by De Folter et al. (2005) are formed between proteins that are colocalized in certain cell types. Figure 4 lists the heterodimers that are most likely present in the central cell, antipodal cells, embryo, and (chalazal) endosperm. These data show that in every cell type, different combinations of closely related proteins result in many possible complexes. However, if all putative redundant complexes are considered the same, the central cell and embryo contain only two types of heterodimers, while the antipodal cells and the (chalazal) endosperm have three. These results suggest that the nonredundant type I complexes fulfill 10 different functions in female gametophyte and seed development. However, especially in the endosperm, proteins from the same clade have probably evolved different functions and more nonredundant complexes may exist. This can be concluded from the facts that the agl23 and agl62 single mutants exhibit different mutant phenotypes (Colombo et al., 2008; Kang et al., 2008) and that several genes, like AGL35, have very particular expression patterns. In addition, several distinctly expressed genes, like AGL29 and AGL92, encode proteins that did not dimerize with other MADS domain proteins in the yeast two-hybrid screen but may be involved in MADS complexes in planta or interact with other transcription factors. The total number of functional nonredundant complexes may thus be higher than 10.
Figure 4.
Heterodimers of type I MADS domain proteins in the embryo sac and developing seed. The different dimeric complexes that potentially can occur in the antipodal cells, central cell, embryo, and (chalazal) endosperm in planta are shown in color code in the schematic drawings of the embryo sac and developing seed. The legend on the right shows the different combinations of paralogous proteins that can probably act redundantly in the same complexes. The numbers indicate the different AGLs (e.g. 80 = AGL80; De Folter et al., 2005). Complexes that are probably more abundant are shown more often.
Chalazal Endosperm Nuclei Can Be Recognized Shortly after Fertilization
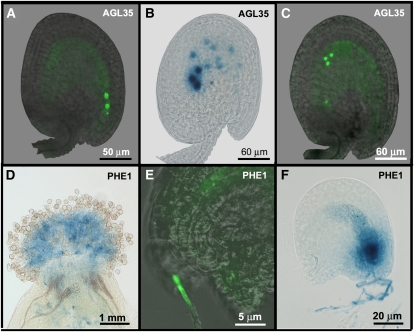
Analysis of the type I expression patterns also revealed interesting information about the development of the endosperm shortly after fertilization. The localization of the Mγ-type protein AGL35 appeared especially remarkable; therefore, we studied it in more detail. AGL35-GFP-GUS was observed for the first time shortly after fertilization in the initial endosperm nuclei positioned at the micropylar end (Fig. 5A). Although the number of endosperm nuclei increases thereafter exponentially, the AGL35-GFP signal remained restricted to three or four nuclei, which appeared to migrate from the micropylar to the chalazal end to form the chalazal endosperm (Figs. 1I and 5C). This suggests that chalazal endosperm identity is already established in the first endosperm nuclei and persists after division only in those nuclei that are destined to become the chalazal endosperm while it becomes lost from the other nuclei. GUS staining of the developing seeds with the pAGL35::AGL35-GFP-GUS construct shows a similar pattern, although a weak signal could also be detected in the other endosperm nuclei due to the higher sensitivity of the method (Fig. 5B). Nevertheless, the AGL35 reporter appears to be a perfect marker in which to study the determination of the chalazal endosperm. We also tested whether AGL35 expression is regulated via imprinting, like the expression of its homolog PHE1 (Köhler et al., 2003b), but reciprocal crosses revealed that the gene is expressed from both the paternal and the maternal transgenes approximately 24 h after pollination.
Figure 5.
Detailed expression patterns of AGL35 and PHE1. A to C, Expression of AGL35 in the developing seed. A, GFP signal in the first two endosperm nuclei and the zygote at 10 h after pollination (hap). B, Strong GUS signal in the nuclei that are migrating to the chalazal end, and weak GUS signal in the peripheral nuclei at 36 hap. C, GFP signal in the nuclei migrating to the chalazal end. d to F, Expression of PHE1 in the pollen tube. D, GUS signal in a wild-type pistil pollinated with pPHE1::PHE1-GFP-GUS pollen. E, GFP signal in a pollen tube that reaches a wild-type embryo sac. F, GUS signal in the synergids of a wild-type embryo released after penetration of a pPHE1::PHE1-GFP-GUS pollen tube.
Marker Lines
In addition to the AGL35 reporter line, several other AGL reporter lines appear very suitable for marker line use. We found that the PHE1 protein is also present in the pollen tube, which was not reported before (Köhler et al., 2003b). To investigate this further, we pollinated wild-type pistils with pPHE1::PHE1-GFP-GUS pollen. The GUS/GFP signal was clearly visible in germinating pollen on the stigma and could subsequently be observed throughout the growing pollen tube (Fig. 5D). After entering the embryo sac via the micropyle, the signal was released in the penetrated synergid cell (Fig. 5, E and F). The GFP signal was more specific than the GUS signal and was concentrated in the growing tip, where the two generative nuclei are located (Fig. 5E). Because the GUS/GFP signal is released in the synergid cell upon the entrance of the pollen tube, the pPHE1::PHE1-GFP-GUS lines are good markers to show that the pollen tube is actually penetrating the embryo sac.
Within the context of the female gametophyte, the DIA, AGL23, AGL28, AGL80, AGL82, and AGL81 reporter constructs can be used to show central cell identity, while the reporters of AGL62, AGL100, AGL57, PHE1, AGL86, and AGL53 are specifically active in the antipodal cells. Also, many chalazal endosperm markers are available, like AGL29, AGL91, AGL40, and AGL62. Finally, we identified the AGL46 reporter to be specific for the peripheral endosperm.
DISCUSSION
We analyzed the expression patterns of 60 type I MADS box genes in Arabidopsis and found that the majority are expressed in the female gametophyte or the developing seed. Our study shows, in contrast to earlier studies (Kofuji et al., 2003; Pařenicová et al., 2003), that many type I genes are distinctly expressed. However, their expression is often very specific and can only be observed in a few cells during a limited time span. Interestingly, there are three cell types in which the type I MADS box genes are predominantly expressed: the central cell and the antipodal cells in the female gametophyte and the chalazal endosperm nuclei in the developing seed. This suggests that the subclass of type I MADS box genes is mainly involved in the maturation of the embryo sac and the development of the endosperm. This idea is supported by the functional characterization of PHE1, AGL80, DIA, and AGL62, which all play a role in either central cell or endosperm development (Köhler et al., 2003b; Portereiko et al., 2006a; Bemer et al., 2008; Kang et al., 2008; Steffen et al., 2008). The type I genes thus seem to have followed a different evolutionary path than the MIKC-type genes, which diverged to fulfill very diverse functions in plant development (Becker and Theissen, 2003; Ferrario et al., 2004).
Type I MADS Box Genes in the Central Cell, Antipodal Cells, and Endosperm
The type I heterodimer AGL80-DIA is essential for central cell development (Portereiko et al., 2006a; Bemer et al., 2008), and mutants in either AGL80 or DIA show a maternal-lethal phenotype due to an impaired central cell. In addition to AGL80 and DIA, we detected expression in the central cell for 15 other type I MADS box genes, mainly Mα- and Mβ-type genes. The fact that the analysis of T-DNA insertion mutants for these genes did not yield mutant phenotypes (Pařenicová et al., 2003) suggests that several of these genes function in a redundant manner. Their roles in central cell functioning remain to be investigated, but it will be interesting to discover whether they are regulated by AGL80-DIA or act independently of this heterodimer. We identified CArG boxes (DNA motif bound by MADS domain proteins) in the promoters of several central cell-expressed genes, suggesting that they may be a target of AGL80-DIA or another MADS dimer active in the central cell.
For 12 type I MADS domain proteins, we observed a GUS signal in the antipodal cells of the embryo sac. It is remarkable that the type I genes seem to be expressed either in the central cell or in the antipodal cells. Expression in both cell types was only occasionally observed for AGL53 and AGL77. Although the antipodal cells have been associated with nutrient transport in several species (Engell, 1994), their function in the Arabidopsis female gametophyte has not been elucidated (Kägi and Gross-Hardt, 2007). Further characterization of the antipodal cell-expressed type I genes will hopefully contribute to a better understanding of the role of the antipodal cells in Arabidopsis. However, also here, redundancy probably complicates the functional analysis, since the phe1 and agl62 single mutants do not exhibit a phenotype in the embryo sac, although they are both strongly expressed in the antipodal cells (Köhler et al., 2003b; Portereiko et al., 2006a).
Recently, several studies provided evidence for the important role of type I MADS box genes in endosperm development. Not only were many genes found to be expressed in 4-d-old proliferating endosperm (Day et al., 2008), but seven type I genes were also revealed to be involved in endosperm hyperproliferation in incompatible hybrid crosses. AGL62, PHE1, PHE2, AGL35, AGL36, AGL40, and AGL90 were found to be down-regulated in wild-type seeds at the transition from syncytial to cellular endosperm growth (Walia et al., 2009). Our data show that their expression becomes restricted to the uncellularized chalazal endosperm at that stage. In developing seeds from incompatible Arabidopsis × A. arenosa crosses, the expression of the genes remained high (and was thus probably not restricted to the chalazal endosperm), concurrent with a lack of endosperm cellularization and endosperm overproliferation (Walia et al., 2009). Walia et al. (2009) found that the Polycomb group complex regulates the suppression of these genes upon cellularization, but it is still unclear if this regulation is direct or indirect. Since the complex has been shown to be involved in the imprinting of PHE1 (Köhler et al., 2003b), it will be interesting to investigate if some of the other genes are imprinted as well. We did not find evidence for imprinting of AGL35, because the transgene was expressed from both the paternal and maternal alleles after reciprocal crosses.
The genes in the study of Walia et al. (2009) belong to the PHE1 and AGL62 clades, the proteins of which were found to interact (De Folter et al., 2005). There are a few genes in these clades that were not identified in the study of Walia et al. (2009). Our data show that two of these genes, AGL23 and AGL28 (AGL62 clade), are also expressed in the chalazal endosperm and may thus play a role in endosperm cellularization as well. This is probably also the case for the PHE1 paralog AGL86, which is expressed in the proliferating endosperm (Day et al., 2008).
Functional Redundancy within the Type I MADS Box Subfamily
The type I MADS box genes in Arabidopsis are subjected to higher birth and death rates than the MIKC-type genes, and many duplication events in the type I subclass appear to have occurred recently (De Bodt et al., 2003b; Nam et al., 2004; Leseberg et al., 2006). Due to this turbulent evolutionary history, a high number of genes probably act in a redundant manner, while others are subjected to nonfunctionalization. This complicates the functional characterization of the type I MADS box genes severely, since single T-DNA insertion mutants rarely show a mutant phenotype. The large-scale expression analysis presented here enables a better prediction of which genes are likely to function in a redundant manner and will thus facilitate the functional analysis of the type I genes. For example, the closely related Mγ-type genes AGL96 and AGL48 are likely to function in a redundant manner in embryo development, while PHE1 and AGL86 may have redundant functions in the antipodal cells and endosperm. In these cases, the analysis of double mutants will probably help to unravel the function of the genes. However, in many other cases, it will be difficult to investigate the functions of the genes by crossing T-DNA insertion mutants, because the genes are located close to each other on the same chromosome. This is especially the case for the Mβ-type genes, which often originated from intrachromosomal duplications and are mainly located in clusters on chromosomes I and V. Also, several Mα- and Mγ-type paralogs, like PHE1 and PHE2, are located in tandem on the Arabidopsis genome. In these cases, transgenic approaches will be required to silence or overexpress the genes and unravel their functions. Suppression approaches like translational fusions with the EAR (for ERF-associated amphiphilic repression) domain (Hiratsu et al., 2003) or artificial microRNAs (Schwab et al., 2006) that target more than a single gene could overcome the redundancy. However, it needs to be taken into account that these methods have a dominant nature and that transgenic seeds may not be obtained if the effects are lethal to the embryo sac or embryo. To tackle this problem, it will be necessary to use inducible transgenes. The prediction of protein functionality and redundancy presented in this study provides an excellent baseline to start these transgenic approaches and unravel the functions of this challenging group of Arabidopsis type I MADS domain proteins.
Evolution of the Mα, Mβ, and Mγ Subclasses
The three subclasses of type I MADS box genes do not share a high sequence similarity and have only the MADS box in common. Phylogenetic analyses have shown that the Arabidopsis Mβ- and Mγ-type proteins are more similar and share a common ancestor, while the Mα-type proteins form a distinct separate group and seem to be more related to the MIKC-type genes (De Bodt et al., 2003b; Pařenicová et al., 2003; Fig. 2). Analyses of the MADS box family in rice (Oryza sativa) and poplar revealed that the Mα- and Mγ-type genes originated before the divergence of monocots and dicots, while the Mβ-type genes were only found in Arabidopsis, indicating that their divergence from the Mγ-type genes is specific for the Brassicaceae (Leseberg et al., 2006; Arora et al., 2007). Our study reveals distinct differences between the expression profiles of the different subclasses and provides more information about their evolutionary histories. A number of recent intrachromosomal duplications in the Mβ-subclass resulted in 21 genes that share a high sequence similarity. The majority of these genes are very weakly expressed in the central cell, suggesting that they originated from one ancestral central cell-expressed gene and currently function in a redundant manner. The fact that many genes are not or only very weakly expressed suggests that several genes are nonfunctional but cannot be recognized as pseudogenes yet. However, several Mβ-type proteins are probably functional and play a role in central cell development together with their Mα-type interaction partners AGL55, AGL83, and AGL99. While the Mβ-type genes are involved in female gametophyte development, the related Mγ-type genes are predominantly expressed during seed development. In contrast to the Mβ-type genes, the Mγ-type genes are distinctly expressed and expression divergence has occurred between the different clades, indicating that the expansion in this subclass occurred earlier. The Mβ- and Mγ-type proteins both interact with Mα-type proteins and together cover all the cell types in which the Mα-type proteins are present. Both subclasses, therefore, could also be considered as one. Because the Mβ-type genes do not form a separate clade in other species (Leseberg et al., 2006), this is probably a better depiction of the situation in all angiosperms.
Nam et al. (2004) estimated that there were four to eight type I genes present in the genome of the most recent common ancestor of Arabidopsis and rice, suggesting that there were only a few type I genes in the genomes of the earliest angiosperms. In line with this, the type I subclass does not appear to be present in gymnosperms, although two paralogous Mγ-type-like sequences are present in the genome of Pinus sylvestris (M. Bemer, unpublished data). Interestingly, one of the unique features of the angiosperms is the double fertilization of the male gametes with the central cell and egg cell, a process of which the evolutionary origin is still puzzling. The discovery that the type I genes are predominantly expressed in the central cell and endosperm suggests, together with their absence in gymnosperms, that this subfamily of MADS box transcription factors has been recruited in basal angiosperms for the evolution of the double fertilization process. Subsequent duplications of the genes may have enabled fine-tuning of this process in different angiosperm lineages. This hypothesis has to be further tested by the functional characterization of type I MADS box genes in other angiosperm species.
MATERIALS AND METHODS
Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were grown in a growth chamber with a 16-h-light/8-h-dark cycle at 22°C. Seeds resulting from floral dip transformation were sterilized for 1 min in 100% ethanol and 5 min in 1% bleach, washed three times in sterile water, and germinated on half-strength Murashige and Skoog selective plates (2.2 g of Murashige and Skoog salts including Gamborg’s B5 vitamins, 0.5 g of MES, 30 mg L−1 kanamycin, or 15 mg L−1 phosphinothricin for Basta selection). After 10 d of incubation in a growth chamber (16 h of light/8 h of dark, 22°C), resistant plants were transferred to soil.
Generation of the Constructs
The reporter constructs were generated using the Gateway system (Invitrogen). Genomic fragments including up to 2 kb of promoter region and terminating just before the stop codon were amplified from Columbia genomic DNA using gene-specific primers with additional nucleotides for integration into the entry vector. The sequences of the primers are listed in Supplemental Table S1. Entry vectors were either constructed using the TOPO system (pENTR/D-TOPO) or via the BP reaction by recombination of the PCR fragments into pDONR221 or pDONR207. The entry vectors were subsequently recombined with the destination vector pKGWFS7 (kanamycin resistance) or pBGWFS7 (Basta resistance; Karimi et al., 2002). The resulting clones were sequenced from both sides with primers 190 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCT-3′) and 191 (5′-GGGGACCACTTTGTACAAGAAAGCTGGGT-3′), and the frame was checked with primer seqGFPrev (5′-AGTCGTGCTGCTTCATGTGGT-3′). The constructs for the Mβ-type genes were completely sequenced. The resulting vectors were transformed into Agrobacterium tumefaciens using freeze-thaw transformation (Chen et al., 1994). Transformation of Arabidopsis wild-type Columbia plants was performed using the floral dip method as described by Clough and Bent (1998).
GUS/GFP Assay
GUS activity was analyzed by staining various tissues 1 to 3 d at 37°C in staining solution (10 mm EDTA, 0.1% Triton X-100, 2 mm Fe2+CN, 2 mm Fe3+CN, 1 mg mL−1 5-bromo-4-chloro-3-indolyl-β-glucuronic acid, and 100 μg mL−1 chloramphenicol in 50 mm phosphate buffer, pH 7.0). If necessary, pistils were incised on both sides for optimal staining of the ovules. Ovules were cleared in 20% lactic acid/20% glycerol and observed on a Zeiss Axiovert 135 microscope using differential interference contrast optics.
For analysis of GFP expression, pistils were dissected on a microscope slide in 50 mm phosphate buffer (pH 7.0) and observed with confocal laser-scanning microscopy using a Leica TCS-SP5 microscope. GFP was excited with an argon laser (488 nm), and emission was detected between 500 and 530 nm.
Neighbor-Joining Tree Construction
The Arabidopsis type I MADS box protein sequences were aligned with ClustalW. The alignment was transferred to BioEdit and adjusted by hand, and the first 240 sites were used to generate a neighbor-joining tree in BioEdit (Accessory Applications > ProtDist). The tree file was opened in TreeView and rooted with the Mα clade.
q-RT-PCR Experiments
Leaf, pistil, and silique samples were harvested from Columbia wild-type Arabidopsis plants. RNA was extracted using the Qiagen RNeasy Plant mini kit, and cDNA was synthesized with the iScript cDNA synthesis kit (Bio-Rad). Real-time RT-PCR was performed with the iQ SYBR Green Supermix from Bio-Rad. The following PCR program was used: 1 min at 95°C, followed by 40 cycles of 10 s at 95°C and 45 s at 57°C. Two biological and two technical replicates were performed. A polyubiquitin gene (UBC) was used as reference gene (Czechowski et al., 2005). The primers are listed in Supplemental Table S1.
The gene names and their Arabidopsis Genome Initiative numbers are listed in Supplemental Table S1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. GFP/GUS signal observed for the different pAGLx::AGLx-GFP-GUS transgenic lines.
Supplemental Table S1. Primers used to generate the pAGLx::AGLx-GFP-GUS constructs (A) and primers used for the q-RT-PCR experiment (B).
Supplemental Table S2. Putative promoter lengths of the type I MADS box genes.
Supplementary Material
Acknowledgments
We thank Marieke Willems and Aline le Rouzic for assisting with the practical work.
References
- Alvarez-Buylla ER, Pelaz S, Liljegren SJ, Gold SE, Burgeff C, Ditta GS, de Pouplana LR, Martinez-Castilla L, Yanofsky MF. (2000) An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Evolution 97: 5328–5333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora R, Agarwal P, Ray S, Singh AK, Singh VP, Tyagi AK, Kapoor S. (2007) MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker A, Theissen G. (2003) The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol Phylogenet Evol 29: 464–489 [DOI] [PubMed] [Google Scholar]
- Bemer M, Gordon J, Weterings K, Angenent GC. (2010) Divergence of recently duplicated Mγ-type MADS-box genes in petunia. Mol Biol Evol 27: 481–495 [DOI] [PubMed] [Google Scholar]
- Bemer M, Wolters-Arts M, Grossniklaus U, Angenent GC. (2008) The MADS domain protein DIANA acts together with AGAMOUS-LIKE80 to specify the central cell in Arabidopsis ovules. Plant Cell 20: 2088–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RC, Lemmon BE, Nguyen H, Olsen OA. (1999) Development of endosperm in Arabidopsis thaliana. Sex Plant Reprod 12: 32–42 [Google Scholar]
- Chaudhury AM, Ming L, Miller C, Craig S, Dennis ES, Peacock WJ. (1997) Fertilization-independent seed development in Arabidopsis thaliana. Proc Natl Acad Sci USA 94: 4223–4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Nelson RS, Sherwood JL. (1994) Enhanced recovery of transformants of Agrobacterium tumefaciens after freeze-thaw transformation and drug selection. Biotechniques 16: 664–670 [PubMed] [Google Scholar]
- Christensen CA, King EJ, Jordan JR, Drews GN. (1997) Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sex Plant Reprod 10: 49–64 [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colombo M, Masiero S, Vanzulli S, Lardelli P, Kater MM, Colombo L. (2008) AGL23, a type I MADS-box gene that controls female gametophyte and embryo development in Arabidopsis. Plant J 54: 1037–1048 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day RC, Herridge RP, Ambrose BA, Macknight RC. (2008) Transcriptome analysis of proliferating Arabidopsis endosperm reveals biological implications for the control of syncytial division, cytokinin signaling, and gene expression regulation. Plant Physiol 148: 1964–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bodt S, Raes J, Florquin K, Rombauts S, Rouzé P, Theissen G, Van de Peer Y. (2003a) Genomewide structural annotation and evolutionary analysis of the type I MADS-box genes in plants. J Mol Evol 56: 573–586 [DOI] [PubMed] [Google Scholar]
- De Bodt S, Raes J, Van de Peer Y, Theissen G. (2003b) And then there were many: MADS goes genomic. Trends Plant Sci 8: 475–483 [DOI] [PubMed] [Google Scholar]
- De Folter S, Immink RGH, Kiefer M, Parenicova L, Henz SR, Weigel D, Busscher M, Kooiker M, Colombo L, Kater MM, et al. (2005) Comprehensive interaction map of the Arabidopsis MADS box transcription factors. Plant Cell 17: 1424–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresselhaus T. (2006) Cell-cell communication during double fertilization. Curr Opin Plant Biol 9: 41–47 [DOI] [PubMed] [Google Scholar]
- Engell K. (1994) Embryology of barley. IV. Ultrastructure of the antipodal cells of Hordeum vulgare L. cv. Bomi before and after fertilization of the egg cell. Sex Plant Reprod 7: 333–346 [Google Scholar]
- Ferrario S, Immink RGH, Angenent GC. (2004) Conservation and diversity in flower land. Curr Opin Plant Biol 7: 84–91 [DOI] [PubMed] [Google Scholar]
- Grossniklaus U, Vielle-Calzada JP, Hoeppner MA, Gagliano WB. (1998) Maternal control of embryogenesis by MEDEA, a Polycomb group gene in Arabidopsis. Science 280: 446–450 [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M. (2003) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J 34: 733–739 [DOI] [PubMed] [Google Scholar]
- Johnston AJ, Meier P, Gheyselinck J, Wuest SE, Federer M, Schlagenhauf E, Becker JD, Grossniklaus U. (2007) Genetic subtraction profiling identifies genes essential for Arabidopsis reproduction and reveals interaction between the female gametophyte and the maternal sporophyte. Genome Biol 8: R204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kägi C, Gross-Hardt R. (2007) How females become complex: cell differentiation in the gametophyte. Curr Opin Plant Biol 10: 633–638 [DOI] [PubMed] [Google Scholar]
- Kang IH, Steffen JG, Portereiko MF, Lloyd A, Drews GN. (2008) The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell 20: 635–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kasahara RD, Portereiko MF, Sandaklie-Nikolova L, Rabiger DS, Drews GN. (2005) MYB98 is required for pollen tube guidance and synergid cell differentiation in Arabidopsis. Plant Cell 17: 2981–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofuji R, Sumikawa N, Yamasaki M, Kondo K, Ueda K, Ito M, Hasebe M. (2003) Evolution and divergence of the MADS-box gene family based on genome-wide expression analyses. J Mol Evol 20: 1963–1977 [DOI] [PubMed] [Google Scholar]
- Köhler C, Hennig L, Bouveret R, Gheyselinck J, Grossniklaus U, Gruissem W. (2003a) Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO J 22: 4804–4814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C, Hennig L, Spillane C, Pien S, Gruissem W, Grossniklaus U. (2003b) The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev 17: 1540–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwee H, Sundaresan V. (2003) The NOMEGA gene required for female gametophyte development encodes the putative APC6/CDC16 component of the anaphase promoting complex in Arabidopsis. Plant J 36: 853–866 [DOI] [PubMed] [Google Scholar]
- Leseberg CH, Li A, Kang H, Duvall M, Mao L. (2006) Genome-wide analysis of the MADS-box gene family in Populus trichocarpa. Gene 378: 84–94 [DOI] [PubMed] [Google Scholar]
- Nam J, Kim J, Lee S, An G, Ma H, Nei M. (2004) Type I MADS-box genes have experienced faster birth-and-death evolution than type II MADS-box genes in angiosperms. Proc Natl Acad Sci USA 101: 1910–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad N, Yadegari R, Margossian L, Hannon M, Michaeli D, Harada JJ, Goldberg RB, Fischer RL. (1996) Mutations in FIE, a WD Polycomb group gene, allow endosperm development without fertilization. Plant Cell 11: 407–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat G, Yu H, Sundaresan V. (2007) Cell-fate switch of synergid to egg cell in Arabidopsis eostre mutant embryo sacs arises from misexpression of the BEL1-like homeodomain gene BLH1. Plant Cell 19: 3578–3592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat GC, Yu HJ, Ngo QA, Rajani S, Mayalagu S, Johnson CS, Capron A, Xie LF, Ye D, Sundaresan V. (2005) Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132: 603–614 [DOI] [PubMed] [Google Scholar]
- Pařenicová L, de Folter S, Kieffer M, Horner DS, Favalli C, Busscher J, Cook HE, Ingram RM, Kater MM, Davies B, et al. (2003) Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15: 1538–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portereiko MF, Lloyd A, Steffen JG, Punwani JA, Otsuga D, Drews GN. (2006a) AGL80 is required for central cell and endosperm development in Arabidopsis. Plant Cell 18: 1862–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portereiko MF, Sandaklie-Nikolova L, Lloyd A, Dever CA, Otsuga D, Drews GN. (2006b) NUCLEAR FUSION DEFECTIVE1 encodes the Arabidopsis RPL21M protein and is required for karyogamy during female gametophyte development and fertilization. Plant Physiol 141: 957–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer PS, Holding DR, Groover A, Yordan C, Martienssen RA. (2000) The essential Mcm7 protein PROLIFERA is localized to the nucleus of dividing cells during the G(1) phase and is required maternally for early Arabidopsis development. Development 127: 1815–1822 [DOI] [PubMed] [Google Scholar]
- Steffen JG, Kang H, Portereiko MF, Lloyd A, Drews GN. (2008) AGL61 interacts with AGL80 and is required for central cell development in Arabidopsis. Plant Physiol 148: 259–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen JG, Kang IH, Macfarlane J, Drews GN. (2007) Identification of genes expressed in the Arabidopsis female gametophyte. Plant J 51: 281–292 [DOI] [PubMed] [Google Scholar]
- Walia H, Josefsson C, Dilkes B, Kirkbride R, Harada J, Comai L. (2009) Dosage-dependent deregulation of an AGAMOUS-LIKE gene cluster contributes to interspecific incompatibility. Curr Biol 19: 1128–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuest SE, Vijverberg K, Schmidt A, Weiss M, Gheyselinck J, Lohr M, Wellmer F, Rahnenführer J, von Mering C, Grossniklaus U. (2010) Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Curr Biol 20: 506–512 [DOI] [PubMed] [Google Scholar]
- Yoo SK, Lee JS, Ahn JH. (2006) Overexpression of AGAMOUS-LIKE 28 (AGL28) promotes flowering by upregulating expression of floral promoters within the autonomous pathway. Biochem Biophys Res Commun 348: 929–936 [DOI] [PubMed] [Google Scholar]
- Yu H, Hogan P, Sundaresan V. (2005) Analysis of the female gametophyte transcriptome of Arabidopsis by comparative expression profiling. Plant Physiol 139: 1853–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.